Abstract
Aims
This study sought to assess the volatile organic compound (VOC) profiles of ampicillin‐resistant and ‐susceptible Escherichia coli to evaluate whether VOC analysis may be utilized to identify resistant phenotypes.
Methods and Results
An E. coli BL21 (DE3) strain and its pET16b plasmid transformed ampicillin‐resistant counterpart were cultured for 6 h in drug‐free, low‐ and high‐concentrations of ampicillin. Headspace analysis was undertaken using thermal desorption‐gas chromatography‐mass spectrometry. Results revealed distinct VOC profiles with ampicillin‐resistant bacteria distinguishable from their susceptible counterparts using as few as six compounds. A minimum of 30 compounds (fold change >2, p ≤ 0.05) were differentially expressed between the strains across all set‐ups. Furthermore, three compounds (indole, acetoin and 3‐methyl‐1‐butanol) were observed to be significantly more abundant (fold change >2, p ≤ 0.05) in the resistant strain compared to the susceptible strain both in the presence and in the absence of drug stress.
Conclusions
Results indicate that E. coli with acquired ampicillin resistance exhibit an altered VOC profile compared to their susceptible counterpart both in the presence and in the absence of antibiotic stress. This suggests that there are fundamental differences between the metabolisms of ampicillin‐resistant and ‐susceptible E. coli which may be detected by means of VOC analysis.
Significance and Impact of the Study
Our findings suggest that VOC profiles may be utilized to differentiate between resistant and susceptible bacteria using just six compounds. Consequently, the development of machine‐learning models using VOC signatures shows considerable diagnostic applicability for the rapid and accurate detection of antimicrobial resistance.
Keywords: ampicillin, antimicrobial resistance, Escherichia coli, gas chromatography‐mass spectrometry, metabolic profiling, volatile organic compounds
INTRODUCTION
The prevalence of antimicrobial resistance (AMR) is growing at an alarming rate and represents a major global public health concern (Antimicrobial Resistance Collaborators, 2022). Methods routinely employed in clinical practice for the detection of resistance involve long incubation steps of up to 24 h and are based on subjective breakpoint criteria rather than direct detection of resistance markers (The European Committee on Antimicrobial Susceptibility Testing, 2021). In addition to being time‐consuming, the disadvantage of these methods is that resistance mechanisms may go undetected if the susceptibility criteria are not met, with potentially negative implications for both patient outcomes and the spread of drug‐resistant strains (Kidd et al., 2020; Wiskirchen et al., 2014). There is an urgent need to develop cost‐effective methods for the detection of AMR which have a rapid turnaround time and high accuracy, for implementation into routine clinical diagnostics.
Escherichia coli are a major cause of nosocomial infections including ventilator‐associated pneumonia, hospital‐acquired urinary tract infections and surgical site infections (Khan et al., 2015; Trubiano & Padiglione, 2015). According to the Centers for Disease Control and Prevention National Healthcare Safety Network between 2015 and 2017, E. coli was the most frequently reported pathogen across all hospital‐acquired infections, accounting for 18% of all infections (Weiner‐Lastinger et al., 2020). The consequence of multidrug resistance within these organisms poses significant therapeutic challenges and is of particular concern.
AmpC β‐lactamase production may arise through the inducible expression of the chromosomal ampC gene via mutations of the promoter or regulatory genes, including ampR, or through transmissible plasmids harbouring ampC genes and enabling high‐level production of AmpC β‐lactamases (Caroff et al., 1999). AmpC production is associated with resistance to a broad range of antibiotics, including third‐generation cephalosporins and reduced inhibition by commonly used β‐lactamase inhibitors such as clavulanic acid and tazobactam (Belley et al., 2021). Furthermore, AmpC β‐lactamases may confer resistance to carbapenems when coupled with porin mutations which reduce outer membrane permeability (Hamzaoui et al., 2018).
Bacteria display species‐specific profiles of volatile organic compounds (VOCs) and these may be used as markers of pathogen infection in some disease states (Ahmed et al., 2017; Bos et al., 2013; Drabińska et al., 2019; Rees, Burklund, et al., 2018; Schulz & Dickschat, 2007; Sethi et al., 2013). In light of these compounds largely arising from upstream metabolic processes, VOCs are representative of metabolic phenotype and offer valuable insight into the cellular state. Recent studies have demonstrated that differences exist in the VOC profiles between susceptible and resistant bacterial strains (Drabińska et al., 2022; Rees, Nasir, et al., 2018; Smart et al., 2019).
In the current study, we analysed the headspace of AmpC‐producing and nonproducing E. coli by means of thermal desorption‐gas chromatography‐mass spectrometry (TD‐GC‐MS) for the purposes of assessing microbial volatile metabolites. Using a metabolomics‐based approach, we sought to examine the differential volatile profiles produced in vitro by ampicillin‐susceptible and ‐resistant E. coli and to assess the influence of drug stress on these bacteria.
METHODS
pET16b plasmid transformation
Fifty microlitres of competent E. coli BL21 (DE3) cells (New England BioLabs) were combined with 5 ng of pET16b plasmid DNA (Addgene) and incubated on ice for 30 min. The pET16b plasmid contained the ampR gene and promoter for inducible expression of the E. coli chromosomal ampC gene. Contents were subjected to heat shock for 10 s at 42°C followed by a 5 min incubation on ice. Nine hundred and fifty microlitres of super optimal broth with catabolite repression media were added, and the transformed cells were incubated at 37°C with 180 rpm shaking for 1 h. The resulting culture was spread onto Luria‐Bertani (LB) agar plates containing 50 μg ml−1 ampicillin sodium (Formedium) and incubated overnight at 37°C for plasmid uptake selection.
Bacterial cultures
Two E. coli nonpathogenic laboratory strains, BL21 (DE3) and pET16b plasmid‐transformed BL21 (DE3) (further referred to as BL21‐AmpC), were subcultured on LB agar plates and incubated at 37°C for 24 h. Single colonies of each strain were removed from plates and inoculated in 5 ml LB broth overnight at 37°C with 180 rpm shaking. Nitrocefin discs (Sigma‐Aldrich) were utilized to confirm β‐lactamase production in the plasmid‐transformed strain and absence in the nontransformed strain. The broth microdilution method was implemented for the determination of the ampicillin minimum inhibitory concentration (MIC) (The European Committee on Antimicrobial Susceptibility Testing, 2021). Each strain was assayed in triplicate, and the method was performed three times across three different days. Results were interpreted using the 2021 European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint criteria (The European Committee on Antimicrobial Susceptibility Testing, 2021).
Headspace sampling from cultures
Overnight cultures of the two E. coli strains were standardized to 0.1 OD600 with LB broth (~1 × 108 colony forming unit ml−1). One hundred microlitres of culture were inoculated in 20 ml headspace vials containing 1 ml supplemented LB broth. Three set‐ups were prepared for each strain: drug‐free (BL21 n = 7, BL21‐AmpC n = 7), low concentration ampicillin (2 μg ml−1; BL21 n = 10, BL21‐AmpC n = 10) and high concentration ampicillin (10 μg ml−1; BL21 n = 7, BL21‐AmpC n = 7). Noninoculated media controls were prepared for comparative purposes and background subtraction. Inert‐coated stainless steel HiSorb™ probes (Markes International) comprising a polydimethylsiloxane sorbent were inserted into the headspace. Headspace was passively collected over a 6 h incubation period at 37°C with 180 rpm shaking after which the probes were removed and inserted into stainless steel tubes for analysis.
