Abstract
Increasing evidence suggests a role of the gut microbiome in the development of colorectal cancer (CRC) and that it can serve as a biomarker for early diagnosis. This review aims to give an overview of the current status of published studies regarding the microbiome as a screening tool for early CRC detection. A literature search was conducted using PubMed and EMBASE in August 2022. Studies assessing the efficacy of microbiome-derived biomarkers based on noninvasive derived samples were included. Not relevant studies or studies not specifying the stage of CRC or grouping them together in the analysis were excluded. The risk of bias for screening tools was performed using the QUADAS-2 checklist. A total of 28 studies were included, ranging from 2 to 462 for CRC and 18 to 665 advanced adenoma patient inclusions, of which only two investigated the co-metabolome as biomarker. The diagnostic performance of faecal bacteria-derived biomarkers had an AUC ranging from 0.28-0.98 for precursor lesions such as advanced adenomas and 0.54-0.89 for early CRC. Diagnostic performance based on the co-metabolome showed an AUC ranging from 0.69 – 0.84 for precursor lesions and 0.65 – 0.93 for early CRC. All models improved when combined with established clinical early detection markers such as gFOBT. A high level of heterogeneity was seen in the number of inclusions and methodology used in the studies. The faecal and oral gut microbiome has the potential to complement existing CRC screening tools, however current evidence suggests that this is not yet ready for routine clinical use.
Key-words: Colorectal cancer, Screening, Biomarker, Microbiome, Metabolomics
Abbreviations: 16S rRNA, 16S ribosomal RNA gene sequencing; AUC, Area Under the Curve; CE-MS, Capillary Electrophoresis Mass Spectrometry; CRC, Colorectal cancer; FIT, Faecal Immunochemical Test; GC-MS, Gas-Chromatography Mass Spectrometry; (g)FOBT, guaiac Faecal Occult Blood Test; OTU, Operational taxonomic unit; qPCR, quantitative real-time Polymerase Chain Reaction; QUADAS, Quality Assessment of Diagnostic Accuracy Studies; U(H)PLC-MS, Ultra-(High)-Performance Liquid Chromatography Mass Spectrometry
Introduction
Colorectal cancer (CRC) is the third commonest cancer worldwide [1] and a leading cause of cancer-associated deaths. Despite this, CRC is often treatable with a high overall survival when detected and treated in its early stages [2]. This has led to the formation of global screening programs for CRC [3], which typically employ faecal biomarkers like the guaiac faecal occult blood test (gFOBT), followed by endoscopy [4]. The gFOBT, is a fast, noninvasive and inexpensive test, however, its diagnostic sensitivity ranges from 7 to 21% for the detection of early cancer (Stage 0 and I) and is unsuitable for the detection of complex adenomas [5,6]. Faecal immunochemical test (FIT) offers a slight improvement in its diagnostic yield for early cancer (maximum sensitivity of 25%), however, with higher associated costs [7]. The FIT-DNA test (or stool DNA test), a test that combines the FIT with DNA markers associated with CRC, shows a further increasement in sensitivity, however at the expense of a decrease in specificity, resulting in more false positive results [8].
Blood-based biomarkers such as Septin 9 and protein-based markers (e.g. CEA, TIMP-1 etc.) have also failed to provide diagnostic utility for early detection [9]. Novel data from recent trials suggest that circulating DNA (cDNA) biomarkers for colorectal cancer have diagnostic and prognostic utility, particularly for the stratification of chemotherapeutic strategies [10] and the prediction of cancer recurrence. However, despite the promise of these approaches for early detection, very limited data exist for the identification of high-risk adenomas.
Increasing evidence points to the gut microbiome as a critical mediator of adenoma formation and CRC risk through parallel processes of co-metabolic dysfunction [11], inflammation, epigenetic programming and DNA damage [12]. Multiple studies have identified the presence of intra-tumoural bacteria in precursors lesions of cancer [13], [14], [15], [16] such as Fusobacterium nucleatum, Escherichia coli, and Bacteroides fragilis [17]. A loss of microbial diversity and increases in the abundance of both mucosal and faecal pathobionts can distinguish healthy persons from patients with CRC [18], [19], [20]. Importantly, microbiome changes can already be observed in very early stages of CRC [21]. It has therefore been hypothesised that the microbiome has value as a biomarker for early cancer detection or to identify those individuals at risk of this disease [22]. This systematic review aims to provide a comprehensive overview of the current state of microbiome-derived biomarkers, based on their genes and their products, for early CRC detection.
Methods
Search strategy
A literature search was carried out from inception to the 3rd of August 2022, using the databases PubMed and EMBASE (via OVID). A combination of the following terms was used: colorectal cancer, screening/early detection, and microbiome. No filters or restrictions were applied. Literature was reviewed following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [23]. The complete search strategy can be found in Supplementary Table S1.
Study selection
Results were screened independently by two authors (F.Z. and H.S.) based on title and abstract for relevance and assessment for inclusion based on full text. Articles were evaluated via Rayyan, developed by Qatar Computing Research Institute, a platform that allows blinded assessment of articles based on title, abstract, and key-words by multiple reviewers [24]. Disagreement of articles screened for possible eligibility based on title and abstract was resolved by discussion. Conflicts over the inclusion of articles for assessment following full-text screening were resolved by consulting a third reviewer (J.K.). If studies used the same datasets, the study most appropriate to our study was selected.
Eligibility criteria
Articles were included for assessment if they assessed the efficacy and/or use of microbiome-derived biomarkers for early CRC detection, including precancerous lesions: complex adenoma > 1 cm, high-grade dysplasia and early cancer: carcinoma in situ (CIS), malignant polyps and stage 0, I or II with N0, M0, according to the American Joint Committee on Cancer (AJCC) TNM Classification of Malignant Tumors [25]. Articles were excluded based on: (I) viral, fungal or solely human-derived biomarkers; (II) investigating prognostic biomarkers; (III) disease other than CRC; (IV) advanced stage CRC; (V) inflammatory bowel disease (IBD)-related CRC or CRC as part of a syndrome; (VI) in vitro or animal studies; (VII) language other than English; (VIII) small sample size (<10); (IX) no full text available; (X) case report or case series; (XI) review; (XII) no clear distinction of the CRC stages assessed; (XIII) all CRC stages grouped in the analysis.
Since this study investigates the use of microbial-derived biomarkers as a screening tool for the early detection of CRC, all sorts of invasive screening studies, for example, studies assessing microbiome composition of the mucosal tissue based on biopsies, were excluded. Studies assessing saliva, urine and blood were regarded as minimally invasive procedures and were included.
Data extraction and assessment
Extraction of data from the eligible articles was performed independently by the two authors (F.Z. and H.S.). Confirmation of the extracted data was performed by a third author (K.Z.). Data extraction included primarily: year of publication, country, number of participants, age, percentage of male, stage of disease, sample origin, biomarker found, technique used for discovery, diagnostic performance of studied biomarker (based on sensitivity, specificity or area under the curve (AUC)) and, if described, comparison with currently used screening tools (e.g. gFOBT, FIT). If any of the data was not described directly, additional information was used to calculate the necessary values if possible. Data was rounded off at either two decimals (AUC) or no decimals (sensitivity, specificity). If stated, specific bacteria or their molecular products and functions per cohort and methodology were reported.
Comparison with healthy controls
Comparison with healthy controls was based, if applicable, via colonoscopy and defined as subjects without prior gastro-intestinal disease(s) or colonoscopic findings with the exception of a few polyps (<5mm) present.
Assessment
Assessment of the strength of overall data regarding individual biomarkers based on their risk of bias and applicability was undertaken using the QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies checklist) [26].
Included studies were independently evaluated for the risk of bias and applicability by two authors (F.Z. and H.S.). The risk of bias was assessed across four domains: patient selection, index test, reference standard, flow and timing (i.e. an appropriate interval between index test and reference as well as the inclusion of all patients in the analysis).
Concerns regarding applicability were assessed on three domains: patient selection, index test and reference standard. Signalling questions of all domains were tailored for this review. Based on these questions, the risk of bias and applicability was classified as low, high or unclear for each domain.
