Abstract
Infectious bronchitis virus (IBV) is a member of the family Coronaviridae, together with viruses such as SARS‐CoV, MERS‐CoV and SARS‐CoV‐2 (the causative agent of the COVID‐19 global pandemic). In this family of viruses, interspecies transmission has been reported, so understanding their pathobiology could lead to a better understanding of the emergence of new serotypes. IBV possesses a single‐stranded, non‐segmented RNA genome about 27.6 kb in length that encodes several non‐structural and structural proteins. Most functions of these proteins have been confirmed in IBV, but some other proposed functions have been based on research conducted on other members of the family Coronaviridae. IBV has variable tissue tropism depending on the strain, and can affect the respiratory, reproductive, or urinary tracts; however, IBV can also replicate in other organs. Additionally, the pathogenicity of IBV is also variable, with some strains causing only mild clinical signs, while infection with others results in high mortality rates in chickens. This paper extensively and comprehensibly reviews general aspects of coronaviruses and, more specifically, IBV, with emphasis on protein functions and pathogenesis. The pathogenicity of the Australian strains of IBV is also reviewed, describing the variability between the different groups of strains, from the classical to the novel and recombinant strains. Reverse genetic systems, cloning and cell culture growth techniques applicable to IBV are also reviewed.
Keywords: avian coronavirus, cloning, genetics, IBV, infectious bronchitis, pathogenesis, protein functions
The natural hosts of many infectious viruses are domestic and wild animals, some of which are able to cross the species barrier and cause disease in humans. Some well recognised examples include the emergence of human immunodeficiency virus (HIV) at the beginning of the 1980s, 1 , 2 Nipah 3 and Hendra viruses 4 , 5 during the 1990s, and the recent outbreaks of Ebolavirus. 6 Each of these viruses has caused considerable loss of human life, and also required significant commitment of resources for the development of more effective treatments, vaccines and methods for prevention. Cases of inter‐species transmission have also been reported among members of the family Coronaviridae. These include the emergence of the severe acute respiratory syndrome coronavirus (SARS‐CoV) at the beginning of the 2000s, 7 , 8 the Middle Eastern respiratory syndrome coronavirus (MERS‐CoV) in 2012, 9 , 10 , 11 and, more recently, SARS‐CoV‐2, the causative agent of the coronavirus pandemic in 2019, known as COVID‐19. 12 SARS‐CoV, MERS‐CoV (Figures 1 and 2) and SARS‐CoV‐2 belong to the genus Betacoronavirus. 14
Figure 1.
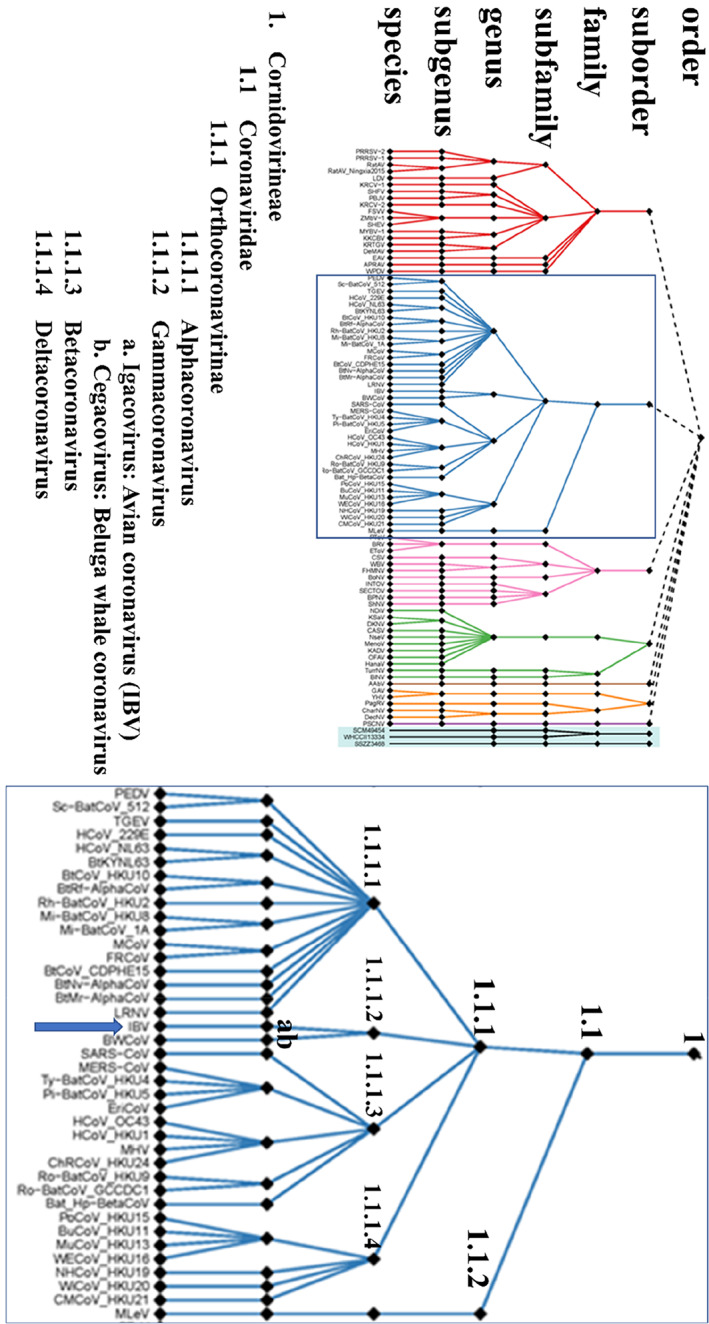
Taxonomy of the members of the order Nidovirales, as recently approved by the international Committee on the Taxonomy of Viruses. On the left is the new classification of the complete order, while on the right the classification of the suborder Cornidovirineae. The blue arrow depicts the position of IBV. Adapted from Siddell et al. 13
Figure 2.
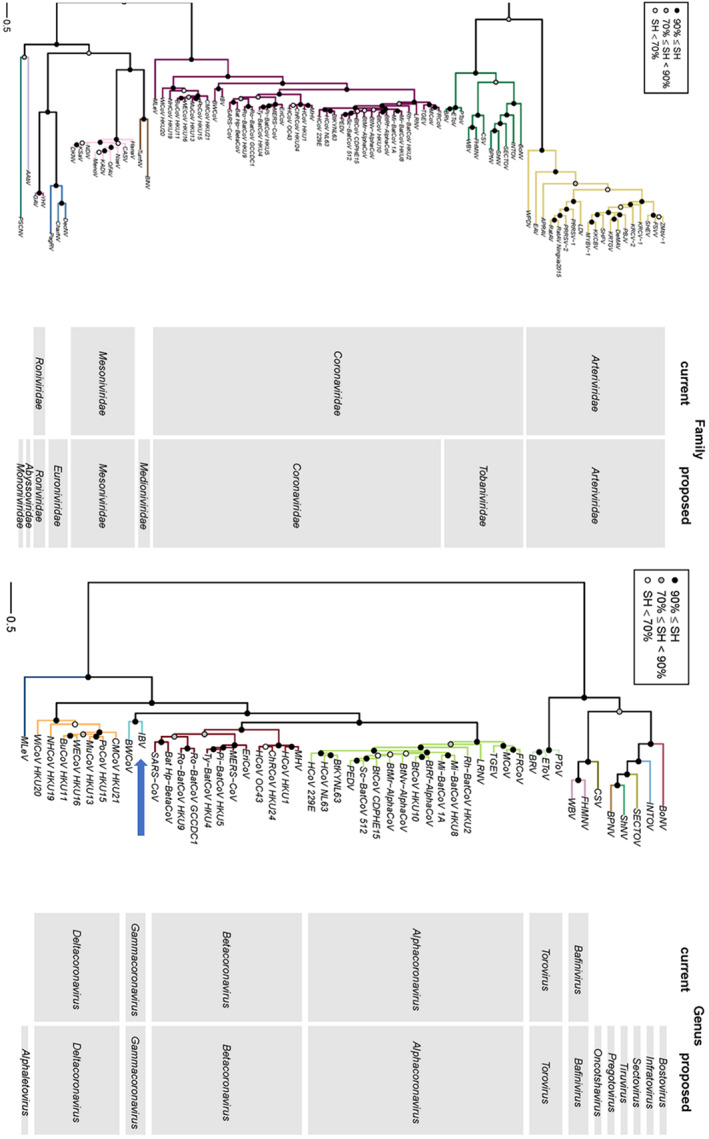
Phylogenetic tree showing the old (current) and new (proposed) classification of families (left) and genera (right) in the order Nidovirales. 13 The tree was inferred by maximum likelihood using concatenated multiple sequence alignments of 3CLpro, NiRAN, RdRp, ZBD and HEL1 domains. The blue arrow depicts the position of IBV. Adapted from Siddell et al. 13
The emergence of SARS‐CoV appears to have resulted from the transmission of the virus from its natural host, the bat. 15 Initially, the virus was also isolated from palm civets (Paguma larvata), 16 which led to large‐scale culling of these animals to control the dissemination of the disease. However, further evidence demonstrated that the civets were not involved in transmission or dissemination of the virus. The virus was subsequently detected in the respiratory tract and faeces of three different species of bats belonging to the genus Miniopterus, especially from M. pusillus. 17 In the case of MERS‐CoV, a very close relative of the virus has been detected in bats in South Africa. 18 Nevertheless, it seems very unlikely that this virus could have crossed the inter‐species barrier from bats to humans, because of the extremely low rate of contact between these insectivorous bats and humans. It appears more likely that an intermediate host was involved. 19 More recent studies have suggested that the intermediate host responsible for the dissemination of MERS‐CoV could have been the dromedary (Camelus dromedarius), because antibodies against MERS‐CoV have been detected in serum samples from these animals. 20 , 21 In the case of SARS‐CoV‐2, pangolins appear to be the most probable intermediate host. 22 A whole‐genome sequencing study of SARS‐CoV‐2 revealed that a coronavirus isolated from Malayan pangolins (Manis javanica) in 2019 23 shared 91.02% nucleotide identity with SARS‐CoV‐2 at whole‐genome level and five key amino acids in the receptor‐binding domain of the S1 glycoprotein. 22
The family Coronaviridae
Members of the family Coronaviridae cause disease in a very wide range of animal species, including humans. Coronaviruses were first described in the 1930s, with the discovery 24 and isolation 25 of infectious bronchitis virus (IBV) in chickens. In the 1940s, a new coronavirus disease, transmissible gastroenteritis, was described in pigs. Under experimental conditions, the disease caused mortality in naïve piglets placed in contact with piglets inoculated with triturated gastrointestinal (GI) tracts of sick pigs (proving its transmissibility). Piglets could also be infected with triturated GI tracts after filtration to eliminate involvement of bacteria. Cross‐species infection was attempted in this study, by feeding a calf with the triturated GI tract of an affected pig. While the calf did not show any clinical signs, piglets administered the same material promptly developed disease. 26 However, it was not until 1970 that the causative agent of transmissible gastroenteritis was identified as a coronavirus, after morphological analysis using electron microscopy was performed. 27 Murine encephalomyelitis virus, which causes demyelination in affected mice, rats and hamsters, was also studied about the same time. Attempts to isolate a bacterial agent were unsuccessful, and bacteria were not observed in tissue sections or direct smears. The agent remained infective after filtration, but did not produce any inclusion bodies and attempts to propagate it in cell cultures or embryonated eggs were unsuccessful. Even though it was not possible to clearly classify this virus, it was concluded that it did not belong to the mouse encephalomyelitis group (i.e., Theiler's virus), as it could not be propagated in embryonated eggs and did not produce myositis in infected animals. 28 Additionally, this virus did not resemble the virus causing lymphocytic choriomeningitis either. Herpesviruses were ruled out as well, due to the absence of inclusion bodies in the tissues affected, and the inability to propagate the agent in embryonated eggs. 28 It was not until the 1960s that coronaviruses were further characterised. In 1967, a virus isolated from cases of common cold in humans was passaged intracranially in mice and it found to be distinct from MHV and another human virus, 229E, which had been recognised as a human coronavirus. 29 , 30 All these human isolates were described as “IBV‐like” viruses. IBV and human coronavirus strain 229E were grouped together in 1967, 26 and murine encephalomyelitis virus was described as a new member of the family in 1969. 31 A wide range of coronaviruses have been found to be causes of clinical disease in humans 8 , 32 and a wide range of animal species, including chickens, 24 , 25 , 33 , 34 , 35 mice, 36 pigs, 37 dogs, cats and wild animals, including bats, the Beluga whale 38 and several small bird species belonging to the order Passeriformes (Figures 1 and 2). The family Coronaviridae is the largest of the order Nidovirales. Formerly, this family was divided into two subfamilies, Coronavirinae and Torovirinae. The subfamily Torovirinae included viruses from cattle, pigs and horses (toroviuses) and the only member known to infect fish (Bafinivirus), 39 while the subfamily Coronavirinae was divided into the genera Alpha, Beta, Gamma and Deltacoronavirus. However, the classification of the viruses belonging to the order Nidovirales was recently changed. The order is now divided into seven suborders, Abnidovirineae, Arnidovirineae, Mesnidovirineae, Monidovirineae, Ronidovirineae, Tornidovirineae and Cornidovirineae. The suborder Cornidovirineae includes only one family, Coronavidae, which is divided into two subfamilies, Letovirinae and Orthocoronavirinae. The subfamily Orthocoronavinae is then divided into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. Two new subgenuses were created, Cegacovirus and Igacovirus. The subgenus Cegacovirus only includes the Beluga whale coronavirus SW1, while the Igacoronavirus subgenus includes the avian coronaviruses 13 (Figures 1 and 2). Based on molecular clock analyses, it has been estimated that the most recent common ancestor of all coronaviruses was present approximately 8,100 years B.C. The most recent common ancestors for each individual coronavirus group were estimated at 2,400 years B.C. for alphacoronaviruses, 3,300 years B.C. for betacoronaviruses, 2,800 years B.C. for gammacoronaviruses and 3,000 years B.C. for deltacoronaviruses. It has also been proposed that bats and birds are ideal natural reservoirs of coronaviruses, and that bats are the natural source for alpha‐ and betacoronaviruses, while wild birds are the natural reservoir of gamma‐ and deltacoronaviruses. 40
Alphacoronavirus
This genus includes viruses affecting humans (such as the human coronaviruses 229E and NL63), bats (Rhinolophus, Scotophilus and Miniopterus bat coronaviruses) and pigs (such as porcine epidemic diarrhoea virus 8 , 39 and transmissible gastroenteritis virus (TGEV)). 41 Based on molecular clock analyses, interspecies transmission has been suggested to have occurred within this group, between the fruit bat Leschenault's rousettes (Rousettus leschenaulti) and the insectivorous bat Pomona leaf‐nosed bat (Hipposideros pomona). 42
Betacoronavirus
