Highlights
-
•
Developed a practical AI model to learn from the actual treatment records with the incorporation of physicians’ logical decision process to mimic physicians’ clinical decision-making (CDM).
-
•
The model was trained intentionally to be institution-specific or physician-specific to address the practice variations in clinical practice.
-
•
This is the first time an AI model has been developed to predict dose prescription based on both image and non-image clinical parameters for CDM of radiotherapy.
-
•
Results demonstrated the effectiveness of the model, which can become a valuable tool for secondary opinion consultation for patients or QA and training of physician practices.
Keywords: Clinical decision-making, Metastases, Deep learning
Abstract
Purpose
AI modeling physicians’ clinical decision-making (CDM) can improve the efficiency and accuracy of clinical practice or serve as a surrogate to provide initial consultations to patients seeking secondary opinions. In this study, we developed an AI network to model radiotherapy CDM and used dose prescription as an example to demonstrate its feasibility.
Materials/Methods
152 patients with brain metastases treated by radiosurgery from 2017 to 2021 were included. CT images and tumor and organ-at-risk (OAR) contours were exported. Eight relevant clinical parameters were extracted and digitized, including age, numbers of lesions, performance status (ECOG), presence of symptoms, arrangement with surgery (pre- or post-surgery radiation therapy), re-treatment, primary cancer type, and metastasis to other sites. A 3D convolutional neural network (CNN) architecture was built using three encoding paths with the same kernel and filters to capture the different image and contour features. Specifically, one path was built to capture the tumor feature, including the size and location of the tumor, another path was built to capture the relative spatial relationship between the tumor and OARs, and the third path was built to capture the clinical parameters. The model combines information from three paths to predict dose prescription. The actual prescription in the patient record was used as ground truth for model training. The model performance was assessed by 19-fold-cross-validation, with each fold consisting of randomly selected 128 training, 16 validation, and 8 testing subjects.
Result
The dose prescriptions of 152 patient cases included 48 cases with 1 × 24 Gy, 48 cases with 1 × 20–22 Gy, 32 cases with 3 × 9 Gy, and 24 cases with 5 × 6 Gy prescribed by 8 physicians. The AI model prescribed correctly for 124 (82 %) cases, including 44 (92 %) cases with 1 × 24 Gy, 36 (75 %) cases with 1 × 20–22 Gy, 25 (78 %) cases with 3 × 9 Gy, and 19 (79 %) cases with 5 × 6 Gy. Analysis of the failed cases showed the potential cause of practice variations across individual physicians, which were not accounted for in the model trained by the group data. Including clinical parameters improved the overall prediction accuracy by 20 %.
Conclusion
To our best knowledge, this is the first study to demonstrate the feasibility of AI in predicting dose prescription in CDM in radiation therapy. Such CDM models can serve as vital tools to address healthcare disparities by providing preliminary consultations to patients in underdeveloped areas or as a valuable quality assurance (QA) tool for physicians to cross-check intra- and inter-institution practices.
1. Introduction
Clinical decision-making (CDM) is involved in every step of the radiation therapy workflow, including initial consultation, patient simulation scanning, target and healthy tissue contouring, dose prescription, treatment planning and review, quality assurance (QA), delivery of radiation therapy, and follow-up care [1]. Consultation with a radiation oncologist is the first step in the clinical workflow. The radiation oncologist reviews detailed patient medical information and discusses with patients about different treatment options to decide upon a treatment plan [2]. Following the initial consultation, the patient will undergo a simulation CT image scan and other imaging modality scans such as MRI as needed. The physician will then use the images to contour the tumor and organs at risk (OAR) and prescribes radiation dose to the tumor with dose constraints for the surrounding OARs. Based on the contours and dose prescription, dosimetrists and physicists will design the treatment plan with different beam arrangements to achieve adequate dose coverage to the tumor while minimizing the radiation dose to OARs. The treatment plan is then reviewed and approved by the physician and verified by the QA process. Once the treatment plan is ready, the patient returns for the actual treatment delivery, which spans from a few days to weeks. After the treatment is completed, follow-up visits and scans are arranged to assess the treatment response and design medical care accordingly [1], [3]. Globally, there is a significant healthcare inequality with physicians in the developing countries or even low-resource areas in the developed countries lacking expertise or experience in CDM, significantly limiting the quality of care patients receive in these regions. Addressing this clinical challenge is especially critical for cancer patients since the physician’s CDM can directly impact the patient’s survival or life expectancy. Over half of all patients with cancer live in low-income or middle-income countries [4]. Workforce and equipment shortages in these resource-constrained settings have left >50 % of patients who are expected to benefit from radiotherapy without access to this treatment, with this value being up to 90 % in some low-income countries [5]. There is also a disparity among the workforce within developed countries with fewer resources and expertise in small rural hospitals [6], [7]. AI has the potential to alleviate some of these workforce shortages and inequality by providing specialized expert knowledge across disease sites and treatment modalities [8]. For example, AI can be trained to model the expert physicians’ decision-making process and act as a surrogate of them to assist the clinical decision process in clinics with limited resources or experiences. AI-assisted clinical decision-making can increase the efficiency, accuracy, and quality of radiation therapy, thus enhancing value-based cancer care delivery in today’s resource-limited healthcare environment.
