Abstract
Adenine base editors (ABEs) can mediate two transition mutations, A-to-G and T-to-C, which are suitable for repairing G·C-to-T·A pathogenic variants, the most significant human pathogenic variant. By combining the protospacer adjacent motif (PAM)less SpRY nuclease with F148A-mutated TadA∗8e deaminase, we developed a new editor, SpRY-ABE8eF148A, in this study, which has narrowed the editing range and enhanced A-to-G editing efficiency in most sites with NR/YN PAMs. Furthermore, compared with SpRY-ABE8e, SpRY-ABE8eF148A significantly decreased the RNA off-target effect. Therefore, this engineered base editor, SpRY-ABE8eF148A, expanded the editing scope and improved the editing precision for G·C-to-T·A pathogenic variants. Besides, we established a bioinformatics tool, adenine base-repairing sgRNA database of pathogenic variant (ARDPM), to facilitate the development of precise editors.
Keywords: MT: RNA/DNA Editing, ABEs, SpRY-ABE8eF148A, PAMless, narrowed window, ARDPM
Graphical abstract
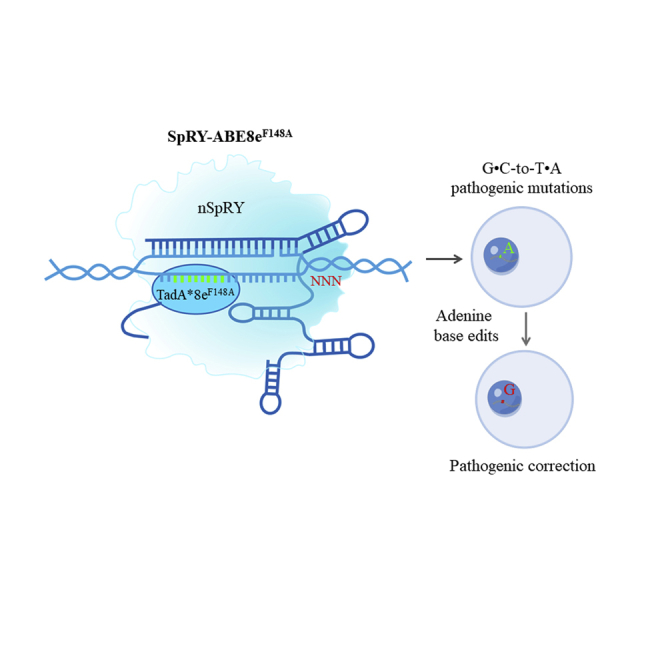
By combining the PAMless SpRY nuclease with F148A-mutated TadA∗8e deaminase, we developed a new editor, SpRY-ABE8eF148A, in this study, which has narrowed the editing window and enhanced A-to-G editing efficiency as well as decreased the RNA off-target effect.
Introduction
Single-nucleotide polymorphisms (SNPs) are the most significant group of human pathogenic variants.1,2,3 According to the Clinvar database (https://www.ncbi.nlm.nih.gov/clinvar/), G·C-to-T·A pathogenic variants constitute about 50% of the known point mutations. Adenine base editors (ABEs) can mediate the conversion of A·T-to-G·C in genomic DNA without double-stranded DNA cleavage.4,5,6 ABEs can mediate two transition mutations, A-to-G and T-to-C, and are potentially applied for repairing G·C-to-T·A pathogenic variants. ABEs perform efficient A-to-G base editing with deamination activity restricted to an ∼5 base pair (bp) window of single-stranded DNA (ssDNA; positions ∼4–8, counting the PAM at positions 21–23) generated by dCas9.4 ABE8e improved the Cas domain compatibility, substantially enhancing editing efficiencies when paired with Cas9 or Cas12 homologs.7 One of the engineered variants of Streptococcus pyogenes Cas9, SpRY, was released from the NGG protospacer adjacent motif (PAM) requirement. Thus, SpRY nuclease and base-editor variants can target many PAMs, exhibiting robust activities on a wide range of sites with NRN PAMs in human cells.8 Based on this development, a new ABE, the SpRY-ABE8e, capable of efficiently editing A-to-G in most sites with NRN (R = A or G) and NYN (Y = C or Y) PAMs was developed.9
The ABE toolbox can repair almost all G·C-to-T·A pathogenic variants, although the bystander edits and potential RNA off-targets restrict the editing accuracy and its therapeutic applications.10,11 ABE engineering showed that an F148A mutation of TadA abolished the enzyme activity in Escherichia coli.12,13 Hence, researchers developed ABE7.10F148A, which can substantially narrow the A-to-G editing range. This development improved DNA base-editing precision.10
In this study, we aimed to develop a new ABE, SpRY-ABEF148A, by engineering the F148A mutation of TadA∗8e to increase the editing precision of SpRY-ABE8e. We found that this SpRY-ABE8eF148A was a functional ABE that precisely repaired G·C-to-T·A pathogenic variants with highly efficient A-to-G base editing and reduced RNA off-target effects.
Results
SpRY-ABE8eF148A activities as a near-PAMless ABE
G·C-to-T·A pathogenic variants account for 46.11% of 99,7743 pathogenic variants in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed February 2022), which is the largest class of human pathogenic variants4,5,6 (Figures S1A and S1B). We designed all gRNA sequences of ABE editors for repairing G·C-to-T·A pathogenic variants. The target A base is located in positions 4, 5, 6, 7, and 8 of the 5 bp editing range. Most of the single guide RNAs (sgRNAs) that repair both C-to-T and G-to-A pathogenic variants did not contain NGG in their PAMs (Figures S1C and S1E) but harbored randomly distributed NAN, NCN, NGN, and NTN PAMs (Figures S1D and S1F). These results demonstrated that PAMless ABEs were inevitable and necessary for repairing more C-to-T and G-to-A pathogenic variants.