TD‐GC‐MS analysis
Samples were dry purged with N2 at 50 ml min−1 for 4 min to remove water residue and analysed by TD‐GC‐MS. A gaseous internal standard (1 ppmV p‐bromofluorobenzene in N2; Thames Restek) was spiked onto each sample prior to desorption. VOCs were thermally desorbed at 280°C for 5 min (TD100; Markes International) and then transferred with split injection (1:10) to a cryofocusing trap maintained at 0°C, which was subsequently flash heated to 280°C for 2 min. VOC separation was performed on an Agilent 7890B GC (Agilent Technologies) using an Agilent DB‐5ms column (30 m × 0.25 mm × 0.25 μm) with constant helium flow (1 ml min−1). The GC column oven was set to a linear temperature ramp programme with an initial temperature of 30°C, increasing at 7.5°C min−1 to 250°C (29.33 min total GC cycle time). After GC separation, VOCs were transferred to an Agilent 7010 MS to obtain mass spectra. The MS utilized an electron ionization (EI+) source set to 70 eV and 150°C, and a triple quadrupole mass analyser in full scan mode across a range of 40–300 m/z with an acquisition rate of 5 Hz.
Data analysis
Spectral deconvolution was performed using MassHunter Quantitative Analysis software (Agilent Technologies) with a retention window size factor of 100 and delta m/z tolerance of 0.3 AMU left/0.7 AMU right. Detected peaks were aligned using a retention time window of ±0.1 min, and tentative peak identifications were performed by comparing mass spectra against the National Institute of Standards and Technology (NIST) mass spectral library (version 2014) (Wallace, 2014). Only compounds with a mass spectral match factor ≥70% were selected, with annotations listed as the compound name of the top match score. Integrated peak areas were normalized against the internal standard, log10 transformed and auto‐scaled. Manual background subtraction was undertaken for each peak by subtracting peak areas found in the media from those of the experimental spectra. Only those compounds which displayed positive peak areas after this subtraction in at least all bar one replicates were considered in the analysis. The ‘all bar one’ criterion was applied to ensure that any biological variation present within our small sample size did not skew results during this early exploratory stage. In the event of the absence of a peak, a nominal value of 1/5 of the minimum positive value was assigned for each variable. Peaks resulting from suspected environmental contaminants and artefacts were removed from the analysis, for example siloxanes and phthalate‐derived compounds. The retention index of each peak was calculated according to the IUPAC's definition of the temperature programmed Kovat's index equation (IUPAC, 1997). Calculated retention indices were compared against those in the literature using the NIST database. Compound identifications with retention indices deviating by more than 5% from the literature were rejected. Multivariate statistical analyses utilizing the unsupervised machine‐learning methods principal component analysis (PCA) and hierarchal clustering analysis (HCA) were performed using R v4.1.2 (R Core Team, 2021). Univariate statistical analysis was performed by Student's t tests (α = 0.05), and volcano plots were constructed in R to assess the differential expression of features.
RESULTS
MIC determination
The BL21‐AmpC strain demonstrated growth in ampicillin concentrations up to and including 128 μg ml−1, confirming the acquisition of ampicillin resistance. The MIC of the non‐AmpC‐producing strain (BL21) was determined to be 4 μg ml−1, classifying the strain as susceptible based on EUCAST breakpoint values (The European Committee on Antimicrobial Susceptibility Testing, 2021). The results from this assay were used to select the experimental ampicillin concentrations for the VOC analysis of ampicillin‐challenged bacteria. A low‐level ampicillin concentration of 2 μg ml−1 was selected, whereas 10 μg ml−1 was chosen for high concentration analysis.
Core volatiles of untreated E. coli strains
The core volatilome was defined as the set of VOCs produced by both strains and included 42 compounds; an additional 14 compounds were detected in the susceptible strain. The pan‐volatilome was defined as the entire set of 66 volatile metabolites. Univariate analysis using Student's t test demonstrated that 31 compounds differed in abundance (p ≤ 0.05) between untreated susceptible and resistant strains. Five compounds were significantly increased (p ≤ 0.05) in the AmpC‐producing strain compared to the susceptible strain, identified as acetoin (retention time [RT] 3.563 min), 3‐methyl‐1‐butanol (RT 3.933 min), an unknown alkane (RT 14.552 min), indole (RT 14.902 min) and an unknown benzene derivative (RT 22.082 min). The detected compounds are detailed in Table 1. Annotations were compared to the published microbial VOC database (mVOC 2.0) (Lemfack et al., 2018).