This genus comprises almost exclusively viruses affecting humans (including SARS‐CoV 43 and SARS‐CoV‐2 14 ) and bats. However, it also includes the murine coronavirus, murine hepatitis virus (MHV), a member of the family that has been used as a model coronavirus. In 1982, Collins, Knobler, Powell and Buchmeier 44 studied the role of some of the proteins of the murine coronavirus strain JMH during host infection using monoclonal antibodies. They found that glycoprotein 1 or GP‐1 (the protein currently referred as the S glycoprotein) conferred the attachment and membrane fusion activities of the virus. It is now known that these activities are conferred by the S1 (attachment) and S2 (membrane fusion) regions of the protein, respectively, a characteristic shared by the other members of the family. MHV was also the model used to determine that the E1 envelope protein was glycosylated by O‐linked glycosylation of serine and threonine residues, and not by conjugating core oligosaccharides from dolichol pyrophosphate intermediates to asparagine residues. 45 This feature was also observed in IBV, and thus proposed to be a characteristic of all members of the family Coronaviridae, and could play an essential role during viral replication. 46 As with alphacoronaviruses, the natural reservoirs of members of this genus are the bats. 40 The genus is subdivided into four subgroups, 2a, 2b, 2c and 2d. 47
Gammacoronavirus
These viruses were formerly known as avian coronaviruses. However, this classification recently changed after the revision of the taxonomy of the coronaviruses, including reorganisation of the entire order Nidovirales. 13 This recent update separated the avian coronaviruses, now referred as the subgenus Igacovirus, from the beluga whale coronavirus, 38 which now belongs to the subgenus Cegacovirus. However, there are some avian coronaviruses that are not members of the subgenus Igacovirus. These coronaviruses have been isolated from wild birds in the order Passeriformes, and have been classified into another genus, Deltacoronavirus.
Igacovirus
The best‐studied members of this subgenus are IBV, turkey coronavirus (TCoV) and pheasant coronavirus (PhCoV). IBV was first detected in 1931 24 in chickens with respiratory disease. The clinical manifestations of the disease caused by IBV now include respiratory signs, such as coughing and tracheal râles, as well as signs reflecting disease in the reproductive, urinary and, in some cases, digestive system, depending on the strain involved. TCoV can cause disease in gallinaceous birds of the species Meleagris gallopavo, and has been linked to transmissible enteritis of turkeys, or Bluecomb disease. Coronavirus‐like particles have been detected in samples from birds with Bluecomb disease using electron microscopy, 48 , 49 providing evidence of the association between this disease and TCoV. TCoV has also been associated with poult enteritis and mortality syndrome (PEMS). The TCoVs that cause Bluecomb disease and PEMS are antigenically related, as demonstrated by immunofluorescence and immunoperoxidase staining of tissue samples from affected birds. 50
Deltacoronavirus
The genus Deltacoronavirus was recently created and includes viruses infecting birds in the order Passeriformes, including munias, thrushes and bulbuls. It has been suggested that other coronaviruses could be assigned to this group, including a porcine coronavirus (PorCoV HKU15), and coronaviruses infecting white‐eyes, sparrows, magpie robins, night herons, wigeons and common moorhens. These wild bird coronaviruses are very distinct from IBV and TCoV, with overall 3‐chymotrypsin‐like protease (3CL‐pro), RNA‐dependent RNA polymerase (RdRp), helicase (Hel), spike (S) and nucleoprotein (N) amino acid sequence similarities below 56.6 and 57.1%, respectively. 40
Infectious bronchitis virus
General aspects of IBV
Infectious bronchitis (IB) is a contagious disease caused by IBV that affects birds belonging to the species Gallus gallus. IBV causes significant economic losses in poultry industries around the globe. It was formerly thought that chickens were the only host of IBV, but it is now apparent that it can infect other avian species, although only chickens appear to develop disease. 51 IBV was first identified in North Dakota, USA, by Schalk and Hawn, 24 isolated in embryonated chicken eggs and named infectious bronchitis virus by Beaudette and Hudson. 25 In Australia, it was initially called uraemia virus, but later recognised as being the same aetiological agent as that causing infectious bronchitis in the USA. 33 , 52
Virus morphology
Avian coronaviruses (IBV and TCoV) are pleomorphic enveloped viruses, but are predominantly round in shape. Their main feature is the presence of club‐shaped projections, or peplomers, on their surface, which gives these viruses the appearance of the corona of the sun, 53 thus explaining the origin of the name of the family. The envelope surrounds a helical nucleocapsid (which is unusual in positive‐stranded RNA animal viruses) that contains the genome. 54 , 55 , 56 The genomes of the avian coronaviruses are single‐stranded, positive‐sense linear RNA molecules of about 27.6 kilobases. They have a guanosine cap at the 5′ end, and a polyadenine (poly A) tail at the 3′ end. 57 , 58 The genome is subdivided into two main regions, the polymerase genes, and the structural and accessory genes.
Polymerase genes
This region comprises two‐thirds of the genome (about 20 kb) and contains only two open‐reading frames (ORFs), 1a and 1ab (Figure 3), which are separated as a result of a minus 1 (−1) ribosomal frameshift. 59 , 60 During the transcription process, the ribosome changes the reading frame in response to the presence of the heptanucleotide “slippery sequence” UUUAAAC, 61 , 62 which is located upstream of a pseudoknot structure in the RNA. 60 , 61 , 63 This ribosomal frameshift allows the ribosome to skip the stop codon of the 1a ORF to translate both 1a and 1b open reading frames as a single 1ab polyprotein, also known as pp1ab. These two ORFs are translated into two large polyproteins pp1a and pp1ab. The polyproteins are post‐translationally cleaved into smaller replicase peptides, or non‐structural proteins (nsp) (Figure 3). In most members of the family Coronaviridae these polyproteins are cleaved into 16 smaller peptides, but Gammacoronavirus and Deltacoronavirus lack nsp1 and instead have a larger nsp2. Several studies have identified or predicted the roles of a number of these peptides. 64 , 65
Nsp1 is a replicase peptide absent in members of the genera Gammacoronavirus (IBV) and Deltacoronavirus, but present in members of the genera Alphacoronavirus and Betacoronavirus. 64 , 65 , 66 , 67 , 68 Although the deletion of this protein from mouse hepatitis virus did not affect its ability to replicate in cell cultures, it caused its attenuation in vivo, suggesting an important role in the pathogenicity of the virus. 69 It is hypothesised that nsp1 suppresses mRNA expression in the host. 67 As this peptide is absent in gammacoronaviruses, the inhibitory role of this protein (host shutoff) in IBV appears to be replaced by a similar function contributed by the accessory protein 5b. 68
Nsp2 has been associated with a proteinase activity and described as a 3‐chymotrypsin‐like (3CLpro) protease in a variant of SARS‐CoV from Singapore. 70 However, this 3CL activity has been associated with nsp5 in HCoV229E, MHV, other SARS‐CoVs and IBV. 65
Nsp3 has a protease activity similar to that of papain and is thus referred as papain‐like protease or PLP. The nsp3 possesses the proteolytic domain glycine–glycine (Gly‐Gly). 65 , 71 , 72 In the first stages of infection, this protease is released by an autocatalytic or in cis process, directed against two highly conserved domains flanking the nsp3 region. 73 It has been hypothesised that changes in these flanking domains could interfere with this autocatalytic release of nsp3. 74 This peptide has also been found to have adenosine diphosphate‐ribose 1″‐phosphatase activity, and transmembrane 71 and putative Cys/His‐rich metal‐binding domains. 58 , 65
Genes nsp4 and nsp6 encode putative transmembrane domains. 65 These domains have been shown to participate in the autocatalytic release of the 3CL protease (nsp5) in in vitro assays. 75 This autocatalytic release of 3CLpro is an essential step during the initial stages of replication. 65 It has been speculated that changes in these domains could alter the pathogenicity of IBV. 74
Gene nsp5 encodes the main protease or 3‐chymotrypsin‐like protease (3CLpro), which post‐translationally cleaves the IBV polyproteins. 65 The 3CLpro is very similar to the 3C protease of picornaviruses, 76 as they share a similar P1—P1 substrate specificity, which is determined in part by His residues. Because of these similarities, the proteases of members of the Nidovirales are referred as 3CL. 58 , 71 During the first stages of replication this protease is released by an autocatalytic process. 65 , 72 , 75 , 77 The cleavage motif for this protease is glutamine‐serine, alanine, asparagine, glycine (Gln‐Ser, Ala, Asn, Gly). 65 , 71
Nsp7 appears to play a role in assisting Nsp8 in its primase activity, forming a supercomplex with it. In SARS‐CoV, nsp7 has a ribbon shape, formed by a helical bundle of three α‐helices, HB1, HB2 and HB3. 78 , 79 Other studies using infectious clones of MHV have demonstrated that nsp7, together with the nsps 8–10, are essential for viral replication, as the deletion of any of them blocks RNA replication and viral infection. 80 A putative function of this peptide has not yet been described in IBV.
Nsp8 in SARS‐CoV has been associated with primase activity, which is required for the correct function of the RNA‐dependent RNA polymerase (RdRp). Primases generate a primer, which is used by the RNA‐dependent RNA polymerase (nsp12) to initiate the formation of the nascent strand. 81 This primase function has also been described in and is shared by other positive‐sense RNA viruses, such as poliovirus. 82 It has been shown that nsp8 can form a cylindrical hexadecameric supercomplex with nsp7. The shape of nsp8 resembles a “golf club” but has two markedly distinct conformations. Even though it has RdRp activity, this appears to be less efficient than that of the primer‐dependant nsp12‐RdRp. 79 , 83 , 84 The hexadecameric structure has not been demonstrated for IBV and the functions of this peptide have not been studied thus far.
