Abstract
Since human lactoferrin (hLF) binds to bacterial products through its highly positively charged N terminus, we investigated which of the two cationic domains is involved in its bactericidal activity. The results revealed that hLF lacking the first three residues (hLF−3N) was less efficient than hLF in killing of antibiotic-resistant Staphylococcus aureus, Listeria monocytogenes, and Klebsiella pneumoniae. Both hLF preparations failed to kill Escherichia coli O54. In addition, hLF−3N was less effective than hLF in reducing the number of viable bacteria in mice infected with antibiotic-resistant S. aureus and K. pneumoniae. Studies with synthetic peptides corresponding to the first 11 N-terminal amino acids, designated hLF(1–11), and fragments thereof demonstrated that peptides lacking the first three N-terminal residues are less effective than hLF(1–11) in killing of bacteria. Furthermore, a peptide corresponding to residues 21 to 31, which comprises the second cationic domain, was less effective than hLF(1–11) in killing of bacteria in vitro and in mice having an infection with antibiotic-resistant S. aureus or K. pneumoniae. Using fluorescent probes, we found that bactericidal hLF peptides, but not nonbactericidal peptides, caused an increase of the membrane permeability. In addition, hLF killed the various bacteria, most probably by inducing intracellular changes in these bacteria without affecting the membrane permeability. Together, hLF and peptides derived from its N terminus are highly effective against infections with antibiotic-resistant S. aureus and K. pneumoniae, and the first two arginines play an essential role in this activity.
Human lactoferrin (hLF) is a major component of the nonspecific defense of mucosal surfaces and neutrophils and is active against a variety of pathogens (reviewed in references 21 and 26). This protein displays antimicrobial properties against gram-positive and gram-negative bacteria by limiting the availability of environmental iron (2). However, since iron-saturated hLF is also able to kill certain bacteria (7), mechanisms other than iron depletion apparently are involved in the antibacterial activity of LF. Recent studies have indicated that peptides obtained after enzymatic hydrolysis of hLF (1, 22, 37) and bovine LF (bLF) (5, 12, 30) are much more effective in killing bacteria than is the intact protein. Amino acid sequence analysis revealed that these antimicrobial peptides comprised (parts of) the N-terminal residues 1 to 47 of hLF (1, 37) or 1 to 48 of bLF (5, 12). Since most natural antimicrobial peptides are cationic (18), it is likely that the N-terminal cationic domains of hLF and bLF play an essential role in their bactericidal activity. hLF contains two N-terminal cationic domains, i.e., RRRR (residues 2 to 5) and RKVR (residues 28 to 31), whereas the bactericidal domain of bLF comprises residues 17 to 42 (12, 30). It has been reported that peptides of bLF origin (12) as well as synthetic peptides that include the second cationic domain of hLF (3) show antibacterial activity resulting from depolarization of the membrane, increased membrane permeability, and metabolic injury. Moreover, some controversy exists as to whether hLF binds to bacterial products, such as endotoxin and glycosaminoglycans, through its first (17, 32, 38) or second (6, 36) cationic domain. Based on these considerations, the present study was undertaken to determine whether the first and/or second cationic domain of natural hLF is important for its antibacterial activity and to delineate the essential N-terminal residues.
MATERIALS AND METHODS
LF and synthetic peptides.
Natural hLF (77,000 Da) was purified from fresh milk of a single donor by cationic exchange chromatography using S-Sepharose (32). The N terminus of this protein was intact, as determined by N-terminal analytical Mono-S chromatography and N-terminal Edman degradation and sequencing. hLF, which was free of contamination with endotoxin and human lysozyme (15) and had been purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, was dialyzed against saline and stored at −70°C at concentrations of approximately 20 mg/ml. An hLF variant lacking the first three N-terminal residues (GRR), referred to below as hLF−3N, was isolated from natural hLF treated with trypsin as described previously (1). As a control, bovine serum albumin (Sigma Chemical Co., St. Louis, Mo.) was applied instead of hLF or hLF−3N. Synthetic peptides corresponding to residues 1 to 11 of hLF (GRRRRSVQWCA, 1,494 Da), referred to below as hLF(1–11), and fragments thereof and a peptide corresponding to residues 21 to 31 of hLF (FQWQRNMRKVR, 1,567 Da), referred to below as hLF(21–31), were prepared and purified as described previously (4). The purity of the synthetic peptides usually exceeded 88%, as determined by reverse-phase high-performance liquid chromatography. Stocks of synthetic peptides at 1 mM in 5% (vol/vol) acetic acid (pH < 6.0) were stored at −20°C and dried in a Speed-Vac (Savant Instruments Inc., Farmingdale, N.Y.) immediately before use. As controls, we applied synthetically prepared protegrin-1 (RGGRLCYCRRRFCVCVVGR, 2,161 Da), a defensin-like molecule with broad-spectrum antimicrobial activity that had been isolated from pig leukocytes (14, 25), as a positive control and peptide 4 (RPVVSTQLLNGSLAEEEVV, 2,171 Da), part of the gp120 protein from human immunodeficiency virus type 1, as a negative control.
Bacteria.
Antibiotic-resistant Staphylococcus aureus strain 2141, a clinical isolate (Department of Infectious Diseases, Leiden University Medical Center [LUMC]), was highly resistant to methicillin (MIC, >256 mg/liter), cloxacillin (MIC, >256 mg/liter), penicillin (MIC, >0.25 mg/liter), gentamicin and netilmycin (MIC of both antibiotics >8 mg/liter), erythromycin (MIC, >2 mg/liter), and tetracycline (MIC, >4 mg/liter) and displayed limited sensitivity (MIC, <2 mg/liter) to teicoplanin and rifampin. Listeria monocytogenes strain EGD, Klebsiella pneumoniae ATCC 43816, and Escherichia coli O54 were purchased from the American Type Culture Collection (Rockville, Md.). Antibiotic-resistant S. aureus, K. pneumoniae, and E. coli were cultured overnight in nutrient broth (Oxoid, Basingstoke, United Kingdom) at 37°C, diluted in tryptase soya broth (Oxoid), and cultured for an additional 2 h in a shaking water bath at 37°C. L. monocytogenes was cultured overnight in brain heart infusion broth (Oxoid) at 37°C, diluted, and cultured for an additional 2 h in a shaking water bath at 37°C.