TABLE 1.
Core VOCs detected for untreated ampicillin‐susceptible and ‐resistant strains of Escherichia coli BL21
| VOC | CAS | RT (min) | Calculated RI | RI NIST | Normalized peak area | p | Found in mVOC 2.0 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m/z | BL21 | BL21‐AmpC | ||||||||||
| Quant | Qual | Mean | SD | Mean | SD | |||||||
| Dimethyl sulfide | 75‐18‐3 | 62.0 | 47.0, 45.0 | 2.244 | 507 | 516 | 260.25 | 120.27 | 54.54 | 22.87 | <0.005 | Y |
| Unknown 1 (2‐ketone) | 51410‐11‐8 | 43.0 | – | 2.542 | 580 | — | 266.69 | 225.94 | 215.86 | 211.33 | ns | – |
| Acetoin | 513‐86‐0 | 45.0 | 88.0 | 3.563 | 704 | 706 | 26.32 | 9.31 | 36.79 | 10.04 | ns | Y |
| 3‐Methyl‐1‐butanol | 123‐51‐3 | 55.0 | 70.0 | 3.933 | 729 | 727 | 846.09 | 141.02 | 1420.57 | 188.86 | <0.005 | Y |
| Dimethyl disulfide | 624‐92‐0 | 93.9 | – | 4.080 | 739 | 748 | 1374.12 | 708.00 | 1588.52 | 671.16 | ns | Y |
| 2‐Methylthiazole | 3581‐87‐1 | 98.9 | 57.7, 57.0 | 5.045 | 803 | 832 | 2.24 | 1.25 | – | – | <0.001 | N |
| 3‐Methylbutanoic acid | 503‐74‐2 | 60.0 | 87.0 | 5.579 | 830 | 835 | 4.92 | 4.95 | 5.31 | 5.52 | ns | Y* |
| Unknown 2 (alcohol) | – | 98.0 | 97.0, 53.0 | 5.927 | 848 | – | 2.85 | 2.18 | – | – | <0.005 | – |
| 1‐Hexanol | 111‐27‐3 | 56.0 | 69.0, 55.0 | 6.249 | 865 | 869 | 17.18 | 3.88 | 19.62 | 3.21 | ns | Y |
| Unknown 3 (benzene derivative) | – | 132.9 | 150.9, 134.9 | 6.775 | 892 | – | 191.38 | 120.00 | 190.35 | 108.15 | ns | ‐ |
| 2,5‐Dimethylpyrazine | 123‐32‐0 | 80.9 | – | 7.118 | 909 | 911 | 33.32 | 17.02 | 28.45 | 13.82 | ns | Y |
| Dimethyl trisulfide | 3658‐80‐8 | 125.8 | 78.9 | 8.377 | 969 | 969 | 429.99 | 217.06 | 375.97 | 164.29 | ns | Y |
| Hexanoic acid | 142‐62‐1 | 60.0 | 73.0 | 8.457 | 973 | 974 | 114.82 | 71.74 | 157.58 | 173.38 | ns | Y* |
| Phenol | 108‐95‐2 | 93.9 | 66.0 | 8.494 | 975 | 976 | 116.80 | 43.26 | 84.71 | 49.26 | ns | Y |
| 2‐Ethyl‐1‐hexanol | 104‐76‐7 | 57.0 | 70.0, 55.0 | 9.580 | 1026 | 1025 | 22.41 | 9.38 | 18.78 | 16.70 | ns | Y |
| Benzyl alcohol | 100‐51‐6 | 107.9 | 78.9, 77.0 | 9.704 | 1032 | 1035 | 79.95 | 15.21 | 83.82 | 20.59 | ns | Y |
| 1‐Ethenoxy‐2‐ethylhexane | 103‐44‐6 | 71.0 | – | 9.772 | 1036 | 1038 | 2.95 | 0.92 | 3.60 | 1.62 | ns | N |
| 1‐Methyl‐2‐pyrrolidinone | 872‐50‐4 | 98.9 | 98.0, 71.0 | 9.781 | 1036 | 1034 | 5.54 | 5.16 | 8.31 | 10.89 | ns | N |
| Tetrahydro‐2H‐pyran‐2‐one | 542‐28‐9 | 99.9 | 57.0, 56.0 | 10.039 | 1048 | 1010 | 1.02 | 0.60 | – | – | <0.001 | N |
| 3‐Acetyl‐1H‐pyrroline | 1072‐82‐8 | 93.9 | 108.9, 66.0 | 10.247 | 1058 | 1061 | 3.15 | 2.29 | – | – | <0.005 | N |
| Acetophenone | 98‐86‐2 | 104.9 | 119.9, 77.0 | 10.388 | 1065 | 1065 | 121.25 | 65.87 | 149.80 | 120.26 | ns | Y |
| 3‐Ethyl‐2,5‐dimethylpyrazine | 13360‐65‐1 | 134.9 | 135.9 | 10.579 | 1074 | 1079 | 9.31 | 4.28 | – | – | <0.001 | Y |
| 2,6‐Diethylpyrazine | 13067‐27‐1 | 135.9 | 134.9 | 10.767 | 1083 | 1085 | 2.21 | 1.60 | – | – | <0.005 | N |
| 2‐Nonanone | 821‐55‐6 | 57.7 | 71.0, 58.6 | 10.883 | 1089 | 1090 | 13.03 | 1.99 | 13.15 | 1.52 | ns | Y |
| 2‐Ethylhexanoic acid | 149‐57‐5 | 88.0 | 73.0, 87.0 | 11.294 | 1109 | 1108 | 40.77 | 20.12 | 22.71 | 18.38 | ns | N |
| Phenylethyl alcohol | 60‐12‐8 | 91.0 | 91.9, 121.9 | 11.352 | 1112 | 1112 | 49.03 | 10.57 | 53.36 | 10.45 | ns | Y |
| Unknown 4 (thiazole derivative) | – | 111.9 | 112.9 | 11.512 | 1120 | – | 2.79 | 1.11 | – | – | <0.001 | – |
| Unknown 5 (benzene derivative) | – | 117.1 | 296.8, 90.1 | 11.845 | 1136 | – | 11.86 | 2.94 | 5.55 | 2.54 | <0.001 | – |
| Unknown 6 (benzaldehyde derivative) | – | 148.9 | 149.9, 131.9 | 12.208 | 1154 | – | 0.74 | 0.41 | – | – | <0.001 | – |
| 1‐Nonanol | 143‐08‐8 | 70.0 | 69.0, 56.0 | 12.515 | 1170 | 1173 | 3.28 | 0.95 | 3.63 | 0.58 | ns | N |
| Naphthalene | 91‐20‐3 | 127.9 | – | 12.890 | 1189 | 1188 | 5.65 | 3.72 | – | – | < 0.005 | N |
| Dodecane | 112‐40‐3 | 71.0 | 57.0, 85.0 | 13.120 | 1200 | 1200 | 3.35 | 2.41 | – | – | < 0.005 | Y* |
| Decanal | 112‐31‐2 | 82.0 | 55.0, 111.9 | 13.215 | 1205 | 1203 | 6.69 | 6.63 | – | – | <0.05 | Y |
| 2,4‐Dimethylbenzaldehyde | 15764‐16‐6 | 132.9 | 105.0, 77.0 | 13.458 | 1218 | 1217 | 3.66 | 0.73 | 2.63 | 1.49 | ns | N |
