Abstract
Based on morphological and molecular analyses, five families have been recognized within the crustose brown algal order Ralfsiales. Our morphological and molecular sequence data were used to assess the establishment and phylogenetic relationship of Sungminia gen. nov. Phylogenies based on rbcL and concatenated rbcL and COI‐5P genes support the recognition of Sungminia composed of three distinct lineages, Sungminia gladiata sp. nov., S. pyriformis sp. nov., and S. asiatica sp. nov. We consider that the Sungminia group is clearly distinct at the family level and propose to place Sungminia in a new family, the Sungminiaceae fam. Nov. Our phylogenetic analyses show that the Sungminiaceae forms a strongly supported monophyletic clade with probable sister relationship to the Mesosporaceae. The Sungminiaceae is characterized by perithallial erect filaments moderately adhered, the rod‐shaped perithallial erect filaments, plurangia terminated with single sterile cell, and unangia terminally inserted on 1–2 celled stalk that is lateral‐basal or sessile to a paraphysis.
Keywords: COI‐5P, phylogeny, Ralfsiales, rbcL, Sungminia asiatica sp. nov., Sungminia gen. Nov., Sungminia gladiata sp. nov., Sungminia pyriformis sp. nov., Sungminiaceae fam. Nov., taxonomy
Abbreviations
- BI
Bayesian inference
- BS
Bootstrap
- COI‐5P
5′ region of the cytochrome oxidase subunit 1 gene
- CUK
herbarium of Chosun University
- GTR
General Time Reversible
- MABIK
Marine Biodiversity Institute of Korea
- ML
Maximum Likelihood
- PP
Posterior probability
- RAxML
Randomized Axelerated Maximum Likelihood
The brown algal order Ralfsiales was first described based on morphological taxonomy (Nakamura 1972). After numerous studies questioned the validity of this order (Tanaka and Chihara 1980, 1982, Nelson 1982, Kawai 1989, Silva and de Reviers 2000), the Ralfsiales has been recognized as a distinct taxonomic group by Lim et al. (2007) based on morphology and molecular data (rbcL gene sequences). The Ralfsiales has been recognized by emended morphological characters: (i) discoid early development of the thallus, (ii) one to several plate‐ or cup‐shaped chloroplasts without pyrenoids, (iii) plurangia with sterile terminal cell(s) and terminal unangia and (iv) presence of crustose gametophytic or sporophytic stages in their life history (Lim et al. 2007). The classification system of the Ralfsiales at the familial level has been added and revised several times (Tanaka and Chihara 1982, Lim et al. 2007, León‐Alvarez et al. 2017, Parente et al. 2021).
The principal morphological characters delineating families within the Ralfsiales are the number of plastids per cell, distinct delineation of cortex and medulla, the number of sterile cells on plurangia, and paraphyses in unangia (Tanaka and Chihara 1982, Lim et al. 2007, León‐Alvarez et al. 2017, Parente et al. 2021). However, due to a lack of robust taxonomic characteristics, especially in vegetative samples, it is difficult to identify crustose species correctly and recognize their taxonomic placement within the Ralfsiales based on traditional morpho‐anatomical characteristics (Parente and Saunders 2019). Recently, molecular analyses have been used to infer phylogenetic relationships among genera within the Ralfsiales and to circumscribe their higher‐level rank (Lim et al. 2007, León‐Alvarez et al. 2017, Parente et al. 2021). The rbcL gene has been considered to have the most suitable resolution for discerning the ordinal and familial phylogenetic relations within the Phaeophyceae (Sasaki et al. 2001, Kawai and Sasaki 2004, Draisma et al. 2010). Currently, five families have been recognized within the Ralfsiales based on morphological and molecular analyses: Hapalospongidiaceae, Mesosporaceae, Neoralfsiaceae, Pseudoralfsiaceae, and Ralfsiaceae (Farlow 1881, Lim et al. 2007, Poong et al. 2013, 2014, 2017, León‐Alvarez et al. 2017, Parente et al. 2021).
Currently, 10 species within the Ralfsiales have been reported from Korea (Lee and Kang 1986, Lee and Kang 2001, Keum 2010, Oteng'o and Won 2020, Oteng'o et al. 2021). Although most crustose brown algal species have been known as species of Ralfsia and Neoralfsia based on their morphology (Lee and Kang 1986, Lee and Kang 2001, Lee 2008, Keum 2010), Oteng'o et al. (2021) recently described two new Endoplura species, E. jejuensis and E. koreana, within the Ralfsiaceae from Korea based on morphological and molecular analyses. We collected some unidentified samples of Ralfsia‐like crustose brown algae along the coastlines of Korea from 2017 to 2020. Using molecular and morphological analyses, we analyze these crusts in comparison with known members of the Ralfsiales to elucidate their taxonomy and relationships.
MATERIALS AND METHODS
Specimen collections
Samples were collected from intertidal areas along the east, west, and south coastlines of Korea from 2017 to 2020. Vouchers were air‐dried with fragments preserved in silica gel for molecular and anatomical analyses. Representative voucher specimens examined in this study were deposited in the herbarium of Chosun University (CUK) and Marine Biodiversity Institute of Korea (MABIK), Korea.
DNA extractions, PCR amplification, and sequencing
Genomic DNA was extracted using the NucleoSpin Plant II Kit (Macherey‐Nagel, Düren, Germany) according to the manufacturer's protocol. The extracted DNA was stored at −20°C and used to amplify the rbcL and COI‐5P genes. All polymerase chain reactions (PCRs) were performed using the HelixAmp Ready‐2x‐Go premix (NanoHelix Co., Ltd., Daejeon, Korea) following the manufacturer's protocol. The rbcL gene was amplified and sequenced in two reactions using the primer pairs NDrbcL2‐DRL1R and DRL2F‐R3A (Kogame et al. 1999, Hwang et al. 2005). The mitochondrial COI‐5P gene was amplified and sequenced using GWSFn and GWSRx (Saunders and McDevit 2012). The PCR amplification for the rbcL gene was performed as described by Oteng'o et al. (2021) while that for the COI‐5P gene was performed as described by Saunders and McDevit (2012). The additional rbcL gene and COI‐5P sequences used in the phylogenetic analysis were selected from GenBank (Table S1 in the supporting information).