AI has pushed the limits of what is possible in the domain of medical image processing, particularly in image registration, detection, segmentation, regression, and classification [9], [10], [11], [12], [13]. Meanwhile, AI has been reported to improve the quality and efficiency of a large variety of tasks in radiation oncology, such as image enhancement, treatment planning, organ segmentation, quality assurance, and treatment response prediction, as shown in many publications including ours [14], [15], [16], [17], [18], [19]. Convolutional neural network (CNN) has been extensively studied and shown to improve prediction performance using large amounts of pre-labeled data [20]. AI is transforming many fields of medicine and has the potential to address many of the challenges faced in radiation therapy and thereby improve the availability and quality of cancer care worldwide. Although novel innovations in AI have enabled the comprehensive analysis of diverse observations such as clinical, imaging, genomics, and treatment features [21], most AI-based applications in medicine focus on diagnosis or treatment optimization, and few are involved with treatment decision making [22]. Clinical decision-making in oncology is often complicated, lacks consensus, and contains uncertainties [23]. Back in 2014, IBM’s artificial intelligence platform, known as IBM Watson, was developed to perform CDM in diagnosis and treatment. Watson aimed to learn from the vast amount of literature, clinical guidelines, treatment records, and outcome data to try to come up with clinical decisions that can even outperform physicians, which is overly ambitious and unrealistic at present due to the limitation of current AI models and various practical challenges in using the medical literature and patient records [24]. Besides, the Watson models ignored the clinical practice variations when training and testing the models in quite different clinical practices in different countries, leading to unsatisfactory performance [25]. To date, it remains challenging and impractical to build an effective universal CDM model that can account for all the variations across different clinical practices.
We propose to take a more practical approach to tackle this issue by developing AI to solely follow the thought process of physicians to mimic their CDM. The goal is to develop a model to reproduce physician CDM as closely as possible. Instead of training the model from literature, we propose to train the model based on the actual treatment records with the incorporation of physicians’ logical decision process in the model. Moreover, the model is trained to be institution-specific using the dataset from a specific institution, removing impact from practice variations across institutions. These practical designs make it more realistic to build an effective model for CDM. In this study, we used dose prescription in radiotherapy as an example to demonstrate the feasibility of such an approach. SRS dose prescription has evolved since RTOG 90-05, which was a dose escalation trial prescribing single fraction radiosurgery for recurrent previously irradiated solitary brain tumors, establishing dose prescriptions based on size. [26] Larger tumors have a higher risk of local failure and radio-necrosis, which led to fractionation for larger lesions to try and improve local control and reduce the risk of radio-necrosis. Dose fractionation is also considered for lesions adjacent to critical structures, such as the brainstem and optic apparatus, to minimize the toxicities. The scope of SRS and experience with SRS has also expanded with its use in the up-front setting and for multiple metastases, while at the same time, advances in systemic therapy improved extracranial control of the disease. SRS dose prescription is a complex decision process involving balancing many factors, including the number of lesions, size of lesions, location of lesions, the total volume of disease, prior treatment, performance status, histology, etc. We developed a three-path three-dimensional CNN model to automatically prescribe doses based on lesion and OARs from CT images and non-image clinical parameters. To our knowledge, this is the first time an AI model has been developed to predict dose prescription in CDM of radiotherapy. Such a CDM model can serve as a surrogate for the physicians it models from to address healthcare disparities by providing preliminary consultations to patients in underdeveloped areas. It can also serve as a valuable QA tool representing a specific institution’s clinical practice for physicians to cross-check intra- and inter-institution practice variations.
2. Materials and methods
2.1. Patient data extraction
The study included 152 patients with brain metastases treated with Stereotactic Radiosurgery (SRS) or Stereotactic Radiation Therapy (SRT) from 2017 to 2021. The study was approved by the institutional review board (IRB) to perform the human subject’s research. All methods to acquire image data were performed following the relevant guidelines and regulations.
2.2. CT data processing
3D CT images and radiation therapy (RT) structures, including target volume and organ at risk (OAR) structures, were extracted from the patient record in the treatment planning system. The target volume used for dose prescription can be a gross tumor target volume (GTV) or a planning target volume (PTV) with margins added to the GTV, depending on the location and size of the target and the preference of the physician. Brainstem structure was exported as the primary OAR structure since it’s the most commonly concerned OAR structure in SRS/SRT dose prescription. In routine clinical practice, physicians typically evaluate the target volume's size, shape, and location and the relative position between the target and OAR to decide the dose prescription. To mimic the physician’s thought process, we extracted the masks of the target volume and OAR to obtain the above information physicians used to decide on dose prescription and used them as inputs to the AI model. To differentiate the target and OAR masks in the model inputs, the mask values were set to 1 and 2 for OAR and target masks, respectively.
2.3. Clinical parameter processing
Clinical parameters are also important factors physicians consider when choosing dose prescriptions. Therefore, in this study, we also extracted clinical parameters and digitized them for use as inputs to the AI model. The list of non-image clinical parameters and their selections for the model inputs based on physicians’ consideration of their relevance to dose prescription design are the following: Age (Y), Number of lesions (Y), ECOG (Y), Primary cancer type (Y), Genetic (N), Re-treatment (Y), Adjuvant chemotherapy (N), Metastasis to other sites (Y), Presence of symptoms (Y), Pre/Post-surgery (Y), Current medication (N). The selected clinical parameters were digitized as follows based on the physician’s input: age (0 for <60 years; 1 for 60–75 years; 2 for >75 years old), number of lesions (0 for 1 lesion; 1 for 2–5 lesions; 2 for >5 lesions), ECOG (0 for fully active; 1 for restricted in physically strenuous activity; 2 for ambulatory and capable of all self-care but unable to carry out any work activity; 3 for capable of only limited self-care; 4 for completely disabled; 5 for death), primary cancer type (1 for melanoma and sarcoma cancer which will be given special consideration in dose prescription based on input from the physician; 0 for others.), re-treatment (0 for no re-treatment; 1 for the re-treatment in a different region; 2 for re-treatment in the same region), metastasis to other sites (0 for no metastatis; 1 for metastasis to other sites), presence of symptoms (0 for no; 1 for yes), pre/post-surgery (0 for no surgery before or after RT; 1 for pre/post-surgery with RT).
