SUMMARY
Gene silencing for functional studies in plants has been largely facilitated by manipulating viral genomes with inserts from host genes to trigger virus‐induced gene silencing (VIGS) against the corresponding mRNAs. However, viral genomes encode multiple proteins and can disrupt plant homeostasis by interfering with endogenous cell mechanisms. To try to circumvent this functional limitation, we have developed a silencing method based on the minimal autonomously‐infectious nucleic acids currently known: viroids, which lack proven coding capability. The genome of Eggplant latent viroid, an asymptomatic viroid, was manipulated with insertions ranging between 21 and 42 nucleotides. Our results show that, although larger insertions might be tolerated, the maintenance of the secondary structure appears to be critical for viroid genome stability. Remarkably, these modified ELVd molecules are able to induce systemic infection promoting the silencing of target genes in eggplant. Inspired by the design of artificial microRNAs, we have developed a simple and standardized procedure to generate stable insertions into the ELVd genome capable of silencing a specific target gene. Analogously to VIGS, we have termed our approach viroid‐induced gene silencing, and demonstrate that it is a promising tool for dissecting gene functions in eggplant.
Keywords: viroids as biotechnological tools, gene silencing, RNA replicon, Avsunviroidae, regulation of plant gene expression, technical advance
Significance Statement
We developed a standardized viroid‐based procedure (termed VdIGS) for targeted silencing of eggplant genes. Modified ELVd variants (with insertions up to 42 nt) are able to replicate and induce systemic infection in host plants. This innovative approach constitutes the first use of biological forms of viroids as a tool for plant biotechnology.
INTRODUCTION
Gene silencing for functional studies in plants has been largely facilitated by manipulating viral genomes with inserts derived from host genes to trigger virus‐induced gene silencing (VIGS) against the corresponding mRNAs (Abrahamian et al., 2020; Lu et al., 2003; Ramegowda et al., 2014). This technology has been adapted for high‐throughput functional genomics as it by‐passes the labor‐extensive plant transformation (Senthil‐Kumar & Mysore, 2011). Therefore, transient silencing of endogenous plant genes, for basic research or biotechnological applications, has been achieved using a wide variety of plant viruses (Lange et al., 2013). However, viral genomes encode multiple proteins that can disrupt plant homeostasis by interfering with endogenous cell mechanisms (Pallas & García, 2011). Furthermore, viral genomes are thousands of bases in length, and thus can produce viral small RNAs targeting several dozens of plant genes (Annacondia & Martinez, 2021). All of that may limit the functional analysis of a targeted gene in an individualized manner. Additionally, the complexity of plant virus genomes can make it challenging to construct efficient infectious clones that are required for VIGS (Pasin et al., 2019).
To attempt to circumvent these issues, we have developed a silencing method based on viroids (Flores et al., 2004; Owens, 2007). These pathogenic RNAs, naturally found infecting higher plants, are characterized by an extremely reduced genome size (246–401 nt) with null or residual protein‐coding capability (Katsarou et al., 2022; Marquez Molins et al., 2021). Its mature form is a covalently closed, single‐stranded RNA molecule with a highly compact structure that protects them against the RNA‐silencing mediated degradation (Elena et al., 2009; Gómez & Pallás, 2007; Itaya et al., 2007). According to the compartment in which replication takes place, viroids have been classified in two families: Pospiviroidae (nucleus) (Di Serio et al., 2021) and Avsunviroidae (chloroplast) (Di Serio et al., 2018). It has been described that viroids of both families trigger plant RNA silencing, and thus viroid‐derived small RNAs (vd‐sRNAs) are produced in infected plants (Itaya et al., 2001; Martinez et al., 2010; Martinez de Alba et al., 2002; Papaefthimiou et al., 2001; Ramesh et al., 2021). Although the involvement of vd‐sRNAs in the downregulation of host transcripts has been demonstrated (Adkar‐Purushothama et al., 2018; Adkar‐Purushothama & Perreault, 2018; Avina‐Padilla et al., 2015; Bao et al., 2019; Navarro et al., 2012; Navarro, Gisel, et al., 2021; Wang et al., 2011), the use of any of these circular RNA replicons as a biotechnological tool for silencing selected plant genes has not been reported yet.
Viroid infection is frequently associated with evident phenotypic alterations in host plants (Adkar‐Purushothama & Perreault, 2020; Marquez‐Molins et al., 2021; Navarro, Flores, & Di Serio, 2021); however, asymptomatic viroid infections have been also reported as, for example, with eggplant latent viroid (ELVd) on eggplant (Daròs, 2016). This viroid is one of the latest members of the family Avsunviroidae formally characterized (Fadda et al., 2003) and as the other members of this family replicates and accumulates in the chloroplast, although it is also specifically trafficked to the nucleus (Gómez & Pallás, 2012). ELVd sequence folds in a branched quasi‐rod‐like structure with two bifurcations (upper left and upper right hairpins) at both ends (Figure 1a). Interestingly, a mutational analysis of ELVd to inspect RNA processing led to the discovery that an eight‐nucleotide insertion in the terminal loop of the right upper hairpin of ELVd was tolerated and partially maintained in the progeny, while all the other assayed mutations impede the infection of its natural host, eggplant (Martínez et al., 2009).
Figure 1.
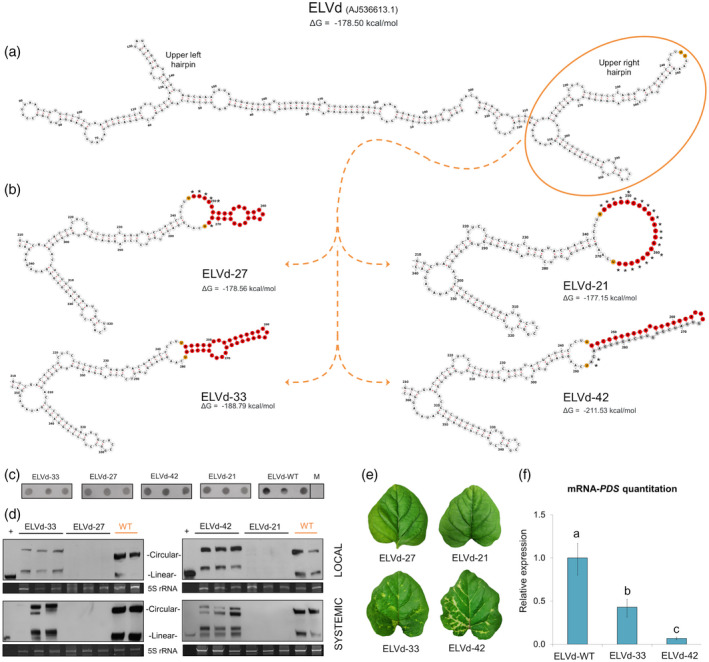
Stability of insertions into the eggplant latent viroid (ELVd) genome and induced silencing of the phytoene desaturase (PDS) gene.
