Abstract
MicroRNA21 (MIR21) abundance in porcine oocytes and cumulus cells increases during in vitro maturation. The mechanism by which MIR21 regulates oocyte maturation and the effect on the developmental competence of subsequent embryos remains unclear. The objective of this study was to assess the function of MIR21 during porcine oocyte maturation and its effect on embryonic development. Treatment with peptide nucleic acid MIR21 inhibitor (MIR21‐PNA), designed to specifically bind to and prevent MIR21 activity during in vitro oocyte maturation, decreased cumulus cell expansion, and the oocyte ability to achieve metaphase II maturation stage when compared to control groups. Following parthenogenetic activation, the cleavage rate at 48 h in the MIR21‐PNA group was decreased (p ≤ 0.03) relative to the control groups. Additionally, liquid chromatography‐mass spectrometry (LC‐MS/MS) of oocyte and cumulus cell total protein following MIR21‐PNA treatment during in vitro maturation identified changes in signaling pathways with primary involvement of glucose metabolism (GM) pathways. Furthermore, there was no difference (p = 0.21) in oocyte maturation of control and MIR21‐PNA treated oocytes when cultured in pyruvate lacking medium. Finally, MIR21‐PNA treatment decreased (p = 0.04) glutathione and increased (p = 0.07) reactive oxygen species production in the oocyte. These data suggest that MIR21 influences porcine oocyte maturation by regulating GM pathways in the cumulus–oocyte complex.
Keywords: glucose metabolic pathways, microRNA21, oocyte maturation, pig, reproduction
1. INTRODUCTION
During oocyte maturation, the oocyte undergoes structural and functional changes, brought on by completion of meiosis involving chromatin transformation and cytoplasmic organelle restructuring (Eppig, 1996). Following progression from prophase I to metaphase II (MII), the maternal ribonucleic acids and proteins in the oocyte are primed for their functions in supporting subsequent embryonic development (Lei et al., 2010). Cumulus cells (CCs) are critical for oocyte maturation (Alam & Miyano, 2020) and removal of oocytes from the microenvironment of CCs decreases the quality of MII‐stage oocytes, and their subsequent ability to develop into embryos once fertilized (Dey et al., 2012; L. Zhang et al., 1995). The bidirectional interface between CCs and the oocyte, created via gap junctions, aids in the transport of essential amino acids and substrates for energy production, which is critical to the maturation process (Pelland et al., 2009; Russell et al., 2016). Glucose is crucial for the oocyte during maturation. However, mammalian oocytes cannot utilize glucose as a substrate and rely on their surrounding CCs to convert glucose into alternative substrates such as pyruvate, lactate, or nicotinamide adenine dinucleotide phosphate (NADPH), and then transfer it through the gap junctions into the oocyte (Cetica et al., 2002). In turn, the oocyte secretes factors, such as growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15), which regulate processes such as differentiation, proliferation, and prevention of apoptosis of CCs (R. B. Gilchrist et al., 2004; McIntosh et al., 2008; McNatty et al., 2005). Therefore, understanding the regulatory mechanisms by which these factors are transferred between the oocyte and CCs is essential for understanding the biology of oocyte maturation.
MicroRNA (miRNA) are small (18–24 nucleotides), noncoding nucleic acid molecules that bind with the 3′ untranslated regions (3′ UTRs) of target mRNA molecules, acting as a molecular switch regulating gene expression and translational repression (Bagga et al., 2005; Bartel, 2004). MicroRNA are highly conserved among eukaryotes and are involved in various physiological and pathological processes through their effects on cell proliferation, differentiation, and apoptosis (Lai & Flynt, 2008) and can regulate up to several hundred genes with well over 30% of mRNAs being subjected to miRNA regulation (Lewis et al., 2005). Microarray analyses of porcine oocytes have identified greater than 400 genes as being expressed differently in post‐maturation MII‐stage oocytes compared to germinal vesicle (GV) stage oocytes (Budna et al., 2017).
While miRNA abundance during oocyte maturation has been characterized in cattle (Abd El Naby et al., 2013; G. C. Gilchrist et al., 2016; Tesfaye et al., 2009), humans (Assou et al., 2013; Battaglia et al., 2016), and pigs (Yang et al., 2012), the function of microRNA‐21 (MIR21) during porcine oocyte maturation is still being explored. Utilizing miRNA microarray and deep sequencing analysis, MIR21 has previously been shown to be more abundant in mature MII‐stage porcine oocytes and CCs than in GV‐stage oocytes (Yang et al., 2012). MIR21 is enriched in murine CCs and acts in an antiapoptotic role, and reduces ovulation when depleted (Carletti et al., 2010). We have also discovered that porcine oocytes treated with an anti‐MIR21 peptide nucleic acid (PNA) during in vitro maturation (IVM) reduced the ability to achieve MII stage maturation and downstream embryo development (Hale et al., 2020). Moreover, overexpression of MIR21 in porcine CCs promotes cell expansion and oocyte maturation via downregulation of TIMP3 expression (Bo & Julang, 2018). Bovine CC expansion is prevented by decreased pri‐miR‐21 expression by inhibiting the STAT3 pathway during IVM (Tscherner et al., 2018). Additionally, up‐ and downregulation of MIR21 expression in murine oocytes reduced and increased cumulus cell apoptosis, respectively (Han et al., 2017).
Hence, the purpose of this study was to investigate the hypothesis that MIR21 function during in vitro maturation of the cumulus–oocyte complex (COC) impacts regulatory events in oocyte and cumulus cells, affecting the ensuing embryo's developmental competence. To do this, a MIR21 inhibitor was added to the IVM medium to discern the molecular pathways affected by MIR21 during porcine oocyte IVM. We utilized parthenogenetic activation (PA) to assess the effect of MIR21 inhibition on early embryo development as we wanted to eliminate any extraneous effect caused inadvertently by the introduction of spermatozoa‐ or somatic cell‐derived factors, as may be observed in embryos produced using in vitro fertilization (IVF) or somatic cell nuclear transfer (SCNT), respectively.
2. RESULTS
2.1. MicroRNA21 inhibitor added to in vitro maturation media decreases oocyte maturation rate and early embryonic development rate
To investigate previously reported temporal changes in MIR21 abundance in porcine oocytes undergoing maturation and early embryo development, we evaluated GV‐ to MII‐stage oocytes and early embryonic stages following IVF, and observed an increase in MIR21 abundance throughout COC maturation which rapidly decreased post‐fertilization, maintaining low levels thereafter until a rapid increase in MIR21 abundance was observed in the blastocyst stage (Figure 1a).
Figure 1.
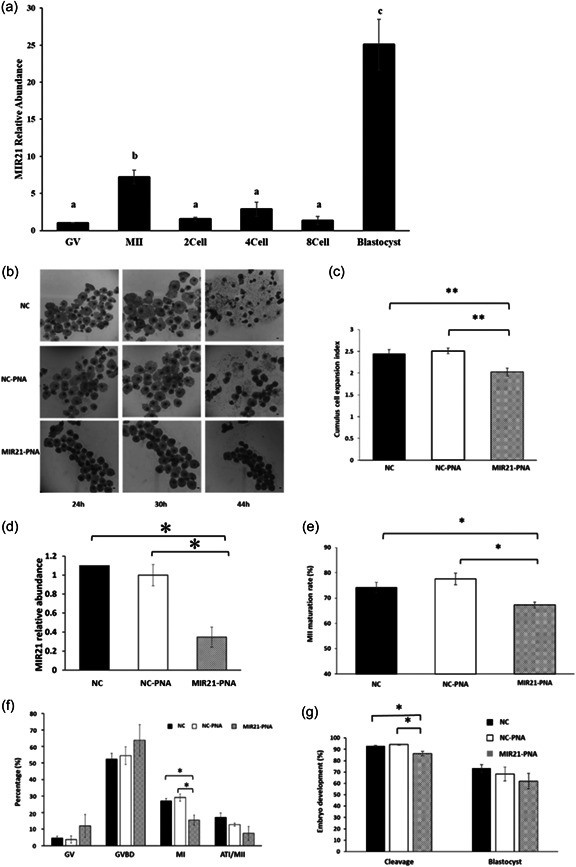
MIR21 inhibitor (MIR21‐PNA) decreases cumulus cell expansion, oocyte maturation, and early embryo development. (a) MIR21 abundance increased during the oocyte maturation phase from germinal vesicle (GV) to MII stage, thereafter decreasing post‐fertilization during early embryo development and then increasing markedly in the blastocyst. Letters indicate a statistical difference (p < 0.05) in MIR21 abundance between development stages. (b) Cumulus cell expansion was assessed after in vitro maturation (IVM) with negative control (NC), negative control‐peptide nucleic acid (NC‐PNA), or MIR21‐inhibitor (MIR21‐PNA) added to IVM medium. (c) Cumulus cell expansion index was calculated after treatments and was decreased in the MIR21‐PNA treated cumulus–oocyte complexes (COCs) compared to the NC and NC‐PNA treated groups. (d) Relative abundance of MIR21 in oocytes, as assessed by qRT‐PCR after IVM with NC‐PNA or MIR21‐PNA added to IVM medium, was reduced (p = 0.01) in MIR21‐PNA treated oocytes compared to the NC‐PNA group, confirming inhibitor effectiveness in oocytes undergoing treatment. (e) Oocyte maturation rate assessed by extrusion of first polar body at MII‐stage after 44 h of maturation and (f) development rate to other meiotic stages, after MIR21‐PNA treatment for 27 h during maturation, demonstrated a decrease in MI‐ and MII‐stage oocytes. (g) Early embryo development evaluated by cleavage and blastocyst development rates in parthenogenetically activated oocytes treated with NC, NC‐PNA, or MIR21‐PNA during IVM determined decreased cleavage rate but no changes in blastocyst development rate due to MIR21‐PNA treatment. All data are mean ± SEM, *p < 0.05, **p < 0.01. Bar = 100 µm.