2.4. Network architecture
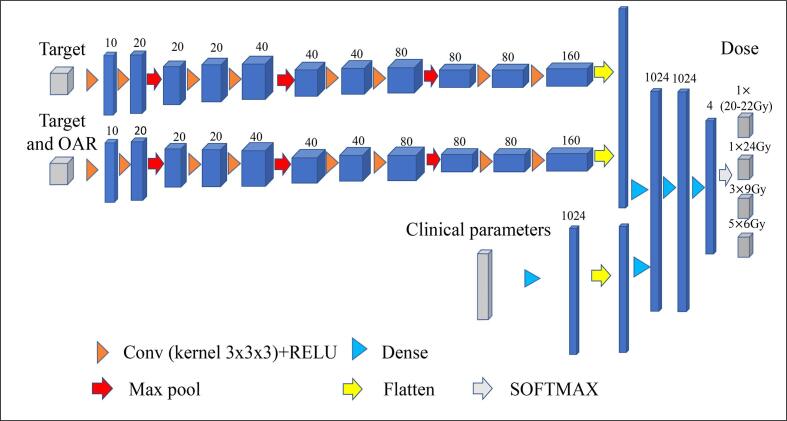
As shown in Fig. 1, a 3D convolutional neural network (CNN) architecture with three encoding paths was built to capture the image and non-image features based on the CT data and clinical parameters. Specifically, the first path was built to capture the target volume feature, including the size and location of the target; the second path was built to capture the relative spatial relationship between the target and brainstem; the third path was built to capture the patient clinical parameters. Both the first and second encoding paths have the same kernel with fixed filters to control the unique feature of each CT image input. Inside each of these two encoding paths, the corresponding kernel convolution is applied twice with a rectified linear unit (RELU), a dropout layer is included between the convolutions with a dropout rate of 0.4, and a 2 × 2 × 2 max-pooling operation is used in each layer [20]. The number of feature channels doubles after the max-pooling operation. Inside the third encoding path for clinical parameters, a dense layer is applied to increase the weighting of the sparse clinical parameters. After encoding, the three paths are processed by 3D convolutional layers for feature extraction and then connected to three fully connected layers. Dropout layers with a rate of 0.4 are applied after each fully connected layer. In the final step, the output from the last fully connected layer feeds a SoftMax, which maps the feature vector to the final classification of dose prescriptions. Four classes of dose prescriptions were used in our study, including 1 × 20-22 Gy, 1 × 24Gy, 3 × 9Gy, and 5 × 6Gy. Note that 20–22 Gy dose prescriptions were combined into one class in this initial study due to their similarity in clinical considerations. Categorical cross-entropy was applied as a loss function. Glorot (Xavier) normal initializer was used for this symmetric CNN based on CT images [27], which drew samples from a truncated normal distribution centered on zero with stdev = sqrt (2/( + )), where and were the numbers of input and output units, respectively, in the weight tensor. Adam optimizer was applied to train this model [28]. The learning rates ranging from 1 × 10−6 to 1 × 10−4 were tested and a learning rate of 2 × 10−5 with 1000 epochs was selected based on the model convergence.
Fig. 1.
The architecture of the three-path three-dimensional CNN model. Each blue cuboid corresponds to a feature map. The number of channels is denoted on the top of the cuboid. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The 3 Paths model above will be called 3P model in the following sections. We also created two other models for comparison: (1). 1 Path (1P) model: one-path three-dimensional CNN model that only uses the second path of the 3P model to capture the target and brainstem information for the input; (2). 2 Path (2P) model: a two-path three-dimensional CNN model that uses the first two paths of the 3P model as input to use only image information without clinical parameters for dose prediction.
2.5. Model training, validation, and testing
Target size and target-to-OAR distance have a major impact on the dose prescription. A high fractional dose is typically given to patients with a small target and large target-to-OAR distance, while a low fractional dose is given to the contrary. Dose prescription for patients with medium target size and target-to-OAR distance is the most challenging to learn for AI models since it’s in the grey areas transitioning from high to low fractional dose. Thus, more training data from these medium cases are needed to train the AI model to handle these challenging situations. To address this need, we selected 24 patients (out of the 152) with medium target size and target-to-OAR distances and performed data augmentation by rotating the images by 90, 180, and 270 degrees. The final model performance was assessed by 19-fold cross-validation, in which each fold consisted of randomly selected 128 training, 16 validation, and 8 testing subjects. Note that the augmented images were only used when the patient was selected for the training data.
2.6. Model performance evaluation and statistical analysis
The performance of the dose prediction model was evaluated using the following quantitative metrics: accuracy, sensitivity, specificity, receiver-operating characteristic (ROC) curve analysis, and the area under the curve (AUC). A confusion matrix was used to compare the predicted prescription with the actual prescription extracted from the patient record [29].
