Abstract
Introduction
Despite advancements in molecular and histopathologic characterization of pediatric low-grade gliomas (pLGGs), there remains significant phenotypic heterogeneity among tumors with similar categorizations. We hypothesized that an unsupervised machine learning approach based on radiomic features may reveal distinct pLGG imaging subtypes.
Methods
Multi-parametric MR images (T1 pre- and post-contrast, T2, and T2 FLAIR) from 157 patients with pLGGs were collected and 881 quantitative radiomic features were extracted from tumorous region. Clustering was performed using K-means after applying principal component analysis (PCA) for feature dimensionality reduction. Molecular and demographic data was obtained from the PedCBioportal and compared between imaging subtypes.
Results
K-means identified three distinct imaging-based subtypes. Subtypes differed in mutational frequencies of BRAF (p < 0.05) as well as the gene expression of BRAF (p<0.05). It was also found that age (p < 0.05), tumor location (p < 0.01), and tumor histology (p < 0.0001) differed significantly between the imaging subtypes.
Conclusion
In this exploratory work, it was found that clustering of pLGGs based on radiomic features identifies distinct, imaging-based subtypes that correlate with important molecular markers and demographic details. This finding supports the notion that incorporation of radiomic data could augment our ability to better characterize pLGGs.
Keywords: Radiomics, Radiogenomics, Pediatric low-grade glioma, Unsupervised machine learning
Introduction
Brain tumors are the most common solid malignancy in pediatric populations and pediatric low-grade gliomas (pLGGs) are the most common type of childhood brain tumor [1]. pLGGs are a large and heterogenous group of tumors which include pilocytic astrocytomas (PAs), pleomorphic xanthoastrocytomas (PXAs), diffuse low grade astrocytomas, and many others [2]. Together, these tumors account for 40% of central nervous system tumors in children [1]. While surgical resection remains a mainstay of pLGG treatment with curative potential in the case of total resection, cases in which this is not possible go on to become chronic and morbid conditions that greatly impact the patient's quality of life [2,3]. While overall survival of patients with pLGGs who undergo standard chemotherapy and radiation therapy are quite high, the 10-year progression-free survival rate is less than 50% [4].
Despite integration of molecular data in the characterization of pLGGs and their use in aiding chemotherapeutic selection, patients with molecularly similar tumors often respond differently to treatments [5]. This in turn results in unnecessary morbidity due to the futility of the agent chosen to treat the disease which necessitates further characterization of tumors beyond the current state of the field. Radiomic data shows promise in filling this knowledge gap. In adult GBM, based on radiomic features extracted from multi-parametric MRI, distinct imaging subtypes that correspond to differences in key disease characteristics including mortality and the underlying molecular subtypes have been found [6]. It is possible that similar relationships exist in pLGGs, lending credence to the idea that radiomic data could independently add to the ability to characterize tumors more comprehensively.
While radiomics has been implemented in pLGGs to work towards pretherapeutic differentiation of molecular subtypes in a supervised fashion [7], to our knowledge there has been no work done to identify radiomic subtypes of pLGGs in an unsupervised approach. An unsupervised radiomic analysis approach allows for the exploration and characterization of the underlying heterogeneity in the structural patterns, reflected in radiographic phenotypes and quantified by radiomic features. These pattern analysis methods have the potential to elucidate the biological processes that form similar or dissimilar imaging phenotypes and clinical presentation of the tumors among patients. In principle, imaging subtypes, similar to molecular subtypes, are characterized through clustering methods that group the most similar tumors together in a subtype, in a way that they are most distinctive from the other subtypes. Such unbiased data-driven approaches have found popularity in several neuroimaging studies and helped in better understanding of the disease biology [8].
We hypothesize that specific imaging subtypes can be identified through a data-driven approach and that certain imaging characteristics are associated with molecular markers. Therefore, by grouping the tumors based on their common imaging characteristics, similar underpinnings of these tumor may be captured as well. Thus, using unsupervised machine learning methods to analyze radiomic data obtained from multi-parametric MRI (mpMRI) modalities collected through standard clinical protocols, we aimed to identify imaging-based subtypes of pLGGs. Additionally, we sought to explore the relationships of these subtypes with key molecular markers and patient demographic information.
Methods
Subjects
The data for this study was retrospectively collected from the Children's Hospital of Philadelphia (CHOP). The patient selection was based on availability of specimen/sequencing and pre-surgical and pre-treatment multi-parametric MRI scans. Raw genomic and imaging data were available through the Children's Brain Tumor Network (www.cbtn.org) and were accessed through the Gabriella Miller Kids First Portal (https://kidsfirstdrc.org/). Processed genomic data was obtained from the Open Pediatric Brain Tumor Atlas (https://github.com/AlexsLemonade/OpenPBTA-analysis). Clinical information was collected through the PedcBioPortal (https://pedcbioportal.kidsfirstdrc.org:443/saml/discovery?entityID=d3b-center.auth0.com&returnIDParam=idp) [9,10].