Secondary structures predicted for the complete unmodified ELVd‐WT (accession AJ536613.1) (a) and the partial ELVd region, orange circle, surrounding the artificial 21‐, 27‐, 33‐ and 42‐nucleotide (nt) insertions (b). Secondary structures were obtained with mfold at 25°C and displayed with forna. The minimal free energy of the complete structures is indicated. The nucleotide positions 245 and 246 of ELVd‐WT (between which the artificial sequences were inserted) are depicted in orange. The nucleotides designed to target the PDS are depicted in red. In the case of the 42‐nt insertion, the complementary nucleotides necessary to form a hairpin‐like structure are in gray. The nucleotides of the artificial insertions that enlarge the terminal loop of ELVd‐WT are marked with an asterisk.
(c) Dot blot hybridization of RNA extracted from inoculated cotyledons of three bio‐replicates per construct at 2 days post‐inoculation (dpi). A mock sample (M) was used as a negative control.
(d) Northern blot of total RNA extracted from inoculated (6 dpi) and apical (25 dpi) tissue. The first lane corresponds to monomeric linear transcript of ELVd used as a positive control (+). The hybridization was performed with a DIG‐labeled riboprobe for detecting ELVd plus strand. An image of the acrylamide gel stained with ethidium bromide is shown below as a loading control. Note the presence, in the modified ELVd chimeras, of circular and linear forms larger than the monomeric ELVd‐WT (circular and linear) or control (only linear).
(e) Phenotype of the apical leaves of plants inoculated with the four modified ELVd variants at 25 dpi.
(f) Histogram showing the relative accumulation of PDS transcripts [estimated by reverse transcriptase‐quantitative polymerase chain reaction (RT‐qPCR)] in the first apical leaves of eggplants inoculated with ELVd‐42 and ELVd‐33 in comparison with ELVd‐WT. Different letters indicate significant differences (P < 0.05), estimated by one‐way analysis of variance (anova) and Fisher's Least Significant Difference (LSD). Error bars represent the standard error values. [Colour figure can be viewed at wileyonlinelibrary.com]
Here, we have analyzed how larger insertions into this terminal loop affect ELVd viability and whether these insertions could cause the silencing of a target gene. Our results show that modified ELVd variants (with insertions of up to 42 nucleotides) are able to replicate and induce systemic infection. Additionally, these manipulated ELVd chimeras can efficiently promote the silencing of selected eggplant genes, as evidenced by phenotypic alterations and transcript quantification. Inspired by the design of artificial microRNAs (Carbonell, 2019; Carbonell et al., 2014), we have developed a standardized cloning procedure into the ELVd genome for facilitating the silencing of a target gene. Analogously to VIGS, we have termed our approach viroid‐induced gene silencing (VdIGS), and demonstrated that it is a promising tool for dissecting gene functions in eggplant.
RESULTS
Chimeric ELVd‐variants with artificial insertions are able to establish a systemic infection and silence a plant gene
In order to test whether larger insertions, with the potential to generate small RNAs specific to a target gene, could be relatively stable in the ELVd genome, we designed four constructs (see Table S1 for detailed information) containing complementary insertions to the phytoene desaturase (PDS) gene and a randomly designed structured sequence (Figure 1a). First, two hairpin‐like insertions of 33‐ (ELVd‐33) and 27‐ nucleotides (ELVd‐27) containing complementary and non‐complementary sequences to PDS, respectively, were obtained (Figure 1b, left part). These constructs were assayed by agroinfection in eggplants, and their transcription in the inoculated tissue was confirmed at 2 days post‐inoculation (dpi) by dot blot assays (Figure 1c). At 6 dpi, mature circular forms (indicative of viroid replication) were only detected in the cotyledons inoculated with the modified ELVd‐33 variant (Figure 1d, left). As expected, mature forms of ELVd‐33 variant were longer than unmodified ELVd‐WT (Figure 1d). Noticeably, viroid mature forms corresponding to ELVd‐33 (larger than ELVd‐WT) were also detected in the non‐inoculated apical leaves of two of the three bio‐replicates at 25 dpi (Figure 1d, left), showing that the modified ELVd‐33 variant is able to establish a systemic infection in eggplant maintaining the artificial insertion.
The PDS gene is a recognized indicator gene for VIGS, which encodes an enzyme that catalyzes an important step in the carotenoid biosynthesis pathway, resulting in a characteristic photo‐bleaching when this gene is silenced. Visual inspection of inoculated and control plants at 25 dpi showed the characteristic whitening attributed to the lack of PDS activity in the apical leaves of plants infected with ELVd‐33 (Figure 1e), suggesting that this variant could be able to induce the silencing of this indicator gene. The decrease in the accumulation of PDS transcripts in ELVd‐33 infected plants measured by reverse transcriptase‐quantitative polymerase chain reaction (RT‐qPCR; Figure 1f) confirmed this. Conversely, plants inoculated with ELVd‐27 variant (unable to develop local and systemic infection) did not render the characteristic phenotype of the PDS silenced plants (Figure 1e, upper left).
As explained above, the 33‐nt insert corresponds to a PDS sequence that folds into a hairpin‐like structure. That could limit the versatility of our system, being restricted to transcripts able to form stable hairpins. To circumvent such methodological restriction, we designed a 21‐nt artificial micro‐RNA (amiRNA) specific to target PDS transcripts. Additionally, a 42‐nt sequence (Figure 1b, lower right) consisting of this specific amiRNA and its corresponding star sequence (Figure 1b, in gray) was designed to be folded into a hairpin. Both 21‐ and 42‐nt sequences were inserted in the identical position of the ELVd‐genome (Figure 1, indicated in orange) to generate the ELVd‐21 and ELVd‐42 variants (Figure 1b, upright part). Similarly, these constructs were agroinfiltrated, and the accumulation of ELVd transcripts in the inoculated tissue was confirmed at 2 dpi by dot blot (Figure 1c). Northern blot assays demonstrated that the ELVd‐42 variant was able to infect (locally and systemically) inoculated eggplants (Figure 1d, right), while mature forms corresponding to the ELVd‐21 variant were not detected in any of the analyzed tissues. In contrast to what was observed for ELVd‐33, mature forms corresponding to ELVd‐42 variants (larger than ELVd‐WT forms) were detected in all the bio‐replicates at 25 dpi but not always as the predominant biological form. However, the plants infected with ELVd‐42 (containing an amiRNA/amiRNA* insert) displayed a more evident whitening (Figure 1e) and a more significant reduction in the accumulation level of PDS transcripts measured by RT‐qPCR (Figure 1f).
From the secondary structural predictions, it was evident that the terminal loop (considering ELVd‐WT) of the modified right upper hairpin contains more additional unpaired nucleotides (marked with asterisks in Figure 1b) in ELVd‐21 (21) and ELVd‐27 (6), than in ELVd‐33 (0) and ELVd‐42 (2), strongly suggesting that an enlarged loop may compromise the viability of the modified ELVd variants. To test this possibility, three new 42‐nt insertions that enlarged the terminal loop in one–three nucleotides were assayed (Figure S1). The results show that mature circular forms (indicative of viroid replication) were detected at 6 dpi in plants inoculated with the variants with one (ELVd‐42 A285U) and two (ELVd‐42 A284U/A285U) extra‐unpaired nucleotides, but not for the variant with three additional unpaired nucleotides in the loop (ELVd‐42 nt G283U/A284U/A285U). According to these results, we inferred that ELVd chimeras with artificial 42‐nt insertions that represent the 12.6% of the ELVd genome (333 nt of ELVd‐WT) maintaining a terminal stem‐loop structure similar (with no more than two extra nucleotides) to that of unmodified ELVd are able to induce systemic infection in eggplant and can trigger the silencing of a target plant gene.
