Abstract
RodZ of rod‐shaped bacteria functions to link MreB filaments to the Rod peptidoglycan (PG) synthase complex that moves circumferentially perpendicular to the long cell axis, creating hoop‐like sidewall PG. Ovoid‐shaped bacteria, such as Streptococcus pneumoniae (pneumococcus; Spn) that lack MreB, use a different modality for peripheral PG elongation that emanates from the midcell of dividing cells. Yet, S. pneumoniae encodes a RodZ homolog similar to RodZ in rod‐shaped bacteria. We show here that the helix‐turn‐helix and transmembrane domains of RodZ(Spn) are essential for growth at 37°C. ΔrodZ mutations are suppressed by Δpbp1a, mpgA(Y488D), and ΔkhpA mutations that suppress ΔmreC, but not ΔcozE. Consistent with a role in PG elongation, RodZ(Spn) co‐localizes with MreC and aPBP1a throughout the cell cycle and forms complexes and interacts with PG elongasome proteins and regulators. Depletion of RodZ(Spn) results in aberrantly shaped, non‐growing cells and mislocalization of elongasome proteins MreC, PBP2b, and RodA. Moreover, Tn‐seq reveals that RodZ(Spn), but not MreCD(Spn), displays a specific synthetic‐viable genetic relationship with aPBP1b, whose function is unknown. We conclude that RodZ(Spn) acts as a scaffolding protein required for elongasome assembly and function and that aPBP1b, like aPBP1a, plays a role in elongasome regulation and possibly peripheral PG synthesis.
Keywords: class A PBP function and regulation, elongasome assembly, peptidoglycan synthesis, synthetic‐viable genetic relationships
This paper establishes RodZ as an essential scaffolding protein required for the assembly and function of the elongasome that synthesizes peripheral peptidoglycan (pPG) in Streptococcus pneumoniae, which lacks an MreB homolog. (Top panel) The assembly hierarchy mediated by RodZ(Spn). (Bottom panel) This paper also reports synthetic‐viable, suppressor relationships between Class A aPBP1b and aPBP1a and components of the core pPG elongasome, suggestive of failsafe bypass mechanisms that restore function of the pPG elongasome and viability.
1. INTRODUCTION
The peptidoglycan (PG) mesh, which consists of peptide‐cross‐linked glycan chains, determines the shape of eubacteria, contributing to their colonization and survival in different environmental niches (Daitch & Goley, 2020; Egan et al., 2020; Kumar et al., 2022; Rohs & Bernhardt, 2021; Young, 2006). PG also protects bacteria from turgor pressure and serves as a scaffold for the attachment of extracellular proteins and exopolysaccharide capsules and wall‐teichoic acids of Gram‐positive bacteria, which lack an outer membrane (Rajagopal & Walker, 2017; Vollmer et al., 2019). PG synthesis has been a major target for many classes of antibiotics, starting with the β‐lactam penicillin (Bush & Bradford, 2016); however, resistance to cell‐wall targeted antibiotics is now a serious, widespread health problem (CDC, 2019; Hakenbeck, 2014; Hakenbeck et al., 2012). Because of its extracellular location, absence in eukaryotic hosts, and many vulnerable enzymatic and regulatory steps, PG synthesis remains a leading target for the discovery and development of new classes of antibiotics (CDC, 2019; den Blaauwen et al., 2014; Lewis, 2020; Ling et al., 2015; Sham et al., 2012).
Formation of ovoid‐shaped (ovococcal) bacteria (Zapun et al., 2008), such as the major respiratory pathogen Streptococcus pneumoniae (pneumococcus; Spn) (Weiser et al., 2018), requires two modes of PG synthesis (reviewed in [Briggs et al., 2021; Massidda et al., 2013; Perez, Boersma, et al., 2021; Vollmer et al., 2019]). Septal PG (sPG) synthesis separates dividing pneumococcal cells at midcell into two daughter cells, whereas peripheral PG (pPG) synthesis is a form of sidewall PG synthesis that also emanates from the midcell division ring of dividing pneumococcal cells. All protein components for both modes of PG synthesis are initially organized by FtsZ, FtsA, and EzrA into a single ring at the equators of predivisional pneumococcal cells (Perez et al., 2019). In the course of division, the sPG synthesis machine moves with the constricting FtsZ ring at the leading edge of the closing septal annulus, separate from the pPG synthesis machine that remains at the outer edge of the septal disk (Briggs et al., 2021). This dual pattern of PG synthesis was recently visualized as two concentric midcell rings by high‐resolution structured‐illumination microscopy (3D‐SIM) (Perez, Boersma, et al., 2021) and direct stochastic optical reconstruction microscopy (dSTORM) (Trouve et al., 2021) microscopy of vertically oriented pneumococcal cells. At the start of division, pPG synthesis likely begins slightly before sPG synthesis, but throughout most of the cell cycle, sPG and pPG synthesis and PG remodeling at midcell are concurrent and highly coordinated (Briggs et al., 2021; Perez, Boersma, et al., 2021; Trouve et al., 2021; Tsui et al., 2014; Wheeler et al., 2011).
Midcell localization of sPG and pPG synthesis in ovococci is fundamentally different in many ways from the patterns of sPG and sidewall PG synthesis used by rod‐shaped bacteria (Rohs & Bernhardt, 2021). In Bacillus subtilis, which like S. pneumoniae is a low‐GC Gram‐positive bacterium, a wall of sPG is synthesized during septal closure without surface constriction between daughter cells that are later separated by PG hydrolases (Errington & Wu, 2017; Straume et al., 2021). Gram‐positive coccoid bacteria, such as Staphylococcus aureus, also synthesize a septal cell wall between daughter cells, which are later separated by a rapid PG hydrolytic popping mechanism (Lund et al., 2018; Saraiva et al., 2020; Straume et al., 2021). In Gram‐negative Escherichia coli, septal closure and cell separation are largely concurrent (Rohs & Bernhardt, 2021), similar to what is observed for S. pneumoniae. However, in E. coli, the regulation of sPG synthesis occurs by a different mechanism than in S. pneumoniae, which lacks FtsN‐mediated activation of essential Class B PBP3 (FtsI) transpeptidase activity required for septal closure in E. coli (Briggs et al., 2021; Pichoff et al., 2019; Rohs & Bernhardt, 2021).
Early in cell division of rod‐shaped bacteria, preseptal PG synthesis pushes sidewall PG outward from the Z‐ring (Aaron et al., 2007; Pazos et al., 2018; van Teeseling, 2021), resembling the pPG synthesis that occurs throughout the pneumococcal cell cycle (Briggs et al., 2021). However, following preseptal PG synthesis, the Rod‐complex elongasome, containing conditionally essential MreB and the sidewall PG synthase complex, assembles along the curved cylindrical body of rod‐shaped cells (Bratton et al., 2018; Hussain et al., 2018; Morgenstein et al., 2015; Rohs & Bernhardt, 2021). MreB is an actin‐like homolog that polymerizes into multiple, short, curved filaments along the cell membrane, perpendicular to the cell long axis, which is the direction of maximum negative Gaussian curvature (Bratton et al., 2018; Hussain et al., 2018; Morgenstein et al., 2015; Rohs & Bernhardt, 2021). MreB filaments are linked to the sidewall PG synthase complex by RodZ, whose cytoplasmic helix‐turn‐helix (HTH) interacts with MreB inside the cell (Ago & Shiomi, 2019; Bendezu et al., 2009; Morgenstein et al., 2015; van den Ent et al., 2010). Bitopic RodZ (see Figures 1 and S1) also forms a complex with the bitopic and polytopic PG synthesis proteins in the elongasome, including MreC and MreD (positive regulators) (Rohs et al., 2018; Rohs et al., 2021; Rohs & Bernhardt, 2021), RodA (SEDS glycosyl transferase) (Meeske et al., 2016; Sjodt et al., 2018; Sjodt et al., 2020), and an essential Class B penicillin‐binding protein (bPBP2 transpeptidase in E. coli) (Rohs & Bernhardt, 2021; Sjodt et al., 2020). The interaction between RodZ and MreB can modulate the density and length of MreB filaments (Bratton et al., 2018; Colavin et al., 2018; Hussain et al., 2018). In E. coli, a limiting amount of bPBP2 seems to bind to PG and recruit the rest of the Rod elongasome (Ozbaykal et al., 2020). Movement of the assembled Rod elongasome is driven by sidewall PG synthesis itself, rather than by ATPase‐dependent treadmilling of MreB (Rohs & Bernhardt, 2021), with the MreB filaments serving as curvature‐sensing rudders (Hussain et al., 2018). Finally, in contrast to the parallel PG synthesis by the Rod elongasome, synthesis of sidewall PG by Class A PBPs appears to be largely non‐ordered, possibly filling in or reinforcing gaps left by the Rod system (Cho et al., 2016; Dion et al., 2019; Lamanna & Maurelli, 2022; Rohs & Bernhardt, 2021).
FIGURE 1.
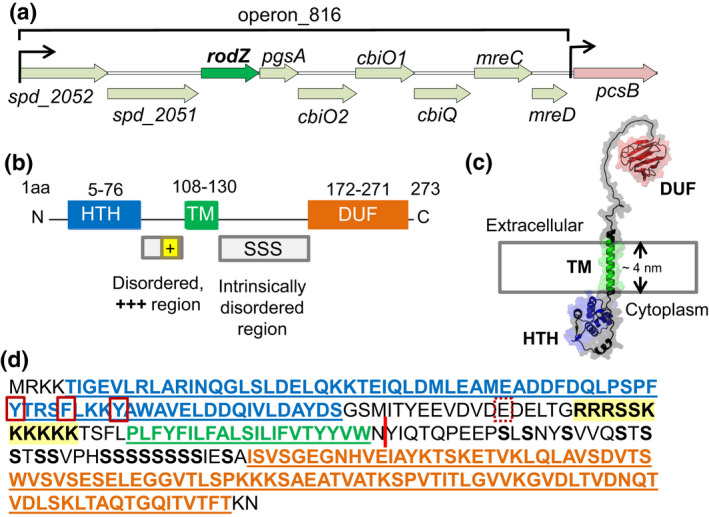
Location and domains of RodZ(Spn). (a) rodZ (spd_2050) is predicted to be a member of operon_816 in the Spn D39 chromosome (Slager et al., 2018). Operon_816 consists of spd_2052 (putative zinc protease), spd_2051 (putative M16 family peptidase), rodZ, pgsA (CDP‐diacylglycerol‐glycerol‐3‐phosphate 3‐phosphatidyltransferase), cbiO2, cbiO1, cbiQ (putative ATPase and transmembrane components of a cobalt ABC transporters), mreC, and mreD. (b) 2D protein structure of RodZ(Spn). Black line indicates residues that are not part of known domains. The intracellular helix‐turn‐helix (HTH) domain, transmembrane (TM), and extracellular domain of unknown function (DUF 4115) are depicted as blue, green, and orange, respectively, and intrinsically disordered regions are represented as gray boxes. The HTH and DUF domains are predicted to be in alpha helices and beta sheets, respectively, by AlphaFold2 (Jumper et al., 2021). TM domain was determined with TMHMM server. The positively charged juxta‐membrane region of the intracellular linker is shown as a yellow box (+++). SSS symbolizes multiple serine residues in the extracellular linker. (c). Predicted 3D structure of RodZ(Spn) generated using the AlphaFold2 webserver. (d) Amino acid sequence of RodZ(Spn). Color coding is as described in (b), except that the multiple serine residues are bolded and the positive juxta‐membrane is both bolded and highlighted in yellow. Y51 and F55 within helix 4 of the HTH domain (red boxes) correspond to the positions of aromatic amino acids that interact with MreB in E. coli (see Figure S1) (van den Ent et al., 2010). E89 (dotted box) corresponds to the position of S85 in RodZ(Bsu) that may be phosphorylated (Sun & Garner, 2020). The red bar between N131 and Y132 marks the first TA site in a TAT (Y132) codon with a Tn‐mariner insertion recovered by Tn‐seq of the WT strain (Figure 2). The junction of the Tn insertion creates a TAA stop codon, indicating that RodZ(M1‐N131) is viable.
In contrast to rod‐shaped bacteria, the pPG elongasome of S. pneumoniae is zonal and confined to the midcell of dividing pneumococcal cells. As division proceeds, the pPG elongasome locates to an outer ring of PG synthesis at the edge of the septal annulus (Briggs et al., 2021; Perez, Boersma, et al., 2021; Trouve et al., 2021). Homologs of MreC, MreD, an essential Class B PBP (bPBP2b), and RodA have been associated with the pneumococcal pPG elongasome by genetic, physiological, and bacterial two‐hybrid (B2H) experiments (Berg et al., 2013; Land & Winkler, 2011; Massidda et al., 2013; Philippe et al., 2014; Stamsas et al., 2017; Straume et al., 2017; Tsui et al., 2014; Zheng et al., 2017). In addition, Class A aPBP1a and CozE have been linked to pneumococcal pPG elongasome function through a synthetic‐viable genetic relationship, in that Δpbp1a or cozE depletion suppresses ΔmreCD (Fenton et al., 2016; Land & Winkler, 2011; Tsui et al., 2016). Complexes containing aPBP1a and pPG elongasome proteins have also been detected by co‐immunoprecipitation (co‐IP) and bacterial two‐hybrid (B2H) assays (Fenton et al., 2016; Stamsas et al., 2017). Similarly, aPBP1a is synthetically viable with muramidase MpgA (formerly MltG(Spn), in that Δpbp1a suppresses ΔmpgA (Taguchi et al., 2021; Tsui et al., 2016). Each of these proteins localizes to the midcell of dividing pneumococcal cells, consistent with the zonal mechanism of pPG elongation (Briggs et al., 2021; Land et al., 2013; Tsui et al., 2014; Tsui et al., 2016).
S. pneumoniae, like most non‐rod‐shaped bacteria, encodes a RodZ homolog (Figures 1 and S1), despite the absence of an MreB homolog. The secondary structure of RodZ(Spn) is remarkably similar to RodZ homologs in rod‐shaped bacteria (Figures 1 and S1) (Ago & Shiomi, 2019; Alyahya et al., 2009; Bendezu et al., 2009; Shiomi et al., 2008). RodZ(Spn) contains a cytoplasmic N‐terminal HTH domain of the XRE family, which contains five helices that often mediate protein interactions (Aravind et al., 2005). The HTH domain is connected to a TM domain by a disordered “juxtamembrane” domain, which is positively charged in RodZ(Spn) and RodZ(Eco) (Figures 1d and S1) (Bendezu et al., 2009). Non‐conserved Ser85 in this linker region of RodZ(Bsu) (Figures 1 and S1b) has been reported to be phosphorylated in a preliminary report (Sun & Garner, 2020). The non‐conserved extracellular linker of RodZ(Spn) contains a large number of repeated Ser residues and connects the TM domain to a domain of unknown function (DUF#4115) that is predicted by AlphaFold2 to fold into a beta‐strand structure (Figure 1c), similar to DUF determined for RodZ(Bsu) (Ago & Shiomi, 2019). rodZ is essential or conditionally essential in E. coli, B. subtilis, and Caulobacter crescentus (Alyahya et al., 2009; Bendezu et al., 2009; Muchova et al., 2013). Structure–function mutagenesis shows that the cytoplasmic HTH and TM domains of RodZ are essential for its function in E. coli and C. crescentus, whereas extracellular domains can be deleted without severe cell growth and morphology phenotypes (Alyahya et al., 2009; Bendezu et al., 2009; Morgenstein et al., 2015). In this regard, RodZ of Rickettsia and Chlamydia species lack an extracellular domain (Kemege et al., 2015). These obligate intracellular pathogens lack FtsZ and use a modified Rod complex consisting of homologs of MreB, RodZ, MreC, a bPBP, and RodA to synthesize midcell PG (Liechti et al., 2016; Ouellette et al., 2020; Ranjit et al., 2020).
In this paper, we demonstrate by Tn‐seq, transformation assays, and protein depletion that RodZ(Spn) is conditionally essential in serotype‐2 D39 strains of S. pneumoniae at 37°C. Tn‐seq and structure–function analyses show that this essentiality requires the HTH and TM domains of RodZ(Spn), but not the extracellular linker or DUF domains. Suppression patterns of ΔrodZ and ΔmreC, but not ΔcozE, mutants phenocopy each other, linking RodZ(Spn) to the pPG elongasome. The conclusion that RodZ(Spn) is a member of the pPG elongasome is supported by interaction studies using co‐IP and B2H assays and by microscopic co‐localization of RodZ(Spn) and MreC(Spn) or aPBP1a(Spn) throughout the pneumococcal cell cycle. Depletion of RodZ(Spn) stops growth and results in viable, rounded, heterogeneous cells with a qualitatively different appearance from cells depleted for MreC or bPBP2b (Berg et al., 2013; Land & Winkler, 2011; Tsui et al., 2014). Depletion of RodZ(Spn) or MreC(Spn) further reveals a hierarchy for pPG elongasome assembly. Finally, Tn‐seq experiments show the unexpected result that Class A aPBP1b, whose function is not known, is in a synthetic‐viable genetic relationship with RodZ, but not MreCD, whereas Class A aPBP1a is in a synthetic‐viable genetic relationship with MreC, MreD, and RodZ. Together, these results show that RodZ(Spn) still acts as an essential scaffold protein through its HTH and TM domains for pneumococcal pPG elongasome assembly and function, despite the absence of MreB. This study also shows that diverse cell morphology and genetic phenotypes result when different members of the pPG elongasome are absent or depleted. Last, this work shows that aPBP1b and aPBP1a play different roles in modulating the function of the S. pneumoniae pPG elongasome and possibly participate in pPG synthesis. These results are discussed in terms of a model in which failsafe mechanisms can bypass or regulate the function of the pneumococcal core RodZ‐MreCD‐bPBP2b‐RodA elongasome to ensure viability.
2. RESULTS
2.1. RodZ is conditionally essential in S. pneumoniae D39 at 37°C
RodZ(Spn) is annotated as “probably not essential” in serotype 2 D39 strains based on recent genomics approaches (see PneumoBrowse site) (Slager et al., 2018). Likewise, a Tn‐seq screen of serotype 4 TIGR4 strains recovered insertions in rodZ that seemed to grow in laboratory media (van Opijnen & Camilli, 2012). ΔrodZ mutants were reported in unencapsulated (Δcps) R6 laboratory strains (Martin‐Galiano et al., 2014; Stamsas et al., 2017; Straume et al., 2017), whose progenitor is strain D39 (Lanie et al., 2007). However, we previously reported that we could not obtain ΔrodZ mutants in an unencapsulated (Δcps) derivative constructed in progenitor strain D39 (Tsui et al., 2016).
