Abstract
Diabetes is a metabolic disease that raises the risk of microvascular and neurological disorders. Insensitivity to insulin is a characteristic of type II diabetes, which accounts for 85-90 percent of all diabetic patients. The fundamental molecular factor of insulin resistance may be impaired cell signal transduction mediated by the insulin receptor (IR). Several cell-signaling proteins, including IR, insulin receptor substrate (IRS), and phosphatidylinositol 3-kinase (PI3K), have been recognized as being important in the impaired insulin signaling pathway since they are associated with a large number of proteins that are strictly regulated and interact with other signaling pathways. Many studies have found a correlation between IR alternative splicing, IRS gene polymorphism, the complicated regulatory function of IRS serine/threonine phosphorylation, and the negative regulatory role of p85 in insulin resistance and diabetes mellitus. This review brings up-to-date knowledge of the roles of signaling proteins in insulin resistance in order to aid in the discovery of prospective targets for insulin resistance treatment.
1. Introduction
Diabetes is a complex metabolic disorder associated with increased incidence of cardiovascular and neurological complications. The global prevalence of diabetes has increased rapidly from 4.7% (1980) to 8.7% (2014) [1]. According to the International Diabetes Federation, the number of diabetics worldwide will rise from 451 million in 2017 to 673 million by 2045 [2]. Type II diabetes mellitus (T2D), previously known as non-insulin-dependent diabetes, accounting for 85-90% of the number of diabetic patients, is characterized by impaired insulin sensitivity, diminished beta-cell function, and increased blood glucose levels [3].
One of the key pathogenesis of T2D is insulin resistance, which is also a critical inducement for early prevention and treatment. Insulin resistance is affected by genetic factors including mutations and polymorphism of insulin receptors, insulin receptor substrates, and signal transduction proteins such as PI3K; the binding of which with activated IRS is the critical step linking IR activation to downstream metabolic functions. Extrinsic factors involve circulating metabolites, inflammatory signals, the gut microbiome, and obesity, which are characterized by chronically elevated free fatty acids and may result in lipotoxicity [4–9].
Mutations in the insulin receptor (IR) gene are associated with metabolic syndromes such as the insulin resistance, which can lead to T2D cardiovascular disorders. More than 50 mutations in IR have been identified that are linked to rare forms of insulin resistance [10]. However, T2D caused by the permanent insulin resistance resulted from mutation of IR is uncommon [11]. Insulin resistance is caused primarily by abnormalities in insulin signal transduction.
Insulin signaling downstream of IR is primarily mediated by the insulin receptor substrate (IRS), which activates the phosphatidylinositol 3-kinase (PI3K)-protein kinase B (PKB/AKT) and ERK/MAPK (mitogen-activation protein kinase) pathways, which crosstalk with other signaling pathways [12]. Activation of IR by insulin or insulin-like growth factor (IGF) leads to autophosphorylation of the beta subunit of IR and subsequently phosphorylates IRS at tyrosine residues. Phosphorylated IRS acts as a docking site for proteins with SH2 domains, such as the PI3K regulatory component p85, causing the PI3K-AKT pathway to be activated. Glucose uptake and metabolism as well as fatty acid and protein synthesis are all aided by these intracellular signaling pathways. Binding of IRS with adapter protein Grb2 mediates activation of the ERK pathway which is involved in regulation of genes related to cell survival, cell growth, and differentiation [13]. Ligand-activated IR is internalized and trafficked to early endosome (EE) for dephosphorylation, after which it is degraded or recycled back to the plasma membrane. In insulin resistance and diabetes circumstances, IR trafficking is changed. IR has a spatial preference in triggering its downstream signaling pathway. When internalized, it tends to activate the ERK pathway, while on the plasma membrane, it initiates the PI3K/AKT pathway [14].
Insulin resistance has been linked to alternative splicing abnormalities in IR and IRS gene polymorphism as well as the negatively regulatory effect of p85 in a number of studies.
Extrinsic factors that contribute to insulin resistance have been updated [6]. This review seeks to provide an update on recent research on the molecular mechanisms driving insulin resistance, with a focus on alternative splicing, gene polymorphism, and IRS and PI3K negative regulation.
2. Alternative Splicing of IR
IR is a transmembrane glycoprotein and a receptor tyrosine kinase that can be activated by insulin and insulin-like growth factor (IGFI and IGFII). IR is encoded by a single gene INSR; however, it forms two functionally related but different isoforms, IR-A (lacking exon 11) and IR-B (including exon 11), due to alternative splicing of exon 11 (Figure 1) [15]. IR-A is predominantly expressed in fetal and tumor cells and is involved in cell proliferation, while IR-B is found in insulin-sensitive tissue such as pancreatic β cells, muscle, kidney, adipose tissue, and liver [16]. There have been reports of functional differences between these two isoforms. IR-B has a high affinity for insulin and is more active in glucose homeostasis, whereas IR-A has a high affinity for IGFII and has reduced tyrosine kinase activity and signaling capacity. IR-A and IR-B activate distinct downstream pathways in pancreatic cells, notably IR-A/PI3K Ia/p70S6 and IR-B/PI3K class II-like/Akt, to regulate insulin transcription and cell survival [17]. Alternative splicing of INSR is linked to insulin and glucose levels [18]. The abnormalities in INSR splicing and postreceptor signaling are associated with insulin resistance and hyperinsulinaemia [19, 20]. As observed in adipose tissue, IR-B is increased in response to weight loss with a strong negative correlation with fasting insulin levels and alternative splicing of INSR correlates with the expression of HNRNPA1, SF3A1, and SFRS7 [21]. HNRNPA1 has been previously identified as a known splicing factor to inhibit exon 11 inclusion [22]. Increased expression of CUG-BP, a regulator of pre-mRNA splicing, caused a switch from IR-B to IR-A in skeletal muscle, resulting in reduced insulin signaling activation and contributing to insulin resistance [23].
Figure 1.
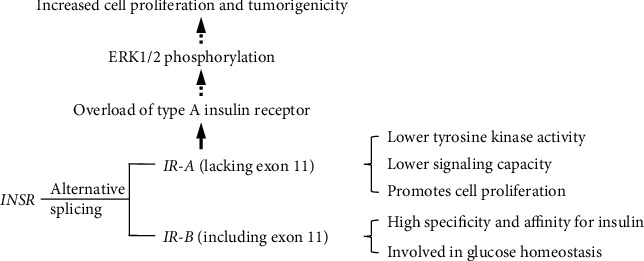
Alternative splicing of INSR and aberrant IR-A/IR-B ratio contributes to insulin resistance.
3. Negative Regulation of IR
The level of tyrosine phosphorylation of IR is crucial for controlling insulin signaling, while activation of IR can be inhibited by various proteins for posttranslational modifications, such as tyrosine phosphatase, the Grb protein family (growth-factor-receptor bound protein), SOCS (suppressor of cytokine signaling), and PC-1 or ENPP1 (plasma-cell-membrane glycoprotein-1, also referred to as ectonucleotide pyrophosphatase phosphodiesterase 1). PTP1B (protein tyrosine phosphatase 1B) is the most studied among the tyrosine phosphatases, which interacts with IR and dephosphorylate key tyrosine residues to limit its activity. PTP1Bgene knockout can increase insulin sensitivity by enhancing IR signaling [24]. Other regulatory proteins, such as SOCS1 and SOCS3, Grb10, Grb14, and PC1, decrease IR binding to IRS or change its kinase activity, thus inhibit IR function [25, 26]. Grb10 or Grb14 overexpression suppresses IRS tyrosine phosphorylation, whereas Grb14 gene knockout improves glucose homeostasis in the liver, white adipose tissues, and heart [27, 28].
SOCS expression has been found to be upregulated in insulin resistance, implying that it plays a role in the feedback control of insulin signaling and the development of diabetes [25]. In the mouse liver, overexpression of SOCS1 reduces insulin sensitivity, whereas suppression of SOCS3 expression improves insulin sensitivity [29]. Furthermore, SOCS directly bind to IR and decrease IRS phosphorylation, reducing insulin signaling, and they further disrupt insulin signaling by promoting ubiquitin-mediated IRS protein degradation [30, 31].