157 subjects were included in the study based on the following criteria: 1) less than 18 years of age at the time of initial imaging; 2) availability of pre-treatment pre- and post-contrast T1-weighted images (T1, T1-Gd), T2-weighted, and T2- fluid-attenuated inversion recovery (T2-FLAIR) MR imaging; 3) confirmed diagnosis of pediatric low-grade glioma (pLGG) as determined by pathology report (Table 1).
Table 1.
Cohort characteristics.
| Age Range (at time of tissue collection) | 0-22* years | ||
|---|---|---|---|
| Sex | Male | 78 | 49.68% |
| Female | 79 | 50.32% | |
| Race | White | 78 | 49.68% |
| Black | 15 | 9.55% | |
| Asian | 4 | 2.55% | |
| American Indian | 1 | 0.64% | |
| Other | 17 | 10.83% | |
| NA | 42 | 26.75% | |
| Tumor Tissue Site | Posterior Fossa | 75 | 47.77% |
| Suprasellar | 34 | 21.66% | |
| Supratentorial (Hemispheric) | 29 | 18.47% | |
| Ventricular | 9 | 5.73% | |
| Other | 10 | 6.37% | |
| Tumor Histologies | Pilocytic Astrocytoma (JPA) | 91 | 57.96% |
| Fibrillary Astrocytoma | 15 | 9.55% | |
| Diffuse Astrocytoma | 10 | 6.37% | |
| Low Grade Glioma (Not otherwise Specified) | 8 | 5.10% | |
| Pilomyxoid Astrocytoma | 7 | 4.46% | |
| Pilocytic/Pilomyxoid Astrocytoma | 6 | 3.82% | |
| Subependymal Giant Cell Astrocytoma | 4 | 2.55% | |
| Pleomorphic Xanthoastrocytomas | 4 | 2.55% | |
| Angiocentric glioma | 3 | 1.91% | |
| Oligodendroglioma | 2 | 1.27% | |
| Ganglioma | 2 | 1.27% | |
| Pilocytic and Diffuse Astrocytoma | 2 | 1.27% | |
| Ganglion Cell Tumor | 1 | 0.64% | |
| Diffuse Infiltrating Astrocytoma | 1 | 0.64% | |
| Glioneuronal Tumor | 1 | 0.64% |
Ages of patients at time of imaging was under 18. Biopsy, for some patients, occurred years after initial imaging.
Molecular analysis
All molecular data was obtained from the PedCBioportal (https://pedcbioportal.kidsfirstdrc.org), which is an open access data visualization platform that hosts multi-institutional data across multiple cancerous and non-cancerous pathologies [9,10]. Gene mutations data on the portal were sourced from clinical gene panels and whole genome sequencing, where available. Expressions data was obtained through RNA sequencing where available.
Initial molecular analysis was focused on 10 genes of interest that play a role in the pathogenesis of pLGG: BRAF, FGFR1, TSC1, TSC2, NF1, MYB, EGFR, ALK, IDH1, and RB1.
Histology data was obtained from review of pathology reports and included all cases that met the WHO grade I/II criteria for Low Grade Glioma.
MRI data acquisition
MR imaging was originally acquired as standard of practice for all subjects and accessed retrospectively for this study. All imaging was acquired on 1.5-3T scanners from Siemens and General Electric manufacturers. T1-weighted pre- (T1) and post-contrast (T1-Gd), T2-weighted, and T2-FLAIR imaging sequences were included in this study. Only images acquired prior to treatment were included. For each patient, the earliest imaging event that included all four modalities were selected.
Pre-processing
mpMRI volumes were pre-processed within the Cancer Imaging Phenomics Toolkit open-source software (CaPTk, https://www.cbica.upenn.edu/captk) [11,12] utilizing methods previously described in detail [13]. In summary, images were co-registered to an atlas [14] and resampled to a 1 × 1 × 1 mm3 resolution utilizing the Greedy tool (https://github.com/pyushkevich/greedy) [15]. Skull stripping was then performed using BrainMaGe [16]. As this method is developed for adult brain tumors and cannot always correctly segment pediatric scans, brain masks were revised manually. The skull-stripped images were then corrected for intensity non-uniformity. Pixels with intensities beyond the 99.9th percentile were removed, and image intensities were rescaled to the range of 0 and 255.
Tumor segmentation
Tumor segmentation was performed in a semi-automatic fashion. Pre-processed MR-images were first segmented automatically by a deep learning segmentation model trained to segment adult brain tumors. [17] Tumor regions of interest (ROIs) were then manually revised using ITK-SNAP [18] (http://www.itksnap.org/) and edited/confirmed by board-certified experienced neuroradiologists (A.N, A.V, J.W) to create final ROIs.