Sequence and phylogenetic analyses
We analyzed the data sets of the rbcL and the concatenated rbcL and COI‐5P gene sequence data including our sequences and sequences from members of all five extant families within the Ralfsiales to resolve phylogenetic relationships within the order (Table S1). The 34 rbcL and 27 COI‐5P gene data sets were aligned using ClustalW (Thompson et al. 1994). Sargassum muticum and Tilopteris mertensii were selected as outgroups. Before performing the phylogenetic analyses, both the best model of nucleotide or protein evolution and the best combination of partitions were computed using PartitionFinder 2.1.1 (Lanfear et al. 2017). Maximum likelihood (ML) analysis was estimated by the General Time‐Reversible (GTR) + Γ + I model with 1000 bootstrap (BS) replications using RAxMLGUI v1.5 (Silvestro and Michalak 2012). Bayesian inference was performed using MrBayes 3.2.6 (Huelsenbeck and Ronquist 2001, Ronquist and Huelsenbeck 2003). Markov chain Monte Carlo runs were conducted for 2,000,000 generations, each with one cold chain and three heated chains, using the GTR + Γ + I evolutionary model and sampling and printing every 1,000 generations. Summary trees were generated using a burn‐in value of 25%. Phylogenetic trees inferred from concatenated rbcL and COI‐5P gene sequence data sets (Table S1) were constructed to delimit the species, generic, and family boundaries within the Ralfsiales. The phylogenetic trees were constructed using the ML method in RAxML and MrBayes and expressed using FigTree ver. 1.4.0 (Rambaut 2012).
Morphological methods
To document their external morphology related to characters, including color, outline, and surface, the fresh samples were photographed with a waterproof digital camera (Nikon COOLPIX AW100, Nikon Corp., Japan). The samples were detached from the substrate using a single‐edged blade before the analysis of anatomical characters. Squashed and microtome‐sectioned preparations were prepared for each sample. For the microtome‐sectioned preparations, samples were embedded in a matrix (OCT; CellPath, Ltd., Newtown, Wales, UK) and sectioned (8–10 μm thickness) using a freezing microtome (Shandon Cryotome FSE; Thermo Shandon, Ltd., Loughborough, UK). Sectioned and squashed samples were stained with a 1:1 mixture of aqueous aniline blue and acetic acid. Sections were mounted in 50% corn syrup and photographed with a DP‐71 camera (Olympus, Tokyo, Japan) mounted on a BX‐51TRF microscope (Olympus). Digitized images were edited for clarity using Adobe Photoshop software ver. 6.1 (Adobe Systems Inc., San Jose, CA, USA).
RESULTS
Sungminia Oteng'o, Won & T.O. Cho gen. nov.
Description
Dimerous thalli are crusts, mucilaginous, firmly attached to substratum without rhizoids. They are composed of hypothallial basal layer and perithallial erect filaments. The hypothallial basal layer is formed by 1–2 prostrate filaments that are composed of cuboid to elongated cells which give rise to perithallial erect filaments. Perithallial erect filaments are straight or slightly curved, simple or sparsely branched, moderately adhering, and rod‐shaped perithallial erect filaments. Apical vegetative cells are larger than the other cells. Tufts of hair in pits arise from the hypothallial basal layer or form lower to middle portions of perithallial erect filaments. Chloroplasts are one per cell without pyrenoid. Plurangia are uniseriate or biseriate and intercalary with a terminal sterile cell. Unangial reproductive thalli produce filaments of two types, both originating from a perithallial erect cell that branches to give rise to a paraphysis and shorter stalk, which supports a terminal unangium. Unangia are sessile and lateral‐basal to a paraphysis or terminally inserted on 1–2 celled stalk lateral‐basal to a paraphysis. Paraphyses are of clavate shape and composed of 7–20 celled rod‐shaped cells.
Etymology
The name “Sungminia” is in honor of Prof. Sung‐Min Boo of Chungnam National University, Daejeon, Korea, for his outstanding contributions to macroalgal taxonomy and phylogeny.
Type species
Sungminia gladiata
Sungminia gladiata Oteng'o, Won & T.O. Cho sp. nov.
Description
Dimerous thalli are small and irregularly expanded epilithic crusts, light to olive brown in color, mucilaginous, firmly attached to the substratum, lacking rhizoids, and growing to 2.3 cm in diameter and 207–456 μm thick (Fig. 1A). Hypothallial basal layers are formed by 1–2 prostrate filaments that are composed of horizontally elongated cells 7–17 μm long and 5–12 μm wide that give rise to erect perithallial filaments. Erect perithallial filaments are straight, sparsely branched below middle portions, rod‐shaped, and laterally adjoined (Fig. 1B) but can be partially separated with pressure. Apical vegetative cells are 1.2–2.0 times larger than other cells of erect perithallial filaments, 9–22 μm long and 4–12 μm wide (Fig. 1C). Tufts of hairs in pits arise from the hypothallial basal layer or the lower to middle portions of erect perithallial filaments (Fig. 1D). There is one chloroplast per cell without a pyrenoid. Plurangia are uniseriate, 60–107 μm long and 3–8 μm wide, intercalary, and terminated by single sword‐shaped sterile cell measuring 14–25 μm long and 1–8 μm wide (Fig. 1, E and F). Unangial reproductive thalli produce filaments of two types, both originating from erect perithallial cells that branch to give rise to a paraphysis and shorter stalk cell that supports a terminal unangium. Unangia are oblong to obovoid, terminally produced on 1–2 celled stalks, 41–70 μm long and 7–30 μm wide (Fig. 1, G and H). Paraphyses are clavate, 104–162 μm long and 4–21 μm wide, composed of 12–20 cells, and distinguished from erect perithallial filaments by their club shape and having rod‐shaped cells while the latter have cells that are mostly wider than long (Fig. 1H).
Fig. 1.