3. Result
3.1. Patient characteristics
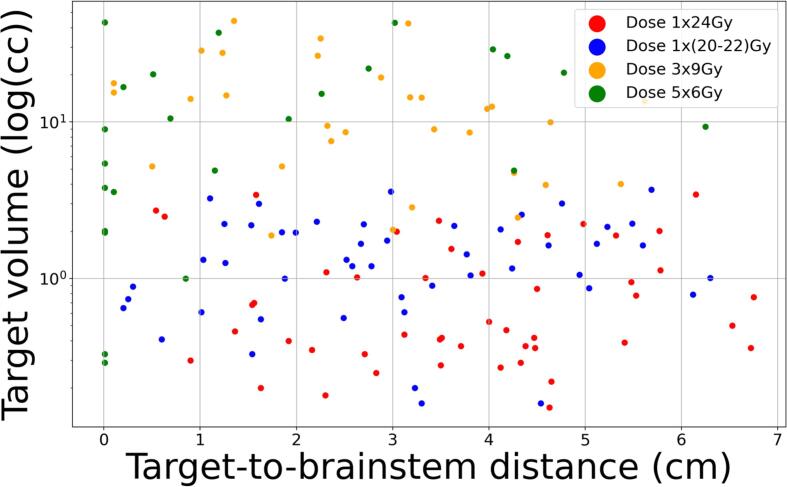
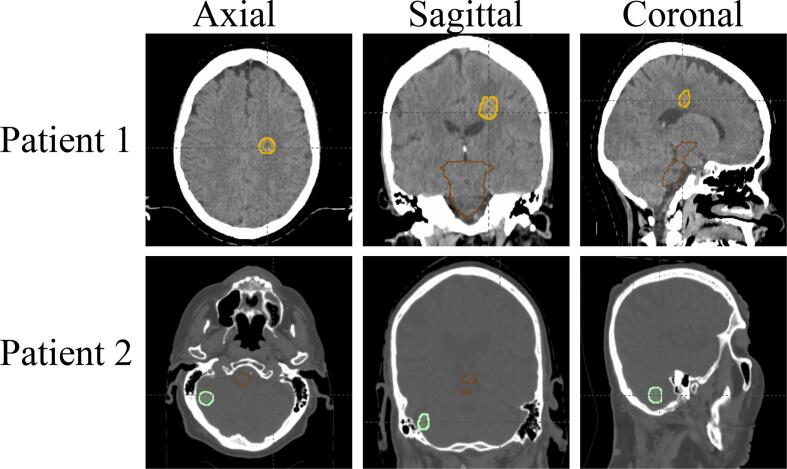
A total of 152 patients with dose prescriptions 1 × 20–22 Gy, 1 × 24 Gy, 3 × 9 Gy, 5 × 6 Gy were evaluated. Of these patients, 48 (31.5 %) were dose prescription 1 × 20–22 Gy, 48 (31.5 %) were dose prescription 1 × 24 Gy, 32 (21.0 %) were dose prescription 3 × 9 Gy, and 24 (16.0 %) were dose prescription 5 × 6 Gy. The clinical parameters of the patient population are shown in Table 1, including age, numbers of lesions, performance status (ECOG), symptomatic, surgery arrangement, re-treatment, primary cancer type, and metastasis to other sites. Fig. 2a shows the scatter plots for the target volume vs target-to-brainstem distances for all patient cases treated with different dose prescriptions. Although Fig. 2a shows a general trend of decreasing target-to-brainstem distances and increasing target volumes from high fractional such as 1 × 24Gy to low fractional doses such as 3 × 9Gy or 5 × 6Gy, there are also “grey zones” where cases with different dose prescriptions are mixed. This scatters plot demonstrates the challenge of predicting dose prescriptions, especially for cases in the grey zone, and the need to incorporate non-image clinical parameters for these predictions. Fig. 2b further shows two example patients with similar target-to-brainstem distances and different tumor sizes. Based on the images, patient 1 had a smaller tumor size and thus should receive a higher fractional dose than patient 2, which is contrary to the actual prescription doses. These examples further demonstrates the need for non-image clinical parameters for dose prescription prediction.
Table 1.
Summary of patient characteristics and treatment parameters.
| Parameters | Patients who received dose 1 × 24 Gy |
Patients who received dose 1 × 20–22 Gy |
Patients who received dose 3 × 9 Gy | Patients who received dose 5 × 6 Gy |
|---|---|---|---|---|
| Total Patient Number | 48 | 48 | 32 | 24 |
| Age (y) | ||||
| Median | 63 | 66 | 64 | 65 |
| Range | 35---84 | 36---84 | 27---90 | 36---85 |
| ECOG | ||||
| 0 | 21 (44 %) | 6 (13 %) | 10 (31 %) | 1 (4 %) |
| 1,2 | 26 (54 %) | 41 (85 %) | 22 (69 %) | 19 (79 %) |
| 3 | 1 (2 %) | 1 (2 %) | 0 (0 %) | 4 (17 %) |
| Number of metastases | ||||
| Single | 25 (52 %) | 21 (44 %) | 20 (62 %) | 13 (54 %) |
| Multiple | 23 (48 %) | 27 (56 %) | 12 (38 %) | 11 (46 %) |
| Presence of symptoms | ||||
| Yes | 10 (21 %) | 18 (38 %) | 10 (31 %) | 12 (50 %) |
| No | 38 (79 %) | 30 (62 %) | 22 (69 %) | 12 (50 %) |
| Primary Cancer type | ||||
| Lung | 28 (58 %) | 24 (50 %) | 17 (53 %) | 16 (67 %) |
| Breast | 9 (19 %) | 7 (15 %) | 4 (13 %) | 2 (8 %) |
| Melanoma | 1 (2 %) | 4 (8 %) | 0 (0 %) | 1 (4 %) |
| others | 10 (21 %) | 13 (27 %) | 11 (34 %) | 5 (21 %) |
| Re-treatment | ||||
| Yes | 4 (8 %) | 22 (46 %) | 2 (6 %) | 9 (38 %) |
| No | 44 (92 %) | 26 (54 %) | 30 (94 %) | 15 (62 %) |
| Pre/Post surgery | ||||
| Yes | 2 (4 %) | 12 (25 %) | 12 (38 %) | 10 (42 %) |
| No | 46 (96 %) | 36 (75 %) | 20 (62 %) | 14 (58 %) |
| Metastasis to other sites | ||||
| Yes | 8 (17 %) | 25 (52 %) | 14 (44 %) | 12 (50 %) |
| No | 40 (83 %) | 23 (48 %) | 18 (56 %) | 12 (50 %) |
Fig. 2.
A. scatter plots of the target volume vs target-to-brainstem distance for all patients treated with different dose prescriptions.. b. Two representative patients. Patient 1 (top) lesion size (2.05 cc), distance from the brainstem (3.03 cm), treated with 3 × 9 Gy. Patient 2 lesion size (2.00 cc), distance from the brainstem (3.00 cm), treated with 1 × 24 Gy.