Assay for antibacterial activity of lactoferrins and related peptides.
An in vitro assay was used to assess the antibacterial activity of hLF and related peptides. In short, bacteria were washed with phosphate-buffered saline (PBS) (pH 7.4) and diluted to approximately 2 × 106 CFU/ml of PBS (pH 6.0) (7) supplemented with 0.1% (vol/vol) Tween 20 (hereafter designated PBS-Tw). We added this detergent to PBS to reduce the binding of hLF to the test tube, since preliminary experiments indicated that assessment of the bactericidal activity of this protein in the presence of Tween 20 is more reliable. Equal volumes of this bacterial suspension and PBS-Tw containing various concentrations of hLF or related peptides were mixed. At various intervals (range, 0 to 4 h) after incubation at 37°C, the number of viable bacteria was determined microbiologically. Bovine serum albumin (Sigma) or no peptide was included as the negative control; protegrin served as the positive control. The detection limit was 4,000 CFU.
Assay for membrane permeability.
Antimicrobial peptides may cause bacterial cell death by inducing an increase in membrane permeability (24, 29); therefore, we monitored changes in the membrane permeability of bacteria on exposure to hLF, hLF−3N, and related peptides by using propidium iodide (PI; Sigma) staining and fluorescence-activated cell sorter (FACS) analysis (3, 19). Stock solutions of PI (1 mg/ml of deionized water) were prepared. Approximately 2 × 106 bacteria/ml of PBS-Tw were incubated for various intervals at 37°C, washed, incubated for 5 min with 1 μg of PI/ml (final concentration) and then analyzed with a FACScan instrument (Becton & Dickinson, San Jose, Calif.) equipped with a argon laser at 488 nm. The photomultiplier voltages were routinely set at 500 V for PI fluorescence intensity in the second channel. Data acquisition and analysis were controlled using Lysis II software.
Assay for nonspecific esterase activity.
An increase in membrane permeability may not be the prime reason for bacterial cell death caused by hLF; therefore, we investigated the effects of hLF, hLF−3N and related peptides in bacteria on the intracellular nonspecific esterase activity of bacteria assessed using 5-sulfofluorescein diacetate (SFDA) (Molecular Probes, Leiden, The Netherlands) and FACS analysis (31). For this purposes, a stock solution of SFDA (1 mg/ml of ethanol) was prepared. Approximately 2 × 106 CFU of bacteria/ml of PBS-Tw was incubated with several concentrations of hLF, hLF−3N, and hLF-related peptides for various intervals at 37°C, washed, incubated with 100 μM SFDA/ml (final ethanol concentration, 20% [vol/vol]) for 20 min at room temperature in the dark, and then analyzed with a FACScan instrument (excitation, 488 nm; emission, 700 nm) (31) using the photomultiplier voltage of the first channel set at 700 V.
Labeling of LF and related peptides with 99mtechnetium.
Lactoferrins and related peptides were labeled with 99mtechnetium (99mTc) as described previously (23, 34, 35). Briefly, 10 μl of a peptide solution (1 mg/ml of acetic acid) was added to 4 μl of an aseptic solution of 0.5 mg of stannous pyrophosphate/ml (Department of Clinical Pharmacy and Toxicology, LUMC). Immediately thereafter, 2 μl of a solution of 10 mg of KBH4 (crystalline; Sigma) per ml of 0.1 M NaOH was added. After addition of 0.1 ml of sodium [99mTc]pertechnetate solution (200 MBq/ml; Mallinckrodt Medical BV, Petten, The Netherlands), the mixture was gently stirred at ambient temperature for 30 min and was then ready for use. Quality control of the labeling was performed by reverse-phase high-performance liquid chromatography analysis using a Sep-Pak C18 cartridge (Waters, Milford, Mass.) in 20 ml of 0.01 M acetic acid. After rinsing with 20 ml of 0.01 M acetic acid, proteins and peptides were eluted with 2 ml of methanol (Sigma). Labeling yields amounted to 88 to 95%. The precise binding site of the radionuclide on hLF and related peptides is not known yet. However, this labeling procedure is very mild to these proteins and peptides since it does not affect their biological activity (23, 34, 35), cationicity (as determined by mono-S chromatography) (32), or binding to receptors (R. Rossin, M. C. Giron, D. Blok, R. Feitsma, U. Mazzi, and E. K. J. Pauwels, Abstract, Nucl. Med. Commun., 21:593–594, 2000).
Experimental thigh muscle infections.
All animal studies were done in compliance with the Leiden Experimental Animal Committee and Dutch laws related to the conduct of animal experiments. Our interest was to assess whether hLF, hLF−3N, and related peptides display antimicrobial activity in vivo. For this purpose, a well-established animal model for experimental thigh infections was used (34, 35). In short, specific-pathogen-free male Swiss mice weighing 20 to 25 g (Broekman Institute, Someren, The Netherlands) were anesthetized with a single intraperitoneal injection of 0.1 ml of saline containing 1 mg of fluanisone and 0.03 mg of fentanyl citrate (Hypnorm; Janssen Pharmaceutics, Tilburg, The Netherlands). Immediately thereafter, 107 CFU of antibiotic-resistant S. aureus or K. pneumoniae in 0.1 ml of saline was injected into the right thigh muscle. At 24 h thereafter, 0.2 ml of saline, containing various amounts of 99mTc-hLF or radiolabeled peptides (0.3 to 30 μmol), was injected intravenously. As controls, mice were injected with 0.2 ml of saline containing human immunoglobulin G (IgG) (34, 35) or saline without any peptide. At 24 h after injection of hLF or related peptides, the mice were sacrificed by an intraperitoneal injection of 0.25 ml of saline containing 12 mg of sodium pentobarbital (Sanofi BV, Div. Algin, Maassluis, The Netherlands). Next, the infected thigh muscles were removed and, after being weighed, homogenized in 4 ml of PBS. Appropriate dilutions of the homogenates were applied to diagnostic sensitivity test plates (Oxoid), and the number of colonies was determined after an overnight culture at 37°C. Results are expressed as the number of CFU per gram of infected tissue. All negative cultures were assigned the value of 100 CFU/ml of homogenate, which is the lower limit of detection.