Nsp9 forms a homodimer that appears to have single‐stranded RNA binding activity in coronaviruses, including SARS‐CoV 85 and IBV. 86 Specific amino acid substitutions can destabilise this homodimer and abolish the transcription of mRNAs and the recovery of infectious viral particles, demonstrating that the homodimeric structure of this protein is essential for viral replication. 86 The deletion of this protein has also been shown to block MHV replication. 80
Nsp10 has been found to be associated with the functions of other peptides, including nsp5 (3CLpro), nsp14 (ExoN) and nsp16 (2'‐O‐MT). 87 The creation of an nsp10 temperature sensitive mutant of MHV altered the protease activity of nsp5. 88 Additionally, it has been shown that nsp10 enhances the exonuclease activity of nsp14 35‐fold. Moreover, nsp10 contains a Cys/His‐rich domain, 65 and the deletion of this protein from MHV impairs viral replication. 80
Nsp12 is the RNA‐dependent RNA polymerase (RdRp), 65 , 89 which is shared by all members of the family Coronaviridae. 65 , 83 , 90 The sequence encoding this polymerase activity occupies two‐thirds of the nsp12 coding region. Even though the RdRp sequence is highly conserved among most of the positive‐sense RNA viruses, the RdRp domain in coronaviruses is substantially different. 65 , 91 , 92 Previous studies have suggested that the nsp12 replicase activity may require nsp5, nsp8 and nsp9 during replication. 93 The polymerase of nsp12 is described as primer‐dependent in SARS‐CoV and, as discussed above, the primase activity can be provided by nsp8. 81 , 94
Nsp13 is the helicase. A single amino acid change (arginine to proline at position 132) in this protein has been shown to eliminate infectivity of IBV in Vero cells. This point mutation blocks transcription of the sub‐genomic RNAs (sgRNAs). 95 The protein has a putative Cys/His‐rich metal‐binding domain at its N‐terminus. 58 , 65 , 96 The helicase activity of nsp13 has been extensively studied in SARS‐CoV, and it belongs to the helicase superfamily 1 and has both DNA and RNA duplex‐unwinding activities. 96 , 97 In SARS‐CoV, this activity operates in a 5′‐to‐3′ direction. It has NTPase activity that uses any nucleotide or deoxynucleotide. It is also involved in the formation of the 5′ guanosine cap on the genome. 96 The helicase has been studied as a target for compounds that could potentially inhibit the growth of coronaviruses. Natural compounds, such as myricetin and scutellarein, can strongly inhibit the NTPase activity of the SARS‐CoV helicase, but do not stop its helicase activity. 98 Another compound referred as SSYA10‐001 was able to inhibit the helicase activity of SARS, 99 MHV and MERS coronaviruses. 100
Nsp14 has 3‐to‐5 exoribonuclease (ExoN) activity, functioning as a proof‐reading mechanism, and guanine‐N7‐methyltransferase (N7‐MTase) activity. Both functions are physically linked and are essential for replication of members of the family Coronaviridae. 86 , 87 , 101 The exoribonuclease belongs to the DEDD superfamily, as it possesses the conserved amino acid residues Asp‐Glu‐Asp‐Asp, which are found in the IBV nsp14 at positions 89, 91, 241 and 271. 87 Nsp14 has three conserved motifs, referred as motifs I, II and III. Studies conducted with SARS‐CoV have demonstrated that changes in motif I impair the fidelity of replication of the virus, increasing the frequency of mutations in progeny viruses after a few passages in cell cultures. 102 Nsp14 has also been linked in MHV coronavirus to the capacity of the virus to evade the innate immune response in the host during replication, as tested in cell culture experiments using an nsp14‐defective MHV. 103
Nsp15 is predicted to be a poly(U)‐specific endoribonuclease that initiates a cascade of poorly‐characterised reactions that may involve the nsp14 exonuclease, leading to the production of mature U16 and U86 small nucleolar RNAs (snoRNAs), which may be utilised in diverse rRNA processing events. 65 It has also been suggested that nsp15 in IBV could play a role assisting the virus to evade the host immune system, inducing host cell shutoff and thus limiting the production of interferon. 68 This shutoff of the host immune system has also been described in other members of the family Coronaviridae, but is effected by nsp1 104 instead.
-
Nsp16 is predicted to be a ribose 2'‐O‐methyltransferase 65 that assists in the methylation of rRNAs. 43 This 2'‐O‐methyltransferase activity methylates the 5′ cap of the mRNA, leading to the recognition of the viral RNA as a host mRNA. Additionally, nsp16 has been shown to interfere with the production of type I interferon. 105
The processing of the polyproteins is catalysed by the main and the accessory proteases. The crystallographic structure of the 3CLpro has been determined for human coronavirus strain 229E and TGEV, and it seems to be highly conserved among the rest of the members of the family Coronaviridae. The 3 CLpro has three domains, with domains I and II forming 6‐stranded antiparallel β barrels, highly similar to the structure of the picornavirus 3Cpro, and the substrate‐binding site is located between these two domains. Domain II and the C‐terminus of domain III are connected by a long loop. Domain III consists of a globular cluster of five helices and appears to be responsible for the proteolytic activity of the enzyme. 65 , 106 , 107 The accessory protease, also known as papain‐like protease (PLP or PLpro), conserves some of the properties of other accessory proteases. PLP recognises one or two sites in the amino‐terminal half of the polyprotein, upstream of the position of the main protease domains, cleaving peptides that all contain at least one small residue at the scissile bond, and has a catalytic dyad consisting of a Cys and a downstream His, and may employ variants of the α + β fold that is conserved in this class of proteases. 71
Figure 3.
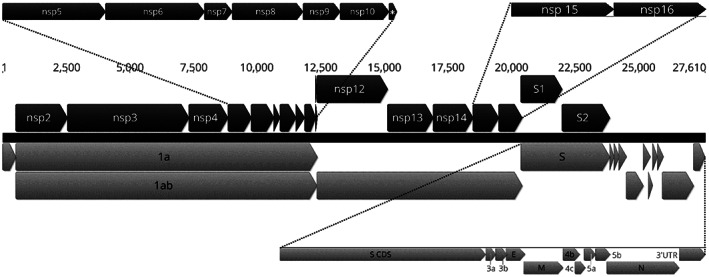
Diagram showing the position of the coding sequences (CDS, grey arrows) and the mature peptides (black arrows) in the IBV genome. The numbers indicate the nucleotide position. The black dotted lines are used to magnify the annotations of the smaller peptides to increase clarity. Nsp, non‐structural protein. *, nsp 11. This genome arrangement is based on the nucleotide sequence of the Australian IBV vaccine strain VicS.
Structural and accessory genes
Five genes comprise the terminal third of the genome, with the coding regions totalling approximately 7.6 kb. Four of the coding regions encode structural proteins, which are integral components of the virion. These are the spike glycoprotein, or Sgp, the membrane glycoprotein, or Mgp, the envelope glycoprotein, or Egp, and the nucleocapsid protein, or Np. The other coding regions encode small peptides known as the accessory proteins. The structural and accessory protein genes are located, from 5′ to 3′, in the viral genome in the following order:
Gene 2 is the largest gene of this section of the genome. It is translated into a protein of about 1,160 amino acids known as the spike protein, the S glycoprotein or Sgp (Figure 3). This glycoprotein has at least eight glycosylation sites, which have been confirmed by mass spectrometric analysis. 108 It is also known as the peplomer, as it projects from the surface of the virion. The S protein is post‐translationally cleaved into two sub‐units, S1 (the amino‐terminal end), also known as the receptor binding unit, and S2 (the carboxyl‐terminal end), also known as the membrane fusion unit (Figure 3). This cleavage occurs at the motifs RRFRR, RRSRR or RRHRR. 109 , 110 , 111 The S1 protein can induce protective antibodies against IBV 112 and recognises the host cell receptors for the virus. 44 , 113 , 114 , 115 S1 has also been implicated in the tissue tropism of IBV, as changes in this protein are correlated with changes in the ability of this virus to replicate in different cell cultures. 116 It has been suggested that the hypervariable regions of the S1 protein could be responsible for tissue tropism and pathogenicity. 117 Because of its inter‐strain sequence variability, the nucleotide sequence of S1 has been used to group IBVs into different genotypes. 118 The S2 sub‐unit anchors the protein to the virion and mediates membrane fusion between the virion envelope and the host cell membrane. 110 A recent study has suggested that the S2 sub‐unit could be responsible for determining the cell tropism of IBV. A recombinant virus derived from the Beaudette strain, which replicates in Vero cells, lost the capacity to replicate in this cell line after a total or partial replacement of its S2 subunit with that of the M41‐CK strain, which only replicates in primary cells. 119
Gene 3 is located immediately downstream of gene 2 and is a polycistronic gene, encoding 3 proteins, 3a, 3b and 3c (Figure 3). The 3a and 3b peptides have not yet been characterised. The 3c protein is also known as the envelope or E glycoprotein and is an integral component of the envelope of the virion. 120 , 121 It has been suggested that none of these proteins are essential for viral replication in vitro, and this has been shown to be the case for MERS‐CoV and IBV. 122 , 123 However, a recent study has shown that the deletion of 3a or 3b in IBV can induce attenuation in vivo and in vitro. 124 Other studies on MERS‐CoV have demonstrated that its absence induces a defect in the capacity of the virus to disseminate in the host. Even though the E protein does not appear to be essential for viral replication, it does appear to be essential for the correct assembly of viral particles. In the absence of this protein, aberrant and abnormally elongated viral particles are produced in MHV. 125 Experiments performed on IBV have shown that mutant viruses with an altered E protein had delayed replication compared to wild‐type viruses, and misshapen virions were also detected by electron microscopy. These defective viral particles also had reduced numbers of projections of the S protein. The quantity of S protein was also reduced in the defective mutant, as assessed by immunoblotting. Defective M proteins lacking the N‐terminal domain were also detected. 126
Gene 4 is also a polycistronic gene, encoding 3 different peptides. The first open reading frame (ORF) encodes the membrane protein, commonly referred as the M glycoprotein or Mgp, the major protein component of the envelope of the virion. The second ORF, which slightly overlaps with the M protein ORF, encodes the 4b peptide. The third ORF encodes the 4c peptide (Figure 3). The function of these two small accessory peptides (4b and c) remains unknown. The M protein ranges in size from 25 to 30 kDa and determines the shape of the virion. The Mgp is embedded in the virion membrane by three transmembrane domains. 115 Mgp can induce a humoral immune response in chickens. However, this response is not sufficient to prevent clinical disease. 127 The cytoplasmic tails of Mgp and Egp appear to interact during viral assembly. 128
Gene 5 does not encode any of the structural proteins, but rather two small accessory peptides, 5a and 5b (Figure 3). ORF 5a has been shown to be nonessential for viral replication in Vero cells, 129 but its replacement with enhanced green fluorescent protein (eGFP) caused reduced replication and smaller plaques in Vero cell cultures. Surprisingly, these replication deficits were reduced after the mutants lost the eGFP gene, even though the ORF 5a deletion remained. However, more recent studies have shown that the deletion of 5a or 5b induced a progeny that exhibited an attenuated phenotype in in vitro, in ovo and in vivo assays. 22 , 124 It has been speculated that protein 5b could play an important role in modulation of the production of interferon in the host (host shutoff). 68
Gene 6 is the last ORF of the IBV genome and encodes the nucleocapsid protein (Figure 3). This is a non‐specific RNA‐binding protein that plays an important multifunctional role in the life cycle of the virus. In TGEV the N protein can act as an RNA chaperone. This role is likely to be shared by the Np of other members of Coronaviridae, including IBV, given their level of nucleotide sequence similarity. 130 RNA chaperones have been described in other organisms, including bacteria, such as Vibrio harveyi and Vibrio cholerae, 131 Escherichia coli 132 and Salmonella Typhimurium, 133 and the +ssRNA viruses HIV 134 and hepatitis C virus. 135 , 136 The role of these chaperone proteins is to bind and rescue RNA molecules from self‐folding states in which they are non‐functional. They also facilitate template switching during viral RNA transcription. 137 Although the Np can induce an antibody‐mediated immune response, these antibodies do not appear to be protective, at least in chickens. 127
Pathogenesis of IBV