PC-1 expression was found to be higher in T2D patients' muscle, fibroblasts, and adipose tissues, while overexpression of PC-1 in established cell lines inhibited IR autophosphorylation and resulted in insulin resistance [32, 33]. Moreover, PC-1 expression is higher in the fibroblasts isolated from nonobese nondiabetic insulin-resistant subjects, suggesting that PC-1 may play a role in the development of insulin resistance. PC-1 inhibits IR autophosphorylation by directly engaging with 485-599 amino acids (AA) of IR, a tyrosine kinase regulatory domain essential for conformational change, and hence down-regulates subsequent downstream signal transduction, according to further studies [34].
The 121st amino acid of PC-1 is critical for its interaction with IR, and a functional missense nucleotide polymorphism resulting in a lysine to glutamine amino acid change has a stronger interaction with IR and is more effective in lowering IR autophosphorylation [35]. Although the PC-1 gene polymorphism (K121Q) may be a predisposing factor for insulin resistance and T2D, the results have been conflicting, as shown in Table 1. Some studies have made a connection between K121Q and insulin resistance and T2D in the populations such as Ukrainians [36], north Indians [37], South Africans of mixed-ancestry [38], Chinese [39, 40], Americans, Europeans, Africans [41], Asians [42], and Zanjans [43], but others have found no link in Pakistani Punjabis [44], Malaysians [45], Lebanese and Tunisians [46], Chinese [47, 48], and Danish Caucasians [49]. To understand its specific molecular significance in insulin resistance, more research with bigger sample sizes is required.
Table 1.
Association with gene polymorphism of PC-1 (K121Q) with insulin resistance or T2D in various populations.
| Association with gene polymorphism of PC-1 (K121Q) | Population | Reference |
|---|---|---|
| Associated with T2D | Ukrainian | [36] |
| Associated with T2D | South African mixed-ancestry population | [38] |
| Associated with insulin resistance | North Indian | [37] |
| Associated with T2D susceptibility | Chinese | [39, 40] |
| Associated with T2D risk | American, European and African | [41] |
| Increased susceptibility to diabetic kidney disease | European and Asian | [42] |
| Enhanced susceptibility to coronary artery disease | South Indian patients with T2D | [50] |
| Associated with T2D | Zanjan | [43] |
| No association with T2D | Lebanese and Tunisian | [46] |
| No association with T2D or obesity | Chinese | [47, 48] |
| No association with insulin resistance in T2D | Pakistani Punjabi | [44] |
| No association with T2D | Malaysian | [45] |
| No association with insulin resistance or T2D | Danish Caucasians | [49] |
4. IRS
IRS is a crucial mediator of insulin action and serves as a major site for both positive and negative control of insulin signaling transduction. IRS is made up of six members, from IRS1 to IRS6, all of which have relatively similar gene sequences and three-dimensional structures.
The pleckstrin-homology (PH) domain, the adjacent phosphotyrosine-binding (PTB) domain, and the C-terminal domain of them are all extremely similar [51, 52]. The PTB domains bind to the NPEpY sequence of IR, and the C-terminal domain has roughly 20 potential tyrosine phosphorylation sites. On activation of IR, these sites can be phosphorylated and bind to proteins with the Src homology domain 2 (SH2), such as the p85 subunit of the PI3K protein, Grb-2 protein, and the tyrosine protein phosphorylase SHP-2 [53].
As a key node of insulin signaling pathways, the loss of each isoform of IRS leads to varied physiologic results. The distribution and function of the IRS isoforms are summarized in Table 2. IRS1 gene knockout causes cell differentiation abnormalities in preadipocytes, whereas IRS2 gene knockout has no effect on cell differentiation but causes nonresponsiveness to insulin-stimulated glucose transport [54]. IRS1 gene knockout mice exhibit insulin deficiency in muscle tissue, whereas IRS2 gene knockout animals have insulin deficiency mostly in the liver and generate growth abnormalities in a few tissues, including neurons and pancreatic cells [55, 56]. IRS1 and IRS2 have complimentary effects on activating the AKT signaling pathway but play distinct roles in regulating gene expression, according to IRS1 and IRS2 tissue-specific knockouts studies in the liver.
Table 2.
Distribution and functions of different IRS isoforms.
| Isoforms | Distribution | Activation | Function |
|---|---|---|---|
| IRS1 | Widely expressed in various tissues | PI3K, SHP2, and Grb2 | Cell differentiation and glucose homeostasis [59] |
| IRS2 | Widely expressed in various tissues | PI3K, SHP2, and Grb2 | Cell growth and differentiation, mainly participate in glucose homeostasis in liver [59] |
| IRS3 | Adipocytes and brain | PI3K and SHP2 | Cell growth of adipocytes [60] |
| IRS4 | Embryonic tissues | PI3K and Grb2 | Cell proliferation and differential and reproductive capacity [61] |
| IRS5 | Mainly in kidney and liver | Undetected | Undefined [62, 63] |
| IRS6 | Skeletal muscle | Undetected | Undefined [63] |
The downregulation of the IRS1 gene causes the expression of genes involved in gluconeogenesis to increase, whereas the downregulation of the IRS2 gene causes the expression of genes involved in abiogenesis to increase [57]. IRS1 controls glucose uptake, while IRS2 is more closely related to MAPK activation, according to research using small interfering RNAs (siRNAs) to suppress the expression of IRS1 or IRS2 genes in L6 myotubes [58]. Hyperinsulinemia can reduce intracellular levels of IRS1 and IRS2 genes in cell culture models and mouse tissues. The following is the specific mechanism of action: at the transcriptional level, hyperinsulinemia causes IRS1 protein degradation and inhibits IRS2 production.
5. Gene Polymorphism of IRSs
The prevalent polymorphism of IRS1 is a glycine to arginine substitution in codon 972 (Gly972Arg), which is located between two potential tyrosine phosphorylation sites involved in binding with p85. This polymorphism has been linked to the development of T2D in obese Caucasian children [64], Egyptian patients with chronic hepatitis C virus infection and T2D [65], Kurdish ethnic, and Saudi and Pakistani populations [66–68]. However, no such association with this genetic variant has been found in Sistan and Baluchistan population of Iran [69], Arab, or Berber and Asian Indian populations [70, 71] with T2D (Table 3). The discrepancy could be attributed to differences in racial and ethnic distribution, sample size, and TM subclassifications as well as the inclusion or exclusion of certain insulin resistance confounders.
Table 3.
Association with gene polymorphism of IRS with insulin resistance or DM in various populations.