MIR21‐PNA treatment during COC maturation was used to further evaluate the specific mechanism by which MIR21 inhibition decreases oocyte maturation rate and early embryo development potential. MicroRNA21‐PNA was added to culture medium during oocyte maturation, using a concentration gradient to determine the appropriate concentration to be utilized for downstream experiments. As a result, 1 and 10 nM doses of MIR21‐PNA did not affect cumulus cell expansion (data not shown), but incubation with 100 nM of MIR21‐PNA decreased cumulus cell expansion after 30 h treatment during IVM, when compared to the negative control group (NC; p = 0.003) and NC supplemented with 100 nM scrambled PNA group (NC‐PNA; p < 0.001; Figure 1b,c). MIR21 abundance in the denuded oocyte after MIR21‐PNA treatment, as assessed by qRT‐PCR, was reduced (63.5± 10.1%; p= 0.01) compared to the NC‐PNA group (Figure 1d). The maturation rate of oocytes incubated in 100 nM MIR21‐PNA for 44 h (Figure 1e) was evaluated and first polar body (PB1) extrusion rates were decreased in the MIR21‐PNA group (67.3 ± 1.2%) compared with the NC (74.1 ± 2.2%; p = 0.028) and NC‐PNA (77.6 ± 2.3%; p = 0.019) groups.
The effect of MIR21‐PNA treatment on meiotic cell cycle progression in porcine oocytes after 27 h in culture was also assessed. Oocytes were classified into GV, GVBD (germinal vesicle breakdown), MI (metaphase I), ATI (anaphase‐telophase I), and MII (metaphase II) stages (Supporting Information: Figure 1), and the proportions of oocytes that progressed to these meiotic stages were assessed (Figure 1f). The percentages of oocytes in the GV, GVBD, and ATI/MII phase of development in the MIR21‐PNA group were not different from the NC and NC‐PNA groups. The MI rate of the MIR21‐PNA oocytes was lower (15.5 ± 3.0%) than that of NC (27.1 ± 1.5%; p = 0.025) and NC‐PNA (29.1 ± 2.2%; p = 0.021) groups.
Following IVM, PA was used to evaluate cytoplasmic maturation of MIR21‐PNA treated oocytes. The cleavage rate (embryos achieving two‐cell stage) was decreased in the MIR21‐PNA group (86.3 ± 1.9%) as compared to the NC (92.8 ± 0.7%; p = 0.032) and NC‐PNA (94.1 ± 0.5%; p = 0.007) groups, whereas blastocyst rate did not differ between treatment groups (Figure 1g). SCNT and IVF‐derived embryos were produced to explore the effect of MIR21‐PNA on early embryo development. Still, no differences between the NC‐PNA and MIR21‐PNA treated oocytes were observed (Supporting Information: Figure 2).
2.2. MicroRNA21 inhibition during oocyte maturation alters proteins associated with GM pathways
To evaluate changes in protein abundance in oocytes and CCs after MIR21 suppression, COCs treated with NC‐PNA and MIR21‐PNA during IVM were collected for proteomic analysis (Figure 2a). The proteomic profiles of the samples revealed that the MIR21‐PNA treatment had more of an effect on the CCs than on the oocytes (Figure 2b–e). A total of 790 proteins were identified in the cumulus cell samples, and 636 proteins were identified in the oocytes, of which 299 were identified in both the cumulus cell and oocyte samples (Figure 3a). The differentially abundant proteins (DAPs) were evaluated and 110 and 30 DAPs were identified in the CCs and oocyte samples, respectively (Figure 3b). Two proteins were differentially abundant in both the CC and oocyte samples as a result of MIR21 inhibition. The MIR21‐PNA treatment resulted in a greater abundance of 106 DAPs and lower abundance of 4 DAPs in the CCs, whereas the oocytes only had 11 DAPs in greater abundance and 19 DAPs in lower abundance, when compared to the NC‐PNA control (Figure 3c). Gene ontology analyses identified the greatest number of altered CC proteins were involved in metabolic, spliceosome, and mRNA surveillance pathways (Figure 3d). In contrast, the DAPs in the oocytes were involved in metabolic, proteasome, glutathione, and cell cycle pathways (Figure 3e).
Figure 2.
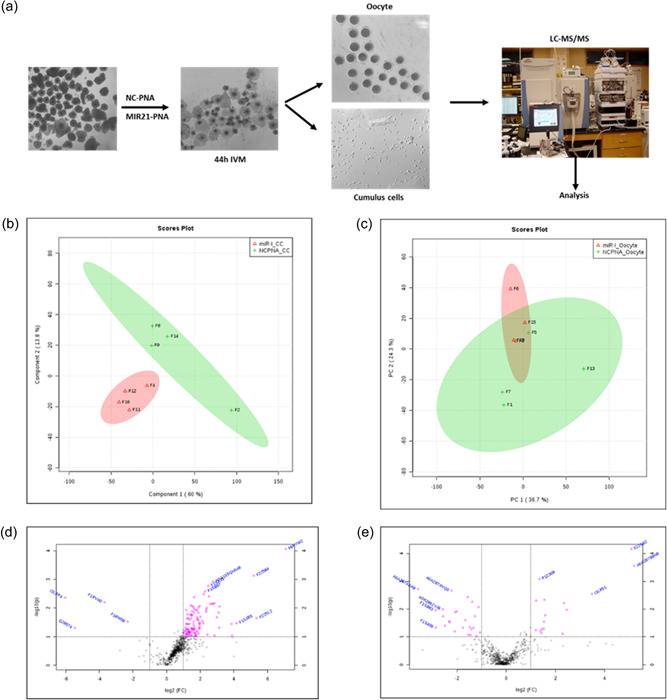
Proteomic analysis of cumulus cells and oocytes treated with negative control‐peptide nucleic acid (NC‐PNA) or MIR21‐PNA during in vitro maturation identified the role of glucose metabolism pathways during oocyte maturation. (a) Cumulus–oocyte complexes (COCs) were treated with NC‐PNA or MIR21‐PNA during in vitro maturation (IVM), after which the oocytes and cumulus cells were collected separately for proteomic analysis. Principal component analysis (PCA) of the proteins identified for (b) cumulus cells and (c) oocytes after NC‐PNA or MIR21‐PNA treatment demonstrated that the MIR21‐PNA treatment generated more differentially abundant proteins (DAPs) in the cumulus cells than in oocytes. Volcano plots depict relative abundance of identified proteins in (d) cumulus cells and (e) oocytes after MIR21‐PNA treatment, with DAPs (adjusted p < 0.05; fold change >10%) indicated in pink.
Figure 3.
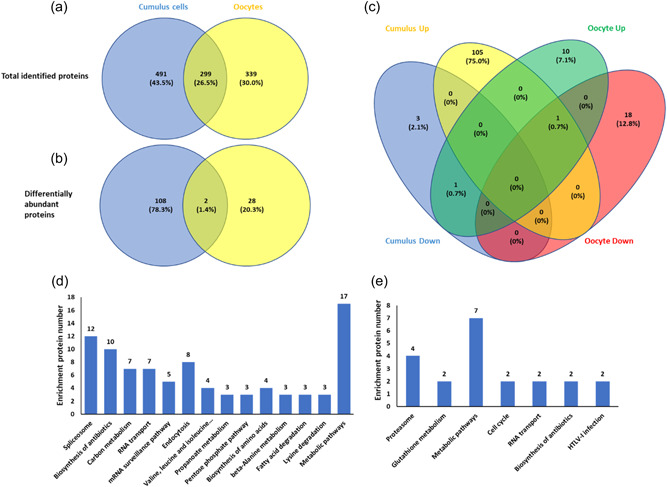
Differentially abundant proteins (DAPs) in cumulus cells and oocytes treated with negative control‐peptide nucleic acid (NC‐PNA) or MIR21‐PNA during in vitro maturation (IVM) show involvement of metabolic pathways in oocyte maturation. (a) Venn diagram indicating the total number of proteins identified in cumulus cells and oocytes as an effect of MIR21‐PNA treatment when compared to the NC‐PNA treated groups. (b) Venn diagram indicating the DAPs in cumulus cells and oocytes as an effect of MIR21‐PNA treatment compared to the NC‐PNA treated group. (c) Venn diagram representing the increased and decreased protein abundances in cumulus cells and oocytes as an effect of MIR21‐PNA treatment compared to the NC‐PNA treated group. KEGG pathway analysis of differentially regulated proteins identified in (d) cumulus cells and (e) oocytes as an effect of MIR21‐PNA treatment compared to the NC‐PNA treated groups.
Three enzymes are involved in the pentose phosphate pathway (PPP), namely glucose phosphate isomerase 1 (GPI1), 6‐phosphogluconolactonase (PGLS), and ribulose‐phosphate‐3‐epimerase (RPE), were in greater abundance in the CCs treated with MIR21‐PNA when compared to the NC‐PNA control (Supporting Information: Table 1). Similarly, two enzymes involved in the GSH metabolism pathway, namely glucose‐6‐phosphate dehydrogenase X‐linked (G6PDX) and glutamate‐cysteine ligase catalytic subunit (GCLC), were in greater abundance in the oocytes treated with MIR21‐PNA when compared to the NC‐PNA control (Supporting Information: Table 2).