IBV has been associated with a variety of clinical signs in its host, the domestic chicken (Gallus gallus). The virus initially replicates in the upper respiratory tract and appears to enter host cells by viropexis. 138 The tissue tropism varies between strains, but the basis of this variation is not well understood. Experiments conducted with recombinant viruses generated under laboratory conditions using the highly attenuated Beaudette strain and the pathogenic strain M41 have suggested that tissue tropism is determined by the S protein. In these experiments, the segment of the genome encoding the S protein of Beaudette was replaced with the homologous segment of M41. The new recombinant, BeauR‐M41 (S), had the same tropism as M41, suggesting that the cell targeting was determined by this region. 116 However, this cell tropism was only assessed in cell cultures (CK, Vero, CEF and BHK‐21 cells), so the role played in tropism in vivo is yet to be confirmed. A limitation of this study was the high level of attenuation of the Beaudette strain. The switch in the cell tropism induced by the modification in the S protein did not alter the pathogenicity of the virus in vitro, 139 so it is possible that factors influencing the virulence of IBV are encoded elsewhere in the genome. 140 Even though it is suspected that those virulence factors could be encoded in the polymerase genes (1a and 1ab), it has been demonstrated, using a Beaudette and M41 in vitro recombinant, that that function is not determined by nsp3, which is a putative ADP‐ribose‐1″‐phosphate or ADRP. 141 IBV initially replicates in the Harderian glands. The histopathological changes that IBV induces in the glands include degeneration of the epithelium of the collecting ducts, lymphocytic infiltration of the septa, subepithelial infiltration of heterophils and exfoliation of epithelial cells inside the collecting tubules. 142 The live attenuated strain H‐120 has been shown to replicate in the Harderian gland tissue, as detected by immunofluorescence using monoclonal antibodies, and by histopathological examination. Immunofluorescently labelled cells were detected from day 3 to day 14 after inoculation, and the histopathological changes in the Harderian glands varied over time. Three days after inoculation, there was an increase in the number of plasma cells in the subepithelium (with Russell bodies), and slight lymphocytic infiltration of the collecting tubules. At 7 days there was an increase in the number of plasma cells, lymphocytes and heterophils. At 10 days, lymphoid infiltration became more evident, with some destruction of the epithelium of the tubules. At 14 days, there was some restoration of the epithelial cells of the collecting tubules. 143 The variation in tissue tropism of IBV contributes to variations in the clinical signs in infected birds. Generally, these differences enable classification of viruses as respiratory, reproductive and nephropathogenic, based on the three main disease manifestations. 24 , 33 , 34 , 51 , 144 , 145 , 146 , 147 Although IBV can replicate in the gastrointestinal tract, and this has not generally been associated with pathological changes, 147 IBV has been isolated from the proventriculus of chickens with proventriculitis. The authors of this study were able to isolate IBV from the proventriculus and to reproduce proventriculitis experimentally. 148 Birds with proventriculitis had ruffled feathers and developed respiratory signs, including tracheal râles, coughing, sneezing and nasal discharge. They also had white to yellow‐creamy faeces, which are commonly associated with diarrhoea and other gastrointestinal tract diseases. At necropsy, the mucosa of the proventriculus appeared thickened, with a white exudate emerging when squeezed (however, this could have been normal gastric acid or pepsinogen stored in the proventricular glands). The whole organ appeared to be enlarged and swollen. At later stages, ulcers and haemorrhages were seen in the papillae of the proventriculus. Lesions were also detected in other parts of the digestive tract, including thinning of the duodenal wall and haemorrhages in the caecal tonsils. 148 A strain isolated in Brazil (IBVPR03) exhibited a particular tropism for both ovaries and testes (gonadotropism) in infected chickens. Interestingly, this strain did not replicate in the trachea, even though it belonged to the Massachusetts genotype. However, full‐genome sequencing of this isolate was not performed, so it is not possible to attribute this change in tissue tropism to any specific peptide change or mutation. 149 Recent studies have been focused on finding a treatment that could inactivate IBV or at least inhibit its growth rate in its host. Astragalus polysaccharides (APS) have been shown to inhibit the growth of IBV in chicken embryo kidney (CEK) cells in a dose‐dependent manner. The reduction in replication of IBV was associated with a down‐regulation of pro‐inflammatory genes, such as those for IL‐1β, IL‐6, IL‐8 and TNF‐α. 150
Respiratory strains of IBV
Even though most strains of IBV primarily replicate in the upper respiratory tract and then disseminate to distal organs, 51 , 110 there are some strains that only replicate and affect the respiratory tract. These strains do not appear to be able to replicate in other organs. Disease is normally characterised by conjunctivitis, tracheal râles, coughing and sneezing, and in some cases is accompanied by nasal discharge. 146 This is usually the mildest form of disease associated with IBV, and the mortality rate is generally low. 151 Nevertheless, disease associated with respiratory strains of IBV can be complicated by secondary and opportunistic pathogens, such as Escherichia coli, 146 , 152 and mortality rates of up to 10% can be seen. 151 Some Australian strains of IBV, including Q1/89 and N3/89, have been found to have tropism for only the respiratory tract and a low pathogenicity. The replication of these strains occurs exclusively in the trachea, with no signs of replication in the kidneys. 151 These strains have been classified historically as subgroup 2 strains, using the Australian nomenclature, but they were recently re‐classified, using a global scheme, as belonging to Group III‐1. 118 While most IBV strains initially replicate in the upper respiratory tract, inducing different levels of respiratory tract disease, some can establish a viraemia and then replicate in other tissues, including those of the reproductive tract and kidneys. 51 , 110 , 147 , 153
Reproductive strains of IBV
IBV can cause lesions in multiple regions of the reproductive tract. The target cells are the ciliated and granular cells of the surface epithelia and the secretory epithelial cells of the tubular glands of both the infundibulum and magnum, although the lesions are more severe in the magnum. 154 In the magnum, infection results in loss of mucopolysaccharides from the epithelial cells, dilation of the tubular glands, and lymphocytic infiltration and oedema of the submucosa. 142 Infection of layer chickens with the Australian vaccine strain VicS resulted in a lighter eggshell colour than infection with other strains, even though the lesions induced by this vaccine strain were very mild. 155 This condition has been referred as bleaching of the eggshell. 35 The first studies of the effect of IBV on the chicken oviduct were conducted in the 1950s. 35 , 144 At the time, most laying hens infected with IBV were shown to suffer a drop in their egg production and about 70% of them did not return to lay after 5 weeks after infection. If hens become infected during the first weeks of life, the effects on the reproductive tract appear to be irreversible and the hens never achieve their full genetic potential as layers. 35 The weight of the laid eggs was also reduced in the infected group, compared to the uninfected control group. The internal quality of the egg was also affected, and the proportion of misshapen eggs increased. The changes induced by IBV in the egg include watery albumen, 154 misshapen eggs and changes in shells, which become thinner, and have a rougher surface and have flattened sides. 35 A quarter of the infected birds laid soft‐shelled eggs and 10% produced eggs with a rough surface. The changes in the albumen could originate from the histopathological changes in the cells of the magnum, which could affect the protein content of the albumen. These histopathological changes include the deciliation of the epithelial cells, and changes in the secretory epithelial cells. 154
Nephropathogenic strains of IBV
Nephropathogenic IBV (NIBV) strains cause nephritis in chickens and are the most pathogenic IBVs, causing higher levels of mortality than respiratory and reproductive strains. Initial viral replication occurs in the respiratory tract, with viraemic spread to the kidneys. 51 , 145 , 156 NIBV was first described in Australia and was referred to as “uraemia syndrome”. 33 The aetiological agent of this syndrome was subsequently found to be IBV 157 and the disease was described as an “aberrant” form of IB. The viral strains involved were shown to be serologically related but not identical to the respiratory strains previously described. 158 Since these initial studies, many strains of IBV around the world have been characterised as nephropathogenic. In Australia, these strains were classified as highly, moderately or lowly pathogenic, and includes almost exclusively strains referred as the “classical” strains, mostly isolated between the 1960s and 1990s. Strain N1/62, also referred as T‐0 or T‐strain, is a highly pathogenic strain and is recognised as the archetypical nephropathogenic strain. The moderate pathogenicity group includes strains Q1/73 and NT2/89, and the low pathogenicity group includes V2/71 and N4/81. 159 , 160 , 161 Several NIBV have been reported from different countries. During the 1960s, the strain Holte, affecting laying hens in North America (US and Canada), was characterised. The reports described a clinical condition in Leghorn laying hens characterised by an increase in the mortality rate. The affected birds also had enlarged kidneys, two to four times their normal size, containing urate crystals and haemorrhages, and urate deposits in the ureters. 157 , 162 Similarly, the nephropathogenic Japanese strain MA‐87 initially replicates in the renal tubules in the medulla of the kidney, resulting in enlarged and pale kidneys, with urate deposits in the renal tubules. 156 Recently, QX‐ and 4/91‐related isolates were found in broilers with damaged kidneys in Western and Central Algeria. A new genotype referred as IBDZ13a has also been isolated from chickens with nephritis, and shares 93% identity with the vaccine strain 4/91. This is the first isolation of a nephropathogenic IBV in Algeria. 163 There is a significant dietary contribution to the incidence of the nephritis syndrome caused by IBV. 51 A layer ration with a high calcium:phosphorus ratio of 7:1, when fed to pullets during their grow out stages, increases the likelihood of urolithiasis, and this likelihood is further increased when the pullets are infected with the IBV strain Gray. The levels of urolithiasis in infected birds fed a regular grower ration (with a calcium:phosphorus ratio of 2:1) did not differ from those of uninfected birds, suggesting that the Gray strain alone may not be capable of inducing urolithiasis. 164 , 165 The macroscopic changes induced in the urinary tract of chickens infected with IBV include pale discolouration of the kidneys, increased kidney size, and deposition of urates inside the tubules and ureters. 145 , 156 Histopathological changes include interstitial nephritis, with extensive damage of the tubular epithelium, consisting of granular degeneration, vacuolation and desquamation. Heterophil infiltration is seen in the early stages of the disease, with most damage seen in the medulla of the kidneys. Over time, the inflammatory cell population of the affected areas changes from heterophils to lymphocytes and plasma cells. The main targets of IBV in the early stages are the cells of the lower nephron and the ducts. The epithelial cells of the collecting ducts, collecting tubules and distal convoluted tubules are the first cells to be infected, followed by the cells in the loop of Henle. The proximal convoluted tubules normally appear to be less affected. 51 , 156 , 166 Young birds (less than 2 weeks of age) are more susceptible to nephropathogenic strains of IBV than older birds. 167 When chickens are affected by a nephropathogenic strain that results in renal damage, there is an increase in serum uric acid levels and a decrease in the packed red cell volume. 168 The pathology of nephropathogenic IBV strains could be mediated by metabolic disorders induced by the virus in the host. Infection of chicken kidney (CK) cells with a nephropathogenic strain of IBV resulted in down‐regulation of xanthine oxidase gene mRNA expression and an increase in the uric acid concentration in the infected cells. 169
Pathogenesis of Australian IBV strains
The classical form of IBV in Australia was characterised by uraemia, 33 , 34 while the classical form of IBV in North America was characterised mainly by upper respiratory tract lesions. 24 , 25 The nephropathogenic T‐strain has been extensively described as a representative Australian strain and as a classical nephropathogenic strain. 34 , 148 , 159 , 167 , 168 , 170 A study of 25 strains of IBV isolated between 1961 and 1994 151 compared the pathogenicity of these strains in specific‐pathogen‐free (SPF) white Leghorn chickens. Twelve strains were found to be nephropathogenic, while ten were classified as respiratory and three were classified as having a mixed pathogenicity. Tropism for the reproductive tract (oviduct) was not evaluated in this study. The severity of the nephropathogenicity was classified as high, moderate or low, and there appeared to be a change in the pathogenicity of the Australian isolates of IBV over the years. Of the twelve strains isolated during the 1960s and 1970s, nine were classified as nephropathogenic, with a mortality rate of 15–90%. Among the thirteen strains isolated in the 1980s and 1990s, only three were classified as nephropathogenic, with a mortality rate ranging from 5 to 37%, while the remaining nine strains were classified as respiratory strains. Interestingly, seven of these nine strains did not replicate in the renal tissues. The emergence of a recombinant strain of IBV was reported by Hewson, Noormohammadi, Devlin, Browning, Schultz and Ignjatovic. 171 This strain, N1/08, was isolated from an IBV outbreak in 2008 on a farm in New South Wales and was found to be a recombinant between subgroup 2 (currently GIII‐1) and subgroup 3 (currently GV‐1) strains. This strain causes mild respiratory signs and tracheal lesions, head shaking, and irritation and swelling of the eyes. The histopathological changes include lesions in the upper and lower trachea, with deciliation and hyperplasia, and inflammatory cell infiltration into the lamina propria. Viral RNA was detected in the caecal tonsil tissue of 5 of 40 infected chickens, and in the renal tissue of 1 of 40 infected chickens, but there were no histopathological changes detected in the kidneys. The reproductive tract was not examined in this study, so the effect of this strain on these tissues remains unknown. A summary of the pathogenesis of Australian IBV strains is provided in Table 1. The nomenclature used in Australia for the designation of new isolates is generally based on the model SI/YY, where S indicates the state in which the strain was isolated (V for Victoria, Q for Queensland, N for New South Wales, NT for Northern Territory, T for Tasmania), I indicates the number of the isolate (if there was more than one that year in that state), and YY indicates the year of isolation (e.g., N1/08 is the first isolate from New South Wales in 2008). 151 , 172 However, the international nomenclature proposed by Jackwood, 173 following that initially proposed by Cavanagh, 174 has the format IBV/bird type/country of origin/genotype or serotype/strain designation/year of isolation. Following this nomenclature, the N1/08 isolate would be referred as IBV/Ck/Aus/GIII‐1/N1/08.