Results
Study selection
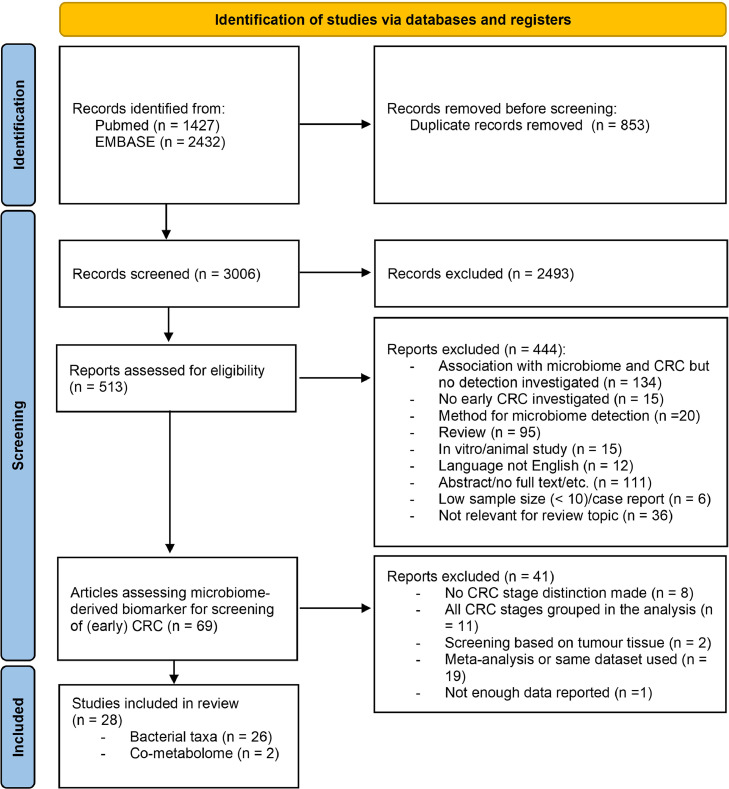
A literature search was performed using the databases PubMed and EMBASE (via OVID). A total of 3859 records were retrieved, 1427 from PubMed and 2432 from EMBASE. After removing duplicates, 3006 records were screened based on title and abstract, resulting in 513 records being evaluated for eligibility based on full-text screening. A total of 69 studies evaluating microbiome-derived indicators for early CRC diagnosis were again carefully screened, resulting in another 41 records to be excluded based on: a lack of CRC stage distinction (n = 8), all stages grouped together in the analysis (n = 11), screening based on tumour tissue (n = 2), meta-analysis or usage of the same dataset in another article already included (n = 19) and insufficient data reported (n = 1). A total of 28 studies remained to be included in this systematic review. Study selection and exclusion were based on the PRISMA guidelines [23]. See Fig. 1 for a detailed overview of the selection process.
Fig. 1.
PRISMA flow diagram of exclusion and included of studies.
Study characteristics
For microbiome-derived biomarkers based on bacteria for the detection of early CRC, 26 articles were found and two articles were found investigating the co-metabolome solely as a biomarker. Seventeen of the studies were conducted in East Asia [19,[27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]], one in South Asia [43], eight in Europe [44], [45], [46], [47], [48], [49], [50], [51] and two in North America [52,53]. The number of inclusions of these studies ranged between 2 and 462 for CRC, 18 and 665 for adenoma and/or polyp and 24 and 788 for healthy controls. Regarding the stage used in the articles for early CRC detection, five studies were based on stage I or II [29,30,37,40,50] and two used stage 0 [19,38]. The other studies investigated a precursor lesion of CRC, either advanced adenomas or polyps (AP) [28,[31], [32], [33], [34], [35], 39,41,[44], [45], [46], [47], [48], [49], [51], [52], [53]]. Only five articles evaluated both precursor lesions of CRC and stages I and II [27,36,38,43,54]. All articles used faecal samples, except for four: two studies used the microbiota collected from the oral mucosa instead of or next to faecal samples [31,46] and two used serum-based samples next to faecal samples [36,54]. Whenever possible, the mean age and percentage of men per group were extracted. The reported mean age ranged from 59 to 73 in the CRC group, from 48 to 68 in the AP group and from 49 to 67 in the healthy controls. The percentages of men in each group ranged from 32 to 67, 19 to 78 and 17 to 56 per cent for CRC, AP and healthy controls, respectively. See Table 1 for the study characteristics as well.
Table 1.
Study characteristics.
| Author, year | Country | Sample | Examined samples (CRC/AP/HC)1 | CRC stage according to TNM ((0)3/I/II/III/IV) | ||
|---|---|---|---|---|---|---|
| (n) | Age (mean (±SD))2 | Gender, m (%) | ||||
| Bacterial taxa | ||||||
| Zhang Y, 2021 [32] | China | Faeces | N/A/268/788 | N/A/61.7 (6.2)/59.9 (6.2) | N/A/74/48 | N/A |
| Zhang S, 2020 [31] | China | Oral mucosa | 161/34/58 | 59.2 (10.7)/51.8 (7.7)/50.7 (11.3) | 67/59/53 | 24/66/60/11 |
| Yachida, 2019 [19] | Japan | Faeces | 258/67/251+40 with history of colorectal surgery | 0 64.8 (7.5), I/II: 63.8 (9.3), III/IV: 59.1 (11) /63.2 (9.1)/ 64.7 (10.6) + 59.2 (12.3) | 0 62, I-II 66, III-IV 58/72/28 + 33 | 73/75/36/52/22 |
| Yao, 2021 [30] | China | Faeces | 206/ N/A /112 | 62.2 (8.6)/ N/A /56.2 (12.8) | 61/ N/A /56 | 98/47/49/12 |
| Xie, 2017 [29] | China | Faeces | 445/288/304 | I-IV 63.5 (10.2), I-II 64 (8.9)/61.9 (8.7)/ 60.1 (8.4) | I-IV 61, I-II 65/63/ 55 | I/II: 142; III/IV: 303 |
| Wei, 2022 [28] | Taiwan | Faeces | 20/67/60 + 40 COB4 | 64 (43–88)/48 (39–60)-/ 61 (31–72) + 52 (35–63) | 35/42/41 + 45 | Not distinguished |
| Mo, 2021 [27] | China | Faeces | 108/18/36 | 58 (26-86) of n = all | 58 of n = all | 20/39/37/12 |
| Liu, 2021 [41] | China | Faeces | 60/37/42 | 64.1 (11.1)/66.5 (3.5)/61.0 (7.9) | 32/19/31 | Not distinguished |
| Liang, 2017 [40] | China | Faeces | 170-2005/97/33-365 | 67.2 (11.6)- 63.4 (9.6)/60.5 (4.7)/59.3 (5.8)-53.2 (12.2) | 59-52/52/39-28 | Stage subset (n=111) 17/42/43/9/111 |
| Liang, 2021 [39] | China | Faeces | 210/115+866/265 | 67.0 (11.3)/61.1 (6.8) + 60.2 (5.0)/58.1 (7.7) | 59/57 + 57/42 | 31/68/73/28 |
| Konishi, 2022 [38] | Japan | Faeces | 462/240/317 | 56/ - /66 | 52 / - /53 | 68/107/77/130/44 |
| Guo, 2018 [37] | China | Faeces | 215/ N/A /156 | 61.2 (12.3)/ N/A / 48.6 (10.3) | 55/ N/A / 45 | 38/59/75/43 |
| Gao, 2022 [36] | China | Faeces | 35/31/34 | 66.1 (10.8)/64.4 (8.4)/57.8 (5.7) | 57/58/17 | 3/21/9/2 |
| Goedert, 2015 [47] | China | Faeces | 2/20/24 | 65 (61–69) of n = all | N/A/60/29 | 1/0/1/0 |
| Gao, 2020 [35] | China | Faeces | 55/85/110 | N/A /63.1 (12.8)/65 (10.4) | N/A /63/29 | N/A |
| Coker, 2022 [34] | China | Faeces | 118/140/128 | 73.2 (10.4)/65.8 (5.5)/64 (6.8) | 54/59/46 | N/A |
| Ai, 2017 [33] | China/France | Faeces | 42/47/52 | 62.9 (1.5)/58.9 (1.5)/52.3 (1.5) | 43/51/40 | I/II 12 III/IV 30 |
| Rezasoltani, 2018 [43] | Iran | Faeces | 20/42/31 | 60.9 (13.5)/58.6 (13.5)/59.8(17) | 56/55/52 | 20/0/0/0 |
| Zeller, 2014 [50] | France | Faeces | 53/42/61 | 70.5-65.0*/62.0-68.0⁎⁎/63.0 | 55/71/46 | 15/7/10/21 |
| Young, 2021 [49] | UK | Faeces | 430/665/6667 | 68.1 (5.0)/66.3 (4.7)/66.6 (4.3) | 67/65/52 | Not distinguished |