Radiomic feature extraction
881 MR-imaging based radiomic features were extracted using CaPTk software from the ROIs across all four imaging modalities. Features included histogram, morphology, and texture-based features. The extracted radiomic features were then z-scored using the normalize function in the preprocessing module of Scikit-learn [19] within Python (Python version 3.9.12).
Unsupervised machine learning
Feature reduction was performed using Principal Component Analysis (PCA) to reduce the dimensionality of the features and reduce noise from the data. PCA components that accounted for 90% of the variance in the data cumulatively were chosen for the clustering step.
K-means clustering was performed on the selected PCA components to identify imaging subtypes that capture similarities in feature profiles between subjects. This clustering was performed in python using Scikit-Learn's KMeans package. A range of 1 to 20 was searched for selecting the optimum number of clusters and the point of inflection of inertia, or within cluster sum of squares, was determined to be the optimal value.
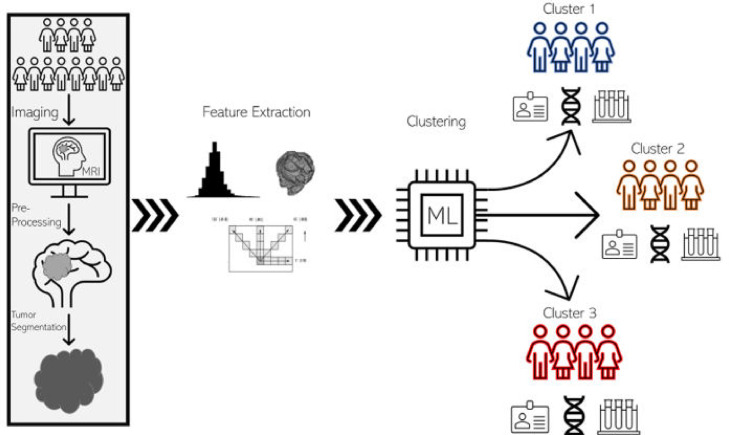
K-means clustering was performed with 10,000 iterations with random initialization to ensure robustness of clustering. A schematic of the entire feature extraction and clustering pipeline is included in Fig. 1.
Fig 1.
Radiomics workflow. The radiomic clustering workflow begins with the feature extraction pipeline, depicted in the rectangle at the left of the schematic. Patients first undergo image acquisition through routine MR imaging protocols. Images are then accessed and pre-processed which involves stripping away extra-axial tissues from the images leaving only the brain and tumor volumes. The tumor is then segmented out leaving only tumor volume. Engineered radiomic features, including texture-based, morphologic, and histogram features, are then extracted from the tumor volumes. The features are then fed into the unsupervised machine learning algorithm, which sorts the patients based on the radiomic features. The end results are distinct groups of patients, categorized based on imaging, that can be compared on their clinical, genetic, and other molecular characteristics.
Statistical analysis
Chi-squared test was used to identify significant differences in categorical variables including mutational frequency between radiomic subtypes, differences in tumor location, and differences in histology. Genes of interest were categorized in a binary fashion (mutated vs wildtype). Histology data obtained from pathology reports was analyzed with each histology as its own category. Histologies that did not occur across all clusters were binned as “other”. Kruskal-Wallis test was used to compare continuous variables including age and gene expressions between clusters. Alpha was set to 0.05 for all analyses.
Assessing feature weights
To provide increased interpretability of the clusters, the weight of each feature on the clustering process was assessed in the following way. Features were normalized using z-score method and the scalar product was found between each feature and PCA component. The absolute value of this product was deemed to be the relative influence of each feature along the vector of the PCA component. These influences were then multiplied by the variance explained by their respective PCA component, resulting in a value that served as a reflection of the influence of the individual feature on the clustering.
Results
Unsupervised learning on radiomic features
PCA produced 48 principal components that explained 90% of the variability in the feature space. Based on inertias, it was found that the optimal number of clusters through K-means analysis on the 48 principal components was 3. Cluster labels were then assigned to each of the subjects based on closest distance to centers of the 3 identified clusters.
