Abstract
Lipopolysaccharide (LPS) derived from the periodontal pathogen Porphyromonas gingivalis has been reported to differ structurally and functionally from enterobacterial LPS. These studies demonstrate that in contrast to protein-free enterobacterial LPS, a similarly purified preparation of P. gingivalis LPS exhibited potent Toll-like receptor 2 (TLR2), rather than TLR4, agonist activity to elicit gene expression and cytokine secretion in murine macrophages and transfectants. More importantly, TLR2 stimulation by this P. gingivalis LPS preparation resulted in differential expression of a panel of genes that are normally induced in murine macrophages by Escherichia coli LPS. These data suggest that (i) P. gingivalis LPS does not signal through TLR4 and (ii) signaling through TLR2 and through TLR4 differs quantitatively and qualitatively. Our data support the hypothesis that the shared signaling pathways elicited by TLR2 and by TLR4 agonists must diverge in order to account for the distinct patterns of inflammatory gene expression.
Lipopolysaccharides (LPS) are among the most potent inflammatory bacterial mediators and have been strongly implicated in the inflammatory response associated with gram-negative sepsis. Most LPS signaling studies have used LPS preparations derived from species within the Enterobacteriaceae, which possess relatively well-conserved lipid A structures (reviewed in reference 36). A convergence of data suggest that these prototypic LPS preparations, when highly purified, elicit LPS responses that are restricted in the use of TLR4 as the principal signal-transducing molecule (reviewed in reference 21), which is strongly supported by the finding that synthetic E. coli lipid A activated Toll-like receptor 4 (TLR4) and not TLR2 transfectants (8). However, the lipid A of nonenterobacterial species, e.g., Porphyromonas gingivalis, which has been implicated in the inflammation associated with chronic periodontitis (reviewed in reference 9), differs both structurally and functionally from enterobacterial lipid A. Specifically, the major species of P. gingivalis lipid A is composed of unique branched fatty acids, with longer carbon chains than in enterobacterial lipid A, the absence of a phosphoryl group at position 4′ of the nonreducing glucosamine, as well as other modifications (Fig. 1) (1). Consistent with these structural differences is the finding that P. gingivalis LPS activity is poorly inhibited by polymyxin B (12), which has been postulated to inactivate LPS by binding electrostatically to negatively charged phosphate groups, leading to a subsequent interaction of polymyxin B with the hydrophobic fatty acids (25, 33). Although P. gingivalis-induced signaling was shown some time ago to be CD14 dependent (34), site-specific mutagenesis of CD14 suggests that the substitution of certain charged amino acids differentially affects the abilities of Escherichia coli and P. gingivalis LPS to bind CD14 (4, 5). In addition, binding of P. gingivalis LPS to LPS binding protein has been reported to be 100-fold less than observed for E. coli LPS (9). In vivo, P. gingivalis LPS has been reported to be much less toxic than other LPS preparations (reviewed in reference 27). P. gingivalis LPS has also been shown to be active on C3H/HeJ macrophages (12, 32), which possess a point mutation in tlr4 that precludes signaling by enterobacterial LPS (24, 26). In contrast, Tabeta et al. (30) reported that human gingival fibroblasts exhibit a slight increase above basal interleukin-6 (IL-6) production upon stimulation with P. gingivalis LPS and that a monoclonal antibody directed against human TLR4 reduced the IL-6 level below that of the medium-treated cells. Differences in cytokine gene expression or secretion by P. gingivalis LPS and enterobacterial LPS preparations have also been reported for both myeloid and nonmyeloid cell types (4, 11, 12, 32).
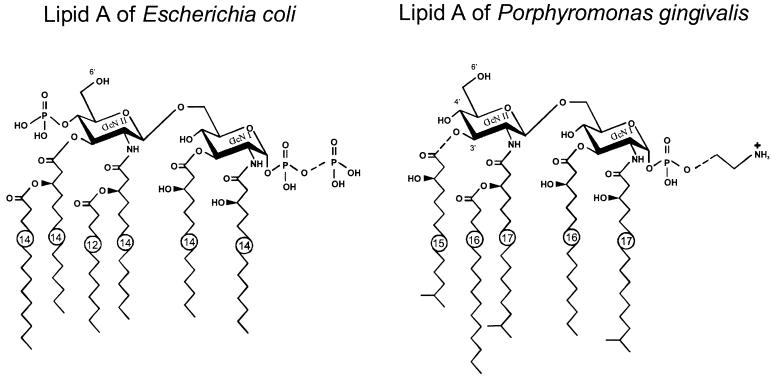
FIG. 1.
(A) Chemical structures of the major species of lipid A produced by E. coli and P. gingivalis. The major structural differences include the nature and number of fatty acids, presence or absence of the second phosphate in position 4′, and substitution of the position 1 phosphate. A more extensive and detailed comparison is presented in reference 36.