| Tarallo, 2019 [48] | Italy | Faeces | 29/27/24 | Not disclosed | Not disclosed | Not distinguished |
| Flemer, 2017 [46] | Ireland | Faeces | 69/23/62 | 65.3 (10.8)/60.4 (13.4)/63.9 (11.1) | 67/78/51 | Not distinguished |
| Oral mucosa | 45/21/25 | 65.7 (10.9)/59.2 (15.1)/51.5 (12.4) | 56/71/38 | |||
| Eklöf, 2017 [45] | Sweden | Faeces | 39/134/65 | N/A | 51/60/54 | 2/21/8/7 |
| Clos-Garcia, 2020 [44] | Spain | Faeces | All: 99/69/77 Analysis: 83/62/74 | 70.2/68/65 (all) | 62/60/45 (all) | 3/22/22/30/6 (analysis) |
| Zackular, 2014 [53] | North America | Faeces | 30/30/30 | 59.4 (11.0)/ 61.3(11.1)55.3 (9.2) | 70/60/37 | Not distinguished |
| Baxter, 2016 [52] | USA/Canada | Faeces | 120/198/172 | 60 (median, range 29-89) | Not disclosed | Not distinguished |
| Co-metabolome (different cohort) | ||||||
| Yachida 2019 [19] | Japan | Faeces | 178/45/149+34 with history of colorectal surgery | 0 63.7 (8.5), I/II 63.6 (8.8), III/IV59.5 (11.5)/64.4 (8.5)/64.1 (10.9) + 60.6 (11.9) | 0 50, I-II 65, III-IV 53/ 78/58 + 47 | 30/51/29/44/24/34 |
| Gao, 2022 [36] | China | Serum | 35/31/34 | 66.1 (10.8)/64.4 (8.4)/57.8 (5.7) | 57/58/17 | 3/21/9/2 |
| Chen, 2022 [54] | China | Serum Faeces | 84/19/53 | 58 (7.2)3/53.8 (7.3) | 723/38 | 8/9/18/20/19 (29 no record) |
| Bosch, 2022 [51] | the Netherlands | Faeces | N/A /19/19 | N/A /73 (6.1)/68 (10.4) | N/A /90/68 | N/A |
CRC = colorectal cancer, AP = adenoma and/or polyp, HC = healthy controls
Unless differently defined
Stage 0 = Intramucosal carcinoma/polypoid adenomas with high-grade dysplasia, pTis/Stage 0 CRC (as defined by Yachida et al. [19])
Colonic occult blood loss
HC and CRC based on two separate cohorts
Asymptomatic and symptomatic patients with AP. Analyses used in this review only on asymptomatic patients.
Normal and non-neoplastic colonoscopy grouped together
Divided in stage I/II and III/IV,
Divided in small and large adenoma
Diagnostic performance of bacteria-derived biomarkers
The overall performance of bacteria-derived biomarkers for the detection of precursor lesions showed an AUC ranging from 0.28-0.98, a sensitivity ranging from 18-100 per cent and specificity ranging from 39-97 per cent. Notably, the high AUC of the overall range for precursor lesions was determined by two studies that used oral mucosa instead of or in conjunction with faecal samples. These studies by Flemer et al. and Zhang S. et al. [31,46], reported the highest AUC found of 0.98 (95% CI 0.95-0.98), with a sensitivity of 88 per cent and AUC of 0.95 (95% CI 0.91-0.99), respectively, suggesting a high accuracy for the use of microbiome-derived biomarkers based on the oral mucosa for precursor lesions. The same study by Flemer et al. evaluated the performance based solely on bacteria from faecal samples with a reported AUC of 0.87 (no 95% CI available) [46].
For early stages of CRC, the overall performance of solely bacteria-derived biomarkers, based on AUC as well, showed a range from 0.54 to 0.89 and sensitivity ranging from 60 to 100 per cent. The studies of Yachida et al. and Konishi et al. were the only ones that included stage 0 in their analyses and reported an AUC of 0.73 and 0.54, respectively [19,38]. For stage I, an AUC range between 0.74 and 0.97 was seen, with a sensitivity ranging from 90 to 100 per cent, and stage I/II or only stage II showed a range between 0.82-0.89, with a sensitivity ranging from 60 to 100 per cent. Although some articles demonstrated very high performance in patients with stage I, performance generally increased with increasing stage. The study of Rezasoltani et al. reported the highest performance, with an AUC of 0.97 (no 95% CI available) and a sensitivity and specificity of 100 and 77, respectively [43]. This was based, however, on a very small sample size of only 20 patients with stage I CRC combined with 43 patients with adenomas.
Most panels were based on multiple bacteria ranging from single up to 50; however, no better performance was seen for the detection of precursor lesions or CRC when more bacteria in the panels were applied. However, the performance of bacterial panels was generally enhanced when combined with other characteristics such as body mass index (BMI), age, sex and ethnicity. Likewise, the addition of other faecal markers like FIT or gFOBT strengthened the accuracy of the models overall. The use of only two bacteria (Fusobacterium nucleatum and Parvimonas micra) combined with FIT, faecal DNA methylation (i.e. Septin9, NDRG4, BMP3) or mutation markers (i.e. KRAS, BRAF, PI3KCA) resulted in a reported increase in AUC from 0.57 to 0.73 for the detection of precursor lesions when compared to the bacterial model alone [27]. However, sensitivity and specificity of respectively 39 and 83 for only the bacteria model changed in respectively 28 and 94 when combined. Liu et al. added Fusobacterium nucleatum and pks+ Escherichia Coli to the gFOBT and two blood-based markers (carcinoembryonic antigen (CEA) and carbohydrate antigen (CA) 19-9) with a reported AUC of 0.85 (95% CI 0.57-1.0) [41]. Again, this was accompanied by a drop in sensitivity from 91 per cent based on a measurement of bacteria alone to 67 per cent when combined. However, there was an increase in specificity from 75 to 90 per cent for the combined model. CEA as an additional marker was also used by Xie et al. with FIT and two bacteria (Clostridium symbiosum and Fusobacterium nucleatum) with a reported AUC of 0.82 (95% CI 0.77–0.87) [29]. A summary of the performance capacity of each study is outlined in Table 2.
Table 2.