Table 1.
Tissue tropism and pathogenicity of the Australian IBV strains studied to date
| Isolate | Genotype | Tissue tropism | Pathogenicity | Mortality rate | Clinical signs | Gross lesions | Histopathological lesions | Reference |
|---|---|---|---|---|---|---|---|---|
| N1/62 | GI‐5 | K | High | 80–90% | Prominent nephritis | Urate deposits in kidneys | Severe in kidneys | Ignjatovic et al 151 |
| K‐RT | High | 90% | Not determined | Not determined | Severe in kidneys, mild in trachea | Sapats et al, 1996 | ||
| Q1/65 | ND | K | High | 80–90% | Prominent nephritis | Urate deposits in kidneys | Severe in kidneys | Ignjatovic et al 151 |
| N8/74 | ND | K | High | 80–90% | Prominent nephritis | Urate deposits in kidneys | Severe in kidneys | Ignjatovic et al 151 |
| Q1/61 | ND | K | Moderate | 30–65% | Moderate nephritis | Irregular or no urate deposits | Mild to severe in kidneys | Ignjatovic et al 151 |
| Q1/63 | ND | K | Moderate | 30–65% | Moderate nephritis | Irregular or no urate deposits | Mild to severe in kidneys | Ignjatovic et al 151 |
| Q1/73 | GI‐6 | K | Moderate | 30–65% | Moderate nephritis | Irregular or no urate deposits | Mild to severe in kidneys | Ignjatovic et al 151 |
| N1/75 | ND | K | Moderate | 30–65% | Moderate nephritis | Irregular or no urate deposits | Mild to severe in kidneys | Ignjatovic et al 151 |
| Q1/76 | GI‐6 | K | Moderate | 30–65% | Moderate nephritis | Irregular or no urate deposits | Mild to severe in kidneys | Ignjatovic et al 151 |
| NT2/89 | ND | K | Moderate | 30–65% | Moderate nephritis | Irregular or no urate deposits | Mild to severe in kidneys | Ignjatovic et al 151 |
| V2/71 | GI‐6 | K | Low | 5–15% | Mild to mod. nephritis | No gross lesions in kidneys | Mild to severe in kidneys | Ignjatovic et al 151 |
| N4/81 | ND | K | Low | 5–15% | Mild to mod. nephritis | No gross lesions in kidneys | Mild to severe in kidneys | Ignjatovic et al 151 |
| N25/87 | ND | K | Low | 5–15% | Mild to mod. nephritis | No gross lesions in kidneys | Mild to severe in kidneys | Ignjatovic et al 151 |
| Vaccine 5 | ND | K | Low | 5–15% | Mild to mod. nephritis | No gross lesions in kidneys | Mild to severe in kidneys | Ignjatovic et al 151 |
| N2/62 | ND | U | Low | No mortality | Mild to trans. nephritis | No gross lesions in kidneys | Mild in kidneys | Ignjatovic et al 151 |
| Q1/64 | ND | U | Low | No mortality | Mild to trans. nephritis | No gross lesions in kidneys | Mild in kidneys | Ignjatovic et al 151 |
| N1/81 | ND | U | Low | No mortality | Mild to trans. nephritis | No gross lesions in kidneys | Mild in kidneys | Ignjatovic et al 151 |
| Vaccine 1 | ND | U | Low | No mortality | Mild to trans. nephritis | No gross lesions in kidneys | Mild in kidneys | Ignjatovic et al 151 |
| V1/71 | GI‐5 | RT (t) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| V4/90 | ND | RT (t) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| V6/92 | GIII‐1 | RT (t) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| N6/88 | ND | RT (at) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | Mild in kidneys | Ignjatovic et al 151 |
| Q1/89 | ND | RT (at) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| N3/89 | ND | RT (at) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| V19/91 | ND | RT (at) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| V2/93 | ND | RT (at) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | Mild in kidneys | Ignjatovic et al 151 |
| V3/93 | ND | RT (at) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| N1/94 | ND | RT (at) | Low | No mortality | No signs of nephritis | No gross lesions in kidneys | No lesions detected | Ignjatovic et al 151 |
| VicS | GI‐6 | K‐RT | Low | No mortality | ND | ND | Mild in kidneys and trachea | Sapats et al, 1996 |
| N3/62 | GI‐5 | K‐RT | Low | No mortality | ND | ND | Mild in trachea | Sapats et al, 1996 |
| N9/74 | GI‐5 | K‐RT | High | 96% | ND | ND | Mild in kidneys and trachea | Sapats et al, 1996 |
| N2/75 | GI‐6 | K‐RT | Moderate | 32% | ND | ND | Moderate in kidneys and trachea | Sapats et al, 1996 |
| N1/88 | GIII‐1 | RT | Low | No mortality | ND | ND | Moderate in trachea | Sapats et al, 1996 |
| Q3/88 | GIII‐1 | RT | Low | No mortality | ND | ND | Moderate in trachea | Sapats et al, 1996 |
| V5/90 | GI‐6 | K‐RT | Low | No mortality | ND | ND | Moderate in trachea | Sapats et al, 1996 |
| V18/91 | GIII‐1 | RT | Low | No mortality | ND | ND | Mild in trachea | Sapats et al, 1996 |
| N1/08 | GIII‐1 | RT | Low | No mortality | Mild respiratory signs | ND | Lesions in the mucosa of trachea | Hewson et al 171 |
Abbreviations: at, atypical; K, kidney; mod, moderate; ND, not determined; RT, respiratory tract; t, typical; trans, transient; U, uncertain/mixed.
Coronaviral replication in cell culture systems
Cell cultures have been used for the propagation of coronaviruses for decades, with differing levels of success, depending on the virus, the conditions used for cell culture, and the cell line used. In one study, cell culture propagation of porcine epidemic diarrhoea virus (PEDV) was attempted 175 using different types of primary and secondary foetal pig cells (intestine, lung, kidney, liver, spleen, brain and skin), and continuous cell lines, including Vero (African green monkey kidney), PD5 (porcine thyroid), PK15 (porcine kidney) and HRT18 (human rectal tumour) cells. The growth medium used was Eagle's minimal essential medium (EMEM) buffered with 30 mM HEPES (N2‐hydroxyethylpiperazine‐N‐2‐ethanesulfonic acid) and supplemented with 10% heat inactivated foetal calf serum (FCS) and antibiotics (100 IU of penicillin and 100 μg of streptomycin per ml). As maintenance medium, either EMEM with 2% FCS or EMEM supplemented with 0.3% tryptose phosphate broth, 0.02% yeast extract and 10 μg trypsin per ml was used. Because trypsin is labile, 80% of the medium was changed daily. In this study, the virus did not replicate in primary or secondary pig cell cultures or any of the cell lines, except Vero cells. Even though there were some signs of virus growth under all the conditions assessed, formation of syncytia and full cytopathic effect were only seen in the presence of a maintenance medium supplemented with trypsin. Replication of PEDV in vitro requires medium supplemented with trypsin. 175 As described above, some coronaviruses require cleavage of the spike protein into its two subunits for successful adsorption to and penetration into the host cells. 175 , 176 Successful replication of the Mebus strain of bovine coronavirus (BCoV) was achieved in human rectal tumour (HRT‐18) cells maintained in serum‐free medium containing 2.5 μg of pancreatin per ml. Pancreatin contains several enzymes, including trypsin. The cytopathic effect was not evident until 7 days post infection, and consisted of granular, swollen or enlarged cells that form syncytia. 177
Coronavirus cell receptors
The presence of cell receptors that are recognised by coronaviruses is a critical requirement for infection. These receptors have been described for some members of the family Coronaviridae. The angiotensin‐converting enzyme has been found to be a receptor for SARS‐CoV and its role has been tested in Vero E6 cells. 178 Mouse hepatitis virus MHV‐59 can use several isoforms of the mouse hepatitis virus receptor (MHVR), a member of the carcinoembryonic antigen glycoprotein family and the immunoglobulin superfamily. 179 If MHV‐resistant human cells are transfected with a construct that expresses MHVR1, they become susceptible to infection with MHV‐59. 180 The receptor for porcine epidemic diarrhoea virus (PEDV), transmissible gastroenteritis virus (TGEV) and human coronavirus 229E (HCoV‐229E) is the aminopeptidase N (APN) receptor. 181 , 182 Studies conducted in IBV suggest that sialic acid is very important in the interaction of the viral particles with the cell receptors. Vero, baby hamster kidney and primary chicken kidney cells become resistant to IBV after a treatment with neuroaminidase. The role of sialic acid in the attachment of IBV has also been tested in erythrocytes. 183 , 184 Additionally, previous studies have demonstrated, by using feline kidney cells infected with IBV, that the feline aminopeptidase N, or fAPN, serves as a receptor for IBV during infection. 185
Foetal bovine serum (FBS) in coronaviral cell cultures
FBS has been shown to interfere with the replication of some viruses in cell cultures. In one study, when FBS was added at 2% to the maintenance medium for Vero cells cultures, PEDV was detected in individual cells by immunofluorescence, but syncytia did not form, and the virus was unable to be passaged. 175 FBS has also been found to have a growth inhibitory effect on human coronavirus OC43, and the factor responsible was not believed to be antibody. 186 The growth of IBV in cell cultures has also been found to be inhibited by FBS. Hopkins 160 found that the inclusion of FBS at 7% in the agar overlay used for plaque counting reduced the viral titres of several different strains of IBV (97, SE 17, JMK, 13721, 46 and 41) by 10‐ to 100‐fold. This inhibition was also believed to be non‐specific and did not involve antibodies.
Growth of IBV in cell cultures
The growth rate of IBV in cell cultures is relatively low and isolation in cell cultures can be difficult, especially with field strains, even for reisolation of vaccine strains from vaccinated birds. In order to succeed, strains must be adapted to grow in cell cultures by serial blind passages, with the number of passages required varying widely. The cells that are more sensitive to IBV are CEK and CK cells. 51 , 187 Electron microscopy of CEK cells infected with IBV (Figures 4 and 5) shows how actively the virus replicates in the cytoplasm of these cells. IBV induces cell death or formation of syncytia (Figure 4, bottom right photograph). The replication of IBV during infection induces cellular membrane rearrangements. 188 These membrane rearrangements occur in both continuous cell lines and also in primary cell cultures. One of the most noticeable changes induced by IBV in infected cells are zippered endoplasmic reticula (ERs) and spherules. These spherules may be connected to the cell cytoplasm by the zippered ERs. The zippered ERs can be seen as early as 48 h after inoculation, coinciding with late‐stage replication of IBV. Membrane rearrangements have also been studied in SARS‐CoV. The viral particles appear to be grouped or packed inside the vesicle packets (VP), 188 , 189 which are seen in the late stages of viral replication, and result from fusion between multiple double‐membrane vesicles (DMV) (Figure 5A,B). In SARS‐CoV, DMVs can be found predominantly in the perinuclear area of the cell. In the later stages of viral replication, virus particles can be found in clusters (Figure 5C,D). The poor growth of IBV in cell cultures has driven the focus of development of live attenuated vaccines towards serial passages of candidate strains in embryonated eggs, as was first reported for the development of the H52 and H120 vaccines in The Netherlands more than 50 years ago. These vaccines needed 52 and 120 serial passages, respectively, in embryonated eggs to become attenuated. 190 Generally, attenuation of a strain of IBV in embryonated eggs requires 75 to over 100 passages, but the number of passages can be reduced to 16 by heat treatment of the virus by incubation at 56°C. After heat treatment and 16 passages in embryonated eggs, strain GA08 had reduced virulence and induced protection against challenge with the homologous strain. However, the factors or genetic mutations contributing to this attenuation remain unknown. 191 Interestingly, it has been found that some strains, such as Holte and SE‐17, produce clearer plaques in cell monolayers when incubated at 40°C instead of 37°C, although the titre is not affected. 187 The changes seen in cell culture monolayers after infection with viruses are referred as the cytopathic effect (CPE). In the case of IBV, the CPE observed is granularity and vacuolation of the cytoplasm, clumping and shrinkage of cells, and eventually detachment from the cell monolayer. In the initial passages, syncytia are rare, but after the adaptation of the strain to growth in cell cultures, they can be readily seen after 24 h of incubation. The amount of CPE increases with serial passage of the virus in cell culture. 187 In Vero cells infected with SARS‐CoV, the CPE has been described as rounding of the cells against a granular background at 5–11 days post inoculation. 70 With bovine coronavirus, the CPE in BEK‐1 cells (from bovine embryonic kidneys) consisted of rounded cells first appearing at 3 days post inoculation, increasing in number as incubation progressed, and subsequently formation of syncytia. The cell sheet was destroyed by 4 to 5 days after inoculation. 192 The morphology of the plaques formed by IBV depends on the strain and the number of times it has been passaged in cell monolayers and varies from small (1–3 mm) to large plaques with irregular or wavy borders. 193
Figure 4.