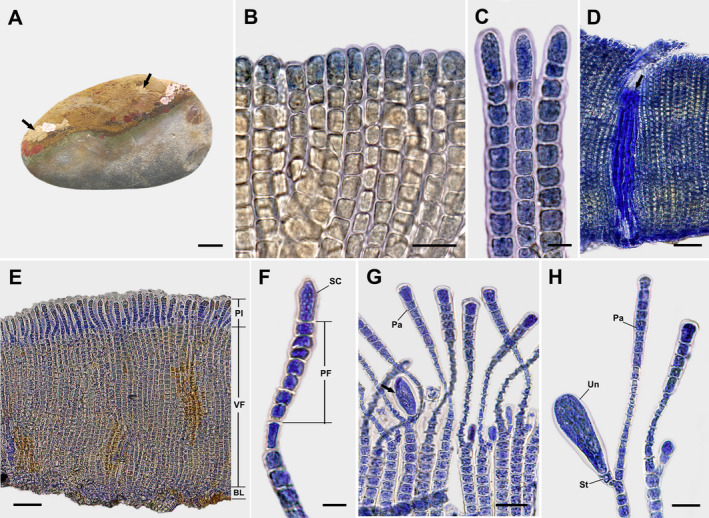
Sungminia gladiata Oteng'o, Won & T.O. Cho sp. nov. (A) Thalli on the rock forming small irregular epilithic olive‐brown crusts (arrows). Scale bar = 2 cm. (B) Longitudinal section view of vegetative thallus showing laterally adjoined erect perithallial filaments. Scale bar = 25 μm. (C) Apical cells of vegetative erect perithallial filaments. Scale bar = 10 μm. (D) Tufts of hairs (arrow) in pits developed from hypothallial basal layer or the lower portions of erect perithallial filaments. Scale bar = 50 μm. (E) Longitudinal section view of reproductive thallus showing plurangia (Pl) and vegetative filaments (VF) developed from the hypothallial basal layer (BL). Scale bar = 50 μm. (F) Plurangium composed of plurangial filament (PF) and terminal sword‐shaped sterile cell (SC). Scale bar = 10 μm. (G) Longitudinal section view of reproductive thallus showing unangia (arrow) and paraphyses (Pa). Scale bar = 25 μm. (H) Unangium (Un) on stalk (St) with paraphysis (Pa). Scale bar = 20 μm.
Holotype (designated here)
MABIK AL00084654 (= CUK19619B), leg. T.O. Cho, S.Y. Jeong, J. Avila, A. Oteng'o, and G.C. Choi, 01 May 2019, Seongsan Ilchul Peak, Jeju Island, Korea, 33°27′38.2″ N, 126°56′05.2″ E, 1–2 m depth.
Isotypes
CUK19594B, CUK19600B, CUK19607B, CUK19630B, CUK19631B.
Etymology
Named for sterile cells that are sword‐shaped (= gladiata L. f., straight or slightly curved with parallel edges and acute apices) and terminate the plurangia.
Other specimen examined
CUK19816B, leg. T.O. Cho & B.Y. Won, 01 August 2019, Oeyeondo, Chungcheongnam‐do, Korea, 36°13′43.4″ N, 126°04′29.1″ E, 1–2 m depth.
Habitat and distribution in Korea
This species is presently known from two locations, Seongsan Ilchul Peak (Jeju Island) and Oeyeondo (Chungcheongnam‐do), growing on rocks and pebbles in the intertidal zone. Plurangia and unangia are found on separate thalli.
Sungminia asiatica Oteng'o, Won & T.O. Cho sp. nov.
Description
Dimerous thalli are irregularly expanded, epilithic crusts, brown to yellowish brown in color, mucilaginous, firmly attached to the substratum, lacking rhizoids, and 0.3–3.3 cm in diameter and 90–611 μm thick (Fig. 2A). The monostromatic hypothallial basal layer is composed of horizontally elongated cells that are 5–16 μm long and 3–12 μm wide and give rise to erect perithallial filaments (Fig. 2, B and E). Perithallial filaments are straight to slightly curved, sparsely branched throughout, rod‐shaped, and laterally adjoined (Fig. 2, B and E) but can be partially separated with pressure. Apical vegetative cells are 1.8–2.5 times larger than other cells of erect perithallial filaments, 10–22 μm long and 4–10 μm wide (Fig. 2C). Tufts of hairs in pits arise from the hypothallial basal layer or the lower to middle portions of erect perithallial filaments (Fig. 2D). There is one chloroplast per cell without a pyrenoid. Plurangia are uniseriate or biseriate, 42–62 μm long and 3–8 μm wide, intercalary, terminated by single dome‐shaped sterile cells measuring 11–16 μm long and 4–7 μm wide (Fig. 2, E and F). Unangial reproductive thalli produce filaments of two types, both originating from erect perithallial cells that branch to give rise to paraphyses and shorter stalk cells that support terminal unangia. Unangia are obovoid to oblong, sessile, and lateral‐basal to a paraphysis or terminally produced on 1–2 celled stalks that are 43–94 μm long and 7–45 μm wide (Fig. 2, G and H). The paraphyses are clavate, 89–157 μm long and 2–18 μm wide, composed of 10–16 cells, and distinguished from erect perithallia filaments by their club shape and having rod‐shaped cells, while the latter have cells mostly wider than long (Fig. 2H).
Fig. 2.
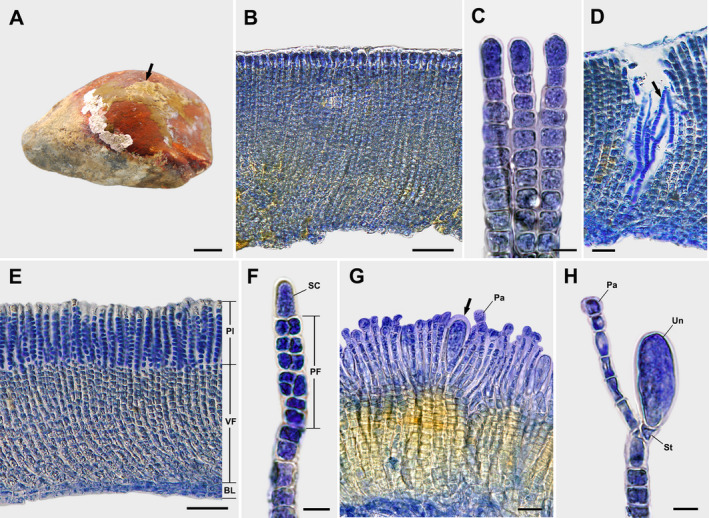
Sungminia asiatica Oteng'o, Won & T.O. Cho sp. nov. (A) Thalli on the rock forming an irregular yellowish‐brown crust (arrow). Scale bar = 2 cm. (B) Longitudinal section view of vegetative thallus showing laterally adjoined erect perithallial filaments. Scale bar = 50 μm. (C) Apical cells of vegetative erect perithallial filaments. Scale bar = 10 μm. (D) Tufts of hairs (arrow) in pits developed from hypothallial basal layer or the lower to middle portions of erect perithallial filaments. Scale bar = 25 μm. (E) Longitudinal section view of a reproductive thallus showing plurangia (Pl) and vegetative filaments (VF) developed from the hypothallial basal layer (BL). Scale bar = 50 μm. (F) Plurangium composed of plurangial filament (PF) and terminal dome‐shaped sterile cell (SC). Scale bar = 10 μm. (G) Longitudinal section view of reproductive thallus showing unangia (arrow) and paraphyses (Pa). Scale bar = 25 μm. (H) Unangium (Un) on stalk (St) with paraphysis (Pa). Scale bar = 10 μm.