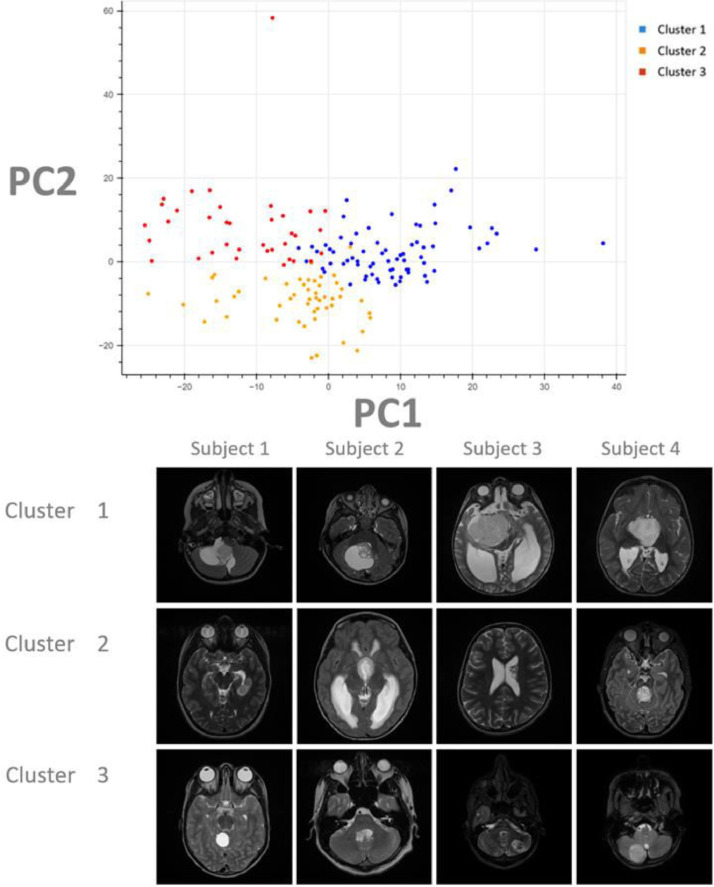
Cluster 1 contained 71 subjects, cluster 2 contained 36 subjects, and cluster 3 contained 50 subjects. These three clusters (subtypes) were utilized for comparisons to molecular status (Fig. 2). Example images were selected from subjects near the center of each cluster and included for illustrative purposes (Fig. 2).
Fig 2.
Clustering projection and illustrative images. In the top chart, the final imaging-based clustering results are depicted here with each point representing a unique subject plotted against the first two principal components (PCs). Each color represents a cluster group (Cluster 1: Blue; Cluster 2: Orange; Cluster 3: Red). In this analysis, the first two PCs explain only 25 percent of the variance in the feature set which may explain the proximity of the clusters on this projection. Although the true clustering is done on 48 dimensions, separation of the subjects can be appreciated even on this two-dimensional projection of the data.
Below the chart, representative images were selected from the T2 Axial MR images from 4 patients in each cluster. These patients were picked from the center-most regions of each cluster and can thus be presumed to be most representative of their groups. Although the full volume of tumor from all 4 modalities (T1 pre-contrast, T1 post-contrast, T2, and FLAIR) was utilized for this work, for illustrative purposes only the axial T2 slice demonstrating the largest diameter of tumor was selected for this figure.
Results are summarized in Table 2
Table 2.
Clustering results.
| Cluster 1 | Cluster 2 | Cluster 3 | p-value | ||
|---|---|---|---|---|---|
| Sex | 0.802 | ||||
| Male | 37 | 18 | 23 | ||
| Female | 34 | 18 | 27 | ||
| Mean Age (years) at Imaging | 10 | 6 | 5 | 0.0334 | |
| Tumor Location | 0.0014 | ||||
| Posterior Fossa | 44 | 15 | 16 | ||
| Suprasellar | 19 | 7 | 8 | ||
| Supratentorial | 4 | 8 | 17 | ||
| Ventricular | 2 | 3 | 4 | ||
| Other | 2 | 3 | 5 | ||
| Tumor Histology | <0.0001 | ||||
| Pilocytic Astrocytoma | 58 | 18 | 17 | ||
| Diffuse Astrocytoma | 1 | 1 | 9 | ||
| Fibrillary Astrocytoma | 3 | 3 | 9 | ||
| Pilomyxoid Astrocytoma | 3 | 2 | 2 | ||
| LGG (Not Otherwise Specified) | 1 | 3 | 4 | ||
| Other | 5 | 9 | 9 | ||
| Mutation Frequencies (percentage of samples altered) | |||||
| BRAF | 20 | 49.25 | 47.22 | 0.0346 | |
| FGFR1 | 4 | 10.45 | 0 | 0.0987 | |
| TSC1 | 28 | 41.79 | 38.89 | 0.479 | |
| TSC2 | 28 | 37.31 | 38.89 | 0.645 | |
| NF1 | 24 | 38.81 | 41.67 | 0.325 | |
| MYB | 0 | 2.99 | 0 | 0.297 | |
| EGFR | 4 | 11.94 | 16.67 | 0.317 | |
| ALK | 4 | 16.42 | 25 | 0.0933 | |
| IDH1 | 8 | 7.46 | 5.56 | 0.916 | |
| RB1 | 24 | 35.82 | 30.56 | 0.544 | |
| Expresion Levels (mean Log2) | |||||
| BRAF | 4.08 | 3.75 | 3.98 | 0.0152 | |
| FGFR1 | 6.34 | 6.35 | 6.56 | 0.459 | |
| TSC1 | 3.61 | 3.43 | 3.58 | 0.0923 | |
| TSC2 | 4.87 | 4.94 | 5.14 | 0.187 | |
| NF1 | 4.95 | 4.83 | 4.85 | 0.707 | |
| MYB | 0.13 | 0.28 | 0.15 | 0.709 | |
| EGFR | 2.19 | 2.55 | 2.07 | 0.308 | |
| ALK | 1.74 | 1.86 | 1.51 | 0.377 | |
| IDH1 | 4.36 | 4.46 | 4.54 | 0.631 | |
| RB1 | 4.63 | 4.58 | 4.7 | 0.521 |
Molecular comparisons of imaging-based subtypes
PedCBioportal's PBTA (provisional) database was queried using subject identifiers from each imaging subtype. From the total 157 pLGG subjects with pre-treatment imaging available, 128 samples had molecular sequencing data available on the portal and were used for exploration of molecular markers.