Included studies assessing microbiome-derived biomarkers for early CRC detection
| Reference | Comparison of interest1 | Biomarker | Performance for early CRC detection | Analytical method2 | |
|---|---|---|---|---|---|
| AUC (95% CI) | Sens/spec | ||||
| Zhang Y, 2021 [32] | AP vs HC | 13 OTU3 | 0.61 (0.55–0.66) | 34 / 80 | 16S |
| AP vs HC | 13 OTU + FIT | 0.64 (0.58–0.69) | 41 / 80 | ||
| AP vs HC | 13 OTU + FIT + APCS4 | 0.71 (0.65-0.75) | 46 / 80 | ||
| Zhang S, 2020 [31] | AP vs HC | 5 OTU | 0.95 (0.91-0.99) | 16S | |
| Yachida, 2019 [19] | Stage 0 vs HC | 29 species | 0.73 | Metagenomics | |
| 16 KO genes | 0.75 | ||||
| 29 species + 24 metabolites + 16 KO genes | 0.78 | ||||
| Yao, 2021 [30] | Stage I/II vs HC | 5 species | N/A | I 62 / -, II 72 / - | qPCR |
| Stage I/II vs HC | 5 species + FIT | N/A | I 70 / -, II 89 / - | ||
| Xie, 2017 [29] | Stage I/II vs HC | Fusobacteria nucleatum | 0.59 (0.54-0.64) | qPCR | |
| Clostridium symbiosum | 0.73 (0.68-0.77) | ||||
| Clostridium symbiosum + FIT | 0.81 (0.75-0.86) | ||||
| Clostridium symbiosum and Fusobacteria nucleatum + FIT + CEA | 0.82 (0.77-0.87) | ||||
| Wei, 2022 [28] | AP vs HC | 5 OTUs | 0.71 (0.63-0.80) | 16S | |
| Mo, 2021 [27] | AP vs HC | Fusobacterium nucleatum and Parvimonas micra | 0.57 | 39 / 83 | qPCR |
| Fusobacterium nucleatum and Parvimonas micra + DNA methylation/mutation5 + FIT | 0.73 | 28 / 94 | |||
| Stage I/II | Fusobacterium nucleatum and Parvimonas micra + DNA methylation/mutation + FIT | N/A | I 60 / -, II 85 / - | ||
| Liu, 2021 [41] | AP vs HC | Fusobacterium nucleatum | 0.74 (0.56-0.91) | 71 / 65 | qPCR |
| pks+ Escherichia coli | 0.82 (0.64-0.98) | 91 / 75 | |||
| Fusobacterium nucleatum, pks+ Escherichia coli + CEA + CA19-9 + FOBT | 0.85 (0.57-1.0) | 67 / 90 | |||
| 4 species + FIT | N/A | 49 / 81 | |||
| Liang, 2017 [40] | Stage I/II vs HC | 4 species + FIT | N/A | I 77 / -, II 100 / - | qPCR |
| Liang, 2021 [39] | AP vs HC | 4 species | 0.67 (0.62-0.72) | 39 / 83 | qPCR |
| 4 species + FIT | N/A | 49 / 81 | |||
| Konishi, 2022 [38] | AP vs HC | 50 OTUs | 0.52 (0.45 – 0.60) | 16S | |
| Stage 0 vs HC | 0.54 (0.45 – 0.62) | ||||
| Stage I vs HC | 0.74 (0.68 – 0.80) | ||||
| Stage II vs HC | 0.86 (0.81 – 0.90) | ||||
| Guo, 2018 [37] | Stage I vs HC | 3 taxa | 0.80 | 90 / 60 | qPCR |
| Stage I/II vs HC | 0.89 | 90 / 76 | |||
| Goedert, 2015 [47] | AP vs HC | 5 taxa + 7 genera | 0.77 | 16S | |
| Gao, 2022 [36] | AP vs HC | 12 species | 0.66 (0.57-0.75) | Metagenomics | |
| Stage I vs HC | 0.88 (0.72-1.00) | ||||
| Stage II vs HC | 0.96 (0.91-1.00) | ||||
| Gao, 2020 [35] | AP vs HC | 18 genera | 0.62 (0.52-0.71) | 84 / 39 | Metagenomics |
| 18 genera + FIT | 0.72 (0.63-0.81) | ||||
| Coker, 2022 [34] | AP vs HC | 14 species | 0.84 (0.80-0.89) | Metagenomics | |
| 14 species + 2 metabolites | 0.88 (0.84-0.92) | ||||
| Ai, 2017 [33] | AP vs HC | 6 species | 0.87 | 16S | |
| Rezasoltani, 2018 [43] | AP + Stage I vs HC | 5 taxa | 0.97 | 100 / 77 | qPCR |
| Zeller, 2014 [50] | CRC vs HC^ | 22 species | 0.84 | Metagenomics | |
| CRC vs HC^ | 22 species + FOBT | 0.87 | |||
| Young, 2021 [49] | AP vs HC* | 15 taxa | 0.72 (0.68-0.75) | 16S | |
| AP vs HC⁎⁎ | 15 taxa | 0.82 (0.79-0.84) | |||
| AP vs HC* | 15 taxa + age + sex | 0.71 (0.67-0.74) | |||
| AP vs HC⁎⁎ | 15 taxa + age + sex | 0.84 (0.82-0.86) | |||
| Tarallo, 2019 [48] | AP vs HC | hsa-miRNA + bsRNA + mbDNA6 | 0.47 | sRNA7/Metagenomics | |
| Flemer, 2017 [46] | AP vs HC | 16 OTUs (faecal) | 0.87 | 16S | |
| 12 OTUs (oral) | 0.89 (0.80-0.89) | 67 / - | |||
| 16 OTUs (faecal) + 12 OTUs (oral) | 0.98 (0.95-0.98) | 88 / 94 | |||
| Eklöf, 2017 [45] | AP vs HC | clbA+ bacteria, Fusobacterium nucleatum | N/A | 47 / 63 | qPCR |
| clbA+ bacteria, Fusobacterium nucleatum + FIT | N/A | 61 / 63 | |||
| Clos-Garcia, 2020 [44] | AP vs HC | 16 genera | 0.28 | 16S | |
| Zackular, 2014 [53] | AP vs HC | 5 OTUs | 0.84 (0.74–0.94) | 16S | |
| 5 OTUs + age + race + BMI | 0.90 (0.82–0.98) | 90 / 80 | |||
| Baxter, 2016 [52] | AP vs HC | 22 OTUs | 0.67 | 18 / 97 | 16S |
| 23 OTUs + FIT | 0.76 | ||||
CRC = colorectal cancer, AP = adenoma and/or polyp, HC = healthy controls
Analytical method: 16S = 16S rRNA gene sequence, qPCR = real-time quantitative PCR, metagenomics
OTU = operational taxonomic unit
APCS = Asia-Pacific Colorectal Screening, a validated risk-stratification tool
Methylated genes: Septin9, NDRG4, BMP3. Mutated genes: KRAS, BRAF, PI3KCA
hsa = Homo sapiens microRNAs, bsRNA = bacterial small RNAs, mbDNA = microbial DNA
sRNA = small RNA sequencing
Based on CRC vs HC, but similar changes in microbial abundance observed in early-stages of CRC
Healthy controls with normal colonoscopy
Healthy controls, blood negative in stool
Analytical methodology of bacteria-derived biomarkers
Biomarkers in faecal samples for early CRC detection were typically based on the relative abundance of bacteria. Relevant bacteria were either preselected from relevant literature or associations were discovered with selected bacteria between the CRC, precursor lesions and healthy controls. (See Supplementary file S4 for the bacteria and the analytical methods used for the detection of early CRC). For the detection of these bacteria, different techniques were used. 11 articles used 16S rRNA gene sequencing [28,[31], [32], [33], 38,44,46,47,49,52,53], whereas nine used qPCR [27,29,30,37,[39], [40], [41], 43,45] and six used metagenomic shotgun sequencing [19,[34], [35], [36], 48,50]. Universal primers were used in most articles using 16S rRNA gene sequence analysis. However, a large range was seen in the variable region used for the analysis per study, ranging from V1 to V4. For qPCR, different primers were sometimes used (See Supplementary file S4). It is important to note that studies using 16S were only capable of giving bacterial taxa, whereas studies employing qPCR or metagenomics were able to identify or use bacteria on their species level for the detection of early CRC. Furthermore, between the included articles, a high heterogeneity was seen between sample collection methods as well as processing kits for DNA extraction which (see Supplementary Table S2 for a summarised description of sample collection and DNA processing kits per article).
Statistical approaches for the classification models for the prediction of early CRC were based on different models. Besides simple logistic regression models (LRM) [27,29,30,35,[37], [38], [39], [40], [41], 43,45], a random forest model (RFM) was often used [28,31,34,36,[47], [48], [49], 52], followed by LASSO logistic regression models [32,44,50] and Bayesian methods [33,53]. Two studies even used two classification models [19,46]. Only the study by Ai et al. investigated on forehand the best classification model, comparing Bayesian methods, RFM, LRM, F measure and Matthews correlation coefficient, finding the best model based on Bayesian methods [33].