Holotype (designated here)
MABIK AL00084653 (= CUK19812B), leg. T.O. Cho & B.Y. Won, (01 August 2019), Oeyeondo, Chungcheongnam‐do, Korea, 36°13′43.4″ N, 126°04′29.1″ E, 1–2 m depth.
Etymology
Named for its geographical distribution in Asia including Korea, its type locality.
Other specimens examined
CUK19175C, T.O. Cho & B.Y. Won, 08 October 2018, Gijang, Busan, Korea, 35°15′29.6″ N, 129°14′06.7″ E, 1–2 m depth; CUK20636A, CUK20631A&B, CUK20656, T.O. Cho, J. Avila, A. Oteng'o & G.C. Choi, 12 August 2020, Dumunjin Port, Baekryeong Island, Korea, 37°58′34.5″ N, 124°37′04.2″ E, 1–2 m depth; CUK20874, T.O. Cho & B.Y. Won, 28 November 2020, Daecheon Port, Chungcheongnam‐do, Korea, 36°19′43.9″ N, 126°30′14.3″ E, 1–2 m depth.
Habitat and distribution in Korea
Known from several locations, Daecheon Port and Oeyeondo (Chungcheongnam‐do), Dumunjin Port (Baekryeong Island), and Gijang (Busan), growing on rocks and pebbles in the intertidal zone. Plurangia and unangia are found on separate thalli.
Sungminia pyriformis Oteng'o, Won & T.O. Cho sp. nov.
Description
Dimerous thalli are light to yellowish brown crusts that are mucilaginous, firmly attached to the substratum without rhizoids, and 0.2–1.3 cm in diameter and 86–243(−392) μm thick (Fig. 3A). Hypothallial basal layers are formed by one to several prostrate filaments that are composed of horizontally elongated cells 6–16 μm long and 3–10 μm wide that give rise to erect perithallial filaments (Fig. 3B). Erect perithallial filaments are straight to slightly curved, simple, rod‐shaped, and laterally adjoined (Fig. 3B) but can be partially separated with pressure. Apical vegetative cells are 1.8–2.4 times larger than other cells of erect perithallial filaments, 12–23 μm long and 5–12 μm wide (Fig. 3C). Tufts of hairs arise in pits from the hypothallial basal layer or the lower to middle portions of erect perithallial filaments (Fig. 3D). There is one chloroplast per cell without a pyrenoid. Plurangia are uniseriate or biseriate, 31–48 μm long and 5–10 μm wide, intercalary, and terminated by single pyriform sterile cells measuring 16–26 μm long and 5–15 μm wide (Fig. 3, E and F). Unangial reproductive thalli produce filaments of two types, both originating from an erect perithallial cell that branches to give rise to a paraphysis and shorter stalk cell that supports a terminal unangium. Unangia are obovoid and terminally produced on 1–2 celled stalks, 80–131 μm long and 7–34 μm wide (Fig. 3, G and H). Paraphyses are clavate, 171–268 μm long and 6–26 μm wide, composed of 7–10 cells, and distinguished from erect perithallial filaments by their clavate shape and having rod‐shaped cells, while the latter have cells mostly wider than long (Fig. 3H).
Fig. 3.
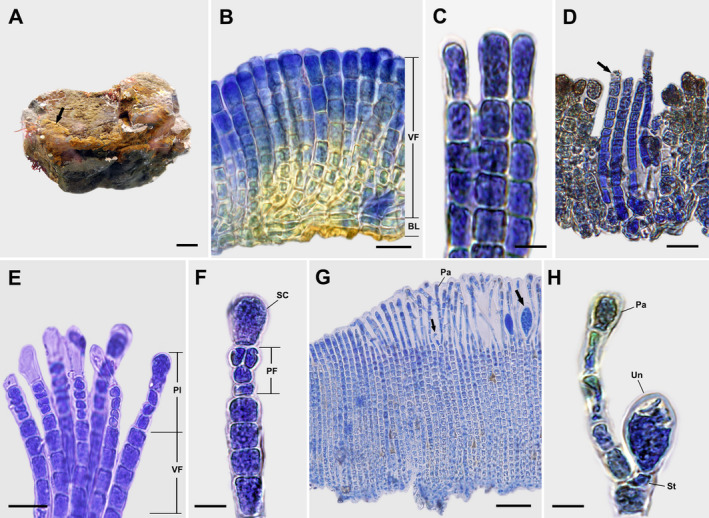
Sungminia pyriformis Oteng'o, Won & T.O. Cho sp. nov. (A) Thallus on the rock forming irregular epilithic yellowish‐brown crusts (arrow). Scale bar = 2 cm. (B) Longitudinal section view of vegetative thallus showing laterally adjoined perithallial erect filaments composed of vegetative filaments (VF) and hypothallial basal layer (BL). Scale bar = 25 μm. (C) Apical cells of vegetative erect perithallial filaments. Scale bar = 10 μm. (D) Tufts of hairs (arrow) in pits developed from the hypothallial basal layer or the lower portions of erect perithallial filaments. Scale bar = 20 μm. (E) Longitudinal section view of reproductive thallus showing plurangia (Pl) and vegetative filaments (VF). Scale bar = 20 μm. (F) Plurangium composed of plurangial filament (PF) and terminal pyriform sterile cell (SC). Scale bar = 10 μm. (G) Longitudinal section view of reproductive thallus showing young and mature unangia (arrows) and paraphyses (Pa). Scale bar = 50 μm. (H) Unangium (Un) on stalk (St) with paraphysis (Pa). Scale bar = 10 μm.