Dynamics of the PDS silencing mediated by the infection with chimeric ELVd
To provide further insights about the stability of the modified ELVd chimeras and characterize the progression of the PDS silencing, we infected plants with the ELVd‐42 chimera in which half of the insertion is designed for targeting the PDS (as an amiRNA) and the other half for structural compensation (star sequence). At 25 dpi, all plants inoculated with ELVd‐42 displayed whitening in systemic leaves, in contrast to the asymptomatic mock plants infected with the wild‐type (WT; Figure 2a). Therefore, we analyzed the apical leaves at 23, 25, 27 and 29 dpi. Northern blot analysis demonstrated that circular and linear forms associated to ELVd‐42 (375 nt) can be detected in apical leaves of infected plants at the four tested time‐points (23, 25, 27 and 29 dpi) (Figure 2b). Mature forms corresponding to the reversion to ELVd‐WT (333 nt) were also observed, which was confirmed by sequencing (Figure S2). Circular and linear forms associated to ELVd‐42 were predominant at 23 and 25 dpi. However, the apparent increasing accumulation of deletion derivatives of ELVd‐42 indicate that the viroid reverts to WT over time. Quantification of transcripts by RT‐qPCR showed a significant reduction of PDS mRNAs in infected plants at all the four time‐points (Figure 2c), showing that in this time‐range the efficiency of the ELVd‐mediated silencing of PDS was a stable and comparable phenomenon.
Figure 2.
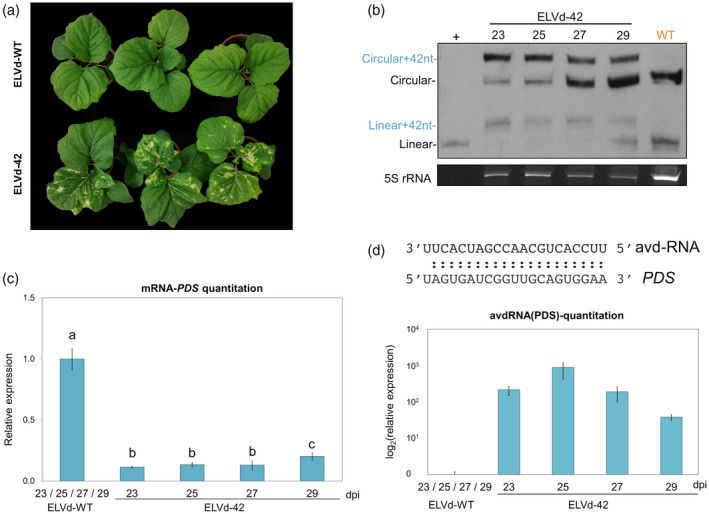
Progression of the silencing of the phytoene desaturase (PDS) induced by an artificial viroid‐derived sRNA.
(a) Phenotype of representative plants inoculated with ELVd‐WT (upper) and ELVd‐42 (lower) at 25 days post‐inoculation (dpi).
(b) Northern blot of RNA extracted from apical leaves of three plants inoculated with ELVd‐42 at 23, 25, 27 and 29 dpi, and an RNA mix of three plants infected with ELVd‐WT at these same time‐points. The first lane corresponds to monomeric linear transcript of ELVd used as a positive control (+). Circular and linear forms are marked in blue (ELVd‐42) and in black (ELVd‐WT). The hybridization was performed with a DIG‐labeled riboprobe for detecting ELVd plus strand. An image of the acrylamide gel stained with ethidium bromide is shown below as a loading control.
(c) Histogram showing the means of the relative accumulation of PDS transcripts [estimated by reverse transcriptase‐quantitative polymerase chain reaction (RT‐qPCR)] in the first apical leaves of three eggplants inoculated with ELVd‐42 at 23, 25, 27 and 29 dpi. The mean of the values obtained from three plants inoculated with WT‐ELVd at comparable dpi was used as reference value.
(d) Graphic representing the quantitation of the artificial viroid‐derived small RNA (avd‐RNA) designed to silence the PDS mRNA in the infected plants by stem‐loop RT‐qPCR. The sequence of the avd‐RNA and the target region in the mRNA of PDS are indicated above. Different letters indicate significant differences (P < 0.05), estimated by one‐way analysis of variance (anova) and Fisher's Least Significant Difference (LSD). Error bars represent the standard error values. [Colour figure can be viewed at wileyonlinelibrary.com]
To analyze up to what extent the chimeric ELVd‐42 persisted in the infected plants, despite its obvious lower fitness in comparison with the WT, we sampled 10 plants at 60 dpi (Figure S3). Although ELVd‐WT was the predominant form, the chimeric ELVd‐42 was detected in 8 out of 10 infected plants (Figure S3a). Furthermore, whitening in leaves was still observed in 9 out of 10 plants (Figure S3b). Overall, these results demonstrate that the chimeric ELVd‐42 is relatively stable in infected plants for a long time period (at least 60 days), being also able to induce the characteristic leaf whitening associated to PDS‐silencing.
In order to efficiently target the eggplant PDS gene, the variant ELVd‐42 contains a sRNA of 21 nt designed as an amiRNA to cleave the PDS mRNA (see “Experimental Procedures” section for details), and thus we denominated it artificial viroid‐derived small RNA (avd‐sRNA). To validate that this avd‐sRNA designed to target the PDS was indeed being produced, a stem‐loop RT‐qPCR was used to quantify its accumulation in the four time‐points mentioned above (Figure 2d). As expected, this avd‐sRNA was not detected in control samples inoculated with ELVd‐WT. In coincidence with that observed for the ELVd‐42 mature forms, the levels of avd‐sRNA peaked at 25 dpi and subsequently decreased at 27 and 29 dpi (Figure 2d).
To determine if the observed phenotypic effects (bleaching) induced by the modified ELVd‐variants could be caused by the vector carrying a control insert, we designed the chimeric ELVd‐GFP construct containing a 21‐nt sequence derived from the GFP transcript and a complementary sequence for structural compensation (Figure S4; Table S2). Mature forms of the viroid containing the GFP fragment were detected in apical leaves of the six plants inoculated with ELVd‐GFP at 25 dpi (Figure S4). This result showed that the ELVd‐GFP was able to establish a systemic infection. However, in contrast to plants inoculated with ELVd‐42, infection with the ELVd‐GFP variant was not associated with bleaching observed in PDS‐silenced plants (Figure S4).
A standardized procedure for VdIGS
We have designed a standardized procedure for VdIGS in which a 21‐nt sequence (designed to silence a specific target gene) is structurally compensated by an additional 21 nt to form a stable hairpin (Figure 3). The design of the 21‐nt sequence specific for silencing the selected target gene is based on that of amiRNAs (Carbonell, 2019; Carbonell et al., 2014). Specifically, the software P‐SAMS was used to design a 21‐nt amiRNA, and the corresponding star sequence (amiRNA*) provided by the P‐SAMS is used to form the hairpin (Fahlgren et al., 2016). For cloning into the ELVd genome the resultant 42‐nt (amiRNA‐amiRNA*) sequence, self‐hybridizing DNA oligonucleotides are designed with CCTT and TTGA overhangs in order to produce a seamless ligation into the ELVd genome between positions 245 and 246. Therefore, the hybridized oligonucleotides can be directly ligated using T4 DNA ligase in the correct orientation into a plasmid harboring the ELVd genome cDNA, which is further dimerized into a binary vector to obtain the infectious clone vector as previously described (Marquez‐Molins et al., 2019; Prol et al., 2021).
