Summary
Fibrosis is a prominent pathological feature of skeletal muscle in Duchenne muscular dystrophy (DMD). The commonly used disease mouse model, mdx5cv, displays progressive fibrosis in the diaphragm but not limb muscles. We use single-cell RNA sequencing to determine the cellular expression of the genes involved in extracellular matrix (ECM) production and degradation in the mdx5cv diaphragm and quadriceps. We find that fibro/adipogenic progenitors (FAPs) are not only the primary source of ECM but also the predominant cells that express important ECM regulatory genes, including Ccn2, Ltbp4, Mmp2, Mmp14, Timp1, Timp2, and Loxs. The effector and regulatory functions are exerted by diverse FAP clusters which are different between diaphragm and quadriceps, indicating their activation by different tissue microenvironments. FAPs are more abundant in diaphragm than in quadriceps. Our findings suggest that the development of anti-fibrotic therapy for DMD should target not only the ECM production but also the pro-fibrogenic regulatory functions of FAPs.
Subject areas: Musculoskeletal medicine, Genetics
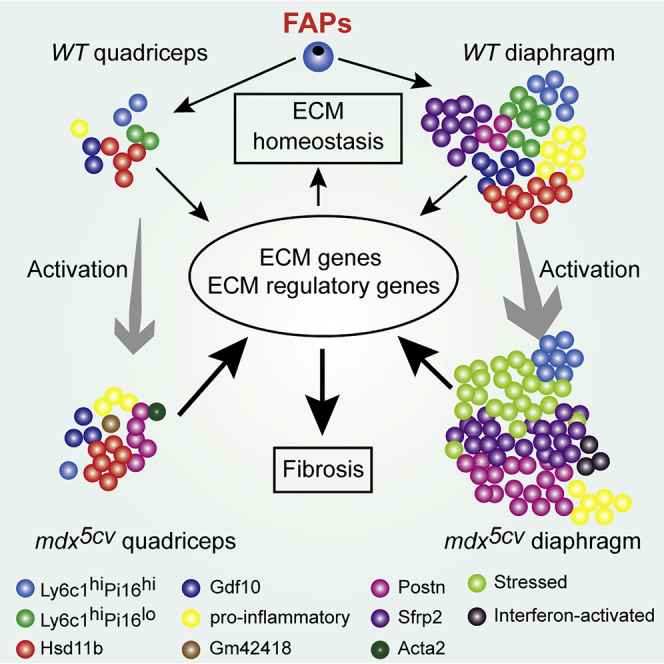
Graphical abstract
Highlights
-
•
scRNAseq supports the effector and regulatory functions of FAPs in muscle fibrosis
-
•
FAPs are more abundant in the diaphragm than in quadriceps
-
•
Diaphragm and quadriceps FAPs contain different clusters in the steady state
-
•
Diaphragm and quadriceps FAPs respond differently to muscular dystrophy
Musculoskeletal medicine; Genetics
Introduction
Duchene muscular dystrophy (DMD) is the most common genetic muscle disease, which is lethal with no cure currently.1,2 It is an X-linked recessive disease caused by the defective dystrophin gene. Fibrosis is a prominent pathologic feature of skeletal muscle, which directly contributes to muscle weakness.3,4 Development of effective anti-fibrotic therapies relies on the understanding of molecular and cellular mechanisms mediating and regulating muscle fibrosis. The commonly used DMD mouse models, mdx and mdx5cv, show progressive fibrosis in the diaphragm but not limb muscles.5,6,7,8 Little is known why limb and respiratory muscles show such a difference, and understanding the cellular and molecular contributions to the difference may benefit the anti-fibrotic therapy development.
Fibrosis is characterized by excessive accumulation of extracellular matrix (ECM) proteins that consist of collagens, fibronectin, and proteoglycans.9,10,11 Fibrosis development and progression in skeletal muscle can be promoted at different levels, including ECM gene upregulation stimulated by pro-fibrogenic factors such as transforming growth factor β1 (TGF-β1),12 connective tissue growth factor (CTGF),13 and osteopontin,14,15,16 reduced matrix metalloproteinases (MMPs)-mediated ECM protein degradation,17 increased tissue inhibitors of matrix metalloproteinases (TIMPs),17 and heightened collagen cross-linking catalyzed by lysyl oxidases (LOXs), which makes ECM stiff and resistant to proteolytic degradation.18,19
Fibro/adipogenic progenitors (FAPs) are muscle interstitial mesenchymal progenitors, expressing mesodermal marker PDGFRα and stem cell markers Sca-1 and CD34. They are bipotent progenitors that can differentiate into fibroblasts and adipocytes.20,21,22 They play important roles in skeletal muscle injury and repair.22,23 Upon injury, they become activated, proliferating and expanding rapidly.20,24,25 They provide a favorable tissue environment to promote satellite cell-mediated muscle regeneration.20,26,27,28,29 As muscle regeneration proceeds, excessive FAPs are cleared from the regenerative niche by apoptosis.25 Failing to do so leads to the pathological accumulation of FAPs, contributing to fibro-fatty replacement of muscle.21,25
In the present study, we performed single-cell RNA sequencing (scRNAseq) to determine the cellular expression of the genes involved in ECM production and degradation during skeletal muscle fibrosis in mdx5cv, and to identify differences between diaphragm and quadriceps. Our findings demonstrate that FAPs are not only the key effector cells that express a high level of ECM genes, but also the predominant regulatory cells that express a high level of Ccn2, Ltbp4, Mmp2, Mmp14, Timp1, Timp2, and Loxs to regulate ECM production and degradation. The effector and regulatory functions are exerted by diverse FAP clusters, which are different between diaphragm and quadriceps, indicating different tissue microenvironments in these two types of skeletal muscles. FAPs are much more abundant in diaphragm than in quadriceps. FAPs are the key effectors and regulators of mdx5cv diaphragm fibrosis.
Results
Fibro/adipogenic progenitors are the most abundant mononuclear cells in diaphragm but not quadriceps of both wild-type and mdx5cv mice
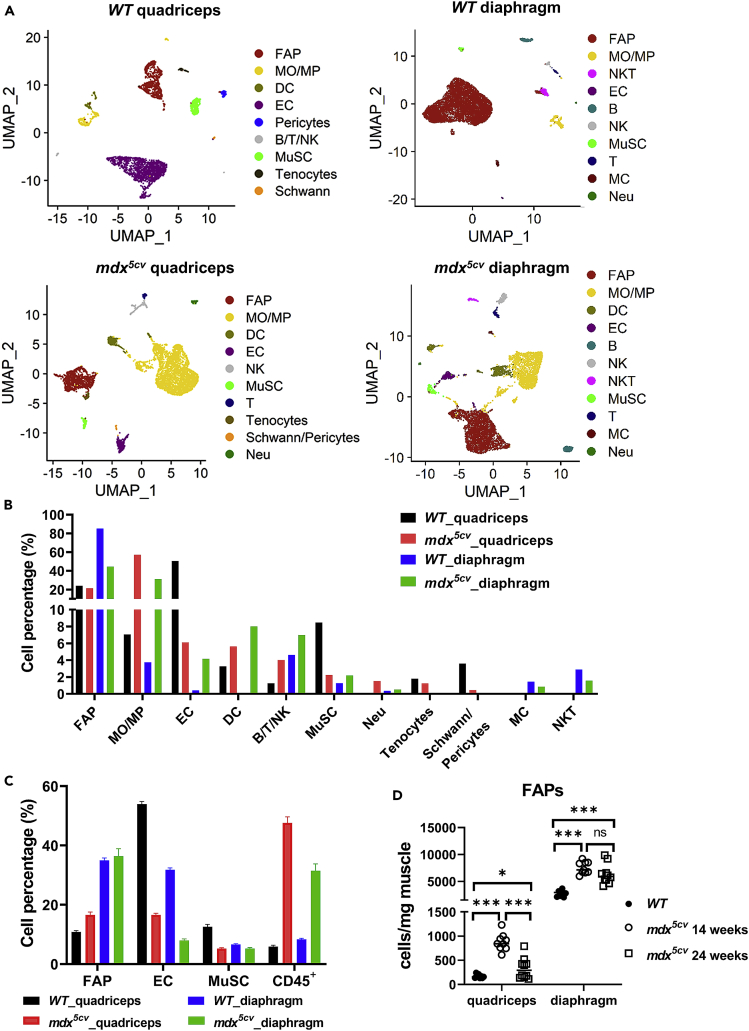
Muscle necrosis, inflammation, and fibrosis are evident in both diaphragm and quadriceps muscles of mdx5cv at 14 weeks of age. However, these changes subside afterward in the quadriceps with progressive fibrosis mainly seen in the diaphragm at 24 weeks of age (Figure S1A).5,30 To uncover the cellular and molecular differences accounting for the divergent fibrosis progression, we performed scRNAseq using single-cell suspensions prepared from diaphragm and quadriceps of mdx5cv and WT mice at 14 weeks of age. The single-cell suspensions contain mononuclear cells but not myofibers, as polynuclear myofibers cannot be included due to their large size. scRNAseq of the four muscle samples, each pooled from five male mice, were performed simultaneously (see Table S1 for quality control). By filtering out the cells of low quality,31 we obtained 16,766 genes from 3,814 cells of WT quadriceps, 17,677 genes from 6,988 cells of WT diaphragm, 18,398 genes from 6,302 cells of mdx5cv quadriceps, and 18,152 genes from 6,089 cells of mdx5cv diaphragm for analysis. Sequencing data were first analyzed using Uniform Mani-fold Approximation and Projection (UMAP) for dimension reduction to generate functionally enriched clusters in each sample. The identities of the generated clusters were determined by the expression of cell type-specific marker genes (Figure S2). Multiple cell types were identified in each sample (Figures 1A and S2, Tables S2 and S3). Most of the cell types were present in all four samples, but mesothelial cells (MCs) and invariant natural killer T cells (NKT) cells were only identified in diaphragm, while Schwann cells, pericytes, and tenocytes were only identified in quadriceps (Figure 1A). Mesothelium covers the surface of diaphragm but not quadriceps,32 which explains the detection of MCs only in diaphragm. The lack of other cell types in diaphragm or quadriceps is likely due to their very low abundance. Clusters with a high-level expression of Pdgfra, Ly6a, and Cd34 and lack of expression of Pecam1, Ptprc, Itga7, Tnmd, and Fmod were identified as FAPs (Figure S2B).
Figure 1.
FAPs are the most abundant among mononuclear cells in diaphragm but not quadriceps of both wild-type (WT) and mdx5cv mice
(A) Uniform Mani-fold Approximation and Projection (UMAP) dimension reduction analysis showing different cell types in WT and mdx5cv quadriceps and diaphragm muscles. FAP: Fibro/adipogenic progenitors, MO/MP: Monocyte/Macrophages, DC: Dendritic cells, EC: Endothelial cells, MuSC: Myogenic stem cells, B: B cells, T: T cells, NK: Natural killer cells, NKT: invariant Natural Killer T Cells, MC: Mesothelial cells, Neu: Neutrophils.
(B) Bar graph showing percentage of each cell type to total cells analyzed by scRNAseq in each indicated muscle sample, quantified by UMAP analysis.
(C) Bar graph showing percentages of FAP, EC, MuSC, and CD45+ cells to total mononuclear cells, quantified by FACS, in each indicated muscle sample (Qua.: quadriceps; Dia.: diaphragm).
(D) Scatterplot showing and comparing cell densities of FAPs in WT quadriceps, WT diaphragm, mdx5cv quadriceps, and mdx5cv diaphragm determined by FACS analysis. Data are represented as mean ± SEM Asterisks indicate significant differences (∗p < 0.05, ∗∗∗p < 0.001, ns: no significance, Kruskal-Wallis test followed by Dunn’s test for multiple comparisons).