3.2. Dose prescription prediction accuracy for different models
To investigate the impact of different model inputs on the prediction accuracy, we evaluated the three models explained in the section 2.4: (1). 1P model: 88 (58 %) patients were predicted correctly, and 64 (42 %) were misclassified based on the contour tumor and OARs in the CT images. (2). 2P model: 102 (67 %) dose prescriptions were predicted correctly, and 50 (33 %) were misclassified. (3). 3P model: 124 (82 %) dose prescriptions were predicted correctly, and 28 (18 %) were misclassified, showing the benefit of adding clinical parameters. The misclassified dose prescriptions for the 3P model include 4 (1 × 24 Gy), 12 (1 × 21 Gy), 7 (3 × 9 Gy), and 5 (5 × 6 Gy). Table 2 shows the detailed model validation results. As indicated by the green numbers, the 3P model outperformed other models for all dose prescription categories. We further asked a designated physician to retrospectively review and prescribe dose to the 28 failed cases by the 3P model. Results showed prescriptions for 14 cases were changed by the physician compared to the treatment record and matched with the AI prediction. This preliminary analysis indicates practice variations across individual physicians can be a cause of failed cases in the model prediction.
Table 2.
Summary of dose prediction performance using different networks. (Green and Red colors highlight the correct and wrong predictions, respectively).
| Dose prescription |
1 × 24 Gy in record (48) |
1 × 20–22 Gy in record (48) |
3 × 9 Gy In record (32) |
5 × 6 Gy In record (24) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model Type (CNN) | 1P | 2P | 3P | 1P | 2P | 3P | 1P | 2P | 3P | 1P | 2P | 3P |
| Predicted 1 × 24 Gy | 29 | 34 | 44 | 12 | 10 |
7 | 2 | 0 | 0 | 2 | 0 | 0 |
| Predicted 1 × 20–22 Gy | 12 | 12 | 2 | 27 | 32 | 36 | 4 | 3 | 1 | 2 | 1 | 1 |
| Predicted 3 × 9 Gy | 3 | 2 | 2 | 6 | 6 | 3 | 16 | 19 | 25 | 6 | 6 | 4 |
| Predicted 5 × 6 Gy | 4 | 0 | 0 | 3 | 2 | 2 | 8 | 10 | 6 | 16 | 17 | 19 |
| Model type | Accuracy mean (range) |
Sensitivity mean (range) |
Specificity mean (range) |
AUC | ||||||||
| 1P | 0.79 (0.74–0.85) | 0.58 (0.50–0.67) | 0.86 (0.83–0.88) | 0.84 | ||||||||
| 2P | 0.84 (0.79–0.88) | 0.67 (0.59–0.71) | 0.89 (0.85–0.91) | 0.88 | ||||||||
| 3P | 0.91 (0.88–0.93) | 0.81 (0.75–0.92) | 0.94 (0.91–0.96) | 0.94 | ||||||||
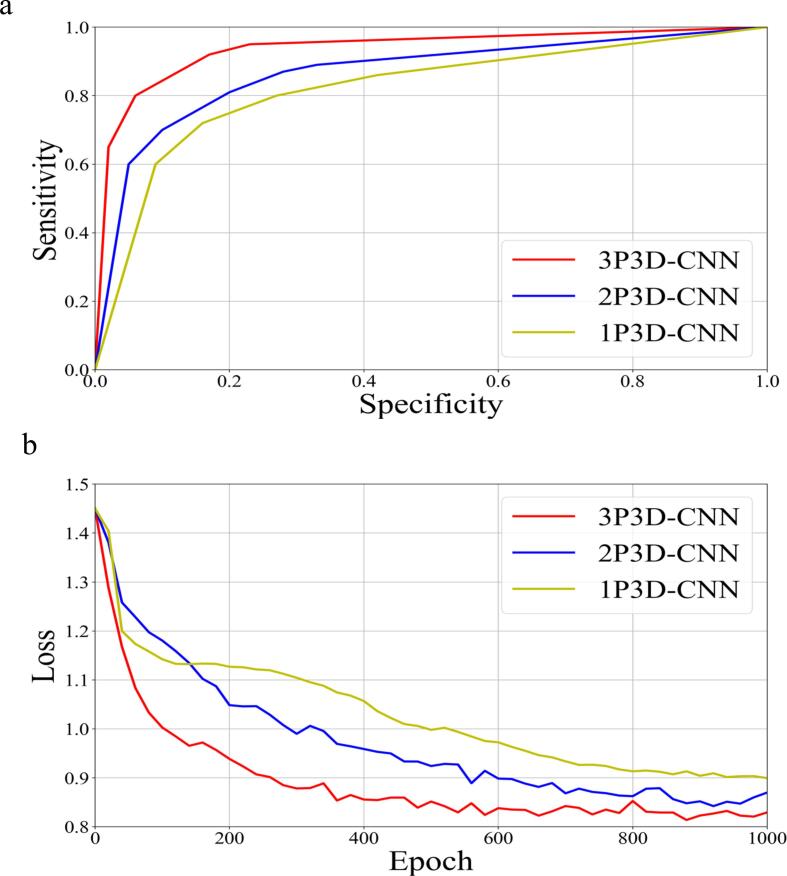
Fig. 3a shows the three trained models’ receiver operating characteristic curves for all the patients tested. The proposed three-path three-dimensional CNN achieved the best classification performance among all three models from Fig. 3a. The mean area under the curve (AUC) values were 0.84 (1P), 0.88 (2P), and 0.94 (3P), respectively. Fig. 3b shows the average loss function convergence for the training data, respectively.
Fig. 3.
A. roc curves from the three models, including one-path three-dimensional cnn (1p3d-cnn), two-path three-dimensional cnn (2p3d-cnn), and three-path three-dimensional cnn (3p3d-cnn). b. the objective loss function along with the epoch for the three cnn models.