Figure 3.
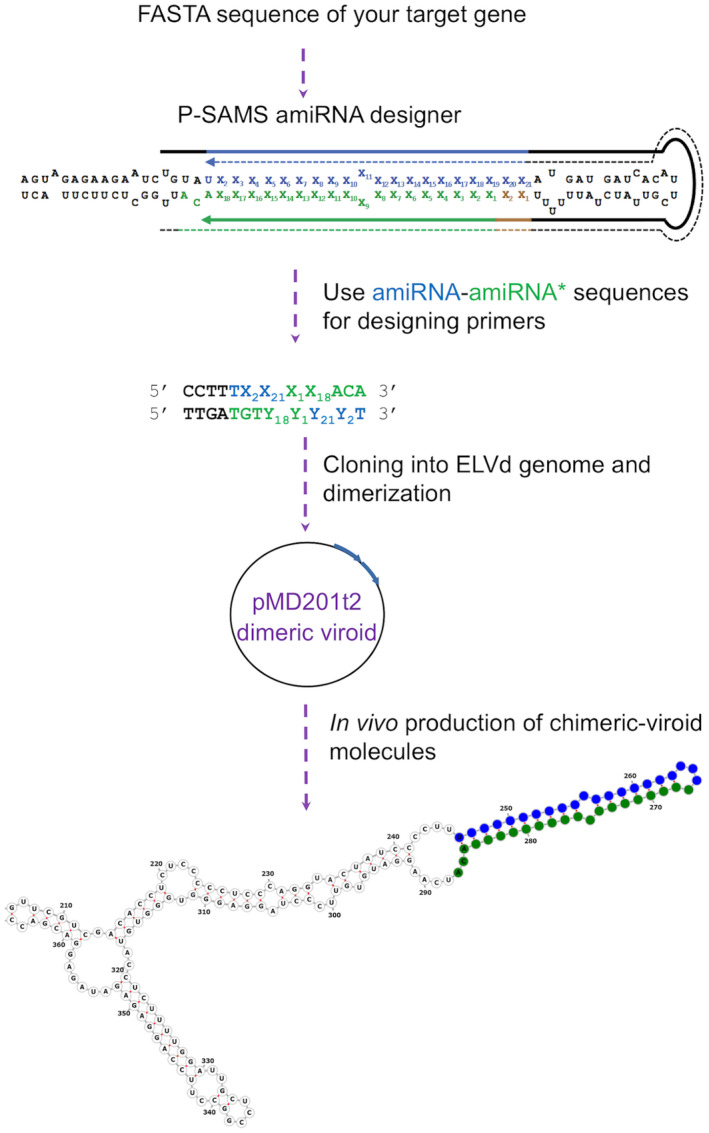
Workflow for generating viroid‐induced gene silencing (VdIGS) constructs.
To design a construct for targeting a desired gene, its sequence is introduced in the P‐SAMS website for artificial microRNA (amiRNA) design. A pair consisting of an amiRNA (blue) and its star sequence (amiRNA*, in green) should be chosen. Xn indicates any nucleotide and Yn the complementary nucleotide to Xn. Subsequently, oligonucleotides with these sequences plus four nucleotides of the viroid (in black) are designed as represented. These oligonucleotides are self‐hybridized and directly cloned into a plasmid harboring the ELVd genome, in which the ccdB gene is excised by BsmbI digestion. Once this construct is obtained, a dimer is generated into the binary vector pMD201t2 as previously reported (Marquez‐Molins et al., 2019). This plasmid is transformed into agrobacterium and the VdIGS RNAs are generated in vivo. [Colour figure can be viewed at wileyonlinelibrary.com]
To evaluate whether, irrespectively of the transcript sequence, the developed strategy can produce stable chimeric‐ELVd variants able to establish a systemic infection and induce the silencing of a target gene, we generated two constructs (ELVd‐CHLI‐a and ELVd‐CHLI‐b) targeting another eggplant indicator gene, the magnesium‐chelatase subunit ChlI. The two constructs target two different regions of the open reading frame of this gene (SMEL_004g204170.1.01) located at positions 114–134 (ELVd‐CHLI‐a) and 896–916 (ELVd‐CHLI‐b). These constructs were designed as described in Figure 3, and the insertions folded into the expected hairpins (Figure 4a). The stability of the ELVd‐CHLI‐a and ELVd‐CHLI‐b variants was analyzed by Northern blot assay at 25 dpi (Figure 4b). ELVd‐CHLI‐b construct seemed to be more stable than ELVd‐CHLI‐a. Consistently, although in both cases the characteristic yellowing produced by the ChlI silencing was observed, ELVd‐CHLI‐b caused a greater phenotypic effect (Figure 4c). The silencing of this gene was corroborated in both cases by a significant decrease in the expression of this gene as measured by RT‐qPCR (Figure 4d). Therefore, these results support that VdIGS is an effective technology for silencing a preselected eggplant gene.
Figure 4.
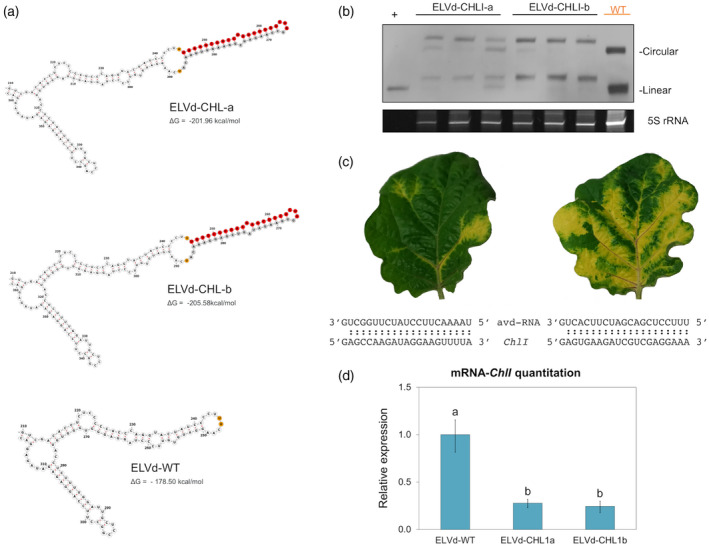
Assessment of the applicability of the viroid‐induced gene silencing (VdIGS) procedure for silencing an additional eggplant gene.
(a) Secondary structure predictions of the two ELVd chimeras (ELVd‐CHLI‐a and ELVd‐CHLI‐b) containing artificial insertions targeting two specific regions of the magnesium‐chelatase subunit (ChlI) mRNA. The corresponding region in the unmodified ELVd‐WT is also shown. The structure predictions were generated with mfold at 25°C, displayed with Forna. Only the ELVd region surrounding the artificial insertions is shown. The minimal free energy of each complete structure is indicated below. The sequence of the two artificial viroid‐sRNAs (avd‐sRNA) is depicted in red while the compensatory sequence is in gray. The nucleotide positions 245 and 246 of ELVd‐WT (between which the artificial sequences were inserted) are depicted in orange.