In both WT and mdx5cv diaphragm, FAPs were the most abundant cell type identified by scRNAseq (Figure 1A). Endothelial cells (ECs) and monocytes/macrophages (MOs/MPs) were the most abundant cell types in WT and mdx5cv quadriceps, respectively (Figures 1A and 1B and Table S3). Since the cell representation could be altered by multiple processing steps of scRNAseq, we further determined the relative abundance of FAPs, ECs, myogenic stem cells (MuSCs), and CD45+ cells (contain MOs/MPs) by flow cytometry analysis (FACS) (Figures 1C and S1B). FACS and scRNAseq revealed similar percentages of these cells, except for the ECs in WT diaphragm, which showed a much lower percentage by scRNAseq than by FACS (Figures 1B and 1C). FAPs are therefore over-represented in WT diaphragm by scRNAseq. But FACS also showed that FAP was the most abundant cell type in WT diaphragm (Figure 1C). The FAP cell density, determined by FACS, was markedly higher in diaphragm than in quadriceps (Figure 1D). It increased dramatically in both quadriceps and diaphragm of mdx5cv as compared with WT at 14 weeks of age, but it was then reduced in mdx5cv quadriceps while remained high in mdx5cv diaphragm at 24 weeks of age (Figure 1D), corresponding to the progressive fibrosis seen in mdx5cv diaphragm but not quadriceps (Figure S1). The findings are consistent with previous studies reporting a higher level of PDGFRα protein expression in diaphragm than in limb muscles.33,34,35
Fibro/adipogenic progenitors are the primary cells expressing extracellular matrix genes in the skeletal muscle of both wild-type and mdx5cv mice
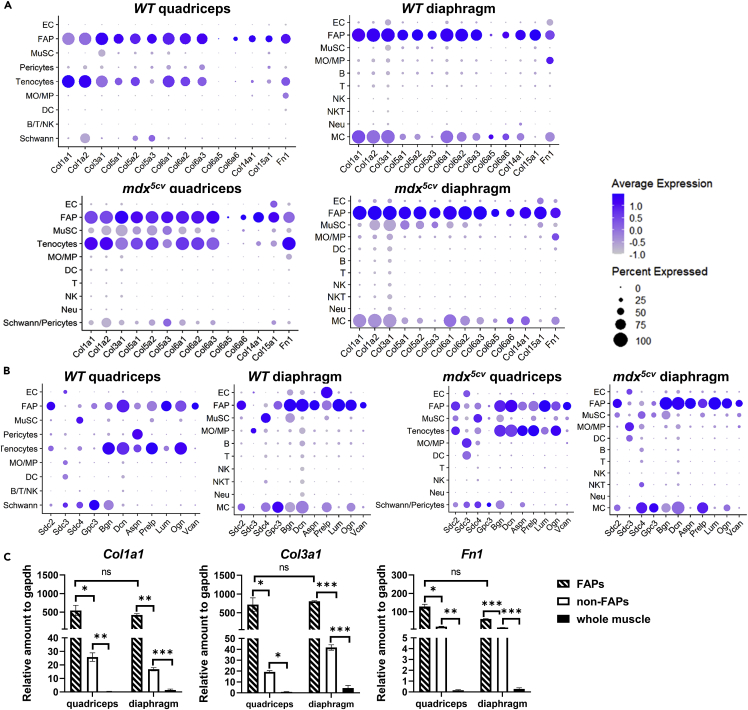
FAPs are known ECM-producing cells.36 To determine the relative contribution of different cell types to ECM production, we compared the cellular expression of the genes encoding collagens, fibronectin, proteoglycans, and synthesizing enzymes for glycosaminoglycans (GAGs) which are the components of many proteoglycans10 (Figures 2, S3, and S4). For all the collagen genes with detectable expression by scRNAseq, the high expression was predominantly detected in FAPs, as well as tenocytes in quadriceps, which include Col1s and Col3s that encode collagen I and collagen III, the two major fibrillary collagen constituents in interstitial tissue of skeletal muscle37 (Figures 2A and S3). FAPs and quadriceps tenocytes were also the major cell types that expressed a high level of fibronectin gene (Fn1) (Figure 2A) and many proteoglycan genes (Figures 2B and S4A). In addition, FAPs moderately expressed several genes encoding the synthesizing enzymes of GAGs (Figure S4B). MCs, Schwann cells, pericytes, and MuSCs also expressed multiple ECM genes but at a lower level (Figures 2A and 2B). Considering the relative low abundance, their contribution to the ECM production is likely limited. To determine the potential contribution by myofibers which cannot be included for scRNAseq, we performed quantitative reverse-transcription PCR (qRT-PCR) to compare FACS-sorted FAPs, FACS-sorted non-FAP cells, and whole muscle tissue samples from mdx5cv mice. The mRNA expression of Col1a1, Col3a1, and Fn1 by the whole muscle tissue was significantly lower than that by the two sorted cell samples (Figure 2C), indicating that myofibers do not significantly contribute to ECM production in dystrophic muscles. Together, our findings are consistent with the notion that FAPs are the key ECM-producing cells in skeletal muscle in both steady state and mdx5cv. Given the persistent high FAP density in mdx5cv diaphragm (Figure 1C), FAPs are the main effector cells mediating the progression of fibrosis in this muscle.
Figure 2.
FAPs are the predominant cells expressing extracellular matrix (ECM) genes in both WT and mdx5cv quadriceps and diaphragm
(A) Dot plot showing the expression of collagen and fibronectin genes by different cell types in different muscles. The dot color intensity reflects the average gene expression level, and the dot size represents the percentage of the cells expressing the gene.
(B) Dot plot showing the expression of proteoglycan genes.
(C) qRT-PCR showing and comparing Col1a1 and Col3a1, and Fn1 expression by FACS-sorted FAPs, FACS-sorted non-FAPs, and whole muscle of mdx5cv quadriceps and diaphragm at 14 weeks of age. Data are represented as mean ± SEM Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns: no significance, Kruskal-Wallis test followed by Dunn’s test for multiple comparisons). N = 5 mice/group.
Fibro/adipogenic progenitors are the predominant cells expressing many regulatory genes involved in skeletal muscle extracellular matrix production and degradation
To address which cell types are the key regulators of skeletal muscle fibrosis, we next analyzed the cellular expression of the genes encoding fibrogenic factors, MMPs, TIMPs, and LOXs.
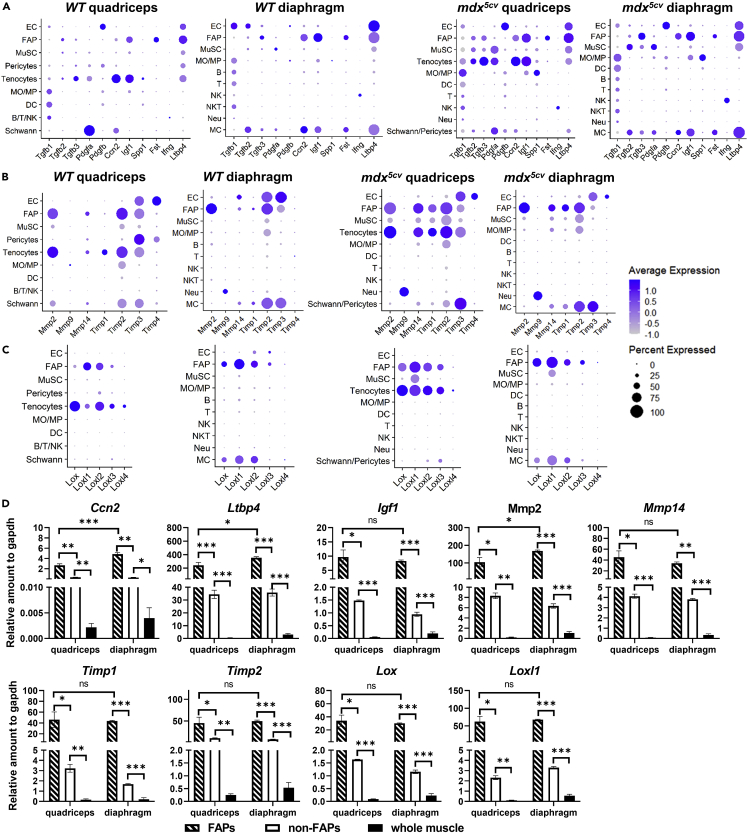
Among the genes that regulate FAP activation and ECM production (Figures 3A and S5A), the CTGF gene (Ccn2), insulin-like growth factor-1 (IGF-1) gene (Igf1), and follistatin gene (Fst) were preferentially expressed by FAPs in both WT and mdx5cv diaphragm. In quadriceps, tenocytes expressed a higher level of Ccn2 and Igf1 than FAPs, while FAPs were the major source of Fst. FAPs also expressed a high level of latent TGFβ binding protein 4 gene (Ltbp4). The FAP expression of these genes has also been reported by other studies.20,24,27,38,39 The findings indicate that FAPs regulate their own differentiation and ECM gene expression. FAPs in diaphragm and tenocytes in quadriceps expressed a high level of Tgfb3. Tgfb1 and Spp1, the two potent pro-fibrogenic genes, were mostly expressed by MOs/MPs in mdx5cv, consistent with the notion that MOs/MPs promote muscle fibrosis in mdx.40,41 MuSCs were the predominant cells expressing Tgfb2 and Pdgfa in mdx5cv but not WT muscles, indicating their acquired fibrogenic potential in mdx5cv. Pdgfb was mainly expressed by ECs in mdx5cv muscles. These findings are consistent with the previous report.42 IFN-γ gene (Ifng) was mostly expressed by NK cells. Taken together, different fibrogenic factor genes are preferentially expressed by different cells, with FAPs, tenocytes, and immune cells being the most important contributors. The contribution of MCs to diaphragm fibrosis is likely limited due to their rarity.
Figure 3.
FAPs are the predominant cells expressing important regulatory genes of ECM production and degradation
(A-C) Dot plots showing the expression of fibrogenic factor genes (A), Mmps (B), Timps (B), and Loxs (C) by different cell types in quadriceps and diaphragm of WT and mdx5cv mice.
(D) qRT-PCR showing and comparing the expression of selected ECM regulatory genes by FACS-sorted FAPs, FACS-sorted non-FAPs, and whole muscle of mdx5cv quadriceps and diaphragm at 14 weeks of age. Data are represented as mean ± SEM Asterisks indicate significant differences (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns: no significance, Kruskal-Wallis test followed by Dunn’s test for multiple comparisons). N = 5 mice/group.
Fibrosis is regulated not only at the level of ECM production but also at the level of ECM degradation. We next addressed which cells expressed Mmps, Timps, and Loxs to regulate ECM degradation (Figures 3B, 3C, and S5B). Among all the Mmps, Mmp2, Mmp9, and Mmp14 were strongly expressed (Figure S5B), with the expression of Mmp2 and Mmp14 mainly by FAPs and tenocytes while the expression of Mmp9 by neutrophils (Figure 3B). Timps were expressed by multiple cell types (Figure 3B), but Timp1 and Timp2 were preferentially expressed by FAPs and tenocytes. ECs were the main cells expressing Timp3 and Timp4, except for WT diaphragm where the Timp4 expression was diminished. Therefore, FAPs appear to play an active role in regulating ECM degradation. The genes that encode cross-linking enzymes, LOXs, were mainly expressed by FAPs and tenocytes (Figure 3C). FAPs may thus also regulate collagen cross-linking.
Taken together, FAPs actively express Ccn2, Ltbp4, Igf1, Fst, Mmp2, Mmp14, Timp1, Timp2, and Lox genes in skeletal muscle, regulating ECM production and degradation. Quantitative RT-PCR with FACS-sorted FAPs and non-FAP cells, as well as whole muscle samples, confirmed the major contribution of FAPs to the ECM regulatory gene expression in mdx5cv quadriceps and diaphragm (Figure 3D). The findings support the notion that FAPs may play a key role not only in mediating but also in regulating skeletal muscle ECM homeostasis in the steady state and fibrosis in muscular dystrophy.21,24,25,33,35,43,44,45
Fibro/adipogenic progenitors in mdx5cv diaphragm and quadriceps display differences in transcriptomes
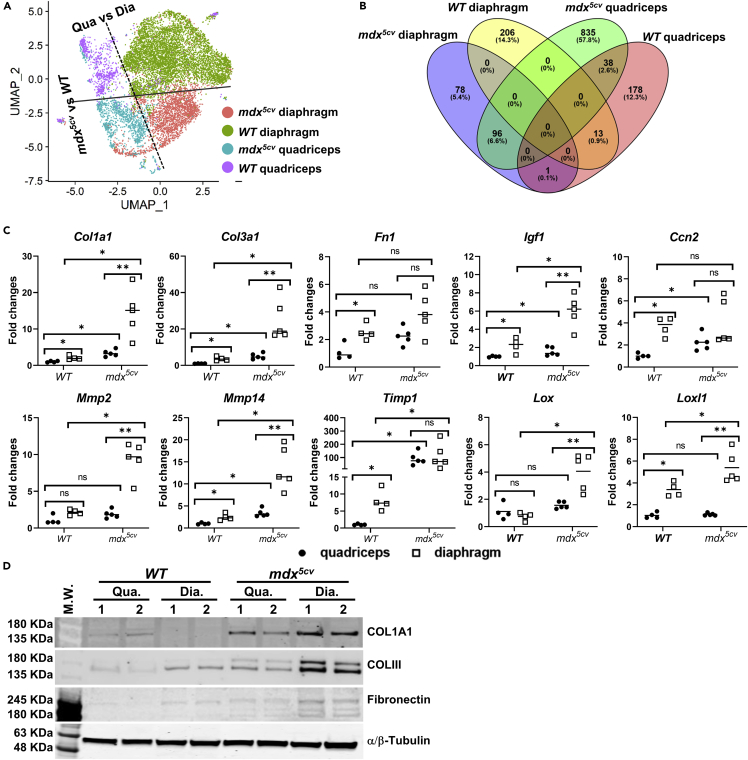
Since FAPs not only produce ECM but also regulate ECM production and degradation, we next determined whether the transcriptomes of FAPs in mdx5cv quadriceps and diaphragm are different to account for the divergent fibrosis progression in these two muscles. To this end, transcriptome data of FAPs from all four muscle samples (WT quadriceps: 846 cells; WT diaphragm: 5,829 cells; mdx5cv quadriceps: 1,140 cell; mdx5cv diaphragm: 2,515 cells) were merged and analyzed for differentially expressed genes (DEGs, log2FC ≥ 0.5 compared to the average expression level). UMAP analysis showed that FAP transcriptomes were different by the muscle type (quadriceps vs. diaphragm) and the disease state (WT vs. mdx5cv) (Figure 4A), with 835 distinct DEGs identified in mdx5cv quadriceps and 78 in mdx5cv diaphragm (Figure 4B and Data S1). The DEGs in mdx5cv quadriceps FAPs were functionally enriched in signaling pathways of TNF-α, IL-4 and IL-13, TGF-β, PDGF, and HGF, and in biological processes of cell migration, apoptosis, and angiogenesis. The DEGs in mdx5cv diaphragm FAPs were functionally enriched in TGF-β response and ECM regulation. It is surprising that most of the ECM genes, fibrogenic factor genes, Mmps, Timps, and Loxs were not present in the distinct DEGs of mdx5cv quadriceps or diaphragm FAPs (Data S1). Likewise, qRT-PCR showed no significant difference in Col1a1, Col3a1, Fn, Igf1, Mmp14, Timp1, Timp2, Lox, and Loxl1 expression between mdx5cv quadriceps and diaphragm FAPs (Figures 2C and 3D). The expression of Ccn2, Ltbp4, and Mmp2 only showed a less than 2-fold increase in mdx5cv diaphragm FAPs as compared with mdx5cv quadriceps FAPs (Figure 3D). Therefore, there is no significant difference in the FAP expression of the ECM or ECM regulatory genes at a single-cell level between the two muscles. However, the gene expression of Col1a1, Col3a1, Igf1, Mmp2, Mmp14, Lox and Loxl1 (Figure 4C) and the protein expression of collagen I, collagen III, and fibronectin (Figures 4D and S3B) at a whole-muscle level was significantly higher in mdx5cv diaphragm than in mdx5cv quadriceps, which is most likely contributed by the higher FAP density in mdx5cv diaphragm.