Holotype (designated here)
MABIK AL00084652 (= CUK19694A), leg. T.O. Cho & B.Y. Won, 06 June 2019, Saegdal‐dong, Jeju Island, Korea, 33°14′28.6″ N, 126°23′52.6″ E, 1–2 m depth.
Isotypes
CUK19697B.
Etymology
Named for the pear‐shaped sterile cells (=pyriformis, L. f.)
Other specimen examined
CUK18425, T.O. Cho, S.Y. Jeong, J. Avila & A. Oteng'o, 03 November 2017, Dala Park, Gyeongsangnam‐do, Korea, 34°46′11.4″ N, 128°23′51.5″ E, 1–2 m depth.
Habitat and distribution in Korea
This species is presently known from two locations, Dala Park (Gyeongsangnam‐do) and Saegdal‐dong (Jeju Island), growing on rocks and pebbles in the intertidal zone. Plurangia and unangia are found on separate thalli.
Phylogenetic analyses
A total of 15 sequences from the rbcL (1279 bp) and COI‐5P (657 bp) loci were obtained from samples collected from Korea (Table S1). Phylogenetic analyses of rbcL gene data set and concatenated data sets showed similar topologies in ML and Bayesian analyses (Fig. 4, Fig. S1 in the Supporting Information). Sungminia gen. nov. formed a monophyletic clade with strong support (100%/1.00 for ML and BPP) with three distinct new species described below (Sungminia asiatica sp. nov., S. gladiata sp. nov., and S. pyriformis sp. nov.; Fig. 4). Sungminia pyriformis was a sister clade to S. gladiata in the rbcL tree (Fig. 4), while it was nested sister to S. asiatica in concatenated tree (Fig. S1). These three Sungminia species were resolved with full support as monophyletic from the remaining genera within the Ralfsiales (Fig. 4). Sequence divergences between Sungminia gen. Nov. and other genera within the Ralfsiales were 9.6–13.9% for the rbcL gene and 30.0–35.4% for the COI‐5P gene (Table 1). Interspecific sequence divergences between S. asiatica and S. gladiata were 3.0% for rbcL and 9.3% for COI‐5P; between S. asiatica and S. pyriformis, they were 2.8–2.9% for rbcL and 8.9% for COI‐5P; between S. pyriformis and S. gladiata, they were 2.2–2.4% for rbcL and 8.8% for COI‐5P.
Fig. 4.
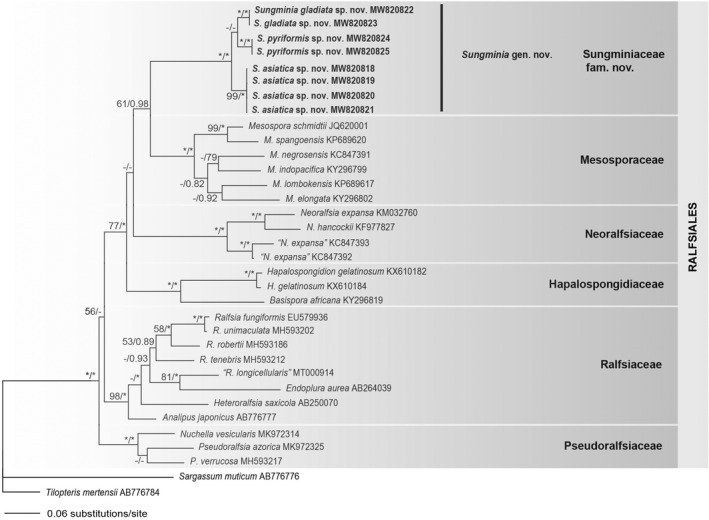
Maximum likelihood tree based on rbcL gene sequence alignment for the Sungminiaceae fam. Nov. and other Ralfsiales species. The value above branches = Maximum likelihood bootstrap values in % > 50, Bayesian posterior probabilities >0.75. Values lower than BS 50 or BPP 0.75 are indicated by hyphens (−). Values of BS 100 or BPP 1.00 are indicated by asterisks (*). Taxa in bold text are described in this paper.
Table 1.
Gene sequence divergences (rbcL/COI‐5P) among families including Sungminiaceae fam. Nov. within Ralfsiales.
| % gene sequence divergence | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 Sungminiaceae fam. Nov. | – | – | – | – | – | – |
| 2 Mesosporaceae | 9.8–11.8/30.0–34.3 | – | – | – | – | – |
| 3 Hapalospongidiaceae | 11.9–13.0/32.8–34.3 | 10.9–12.8/23.2–26.2 | – | – | – | – |
| 4 Neoralfsiaceae | 10.4–13.9/32.9–34.1 | 10.9–13.7/22.8–24.7 | 11.3–14.0/21.2–22.2 | – | – | – |
| 5 Ralfsiaceae | 9.6–13.5/31.5–34.4 | 9.1–12.8/22.2–26.8 | 11.3–13.6/20.2–21.9 | 10.0–13.6/20.0–23.5 | – | – |
| 6 Pseudoralfsiaceae | 10.4–12.6/33.3–35.4 | 9.4–11.1/23.8–28.8 | 11.8–13.8/22.2–24.8 | 11.1–13.5/22.2–25.0 | 7.5–11.1/21.9–25.6 | – |
DISCUSSION
We propose a new genus, Sungminia gen. nov., containing three new species based on morphological and molecular analyses. The sequences of rbcL and COI‐5P from the specimens collected from Korea were nested as a distinct clade, Sungminia, composed of S. gladiata, S. asiatica, and S. pyriformis. Sungminia is characterized by (i) thalli with perithallial erect filaments moderately adhered, (ii) the rod‐shaped perithallial erect filaments, (iii) hairs in tufts in pits, (iv) single chloroplasts per cell without pyrenoid, (v) plurangia with one apical sterile cell and (vi) unangia are sessile or terminally inserted on a stalk lateral‐basal to a paraphysis. Sungminia gladiata sp. nov. is distinguished from the other two species by having long uniseriate plurangia terminated with sword‐shaped sterile cells. Sungminia pyriformis sp. nov. is distinguished from the other two species by having simple perithallial erect filaments and plurangia with terminal pyriform sterile cells. Sungminia asiatica sp. nov. differs from the former two new species by having a monostromatic hypothallial basal layer, and plurangia terminated by dome‐shaped sterile cells. The shape of sterile cells may be recognized one of key characteristics for distinguishing each species of this genus: sword‐shaped in S. gladiata, pear‐shaped in S. pyriformis, and dome‐shaped in S. asiatica.