2.3. In the absence of pyruvate during IVM, microRNA21 inhibition did not affect oocyte maturation but did influence embryo development
MIR21 involvement in the regulation of glucose and pyruvate production during denuded oocyte IVM was evaluated in light of the DAP identified in the proteomics analysis. Media lacking pyruvate for IVM with NC‐PNA and MIR21‐PNA treatments was employed and the maturation rate in NC‐PNA and MIR21‐PNA treated oocytes was not different (p = 0.21; Figure 4a). After PA, the embryo cleavage rate was decreased (p = 0.02) in the MIR21‐PNA treated group (75.1 ± 3.6%) compared with the NC‐PNA group (87.8 ± 2.4%; Figure 4b). Though the blastocyst development rate was not significantly (p = 0.06) different between treatment groups, a numerical decrease was observed in the MIR21‐PNA group (37.5 ± 2.8%) compared to the NC‐PNA group (52.2 ± 6.9%), which may still be biologically relevant (Figure 4b). The blastocyst cell number was not different (p = 0.72) between the two treatment groups (Figure 4c).
Figure 4.
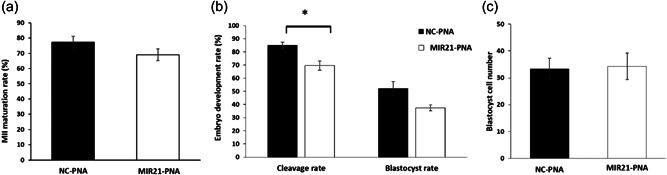
MIR21 inhibition did not affect oocyte maturation in the absence of pyruvate in the in vitro maturation (IVM) medium. (a) Addition of MIR21‐PNA in a pyruvate‐free IVM medium did not affect oocyte maturation rate compared to the negative control‐peptide nucleic acid (NC‐PNA)‐treated oocytes. (b) Early embryo development after parthenogenetic activation of the above MIR21‐PNA treated oocytes reduced cleavage rate, but there was no change in blastocyst development rate when compared to the NC‐PNA treated group. (c) No differences were observed in blastocyst cell number between the two treatment groups. All data are mean ± SEM, *p < 0.05.
2.4. MicroRNA21 inhibition during in vitro maturation influences mitochondrial membrane potential and abundance of specific antioxidant and apoptosis‐related transcripts and proteins
The abundance of GCLC in oocytes was quantified using immunofluorescence (Figure 5a). After MIR21‐PNA treatment, GCLC protein expression was greater (p = 0.037) than in the NC‐PNA treated group (Figure 5b). Mitochondrial membrane potential (ΔΨm) was evaluated and the average mitochondrial ΔΨm from the MIR21‐PNA treated oocytes was decreased (p = 0.001) as compared to the NC‐PNA group (Figure 5c,d).
Figure 5.
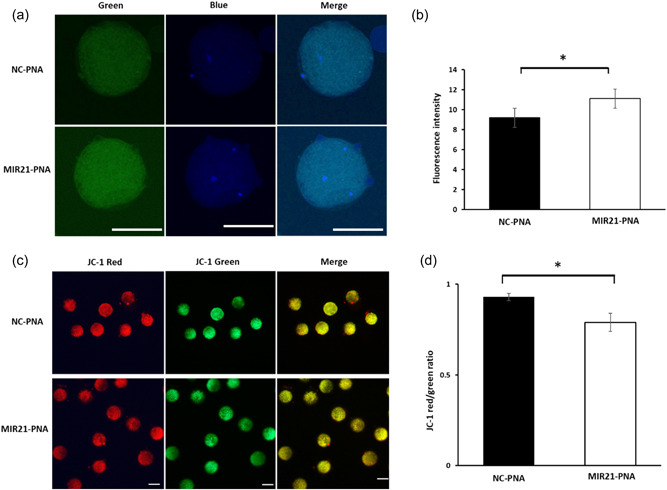
Glutamate‐cysteine ligase, catalytic unit (GCLC) expression and mitochondrial membrane potential are changed in oocytes treated with MIR21‐PNA during in vitro maturation (IVM). (a) GCLC protein expression (in green) as assessed by immunofluorescence in oocytes treated with MIR21‐PNA compared to the negative control‐peptide nucleic acid (NC‐PNA) treated oocytes. (b) Quantification of fluorescence intensity indicates increased GCLC expression in oocytes treated with MIR21‐PNA compared to the NC‐PNA treated oocytes. (c) NC‐PNA and MIR21‐PNA treated oocytes were labeled with JC‐1 and evaluated for J‐monomers with Fluorescein (FITC, green) and J‐aggregates with Rhodamine (RITC, red) channels. (d) Quantification of the fluorescence ratio of RITC/FITC (red/green) indicated a reduction in the average value of mitochondrial membrane potential (ΔΨm) in MIR21‐PNA treated oocytes compared to the NC‐PNA treated oocytes. All data are mean ± SEM, *p < 0.05, scale bar = 100 µm.
The levels of both glutathione (GSH; Figure 6a) and reactive oxygen species (ROS; Figure 6c) were determined in oocytes after MIR21‐PNA and NC‐PNA treatment. Abundance of GSH was lower (p = 0.04) in the MIR21‐PNA group (69.35 ± 1.14 pixels/oocyte) as compared to the NC‐PNA group (91.78 ± 1.48 pixels/oocyte; Figure 6b). Relative ROS fluorescence intensity (Figure 6d) in oocytes treated with MIR21‐PNA (70.88 ± 15.21 pixels/oocyte) was numerically higher compared to the NC‐PNA group (48.41 ± 10.24 pixels/oocyte), though not significantly different (p = 0.07).
Figure 6.
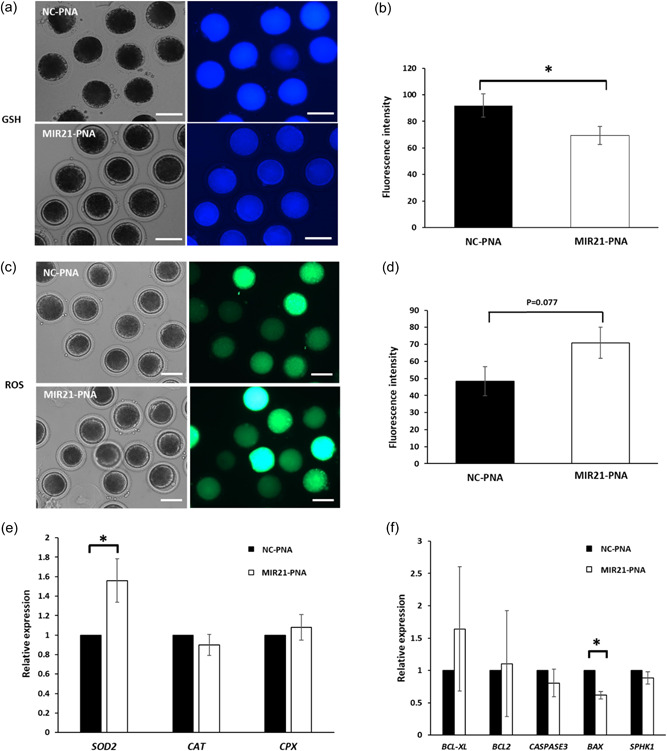
MIR21‐PNA affects reactive oxygen species (ROS) and Glutathione (GSH) expression and apoptosis‐related gene abundance in oocytes compared to the negative control‐peptide nucleic acid (NC‐PNA) treatment. (a) Representative intracellular GSH expression in oocytes treated with MIR21‐PNA or NC‐PNA. (b) Quantification of fluorescence intensity in oocytes determined that MIR21‐PNA treatment reduced GSH expression compared to that in the NC‐PNA treated oocytes. (c) Representative intracellular ROS expression in oocytes treated with MIR21‐PNA or NC‐PNA. (d) Quantification of fluorescence intensity in oocytes demonstrated that MIR21‐PNA treatment increased ROS expression as compared to that in the NC‐PNA treated oocytes. Quantitative RT‐PCR analysis of the relative abundance of antioxidant‐related genes (e) SOD2, CAT, CPX, and apoptosis‐related genes (f) BCL‐XL, BCL2, CASPASE3, BAX, SPHK1 in oocytes treated with MIR21‐PNA or NC‐PNA determined an effect of MIR21‐PNA treatment on SOD2 and BAX mRNA abundance. All data are mean ± SEM, *p < 0.05, scale bar = 100 µm.
Relative mRNA abundance of antioxidant and apoptosis‐related genes in the oocytes treated with MIR21‐PNA or NC‐PNA for 44 h was analyzed (Figure 6e,f; Table 1). Abundance of SOD2 was higher (p = 0.032), and BAX abundance was lower (p = 0.048) in oocytes treated with MIR21‐PNA compared with NC‐PNA treated group, whereas other genes evaluated did not differ between treatment groups (Figure 6e,f; Table 1).
Table 1.