Pharmacology.
The accumulation of proteins and peptides at the site of infection, as well as the pharmacology of the radiolabeled hLF preparations and related peptides in mice infected with antibiotic-resistant S. aureus or K. pneumoniae, was assessed by performing scintigraphy. The mice were placed in a supine position on a planar gamma camera (GCA 7100/UI; Toshiba, Tokyo, Japan) with both hind legs spread out and fixed with surgical tape. Continuous whole-body acquisitions of every 60 s during the first hour after injection of the tracer were made with the gamma camera, equipped with a low-energy general-purpose parallel-hole collimator. The camera was connected to a computer (GMS 5500 UI; Toshiba), and high-resolution images of the animals were stored in a 256-by-256 matrix. The energy peak was set at 140 keV with a window of 20%. On the scintigrams, anatomically adjusted regions of interest (ROI) were drawn over the heart and both thighs, providing data about the clearance of 99mTc-peptides and accumulation at the site of infection. The clearance of 99mTc-labeled hLF or related peptides from the circulation during the first hour after injection was assessed by determining the half-life (t1/2) of radioactivity in ROI drawn over the heart at various time intervals. The amount of radioactivity in the heart was corrected for decay and expressed as a percentage of injected dose (ID). Accumulation of hLF and related peptides at the site of infection is expressed as the ratio of the radioactivity counts in identical regions drawn over the infected (target) and the noninfected thigh muscle (nontarget), referred to below as the T/NT ratio (34, 35).
Statistical analysis.
Differences between the values for hLF, hLF−3N, and bovine serum albumin, as well as for the various hLF-related peptides and the negative control peptide, were analyzed using the Mann-Whitney U test. The level of significance was set at P < 0.05.
RESULTS
Antibacterial activity of hLF, hLF−3N, and related synthetic peptides.
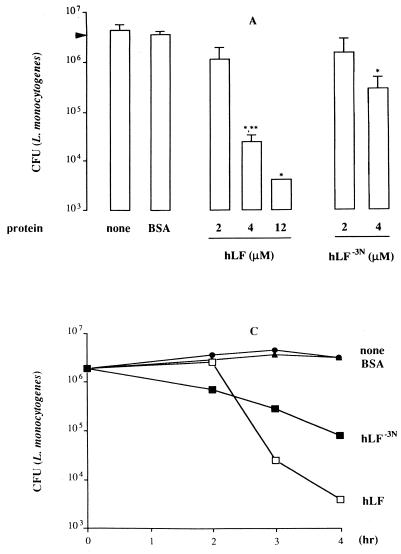
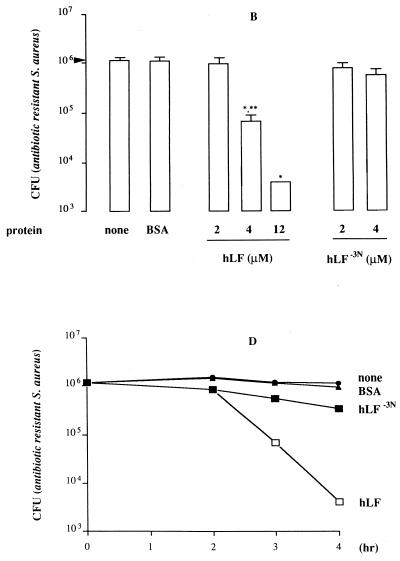
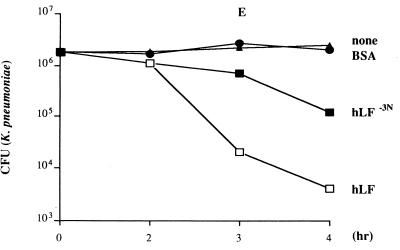
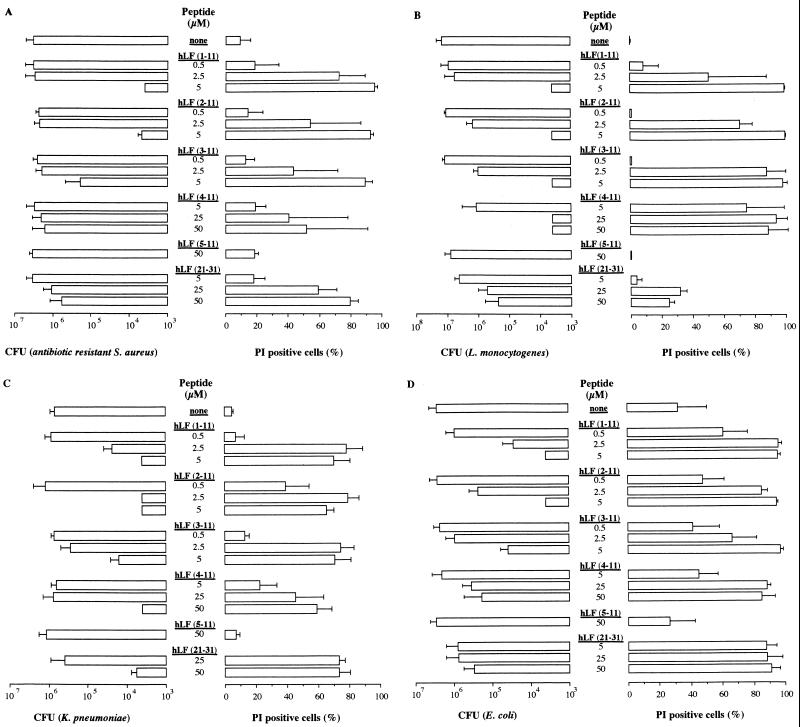
Based on the observation that the first cationic domain of hLF is essential for its interaction with various molecules, such as endotoxin and heparin (17, 32, 38), we compared the antibacterial activity of hLF−3N and natural hLF. The results revealed that both forms of hLF killed antibiotic-resistant S. aureus (Fig. 1A), L. monocytogenes (Fig. 1B) and K. pneumoniae (results not shown) in a dose-dependent fashion, with hLF being more potent (P < 0.05) than hLF−3N. In addition, natural hLF acted faster (P < 0.05) than hLF−3N in killing of these bacteria (Fig. 1C to E). In contrast, E. coli was not killed by hLF or hLF−3N even at concentrations up to 12 μM (results not shown). To find which N-terminal amino acids of hLF are essential for killing of bacteria, we determined the in vitro bactericidal effects of synthetic peptides corresponding to the first 11 N-terminal residues of hLF and fragments thereof against various bacteria. The results revealed that hLF(1–11), hLF(2–11), and hLF(3–11) displayed bactericidal activity against the various bacteria whereas hLF(5–11) was ineffective (Fig. 2). Dose-response studies revealed that hLF(1–11) and hLF(2–11) were considerably (P < 0.05) more efficient than hLF(3–11) in killing of antibiotic-resistant S. aureus (Fig. 2A), K. pneumoniae (Fig. 2C), and E. coli (Fig. 2D) but not L. monocytogenes (Fig. 2B). In addition, hLF(4–11) killed L. monocytogenes (Fig. 2B), K. pneumoniae (Fig. 2C), and E. coli (Fig. 2D) only at high concentrations but did not kill antibiotic-resistant S. aureus (Fig. 2A). Together, these data indicate that the first two arginines are essential for the bactericidal activity of hLF against these bacteria. Next, we compared the bactericidal activities of hLF(21–31), which includes the second cationic domain of hLF and is known to display bactericidal activity (22), and hLF(1–11) against antibiotic-resistant S. aureus, L. monocytogenes, K. pneumoniae, and E. coli. The results revealed that hLF(1–11) is at least 10 times more (P < 0.05) efficient than hLF(21–31) (Fig. 2).