(b) Northern blot of total RNA extracted from apical leaves of plants inoculated with the ELVd‐CHLI‐a, ELVd‐CHLI‐b and ELVd‐WT at 25 days post‐inoculation (dpi). The first lane corresponds to monomeric linear transcript of ELVd used as a positive control (+). Note the presence, in the plants inoculated with the modified variants, of circular and linear forms larger than the monomeric ELVd‐WT (circular and linear) or control (only linear). The hybridization was performed with a DIG‐labeled riboprobe for detecting ELVd plus strand. An image of the acrylamide gel stained with ethidium bromide is shown below as a loading control.
(c) Phenotype of representative apical leaves of plants inoculated with ELVd‐CHLI‐a (left) and ELVd‐CHLI‐b (right) at 25 dpi. The sequence of the two designed avd‐RNAs and their predicted target region in the Chll mRNA are indicated below.
(d) Histogram showing the relative accumulation of Chll transcripts in apical leaves of plants inoculated with ELVd‐CHLI‐a and ELVd‐CHLI‐b at 25 dpi measured by reverse transcriptase‐quantitative polymerase chain reaction (RT‐qPCR). Different letters indicate significant differences (P < 0.05), estimated by one‐way analysis of variance (anova) and Fisher's Least Significant Difference (LSD). Error bars represent the standard error values. [Colour figure can be viewed at wileyonlinelibrary.com]
DISCUSSION
The possibility to silence endogenous genes in plants has revolutionized plant biology. Currently one of the most popular procedures to study the functions of host genes in plants relies on the virus‐induced gene silencing methodology. The virus amplification capacity allows the exponential expression of partial sequences of the host gene of interest. However, the requirement of various proteins for this amplification (replicase proteins) and its movement through the plant (movement proteins) can mask or interfere with the desired effects. These undesirable interferences could be avoided with the use of replicative nucleic acids without coding capacity. Viral satellite DNA associated with tomato yellow leaf curl China virus (TYLCCNV) has been engineered to function as an apparent non‐protein‐coding silencing vector allowing the transcriptional and/or posttranscriptional regulation of diverse genes in nicotiana, tomato and petunia (Ju et al., 2016, 2017; Tao et al., 2006; Tao & Zhou, 2004). Although a satellite DNA has the advantage of allowing the stable insertion of bigger sequences (170–521 nt), it has the important disadvantage of being a non‐autonomous molecule. Therefore, satellites require a helper virus to replicate and accumulate in the plant.
In this work, we have used diverse artificial insertions into an asymptomatic autonomously replicating viroid (ELVd) to study the viability and stability of the modified genomes to establish a systemic infection and the potential use of these chimeric ELVd to promote the silencing of a host gene of choice. We found that modified ELVd genomes with a 42‐nt insertion folded into a hairpin are relatively stable, being the predominant form in non‐inoculated tissues at least up to 25 dpi. Moreover, our results show that artificial sRNAs derived from modified variants (avd‐sRNAs) can efficiently induce the silencing of previously selected eggplant genes. This technique named as VdIGS combines the target specificity of 21‐nt small RNAs and the systemic movement of an asymptomatic viroid, being an innovative tool for plant biotechnology. The specific gene silencing induced by chimeric ELVd variants exhibits its maximum level between 23 and 29 dpi, but the phenotypic alteration can endure up to 60 dpi, providing a presumably sufficient time‐range for the functional analysis of the silenced genes. However, the utility of VdIGS at later time‐points (60 dpi) would be subject to a lot more variability (likely due to the increasing reversion to WT forms).
The VdIGS can be primarily used for studying gene function in eggplant, which is among the top five most important vegetables in the Mediterranean and Asia (Taher et al., 2017) and its genome has been recently re‐sequenced (Barchi et al., 2019; Gramazio et al., 2019), demonstrating that it is a well‐studied crop with on‐going attention. Another advantage is the cloning procedure that does not require the amplification of the target gene as in VIGS (typical insert size is 300 nt) because oligonucleotides (46 nt in length) are self‐hybridized and cloned into the ELVd genome.
It is worth noting that significant silencing of PDS and Chll transcripts is not always accompanied by homogenous phenotypic alterations in systemically infected eggplant leaves (Figures 2 and 4). This observation is consistent with previous results showing that VIGS by short PDS‐insertions (below 120 nt) triggered minimum or no bleaching phenotype in plants infected with modified brome mosaic virus (Wang et al., 2021), or barley stripe mosaic virus (Bruun‐Rasmussen et al., 2007), although measurements of the respective transcripts levels showed significant reductions. In one of these cases (Bruun‐Rasmussen et al., 2007), it was suggested that to trigger a visible bleaching phenotype PDS transcripts levels should be lower than a specific threshold at a certain point in leaf development. However, the observation that plants infected with ELVd‐42 had an approximately 90% reduction in PDS mRNA accumulation and exhibited irregular bleaching (Figure 2) indicates that this threshold was reached. To better understand the patchiness, it may be interesting to test for the presence of the insert and the target gene silencing in different portions of leaves showing patches of visible silencing to help explain these phenotypic inconsistencies (Wang et al., 2021).
Additionally, our results might have implications for understanding viroid biology and evolution. We show that an enlarged loop abolished the accumulation of mature forms and only hairpin structures were stable. That observation explains why inserts with roughly the same size (40 nt) but with 16 nt folding into a loop failed to infect eggplant (Daròs et al., 2018). Moreover, chimeric ELVd reverted to the WT over time, but there was an apparent absence of intermediary forms, suggesting that the maintenance of a compact secondary structure should be essential for this viroid. The incorporation of novel sequences into the viroid genome could be thus constrained by important structural limitations, and the insertions must not impede the transcription, circularization and movement of these RNA replicons.
Regarding the fundamental aspect of viroid–host interactions, our results reinforce and expand previous findings (using Peach latent mosaic viroid as a model) showing that the small RNAs derived from chloroplast‐replicating viroids can be functional and promote the silencing of endogenous host genes (Delgado et al., 2019; Navarro et al., 2012).
In conclusion, here we have developed a simple, fast, cost‐effective and standardized procedure based on the use of minimal replicating systemic RNAs able to generate tailored sRNAs for efficient silencing of predetermined eggplant genes. This is a conceptual novelty that further expands the biotechnological applications of viroids in general, and ELVd in particular (Daròs et al., 2018; Ortolá et al., 2021). In the future, it will be very interesting to analyze the potential to transfer this technological approach to other viroids with broader host ranges, such as hop stunt viroid (HSVd; Marquez‐Molins et al., 2021), to expand the utility of VdIGS to other economically important crops.