Microbiome-derived biomarkers based on co-metabolome
The metabolome may provide an alternative source of novel biomarkers. The metabolome, defined as the collection of all small low-molecular-weight compounds (<1kDa) in a biological sample, is a time-dependent, multiparametric analysis of the functional output of a biological system and its exposome [55]. The metabolome expression will therefore vary with subtle or early changes in a given disease state [56,57]. The co-metabolome, defined as the set of compounds which are the product of more than one genome interacting in a symbiotic system, is of particular pertinence in the diagnosis of colorectal cancer [55]. Five articles investigated the diagnostic performance of the faecal co-metabolome next to, or in conjunction with, bacteria-derived biomarkers [19,28,34,36,44]. Only two articles were found that solely investigated the diagnostic performance of the co-metabolome for the detection of early CRC [51,54]. The overall performance for precursor lesions based on the co-metabolome reported a range in AUC from 0.69 to 0.84, whereas stage I and II of CRC ranged between 0.65 to 0.93. Chen et al. demonstrated the best performance for precursor lesions and stage I and II of CRC of 0.84 (sensitivity 63 per cent, specificity 85 per cent) and 0.93 (sensitivity 88 per cent, specificity 85 per cent), respectively, based on a model of eight gut-microbiome associated serum metabolites (GMSM-panel) in a matched faeces and serum cohort [54]. Their panel, based on bacterial metabolism, emphasised on secondary bile acids and described an upregulation in serum concentrations of unconjugated primary bile acids (CA) and deoxycholic acid (DCA) in CRC patients (see Supplementary file S4 for all specific metabolites used). Interestingly, the study of Yachida et al. [19] reported an alteration in bile acid metabolism as well, with a significant increase in DCA in patients with stage 0 CRC. These findings suggest a role for bacterial metabolism, especially bile acid metabolism, in early CRC. Four articles described the performance of metabolites compared with bacterial species. Yachida et al. [19] reported an increase in performance, based on AUC, from 0.73 based on 29 bacterial species and 0.65 based on metabolites to 0.78 when combined. Gao et al. [36], interestingly the only study who looked solely at the serum metabolomic profile instead of the faecal metabolomic profile, reported an AUC of 0.91 when combined with bacterial species for the detection of precursor lesions. Coker et al. [34] reported an AUC of 0.69 based on metabolites and an AUC of 0.84 based on 14 bacterial species. They reported an AUC of 0.88 when these 14 species were combined with the two most distinctive metabolites (L-Asparagine and Phenyllactic acid). Finally, the study of Clos-Garcia et al. [44] reported a low AUC of 0.30 when metabolites and bacteria were combined. This study showed the lowest AUC as well based only on bacteria of 0.28. A summary of the results is presented in Table 3.
Table 3.
Co-metabolome as screening tool for early CRC detection.
| Reference | Comparison of interest | Biomarker | Performance for early CRC detection | Analytical method1 | |
|---|---|---|---|---|---|
| wAUC (95% CI) | Sens/Spec | ||||
| Yachida, 2019 [19] | Stage 0 vs HC | 24 metabolites | 0.65 | CE-TOFMS | |
| Wei, 2022 [28] | AP vs HC | 4 metabolites | 0.73 (0.63-0.82) | UPLC-MS/MS | |
| 4 OTUs + 5 metabolites | 0.90 (0.85-0.96) | ||||
| Gao, 2022 [36] | Stage I vs HC | 3 metabolites | 0.82 (0.68-0.97) | UPLC-MS/MS | |
| Stage II vs HC | 0.87 (0.79-0.95) | ||||
| Stage I/II vs HC | 0.85 (0.76-0.93) | ||||
| AP vs HC | 12 OTUs + 3 metabolites | 0.91 (0.83-0.99) | |||
| Coker, 2022 [34] | AP vs HC | 11 metabolites | 0.69 (0.62-0.81) | GC-TOFMS | |
| 11 metabolites + age + gender + obesity | 0.75 (0.69-0.81) | ||||
| 4 metabolites + 6 species | 0.75 (0.69-0.81) | ||||
| Clos-Garcia, 2020 [44] | AP vs HC | 16 genera + 6 metabolites | 0.30 | UHPLC-MS | |
| Chen, 2022 [54] | AP vs HC | 8 metabolites | 0.84 | 63 / 85 | UPLC-MS |
| Stage I/II vs HC | 0.93 | 88 / 85 | |||
| Bosch, 2022 [51] | AP vs HC | 3 metabolites | 0.79 (0.64–0.94) | 79 / 74 | UPLC-MS |
CE-TOFMS = capillary electrophoresis time-of-flight mass spectrometry, U(H)PLC-MS/MS = Ultra-(High)-Performance Liquid Chromatography Tandem Mass Spectrometry
Analytical methodology of co-metabolomic markers
The methods used to detect metabolites were all based on mass spectrometry (MS), with variations in chromatography techniques. Most used liquid chromatography MS [28,36,44,51,54], and only one used gas-chromatography GS-MS [34], and one applied capillary electrophoresis MS [19]. Most studies investigated the faecal co-metabolome [19,28,34,44,51], with only two studies investigating the serum co-metabolome either solely or in conjunction with the faecal metabolome [36,54].
It has been reported that volatile components of the co-metabolome are more prone to variations in storage conditions and sampling methodologies than the microbiome [58,59]. The sampling methodologies were extracted per study, although not all studies disclosed details of sample collection, showing a large heterogeneity between trial designs (see Supplementary Table S2 for a detailed description). The study of Yachida et al. was the only study to collect faeces on site and immediately stored for analysis under appropriate conditions (dry ice and -80°C for long-term storage) [19]. Two studies let the patient collect the faeces at home and stored in their own freezer, leaving time until long-term storage unclear [44,51].
Risk of bias and applicability
The QUADAS-2 tool was used to assess the risk of bias and applicability. The results are summarised in Supplementary Table S3 as well as presented in Supplementary Fig. S1.
Overall the risk of bias for the first domain, 'patient selection,' was low in 17 studies, high in 10 and unclear in one since most of the studies were cohort studies. For the second domain ‘index test', the risk of bias was very high, with 23 studies high risk, zero low and five unclear. The third domain 'reference test' showed a low risk of bias with zero high, one unclear and 27 low risk. For the last domain 'flow and timing', low risk was seen in 19 studies, high risk in two and unclear in seven studies. Since we preselected articles only applicable to the detection or description of early CRC, all studies showed a high degree of applicability.
Discussion
A total of 28 studies assessed microbiome-derived biomarkers for the detection of early CRC. Despite this, a large degree of heterogeneity was observable in cohort sizes, demographic properties, microbiome sequencing analyses and statistical models used, making a robust interpretation of these data challenging and preventing meta-analysis. Therefore, results were only reported in a descriptive manner, summarising the diagnostic performance of microbiome-derived biomarkers for early CRC detection. The biomarkers identified in these analyses are also yet to be externally validated. Despite this, it appears that the faecal and oral microbiome may have some diagnostic utility for the early detection of CRC.
Diagnostic performance of microbiome-derived biomarkers
The diagnostic performance demonstrated a wide confidence interval across the reported studies for the measurement of the relative abundance of faecal bacteria (0.28 – 0.98 for precursor lesions and a range of 0.54-0.89 for early CRC, based on AUC). Diagnostic performance increased when the bacterial panel was added to existing screening tests, like the gFOBT or FIT, or added to DNA-makers for methylated (Septin9, NDRG4, BMP3) or mutated genes (KRAS, BRAF, PI3KCA), suggesting that bacteria-derived biomarkers could be used to augment current screening programs. Diagnostic performance did not significantly vary in those studies that used small numbers of bacteria as compared to studies using a multitude of bacteria in their panels. This is in contrast to recent literature investigating the microbiome as a biomarker in (all stages of) CRC [60,61]. Many of these studies were not prospective or powered to a primary diagnostic endpoint, or performed in clinically representative populations (e.g. within population demographics typically called for screening).. Moreover, many did not make a distinction between stage or anatomical location (e.g. right or left colon) of CRC in their analysis [62], [63], [64], [65]. Similarly, some studies simply group cancer stages together to produce summary diagnostic statistics [66], [67], [68], [69], [70], [71]. As expected, studies that did compare the performance of the gut microbiome across AJCC different stages often report an increase in performance in stage III and IV cancers [36], [37], [38].