| Gene polymorphism | Results | Population | Reference |
|---|---|---|---|
| IRS1Arg972Gly | Increased insulin resistance | Obese Caucasian children | [64] |
| IRS1 Gly972Arg and Ala512Pro | Not associated with T2D | Sistan and Baluchistan population of Iran | [69] |
| IRS1 Gly972Arg | Not associate with prediabetes | Northern Vietnamese women | [84] |
| IRS1 Gly972Arg | Contributing risk factor for the development of T2D | Egyptian patients with chronic hepatitis C virus infection and T2D | [65] |
| IRS1Gly972Arg | Involved with GDM | Saudi, Iraq, Greek, and Egyptian population | [72–75] |
| IRS1Gly972Arg | Associate with risk of obesity | Obese Polish pregnant women | [81] |
| IRS1Gly972Arg | Associated with T2D | Kurdish ethnic, Saudi, and Pakistani population | [66–68, 85] |
| IRS1Gly972Arg | Correlated with newly diagnosed diabetic patients | Iranian | [86] |
| IRS1Gly972Arg and Ala513Pro | Not associated with T2D | Arab or Berber and Asian Indian populations | [70, 71] |
| IRS2Gly1057Asp | Not associated with T2D | Arab or Berber and Tunisian population | [70, 87] |
| IRS2 Gly1057Asp | Associate with T2D only in female | Bangladeshi population | [80] |
| IRS2 Gly1057Asp | No association with obesity | Obese Polish pregnant women | [81] |
| IRS2Gly1057Asp | Associated with T2D | Kurdish ethnic | [66] |
| IRS2 1057G/D | Associated with DM | Iranian | [77] |
| IRS2Asp1057Gly | Increased insulin resistance | Obese Caucasian children | [64] |
| IRS2Gly1057Asp | Associated with GDM | Turkish women | [79] |
| IRS2Gly1057Asp | Related with coronary artery disease | Taiwanese | [88] |
| IRS2Gly1057Asp | Increases susceptibility to T2D | Asian Indian | [89] |
| IRS4 Val1100Ile, His879Asp, His879Tyr, Ser439Cys, Arg411Glu, and Leu34Phe | Associated with body mass index in patient with schizophrenia | Caucasians | [82] |
| IRS4Leu34Phe, Arg411Gly, Gly584Cys, His879Asp, and Lys883Thr | Not associated with T2D or insulin resistance | Danish Caucasians | [83] |
Gestational diabetes mellitus (GDM), which affects 2-22 percent of all pregnancies, has a genetic background that is similar toT2D, including decreased insulin production and insulin resistance. As a result, similar genetic variations linked to T2D could be applied to predict the risk of GDM. In Saudi [72], Egyptian [73], Iraq [74], and Greek population [75], the association with IRS1 Gly972Arg GDM has been observed. The Gly1057Asp variation of the IRS2 gene has been widely documented, and it is thought to be linked to insulin resistance, T2D, and obesity. Gly1057Asp inside IRS2 has been linked to greater insulin resistance and T2D in obese Caucasian youngsters, Kurdish ethnics, Iranians, and Asian Indians [66, 76–78], while research in a Turkish women's population suggests that this genetic variation may be linked to GDM as well [79]. However, the genetic variant of IRS2 Gly1057Asp was significantly associated with risk of T2D only in female Bangladeshi population, not males, and the authors were unable to replicate the association of IRS2 Gly1057Asp with obesity [80], while no association with obesity has been found in obese Polish pregnant women [81], implying more appropriate controls for related confounding factors, and a larger sample size is required to confirm this finding.
Six novel IRS4 SNPs have been reported to be linked with BMI in schizophrenic patients [82], while another investigation found no link between IRS4 gene polymorphism and insulin resistance or T2D [83]. The differing results of various researches may be caused by population stratification; adoption of the case-control design would limit the link between allelic polymorphisms in candidate genes and diabetes, which may be related to ethnic or environmental factors produced by population stratification. In addition, the efficacy of different approaches for detecting gene polymorphism varies. To elucidate the significance of gene variation in distinct IRS isoforms in insulin resistance or diabetes mellitus, more research with a larger population and a reasonable design is required.
6. Elevated Serine Phosphorylation of IRSs
There are more than 70 potential serine/threonine phosphorylation sites in IRS protein, in addition to tyrosine phosphorylation sites, which can be induced by a variety of factors, including tumor necrosis factor-α (TNF-α) [90, 91], c-Jun-amino-terminal kinase (JNK) [92], protein kinase C (PKC) [93], glycogen synthase kinase-3 (GSK-3) [94], SOCS-3 [95], and mitochondrial dysfunction [96]. Serine phosphorylation of IRS1 increases with insulin resistance, and serine hyperphosphorylation of IRS1 is thought to be a negative regulator of insulin signal transduction in general (Figure 2) [97, 98]. Increased serine phosphorylation levels of IR and IRS, as a result of increased circulating fatty acids and ectopic lipid accumulation in muscle and liver, contribute to insulin resistance. Furthermore, elevated levels of circulating fatty acids caused by malnutrition downregulate the insulin signaling pathway by activating serine/threonine phosphorylation kinases including JNK and PKC, as well as impact IRS tyrosine phosphorylation levels by increasing transcription of SOCS proteins [99–101]. IRS1 subcellular localization, trafficking, and degradation as well as interaction with other signaling molecules are all part of the negative regulatory function of serine/threonine phosphorylation. IRS1 phosphorylation at Ser636/639 and Ser307 inhibits IRS1 binding to IR and elevated levels of phosphorylation of serine/threonine in IRS1 reduce IRS1 affinity with the p85 regulatory subunit of PI3K, weakening insulin signal transduction and leading to the symptom of insulin resistance, according to studies [102, 103].
Figure 2.
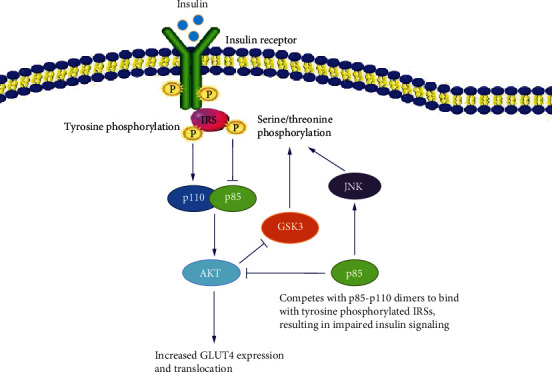
Schematic diagram of potential negative regulation of insulin signal transduction.
In fact, serine phosphorylation of IRS1 has been shown to have a positive regulatory role, promoting tyrosine phosphorylation while inhibiting serine/threonine phosphorylation at other sites. For example, phosphorylation of IRS1 at Ser629 causes phosphorylation at Ser636 to decrease, adversely regulating IRS1 and increasing insulin activity [104]. While a mutation study found that phosphorylation of hSer1223/m1214 (human and rat IRS1) interferes with IRS1's interaction with SHP-2, a negative modulator of IRS1 tyrosine phosphorylation, most likely due to steric hindrance, implying a positive regulatory function [105]. Basal phosphorylation at the serine/threonine of IRS1 potentiates insulin-stimulated tyrosine phosphorylation, whereas hyperphosphorylation inhibits phosphorylation produced by the insulin receptor tyrosine kinase [106, 107].
Serine/threonine phosphorylation at specific sites on IRS provides negative charge, altering protein interactions and reducing downstream cell signaling. By inducing conformational changes in IRS, phosphorylation at certain sites promotes protein interaction and enhances downstream signaling. Furthermore, depending on the time course of insulin action, the interaction status, and the implications of temporal variations in IRS phosphorylation, phosphorylation at specific serine/threonine residues may have varied effects [108]. Phosphorylation of Ser-302/318 is associated with enhanced insulin signaling during the early stages of insulin action, but it is also necessary for attenuating insulin actions during the late stages [109]. Studies into the regulatory effects of serine/threonine phosphorylation patterns on IRS1 activity will aid in understanding insulin resistance.
7. PI3K
PI3K inhibitors inhibit practically all of insulin's metabolic activities, including glucose transport, glycogen synthesis, lipid synthesis, and adipocyte differentiation, demonstrating that PI3K plays an important role in insulin signaling [12]. Class I PI3K is a heterodimeric protein consisting of the regulatory subunit p85 and the catalytic subunit p110. It has dual enzymatic activities of both serine/threonine protein kinase and phospholipid kinase. The catalytic subunit p110 is usually in conjunction with the regulatory subunit, and the free p110 is unstable and readily degraded. p85 not only stabilizes p110 but also limits its enzymatic activity by inducing conformational changes [110]. After the regulatory subunit p85 is phosphorylated at Tyr688, the inhibition is abolished. Phosphorylation of Ser608 of p85 by the serine kinase activity of p110 decreases the heterodimer's lipid kinase activity [111]. Additionally, it was discovered in cell culture models that PI3K modulates IRS1 phosphorylation at serine sites and inhibits IRS1 signaling [112].
Insulin sensitivity was increased by a mutation in p85, suggesting that p85 plays a negative function in insulin signal transduction [113, 114]. There are three possible mechanisms which p85 suppresses insulin signaling. The reduction of free monomers in the PI3K-regulatory subunit p85 is the first regulatory mechanism. The amount of p85 is more than p110 and the phosphorylated IRS protein under normal conditions, and there is a balance between the p85-p110 dimer and the free p85 monomers of PI3K. Tyrosine phosphorylation is competed with by free p85. As a consequence of reduced level of free p85, p85-p110 dimers can interact with more phosphorylated IRSs, improving insulin signal transduction [115]. In addition to IRS serine phosphorylation, several investigations have revealed that p85 overexpression is an important molecular mechanism that causes insulin resistance [116]. The isolation of PI3K via the formation of an isolation complex between the p85 monomer and IRS1 to downregulate IR signaling is the second mechanism [117]. Crosstalk between the p85 subunit and the JNK pathway is the third negative regulatory mechanism. Studies have shown that p85 is essential for insulin-stimulated JNK activation. JNK activity is regulated by p85, according to recombination experiments [115]. JNK has been identified in a number of studies to have an important role in metabolism and the development of impaired glucose tolerance and insulin resistance as a result of obesity. Activated JNK phosphorylates the Ser307 residue in IRS1's PTB domain, decreasing its tyrosine phosphorylation and inhibiting the IRS1-PI3K signaling pathway. The phosphorylation level of IRS1 Ser307 induced by JNK was significantly lower in JNK1 gene knockout mice fed a high fat diet (HFD), suggesting that it may have a protective role in the occurrence of impaired glucose tolerance and insulin resistance [118].