| Unknown 7 (fatty acid methyl ester) | – | 87.0 | 88.0, 71.0 | 13.535 | 1222 | – | 14.94 | 4.40 | 11.95 | 3.05 | ns | – |
| 3‐Phenylfuran | 13679‐41‐9 | 143.9 | 114.9 | 13.578 | 1224 | 1224 | 6.68 | 2.50 | – | – | <0.001 | N |
| Benzothiazole | 95‐16‐9 | 134.9 | 107.9, 73.0 | 13.662 | 1229 | 1228 | 13.98 | 9.32 | – | – | <0.005 | N |
| Unknown 8 | – | 58.6 | 102.9, 59.3 | 13.832 | 1237 | – | 0.75 | 0.65 | – | – | < 0.01 | – |
| 3‐Butyl‐2,5‐dimethylpyrazine | 40790‐29‐2 | 121.9 | – | 14.132 | 1253 | 1263 | 3.51 | 1.56 | – | – | < 0.001 | Y* |
| Unknown 9 (alkane) | – | 71.0 | 85.0, 57.0 | 14.552 | 1275 | – | 0.92 | 0.82 | 3.81 | 4.95 | ns | – |
| Indole | 120‐72‐9 | 116.9 | 89.9, 88.9 | 14.902 | 1294 | 1293 | 60531.32 | 9898.92 | 78004.51 | 13472.80 | < 0.05 | Y |
| Unknown 10 (pyridine derivative) | – | 119.9 | 134.9, 92.0 | 15.066 | 1303 | – | 8.92 | 2.36 | 5.63 | 2.71 | < 0.05 | – |
| Undecanal | 112–44–7 | 67.0 | 82.0, 68.0 | 15.141 | 1307 | 1309 | 2.83 | 2.15 | 2.07 | 1.77 | ns | N |
| Unknown 11 (ester) | – | 144.9 | 102.9 | 15.652 | 1335 | – | 1.92 | 1.50 | 3.99 | 5.84 | ns | – |
| 4‐Methylquinoline | 491‐35‐0 | 142.9 | 141.9, 114.9 | 16.495 | 1382 | 1384 | 2.73 | 2.44 | 1.62 | 1.17 | ns | N |
| Tetradecane | 629‐59‐4 | 57.0 | 71.0, 85.0 | 16.812 | 1400 | 1400 | 8.65 | 5.41 | – | – | <0.001 | Y |
| Unknown 12 (ketone) | – | 82.0 | 109.0, 96.0 | 16.957 | 1409 | – | 5.07 | 2.99 | – | – | <0.001 | – |
| 2,6‐Bis(1,1‐dimethylethyl)‐2,5‐cyclohexadiene‐1,4‐dione | 719‐22‐2 | 176.9 | 134.9, 204.9 | 17.897 | 1464 | 1458 | 1.33 | 0.78 | – | – | < 0.001 | N |
| Unknown 13 (alkene) | – | 109.9 | 96.0, 178.9 | 17.986 | 1469 | – | 9.13 | 1.92 | 7.86 | 0.88 | ns | – |
| Unknown 14 (branched alkene) | – | 55.0 | – | 18.049 | 1473 | – | 1.51 | 1.02 | – | – | <0.005 | – |
| 2‐Tridecanone | 593‐08‐8 | 57.7 | 71.0, 58.6 | 18.396 | 1494 | 1498 | 25.64 | 3.23 | 27.73 | 2.92 | ns | Y |
| 2,4‐Di‐tert‐butylphenol | 96‐76‐4 | 190.9 | – | 18.593 | 1505 | 1518 | 175.87 | 103.49 | 62.08 | 20.83 | < 0.05 | Y* |
| Tridecanal | 10486‐19‐8 | 82.0 | 96.0, 94.9 | 18.676 | 1510 | 1510 | 2.65 | 2.53 | – | – | < 0.05 | N |
| Dibenzofuran | 132‐64‐9 | 167.9 | 138.9 | 18.887 | 1523 | 1521 | 1.51 | 1.13 | 1.48 | 2.05 | ns | N |
| Unknown 15 (furan derivative) | – | 101.9 | 173.8 | 19.089 | 1535 | – | 13.23 | 19.38 | 58.82 | 83.07 | ns | – |
| Hexadecane | 544‐76‐3 | 71.0 | 57.0, 85.0 | 20.110 | 1600 | 1600 | 2.61 | 1.79 | – | – | < 0.005 | Y* |
| (1‐Pentylhexyl)‐benzene | 4537‐14‐8 | 91.0 | 160.9 | 20.546 | 1629 | 1620 | 1.16 | 0.78 | 0.96 | 0.78 | ns | N |
| (1‐Ethylnonyl)‐benzene | 4536‐87‐2 | 91.0 | 118.9 | 21.100 | 1667 | 1656 | 0.91 | 0.70 | – | – | < 0.005 | N |
| 2‐Pentadecanone | 2345‐28‐0 | 57.7 | 71.0, 58.6 | 21.582 | 1650 | 1697 | 38.77 | 6.17 | 36.93 | 6.35 | ns | Y |
| Unknown 16 (branched alkane) | – | 71.0 | 57.0, 85.0 | 21.632 | 1703 | – | 3.51 | 1.79 | 3.51 | 1.57 | ns | – |
| (1‐Methyldecyl)‐benzene | 4536‐88‐3 | 104.9 | 220.9, 91.0 | 21.684 | 1706 | 1692 | 1.59 | 1.00 | – | – | < 0.001 | ‐ |
| Unknown 17 (benzene derivative) | – | 147.9 | 162.9, 91.0 | 22.082 | 1733 | – | 24.09 | 19.04 | 46.19 | 17.07 | < 0.05 | – |
| Octadecane | 593‐45‐3 | 71.0 | 85.0, 57.0 | 23.078 | 1800 | 1800 | 1.87 | 1.29 | – | – | < 0.005 | N |
| 2‐Heptadecanone | 2922‐51‐2 | 71.0 | 57.7, 85.0 | 24.453 | 1893 | 1899 | 20.92 | 22.76 | 8.64 | 1.69 | ns | N |
| 5‐Dodecyldihydro‐2(3H)‐furanone | 730‐46‐1 | 85.0 | 83.0 | 27.227 | 2080 | 2104 | 28.47 | 7.37 | 23.60 | 13.41 | ns | N |
| Unknown 18 (alcohol) | – | 71.0 | 85.0, 57.0 | 27.641 | 2107 | – | 63.33 | 53.50 | 54.77 | 26.81 | ns | – |
Note: ns, not significant (p > 0.05).
Abbreviations: CAS, chemical abstracts service number; NIST, National Institute of Standards and Technology; RI, retention index; RT, retention time; VOC, volatile organic compound.
Listed in mVOC 2.0 but not for E. coli.
Effect of treatment with 10 μg ml−1 ampicillin on the volatilome
The volcano plot in Figure 1 depicts the differential VOC expression of the BL21 susceptible strain in the presence of 10 μg ml−1 ampicillin compared to the untreated condition. Of the 66 pan‐volatilome compounds, 57 were downregulated (fold change >2, p ≤ 0.05), with all but 14 compounds less than the limit of detection.
FIGURE 1.
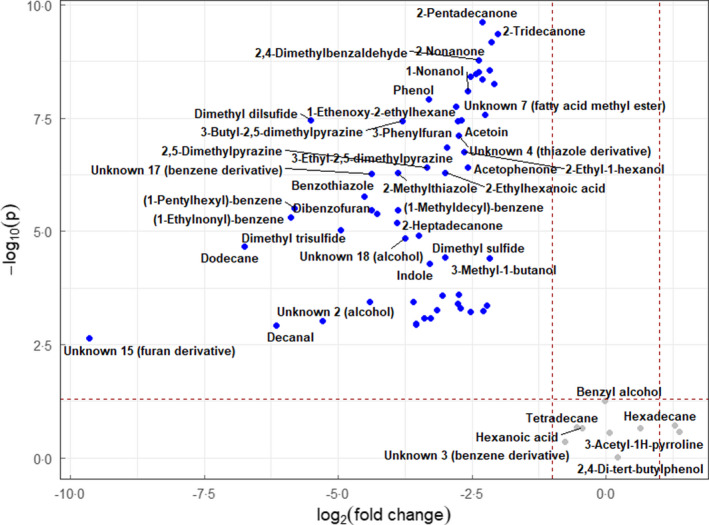
Volcano plot of volatile organic compounds detected in Escherichia coli BL21 strain in 10 μg ml−1 ampicillin‐supplemented media compared with drug‐free control with fold change threshold (x) 2 and t test threshold (y) 0.05. Blue (downregulated), grey (nonsignificant).