SpRY nuclease and base-editor variants can target PAMs, exhibiting robust activities on many sites with NRN PAMs in human cells.8 SpRY-ABE8e can efficiently perform A-to-G editing in NRN PAM sites.9,14 Bystander edits in the editing range are the main constraint for precisely repairing pathogenic variants by DNA base editors. A previous study showed that ABE7.10F148A narrowed the editing range and achieved precise DNA base editing.10
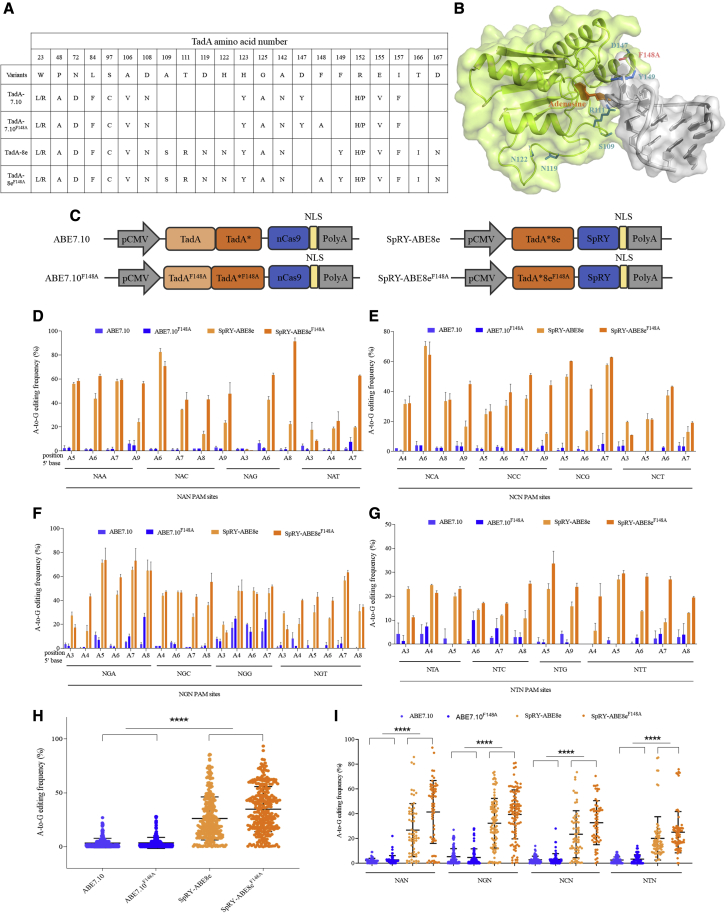
To determine whether TadA∗8eF148A could improve the precision of the SpRY-ABE8e editor, we constructed a new DNA base editor, SpRY-ABE8e F148A, by introducing an F148A mutation in the TadA∗8e deaminase (Figures 1A and 1B). To assess the editing efficiency of different TadA variants with different PAMs, we constructed four ABEs, ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A (Figure 1C). Then, we evaluated the editing efficiency of these four ABEs in the endogenous site VISTA enhancer hs267 (NG_053265.1) with NAN, NCN, NGN, and NTN PAMs. We found that ABE7.10 and ABE7.10F148A showed no editing efficiency in most PAMs, except for the NGG PAM (Figure S1). Comparatively, SpRY-ABE8eF148A achieved 62.90% editing efficiency in NAN PAM (Figure S1A), 27.38% in NCN PAM (Figure S1B), 37.28% in NGN PAM (Figure S1C), and 27.31% in NTN PAM (Figure S1D), respectively.
Figure 1.
Comparison of ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A base editors’ activities across NR/YN PAM sites in HEK293T cells
(A) Conserved mutations and genotypes of four TadA variants. (B) The structure of TadA∗8eF148A variant. The side chains of F148A of TadA∗ are shown as red sticks. (C) The architecture of ABE7.10, ABE7.10F148A, ABE8e, and ABE8eF148A. (D) A-to-G base editing of 12 endogenous sites in HEK293T cells bearing NAN PAM. Editing efficiency is shown as mean ± SEM, n = 3 independent experiments. Edited adenines were counted with setting the base distal to PAM as position 1. (E) A-to-G base editing of 12 endogenous sites in HEK293T cells bearing NCN PAM. (F) A-to-G base editing of 12 endogenous sites in HEK293T cells bearing NGN PAM. (G) A-to-G base editing of 12 endogenous sites in HEK293T cells bearing NTN PAM. (H) Editing efficiencies across target sites with NR/YN PAMs in HEK293T cells. p < 0.0001 for comparison of ABE7.10 and ABE7.10F148A with SpRY-ABE8e and SpRY-ABE8eF148A. (I) Editing efficiencies across target sites with NAN, NGN, NCN, and NTN in HEK293T cells, respectively.
To further illustrate the efficiency of the new ABE, we compared the A-to-G base-editing efficiency among ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A across 48 sites (12 sites harboring NAN, 12 sites harboring NGN, 12 sites harboring NCN, and 12 sites harboring NTN) in both HEK293T and HeLa cells. The results showed that SpRY-ABE8e and SpRY-ABE8eF148A markedly increased editing efficiency at nearly all tested sites (Figures 1H, 1I, S3E, and S3F) in HEK293T and HeLa cells, which offered nearly ten times higher efficiency than ABE7.10 and ABE7.10F148A. For 12 NAN PAM sites, SpRY-ABE8eF148A achieved 50.73% editing efficiency in HEK293T cells (Figures 1D) and 42.57% in HeLa cells (Figure S3A); for 12 NCN PAM sites, SpRY-ABE8eF148A achieved 40.59% editing efficiency in HEK293T cells (Figures 1E) and 34.74% in HeLa cells (Figure S3B); for 12 NGN PAM sites, SpRY-ABE8eF148A achieved 46.43% editing efficiency in HEK293T cells (Figure 1E) and 36.85% in HeLa cells (Figure S3C); and for 12 NTN PAM sites, SpRY-ABE8eF148A achieved 22.5% editing efficiency in HEK293T cells (Figure 1F) and 36.68% in HeLa cells (Figure S3D). Therefore, this new DNA base editor, SpRY-ABE8eF148A, efficiently performed A-to-G editing in most NR/YN PAMs.