Primer sequences for qPCR
| Gene ID | Accession number | Primer sequences (5′−3′) | Product size (bp) | References |
|---|---|---|---|---|
| BAX a | XM_003127290.4 | F: GCCGAAATGTTTGCTGACG | 157 | Song et al. (2017) |
| R: GCAGCCGATCTCGAAGGA | ||||
| BCL2 b | XM_021099602.1 | F: CGTCCCAGCTCCACATCACC | 130 | Song et al. (2017) |
| R: AGTGCCCCACCGAAGGAGAA | ||||
| BCL2L1 c | XM_021077300.1 | F: ACTGAATCAGAAGCGGAAAC | 294 | J. Liu et al. (2015) |
| R: AAAGCTCTGATACGCTGTCC | ||||
| CASP3 d | XM_005671704 | F: TTTGCGTGCTTCTAAGCCAT | 147 | Song et al. (2017) |
| R: GGCAGGCCTGAATTATGAAA | ||||
| CAT e | XM_003993157.4 | F: ACATGGTCTGGGATTCTGG | 172 | Li et al. (2015) |
| R: TCATGTGCCTGTGCCATCT | ||||
| CPXCR1 f | XM_021079849.1 | F: CACCCAGATGAATGAGCTGC | 163 | Y. Zhang et al. (2018) |
| R: CATGAAGTTGGGCTCGAACC | ||||
| EF1A1 g | NM_001097418.2 | F: AATGCGGTGGGATCGACAAA | 120 | Li et al. (2018) |
| R: CACGCTCACGTTCAGCCTTT | ||||
| SOD2 h | NM_214127.2 | F: TCAAGGAGAAGTTGACCGCT | 181 | Y. Zhang et al. (2018) |
| R: AGGTAATACGCATGCTCCCA | ||||
| SPHK1 i | XM_021066593.1 | F: CTACGAGCAGGTGACGAATG | 132 | Wang et al. (2017) |
| R: AGGCTGAGCACGGAGAAGAC |
BCL2‐associated X protein.
BCL2 apoptosis regulator.
BCL2 like 1.
Caspase 3, apoptosis‐related cysteine peptidase.
Catalase.
CPX chromosome region candidate 1.
Eukaryotic translation elongation factor 1 alpha 1.
Superoxide Dismutase 2, mitochondrial.
Sphingosine kinase 1.
3. DISCUSSION
MicroRNA21 abundances in MII‐stage oocytes and CCs are greater than in GV‐stage oocytes (Wright et al., 2016; Yang et al., 2012), suggesting a potential involvement in porcine oocyte maturation. This study has discovered that inhibition of MIR21 function via MIR21‐PNA treatment during oocyte maturation influences CC expansion, oocyte maturation, and early embryonic development. Moreover, proteomics analysis indicated that oocyte and CC glucose metabolism (GM) pathways are impacted by MIR21 inhibition. Further assessment revealed that MIR21 may also affect the oocyte nuclear and cytoplasmic maturation, potentially altering ATP, ROS, and GSH production in oocytes.
MicroRNA function is thought to be repressed during the oocyte to zygotic genome activation (ZGA) stage, and many maternal miRNAs are degraded during this transition (García‐López & del Mazo, 2012; Tang et al., 2007). Conversely, it has also been hypothesized that certain pri‐miRNA may be transcriptionally active during oocyte maturation (G. C. Gilchrist et al., 2016). In the current study, MIR21 abundance was decreased after fertilization, from the MII‐stage to the eight‐cell embryonic stage, potentially indicating that MIR21 may not be as actively functional during early embryonic development stages as it is during the COC maturation period. This could explain the significant deficits in cleavage rate reported in this study when COCs were treated with MIR21‐PNA during IVM. Interestingly, MIR21 abundance was highest in the blastocyst stage embryo. Upon ZGA, the sudden increase in MIR21 abundance observed potentially suggests its involvement during the blastocyst stage of development. MIR21 is increased at the implantation site and influences matrix metallopeptidase 2 (MMP2) and matrix metallopeptidase 9 (MMP9) abundance during implantation in mice (Hu et al., 2008). These metalloproteinases are expressed by the porcine blastocyst during the peri‐implantation period (Menino et al., 1997; Rashev, 2009) suggesting a potential function for MIR21 during the blastocyst stage and supports the observation of considerably higher abundance in these later stages of porcine embryos in this study.
Knockdown of MIR21 during porcine COC maturation has decreased CC expansion and oocyte maturation, and inversely, increased MIR21 enhanced cumulus cell expansion and oocyte maturation (Bo & Julang, 2018). Treatment of bovine COCs with a STAT3 pathway inhibitor markedly decreased pri‐miR‐21 abundance, in turn preventing CC expansion (Tscherner et al., 2018). The findings of this current study support that CC expansion and oocyte maturation are decreased after MIR21‐PNA treatment during IVM. During normal porcine embryo development, we have shown that MIR21 abundance is reduced as the embryo develops from the two‐cell to eight‐cell stages. Interestingly, when the MIR21 inhibitor was injected into MII stage oocytes, there was no effect on the developmental ability of PA embryos (data not shown), as we are essentially mimicking what the embryo does normally during this phase (low MIR21 abundance during two‐cell to eight‐cell stages). This indicates that MIR21 inhibition during IVM is the critical factor for embryo quality and maturation to MII‐stage, even though it might not affect later development significantly, further demonstrating the stage‐specific role of MIR21 during porcine oocyte maturation.
Cumulus cell expansion observed during oocyte IVM involves the synthesis of extracellular matrix (ECM) proteins by CCs in response to epidermal growth factor (EGF) or follicle‐stimulating hormone (FSH) stimulation (Buccione et al., 1990). Hyaluronic acid is the major structural backbone of cumulus cell‐derived ECM, and both glucose and glucosamine are major substrates for hyaluronic acid synthesis (Salustri et al., 1989). The proteomics data presented in this study identified the hyaluronic acid synthesis pathway‐related gene, glucosamine‐6‐phosphate N‐acetyltransferase (GNPNAT1/GNA1), decreased in CCs after MIR21‐PNA treatment during IVM. GNA1 converts glucosamine‐6‐phosphate into N‐acetylglucosamine‐6‐phosphate, which is dephosphorylated and secreted into the growth medium. Hence, it is possible that MIR21 inhibition affects hyaluronic acid production, thus potentially suppressing CC expansion.
As previously reported, this study validated porcine oocyte maturation rate disruption after MIR21‐PNA treatment during IVM (Bo & Julang, 2018; Tscherner et al., 2018). Using a global proteomics assessment of treated oocytes and CCs, proteins involved in the GM pathway were identified to be influenced by MIR21‐PNA treatment. It is documented that GM is essential for oocyte nuclear and cytoplasmic maturation during the transition to the MII‐stage (Alam & Miyano, 2020). However, the oocyte itself has a poor capacity to utilize glucose (Cetica et al., 2002). Therefore, the CCs metabolize available glucose and transfer these metabolic intermediates to the oocyte (Biggers et al., 1967; Sutton‐McDowall et al., 2010). This COC metabolic strategy was supported by the current findings demonstrating that oocytes devoid of CCs were unable to take full advantage of the glucose present in the culture medium, and only the addition of pyruvate increased the ability to achieve MII‐stage maturation and subsequent embryonic developmental ability (Yuan et al., 2016). Also, treatment with MIR21‐PNA decreased the oocyte maturation rate during IVM with glucose and pyruvate in the maturation medium. However, during pyruvate deprivation, oocyte maturation was unaffected by MIR21‐PNA suggesting MIR21 may affect oocyte maturation through regulation of pyruvate uptake and utilization in porcine oocytes. Indeed, MIR21 can regulate the GM pathway in many cell types. For example, MIR21 decreased the abundance of phosphoenolpyruvate carboxykinase (PEPCK) and glucose‐6‐phosphatase (G6Pase) by targeting Forkhead Box O1 (FOXO1) in primary mouse hepatocytes (Luo et al., 2017). Further, in non‐small cell lung cancer, MIR21 promoted glycolysis and increased lactate generation by targeting fructose‐1,6‐biphosphatase (FBP1; Q. Dai et al., 2017).
Four important GM pathways; glycolysis, PPP, hexosamine biosynthesis pathway, and the polyol pathway, are involved in pig COC maturation. In the PPP, NADPH (important for the reduction of antioxidant GSH) and phosphoribosyl pyrophosphate (PRPP, a substrate for de novo purine synthesis important for meiotic regulation) are the main products in CCs, which are then transferred into the oocytes (Biggers et al., 1967; Sutton‐McDowall et al., 2010). The PPP plays a more important role than glycolysis during mouse oocyte maturation (Jimenez et al., 2013; Xie et al., 2016). However, GM enzyme activity inhibitors during IVM and RNAi gene silencing in CCs determined that in porcine CCs, GM promoted oocyte maturation by releasing metabolites derived via PPP and glycolysis (Wen et al., 2020).
Both glycolysis and the PPP promote porcine oocyte cytoplasmic maturation by supplying energy and regulating maternal gene expression (Yuan et al., 2016). Inhibition of the PPP reduced ATP abundance, increased ROS abundance, and reduced NADPH levels in porcine oocytes (Yuan et al., 2016). The current study discovered that the PPP proteins GPI1, 6PGL, and RPE were in greater abundance in CCs due to MIR21‐PNA treatment during IVM. GPI1 converts glucose‐6‐phosphate to fructose‐6‐phosphate and is involved in the glycolytic and PPP pathways (Keighren et al., 2016). 6PGL catalyzes the conversion of 6‐phosphogluconolactone to 6‐phosphogluconic acid, both intermediates in the oxidative phase of the PPP, during which process glucose is converted into ribulose 5‐phosphate. This is processed further by the cell during the non‐oxidative phase of the PPP to synthesize nucleotides, ATP molecules, and coenzyme A (Tran et al., 2018).