Gene polymorphism of PI3KR1, the coding gene of the PI3K p85 subunit, may be associated with GDM [119], T2D [120], insulin resistance, obesity and numerous cancers. A study conducted in Italy found no link between PI3KR1 and GDM, although the sample size was small, with only 38 pregnant women and 240 controls included [121]. Another study made in China including 334 cases and 367 controls found that PI3KR1 was involved in abnormal glucose metabolism [119]. A similar finding was found in a Turkish population with 427 diabetic patients and 413 controls, supporting the idea that PI3KR1 is linked to T2D and accompanying symptoms [120].
8. Targeting PTP1B in Insulin Resistance Treatment
Tyrosine residues of IR and IRS are dephosphorylated by PTP1B, which is one of the most well-studied tyrosine phosphatases, acting as a crucial negative regulator of the insulin signaling pathways [122]. PTP1B modulation may be valuable as a prospective therapeutic target for the treatment of T2D, and new PTP1B-targeting inhibitors or medications have been developed [123–128]. Due to comparatively lower toxicity, plant-derived medicines have become more prominent. Rampadarath et al. suggested flavonoid C glycosides, particularly orientin, as a possible therapeutic agent in the management of T2D [124]. Low-molecular-weight polymannuronic acid phosphate (LPMP), according to studies by Li et al., may be a promising candidate for an antidiabetic medication since it can reduce oxidative stress and improve insulin sensitivity [127]. The use of accessible synthetic medications, phytochemicals, and potential underlying mechanisms should be highlighted in depth in order to provide management suggestions for insulin resistance in the future [128].
9. Conclusion
Since the prevalence of diabetes has increased considerably around the world, understanding the molecular basis of insulin resistance is both theoretically and practically important. More research into the molecular processes of insulin signal transduction, particularly aberrant IR splicing, IRS gene polymorphism, regulatory effects of IRS phosphorylation, and the regulatory function of p85 on insulin activities, is valuable and still needed.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (32160214), the High-Level Talent Project of the Natural Science Foundation of Hainan Province (821RC1053) to Dayong Wang, and the Key Research and Development Project of Hainan Province (ZDYF2017090) to Baochun Wang.
Contributor Information
Baochun Wang, Email: 1387618006@163.com.
Dayong Wang, Email: wangdy@hainanu.edu.cn.
Conflicts of Interest
The author declares that there is no conflict of interest that could compromise the review's objectivity.
Authors' Contributions
All authors have contributed to the planning and writing of this review.
References
- 1.Zhou B., Lu Y., Hajifathalian K., et al. Worldwide trends in diabetes since 1980: a pooled analysis of 751 population- based studies with 4∗4 million participants. The Lancet . 2016;387(10027):1513–1530. doi: 10.1016/S0140-6736(16)00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho N. H., Shaw J. E., Karuranga S., et al. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Research & Clinical Practice . 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 3.Ribbing J., Hamrén B., Svensson M. K., Karlsson M. O. A model for glucose, insulin, and beta-cell dynamics in subjects with insulin resistance and patients with type 2 diabetes. Journal of Clinical Pharmacology . 2010;50(8):861–872. doi: 10.1177/0091270009349711. [DOI] [PubMed] [Google Scholar]
- 4.Wang H., Gou W., Su C., et al. Association of gut microbiota with glycaemic traits and incident type 2 diabetes, and modulation by habitual diet: a population-based longitudinal cohort study in Chinese adults. Diabetologia . 2022;65(7):1145–1156. doi: 10.1007/s00125-022-05687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seah J., Hong Y., Cichońska A., et al. Circulating metabolic biomarkers are consistently associated with type 2 diabetes risk in Asian and European populations. The Journal of Clinical Endocrinology and Metabolism . 2022;107(7):e2751–e2761. doi: 10.1210/clinem/dgac212. [DOI] [PubMed] [Google Scholar]
- 6.Batista T., Haider N., Kahn C. Defining the underlying defect in insulin action in type 2 diabetes. Diabetologia . 2021;64(5):994–1006. doi: 10.1007/s00125-021-05415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalid M., Alkaabi J., Khan M., Adem A. Insulin signal transduction perturbations in insulin resistance. International Journal of Molecular Sciences . 2021;22(16, article 8590) doi: 10.3390/ijms22168590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benito-Vicente A., Jebari-Benslaiman S., Galicia-Garcia U., Larrea-Sebal A., Uribe K., Martin C. Molecular mechanisms of lipotoxicity-induced pancreatic β-cell dysfunction. International Review of Cell and Molecular Biology . 2021;359:357–402. doi: 10.1016/bs.ircmb.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Daziano G., Blondeau N., Béraud-Dufour S., et al. Sortilin-derived peptides promote pancreatic beta-cell survival through CREB signaling pathway. Pharmacological Research . 2021;167, article 105539 doi: 10.1016/j.phrs.2021.105539. [DOI] [PubMed] [Google Scholar]
- 10.Rojek A., Niedziela M. Insulin receptor and its relationship with different forms of insulin resistance. Advances in Cell Biology . 2010;2(2):59–90. doi: 10.2478/v10052-010-0004-8. [DOI] [Google Scholar]
- 11.Iwanishi M., Toru K., Choka A., et al. Clinical characteristics in two patients with partial lipodystrophy and type A insulin resistance syndrome due to a novel heterozygous missense mutation in the insulin receptor gene. Diabetes Research and Clinical Practice . 2019;152:79–87. doi: 10.1016/j.diabres.2019.04.034. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty C., Doss C. G. P., Bhatia R., Agoramoorthy G. Profiling of phosphatidylinositol 3-kinase (PI3K) proteins in insulin signaling pathway. Applied Biochemistry and Biotechnology . 2015;175(7):3431–3446. doi: 10.1007/s12010-015-1515-4. [DOI] [PubMed] [Google Scholar]
- 13.Pei J., Xiao Z., Gou Z., et al. Sustained Stimulation of β2AR Inhibits Insulin Signaling in H9C2 Cardiomyoblast Cells Through the PKA-Dependent Signaling Pathway. Diabetes, metabolic syndrome and obesity : targets and therapy . 2020;13:3887–3898. doi: 10.2147/DMSO.S268028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Huang L., Qi X., Chen C. Insulin receptor trafficking: consequences for insulin sensitivity and diabetes. International Journal of Molecular Sciences . 2019;20(20):p. 5007. doi: 10.3390/ijms20205007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chifei K., Gadi S., Annie J. SUN-131 the roles of two insulin receptor isoforms in triple negative breast cancer growth. Journal of the Endocrine Society . 2020;4(Supplement 1) doi: 10.1210/jendso/bvaa046.1817. [DOI] [Google Scholar]
- 16.Escribano O., Beneit N., Rubio-Longás C., López-Pastor A., Gómez-Hernández A. The role of insulin receptor isoforms in diabetes and its metabolic and vascular complications. Journal Diabetes Research . 2017;2017, article 1403206:1–12. doi: 10.1155/2017/1403206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbara L., Ingo L., Tilo M. Selective insulin signaling through A and B insulin receptors regulates transcription of insulin and Glucokinase genes in pancreatic β cells. Molecular Cell . 2001;7(3):559–570. doi: 10.1016/s1097-2765(01)00203-9. [DOI] [PubMed] [Google Scholar]