We performed a series of experiments to reconcile these conflicting previous results, leading to the conclusion that rodZ is essential for growth at 37°C in unencapsulated and encapsulated D39 strains, although poor growth occurs at 32°C (Figure S2). Tn‐seq analysis showed that insertions occur in the non‐essential carboxyl‐terminal DUF‐domain half of rodZ(Spn), but are not recovered in the essential transmembrane and amino‐terminal domains (Figures 1d and 2a [WT]). This pattern of insertions relative to RodZ(Spn) domain functions is taken up further below and underlies why a substantial number of non‐essential rodZ insertions were detected in previous Tn‐seq experiments. In addition, the Tn‐seq profile here (Figure 2a) confirms previous Tn‐seq (Fenton et al., 2016) and complementation results (see below; [Rued et al., 2017]) showing that mreC is essential in S. pneumoniae D39, contrary to a conclusion in (Straume et al., 2017).
FIGURE 2.
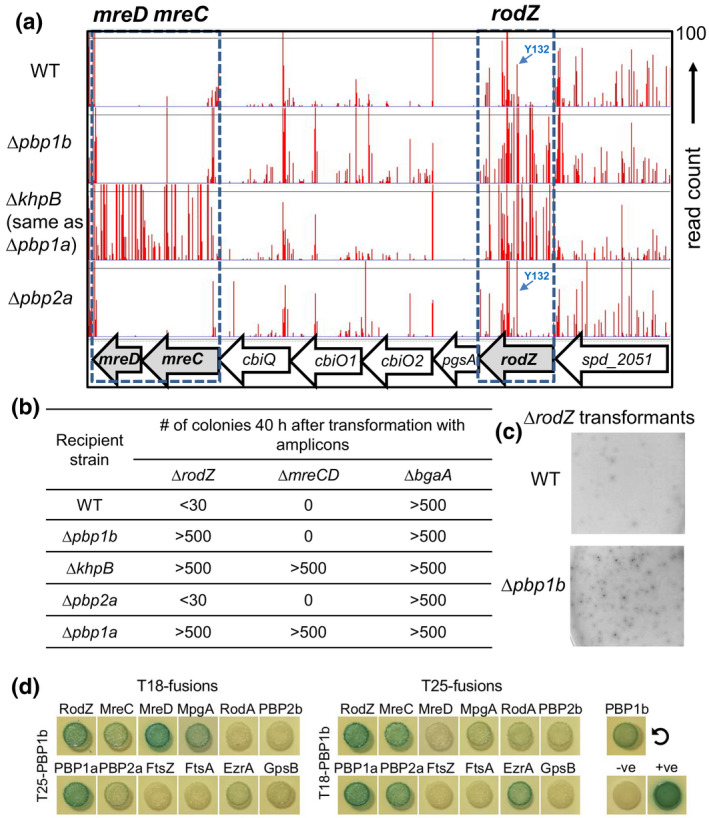
Tn‐seq analysis reveals suppression of ∆rodZ, but not ∆mreCD, lethality by ∆pbp1b deletion. (a) Tn‐seq transposon insertion profile for the genome region covering spd_2051, rodZ, pgsA, cbiO2, cbiO1, cbiQ, mreC, and mreD of mini‐mariner Malgellan6 transposon (Tn) into the genomes of WT (D39 ∆cps rpsL1, IU1824), ∆pbp1b (IU14697), ∆khpB (IU10592), or ∆pbp2a (IU13256) strains. In vitro transposition reactions, containing purified genomic DNA, Magellan6 plasmid DNA, and purified MarC9 mariner transposase, transformation, harvesting of transposon‐inserted mutants, NextSeq 75 high‐output sequencing, and analysis were performed as described in Experimental procedures. (b) Transformation assay confirming that Δpbp1b suppresses RodZ, but not MreCD, essentiality. Results were obtained 40 h after transformation of WT, ∆pbp1b, ∆khpB, ∆pbp2a, or ∆pbp1a (IU6741) strains with linearized ∆rodZ::Pc‐aad9, ∆mreCD<>aad9, or positive control ∆bgaA::Pc‐erm amplicons. Numbers of transformants were normalized to correspond to 1 ml of transformation mixture. Similar results were obtained in two or more independent experiments. Similar results were obtained after 24 h of incubation, except that colonies ∆rodZ ∆pbp1b transformants were fainter in appearance than at 40 h, and <10 ∆rodZ colonies were obtained in the WT and ∆pbp2a backgrounds (data not shown). (c) Appearance of colonies of the WT or ∆pbp1b strain 40 h after transformation with the ∆rodZ::Pc‐aad9 amplicon. (d) aPBP1b interacts with RodZ, MreC, MreD, EzrA, MpgA, aPBP1a, and aPBP2a as well as with itself (circular arrow) in B2H assays. Agar plates were photographed after 40 h at 30°C. B2H assays were performed as described in Experimental procedures.
Transformation assays confirmed that ΔrodZ was not viable in unencapsulated WT D39 strains at 37°C, unless it was complemented in a merodiploid strain by an ectopic copy of rodZ + that was under control of a zinc‐inducible promoter (Table 1, rows 1–4). At the lower temperature of 32°C, we did observe small colonies of ΔrodZ mutants in merodiploid strains lacking the −Zn inducer for rodZ + expression (Figure S2a) or in transformation assays (Figure S2c, line 3). However, we found that slow growth at 32°C is a phenotype of mutants lacking other components of the pneumococcal pPG elongasome, including MreC, bPBP2b, or RodA (Figures S2b,c, lines 4–6). In contrast to the unencapsulated D39 background, ΔrodZ transformed into the R6 laboratory strain (Table 1, line 17), as reported previously (Stamsas et al., 2017; Straume et al., 2017); however, R6 derivatives contain mutations in pbp1a that suppress mutations in genes encoding the peripheral PG (pPG) synthesis elongasome machine (Land & Winkler, 2011; Tsui et al., 2016), including ΔmreC (Table 1, line 17). Finally, comparable experiments at 37°C showed that ΔrodZ could not be transformed into encapsulated D39 progenitor strains (Table 1, lines 18 and 19). Numerous mutations suppress ΔrodZ in S. pneumoniae (see next section) and likely account for the small number of colonies that arose in the D39 strains transformed with ΔrodZ amplicons. The essentiality of rodZ in the encapsulated D39 strains was corroborated in merodiploid strains, where hundreds of colonies were obtained only in the complementation strain (+Zn), but not in the depleted ΔrodZ strain (−Zn; Table 1, lines 20 and 21). We conclude that rodZ is essential for growth of unencapsulated or encapsulated D39 strains of S. pneumoniae at the optimal culture temperature of 37°C. Because capsule partially masks primary phenotypes of PG synthesis mutants in S. pneumoniae and complicates microscopy due to cell chaining (Barendt et al., 2009), the rest of these studies of rodZ physiology and function were performed in the D39 unencapsulated genetic background.
TABLE 1.
∆rodZ has a similar suppression pattern as ∆mreC, but not to ΔcozE, in transformation assays at 37°C a
| Recipient strain and condition | Genotype | Number of colonies at 22 h after transformation with deletion amplicons b | ||
|---|---|---|---|---|
| ∆rodZ | ∆mreC | ∆cozE | ||
| IU1824 (D39 ∆cps rpsL1) unencapsulated genetic background | ||||
| 1. IU1824 −Zn | WT | 0 c | 0 (3) | >500 tiny (4) |
| 2. IU1824 +Zn d , e | 0 f (3) | 0 (2) | >500 tiny (2) | |
| 3. IU9613 −Zn |
rodZ +// ΔbgaA::PZn‐rodZ |
0 c (6) | 0 (3) | >500 tiny (1) |
| 4. IU9613 +Zn d | >500 (6) | 0 (3) | >500 tiny (1) | |
| 5. IU10220 −Zn |
mreC +// ΔbgaA::PZn‐mreC |
0 c (3) | 0 (4) | >500 tiny (1) |
| 6. IU10220 +Zn d | 0 f (3) | >500 (4) | >500 tiny (1) | |
| 7. IU12681 −Zn |
cozE +// ΔbgaA::PZn‐cozE |
0 c (1) | 0 (1) | >500 tiny (2) |
| 8. IU12681 +Zn d | 0 f (1) | 0 (1) | >500 normal (2) | |
| 9. IU6741 | ∆pbp1a | >500 (2) | >500 (2) | >500 tiny (2) |
| 10. IU9760 |
mpgA(Y488D) (formerly mltG) |
>500 (2) | >500 (2) | <300 tiny (2) |
| 11. IU9036 | ∆khpA | >500 (2) | >500 (3) | >500 tiny (2) |
| 12. IU12719 |
ftsA +// ΔbgaA::P ftsA ‐ftsA |
>500 g (2) | >500 (2) | >500 tiny (1) |
| 13. IU12310 −Zn |
ftsA +// ΔbgaA::PZn‐ftsA |
0 c (1) | 0 h (2) | nd |
| 14. IU12310 +Zn e | >500 g (2) | >500 h (2) | nd | |
| 15. IU12286 −Zn |
ftsZ +// ΔbgaA::PZn‐ftsZ |
0 c (3) | 0 (2) | >500 tiny (1) |
| 16. IU12286 +Zn e | 0 f (3) | 0 (2) | >500 tiny (1) | |
| R6 unencapsulated laboratory strain background | ||||
| 17. R6 (EL59) | WT | >500 (2) | >500 (1) | >500 small (2) |
| D39 cps + encapsulated background | ||||
| 18. IU1690 | cps + | <12 colonies i (5) | <2 (3) | ≈300 small (2) |
| 19. IU1781 | cps + rpsL1 | ≈40 colonies i ; various sizes (7) | <5 (2) | ≈300 small (2) |
| Δcps and cps + ΔrodZ//P Zn ‐rodZ + merodiploid strains depleted or containing RodZ | ||||
| Number of colonies 22 h after transformation with markerless rodZ Δ(21–257 aa) amplicon j | ||||
| Transformation condition | ||||
| Recipient strain | Genotype | −Zn | +Zn | |
| 20. IU12515 | D39 Δcps rpsL1 ΔrodZ::Pc ‐[kan‐rpsL +]//ΔbgaA::PZn‐rodZ | <5; various sizes (2) | >300 WT size (2) | |
| 21. IU15645 | D39 cps + rpsL1 ΔrodZ::Pc ‐[kan‐rpsL +]//ΔbgaA::PZn‐rodZ | <16; various sizes (2) | ≈300 WT size (2) | |
Recipient strains were constructed as described in Table S1. Transformations and visualization of colonies normalized to 1 ml of transformation mixture were performed as described in Experimental procedures. All transformation experiments were performed with no added DNA as the negative control and with a Δpbp1b amplicon containing the same antibiotic selection as the positive control. 30 ng of purified amplicons were used for each transformation. The volumes of transformation mixture plated (100–500 μl) were adjusted to provide ≈150–300 colonies with Δpbp1b amplicons. Transformations with control Δpbp1b or Δspd_1874 amplicons yielded >500 colonies for Δcps strains per 1 ml of transformation mixture. Transformants were confirmed by PCR reactions.
Results are presented as the number and appearance of colonies (amplicon used, number of biological replicates [n]). Unless specified, the sizes of colonies were similar to those obtained with the Δpbp1b amplicon. Amplicons generated with primers and genomic DNA templates listed in Tables S1 and S4 were: ΔrodZ::Pc ‐aad9, ΔrodZ::Pc ‐erm, ΔrodZ::Pc ‐[kan‐rpsL +], ΔmreC::Pc ‐erm, ΔcozE::Pc ‐erm, and ΔcozE::Pc ‐cat. Similar results were obtained with amplicons with different antibiotics markers when transformed into D39 ∆cps rpsL1 background strains.
Fast‐growing suppressors (<5 colonies per plate) were occasionally seen for the WT or merodiploid strains under the non‐inducing condition.
Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4) was added to BHI growth media to induce RodZ, MreC, or CozE expression in merodiploid strains before transformation, and in the transformation mixes, which were then divided into soft agar and TSAII‐BA plates containing or lacking Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4).
0.5 mM ZnCl2 + 0.05 mM MnSO4 (Zn) was used in the transformations to induce FtsA or FtsZ expression.
Faint colonies were present when transformation was carried out in the presence of Zn. However, these colonies are not viable when re‐streaked on TSAII‐BA plates containing antibiotics.
No transformants were obtained previously when a promoterless ∆rodZ<>aad9 amplicon was used to transform IU12310 (D39 Δcps rpsL1 ΔbgaA::P Zn ‐ftsA) in the presence of Zn inducer (Zheng et al., 2017). We reprised this transformation with amplicons that contain a promoter in front of antibiotic‐resistance genes (ΔrodZ::Pc ‐aad9 or ΔrodZ::Pc ‐erm) and obtained >500 transformants (row 15). In addition, we obtained >500 transformants of promoter‐containing amplicons into strains that constitutively express ftsA (rows 13). Two reasons could explain these different outcomes. First, the lack of transformants obtained before could be due to insufficient expression of the in‐frame antibiotic resistance gene inserted into the rodZ reading frame. Alternatively, ΔrodZ::Pc ‐aad9 and ΔrodZ::Pc ‐erm constructs may be polar and increase transcription of the downstream genes (pgsA, cbiO1, cbiO2, cbiQ, mreC and mreD; Figure 1a), which may somehow cause ftsA overexpression to suppress the lethality of rodZ deletion. We did not distinguish between these two explanations in this study.
ΔmreCD<>aad9 amplicon was used instead of ΔmreC::Pc‐erm amplicon for these transformations.
ΔrodZ::Pc ‐aad9 amplicons used in these transformations.
2.2. ∆rodZ has a similar suppression pattern as ∆mreC, but not to Δ cozE, in transformation assays at 37°C
Mutations in essential genes of the pneumococcal PG elongasome complex, including mreC, pbp2b, and rodA, are suppressed by several kinds of mutations (Land & Winkler, 2011; Stamsas et al., 2017; Tsui et al., 2016; Zheng et al., 2017). For example, ΔmreC is suppressed by Δpbp1a (Class A aPBP1a), mpgA(Y488D) (reduced activity of MpgA (formerly MltG(Spn)) muramidase, ΔkhpA (RNA binding regulator), and overexpression of FtsA (division actin‐homolog) (Table 1, lines 9–14) (Land & Winkler, 2011; Stamsas et al., 2017; Taguchi et al., 2021; Tsui et al., 2016; Zheng et al., 2017). To link RodZ to MreC and pPG synthesis, we showed that ΔrodZ has the same suppression pattern as ΔmreC. ΔrodZ or ΔmreC is complemented in merodiploid strains by ectopic expression (+Zn) of rodZ + or mreC +, respectively (Table 1, lines 4 and 6). Like ΔmreC, ΔrodZ is suppressed by Δpbp1a, mpgA(Y488D), ΔkhpA, and overexpression of FtsA (Table 1, lines 9–14). This identical suppression pattern provides strong evidence that RodZ functions in the pPG synthesis elongasome, along with MreC, bPBP2b, and RodA. In addition, overexpression of MreC or FtsZ in merodiploid strains did not bypass the requirement for RodZ (Table 1, lines 6 and 16). Likewise, the requirement for MreC was not bypassed by overexpression of RodZ or FtsZ (Table 1, lines 4 and 16).
We used transformation assays to study two other aspects of RodZ function. CozE was discovered in a Tn‐seq of essential genes that become dispensable in a mutant lacking aPBP1a (Fenton et al., 2016). Localization and interaction studies indicated that CozE is a member of the MreCD complex in the pneumococcal elongasome. Therefore, we fully expected ΔcozE mutants to show the same suppression patterns as ΔmreC and ΔrodZ mutants. Unexpectedly, we found that ΔcozE significantly reduced, but did not abolish, colony growth, indicating that cozE is dispensable under the conditions used here (Table 1, line 1 and 2). This non‐essentiality was recapitulated in Tn‐seq experiments showing that insertion in cozE are recovered in the WT strain propagated in BHI broth (see below; data not shown). The colony growth defect of a ΔcozE mutant was fully complemented by ectopic expression of cozE + in a merodiploid strain (Table 1; line 8). However, the ΔcozE colony growth was not ostensibly improved by any of the mutations that suppressed both ΔmreC and ΔrodZ, including Δpbp1a (Table 1, lines 9–16). Thus, loss of CozE is not equivalent to loss of MreC or RodZ under some growth conditions, suggesting different functions in the elongasome and/or cell growth. This difference was not studied further here.
Last, rodZ is immediately upstream of essential pgsA, which encodes phosphatidylglycerol phosphate synthase, required for phospholipid synthesis (Figure 1a). In B. subtilis, insertion mutations in rodZ can have polar effects on pgsA expression (van Beilen et al., 2016). In this study of S. pneumoniae, rodZ mutant growth and cell morphology phenotypes are complemented by expression of an ectopic copy of rodZ + and markerless ΔrodZ mutations are used for most experiments (Table 1, lines 4, 20, and 21; Figures 3 and 4). Conversely, ectopic expression of pgsA + (+Zn) complemented ΔpgsA for growth in a ΔpgsA//PZn‐pgsA + merodiploid (Table S5, line 11), but overexpression of PgsA in a pgsA + //PZn‐pgsA + merodiploid strain (+Zn) did not allow growth of ΔrodZ transformants (Table 1, line 9). We conclude from genetic complementation along with the use of markerless alleles that phenotypes attributed to mutations in rodZ in this study are not caused by polarity on pgsA or other downstream genes.
FIGURE 3.