3.3. Impact of including clinical parameters on the model performance
Results in Table 2 showed that including clinical parameters in the 3P model achieved the total accurate dose predictions for 124 patients compared to the 102 patients achieved by the 2P model without clinical parameters, indicating an increase of 20 % in prediction accuracy. The 3P model maintained accurate predictions for patients that were predicted correctly by the 2P model and corrected the wrong predictions by the 2P model for 22 patients. These 22 improved patient predictions based on clinical parameters included 10 patients for 1 × 24 Gy, 5 patients for 1 × 21 Gy, 5 patients for 3 × 9 Gy, and 2 patients for 5 × 6 Gy. The most prediction improvements were seen in the highest dose prescription category of 1 × 24 Gy, showing the important role of using clinical parameters. As shown in Table 1, patients in this dose prescription group (1 × 24 Gy) had the best clinical performance status represented by ECOG, which became a vital factor for physicians to consider prescribing high fractional doses. Therefore, including these clinical parameters was crucial for increasing the model's prediction accuracy. Fig. 4a shows an example of a patient with head and neck primary cancer and metastases to the lung and brain, who received the SRS to 4 brain lesions two months ago. This patient had the following characteristics: age (53), ECOG (1), number of lesions (1), volume (0.87 cc), and a small amount of edema surrounding the lesion. 2P model predicted the dose to be 1 × 24 Gy just based on the image since this is a small tumor far away from OAR. However, 3P model predicted the dose to be 1 × 20–22 Gy based on both image and non-image clinical information. The lower dose prescription by 3P model was due to consideration of poor patient prognosis and retreatment recorded in the clinical information. This patient was indeed treated by 1 × 21 Gy according to the record.
Fig. 4.
A. example patient showing the benefit of including clinical parameters for prediction. b. c. simulated cases to interpret the model prediction.
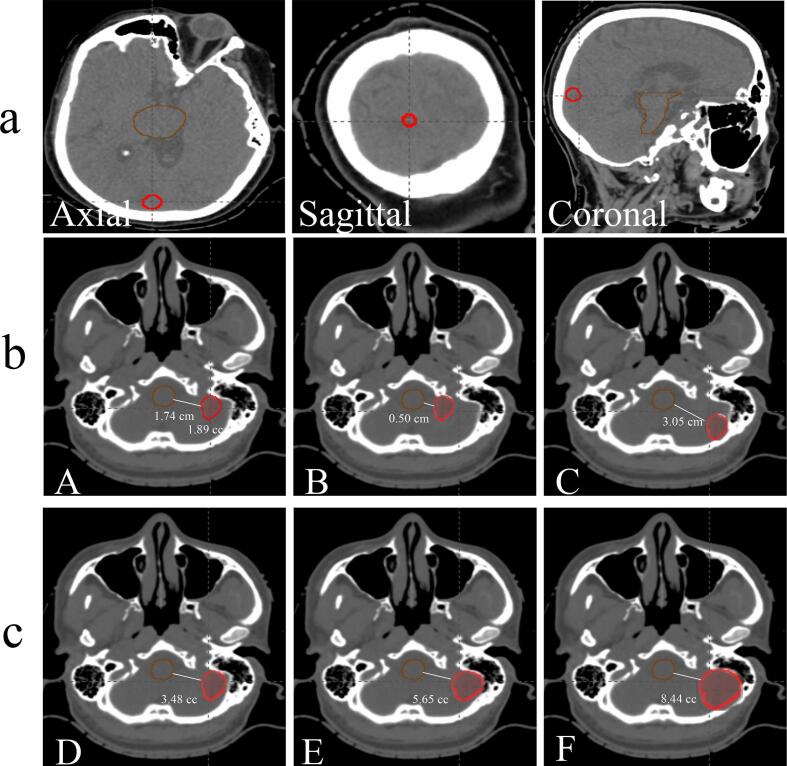
3.4. Model interpretability
To interpret the performance of our three-path three-dimensional CNN model, we studied a representative patient with a dose prescription of 3 9 Gy, a target volume of 1.89 cc, a target to brainstem distance of 1.74 cm, ECOG (1), number of lesions (1), age (47), primary cancer type (lung), re-treatment (Yes), metastasis to other sites (Yes), presence of symptoms (No), Pre/Post-surgery (No). This patient’s dose prescription was successfully predicted to be 3 × 9 Gy by our 3P model. To analyze and understand how the model made the prediction based on the various input factors, we did simulation studies to investigate the impact of individual input on the final prediction. To study the impact of the target-to-brainstem distance, we changed the target-to-brainstem distances from 1.74 cm to 0.50 cm and 3.05 cm while maintaining the target volume and re-tested the model with these new cases (Fig. 4b A-C). The model predicted 5 × 6 Gy and 1 × 21 Gy for the cases with distances of 0.50 cm and 3.05 cm, respectively, showing it learned to reduce the fractional dose for shorter distances. To study the impact of target size, we increased the target volume from 1.89 cc to 3.48 cc, 5.65 cc, and 8.44 cc while maintaining the target-to-brainstem minimal distance (Fig. 4c D-F), and re-tested the model with these three new cases. The probability of dose prescription of 5 × 6 Gy of these three volumes was 0.6, 0.8, and 1.0, respectively, showing the model learned to reduce the fractional dose when tumor size increases. To study the impact of clinical parameters, we changed all the clinical parameters (ECOG, age, primary cancer type, number of lesions, presence of symptoms, Pre/Post-surgery) to the worst situation. The probability of dose prescription of 5 × 6 Gy increased from 0.1 to 0.6 for the target volume of 1.89 cc when the clinical parameters deteriorated. This shows the model is able to reduce the fractional dose for patients with poor conditions, which is consistent with our clinical practice. Moreover, we increased the target volume from 1.89 cc to 3.48 cc and kept the clinical parameters in the worst situation, the probability of dose prescription of 5 × 6 Gy increased from 0.6 to 0.96, showing it learned to reduce the fractional dose for worse clinical parameters. These results demonstrated the model’s capability to learn the physicians’ general rules of prescribing higher fractional doses for smaller tumors or tumors far away from the brainstem and lower fractional doses for larger tumors or tumors close to the brainstem.