- 18.Malakar P., Chartarifsky L., Hija A., et al. Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival. Scientific reports . 2016;6:1–14. doi: 10.1038/srep31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renna L. V., Bose F., Brigonzi E., Fossati B., Meola G., Cardani R. Aberrant insulin receptor expression is associated with insulin resistance and skeletal muscle atrophy in myotonic dystrophies. PLoS One . 2019;14(3, article e0214254) doi: 10.1371/journal.pone.0214254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renna L., Bosè F., Iachettini S., et al. Receptor and post-receptor abnormalities contribute to insulin resistance in myotonic dystrophy type 1 and type 2 skeletal muscle. PLoS One . 2017;12(9, article e0184987) doi: 10.1371/journal.pone.0184987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminska D., Hämäläinen M., Cederberg H., et al. Adipose tissue INSR splicing in humans associates with fasting insulin level and is regulated by weight loss. Diabetologia . 2014;57(2):347–351. doi: 10.1007/s00125-013-3097-4. [DOI] [PubMed] [Google Scholar]
- 22.Talukdar I., Sen S., Urbano R., Thompson J., Yates J. R., Webster N. J. hnRNP A1 and hnRNP F modulate the alternative splicing of exon 11 of the insulin receptor gene. PLoS One . 2011;6(11, article e27869) doi: 10.1371/journal.pone.0027869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savkur R., Philips A. V., Cooper T. A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nature Genetics . 2001;29(1):40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 24.Xue B., Kim Y. B., Lee A., et al. Protein-tyrosine phosphatase 1B deficiency reduces insulin resistance and the diabetic phenotype in mice with polygenic insulin resistance. Journal of Biological Chemistry . 2007;282(33):23829–23840. doi: 10.1074/jbc.M609680200. [DOI] [PubMed] [Google Scholar]
- 25.Ueki K., Kondo T., Kahn C. R. Suppressor of Cytokine Signaling 1 (SOCS-1) and SOCS-3 Cause Insulin Resistance through Inhibition of Tyrosine Phosphorylation of Insulin Receptor Substrate Proteins by Discrete Mechanisms. Molecular & Cellular Biology . 2004;25(19):5434–5446. doi: 10.1128/mcb.24.12.5434-5446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carré N., Goenaga D., Burnol A. F. Modulation of insulin sensitivity by the Grb7 family of molecular adaptors. Obésité . 2011;6(2):114–122. doi: 10.1007/s11690-011-0280-y. [DOI] [Google Scholar]
- 27.Ding X., Iyer R., Novotny C., Metzger D., Zhou Y. Inhibition of Grb14, a negative modulator of insulin signaling, improves glucose homeostasis without causing cardiac dysfunction. Scientific Reports . 2020;10(1, article 3417) doi: 10.1038/s41598-020-60290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto Y., Ishino F., Kaneko-Ishino T., et al. Type 2 diabetes mellitus in a non-obese mouse model induced by Meg1/Grb10 overexpression. Experimental Animals . 2008;57(4):385–395. doi: 10.1538/expanim.57.385. [DOI] [PubMed] [Google Scholar]
- 29.Shi H., Tzameli I., Bjorbaek C., Flier J. S. Suppressor of cytokine signaling 3 is a physiological regulator of adipocyte insulin signaling. Journal of Biological Chemistry . 2004;279(33):34733–34740. doi: 10.1074/jbc.M403886200. [DOI] [PubMed] [Google Scholar]
- 30.Rui L. Y., Yuan M. S., Frantz D., Shoelson S., White M. Socs1 induces insulin resistance by promoting ubiquitin-mediated degradation of IRS1 and IRS2. Diabetes . 2002;51:A57–A58. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 31.Rui L., Yuan M., Frantz D., Shoelson S., White M. F. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. Journal of Biological Chemistry . 2002;277(44):42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 32.Dong H., Maddux B. A., Altomonte J., et al. Increased hepatic levels of the insulin receptor inhibitor, PC-1/NPP1, induce insulin resistance and glucose intolerance. Diabetes . 2005;54(2):367–372. doi: 10.2337/diabetes.54.2.367. [DOI] [PubMed] [Google Scholar]
- 33.Maddux B. A., Chang Y. N., Accili D., Mcguinness O. P., Goldfine I. D. Overexpression of the insulin receptor inhibitor PC-1/ENPP1 induces insulin resistance and hyperglycemia. American Journal of Physiology. Endocrinology and Metabolism . 2006;290(4):E746–E749. doi: 10.1152/ajpendo.00298.2005. [DOI] [PubMed] [Google Scholar]
- 34.Maddux B. A., Goldfine I. D. Membrane glycoprotein PC-1 inhibition of insulin receptor function occurs via direct interaction with the receptor alpha-subunit. Diabetes . 2000;49(1):13–19. doi: 10.2337/diabetes.49.1.13. [DOI] [PubMed] [Google Scholar]
- 35.Costanzo B. V., Trischitta V., Paola R. D., et al. The Q allele variant (GLN121) of membrane glycoprotein PC-1 interacts with the insulin receptor and inhibits insulin signaling more effectively than the common K allele variant (LYS121) Diabetes . 2001;50(4):831–836. doi: 10.2337/diabetes.50.4.831. [DOI] [PubMed] [Google Scholar]
- 36.Marchenko I. V., Dubovyk Y. I., Matlai O. I., Biesiedina A. A., Kniazkova P. V., Harbuzova Y. A. The analysis of association between ENPP1 K121Q polymorphism and risk factors of type 2 diabetes mellitus in ukrainian population. Wiadomosci Lekarskie . 2018;71(4):815–820. [PubMed] [Google Scholar]
- 37.Jai P., Balraj M., Shally A., Agarwal C. G., Neena S. K121Q ENPP1/PC-1 gene polymorphism is associated with insulin resistance in a north Indian population. Journal of Genetics . 2013;92(3):571–576. doi: 10.1007/s12041-013-0287-2. [DOI] [PubMed] [Google Scholar]
- 38.Yako Y. Y., Madubedube J. H., Kengne A. P., Erasmus R. T., Pillay T. S., Matsha T. E. Contribution of ENPP1, TCF7L2, and FTO polymorphisms to type 2 diabetes in mixed ancestry ethnic population of South Africa. African Health Sciences . 2015;15(4):1149–1160. doi: 10.4314/ahs.v15i4.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsiao T. J., Lin E. The ENPP1 K121Q polymorphism is associated with type 2 diabetes and related metabolic phenotypes in a Taiwanese population. Molecular and Cellular Endocrinology . 2016;433:20–25. doi: 10.1016/j.mce.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Li Y. Y. ENPP1 K121Q polymorphism and type 2 diabetes mellitus in the Chinese population: a meta-analysis including 11 855 subjects. Metabolism-Clinical & Experimental . 2012;61(5):625–633. doi: 10.1016/j.metabol.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Fajar J. K. The association of ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) K121Q gene polymorphism with the risk of type 2 diabetes mellitus in European, American, and African populations: a meta-analysis. Journal of Health Sciences . 2016;6(2):76–86. doi: 10.17532/jhsci.2016.358. [DOI] [Google Scholar]
- 42.Alves S. D., Piucco B. M., Marmontel S. B., et al. Association between the ENPP1 K121Q polymorphism and risk of diabetic kidney disease: a systematic review and meta-analysis. PLoS One . 2015;10(3, article e0118416) doi: 10.1371/journal.pone.0118416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golbon P., Esmaeilzadeh A., Mahmazi S. Association of ENPP1 (K121Q rs 1044498) and TCF7L2 (C/T rs7903146) gene polymorphisms with Type2 diabetes in Zanjan population (northwest, Iran) Journal of Advances in Medical and Biomedical Research . 2018;26(118):9–14. doi: 10.30699/jambs.26.118.9. [DOI] [Google Scholar]
- 44.Albegali A. A., Shahzad M., Ullah M. I., Mahmood S., Rashid M. Association of genetic polymorphism of PC-1 gene (rs1044498 Lys121Gln) with insulin-resistant type 2 diabetes mellitus in Punjabi population of Pakistan. Molecular Genetics & Genomic Medicine . 2019;7(8):e775–e775. doi: 10.1002/mgg3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasudevan R., Ismail P., Ali A., Mansor M. S. No association of TCF7L2 and ENPP1 gene polymorphisms in Malaysian type 2 diabetes mellitus with or without hypertension. Research Journal of Biological Sciences . 2009;6:703–709. [Google Scholar]