Figure 4.
FAPs in WT and mdx5cv diaphragm and quadriceps display differences in transcriptomes
(A) UMAP dimension reduction analysis showing different FAP transcriptomes in different muscle samples.
(B) Venn diagram depicting differentially expressed genes (DEGs) shared by or unique in FAPs of different muscles.
(C) qRT-PCR showing and comparing the expression of selected ECM and ECM regulatory genes by different muscles. Data are represented as mean ± SEM Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01; ∗∗∗p < 0.001, ns: no significance, Kruskal-Wallis test followed by Dunn’s test for multiple comparisons). N = 5 mice/group.
(D) Western blot showing and comparing the protein expression of collagen 1α1 (COL1A1), collagen 3α1 (COL3A1), and fibronectin among different muscles. Results showed represent 3 independent experiments with a total of 6 mice for each group.
Fibro/adipogenic progenitors consist of diverse clusters which are different between quadriceps and diaphragm in wild-type and mdx5cv mice
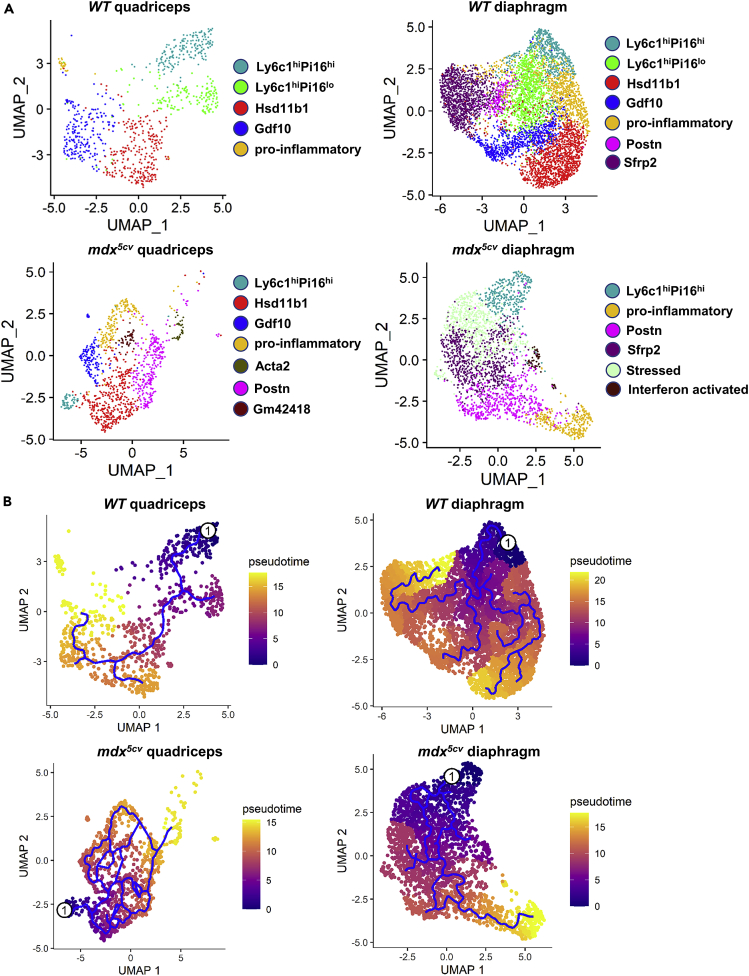
FAPs are functionally heterogeneous in both steady state and disease state.22,36,44,46,47,48,49 There are three types of tissue fibroblast clusters: universal clusters that are shared by all tissue fibroblasts in the steady state, specialized clusters that are induced by the tissue-specific microenvironment in the steady state, and activated clusters that are induced by a disease.50 We next addressed whether quadriceps and diaphragm have different specialized FAP clusters in the steady state and different activated FAP clusters in mdx5cv and whether different FAP clusters exert different effector and regulatory functions. To this end, we extracted transcriptome data of FAPs and re-clustered by UMAP analysis. Multiple clusters with differential transcriptomes were identified in each sample (Figures 5A and S6A).
Figure 5.
FAPs consist of diverse clusters which are different among quadriceps and diaphragm in WT and mdx5cv mice
(A) UMAP clustering identifying FAP clusters in each muscle sample.
(B) Pseudotime analysis showing the trajectory of FAP differentiation starting from the Ly6c1hiP16hi cluster (labeled as 1) in each muscle sample.
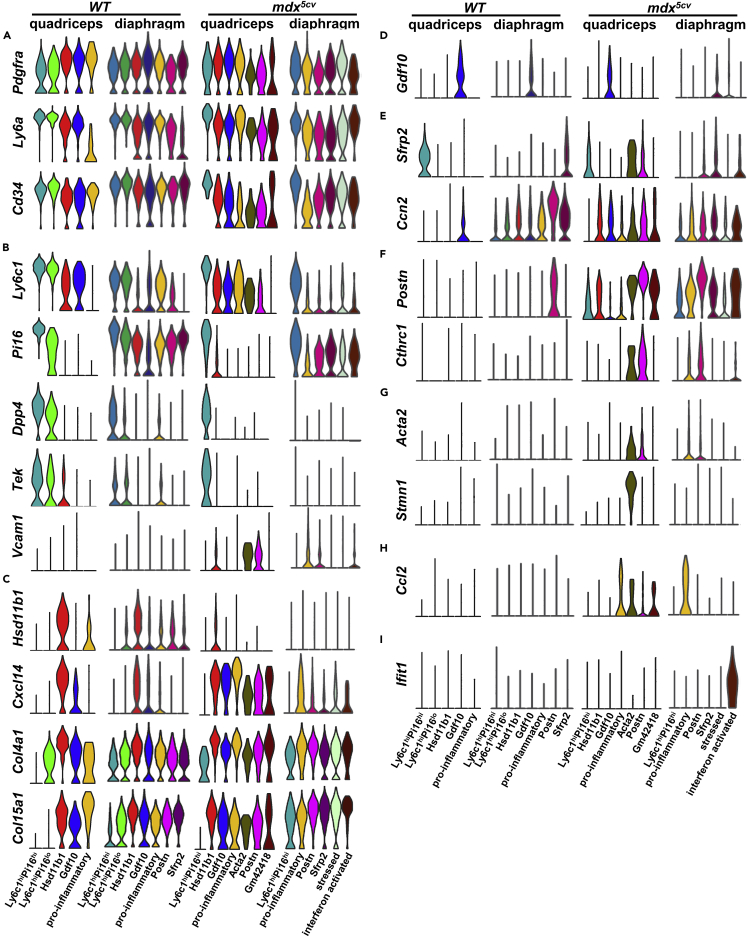
Five FAP clusters were identified in WT quadriceps. The two universal clusters identified by the cross-tissue transcriptome analysis of fibroblasts,50 including the adventitial stromal cell-like fibroblasts featured by Ly6c1, Pi16, Dpp4, and CD55 and the basement membrane-associated fibroblasts featured by Hsd11b1, Cxcl14, Col4a1, Col15a1, and Hspg2,50 were identified in our WT quadriceps FAPs and named Ly6c1hi and Hsd11b1 clusters, respectively (Figures 5A, 6A-6C, and S6A.1; Table S4). Similar FAP clusters were also identified by two previous transcriptome studies36,46 and named Dpp4+ and Cxcl14+ clusters.46 The findings confirm the reliability of our analysis. We named the clusters with different featured genes to best distinguish them from the other clusters in all four samples. Interestingly, we identified two adventitial Ly6c1hi FAP clusters, which were distinguished by the expression level of Pi16: Ly6c1hiPi16hi and Ly6c1hiPi16lo (Figures 5A and 6B; Table S4). Compared to the Ly6c1hiPi16hi cluster, the Ly6c1hiPi16lo cluster expressed a lower level of Dpp4 but a higher level of Col4a1 (Figures 6B and 6C), suggesting that this is an intermediate cluster between the adventitia stromal cell-like and basement membrane-associated clusters. The Ly6c1hi clusters expressed the highest level of mesenchymal progenitor markers Cd34 and Ly6a/Sca-1 (Figure 6A), indicating their higher stemness than the other clusters. Pseudotime analysis confirmed the progenitor status of the Ly6c1hiPi16hi cluster (Figure 5B), consistent with the finding by others.50 The Ly6c1hi clusters also expressed the highest level of TEK tyrosine kinase gene (Tek) (Figure 6B), an important molecule involved in vascular maintenance and homeostasis.44,51 The Hsd11b1 cluster expressed a high level of homeostatic chemokine genes, Cxcl14 and Ccl11 (Figure 6C and Table S4), presumably helping maintain tissue homeostasis.52,53 The two other clusters are specialized (Figures 5A, 6D, and S6A.1; Table S4). The Gdf10 cluster was enriched for genes involved in chondrogenesis, such as Gdf10, Mgp, Prg4, and Cilp (Figure 6D and Table S4). The pro-inflammatory cluster was very small (Figure S6B) and enriched for genes upregulated during inflammation, such as Cxcl1, Ccl7, and Spp1 (Table S4). As skeletal muscle suffers constant micro-injuries due to mechanical stretch, the pro-inflammatory cluster may represent the FAPs that respond to micro-injuries. To infer the progression trajectories among FAP clusters, we did Monocle pseudotime trajectory analysis (Figure 5B). The Ly6c1hiPi16lo cluster bridged the progression from the Ly6c1hiPi16hi cluster to the Hsd11b1 cluster, and the two specialized clusters were directly from the Hsd11b1 cluster. The findings are consistent with the analysis of cross-tissue fibroblasts.50
Figure 6.
FAP clusters show diverse gene expression depending on the muscle type and disease state
Violin plots showing gene expression by different FAP clusters in different muscle samples.
(A) FAP marker genes.
(B) Featured genes of Ly6c1hi cluster.
(C) Featured genes of Hsd11b1 cluster.
(D) Gdf10.
(E) Featured genes of Sfrp2 cluster.
(F) Featured genes of Postn cluster.
(G) Featured genes of Acta2 cluster.
(H) Ccl2.
(I) Ifit1.
Seven FAP clusters were identified in WT diaphragm, of which five were shared by WT quadriceps, including Ly6c1hiPi16hi, Ly6c1hiPi16lo, Hsd11b1, Gdf10, and pro-inflammatory clusters (Figures 5A, 6A-6D, and S6A.2; Table S5). But they all showed some differences in featured gene expression compared to their counterparts in WT quadriceps (Tables S4 and S5). The differential FAP specialization was further demonstrated by the presence of two additional FAP clusters in WT diaphragm: the Sfrp2 cluster and the Postn cluster. The Sfrp2 cluster featured Sfrp2 expression and enriched pro-fibrotic genes, including Ccn2, Eln, and Mfap4 (Figure 6E and Table S5). The Postn cluster was also enriched for pro-fibrotic genes, including Ccn2, Postn, and C1qtnf3 (Figures 6E and 6F and Table S5). Interestingly, Sfrp2 was selectively expressed by the Ly6c1hiPi16hi cluster in quadriceps while by the Sfrp2 cluster in diaphragm, (Figure 6E). Sfrp2 gene encodes secreted frizzled-related protein 2 (SFRP2), which is an inhibitor of Wnt signaling and may regulate cell proliferation54 and tissue fibrosis.55,56 These findings suggest a generally higher pro-fibrotic microenvironment in diaphragm than in quadriceps, consistent with the higher FAP density in diaphragm than in quadriceps. In addition, the percentage of pro-inflammatory cluster increased dramatically in WT diaphragm as compared to WT quadriceps (Figure S6B), suggesting an increased level of micro-injuries in the steady state diaphragm. Pseudotime analysis showed that all the specialized clusters, in parallel with the Hsd11b1 cluster, were progressed from the Ly6c1hiPi16lo cluster which was directly from the Ly6c1hiPi16hi progenitor (Figure 5B).