Having purified P. gingivalis LPS by the same protocol as used to demonstrate the restricted usage of TLR4 by enterobacterial LPS to mediate signaling (8), we demonstrate herein that a preparation of P. gingivalis LPS utilizes TLR2, not TLR4, to mediate inflammatory signaling. In addition, P. gingivalis LPS differentially activates a panel of genes relative to E. coli LPS, suggesting that TLR2- and TLR4-mediated signaling pathways diverge.
MATERIALS AND METHODS
Mice.
C3H/OuJ and C3H/HeJ mice were purchased from the Jackson Laboratory, (Bar Harbor, Maine). Thioglycolate-elicited peritoneal exudate macrophages were cultured as described previously (29).
LPS preparations.
E. coli K235 LPS was prepared by a modification of the phenol water extraction method of McIntire et al. (20) (<0.008% protein). E. coli J5 (Rc) LPS was purchased from List Biological Laboratories (Campbell, Calif.) and was subjected to phenol reextraction (repurified) by a method that results in the elimination of contaminants that are active on C3H/HeJ macrophages (17, 19). This reextraction method, detailed more recently by Hirschfeld et al. (8), results in enterobacterial LPS preparations that utilize TLR4, and not TLR2, for signaling. Briefly, 5 mg of Rc LPS was resuspended in 1 ml of room temperature, endotoxin-free water containing 0.2% triethylamine (TEA). The sample was split into two 500-μl aliquots, and one aliquot was stored at 4°C without further manipulation (unpurified LPS). Deoxycholate was added to the remaining aliquot to a final concentration of 0.5%, followed by the addition of 500 μl of water-saturated phenol. The sample was vortexed intermittently for 5 min, and the phases were allowed to separate at room temperature for 5 min. The sample was placed on ice for 5 min and then centrifuged at 4°C for 2 min at 10,000 × g. The top aqueous layer was transferred to a new tube, and the phenol phase was subjected to reextraction with 500 μl of 0.2% TEA–0.5% deoxycholate. The aqueous phases were pooled and reextracted with 1 ml of water-saturated phenol. The pooled aqueous phases were adjusted to 75% ethanol and 30 mM sodium acetate and were allowed to precipitate at −20°C for 1 h. The precipitate was centrifuged at 4°C for 10 min at 10,000 × g, washed in 1 ml of cold 100% ethanol, and air dried. The precipitate was resuspended in the original volume (500 μl) of 0.2% TEA. One hundred percent recovery was assumed for the purified LPS sample (19), which will be referred to as purified LPS. P. gingivalis 33277 LPS was purified by two rounds of hot phenol extraction (22), followed by phenol reextraction (8, 17, 19). Colloidal gold staining was carried out using a kit (Enhanced Colloidal Gold; Bio-Rad, Hercules, Calif.), which has a lower limit of sensitivity of 10 to 100 pg protein. Twenty micrograms of P. gingivalis LPS was analyzed by thin-layer chromatography (TLC) using silica gel H plates, developed with chloroform-methanol-water-ammonium hydroxide (50:25:4:2). The plate was sprayed with dichromate solution and charred. Most of the LPS chromatographed at the origin. When the LPS was acid hydrolyzed (0.1 N HCl at 100°C for 25 min) and analyzed by TLC, most of the lipid A migrated off the origin, consistent with an LPS preparation that is >95% pure.
Isolation of total cellular RNA and RT-PCR.
All procedures for detection of cytokine and chemokine mRNA by semiquantitative RT-PCR have been detailed previously (18, 28). Briefly, total cellular RNA was extracted from macrophage cultures and was reverse transcribed. PCR amplifications were performed on the resultant cDNA for the gene of interest, using specific sense and antisense primers for cytokine mRNA, i.e., IL-1β (35 cycles), tumor necrosis factor alpha (TNF-α) (31 cycles), IL-6 (30 cycles), gamma interferon (IFN-γ) (35 cycles), IL-12 p35 (31 cycles), IL-12 p40 (22 cycles), and for chemokine mRNA, i.e., MIP-1α (27 cycles), MIP-2 (26 cycles), IP-10 (30 cycles), JE (32 cycles), and MCP-5 (29 cycles). The gene encoding hypoxanthine-guanine phosphoribosyltransferase (HPRT) (24 cycles) was included as a housekeeping gene to control for differences in cDNA for each treatment during the amplification reaction. PCR amplification products were electrophoresed on a 1% agarose gel and blotted overnight onto a Nytran membrane. The DNA was then UV cross-linked onto the membrane and baked at 80°C for 2 h. The amplified PCR products were detected by Southern blot analysis using gene-specific oligonucleotide probes labeled with the Amersham 3-oligolabeling and detection systems (Amersham International, Buckinghamshire, England).
Cell lines and transfections.