- 46.Mtiraoui N., Turki A., Nemr R., et al. Contribution de variants frequents de ENPP1 , IGF2BP2 , KCNJ11 , MLXIPL , PPARγ, SLC30A8 et TCF7L2 au risque de diabete de type 2 dans des populations arabes libanaise et tunisienne. Diabetes & Metabolism . 2012;38(5):444–449. doi: 10.1016/j.diabet.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Shi X., Wang L., Jin F., et al. The ENPP1 K121Q polymorphism is not associated with type 2 diabetes in northern Chinese. Acta Diabetologica . 2011;48(4):303–310. doi: 10.1007/s00592-011-0281-1. [DOI] [PubMed] [Google Scholar]
- 48.Zhao T., Liu Z., Zhang D., et al. The ENPP1 K121Q polymorphism is not associated with type 2 diabetes or obesity in the Chinese Han population. Journal of Human Genetics . 2011;56(1):12–16. doi: 10.1038/jhg.2010.124. [DOI] [PubMed] [Google Scholar]
- 49.Rasmussen S. K., Urhammer S. A., Pizzuti A., et al. The K121Q variant of the human PC-1 gene is not associated with insulin resistance or type 2 diabetes among Danish Caucasians. Diabetes . 2000;49(9):1608–1611. doi: 10.2337/diabetes.49.9.1608. [DOI] [PubMed] [Google Scholar]
- 50.Sumi S., Ramachandran S., Ramankutty V., et al. ENPP1 121Q functional variant enhances susceptibility to coronary artery disease in South Indian patients with type 2 diabetes mellitus. Molecular and Cellular Biochemistry . 2017;435(1-2):67–72. doi: 10.1007/s11010-017-3057-2. [DOI] [PubMed] [Google Scholar]
- 51.Chakraborty C., Doss C. G. P., Bandyopadhyay S., Sarkar B. K. Mapping the structural topology of IRS family cascades through computational biology. Cell Biochemistry & Biophysics . 2013;67(3):1319–1331. doi: 10.1007/s12013-013-9664-y. [DOI] [PubMed] [Google Scholar]
- 52.Chiranjib C., Govindasamy A., Hsu M. J. Exploring the evolutionary relationship of insulin receptor substrate family using computational biology. PLoS One . 2011;6(2, article e16580) doi: 10.1371/journal.pone.0016580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taniguchi C., Emanuelli B., Kahn C. Critical nodes in signalling pathways: insights into insulin action. Nature Reviews Molecular Cell Biology . 2006;7(2):85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 54.Tseng Y., Butte A., Kokkotou E., et al. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nature Cell Biology . 2005;7(6):601–611. doi: 10.1038/ncb1259. [DOI] [PubMed] [Google Scholar]
- 55.Araki E., Lipes M. A., Patti M. E., et al. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature . 1994;372(6502):186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 56.Kubota N., Tobe K., Terauchi Y., et al. Disruption of insulin receptor substrate 2 causes type 2 diabetes because of liver insulin resistance and lack of compensatory beta-cell hyperplasia. Diabetes . 2000;49(11):1880–1889. doi: 10.2337/diabetes.49.11.1880. [DOI] [PubMed] [Google Scholar]
- 57.Taniguchi C., Ueki K., Kahn C. Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. The Journal of Clinical Investigation . 2016;126(11):p. 4387. doi: 10.1172/JCI90689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang C., Thirone A., Huang X., Klip A. Differential contribution of insulin receptor substrates 1 versus 2 to insulin signaling and glucose uptake in L6 myotubes. The Journal of Biological Chemistry . 2005;280(19):19426–19435. doi: 10.1074/jbc.M412317200. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y., Zhu S., Cai C., et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Science Foundation in China . 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 60.Kabuta T., Take K., Kabuta C., Hakuno F., Takahashi S. I. Differential subcellular localization of insulin receptor substrates depends on C-terminal regions and importin β. Biochemical & Biophysical Research Communications . 2008;377(3):741–746. doi: 10.1016/j.bbrc.2008.09.106. [DOI] [PubMed] [Google Scholar]
- 61.Ikink G. J., Boer M., Bakker E., Hilkens J. IRS4 induces mammary tumorigenesis and confers resistance to HER2-targeted therapy through constitutive PI3K/AKT-pathway hyperactivation. Nature Communications . 2016;7(1, article 13567) doi: 10.1038/ncomms13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gray S. G., Al-Sarraf N., Baird A. M., Gately K., O’Byrne K. J. Transcriptional regulation of IRS5/DOK4 expression in non -small-cell lung cancer cells. Clinical Lung Cancer . 2008;9(6):367–374. doi: 10.3816/CLC.2008.n.053. [DOI] [PubMed] [Google Scholar]
- 63.Cai D., Dhe-Paganon S., Melendez P. A., Lee J., Shoelson S. E. Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. Journal of Biological Chemistry . 2003;278(28):25323–25330. doi: 10.1074/jbc.M212430200. [DOI] [PubMed] [Google Scholar]
- 64.Le F. S., Le S. C., Bougnères P. Increased insulin resistance in obese children who have both 972 IRS-1 and 1057 IRS-2 polymorphisms. Diabetes . 2002;51(Supplement 3):S304–S307. doi: 10.2337/diabetes.51.2007.s304. [DOI] [PubMed] [Google Scholar]
- 65.Bedair R. N., Magour G. M., Ooda S. A., Amar E. M., Awad A. M. Insulin receptor substrate-1 G972R single nucleotide polymorphism in Egyptian patients with chronic hepatitis C virus infection and type 2 diabetes mellitus. Egyptian Liver Journal . 2021;11(1) doi: 10.1186/s43066-020-00069-1. [DOI] [Google Scholar]
- 66.Haghani K., Bakhtiyari S. The study on the relationship between IRS-1 Gly972Arg and IRS-2 Gly1057Asp polymorphisms and type 2 diabetes in the Kurdish ethnic group in West Iran. Genetic Testing and Molecular Biomarkers . 2012;16(11):1270–1276. doi: 10.1089/gtmb.2012.0160. [DOI] [PubMed] [Google Scholar]
- 67.Alsalman H. A., Kaabi Y. A. Lack of association between the insulin receptor substrates-1 Gly972Arg polymorphism and type-2 diabetes mellitus among Saudis from Eastern Saudi Arabia. Saudi Medical Journal . 2015;36(12):1420–1424. doi: 10.15537/smj.2015.12.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Albegali A. A., Shahzad M., Mahmood S., Ullah M. I. Genetic association of insulin receptor substrate-1 (IRS-1, rs1801278) gene with insulin resistant of type 2 diabetes mellitus in a Pakistani population. Molecular Biology Reports . 2019;46(6):6065–6070. doi: 10.1007/s11033-019-05041-w. [DOI] [PubMed] [Google Scholar]
- 69.Ghalehnoo Z. R. Investigating the association of Gly972Arg and Ala512Pro polymorphism of IRS1 gene with type 2 diabetes in Sistan and Baluchistan province. Journal of Kerman University of Medical Sciences . 2021;5(2):56–66. [Google Scholar]
- 70.Fadiel A., Hamza O. B. Polymorphism study of the insulin receptor substrate IRS1 and IRS2 genes associated with type 2 diabetes in ethnic groups of Djerba island. Clinical Medicine Reviews in Vascular Health . 2010;2(2):185–190. doi: 10.4137/CMRVH.S3378. [DOI] [Google Scholar]
- 71.Bodhini D., Radha V., Mohan V. Association study ofIRS1gene polymorphisms with type 2 diabetes in South Indians. Diabetes Technology & Therapeutics . 2011;13(7):767–772. doi: 10.1089/dia.2011.0017. [DOI] [PubMed] [Google Scholar]
- 72.Alharbi K. K., Khan I. A., Abotalib Z., Al-Hakeem M. M. Insulin receptor substrate-1 (IRS-1) Gly927Arg: correlation with gestational diabetes mellitus in Saudi women. BioMed Research International . 2014;2014:5. doi: 10.1155/2014/146495.146495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barseem N. F., Khattab E., Dawood R., Mohamed S. GST T1, M1, and IRS-1 G972R genetic variants association to gestational diabetes mellitus (GDM) in Egyptian women: linkage to maternal hyperglycemia. Current Diabetes Reviews . 2022;18(2) doi: 10.2174/1573399817666210219124628. [DOI] [PubMed] [Google Scholar]