The optimal diagnostic performance across all studies showed an AUC of 0.98, with a sensitivity and specificity of 88 and 94 per cent, and employed an oral mucosa-based detection of bacterial strains [46]. Bacterial taxa statistically associated with CRC, such as Fusobacterium, Peptostreptococcus, Porphyromonas, and Parvimonas, were consistently detected in the oral cavity of those patients with CRC [46]. However, this observation is not CRC-specific, as these bacteria have also been observed to be enriched in other gastro-intestinal (GI) tract cancers, such as oesophageal [42] and pancreatic cancer [72]. Interestingly, these bacteria were however abundant in precursor lesions for bowel cancer. This is important as colonic adenoma detection rates range from 26 to 47% in the screening populations [73] and prevalence rates of cancer within these polyps have been reported in 0.9–2.8 % of polyps ≤5 mm and 5.3–15.5 % of polyps between 6 and 9 mm [74]. The risk of adenoma progression to cancer is dependent on their histological subtype and anatomical location (right vs left) but also on environmental factors (e.g. diet or obesity) that are co-regulated by the gut microbiome [75]. A recent prospective analysis of 231 patients undergoing screening colonoscopy analysed faecal swabs using 16S RNA and demonstrated a classification accuracy for adenomas >75% for Naïve Bayes and Neural Network models using informative OTUs [76]. Previous analysis of faecal samples by 16 sRNA of patients with conventional adenomas is depleted in a network of Clostridia operational taxonomic units from families Ruminococcaceae, Clostridiaceae, and Lachnospiraceae and enriched in the classes Bacilli and Gammaproteobacteria, order Enterobacteriales, and genera Actinomyces and Streptococcus [15].
Of all the screened articles, only seven investigated the potential of the co-metabolome as a diagnostic tool for early CRC detection. A similar effect was reported with an AUC ranging from 0.69 – 0.84 for precursor lesions and 0.65 – 0.93 for stages I and II of CRC. However, there were insufficient articles on the efficacy of the co-metabolome for early CRC detection to form any proper conclusions. Oncogenic co-metabolites produced by luminal and mucosal gut microbiota (e.g. bile acids) are over-abundant in those populations at high risk of colonic cancer, while beneficial microbial co-metabolites, like short-chain fatty acids (SCFAs), are often decreased [76,77]. Changes in metabolic profiles may therefore reflect an early pro-oncogenic environment making them valuable in screening programs, however this hypothesis is yet to be tested at scale.
Currently, most research regarding the investigation of a microbiome-derived biomarker for (early) CRC is based on differences in taxonomic variation. However, some studies suggest a higher sensitivity for the detection of CRC based on bacterial genes [78]. A direct comparison of bacterial taxa vs bacterial genes for the detection of CRC has been recently investigated by Norouzi-Beirami et al. [79]. This study noticed a small increase in sensitivity and specificity for the detection of CRC based on genes in comparison to bacterial taxa. However, the increase was almost gone when tested in an independent cohort. Superior accuracy was however detected based on functional features (AUC 0.71) compared to taxa or gene level (with a corresponding AUC of both of 0.59). Most studies have been referring to bacterial taxonomy because this is the most standardised technique in this field, mainly based on only one bacterial gene, the 16S rRNA gene. To use other bacterial genes as biomarkers requires standardised procedures and bioinformatic pipelines as well as the establishment of universally usable primers for these specific genes, which are also very challenging at this specific moment considering the advances in whole genome shotgun sequencing. Nevertheless, one major limitation of taxonomic identification as well as gene level, especially in regard to early CRC, is that it does not give any insight into, or is necessarily associated with, functional variation of the genetic content. When investigating a more accurate and sensitive biomarker for the prediction of early CRC, it is the functional level rather than bacterial taxa or genes that could provide insight into microbiome changes associated with the development of CRC and therefore be used as a possible biomarker for the detection of early CRC.
Limitations of microbiome-derived biomarkers
The gut microbiome demonstrates significant inter-individual variation and macro and microscopic anatomical heterogeneity, governed by factors such as age, gender, BMI, diet and antibiotic use. When comparing healthy controls with colorectal patients, these factors should ideally be incorporated in the analyses via matching of the groups of interest. Unfortunately, most of the included articles only reported possible influencing factors on the microbiome composition without any matching between the two groups. Pilot studies in CRC have also identified significant variation between cancers [80], in part because the gut microbiome is dynamic and evolves with the pathology, in part because it is subject to confounding environmental factors, such as diet, medication, smoking and other lifestyle factors [81]. These were rarely accounted for in the analyses, and most were often not reported. The heterogeneity in the diversity of the faecal microbiome exists at both a phyla and strain level between geographically discrete populations and across countries [82,83]. The majority of included articles were performed in Asia, potentially limiting the translation to other regions, and it is unclear therefore how many of these studies are translatable.
One other major limitation in these studies has been the lack of standardisation in both sample collection and processing. Sample collection showed a wide heterogeneity between studies (e.g. before or after bowel-cleansing agents required for the colonoscopy) as well as temperature and time until long-term storage at -80°C. These factors are known to influence microbiome composition [84,85]. Furthermore, DNA extraction was carried out using a range of DNA extraction kits, adding an additional variable influencing the outcome of microbiome composition (see Supplementary Table S2 for a summary of sample collection and DNA processing of the included studies). A lack of standardisation was also seen in the analytical approaches employed by the included studies. All studies used different analytical methods to identify bacteria, either 16S rRNA, qPCR or metagenomics. 16S rRNA analysis is largely restricted to taxonomic analysis with an inferred functional interpretation. Metagenomics permits species-level identification and deep functional insights, bypassing some of these challenges [86], although it is not yet affordable for population-level analyses and its interpretation requires significant computing power. Within these methods, there was again a large heterogeneity in the choice of primers used, leading to potential biases and making comparisons between them almost impossible [87].
Research priorities and the ideal microbiome biomarker study
If the microbiome is to be realistically mined for its biomarker potential in CRC, there is an urgent need to move away from small-scale pilot studies into prospectively, adequately powered, multicentred trials that are quality assured from a sampling and analytical methodology. Currently, there is a lack of consensus on the most robust method of either qualitatively or quantitively analysing the gut microbiome for early cancer detection in terms of its function, absolute or relative abundance, its diversity or species richness. Given the significant inter-individual variation in the global microbiome and its evolutionary instability within and on CRC, it is not yet clear if the microbiome is a viable biomarker for early cancer. Therefore future studies must seek to test biomarkers that are mechanistically linked to established molecular pathways and which are established to exist across vulnerable populations. However, this is potentially where the microbiome has its greatest potential, as it can provide future clinicians with noninvasive, actional information on the anatomical location or progression of the disease.
Conclusion
Gut microbial-derived biomarkers could be leveraged to enhance current screening programs for CRC. However, significant barriers must be overcome before this can be achieved. Current analyses cannot be meta-analysed due to large observed variations in study design and analytical precision. Future CRC microbiome studies therefore require precision and must account for environmental and sampling confounding and bias, and provide methodological quality assurance and establish cost-effectiveness.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Supplementary materials
Supplementary material of this systematic review can be found in the online version. The following supplementary material of this systematic review can be found in the online version. Fig. S1. QUADAS-2 overview, Table S1. Search strategy, Table S2. Sample collection and DNA processing, Table S3. QUADAS-2 results, Excel file S4. Additional characteristics of analytical method and biomarker used.
CRediT authorship contribution statement
Florine H. Zwezerijnen-Jiwa: Conceptualization, Writing – original draft. Hugo Sivov: Data curation. Petra Paizs: Writing – review & editing. Konstantina Zafeiropoulou: Visualization, Writing – review & editing. James Kinross: Supervision, Writing – review & editing.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100868.