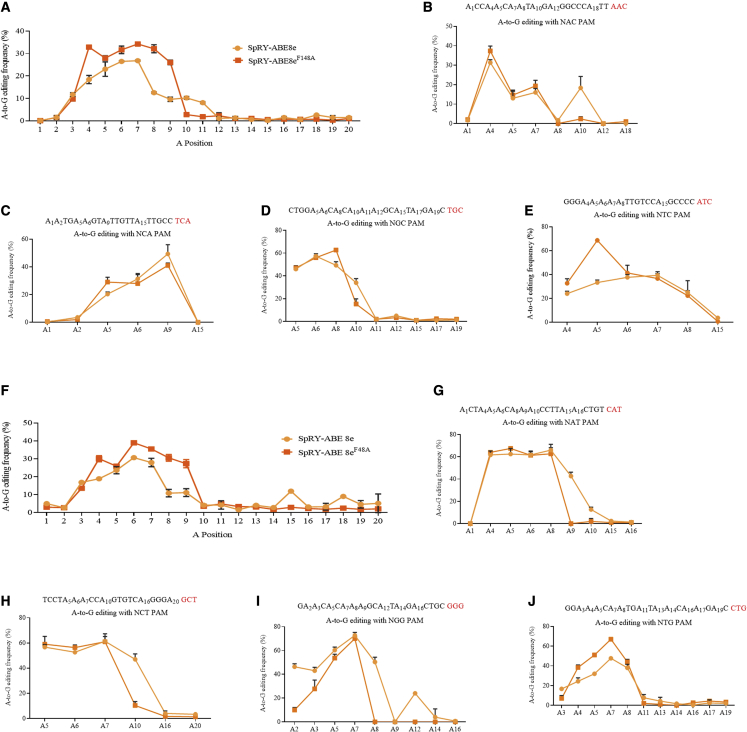
SpRY-ABE8eF148A narrows the editing range
We analyzed the editable positions to evaluate whether the new SpRY-ABE8eF148A editor reduced bystander A edits. Results showed that the editing range of SpRY-ABE8e spanned from positions 3 to 11 in HEK293T cells. In contrast, the editing range of the SpRY-ABE8eF148A editor mainly appeared in 3–10 bp (Figure 2A).
Figure 2.
TadA∗8eF148A variant capable of narrowing the width of the editing range
(A) Editing range of SpRY-ABE8eF148A variant in HEK293T cells. Edited adenines were counted with setting the base distal to PAM as position 1; each data point is shown as mean ± SEM. (B) Editing range of SpRY-ABE8eF148A variant bearing NAN PAM in HEK293T cells. (C) Editing range of SpRY-ABE8eF148A variant bearing NCN PAM in HEK293T cells. (D) Editing range of SpRY-ABE8eF148A variant bearing NGN PAM in HEK293T cells. (E) Editing range of SpRY-ABE8eF148A variant bearing NTN PAM in HEK293T cells. (F) Editing range of SpRY-ABE8eF148A variants in HeLa cells. (G) Editing range of SpRY-ABE8eF148A variant bearing NAN PAM in HeLa cells. (H) Editing range of SpRY-ABE8eF148A variant bearing NCN PAM in HeLa cells. (I) Editing range of SpRY-ABE8eF148A variant bearing NGN PAM in HeLa cells. (J) Editing range of SpRY-ABE8eF148A variant bearing NTN PAM in HeLa cells.
SpRY-ABE8eF148A had a narrower editing range in 20 bp sgRNAs with NAN PAM (Figure 2B), NCN PAM (Figure 2C), NGN PAM (Figure 2D), and NTN PAM in HEK293T cells (Figure 2E). As for HeLa cells, the editing range of SpRY-ABE8e mainly spanned from position 3 to 11 and appeared in distal A edits, such as A15 and A18 (Figure 2F). The editing range of the SpRY-ABE8eF148A editor mainly appeared in 3–10 bp (Figure 2F). Besides, SpRY-ABE8eF148A also had a narrower editing range in 20 bp sgRNAs with NAN PAM (Figure 2G), NCN PAM (Figure 2H), NGN PAM (Figure 2I), and NTN PAM in HeLa cells (Figure 2J).
As mentioned above, the SpRY-ABE8eF148A editor narrows the editing range, which could be a good candidate for a precise ABE.