The human astrocytoma cell lines U87 and U373, were obtained from the American Type Culture Collection (Manassas, Va.). The subclone of the human embryonic kidney epithelial cell line HEK 293 and the constructs for FLAG-tagged human TLR1, TLR2, TLR3, and TLR4, pFLAG control vector, the ELAM-1 luciferase reporter construct, and Rous sarcoma virus–β-galactosidase (RSV-β-Gal) were provided by Tularik (South San Francisco, Calif.) (13). Conditions for transfection of cells have been detailed elsewhere (7, 8). Briefly, 293 cells were cotransfected in six-well plates using a calcium phosphate kit (Clontech, Palo Alto, Calif.) with 2, 0.5, and 0.5 μg of the TLR expression construct, the ELAM-1 luciferase reporter construct, and the RSV-β-Gal construct, respectively, to normalize for transfection efficiency. Cells were grown for 36 h and stimulated with the indicated agonist for an additional 6 h. U87 cells were transfected in 12-well plates using pFx-2 (Invitrogen, Carlsbad, Calif.) with 2 μg of either TLR2 or TLR4 expression construct. Cells were then grown for 24 h in Dulbecco modified Eagle medium (DMEM) with Nutridoma-HU (Boehringer Mannheim, Indianapolis, Ind.) followed by stimulation with agonist for an additional 24 h in DMEM–Nutridoma-HU containing 2% human serum. U373 cells were grown in 24-well plates for 24 h in DMEM with Nutridoma-HU followed by stimulation with agonist for an additional 24 h in DMEM–Nutridoma-HU containing 2% human serum.
Luciferase and cytokine assays.
IL-6 (U87 and U373 cells) and IL-8 (HEK 293) levels were measured by enzyme-linked immunosorbent assay (ELISA; Endogen, Woburn, Mass.). ELISAs specific for total and bioactive murine IL-12 have been detailed elsewhere (29). Murine TNF-α was also measured by ELISA (Genzyme, Cambridge, Mass.). To assay for NF-κB-dependent luciferase activity, transfected 293 cells were lysed using reporter lysis buffer (Promega, Madison, Wis.), and 20 μl of lysate was assayed for both luciferase and β-galactosidase activities using a Dynatec MLX luminometer after incubation in luciferase assay reagent (Promega) and Galacto-Light with light emission accelerator (Tropix, Bedford, Mass.), respectively.
RESULTS AND DISCUSSION
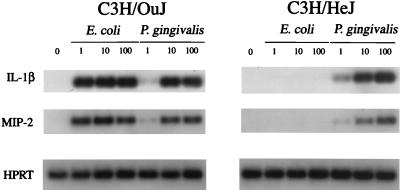
Previous reports that P. gingivalis LPS (i) activates C3H/HeJ macrophages, (ii) differs from enterobacterial LPS in its capacity to elicit a variety of responses, and (iii) possesses a lipid A with a markedly distinct structure led us to evaluate this LPS further upon hot phenol water extraction (22), followed by repurification using a method demonstrated to eliminate TLR2-dependent ligands from several enterobacterial LPS preparations (8). The preparation of P. gingivalis LPS used in this study was confirmed to be essentially protein free, as evidenced by a lack of detectable bands in colloidal gold-stained blots of 10 μg of P. gingivalis LPS preparation (Fig. 2); i.e., the protein concentration of this preparation is <100 pg of protein/10 μg of LPS, or <0.001%, in contrast to 5 μg of the commercial E. coli Rc LPS, which contained clearly detectable protein bands that were eliminated by repurification. Figure 3 illustrates that under conditions where protein-free E. coli K235 LPS completely discriminates between LPS-normoresponsive C3H/OuJ and TLR4-defective C3H/HeJ macrophages with respect to induction of IL-1β and MIP-2 mRNA, P. gingivalis LPS elicits very comparable levels of dose-dependent induction of both genes in macrophages from both strains. Thus, these data confirm previous evidence supporting the hypothesis that stimulation of murine macrophages with P. gingivalis LPS is TLR4 independent (12, 32). Moreover, the potency of the E. coli LPS is clearly greater than that of the P. gingivalis LPS, as evidenced by a clear diminution of signal in those samples stimulated by P. gingivalis versus E. coli LPS at 1 ng/ml.
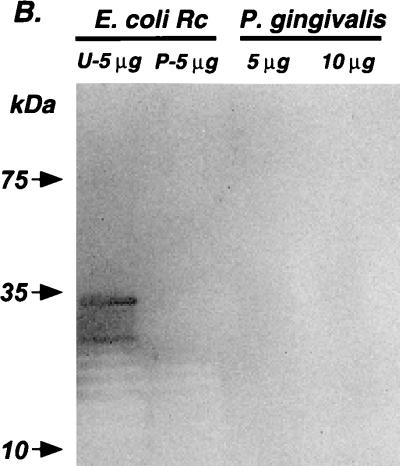
FIG. 2.
Repurified P. gingivalis LPS is not contaminated with endotoxin protein. A commercial preparation of E. coli J5 (Rc) LPS and a preparation of P. gingivalis LPS isolated according to Millar et al. (22) were subjected to a modified phenol reextraction protocol previously shown to eliminate trace endotoxin protein contamination (8, 17, 19). Unpurified (U) or repurified (P) Rc LPS samples are indicated. Five micrograms of both Rc LPS samples and 5 and 10 μg of repurified P. gingivalis LPS were submitted to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Samples were resolved on a 4 to 20% gradient gel and then transferred to a polyvinylidene membrane. Membranes were subsequently stained with colloidal gold.