The volcano plot in Figure 2 shows differentially expressed VOCs of the BL21‐AmpC strain in the presence and absence of 10 μg ml−1 ampicillin. A total of 26 altered VOCs (fold change >2, p ≤ 0.05) were observed when the AmpC‐producing strain was exposed to 10 μg ml−1 ampicillin compared to when the strain was grown in LB only. Compounds putatively identified as octanoic acid (RT 12.408 min) and tetradecanoic acid (RT 22.623 min) were detected in the headspace of the AmpC‐producing strain but were not previously observed in the pan‐volatilome. Two compounds which exhibited significantly higher levels (p ≤ 0.05) in the treated state (3‐acetyl‐1H‐pyrroline [RT 10.247 min] and an unknown thiazole derivative [RT 11.512 min]) were not previously observed in the volatilome of the untreated resistant strain, although they were detected in the untreated susceptible strain and formed part of the pan‐volatilome.
FIGURE 2.
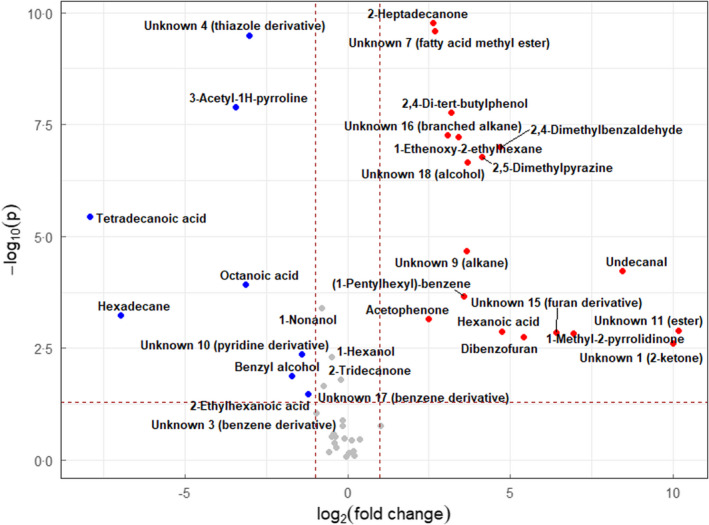
Volcano plot of volatile organic compounds detected in AmpC‐producing Escherichia coli BL21 strain in 10 μg ml−1 ampicillin‐supplemented media compared with drug‐free control with fold change threshold (x) 2 and t tests threshold (y) 0.05. Blue (downregulated), red (upregulated) and grey (nonsignificant).
Principal component analysis was implemented to determine whether the VOC profiles of the resistant strain in the presence and absence of 10 μg ml−1 ampicillin could be distinguished. Figure 3 demonstrates that the 10 μg ml−1 ampicillin‐stressed and unstressed BL21‐AmpC‐producing isolates separate across the first principal component (total explained variance = 50.9%).
FIGURE 3.
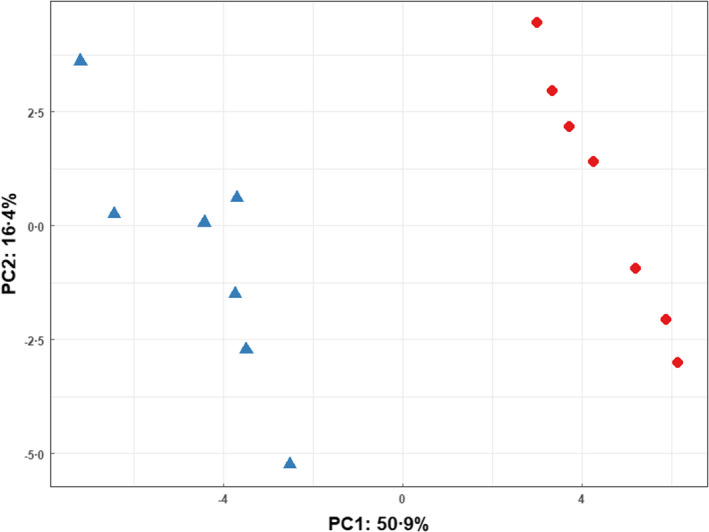
Principal component analysis scores plot comparing AmpC‐producing Escherichia coli BL21 strain in 10 μg ml−1 ampicillin‐supplemented media ( ) with drug‐free media (
) with drug‐free media ( ).
).
Treatment with 2 μg ml−1 ampicillin
The volcano plot (Figure 4) indicates 40 significantly altered VOCs (fold change >2, p ≤ 0.05) between the AmpC‐producing and nonproducing BL21 strains after incubation with 2 μg ml−1 ampicillin. Similar numbers of alcohols, heterocyclics, and aldehydes were detected between the two strains. The resistant strain showed a greater abundance of hydrocarbons (75%), whereas VOCs of the susceptible strain were enriched for ketones (64%), fatty acids (100%), esters (78%), and benzene‐derivatives (80%).
FIGURE 4.
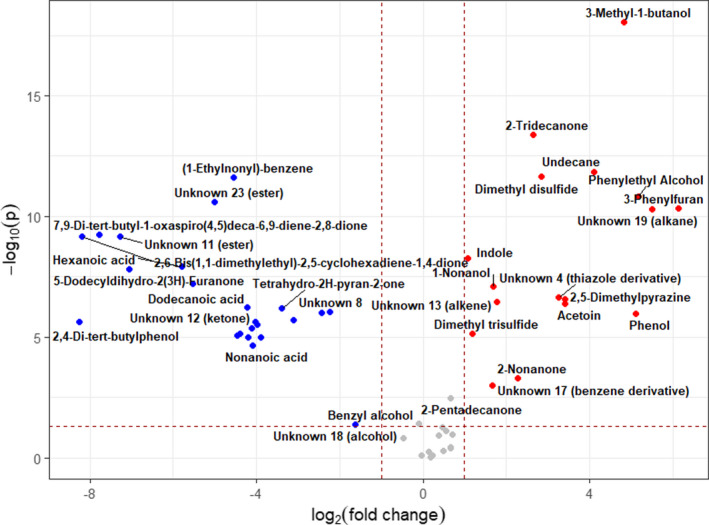
Volcano plot of volatile organic compounds detected in Escherichia coli BL21‐AmpC strain compared with E. coli BL21 strain in 2 μgml−1 ampicillin‐supplemented media with fold change threshold (x) 2 and t tests threshold (y) 0.05. Blue (downregulated), red (upregulated) and grey (nonsignificant).
The heat map generated by HCA utilizing all detected compounds in both treated and untreated conditions (Figure S1) revealed distinct clustering patterns and differences between the four groups (BL21 untreated n = 7, BL21‐AmpC untreated n = 7, BL21 + 2 μg ml−1 ampicillin n = 10, BL21‐AmpC + 2 μg ml−1 ampicillin n = 10). The presence of drug stress in both strains correlated with higher abundances of pyrazine‐related compounds and lower abundances of several core volatiles such as phenol and 2‐pentadecanone. The BL21‐AmpC strain exhibited higher levels of other core volatiles including indole, 3‐methyl‐1‐butanol, dimethyl disulfide and dimethyl trisulfide under drug stress. Alkanes appeared enriched in the untreated BL21 group, whereas fatty acids were seen in greater abundance in the treated BL21 group.