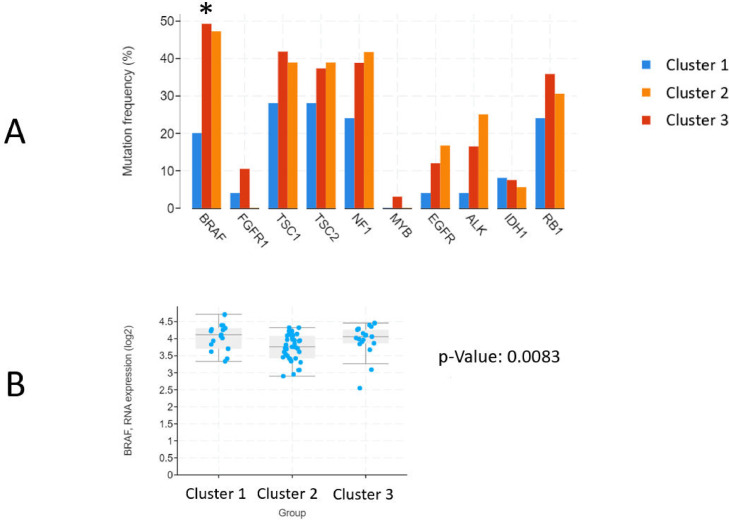
Of the 10 genes of interest described in the above section, only BRAF was found to have significant differences in mutational frequency between the three imaging subtypes (X2 [2, N = 128] = 6.73, p = 0.0346). Cluster 1 had 20%, Cluster 2 had 49.3% and Cluster 3 had 47.2% of samples carrying a BRAF mutation. Of note, this does not include data on structural variants (ie. gene fusions).
Kruskal-Wallis test on expressions data further revealed expressions differences in BRAF between imaging subtypes as well (H = 9.59 [2, N = 73], p = 0.0083). The mean log2 expression of BRAF was 4.08, 3.75, and 3.98 in Clusters 1, 2, and 3 respectively. The remaining genes showed no differences (p > 0.05) in alterations or expressions between subtypes (Fig. 3).
Fig 3.
Molecular analysis. (A) Mutation frequencies of each of the 10 genes of interest are reported. The X-axis contains each of 10 genes of interest. The Y-axis represents percentage of patients in each cluster that contain a mutation in the respective gene. Genes that had significant differences in mutation frequencies between clusters were marked with an asterisk (*) next to the gene symbol on the X-axis. (B) Box-and-whisker plot for BRAF expressions between clusters. Kruskal-Wallis test was used to determine significance of differences observed between clusters, p-value from this analysis is included to the right of the chart.
Additional differences between image-based subtypes
Tissue histology was also compared between imaging subtypes through review of clinical pathology free-text reports generated at the time of initial tissue collection. Chi-squared test suggested that there were significant differences in histology between imaging-based clusters (X2 [10, N = 157] = 39.97, p < 0.0001).
Age at the time of tissue collection was found to be significantly different between clusters based on Kruskal-Wallis test (p = 0.0334) with cluster 1 being the oldest (median = 10 years), as compared to cluster 2 (median = 6 years) and cluster 3 (median = 5 years).
Finally, tumor tissue site was also significantly different between imaging subtypes based on chi-square test (X2 [8, N = 157] = 25.28, p = 0.0014).