This study determined that both ATP and GSH abundance decreased and ROS abundance increased in oocytes treated with MIR21‐PNA, suggesting MIR21 may alter other energy metabolism genes in the COC. MIR21‐5p directly targets pyruvate dehydrogenase A1 (PDHA1), a pyruvate dehydrogenase enzyme complex component, which links glycolysis and the tricarboxylic acid cycle to regulate glycolysis in gastric cancer cells (Z. Liu et al., 2018). Taken together, the findings of this study support the position that MIR21 influences porcine oocyte maturation by regulating the GM pathway.
In summary, the data from this study suggest that MIR21 affects porcine oocyte nuclear maturation but not necessarily cytoplasmic maturation, putatively by regulating genes and/or proteins involved in GM in CCs during IVM. Moreover, this study has further validated that MIR21 is differentially abundant in different oocyte maturation and early embryonic development stages, further implicating its potential role(s) in regulating oocyte maturation and early embryonic development.
4. MATERIALS AND METHODS
Unless otherwise stated, all chemicals and antibodies used in this study were purchased from Sigma‐Aldrich.
4.1. Oocyte collection and in vitro maturation
Ovaries from mature female pigs were obtained from a local abattoir, transported to the laboratory in a thermos maintained at approximately 30–35°C, for isolation of COCs that were subjected to IVM as previously described (Wright et al., 2016; Yang et al., 2012) with minor modifications. Briefly, follicular fluid from 3 to 5 mm antral follicles was aspirated using an 18‐gauge needle attached to a 10‐ml disposable syringe.
The COCs or GV‐stage denuded oocytes were cultured in a chemically defined IVM medium (TCM‐199 with Hepes, containing 0.1% [w/v] polyvinyl alcohol, 3.05 mM d‐glucose, 0.91 mM sodium pyruvate, 0.57 mM l‐cysteine, 0.5 μg/ml FSH, 0.5 μg/ml luteinizing hormone, 10 ng/ml EGF, 25 ng/ml gentamicin) for 44 h at 38.5°C in 5% CO2. GV‐stage denuded oocytes were obtained by incubating COCs in 0.1% hyaluronidase in TCM‐199 with Hepes for 5 min at 38.5°C, followed by gentle vortexing for 5 min in the same medium.
Cleavage rate was calculated as the percentage of embryos that cleaved to the two‐cell stage. Blastocyst cell number was evaluated by staining blastocysts with 20 μg/ml of Hoechst 33342 for 10 min, mounting them onto glass slides, and applying a cover slip. The blastocysts thus stained were visualized under UV light and images captured using an inverted microscope (Olympus). Total cell number per blastocyst was counted using the NIH ImageJ blast software.
For the experiments evaluating the effect of MIR21 on pyruvate production during IVM, the oocytes were cultured in IVM medium as described above but without the addition of sodium pyruvate.
4.2. Evaluation of cumulus cell expansion
After 30 h of in vitro culture, the porcine COCs were assessed for degree of expansion using a stereoscope, as previously described (Eva et al., 2000). Briefly, the CC expansion was assessed by a subjective scoring system from 0 to 4, wherein 0 indicated no expansion, 1 indicated minimal changes observed in the outermost layer of CCs, 2 indicated expansion involving the peripheral layers of the cumulus oophorus, 3 indicated CC expansion up to, but not including, the corona radiata, and 4 indicated complete CC expansion including the corona radiata cells. At least 20 COCs for each treatment group were evaluated, and three independent replicates were performed. Expanded COCs with a score of 2 or higher were included in all the experiments conducted hereafter.
4.3. Incubation of cumulus–oocyte complexes in IVM medium with MIR21‐PNA
An artificially constructed oligonucleotide sequence, also known as PNA (Panagene Inc.), was designed to bind to and prevent MIR21 activity specifically. A scrambled PNA with no predicted targets was used as a negative control (NC‐PNA). PNA oligonucleotides were diluted in water at a stock concentration of 100 µM and then added to the maturation medium to acquire a final concentration of 1, 10, 100, and 200 nM. The 1 and 10 nM doses did not influence COC maturation in either control or MIR21‐PNA treated groups, and hence were not used. The 100 nM dose did not influence COC maturation rate in the control group while showing a significant change in the MIR21‐PNA treated group. Moreover, the 200 nM dose was observed to damage the maturing oocytes, in the NC‐PNA group. Hence the 100 nM dose was utilized for all downstream experiments. A control group without PNA was used to evaluate potential PNA toxicity. At least 20 COCs were included in each treatment group, and three independent replicates were performed.
4.4. Parthenogenetic activation, somatic cell nuclear transfer, and in vitro fertilization
PA was conducted on MII‐stage oocytes in activation medium (0.3 M mannitol, 0.1 mM CaCl2, 0.1 mM MgCl2, and 0.5 mM Hepes) with two DC pulses (1‐s interval) at 1.2 kV/cm for 30 μs using a BTX Electro Cell Manipulator (Harvard Apparatus). Oocytes were incubated in embryo activation medium (10 μg/ml cycloheximide and 10 μg/ml cytochalasin B) for 4 h, and switched to porcine zygote medium 3 (PZM3; 0.6312 g NaCl, 0.2106 g NaHCO3, 0.0746 g KCl, 0.0048 g KH2PO4, 0.0022 g sodium pyruvate, 0.0146 g l‐glutamate, 0.0546 g hypotaurine, 0.0617 g calcium lactate, 0.001 g gentamicin, 2.0 ml BME essential amino acid, 1.0 ml MEM non‐essential amino acid, and 0.3 g BSA in every 100 ml of medium) in four‐well Nunclon dishes (Nunc) at 38.5°C in 5% CO2 for embryo culture.
For SCNT, MII‐stage oocytes were held in manipulation medium (TCM199 with HEPES supplemented with 7 μg/ml cytochalasin B) to allow equilibration. The polar body along with a portion of the adjacent cytoplasm, presumably containing the metaphase II plate, was extracted, and a single porcine somatic cell was inserted in the perivitelline space using a thin glass capillary. Reconstructed embryos were then held in fusion medium (0.3 M mannitol, 0.1 mM CaCl2, 0.1 mM MgCl2, and 0.5 mM HEPES) and treated with two DC pulses (1‐s interval) at 1.2 kV/cm for 30 μs using a BTX Electro Cell Manipulator (Harvard Apparatus). Embryos were then incubated in activation medium (10 μg/ml cycloheximide and 10 μg/ml cytochalasin B) for 4 h, then immediately moved to PZM‐3 for embryo culture at 38.5°C in 5% CO2.
IVF was performed as previously described (Yang et al., 2012). Briefly, approximately 25 MII‐stage oocytes were rinsed and transferred into equilibrated 50 μl droplets of modified Tris‐buffered medium (mTBM; 113.1 mM NaCl, 3 mM KCl, 7.5 mM CaCl2, 11 mM glucose, 20 mM Tris, 5 mM sodium pyruvate, 2 mM caffeine, and 2 mg/ml bovine serum albumin [BSA]). Boar semen was washed with Dulbecco's phosphate‐buffered saline (DPBS) supplemented with 0.1% BSA, 25 ng/ml gentamicin, and centrifuged at 100 g for 3 min. A 100 μl aliquot of semen was diluted in 3 ml DPBS supplemented with 0.1% BSA, layered over 60% Percoll, and centrifuged at 650g for 20 min. After discarding the supernatant, the sperm pellet was resuspended in mTBM and centrifuged for 10 min. The sperm pellet was resuspended in mTBM to achieve a concentration of 1 × 106 sperm/ml. About 50 μl of this sperm suspension was added into the droplets with MII‐stage oocytes to obtain a final sperm concentration of 0.5 × 106 sperm/ml. After 4–6 h of incubation, putative zygotes were rinsed and transferred into 500 μl PZM3 in four‐well Nunclon dishes (Nunc) and incubated at 38.5°C in 5% CO2.
4.5. Quantification of MIR21 abundance in oocytes and early stage embryos
Pools of exactly 25 denuded oocytes or embryos were collected from three independent replicates for each development stage and treatment group in a minimal amount of PBS‐PVA, and utilized for downstream procedures as previously established (Adur et al., 2017; Wright et al., 2016; Yang et al., 2012). TaqMan® Gene Expression Cells‐to‐CT™ Kit (Ambion®) was used to lyse oocytes or embryos and prepare samples for quantitative real‐time polymerase chain reaction (qRT‐PCR). Lysis solution and DNase from the Cells‐to‐CT™ kit were added to each pool at 4.95 and 0.05 µl, respectively, and incubated at room temperature for 5 min. Stop solution (0.5 µl) was added, and the samples were incubated for 2 min.
MIR21 was quantified using a TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems) followed by qRT‐PCR with TaqMan® MicroRNA Assay using hsa‐MIR21 (Cat# 4427975; Applied Biosystems), which has sequence similarity to the mature miR21 sequence for porcine species. Briefly, the reverse transcription (RT) reaction of 7 µl master mix (0.15 μl of 100 mM dNTPs, 1 μl of MultiScribe™ Reverse Transcriptase, 1.5 μl of 10× Reverse Transcription buffer, 0.19 μl of RNase inhibitor, and 4.16 μl of nuclease‐free water), 3 µl primers (5×), and 1 µl sample lysate, made up to 15 μl with nuclease‐free water, was subjected to reverse transcription conditions of 16°C for 30 min, 42°C for 30 min and 85°C for 5 min. Subsequently, the qRT‐PCR reaction of 20 µl included 1.33 µl of the RT product, 1 µlL TaqMan® MicroRNA Assay (20×), 10 µl TaqMan® Universal PCR Master Mix II (2×), and 7.67 µl nuclease‐free water. Thermal cycler conditions for the TaqMan® MicroRNA qRT‐PCR were 95°C for 10 min, followed by 50 cycles of 95°C for 15 s, and 60°C for 60 s. Fluorescent data acquisition was conducted during the 60°C extension step. Quantification of MIR21 abundance in oocytes and embryos was conducted as previously reported (Yang et al., 2012). Briefly, ΔC t was calculated by subtracting the single greatest sample C t value (i.e., the sample with the lowest abundance) from all other sample C t values. Assuming that each cycle difference is equivalent to a twofold difference, the relative fold‐change for each sample was calculated by applying the equation 2−∆C t .