The top six discriminatory compounds identified by HCA were putatively identified as an unknown ester (RT 23.389 min), 3‐methyl‐1‐butanol, undecane (RT 11.112 min), an unknown fatty acid methyl ester (RT 13.535 min), 2‐tridecanone (RT 18.396 min), and 3‐phenylfuran (RT 13.578 min). HCA exclusively employing these six compounds (Figure 5) demonstrated clear differences between the groups.
FIGURE 5.
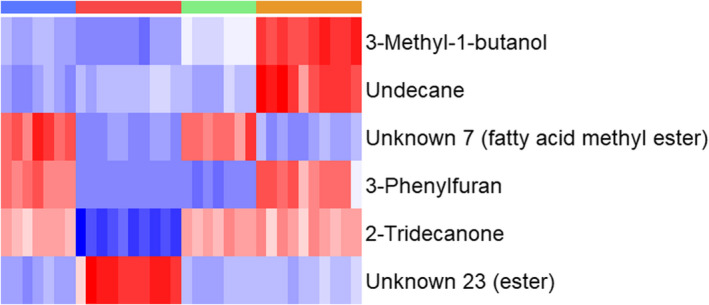
Heat map generated using hierarchical clustering analysis for the differentiation between AmpC‐producing and nonproducing Escherichia coli BL21 strains when treated with and without 2 μg ml−1 ampicillin using the top six identified compounds. Groups are represented in columns left to right by blue: BL21 (untreated), red: BL21 (2 μg ml−1 ampicillin), green: BL21‐AmpC (untreated) and orange: BL21‐AmpC (2 μg ml−1 ampicillin). Coloured cells correspond to compound peak intensities, with relative content for a given compound shown in red or blue to signify high and low values, respectively.
The PCA scores plot (Figure 6) of the 2 μg ml−1 ampicillin‐treated and untreated strains showed excellent separation across the first and second PCs. The untreated strains did not exhibit substantial separation from each other.
FIGURE 6.
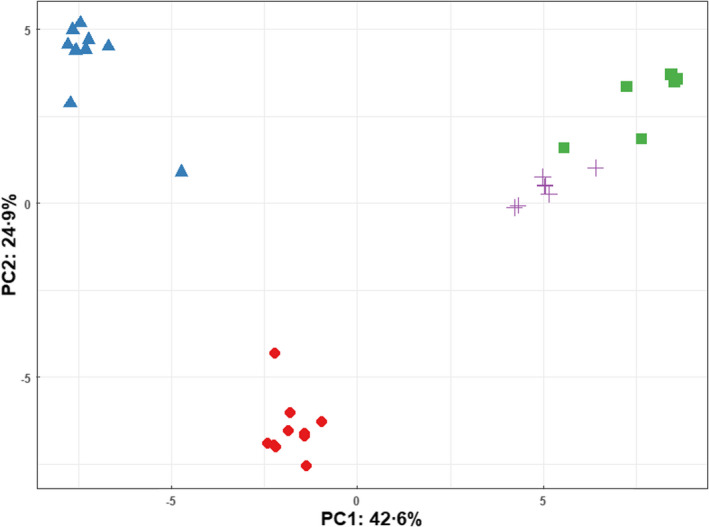
Principal component analysis scores plot comparing Escherichia coli BL21 and BL21‐AmpC strains in the presence and absence of 2 μg ml−1 ampicillin‐supplemented media.  BL21 (2 μg ml−1 ampicillin), + BL21 (untreated),
BL21 (2 μg ml−1 ampicillin), + BL21 (untreated),  BL21‐AmpC (2 μg ml−1 ampicillin),
BL21‐AmpC (2 μg ml−1 ampicillin),  BL21‐AmpC (untreated).
BL21‐AmpC (untreated).
We also sought to evaluate the effect of ampicillin concentration on the number of VOCs detected in the headspace of the two strains. In the untreated condition, the susceptible and resistant strains reported 66 and 42 compounds, respectively. Higher concentrations of ampicillin were correlated with lower numbers of detected VOCs in the susceptible strain (2 μg ml−1 n = 47, 10 μg ml−1 n = 14). However, while the number of detected VOCs decreased overall after subjecting the resistant strain to drug stress compared to the untreated condition, there was no significant difference in the detected number of VOCs between the 2 μg ml−1 (n = 32) and 10 μg ml−1 (n = 30) ampicillin set‐ups.
DISCUSSION
We employed TD‐GC‐MS to assess the VOC profiles associated with AmpC‐producing and nonproducing E. coli, in order to evaluate any differences in the VOC profiles of ampicillin‐resistant and ‐susceptible bacteria under drug stress conditions. We identified several compounds which may be markers of AmpC production and by using machine‐learning algorithms demonstrated that the VOC profiles of ampicillin‐resistant and ‐susceptible bacteria are different. These findings indicate that bacteria with acquired resistance exhibit an altered metabolic phenotype compared to their susceptible counterpart and suggest that VOC profiles may be utilized to differentiate between resistant and susceptible strains of bacteria.
In the absence of drug stress, differences between the volatilomes of the resistant and susceptible strains were observed. Most compounds were found to be present or significantly increased in the susceptible strain and absent or significantly decreased in the resistant strain. However, five compounds were found to be significantly higher in the resistant strain suggesting that the metabolism of this strain is not completely downregulated in comparison. This is indicative of a fundamental biological difference in the metabolisms between the two strains.
Similar to previous findings, many of the compounds which were found to be significantly different between the untreated strains were no longer significantly different after the addition of a sublethal dose of ampicillin (Smart et al., 2019). The acquisition of resistance is known to confer a metabolic burden (Martinez & Rojo, 2011; Millan & Maclean, 2019). Thus, the absence of selection pressures for resistance reduces the competitiveness of the organism harbouring the resistance determinants. This may explain the lower abundance of VOCs detected in the untreated resistant strain as well as the detection of compounds under drug stress which were previously observed only in the untreated susceptible strain.
Three compounds (indole, acetoin and 3‐methyl‐1‐butanol) were observed to be significantly more abundant in the ampicillin‐resistant strain compared to the susceptible strain both in the presence and in the absence of drug stress. As well as having a role in tryptophan biosynthesis, indole has been implicated as a signalling molecule in the bacterial stress response as well as in the regulation of biofilm production (Di Martino et al., 2003; Kuczynska‐Wisnik et al., 2010). Indole signalling has also been shown to induce persistence in E. coli exposed to antibiotics and increase AMR (Vega et al., 2012). Acetoin has been shown to have a regulatory effect on motility and biofilm formation in bacteria (Létoffé et al., 2014). It is produced through the conversion of pyruvate and has been observed to prevent intracellular pH decreasing excessively (Vivijs et al., 2014). Modifications to the amino acid‐derived starter units for fatty acid biosynthesis may yield branched chain compounds, of which branched chain aldehydes are intermediates. For example, the metabolism of leucine may give rise to 3‐methylbutanal. The subsequent reduction by alcohol dehydrogenase may lead to the production of methyl alcohols, for example 3‐methyl‐1‐butanol (Schulz & Dickschat, 2007). Owing to the fact that LB broth contains tryptone and thus a range of amino acids, it is plausible that the high levels of 3‐methyl‐1‐butanol in the resistant strain have arisen due to the metabolism of culture medium amino acids (Filipiak et al., 2013). Our study showed that the two strains had significantly altered levels of VOCs involved in fatty acid biosynthesis and metabolism. The increased prevalence of fatty acids, for example octanoic and tetradecanoic acid in the headspace of the AmpC‐producing strain under drug stress may be explained by the biosynthesis of fatty acids. Conversely, the presence of smaller chain fatty acids in the susceptible strain under drug stress is likely resultant of fatty acid metabolism via the β‐oxidation pathway (Schulz & Dickschat, 2007). A multitude of other compounds are also produced along this pathway including 1‐alcohols, alkanes and 2‐substituted compounds.