EXPERIMENTAL PROCEDURES
Molecular cloning
The lethal gene ccdB flanked by BsmBI sites was inserted into positions 245 and 246 of Eggplant latent viroid accession AJ536613.1 and cloned into a pBluescript SK plasmid using high‐fidelity PrimeSTAR HS DNA Polymerase (Takara, Kusatsu, Japan). This plasmid linearized with BsmBI was used to ligate self‐hybridized oligonucleotides into positions 245 and 246 of ELVd. The oligonucleotides were self‐hybridized by denaturing for 5 min at 95°C and cooling down (0.05°C sec−1) to 25°C and then ligated using T4 DNA ligase (Thermo Scientific™, Waltham, MA, USA). The resulting plasmids were used as a template for generating the dimeric viroid sequences into a binary vector as has been previously described (Marquez‐Molins et al., 2019). The oligonucleotides used to generate each insert are listed in Table S1.
RNA secondary structure predictions
ELVd (AJ536613.1) secondary structure of the plus polarity had been previously elucidated and the in vivo data obtained by high‐throughput selective 2′‐hydroxyl acylation analyzed by primer extension (hSHAPE) support the prediction of Mfold software (López‐Carrasco et al., 2016). For our analysis, the minimal free energy was calculated considering that these are circular molecules and a temperature of 25°C using Mfold (Zuker, 2003). RNA secondary structures files were generated in Vienna format and were displayed using forna (Kerpedjiev et al., 2015).
Viroid inoculation and sample collection
Plants at the cotyledon stage (2 weeks after sowing) of Solanum melongena cv. Black beauty were inoculated by agroinfiltration with a culture of Agrobacterium tumefaciens strain C58 harboring the pMD201t2 binary vector with the correspondent ELVd‐modified construct (Table S2). The overnight‐grown bacterial culture was diluted in infiltration buffer (MES 0.1 m, MgCl2 1 m) up to an optical density at 600 nm of 1 and infiltrated on the abaxial side of one cotyledon using a needle‐less syringe. Plants were kept in a photoperiod of 16 h under visible light and 25°C (light)/22°C (darkness). Plants inoculated with an unmodified ELVd‐construct (WT) were used as controls.
To analyze viroid processing in local tissue, agroinfiltrated cotyledons were collected at 2 or 6 dpi. Three biological replicates were performed for each construct. Each analyzed replicate corresponds to a pool of six cotyledons recovered from three inoculated plants.
Systemically‐infected tissue was collected from the first expanded leaf below the apex of agroinfiltrated plants and analyzed individually. The number of biological replicates for each assay and the dpi of those samples were collected and are detailed specifically for each experiment in the Results section.
RNA extraction and Northern/dot blot
Total RNA was extracted from systemic leaves using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For Northern blot analysis, 3 μg of total RNA per sample was mixed with 1 volume of formamide and loading buffer 6 ×, denatured for 10 min at 95°C and then loaded into a polyacrylamide gel electrophoresis 5% urea 8 m and TBE 89 mm gel. RNA electrophoresis was performed at 200 V for 1 h, and then RNA was transferred to a nylon membrane using a MiniProtean 3 system (BioRad, Hercules, CA, USA). Transfer conditions were 100 V for 1 h at 4°C in TBE buffer 1 ×. Nucleic acids transferred to the membrane were covalently fixed using ultraviolet light (700 × 100 J cm−2). Dot‐blot samples were directly applied onto the membrane. Hybridization and chemiluminescent detection were performed as previously described (Herranz et al., 2005) using a DIG‐labeled probe to detect the plus polarity strand of ELVd.
Small RNA purification and stem‐loop RT
Low‐molecular weight RNA (< 200 nt) were isolated from total RNA using REALTOTAL microRNA Kit RBMER14 (Durviz, Paterna, Valencia, Spain) according to the manufacturer's instructions. Stem‐loop‐specific reverse transcription for small RNA detection was performed as previously described (Bustamante et al., 2018).
RT‐PCR and qPCR
First‐strand cDNA was synthesized by reverse transcription using RevertAid cDNA Synthesis Kit (Thermo Scientific™). ELVd molecules were amplified with the specific primers Fw ELVd and Rv ELVd (Table S1), cloned into pJET 1.2 (Thermo Scientific) and sequenced by Sanger at Eurofins Genomics using Mix2Seq. RT‐qPCR assays were performed using PyroTaq EvaGreen mix Plus (ROX; CulteK Molecular Bioline, Madrid, Spain) according to the manufacturer's instructions. All analyses were performed in triplicate on an ABI 7500 Fast‐Real Time qPCR instrument (Applied Biosystems, Waltham, MA, USA) as previously described (Sanz‐Carbonell et al., 2020). The efficiency of PCR amplification was derived from a standard curve generated by four 10‐fold serial dilution points of cDNA obtained from a mix of all the samples. The sequences of all primers are listed in Table S1. Relative RNA accumulation was determined by using the comparative ΔΔCT method (Livak & Schmittgen, 2001) and normalized to the geometric mean of cyclophilin (SMEL_001g116150.1.01) and F‐Box (SMEL_002g166030.1.01), as references for mRNA, whereas the small nucleolar U6 RNA (SMEL3Ch05) was used for normalizing sRNA‐quantitation. The statistical analysis was performed with Statgraphics (https://www.statgraphics.com/). First, replicate populations were verified to have normal variances. Next one‐way analysis of variance (anova) and Fisher's Least Significant Difference (LSD) were used to obtain the significance.
AUTHOR CONTRIBUTIONS
JM‐M designed the strategy. JM‐M, AGH‐A and MU‐P performed the experiments. JM‐M, VP and GG analyzed and discussed the results. JM‐M and GG conceived the project and wrote the main manuscript text. All authors read, revised and approved the final manuscript.
CONFLICT OF INTEREST
The author(s) declare no competing interests.
Supporting information
Figure S1. Enlarged loops affect the viability of chimeric ELVd variants.
Figure S2. Modified ELVd‐42 reverts to the original unmodified form.
Figure S3. Analysis of ELVd‐42 60 days after inoculation.
Figure S4. The stuffer control ELVd‐GFP with a 42‐nucleotide insertion does not produce phenotypic alterations.
Table S1. Detail of the primers used in this study
Table S2. Detail of the sequences inserted in the constructs used in this work
ACKNOWLEDGEMENTS
This work was supported by the Spanish Agencia Estatal de Investigacion (co‐supported by FEDER) Grants PID2019‐104126RB‐I00 (GG) and PID2020‐115571RB‐I00 (VP). JM‐M and AGH‐A were recipients of a pre‐doctoral contract from the Generalitat Valenciana (ACIF‐2017‐114 and ACIF‐2021‐202). The funders had no role in the experiment design, data analysis, decision to publish, or preparation of the manuscript. The authors want to acknowledge the valuable comments related to new control experiments made by two anonymous reviewers.
Contributor Information
Joan Marquez‐Molins, Email: j.molins@csic.es.
Gustavo Gomez, Email: gustavo.gomez@csic.es.
DATA AVAILABILITY STATEMENT
All the biological and chemical materials not commercially available that are used for the experiments reported here are available from gustavo.gomez@csic.es upon reasonable request.