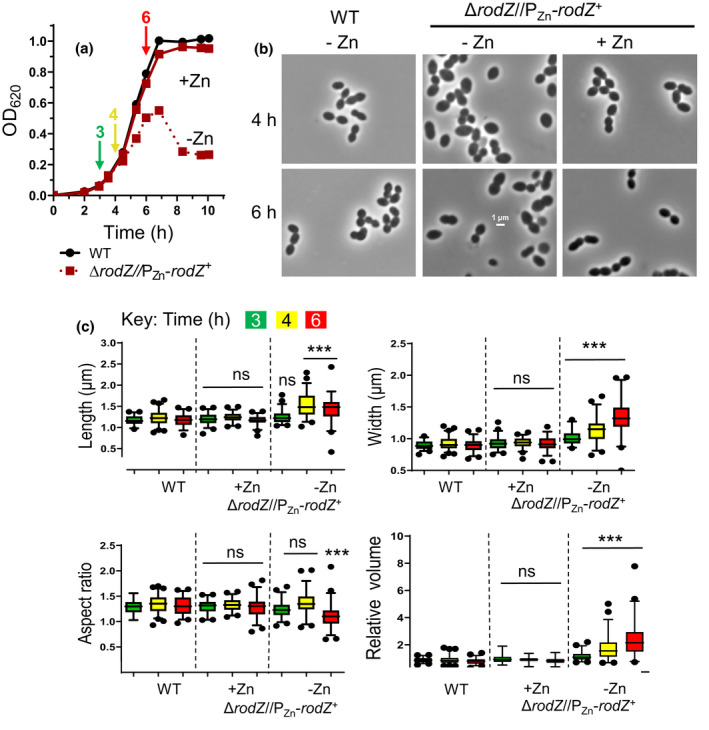
Depletion of RodZ results in cell rounding, indicative of a defect in peripheral PG synthesis. (a). Representative growth curves of IU1824 (WT) and depletion/complementation strain IU12738 (ΔrodZ//PZn‐rodZ +) with or lacking Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4). IU1824 or IU12738 was grown overnight in BHI broth at 37°C lacking or with Zn inducer, respectively. Samples were re‐suspended in fresh BHI ± Zn inducer to an OD620 ≈ 0.003. Arrows indicate times (3, 4, and 6 h) at which samples were taken for phase‐contrast microscopy. (b) Representative micrographs of IU1824 (WT) and IU12738 (ΔrodZ//PZn‐rodZ +) grown in the presence or absence of Zn inducer and sampled after 4 or 6 h. all images are at the same magnification (scale bar = 1 μm). (c) Box‐and‐whiskers plots (5 to 95 percentile) of cell length, width, aspect ratio, and relative volume measured for IU1824 grown in the absence of Zn inducer, and IU12738 grown in the presence or absence of Zn inducer for 3, 4, and 6 h. for each time point, ≈50–80 cells per sample were measured, and statistical analysis was conducted using the non‐parametric, one‐way ANOVA Kruskal–Wallis test in GraphPad prism. Statistical comparisons were carried out for IU12738 grown in the presence or absence of Zn inducer compared with the WT control at the respective time points. ***, p < 0.001; ns, non‐significant. Results shown are representative from one of at least three independent biological replicates.
FIGURE 4.
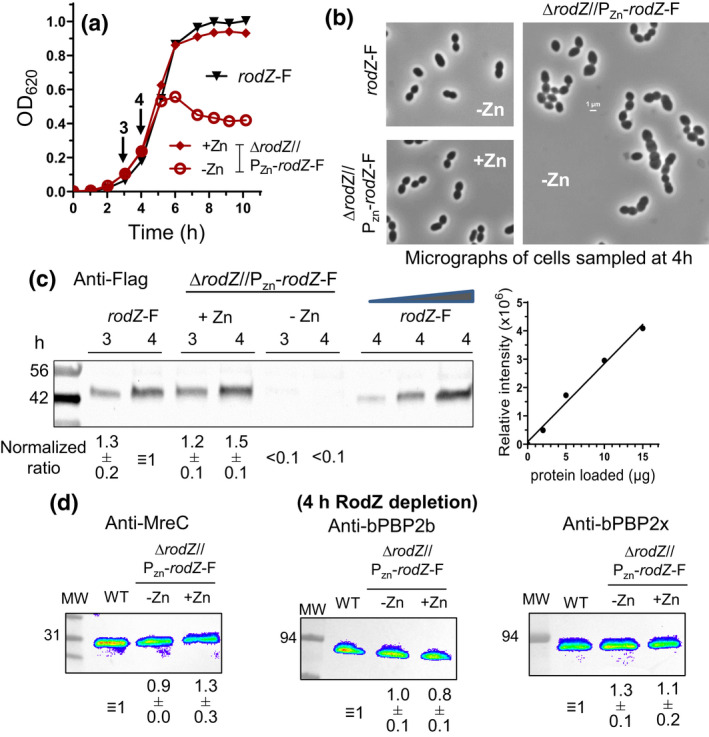
RodZ levels decrease to an undetectable level upon depletion for 3 h. (a) Representative growth curves of rodZ‐FLAG (IU14594) and depletion strain ΔrodZ//PZn‐ rodZ‐FLAG (IU10947) with or lacking Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4), where “F” is used as an abbreviation for the FLAG tag. IU14594 or IU10947 was grown overnight in BHI lacking or with Zn inducer, respectively, and diluted into BHI with no Zn for IU14594, and into BHI with or lacking Zn for IU10947. Cultures were sampled at 3 or 4 h for Western analysis (arrows). (b) Representative micrographs of IU14594 (rodZ‐F; −Zn) and IU 10947 (ΔrodZ//PZn‐rodZ‐F; +Zn or −Zn) sampled at 4 h. scale bar = 1 μm (all images are at the same magnification). (c) Representative quantitative Western blot showing RodZ‐F amount expressed from the native chromosomal site in IU14594 or from the ectopic site in the presence or absence of Zn inducer in IU10947 (ΔrodZ//PZn‐ rodZ +‐F) sampled at 3 and 4 h. 10 μg of crude cell lysates were loaded in the left 6 lanes, and 2, 5, or 15 μg were loaded in the right three lanes to generate a standard curve for quantitation. SDS‐PAGE and western blotting were carried out as described in Experimental procedures using Licor IR Dye800 CW secondary antibody detected with an Azure Biosystem 600. Signals obtained with anti‐FLAG antibody were normalized for total protein in each lane using Totalstain Q‐NC (Azure Scientific). Normalized ratios indicate RodZ‐F protein amounts (mean ± SEM) from 3 or 4 h samples for IU10947 relative to IU14594 at 4 h. (D) MreC, bPBP2b, and bPBP2x protein levels are not altered by RodZ depletion. Protein samples were obtained from IU14594 (rodZ‐F WT), or IU10947 (ΔrodZ//PZn‐ rodZ‐F) grown in the presence or absence of Zn inducer for 4 h. 3 μg of crude cell lysates were loaded in each lane. SDS‐PAGE and Western blotting were carried out with primary antibodies to MreC, bPBP2b, or bPBP2x. Chemiluminescence signals obtained with secondary HRP‐conjugated antibodies were detected using an IVIS imaging system. Ratios indicate protein amounts (average ± SEM) in IU10947 (ΔrodZ//PZn‐ rodZ +‐F) relative to those in IU14594 (WT) from two independent biological replicates.
2.3. RodZ is required for normal cell shape and morphology of S. pneumoniae
To study primary rodZ mutant phenotypes without accumulating suppressors (Table 1), RodZ was depleted in ΔrodZ//PZn‐rodZ + and ΔrodZ//PZn‐rodZ‐FLAG merodiploid mutants (−Zn), and cultures were sampled at various times after depletion (Figures 3 and 4, where F is used as an abbreviation for the FLAG tag here and elsewhere). RodZ depletion causes a decrease in apparent growth rate after ≈4.5 h, followed by a decrease in culture density at ≈7 h (Figures 3a and 4a). In controls for depletion experiments, cells expressing functional RodZ‐FLAG from the native locus grew at the same rate in BHI broth lacking or containing the Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4), where 1/10 MnSO4 was added to lessen Zn2+ toxicity (Figure S3a) (Jacobsen et al., 2011; Perez, Villicana, et al., 2021; Tsui et al., 2016), and cell morphology and cellular RodZ‐FLAG amount were not appreciably altered by Zn addition (Figure S3b,c). By 4 h of RodZ depletion, cells tended to become larger and more heterogeneous in shape, with some cells becoming round and others exhibiting pointed ends (Figures 3b,c and 4b). By 6 h of RodZ depletion, cell width and size increased significantly, and cells became more spherical with an average aspect ratio approaching 1 (Figure 3b,c). Quantitative western blotting showed that RodZ‐FLAG was not detectable by 3 h of depletion and that RodZ depletion for 4 h did not alter the cellular amounts of MreC, bPBP2b, or bPBP2x (Figure 4d). Depletion of RodZ also led to a moderate increase in the number of cells in chains (Figure S4).
Depletion of RodZ for 4 or 6 h is bacteriostatic and did not lead to a loss of cell viability, as determined by live‐dead staining (Figure 5) or by recovery of CFUs following RodZ depletion for at least 7 h (Figure S5). Finally, we tested whether overexpression of RodZ affected growth or cell morphology of S. pneumoniae, as happens in E. coli and C. crescentus (Alyahya et al., 2009; Bendezu et al., 2009; Shiomi et al., 2008). Overexpression of RodZ‐FLAG by ≈2.5 fold (Figure S6c) did not have an appreciable effect on pneumococcal growth (Figure S6a) or cell morphology (Figure S6b,d), although more lysed cells were observed when RodZ‐FLAG was overexpressed. Altogether, these results indicate that RodZ is required for normal morphology of pneumococcal cells. Notably, cell shape and size tend to be more heterogeneous for RodZ depletion than for depletion of other pneumococcal elongasome components, such as MreC, bPBP2b, or FtsEX, which results in chains of relatively uniform, spherical cells at 4 h (Berg et al., 2013; Sham et al., 2013; Tsui et al., 2014).
FIGURE 5.
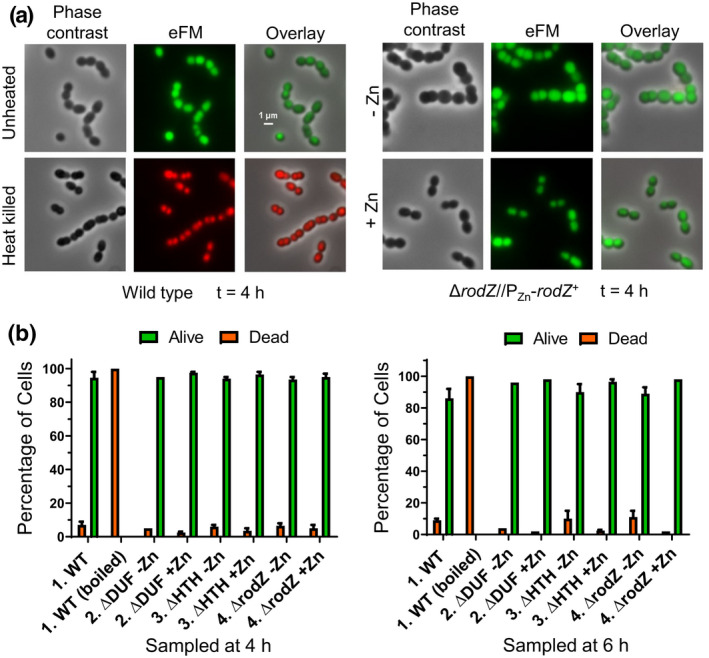
Cells depleted of RodZ(Spn) for 6 h remain viable. (a) Representative phase‐contrast and 2D epifluorescence microscopy (eFM) images of cells of WT (IU1824; left panels) or the ΔrodZ//PZn‐rodZ + merodiploid strain (IU12738; right panels) depleted for RodZ (−Zn) or replete with RodZ (+Zn) for 4 h and stained for live (green) and dead (red; indicative of membrane permeability) cells. Cells were grown as described in Figure 3a and stained with the live‐dead BacLight bacterial viability kit (Syto9 and propidium iodide) as described in Experimental procedures. Most exponentially growing WT cells are alive (green), while WT cells heat‐killed by boiling for 5 min at 95°C are dead (red). RodZ depleted (−Zn) or RodZ replete (+Zn) cells are mostly alive (green) at 4 h. All images are at the same magnification (scale bar = 1 μm). (b) Quantitation of the percentage of live or dead WT (IU1824) cells growing exponentially (−Zn) or boiled (control), and rodZ(ΔDUF)//PZn‐rodZ (IU12699), rodZ (ΔHTH)//PZn‐rodZ (IU12696), or ΔrodZ//PZn‐rodZ (IU12738) cells grown in the presence (+Zn; RodZ+) or absence (−Zn; mutant RodZ and/or RodZ depleted) of inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4) for 4 or 6 h. Two hundred cells were examined and scored for each sample. Except for the boiled control cells, most cells remained viable. Data are averaged (± SEM) from 2 independent experiments, except for the 6 h time points of IU12699 and IU12738, which are from a single experiment.
2.4. The RodZ(Spn) HTH and TM domains, but not the DUF domain, are required for normal growth
RodZ(Spn) has the same overall domain structure as RodZ from bacteria that express MreB homologs (Introduction; Figures 1 and S1). Tn‐seq of the WT strain demonstrated that viable insertions are obtained in the C‐terminus of rodZ(Spn) starting with TA in the TAT (Y132) codon, resulting in a stop codon after AAC (N131) (Figures 1d and 2a). Thus, the entire extracellular region of RodZ(Spn) from aa Y132‐N273, including DUF4115 and the disordered extracellular domain proximal to the membrane are dispensable for growth in BHI broth at 37°C. This conclusion was confirmed by deletion mutations constructed in a rodZ//PZn‐rodZ + merodiploid strain (Table 2, lines 6 and 7; Figure 6). RodZ(M1‐Q134) and RodZ(M1‐T135) mutants form normal‐sized colonies on TSAII‐BA plates, as did other deletion mutants of the extracellular domains (Table 2; Figure 6a). Cells of RodZ(ΔDUF) and RodZ(M1‐Q195; lacking DUF and the C‐terminal region) resemble WT cells in BHI broth at 37°C (Figures 6b and S7). By contrast, the RodZ(M1‐Q134) mutant forms wider, bigger cells than WT, indicative of partial RodZ function or instability, whereas RodZ(M1‐T135) cells are mostly WT, with infrequent larger, wider cells (Figure 6b). C‐terminal FLAG‐tagged WT RodZ was readily detected by western blotting (Figure 4c). Curiously, truncated RodZ(M1‐Q195) with a C‐terminal FLAG‐tag could not be detected by western blotting (Figure 6), despite not showing growth or morphology phenotypes, suggesting C‐terminal degradation in the absence of the DUF domain.
TABLE 2.
The RodZ(Spn) HTH and TM domains are essential, while the extracellular domain, including DUF, is dispensable
| Condition | Results 20–24 h | |||
|---|---|---|---|---|
|
Initial transformation a Number of colonies and size |
Re‐streak Growth and colony size |
|||
| −Zn | +Zn | −Zn | +Zn | |
| Recipient strain: ΔrodZ::Pc ‐[kan‐rpsL +]//PZn‐rodZ + | ||||
| Amplicon | ||||
| 1. rodZ + (1‐273 aa) b positive control | >300, WT | >300, WT | WT | WT |
| 2. No DNA negative control | 0 | 0 | ND c | ND c |
| 3. rodZ (1‐261 aa) | >300, WT | >300, WT | WT | WT |
| 4. rodZ (1‐195 aa) | >300, WT | >300, WT | WT | WT |
| 5. rodZ Δ(196‐261 aa) = ∆DUF | >300, WT | >300, WT | WT | WT |
| 6. rodZ (1‐135 aa) | >300, WT | >300, WT | WT | WT |
| 7. rodZ (1‐134 aa) | >300, WT | >300, WT | WT | WT |
| 8. rodZ ∆(1‐103 aa) | <5 d | >300, WT | ≈10–20 Heterogeneous μcolonies; green sheen | WT |
| 9. rodZ (1‐72 aa) | <5 d | >300, WT | ≈10–20 Heterogeneous μcolonies; green sheen | WT |
| 10. rodZ Δ(21‐257 aa) e = ∆RodZ | <5 d | >300, WT | <5 d | WT |
| 11. rodZ ∆(4‐68 aa) = ∆HTH | <5 d | >300, WT | <5 d | WT |
| 12. rodZ + ‐FLAG | >300, WT | >300, WT | WT | WT |
| 13. rodZ (Y51A F55A Y59A)‐FLAG | >300, WT | >300, WT | WT | WT |
Amplicons were transformed into merodiploid strain ΔrodZ::Pc ‐[kan‐rpsL +]//PZn‐rodZ + (IU12515), replacing the ΔrodZ::Pc ‐[kan‐rpsL +] cassette at the native chromosomal locus. Prior to transformation, ΔrodZ::Pc ‐[kan‐rpsL +]//PZn‐rodZ + (IU12515) was grown in BHI broth containing Zn inducer (0.4 mM ZnCl2 and 0.04 mM MnSO4). Transformation experiments were first performed with fusion amplicons and repeated one or more times with sequence‐verified amplicons obtained from strains: IU12696 (∆HTH), IU12699 (∆DUF), IU12738 (∆rodZ), IU12792 (rodZ(1‐72)), IU12794 (rodZ(1‐261)), IU12797 (rodZ(1‐195)), IU12799 (rodZ(1‐135)), IU12800 (rodZ(1‐103)), IU12803 (rodZ(1‐134)), or IU15628 (rodZ(Y51A F55A Y59A)‐F). Transformation mixtures were added to soft agar ±Zn inducer and subsequently plated onto TSAII‐BA plates ±Zn inducer. As the positive control, a rodZ + amplicon was transformed into the recipient ‐Zn inducer. Negative controls lacked amplicon DNA in transformation mixtures. Plates were incubated at 37°C in the presence of 5% CO2 for 20–24 h before being scored for colony number and morphology. The number of colonies is normalized to 1 ml of transformation mixture. “μcolonies” (micro‐colonies) were barely visible by eye, but observed with a dissecting microscope. “green‐sheen” refers to shiny green coloration observed on top of the blood agar, possibly due to partial hemolysis. Transformations were done at least twice independently with similar results.
aa, amino acid.
ND, not determined.
<5, indicates 0–4 colonies of different sizes.
Similar Zn inducer‐dependent growth occurred when the rodZ Δ(21‐257aa) amplicon was transformed into encapsulated strain IU15645 (D39 cps + rpsL1 ΔrodZ::Pc ‐[kan‐rpsL +]// ΔbgaA::PZn‐rodZ) ± Zn inducer.
FIGURE 6.