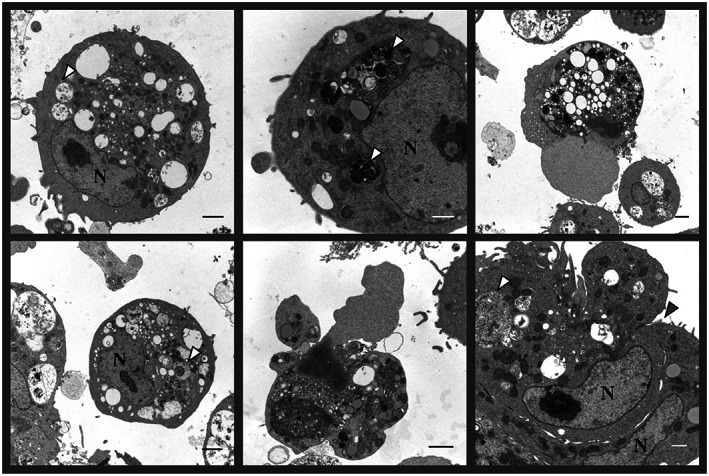
Electron micrographs of chicken embryo kidney (CEK) cells infected with the IBV strain VicS‐v for 48 h. The white arrow heads indicate the location of viral particles in some of the vesicular packets. The grey arrow head indicates the location of the junction of two adjacent cells, which are forming a syncytium. N, nuclei. Scale bars represent 500 nm (white) and 1 μm (black). Photograph, courtesy of Ms Liliana Tatarczuch, Melbourne Veterinary School, Faculty of Veterinary and Agricultural Sciences, The University of Melbourne.
Figure 5.
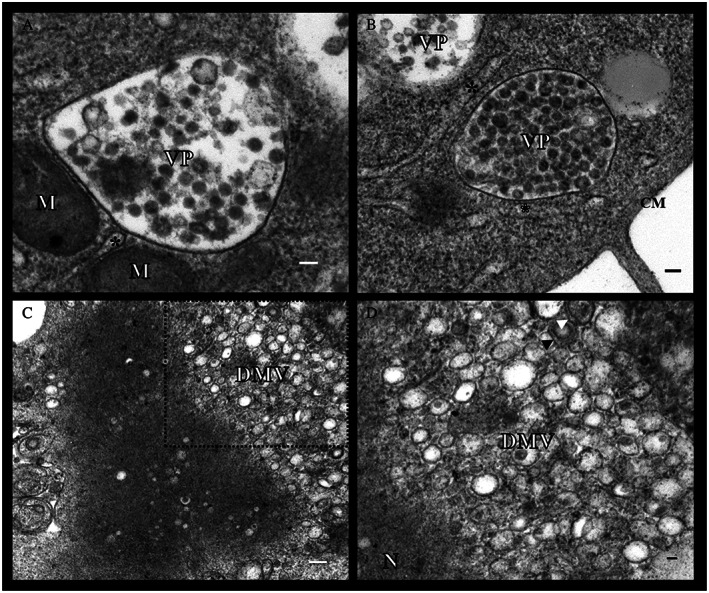
Details of the electron micrographs of chicken embryo kidney (CEK) cells infected with the IBV strain VicS‐v for 48 h. VP, vesicular packets; CM, cellular membrane; DMV, double‐membrane vesicles; M, mitochondria. Examples of DMVs are indicated by the black and white arrow heads. The reticular membranes associated with the vesicles are marked with an asterisk. Scale bars represent 100 nm (A), (B) and (D) and 200 nm (C). Photograph, courtesy of Ms Liliana Tatarczuch, Melbourne Veterinary School, Faculty of Veterinary and Agricultural Sciences, The University of Melbourne.
Conclusions
This review provides background about the family Coronaviridae, and a comprehensive overview of current understanding about the avian coronaviruses. It should enable the reader to appreciate the complexity of the molecular biology and genetics of these viruses, as well as the pathogenesis of the diseases they cause. It should also be a useful source for those interested in the ecology of Australian infectious bronchitis viruses, and in the development of more effective tools to control the diseases they cause in Australia.
Conflict of interest and sources of funding
Glenn F. Browning is an Editorial Board member of the journal and co‐author of this article. He was excluded from the peer‐review process and all editorial decisions related to the acceptance and publication of this article. Peer review was handled independently by members of the Editorial Board to minimise bias.
Acknowledgments
The authors acknowledge the contribution of Ms Liliana Tatarczuch, Melbourne Veterinary School, Faculty of Veterinary and Agricultural Sciences, The University of Melbourne, for her technical contribution during electron microscopy. The authors also acknowledge the Government of Chile through the “Becas Chile” scholarship program, and the Melbourne Veterinary School, Faculty of Veterinary and Agricultural Sciences at The University of Melbourne, which supported this doctoral candidature. Open access publishing facilitated by The University of Melbourne, as part of the Wiley ‐ The University of Melbourne agreement via the Council of Australian University Librarians.
Quinteros, JA. , Noormohammadi, AH. , Lee, SW. , Browning, GF. and Diaz‐Méndez, A. , Genomics and pathogenesis of the avian coronavirus infectious bronchitis virus. Aust Vet J. 2022;100:496–512. 10.1111/avj.13197
References
- 1. Beral V, Peterman TA, Berkelman RL et al. Kaposi's sarcoma among persons with AIDS: A sexually transmitted infection? Lancet 1990;335:123–128. [DOI] [PubMed] [Google Scholar]
- 2. Hymes K, Greene J, Marcus A et al. KAPOSI'S sarcoma in homosexual men—A report of eight cases. Lancet 1981;318:598–600. [DOI] [PubMed] [Google Scholar]
- 3. Chua KB, Bellini WJ, Rota PA et al. Nipah virus: A recently emergent deadly paramyxovirus. Science 2000;288:1432–1435. [DOI] [PubMed] [Google Scholar]
- 4. Field H, Young P, Yob JM et al. The natural history of Hendra and Nipah viruses. Microbes Infect 2001;3:307–314. [DOI] [PubMed] [Google Scholar]
- 5. Murray K, Selleck P, Hooper P et al. A morbillivirus that caused fatal disease in horses and humans. Science 1995;268:94–97. [DOI] [PubMed] [Google Scholar]
- 6. Pourrut X, Kumulungui B, Wittmann T et al. The natural history of Ebola virus in Africa. Microbes Infect 2005;7:1005–1014. [DOI] [PubMed] [Google Scholar]
- 7. May RM, McLean AR, Pattison J et al. Viral evolution and the emergence of SARS coronavirus. Philos Trans R Soc Lond Ser B Biol Sci 2004;359:1059–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peiris JSM, Lai ST, Poon LLM et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coleman CM, Frieman MB. Emergence of the Middle East respiratory syndrome coronavirus. PLoS Pathog 2013;9:e1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raj VS, Osterhaus ADME, Fouchier RAM et al. MERS: Emergence of a novel human coronavirus. Curr Opin Virol 2014;5:58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zaki AM, van Boheemen S, Bestebroer TM et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814–1820. [DOI] [PubMed] [Google Scholar]
- 12. Lai C‐C, Shih T‐P, Ko W‐C et al. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and coronavirus disease‐2019 (COVID‐19): The epidemic and the challenges. Int J Antimicrob Agents 2020;55:105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Siddell SG, Walker PJ, Lefkowitz EJ et al. Additional changes to taxonomy ratified in a special vote by the international committee on taxonomy of viruses (October 2018). Arch Virol 2019;164:943–946. [DOI] [PubMed] [Google Scholar]
- 14. Jaimes JA, André NM, Chappie JS et al. Phylogenetic analysis and structural modeling of SARS‐CoV‐2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 2020;432:3309–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W, Shi Z, Yu M et al. Bats are natural reservoirs of SARS‐like coronaviruses. Science 2005;310:676–679. [DOI] [PubMed] [Google Scholar]
- 16. Guan Y, Zheng BJ, He YQ et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003;302:276–278. [DOI] [PubMed] [Google Scholar]
- 17. Poon LLM, Chu DKW, Chan KH et al. Identification of a novel coronavirus in bats. J Virol 2005;79:2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ithete NL, Stoffberg S, Corman VM et al. Close relative of human middle east respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis 2013;19:1697–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Groot RJ, Baker SC, Baric RS et al. Middle East respiratory syndrome coronavirus (MERS‐CoV): Announcement of the coronavirus study group. J Virol 2013;87:7790–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chastel C. Middle East respiratory syndrome (MERS): Bats or dromedary, which of them is responsible? Bull Soc Pathol Exot 1990;2014(107):69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benjamin M, Marcel AM, Victor MC et al. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis J 2014;20:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Y, Cheng J, Yan S et al. S gene and 5a accessory gene are responsible for the attenuation of virulent infectious bronchitis coronavirus. Virology 2019;533:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu P, Chen W, Chen J‐P. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica). Viruses 2019;11:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schalk A, Hawn M. An apparently new respiratory disease of baby chicks. J Am Vet Med Assoc 1931;78:19. [Google Scholar]
- 25. Beaudette F, Hudson C. Cultivation of the virus of infectious bronchitis. J Am Vet Med Assoc 1937;90:51–60. [Google Scholar]
- 26. Doyle LP, Hutchings LM. A transmissible gastroenteritis in pigs. J Am Vet Med Assoc 1946;108:257–259. [PubMed] [Google Scholar]
- 27. Tajima M. Morphology of transmissible gastroenteritis virus of pigs. Arch Gesamte Virusforsch 1970;29:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheever FS, Daniels JB, Pappenheimer AM et al. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin: I. isolation and biological properties of the virus. J Exp Med 1949;90:181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McIntosh K, Becker WB, Chanock RM. Growth in suckling‐mouse brain of "IBV‐like" viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci 1967;58:2268–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McIntosh K, Dees JH, Becker WB et al. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci 1967;57:933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McIntosh K, Kapikian AZ, Hardison KA et al. Antigenic relationships among the coronaviruses of man and between human and animal coronaviruses. J Immunol 1969;102:1109–1118. [PubMed] [Google Scholar]
- 32. Stadler K, Masignani V, Eickmann M et al. SARS‐beginning to understand a new virus. Nat Rev Microbiol 2003;1:209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cumming RB. The etiology of “uraemia” of chickens. Aust Vet J 1962;38:554. [Google Scholar]
- 34. Cumming RB. Infectious avian nephrosis (uraemia) in Australia. Aust Vet J 1963;39:360. [Google Scholar]
- 35. Sevoian M, Levine PP. Effects of infectious bronchitis on the reproductive tracts, egg production, and egg quality of laying chickens. Avian Dis 1957;1:136–164. [Google Scholar]
- 36. Parker JC, Cross SS, Rowe WP. Rat coronavirus (RCV): A prevalent, naturally occurring pneumotropic virus of rats. Arch Gesamte Virusforsch 1970;31:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pensaert MB, de Bouck P. A new coronavirus‐like particle associated with diarrhea in swine. Arch Virol 1978;58:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mihindukulasuriya KA, Wu G, St. Leger J et al. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol 2008;82:5084–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masters PS, Perlman S. Chapter 28: Coronaviridae. In: Knipe DM, Howley PM, editors. Fileds Virology. 6th edn. Lippincott William & Wilkins, Two Commerce Square, 2001 Market Street, Philadelphia, PA 19103 USA, 2013;825–858. [Google Scholar]
- 40. Woo PCY, Lau SKP, Lam CSF et al. Discovery of seven novel mammalian and avian coronaviruses in Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J Virol 2012;86:3995–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang C, Lokugamage KG, Rozovics JM et al. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell‐free HeLa cell extracts but not in rabbit reticulocyte lysate. J Virol 2011;85:638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lau SKP, Li KSM, Tsang AKL et al. Recent transmission of a novel alphacoronavirus, bat coronavirus HKU10, from Leschenault's rousettes to Pomona leaf‐nosed bats: First evidence of interspecies transmission of coronavirus between bats of different suborders. J Virol 2012;86:11906–11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Snijder EJ, Bredenbeek PJ, Dobbe JC et al. Unique and conserved features of genome and proteome of SARS‐coronavirus, an early split‐off from the coronavirus group 2 lineage. J Mol Biol 2003;331:991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Collins AR, Knobler RL, Powell H et al. Monoclonal antibodies to murine hepatitis virus‐4 (strain JHM) define the viral glycoprotein responsible for attachment and cell‐cell fusion. Virology 1982;119:358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holmes KV, Doller EW, Sturman LS. Tunicamycin resistant glycosylation of a coronavirus glycoprotein: Demonstration of a novel type of viral glycoprotein. Virology 1981;115:334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stern DF, Sefton BM. Coronavirus proteins: Structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins. J Virol 1982;44:804–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chan JFW, Li KSM, To KKW et al. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV‐EMC) the beginning of another SARS‐like pandemic? J Infect 2012;65:477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Panigrahy B, Naqi SA, Hall CF. Isolation and characterization of viruses associated with transmissible enteritis (Bluecomb) of turkeys. Avian Dis 1973;17:430–438. [PubMed] [Google Scholar]
- 49. Ritchie AE, Deshmukh DR, Larsen CT et al. Electron microscopy of coronavirus‐like particles characteristic of Turkey Bluecomb disease. Avian Dis 1973;17:546–558. [PubMed] [Google Scholar]
- 50. Guy JS, Barnes HJ, Smith LG et al. Antigenic characterization of a Turkey coronavirus identified in Poult enteritis‐ and mortality syndrome‐affected turkeys. Avian Dis 1997;41:583–590. [PubMed] [Google Scholar]
- 51. Jackwood MW, de Witt JJ. Infectious bronchitis. In: Swayne DE, Glisson JR, McDougald LR, et al., editors. Diseases of poultry. 13th edn. Ames: John Wiley & Sons, Inc, 2013;139–159. [Google Scholar]
- 52. Faragher JT. Infectious bronchitis ‐ the disease in Australia. In: McFerran JB, McNulty MS, editors. Virus infections of birds. Amsterdam, Netherlands: Elsevier, 1993;268–270. [Google Scholar]
- 53. Tyrell DAJ. Coronaviruses. Nature 1968;220:650. [Google Scholar]
- 54. Abdel‐Moneim AS. Coronaviridae: Infectious bronchitis virus. In: Bayry J, editor. Emerging and re‐emerging infectious diseases of livestock. Springer International Publishing, Cham, 2017;133–166. [Google Scholar]
- 55. Davies HA, Dourmashkin RR, Macnaughton MR. Ribonucleoprotein of avian infectious bronchitis virus. J Gen Virol 1981;53:67–74. [DOI] [PubMed] [Google Scholar]