Seven FAP clusters were identified in mdx5cv quadriceps. Compared to WT quadriceps FAPs, the mdx5cv quadriceps FAPs lost the Ly6c1hiPi16lo intermediate universal cluster but acquired three new activated clusters: Postn, Acta2, and Gm42418 clusters (Figures 5A, 6A-6G, and S6A.3; Table S6). The Postn cluster was also enriched for Ccn2, Postn, and C1qtnf3 as seen in WT diaphragm (Figures 6E and 6F and Table S6). The unique small Acta2 cluster featured the expression of Acta2, which encodes α-smooth muscle actin that is considered a marker of activated myofibroblasts.57 This cluster was also enriched for genes involved in cell proliferation, including Stmn1, Hmgb2, and S100a4 but not Cdk genes (Figure 6G and Table S6). Therefore, it may represent a cycling population of activated fibroblasts. The small Gm42418 cluster featured high expression of Gm42418, Cd74, and several ribosome protein genes (Table S6). Pro-fibrotic Cthrc1, which was not detectable in WT FAPs, was enriched in the Postn and Acta2 clusters (Figure 6F). The FAP expression of Postn and Cthrc1 was also reported in acutely injured leg muscle.36,46 Vcam1, a marker for pro-fibrotic FAPs,44 was detected in the Hsd11b, Postn, and Acta2 clusters but not in WT FAPs (Figure 6B). The Ly6c1hiPi16hi, Hsd11b1, Gdf10, and pro-inflammatory clusters in mdx5cv quadriceps also showed profound changes in transcriptomes when compared to their counterparts in WT quadriceps (Tables S4 and S6). For instance, Ccl2 expression was observed in the pro-inflammatory FAP cluster in mdx5cv quadriceps but not in WT quadriceps or diaphragm (Figure 6H), and CCL2 mediates MO/MP infiltration in injured skeletal muscle.58 Likewise, a subpopulation of FAPs in acutely injured muscle also expressed Ccl2.46 Importantly, the relative abundance of the pro-inflammatory cluster was significantly increased, while the Ly6c1hiPi16hi progenitor cluster was significantly decreased (Figure S6B). Along with the loss of the Ly6c1hiPi16lo cluster, these changes indicate that the universal Ly6c1hi clusters are greatly perturbed by muscular dystrophy, and they are differentiated into pro-inflammatory and pro-fibrotic clusters, corresponding to the inflammation and fibrosis seen in mdx5cv quadriceps at 14 weeks of age (Figure S1). Pseudotime analysis showed that the Hsd11b1 cluster, progressed from the Ly6c1hiPi16hi cluster, is the source of all the specialized and activated clusters (Figure 5B).
Mdx5cv diaphragm FAPs contained six clusters, four of which were also present in WT diaphragm FAPs, including the Ly6c1hiPi16hi, Sfrp2, Postn, and pro-inflammatory clusters (Figures 5A, 6, and S6A.4; Table S7). The Postn and pro-inflammatory clusters acquired the expression of pro-fibrotic Vcam1and Cthrc1 (Figures 6B and 6F). As seen in mdx5cv quadriceps, FAPs in mdx5cv diaphragm did not contain the Ly6c1hiPi16lo cluster. Compared to the other three samples, FAPs in mdx5cv diaphragm showed substantial changes in clustering, including the loss of the universal Hsd11b1 cluster and the specialized Gdf10 cluster, and the presence of two distinct activated clusters: the stressed cluster and the interferon-activated cluster (Figures 5A, 6I, and S6A.4; Table S7). The stressed cluster was enriched for stress-response genes, including Apod, Egr1, Jun, and Hspa1b (Table S7), a sign of increased tissue stress in mdx5cv diaphragm. The interferon-activated cluster uniquely expressed interferon-response genes, including Ifit1, Ifit3, Ligp1, and Cxcl9 (Figure 6I and Table S7), which was likely related to the significant Ifng expression by NK cells in mdx5cv diaphragm (Figure 3A). Although this cluster is very small (Figure S6B), its unique presence suggests a different inflammatory microenvironment in mdx5cv diaphragm than in mdx5cv quadriceps. Pseudotime analysis showed a sequential progression: Ly6c1hiPi16hi → Stressed → Sfrp2 → Postn → pro-inflammatory clusters, with the interferon-activated cluster progressed from the stressed cluster (Figure 5B).
Taken together, quadriceps and diaphragm FAPs are transcriptionally different, each containing unique specialized clusters in the steady state and distinct activated clusters in mdx5cv, with more pro-fibrotic features present in the diaphragm FAPs than in the quadriceps FAPs.
Extracellular matrix and extracellular matrix regulatory genes are expressed by multiple Fibro/adipogenic progenitor clusters in mdx5cv skeletal muscles
To further address the contributions of individual FAP clusters to the effector and regulatory functions related to skeletal muscle fibrosis in mdx5cv, we analyzed the expression of the genes involved in ECM production and degradation by different FAP clusters in mdx5cv diaphragm and quadriceps.
The Postn cluster expressed the highest level of most of the collagen genes in both quadriceps and diaphragm, including Col1s and Col3s (Figure 7A). Fn1, however, was preferentially expressed by the Ly6c1hiPi16hi cluster (Figure 7A). Different proteoglycan genes were preferentially expressed by different FAP clusters (Figure 7B). Ccn2 was preferentially expressed by the Sfrp2 cluster in diaphragm while was similarly expressed by several clusters in quadriceps (Figure 7C). The Hsd11b1 cluster in quadriceps and the Sfrp2 cluster in diaphragm expressed the highest level of Mmp2 (Figure 7D). The quadriceps Acta2 cluster expressed a relatively high level of Loxs, especially Loxl3 (Figure 7F). The Postn cluster in both muscles expressed the highest level of Igf1, Mmp14, and Loxl1, as well as a moderate level of Lox, Loxl2, and Loxl3 (Figures 7C-7F). The pro-inflammatory cluster expressed the highest level of Fst and Timp1 in both muscles (Figures 7C and 7E) and the highest level of Lox in diaphragm (Figure 7F). Timp2 and Timp3 were highly expressed by the Ly6c1hiPi16hi cluster (Figure 7E). These findings suggest that the ECM production and degradation in mdx5cv skeletal muscle may be regulated by multiple specialized and activated FAP clusters, and the Postn cluster appears the most pro-fibrotic one.
Figure 7.
ECM genes and ECM regulatory genes are expressed by multiple FAP clusters
(A–F) Dot plot showing the expression of collagen and fibronectin genes (A), proteoglycan genes (B), fibrogenic factor genes (C), Mmps (D), Timps (E), and Loxs (F) by FAP clusters of mdx5cv quadriceps and diaphragm.
Discussion
Fibrosis is a prominent pathological feature of skeletal muscle in patients with DMD and animal models, which directly contributes to muscle weakness.3,4 FAPs are the main fibrogenic cells in skeletal muscle.20,21 By scRNAseq analysis of intramuscular mononuclear cells derived from limb (quadriceps) and respiratory (diaphragm) muscles of both WT and mdx5cv mice, our present study has generated several important findings, which deepens our understanding of the cellular and molecular mechanisms mediating and regulating skeletal muscle fibrosis.
First, despite more severe and progressive fibrosis seen in mdx5cv diaphragm than in mdx5cv quadriceps, the expression of most ECM genes by FAPs at a single-cell level is not different between these two muscles. But the ECM gene and protein expression at a whole-muscle level is significantly higher in mdx5cv diaphragm than in mdx5cv quadriceps. Since FAPs are the primary source of ECM in skeletal muscle, the relatively high ECM gene expression in mdx5cv diaphragm is most likely contributed by the high abundance of FAPs in this muscle. It remains possible that the ECM protein degradation may also be different between mdx5cv diaphragm and quadriceps.
The difference in the FAP density between mdx5cv quadriceps and diaphragm is prominent. It could be in part intrinsic to the muscle type, as the FAP density is much higher in WT diaphragm than in WT quadriceps. Interestingly, the percentages of proliferating and apoptotic FAPs are both higher in mdx5cv quadriceps than in mdx5cv diaphragm (Figures S7A and S7B), indicating a higher FAP turnover in mdx5cv quadriceps than in mdx5cv diaphragm. The expression of the FAP senescence genes, including Cdkn1a, Cdkn2a, Trp53, and Fas,59 is low in both mdx5cv quadriceps and diaphragm FAPs (Figure S7C). Apoptosis, proliferation, and fibrogenic function of FAPs are regulated by TNF-α and TGF-β signaling. While TNF-α signaling induces FAP apoptosis, TGF-β signaling inhibits FAP apoptosis,25 and it also promotes FAP proliferation and fibrogenic differentiation.34,35,60 While TGF-β signaling is enriched in the DEGs of both mdx5cv diaphragm and quadriceps FAPs, TNF-α signaling is enriched only in the mdx5cv quadriceps FAPs. The difference might contribute, in part, to the persistent high number of FAPs in mdx5cv diaphragm.
Second, our study supports the notion that FAPs are not only important effectors but also the master regulators of ECM homeostasis and fibrosis in skeletal muscle. FAPs are known primary ECM producers. Our study further suggests that FAPs may also be the critical regulators of ECM production and degradation, as they may regulate many aspects of skeletal muscle fibrogenesis by expressing Ccn2, Ltbp4, Mmp2, Mmp14, Timp1, Timp2, and Loxs.
Fibrosis is the end result of chronic inflammation, and the two potent pro-fibrogenic genes, Tgfb1 and Spp1, are mainly expressed by MOs/MPs in mdx5cv muscles. But Ltbp4, an important regulatory gene of TGF-β bioavailability61 and a genetic modifier of DMD mice and humans,62,63,64,65 is predominantly expressed by FAPs. FAPs also express TGF-β1, TGF-β2, and TGF-β3, which can promote their own fibrogenic differentiation.24 Another important pro-fibrogenic growth factor, CTGF, is highly expressed by FAPs. CTGF stimulates Col1, Col3, and Fn expression. Ccn2 overexpression in mice caused dystrophic changes,66 while CTGF inhibition in mdx reduced muscle fibrosis without affecting TGF-β signaling.67 Interestingly, a recent study showed that CTGF, derived from myofibers but not fibroblasts, affected collagen content and organization, contributing to muscular dystrophy in δ-sarcoglycan-null mice.38 The role of FAP-derived CTGF in mdx5cv needs to be further determined. FAPs are also the major cells that express the IGF-1 and follistatin genes, regulating not only muscle fibrosis but also muscle regeneration.
MMPs and TIMPs are multifunctional proteins, regulating inflammation, fibrogenesis, myogenesis, and angiogenesis in skeletal muscle via controlling the degradation of ECM and the activation of growth factors and cell adhesion molecules.17,68 Of the three Mmps that are highly expressed by skeletal muscle mononuclear cells, two are highly expressed by FAPs, including Mmp2 and Mmp14. Mmp9 is mainly expressed by neutrophils. These MMPs play a key role in collagen and fibronectin degradation, which can limit fibrosis. On the other hand, MMP-9 and MMP-14 can also promote fibrosis by cleaving and activating latent TGF-β1 and CTGF.69,70 They are double-edged swords. Although MMP-14 has been implicated in promoting adipogenic differentiation of FAPs,71,72 the expression of the adipogenic genes, including Pparg, Fabp4, Adipog, and Cebpa, is barely detectable in mdx5cv FAPs (Figure S7D). MMP-14, along with TIMP2, activates MMP-2, and both MMPs are critically involved in skeletal muscle maturation.17,68,73,74,75 Ablation of MMP-2 in mdx impaired muscle regeneration and angiogenesis.76 The role of MMP-14 in mdx has not been well studied. Given the diverse functions of this membrane-bound MMP, the overall role of the FAP-derived MMP-14 in mdx5cv is difficult to predict and needs to be further determined.
TIMPs inhibit the proteolytic activity of MMPs. Among the four TIMP genes, Timp1 and Timp2 are mainly expressed by FAPs. FAPs are thus likely the significant contributors to the elevated TIMP-1 and TIMP-2 levels in the plasma and skeletal muscles of patients with DMD and animal models.77,78,79,80 TIMP-1 and TIMP-2 can promote fibrosis not only by reducing ECM proteolysis but also by stimulating fibrogenic cell proliferation and collagen production.17 FAP-derived TIMPs may thus contribute significantly to the skeletal muscle fibrosis in mdx5cv. Being the major cells that express Timp3 and Timp4, ECs may regulate FAP differentiation, as Timp3 inhibits adipogenic differentiation of FAPs.72,81,82
The important regulatory functions of FAPs are further exemplified by their significant expression of Loxs for collagen cross-linking. FAPs, therefore, may regulate the mechanical properties of ECM and contribute to the increased collagen cross-linking detected in skeletal muscles of patients with DMD and animal models.19 Taken together, FAPs appear critically involved in the regulation of skeletal muscle fibrosis in mdx5cv. FAPs may also regulate inflammation, regeneration, and angiogenesis associated with muscular dystrophy via their predominant expression of important growth factors, Mmps, and Timps.
Third, our study demonstrates the presence of FAP clusters specialized by muscle type in the steady state, limb (quadriceps) muscle vs. respiratory (diaphragm) muscle. While FAPs in both muscles contain the universal adventitia-associated Ly6c1hi clusters and basement membrane-associated Hsd11b1 cluster and share the specialized Gdf10 and pro-inflammatory clusters, the diaphragm FAPs also contains the uniquely specialized Postn and Sfrp2 clusters. Based on the enriched genes in the muscle type-specific FAP clusters, the diaphragm appears to have an increased level of micro-injury and a generally high pro-fibrotic microenvironment compared to the quadriceps. Likewise, muscle type-specific clusters have also been identified in the steady state resident macrophages, which also suggests an increased level of micro-injury in diaphragm as compared to quadriceps.83 The intrinsic differences in tissue microenvironment may be related to the different structural origins that limb and diaphragm muscles arise from during embryonic development,22 and they may contribute, in part, to the differential responses to a variety of muscle diseases. While some muscle diseases affect both limb and respiratory muscles, others preferentially affect limb or respiratory muscles.