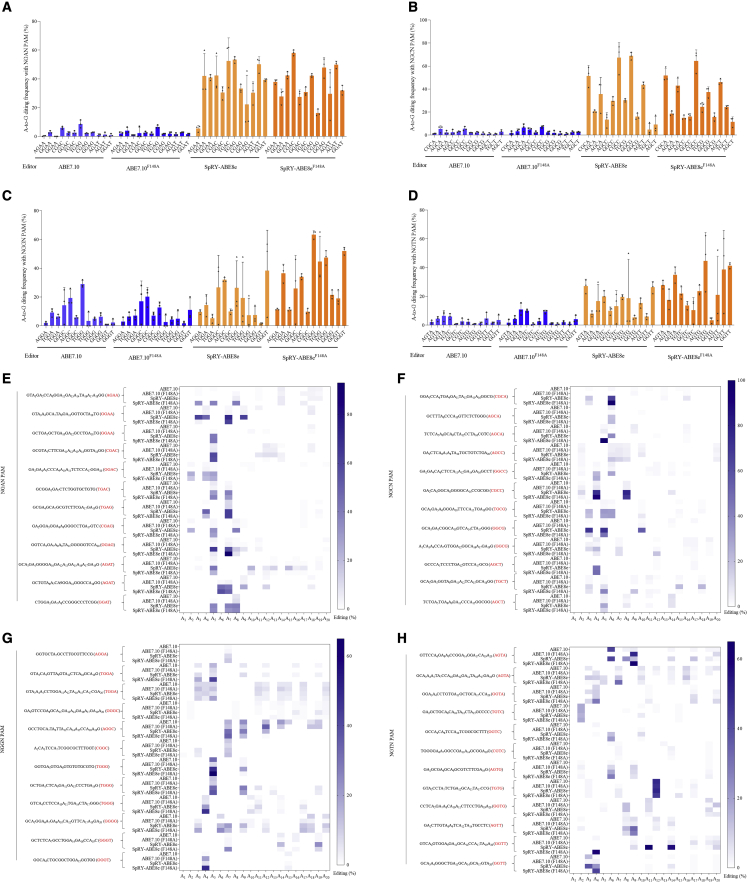
SpRY-ABE8eF148A performs highly efficient editing in NGN PAM sites
Most ABEs, including the two ABEs used in this study, ABE7.10 and ABE7.10F148A, mediated A-to-G base edits in NGN PAMs. Research showed that the R1333P substitution in SpRY might also enable targeting of any base in the second PAM position (including thus far unexamined NYN PAMs).8 We assessed the activities of SpRY-ABE8e, SpRY-ABE8EF148A, ABE7.10, and ABE7.10F148A on 16 types of NGNN PAMs, and these NGNN PAMs contain NGAN (NGAA, NGAC, NGAG, NGAT); NGCN (NGCA, NGCC, NGCG, NGCT); NGGN (NGGA, NGGC, NGGG, NGGT); and NGTN (NGTA, NGTC, NGTG, NGTT) (Table S1). We statistically evaluated the average editing efficiency of ABE7.10 and ABE7.10F148A bearing the 3–7 bp editing range and the average editing efficiency of SpRY-ABE8e and SpRY-ABE8EF148A bearing the 3–10 bp editing range.
Results illustrated that SpRY-ABE8eF148A achieved 36.83% editing efficiency in NGAN PAM, 35.96% in NGAA, 38.78% in NGAC, 35.46% in NGAG, and 37.14% in NGAT; SpRY-ABE8e showed 36.87% editing efficiency in NGAN PAM, 29.50% in NGAA, 41.78% in NGAC, 36.27% in NGAG, and 39.93% in NGAT (Figure 3A); SpRY-ABE8eF148A achieved 30.69% editing efficiency in NGCN PAM, 37.83% in NGCA, 31.79% in NGCC, 25.82% in NGCG, and 27.31% in NGCT; SpRY-ABE8e showed 32.60% editing efficiency in NGCN, 35.94% in NGCA, 36.80% in NGCC, 38.44% in NGCG, and 19.22% in NGCT(Figure 3B); SpRY-ABE8eF148A achieved 31.38% editing efficiency in NGGN PAM, 19.66% in NGGA, 23.28% in NGGC, 51.78% in NGGG, and 30.82% in NGGT; SpRY-ABE8e showed 16.56% editing efficiency in NGGN PAM, 9.80% in NGGA, 22.89% in NGGC, 17.67% in NGGG, and 15.87% in NGGT (Figure 3C); SpRY-ABE8eF148A achieved 24.94% editing efficiency in NGTN PAM, 26.81% in NGCA, 15.39% in NGTC, 23.89% in NGTG, and 33.67% in NGTT; and SpRY-ABE8e achieved 15.53% editing efficiency in NGTN PAM, 17.27% in NGTA, 14.39% in NGTC, 14.59% in NGTG, and 15.89% in NGTT (Figure 3D).
Figure 3.
TadA∗8eF148A variant capable of efficient A-to-G editing with NGNN PAM
(A) Mean editing efficiency mediated by ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A with NGAN PAM. Edited adenines were counted with setting the base distal to PAM as position 1; each data point is shown as mean ± SEM. (B) Mean editing efficiency mediated by ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A with NGCN PAM. (C) Mean editing efficiency mediated by ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A with NGGN PAM. (D) Mean editing efficiency mediated by ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A with NGTN PAM. (E) A-to-G editing efficiency of all A sites with NGAN PAM. (F) A-to-G editing efficiency of all A sites with NGCN PAM. (G) A-to-G editing efficiency of all A sites with NGGN PAM. (H) A-to-G editing efficiency of all A sites with NGTN PAM.
Comparatively, ABE7.10 and ABE7.10F148A had poor editing efficiency in NGAN, NGCN, and NGTN PAM sites (Figures 3A, 3B, and 3D). ABE7.10 achieved 8.67% editing efficiency in NGGN PAM, 5.87% in NGGA, 13.17% in NGGC, 12.44% in NGGG, and 3.19% in NGGT, respectively, and ABE7.10F148A showed 8.19% editing efficiency in NGGN PAM, 5.53% in NGGA, 14.78% in NGGC, 6.56% in NGGG, and 5.89% in NGGT, respectively (Figure 3C).
We also statistically assessed A-to-G base-editing efficiency of all the A sites in the sgRNA. Results clearly illustrated that SpRY-ABE8e and SpRY-ABE8eF148A were more efficient than ABE7.10 and ABE7.10F148A. Compared with SpRY-ABE8e, SpRY-ABE8eF148A activated mainly in the 5–8 bp window, which showed a narrower editing range than the NGAN, NGCN, NGGN, and NGTN PAM sites (Figures 3E–3H). These results indicated that SpRY-ABE8eF148A efficiently performed A-to-G editing in most NGNN PAM sites and narrowed the editing range.