4.6. Real‐time quantitative polymerase chain reaction
Pools of 20 MII‐stage oocytes were collected from three independent experimental replicates to quantify transcript abundance of specific genes in porcine oocytes after MIR21‐PNA treatment during the IVM process. Total RNA was extracted as per the manufacturer's instructions using the RNeasy Micro Kit (Qiagen) and reverse‐transcribed using the QuantiTect Reverse Transcription Kit (Qiagen). Thereafter, qRT‐PCR was conducted using SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol on an Eppendorf MasterCycler (Eppendorf). The primer sequences (Table 1) were synthesized by Integrated DNA Technologies (IDT). Eukaryotic translation elongation factor 1 alpha 1 (EF1A1) was used as an endogenous normalization control, as it was observed to be unchanged between control and treated oocytes. Reaction conditions were 95°C for 10 min, followed by 35 cycles of 95°C for 15 s, and 60°C for 60 s. Target gene abundance was quantified using the comparative cycle threshold (C t ) method, as previously established (Yang et al., 2012). Briefly, the ΔC t values were obtained for each sample by subtracting the EF1A1 C t value from its respective C t value for the target gene. Then ΔΔC t was calculated using the sample with the highest ΔC t value as an arbitrary constant to subtract from all other sample ΔC t values. Assuming an amplification efficiency of 2 during each cycle, relative mRNA abundance was quantified using the equation. Statistical analysis for each assay was performed on the ΔC t values.
4.7. Proteomic analysis of cumulus cells and oocytes after MIR21‐PNA and NC‐PNA treatment
After 42 h of MIR21‐PNA and NC‐PNA treatment during IVM, pools of 100 oocytes and their corresponding CCs (four independent replicates), were collected in separate microtubes. Total protein from the oocyte and CC samples was extracted in 10 µL lysis buffer (50 mM Tris‐HCl, 1 mM EDTA, pH 8.5), homogenized by vortexing, and protein concentration was quantified using the Pierce Micro BCA™ Assay Kit (Thermo Fisher Scientific). Oocyte and CC samples were diluted to 1 µg/µl and submitted for label‐free relative quantification of peptides using Liquid Chromatography‐Mass Spectrometry (LC‐MS/MS) analysis, conducted at the Protein Facility (Iowa State University). Protein extracts were digested with trypsin/Lys‐C and a Peptide Retention Time Calibration (PRTC; Pierce part #88320) standard was spiked into the sample to serve as an internal control. The sample was then injected into a chromatography column (Agilent Zorbax SB‐C18, 5 microns; part#820966‐922), peptides separated using an EASY nLC‐1200 (Thermo Fisher Scientific) coupled to a Nanospray FlexIon source (Thermo Fisher Scientific), and analyzed using a Q Exactive Hybrid Quadrupole‐Orbitrap Mass Spectrometer with an HCD fragmentation cell. The intact and fragmentation patterns thus obtained were compared to a theoretical fragmentation pattern (from either MASCOT or Sequest HT) to identify the peptides that could in turn be used to identify the native protein, based on the areas of the top three unique peptides for that specific protein. Since each sample had the same amount of PRTC spiked in, the PRTC areas were used to normalize the data between samples. Spectra thus obtained were analyzed in Proteome discoverer (2.1) software against Sus scrofa database using only medium and high confidence peptides with a global false discovery rate (FDR) of less than 5% based on a target‐decoy approach, and normalization was done by the constant algorithm.
Proteome analysis was conducted using MetaboAnalyst 3.0 (Xia et al., 2015) at the Genome Informatics Facility (Iowa State University). Data integrity was confirmed (no peptide with more than 50% missing replicates and positive values for the area), and missing value estimation was imputed using the Singular Value Decomposition (SVD) method. Non‐informative values were identified using interquartile range (IQR) estimation method and deleted, after which data transformation was performed using generalized logarithm transformation (glog) to make individual features more comparable between treatments. The control and treatment samples were compared by Student's t‐test and only p values less than 0.1 were considered statistically significant. Fold change (FC) analysis between the NC‐PNA control and MIR21‐PNA treated groups was performed with a threshold of two, and a volcano plot was created to depict this change against the t‐test analysis. Principal component analysis (PCA) was conducted using the prcomp package and pairwise score plots with an overview of the various separation patterns among the most significant components were created. Partial least squared (PLS) regression was conducted using the plsr function provided by R pls package and PLS‐DA models were built to classify and cross‐validate PLS using the caret package. For all proteins that were significant, S. scrofa orthologs were retrieved from Uniprot (Bateman et al., 2021) and mapped to KEGG (Kanehisa, 2000) pathways to assess the biological pathway association of the proteins.
4.8. Immunofluorescence assessment of glutamate‐cysteine ligase, catalytic unit in the oocyte
Pools of 20 MII‐stage oocytes (three independent replicates) were incubated in rabbit anti‐human GCLC antibody (1:400; Sigma‐Aldrich, AV54576) for 2h at room temperature, followed by incubation in the secondary antibody (1:1000) for 1 h at room temperature. After three washes for 15 min each, oocytes were incubated in Hoechst 33342 (10 μg/ml in PBS) for 10 min at room temperature. Finally, samples were mounted on glass slides and examined with a Leica SP5X MP confocal laser‐scanning microscope. The fluorescence intensity of GCLC expression was quantified using ImageJ software (Version 1.50; National Institutes of Health).
4.9. Measurement of mitochondrial membrane potential (ΔΨm) by JC‐1 staining
Mitochondrial membrane potential (ΔΨm) of the oocytes was evaluated using the lipophilic cationic dye JC‐1 (T3168, Thermo Fisher Scientific), as previously established (J. Dai et al., 2015). Briefly, 20 MII‐stage oocytes (three independent replicates) were incubated in 2 µM JC‐1 in TCM‐199, for 30 min at 38.5°C in 5% CO2. They were then washed with PBS to eliminate nonspecific surface fluorescence and mounted on glass slides for evaluation. Samples were visualized using a Leica SP5X MP confocal microscope using the rhodamine isothiocyanate (RITC, red) channel to identify J‐aggregates and fluorescein isothiocyanate (FITC, green) channel to identify the monomeric form of JC‐1, inside the inner mitochondrial membrane. Images of double fluorescence were recorded in oocytes with the largest diameter and analyzed using Image J software (Version 1.50; National Institutes of Health) to quantify the signal intensity of red and green fluorescence. The ratio of RITC (J‐aggregate) to FITC (J‐monomer) for each oocyte was determined and reported as the ΔΨm.
4.10. Measurement of ROS and GSH levels in MII‐stage oocytes
To detect ROS levels, a pool of 10 MII‐stage oocytes was incubated in 10 μM 2′,7′‐dichlorodihydrofluorescein diacetate (H2DCFDA; Thermo Fisher Scientific) for 15 min, followed by spectroscopy for green fluorescence at 490 nm (Yuan et al., 2016). To detect the levels of GSH, a pool of 10 MII‐stage oocytes was incubated in 10 μM 4‐chloromethyl‐6,8‐difluoro‐7‐hydroxycoumarin (CMF2HC) Cell Tracker Blue dye (Thermo Fisher Scientific) for 20 min, followed by spectroscopy for blue fluorescence at 370 nm (Yuan et al., 2016). Three independent replicates were conducted, and oocytes from the different treatment groups were processed identically with respect to incubation, washing, mounting on glass slides, and imaging. ImageJ software (Version 1.50; National Institutes of Health) was used to analyze the fluorescence intensity for ROS and GSH abundance in the oocytes.
4.11. Statistical analysis
Statistical analyses for all oocyte and embryo cultures, immunofluorescence, and PCR experimental results were performed using SPSS software (version 17.0, SPSS). Shapiro–Wilk test for normality check was utilized to evaluate all data generated, and when needed data were log‐transformed before analysis. Differences between two treatment groups were evaluated using the independent‐samples t‐test, whereas multiple comparison tests were analyzed using one‐way analysis of variance (one‐way ANOVA). Data are expressed as mean ± SEM, and a p value less than 0.05 was considered statistically significant.
AUTHOR CONTRIBUTIONS
Yunsheng Li and Malavika K. Adur contributed equally to this study. Yunsheng Li and Jason W. Ross designed the study. Yunsheng Li and Malavika K. Adur conducted experiments and wrote the manuscript. Jason W. Ross, Aileen F. Keating, and Steven M. Lonergan provided intellectual input contributing to the design and/or execution of experiments in addition to editing the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information.
ACKNOWLEDGMENT
This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2017‐67015‐26459 from the USDA National Institute of Food and Agriculture. Open access funding provided by the Iowa State University Library.