- 74.Yenzeel J., Hussain Z. K., Hassani H. H. Genetic variation of IRS1 gene in women with gestational diabetes mellitus in third trimester stage in Iraq. Iraqi Journal of Science . 2018;59(3A):1176–1182. doi: 10.24996/ijs.2018.59.3A.4. [DOI] [Google Scholar]
- 75.Pappa K. I., Gazouli M., Economou K., et al. Gestational diabetes mellitus shares polymorphisms of genes associated with insulin resistance and type 2 diabetes in the Greek population. Gynecological Endocrinology . 2011;27(4):267–272. doi: 10.3109/09513590.2010.490609. [DOI] [PubMed] [Google Scholar]
- 76.Le Fur S., Le Stunff C., Bougnères P. Increased insulin resistance in obese children who have both 972IRS-1and 1057IRS-2polymorphisms. Diabetes . 2002;51(Supplement 3):S304–S307. doi: 10.2337/diabetes.51.2007.S304. [DOI] [PubMed] [Google Scholar]
- 77.Mehrnoosh K., Samaneh E., Ozra T. M., Amoli M. M. Association between genetic variants and diabetes mellitus in Iranian populations: a systematic review of observational studies. Journal of Diabetes Research . 2015;2015:21. doi: 10.1155/2015/585917.585917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhatt S., Guleria R. Association of IRS1 (Gly972Arg) and IRS2 (Gly1057Asp) genes polymorphisms with OSA and NAFLD in Asian Indians. PloS one . 2021;16(2, article e0245408) doi: 10.1371/journal.pone.0245408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ayaz L., Celik S. K., Cayan F. The G1057D polymorphism of insulin receptor substrate-2 associated with gestational diabetes mellitus. Gynecological Endocrinology . 2014;30(2):165–168. doi: 10.3109/09513590.2013.871516. [DOI] [PubMed] [Google Scholar]
- 80.Bappy H., Goswami A., Huda N., Hosen M. I., Nabi A. Gender specific association of missense variant rs1805097 of IRS-2 and noncoding variant rs841853 of GLUT-1 genes with susceptibility to type 2 diabetes in Bangladeshi population. Gene Reports . 2020;21, article 100866 doi: 10.1016/j.genrep.2020.100866. [DOI] [Google Scholar]
- 81.Ag A., Mw A., Bca B., et al. Polymorphism analysis of the Gly972Arg IRS-1 and Gly1057Asp IRS-2 genes in obese pregnant women. Reproductive Biology . 2020;20(3):365–370. doi: 10.1016/j.repbio.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 82.Melkersson K., Persson B. P-709 - association between body mass index and insulin receptor substrate-4 (IRS-4) gene polymorphisms in patients with schizophrenia. Neuro Endocrinology Letters . 2012;27(5):1–1. doi: 10.1016/S0924-9338(12)74876-9. [DOI] [PubMed] [Google Scholar]
- 83.Almind K., Frederiksen S. K., Ahlgren M. G., et al. Common amino acid substitutions in insulin receptor substrate-4 are not associated with type II diabetes mellitus or insulin resistance. Diabetologia . 1998;41(8):969–974. doi: 10.1007/s001250051015. [DOI] [PubMed] [Google Scholar]
- 84.Trung T., Binh T. Q. The Association between the Gly972Arg Polymorphism in IRS1 Gene and the Risk of Prediabetes among Vietnamese Women. VNU Journal of Science: Medical and Pharmaceutical Sciences . 2018;34(2) doi: 10.25073/2588-1132/vnumps.4129. [DOI] [Google Scholar]
- 85.Alharbi K. K., Khan I. A., Munshi A., Alharbi F. K., Al-Sheikh Y., Alnbaheen M. S. Association of the genetic variants of insulin receptor substrate 1 (IRS-1) with type 2 diabetes mellitus in a Saudi population. Endocrine . 2014;47(2):472–477. doi: 10.1007/s12020-014-0177-2. [DOI] [PubMed] [Google Scholar]
- 86.Shakeri H., Khoshi A., Kaffash Bajestani M., et al. Association of IRS1 Gly971Arg gene polymorphism with insulin resistance in Iranian newly diagnosed diabetic adults. Acta Endocrinologica . 2019;15(3):317–322. doi: 10.4183/aeb.2019.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ouederni T. B., Sanchez-Corona J., Martinez S., et al. The G1057D polymorphism of IRS-2 gene is not associated with type 2 diabetes and obese patients among ethnic groups in Tunisian population. Clinical Biochemistry . 2009;42(10-11):1169–1173. doi: 10.1016/j.clinbiochem.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 88.Chan S. H., Chen J. H., Li Y. H., Tsai L. M. Gly1057Asp polymorphism of insulin receptor substrate-2 is associated with coronary artery disease in the Taiwanese population. Journal of Biomedical Science . 2012;19(1):p. 100. doi: 10.1186/1423-0127-19-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bodhini D., Radha V., Deepa R., et al. The G1057D polymorphism of IRS-2 gene and its relationship with obesity in conferring susceptibility to type 2 diabetes in Asian Indians. International Journal of Obesity . 2007;31(1):97–102. doi: 10.1038/sj.ijo.0803356. [DOI] [PubMed] [Google Scholar]
- 90.Alipourfard I., Datukishvili N., Mikeladze D. TNF-α downregulation modifies insulin receptor substrate 1 (IRS-1) in metabolic signaling of diabetic insulin-resistant hepatocytes. Mediators of Inflammation . 2019;2019:6. doi: 10.1155/2019/3560819.3560819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shibata T., Takaguri A., Ichihara K., Satoh K. Inhibition of the TNF-α–Induced serine phosphorylation of IRS-1 at 636/639 by AICAR. Journal of Pharmacological Sciences . 2013;122(2):93–102. doi: 10.1254/jphs.12270FP. [DOI] [PubMed] [Google Scholar]
- 92.Sharfi H., Eldar-Finkelman H. Sequential phosphorylation of insulin receptor substrate-2 by glycogen synthase kinase-3 and c-Jun NH2-terminal kinase plays a role in hepatic insulin signaling. American Journal of Physiology Endocrinology & Metabolism . 2008;294(2):E307–E315. doi: 10.1152/ajpendo.00534.2007. [DOI] [PubMed] [Google Scholar]
- 93.Ziva L. Coordinated phosphorylation of insulin receptor substrate-1 by glycogen synthase kinase-3 and protein kinase CβII in the diabetic fat tissue. American Journal of Physiology Endocrinology & Metabolism . 2008;294(6):E1169–E1177. doi: 10.1152/ajpendo.00050.2008. [DOI] [PubMed] [Google Scholar]
- 94.Liberman Z., Eldar-Finkelman H. Serine 332 phosphorylation of insulin receptor substrate-1 by glycogen synthase kinase-3 attenuates insulin signaling. Journal of Biological Chemistry . 2005;280(6):4422–4428. doi: 10.1074/jbc.M410610200. [DOI] [PubMed] [Google Scholar]
- 95.Yang S. J., Xu C. Q., Wu J. W., Yang G. S. SOCS3 inhibits insulin signaling in porcine primary adipocytes. Molecular and Cellular Biochemistry . 2010;345(1-2):45–52. doi: 10.1007/s11010-010-0558-7. [DOI] [PubMed] [Google Scholar]
- 96.Parish R., Petersen K. F. Mitochondrial dysfunction and type 2 diabetes. Current Diabetes Reports . 2005;5(3):177–183. doi: 10.1007/s11892-005-0006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Werner E. D., Lee J., Hansen L., Yuan M., Shoelson S. E. Insulin Resistance Due to Phosphorylation of Insulin Receptor Substrate-1 at Serine 302. Journal of Biological Chemistry . 2004;279(34):35298–35305. doi: 10.1074/jbc.M405203200. [DOI] [PubMed] [Google Scholar]
- 98.Ragheb R., Shanab G., Medhat A. M., Seoudi D. M., Adeli K., Fantus I. G. Free fatty acid-induced muscle insulin resistance and glucose uptake dysfunction: evidence for PKC activation and oxidative stress-activated signaling pathways. Biochemical & Biophysical Research Communications . 2009;389(2):211–216. doi: 10.1016/j.bbrc.2009.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lyu K., Zhang Y., Zhang D., et al. A Membrane-Bound Diacylglycerol Species Induces PKCϵ-Mediated Hepatic Insulin Resistance. Cell Metabolism . 2020;32(4):654–664.e5. doi: 10.1016/j.cmet.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Glass C., Olefsky J. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metabolism . 2012;15(5):635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson A. F., Olefsky J. The origins and drivers of insulin resistance. Cell . 2013;152(4):673–684. doi: 10.1016/j.cell.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 102.Pende M., Kozma S. C., Jaquet M., et al. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature . 2000;408(6815):994–997. doi: 10.1038/35050135. [DOI] [PubMed] [Google Scholar]