The Ralfsiales is an order of crustose brown algae containing five families: Hapalospongidiaceae, Mesosporaceae, Neoralfsiaceae, Pseudoralfsiaceae, and Ralfsiaceae (Farlow 1881, Tanaka and Chihara 1982, Lim et al. 2007, León‐Alvarez et al. 2017, Parente et al. 2021). Molecular analyses of crustose brown algae have expanded our understanding of the phylogenetic affinities between related families of the Ralfsiales and have helped us infer their taxonomic positions. Our phylogenetic analyses of rbcL and concatenated rbcL and COI‐5P gene sequences show that Sungminia forms a strongly supported monophyletic clade and a probable sister relationship with the Mesosporaceae. Our multigene sequence analyses revealed the genetic differences in rbcL gene sequences (9.8–11.8%) and COI‐5P gene (30.0–34.3%) between Sungminia and Mesospora, while in rbcL gene sequences (9.6–13.9%) and COI‐5P gene (31.5–35.4%) between Sungminia and other genera within the Ralfsiales, respectively. As a comparison, families recognized within the Ralfsiales differed by ≧9.7% in the rbcL gene (León‐Alvarez et al. 2017, Parente et al. 2021). We propose that Sungminia can be recognized as the basis for a new, higher‐level taxon within the Ralfsiales.
While molecular data can provide compelling evidence for erecting a new taxon, they should not be the sole basis for determining higher‐level classification to the exclusion of vegetative and reproductive morphology. We have revisited the morphological descriptions and compared the most common characteristics of Sungminia with genera of the other families in the Ralfsiales (Table 2). Sungminia and Mesospora (Mesosporaceae) share morphological features of the absence of medullary layer and rhizoids, and cells with a single chloroplast lacking pyrenoids. However, Sungminia is distinguished from Mesospora by having unangia developed on short 1–2 celled stalk‐like filaments, paraphyses in unangial reproductive thalli, and erect perithallial filaments moderately adhered. Sungminia is also distinguished from Hapalospongidion (Hapalospongidiaceae) which has unangia developed on long 7–19(−31) celled stalk‐like filaments without paraphyses (León‐Alvarez et al. 2017), from Neoralfsia (Neoralfsiaceae) which has distinct delineation of cortex and medulla and rhizoids on the base of thallus (Lim et al. 2007), and from Pseudoralfsia (Pseudoralfsiaceae) and the crustose genera (Ralfsia, Endoplura, and crustose phase of Heteroralfisa) (Ralfsiaceae) which have firmly adhering erect filaments (Farlow 1881, Hollenberg 1969, Parente et al. 2021; Table 2).
Table 2.
Comparison of morphological features among Sungminia of Sungminiaceae fam. Nov. and genera of the other families within Ralfsiales.
| Taxa/Characteristics | Sungminiaceae fam. Nov. (Sungmina gen. Nov.) | Hapalospongidiaceae (Hapalospongidion) | Mesosporaceae (Mesospora) | Neoralfsiaceae (Neoralfsia) | Pseudoralfsiaceae (Pseudoralfsia, Nuchella) | Ralfsiaceae (Ralfsia, Endoplura, Heteroralfsia) |
|---|---|---|---|---|---|---|
| Erect perithallial erect filaments | Moderately adhered | Loosely adhered | Loosely adhered | Firmly adhered | Firmly adhered | Firmly adhered |
| Shape of erect perithallial filaments | Rod‐shaped | Tapered downward | Tapered downward | Tapered downward | Rod‐shaped | Tapered upward or rod‐shaped |
| Distinct delineation of cortex and medullar layer | No | No | No | Yes | No | No/yes |
| Form of hairs | Tuft | Single/tuft | Tuft | Hair pit | Hair pit | – |
| No. of plastids per cell | One | One to several | One to several | One | One | One to several |
| No. of sterile cell on plurangia | One | One to several | One or three | One | One or two | One to several |
| Unangia | Sessile or on stalk‐like filament | On stalk‐like filament | On stalk‐like filament | On stalk‐like filament | Sessile or on stalk‐like filament | Sessile or on stalk‐like filament |
| No. of cells in stalk‐like filaments | 1–2 | 7–19 (31) | 5–12 | 3–6 | 1–3 | 1–6 |
| Paraphyses with unangium | Present | Absent | Absent | Present | Present | Present |
| No. of cells in paraphyses | 7–20 | – | – | – | Up to 13 | (6)–8–12 |
| Rhizoids | Absent | Absent | Absent | Present | Present or absent | Present or absent |
| Reference | This study | León‐Alvarez et al. 2017 | Weber‐van Bosse 1911, Tanaka and Chihara 1982 | Lim et al. 2007 | Parente et al. 2021 | Farlow 1881, Hollenberg 1969 |
The reproductive structure of unangia has been used to distinguish the families within the Ralfsiales (Tanaka and Chihara 1982, Lim et al. 2007, León‐Alvarez et al. 2017, Parente et al. 2021). There are two types: unangia with and without paraphyses. Unangia without paraphyses have been known to occur in the Hapalospongidiaceae and Mesosporaceae (Tanaka and Chihara 1982, León‐Alvarez et al. 2017). However, in the Hapalospongidiaceae, unangia originate on the stalk‐like filaments laterally developed from hypothallial basal layer, whereas in the Mesosporaceae, unangia originate on the stalk‐like filaments laterally developed from a basal cell of perithallial erect filament. Unangia with paraphyses have been known to occur in the Neoralfsiaceae, Pseudoralfsiaceae, and Ralfsiaceae (Hollenberg 1969, Lim et al. 2007, Parente et al. 2021) along with the Sungminiaceae, proposed below. However, unangia in the Sungminiaceae differ from unangia in the Neoralfsiaceae (3–6 celled), Pseudoralfsiaceae (1–3 celled), and Ralfsiaceae (1–6 celled) in that they develop from 1–2 celled short stalk‐like filaments (Table 2). Also, because the detailed morphologies of unangia (e.g., stalk‐like filaments, paraphyses, position of unangia) vary within families, the combinations of these characteristics may be important in taxonomy for each group of different levels within the Ralfsiales. Our morpho‐anatomical observations and phylogenetic analyses support monophyly of the Sungminia clade within the order Ralfsiales as a probable sister taxon to the Mesosporaceae. We consider that the Sungminia group is clearly distinct at the family level and therefore propose to place the Sungminia in a new family designated here:
Sungminiaceae Oteng'o, Won & T.O. Cho fam. Nov.