Appendix. Supplementary materials
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Toes-Zoutendijk E, Bonfrer JMG, Ramakers C, Thelen M, Spaander MCW, Dekker E, et al. Quality monitoring of a FIT-based colorectal cancer screening program. Clin. Chem. 2019;65(3):419–426. doi: 10.1373/clinchem.2018.294041. [DOI] [PubMed] [Google Scholar]
- 4.Davila RE, Rajan E, Baron TH, Adler DG, Egan JV, Faigel DO, et al. ASGE guideline: colorectal cancer screening and surveillance. Gastrointest Endosc. 2006;63(4):546–557. doi: 10.1016/j.gie.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Lin JS, Perdue LA, Henrikson NB, Bean SI, Blasi PR. Screening for colorectal cancer: updated evidence report and systematic review for the US preventive services task force. JAMA. 2021;325(19):1978–1998. doi: 10.1001/jama.2021.4417. [DOI] [PubMed] [Google Scholar]
- 6.Gupta S. Screening for colorectal cancer. Hematol. Oncol. Clin. North Am. 2022;36(3):393–414. doi: 10.1016/j.hoc.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton KF, Newman W, Hill J. Review of biomarkers in colorectal cancer. Colorectal Dis. 2012;14(1):3–17. doi: 10.1111/j.1463-1318.2010.02439.x. [DOI] [PubMed] [Google Scholar]
- 8.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Tanaka M, Tanaka T, Ishigamori R. Biomarkers for colorectal cancer. Int. J. Mol. Sci.. 2010;11(9):3209–3225. doi: 10.3390/ijms11093209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB, 3rd, Boland P, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat. Rev. Clin. Oncol. 2020;17(12):757–770. doi: 10.1038/s41571-020-0392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8):e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71(5):1020. doi: 10.1136/gutjnl-2021-326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knippel RJ, Sears CL. The microbiome colorectal cancer puzzle: initiator, propagator, and avenue for treatment and research. J. Natl. Compr. Canc. Netw. 2021;19(8):986–992. doi: 10.6004/jnccn.2021.7062. [DOI] [PubMed] [Google Scholar]
- 15.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4(1):69. doi: 10.1186/s40168-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinross J, Mirnezami R, Alexander J, Brown R, Scott A, Galea D, et al. A prospective analysis of mucosal microbiome-metabonome interactions in colorectal cancer using a combined MAS 1HNMR and metataxonomic strategy. Sci. Rep. 2017;7(1):8979. doi: 10.1038/s41598-017-08150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Chang Y, Zheng Q, Zhang R, Hu C, Jia W. Altered intestinal microbiota associated with colorectal cancer. Front Med. 2019;13(4):461–470. doi: 10.1007/s11684-019-0695-7. [DOI] [PubMed] [Google Scholar]
- 18.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019;25(6):968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 20.Xu K, Jiang B. Analysis of mucosa-associated microbiota in colorectal cancer. Med. Sci. Monit. 2017;23:4422–4430. doi: 10.12659/MSM.904220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493(7430):45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott AJ, Alexander JL, Merrifield CA, Cunningham D, Jobin C, Brown R, et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut. 2019;68(9):1624–1632. doi: 10.1136/gutjnl-2019-318556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan—a web and mobile app for systematic reviews. Syst. Rev. Pharm. 2016;5(1):210. doi: 10.1186/s13643-016-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiser MR. AJCC 8th Edition: colorectal cancer. Ann. Surg. Oncol. 2018;25(6):1454–1455. doi: 10.1245/s10434-018-6462-1. [DOI] [PubMed] [Google Scholar]
- 26.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 27.Mo S, Wang H, Han L, Xiang W, Dai W, Zhao P, et al. Fecal multidimensional assay for non-invasive detection of colorectal cancer: fecal immunochemical test, stool DNA mutation, methylation, and intestinal bacteria analysis. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.643136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei PL, Wu MS, Huang CK, Ho YH, Hung CS, Lin YC, et al. Exploring gut microenvironment in colorectal patient with dual-omics platform: a comparison with adenomatous polyp or occult blood. Biomedicines. 2022;10(7) doi: 10.3390/biomedicines10071741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie YH, Gao QY, Cai GX, Sun XM, Sun XM, Zou TH, et al. Fecal clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMedicine. 2017;25:32–40. doi: 10.1016/j.ebiom.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y, Ni H, Wang X, Xu Q, Zhang J, Jiang L, et al. A new biomarker of fecal bacteria for non-invasive diagnosis of colorectal cancer. Front. Cell Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.744049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Kong C, Yang Y, Cai S, Li X, Cai G, et al. Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics. 2020;10(25):11595–11606. doi: 10.7150/thno.49515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Lu M, Lu B, Liu C, Ma Y, Liu L, et al. Leveraging fecal microbial markers to improve the diagnostic accuracy of the fecal immunochemical test for advanced colorectal adenoma. Clin. Transl. Gastroenterol. 2021;12(8):E00389. doi: 10.14309/ctg.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ai L, Tian H, Chen Z, Chen H, Xu J, Fang JY. Systematic evaluation of supervised classifiers for fecal microbiota-based prediction of colorectal cancer. OncoTargets Ther. 2017;8(6):9546–9556. doi: 10.18632/oncotarget.14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, et al. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. 2022;10(1):35. doi: 10.1186/s40168-021-01208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao R, Wang Z, Li H, Cao Z, Gao Z, Chen H, et al. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. J. Gastroenterol. Hepatol. 2020;35(12):2109–2121. doi: 10.1111/jgh.15077. [DOI] [PubMed] [Google Scholar]
- 36.Gao R, Wu C, Zhu Y, Kong C, Zhu Y, Gao Y, et al. Integrated analysis of colorectal cancer reveals cross-cohort gut microbial signatures and associated serum metabolites. Gastroenterology. 2022;163(4):1024–1037. doi: 10.1053/j.gastro.2022.06.069. [DOI] [PubMed] [Google Scholar]
- 37.Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, et al. A simple and novel fecal biomarker for colorectal cancer: Ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin. Chem. 2018;64(9):1327–1337. doi: 10.1373/clinchem.2018.289728. [DOI] [PubMed] [Google Scholar]
- 38.Konishi Y, Okumura S, Matsumoto T, Itatani Y, Nishiyama T, Okazaki Y, et al. Development and evaluation of a colorectal cancer screening method using machine learning-based gut microbiota analysis. Cancer Med. 2022;11(16):3194–3206. doi: 10.1002/cam4.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang JQ, Wong SH, Szeto CH, Chu ES, Lau HC, Chen Y, et al. Fecal microbial DNA markers serve for screening colorectal neoplasm in asymptomatic subjects. J. Gastroenterol. Hepatol. 2021;36(4):1035–1043. doi: 10.1111/jgh.15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Q, Chiu J, Chen Y, Huang Y, Higashimori A, Fang J, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin. Cancer Res. 2017;23(8):2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 41.Liu K, Yang X, Zeng M, Yuan Y, Sun J, He P, et al. The role of fecal fusobacterium nucleatum and pks(+) escherichia coli as early diagnostic markers of colorectal cancer. Dis. Markers. 2021;2021 doi: 10.1155/2021/1171239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Winckler B, Lu M, Cheng H, Yuan Z, Yang Y, et al. Oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0143603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rezasoltani S, Sharafkhah M, Asadzadeh Aghdaei H, Nazemalhosseini Mojarad E, Dabiri H, Akhavan Sepahi A, et al. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J. Microbiol. Methods. 2018;155:82–88. doi: 10.1016/j.mimet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Clos-Garcia M, Garcia K, Alonso C, Iruarrizaga-Lejarreta M, D'Amato M, Crespo A, et al. Integrative analysis of fecal metagenomics and metabolomics in colorectal cancer. Cancers. 2020;12(5):1142. doi: 10.3390/cancers12051142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer. 2017;141(12):2528–2536. doi: 10.1002/ijc.31011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goedert JJ, Gong Y, Hua X, Zhong H, He Y, Peng P, et al. Fecal microbiota characteristics of patients with colorectal adenoma detected by screening: a population-based Study. EBioMedicine. 2015;2(6):597–603. doi: 10.1016/j.ebiom.