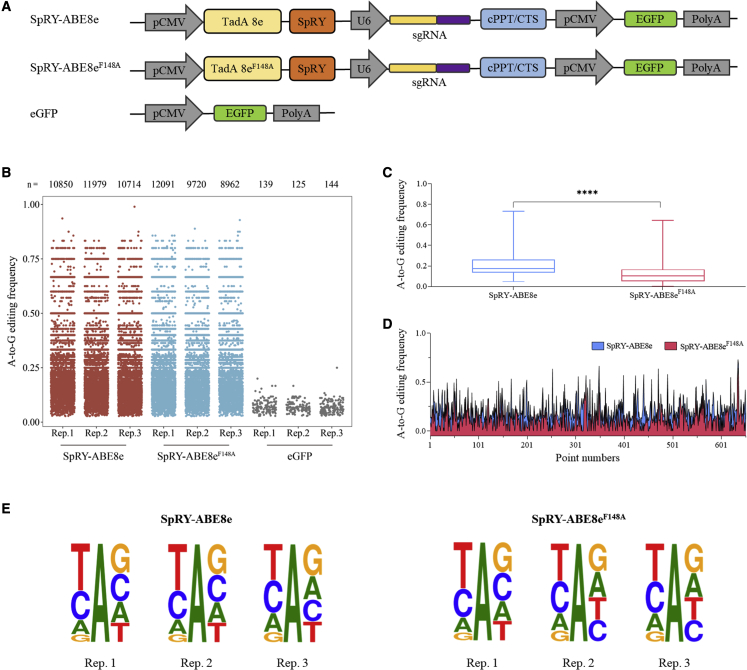
SpRY-ABE8eF148A reduces RNA off-target effect
The potential off-target effect limits the biomedical application of base editors.10,11 To evaluate the RNA off-target effect, we assessed the RNA off-target single-nucleotide variation (SNV) numbers and RNA off-target editing efficiency of SpRY-ABE8eF148A and SpRY-ABE8e by RNA sequencing. We established three experiment groups: the SpRY-ABE8e editing group, the SpRY-ABE8eF148A editing group, and an EGFP group as the control (Figure 4A). RNA off-target SNVs showed that SpRY-ABE8eF148A and SpRY-ABE8e had equal A-to-G RNA off-target SNV numbers. The A-to-G RNA off-target SNV numbers of three repeats of SpRY-ABE8eF148A were 10,752, 8,478, and 7,740; however, the A-to-G RNA off-target SNV numbers of three repeats of SpRY-ABE8e were 9,652, 10,667, and 9,570, respectively (Figure 4B). To assess whether SpRY-ABE8eF148A narrowed the editing range, we calculated the average off-target A-to-G editing efficiency of SpRY-ABE8eF148A and SpRY-ABE8e. Compared with that of SpRY-ABE8e, the A-to-G off-target editing efficiency of SpRY-ABE8eF148A was significantly reduced (p < 0.0001) (Figures 4C and 4D). In addition, we found that the off-target editing motifs of SpRY-ABE8eF148A and SpRY-ABE8e were unbiased (Figure 4E). Although SpRY-ABE8eF148A slightly reduced the total RNA off-target SNV numbers, it significantly decreased the A-to-G RNA off-target editing frequency.
Figure 4.
Elimination of off-target RNA SNVs by engineered of TadA∗8eF148A deaminase
(A) Schematics of off-target detection groups. (B) Representative distributions of off-target RNA SNVs on human chromosomes for SpRY-ABE8e, SpRY-ABE8eF148A, and EGFP. (C) Mean off-target editing efficiency of SpRY-ABE8e and SpRY-ABE8eF148A. (D) Specific off-target editing efficiency of SpRY-ABE8e and SpRY-ABE8eF148A. Plots represent all A-to-G sites. Each plot represents the frequency of A-to-G. (E) Sequence logos from off-target edited adenines identified in each RNA-seq replicate.
Expanding the application of SpRY-ABE8eF148A in correcting disease-relevant loci
Currently, very few reports on sgRNA designers or tools for DNA base editing are presented. Hence, we established an adenine base-repairing sgRNA database of pathogenic variants (ARDPM; http://47.92.172.28:12026/). ARDPM is a database of G·C-to-A·T pathogenic variant-repairing sgRNAs achieved via A-to-G base editing by PAMless ABEs. This database provides all available sgRNAs of PAMless-ABEs that can repair G·C-to-A·T pathogenic variants from the ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed February 2022). Each sgRNA shows sufficient editing-tool information, including the position of the target edit base, the bystander edit number, the PAM, and the sgRNA sequence. Researchers can select the appropriate sgRNA for repairing specific G·C-to-A·T pathogenic variants.