Feature weights and informative radiomic features
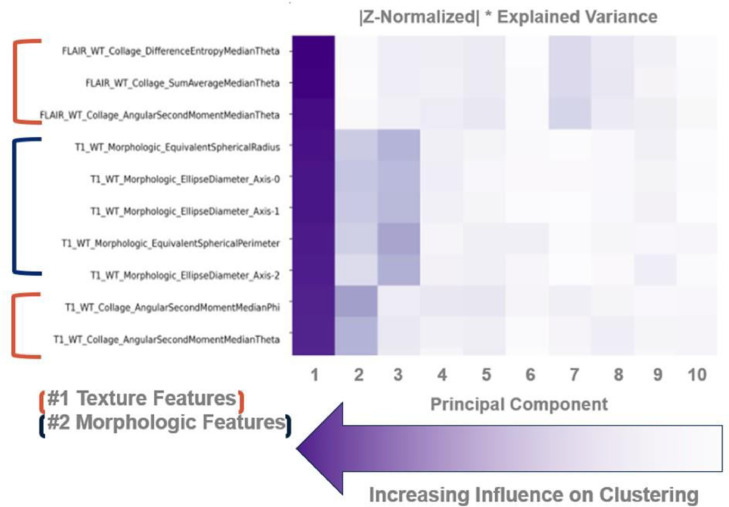
On assessment of feature weights, it was found that texture-based (collage) features and morphologic features, namely those describing the size of the tumors, were most influential in the creation of imaging subtypes. Furthermore, all top ten weighted features were obtained from the morphology of the tumor masks and the FLAIR sequences (Fig. 4).
Fig 4.
Feature weight matrix. This heatmap demonstrates the relative influence of each feature on the overall clustering attempt. Darker cells depict higher values of influence as calculated by the scalar product between the feature vector and the principal component vector multiplied by the explained variance of the respective principal component. By performing this operation, the relative importance of certain types of features and imaging sequences can be understood. Feature names describe the attribute that they measure in the following way: ImagingSequence_FeatureType_Metric.
Discussion
This work, to the best of our knowledge, is the first application of unsupervised machine learning methods in identifying imaging-based subtypes of pediatric low-grade gliomas. The principal contribution of this work is a demonstration that an unbiased approach to high-throughput image analysis can identify imaging-based subtypes that may have an underlying biological basis.
In this work, the unsupervised approach was undertaken to explore the utility of radiomics in explaining phenotypic differences within pLGGs. By focusing primarily on the intra-tumoral radiomic feature-space, this work identifies imaging subtypes without any a priori sub-categorization. In doing so, it allows for flexibility in the classification of these tumors beyond existent categorizations and groupings that are based on molecular information alone. A major advantage of such an approach is its ability to reveal relatively homogenous subtypes from a large group of samples in an unbiased way [20]. Information extracted as a result of such work can add to the metrics, we use to define specific tumors. The current trajectory of tumor classifications supports this type of data layering, with the most recent WHO guidelines incorporating layers of molecular and histologic data in tumor classification [21]. It is possible that an additional layer of radiomic information could be valuable in further refining our understanding of tumors and augmenting our ability to create integrated diagnoses. [22] In other sects of neuroscience, similar approaches have been instrumental in furthering our understanding of adult neuro oncology, neurodegenerative diseases, psychiatric conditions, and autism spectrum disorders [20].
The tumors comprising pLGGs are diverse in their histopathology. On a molecular scale they converge on an upregulation of the mitogen-activated protein kinase (MAPK) pathway through a variety of genomic aberrations [23,24]. Mutations or fusions involving the B-Raf proto-oncogene, serine/threonine kinase (BRAF) gene are the most common alterations found in pLGGs; namely, the BRAF V600E mutation (p.V600E) and KIAA1549-BRAF fusion gene are the two major BRAF alterations that hold clinical significance and are used to guide treatment selection [24,25]. Other common genetic alterations in the MAPK pathway include those in EGFR, FGFR1, MYB, MYBL1, and NTRK2 [24]. Patients with Neurofibromatosis type 1 and tuberous sclerosis also have a predisposition for developing pLGGs due to congenital aberrations in NF1 and TSC1/TSC2, respectively. NF1 is a tumor suppressor gene that encodes neurofibromin 1, a negative regulator of RAS, which is a signal transducer in the MAPK pathway. TSC1 and TSC2 encode for hamartin and tuberin, respectively, which are components of the mTOR pathway downstream of RAS [24]. This background is what informed the selection of the genes of interest in this study.
A major, general criticism of unsupervised tasks is the difficulty in interpreting the clinical significance of their classifications. This work addresses these concerns by analyzing key molecular markers, histologic diagnoses, and demographic information that underlie the image-based subtypes. Interestingly, the analysis found that there were significant differences in both the mutations and expressions of BRAF, a key gene identified in the literature review that holds clinical significance in treatment selection. This finding suggests that the imaging-based classification has true biologic basis. Furthermore, it supports the hypothesis that molecular foundations underlie imaging phenotypes and suggests that by grouping based on imaging features, we can gain insight into the molecular landscapes that inform them. However, the work also demonstrated that mutation and expression differences alone did not explain the classifications of the images. This may point towards the idea that radiomics is capturing currently unleveraged information, although such conclusions cannot be drawn from this work alone.