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarallo S, Ferrero G, Gallo G, Francavilla A, Clerico G, Realis Luc A, et al. Altered fecal small RNA profiles in colorectal cancer reflect gut microbiome composition in stool samples. mSystems. 2019;4(5) doi: 10.1128/mSystems.00289-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young C, Wood HM, Balaguer AF, Bottomley D, Gallop N, Wilkinson L, et al. Microbiome analysis of more than 2,000 NHS bowel cancer screening programme samples shows the potential to improve screening accuracy. Clin. Cancer Res. 2021;27(8):2246–2254. doi: 10.1158/1078-0432.CCR-20-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10(11):766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bosch S, Acharjee A, Quraishi MN, Rojas P, Bakkali A, Jansen EEW, et al. The potential of fecal microbiota and amino acids to detect and monitor patients with adenoma. Gut Microbes. 2022;14(1) doi: 10.1080/19490976.2022.2038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baxter NT, Ruffin MT, Rogers MA. Schloss PD. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8(1):37. doi: 10.1186/s13073-016-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zackular JP, Rogers MAM, Ruffin MT. Schloss PD. the human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014;7(11):1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen F, Dai X, Zhou CC, Li KX, Zhang YJ, Lou XY, et al. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. 2022;71(7):1315–1325. doi: 10.1136/gutjnl-2020-323476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134(5):714–717. doi: 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 56.Han S, Gao J, Zhou Q, Liu S, Wen C, Yang X. Role of intestinal flora in colorectal cancer from the metabolite perspective: a systematic review. Cancer Manag Res. 2018;10:199–206. doi: 10.2147/CMAR.S153482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batty CA, Cauchi M, Lourenço C, Hunter JO, Turner C. Use of the analysis of the volatile faecal metabolome in screening for colorectal cancer. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Spiegeleer M, De Graeve M, Huysman S, Vanderbeke A, Van Meulebroek L, Vanhaecke L. Impact of storage conditions on the human stool metabolome and lipidome: Preserving the most accurate fingerprint. Anal. Chim. Acta. 2020;1108:79–88. doi: 10.1016/j.aca.2020.02.046. [DOI] [PubMed] [Google Scholar]
- 59.Gratton J, Phetcharaburanin J, Mullish BH, Williams HR, Thursz M, Nicholson JK, et al. Optimized sample handling strategy for metabolic profiling of human feces. Anal. Chem. 2016;88(9):4661–4668. doi: 10.1021/acs.analchem.5b04159. [DOI] [PubMed] [Google Scholar]
- 60.Yu L, Zhao G, Wang L, Zhou X, Sun J, Li X, et al. A systematic review of microbial markers for risk prediction of colorectal neoplasia. Br. J. Cancer. 2022;126(9):1318–1328. doi: 10.1038/s41416-022-01740-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erben V, Bhardwaj M, Schrotz-King P, Brenner H. Metabolomics biomarkers for detection of colorectal neoplasms: a systematic review. Cancers. 2018;10(8) doi: 10.3390/cancers10080246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, Geng R, Liu L, Jin X, Yan W, Zhao F, et al. Gut microbiota-based algorithms in the prediction of metachronous adenoma in colorectal cancer patients following surgery. Front. Microbiol. 2020;11:1106. doi: 10.3389/fmicb.2020.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma C, Chen K, Wang Y, Cen C, Zhai Q, Zhang J. Establishing a novel colorectal cancer predictive model based on unique gut microbial single nucleotide variant markers. Gut. Microbes. 2021;13(1):1–6. doi: 10.1080/19490976.2020.1869505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J, McDowell A, Kim EK, Seo H, Lee WH, Moon CM, et al. Development of a colorectal cancer diagnostic model and dietary risk assessment through gut microbiome analysis. Exp. Mol. Med. 2019;51(10):117. doi: 10.1038/s12276-019-0313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan B, Ma B, Yu J, Meng Q, Du T, Li H, et al. Fecal bacteria as non-invasive biomarkers for colorectal adenocarcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.664321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Löwenmark T, Löfgren-Burström A, Zingmark C, Eklöf V, Dahlberg M, Wai SN, et al. Parvimonas micra as a putative non-invasive faecal biomarker for colorectal cancer. Sci. Rep. 2020;10(1):15250. doi: 10.1038/s41598-020-72132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magat EM, Balanag GA, CariÑo AM, Fellizar A, Ortin TS, Guevarra L, Jr., et al. Clostridioides difficile antibody response of colorectal cancer patients versus clinically healthy individuals. Biosci. Microbiota Food Health. 2020;39(3):123–127. doi: 10.12938/bmfh.2020-010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mutignani M, Penagini R, Gargari G, Guglielmetti S, Cintolo M, Airoldi A, et al. Blood bacterial DNA load and profiling differ in colorectal cancer patients compared to tumor-free controls. Cancers. 2021;13(24) doi: 10.3390/cancers13246363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Li D, Yang Z, Dai W, Feng X, Liu Y, et al. Establishing high-accuracy biomarkers for colorectal cancer by comparing fecal microbiomes in patients with healthy families. Gut. Microbes. 2020;11(4):918–929. doi: 10.1080/19490976.2020.1712986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu J, Feng Q, Wong SH, Zhang D, Yi Liang Q, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 71.Zhang C, Hu A, Li J, Zhang F, Zhong P, Li Y. Combined non-invasive prediction and new biomarkers of oral and fecal microbiota in patients with gastric and colorectal cancer. Front. Cell Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.830684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahmed S, Naumann DN, Karandikar S. Differences in screening vs. non-screening colonoscopy: scope for improvement? Colorectal Dis. 2016;18(9):903–909. doi: 10.1111/codi.13291. [DOI] [PubMed] [Google Scholar]
- 74.Din S, Ball AJ, Taylor E, Rutter M, Riley SA, Johal S. Polypectomy practices of sub-centimeter polyps in the English Bowel Cancer Screening Programme. Surg. Endosc. 2015;29(11):3224–3230. doi: 10.1007/s00464-015-4064-6. [DOI] [PubMed] [Google Scholar]
- 75.Harewood R, Wooldrage K, Robbins EC, Kinross J, von Wagner C, Cross AJ. Adenoma characteristics associated with post-polypectomy proximal colon cancer incidence: a retrospective cohort study. Br J Cancer. 2022;126(12):1744–1754. doi: 10.1038/s41416-022-01719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang G, Yu Y, Wang YZ, Wang JJ, Guan R, Sun Y, et al. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J. Cell. Physiol. 2019;234(10):17023–17049. doi: 10.1002/jcp.28436. [DOI] [PubMed] [Google Scholar]
- 78.Wu Y, Jiao N, Zhu R, Zhang Y, Wu D, Wang A-J, et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat. Commun. 2021;12(1):3063. doi: 10.1038/s41467-021-23265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Norouzi-Beirami MH, Marashi SA, Banaei-Moghaddam AM, Kavousi K. Beyond taxonomic analysis of microbiomes: a functional approach for revisiting microbiome changes in colorectal cancer. Front. Microbiol. 2019;10:3117. doi: 10.3389/fmicb.2019.03117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Artemev A, Naik S, Pougno A, Honnavar P, Shanbhag NM. The association of microbiome dysbiosis with colorectal cancer. Cureus. 2022;14(2):e22156. doi: 10.7759/cureus.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7(1):17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mancabelli L, Milani C, Lugli GA, Turroni F, Ferrario C, van Sinderen D, et al. Meta-analysis of the human gut microbiome from urbanized and pre-agricultural populations. Environ. Microbiol. 2017;19(4):1379–1390. doi: 10.1111/1462-2920.13692. [DOI] [PubMed] [Google Scholar]
- 84.Jalanka J, Salonen A, Salojärvi J, Ritari J, Immonen O, Marciani L, et al. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64(10):1562–1568. doi: 10.1136/gutjnl-2014-307240. [DOI] [PubMed] [Google Scholar]
- 85.Choo JM, Leong LEX, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015;5(1):16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Durazzi F, Sala C, Castellani G, Manfreda G, Remondini D, De Cesare A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021;11(1):3030. doi: 10.1038/s41598-021-82726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Breitwieser FP, Lu J, Salzberg SL. A review of methods and databases for metagenomic classification and assembly. Briefings Bioinf. 2019;20(4):1125–1136. doi: 10.1093/bib/bbx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.