Fourth, mdx5cv diaphragm and quadriceps FAPs respond differently to muscular dystrophy, with more profound changes seen in the FAP clustering in the diaphragm than in the quadriceps. Both universal and specialized clusters are greatly perturbed by the muscular dystrophy in mdx5cv. While FAPs in both muscles lose the intermediate Ly6c1hiPi16lo cluster, FAPs in mdx5cv diaphragm also lose the universal Hsd11b1 cluster and the specialized Gdf10 cluster. Hsd11b1 encodes 11b-Hydroxysteroid Dehydrogenase 1, which has been implicated in suppressing fibrosis, as deficiency or inhibition of this enzyme promotes fibroblast activation and enhances fibrosis in liver and skin.84,85 The Hsd11b1 cluster is thus likely differentiated into more pro-fibrotic clusters in mdx5cv diaphragm. The selective loss of the Hsd11b1 cluster in mdx5cv diaphragm also suggests more severe perturbation of tissue homeostasis in mdx5cv diaphragm than in mdx5cv quadriceps, as this cluster is enriched for homeostatic chemokine genes. PDGFRα+ mesenchymal progenitors-derived GDF10 has been shown to maintain skeletal muscle integrity86 and suppress adipogenic differentiation of FAPs.49 The loss of this cluster may suggest its differentiation into pathogenic clusters to contribute to muscular dystrophy in mdx5cv diaphragm. Mdx5cv diaphragm acquires two uniquely activated clusters: the stressed cluster and the interferon-activated cluster. The presence of the stressed cluster suggests exceptionally high stress in the microenvironment of mdx5cv diaphragm. Since many stress-responsive genes are also potent pro-inflammatory genes, such as Egr1 and Jun,87,88 the presence of the stressed cluster also indicates a high pro-inflammatory microenvironment in mdx5cv diaphragm. The unique presence of the interferon-activated FAP cluster further supports a special inflammatory microenvironment in mdx5cv diaphragm. These two uniquely activated clusters likely contribute to the persistent inflammation and progressive fibrosis in mdx5cv diaphragm.
In summary, our study supports the critical roles of FAPs in skeletal muscle ECM homeostasis and fibrosis, both as predominant effectors and crucial regulators. The persistent high abundance of FAPs appears the main contributor to the progressive fibrosis in mdx5cv diaphragm. Limb and diaphragm muscles have different embryonic origins, locate at different anatomical positions, and work under different mechanical stress. These differences likely drive the differential FAP specialization in the steady state and differential FAP activation in muscular dystrophy. The high pro-fibrotic microenvironment likely contributes to the persistent high FAP abundance and progressive fibrosis in mdx5cv diaphragm. The anti-fibrotic therapy development for DMD should target not only the ECM production but also the pro-fibrogenic regulatory functions of FAPs.
Limitations of the study
Since polynuclear myofibers cannot be included in single-cell suspensions for scRNAseq or FACS sorting due to their large size, their expression of the ECM and ECM regulatory genes cannot be directly assessed or compared with FAPs. Future snRNAseq can help circumvent the difficulty. scRNAseq cannot assess locations of FAPs in dystrophic muscle, while spatial transcriptomics can (https://www.biorxiv.org/content/10.1101/2022.03.17.484699v1.full.pdf). Future spatial transcriptomics studies will be helpful to localize different FAP clusters and correlate with histopathological changes in dystrophic muscle. Our scRNAseq is performed using the mice at 14 weeks of age, after which mdx5cv diaphragm and limb muscles undergo divergent fibrosis progression. Future studies at later time points may help identify additional differences in FAP transcriptomes that underlie the divergent fibrosis progression. Our study has identified several regulatory genes of fibrosis that are predominantly expressed by FAPs. Future studies with FAP-specific deletion are needed to verify the functional importance of the FAP expression of these genes in muscular dystrophy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti- a/b-tubulin | Cell Signaling Technology | Cat# 2148S; RRID: AB_2288042 |
| Rabbit anti-collagen 1a1 | Cell Signaling Technology | Cat# 72026S; RRID: AB_2904565 |
| Rabbit anti-fibronectin | Cell Signaling Technology | Cat# 63779S |
| Rabbit anti-collagen III | Abcam | Cat# ab7778; RRID:AB_306066 |
| IRDye 800CW anti-rabbit IgG | LI-COR Biosciences | Cat# 926-32211; RRID: AB_621843 |
| PerCP-Cy5.5 Rat anti-mouse CD45 | BD Biosciences | Cat# 550994; RRID:AB_394003 |
| PE Rat anti-mouse CD45 | BD Biosciences | Cat# 553081; RRID:AB_394611 |
| APC Rat anti-mouse CD31 | BD Biosciences | Cat# 551262; RRID:AB_398497 |
| PE Rat anti-mouse CD31 | BD Biosciences | Cat# 553373; RRID:AB_394819 |
| PE-Cy7 Rat anti-mouse Ly6-A/E (Sca-1) | BD Biosciences | Cat# 558162; RRID:AB_647253 |
| BV421 Rat anti-mouse CD140A (PDGFRa) | BD Biosciences | Cat# 562774; RRID:AB_2728781 |
| PE Rat anti-mouse Integrin α7 | R&D Systems | Cat# FAB3518P; RRID:AB_2249296 |
| APC Rat IgG2a, κ Isotype Control | BD Biosciences | Cat# 551139; RRID: AB_10054921 |
| PE Rat IgG2a, κ Isotype Control | BD Biosciences | Cat# 559317; RRID: AB_10050484 |
| PE-Cy7 Rat IgG2a, κ Isotype Control | BD Biosciences | Cat# 552784; RRID: AB_394465 |
| BV421 Rat IgG2a, k Isotype Control | BD Biosciences | Cat# 562602; RRID:AB_11153860 |
| PerCP-Cy5.5 Rat IgG2b, κ Isotype Control | BD Biosciences | Cat# 550764; RRID: AB_393874 |
| PE Rat IgG2b, k Isotype Control | BD Biosciences | Cat# 555848; RRID:AB_396171 |
| Chemicals, peptides, and recombinant proteins | ||
| Mayer’s Hematoxylin solution | Millipore Sigma | Cat# MHS16 |
| Eosin Y solution | Millipore Sigma | Cat# HT110116 |
| 4% paraformaldehyde solution | Thermo Scientific | Cat# J19943.K2 |
| Poly Mount Xylene | Polysciences | Cat# 24716-120 |
| Collagenase B | Roche Diagnostics | Cat# 11088831001 |
| Dispase II | Roche Diagnostics | Cat# 04942078001 |
| Fetal bovine serum | Corning | Cat# 35-011-CV |
| Bovine serum albumin | Millipore Sigma | Cat# A7030 |
| Normal mouse serum | Millipore Sigma | Cat# S7273 |
| Lympholyte-M solution | Cedarlane | Cat# CL5035 |
| TRIzol reagent | Invitrogen | Cat# 15596026 |
| RPMI 1640 | Gibco | Cat# 11-875-093 |
| RIPA buffer | Pierce Biotechnology | Cat# 89901 |
| cOmplete (protease inhibitor) | Roche Diagnostics | Cat# 04.693.116.001 |
| Bolt 4–12% Bis-Tris mini-gel | Invitrogen | Cat# NW04122 |
| Immobilon PVDF membrane | Millipore | Cat#I SEQ00010 |
| Acridine Orange and Propidium Iodide staining solution | Nexcelom | Cat# CS2-0106 |
| Critical commercial assays | ||
| Vybrant™ DyeCycle™ Violet/SYTOX™ AADvanced™ Kit | Invitrogen | Cat# A35135 |
| EdU Click 647 Kit + EdU | Baseclick GmbH | Cat# BCK647 |
| Rneasy Micro Kit | Qiagen | Cat# 74004 |
| Super-Script III Reverse Transcriptase Kit | Invitrogen | Cat# 18080051 |
| PowerTrack™ SYBR Green Master Mix | Applied Biosystems | Cat# A46109 |
| BCA protein assay Kit | Pierce Biotechnology | Cat# 23225 |
| Single Cell 3′ Reagents Kits V3.1 | 10x Genomics | https://assets.ctfassets.net/an68im79xiti/1eX2FPdpeCgnCJtw4fj9Hx/7cb84edaa9eca04b607f9193162994de/CG000204_ChromiumNextGEMSingleCell3_v3.1_Rev_D.pdf |
| Deposited data | ||
| Single cell-based RNA sequencing data | https://www.ncbi.nlm.nih.gov/geo/ | GEO:GSE218201 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | The Jackson Laboratory | |
| mdx5cv mice | The Jackson Laboratory; Chapman VM, et al. PNAS 86(4): 1292–6. | |
| Oligonucleotides | ||
| Gapdh (Forward): 5′-GTGTTCCTACCCCCAATGTGTC-3′ | IDT DNA Oligos | N/A |
| Gapdh (Reverser): 5′-GTAGCCCAAGATGCCCTTCAGT-3′ | IDT DNA Oligos | N/A |
| Col1a1 (Forward): 5′-GCTCCTCTTAGGGGCCACT-3′ | IDT DNA Oligos | N/A |
| Col1a1 (Reverse): 5′-CCACGTCTCACCATTGGGG-3′ | IDT DNA Oligos | N/A |
| Col3a1 (Forwad): 5′-AACCTGGTTTCTTCTCACCCTTC-3′ | IDT DNA Oligos | N/A |
| Col3a1 (Reverse): 5′-ACTCATAGGACTGACCAAGGTGG-3′ | IDT DNA Oligos | N/A |
| Fn1 (Forward): 5′-AAACTTGCATCTGGAGGCAAACCC-3′ | IDT DNA Oligos | N/A |
| Fn1 (Reverse): 5′-AGCTCTGATCAGCATGGACCACTT-3′ | IDT DNA Oligos | N/A |
| Igf1 (Forward): 5′-CTACAAAAGCAGCCCGCTCT-3′ | IDT DNA Oligos | N/A |
| Igf1 (Reverse): 5′-CTTCTGAGTCTTGGGCATGTCA-3′ | IDT DNA Oligos | N/A |
| Ccn2 (Forward): 5′-AGGACTGCAGCGCGCAATGT-3′ | IDT DNA Oligos | N/A |
| Ccn2 (Reverse): 5′-GAGGCCCTTGTGTGGGTCGC-3′ | IDT DNA Oligos | N/A |
| Mmp2 (Forward): 5′-GCCCCGAGACCGCTATGTCCACT-3′ | IDT DNA Oligos | N/A |
| Mmp2 (Revers): 5′-GCCCCACTTCCGGTCATCATCGTA-3′ | IDT DNA Oligos | N/A |
| Timp1 (Forward): 5′-ACTCGGACCTGGTCATAAGGGC-3′ | IDT DNA Oligos | N/A |
| Timp1 (Reveres): 5′-TTCCGTGGCAGGCAAGCAAAGT-3′ | IDT DNA Oligos | N/A |
| Timp2 (Forward): 5′-GGCAACCCCATCAAGAGGA-3′ | IDT DNA Oligos | N/A |
| Timp2 (Reverse): 5′-CCTTCTGCCTTTCCTGCAATTAG-3′ | IDT DNA Oligos | N/A |
| Mmp14 (Forward): 5′-CTTCAAAGGAGATAAGCACTG-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Mmp14 (Reverse): 5′-AGAAGTAGGTCTTCCCATTG-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Ltbp4 (Forward): 5′- CCCATTCTTCGAAATATCACC-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Ltbp4 (Reverse): 5′- GAAAACCCTCTGAACCATAAG-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Lox (Forward): 5′-CACCGTATTAGAAAGAAGCC-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Lox (Reverse): 5′-GTCCTTCCTACTTAAGCTAATC-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Loxl1 (Forward): 5′-TGCTATGACACATACAATGC-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Loxl1 (Reverse): 5′-GAACAATGTACTTGGGGTTC-3′ | Millipore Sigma | KiCqStart® SYBR® Green Primers: Cat# KSPQ12012 |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo V10 | BD Biosciences | https://www.bdbiosciences.com/en-us/products/software/flowjo-v10-software |
| Image Studio software | LI-COR Biosciences | https://www.licor.com/bio/image-studio/ |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| R package Seurat version 4.0 | Satija Lab at NYGC | https://satijalab.org/seurat/ |
| g:Profiler | https://biit.cs.ut.ee/gprofiler/gost | |
| Monocle3 | https://cole-trapnell-lab.github.io/monocle3/ | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xingyu Wang (wangx20@bu.edu).
Materials availability
This study did not generate unique new reagents or mouse lines.
Experimental model and subject details
Animals
C57BL/6J (WT) and mdx5cv mice (B6 background) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). Only males were used in this study because DMD only affects males. For scRNAseq analysis, WT and mdx5cv mice used were at 14 weeks of ages with body weight between 25 and 30 grams. For FACS analysis determining FAP density, an additional time point at 24 weeks of age was used for mdx5cv mice with body weight between 30 and 35 grams. Our study protocols were approved by the Institutional Animal Care and Use Committee at the Boston University School of Medicine (Boston, MA, USA.).