FIG. 3.
Induction of macrophage gene expression by E. coli K235 LPS and P. gingivalis LPS in C3H/OuJ and C3H/HeJ macrophages. Total macrophage RNA was subjected to RT-PCR with Southern blotting for the detection of IL-1β, MIP-2, and HPRT mRNA as described in the text. HPRT served as the housekeeping gene in these experiments. The data are representative one of three separate experiments.
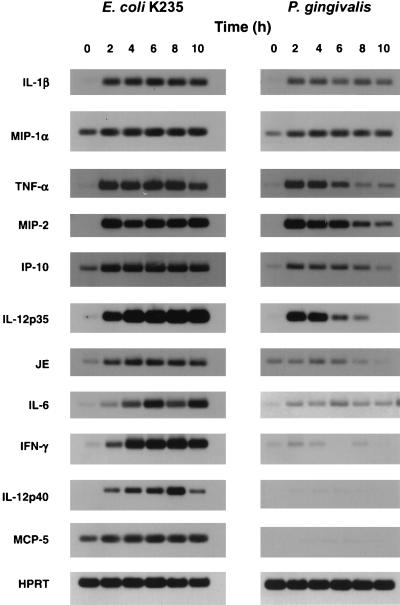
Using 100 ng/ml, a concentration that was superoptimal for induction of expression of both IL-1β and MIP-2 genes by both LPS preparations, we carried out a kinetic analysis and compared induction of 11 cytokine genes in C3H/OuJ macrophages (Fig. 4). Induction of IL-1β and MIP-1α mRNA expression was quite comparable over the time course examined. A second subset of genes, i.e., TNF-α, MIP-2, IP-10, and IL-12 p35, were inducible by the P. gingivalis LPS preparation, but steady-state mRNA levels declined more rapidly than when cells were stimulated with E. coli LPS. Induction of JE and IL-6 mRNA was poor and steady-state levels declined rapidly, while IFN-γ, IL-12 p40, and MCP-5 gene expression was strongly induced by E. coli LPS but not by P. gingivalis LPS. Even at a dose of 1 μg/ml, the P. gingivalis preparation induced MCP-5 only minimally (data not shown). Secretion of TNF-α, total IL-12 (IL-12 p40 plus IL-12 p70), and bioactive IL-12 (IL-12 p70) was markedly attenuated in P. gingivalis- versus E. coli-stimulated cultures (Table 1). Therefore, not only is the P. gingivalis-induced gene expression TLR4 independent, but also such differential gene expression and cytokine secretion implies that there must be qualitative differences in signaling between TLR2 and TLR4.
FIG. 4.
Differential induction of macrophage gene expression by P. gingivalis LPS for a panel of genes normally induced by E. coli K235 LPS. Total RNA was subjected to RT-PCR with Southern blotting for the detection of the specific mRNA species as described in the text. The data represent one of two representative experiments carried out in duplicate. HPRT was used as the housekeeping gene.
TABLE 1.
Induction of cytokines in C3H/OuJ macrophages stimulated with E. coli LPS or with P. gingivalis LPSa
| Cytokine | Level (pg/ml)b
|
||
|---|---|---|---|
| Medium only | E. coli K235 LPS | P. gingivalis LPS | |
| TNF-α (6 h) | <23 | 12,713 ± 3,456 | 757 ± 62 |
| IL-12 p40 + p70 | |||
| 6 h | <15 | 2,701 ± 127 | 150 ± 32 |
| 24 h | <15 | 7,065 ± 438 | 328 ± 25 |
| IL-12 p70 | |||
| 6 h | <15 | 143 ± 2 | <15 |
| 24 h | <15 | 279 ± 20 | <15 |
C3H/OuJ macrophages were cultured at a final density of 4 × 106/well in six-well plates and treated with 2 ml of medium only or medium containing 100 ng of E. coli K235 LPS or P. gingivalis LPS per ml. Supernatants were harvested at 6 or 24 h and assayed by ELISA for TNF-α, total IL-12 (IL-12 p40 plus p70), or bioactive IL-12 p70 as described previously (29).
Arithmetic mean ± standard deviation of duplicate samples.