FIG. 1.
Antibacterial activity of natural hLF and hLF−3N against antibiotic-resistant S. aureus, L. monocytogenes, and K. pneumoniae. (A and B) Dose-dependent reduction of the number of viable antibiotic-resistant S. aureus (A) and L monocytogenes (B) cells by natural hLF and hLF−3N. Approximately 1 × 106 to 2 × 106 CFU of bacteria was exposed to different concentrations of natural hLF or hLF−3N for 3 h at 37°C, and then the number of viable bacteria was determined microbiologically. The detection limit was 4,000 CFU. Results are means and standard deviations of at least three separate experiments. (C to E) Time-dependent reduction of the number of viable antibiotic-resistant S. aureus (C), L. monocytogenes (D), and K. pneumoniae (E) by natural hLF and hLF−3N. Approximately 1 × 106 to 2 × 106 CFU of bacteria was exposed to 4 μM hLF or hLF−3N for various intervals at 37°C, and then the number of viable bacteria was determined microbiologically. Results are means and standard deviations of at least three separate experiments. ∗, P < 0.05 compared to bacteria exposed to bovine serum albumin; ∗∗, P < 0.05 compared to bacteria exposed to 4 μM hLF.
FIG. 2.
Killing of antibiotic-resistant S. aureus (A), L. monocytogenes (B), K. pneumoniae (C), and E. coli (D) by hLF(1–11) as well as fragments thereof and hLF(21–31). Approximately 2 × 106 CFU of the various bacteria was exposed to various concentrations of hLF(1–11) or fragments thereof or hLF(21–31) for 1 h at 37°C, and then the number of viable bacteria was determined microbiologically. The detection limit was 4,000 CFU. Results are means and standard deviations of at least three separate experiments. In addition, the effects of the peptides on the membrane permeability of the various bacteria was assessed using 1 μg of PI/ml and FACS analysis. The results, expressed as mean and standard deviation of the percentage of PI-positive bacteria, are from at least three separate experiments.
Effect of natural hLF, hLF−3N, and related peptides on the membrane permeability as well as the intracellular nonspecific esterase activity of bacteria.
To gain insight into the mechanisms underlying the antibacterial activity of hLF as well as related peptides, we determined their effect on the membrane permeability of bacteria. The results revealed that incubation of the various bacteria with natural hLF (and hLF−3N) was not followed by a significant change of the membrane permeability during the period of analysis, i.e., 4 h after addition of these proteins. Based on these results, we investigated the effects of natural hLF and hLF−3N on the intracellular nonspecific esterase activity of bacteria. We chose this enzyme because its activity correlates with cell viability (31). The results revealed that hLF and hLF−3N inhibited the intracellular nonspecific esterase activity of antibiotic-resistant S. aureus, but not of E. coli, within 1 to 2 h after addition of these proteins (Fig. 3), suggesting that killing of bacteria by hLF (and hLF−3N) could be preceded by intracellular effects in antibiotic-resistant S. aureus without disruption of the membrane integrity. In agreement with this possibility, we found that hLF, as well as hLF−3N, at 4°C did not affect the number of viable bacteria as well as the nonspecific esterase activity whereas the defensin-like peptide protegrin-1 (14, 25) did (results not shown). The protegrin-1 peptide kills bacteria through another mechanism, i.e., increasing the membrane permeability. In contrast to natural hLF and hLF−3N, the various peptides, except hLF(5–11), influenced the membrane permeability in a dose- and time-dependent fashion (Fig. 2). When studied at concentrations that did not increase the membrane permeability, these peptides did not affect the nonspecific esterase activity of bacteria (results not shown). Finally, dose-response studies revealed that hLF(1–11) was considerably (P < 0.05) more efficient than hLF (21–31) in increasing the membrane permeability of bacteria (Fig. 2).