4. Discussion
The study is innovative on several levels. To our best knowledge, this is the first study to demonstrate the feasibility of AI in predicting dose prescription in CDM in radiation therapy. Different from previous methods, we developed the model based on the actual treatment records with the incorporation of physicians’ logical decision process to reproduce physicians’ CDM. Our development of a multi-path approach to incorporate both image information and non-image clinical parameters for CDM prediction is novel and effective in improving prediction accuracy. It also provides the potential to elucidate the impact of each factor in decision-making. Note that combining the imaging and non-imaging features is not trivial. Adding the non-imaging features to the model directly will not be effective because the number of clinical parameters is much less than that of the image features, making clinical parameters easily outweighed by other inputs in the model prediction. Therefore, a technical contribution of the current study is the use of a novel clinical parameters design, where a dense layer allows optimization of the weighting between clinical parameters and CT images. Results showed that our 3P model was able to assign the correct dose prescription to 82 % of patients without considering the practice variations across individual physicians.
Comparison studies showed that the 3P model consistently outperformed the 1P and 2P models in predicting all dose prescription categories. The 1P model only uses one path to capture the information of the target and OAR altogether, and therefore it may not allocate enough attention to the target size, shape, and location, which is crucial for determining the dose prescription. As a result, the 1P model achieved worse performance among all models. The 2P model alleviates the problem in the 1P model by adding a second path using the target image as the input to force the model to learn more from the target information that is vital for dose prescription prediction. Thus, the 2P model achieved superior performance to the 1P model. However, as the 2P model doesn’t use any non-image clinical parameters as input, it can’t predict dose prescriptions correctly for cases where clinical parameters play a major role in the dose prescription selection. The 3P model addressed the limitations of 1P and 2P models by incorporating both image and non-image clinical information for the model prediction and achieved the best performance in terms of prediction accuracy, sensitivity, specificity, and AUC, as shown in Table 3. Furthermore, the model interpretability study demonstrated that the 3P model was able to predict the dose prescription by considering the changes in the target size and target-to-OAR distances, mimicking the physician’s thought process. To validate the efficacy of the proposed CNN model against benchmark machine learning models, a random forest model was constructed to use CT image features (volume, distance from the brainstem), and eight clinical features to predict dose prescription. The random forest model misclassified 60 (39 %) patients, indicating the superiority of the proposed CNN model (misclassification of 18 % in the 3P model) [30].
There are several limitations to this study. First, our study is limited by the sample size of 152 patients. As a result, our study used 19-fold validation for model training and testing. In future work, we will accrue more patient data to further train and validate our model’s performance in different situations. Second, the current study hasn’t considered practice variations across individual physicians, which can impact the dose prescription as shown in the preliminary analysis of the failed cases in the 3P model in section 3.2. To address this limitation, the physician-specific model will be explored to model individual physician’s practices in the future. Moreover, the prediction accuracy would be improved by training a physician-specific model since clinical practice can vary across physicians due to variations in physicians’ knowledge, experiences, preferences, etc. Third, in this study, we only used the brainstem as the OAR for dose prescription because the brainstem is the most important and commonly concerned structure when determining dose prescription. Another important constraint is V12 (volume receiving 12 Gy or higher) of the whole brain excluding GTV. This constraint is inherently considered when the CT images with tumor masks were input into the model since V12 is very much dependent on the tumor size and location. OARs that haven’t been considered include optic nerves, chiasms, and cochlea. These structures can be important when the target is close to them. However, such scenarios account for only a small portion of the patient cases and therefore weren’t included in our study due to the lack of sufficient cases to train the model. We will include these OARs in future model training when sufficient cases are accrued.
This study demonstrated the feasibility of building an AI model to mimic the physician’s decision process for dose prescription. The development will serve as a stepping stone for further expanding the network to model other decision processes in the radiation therapy workflow, eventually building a suite of AI agents for end-to-end CDM support in radiotherapy. Such institution- or physician-specific AI tools can provide preliminary initial or secondary opinion consultations for patients to efficiently and cost-effectively survey the potential treatment options from different institutions/physicians so they can choose and pursue further consultations with the selected physicians afterward. It can also serve as a QA tool for physicians to cross-check practice variations or as a training tool for junior physicians or medical residents.
5. Conclusion
A three-dimensional CNN model with three encoding paths from CT images and non-image clinical parameters was successfully developed to predict dose prescription for brain metastases patients treated by radiotherapy. To our best knowledge, this is the first study to demonstrate the feasibility of AI in predicting dose prescription in CDM. Such CDM models can serve as vital tools to address healthcare disparities by providing preliminary initial or secondary opinion consultations to patients in underdeveloped areas or as a valuable QA tool for physicians to cross-check intra- and inter-institution practice variations.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by NIH R01EB028324.
References
- 1.Fraass B., Doppke K., Hunt M., Kutcher G., Starkschall G., Stern R., et al. American association of physicists in medicine radiation therapy committee task group 53: quality assurance for clinical radiotherapy treatment planning. Med Phys. 1998;25(10):1773–1829. doi: 10.1118/1.598373. [DOI] [PubMed] [Google Scholar]
- 2.Huynh E., Hosny A., Guthier C., Bitterman D.S., Petit S.F., Haas-Kogan D.A., et al. Artificial intelligence in radiation oncology. Nat Rev Clin Oncol. 2020;17(12):771–781. doi: 10.1038/s41571-020-0417-8. [DOI] [PubMed] [Google Scholar]
- 3.Ezzell G.A., Galvin J.M., Low D., Palta J.R., Rosen I., Sharpe M.B., et al. Guidance document on delivery, treatment planning, and clinical implementation of IMRT: report of the IMRT Subcommittee of the AAPM radiation therapy committee. Med Phys. 2003;30(8):2089–2115. doi: 10.1118/1.1591194. [DOI] [PubMed] [Google Scholar]
- 4.Barton M.B., Frommer M., Shafiq J. Role of radiotherapy in cancer control in low-income and middle-income countries. Lancet Oncol. 2006;7(7):584–595. doi: 10.1016/S1470-2045(06)70759-8. [DOI] [PubMed] [Google Scholar]
- 5.Zubizarreta E.H., Fidarova E., Healy B., Rosenblatt E. Need for radiotherapy in low and middle income countries–the silent crisis continues. Clin Oncol. 2015;27(2):107–114. doi: 10.1016/j.clon.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Wang L, Zhang Z, Wang D, Zhu H, Gao Y, et al. “Brilliant AI Doctor” in Rural Clinics: Challenges in AI-Powered Clinical Decision Support System Deployment. Proc. 2021 CHI Conf. Hum. Factors Comput. Syst., 2021, p. 1–18.