This work has several limitations. Primarily, it is a retrospective analysis, and its findings are limited to the purpose of hypothesis generation. Furthermore, while the sample size was larger than other radiomic studies on pLGG, more data is needed for a comprehensive exploration of imaging-genomics relationships. pLGGs encompass a wide variety of tumor histologies, of which only the major types represented by this dataset despite its relatively large size compared to other works involving the application of radiomics in pediatric neuro-oncology.
Furthermore, not all subjects had molecular data available in the publicly accessible repository and so the molecular analysis is prone to the influence of sampling biases. Addition of data, both in terms of detail and availability, would allow for a more rigorous analysis of the imaging subtypes. Differences in image acquisition in terms of MR machine brand and magnet strength was present in this study. Steps were taken to minimize the effect of differences in acquisition as elaborated in the Pre-processing sub-section of the Methods. Despite these steps, we can't exclude the confounding effect of variations in acquisitions and in future work. Notwithstanding these limitations, the work does accomplish its exploratory goal of furthering our understanding of the relationships between imaging phenotypes and biological drivers of disease.
Future works should thus focus on the expansion of cohorts both in terms of number of subjects and diversity of tumors. Cohort size is a universal limitation in radiomic works in pediatric brain tumor populations [26]. Multi-institutional collaboration is the ideal solution to this challenge and is indeed the model upon which open-access data repositories, such as those available via PedCBioportal, are built. By incorporating data from a wider array of institutions, the generalizability of findings will improve, however, this would also necessitate the use of rigorous normalization methods to account for the diversity in imaging acquisition protocols and equipment between institutions. Finally, utilization of a wider range of markers, both clinical and molecular, should be emphasized in future works to better support image subtype identities.
Conclusion
This work demonstrated that readily available imaging sequences can be utilized to create novel radiomic subtypes of pLGG that correlate with key molecular markers and demographic information. The findings suggest that important molecular aberrations are manifested in radiologic phenotypes and that non-invasive imaging can lend insights into the biological underpinnings of pLGGs. The work paves the way for future work in the field that can build towards robust and generalizable imaging subtypes of tumor that may aid in treatment selection and disease prognostication.
Funding
This work was supported by funding from the National Center for Advancing Translational Sciences of the National Institutes of Health [TL1TR001880]
CRediT authorship contribution statement
Debanjan Haldar: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. Anahita Fathi Kazerooni: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Sherjeel Arif: Data curation, Investigation, Project administration, Software, Visualization, Writing – review & editing. Ariana Familiar: Data curation, Investigation, Methodology, Resources, Software, Visualization, Writing – review & editing. Rachel Madhogarhia: Data curation, Investigation, Visualization, Writing – review & editing. Nastaran Khalili: Data curation, Investigation, Visualization, Writing – review & editing. Sina Bagheri: Data curation, Investigation, Visualization, Writing – review & editing. Hannah Anderson: Data curation, Investigation, Project administration, Visualization, Writing – review & editing. Ibraheem Salman Shaikh: Data curation, Investigation, Visualization, Writing – review & editing. Aria Mahtabfar: Data curation, Investigation, Visualization, Writing – review & editing. Meen Chul Kim: Data curation, Investigation, Resources, Visualization, Writing – review & editing. Wenxin Tu: Data curation, Investigation, Visualization, Writing – review & editing. Jefferey Ware: Data curation, Investigation, Supervision, Validation, Visualization, Writing – review & editing. Arastoo Vossough: Data curation, Investigation, Supervision, Validation, Visualization, Writing – review & editing. Christos Davatzikos: Writing – review & editing. Phillip B. Storm: Funding acquisition, Writing – review & editing. Adam Resnick: Conceptualization, Funding acquisition, Methodology, Writing – review & editing. Ali Nabavizadeh: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Validation, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Yanapong Chaisithkarnkha, Kevin Guo, Kelly Garcia-Ramos, Kevin Huang, Shuvanjan Haldar, Rachel K. Patel, Leeya Addisu, Isabella Song, Georgia Georgostathi
References