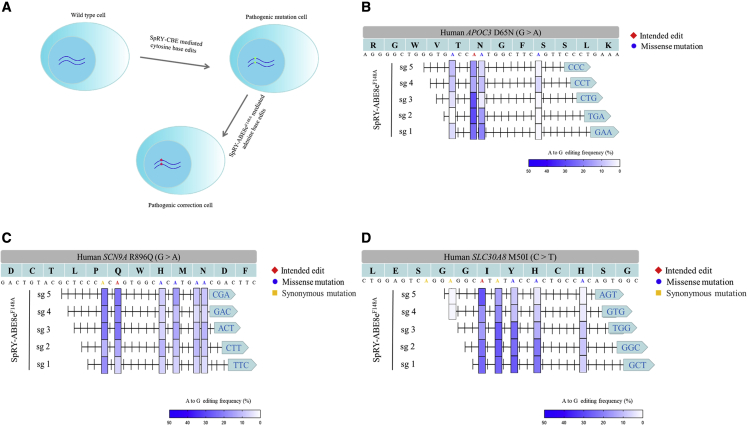
Then, we systematically evaluated the potential of SpRY-ABE8eF148A to correct disease-associated mutations in human cells that lack nearby canonical NGG PAMs. We first established three mutant cell lines harboring three different pathogenic variants, including APOC3 (p.D65N, c.G2871A), SCN9A (p.R896Q, c.G98851A), and SLC30A8 (p.M50I, c.G196816A). We successfully constructed the pathogenic mutation cell lines using the SpRY-CBE cytosine base editor15 with the mutant (mut)-sgRNA (Figures 5A and S4). Five sgRNAs with different PAMs and intended edit positions were designed for each pathogenic variant site.
Figure 5.
Expanded capabilities of SpRY-ABE8eF148A to generate protective genetic variants
(A) Schematic diagram of simulating and correcting the pathogenic variant in HEK293T cells. The disease cell model was established by SpRY-CBE under the guide of mut-sgRNA and then was corrected by SpRY-ABE8eF148A. (B) APOC3 (D65N) pathogenic variant was repaired by SpRY-ABE8eF148A under five different sgRNAs. (C) SCN9A (R896Q) pathogenic variant was repaired by SpRY-ABE8eF148A under five different sgRNAs. (D) SLC30A8 (M50I) pathogenic variant was repaired by SpRY-ABE8eF148A under five different sgRNAs.
For APOC3, there were 3 missense mutations in the editing range of the 5 sgRNAs. sgRNA2 and sgRNA3 mediated the higher A-to-G conversion with efficiencies of 29.33% and 28.67%, respectively; in contrast, the other three sgRNAs (sg1, sg4, and sg5) generated average 23.67%, 13.67%, and 17% editing efficiencies, respectively. Meanwhile, the five sgRNAs (sg1–5) generated average efficiencies of 8.78%, 5.56%, 2.89%, 8.22%, and 5.78% potential missense edits, respectively (Figure 5B). Among the five selected sgRNAs, sgRNA3 might be the candidate sgRNA in correcting the APOC3 (D65N) pathogenic variant.
For SCN9A (R896Q), except for the intended edit, there was one synonymous mutation and four potential missense mutations in the five designed sgRNAs. The intended editing efficiencies of the five sgRNAs (sg1–5) were 6.67%, 13.67%, 21.67%, 20%, and 22.33%, respectively. The five sgRNAs (sg1–5) generated average efficiencies of 6.67%, 9.92%, 10%, 7%, and 10.25% potential missense edits (Figure 5C). Thus, sgRNA4 was selected for the SCN9A (R896Q) pathogenic variant.
For SLC30A8 (M50I), the intended editing efficiencies of the five sgRNAs (sg1–5) were 24.67%, 23%, 15.67%, 17.33%, and 27.67%, respectively. The five sgRNAs (sg1–5) generated average efficiencies of 18.78%, 18%, 15%, 7%, and 4.56% potential missense edits, respectively (Figure 5D). Comparatively, sgRNA5 might be the ideal sgRNA in a correction the SLC30A8 (M50I).
Therefore, these results demonstrated that SpRY-ABE8eF148A could be one potential editor to correct disease-associated mutations in human cells.
Discussion
Base editors hold promise for repairing pathogenic variants as they do not require DSBs, donor DNA templates, or homology-directed repair.16,17,18,19,20 Typical base editors are developed by fusing inactive CRISPR-Cas nuclease or nickase to a deaminase. So far, two main classes of base editors have been identified: cytosine base editors (CBEs), which mediate the C-to-T base conversion,21 and ABEs, which catalyze A-to-G conversion.4 CBEs and ABEs can efficiently repair approximately 1/3 of the currently annotated pathogenic points, representing the four possible transition mutations: C-to-T, A-to-G, T-to-C, and G-to-A.1 In particular, G·C-to-T·A pathogenic variants are the largest class of human pathogenic variants.
ABEs, as a toolbox, have been developed to repair these G·C-to-T·A mutations. For instance, ABE7.104, ABEmax,22 and ABE8e7 mediate A-to-G base editing efficiently. ABE-NG,22 ABEmax-NG,22 and SpRY-ABE8e9,14 expand the related editing scope. The BEs dCpf1-eBE-YE23 and ABE7.10F148A narrow the editing range to improve editing precision. Here, we developed a new ABE, SpRY-ABE8eF148A, that expanded the editing scope and narrowed the editing range. SpRY-ABE8e F148A could be one of the widely targeting and precisely editing ABEs for G·C-to-T·A pathogenic variants. SpRY-ABE8eF148A mediated efficient A-to-G base editing with NAC, NAG, NAT, NCA, NCT, NGC, NGG, NGT, NTC, and NTG (N = “A, T, C, or G”) PAMs. Compared with SpRY-ABE8e, SpRY-ABE8eF148A narrowed the width of the editing range and significantly decreased the A-to-G RNA off-target SNV.