FIG. 3.
Effect of natural hLF, hLF−3N, and protegrin-1 on nonspecific esterase activity of antibiotic-resistant S. aureus. Approximately 2 × 106 CFU of bacteria were incubated with 4 μM natural hLF or hLF−3N in PBS-Tw for 2 h at 37°C and then incubated with 100 μM SFDA in 20% (vol/vol) ethanol for 20 min at room temperature in the dark and washed, and then the fluorescence intensity was measured by flow cytometry. It should be realized that the formation of fluorescein resulting from the degradation of SFDA by nonspecific esterase activity is measured by FACS analysis. Consequently, low mean fluorescence intensities indicate little esterase activity, provided that loss of fluorescein due to an increased membrane permeability can be excluded as an explanation. As controls, bacteria were exposed to 4 μM bovine serum albumin (BSA) (negative control) for 2 h at 37°C or protegrin-1 (positive control) for 30 min at 37°C instead of hLF of hLF−3N. Results are from a representative experiment of three independent experiments.
Antibacterial activity of natural hLF, hLF−3N, and related peptides in experimental thigh muscle infections.
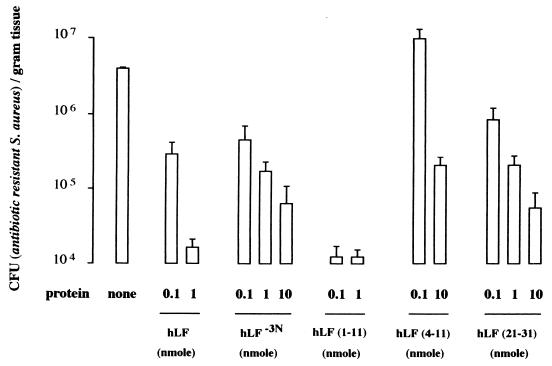
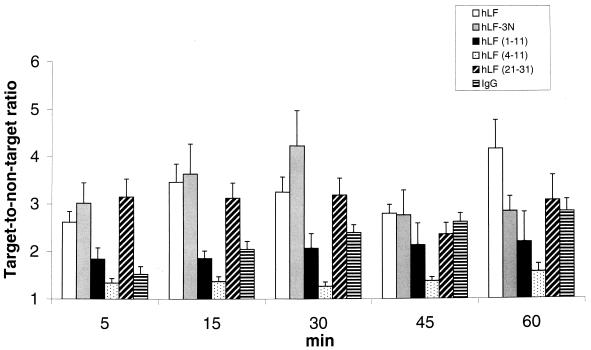
Within 24 h after injection of natural hLF, hLF−3N, hLF(1–11), hLF(4–11), or hLF(21–31), the number of viable bacteria in mice infected with antibiotic-resistant S. aureus was reduced in a dose-dependent fashion; maximum effects were seen with 1 nmol of hLF, 10 nmol of hLF−3N, 0.1 nmol of hLF(1–11), and 10 nmol of hLF(21–31) (Fig. 4). It should be noted that at the lowest level tested (0.1 nmol), the synthetic peptide hLF(1–11) resulted in approximately a 100-fold-higher reduction in the number of viable bacteria than did natural hLF. The highest level of hLF(4–11) exerted some antibacterial activity in mice infected with antibiotic-resistant S. aureus (Fig. 4). Similar results were found (2- to 3-log-unit reduction) in mice infected with K. pneumoniae (results not shown). Pharmacological studies revealed that natural hLF, hLF−3N, hLF(1–11), and hLF(4–11), and hLF(21–31) were rapidly removed from the circulation of these mice with mean t1/2 values of approximately 19, 22, 9, 1, and 2 min, respectively (n = 3). Already within the first 1 min after injection of 1 nmol of radiolabeled natural hLF, hLF−3N, hLF(1–11), hLF(4–11), or hLF(21–31) into antibiotic-resistant S. aureus-infected mice, a significant amount of radiolabeled peptide, i.e., approximately 1 to 1.5% of ID, was observed at the site of infection. Moreover, the amount of radiolabeled hLF or related peptide at the site of infection remained constant during the period of analysis, i.e., the first 60 min after injection of the radiolabeled protein or peptide (Fig. 5), indicating that natural hLF, hLF−3N, and related peptides quickly reached and accumulated at the site of infection.
FIG. 4.
Bactericidal activity of natural hLF, hLF−3N, and various related peptides in antibiotic-resistant S. aureus-infected mice. Mice were intramuscularly infected with approximately 107 CFU of antibiotic-resistant S. aureus in 0.1 ml of saline, and 24 h later 0.2 ml of saline containing various amounts (range, 0.1 to 10 nmol) of natural hLF, hLF−3N, hLF(1–11), hLF(4–11), or hLF(21–31) was injected intravenously. At 24 h after injection of radiolabeled peptide the mice were killed by intraperitoneal injection with sodium pentobarbital, and the thigh muscle was removed, weighed, and then homogenized. Next, serial fold dilutions of these homogenates were pipetted onto plates and the number of CFU was determined microbiologically. It should be kept in mind that hLF(1–11) is much more effective than natural hLF in eliminating bacteria in mice with an experimental infection. Values are means and standard deviations of CFU of antibiotic-resistant S. aureus per gram of thigh muscle (n = 3 experiments; at least 3 mice in each experiment).