- 103.Tanti J. F., Jager J. Cellular mechanisms of insulin resistance: role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Current Opinion in Pharmacology . 2009;9(6):753–762. doi: 10.1016/j.coph.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 104.Luo M., Paul L., Yi Z., et al. Phosphorylation of human insulin receptor substrate-1 at serine 629 plays a positive role in insulin signaling. Endocrinology . 2007;148(10):4895–4905. doi: 10.1210/en.2007-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo M., Sara R., Wang L., et al. Identification of insulin receptor substrate 1 serine/threonine phosphorylation sites using mass spectrometry analysis: regulatory role of serine 1223. Endocrinology . 2005;146(10):4410–4416. doi: 10.1210/en.2005-0260. [DOI] [PubMed] [Google Scholar]
- 106.Greene M. W., Garofalo R. S. Positive and negative regulatory role of insulin receptor substrate 1 and 2 (IRS-1 and IRS-2) serine/threonine phosphorylation. Biochemistry . 2002;41(22):7082–7091. doi: 10.1021/bi015992f. [DOI] [PubMed] [Google Scholar]
- 107.Giraud J., Leshan R., Lee Y. H., White M. F. Nutrient-dependent and insulin-stimulated phosphorylation of insulin receptor substrate-1 on serine 302 correlates with increased insulin signaling. Journal of Biological Chemistry . 2004;279(5):3447–3454. doi: 10.1074/jbc.M308631200. [DOI] [PubMed] [Google Scholar]
- 108.Weigert C., Kron M., Kalbacher H., et al. Interplay and effects of temporal changes in the phosphorylation state of serine-302, -307, and -318 of insulin receptor substrate-1 on insulin action in skeletal muscle cells. Molecular Endocrinology . 2008;22(12):2729–2740. doi: 10.1210/me.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weigert C., Hennige A., Brischmann T., et al. The phosphorylation of Ser318 of insulin receptor substrate 1 is not per se inhibitory in skeletal muscle cells but is necessary to trigger the attenuation of the insulin-stimulated signal. Journal of Biological Chemistry . 2005;280(45):37393–37399. doi: 10.1074/jbc.M506134200. [DOI] [PubMed] [Google Scholar]
- 110.Chuan-Hsiang H., Diana M., Oleg S. K., et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science . 2007;318(5857):1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 111.Lazaros C., Foukas C., Foukas L. C., et al. Regulation of phosphoinositide 3-kinase by its intrinsic serine kinase activity in vivo. Molecular and Cellular Biology . 2004;24(3):966–975. doi: 10.1128/MCB.24.3.966-975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Copps K. D., White M. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia . 2012;55(10):2565–2582. doi: 10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mauvais-Jarvis F., Ueki K., Fruman D. A., et al. Reduced expression of the murine p85α subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. Journal of Clinical Investigation . 2002;109(1):141–149. doi: 10.1172/JCI0213305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ueki K., Yballe C. M., Brachmann S. M., et al. Increased insulin sensitivity in mice lacking p85β subunit of phosphoinositide 3-kinase. Proceedings of the National Academy of Sciences of the United States of America . 2002;99(1):419–424. doi: 10.1073/pnas.012581799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ueki K., Fruman D. A., Yballe C. M., et al. Positive and negative roles of p85α and p85β regulatory subunits of phosphoinositide 3-kinase in insulin signaling. Journal of Biological Chemistry . 2017;292(13):5608–5608. doi: 10.1074/jbc.A117.305602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barbour L. A., Mizanoor Rahman S., Gurevich I., et al. Increased P85α is a potent negative regulator of skeletal muscle insulin signaling and induces in vivo insulin resistance associated with growth hormone excess. Journal of Biological Chemistry . 2005;280(45):37489–37494. doi: 10.1074/jbc.M506967200. [DOI] [PubMed] [Google Scholar]
- 117.Luo J., Field S., Lee J., Engelman J., Cantley L. The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex. The Journal of Cell Biology . 2005;170(3):455–464. doi: 10.1083/jcb.200503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hirosumi J., Tuncman G., Chang L., et al. A central role for JNK in obesity and insulin resistance. Nature . 2002;420(6913):333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 119.Hu S., Ma S., Xun L., Tian Z., Tan H. Relationships of SLC2A4, RBP4, PCK1, and PI3K Gene Polymorphisms with Gestational Diabetes Mellitus in a Chinese Population. BioMed Research International . 2019;2019:9. doi: 10.1155/2019/7398063.7398063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karadoğan A., Arikoglu H., Göktürk F., İşçioğlu F., İpekçi S. PIK3R1 gene polymorphisms are associated with type 2 diabetes and related features in the Turkish population. Advances in Clinical and Experimental Medicine . 2018;27(7):921–927. doi: 10.17219/acem/68985. [DOI] [PubMed] [Google Scholar]
- 121.Tarquini F., Picchiassi E., Centra M., et al. Body mass index associated to rs2021966 ENPP1 polymorphism increases the risk for gestational diabetes mellitus. Gynecological Endocrinology . 2015;31(1):83–86. doi: 10.3109/09513590.2014.958994. [DOI] [PubMed] [Google Scholar]
- 122.Ito Y., Sun R., Yagimuma H., et al. Protein tyrosine phosphatase 1B deficiency improves glucose homeostasis in type 1 diabetes treated with leptin. Diabetes . 2022;71(9):1902–1914. doi: 10.2337/db21-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ali M., Jannat S., Jung H., Choi J. Structural bases for hesperetin derivatives: inhibition of protein tyrosine phosphatase 1B, kinetics mechanism and molecular docking study. Molecules . 2021;26(24):p. 7433. doi: 10.3390/molecules26247433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rampadarath A., Balogun F. O., Pillay C., Sabiu S., Campesi I. Identification of flavonoid C-glycosides as promising antidiabetics targeting protein tyrosine phosphatase 1B. Journal of Diabetes Research . 2022;2022:11. doi: 10.1155/2022/6233217.6233217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen L., Zeng H., Qin H., Ruan X., Yang P. CD36 gene deletion reduces muscle insulin sensitivity in mice by up-regulating PTP1B expression. Journal of Southern Medical University . 2022;42(3):392–398. doi: 10.12122/j.issn.1673-4254.2022.03.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rath P., Ranjan A., Ghosh A., et al. Potential therapeutic target protein tyrosine phosphatase-1B for modulation of insulin resistance with polyphenols and its quantitative structure-activity relationship. Molecules . 2022;27(7):p. 2212. doi: 10.3390/molecules27072212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li D., Zhang S., Yang C., et al. A novel PTP1B inhibitor-phosphate of polymannuronic acid ameliorates insulin resistance by regulating IRS-1/Akt signaling. International Journal of Molecular Sciences . 2021;22(23):p. 12693. doi: 10.3390/ijms222312693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rath P., Ranjan A., Chauhan A., et al. A critical review on role of available synthetic drugs and phytochemicals in insulin resistance treatment by targeting PTP1B. Applied Biochemistry and Biotechnology . 2022;194(10):4683–4701. doi: 10.1007/s12010-022-04028-x. [DOI] [PubMed] [Google Scholar]