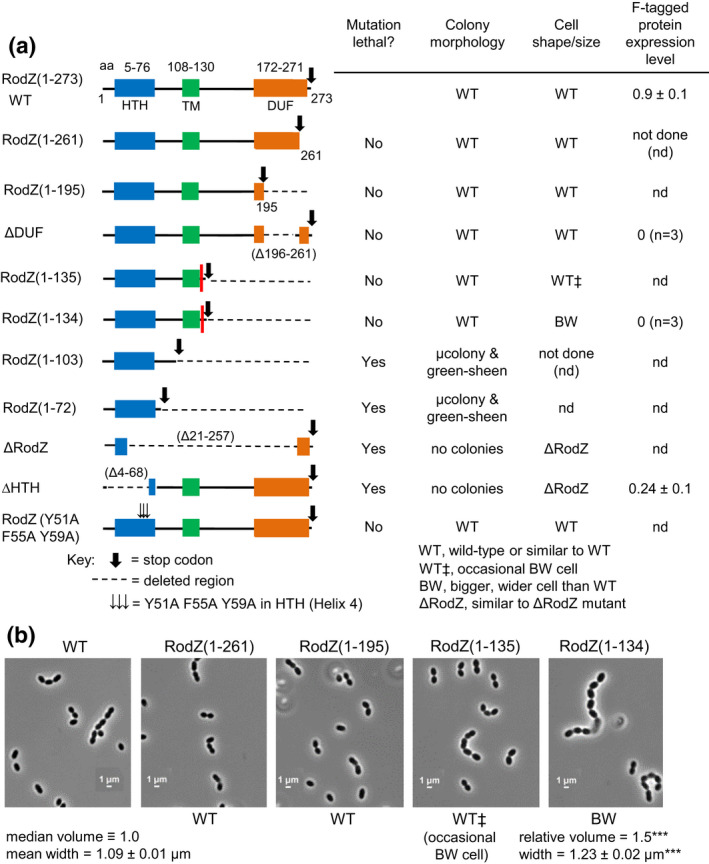
Amino acids 1–131 of RodZ are required for growth of Spn. (a) Amplicons harboring rodZ truncation or codon‐changing alleles were transformed into merodiploid strain IU12515 (ΔrodZ::Pc ‐[kan‐rpsL +] //PZn‐rodZ + ) to replace the Janus cassette (ΔrodZ::Pc ‐[kan‐rpsL +]) as described in experimental procedures and Table 2. Effects of RodZ truncations were determined by transformation assays on TSAII‐blood agar plates with or lacking Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4). Colony numbers, sizes, and morphologies were evaluated compared with rodZ + transformants after 20–24 h incubation at 37°C (see legend to Table 1 for experimental details). “μcolonies” (micro colonies) were barely visible by eye, but observed using a low power microscope. “Green‐sheen” refers to a shiny green pattern observed on top of the blood agar that may be due to partial hemolysis. Similar results were obtained in two independent transformation experiments (Table 2). The red bar between N131 and Y132 in the RodZ(1–135) and RodZ(1–134) entries marks the first TA site with a TnMariner insertion recovered by Tn‐seq of the WT strain (see Figure 2). Cell shapes and sizes were determined for WT and merodiploid mutants depleted for RodZ (see Figures 3 and S7; panel (b), below; Figures S8 and S9). Relative amounts of corresponding truncated RodZ proteins fused to a C‐terminus FLAG tag were determined by quantitative western blotting probed with anti‐FLAG antibody as described in Experimental procedures. Proteins samples were obtained from strains IU14594 (rodZ‐F at native chromosomal locus), IU13457 (rodZ‐F//PZn‐rodZ +), IU13655 (rodZ(ΔDUF)‐F//PZn‐rodZ +), IU13660 ((rodZ(1–134)‐F//PZn‐rodZ +), and IU13705 (rodZ(ΔHTH)‐F//PZn‐rodZ +) (see Table S1). Strains were grown in BHI broth +Zn inducer overnight, followed by growth for 4 h in BHI media lacking or containing Zn inducer as described in Figure 3. Values in the last column are amounts of truncated F‐tagged RodZ variants grown −Zn relative to the amount of RodZ‐F in IU14594. Although IU13655 (rodZ(ΔDUF)‐F//PZn‐rodZ +) and IU13660 ((rodZ(1–134)‐F//PZn‐rodZ +) were viable −Zn inducer, RodZ(ΔDUF)‐F and RodZ(1–134)‐F proteins were not detected in samples grown ± Zn, consistent with cleavage of the FLAG tag off the truncated RodZ variants lacking the C‐terminal DUF domain. (b) Representative micrographs of IU1824 (WT parent), and rodZ truncation mutants IU12794 (rodZ[1–261]//PZn‐rodZ +), IU12797 (rodZ[1–195]//PZn‐rodZ +), IU12799 (rodZ(1–135)//PZn‐rodZ +), and IU12803 (rodZ(1–134)//PZn‐rodZ +), which grow in the absence of Zn inducer. Cells were imaged during exponential growth at an OD620 ≈ 0.1–0.15 after ≈2.5–3.0 h of growth. Representative growth curves of truncated RodZ variants are shown in Figure S9d. Shapes and sizes were categorized as described for panel (a), above. Only the RodZ(M1‐Q134) mutant showed significant changes in relative median cell volume and average width (± SEM) compared with WT (n = 50 cells for each strain). ***, p < 0.001 by the non‐parametric, one‐way ANOVA Kruskal–Wallis test in GraphPad Prism. RodZ(M1‐T135) mutant cells resembled WT, except for an occasional bigger, wider cell.
Transformation assays and growth characterization indicated that mutants lacking the TM, HTH, or the cytoplasmic linker domain of RodZ(Spn) are not viable (Table 2; Figures 2a, 6a, and S8). Reduced RodZ(ΔHTH) mutant protein with a C‐terminal FLAG‐tag was detected in 4‐h depletion experiments (Figure 6a), consistent with lack of RodZ(ΔHTH) function and possibly decreased protein amount underlying its null mutant phenotype. Consistent with loss of function, the RodZ(HTH) domain mediates protein–protein interactions in B2H assays (see below). Changes of aromatic amino acids in RodZ(Spn) at three positions in Helix 4 of the HTH domain, which correspond to the MreB interaction interface in E. coli (van den Ent et al., 2010), do not show growth phenotypes in S. pneumoniae (Figures 6a and S9). In addition, amino acid changes in the membrane‐proximal pedestal region of bPBP2b failed to suppress ΔrodZ lethality in S. pneumoniae (Table S6), unlike the corresponding amino acid changes in E. coli bPBP2 that did suppress ΔrodZ phenotypes (Rohs et al., 2018). Finally, phosphorylation of S85 in RodZ(Bsu) was proposed to increase MreB filament density and growth (Sun & Garner, 2020). S85 of RodZ(Bsu) corresponds in alignment to E89 of RodZ(Spn) (dotted box, Figures 1d and S1), which is not immediately adjacent to other serine or threonine residues. Phostag‐PAGE analysis failed to detect phosphorylation of functional RodZ‐HA3 in S. pneumoniae (Figure S10). From these combined results, we conclude that the cytoplasmic HTH and linker and the TM domain, but not the extracellular domains, of RodZ(Spn) are required for growth at 37°C under the conditions tested here and that amino acids important for RodZ function in E. coli and B. subtilis are not required in S. pneumoniae (see Discussion).
2.5. RodZ(Spn) localizes with known pPG elongasome proteins throughout the pneumococcal cell cycle
The identical suppression pattern of ΔrodZ and ΔmreC in S. pneumoniae (Table 1) supports the hypothesis that RodZ is a member of the pPG elongasome. This hypothesis is further corroborated by protein co‐localization analyses using immunofluorescence microscopy (IFM) as described in Experimental procedures. Strains expressing RodZ‐FLAG3 constructs or other epitope‐tagged proteins from their native chromosomal loci were functional and did not exhibit aberrant growth or cell morphologies (Figure S11). We used a previously published method to compare the average locations of two fluorescent epitope‐tagged proteins relative to DAPI‐stained nucleoids at four stages of division in pneumococcal cells growing exponentially in BHI broth at 37°C (Figure 7) (Land et al., 2013; Tsui et al., 2014). This method also allows statistical comparisons of average midcell widths at different cell division stages as described in Experimental procedures.
FIGURE 7.
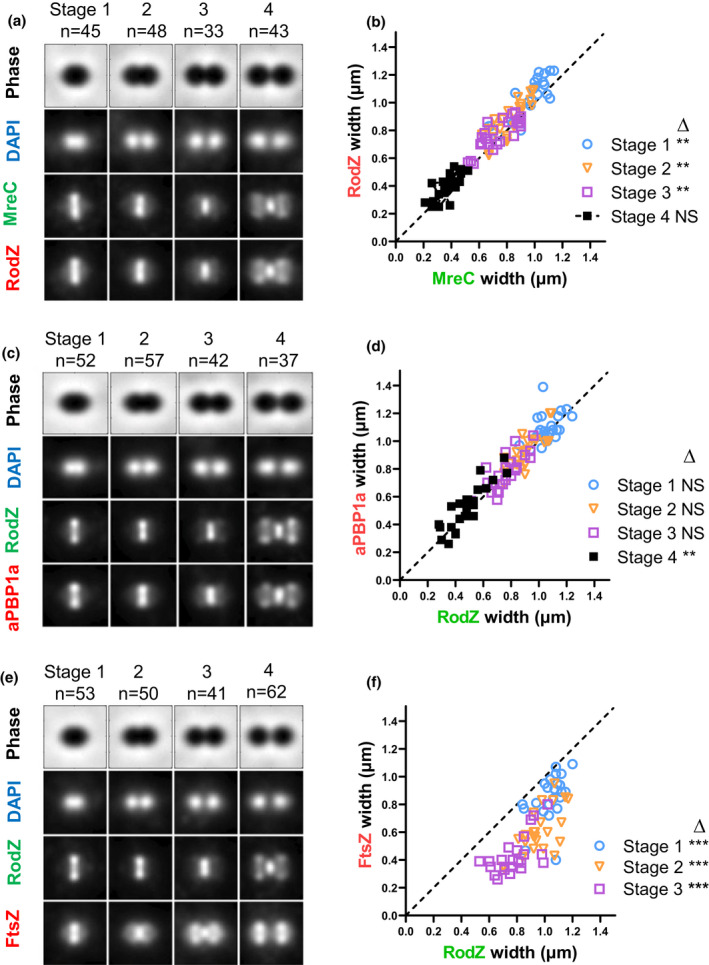
RodZ localizes with MreC and aPBP1a of the peripheral PG synthesis machine. (a) Composite image displaying localization patterns of MreC and RodZ through four stages of pneumococcal growth and division. Images were obtained by dual‐labeling immunofluorescence microscopy (IFM). To construct composite images, n > 30 cells from each division stage were averaged and quantified as described in Experimental procedures. (a) IU7113 (mreC‐L‐F3 rodZ‐Myc) IFM was probed with DAPI (DNA) and anti‐FLAG and anti‐Myc antibodies as detailed in Experimental procedures. (b) Scatter plot of the paired widths of RodZ compared with MreC constructed using the IMA‐GUI program described in Experimental procedures. The dotted line intercepts the origin with slope = 1 and indicates the expected distribution if RodZ and MreC widths are identical. Differences between paired widths were calculated for cells at each division stage, and one‐sample student's t‐tests were performed to determine whether mean differences in widths were significantly different from the null hypothesis value of zero (NS, not significant; **, p < 0.01) (Tsui et al., 2014). (c) Composite image of RodZ and aPBP1a localization in IFM of IU7515 (pbp1a‐L‐F3 rodZ‐Myc) probed with DAPI and anti‐Myc and anti‐FLAG antibodies. (d) Scatter plot of paired width analysis of aPBP1a compared with RodZ. (e) Composite image of RodZ and FtsZ localization in IFM of IU7072 (rodZ‐L‐F3 ftsZ‐Myc) probed with DAPI and anti‐FLAG and anti‐Myc antibodies. (f) Scatter plot of paired width analysis of FtsZ compared with RodZ. ***p value <0.001. Data were obtained from two independent experiments for each comparison.
By this analysis, RodZ co‐localizes throughout the cell cycle with MreC and aPBP1a (Figure 7a–d), which have been implicated in pPG elongation in S. pneumoniae (Briggs et al., 2021; Fenton et al., 2016; Land & Winkler, 2011; Philippe et al., 2014; Straume et al., 2017; Tsui et al., 2016). All three proteins localize at the midcell equator in Stage 1 cells, remain at the midcell septum in Stage 2 and 3 cells, and only appear at the new equators of daughter cells late in division at Stage 4. There is a very small displacement of midcell widths of RodZ‐Myc from those of MreC‐L‐F3 on the diagonal reference line (slope = 1), accompanied by statistically significant width differences for stage 1–3 cells (Figure 7b). These slight differences likely indicate that the C‐terminal epitope tag of RodZ extends further from the cell membrane than that of MreC (see below), with this apparent displacement possibly enhanced by the lengths of the two antibodies used in IFM (Perez, Villicana, et al., 2021). In contrast to RodZ, MreC, and aPBP1a, nascent FtsZ‐ rings move outward toward the future sites of the new equators throughout division and largely leave the septum in Stage 3 and 4 cells (Figure 7e,f) (Perez et al., 2019). We conclude that RodZ co‐localizes with components of the pPG elongasome, which overlaps FtsZ localization in Stage 1 cells, but is different in later stages of the cell cycle. This conclusion is corroborated independently by high‐resolution 3D‐SIM of RodZ and FtsZ in cells at different division stages (Figure S12).
2.6. RodZ(Spn) forms complexes and interacts with proteins in the septal PG and pPG synthesis machines and with PG synthesis regulators
To gain more information about RodZ function in S. pneumoniae, we performed pairwise co‐IP experiments using RodZ‐FLAG and RodZ‐FLAG3 as bait proteins that were probed in western blots for complex formation with proteins involved in PG elongation, septation, or cell division. Representative co‐IP results are shown in Figure 8a, quantitated in Table 3, and summarized in Figure 8b,c. Additional supporting data are in Figures S13 and S14. Strong complex formation was detected between RodZ and pPG elongasome proteins MreC and bPBP2b at some stage of division in non‐synchronized cell cultures (Table 3). The experiment to probe for complexes between RodZ and RodA could not be performed, because cells expressing RodZ‐FLAG3 and HaloTag‐RodA (HT‐RodA) showed a synthetic lysis phenotype not observed in cells separately expressing the fusion proteins (Figure S14a,b). Complexes were also detected between RodZ and protein regulators of PG synthesis (GpsB; StkP [Ser/Thr protein kinase]; DivIVA), Class A PBPs (aPBP1a; aPBP2a), and MpgA (PG muramidase) (Briggs et al., 2021; Massidda et al., 2013). Consistent with these results, MreC, MpgA, or aPBP1a, each of which has been linked to pPG elongation in S. pneumoniae (Briggs et al., 2021; Fenton et al., 2016; Land & Winkler, 2011; Massidda et al., 2013; Philippe et al., 2014; Taguchi et al., 2021; Tsui et al., 2016), pulled down the same set of proteins (Table 3). RodZ was also detected in complexes with the sPG synthesis proteins bPBP2x (Figure 8a) and FtsW (Figure S14c) at some stage of cell division, possibly the initial equatorial ring of newly divided cells (Briggs et al., 2021) or the outer pPG synthesis ring that also contains some bPBP2x, and presumably FtsW (Perez, Boersma, et al., 2021). In contrast, marginal or no complexes were detected between RodZ and FtsA, FtsZ, PhpP (protein phosphatase), or KhpAB (RNA‐binding regulator) (Figures 8 and S13c; Table 3) (Massidda et al., 2013; Mura et al., 2017; Perez et al., 2019; Rued et al., 2017; Stamsas et al., 2017; Zheng et al., 2017). Thus, an in vivo complex containing RodZ and KhpB was not detected, despite a previous report of an interaction in a B2H assay (Winther et al., 2021).
FIGURE 8.
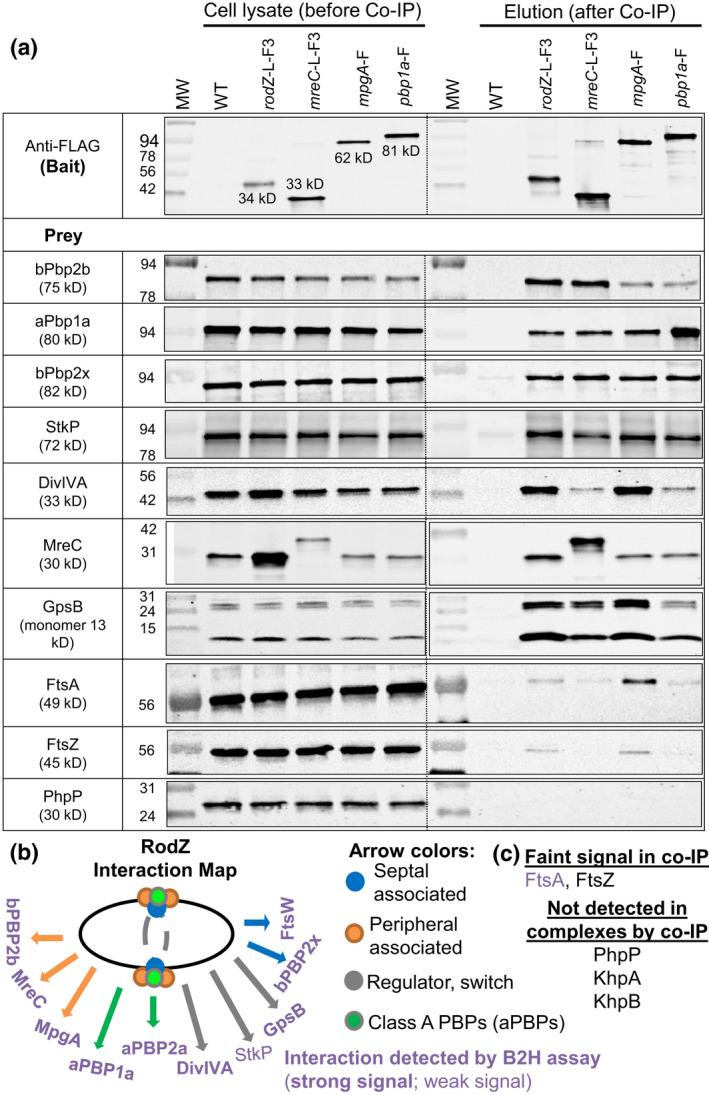
RodZ, MreC, MpgA (formerly MltG), and aPbp1a are in complexes with components of the peripheral and septal PG machines, class a PBPs, and cellular regulators StkP, GpsB, and DivIVA. Co‐IP experiments using non‐FLAG‐tagged WT strain (IU1945) or FLAG‐tagged strains RodZ‐L‐F3 (IU6291), MreC‐L‐F3 (IU4970), MpgA‐F (IU7403), or PBP1a‐F (IU5840) as bait were probed with native antibodies to detect prey proteins bPBP2b, aPBP1a, bPBP2x, StkP, DivIVA, MreC, GpsB, FtsA, FtsZ and PhpP as described in Experimental procedures. Prey proteins were detected in all cell lysates (input; left lanes). In elution output samples (right lanes), prey proteins are undetectable for the WT non‐FLAG‐tagged control strain but are present in different relative amounts in samples of the FLAG‐tagged strains. The top blot was probed with anti‐FLAG primary antibody for detection bait proteins. For most blots, 4 μl (4–6 μg) of each lysate sample (input) were loaded on the left lanes, while 15 μl of each elution output sample (after mixing 1:1 2× Laemmli buffer) were loaded on the right lanes. For detection of GpsB, 6 μl (6 μg) of lysate sample and 25 μl of output were loaded. Two bands are detected with anti‐GpsB antibody in the input and output samples, possibly due to failure of heating to reverse cross‐linking of GpsB monomers. The bottom band corresponds to GpsB monomer (≈13 kDa), whereas the top band is likely a GpsB dimer (≈26 kDa). Bands detected with anti‐MreC or anti‐aPBP1a in MreC‐L‐F3 or Pbp1a‐F strains were F‐tagged bait proteins. For detection of MreC‐L‐F3 or Pbp1a‐F in output elution samples, 3 μl of samples were loaded to each lane. The relative amount of MreC was 5–9‐fold higher in the input lysate of rodZ‐L‐F3‐Pc erm (IU4970; MreC row) compared with that from the untagged WT strain (shown in adjacent lane) or lysate obtained from the markerless rodZ‐F strain (IU14594, data not shown), suggesting that the Pc promoter present in the rodZ‐L‐F3‐Pc erm construct leads to overexpression of downstream genes, including mreC. Nevertheless, the Co‐IP results using the rodZ‐F markerless strain lacking an antibiotic‐resistance cassette (IU14594) were similar to those for the rodZ‐L‐F3‐Pc‐ erm strain (IU4970) (data not shown). Co‐IP experiments were performed 2–6 times with similar results (see Table 3 for quantitation). (b) Interaction map of RodZ in cells detected by co‐IP. (c) Proteins that were weakly or not detected in complex with RodZ by co‐IP.