Description
Dimerous thalli are mucilaginous crusts firmly attached to the substratum without rhizoids. A hypothallial basal layer with one to several cells gives rise to erect perithallial filaments. Erect perithallial filaments are moderately adjoined and rod‐shaped, and their and terminal cells are larger than other cells. Tufts of hair arise in pits arise from the hypothallial basal layer or form in lower to middle portions of erect perithallial filaments Chloroplasts are one per cell and lack a pyrenoid. Plurangia are intercalary and terminated with single sterile cell. Unangial reproductive thalli produce filaments of two types, both originating from an erect perithallial cell that branches to give rise to a paraphysis and shorter stalk that supports a terminal unangium. Unangia are sessile or lateral‐basal to a paraphysis or terminally produced on 1–2 celled stalk or lateral‐basal to a paraphysis. Paraphyses are distinguished from erect perithallial filaments by their clavate shape and having rod‐shaped cells.
Type genus
Sungminia Oteng'o, Won & T.O. Cho gen. Nov.
In our study, we confirmed that all of the known species in the Sungminiaceae are presently distributed only along the coastline of Korea. Future studies based on more extensive collections worldwide are necessary to understand the geographic ranges of species in the Sungminiaceae and to further investigate the relationships of this distinct group of crustose algae.
AUTHOR CONTRIBUTIONS
A.O. Oteng'o: Conceptualization (equal); data curation (equal); formal analysis (equal); methodology (equal); writing – original draft (equal). B.Y. Won: Data curation (equal); formal analysis (equal); project administration (equal); software (equal); writing – review and editing (equal). T.O. Cho: Funding acquisition (equal); project administration (equal); supervision (equal); writing – review and editing (equal).
Supporting information
Figure S1. Maximum likelihood tree based on concatenated DNA sequence (plastid rbcL and mitochondrial COI‐5P) alignments for the Sungminiaceae fam. Nov. and other Ralfsiales species. The value above branches = Maximum likelihood bootstrap values in % >50, Bayesian posterior probabilities >0.75. Values lower than BS 50 or BPP 0.75 are indicated by hyphens (−). Values of BS 100 or BPP 1.00 are indicated by asterisks (*). Taxa in bold text are described in this paper.
Table S1. List of specimen information used in molecular analyses (rbcL, COI‐5P). “–” indicates no sequence obtained.
We thank to Dr. Craig Schneider for many constructive suggestions to improve the manuscript. This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A2059577), by the Ministry of Ocean and Fisheris (MarineBiotics Project, 20210469), and by the National Marine Biodiversity Institute of Korea (2022M01100) to Tae Oh Cho. This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A1A01051909) to Boo Yeon Won.
Editorial Responsibility: C. Schneider (Associate Editor)
[Correction added on 19 September 2022, after first online publication: The copyright line was changed.]
References
- Draisma, S. G. , Prud'homme Van Reine, W. F. & Kawai, H. 2010. A revised classification of the Sphacelariales (Phaeophyceae) inferred from a psbC and rbcL based phylogeny. Eur. J. Phycol. 45:308–26. [Google Scholar]
- Farlow, W. G. 1881. Marine Algae of New England and Adjacent Coast. US Government Printing Office, Washington, 210 pp. [Google Scholar]
- Hollenberg, G. J. 1969. An account of the Ralfsiaceae (Phaeophyta) of California. J. Phycol. 5:290–301. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P. & Ronquist, F. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–5. [DOI] [PubMed] [Google Scholar]
- Hwang, I. K. , Kim, H. S. & Lee, W. J. 2005. Polymorphism in the brown alga Dictyota dichotoma (Dictyotales, Phaeophyceae) from Korea. Mar. Biol. 147:999–1015. [Google Scholar]
- Kawai, H. 1989. Life history and systematic position of Heteroralfsia saxicola gen. et comb. nov. (Ralfsiaceae, Phaeophyceae). Phycologia 28:243–51. [Google Scholar]
- Kawai, H. & Sasaki, H. 2004. Morphology, life history, and molecular phylogeny of Stschapovia flagellaris (Tilopteridales, Phaeophyceae) and the erection of the Stschapoviaceae fam. nov. J. Phycol. 40:1156–69. [Google Scholar]
- Keum, Y. S. 2010. Sphacelariales, Cutleriales, Ralfsiales. In Sook, S. [Ed.] Algal Flora of Korea. Volume 2, number2. Heterokontophyta: Phaeophyceae: Ishigeales, Dictyotales, Desmarestiales, Sphacelariales, Cutleriales, Ralfisales, Laminariales. National Institute of Biological Resources, Incheon, South Korea, pp. 73–108. [Google Scholar]
- Kogame, K. , Horiguchi, T. & Masuda, M. 1999. Phylogeny of the order Scytosiphonales (Phaeophyceae) based on DNA sequences of rbcL, partial rbcS, and partial LSU nrDNA. Phycologia 38:496–502. [Google Scholar]
- Lanfear, R. , Frandsen, P. B. , Wright, A. M. , Senfeld, T. & Calcott, B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34:772–3. [DOI] [PubMed] [Google Scholar]
- Lee, I. K. & Kang, J. W. 1986. A check list of marine algae in Korea. Algae 1:311–25. [Google Scholar]
- Lee, Y. P. & Kang, S. Y. 2001. A Catalogue of the Seaweeds in Korea. Cheju National University Press, Jeju, South Korea, 662 pp. [Google Scholar]