REFERENCES
- Abrahamian, P. , Hammond, R.W. & Hammond, J. (2020) Plant virus‐derived vectors: applications in agricultural and medical biotechnology. Annual Review of Virology, 7, 513–535. 10.1146/annurev-virology-010720-054958 [DOI] [PubMed] [Google Scholar]
- Adkar‐Purushothama, C.R. & Perreault, J.‐P. (2018) Alterations of the viroid regions that interact with the host defense genes attenuate viroid infection in host plant. RNA Biology, 15, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkar‐Purushothama, C.R. & Perreault, J.P. (2020) Current overview on viroid–host interactions. Wiley Interdisciplinary Reviews: RNA, 11, e1570. [DOI] [PubMed] [Google Scholar]
- Adkar‐Purushothama, C.R. , Sano, T. & Perreault, J.‐P. (2018) Viroid‐derived small RNA induces early flowering in tomato plants by RNA silencing. Molecular Plant Pathology, 19, 2446–2458. 10.1111/mpp.12721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annacondia, M.L. & Martinez, G. (2021) Reprogramming of RNA silencing triggered by cucumber mosaic virus infection in Arabidopsis. Genome Biology, 22, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avina‐Padilla, K. , Martinez de la Vega, O. , Rivera‐Bustamante, R. , Martinez‐Soriano, J.P. , Owens, R.A. , Hammond, R.W. et al. (2015) In silico prediction and validation of potential gene targets for pospiviroid‐derived small RNAs during tomato infection. Gene, 564, 197–205. [DOI] [PubMed] [Google Scholar]
- Bao, S. , Owens, R.A. , Sun, Q. , Song, H. , Liu, Y. , Eamens, A.L. et al. (2019) Silencing of transcription factor encoding gene StTCP23 by small RNAs derived from the virulence modulating region of potato spindle tuber viroid is associated with symptom development in potato. PLoS Pathogens, 15, e1008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi, L. , Pietrella, M. , Venturini, L. , Minio, A. , Toppino, L. , Acquadro, A. et al. (2019) A chromosome‐anchored eggplant genome sequence reveals key events in Solanaceae evolution. Scientific Reports, 9, 11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun‐Rasmussen, M. , Madsen, C.T. , Jessing, S. & Albrechtsen, M. (2007) Stability of barley stripe mosaic virus–induced gene silencing in barley. Molecular Plant–Microbe Interactions, 20, 1323–1331. 10.1094/MPMI-20-11-1323 [DOI] [PubMed] [Google Scholar]
- Bustamante, A. , Marques, M.C. , Sanz‐Carbonell, A. , Mulet, J.M. & Gomez, G. (2018) Alternative processing of its precursor is related to miR319 decreasing in melon plants exposed to cold. Scientific Reports, 8, 15538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell, A. (2019) Design and high‐throughput generation of artificial small RNA constructs for plants. Methods in Molecular Biology, 1932, 247–260. [DOI] [PubMed] [Google Scholar]
- Carbonell, A. , Takeda, A. , Fahlgren, N. , Johnson, S.C. , Cuperus, J.T. & Carrington, J.C. (2014) New generation of artificial MicroRNA and synthetic trans‐acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiology, 165, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs, J.A. (2016) Eggplant latent viroid: a friendly experimental system in the family Avsunviroidae. Molecular Plant Pathology, 17, 1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs, J.A. , Aragonés, V. & Cordero, T. (2018) A viroid‐derived system to produce large amounts of recombinant RNA in Escherichia coli . Scientific Reports, 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado, S. , Navarro, B. , Serra, P. , Gentit, P. , Cambra, M.Á. , Chiumenti, M. et al. (2019) How sequence variants of a plastid‐replicating viroid with one single nucleotide change initiate disease in its natural host. RNA Biology, 16, 906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Serio, F. , Li, S.F. , Matoušek, J. et al. (2018) ICTV virus taxonomy profile: Avsunviroidae. The Journal of General Virology, 99, 611–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Serio, F. , Owens, R.A. , Li, S.F. et al. (2021) ICTV virus taxonomy profile: Pospiviroidae. The Journal of General Virology, 102, 1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S.F. , Gómez, G. & Daròs, J.A. (2009) Evolutionary constraints to viroid evolution. Viruses, 1, 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda, Z. , Daròs, J.A. , Fagoaga, C. , Flores, R. & Duran‐Vila, N. (2003) Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). Journal of Virology, 77, 6528–6532. http://jvi.asm.org/content/77/11/6528.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N. , Hill, S.T. , Carrington, J.C. & Carbonell, A. (2016) P‐SAMS: a web site for plant artificial microRNA and synthetic trans‐acting small interfering RNA design. Bioinformatics, 32, 157–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R. , Delgado, S. , Gas, M.E. , Carbonell, A. , Molina, D. , Gago, S. et al. (2004) Viroids: the minimal non‐coding RNAs with autonomous replication. FEBS Letters, 567, 42–48. [DOI] [PubMed] [Google Scholar]
- Gómez, G. & Pallás, V. (2007) A peptide derived from a single‐modified viroid‐RNA can be used as an “in vivo” nucleolar marker. Journal of Virological Methods, 144, 169–171. [DOI] [PubMed] [Google Scholar]
- Gómez, G. & Pallás, V. (2012) Studies on subcellular compartmentalization of plant pathogenic noncoding RNAs give new insights into the intracellular RNA‐traffic mechanisms. Plant Physiology, 159, 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramazio, P. , Yan, H. , Hasing, T. , Vilanova, S. , Prohens, J. & Bombarely, A. (2019) Whole‐genome resequencing of seven eggplant (Solanum melongena) and one wild relative (S. incanum) accessions provides new insights and breeding tools for eggplant enhancement. Frontiers in Plant Science, 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz, M.C. , Sanchez‐Navarro, J.A. , Aparicio, F. & Pallás, V. (2005) Simultaneous detection of six stone fruit viruses by non‐isotopic molecular hybridization using a unique riboprobe or polyprobe. Journal of Virological Methods, 124, 49–55. [DOI] [PubMed] [Google Scholar]
- Itaya, A. , Folimonov, A. , Matsuda, Y. , Nelson, R.S. & Ding, B. (2001) Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Molecular Plant‐Microbe Interactions, 14, 1332–1334. [DOI] [PubMed] [Google Scholar]
- Itaya, A. , Zhong, X. , Bundschuh, R. , Qi, Y. , Wang, Y. , Takeda, R. et al. (2007) A structured viroid RNA serves as a substrate for dicer‐like cleavage to produce biologically active small RNAs but is resistant to RNA‐induced silencing complex‐mediated degradation. Journal of Virology, 81, 2980–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, Z. , Wang, L. , Cao, D. , Zuo, J. , Zhu, H. , Fu, D. et al. (2016) A viral satellite DNA vector‐induced transcriptional gene silencing via DNA methylation of gene promoter in Nicotiana benthamiana . Virus Research, 223, 99–107. [DOI] [PubMed] [Google Scholar]