To the best of our knowledge, this is the first time that sorptive extraction coupled with GC‐MS has been employed for the analysis of headspace volatiles of AmpC‐producing E. coli. Our experimental design aimed to control for confounding biological factors implicated in clinical isolates such as genetic determinants by utilizing plasmid transformation for inducible ampicillin resistance in BL21 (DE3) cells. Differences in the volatile profiles between the resistant and the susceptible strain may thus be attributed to the expression of ampC rather than a complex interplay of multiple factors as may be seen in clinical isolates. Furthermore, the application of ampicillin at a lower concentration permitted direct comparison between the susceptible and resistant strains without the influence of widespread cell death. This ensured that the experimental methodology detected VOC changes associated with AMR rather than detecting alterations correlated with inhibited metabolism and bacterial death as was observed when the susceptible strain was treated with a high concentration of ampicillin. Nonetheless, experimental groups assessing both low and high antibiotic concentrations provide insight into the effect of antibiotic concentration on the VOC profiles of bacteria. VOC analysis of the bacterial headspace at both levels showed significant differences between the VOC profiles and demonstrated potential applicability for the measurement of antimicrobial susceptibility. This is supported by previous studies which have monitored the growth and susceptibility of resistant and susceptible strains using VOCs (Allardyce et al., 2006; Wiesner et al., 2014).
Our findings suggest that ampicillin‐resistant bacteria may be distinguished from their susceptible counterparts using as few as six compounds within a time frame of <7 h. The development of computational models based on VOC signatures for the detection of AMR shows considerable promise for use in routine clinical diagnostics with little sample preparation and technical expertise required. Given the small sample size, future studies should look to expand the number of strains evaluated and assess other means of ampicillin resistance such as plasmid AmpC or alternative AmpC‐derived enzymes (e.g. CMY‐2). In addition, consideration should be given to clinical isolates as well as biological matrices to ascertain whether the same volatile signatures may also be detected in these typically complex samples. Validation of identified biomarkers will facilitate the development of targeted and rapid detection methods, reducing the incubation and analysis time currently required and promoting the timely identification of AMR‐associated infections.
FUNDING INFORMATION
This work was supported by the Biotechnology and Biological Sciences Research Council (grant numbers BB/T008725/1 and BB/M011208/1). WMA, TF and SJF are supported by the NIHR Manchester Biomedical Research Centre.
CONFLICTS OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
Figure S1
Dixon, B. , Ahmed, W.M. , Mohamed, A.A. , Felton, T. & Fowler, S.J. (2022) Metabolic phenotyping of acquired ampicillin resistance using microbial volatiles from Escherichia coli cultures. Journal of Applied Microbiology, 133, 2445–2456. Available from: 10.1111/jam.15716
REFERENCES
- Ahmed, W.M. , Lawal, O. , Nijsen, T.M. , Goodacre, R. & Fowler, S.J. (2017) Exhaled volatile organic compounds of infection: a systematic review. ACS Infectious Diseases, 3, 695–710. Available from: 10.1021/acsinfecdis.7b00088 [DOI] [PubMed] [Google Scholar]
- Allardyce, R.A. , Hill, A.L. & Murdoch, D.R. (2006) The rapid evaluation of bacterial growth and antibiotic susceptibility in blood cultures by selected ion flow tube mass spectrometry. Diagnostic Microbiology and Infectious Disease, 55, 255–261. Available from: 10.1016/j.diagmicrobio.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Antimicrobial Resistance Collaborators . (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet, 399, 629–655. Available from: 10.1016/s0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belley, A. , Morrissey, I. , Hawser, S. , Kothari, N. & Knechtle, P. (2021) Third‐generation cephalosporin resistance in clinical isolates of Enterobacterales collected between 2016–2018 from USA and Europe: genotypic analysis of β‐lactamases and comparative in vitro activity of cefepime/enmetazobactam. Journal of Global Antimicrobial Resistance, 25, 93–101. Available from: 10.1016/j.jgar.2021.02.031 [DOI] [PubMed] [Google Scholar]
- Bos, L.D.J. , Sterk, P.J. & Schultz, M.J. (2013) Volatile metabolites of pathogens: a systematic review. PLoS Pathogens, 9, 1–8. Available from: 10.1371/journal.ppat.1003311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroff, N. , Espaze, E. , Bérard, I. , Richet, H. & Reynaud, A. (1999) Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum β‐lactamase production. FEMS Microbiology Letters, 173, 459–465. Available from: 10.1016/S0378-1097(99)00111-1 [DOI] [PubMed] [Google Scholar]
- Di Martino, P. , Fursy, R. , Bret, L. , Sundararaju, B. & Phillips, R.S. (2003) Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole‐producing bacteria. Canadian Journal of Microbiology, 49, 443–449. Available from: 10.1139/W03-056 [DOI] [PubMed] [Google Scholar]
- Drabińska, N. , de Lacy Costello, B. , Hewett, K. , Smart, A. & Ratcliffe, N. (2019) From fast identification to resistance testing: volatile compound profiling as a novel diagnostic tool for detection of antibiotic susceptibility. Trends in Analytical Chemistry, 115, 1–12. Available from: 10.1016/j.trac.2019.03.019 [DOI] [Google Scholar]
- Drabińska, N. , Hewett, K. , White, P. , Avison, M.B. , Persad, R. , Ratcliffe, N.M. et al. (2022) Application of a solid‐phase microextraction‐gas chromatography‐mass spectrometry/metal oxide sensor system for detection of antibiotic susceptibility in urinary tract infection‐causing Escherichia coli – a proof of principle study. Advances in Medical Sciences, 67, 1–9. Available from: 10.1016/j.advms.2021.09.001 [DOI] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing . (2021) Breakpoint tables for interpretation of MICs and zone diameters, version 11.0.