TABLE 3.
RodZ, MreC, MpgA, and aPBP1a are in complexes with key components of the peripheral and septal PG synthesis machines and division regulators StkP, GpsB, and DivIVA
| Prey proteins tested | Primary antibodies | Ratio of prey protein band/background a | ||||
|---|---|---|---|---|---|---|
| Bait protein | ||||||
| RodZ‐F b | RodZ‐L‐F3 c | MreC‐L‐F3 | MpgA‐F | PBP1a‐F | ||
| MreC | Anti‐MreC | 24 ± 1 (2) | 100 ± 11 (3) | NA d | 22 ± 9 (3) | 19 ± 8 (3) |
| bPBP2b | Anti‐PBP2b | 26 ± 24 (2) | 63 ± 28 (5) | 47 ± 17 (6) | 13 ± 10 (3) | 13 ± 11 (3) |
| aPBP1a | Anti‐PBP1a | 11 ± 8 (2) | 61 ± 28 (6) | 41 ± 15 (6) | 22 ± 10 (4) | NA d |
| aPBP2a | Anti‐PBP2a | 29 ± 5 (2) | 34 ± 10 (4) | 20 ± 6 (4) | ND e | ND e |
| GpsB | Anti‐GpsB | 24 ± 6 (2) | 51 ± 20 (5) | 26 ± 10 (4) | 38 ± 22 (3) | 16 ± 9 (2) |
| StkP | Anti‐StkP | 20 (1) | 23 ± 3 (3) | 14 ± 1 (4) | 34 ± 13 (3) | 28 ± 12 (3) |
| DivIVA | Anti‐DivIVA | 7.9 (1) | 18 ± 1.5 (3) | 4.3 ± 0.9 (3) | 53 ± 30 (2) | 6.8 ± 2.3 (2) |
| bPBP2x | Anti‐PBP2x | 4.8 ± 2.5 (2) | 8.7 ± 2.3 (4) | 10 ± 4 (4) | 12 ± 4 (4) | 15 ± 7 (3) |
| MpgA‐HA | Anti‐HA | ND e | 8.6 ± 3.1 (3) | ND e | ND e | ND e |
| FtsW‐L‐GFP | Anti‐GFP | ND e | 4.5 ± 1.1 (3) | ND e | ND e | ND e |
| FtsA | Anti‐FtsA | 2.0 ± 0.0 (2) | 1.9 ± 0.4 (3) | 1.2 ± 0.1 (4) | 4.5 ± 0.7 (3) | 1.6 ± 0.1 (3) |
| FtsZ | Anti‐FtsZ | 1.1 (1) | 1.7 ± 0.3 (3) | 1.0 ± 0.0 (4) | 2.0 ± 0.6 (3) | 1.1 ± 0.1 (3) |
| PhpP | Anti‐PhpP | 0.9 (1) | 1.0 ± 0.0 (3) | 1.1 ± 0.1 (4) | 1.1 ± 0.0 (3) | 1.2 ± 0.1 (3) |
| Prey proteins tested | Primary antibody | Bait protein | ||
|---|---|---|---|---|
| PBP1a‐F | KhpA‐L‐F3 | KhpB‐L‐F3 | ||
| RodZ‐HA3 | Anti‐HA | 8.6 ± 0.3 (2) | 1.2 ± 0.0 (2) | 1.4 ± 0.0 (2) |
Quantification and summary of RodZ interactions investigated in S. pneumoniae using Co‐IP assay. Interaction ratios were obtained from co‐IP experiments and western blot analysis as described in Experimental procedures. The interaction ratio is the mean ± SEM ([n], number of biological replicates) value of the prey band signal divided by that of the signal obtained from a non‐FLAG tagged sample. A mean ratio of band intensity ≥2 between FLAG‐tagged and non‐FLAG‐tagged samples was considered indicative of protein interaction. Co‐IP experiments are performed with lysates obtained from non‐FLAG‐tagged WT strain (IU1945), and isogenic FLAG‐tagged strains rodZ‐L‐F3 (IU6291), which contains three tandem FLAG tags, mreC‐L‐F3 (IU4970), mpgA‐F (IU7403), or pbp1a‐F (IU5840) as bait, or from non‐FLAG‐tagged WT parent strain (IU1824), and isogenic rodZ‐F markerless (IU14594). Co‐IP elution samples were probed with native antibodies to detect prey proteins bPBP2b, aPBP1a, bPBP2x, StkP, DivIVA, MreC, GpsB, FtsA, FtsZ and PhpP (Figure 8). RodZ/aPBP2a and MreC/aPBP2a complexes were detected with IU1945 (WT), K166 (Δpbp2a), IU4970 (mreC‐L‐F3), IU17817 (mreC‐L‐F3 Δpbp2a), IU6291 (rodZ‐L‐F3), and IU17821 (rodZ‐L‐F3 Δpbp2a) (Figure S13a). RodZ/MpgA complex was examined with strains IU7399 (mpgA‐HA) and IU7584 (rodZ‐L‐F3 mpgA‐HA), and anti‐HA antibody to detect MpgA‐HA as prey (Figure S13b). Strains used for examining complexes of aPBP1a/RodZ, KhpA/RodZ, and KhpB/RodZ interaction using PBP1a‐F, KhpA‐L‐F3, and KhpB‐L‐F3 as bait were IU11828 (rodZ‐HA3), IU11925 (rodZ‐HA3 pbp1a‐F), IU17873 (rodZ‐HA3 khpA‐L‐F3) and IU17877 (rodZ‐HA3 khpB‐L‐F3), with the use of anti‐HA to detect the prey protein RodZ‐HA3 (Figure S13c). Strains IU8918 (ftsW‐L‐gfp), IU16026 (rodZ‐L‐F3 ftsW‐L‐gfp), and an anti‐GFP antibody were used for detection of RodZ/FtsW interaction (Figure S14c).
rodZ‐F markerless (IU14594) construct has no polar effect on the expression of the downstream gene mreC.
IU6291 (rodZ‐L‐F3‐Pc erm) strain showed polar effect of increased expression (5–9‐fold) of mreC, which is downstream of rodZ. IU6291 shows no growth or morphological phenotypes compared with its rodZ + WT parent (Figure S11).
NA, not applicable, since MreC‐L‐F3 or PBP1a‐F were used as bait.
ND, not determined.
B2H assays were performed in E. coli to test for direct interactions between WT RodZ (Spn) or truncated constructs lacking the HTH or DUF domain of RodZ(Spn) and the set of proteins mentioned above, as well as additional proteins not analyzed by co‐IP (Figures 9 and S15; Table S7). B2H assays revealed RodZ(Spn) self‐interaction and a strong signal of interactions, usually bidirectional, between RodZ(Spn) and GpsB, MreC, MreD, MpgA, bPBP2b, RodA, aPBP1a, aPBP2a, bPBP2x, FtsW, EzrA, DivIVA, or aPBP1b (Figures 2d, 8c and 9a). Weaker signals of unidirectional interaction or no interaction were detected by B2H between RodZ(Spn) and StkP, FtsA, or FtsZ (Figure 9a). For comparison, B2H assays were performed to determine direct interactions of pPG elongasome proteins MreC(Spn) or MreD(Spn) with Class A PBPs. MreC interacts with itself and shows bidirectional interactions with aPBP1a, aPBP2a, or aPBP1b, whereas MreD also self‐interacts and shows bidirectional interactions with aPBP1a, but unidirectional interactions with aPBP2a or aPBP1b (Figures 2d and 9b).
FIGURE 9.
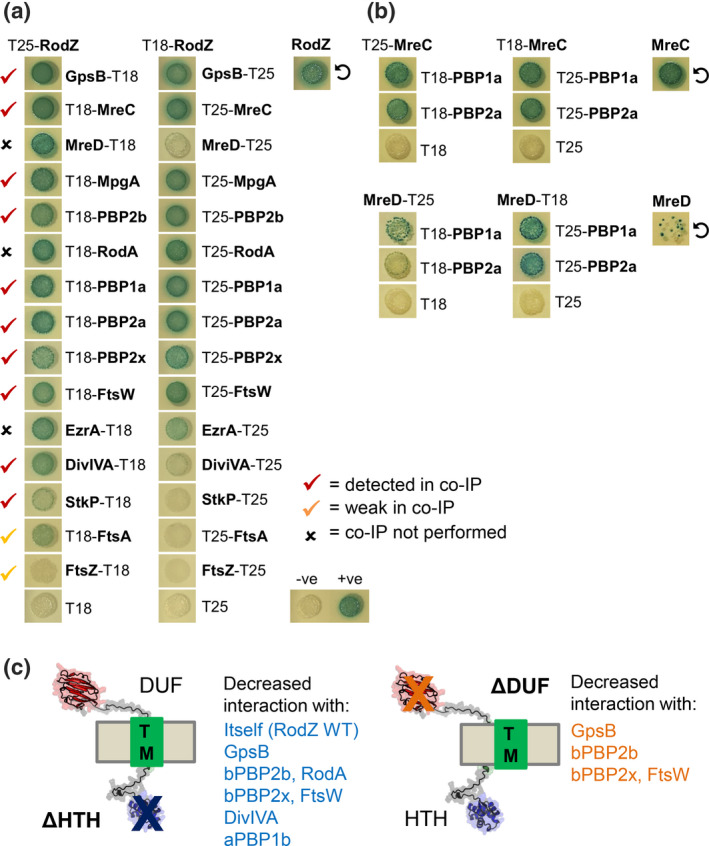
RodZ interacts with numerous cell elongation and division proteins as well as with itself in B2H assays. (a) RodZ interacts with GpsB, MreC, MreD, MpgA, bPBP2b, RodA, aPBP1a, aPBP2a, bPBP2x, FtsW, EzrA, and DivIVA in both directions, and with StkP and FtsA with a lower signal and only in one direction. Also, RodZ self‐interaction is shown (circular arrow). B2H assays were performed as described in Experimental procedures. Agar plates were photographed after 40 h at 30°C. See Figure S15 for earlier time points at 24, 30, and 36 h. Control experiments showed that all tested proteins exhibited self‐interaction, indicative of functional intactness for interaction (data not shown). (b) MreC and MreD interact with aPBP1a and aPBP2a and also self‐interact (circular arrows). Agar plates were photographed after 40 h at 30°C. The punctate appearance of the spot showing MreD self‐interaction is likely due to high toxicity of the S. pneumoniae mreD hybrid constructs in E. coli. (c) Summary of decreased interactions of Spn RodZ(ΔHTH) and RodZ(ΔDUF) compared with RodZ WT with certain PG synthesis and division proteins in B2H assays. Data are shown in Figure S15.
Finally, B2H assays were used to determine whether the absence of the HTH or DUF domain reduces binding to the abovementioned proteins. Compared with WT RodZ(Spn), the absence of the HTH or DUF domain does not completely abolish the interactions between the truncated RodZ variants and any of the numerous partners tested (Figure S15; Table S7). For many of them, including MreC, MreD, MpgA, aPBP2a, and EzrA, the signal was unchanged compared with WT RodZ(Spn) at the endpoint of the assay (data not shown). Yet, the absence of the HTH domain significantly reduces the interactions of RodZ(Spn) with itself and several proteins, including GpsB, bPBP2b, RodA, bPBP2x, FtsW, DivIVA, and aPBP1b (Figure 9c and S15; Table S7). Although not essential in S. pneumoniae (Figures 2 and 6; Table 2), the absence of the extracellular DUF domain also reduces the interactions with GpsB, bPBP2b, bPBP2x, or FtsW in the B2H assay (Figures 9c and S15; Table S7). Together, we conclude that RodZ(Spn) is in complexes with numerous pPG elongasome proteins, PG synthesis regulatory proteins, and a few sPG synthesis proteins, possibly through direct interactions in some cases. Implications of these complexes to pneumococcal PG synthesis and division are considered further in Discussion.
2.7. Depletion of RodZ(Spn) mislocalizes MreC, bPBP2b, and RodA, but not other pPG and sPG synthesis proteins
We tested the hypothesis that RodZ(Spn) organizes the assembly of pPG elongasome proteins. We first determined whether incorporation of a fluorescent d‐amino acid (FDAA) changes in a rodZ(ΔDUF), rodZ(ΔHTH), or ΔrodZ merodiploid mutant after ectopically expressed WT RodZ is depleted (Figure S16). FDAA incorporation indicates regions of active PBP transpeptidase activity during PG synthesis (Boersma et al., 2015; Tsui et al., 2014), but does not distinguish between sPG and pPG synthesis at the midcell of S. pneumoniae cells (Perez, Boersma, et al., 2021). As expected, FDAA labeling in ΔrodZ(DUF) cells depleted for RodZ (−Zn) is the same as that of cells expressing RodZ (+Zn or WT). FDAA is also similar in ΔrodZ(HTH) or ΔrodZ cells depleted of RodZ (−Zn inducer), although the RodZ depletion changes the cell size and morphology (Figure S16). Results presented next show that RodZ depletion disrupts normal localization of MreC and the bPBP2b:RodA pPG synthase. Therefore, we interpret the FDAA labeling at midcell and equators of ΔrodZ(HTH) or ΔrodZ cells depleted of RodZ to reflect sPG synthesis, which is not disrupted by RodZ depletion. We conclude that RodZ depletion does not lead to widespread mislocalization of sPG synthesis, as occurs upon FtsZ, FtsA, or EzrA depletion, where EzrA(Spn) acts as a positive regulator of FtsZ‐ring formation in pneumococcus (Mura et al., 2017; Perez, Villicana, et al., 2021).
We next constructed ΔrodZ//PZn‐rodZ + merodiploid strains expressing from native chromosomal loci 12 other PG synthesis and division proteins fused to epitope tags, fluorescent reporter proteins, or a HaloTag (HT) (Figures S17–S22). Apart from three exceptions, we did not observe pronounced fusion‐associated phenotypes that suppressed or exacerbated growth defects upon RodZ depletion (−Zn) in these strains. A sfGFP‐MpgA fusion suppressed ΔrodZ lethality (Figure S18a,b), likely because of reduced MpgA enzymatic activity, which is known to bypass the requirement for the pPG elongasome in S. pneumoniae (Taguchi et al., 2021; Tsui et al., 2016). Conversely, GFP‐MpgA or HT‐bPBP2x fusion exacerbated the drop in OD620 upon RodZ depletion, without overtly changing localization of the fusion proteins after 4 h of RodZ depletion (Figures S18c and S22a).
Of the 12 proteins tested, aberrant localization upon RodZ depletion was only observed for MreC (Figures 10a, 11, and S21), bPBP2b (Figures 10b, S21, and S22b), and RodA (Figure S22c) (summarized in Figure 12). Mislocalization of MreC, bPBP2b, and RodA upon RodZ depletion was demonstrated by demographic analysis (Figures 11, S21, and S22) and confirmed independently by IFM for MreC and bPBP2b (Figure 10). By contrast, MpgA (Figure S18) and aPBP1a (Figure S20) (pPG synthesis); bPBP2x (Figures S19 and S22d) (sPG synthesis); FtsZ (Figure S19), MapZ (Figure S19), EzrA (Figure S19), and FtsA (Figure S20) (Z‐ring organization); and StkP (Figure S20) and DivIVA (Figure S19) (pPG and sPG synthesis) localize normally at midcell upon RodZ depletion (Figure 12) (for functions, see [Briggs et al., 2021, Massidda et al., 2013, Straume et al., 2021]). We conclude that RodZ(Spn) is required for normal assembly and localization of MreC, bPBP2b, and RodA in the pPG elongasome.
FIGURE 10.