Method details
Muscle sample collection
Quadriceps and diaphragm were collected from 14-week-old male WT mice, and 14-week-old and 24-week-old male mdx5cv mice. Tendons were removed from the tissue specimens. Collected muscle samples were thoroughly washed in 1× phosphate-buffered saline (PBS, pH 7.4) to remove blood contamination.
Histopathological analysis
Collected muscle samples were frozen in liquid nitrogen-chilled isopentane for 30 seconds. Cross sections were then obtained on a Leica cryostat at a thickness of 10 μm. Sections were air-dried for 30 minutes at room temperature and then fixed in 4% (w/v) of paraformaldehyde (Thermo Scientific) for 10 minutes followed by washing in 1× PBS for 5 minutes. Hematoxylin and Eosin (H&E) staining was then performed as following: 1) stained in Mayer’s Hematoxylin solution (Millipore Sigma, Burlington, MA, USA.) for 10 minutes, 2) rinsed in warm running tap water for 15 minutes, 3) washed in distilled water for 30 seconds, 4) placed in 95% (v/v) reagent alcohol for 30 seconds, 5) stained in Eosin Y solution (Alcoholic, Millipore Sigma) for 1 minute, 6) dehydrated and cleared through 2 changes each of 95% reagent alcohol, absolute reagent alcohol, and xylene for 2 minutes each, 7) mounted with Poly Mount Xylene (Polysciences, Warrington, PA, USA). Stained sections were viewed and photographed under a bright-field microscope (Nikon, Y-THR-L).
Muscle single-cell suspension preparation
Single-cell suspension was prepared by collagenase/dispase digestion.83 Quadriceps and diaphragm samples (less than 250 mg) were minced in 2.5 mL digestion solution (1 U/ml collagenase B and 1 U/ml dispase II (Roche Diagnostics, Indianapolis, IN, USA) in 1× PBS) and incubated at 37°C for 1 hour. The reaction was terminated by adding 10 mL PBS containing 10% (v/v) fetal bovine serum (FBS, Corning, Glendale, AZ, USA). The mixture was then filtered through a 70-μm cell strainer and centrifuged at 250 g for 5 minutes. The pellet was collected, and the supernatant was centrifuged again at 250 g for 5 minutes at 4°C. The pellet was combined with the pellet from the first centrifugation, washed with PBS, and centrifuged at 670 g for 10 minutes at 4°C. The pellet was re-suspended in 3 mL PBS and filtered through a 40-μm cell strainer. Cell suspension was layered on equal volume of the Lympholyte-M solution (Cedarlane, Burlington, NC, USA) and centrifuged at 2,100 g for 45 minutes at room temperature. Cells at the interface were collected in 10 mL PBS containing 10% FBS, pelleted by centrifugation at 670 g for 10 minutes at 4°C, and re-suspended in appropriated buffer for following analysis.
Flow cytometry analysis and cell sorting
Single-cell pellets were washed once in ice-cold FACS buffer (1× PBS, 0.5% (w/v) bovine serum albumin (BSA, Millipore Sigma), 0.1% (w/v) sodium azide), centrifuged at 500 g for 3 minutes at 4°C, and re-suspended in ice-cold FACS staining buffer (1× PBS, 2% (w/v) BSA, 2% (v/v) normal mouse serum (Millipore Sigma).
For FACS analysis of different cell types, cells were stained on ice for 20 minutes with fluorescence-labelled antibodies targeting cell-type specific markers: Fibro/Adipogenic progenitors (FAPs) were identified as Sca-1+PDGFRα+CD45−CD31-α7-integrin- cells, endothelial cells (ECs) were identified as CD31+CD45− cells, and muscle satellite cells (MuSCs) were identified as α7-integrin+CD45−CD31−Sca-1- cells. Fluorescence-labelled corresponding normal IgG isotypes were included as negative controls for gating. All antibodies were from commercial providers (See key resources table for details) and diluted for staining following manufacturer’s recommendations. After staining, cells were washed twice with FACS buffer and analyzed on a LSR II (BD Bioscience, San Jose, CA, USA) with BD FACSDiva™ software. Collected data were then analyzed using FlowJo software (Tree Star, Inc., Ashland, OR, USA).
To clean up single cell suspension for subsequent single-cell based analysis, cell sorting based on FSC/SSC was performed to exclude dead cells, cell duplexes, and tissue debris. Sorting FAPs and non-FAPs for qRT-PCR analysis were performed with antibodies listed above. Cell sorting was done by the Flow Cytometry Core of Boston University School of Medicine. Collected cells from 5 individual male mice were combined, and pelleted by centrifuging at 300 g for 10 minutes. For scRNAseq analysis, pellets were re-suspended in FBS supplemented with 10% of DMSO, and cryopreserved in liquid nitrogen. For qRT-PCR analysis, pellets were dissolved in TRIzol reagent (Invitrogen, Waltham, MA, USA) for RNA preparation.
FAP apoptosis assay
Muscle single-cell suspension was first stained with antibodies identifying FAPs as described in the section of flow cytometry analysis and cell sorting. After being washed twice with FACS buffer, cells were again re-suspended in FACS buffer and stained with Vybrant™ DyeCycle™ Violet/SYTOX™ AADvanced™ reagents (Invitrogen) following manufacturer’s instruction. Stained cells were analyzed by flow cytometry immediately. Apoptotic FAPs were identified as Vybrant+SYTOX−.
FAP proliferation assay
Mdx5cv mice at 14 weeks of age received intraperitoneal injection of EdU (100 mg/mouse, Baseclick GmbH, Germany). Quadriceps and diaphragm were then collected 24 hours later and single-cell suspension was prepared as described in the section of muscle single-cell suspension preparation. FAP proliferation was then analyzed by determining the incorporation of EdU using EdU Click 647 Kit (Baseclick GmbH, Germany) following manufacturer’s instruction, combined with FACS staining and analysis of FAPs as described in the section of flow cytometry analysis and cell sorting.
RNA preparation and qRT-PCR
Total RNA samples were prepared using TRIzol reagent (Invitrogen). Samples of FAPs and non-FAPs were prepared by cell sorting as stated above and dissolved in TRIzol reagent. To prepare whole-muscle RNA, quadriceps and diaphragm, collected from 14-week-old male WT and mdx5cv mice, were homogenized in TRIzol reagent using Bead Mill 24 homogenizer (Fisher Scientific). Total RNA was then prepared following manufacturer’s instruction. TRIzol-prepared total RNA was further cleaned up using RNeasy Micro Kit (Qiagen, Germantown, MD, USA) following manufacturer’s instruction. Concentration of cleaned RNA was measured using NanoDrop 2000 Spectrophotometer (Thermo Scientific). 1 μg of total RNA was then reverse-transcribed into cDNA using Super-Script III Reverse Transcriptase system (Invitrogen) following manufacturer’s instruction. Expression of genes at mRNA level was then analyzed by real-time PCR with PowerTrack™ SYBR Green Master Mix (Applied Biosystems, Waltham, MA, USA) using StepOne™ Real Time PCR System (Applied Biosystems). Sequences and providers of the primers used are listed in key resources table. The sequences and target genes of the primers were listed in key resources table. Data was calculated by ΔΔCt method. For sorted cell analysis, data was presented as bar graph with error bar showing standard deviation of triplicated PCR results. For whole muscle analysis, data were presented as scatter plot showing results of each individual samples.
Western blot
Quadriceps and diaphragm, collected from 14-week-old male WT and mdx5cv mice, were homogenized in RIPA buffer (Pierce Biotechnology, Waltham, MA, USA) supplemented with cOmplete protease inhibitor cocktail (Roche Diagnostic). After centrifugation (10,000 ×g for 15 minutes), the protein concentration was quantitated by Pierce BCA protein assay kit (Pierce Biotechnology). 50 μg of protein lysate were run on 4–12% Bis-Tris mini-gel (Invitrogen) under reducing condition then transferred to PVDF membranes (Immobilon). Blots were blocked with 5% (w/v) of non-fat milk in TBS-T (1× Tris-buffered saline (pH7.6) with 0.1% (v/v) Tween 20) and then incubated with primary antibodies include α/β-tubulin (1:1000 dilution), collagen 1α1 (1:500), fibronectin (1:250) (all purchased from Cell Signaling Technology, Danvers, MA, USA), and collagen III (1:500) (Abcam, Waltham, MA, USA). Signals were detected by IRDye 800 or 680 (LI-COR Biosciences, Lincoln, NE)-conjugated secondary antibodies with appropriate species specificity. Immunoblots were visualized by the Image Studio software (LI-COR Biosciences) that accompanies the LI-COR Odyssey infrared system (LI-COR Biosciences).
Single-cell cDNA library preparation and sequencing
Cryopreserved cells were thawed in a 37°C water bath for two minutes. The thawed content was added dropwise into 9 mL of warm freshly prepared RPMI 1640 (Gibco) with 20% of FBS, spun 300 g for 5 minutes, and re-suspended in 5 mL of RPMI with10% of FBS. The cells were counted, and viability was assessed using Acridine Orange and Propidium Iodide staining followed by counting on a K2 Cellometer (Nexcelome, Lawrence, MA, USA). Next, scRNAseq was performed following the Single Cell 3′ Reagents Kits V3.1 User Guidelines (10x Genomics, Pleasanton, CA, USA). All samples were above 70% viability. Cells were loaded to target 5,000 cells final for generating the single-cell emulsion (Chromium Single Cell 3′ Chip kit A v2 PN-12036 or v3 chip kit B PN-2000060). Reverse-transcription (RT) was performed in the emulsion, cDNA amplified, and library constructed with v3.1 chemistry. Libraries were indexed for multiplexing (Chromium i7 Multiplex kit PN-12062). Following preparation, libraries were quantified by Qubit 3 fluorometer (Invitrogen) and quality was assessed by Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Equivalent molar concentrations of libraries were pooled, and the reads were adjusted after sequencing the pools in a Miseq (Illumina, San Diego, CA, USA). The libraries were then sequenced in a Novaseq 6000 (Illumina) at the New York Genome Center (NYGC, New York, NY, USA) following 10× Genomics recommendations.
Single cell-based mRNA sequencing analysis
The gene × cell expression matrices were loaded to the R package Seurat version 4.0 (https://cran.r-project.org/web/packages/Seurat/index.html) for downstream analyses.31 Cells of low quality, defined by the detectable expression of fewer than 200 genes per 1000 Unique Molecular Identifiers (UMI) and the presence of more than 7.5% mitochondria content, were filtered out.31 The Seurat default global-scaling normalization method LogNormalize was used to normalize the feature expression measurements for each cell by the total expression, multiplies this by a scale factor and log-transforms the result. The Seurat default global-scaling normalization method, “LogNormalize”, was used for scaling the data.89 The method Vst was used to identify highly variable features in function FindVariableFeatures.
A shared-nearest neighbor (SNN) graph–based clustering method was constructed to identify discrete cell populations. Then the obtained clusters were optimized by using the “FindClusters” function.
Linear dimensional reduction PCA was performed on variable genes. A clear Elbow in the plot of the PC standard deviation was used to define the significant PCs. Based on the identified PCs above, non-linear dimensional reduction technique UMAP was performed.89,90,91 Differentially Expressed Genes (DEGs) were calculated and obtained by the function of FindAllMarkers based on the Wilcoxon rank sum test.31 Top marker genes in each cluster shown in Heatmap were ordered by log fold-change of the average expression. Differential gene expression analysis was performed in each of the cell clusters, and the log transformed fold changes (log2FC) were used to perform hierarchical clustering between genes.
Functional enrichment analysis
We used g:Profiler webserver (https://biit.cs.ut.ee/gprofiler/gost) to perform functional enrichment analysis for the identified DEGs in individual clusters or subclusters. Top DEGs (adj. p-value < 0.05) were tested for enrichment of biological pathways with Gene Ontology (GO), Reactome, CORUM and KEGG. Results from different databases were compared to ensure the reliability of the analysis.
Pseudotime trajectory analysis
The cells of FAP clusters were further analyzed using Monocle for pseudotime trajectory.92 Seurat objects were converted to CellDataSet objects using the as.cell_data_set function from SeuratWrappers. The trajectories were built using Monocle 3 (https://cole-trapnell-lab.github.io/monocle3/docs/citations/, https://satijalab.org/signac/articles/monocle.html). Each sample was analyzed separately by the same procedure. Interactive function order cells was used to select cells as the root nodes in the graph. The function get_earliest_principal_node was applied to identify the root principal points.
Venn diagram
Differential gene expression among FAPs of 4 muscles (WT diaphragm, mdx5cv diaphragm, WT quadriceps, and mdx5cv quadriceps) were determined by Seurat FindMarkers function, using a log fold-change threshold of 0.5 and an adjusted p value cutoff of 0.05. Venn diagram was prepared using Venny2.1.
Quantification and statistical analysis
Data were analyzed using GraphPad Prism 8 software (GraphPad Software, San Diego, CA, USA). The Mann-Whitney test was performed to compare between two groups; the Kruskal-Wallis test followed by Dunn’s test was performed to compare multiple (>=3) groups. A p value of <0.05 was considered statistically significant.