- Filipiak, W. , Sponring, A. , Filipiak, A. , Baur, M. , Troppmair, J. , Amann, A. , et al. (2013) Volatile organic compounds (VOCs) released by pathogenic microorganisms in vitro: potential breath biomarkers for early‐stage diagnosis of disease. In: Davis, C. & Beauchamp, J. (Eds.) Volatile biomarkers: non‐invasive diagnosis in physiology and medicine. Amsterdam, The Netherlands: Elsevier, pp. 463–512. Available from: 10.1016/B978-0-44-462613-4.00023-4 [DOI] [Google Scholar]
- Hamzaoui, Z. , Ocampo‐Sosa, A. , Fernandez Martinez, M. , Landolsi, S. , Ferjani, S. , Maamar, E. et al. (2018) Role of association of OmpK35 and OmpK36 alteration and bla ESBL and/or bla AmpC genes in conferring carbapenem resistance among non‐carbapenemase‐producing Klebsiella pneumoniae . International Journal of Antimicrobial Agents, 52, 898–905. Available from: 10.1016/j.ijantimicag.2018.03.020 [DOI] [PubMed] [Google Scholar]
- IUPAC . (1997) In: McNaught, A.D. & Wilkinson, A. (Eds.) Compendium of chemical terminology (the ‘gold book’), 2nd edition. Oxford: Blackwell Scientific Publications. Online version (2019‐) created by Chalk, S. J. Available from: 10.1351/goldbook.r05360 [DOI] [Google Scholar]
- Khan, H.A. , Ahmad, A. & Mehboob, R. (2015) Nosocomial infections and their control strategies. Asian Pacific Journal of Tropical Biomedicine, 5, 509–514. Available from: 10.1016/j.apjtb.2015.05.001 [DOI] [Google Scholar]
- Kidd, J.M. , Livermore, D.M. & Nicolau, D.P. (2020) The difficulties of identifying and treating Enterobacterales with OXA‐48‐like carbapenemases. Clinical Microbiology and Infection, 26, 401–403. Available from: 10.1016/j.cmi.2019.12.006 [DOI] [PubMed] [Google Scholar]
- Kuczynska‐Wisnik, D. , Matuszewska, E. , Furmanek‐Blaszk, B. , Leszczyńska, D. , Grudowska, A. , Szczepaniak, P. , et al. (2010) Antibiotics promoting oxidative stress inhibit formation of Escherichia coli biofilm via indole signalling. Research in Microbiology, 161(10), 847–853. Available from: 10.1016/j.resmic.2010.09.012 [DOI] [PubMed] [Google Scholar]
- Lemfack, M.C. , Gohlke, B.O. , Toguem, S.M.T. , Preissner, S. , Piechulla, B. & Preissner, R. (2018) mVOC 2.0: a database of microbial volatiles. Nucleic Acids Research, 46, D1261–D1265. Available from: 10.1093/nar/gkx1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létoffé, S. , Audrain, B. , Bernier, S.P. , Delepierre, M. & Ghigo, J. (2014) Aerial exposure to the bacterial volatile compound trimethylamine modifies antibiotic resistance of physically separated bacteria by raising culture medium pH. mBio, 5, e00913. Available from: 10.1128/mBio.00944-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J.L. & Rojo, F. (2011) Metabolic regulation of antibiotic resistance. FEMS Microbiology Reviews, 35, 768–789. Available from: 10.1111/j.1574-6976.2011.00282.x [DOI] [PubMed] [Google Scholar]
- Millan, A.S. & Maclean, R.C. (2019) Fitness costs of plasmids: a limit to plasmid transmission. Microbiology Spectrum, 5, MTBP0016‐2017. Available from: 10.1128/microbiolspec.MTBP-0016-2017 [DOI] [PubMed] [Google Scholar]
- R Core Team . (2021) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rees, C.A. , Burklund, A. , Stefanuto, P.H. , Schwartzman, J.D. & Hill, J.E. (2018) Comprehensive volatile metabolic fingerprinting of bacterial and fungal pathogen groups. Journal of Breath Research, 12, 026001. Available from: 10.1088/1752-7163/aa8f7f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, C.A. , Nasir, M. , Smolinska, A. , Lewis, A.E. , Kane, K.R. , Kossmann, S.E. et al. (2018) Detection of high‐risk carbapenem‐resistant Klebsiella pneumoniae and Enterobacter cloacae isolates using volatile molecular profiles. Scientific Reports, 8, 13297. Available from: 10.1038/s41598-018-31543-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, S. & Dickschat, J.S. (2007) Bacterial volatiles: the smell of small organisms. Natural Product Reports, 24, 814–842. Available from: 10.1039/b507392h [DOI] [PubMed] [Google Scholar]
- Sethi, S. , Nanda, R. & Chakraborty, T. (2013) Clinical application of volatile organic compound analysis for detecting infectious diseases. Clinical Microbiology Reviews, 26, 462–475. Available from: 10.1128/CMR.00020-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, A. , de Lacy Costello, B. , White, P. , Avison, M. , Batty, C. , Turner, C. et al. (2019) Sniffing out resistance – rapid identification of urinary tract infection‐causing bacteria and their antibiotic susceptibility using volatile metabolite profiles. Journal of Pharmaceutical and Biomedical Analysis, 167, 59–65. Available from: 10.1016/j.jpba.2019.01.044 [DOI] [PubMed] [Google Scholar]
- Trubiano, J.A. & Padiglione, A.A. (2015) Nosocomial infections in the intensive care unit. Anaesthesia and Intensive Care Medicine, 16, 598–602. Available from: 10.1016/j.mpaic.2015.09.010 [DOI] [Google Scholar]
- Vega, N.M. , Allison, K.R. , Khalil, A.S. & Collins, J.J. (2012) Signaling‐mediated bacterial persister formation. Nature Chemical Biology, 8, 431–433. Available from: 10.1038/nchembio.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivijs, B. , Moons, P. , Aertsen, A. & Michiels, C.W. (2014) Acetoin synthesis acquisition favors Escherichia coli growth at low pH. Applied and Environmental Microbiology, 80, 6054–6061. Available from: 10.1128/AEM.01711-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, W.E. (2014) NIST mass spectral library (NIST 14). In: Linstrom, P.J. & Mallard, W.G. (Eds.) NIST chemistry webbook, NIST standard reference database. Gaithersburg, MD: National Institute of Standards and Technology. Available from: 10.18434/T4H594 [DOI] [Google Scholar]
- Weiner‐Lastinger, L.M. , Abner, S. , Edwards, J.R. , Kallen, A.J. , Karlsson, M. , Magill, S.S. et al. (2020) Antimicrobial‐resistant pathogens associated with adult healthcare‐associated infections: summary of data reported to the National Healthcare Safety Network, 2015–2017. Infection Control & Hospital Epidemiology, 41, 1–18. Available from: 10.1017/ice.2019.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner, K. , Jaremek, M. , Pohle, R. , Von Sicard, O. & Stuetz, E. (2014) Monitoring of bacterial growth and rapid evaluation of antibiotic susceptibility by headspace gas analysis. Procedia Engineering, 87, 332–335. Available from: 10.1016/j.proeng.2014.11.750 [DOI] [Google Scholar]
- Wiskirchen, D.E. , Nordmann, P. , Crandon, J.L. & Nicolau, D.P. (2014) Efficacy of humanized carbapenem and ceftazidime regimens against Enterobacteriaceae producing OXA‐48 carbapenemase in a murine infection model. Antimicrobial Agents and Chemotherapy, 58, 1678–1683. Available from: 10.1128/AAC.01947-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1