- 7.Kompa B., Snoek J., Beam A.L. Second opinion needed: communicating uncertainty in medical machine learning. NPJ Digit Med. 2021;4:1–6. doi: 10.1038/s41746-020-00367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosny A., Aerts H.J.W.L. Artificial intelligence for global health. Science. 2019;366(6468):955–956. doi: 10.1126/science.aay5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol (Berl) 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 10.Frid-Adar M., Diamant I., Klang E., Amitai M., Goldberger J., Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321:321–331. [Google Scholar]
- 11.Cao Y., Vassantachart A., Jason C.Y., Yu C., Ruan D., Sheng K., et al. Automatic detection and segmentation of multiple brain metastases on magnetic resonance image using asymmetric UNet architecture. Phys Med Biol. 2021;66:015003. doi: 10.1088/1361-6560/abca53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassantachart A., Cao Y., Gribble M., Guzman S., Ye J.C., Hurth K., et al. Automatic differentiation of Grade I and II meningiomas on magnetic resonance image using an asymmetric convolutional neural network. Sci Rep. 2022;12(1) doi: 10.1038/s41598-022-07859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Z., Yin F.-F., Ge Y., Ren L. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration. Phys Med Biol. 2020;65:015011. doi: 10.1088/1361-6560/ab5da0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohammadi R., Shokatian I., Salehi M., Arabi H., Shiri I., Zaidi H. Deep learning-based Auto-segmentation of Organs at Risk in High-Dose Rate Brachytherapy of Cervical Cancer. Radiother Oncol. 2021;159:231–240. doi: 10.1016/j.radonc.2021.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y., Hosny A., Zeleznik R., Parmar C., Coroller T., Franco I., et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res. 2019;25:3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z., Huang M.i., Jiang Z., Chang Y., Lu K.e., Yin F.-F., et al. Patient-specific deep learning model to enhance 4D-CBCT image for radiomics analysis. Phys Med Biol. 2022;67(8):085003. doi: 10.1088/1361-6560/ac5f6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Z., Zhang Z., Chang Y., Ge Y., Yin F.-F., Ren L. Enhancement of Four-dimensional Cone-beam Computed Tomography (4D-CBCT) using a Dual-encoder Convolutional Neural Network (DeCNN) IEEE Trans Radiat Plasma Med Sci. 2021 doi: 10.1109/trpms.2021.3133510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Z., Yin F.-F., Ge Y., Ren L. Enhancing digital tomosynthesis (DTS) for lung radiotherapy guidance using patient-specific deep learning model. Phys Med Biol. 2021;66:035009. doi: 10.1088/1361-6560/abcde8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Z., Chen Y., Zhang Y., Ge Y., Yin F.-F., Ren L. Augmentation of CBCT reconstructed from under-sampled projections using deep learning. IEEE Trans Med Imaging. 2019;38(11):2705–2715. doi: 10.1109/TMI.2019.2912791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. ArXiv Prepr ArXiv14091556 2014.
- 21.Ahmad Z., Rahim S., Zubair M., Abdul-Ghafar J. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn Pathol. 2021;16:1–16. doi: 10.1186/s13000-021-01085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang F., Jiang Y., Zhi H., Dong Y.i., Li H., Ma S., et al. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2(4):230–243. doi: 10.1136/svn-2017-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeBlanc T.W., Fish L.J., Bloom C.T., El-Jawahri A., Davis D.M., Locke S.C., et al. Patient experiences of acute myeloid leukemia: a qualitative study about diagnosis, illness understanding, and treatment decision-making. Psychooncology. 2017;26(12):2063–2068. doi: 10.1002/pon.4309. [DOI] [PubMed] [Google Scholar]
- 24.Strickland E. IBM Watson, heal thyself: how IBM overpromised and underdelivered on AI health care. IEEE Spectr. 2019;56(4):24–31. [Google Scholar]
- 25.Lee W.-S., Ahn S.M., Chung J.-W., Kim K.O., Kwon K.A., Kim Y., et al. Assessing concordance with Watson for Oncology, a cognitive computing decision support system for colon cancer treatment in Korea. JCO Clin Cancer Inform. 2018;2:1–8. doi: 10.1200/CCI.17.00109. [DOI] [PubMed] [Google Scholar]
- 26.Shaw E., Scott C., Souhami L., Dinapoli R., Kline R., Loeffler J., et al. Single dose radiosurgical treatment of recurrent previously irradiated primary brain tumors and brain metastases: final report of RTOG protocol 90–05. Int J Radiat Oncol Biol Phys. 2000;47(2):291–298. doi: 10.1016/s0360-3016(99)00507-6. [DOI] [PubMed] [Google Scholar]
- 27.Glorot X., Bengio Y. Understanding the difficulty of training deep feedforward neural networks. Proc Thirteen Int Conf Artif Intell Stat. 2010:249–256. [Google Scholar]
- 28.Kingma DP, Ba J. Adam: A method for stochastic optimization. ArXiv Prepr ArXiv14126980 2014.
- 29.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27(8):861–874. [Google Scholar]
- 30.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]