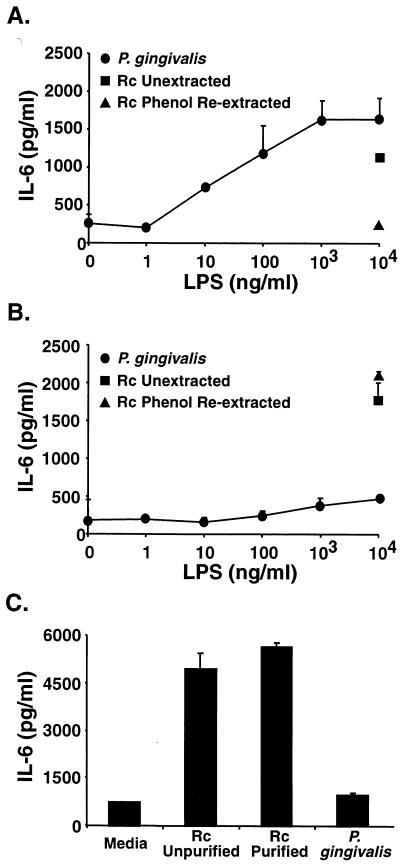
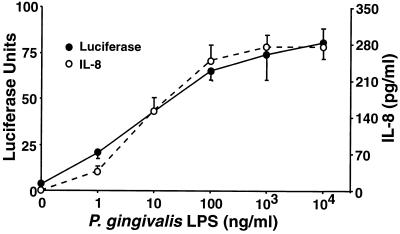
We recently demonstrated that the apparent TLR2 dependency of many LPS preparations is eliminated by phenol reextraction, while TLR4 dependency is retained (8, 17, 19). In contrast, our P. gingivalis preparation, which was confirmed to be equivalently protein free and exhibited an electrophoretic mobility in TLC consistent with LPS, did not appear to stimulate gene expression through TLR4 since IL-1β and MIP-2 genes were expressed in very comparable, dose-dependent fashions in both normal and TLR4-defective macrophages (Fig. 3). Therefore, to confirm the TLR4 independence of the P. gingivalis LPS preparation, the U87 astrocytoma cell line was transiently transfected with either human TLR4 or TLR2 constructs and then stimulated with either E. coli Rc LPS (as purchased or repurified) or the P. gingivalis LPS preparation, repurified identically to the E. coli Rc preparation. Figure 5 illustrates that the P. gingivalis LPS induces IL-6 production in U87 cells that overexpress TLR2 (Fig. 5A) but not TLR4 (Fig. 5B). As recently reported (8), the commercially prepared and repurified E. coli Rc preparations both stimulated TLR4 transfectants, but only the unpurified preparation was active in TLR2 transfectants. These findings were confirmed using a similar astrocytoma cell line, U373, that has been reported to be responsive to E. coli LPS but not to bacterial lipoproteins and to express endogenous TLR4 but not TLR2 mRNA (7). U373 cells secreted IL-6 in response to unpurified or purified E. coli Rc LPS but not in response to the P. gingivalis LPS preparation (Fig. 5C). TLR2 transfection of HEK 293 cells also conferred sensitivity to P. gingivalis LPS, as measured by ELAM-1 luciferase reporter gene expression or IL-8 secretion (Fig. 6), with a dose dependency that closely parallels that seen in the primary macrophages (Fig. 3). In contrast to the P. gingivalis sensitivity conferred upon HEK 293 cells by TLR2 overexpression, transfection with a pFLAG control vector or a TLR1, TLR3, or TLR4 construct failed to render 293 cells sensitive to P. gingivalis LPS (data not shown). Finally, Chinese hamster ovary (CHO) cells engineered to express an ELAM.Tac (CD25) reporter construct and CD14 only (14, 15) failed to respond to this same preparation of P. gingivalis LPS or to the synthetic TLR2 agonist, tripalmitoyl-S-glycerylcysteine-modified Ser Lys4 peptide, but did respond to E. coli lipid A, presumably via endogenous hamster TLR4. In contrast, CHO cells engineered to express the reporter construct, CD14, and human TLR2 (14, 15) responded to all three stimuli to express Tac antigen (H. Heine, personal communication). Thus, in four separate cell lines (transiently or stably transfected) and in primary macrophages, where functional TLR2 and TLR4 expression levels differ, this P. gingivalis LPS preparation initiates intracellular signaling through TLR2, while E. coli LPS selectively utilizes TLR4. That our preparation of P. gingivalis LPS signals via TLR2 is also consistent with two previous reports that Rhodobacter sphaeroides lipid A, which blocks E. coli LPS-induced signaling in a TLR4-dependent fashion (15), fails to block P. gingivalis LPS-induced TNF production (1, 12).
FIG. 5.
TLR2, but not TLR4, confers responsiveness to P. gingivalis LPS. U87 cells transiently transfected with either human TLR2 (A) or human TLR4 (B) or untransfected U373 cells (C) were stimulated for 24 h with unpurified Rc LPS repurified Rc LPS, or repurified P. gingivalis LPS at the indicated concentrations. Supernatants were collected and assayed for IL-6 production by ELISA.
FIG. 6.
Transfection of TLR2 confers responsiveness to P. gingivalis LPS in HEK 293 cells. HEK 293 cells were transiently transfected with human TLR2 plus the ELAM-1 luciferase reporter construct. Cells were stimulated for 6 h with increasing doses of repurified P. gingivalis LPS. Supernatants were analyzed for IL-8 secretion, and cell lysates were analyzed for NF-κB nuclear translocation, expressed in luciferase units.