- Lee, Y. P. 2008. Marine Algae of Jeju. Academy Publication, Seoul, Korea, 177 pp. [Google Scholar]
- León‐Alvarez, D. , Reyes‐Gómez, V. P. , Wynne, M. J. , Ponce‐Márquez, M. E. & Quiróz‐González, N. 2017. Morphological and molecular characterization of Hapalospongidion gelatinosum, Hapalospongidiaceae fam. nov. (Ralfsiales, Phaeophyceae) from Mexico. Bot. Mar. 60:567–81. [Google Scholar]
- Lim, P. E. , Sakaguchi, M. , Hanyuda, T. , Kogame, K. , Phang, S. M. & Kawai, H. 2007. Molecular phylogeny of crustose brown algae (Ralfsiales, Phaeophyceae) inferred from rbcL sequences resulting in the proposal for Neoralfsiaceae fam. nov. Phycologia 46:456–66. [Google Scholar]
- Nakamura, Y. 1972. A proposal on the classification of the Phaeophyta. In Abbott, I. A. & Kurogi, M. [Eds.] Contributions to the Systematics of the Benthic Marine Algae of the North Pacific. Japanese Society of Phycology, Kobe, Japan, pp. 147–56. [Google Scholar]
- Nelson, W. A. 1982. A critical review of the order Ralfsiales, Ralfsiaceae and the taxonomic position of Analipus japonicus (Harv.) Wynne (Phaeophyta). Brit. Phycol. J. 17:311–20. [Google Scholar]
- Oteng'o, A. O. & Won, B. Y. 2020. Ralfsia longicellularis (Ralfsiales, Phaeophyceae): a Far East Asian endemic brown alga from Korea. Korean J. Environ. Biol. 38:101–5. [Google Scholar]
- Oteng'o, A. O. , Cho, T. O. & Won, B. Y. 2021. Endoplura jejuensis sp. nov. and Endoplura koreana sp. nov. (Ralfsiales, Phaeophyceae) from Korea based on molecular and morphological analyses. Algae 36:155–63. [Google Scholar]
- Parente, M. I. & Saunders, G. W. 2019. A molecular survey of Ralfsia sensu stricto (Ralfsiales, Phaeophyceae) in Canada uncovers three new species: R. robertii sp. nov., R. tenebris sp. nov., and R. unimaculata sp. nov. Botany 97:135–47. [Google Scholar]
- Parente, M. I. , Fletcher, R. L. , Costa, F. O. & Saunders, G. W. 2021. Taxonomic investigation of Ralfsia‐like (Ralfsiales, Phaeophyceae) taxa in the North Atlantic Ocean based on molecular and morphological data, with descriptions of Pseudoralfsiaceae fam. nov., Pseudoralfsia azorica gen. et sp. nov. and Nuchella vesicularis gen. et sp. nov. Eur. J. Phycol. 56:12–23. [Google Scholar]
- Poong, S. W. , Lim, P. E. , Phang, S. M. , Gerung, G. S. & Kawai, H. 2013. Mesospora elongata sp. nov. (Ralfsiales, Phaeophyceae), a new crustose brown algal species from the Indo‐Pacific region. Phycologia 52:74–81. [Google Scholar]
- Poong, S. W. , Lim, P. E. , Phang, S. M. , Sunarpi, H. , West, J. A. & Kawai, H. 2014. A molecular‐assisted floristic survey of crustose brown algae (Phaeophyceae) from Malaysia and Lombok Island, Indonesia based on rbcL and partial cox1 genes. J. Appl. Phycol. 26:1231–42. [Google Scholar]
- Poong, S. W. , Lim, P. E. , Phang, S. M. , Sunarpi, H. , West, J. A. , Miller, K. A. , Nelson, W. & Kawai, H. 2017. Two new species of Mesospora (Ralfsiales, Phaeophyceae) from the subtropical Indo‐Pacific region. Phycologia 56:487–98. [Google Scholar]
- Rambaut, A. 2012. FigTree, tree figure drawing tool v1.4.0. Institute of Evolutionary Biology, University of Edinburgh. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (last accessed 02 February 2021).
- Ronquist, F. & Huelsenbeck, J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–4. [DOI] [PubMed] [Google Scholar]
- Sasaki, H. , Flores‐Moya, A. , Henry, E. C. , Müller, D. G. & Kawai, H. 2001. Molecular phylogeny of Phyllariaceae, Halosiphonaceae and Tilopteridales (Phaeophyceae). Phycologia 40:123–34. [Google Scholar]
- Saunders, G. W. & McDevit, D. C. 2012. Methods for DNA barcoding photosynthetic protists emphasizing the macroalgae and diatoms. Methods Mol. Biol. 858:207–22. [DOI] [PubMed] [Google Scholar]
- Silva, P. C. & de Reviers, B. 2000. Ordinal names in the Phaeophyceae. Cryptogamie Algol. 21:49–58. [Google Scholar]
- Silvestro, D. & Michalak, I. 2012. raxmlGUI: a graphical front‐end for RaxML. Org. Divers. Evol. 12:335–7. [Google Scholar]
- Tanaka, J. & Chihara, M. 1980. Taxonomic study of the Japanese crustose brown algae. General account and the order of Ralfsiales. J. Jap. Bot. 55:193–201. [Google Scholar]
- Tanaka, J. & Chihara, M. 1982. Morphology and taxonomy of Mesospora schmidtii Weber van Bosse, Mesosporaceae fam. nov. (Ralfsiales, Phaeophyceae). Phycologia 21:382–9. [Google Scholar]
- Thompson, J. D. , Higgins, D. G. & Gibson, T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber‐van Bosse, A. 1911. Notice sur quelques genres nouveaux d'algues de l'Archipel Malaisien. Ann. Jard. Bot. Buitenzorg. 24:25–33. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Maximum likelihood tree based on concatenated DNA sequence (plastid rbcL and mitochondrial COI‐5P) alignments for the Sungminiaceae fam. Nov. and other Ralfsiales species. The value above branches = Maximum likelihood bootstrap values in % >50, Bayesian posterior probabilities >0.75. Values lower than BS 50 or BPP 0.75 are indicated by hyphens (−). Values of BS 100 or BPP 1.00 are indicated by asterisks (*). Taxa in bold text are described in this paper.
Table S1. List of specimen information used in molecular analyses (rbcL, COI‐5P). “–” indicates no sequence obtained.