Li, Y. , Adur, M. K. , Lonergan, S. M. , Keating, A. F. , & Ross, J. W. (2022). MicroRNA21 inhibition affects porcine oocyte maturation and alters protein expression critical for metabolic pathway function. Molecular Reproduction and Development, 89, 443–458. 10.1002/mrd.23641
Yunsheng Li and Malavika K. Adur contributed equally to this study.
REFERENCES
- Abd El Naby, W. S. , Hagos, T. H. , Hossain, M. M. , Salilew‐Wondim, D. , Gad, A. Y. , Rings, F. , Cinar, M. U. , Tholen, E. , Looft, C. , Schellander, K. , Hoelker, M. , & Tesfaye, D. (2013). Expression analysis of regulatory microRNAs in bovine cumulus oocyte complex and preimplantation embryos. Zygote, 21(1), 31–51. 10.1017/S0967199411000566 [DOI] [PubMed] [Google Scholar]
- Adur, M. K. , Hale, B. J. , & Ross, J. W. (2017). Detection of miRNA in mammalian oocytes and embryos. In Lee K. (Ed.), Zygotic genome activation: Methods and protocols (pp. 63–81). Springer. 10.1007/978-1-4939-6988-3_5 [DOI] [PubMed] [Google Scholar]
- Alam, M. H. , & Miyano, T. (2020). Interaction between growing oocytes and granulosa cells in vitro. Reproductive Medicine and Biology, 19(1), 13–23. 10.1002/rmb2.12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou, S. , Al‐edani, T. , Haouzi, D. , Philippe, N. , Lecellier, C. H. , Piquemal, D. , Commes, T. , Aït‐Ahmed, O. , Dechaud, H. , & Hamamah, S. (2013). MicroRNAs: New candidates for the regulation of the human cumulus–oocyte complex. Human Reproduction, 28(11), 3038–3049. 10.1093/humrep/det321 [DOI] [PubMed] [Google Scholar]
- Bagga, S. , Bracht, J. , Hunter, S. , Massirer, K. , Holtz, J. , Eachus, R. , & Pasquinelli, A. E. (2005). Regulation by let‐7 and lin‐4 miRNAs results in target mRNA degradation. Cell, 122(4), 553–563. 10.1016/j.cell.2005.07.031 [DOI] [PubMed] [Google Scholar]
- Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bateman, A. , Martin, M.‐J. , Orchard, S. , Magrane, M. , Agivetova, R. , Ahmad, S. , Alpi, E. , Bowler‐Barnett, E. H. , Britto, R. , Bursteinas, B. , Bye‐A‐Jee, H. , Coetzee, R. , Cukura, A. , Da Silva, A. , Denny, P. , Dogan, T. , Ebenezer, T. , Fan, J. , Castro, L. G. , … Teodoro, D. (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research, 49(D1), D480–D489. 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia, R. , Vento, M. E. , Ragusa, M. , Barbagallo, D. , La Ferlita, A. , Di Emidio, G. , Borzí, P. , Artini, P. G. , Scollo, P. , Tatone, C. , Purrello, M. , & Di Pietro, C. (2016). MicroRNAs are stored in human MII oocyte and their expression profile changes in reproductive aging. Biology of Reproduction, 95(6), 131. 10.1095/biolreprod.116.142711 [DOI] [PubMed] [Google Scholar]
- Biggers, J. D. , Whittingham, D. G. , & Donahue, R. P. (1967). The pattern of energy metabolism in the mouse oöcyte and zygote. Proceedings of the National Academy of Sciences of the United States of America, 58(2), 560–567. 10.1073/pnas.58.2.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo, P. , & Julang, L. (2018). MicroRNA‐21 up‐regulates metalloprotease by down‐regulating TIMP3 during cumulus cell‐oocyte complex in vitro maturation. Molecular and Cellular Endocrinology, 477, 29–38. 10.1016/j.mce.2018.05.011 [DOI] [PubMed] [Google Scholar]
- Buccione, R. , Vanderhyden, B. C. , Caron, P. J. , & Eppig, J. J. (1990). FSH‐induced expansion of the mouse cumulus oophorus in vitro is dependent upon a specific factor(s) secreted by the oocyte. Developmental Biology, 138(1), 16–25. 10.1016/0012-1606(90)90172-F [DOI] [PubMed] [Google Scholar]
- Budna, J. , Rybska, M. , Ciesiółka, S. , Bryja, A. , Borys, S. , Kranc, W. , Wojtanowicz‐Markiewicz, K. , Jeseta, M. , Sumelka, E. , Bukowska, D. , Antosik, P. , Brüssow, K. P. , Bruska, M. , Nowicki, M. , Zabel, M. , & Kempisty, B. (2017). Expression of genes associated with BMP signaling pathway in porcine oocytes before and after IVM—A microarray approach. Reproductive Biology and Endocrinology, 15(1), 43. 10.1186/s12958-017-0261-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti, M. Z. , Fiedler, S. D. , & Christenson, L. K. (2010). MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biology of Reproduction, 83(2), 286–295. 10.1095/biolreprod.109.081448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetica, P. , Pintos, L. , Dalvit, G. , & Beconi, M. (2002). Activity of key enzymes involved in glucose and triglyceride catabolism during bovine oocyte maturation in vitro. Reproduction, 124(5), 675–681. 10.1530/rep.0.1240675 [DOI] [PubMed] [Google Scholar]
- Dai, J. , Wu, C. , Muneri, C. W. , Niu, Y. , Zhang, S. , Rui, R. , & Zhang, D. (2015). Changes in mitochondrial function in porcine vitrified MII‐stage oocytes and their impacts on apoptosis and developmental ability. Cryobiology, 71(2), 291–298. 10.1016/j.cryobiol.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Dai, Q. , Li, N. , & Zhou, X. (2017). Increased miR‐21a provides metabolic advantages through suppression of FBP1 expression in non‐small cell lung cancer cells. American Journal of Cancer Research, 7(11), 2121–2130. [PMC free article] [PubMed] [Google Scholar]
- Dey, S. R. , Deb, G. K. , Ha, A. N. , Lee, J. I. , Bang, J. I. , Lee, K. L. , & Kong, I. K. (2012). Coculturing denuded oocytes during the in vitro maturation of bovine cumulus oocyte complexes exerts a synergistic effect on embryo development. Theriogenology, 77(6), 1064–1077. 10.1016/j.theriogenology.2011.10.009 [DOI] [PubMed] [Google Scholar]
- Eppig, J. J. (1996). Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reproduction, Fertility, and Development, 8(4), 485–489. 10.1071/rd9960485 [DOI] [PubMed] [Google Scholar]
- Eva, N. , Barbara, C. V. , & Radek, P. Z. (2000). Secretion of paracrine factors enabling expansion of cumulus cells is developmentally regulated in pig oocytes. Biology of Reproduction, 63(4), 1149–1156. 10.1095/biolreprod63.4.1149 [DOI] [PubMed] [Google Scholar]
- García‐López, J. , & del Mazo, J. (2012). Expression dynamics of microRNA biogenesis during preimplantation mouse development. Biochimica et Biophysica Acta, Gene Regulatory Mechanisms, 1819(8), 847–854. 10.1016/j.bbagrm.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Gilchrist, G. C. , Tscherner, A. , Nalpathamkalam, T. , Merico, D. , & LaMarre, J. (2016). MicroRNA expression during bovine oocyte maturation and fertilization. International Journal of Molecular Sciences, 17(3), 396. 10.3390/ijms17030396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, R. B. , Ritter, L. J. , & Armstrong, D. T. (2004). Oocyte–somatic cell interactions during follicle development in mammals. Animal Reproduction Science, 82, 431–446. 10.1016/j.anireprosci.2004.05.017 [DOI] [PubMed] [Google Scholar]
- Hale, B. J. , Li, Y. , Adur, M. K. , & Ross, J. W. (2020). Inhibition of germinal vesicle breakdown using IBMX increases microRNA‐21 in the porcine oocyte. Reproductive Biology and Endocrinology, 18(1), 39. 10.1186/s12958-020-00603-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X. , Xue, R. , Yuan, H.‐J. , Wang, T.‐Y. , Lin, J. , Zhang, J. , Liang, B. , & Tan, J.‐H. (2017). MicroRNA‐21 plays a pivotal role in the oocyte‐secreted factor‐induced suppression of cumulus cell apoptosis. Biology of Reproduction, 96(6), 1167–1180. 10.1093/biolre/iox044 [DOI] [PubMed] [Google Scholar]
- Hu, S.‐J. , Ren, G. , Liu, J.‐L. , Zhao, Z.‐A. , Yu, Y.‐S. , Su, R.‐W. , Ma, X.‐H. , Ni, H. , Lei, W. , & Yang, Z.‐M. (2008). MicroRNA expression and regulation in mouse uterus during embryo implantation. Journal of Biological Chemistry, 283(34), 23473–23484. 10.1074/jbc.M800406200 [DOI] [PubMed] [Google Scholar]