Our data extend significantly previous observations indicating that P. gingivalis LPS activates macrophages differently from enterobacterial LPS preparations. Clearly, the data presented herein indicate that P. gingivalis LPS does not signal through TLR4, likely accounting for the comparability of induction of IL-1β and MIP-2 mRNA in C3H/OuJ and C3H/HeJ macrophages (Fig. 3). However, we cannot be certain that the TLR2 stimulatory activity detected in the P. gingivalis LPS preparation is attributable to the LPS, despite the fact that the LPS represents the predominant species (>95%). It is possible that a minor contaminant contained within the P. gingivalis LPS preparation is responsible for the TLR2 agonist activity of this preparation. Future experiments using synthetic P. gingivalis lipid A will be required to confirm whether or not P. gingivalis LPS is the true TLR2 agonist.
Faure et al. (6) reported recently that human dermal microvessel or umbilical vein endothelial cells express predominantly TLR4 but very weakly TLR2 and respond vigorously to E. coli LPS but not to Mycobacterium tuberculosis 19-kDa lipoprotein, a TLR2 ligand. Thus, the earlier observation of Cunningham et al. (4) that human endothelial cells responded to produce E-selectin upon stimulation with E. coli but not P. gingivalis LPS are likely explained by our observation that P. gingivalis requires TLR2, and not TLR4, for activation. However, Cunningham et al. (4) also demonstrated that P. gingivalis LPS blocked E. coli LPS-induced E-selectin expression in human umbilical vein endothelial cell cultures in a CD14-independent fashion. Thus, it is possible that P. gingivalis LPS, like R. sphaeroides LPS, is an inactive antagonist of E. coli LPS at the level of TLR4. A second possible mechanism for this inhibition is that engagement of TLR2 by the agonist contained within the P. gingivalis LPS preparation sequesters or exhausts shared signaling molecules (MyD88, etc.) such that subsequent stimulation by E. coli LPS through TLR4 is precluded.
The finding of diminished potency of P. gingivalis versus E. coli LPS for the induction of IL-1β and MIP-2 in C3H/OuJ macrophages (Fig. 3), coupled with differential gene expression (Fig. 4), implies that signaling pathways through TLR2 and TLR4 are quantitatively and/or qualitatively different. One possibility is that the utilization of TLR2 rather than TLR4 results in a more limited capacity to generate intracellular signals required for the expression of certain genes, either through differences related to the strength of signaling through TLR2 versus TLR4 or, perhaps secondarily, through the difference in capacity of TLR2 and TLR4 to recruit additional signaling molecules to the LPS signaling complex. In this regard, it is possible that TLR2 interacts with other TLRs to elicit signaling by our P. gingivalis preparation, as has been recently observed for signaling by Neisseria meningitidis (35). Different affinities of P. gingivalis LPS and enterobacterial LPS for CD14 (5) could also alter the interaction of CD14, once engaged by a particular LPS, with specific TLRs. Regardless of the mechanism, there appears to be a divergence of signaling pathways that results in differential gene expression distal to or distinct from the engagement of shared upstream signaling molecules. This conclusion is also strengthened by our recent observation that another TLR2 agonist, soluble tuberculosis factor (kindly provided by Matthew Fenton) (21), induces IL-1β but not MCP-5 mRNA (V. Toshchakov, unpublished observations).
It is tempting to speculate that the mitigated toxicity of P. gingivalis compared with that of enterobacterial LPS preparations in vivo is secondary to mitigated production of cytokines that have been implicated in endotoxicity (e.g., TNF-α, IL-12, and IFN-γ [reviewed in reference 29). While P. gingivalis can invade epithelial cells and replicate intracellularly (14), the failure of this P. gingivalis LPS preparation to induce IL-12 and IFN-γ mRNA may also contribute to the chronicity of this agent in the pathogenicity of periodontitis, since IL-12 and IFN-γ are necessary for the elimination of many intracellular pathogens, such as Mycobacterium and Listeria species (10). Other bacterial pathogens that produce potent proinflammatory molecules that act through TLR2 but not TLR4 have been identified. These include Borrelia burgdorferi, the agent of Lyme disease, which causes a chronic infection in mice and humans characterized by inflammatory arthritis and produces numerous distinct tripalmitoyl-S-glycerylcysteine-bearing lipoproteins that are TLR2 ligands (2, 3, 7, 16). Several Mycoplasma species, which are also associated with chronic infections and arthritis, produce extremely potent diacylated lipoproteins that interact with TLR2 (23, 31). Although both B. burgdorferi and Mycoplasma species cause chronic diseases, chronicity cannot be attributed solely to signaling through TLR2 since administration of high doses of purified TLR2-dependent bacterial lipoproteins has been associated with acute, toxic-type syndromes (37).
ACKNOWLEDGMENTS
We thank Ulrich Zähringer, Research Center Borstel, Borstel, Germany, for providing Fig. 1 and for helpful discussions about differences in E. coli and P. gingivalis lipid A structures. We also acknowledge Carsten J. Kirschning, Ralf Schwandner, and Holger Wesche for providing the HEK 293 cells and expression constructs of human TLRs, ELAM-1 luciferase, and RSV-β-Gal. Finally, we thank Holger Heine for sharing unpublished data on the activity of our P. gingivalis LPS preparation in CHO cell transfectants.