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis And Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number 1R01AR074428 (LZ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
XW: Conducted the experiments, analyzed the data, made the figures, and wrote the article. JC: Analyzed the scRNAseq data, conducted the experiments, and made the figures. SH: Conducted the experiments. YW: Conducted the experiments. GRS: Analyzed the scRNAseq data. FRZ: Conducted the scRNAseq experiment. SCS: Supervised the scRNAseq experiment. LZ: Designed the study, analyzed the data, and wrote the article.
Declaration of interests
All the authors have no conflict of interest to report.
Published: January 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105775.
Supplemental information
Data and code availability
Single-cell based RNA sequencing (scRNAseq) data have been deposited to GEO repository (GEO: GSE218201) and are publicly available as of the date of publication. The accession number is also listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Duan D., Goemans N., Takeda S., Mercuri E., Aartsma-Rus A. Duchenne muscular dystrophy. Nat. Rev. Dis. Primers. 2021;7:13. doi: 10.1038/s41572-021-00248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryder S., Leadley R.M., Armstrong N., Westwood M., de Kock S., Butt T., Jain M., Kleijnen J. The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: an evidence review. Orphanet J. Rare Dis. 2017;12:79. doi: 10.1186/s13023-017-0631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desguerre I., Mayer M., Leturcq F., Barbet J.P., Gherardi R.K., Christov C. Endomysial fibrosis in Duchenne muscular dystrophy: a marker of poor outcome associated with macrophage alternative activation. J. Neuropathol. Exp. Neurol. 2009;68:762–773. doi: 10.1097/NEN.0b013e3181aa31c2. [DOI] [PubMed] [Google Scholar]
- 4.Klingler W., Jurkat-Rott K., Lehmann-Horn F., Schleip R. The role of fibrosis in Duchenne muscular dystrophy. Acta Myol. 2012;31:184–195. [PMC free article] [PubMed] [Google Scholar]
- 5.Beastrom N., Lu H., Macke A., Canan B.D., Johnson E.K., Penton C.M., Kaspar B.K., Rodino-Klapac L.R., Zhou L., Janssen P.M.L., Montanaro F. mdx((5)cv) mice manifest more severe muscle dysfunction and diaphragm force deficits than do mdx Mice. Am. J. Pathol. 2011;179:2464–2474. doi: 10.1016/j.ajpath.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupont-Versteegden E.E., McCarter R.J. Differential expression of muscular dystrophy in diaphragm versus hindlimb muscles of mdx mice. Muscle Nerve. 1992;15:1105–1110. doi: 10.1002/mus.880151008. [DOI] [PubMed] [Google Scholar]
- 7.Stedman H.H., Sweeney H.L., Shrager J.B., Maguire H.C., Panettieri R.A., Petrof B., Narusawa M., Leferovich J.M., Sladky J.T., Kelly A.M. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352:536–539. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 8.Zhou L., Porter J.D., Cheng G., Gong B., Hatala D.A., Merriam A.P., Zhou X., Rafael J.A., Kaminski H.J. Temporal and spatial mRNA expression patterns of TGF-beta1, 2, 3 and TbetaRI, II, III in skeletal muscles of mdx mice. Neuromuscul. Disord. 2006;16:32–38. doi: 10.1016/j.nmd.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Csapo R., Gumpenberger M., Wessner B. Skeletal muscle extracellular matrix - what do we know about its composition, regulation, and physiological roles? A narrative review. Front. Physiol. 2020;11:253. doi: 10.3389/fphys.2020.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillies A.R., Lieber R.L. Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve. 2011;44:318–331. doi: 10.1002/mus.22094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W., Liu Y., Zhang H. Extracellular matrix: an important regulator of cell functions and skeletal muscle development. Cell Biosci. 2021;11:65. doi: 10.1186/s13578-021-00579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ismaeel A., Kim J.S., Kirk J.S., Smith R.S., Bohannon W.T., Koutakis P. Role of transforming growth factor-beta in skeletal muscle fibrosis: a review. Int. J. Mol. Sci. 2019;20:2446. doi: 10.3390/ijms20102446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebolledo D.L., Lipson K.E., Brandan E. Driving fibrosis in neuromuscular diseases: role and regulation of connective tissue growth factor (CCN2/CTGF) Matrix Biol. 2021;11:100059. doi: 10.1016/j.mbplus.2021.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagel C.N., Wasgewatte Wijesinghe D.K., Taghavi Esfandouni N., Mackie E.J. Osteopontin, inflammation and myogenesis: influencing regeneration, fibrosis and size of skeletal muscle. J. Cell Commun. Signal. 2014;8:95–103. doi: 10.1007/s12079-013-0217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramerova I., Kumagai-Cresse C., Ermolova N., Mokhonova E., Marinov M., Capote J., Becerra D., Quattrocelli M., Crosbie R.H., Welch E., et al. Spp1 (osteopontin) promotes TGFbeta processing in fibroblasts of dystrophin-deficient muscles through matrix metalloproteinases. Hum. Mol. Genet. 2019;28:3431–3442. doi: 10.1093/hmg/ddz181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vetrone S.A., Montecino-Rodriguez E., Kudryashova E., Kramerova I., Hoffman E.P., Liu S.D., Miceli M.C., Spencer M.J. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J. Clin. Invest. 2009;119:1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alameddine H.S., Morgan J.E. Matrix metalloproteinases and tissue inhibitor of metalloproteinases in inflammation and fibrosis of skeletal muscles. J. Neuromuscul. Dis. 2016;3:455–473. doi: 10.3233/JND-160183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afratis N.A., Klepfish M., Karamanos N.K., Sagi I. The apparent competitive action of ECM proteases and cross-linking enzymes during fibrosis: applications to drug discovery. Adv. Drug Deliv. Rev. 2018;129:4–15. doi: 10.1016/j.addr.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Smith L.R., Hammers D.W., Sweeney H.L., Barton E.R. Increased collagen cross-linking is a signature of dystrophin-deficient muscle. Muscle Nerve. 2016;54:71–78. doi: 10.1002/mus.24998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joe A.W.B., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M.V. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uezumi A., Ito T., Morikawa D., Shimizu N., Yoneda T., Segawa M., Yamaguchi M., Ogawa R., Matev M.M., Miyagoe-Suzuki Y., et al. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J. Cell Sci. 2011;124:3654–3664. doi: 10.1242/jcs.086629. [DOI] [PubMed] [Google Scholar]
- 22.Contreras O., Rossi F.M.V., Theret M. Origins, potency, and heterogeneity of skeletal muscle fibro-adipogenic progenitors-time for new definitions. Skelet. Muscle. 2021;11:16. doi: 10.1186/s13395-021-00265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theret M., Rossi F.M.V., Contreras O. Evolving roles of muscle-resident fibro-adipogenic progenitors in Health, regeneration, neuromuscular disorders, and aging. Front. Physiol. 2021;12:673404. doi: 10.3389/fphys.2021.673404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Contreras O., Cruz-Soca M., Theret M., Soliman H., Tung L.W., Groppa E., Rossi F.M., Brandan E. Cross-talk between TGF-beta and PDGFRalpha signaling pathways regulates the fate of stromal fibro-adipogenic progenitors. J. Cell Sci. 2019;132:jcs232157. doi: 10.1242/jcs.232157. [DOI] [PubMed] [Google Scholar]
- 25.Lemos D.R., Babaeijandaghi F., Low M., Chang C.K., Lee S.T., Fiore D., Zhang R.H., Natarajan A., Nedospasov S.A., Rossi F.M.V. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med. 2015;21:786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 26.Heredia J.E., Mukundan L., Chen F.M., Mueller A.A., Deo R.C., Locksley R.M., Rando T.A., Chawla A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mozzetta C., Consalvi S., Saccone V., Tierney M., Diamantini A., Mitchell K.J., Marazzi G., Borsellino G., Battistini L., Sassoon D., et al. Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Mol. Med. 2013;5:626–639. doi: 10.1002/emmm.201202096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wosczyna M.N., Konishi C.T., Perez Carbajal E.E., Wang T.T., Walsh R.A., Gan Q., Wagner M.W., Rando T.A. Mesenchymal stromal cells are required for regeneration and homeostatic maintenance of skeletal muscle. Cell Rep. 2019;27:2029–2035.e5. doi: 10.1016/j.celrep.2019.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urciuolo A., Quarta M., Morbidoni V., Gattazzo F., Molon S., Grumati P., Montemurro F., Tedesco F.S., Blaauw B., Cossu G., et al. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun. 2013;4:1964. doi: 10.1038/ncomms2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao W., Wang X., Ransohoff R.M., Zhou L. CCR2 deficiency does not provide sustained improvement of muscular dystrophy in mdx5cv mice. FASEB J. 2017;31:35–46. doi: 10.1096/fj.201600619R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mutsaers S.E. Mesothelial cells: their structure, function and role in serosal repair. Respirology. 2002;7:171–191. doi: 10.1046/j.1440-1843.2002.00404.x. [DOI] [PubMed] [Google Scholar]
- 33.Contreras O., Rebolledo D.L., Oyarzún J.E., Olguín H.C., Brandan E. Connective tissue cells expressing fibro/adipogenic progenitor markers increase under chronic damage: relevance in fibroblast-myofibroblast differentiation and skeletal muscle fibrosis. Cell Tissue Res. 2016;364:647–660. doi: 10.1007/s00441-015-2343-0. [DOI] [PubMed] [Google Scholar]
- 34.Contreras O., Rossi F.M., Brandan E. Adherent muscle connective tissue fibroblasts are phenotypically and biochemically equivalent to stromal fibro/adipogenic progenitors. Matrix Biol. 2019;2:100006. doi: 10.1016/j.mbplus.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Contreras O., Soliman H., Theret M., Rossi F.M.V., Brandan E. TGF-beta-driven downregulation of the transcription factor TCF7L2 affects Wnt/beta-catenin signaling in PDGFRalpha(+) fibroblasts. J. Cell Sci. 2020;133:jcs242297. doi: 10.1242/jcs.242297. [DOI] [PubMed] [Google Scholar]
- 36.Scott R.W., Arostegui M., Schweitzer R., Rossi F.M.V., Underhill T.M. Hic1 defines quiescent mesenchymal progenitor subpopulations with distinct functions and fates in skeletal muscle regeneration. Cell Stem Cell. 2019;25:797–813.e9. doi: 10.1016/j.stem.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahdy M.A.A. Skeletal muscle fibrosis: an overview. Cell Tissue Res. 2019;375:575–588. doi: 10.1007/s00441-018-2955-2. [DOI] [PubMed] [Google Scholar]
- 38.Petrosino J.M., Leask A., Accornero F. Genetic manipulation of CCN2/CTGF unveils cell-specific ECM-remodeling effects in injured skeletal muscle. FASEB J. 2019;33:2047–2057. doi: 10.1096/fj.201800622RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reggio A., Rosina M., Palma A., Cerquone Perpetuini A., Petrilli L.L., Gargioli C., Fuoco C., Micarelli E., Giuliani G., Cerretani M., et al. Adipogenesis of skeletal muscle fibro/adipogenic progenitors is affected by the WNT5a/GSK3/beta-catenin axis. Cell Death Differ. 2020;27:2921–2941. doi: 10.1038/s41418-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tidball J.G., Welc S.S., Wehling-Henricks M. Immunobiology of inherited muscular dystrophies. Compr. Physiol. 2018;8:1313–1356. doi: 10.1002/cphy.c170052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Juban G., Saclier M., Yacoub-Youssef H., Kernou A., Arnold L., Boisson C., Ben Larbi S., Magnan M., Cuvellier S., Théret M., et al. AMPK activation regulates LTBP4-dependent TGF-beta1 secretion by pro-inflammatory macrophages and controls fibrosis in Duchenne muscular dystrophy. Cell Rep. 2018;25:2163–2176.e6. doi: 10.1016/j.celrep.2018.10.077. [DOI] [PubMed] [Google Scholar]
- 42.Contreras O., Córdova-Casanova A., Brandan E. PDGF-PDGFR network differentially regulates the fate, migration, proliferation, and cell cycle progression of myogenic cells. Cell. Signal. 2021;84:110036. doi: 10.1016/j.cellsig.2021.110036. [DOI] [PubMed] [Google Scholar]