Although prime editor (PE) and base editor (BE) are promising tools for molecular therapy in biomedical applications, efforts are still inevitably required to develop simpler PEs and precise BEs that can enhance editing capabilities. PEs substantially expand the scope and capabilities of genome editing. PEs can correct up to 89% of known genetic variants associated with human diseases.24,25,26,27,28,29 Improvements in the stability of editing efficiency, nCas9-MMLV framework, pegRNA, and nicking gRNA are still required to ensure the practical application of ABEs. Furthermore, the editing precision of BEs is the key issue that has to be sorted for specific therapeutic applications. For in vivo delivery, due to the package size of adeno-associated viruses (AAVs), it is difficult to pack the ABE and sgRNA into one AAV. It still needs to package the reagents of BEs into dual AAVs using a split-intein delivery system.30
In summary, we engineered a new BE, SpRY-ABE8eF148A, that mainly generates 3–10 bp editing. Compared with SpRY-ABE8e, in some sites, such as the NAN PAM or NGN PAM sites, SpRY-ABE8eF148A has less bystander edits and a narrower editing range, so it has lower mean off-target editing frequency. SpRY-ABE8eF148A achieved efficient A-to-G editing with RNA off-target elimination. In addition, we established ARDPM, a database of sgRNAs that can repair pathogenic variants by using ABEs. This toolbox, along with SpRY-ABE8eF148A, will contribute to the development of more efficient and precise editors.
Materials and methods
Construction of expression plasmids
EGFP, pCMV-ABE7.10, pCMV-T7-ABEmax(7.10)-SpRY-P2A-EGFP, ABE8e plasmids, and PGL3-U6-GFP were obtained from Addgene (#176015, #102919, #140003, #138489, and #107721). The ABE7.10F148A expression plasmid was constructed based on pCMV-ABE7.10. SpRY-ABE8e and SpRY-ABE8eF148A were constructed based on pCMV-T7-ABEmax(7.10)-SpRY-P2A-EGFP and ABE8e. The TadA∗F148A and TadA∗8eF148A variants were obtained by plasmid-site-directed mutagenesis using a fast-site-directed mutagenesis kit (Tiangen). PGL3-U6-GFP was digested using BsaI, and the linearized vector was used for sgRNA expression vector construction. All the plasmid sequences mentioned above are listed in Table S3, and all gRNA sequences are provided in Table S1.
Cell culture and transfection
In this study, all experiments were performed in two human cell lines, HEK293T and HeLa (ATCC). Cells were grown in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, #11965092) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, #30067334) and 1× penicillin-streptomycin (Thermo Fisher Scientific, #10378016).
At about 12–16 h prior to transfection, approximately 50,000 cells per well in 24-well plates were coated with 1× poly-D-lysine (Thermo Fisher Scientific, #A3890401). The cells were transfected to about 80%–90% confluence. Transfection experiments were performed using the EZ Cell transfection reagent (Shanghai Life iLab Bio Technology, #AC04L092). For each well, 1 μg plasmids were combined with 40 μL Opti-MEM for transfection (Thermo Fisher Scientific, #11058021). Separately, 3 μL EZ Cell transfection reagent was combined with 40 μL Opti-MEM to form the EZ-trans reagent. The plasmids and EZ-trans reagent were then combined and incubated for 15 min to form the plasmid-EZ reagents, which were added to the cells. After 6 h, the culture medium was replaced with fresh medium.
A-to-G base editing in mammalian cells
To assess the activities of ABE7.10, ABE7.10F148A, SpRY-ABE8e, and SpRY-ABE8eF148A in mammalian cells, 660 ng BE vectors and 330 ng sgRNA expression plasmids were transfected into cells in 24-well plates. After 48 h of transfection, 10,000 cells were collected by fluorescence-activated cell sorting (FACs). DNA from cells was harvested using cell lysis solution under the following conditions: 65°C for 1 h, 95°C for 10 min. The cell lysate was then subjected to polymerase chain reaction (PCR) to obtain the targeted PCR fragments under the following conditions: 95°C for 3 min; 95°C for 15 s, 58°C for 15 s, 72°C for 20 s (30 cycles); 72°C for 5 min. PCR primers are shown in Table S2. The PCR fragments were then subjected to Sanger sequencing. A-to-G base-editing efficiency was evaluated using the web tool EditR (https://moriaritylab.shinyapps.io/editr_v10/).31
RNA sequencing
Three experimental groups were set for detecting RNA off-targets, namely, the SpRY-ABE8e editing group, the SpRY-ABE8eF148A editing group, and the EGFP control group. The SpRY-ABE8e + target sgRNA, SpRY-ABE8eF148A + target sgRNA, and EGFP plasmids were separately transfected in HEK293T cells and cultured in a 60 mm dish with three replicates per group. After 48 h, about 500,000 cells (top 5% GFP signal) were collected by using FACS. Total RNA of the collected cells was extracted according to the standard protocol. Nine mRNA samples were prepared for RNA sequencing (RNA-seq).
RNA-seq
RNA-seq was performed on the Illumina sequencing platform of Novogene (https://en.novogene.com/).
Off-target RNA-editing analysis
SNVs were called following the best practices from GATK (v.4.1.9) for RNA-seq data. Clean reads were subjected to local realignment, coordinate sorting, base quality score recalibration, and insertion or deletion (indel) realignment. We performed SNV discovery and used multi-sample variant calling to distinguish between a homozygous reference genotype and a missing genotype among the analyzed samples. SNVs were annotated using the transcript set from the Homo_sapiens_Ensemble_94 version. All the A-to-G RNA off targets and their off-target editing efficiency were calculated.
Statistical analysis
All the experimental data in this study are derived from at least three individual replicates, and data are expressed as mean ± SEM. GraphPad Prism software 8.0.1 was used to analyze data using the two-sided t test. The statistical significance was set at p < 0.05, and asterisks show difference at the following levels: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
This work is supported by the ZJU-Hangzhou Global Scientific and Technological Innovation Center; Zhejiang University (02020200-K02013008); the National Natural Science Foundation of China (32071347); and a research fund from a Joint laboratory grant from Jiangsu Wuzhong Aesthetics Biotech Co., Ltd. We also would like to thank the iBiofoundary and Core Facility of the Institute for Intelligent Bio/Chem Manufacturing, ZJU-Hangzhou Global Scientific and Technological Innovation Center (China) for fabrication and analysis.