This work was supported by NIH grants AI-32223 (J.J.W.) and 5P30-CA-42014 (University of Utah Core facilities), DE-08228 (S.M.M.), GM-50870 (N.Q.), and AI-18797 (S.N.V.), and USUHS protocol R07338 (S.N.V.).
REFERENCES
- 1.Aida Y, Kusumoto K, Nakatomi K, Takada H, Pabst M J, Maeda K. An analogue of lipid A and LPS from Rhodobacter sphaeroides inhibits neutrophil responses to LPS by blocking receptor recognition of LPS and by depleting LPS-binding protein in plasma. J Leukoc Biol. 1995;58:675–682. doi: 10.1002/jlb.58.6.675. [DOI] [PubMed] [Google Scholar]
- 2.Aliprantis A O, Yang R B, Mark M R, Suggett S, Devaux B, Radolf J D, Klimpel G R, Godowski P, Zychlinsky A. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 3.Brightbill H D, Libraty D H, Krutzik S R, Yang R B, Belisle J T, Bleharski J R, Maitland M, Norgard M V, Plevy S E, Smale S T, Brennan P J, Bloom B R, Godowski P J, Modlin R L. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham M D, Bajorath J, Somerville J E, Darveau R P. Escherichia coli and Porphyromonas gingivalis lipopolysaccharide interactions with CD14: implications for myeloid and nonmyeloid cell activation. Clin Infect Dis. 1999;28:497–504. doi: 10.1086/515158. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham M D, Shapiro R A, Seachord C, Ratcliffe K, Cassiano L, Darveau R P. CD14 employs hydrophilic regions to “capture” lipopolysaccharides. J Immunol. 2000;164:3255–3263. doi: 10.4049/jimmunol.164.6.3255. [DOI] [PubMed] [Google Scholar]
- 6.Faure E, Equils O, Sieling P A, Thomas L, Zhang F X, Kirschning C J, Polentarutti N, Muzio M, Arditi M. Bacterial lipopolysaccharide activates NF-kappaB through Toll-like receptor 4 (TLR-4) in cultured human dermal endothelial cells. Differential expression of TLR-4 and TLR-2 in endothelial cells. J Biol Chem. 2000;275:11058–11063. doi: 10.1074/jbc.275.15.11058. [DOI] [PubMed] [Google Scholar]
- 7.Hirschfeld M, Kirschning C J, Schwandner R, Wesche H, Weis J H, Wooten R M, Weis J J. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by Toll-like receptor 2. J Immunol. 1999;163:2382–2386. [PubMed] [Google Scholar]
- 8.Hirschfeld M, Ma Y, Weis J H, Vogel S N, Weis J J. Cutting edge: repurification of LPS eliminates signaling through both human and murine Toll-like receptor 2. J Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- 9.Holt S C, Kesavalu L, Walker S, Genco C A. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann S H E. Immunity to intracellular bacteria. In: Paul W E, editor. Fundamental immunology. 4th ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1999. pp. 1335–1371. [Google Scholar]
- 11.Kent L W, Rahemtulla F, Hockett R D, Gilleland R C, Michalek S M. Effect of lipopolysaccharide and inflammatory cytokines on interleukin-6 production by healthy human gingival fibroblasts. Infect Immun. 1998;66:608–614. doi: 10.1128/iai.66.2.608-614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirikae T, Nitta T, Kirikae F, Suda Y, Kusumoto S, Qureshi N, Nakano M. Lipopolysaccharides (LPS) of oral black-pigmented bacteria induce tumor necrosis factor production by LPS-refractory C3H/HeJ macrophages in a way different from that of Salmonella LPS. Infect Immun. 1999;67:1736–1742. doi: 10.1128/iai.67.4.1736-1742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschning C J, Wesche H, Ayrers T M, Rothe M. Human Toll-like receptor 2 confers responsiveness to bacterial lipipolysaccharide. J Exp Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamont R J, Chan A, Belton C M, Izutsu K T, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lien E, Means T K, Heine H, Yoshimura A, Kusumoto S, Fukase K, Fenton M J, Oikawa M, Qureshi N, Monks B, Finberg R W, Ingalls R R, Golenbock D T. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J Clin Investig. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lien E, Sellati T J, Yoshimura A, Flo T H, Rawadi G, Finberg R W, Carroll J D, Espevik T, Ingalls R R, Radolf J D, Golenbock D T. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 17.Manthey C L, Perera P-Y, Henricson B E, Hamilton T A, Qureshi N, Vogel S N. Endotoxin-induced early gene expression in C3H/HeJ (Lpsd) macrophages. J Immunol. 1994;153:2653–2663. [PubMed] [Google Scholar]
- 18.Manthey C L, Perera P-Y, Salkowski C A, Vogel S N. Taxol provides a second signal for murine macrophage tumoricidal activity. J Immunol. 1994;152:825–831. [PubMed] [Google Scholar]
- 19.Manthey C L, Vogel S N. Elimination of trace endotoxin protein from rough chemotype LPS. J Endotoxin Res. 1994;1:84–92. [Google Scholar]