FIG. 5.
Accumulation of 99mTc-hLF or related peptides at the site of infection in mice experimentally infected with antibiotic-resistant S. aureus. Accumulation of radiolabeled hLF, hLF−3N, hLF(1–11), hLF(4–11), or hLF(21–31) and, as a control, IgG at the site of infection in mice with an experimental thigh muscle infection with antibiotic-resistant S. aureus at various intervals after injection of radiolabeled protein or peptide is represented by the ratio of the radioactivity counts in identical ROI drawn over the infected (target, T) and the noninfected thigh muscle (nontarget, NT). Results (T/NT ratios) are means and standard deviations for at least three mice.
DISCUSSION
The main conclusion to be drawn from the present results is that the N-terminal arginines 2 and 3 of hLF play an important role in its bactericidal activity. This conclusion is based on the following findings. First, the bactericidal activity of the physiologically relevant truncated form hLF−3N (13, 15, 32), i.e., hLF lacking the first 3 amino acids, was significantly lower than that of natural hLF both in in vitro studies and in animals with an experimental thigh muscle infection with antibiotic-resistant S. aureus (Fig. 4) or K. pneumoniae (results not shown). It should be kept in mind that the amounts of hLF and hLF−3N at the site of infection did not differ (Fig. 5), excluding the possibility that differences in the bactericidal activity of these proteins in mice can be attributed to differences in the amount of protein at the site of infection. Second, comparison of the bactericidal activities of hLF(1–11) and peptides lacking one or more of the first 5 residues revealed that the first 2 arginines are essential for killing of bacteria. In agreement with these in vitro data, we found that hLF(1–11) was much more effective than hLF(4–11) in reducing the number of viable bacteria in antibiotic-resistant S. aureus-infected as well as K. pneumoniae-infected mice.
Our second conclusion pertains to mechanisms underlying the bactericidal activity of hLF and related peptides. Natural hLF or hLF−3N interacts with bacteria possibly through specific binding sites (27), porins (9), endotoxin (6, 32) or lipoteichoic acid (C. Ødegard, unpublished data). Interaction of hLF and hLF−3N with bacteria (35) inhibited the nonspecific esterase activity without affecting their membrane permeability. These observations suggest that hLF is taken up by bacteria, where it exerts its effects, such as interaction with ATP (28) and/or mitochondria in Candida albicans, as has recently been reported for histatin (11). These intracellular actions of hLF could result in disruption of the metabolic activity and subsequently the death of the bacteria. In agreement with this possibility, we found that at 4°C hLF is not able to kill bacteria or to affect the nonspecific esterase activity of the bacteria whereas protegrin-1 can do so. These data indicate that the bactericidal action of hLF requires metabolically active bacteria. The sequence of events initiated by synthetic peptides derived from hLF and resulting in the death of the bacteria is difficult to unravel because the bacteria are rapidly killed by these peptides. For example, we could not establish conditions that allowed us to measure the effects of hLF(1–11) and related peptides on the nonspecific esterase activity of bacteria without affecting the membrane permeability. Furthermore, for certain hLF-derived peptides, e.g., hLF(21–31), no clear correlation between reduction of the number of viable bacteria and the percentage of PI-positive bacteria was found. This observation may indicate that enhanced membrane permeability by antimicrobial peptides does not invariably lead to bacterial cell death. Although we cannot draw a definitive conclusion about the mechanisms underlying the bactericidal action of synthetic hLF-derived peptides, it could be that the mechanisms differ between synthetic peptides and natural peptides derived from the N terminus of hLF. It should also be kept in mind that the size and tertiary structure of the present synthetic peptides may differ considerably from those of natural peptides derived from hLF. Moreover, it is highly likely that potent bactericidal peptides are formed from natural hLF in reponse to local conditions at sites where the bactericidal action is required without exposing other anatomical localizations to the adverse effects of these potent peptides. This feature could be an important advantage of hLF when used to treat patients with infections.
A third striking finding in the present study is that hLF-derived peptides are much more potent in mice with experimental infections with bacteria than in in vitro assays for antibacterial activity. Possible explanations for these findings include (i) synergism between hLF or related peptides and other antimicrobial proteins/peptides, such as lysozyme (16), or other local factors, such as pH and Ca2+ and Zn2+ concentrations, and (ii) interactions with host cells, leading to enhanced antibacterial activities of the cells (20).
Other important findings in the present study concern the role of the second cationic domain of hLF in its bactericidal activity. It has been reported that peptides comprising the second cationic domain of hLF display antibacterial activity (3). However, such a peptide, i.e., hLF(21–31), was considerably less efficient in killing of bacteria than hLF(1–11), which may suggest that the first cationic domain contributes more than the second to killing of bacteria. In agreement with this conclusion, we found that hLF(21–31) was less effective than hLF(1–11) in reducing the number of viable bacteria in the thigh muscle of mice infected with antibiotic-resistant S. aureus or K. pneumoniae.
Our observation that natural hLF is not bactericidal for E. coli O54 contrasts with earlier reports by others (7, 33) using different strains of E. coli. Although we cannot offer a definitive explanation for this difference, technical differences, e.g., the use of different E. coli strains, impurity of hLF preparations, pH 2 treatment of the protein (33), and the assay conditions (7), are clearly important. In connection with the last suggestion, we noted that in vitro hLF and hLF−3N displayed optimal bactericidal activity when investigated in PBS-Tw whereas its bactericidal activity in 10 mM sodium phosphate buffer supplemented with 1% tryptase soya broth was rather poor (results not shown). Of course, our in vitro observation that hLF and hLF−3N are not able to kill E. coli does not exclude the posibility that this protein can be used to successfully treat infections with this bacterium, as has recently been reported by others (10).