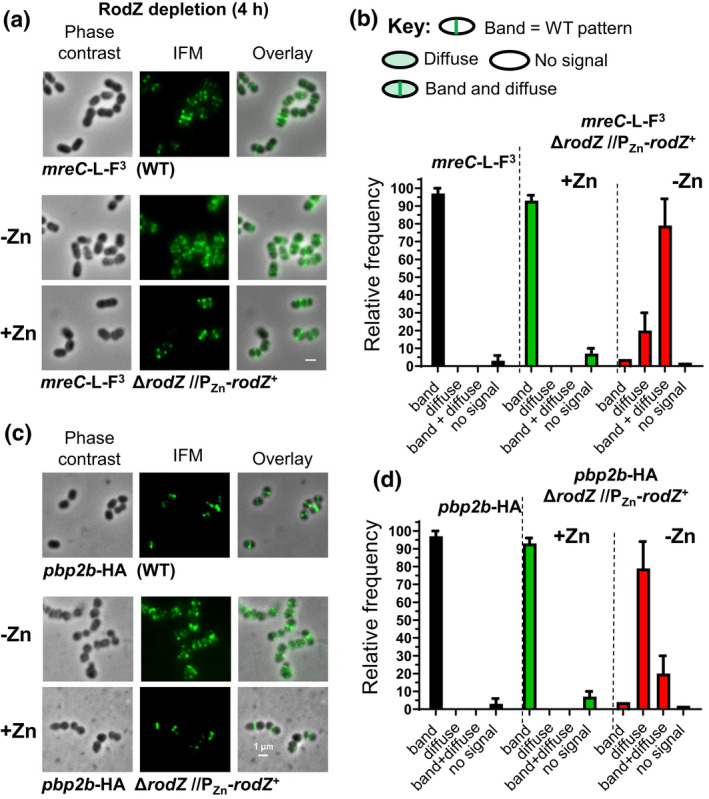
Depletion of RodZ leads to the mislocalization of MreC and bPBP2b detected by immunofluorescence microscopy (IFM). Representative images showing localization of MreC (a) or bPbp2b (c) after depletion of RodZ for 4 h of growth, which reduced RodZ to an undetectable amount (Figure 3c). Phase contrast and 2D IFM was performed as described in experimental procedures using antibody to the FLAG or HA tags. Strains used: (a) WT IU14458 (mreC‐L‐F3) and merodiploid strain IU14158 (mreC‐L‐F3 ΔrodZ//PZn ‐rodZ +); (c): WT IU14455 (pbp2B‐HA) and merodiploid strain IU14131 (pbp2B‐HA ΔrodZ//PZn ‐rodZ +). Quantification of localization patterns of MreC (b) and bPBP2b (d) observed at 4 h in the WT and after RodZ depletion. For each sample and condition, 100 cells were manually examined and scored according to the key. Data are averaged (± SEM) from 2 independent experiments.
FIGURE 11.
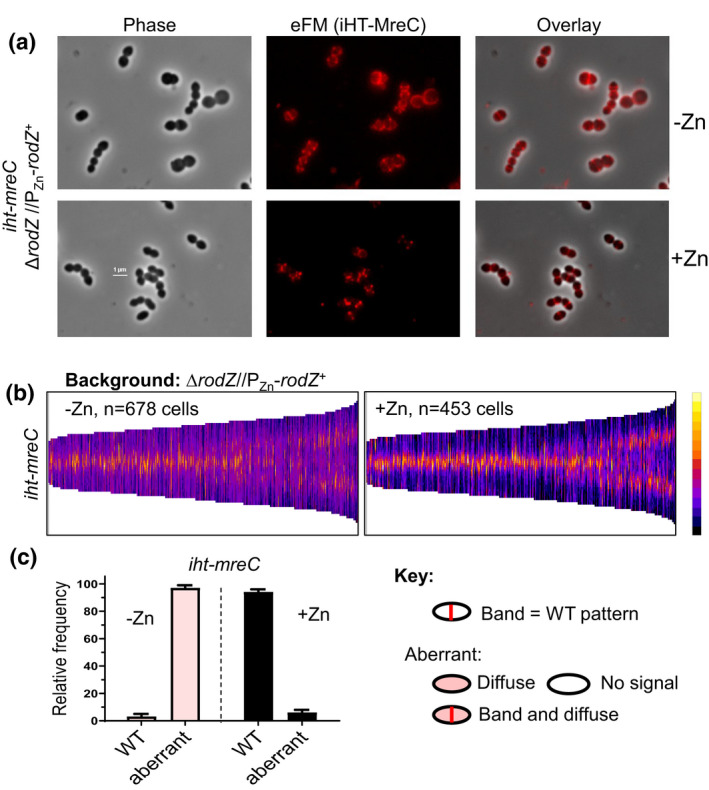
Depletion of RodZ leads to the mislocalization of MreC detected by 2D‐eFM. IU16920 (iht‐mreC ΔrodZ//PZn‐rodZ +; where iht refers to the i‐tag‐HaloTag (Perez et al., 2019)) was grown overnight in the presence of Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4) and diluted into fresh medium to OD620 ≈ 0.003 containing (complementation) or lacking (depletion) Zn inducer. At 4 h, localization of iHT‐MreC was determined following saturation labeling of the iHT domain with HT‐TMR ligand by 2D epifluorescence microscopy (eFM) as described in experimental procedures. (a) Representative micrographs showing iHT‐MreC localization. (b) Demographs displaying fluorescence intensity of iHT‐MreC localization in the absence (−Zn; RodZ depletion) or presence (+Zn; RodZ present) of inducer. N, number of cells aligned and displayed in each demograph. Microscopy and demographs are representative of three independent biological replicates. (c) Bar graph displaying iHT‐MreC localization patterns. For each sample and condition, 100 cells were manually examined and scored according to the key. Data are averaged (± SEM) from two independent experiments.
FIGURE 12.
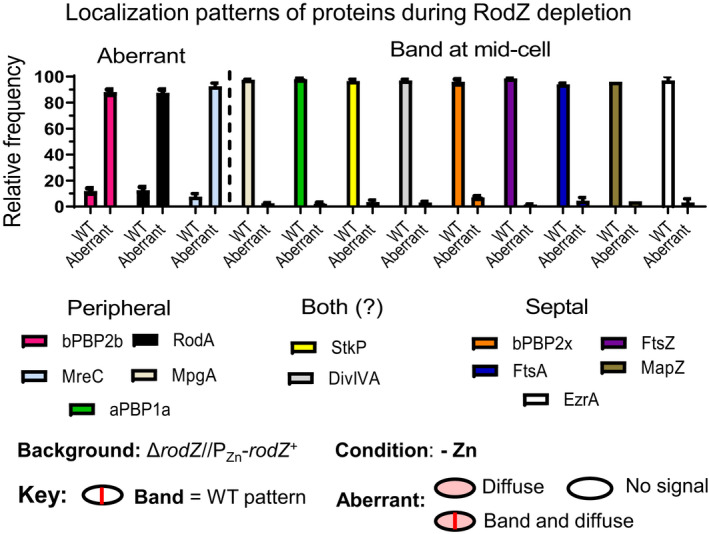
Summary of localization patterns of PG synthesis and division proteins after RodZ depletion (−Zn) for 4 h. Among the peripheral PG synthesis machine components, the morphogenic protein MreC and the PG synthase components bPBP2b (TPase) and RodA (GTase) require RodZ for localization, while the localization of MpgA (formerly MltG(Spn) muramidase and Class A PBP1a was unchanged by RodZ depletion. MreC, bPBP2b, and RodA localized normally in the presence of Zn inducer (Figures 10, 11, and S22). Localization of other cell division and PG synthesis proteins (bPBP2x, FtsZ, FtsA, MapZ, EzrA, StkP and DivIVA) were unaffected by RodZ depletion. Representative micrographs of localization studies are shown in Figures 10, 11, S18, S19, S20, and S22. Hundred cells were scored by eye within a given field in each experiment using the indicated key. Data are averaged (± SEM) from two or more independent experiments of each strain. Strains used: IU16058 (iht‐pbp2b), IU16060 (iht‐rodA), IU16920 (iht‐mreC), IU14433 (gfp‐mpgA), IU14496 (pbp1a‐FLAG), IU14160 (stkP‐FLAG2), IU12993 (ftsZ‐sfgfp), IU13061 (divIVA‐gfp), IU13062 (gfp‐mapZ), IU13058 (ezrA‐sfgfp), IU13000 (isfgfp‐pbp2x), and IU17022 (FLAG‐ftsA) in the ΔrodZ//PZn‐rodZ + background (see Table S1), which was depleted for RodZ as in Figure 3.
To test this notion and further establish the assembly hierarchy, we determined protein localization upon MreC depletion. We first established that Zn inducer (0.4 mM ZnCl2 + 0.4 mM MnSO4) does not affect growth or MreC amount in WT cells (Figure S23). We then showed that MreC depletion for 4 h in a ΔmreC//PZn‐mreC + merodiploid strain reduces MreC cellular amount to ≈10% of WT, but does not alter bPBP2b or bPBP2x cellular amount (Figure S24). Depletion of MreC resulted in mislocalization of bPBP2b detected by demographic analysis of HaloTag‐bPBP2b (Figure 13) and by IFM (Figure S25c). Likewise, demographic analysis showed that RodA mislocalizes upon MreC depletion (Figure S26b). In contrast, RodZ (Figure S25a), aPBP1a (Figure S25b), and bPBP2x (Figure S26c) remained mostly at their normal midcell positions upon MreC depletion. Together, these results support an assembly hierarchy wherein RodZ is required for MreC midcell localization, which in turn, is required for midcell localization of bPBP2b and RodA (Figure 14a).
FIGURE 13.
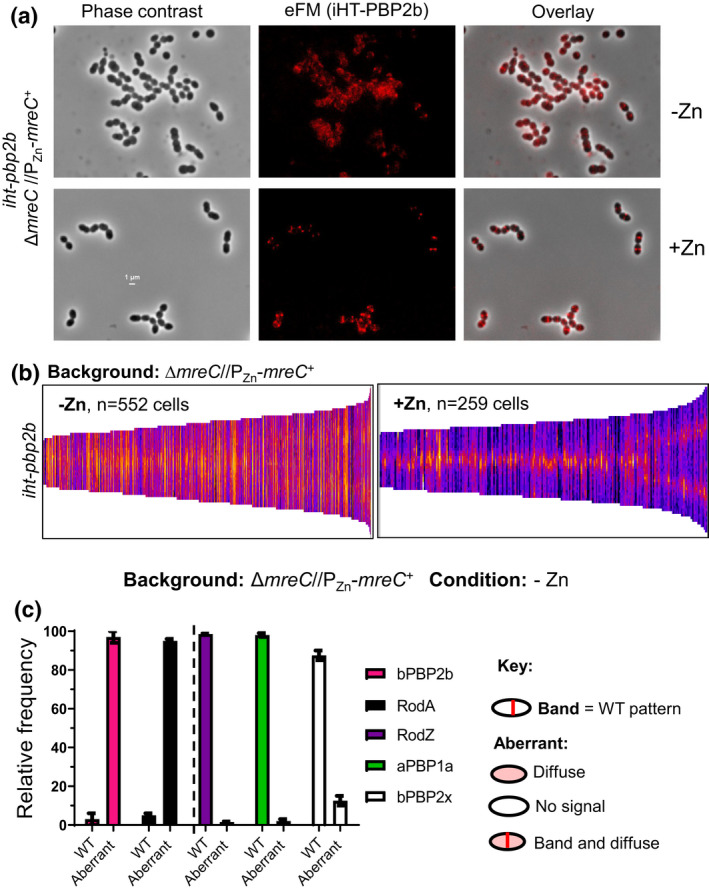
Depletion of MreC leads to mislocalization of bPBP2b and RodA, but not RodZ. For localization of bPBP2b, IU16281 (iht‐pbp2b ΔmreC //PZn ‐mreC +) was grown overnight in the presence of Zn inducer (0.4 mMCl2 Zn + 0.04 mM MnSO4) and diluted into fresh medium containing (complementation) or lacking (depletion) Zn inducer to OD620 ≈ 0.003. After 4 h, iHT‐PBP2b was labeled with a saturating concentration of a HT‐TMR ligand, and localized in cells by 2D epifluorescence microscopy (eFM) as described in Experimental procedures. (a) Representative micrographs of iHT‐PBP2b localization under MreC complementation or depletion conditions. (b) Demographs displaying fluorescence intensity of iHT‐PBP2b localization upon MreC depletion (−Zn) or in the presence of MreC (+Zn) for the number of cells (n) aligned and displayed in each demograph. Microscopy and demographs are representative of three independent biological replicates. (c) Bar graph displaying localization patterns of bPBP2b, RodA, RodZ, aPBP1a, and bPBP2x after MreC depletion (−Zn). For each sample and condition, 100 cells were manually examined and scored according to the key. Data are averaged (± SEM) from two independent experiments. Strains used: IU16281 (iht‐pbp2b), IU16283 (iht‐rodA), IU14598 (rodZ‐FLAG), IU15901 (pbp1a‐FLAG), and IU16326 (iht‐pbp2x) in the ΔmreC//PZn‐mreC + background (see Table S1). Representative micrographs of proteins other than bPBP2b are in Figures S21, S25, and S26.
FIGURE 14.
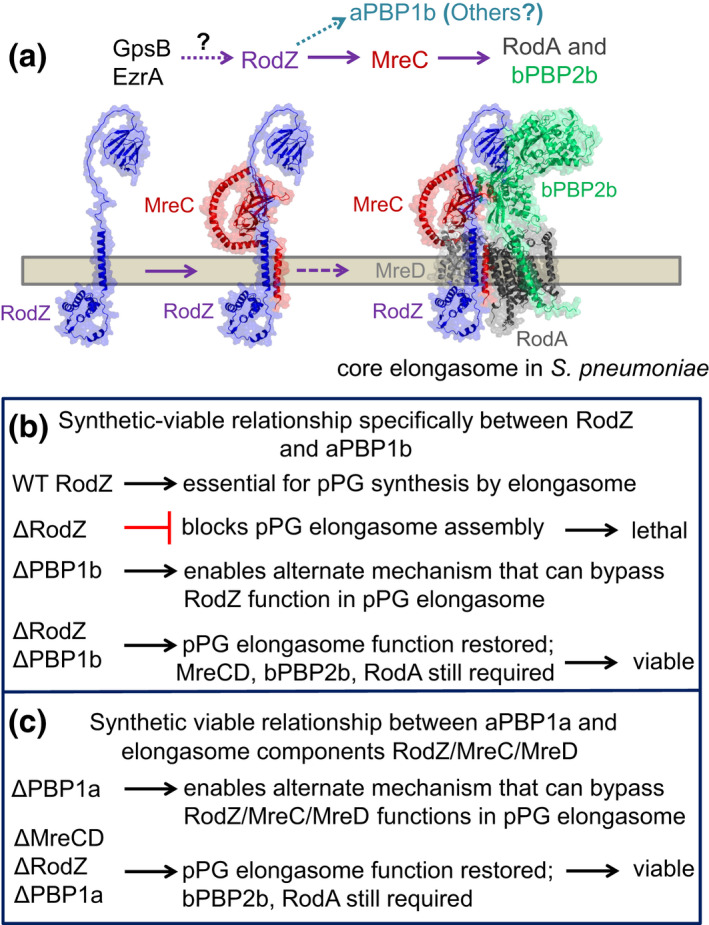
Models of (a) the assembly hierarchy of the pPG core elongasome mediated by RodZ(Spn) and (b and c) bypass pPG synthesis to account for the synthetic‐viable genetic relationships between Class A PBPs and pPG elongasome components in S. pneumoniae. (a) Results presented here establish RodZ(Spn) as an essential scaffolding protein required for the assembly and function of the pPG elongasome. The assembly hierarchy is based on RodZ depletion experiments, protein interaction assays, and genetic relationships described in Results. Depletion of RodZ leads to mislocalization of bPBP2b, RodA, and MreC, which are members of the core pPG elongasome, but not aPBP1a, StkP, FtsA, PBP2x, or MpgA (formerly MltG(Spn)). In turn, depletion of MreC leads to mislocalization of bPBP2b and RodA, but not RodZ or bPBP2x. Hence, depletion of RodZ results in incomplete assembly of the pPG elongasome. Structures of pneumococcal RodZ:MreC, RodZ:MreC:MreD, bPBP2b:RodA, and bPBP2b:RodZ complexes were predicted by AlphaFold‐Multimer (Evans et al., 2021) and aligned by PyMOL, version 2.0 (Schrödinger, LLC) into a model of the core elongasome in S. pneumoniae. Synthetic‐viable genetic relationship between RodZ(Spn) and aPBP1b and interaction experiments described in results implicate aPBP1b in pPG elongasome regulation and possibly in pPG synthesis. Interaction experiments show that RodZ(Spn) interacts with GpsB and EzrA, which have been proposed to play roles in the interface between cell division and PG synthesis in S. pneumoniae (Cleverley et al., 2019; Perez, Villicana, et al., 2021; Rued et al., 2017). See text for additional details. (b) Δpbp1b suppresses ΔrodZ, but not ΔmreCD, and MreCD, bPBP2b, and RodA are still required for viability. (c) Δpbp1a suppresses ΔrodZ, ΔmreC, or ΔmreCD, and bPBP2b and RodA are still required for viability. A favored model postulates that some form of pPG synthesis is required for pneumococcal viability because of the proposed role of pPG synthesis in positioning future equatorial Z‐rings in daughter cells. When functions of the WT RodZ‐MreCD‐bPBP2b‐RodA core elongasome (panel (a), above) are impaired, failsafe mechanisms bypass or modulate the function of the pPG elongasome as indicated and restore division and growth. See text for additional details and alternative models for these synthetic‐viable phenotypes.
2.8. RodZ(Spn), but not MreCD(Spn), displays a synthetic‐viable genetic relationship with aPBP1b
Tn‐seq analysis indicates the essentiality of the cytoplasmic N‐terminal HTH and TM domains of RodZ(Spn) (above; Figure 2a, row 1) and confirmed the essentiality of MreC(Spn) and MreD(Spn) (see above). Unexpectedly, Tn‐seq analysis of a Δpbp1b mutant, which lacks aPBP1b of unknown function, indicates suppression of rodZ essentiality (i.e., insertions throughout the HTH and TM domains), but not mreCD essentiality (Figure 2a, row 2). Previous results and those reported here show that Δpbp1a suppresses the requirement for MreC, MreD, and RodZ (Table 1) (Fenton et al., 2016; Land & Winkler, 2011; Tsui et al., 2016). Likewise, ΔkhpA and ΔkhpB mutations, which result in the absence of the major KhpAB RNA‐binding regulatory protein (Hor et al., 2020; Olejniczak et al., 2022), suppress ΔmreCD and ΔrodZ mutations (Table 1) (Zheng et al., 2017). These results are reiterated by Tn‐seq analysis (Figure 2a, row 3). Finally, in contrast to Δpbp1b or Δpbp1a, Tn‐seq analysis shows that Δpbp2a, which lacks aPBP2a, fails to suppress the essentiality of mreC, mreD, or rodZ in S. pneumoniae (Figure 2a, row 4). We conclude that there is an unanticipated synthetic‐viable genetic relationship between null mutations of pbp1b and rodZ, but not between pbp1b and mreCD (Figure 14b). By contrast, there is a different synthetic‐viable genetic relationship between null mutations of pbp1a and mreC, mreD, or rodZ (Figure 14c; Discussion).