Author contributions
G.L. and Y.Y. conceived and designed the project. G.L. and Y.C. performed most experiments with the assistance of Y.L. H.M., S.L., and Y.Q. analyzed the RNA-seq data. Z.P. predicted the protein structure of TadA∗8eF148A variant. Y.Y. and X.H. supervised the project.
Declaration of interests
The authors declare that they have no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.12.001.
Supplemental information
Data availability
Base-editing efficiency statistics are available at EditR (https://moriaritylab.shinyapps.io/editr_v10/). The SNVs annotated used Homo_sapiens_Ensemble_94 version. SNV callings were done using GATK (v.4.1). Raw data filtering was done with Fastp (v.0.20.1). We also used GraphPad Prism software 8.0.1. The RNA-seq data will be deposited at NCBI Bioproject (NCBI: PRJNA818868).
References
- 1.Landrum M.J., Chitipiralla S., Brown G.R., Chen C., Gu B., Hart J., Hoffman D., Jang W., Kaur K., Liu C., et al. ClinVar: improvements-to-accessing data. Nucleic Acids Res. 2020;48:D835–D844. doi: 10.1093/nar/gkz972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A·T-to-G·C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees H.A., Liu D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 7.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E., et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. https://www.science.org/doi/10.1126/science.aba8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren Q., Sretenovic S., Liu S., Tang X., Huang L., He Y., Liu L., Guo Y., Zhong Z., Liu G., et al. PAMless plant genome editing using a CRISPR-SpRY toolbox. Nat. Plants. 2021;7:25–33. doi: 10.1038/s41477-020-00827-4. [DOI] [PubMed] [Google Scholar]
- 10.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R., et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 11.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. https://www.science.org/doi/10.1126/science.aaw7166 [DOI] [PubMed] [Google Scholar]
- 12.Xiang S., Short S.A., Wolfenden R., Carter C.W., Jr. The structure of the cytidine deaminase-product complex provides evidence for efficient proton transfer and ground-state destabilization. Biochemistry. 1997;36:4768–4774. doi: 10.1021/bi963091e. https://pubs.acs.org/doi/10.1021/bi963091e [DOI] [PubMed] [Google Scholar]
- 13.Kim J., Malashkevich V., Roday S., Lisbin M., Schramm V.L., Almo S.C. Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase. Biochemistry. 2006;45:6407–6416. doi: 10.1021/bi0522394. https://pubs.acs.org/doi/10.1021/bi0522394 [DOI] [PubMed] [Google Scholar]
- 14.Tao W., Liu Q., Huang S., Wang X., Qu S., Guo J., Ou D., Li G., Zhang Y., Xu X., Huang X. CABE-RY: a PAM-flexible dual-mutation base editor for reliable modeling of multi-nucleotide variants. Mol. Ther. Nucleic Acids. 2021;26:114–121. doi: 10.1016/j.omtn.2021.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao X., Guo J., Huang S., Yu W., Li G., An L., Li X., Tao W., Liu Q., Huang X., et al. Engineering of near-PAMless adenine base editor with enhanced editing activity and reduced off-target. Mol. Ther. Nucleic Acids. 2022;28:732–742. doi: 10.1016/j.omtn.2022.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;169:559–636. doi: 10.1016/j.cell.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y., et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. https://www.science.org/doi/10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- 19.Molla K.A., Yang Y. CRISPR/Cas-Mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Hess G.T., Tycko J., Yao D., Bassik M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell. 2017;68:26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua K., Tao X., Han P., Wang R., Zhu J.K. Genome engineering in rice using Cas9 variants that Recognize NG PAM sequences. Mol. Plant. 2019;12:1003–1014. doi: 10.1016/j.molp.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Li X., Wang Y., Liu Y., Yang B., Wang X., Wei J., Lu Z., Zhang Y., Wu J., Huang X., et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 24.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Q., Jin S., Zong Y., Yu H., Zhu Z., Liu G., Kou L., Wang Y., Qiu J.L., Li J., Gao C. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021;39:923–927. doi: 10.1038/s41587-021-00868-w. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y., Yang G., Huang S., Li X., Wang X., Li G., Chi T., Chen Y., Huang X., Wang X. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 2021;31:1134–1136. doi: 10.1038/s41422-021-00520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson J.W., Randolph P.B., Shen S.P., Everette K.A., Chen P.J., Anzalone A.V., An M., Newby G.A., Chen J.C., Hsu A., Liu D.R. Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 2022;40:402–410. doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P.J., Hussmann J.A., Yan J., Knipping F., Ravisankar P., Chen P.F., Chen C., Nelson J.W., Newby G.A., Sahin M., et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635–5652.e29. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zong Y., Liu Y., Xue C., Li B., Li X., Wang Y., Li J., Liu G., Huang X., Cao X., Gao C. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 2022;40:1394–1402. doi: 10.1038/s41587-022-01254-w. [DOI] [PubMed] [Google Scholar]
- 30.Yeh W.H., Shubina-Oleinik O., Levy J.M., Pan B., Newby G.A., Wornow M., Burt R., Chen J.C., Holt J.R., Liu D.R. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aay9101. https://www.science.org/doi/10.1126/scitranslmed.aay9101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: a method-to-quantify base editing from sanger sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. https://www.liebertpub.com/doi/10.1089/crispr.2018.0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Base-editing efficiency statistics are available at EditR (https://moriaritylab.shinyapps.io/editr_v10/). The SNVs annotated used Homo_sapiens_Ensemble_94 version. SNV callings were done using GATK (v.4.1). Raw data filtering was done with Fastp (v.0.20.1). We also used GraphPad Prism software 8.0.1. The RNA-seq data will be deposited at NCBI Bioproject (NCBI: PRJNA818868).