- Jimenez, P. T. , Frolova, A. I. , Chi, M. M. , Grindler, N. M. , Willcockson, A. R. , Reynolds, K. A. , Zhao, Q. , & Moley, K. H. (2013). DHEA‐mediated inhibition of the pentose phosphate pathway alters oocyte lipid metabolism in mice. Endocrinology, 154(12), 4835–4844. 10.1210/en.2012-2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28(1), 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keighren, M. A. , Flockhart, J. H. , & West, J. D. (2016). Survival of glucose phosphate isomerase null somatic cells and germ cells in adult mouse chimaeras. Biology Open, 5(5), 596–610. 10.1242/bio.017111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E. C. , & Flynt, A. S. (2008). Biological principles of microRNA‐mediated regulation: Shared themes amid diversity. Nature Reviews Genetics, 9(11), 831–842. 10.1038/nrg2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, L. , Ping, Z. , & Jurrien, D. (2010). Maternal control of early mouse development. Development, 137(6), 859–870. 10.1242/dev.039487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B. P. , Burge, C. B. , & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120(1), 15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Liu, X. , Chen, Z. , Song, D. , Yang, J. , Zuo, X. , Cao, Z. , Liu, Y. , & Zhang, Y. (2018). Effect of follistatin on pre‐implantational development of pig parthenogenetic embryos. Animal Science Journal, 89(2), 316–327. 10.1111/asj.12936 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang, Z. , He, C. , Zhu, K. , Xu, Z. , Ma, T. , Tao, J. , & Liu, G. (2015). Melatonin protects porcine oocyte in vitro maturation from heat stress. Journal of Pineal Research, 59(3), 365–375. 10.1111/jpi.12268 [DOI] [PubMed] [Google Scholar]
- Liu, J. , Wang, Q.‐C. , Han, J. , Xiong, B. , & Sun, S.‐C. (2015). Aflatoxin B1 is toxic to porcine oocyte maturation. Mutagenesis, 30(4), 527–535. 10.1093/mutage/gev015 [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Yu, M. , Fei, B. , Fang, X. , Ma, T. , & Wang, D. (2018). miR‑21‑5p targets PDHA1 to regulate glycolysis and cancer progression in gastric cancer. Oncology Reports, 40(5), 2955–2963. 10.3892/or.2018.6695 [DOI] [PubMed] [Google Scholar]
- Luo, A. , Yan, H. , Liang, J. , Du, C. , Zhao, X. , Sun, L. , & Chen, Y. (2017). MicroRNA‐21 regulates hepatic glucose metabolism by targeting FOXO1. Gene, 627, 194–201. 10.1016/j.gene.2017.06.024 [DOI] [PubMed] [Google Scholar]
- McIntosh, C. J. , Stan, L. , Steve, L. , Andrea, H. W. , Kenneth, P. M. , & Jennifer, L. J. (2008). The proregion of mouse BMP15 regulates the cooperative interactions of BMP15 and GDF91. Biology of Reproduction, 79(5), 889–896. 10.1095/biolreprod.108.068163 [DOI] [PubMed] [Google Scholar]
- McNatty, K. P. , Juengel, J. L. , Reader, K. L. , Lun, S. , Myllymaa, S. , Lawrence, S. B. , Western, A. , Meerasahib, M. F. , Mottershead, D. G. , Groome, N. P. , Ritvos, O. , & Laitinen, M. P. E. (2005). Bone morphogenetic protein 15 and growth differentiation factor 9 co‐operate to regulate granulosa cell function. Reproduction, 129(4), 473–480. 10.1530/rep.1.0511 [DOI] [PubMed] [Google Scholar]
- Menino, Jr., A. R. , Hogan, A. , Schultz, G. A. , Novak, S. , Dixon, W. , & Foxcroft, G. H. (1997). Expression of proteinases and proteinase inhibitors during embryo‐uterine contact in the pig. Developmental Genetics, 21(1), 68–74. [DOI] [PubMed] [Google Scholar]
- Pelland, A. M. D. , Corbett, H. E. , & Baltz, J. M. (2009). Amino acid transport mechanisms in mouse oocytes during growth and meiotic maturation. Biology of Reproduction, 81(6), 1041–1054. 10.1095/biolreprod.109.079046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashev, P. (2009). Localization of matrix metalloproteinases‐2 and ‐9 in pig trophoblast and endometrium during early pregnancy. Comptes rendus de l'Académie bulgare des Sciences, 62(8), 949–952. [Google Scholar]
- Russell, D. L. , Gilchrist, R. B. , Brown, H. M. , & Thompson, J. G. (2016). Bidirectional communication between cumulus cells and the oocyte: Old hands and new players? Theriogenology, 86(1), 62–68. 10.1016/j.theriogenology.2016.04.019 [DOI] [PubMed] [Google Scholar]
- Salustri, A. , Yanagishita, M. , & Hascall, V. C. (1989). Synthesis and accumulation of hyaluronic acid and proteoglycans in the mouse cumulus cell‐oocyte complex during follicle‐stimulating hormone‐induced mucification. Journal of Biological Chemistry, 264(23), 13840–13847. 10.1016/S0021-9258(18)80077-1 [DOI] [PubMed] [Google Scholar]
- Song, Z.‐Q. , Li, X. , Wang, Y.‐K. , Du, Z.‐Q. , & Yang, C.‐X. (2017). DMBA acts on cumulus cells to desynchronize nuclear and cytoplasmic maturation of pig oocytes. Scientific Reports, 7(1), 1687. 10.1038/s41598-017-01870-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton‐McDowall, M. L. , Gilchrist, R. B. , & Thompson, J. G. (2010). The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction, 139(4), 685–695. 10.1530/REP-09-0345 [DOI] [PubMed] [Google Scholar]
- Tang, F. , Kaneda, M. , O'Carroll, D. , Hajkova, P. , Barton, S. C. , Sun, Y. A. , Lee, C. , Tarakhovsky, A. , Lao, K. , & Surani, M. A. (2007). Maternal microRNAs are essential for mouse zygotic development. Genes and Development, 21(6), 644–648. 10.1101/gad.418707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye, D. , Worku, D. , Rings, F. , Phatsara, C. , Tholen, E. , Schellander, K. , & Hoelker, M. (2009). Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Molecular Reproduction and Development, 76(7), 665–677. 10.1002/mrd.21005 [DOI] [PubMed] [Google Scholar]
- Tran, A. T. , Sadet, A. , Calligari, P. , Lopes, P. , Ouazzani, J. , Sollogoub, M. , Miclet, E. , & Abergel, D. (2018). Targeting the pentose phosphate pathway: Characterization of a new 6PGL inhibitor. Biophysical Journal, 115(11), 2114–2126. 10.1016/j.bpj.2018.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherner, A. , Brown, A. C. , Stalker, L. , Kao, J. , Dufort, I. , Sirard, M.‐A. , & LaMarre, J. (2018). STAT3 signaling stimulates miR‐21 expression in bovine cumulus cells during in vitro oocyte maturation. Scientific Reports, 8(1), 11527. 10.1038/s41598-018-29874-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. , Gao, Y.‐Y. , Chen, L. , Nie, Z.‐W. , Cheng, W. , Liu, X. , Schatten, H. , Zhang, X. , & Miao, Y.‐L. (2017). Melatonin prevents postovulatory oocyte aging and promotes subsequent embryonic development in the pig. Aging, 9(6), 1552–1564. 10.18632/aging.101252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, J. , Wang, G.‐L. , Yuan, H.‐J. , Zhang, J. , Xie, H.‐L. , Gong, S. , Han, X. , & Tan, J.‐H. (2020). Effects of glucose metabolism pathways on nuclear and cytoplasmic maturation of pig oocytes. Scientific Reports, 10(1), 2782. 10.1038/s41598-020-59709-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, E. C. , Hale, B. J. , Yang, C.‐X. , Njoka, J. G. , & Ross, J. W. (2016). MicroRNA‐21 and PDCD4 expression during in vitro oocyte maturation in pigs. Reproductive Biology and Endocrinology, 14(1), 21. 10.1186/s12958-016-0152-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J. , Sinelnikov, I. V. , Han, B. , & Wishart, D. S. (2015). MetaboAnalyst 3.0‐making metabolomics more meaningful. Nucleic Acids Research, 43(W1), W251–W257. 10.1093/nar/gkv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, H.‐L. , Wang, Y.‐B. , Jiao, G.‐Z. , Kong, D.‐L. , Li, Q. , Li, H. , Zheng, L.‐L. , & Tan, J.‐H. (2016). Effects of glucose metabolism during in vitro maturation on cytoplasmic maturation of mouse oocytes. Scientific Reports, 6(1), 20764. 10.1038/srep20764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. X. , Du, Z. Q. , Wright, E. C. , Rothschild, M. F. , Prather, R. S. , & Ross, J. W. (2012). Small RNA profile of the cumulus‐oocyte complex and early embryos in the pig. Biology of Reproduction, 87(5), 117. 10.1095/biolreprod.111.096669 [DOI] [PubMed] [Google Scholar]
- Yuan, B. , Liang, S. , Kwon, J.‐W. , Jin, Y.‐X. , Park, S.‐H. , Wang, H.‐Y. , Sun, T.‐Y. , Zhang, J.‐B. , & Kim, N.‐H. (2016). The role of glucose metabolism on porcine oocyte cytoplasmic maturation and its possible mechanisms. PLoS One, 11(12), e0168329. 10.1371/journal.pone.0168329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Jiang, S. , Wozniak, P. J. , Yang, X. , & Godke, R. A. (1995). Cumulus cell function during bovine oocyte maturation, fertilization, and embryo development in vitro. Molecular Reproduction and Development, 40(3), 338–344. 10.1002/mrd.1080400310 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Wang, T. , Lan, M. , Zang, X.‐W. , Li, Y.‐L. , Cui, X.‐S. , Kim, N.‐H. , & Sun, S.‐C. (2018). Melatonin protects oocytes from MEHP exposure‐induced meiosis defects in porcine. Biology of Reproduction, 98(3), 286–298. 10.1093/biolre/iox185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