We confirmed these synthetic‐viable relationships detected by Tn‐seq by independent transformation assays, in which ΔrodZ or ΔmreCD amplicons were transformed into deletion mutants of recipient strains (Figure 2b,c). Consistent with the Tn‐seq results, Δpbp1b suppresses ΔrodZ, but not ΔmreCD, Δpbp1a or ΔkhpB suppresses ΔrodZ or ΔmreCD, and Δpbp2a does not suppress ΔrodZ or ΔmreCD (Figure 2b,c). This synthetic‐viable genetic relationship between aPBP1b and RodZ could reflect a direct interaction between the proteins. To test this idea, we performed B2H assays. Indeed, aPBP1b interacts with itself and shows bidirectional interactions with RodZ, MreC, or aPBP1a (Figure 2d), as well as unidirectional interactions with MreD, MpgA, aPBP2a, or EzrA. Altogether, these results implicate aPBP1b in the regulation of the pPG elongasome, possibly through direct interaction with RodZ, as discussed below.
3. DISCUSSION
The essentiality of RodZ(Spn) has been controversial (Stamsas et al., 2017; Straume et al., 2017; Tsui et al., 2016). ΔrodZ mutants can be constructed in laboratory strain R6 and its derivatives (Martin‐Galiano et al., 2014; Straume et al., 2017); however, these R6‐derived strains contain dozens of mutations compared with progenitor D39 strain, including mutations in pbp1a (Land & Winkler, 2011; Lanie et al., 2007). Moreover, full suppression of essential mutations in R6‐strains requires additional unspecified mutations, besides those known in pbp1a (Land & Winkler, 2011), making genetic deconvolution problematic. Because of this genetic variance, R6‐derived strains have significant differences in shape and the timing of cell division compared with progenitor D39 strains (Trouve et al., 2021), indicating that R6‐derived strains are not an exact model for mechanisms of PG synthesis and cell division that occur in their virulent progenitor D39 strain. Yet, rodZ has also been classified as non‐essential in several Tn‐seq studies of different S. pneumoniae serotype strains (Slager et al., 2018; van Opijnen & Camilli, 2012). Tn‐seq and deletion analyses (Figures 2 and 6) presented here show that the entire extracellular domain corresponding to about 48% of rodZ(Spn) is dispensable, which may account for these previous misclassifications. The essentiality of rodZ and other members of the pPG elongasome, including mreC, mreD, pbp2b, and rodA, was confirmed by transformation assays of unencapsulated (Δcps) S. pneumoniae D39 grown at 37°C (Table 1; Figure S2c). rodZ was also confirmed to be essential by transformation assays of encapsulated D39 strains (Table 1) and by depletion assays of unencapsulated strains (Figures 3, 4, 5). Analogous to the case in E. coli (Bendezu & de Boer, 2008), this essentiality is conditional in unencapsulated D39 strains. Mutants lacking components of the pneumococcal pPG elongasome (MreCD, bPBP2b, RodA, or RodZ) grow poorly at the lower temperature of 32°C under the growth conditions tested (Figure S2). It remains to be tested whether this residual growth at 32°C, which is close to the that of the nasopharynx, is of physiological significance in vivo, because insertions in mreC, pbp2b, or rodA are not recovered in Tn‐seq screens of a murine nasal colonization model (van Opijnen & Camilli, 2012).
In rod‐shaped bacteria, RodZ acts as the linker between cytoplasmic MreB filaments and the largely extracellular Rod complex containing regulators MreC and MreD and the PG synthase complex consisting of an essential bPBP and SEDS‐protein RodA (reviewed in (Rohs & Bernhardt, 2021)). Despite the absence of MreB, the overall structure of RodZ(Spn) is remarkably similar to that of RodZ in rod‐shaped bacteria that have MreB (Figures 1 and S1) (Ago & Shiomi, 2019; Alyahya et al., 2009; Bendezu et al., 2009; Shiomi et al., 2008). Nevertheless, the TM and cytoplasmic domains, including the HTH domain, are essential for RodZ(Spn) function (Figures 2 and 6; Table 2), similar to their importance in rod‐shaped bacteria (Alyahya et al., 2009; Bendezu et al., 2009; Morgenstein et al., 2015; Shiomi et al., 2008). In rod‐shaped bacteria, the HTH domain of RodZ binds to MreB, and changes in amino acids in one helix (Figures 1 and S9) alter this interaction (Bendezu et al., 2009; Morgenstein et al., 2015; van den Ent et al., 2010). However, amino acid changes at corresponding positions in RodZ(Spn) do not cause a detectable phenotype (Table 2; Figure 6). In contrast to the HTH and TM domains, the RodZ(Spn) β‐strand DUF domain and most of the extracellular unstructured linker (beyond amino acid 132) are dispensable for growth, although cell morphology is altered by truncation ending at amino acid Q134 (Figures 2 and 6; Table 2). Deletion of the RodZ(Spn) DUF domain lessens interactions with a couple of proteins in B2H assays, including bPBP2b (Figures 9c and S15a); however, these interactions, if they exist in pneumococcal cells, are dispensable.
Several lines of evidence presented here strongly link RodZ(Spn) to the elongasome that carries out pPG synthesis at the middle of dividing pneumococcal cells (Briggs et al., 2021; Massidda et al., 2013; Perez, Boersma, et al., 2021; Philippe et al., 2014; Tsui et al., 2014). Transformation assays show that ΔrodZ(Spn) is suppressed by the same mutations that suppress ΔmreC, including Δpbp1a, mpgA(Y488D), ΔkhpA, insertions in khpB, and overexpression of FtsA (Figure 2; Table 1) (Land & Winkler, 2011; Tsui et al., 2016; Zheng et al., 2017). MreC and MpgA are members of the pPG elongasome (Briggs et al., 2021; Fenton et al., 2016; Land & Winkler, 2011; Philippe et al., 2014; Straume et al., 2017; Tsui et al., 2016), and KhpAB and FtsA amounts regulate the requirement for pPG elongasome in S. pneumoniae (Zheng et al., 2017). Although CozE(Spn) also associates with proteins in the pPG elongasome (Fenton et al., 2016), it is neither essential, nor is the poor growth of ΔcozE mutants suppressed by Δpbp1a, mpgA(Y488D), or other mutations that suppress ΔmreC and ΔrodZ in transformation assays (Table 1). These results suggest different functions of MreC and RodZ from CozE in pPG elongation under the conditions tested here.
In addition, throughout the cell cycle, RodZ(Spn) colocalizes with pPG elongasome members MreC and aPBP1a (Figure 7), which are genetically linked to pPG synthesis (see below) (Briggs et al., 2021; Fenton et al., 2016; Land & Winkler, 2011; Massidda et al., 2013; Tsui et al., 2014). These colocalization experiments were performed by IFM of exponentially growing cells expressing C‐terminal RodZ‐Myc, RodZ‐FLAG, or RodZ‐L‐FLAG3 fusions from the rodZ native locus. These constructs did not cause observable growth or morphology defects (Figures S11 and S12). Attempts to construct numerous RodZ(Spn) N‐terminal, C‐terminal, or internal sandwich‐fusions to different fluorescent‐protein reporters were unsuccessful, because the fusions were lethal, caused cell morphology defects, and/or showed diffuse fluorescence over entire cells (data not shown). Finally, depletion experiments of RodZ(Spn) or MreC(Spn) demonstrate that RodZ is required for MreC, bPBP2b, and RodA localization, whereas MreC is required for bPBP2b and RodA localization, but not for RodZ localization (Figures 10, 11, 12, 13, 14). Together, these transformation assays, colocalization, and assembly hierarchy results establish that RodZ is a member of the pPG elongasome of S. pneumoniae.
RodZ(Spn) acting as a scaffold for the MreC/bPBP2b/RodA complex is analogous to its function in rod‐shaped bacteria, where an assembly hierarchy has not yet been reported (Ago & Shiomi, 2019; Bendezu et al., 2009; Morgenstein et al., 2015; Rohs & Bernhardt, 2021). However, there are some specific differences in phenotypes that may reflect the absence of an MreB‐dependent mechanism in S. pneumoniae. Depletion of RodZ(Spn) is bacteriostatic, halts growth, and leads to cell rounding, increased size, and slight chaining (Figures 3, 4, 5 and S4–S5). Nonetheless, cells depleted for RodZ(Spn) are more heterogenous in shape and form shorter chains than cells depleted of MreC(Spn) (Land & Winkler, 2011), bPBP2b(Spn) (Berg et al., 2013; Tsui et al., 2014), or the regulated FtsEX:PcsB(Spn) PG hydrolase (Sham et al., 2011; Sham et al., 2013), which also localizes with the pPG elongasome (Perez, Boersma, et al., 2021) and may act as a remodeling space maker during PG synthesis (Bartual et al., 2014; Briggs et al., 2021; Rued et al., 2019; Sham et al., 2011; Trouve et al., 2021). These results suggest that RodZ(Spn) may have some functions distinct from MreC(Spn), as discussed below for aPBP1b (see Figure 14b). In another example, activated mutants of bPBP2(Eco) largely bypass the requirement for regulators MreC, MreD, and RodZ, suggesting that MreB, bPBP2, and RodA form the core of the E. coli elongasome (Rohs et al., 2018). In contrast, corresponding amino acid changes in the pedestal region of bPBP2b(Spn) fail to suppress ΔrodZ mutations (Table S6), raising the possibility that bPBP2(Eco) activation is dependent on MreB. Other differences in phenotypes suggest functional differences between RodZ in S. pneumoniae and E. coli. Overexpression of FtsZ(Spn) does not suppress ΔrodZ(Spn) (Table 1), and overexpression of RodZ(Spn) does not alter cell shape (Figure S6), in contrast to overexpression phenotypes reported in E. coli and C. crescentus (Alyahya et al., 2009; Bendezu et al., 2009; Shiomi et al., 2008).
To further define the role of RodZ(Spn) in pPG synthesis by the elongasome, we performed co‐IP and B2H assays (summarized in Figures 8 and 9). B2H and bimolecular fluorescence complementation assays and genetic approaches have indicated that RodZ of rod‐shaped bacteria interacts with MreB and with other components of the Rod complex, including MreC, MreD, bPBP2, and RodA (Ago & Shiomi, 2019; Alyahya et al., 2009; Beilharz et al., 2012; Morgenstein et al., 2015; Shiomi et al., 2013; van den Ent et al., 2010). Likewise, RodZ(Spn) is found in complexes in unsynchronized pneumococcal cells with tested proteins bPBP2b, MreC, MpgA, and aPBP1a, which have been implicated in pPG elongation (Figure 8; Table 3) (Briggs et al., 2021; Philippe et al., 2014; Straume et al., 2021). Direct interactions were detected in B2H assays between RodZ(Spn) and these proteins and with other elongasome proteins MreD and RodA (Figure 9), which were not tested in co‐IP assays (Figure 8). In addition, complexes and direct interactions were detected for RodZ(Spn) and other aPBPs (aPBP2a and aPBP1b), PG synthesis regulators (GpsB and DivIVA), the sPG synthase components (bPBP2x and FtsW), and EzrA (Figures 2, 8, and 9; Table 3). Based on B2H assays, many of these interactions are mediated at some level by the cytoplasmic HTH domain of RodZ(Spn), but others are not (Figures 9 and S15; Table S7). Phosphorylation of RodZ(Spn) by StkP was not detected by western blotting (Figure S10) or by phosphoryl‐proteomic analysis (Ulrych et al., 2021).
The functions and timing of these putative interactions of RodZ(Spn) with proteins outside of the canonical elongasome remain to be determined. Pneumococcal GpsB is found in complexes with EzrA, MreC, aPBP2a, bPBP2b, and StkP, which itself is detected in complexes with MreC, bPBP2b, and bPBP2x, at currently unknown stages of cell division (Cleverley et al., 2019; Rued et al., 2017). The interactions and phenotypes of gpsB mutants suggest a model wherein GpsB activates protein phosphorylation by StkP and also balances sPG and pPG synthesis at the midcell of dividing pneumococcal cells (Cleverley et al., 2019; Fleurie, Manuse, et al., 2014; Rued et al., 2017). Moreover, a low level of bPBP2x is detected in the outer midcell pPG synthesis ring of S. pneumoniae (Briggs et al., 2021; Perez, Boersma, et al., 2021; Tsui et al., 2014). On the other hand, strong complexes containing RodZ(Spn) and FtsZ were not detected by co‐IP (Figure 8; Table 3) and no interactions between RodZ(Spn) and FtsZ were detected by B2H assays (Figure 9), in contrast to what was reported in E. coli (Ago & Shiomi, 2019; Yoshii et al., 2019). In this regard, S. pneumoniae lacks a homolog of newly characterized ZapG, which interacts with elongasome proteins, including RodZ(Eco), and divisome proteins (Mehla et al., 2021). At best, only marginal complexes were detected containing RodZ(Spn) and FtsA, which is an actin homolog like MreB (Figure 8). Altogether, the detected interactions and assembly patterns are consistent with RodZ(Spn) acting as a scaffold protein that may link GpsB and EzrA to the assembly and function of the pneumococcal pPG elongasome (Figure 14a).
A main finding from this study is the synthetic‐viable genetic relationship specifically between RodZ and aPBP1b, but not between RodZ and MreCD in S. pneumoniae (Figures 2 and S15). This is one of the first phenotypes that has been associated with pneumococcal aPBP1b, about which little is known (Briggs et al., 2021; Ducret & Grangeasse, 2017; Straume et al., 2021). The regulation and functions of Class A PBPs are not generally well understood in S. pneumoniae and other bacteria, but likely play roles during normal growth and stress conditions (Briggs et al., 2021; Pazos & Vollmer, 2021; Rohs & Bernhardt, 2021; Straume et al., 2021; Vigouroux et al., 2020). aPBP1a and aPBP2a have a synthetic‐lethal relationship, such that Δpbp1a Δpbp2a mutants are inviable (Paik et al., 1999; Straume et al., 2021). In addition, aPBP1a was shown previously to have a synthetic‐viable relationship with the elongasome components MreC, MreD, and RodZ, such that Δpbp1a suppresses and allows growth of strains containing normally lethal ΔmreC, ΔmreCD, or ΔrodZ mutations (Figure 2; Table 1) (Fenton et al., 2016; Land & Winkler, 2011; Tsui et al., 2016). Notably, Δpbp1a does not suppress Δpbp2b or ΔrodA, indicating that aPBP2b and RodA are still required for viability in the absence of MreCD, RodZ, and aPBP1a (Tsui et al., 2016). In contrast to aPBP1a, aPBP1b displays a synthetic‐viable relationship only with RodZ, but not with MreCD, such that Δpbp1b suppresses ΔrodZ, but does not suppress ΔmreCD (Figure 2a). This synthetic viable relationship was confirmed by transformation assays (Figure 2b,c), and B2H assays indicate direct interactions between aPBP1b and RodZ, MreC, or aPBP1a (Figure 2d). Tn‐seq data confirms that Δpbp1b also does not suppress knock‐out insertions in pbp2b or rodA (data not shown).
Different models can account for the synthetic‐viable relationships of aPBP1b and aPBP1a with components of the pPG elongasome. Our favored model postulates that some form of pPG synthesis is essential for pneumococcal viability and that failsafe mechanisms can bypass or modulate the function of the core RodZ‐MreCD‐bPBP2b‐RodA elongasome (Figure 14b,c). pPG synthesis may be essential, because it drives the composite MapZ/FtsZ/FtsA/EzrA nascent equatorial rings from midcell to the middle of daughter cells (Briggs et al., 2021; Fleurie, Lesterlin, et al., 2014; Holeckova et al., 2014; Perez et al., 2019). The absence of aPBP1b induces the pneumococcal WalRK two‐component system (TCS) regulon (Zheng et al., 2017), which responds to cell wall stresses (Gutu et al., 2010; Tsui et al., 2016; Wayne et al., 2012). Induction of the WalRK TCS increases transcript amounts of genes encoding PG hydrolases and PG‐binding proteins (Ng et al., 2005). Another possible change that may occur in the absence of aPBP1b is altered activity and/or interactions of aPBP1a, which associates with the core pPG elongasome and can provide alternate TP and GT activities (Figures 8 and 9) (Briggs et al., 2021; Land & Winkler, 2011; Philippe et al., 2014; Tsui et al., 2016). In a ΔrodZ Δpbp1b mutant, these alternate activities and/or interactions are proposed to bypass the defects in pPG elongasome assembly in the absence of RodZ and allow the aPBP2b:RodA PG synthase and perhaps aPBP1a to carry out sufficient pPG synthesis for division and growth (Figure 14b).
The absence of aPBP1a also induces the WalRK regulon (Zheng et al., 2017), changes cell shape in culture (Land & Winkler, 2011; Tsui et al., 2016), and possibly alters the interactions and/or functions aPBP1b, which is associated with the pneumococcal pPG elongasome (Figure 2). In a Δpbp1a ΔmreCD ΔrodZ mutant, alternate activities and/or interactions would bypass the defects in pPG elongasome assembly in the absence of RodZ and MreCD and allow the bPBP2b:RodA PG synthase and perhaps aPBP1b to carry out sufficient pPG synthesis for division and growth (Figure 14c). Another putative component of failsafe, bypass mechanisms for pPG synthesis is the sPG synthase bPBP2x:FtsW. A majority of bPBP2x migrates in the inner ring of sPG synthesis at the leading edge of the closing septal annulus (Briggs et al., 2021; Perez, Boersma, et al., 2021; Tsui et al., 2014). However, some bPBP2x, and presumably FtsW, remains in the outer pPG synthesis ring and may provide an alternate pathway of pPG synthesis. Consistent with this notion, RodZ(Spn) associates with bPBP2x and FtsW (Figures 8 and 9; Table 3). Finally, although aPBP2a also associates with the pPG elongasome (Figures 8 and 9; Table 3), there is no genetic evidence that implicates aPBP2a in a bypass mechanism of pPG synthesis (Figure 2), and Δpbp2a does not induce the WalRK TCS (Zheng et al., 2017).