Originally, these studies were designed to investigate whether hLF and related peptides can be used to treat experimental infections with bacteria, such as antibiotic-resistant S. aureus. It should be realized that treatment of patients infected with antibiotic-resistant bacteria is an increasingly important problem in hospitals. Currently, there are over 60,000 deaths per year in the United States from hospital-acquired infections. Nearly 40% of hospital-acquired infections with S. aureus are resistant to all antibiotics except methicillin. A major finding in the present structure-function analysis is that hLF(1–11) is highly effective against infections with antibiotic (methicillin)-resistant S. aureus. It should be kept in mind that hLF(1–11) is much more effective than natural hLF in eliminating bacteria in mice with an experimental infection. In addition, during the course of our experiments in mice, we noted that radiolabeled hLF and related peptides rapidly reached the site of infection and remained there throughout for the period of analysis (2 to 4 h). This prompted us to compare the abilities of radiolabeled hLF and related peptides with an established marker of infection or inflammation, i.e., radiolabeled polyclonal human IgG (34, 35), to image of a bacterial infection. The results clearly indicated that radiolabeled hLF and some related peptides visualized the infection much faster than did radiolabeled IgG and that in this respect hLF was superior to the related peptides. The main drawback of radiolabeled hLF for imaging of infections is its ability to bind to leukocytes (21, 26), making this tracer unsuitable for discrimination between sterile inflammatory processes and infectious sites.
ACKNOWLEDGMENTS
The secretarial assistance of M. H. Luijnenburg-Oostdam and E. van Ballegoyen is greatly appreciated.
REFERENCES
- 1.Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K, Tomita M. Identification of the bactericidal domain of lactoferrin. Biochim Biophys Acta. 1992;1121:130–136. doi: 10.1016/0167-4838(92)90346-f. [DOI] [PubMed] [Google Scholar]
- 2.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 3.Chapple D S, Mason D J, Joannou C L, Odell E W, Gant V, Evans R W. Structure-function relationship of antibacterial synthetic peptides homologous to a helical surface region on human lactoferrin against Escherichia coli serotype O111. Infect Immun. 1998;66:2434–2440. doi: 10.1128/iai.66.6.2434-2440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Koster H S, Amons R, Benckhuijsen W E, Feijlbrief M, Schellekens G A, Drijfhout J W. The use of dedicated peptide libraries permits the discovery of high affinity binding peptides. J Immunol Methods. 1995;197:179–188. doi: 10.1016/0022-1759(95)00182-a. [DOI] [PubMed] [Google Scholar]
- 5.Dionysius D A, Milne J M. Antibacterial peptides of bovine lactoferrin: purification and characterization. J Dairy Sci. 1997;80:667–674. doi: 10.3168/jds.S0022-0302(97)75985-X. [DOI] [PubMed] [Google Scholar]
- 6.Elass-Rochard E, Roseanu A, Legrand D, Trif M, Salmon V, Motas C, Montreuil J, Spik G. Lactoferrin-lipopolysaccharide interactions: involvement of the 28–34 loop region of human lactoferrin in the high affinity binding of Escherichia coli 055B5 lipopolysaccharide. Biochem J. 1995;312:839–848. doi: 10.1042/bj3120839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellison R T, Giehl T J, LaForce M M. Damage of the outer membrane of enteric gram-negative bacteria by lactoferrin and transferrin. Infect Immun. 1988;56:2741–2781. doi: 10.1128/iai.56.11.2774-2781.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison R T., III The effects of lactoferrin on Gram-negative bacteria. Adv Exp Med Biol. 1994;357:71–90. doi: 10.1007/978-1-4615-2548-6_8. [DOI] [PubMed] [Google Scholar]
- 9.Erdei J, Forsgren A, Naidu A S. Lactoferrin binds to porins OmpF and OmpC in Escherichia coli. Infect Immun. 1994;62:1236–1240. doi: 10.1128/iai.62.4.1236-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haverson L A, Engberg I, Baltzer L, Dolphin G, Hanson L A, Mattsby-Baltzer I. Human lactoferrin and peptides derived from a surface-exposed helical region reduce experimental Escherichia coli urinary tract infection in mice. Infect Immun. 2000;68:5816–5823. doi: 10.1128/iai.68.10.5816-5823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmerhorst E J, Breeuwe P, van het Hof W, Walgreen E, Oomen L C J M, Veerman E C I, van Nieuw Amerongen A, Abee T. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J Biol Chem. 1999;274:7286–7291. doi: 10.1074/jbc.274.11.7286. [DOI] [PubMed] [Google Scholar]
- 12.Hoek K S, Milne J M, Grieve P A, Dionysius D A, Smith R. Antibacterial activity of bovine lactoferrin-derived peptides. Antimicrob Agents Chemother. 1997;41:54–59. doi: 10.1128/aac.41.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchens T W, Henry J F, Yip T T. Structurally intact (78-kDa) form of maternal lactoferrin purified from urine of preterm infants fed human milk: identification of trypsin-like proteolytic cleavage event in vivo that does not result in fragment dissociation. Proc Natl Acad Sci USA. 1991;15:2994–2998. doi: 10.1073/pnas.88.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokryakov V N, Harwig S S L, Panyutich E A, Shevchenko A A, Aleshina G M, Shamova O V, Korneva H A, Lehrer R I. Protegrins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 15.Legrand D, van Berkel P H, Salmon V, van Veen H A, Slomianny M C, Nuijens J H, Spik G. The N-terminal Arg2, Arg3, Arg4 of human lactoferrin interact with sulphated molecules but not with the receptor present on Jurkat human lymphoblastic T-cells. Biochem J. 1997;327:841–846. doi: 10.1042/bj3270841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leitch E C, Wilcox M D. Elucidation of the antistaphylococcal action of lactoferrin and lysozyme. J Med Microbiol. 1999;48:867–871. doi: 10.1099/00222615-48-9-867. [DOI] [PubMed] [Google Scholar]