- 56. Emmott E, Munday D, Bickerton E et al. The cellular Interactome of the coronavirus infectious bronchitis virus Nucleocapsid protein and functional implications for virus biology. J Virol 2013;87:9486–9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boursnell M, Brown T, Foulds I et al. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol 1987;68:57–77. [DOI] [PubMed] [Google Scholar]
- 58. Gorbalenya AE, Koonin EV, Donchenko AP et al. Coronavirus genome: Prediction of putative functional domains in the non‐structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res 1989;17:4847–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brierley I, Boursnell M, Binns M et al. An efficient ribosomal frame‐shifting signal in the polymerase‐encoding region of the coronavirus IBV. EMBO J 1987;6:3779–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell 1989;57:537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the “slippery‐sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol 1992;227:463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jacks T, Madhani HD, Masiarz FR et al. Signals for ribosomal frameshifting in the rous sarcoma virus gag‐pol region. Cell 1988;55:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pleij C, Bosch L. RNA pseudoknots: Structure, detection, and prediction. Methods Enzymol 1989;180:289–303. [DOI] [PubMed] [Google Scholar]
- 64. Narayanan K, Ramirez SI, Lokugamage KG et al. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res 2015;202:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ziebuhr J. The coronavirus replicase. In: Enjuanes L, editor. Coronavirus replication and reverse genetics. Springer, Berlin, Germany, 2005;57–94. [Google Scholar]
- 66. Cao J, Wu CC, Lin TL. Complete nucleotide sequence of polyprotein gene 1 and genome organization of Turkey coronavirus. Virus Res 2008;136:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jansson AM. Structure of alphacoronavirus TGEV nsp1 has implications for coronavirus nsp1 function and evolution. J Virol 2012;87(5):2949–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kint J, Langereis MA, Maier HJ et al. Infectious bronchitis coronavirus limits interferon production by inducing a host shutoff that requires accessory protein 5b. J Virol 2016;90:7519–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Züst R, Cervantes‐Barragán L, Kuri T et al. Coronavirus non‐structural protein 1 is a major pathogenicity factor: Implications for the rational Design of Coronavirus Vaccines. PLoS Pathog 2007;3:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ruan Y, Wei CL, Ling AE et al. Comparative full‐length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 2003;361:1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus‐encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 2000;81:853–879. [DOI] [PubMed] [Google Scholar]
- 72. Ziebuhr J, Thiel V, Gorbalenya AE. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain‐like proteases that cleave the same peptide bond. J Biol Chem 2001;276:33220–33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Johnson MA, Pooley C, Ignjatovic J et al. A recombinant fowl adenovirus expressing the S1 gene of infectious bronchitis virus protects against challenge with infectious bronchitis virus. Vaccine 2003;21:2730–2736. [DOI] [PubMed] [Google Scholar]
- 74. Quinteros JA, Markham PF, Lee S‐W et al. Analysis of the complete genomic sequences of two virus subpopulations of the Australian infectious bronchitis virus vaccine VicS. Avian Pathol 2015;44:182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tibbles KW, Brierley I, Cavanagh D et al. Characterization in vitro of an autocatalytic processing activity associated with the predicted 3C‐like proteinase domain of the coronavirus avian infectious bronchitis virus. J Virol 1996;70:1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Palmenberg AC. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol 1990;44:603–623. [DOI] [PubMed] [Google Scholar]
- 77. Piñón JD, Mayreddy RR, Turner JD et al. Efficient autoproteolytic processing of the MHV‐A59 3C‐like proteinase from the flanking hydrophobic domains requires membranes. Virology 1997;230:309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Peti W, Johnson MA, Herrmann T et al. Structural genomics of the severe acute respiratory syndrome coronavirus: Nuclear magnetic resonance structure of the protein nsP7. J Virol 2005;79:12905–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhai Y, Sun F, Li X et al. Insights into SARS‐CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nat Struct Mol Biol 2005;12:980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Deming DJ, Graham RL, Denison MR et al. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J Virol 2007;81:10280–10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Imbert I, Guillemot J‐C, Bourhis J‐M et al. A second, non‐canonical RNA‐dependent RNA polymerase in SARS coronavirus. EMBO J 2006;25:4933–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Paul AV, van Boom JH, Filippov D et al. Protein‐primed RNA synthesis by purified poliovirus RNA polymerase. Nature 1998;393:280–284. [DOI] [PubMed] [Google Scholar]
- 83. Sawicki SG. Coronavirus genome replication. In: Viral Genome Replication. New York, NY: Springer, 2009;25–39. [Google Scholar]
- 84. te Velthuis AJW, van den Worm SHE, Snijder EJ. The SARS‐coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res 2012;40:1737–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Egloff M‐P, Ferron F, Campanacci V et al. The severe acute respiratory syndrome‐coronavirus replicative protein nsp9 is a single‐stranded RNA‐binding subunit unique in the RNA virus world. Proc Natl Acad Sci U S A 2004;101:3792–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chen Y, Cai H, Ja P et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 2009;106:3484–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Smith EC, Denison MR. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Curr Opin Virol 2012;2:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Donaldson EF, Graham RL, Sims AC et al. Analysis of murine hepatitis virus strain A59 temperature‐sensitive mutant TS‐LA6 suggests that nsp10 plays a critical role in polyprotein processing. J Virol 2007;81:7086–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ammayappan A, Upadhyay C, Gelb J Jr et al. Complete genomic sequence analysis of infectious bronchitis virus ark DPI strain and its evolution by recombination. Virol J 2008;5:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol 2007;81:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Thor SW, Hilt DA, Kissinger JC et al. Recombination in avian gamma‐coronavirus infectious bronchitis virus. Viruses 2011;3:1777–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Woo PCY, Lau SKP, Huang Y et al. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med 2009;234:1117–1127. [DOI] [PubMed] [Google Scholar]
- 93. Brockway SM, Clay CT, Lu XT et al. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA‐dependent RNA polymerase. J Virol 2003;77:10515–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. te Velthuis AJW, Arnold JJ, Cameron CE et al. The RNA polymerase activity of SARS‐coronavirus nsp12 is primer dependent. Nucleic Acids Res 2010;38:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fang S, Chen B, Tay FPL et al. An arginine‐to‐proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology 2007;358:136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ivanov KA, Thiel V, Dobbe JC et al. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol 2004;78:5619–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ivanov KA, Ziebuhr J. Human coronavirus 229E nonstructural protein 13: Characterization of duplex‐unwinding, nucleoside Triphosphatase, and RNA 5′‐Triphosphatase activities. J Virol 2004;78:7833–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yu M‐S, Lee J, Lee JM et al. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg Med Chem Lett 2012;22:4049–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Adedeji AO, Singh K, Calcaterra NE et al. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob Agents Chemother 2012;56:4718–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Adedeji AO, Singh K, Kassim A et al. Evaluation of SSYA10‐001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob Agents Chemother 2014;58:4894–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chen Y, Tao J, Sun Y et al. Structure‐function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine‐N7‐methyltransferase. J Virol 2013;87:6296–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Eckerle LD, Becker MM, Halpin RA et al. Infidelity of SARS‐CoV nsp14‐exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 2010;6:e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Case JB, Li Y, Elliott R et al. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J Virol 2018;92:e01531‐01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Narayanan K, Huang C, Lokugamage K et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 2008;82:4471–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Züst R, Cervantes‐Barragan L, Habjan M et al. Ribose 2′‐O‐methylation provides a molecular signature for the distinction of self and non‐self mRNA dependent on the RNA sensor Mda5. Nat Immunol 2011;12:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Anand K, Palm GJ, Mesters JR et al. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α‐helical domain. EMBO J 2002;21:3213–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Anand K, Ziebuhr J, Wadhwani P et al. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti‐SARS drugs. Science 2003;300:1763–1767. [DOI] [PubMed] [Google Scholar]
- 108. Zheng J, Yamada Y, Fung TS et al. Identification of N‐linked glycosylation sites in the spike protein and their functional impact on the replication and infectivity of coronavirus infectious bronchitis virus in cell culture. Virology 2018;513:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bosch BJ, van der Zee R, de Haan CAM et al. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J Virol 2003;77:8801–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res 2007;38:281–297. [DOI] [PubMed] [Google Scholar]
- 111. Song CS, Lee YJ, Lee CW et al. Induction of protective immunity in chickens vaccinated with infectious bronchitis virus S1 glycoprotein expressed by a recombinant baculovirus. J Gen Virol 1998;79:719–723. [DOI] [PubMed] [Google Scholar]
- 112. Cavanagh D, Davis PJ, Darbyshire JH et al. Coronavirus IBV: Virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus‐neutralizing or haemagglutination‐inhibiting antibody, or induce chicken tracheal protection. J Gen Virol 1986;67:1435–1442. [DOI] [PubMed] [Google Scholar]
- 113. Kant A, Koch G, van Roozelaar DJ et al. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J Gen Virol 1992;73:591–596. [DOI] [PubMed] [Google Scholar]
- 114. Mardani K, Noormohammadi AH, Hooper P et al. Infectious bronchitis viruses with a novel genomic organization. J Virol 2008;82:2013–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Masters PS. The molecular biology of coronaviruses. In: Karl M, Aaron JS, editors. Adv Virus Res. Champaign, IL: Academic Press, 2006;193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Casais R, Dove B, Cavanagh D et al. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol 2003;77:9084–9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Shan D, Fang S, Han Z et al. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res 2018;250:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Valastro V, Holmes EC, Britton P et al. S1 gene‐based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect Genet Evol 2016;39:349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bickerton E, Maier HJ, Stevenson‐Leggett P et al. The S2 subunit of infectious bronchitis virus Beaudette is a determinant of cellular tropism. J Virol 2018;92:e01044‐01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Liu DX, Cavanagh D, Green P et al. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology 1991;184:531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Liu DX, Inglis SC. Internal entry of ribosomes on a tricistronic mRNA encoded by infectious bronchitis virus. J Virol 1992;66:6143–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Almazán F, DeDiego ML, Sola I et al. Engineering a replication‐competent, propagation‐defective middle east respiratory syndrome coronavirus as a vaccine candidate. MBio 2013;4(5):e00650‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hodgson T, Britton P, Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J Virol 2006;80:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Laconi A, van Beurden SJ, Berends AJ et al. Deletion of accessory genes 3a, 3b, 5a or 5b from avian coronavirus infectious bronchitis virus induces an attenuated phenotype both in vitro and in vivo. J Gen Virol 2018;99:1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Fischer F, Stegen CF, Masters PS et al. Analysis of constructed E gene mutants of mouse hepatitis: Virus confirms a pivotal role for E protein in coronavirus assembly. J Virol 1998;72:7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Machamer CE, Youn S. The transmembrane domain of the infectious bronchitis virus E protein is required for efficient virus release. In: Perlman S, Holmes KV, editors. The Nidoviruses: Toward control of SARS and other Nidovirus diseases. Springer, US, Boston, MA, 2006;193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ignjatovic J, Galli L. The S1 glycoprotein but not the N or M proteins of avian infectious bronchitis virus induces protection in vaccinated chickens. Arch Virol 1994;138:117–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Corse E, Machamer CE. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 2003;312:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Youn S, Leibowitz JL, Collisson EW. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology 2005;332:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zúñiga S, Sola I, Moreno JL et al. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 2007;357:215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lenz DH, Mok KC, Lilley BN et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and vibrio cholerae. Cell 2004;118:69–82. [DOI] [PubMed] [Google Scholar]