- 1.Ostrom Q.T., Gittleman H., Liao P., et al. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro. Oncol. 2017;19(5):v1–v88. doi: 10.1093/neuonc/nox158. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sturm D., Pfister S.M., Jones D.T.W. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J. Clin. Oncol. 2017;35(21):2370–2377. doi: 10.1200/JCO.2017.73.0242. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong G.T., Conklin H.M., Huang S., et al. Survival and long-term health and cognitive outcomes after low-grade glioma. Neuro. Oncol. 2011;13(2):223–234. doi: 10.1093/neuonc/noq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lassaletta A., Zapotocky M., Mistry M., et al. Therapeutic and prognostic implications of BRAF V600E in pediatric low-grade gliomas. J. Clin. Oncol. 2017;35(25):2934. doi: 10.1200/JCO.2016.71.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selt F., van Tilburg C.M., Bison B., et al. Response to trametinib treatment in progressive pediatric low-grade glioma patients. J. Neurooncol. 2020;149(3):499–510. doi: 10.1007/s11060-020-03640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazerooni A.F., Bakas S., Rad H.S., Davatzikos C. Imaging signatures of glioblastoma molecular characteristics: a radiogenomics review. J. Magn. Reson. Imaging. 2020;52(1):54–69. doi: 10.1002/jmri.26907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner M.W., Hainc N., Khalvati F., et al. Radiomics of pediatric low-grade gliomas: toward a pretherapeutic differentiation of BRAF-mutated and BRAF-fused tumors. Am. J. Neuroradiol. 2021;42(4):759–765. doi: 10.3174/ajnr.A6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davatzikos C. Machine learning in neuroimaging: progress and challenges. Neuroimage. 2019;197:652–656. doi: 10.1016/j.neuroimage.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerami E., Gao J., Dogrusoz U., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pati S., Singh A., Rathore S., et al. Springer International Publishing; 2020. The Cancer Imaging Phenomics Toolkit (CaPTk): Technical Overview; pp. 380–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davatzikos C., Rathore S., Bakas S., et al. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging. 2018;5(1) doi: 10.1117/1.JMI.5.1.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fathi Kazerooni A., Saxena S., Toorens E., et al. Clinical measures, radiomics, and genomics offer synergistic value in AI-based prediction of overall survival in patients with glioblastoma. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-12699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohlfing T., Zahr N.M., Sullivan E.V., Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp. 2009;31(5):798–819. doi: 10.1002/hbm.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.IC-P-174: Fast Automatic Segmentation of Hippocampal Subfields and Medial Temporal Lobe Subregions In 3 Tesla and 7 Tesla T2-Weighted MRI - Yushkevich - 2016 - Alzheimer's & Dementia - Wiley Online Library. Accessed July 2, 2022. https://alz-journals.onlinelibrary.wiley.com/doi/full/10.1016/j.jalz.2016.06.205
- 16.Thakur S., Doshi J., Pati S., et al. Brain extraction on MRI scans in presence of diffuse glioma: Multi-institutional performance evaluation of deep learning methods and robust modality-agnostic training. Neuroimage. 2020;220 doi: 10.1016/j.neuroimage.2020.117081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madhogarhia R., Fathi Kazerooni A., Arif S., et al. Vol. 12033. SPIE; 2022. (Automated Segmentation of Pediatric Brain Tumors Based on Multi-Parametric MRI and Deep Learning). Vol. [Google Scholar]
- 18.ITK-SNAP Home. Accessed July 2, 2022. http://www.itksnap.org/pmwiki/pmwiki.php?n=Main.HomePage.
- 19.Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-learn: machine learning in python. J. Mach. Lear. Res. 2011;12(85):2825–2830. [Google Scholar]
- 20.Wen J., Varol E., Yang Z., et al. Subtyping brain diseases from imaging data. arXiv preprint arXiv:220210945. Published online 2022.
- 21.Louis D.N., Perry A., Wesseling P., et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro. Oncol. 2021;23(8):1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fathi Kazerooni A., Bagley S.J., Akbari H., et al. Applications of radiomics and radiogenomics in high-grade gliomas in the era of precision medicine. Cancers. 2021;13(23):5921. doi: 10.3390/cancers13235921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryall S., Tabori U., Hawkins C. Pediatric low-grade glioma in the era of molecular diagnostics. Acta Neuropathol. Commun. 2020;8(1):30. doi: 10.1186/s40478-020-00902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.AlRayahi J., Zapotocky M., Ramaswamy V., et al. Pediatric brain tumor genetics: what radiologists need to know. Radiographics. 2018;38(7):2102–2122. doi: 10.1148/rg.2018180109. [DOI] [PubMed] [Google Scholar]
- 25.Chalil A., Ramaswamy V. Low grade gliomas in children. J. Child Neurol. 2016;31(4):517–522. doi: 10.1177/0883073815599259. [DOI] [PubMed] [Google Scholar]
- 26.Madhogarhia R., Haldar D., Bagheri S., et al. Radiomics and radiogenomics in pediatric neuro-oncology: a review. Neuro-Oncol. Adv. 2022 doi: 10.1093/noajnl/vdac083. Published online May 27vdac083. [DOI] [PMC free article] [PubMed] [Google Scholar]