- 17.Mann D M, Romm E, Migliorini M. Delineation of the glycosaminoglycan-binding site in the human inflammatory response protein lactoferrin. J Biol Chem. 1994;269:23661–23667. [PubMed] [Google Scholar]
- 18.Martin E, Ganz T, Lehrer R I. Defensins and other endogenous peptide antibiotics of vertebrates. J Leukoc Biol. 1995;58:128–137. doi: 10.1002/jlb.58.2.128. [DOI] [PubMed] [Google Scholar]
- 19.Mason D J, Dybowski R, Larrick J W, Gant V A. Antimicrobial action of rabbit leukocyte CAP18106–137. Antimicrob Agents Chemother. 1997;41:624–629. doi: 10.1128/aac.41.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myauchi H, Hashimoto S, Nakajima M, Shinoda I, Fukuwatari Y, Hayasawa H. Bovine lactoferrin stimulates the phagocytic activity of human neutrophils: identification of its active domain. Cell Immunol. 1998;187:34–37. doi: 10.1006/cimm.1997.1246. [DOI] [PubMed] [Google Scholar]
- 21.Nuijens J H, van Berkel P H C, Schanbacher F L. Structure and biological actions of lactoferrin. J Mammary Gland Biol Neoplasia. 1996;1:285–295. doi: 10.1007/BF02018081. [DOI] [PubMed] [Google Scholar]
- 22.Odell E W, Sarra R, Foxworthy M, Chapple D S, Evans R W. Antibacterial activity of peptides homologous to a loop region in human lactoferrin. FEBS Lett. 1996;382:175–178. doi: 10.1016/0014-5793(96)00168-8. [DOI] [PubMed] [Google Scholar]
- 23.Pauwels E K J, Welling M M, Feitsma R I J, Atsma D E, Nieuwenhuizen W. The labelling of proteins and LDL with 99mTc: a new direct method employing KBH4 and stannous chloride. Nucl Med Biol. 1993;20:825–833. doi: 10.1016/0969-8051(93)90148-n. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Playa E, Houghton R A, Blondelle S E. The role of amphipathicity in the folding, self-association, and biological activity of multiple subunit small proteins. J Biol Chem. 1995;270:1048–1056. doi: 10.1074/jbc.270.3.1048. [DOI] [PubMed] [Google Scholar]
- 25.Qu X-D, Harwig S S L, Oren A, Schafer W H, Lehrer R I. Susceptibility of Neisseria gonorrhoeae to protegrins. Infect Immun. 1996;64:1240–1245. doi: 10.1128/iai.64.4.1240-1245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez L, Calvo M, Brock J H. Biological role of lactoferrin. Arch Dis Child. 1992;67:657–661. doi: 10.1136/adc.67.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrijvers A, Bonnah R, Wong H, Retzer M. Bacterial lactoferrin receptors. Adv Exp Med Biol. 1998;443:123–133. doi: 10.1007/978-1-4757-9068-9_15. [DOI] [PubMed] [Google Scholar]
- 28.Semenov D V, Kanyshkova T G, Akimzhanov A M, Buneva V N, Nevinsky G A. Interaction of human milk lactoferrin with ATP. Biochemistry (Moscow) 1998;63:944–951. [PubMed] [Google Scholar]
- 29.Shai Y. Mechanism of the binding, insertion, and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 30.Tomita M, Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K. Potent antibacterial peptides generated by pepsin digestion of bovine lactoferrin. J Dairy Sci. 1991;74:4137–4142. doi: 10.3168/jds.S0022-0302(91)78608-6. [DOI] [PubMed] [Google Scholar]
- 31.Tsuji T, Kawasaki Y, Takeshima S, Sekiya T, Tanaka S. A new fluorescence staining assay for visualizing living micro-organisms in soil. Appl Environ Microbiol. 1995;61:3415–3421. doi: 10.1128/aem.61.9.3415-3421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Berkel P H C, Geerts M E J, van Veen H A, Mericskay M, de Boer H A, Nuijens J H. N-terminal stretch Arg2, Arg3, Arg4, and Arg5 of human lactoferrin is essential for binding to heparin, bacterial lipopolysaccharide, human lysozyme and DNA. Biochem J. 1997;328:145–151. doi: 10.1042/bj3280145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward P P, Piddington C S, Cunningham G A, Zhou X, Wyatt R D, Conneely O M. A system for production of commercial quantities of human lactoferrin: a broad spectrum natural antibiotic. Bio/Technology. 1995;13:498–503. doi: 10.1038/nbt0595-498. [DOI] [PubMed] [Google Scholar]
- 34.Welling M M, Hiemstra P S, van den Barselaar M T, Paulusma-Annema A, Nibbering P H, Pauwels E K J, Calame W. Antibacterial activity of human neutrophil defensins in experimental infections in mice is accompanied by increased leukocyte accumulation. J Clin Investig. 1998;8:1583–1589. doi: 10.1172/JCI3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welling M M, Paulusma-Annema A, Pauwels E K J, Nibbering P H. 99mTechnetium labelled antimicrobial peptides that discriminate between bacterial infections and sterile inflammations. Eur J Nucl Med. 2000;27:292–301. doi: 10.1007/s002590050036. [DOI] [PubMed] [Google Scholar]
- 36.Wu H D, Monroe M, Church F C. Characterization of the glycosaminoglycan-binding region of lactoferrin. Arch Biochem Biophys. 1995;317:85–92. doi: 10.1006/abbi.1995.1139. [DOI] [PubMed] [Google Scholar]
- 37.Yamauchi K, Tomita M, Giehl T J, Ellison R T. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect Immun. 1993;61:719–728. doi: 10.1128/iai.61.2.719-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang G H, Mann D M, Tsai C M. Neutralization of endotoxin in vitro and in vivo by a human lactoferrin-derived peptide. Infect Immun. 1999;67:1353–1358. doi: 10.1128/iai.67.3.1353-1358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]