- 132. Jiang W, Hou Y, Inouye M. CspA, the major cold‐shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem 1997;272:196–202. [DOI] [PubMed] [Google Scholar]
- 133. Sittka A, Pfeiffer V, Tedin K et al. The RNA chaperone Hfq is essential for the virulence of salmonella typhimurium. Mol Microbiol 2007;63:193–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Feng Y‐X, Campbell S, Harvin D et al. The human immunodeficiency virus type 1 gag polyprotein has nucleic acid chaperone activity: Possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol 1999;73:4251–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Gl C, Ivanyi‐Nagy R, Gabus C et al. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res 2004;32:2623–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Mir MA, Panganiban AT. The bunyavirus nucleocapsid protein is an RNA chaperone: Possible roles in viral RNA panhandle formation and genome replication. RNA 2006;12:272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zúñiga S, Cruz JLG, Sola I et al. Coronavirus Nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol 2010;84:2169–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Patterson S, Bingham RW. Electron microscope observations on the entry of avian infectious bronchitis virus into susceptible cells. Arch Virol 1976;52:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hodgson T, Casais R, Dove B et al. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J Virol 2004;78:13804–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Armesto M, Cavanagh D, Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS One 2009;4:e7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Keep S, Bickerton E, Armesto M et al. The ADRP domain from a virulent strain of infectious bronchitis virus is not sufficient to confer a pathogenic phenotype to the attenuated Beaudette strain. J Gen Virol 2018;99:1097–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Chousalkar KK, Roberts JR, Reece R. Comparative histopathology of two serotypes of infectious bronchitis virus (T and N1/88) in laying hens and cockerels. Poult Sci 2007;86:50–58. [DOI] [PubMed] [Google Scholar]
- 143. Toro H, Godoy V, Larenas J et al. Avian infectious bronchitis: Viral persistence in the Harderian gland and histological changes after eyedrop vaccination. Avian Dis 1996;40:114–120. [PubMed] [Google Scholar]
- 144. Broadfoot DI, Smith JWM. Effects of infectious bronchitis in laying hens on egg production, percent Unsettable eggs and hatchability. Poult Sci 1954;33:653–654. [Google Scholar]
- 145. Demcenco B, Balan I, Petcu I et al. Features of avian infectious bronchitis and quality of poultry products. Moldova: Institutional Repository in Agricultural Sciences of the State Agrarian University of Moldova; 2019. [Google Scholar]
- 146. Ignjatovic J, Sapats SI. Avian infectious bronchitis virus. Rev Sci Tech 2000;19:493–508. [DOI] [PubMed] [Google Scholar]
- 147. Raj GD, Jones RC. Infectious bronchitis virus: Immunopathogenesis of infection in the chicken. Avian Pathol 1997;26:677–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yu L, Jiang Y, Low S et al. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis 2001;45:416–424. [PubMed] [Google Scholar]
- 149. Pereira NA, Alessi AC, Montassier HJ et al. Gonadal pathogenicity of an infectious bronchitis virus strain from the Massachusetts genotype. Braz J Microbiol 2019;50:313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zhang P, Liu X, Liu H et al. Astragalus polysaccharides inhibit avian infectious bronchitis virus infection by regulating viral replication. Microb Pathog 2018;114:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ignjatovic J, Ashton DF, Reece R et al. Pathogenicity of Australian strains of avian infectious bronchitis virus. J Comp Pathol 2002;126:115–123. [DOI] [PubMed] [Google Scholar]
- 152. Fabricant J, Levine PP. Experimental production of complicated chronic respiratory disease infection ("air sac" disease). Avian Dis 1962;6:13–23. [Google Scholar]
- 153. Benyeda Z, Mató T, Süveges T et al. Comparison of the pathogenicity of QX‐like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathol 2009;38:449–456. [DOI] [PubMed] [Google Scholar]
- 154. Chousalkar KK, Roberts JR. Ultrastructural study of infectious bronchitis virus infection in infundibulum and magnum of commercial laying hens. Vet Microbiol 2007;122:223–236. [DOI] [PubMed] [Google Scholar]
- 155. Chousalkar KK, Cheetham BF, Roberts JR. Effects of infectious bronchitis virus vaccine on the oviduct of hens. Vaccine 2009;27:1485–1489. [DOI] [PubMed] [Google Scholar]
- 156. Chen BY, Hosi S, Nunoya T et al. Histopathology and immunohistochemistry of renal lesions due to infectious bronchitis virus in chicks. Avian Pathol 1996;25:269–283. [DOI] [PubMed] [Google Scholar]
- 157. Winterfield RW, Hitchner SB. Etiology of an infectious nephritis‐nephrosis syndrome of chickens. Am J Vet Res 1962;23:1273–1279. [PubMed] [Google Scholar]
- 158. Siller WG, Cumming RB. The histopathology of an interstitial nephritis in the fowl produced experimentally with infectious bronchitis virus. J Pathol 1974;114:163–173. [DOI] [PubMed] [Google Scholar]
- 159. Chong KT, Apostolov K. The pathogenesis of nephritis in chickens induced by infectious bronchitis virus. J Comp Pathol 1982;92:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Hopkins SR. Serological comparisons of strains of infectious bronchitis virus using plaque‐purified isolants. Avian Dis 1974;18:231–239. [PubMed] [Google Scholar]
- 161. Mardani K, Browning GF, Ignjatovic J et al. Rapid differentiation of current infectious bronchitis virus vaccine strains and field isolates in Australia. Aust Vet J 2006;84:59–62. [DOI] [PubMed] [Google Scholar]
- 162. Julian RJ, Willis NG. The nephrosis‐nephritis syndrome in chickens caused by a Holte strain of infectious bronchitis virus. Can Vet J 1969;10:18–19. [PMC free article] [PubMed] [Google Scholar]
- 163. Lounas A, Oumouna‐Benachour K, Medkour H et al. The first evidence of a new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and unvaccinated broiler flocks in Algeria. Vet World 2018;11:1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Glahn RP, Wideman JRF, Cowen BS. Order of exposure to high dietary calcium and gray strain infectious bronchitis virus alters renal function and the incidence of Urolithiasis1. Poult Sci 1989;68:1193–1204. [DOI] [PubMed] [Google Scholar]
- 165. Wideman JRF, Closser JA, Roush WB et al. Urolithiasis in pullets and laying hens: Role of dietary calcium and Phosphorus1. Poult Sci 1985;64:2300–2307. [DOI] [PubMed] [Google Scholar]
- 166. Chen BY, Itakura C. Cytopathology of chick renal epithelial cells experimentally infected with avian infectious bronchitis virus. Avian Pathol 1996;25:675–690. [DOI] [PubMed] [Google Scholar]
- 167. Albassam MA, Winterfield RW, Thacker HL. Comparison of the Nephropathogenicity of four strains of infectious bronchitis virus. Avian Dis 1986;30:468–476. [PubMed] [Google Scholar]
- 168. Afanador G, Roberts JR. Effect of nephropathogenic infectious bronchitis viruses on renal function in young male broiler chickens. Br Poult Sci 1994;35:445–456. [DOI] [PubMed] [Google Scholar]
- 169. Liu W, Liu P, Wang T et al. Changes of antioxidant function and the mRNA expression levels of xanthine oxidase in primary chick kidney cell culture caused by nephropathogenic infectious bronchitis virus infection. Pak Vet J 2018;38:51–55. [Google Scholar]
- 170. Cavanagh D, Gelb J Jr. Infectious bronchitis. In: Saif YM, Fadley AM, Glisson JR, et al., editors. Diseases of poultry. 12th edn. Chişinău, Moldova: Blackwell Publishing, 2008;117–130. [Google Scholar]
- 171. Hewson KA, Noormohammadi AH, Devlin JM et al. Evaluation of a novel strain of infectious bronchitis virus emerged as a result of spike gene recombination between two highly diverged parent strains. Avian Pathol 2014;43:249–257. [DOI] [PubMed] [Google Scholar]
- 172. Ignjatovic J, Sapats SI, Ashton F. A long‐term study of Australian infectious bronchitis viruses indicates a major antigenic change in recently isolated strains. Avian Pathol 1997;26:535–552. [DOI] [PubMed] [Google Scholar]
- 173. Jackwood MW. Review of infectious bronchitis virus around the world. Avian Dis 2012;56:634–641. [DOI] [PubMed] [Google Scholar]
- 174. Cavanagh D. A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol 2001;30:109–115. [DOI] [PubMed] [Google Scholar]
- 175. Hofmann M, Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol 1988;26:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Sturman LS, Ricard CS, Holmes KV. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: Activation of cell‐fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J Virol 1985;56:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Benfield DA, Saif LJ. Cell culture propagation of a coronavirus isolated from cows with winter dysentery. J Clin Microbiol 1990;28:1454–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Li W, Moore MJ, Vasilieva N et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003;426:450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Dveksler GS, Dieffenbach CW, Cardellichio CB et al. Several members of the mouse carcinoembryonic antigen‐related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus‐A59. J Virol 1993;67:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Dveksler GS, Pensiero MN, Cardellichio CB et al. Cloning of the mouse hepatitis virus (MHV) receptor: Expression in human and hamster cell lines confers susceptibility to MHV. J Virol 1991;65:6881–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Delmas B, Gelfi J, L'Haridon R et al. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature 1992;357:417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Li BX, Ge JW, Li YJ. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007;365:166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Schultze B, Cavanagh D, Herrler G. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid‐containing receptors on erythrocytes. Virology 1992;189:792–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Winter C, Schwegmann‐Weßels C, Cavanagh D et al. Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J Gen Virol 2006;87:1209–1216. [DOI] [PubMed] [Google Scholar]
- 185. Miguel B, Pharr GT, Wang C. The role of feline aminopeptidase N as a receptor for infectious bronchitis virus. Arch Virol 2002;147:2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Gerna G, Cattaneo E, Cereda PM et al. Human coronavirus OC43 serum inhibitor and neutralizing antibody by a new plaque‐reduction assay. Proc Soc Exp Biol Med 1980;163:360–366. [DOI] [PubMed] [Google Scholar]
- 187. Gillette KG. Plaque formation by infectious bronchitis virus in chicken embryo kidney cell cultures. Avian Dis 1973;17:369–378. [PubMed] [Google Scholar]
- 188. Maier HJ, Hawes PC, Cottam EM et al. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. MBio 2013;4(5):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Knoops K, Kikkert M, Worm SHE et al. SARS‐coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 2008;6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Bijlenga G, Cook JKA, Gelb JJ et al. Development and use of the H strain of avian infectious bronchitis virus from The Netherlands as a vaccine: A review. Avian Pathol 2004;33:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Jackwood MW, Hilt DA, Sellers HS et al. Rapid heat‐treatment attenuation of infectious bronchitis virus. Avian Pathol 2010;39:227–233. [DOI] [PubMed] [Google Scholar]
- 192. Inaba Y, Sato K, Kurogi H et al. Replication of bovine coronavirus in cell line BEK‐1 culture. Arch Virol 1976;50:339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Sapats S, Ashton F, Wright P, Ignjatovic J. Novel variation in the N protein of avian infectious bronchitis virus. Virology 1996;226(2):412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]