- 43.Uezumi A., Ikemoto-Uezumi M., Tsuchida K. Roles of nonmyogenic mesenchymal progenitors in pathogenesis and regeneration of skeletal muscle. Front. Physiol. 2014;5:68. doi: 10.3389/fphys.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malecova B., Gatto S., Etxaniz U., Passafaro M., Cortez A., Nicoletti C., Giordani L., Torcinaro A., De Bardi M., Bicciato S., et al. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nat. Commun. 2018;9:3670. doi: 10.1038/s41467-018-06068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mázala D.A., Novak J.S., Hogarth M.W., Nearing M., Adusumalli P., Tully C.B., Habib N.F., Gordish-Dressman H., Chen Y.W., Jaiswal J.K., Partridge T.A. TGF-beta-driven muscle degeneration and failed regeneration underlie disease onset in a DMD mouse model. JCI Insight. 2020;5:e135703. doi: 10.1172/jci.insight.135703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oprescu S.N., Yue F., Qiu J., Brito L.F., Kuang S. Temporal dynamics and heterogeneity of cell populations during skeletal muscle regeneration. iScience. 2020;23:100993. doi: 10.1016/j.isci.2020.100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giuliani G., Vumbaca S., Fuoco C., Gargioli C., Giorda E., Massacci G., Palma A., Reggio A., Riccio F., Rosina M., et al. SCA-1 micro-heterogeneity in the fate decision of dystrophic fibro/adipogenic progenitors. Cell Death Dis. 2021;12:122. doi: 10.1038/s41419-021-03408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stumm J., Vallecillo-García P., Vom Hofe-Schneider S., Ollitrault D., Schrewe H., Economides A.N., Marazzi G., Sassoon D.A., Stricker S. Odd skipped-related 1 (Osr1) identifies muscle-interstitial fibro-adipogenic progenitors (FAPs) activated by acute injury. Stem Cell Res. 2018;32:8–16. doi: 10.1016/j.scr.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Camps J., Breuls N., Sifrim A., Giarratana N., Corvelyn M., Danti L., Grosemans H., Vanuytven S., Thiry I., Belicchi M., et al. Interstitial cell remodeling promotes aberrant adipogenesis in dystrophic muscles. Cell Rep. 2020;31:107597. doi: 10.1016/j.celrep.2020.107597. [DOI] [PubMed] [Google Scholar]
- 50.Buechler M.B., Pradhan R.N., Krishnamurty A.T., Cox C., Calviello A.K., Wang A.W., Yang Y.A., Tam L., Caothien R., Roose-Girma M., et al. Cross-tissue organization of the fibroblast lineage. Nature. 2021;593:575–579. doi: 10.1038/s41586-021-03549-5. [DOI] [PubMed] [Google Scholar]
- 51.Fiedler U., Augustin H.G. Angiopoietins: a link between angiogenesis and inflammation. Trends Immunol. 2006;27:552–558. doi: 10.1016/j.it.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Marichal T., Mesnil C., Bureau F. Homeostatic eosinophils: characteristics and functions. Front. Med. 2017;4:101. doi: 10.3389/fmed.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kouzeli A., Collins P.J., Metzemaekers M., Meyrath M., Szpakowska M., Artinger M., Struyf S., Proost P., Chevigne A., Legler D.F., et al. CXCL14 preferentially synergizes with homeostatic chemokine receptor systems. Front. Immunol. 2020;11:561404. doi: 10.3389/fimmu.2020.561404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Loon K., Huijbers E.J.M., Griffioen A.W. Secreted frizzled-related protein 2: a key player in noncanonical Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev. 2021;40:191–203. doi: 10.1007/s10555-020-09941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi K., Luo M., Zhang Y., Wilkes D.C., Ge G., Grieskamp T., Yamada C., Liu T.C., Huang G., Basson C.T., et al. Secreted Frizzled-related protein 2 is a procollagen C proteinase enhancer with a role in fibrosis associated with myocardial infarction. Nat. Cell Biol. 2009;11:46–55. doi: 10.1038/ncb1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akpulat U., Onbaşılar İ., Kocaefe Y.Ç. Tenotomy immobilization as a model to investigate skeletal muscle fibrosis (with emphasis on Secreted frizzled-related protein 2) Physiol. Genom. 2016;48:397–408. doi: 10.1152/physiolgenomics.00010.2016. [DOI] [PubMed] [Google Scholar]
- 57.Hinz B., Phan S.H., Thannickal V.J., Prunotto M., Desmoulière A., Varga J., De Wever O., Mareel M., Gabbiani G. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am. J. Pathol. 2012;180:1340–1355. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu H., Huang D., Ransohoff R.M., Zhou L. Acute skeletal muscle injury: CCL2 expression by both monocytes and injured muscle is required for repair. FASEB J. 2011;25:3344–3355. doi: 10.1096/fj.10-178939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saito Y., Chikenji T.S., Matsumura T., Nakano M., Fujimiya M. Exercise enhances skeletal muscle regeneration by promoting senescence in fibro-adipogenic progenitors. Nat. Commun. 2020;11:889. doi: 10.1038/s41467-020-14734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theret M., Low M., Rempel L., Li F.F., Tung L.W., Contreras O., Chang C.K., Wu A., Soliman H., Rossi F.M.V. In vitro assessment of anti-fibrotic drug activity does not predict in vivo efficacy in murine models of Duchenne muscular dystrophy. Life Sci. 2021;279:119482. doi: 10.1016/j.lfs.2021.119482. [DOI] [PubMed] [Google Scholar]
- 61.Todorovic V., Rifkin D.B. LTBPs, more than just an escort service. J. Cell. Biochem. 2012;113:410–418. doi: 10.1002/jcb.23385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bello L., Kesari A., Gordish-Dressman H., Cnaan A., Morgenroth L.P., Punetha J., Duong T., Henricson E.K., Pegoraro E., McDonald C.M., et al. Genetic modifiers of ambulation in the cooperative international neuromuscular research group Duchenne natural history study. Ann. Neurol. 2015;77:684–696. doi: 10.1002/ana.24370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flanigan K.M., Ceco E., Lamar K.M., Kaminoh Y., Dunn D.M., Mendell J.R., King W.M., Pestronk A., Florence J.M., Mathews K.D., et al. LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann. Neurol. 2013;73:481–488. doi: 10.1002/ana.23819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heydemann A., Ceco E., Lim J.E., Hadhazy M., Ryder P., Moran J.L., Beier D.R., Palmer A.A., McNally E.M. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J. Clin. Invest. 2009;119:3703–3712. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van den Bergen J.C., Hiller M., Böhringer S., Vijfhuizen L., Ginjaar H.B., Chaouch A., Bushby K., Straub V., Scoto M., Cirak S., et al. Validation of genetic modifiers for Duchenne muscular dystrophy: a multicentre study assessing SPP1 and LTBP4 variants. J. Neurol. Neurosurg. Psychiatry. 2015;86:1060–1065. doi: 10.1136/jnnp-2014-308409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morales M.G., Cabello-Verrugio C., Santander C., Cabrera D., Goldschmeding R., Brandan E. CTGF/CCN-2 over-expression can directly induce features of skeletal muscle dystrophy. J. Pathol. 2011;225:490–501. doi: 10.1002/path.2952. [DOI] [PubMed] [Google Scholar]
- 67.Morales M.G., Gutierrez J., Cabello-Verrugio C., Cabrera D., Lipson K.E., Goldschmeding R., Brandan E. Reducing CTGF/CCN2 slows down mdx muscle dystrophy and improves cell therapy. Hum. Mol. Genet. 2013;22:4938–4951. doi: 10.1093/hmg/ddt352. [DOI] [PubMed] [Google Scholar]
- 68.Bassiouni W., Ali M.A.M., Schulz R. Multifunctional intracellular matrix metalloproteinases: implications in disease. FEBS J. 2021;288:7162–7182. doi: 10.1111/febs.15701. [DOI] [PubMed] [Google Scholar]
- 69.Tatti O., Vehviläinen P., Lehti K., Keski-Oja J. MT1-MMP releases latent TGF-beta1 from endothelial cell extracellular matrix via proteolytic processing of LTBP-1. Exp. Cell Res. 2008;314:2501–2514. doi: 10.1016/j.yexcr.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 70.Yu Q., Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–176. [PMC free article] [PubMed] [Google Scholar]
- 71.Lin Y., Wen-Jie Z., Chang-Qing L., Sheng-Xiang A., Yue Z. mir-22-3p/KLF6/MMP14 axis in fibro-adipogenic progenitors regulates fatty infiltration in muscle degeneration. FASEB J. 2020;34:12691–12701. doi: 10.1096/fj.202000506R. [DOI] [PubMed] [Google Scholar]
- 72.Kopinke D., Roberson E.C., Reiter J.F. Ciliary hedgehog signaling restricts injury-induced adipogenesis. Cell. 2017;170:340–351.e12. doi: 10.1016/j.cell.2017.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S.A., Mankani M., Robey P.G., Poole A.R., Pidoux I., et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 74.Oh J., Takahashi R., Adachi E., Kondo S., Kuratomi S., Noma A., Alexander D.B., Motoda H., Okada A., Seiki M., et al. Mutations in two matrix metalloproteinase genes, MMP-2 and MT1-MMP, are synthetic lethal in mice. Oncogene. 2004;23:5041–5048. doi: 10.1038/sj.onc.1207688. [DOI] [PubMed] [Google Scholar]
- 75.Snyman C., Niesler C.U. MMP-14 in skeletal muscle repair. J. Muscle Res. Cell Motil. 2015;36:215–225. doi: 10.1007/s10974-015-9414-4. [DOI] [PubMed] [Google Scholar]
- 76.Miyazaki D., Nakamura A., Fukushima K., Yoshida K., Takeda S., Ikeda S.i. Matrix metalloproteinase-2 ablation in dystrophin-deficient mdx muscles reduces angiogenesis resulting in impaired growth of regenerated muscle fibers. Hum. Mol. Genet. 2011;20:1787–1799. doi: 10.1093/hmg/ddr062. [DOI] [PubMed] [Google Scholar]
- 77.Fukushima K., Nakamura A., Ueda H., Yuasa K., Yoshida K., Takeda S., Ikeda S.i. Activation and localization of matrix metalloproteinase-2 and -9 in the skeletal muscle of the muscular dystrophy dog (CXMDJ) BMC Musculoskelet. Disord. 2007;8:54. doi: 10.1186/1471-2474-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nadarajah V.D., van Putten M., Chaouch A., Garrood P., Straub V., Lochmüller H., Ginjaar H.B., Aartsma-Rus A.M., van Ommen G.J.B., den Dunnen J.T., 't Hoen P.A.C. Serum matrix metalloproteinase-9 (MMP-9) as a biomarker for monitoring disease progression in Duchenne muscular dystrophy (DMD) Neuromuscul. Disord. 2011;21:569–578. doi: 10.1016/j.nmd.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 79.Sun G., Haginoya K., Chiba Y., Uematsu M., Hino-Fukuyo N., Tanaka S., Onuma A., Iinuma K., Tsuchiya S. Elevated plasma levels of tissue inhibitors of metalloproteinase-1 and their overexpression in muscle in human and mouse muscular dystrophy. J. Neurol. Sci. 2010;297:19–28. doi: 10.1016/j.jns.2010.06.031. [DOI] [PubMed] [Google Scholar]
- 80.von Moers A., Zwirner A., Reinhold A., Brückmann O., van Landeghem F., Stoltenburg-Didinger G., Schuppan D., Herbst H., Schuelke M. Increased mRNA expression of tissue inhibitors of metalloproteinase-1 and -2 in Duchenne muscular dystrophy. Acta Neuropathol. 2005;109:285–293. doi: 10.1007/s00401-004-0941-0. [DOI] [PubMed] [Google Scholar]
- 81.Qi J.H., Ebrahem Q., Moore N., Murphy G., Claesson-Welsh L., Bond M., Baker A., Anand-Apte B. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med. 2003;9:407–415. doi: 10.1038/nm846. [DOI] [PubMed] [Google Scholar]
- 82.Fernández C.A., Moses M.A. Modulation of angiogenesis by tissue inhibitor of metalloproteinase-4. Biochem. Biophys. Res. Commun. 2006;345:523–529. doi: 10.1016/j.bbrc.2006.04.083. [DOI] [PubMed] [Google Scholar]
- 83.Wang X., Sathe A.A., Smith G.R., Ruf-Zamojski F., Nair V., Lavine K.J., Xing C., Sealfon S.C., Zhou L. Heterogeneous origins and functions of mouse skeletal muscle-resident macrophages. Proc. Natl. Acad. Sci. USA. 2020;117:20729–20740. doi: 10.1073/pnas.1915950117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Terao M., Tani M., Itoi S., Yoshimura T., Hamasaki T., Murota H., Katayama I. 11beta-hydroxysteroid dehydrogenase 1 specific inhibitor increased dermal collagen content and promotes fibroblast proliferation. PLoS One. 2014;9:e93051. doi: 10.1371/journal.pone.0093051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zou X., Ramachandran P., Kendall T.J., Pellicoro A., Dora E., Aucott R.L., Manwani K., Man T.Y., Chapman K.E., Henderson N.C., et al. 11Beta-hydroxysteroid dehydrogenase-1 deficiency or inhibition enhances hepatic myofibroblast activation in murine liver fibrosis. Hepatology. 2018;67:2167–2181. doi: 10.1002/hep.29734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uezumi A., Ikemoto-Uezumi M., Zhou H., Kurosawa T., Yoshimoto Y., Nakatani M., Hitachi K., Yamaguchi H., Wakatsuki S., Araki T., et al. Mesenchymal Bmp3b expression maintains skeletal muscle integrity and decreases in age-related sarcopenia. J. Clin. Invest. 2021;131:e139617. doi: 10.1172/JCI139617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pawlinski R., Pedersen B., Kehrle B., Aird W.C., Frank R.D., Guha M., Mackman N. Regulation of tissue factor and inflammatory mediators by Egr-1 in a mouse endotoxemia model. Blood. 2003;101:3940–3947. doi: 10.1182/blood-2002-07-2303. [DOI] [PubMed] [Google Scholar]
- 88.Schonthaler H.B., Guinea-Viniegra J., Wagner E.F. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 2011;70(Suppl 1):i109–i112. doi: 10.1136/ard.2010.140533. [DOI] [PubMed] [Google Scholar]
- 89.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell based RNA sequencing (scRNAseq) data have been deposited to GEO repository (GEO: GSE218201) and are publicly available as of the date of publication. The accession number is also listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.