- Ju, Z. , Cao, D. , Gao, C. , Zuo, J. , Zhai, B. , Li, S. et al. (2017) A viral satellite DNA vector (TYLCCNV) for functional analysis of miRNAs and siRNAs in plants. Plant Physiology, 173, 1940–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsarou, K. , Adkar‐Purushothama, C.R. , Tassios, E. , Samiotaki, M. , Andronis, C. , Lisón, P. et al. (2022) Revisiting the non‐coding nature of Pospiviroids. Cells, 11, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev, P. , Hammer, S. & Hofacker, I.L. (2015) Forna (force‐directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics, 31, 3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, M. , Yellina, A.L. , Orashakova, S. & Becker, A. (2013) Virus‐induced gene silencing (VIGS) in plants: an overview of target species and the virus‐derived vector systems. Methods in Molecular Biology, 975, 1–14. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. & Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- López‐Carrasco, A. , Gago‐Zachert, S. , Mileti, G. , Minoia, S. , Flores, R. & Delgado, S. (2016) The transcription initiation sites of eggplant latent viroid strands map within distinct motifs in their in vivo RNA conformations. RNA Biology, 13, 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Martin‐Hernandez, A.M. , Peart, J.R. , Malcuit, I. & Baulcombe, D.C. (2003) Virus‐induced gene silencing in plants. Methods, 30, 296–303. [DOI] [PubMed] [Google Scholar]
- Marquez Molins, J. , Navarro, J.A. , Seco, L.C. , Pallas, V. & Gomez, G. (2021) Might exogenous circular RNAs act as protein‐coding transcripts in plants? RNA Biology, 18, 98–107. 10.1080/15476286.2021.1962670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez‐Molins, J. , Navarro, J.A. , Pallas, V. & Gomez, G. (2019) Highly efficient construction of infectious viroid‐derived clones. Plant Methods, 15, 87. 10.1186/s13007-019-0470-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez‐Molins, J. , Gomez, G. & Pallas, V. (2021) Hop stunt viroid: a polyphagous pathogenic RNA that has shed light on viroid–host interactions. Molecular Plant Pathology, 22, 153–162. 10.1111/mpp.13022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Alba, A.E. , Flores, R. & Hernandez, C. (2002) Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. Journal of Virology, 76, 13094–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, F. , Marqués, J. , Salvador, M.L. & Darós, J.A. (2009) Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. The Journal of General Virology, 90, 3057–3065. [DOI] [PubMed] [Google Scholar]
- Martinez, G. , Donaire, L. , Llave, C. , Pallas, V. & Gomez, G. (2010) High‐throughput sequencing of hop stunt viroid‐derived small RNAs from cucumber leaves and phloem. Molecular Plant Pathology, 11, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, B. , Gisel, A. , Rodio, M.E. , Delgado, S. , Flores, R. & Di Serio, F. (2012) Small RNAs containing the pathogenic determinant of a chloroplast‐replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant Journal, 70, 991–1003. 10.1111/j.1365-313X.2012.04940.x [DOI] [PubMed] [Google Scholar]
- Navarro, B. , Flores, R. & Di Serio, F. (2021) Advances in viroid‐host interactions. Annual Reviews in Virology, 8, 305–325. 10.1146/annurev-virology-091919-092331 [DOI] [PubMed] [Google Scholar]
- Navarro, B. , Gisel, A. , Serra, P. , Chiumenti, M. & Di Serio, F. (2021) Degradome analysis of tomato and Nicotiana benthamiana plants infected with potato spindle tuber viroid. International Journal of Molecular Sciences, 22, 3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolá, B. , Cordero, T. , Hu, X. & Daròs, J.‐A. (2021) Intron‐assisted, viroid‐based production of insecticidal circular double‐stranded RNA in Escherichia coli . RNA Biology, 18, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, R.A. (2007) Potato spindle tuber viroid: the simplicity paradox resolved? Molecular Plant Pathology, 8, 549–560. [DOI] [PubMed] [Google Scholar]
- Pallas, V. & García, J.A. (2011) How do plant viruses induce disease? Interactions and interference with host components. The Journal of General Virology, 92, 2691–2705. [DOI] [PubMed] [Google Scholar]
- Papaefthimiou, I. , Hamilton, A. , Denti, M. , Baulcombe, D. , Tsagris, M. & Tabler, M. (2001) Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post‐transcriptional gene silencing. Nucleic Acids Research, 29, 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasin, F. , Menzel, W. & Daròs, J.A. (2019) Harnessed viruses in the age of metagenomics and synthetic biology: an update on infectious clone assembly and biotechnologies of plant viruses. Plant Biotechnology Journal, 17, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prol, F.V. , Márquez‐Molins, J. , Rodrigo, I. , López‐Gresa, M.P. , Bellés, J.M. , Gómez, G. et al. (2021) Symptom severity, infection progression and plant responses in Solanum plants caused by three pospiviroids vary with the inoculation procedure. International Journal of Molecular Sciences, 22, 6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramegowda, V. , Mysore, K.S. & Senthil‐Kumar, M. (2014) Virus‐induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic‐stress‐responsive genes in crop plants. Frontiers in Plant Science, 5, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh, S.V. , Yogindran, S. , Gnanasekaran, P. , Chakraborty, S. , Winter, S. & Pappu, H.R. (2021) Virus and viroid‐derived small RNAs as modulators of host gene expression: molecular insights into pathogenesis. Frontiers in Microbiology, 11, 3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz‐Carbonell, A. , Marques, M.C. , Martinez, G. & Gomez, G. (2020) Dynamic architecture and regulatory implications of the miRNA network underlying the response to stress in melon. RNA Biology, 17, 292–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. & Mysore, K.S. (2011) New dimensions for VIGS in plant functional genomics. Trends in Plant Science, 16, 656–665. [DOI] [PubMed] [Google Scholar]
- Taher, D. , Solberg, S. , Prohens, J. , Chou, Y.Y. , Rakha, M. & Wu, T.H. (2017) World vegetable center eggplant collection: origin, composition, seed dissemination and utilization in breeding. Frontiers in Plant Science, 8, 1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, X. & Zhou, X. (2004) A modified viral satellite DNA that suppresses gene expression in plants. The Plant Journal, 38, 850–860. [DOI] [PubMed] [Google Scholar]
- Tao, X. , Qian, Y. & Zhou, X. (2006) Modification of a viral satellite DNA‐based gene silencing vector and its application to leaf or flower color change in Petunia hybrida . Chinese Science Bulletin, 51, 2208–2213. [Google Scholar]
- Wang, Y. , Shibuya, M. , Taneda, A. , Kurauchi, T. , Senda, M. , Owens, R.A. et al. (2011) Accumulation of potato spindle tuber viroid‐specific small RNAs is accompanied by specific changes in gene expression in two tomato cultivars. Virology, 413, 72–83. https://www.sciencedirect.com/science/article/pii/S0042682211000353 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Chai, C. , Khatabi, B. , Scheible, W.‐R. , Udvardi, M.K. , Saha, M.C. et al. (2021) An efficient brome mosaic virus‐based gene silencing protocol for Hexaploid wheat (Triticum aestivum L.). Frontiers in Plant Science, 12, 685187. 10.3389/fpls.2021.685187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Enlarged loops affect the viability of chimeric ELVd variants.
Figure S2. Modified ELVd‐42 reverts to the original unmodified form.
Figure S3. Analysis of ELVd‐42 60 days after inoculation.
Figure S4. The stuffer control ELVd‐GFP with a 42‐nucleotide insertion does not produce phenotypic alterations.
Table S1. Detail of the primers used in this study
Table S2. Detail of the sequences inserted in the constructs used in this work
Data Availability Statement
All the biological and chemical materials not commercially available that are used for the experiments reported here are available from gustavo.gomez@csic.es upon reasonable request.