- 20.McIntire F C, Sievert H W, Barlow G H, Finley R A, Lee A Y. Chemical, physical, biological properties of a lipopolysaccharide from Escherichia coli K-235. Biochemistry. 1967;6:2363–2372. doi: 10.1021/bi00860a011. [DOI] [PubMed] [Google Scholar]
- 21.Means T K, Wang S, Lien E, Yoshimura A, Golenbock D T, Fenton M J. Human Toll-like receptors mediate cellular activation by Mycobacterium tuberculosis. J Immunol. 1999;163:3920–3927. [PubMed] [Google Scholar]
- 22.Millar S J, Goldstein E G, Levine M J, Hausmann E. Modulation of bone metabolism by two chemically distinct lipopolysaccharide fractions from Bacteroides gingivalis. Infect Immun. 1986;51:302–306. doi: 10.1128/iai.51.1.302-306.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muhlradt P F, kiess M, Meyer H, Sussmuth S, Jung G. Structure and specific activity of macrophage-stimulating lipopeptides from Mycoplasma hyorhinis. Infect Immun. 1998;66:4804–4810. doi: 10.1128/iai.66.10.4804-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in the Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 25.Pristovsek P, Kidric J. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J Med Chem. 1999;42:4604–4613. doi: 10.1021/jm991031b. [DOI] [PubMed] [Google Scholar]
- 26.Qureshi S T, Lariviere L, Leveque G, Clermont S, Moore K J, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) J Exp Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts F A, Richardson G J, Michalek S M. Effects of Porphyromonas gingivalis and Escherichia coli lipopolysaccharides on mononuclear phagocytes. Infect Immun. 1997;65:3248–3254. doi: 10.1128/iai.65.8.3248-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salkowski C A, Detore G, Franks A, Falk M C, Vogel S N. Pulmonary and hepatic gene expression following cecal ligation and puncture: monophosphoryl lipid A prophylaxis attenuates sepsis-induced cytokine and chemokine expression and neutrophil infiltration. Infect Immun. 1998;66:3569–3578. doi: 10.1128/iai.66.8.3569-3578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salkowski C A, Kopydlowski K, Blanco J, Cody M J, McNally R, Vogel S N. IL-12 is dysregulated in macrophages from IRF-1 and IRF-2 knockout mice. J Immunol. 1999;163:1529–1536. [PubMed] [Google Scholar]
- 30.Tabeta K, Yamazaki K, Akashi S, Miyake K, Kumada H, Umemoto T, Yoshie H. Toll-like receptors confer responsiveness to lipopolysaccharide from Porphyromonas gingivalis in human gingival fibroblasts. Infect Immun. 2000;68:3731–3735. doi: 10.1128/iai.68.6.3731-3735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi O, Kaufmann A, Grote K, Kawai T, Hoshino K, Morr M, Mühlradt P F, Akira S. Cutting edge: preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a Toll-like receptor-2- and MyD88-dependent signaling pathway. J Immunol. 2000;164:554–557. doi: 10.4049/jimmunol.164.2.554. [DOI] [PubMed] [Google Scholar]
- 32.Tanamoto K, Azumi S, Haishima Y, Kumada H, Umemoto T. The lipid A moiety of Porphyromonas gingivalis lipopolysaccharide specifically mediates the activation of C3H/HeJ mice. J Immunol. 1997;158:4430–4436. [PubMed] [Google Scholar]
- 33.Vaara M, Vaara T, Jensen M, Helander I, Nurminen M, Rietschel E T, Makela P H. Characterization of the lipopolysaccharide from the polymyxin-resistant pmrA mutants of Salmonella typhimurium. FEBS Lett. 1981;129:145–149. doi: 10.1016/0014-5793(81)80777-6. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe A, Takeshita A, Kitano S, Hanazawa S. CD14-mediated signal pathway of Porphyromonas gingivalis lipopolysaccharide in human gingival fibroblasts. Infect Immun. 1996;64:4488–4494. doi: 10.1128/iai.64.11.4488-4494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyllie D H, Smith S C, Visintin A, Kiss-Toth E. E, Boussouf S, Segal D M, Duff G W, Dower S K. Evidence for an accessory protein function for TLR-1 in anti-bacterial responses. J Immunol. 2000;165:7125–7132. doi: 10.4049/jimmunol.165.12.7125. [DOI] [PubMed] [Google Scholar]
- 36.Zähringer U, Lindner B, Rietschel E T. Chemical structure of lipid A: recent advances in structural analysis of biologically active molecules. In: Brade H, Opal S M, Vogel S N, Morrison D C, editors. Endotoxin in health and disease. New York, N.Y: Marcel Dekker, Inc; 1999. pp. 93–114. [Google Scholar]
- 37.Zhang H, Peterson J W, Niesel D W, Klimpel G R. Bacterial lipoprotein and lipopolysaccharide act synergistically to induce lethal shock and proinflammatory cytokine production. J Immunol. 1997;159:4868–4878. [PubMed] [Google Scholar]