Other genetic patterns strongly support the idea of alternate, bypass mechanisms to maintain pPG synthesis in S. pneumoniae. The pPG elongasome‐associated muramidase MpgA (formerly MltG(Spn)) (Figures 8 and 9) is essential; yet, ΔmpgA is suppressed by Δpbp1a (Taguchi et al., 2021; Tsui et al., 2016). Furthermore, a triple Δpbp1a ΔmpgA Δpbp2b mutant lacks the core pPG elongasome, but is viable and forms elongated cells (Tsui et al., 2016). In addition, a mpgA(Y488D) mutant, which expresses an MpgA with greatly reduced enzymatic activity (Taguchi et al., 2021; Tsui et al., 2016), bypasses the requirement for the core pPG elongasome, in that mpgA(Y488D) Δpbp2b and mpgA(Y488D) ΔrodA mutants are viable and form elongated cells (Tsui et al., 2016). Yet, the mpgA(Y488D) Δpbp2b Δpbp1a mutant is now inviable, which appears at odds with the phenotype of the Δpbp1a ΔmpgA Δpbp2b mutant. This apparent discrepancy can be explained if aPBP1b mediates pPG bypass synthesis in the Δpbp1a ΔmpgA Δpbp2b mutant, while aPBP1a mediates pPG bypass synthesis in the mpgA(Y488D) Δpbp2b mutant, where aPBP1a bypass activity is dependent on the physical presence, but not the activity, of MpgA(Y488D). Finally, the shape and size of WT and Δpbp1b cells grown in BHI broth are similar to those of Δpbp1b ΔrodZ mutants, with some heterogeneity, whereas Δpbp1a ΔrodZ cells have the distinctive shorter, narrower shape of Δpbp1a mutant cells compared with WT (data not shown) (Land & Winkler, 2011; Tsui et al., 2016). These data indicate that suppression of ΔrodZ by Δpbp1b or Δpbp1a are not equivalent, consistent with different mechanisms. Taken together, these results support the hypothesis of multiple bypass pathways for essential pPG synthesis when the core pPG elongasome is incomplete or absent.
Another model for the synthetic‐viable relationships of aPBP1b and aPBP1a with components of the pPG elongasome invokes direct regulation of aPBP activity, interactions, and/or localization (Figure S27) (Land & Winkler, 2011; Tsui et al., 2016). In this model, RodZ(Spn) acts as a negative regulator of aPBP1b activity, interactions, and/or mislocalization. The absence of RodZ causes aPBP1b misregulation that contributes to cell lethality (Figure S27a). Likewise, MreC, MreD, and RodZ would negatively regulate aPBP1a activity, interactions, and/or mislocalization. In the absence of MreC, MreD, or RodZ, aPBP1a misregulation contributes to cell lethality (Figure S27b).
There are issues with this alternative model. First, to date, PBPs have been found to be positively regulated, rather than negatively regulated. For example, in E. coli the activities of FtsI(bPBP3), bPBP2, aPBP1a, and aPBP1b are positively activated by FtsN, MreC, LpoA, and LpoB, respectively (Pazos & Vollmer, 2021; Pichoff et al., 2019; Rohs & Bernhardt, 2021). In S. pneumoniae, aPBP2a is positively regulated by MacP and GpsB (Cleverley et al., 2019; Fenton et al., 2018). Second, aPBP misregulation alone is insufficient to account for cell lethality, because function of the bPBP2b:RodA pPG synthase is still essential (Figure 14b,c), despite suppression of ΔrodZ or ΔmreCD ΔrodZ by Δpbp1b or Δpbp1a, respectively (Figure 2). Third, aPBP1a localizes normally when RodZ or MreC is depleted (Figures 12 and 13c). This result contrasts with a previous conclusion that aPBP1a mislocalizes in the absence of MreC (Fenton et al., 2016). The different results may reflect the ectopic induction of potentially high levels of active GFP‐aPBP1a in a ΔmreC mutant, which is not tolerated, as opposed to the MreC depletion used here in a strain expressing epitope‐tagged aPBP1a‐FLAG from its chromosomal locus (Figure S25b). Moreover, moderate (≈2‐fold) overexpression of aPBP1a in a WT strain does not overtly affect normal cell morphology or growth (Averi McFarland; unpublished result). Finally, because the synthetic‐viable relationships of the aPBPs with components of the pPG elongasome are different, it is difficult to reconcile a model postulating that absence of aPBP1b or aPBP1a in the ΔrodZ or ΔrodZ ΔmreCD mutant, respectively, solely decreases competition or interference with residual bPBP2b:RodA pPG synthase activity. For this type of model to work, there still needs to be differential regulation of aPBP1b or bPBP1a expression, activity, and/or interactions allowing bypass pPG synthesis. Therefore, current data supports alternate mechanisms leading to pPG bypass synthesis more than other models for these synthetic‐viable genetic relationships.
The different synthetic‐viable relationships of aPBP1b and aPBP1a with components of the pPG elongasome indicates functional fungibility during pPG synthesis in S. pneumoniae. This flexibility points to altered protein interactions and/or regulatory pathways that enable alternate pPG synthesis pathways. Some of these outcomes may be through direct interactions, while others may be indirect through additional proteins induced or regulated by stress responses. Along the same lines, CozE does not have a strong synthetic viable relationship with aPBP1a in transformation assays or in Tn‐seq analyses (data not shown) under the conditions used here, and ΔcozE was not suppressed by the array of mutations that suppress ΔrodZ and ΔmreC (Table 1). These shared and different functions and interactions of pPG elongasome members RodZ, MreC, MreD, and CozE require further study at different stages of the pneumococcal cell cycle. Taken together, we conclude that both aPBP1a and aPBP1b play roles in the regulation of the pPG elongasome and possibly participate in pPG synthesis in S. pneumoniae. The action of RodZ(Spn) in assembly and function of the pPG elongasome and the roles of the aPBPs in WT and bypass pPG synthesis are important topics for future studies.
4. EXPERIMENTAL PROCDURES
4.1. Strain construction and growth conditions
Bacterial strains used in this study are derivatives of the unencapsulated, S. pneumoniae serotype 2 strain D39W (Lanie et al., 2007; Slager et al., 2018), and are listed in Table S1. For strains containing antibiotic markers, linear DNA amplicons synthesized by fusion PCR were transformed into competent pneumococcal cells as described in Land et al. (2013); Tsui et al. (2010); Tsui et al. (2014). For antibiotic selection Trypticase soy agar II (modified; Becton‐Dickinson) and 5% (vol/vol) defibrinated sheep blood (TSAII‐BA) plates were supplemented with the following final concentrations of antibiotics: 250 μg kanamycin/ml, 150 μg spectinomycin/ml, 0.3 μg erythromycin/ml, 200 μg streptomycin/ml, or 0.25 μg tetracycline/ml. Strains containing markerless mutations or insertions at the native site in the chromosome, e.g., iht‐pbp2b markerless (i‐tag‐HaloTag (iHT) fused to bPBP2b (Perez et al., 2019)), were constructed through two rounds of transformation via the Janus method, as described in Sung et al. (2001). Linkers used in construction of fluorochrome or epitope‐tagged fusion proteins are listed in Table S2. All primers and templates used in this study are listed in Table S4. All strains were confirmed via PCR and sequencing. For overnight growth, BHI broth was inoculated with frozen glycerol stocks, serially diluted and propagated overnight for 12–13 h at 37°C in an atmosphere of 5% CO2. Antibiotics were not added to the media. To start experiments, overnight cultures with an OD620 of 0.1–0.4 (exponentially growing) were diluted to an OD620 of 0.003 in fresh BHI, lacking antibiotics.
4.2. Tn‐seq transposon library generation and insertion sequencing
Tn‐seq was carried out using protocols in (Fenton et al., 2016; van Opijnen et al., 2015) with the following modifications. A transposon insertion library was generated for each of the following strains: WT D39 ∆cps rpsL1 (IU1824), isogenic ∆pbp1b (IU14697), ∆khpB (IU10592), and ∆pbp2a (IU13256). Approximately 200,000 (WT, ∆pbp1b and ∆khpB) or 300,000 (Δpbp2a) transformants were pooled for each library (see Appendix S1). Genomic DNA preparations were modified from the instructions provided by Qiagen for Gram‐positive bacteria using a DNeasy blood and tissue kit (Qiagen 69,504). 5 ml of cultures at OD620 = 0.4 were centrifuged for 10 min at 5000× g at room temperature and suspended in 180 μl of enzymatic lysis buffer containing 20 mg/ml lysozyme, and incubated for 30 min at 37°C. 10 μl of RNase A (Qiagen 19,101,100 mg/ml) was added, followed by a 5‐min incubation at room temperature. Subsequent steps were as specified by the Qiagen manual, except that DNA was eluted with 100 μl of water. Eluted genomic DNA and pMagellan6 DNA prepared with a QIAprep Spin Miniprep kit (27104) were concentrated by ethanol precipitation to concentrations of more than 0.3 μg/μl in ultrapure distilled water. In vitro transposition reactions were performed as described previously (van Opijnen et al., 2015) with genomic DNA obtained from WT or mutant strains with the following modifications. A reaction mixture of 1 μg genomic DNA, 1 μg pMagellan6 DNA, and 3 μl of purified MarC9 transposase prepared as specified in (van Opijnen et al., 2015) was incubated at 30°C for 4 h. Transposon junctions were repaired by using 1 μl of 3 U/μl T4 DNA polymerase at 12°C for 30 min. All incubation steps were performed in a thermocycler. Ten independent 20 μl‐transposition products were prepared each time and stored at −20°C. Starter cultures for transformation were prepared by growing frozen stocks of respective strains in 4 ml of BHI broth at 37°C with CO2 to OD620 ≈ 0.15. The cultures were centrifuged for 3 min at 16,000× g at room temperature, and pellets were resuspended in 400 μl BHI broth mixed with 600 μl 25% glycerol. 50‐μl aliquots of starter cultures were stored at −80°C.
Transposed DNA was transformed into CSP‐1 induced competent WT or mutant strains and plated onto TSAII agar plates containing spectinomycin and catalase. On the day of transformation, recipient strains were grown from frozen starter cultures in 5 ml of BHI broth to OD620 ≈ 0.03–0.04. TSAII agar (BBL 212305) plates containing spectinomycin and catalase were prepared by pouring 17 ml of warm TSAII agar containing 150 μg/ml spectinomycin into each 100 × 15 mm plate. After solidification, 396 μl (13,000 to 15,000 U) of catalase solution (Worthington, CAT # LS001896) were spread on the surface of each plate and dried for 30 min in a sterile hood. Transformation mixes were prepared by addition of 40 μl CSP‐1 (50 ng/μl), 1 ml heat inactivated horse serum, 45 μl 40% glucose, to 9 ml BHI broth. 300 μl of cell culture at OD620 ≈ 0.03–0.04 were mixed with 700 μl of transformation mix. After incubation at 37°C for 10 min, 3–8 μl of respective transposed DNA were added to each transformation, and the mixtures were incubated at 37°C for 1 h. Transformations containing no DNA or 27 ng of genomic DNA obtained from IU2072 containing spxR::Mariner (Ramos‐Montanez et al., 2008) were used as negative or positive controls. 200 μl of transformed cell culture were spread on the surface of each prepared TSAII/spectinomycin/catalase plate, which were incubated at 37°C in 5% CO2 for 20 h.
After 20 h of incubation, colonies were scraped from 50 plates and collected in 20 ml of BHI broth. Cell suspensions were centrifuged for 8 min at 3000× g at room temperature, and the pellets were resuspended in 3 ml BHI mixed with 2 ml 25% glycerol. From each transformation, 20 250‐μl of transposon library starter culture aliquots were stored at −80°C. For WT, ∆pbp1b, ∆khpB strains, transposon library starter cultures were obtained from ≈200,000 colonies with 11, 2, and 5 independent transformations, respectively. For ∆pbp2a strain, transposon library starter cultures were obtained from ≈300,000 colonies with 4 independent transformations. Transposon library starter cultures from different transformations were thawed and mixed together in proportion to the numbers of transformants obtained from each transformation. The combined starter cultures were diluted to OD620 ≈ 0.005 in 5 ml of BHI broth containing 180 μg/ml spectinomycin and 5 μl/ml Ec‐oxyrase (Oxyrase, EC0005), and were grown at 37°C with 5% CO2 to OD620 ≈ 0.4. 5 ml of culture at OD620 ≈ 0.4 were used to extract genomic DNA using DNeasy blood and tissue kit. 3 μg of DNA from each sample was used for MmeI digestion, followed by ligation to adaptors described in van Opijnen et al. (2015). The samples were further processed according to (Fenton et al., 2016) and sequenced on the Illumina NextSeq 500 using a NextSeq 75 high sequencing kit at the Center for Genomics and Bioinformatics, Indiana University Bloomington. Sequencing reads were de‐multiplexed and trimmed using the QIAGen CLC genomics workbench (version 11.0.1). Data were mapped and analyzed as described in Fenton et al. (2016). Insertion data were visualized graphically using the Artemis genome browser (version 10.2) (Carver et al., 2012). Tn‐seq primary data for the region between mreD (spd_2044) and spd_2051, the gene upstream rodZ (spd_2050), are contained in Appendix S1, including run summaries, number of reads per TA site in each gene, and count ratios for each gene in the indicated mutants compared with WT. p values for comparisons of the number of reads per TA site in each gene were calculated by the Mann–Whitney test using GraphPad Prism (9.2.0).
4.3. Growth of Zn‐dependent depletion and merodiploid strains
Ectopic expression of rodZ or mreC was achieved from a zinc‐inducible promoter (PZn) in the bgaA site (Tsui et al., 2014, 2016). 0.4 mM of ZnCl2 and corresponding 1/10 concentration of MnSO4 were added to TSAII‐BA plates or BHI broth for inducing conditions. Mn2+ was added to Zn2+ conditions to prevent zinc toxicity (Jacobsen et al., 2011; Perez, Villicana, et al., 2021; Tsui et al., 2016). Depletion strains requiring ZnCl2 for growth were grown overnight in BHI broth in the presence of Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4). For depletion/complementation experiments, overnight cultures (OD620 of 0.1–0.4) supplemented with inducer were diluted to an OD620 of 0.003 in 5 ml fresh BHI with or without inducer. Growth was monitored every 1 h using a Genesys 2 spectrophotometer (Thermo Scientific) for 10 h. For these experiments, the point of resuspension serves as time zero, (T = 0). All growth curves and microscopy experiments were performed two or more times with similar results.
4.4. Transformation assays
Transformations were performed as detailed in (Rued et al., 2017; Tsui et al., 2016). All amplicons (experimental and control) contained ≈1 kb flanking region and were obtained from PCR reactions using primer pairs and templates listed in Table S4. Recipient strains were grown to OD620 ≈ 0.03 from glycerol ice stock and 100 μl was added to 900 μl of transformation mix containing 10% (wt/vol) heat‐treated horse serum, 0.18% (wt/vol) glucose, 100 ng CSP‐1 (competence stimulatory peptide, type 1) ml−1 and 9 ml of BHI. The mixture was incubated for 10 min at 37°C in the presence of 5% CO2. 30 or 100 ng of purified amplicon (for unencapsulated and encapsulated strains, respectively) was added to the transformation mixture and incubated for 1 h at 37°C in the presence of 5% CO2. A fraction or the entire final transformation mixture was added to 3 ml of soft agar containing the appropriate antibiotic (72 μl of 0.1 mg erythromycin ml−1, 36 μl of 0.1 mg spectinomycin ml−1, 60 μl of 100 mg streptomycin ml−1, or 120 μl of 50 mg kanamycin ml−1) and plated onto TSAII‐BA plates. Unless explicitly stated, the numbers of colonies listed are normalized to 1 ml of transformation mixture. Transformants were incubated overnight in the presence of 5% CO2 for 20–24 h, at which time colony numbers and morphology were counted and evaluated.
4.5. Viable count (CFU) assays
At various time points (3, 4, 5, 6, and 7.5 h) in depletion and complementation experiments, 100 μl of culture was suspended in 900 μl of 1× PBS and serially diluted from 10−1 to 10−7. 100 μl of three selected dilutions were suspended in 3 ml of molten soft agar and poured onto TSAII‐BA plates with or without inducer, 0.4 mM Zn + 0.04 mM Mn. Solidified plates were incubated at 37°C in the presence 5% CO2 for 20–24 h. Plates containing 30–300 CFUs were scored and counted with respect to colony number and size. CFU/ml values were calculated for each strain and condition.
4.6. Image acquisition and processing
For 2D epifluorescence microscopy (eFM), 2D IFM), and phase‐contrast microscopy (PCM) experiments, images were taken using a Nikon E‐400 epifluorescence phase‐contrast microscope and 100× Nikon Plan Apo oil‐immersion objective (numerical aperture, 1.40) connected to a CoolSNAP HQ2 charged‐coupled device (CCD) camera (Photometrics). Images were analyzed with NIS‐Elements AR software (Nikon). Micrographs were assembled using Adobe Illustrator and all images are to scale.
4.7. Cell length and width measurements
Cell lengths and widths of strain growing exponentially in BHI broth were measured as previously described (Perez, Villicana, et al., 2021, Tsui et al., 2016). For all strains, only ovoid‐shape pre‐divisional cells were measured. Unless indicated in the figure legends, more than 50 cells from one experiment were measured, and plotted with box and whiskers plot (5 to 95 percentile whiskers). p values were obtained by one‐way ANOVA analysis by using the Kruskal–Wallis test in GraphPad Prism program.
4.8. Live‐dead staining and epifluorescence microscopy
Live/dead staining was performed using the BacLight Bacterial Viability kit (Syto9 and Propidium Iodide), according to the manufacturer's instructions (ThermoFisher Scientific, Cat. #L7007) and (Perez, Villicana, et al., 2021; Sham et al., 2013). Briefly, 500 μl of cultures were harvested at 4 and 6 h of growth and centrifuged at 12,000× g for 2.5 min at 25°C. Pellets were re‐suspended in 50 μl of BHI broth plus 2 μl of a 1:1 (v/v) mixture of Syto‐9 and propidium iodide, and incubated for 5 min in the dark at 22°C. As a control, heat killed cells (95°C for 5 min) were stained as described above for comparison. After incubation, samples were immediately imaged using both the Alexa 488 (EX 460–500, DM 505, and BA 510–560) and Alexa 568 (EX 532–587, DM 595, and BA 608–682) filters. The staining pattern of the sample was revealed by superimposing the corresponding Alexa 488 and Alexa 568 images upon one another within the NIS‐Elements AR software (Nikon). A total of 200 cells were categorized based on the staining pattern for quantification of percentage of live vs dead cells.
4.9. Fluorescent d‐amino acid short‐pulse labeling and epifluorescence microscopy
The FDAA TADA (tetramethylrhodamine 3‐amino‐d‐alanine) synthesized as described in Kuru et al. (2012) was obtained from Michael VanNieuwenhze. Samples were processed as described in Boersma et al. (2015); Perez, Boersma, et al. (2021); Tsui et al. (2014) with minor changes. Briefly, at 4 h of growth, 500 μl of culture was harvested and centrifuged at 16,000× g for 5 min at room temperature. Cultures were washed twice with 500 μl ice cold 1× PBS. Pelleted via centrifugation (16,000× for 5 min) and re‐suspended in 250 μl of BHI broth containing TADA at a final concentration of 500 μM. Working solutions of TADA in BHI were diluted from 500 mM stocks in DMSO, which were stored at −20°C. Samples were incubated for 5 min at 37°C. Cells were centrifuged for 2.5 min at 16 000× g at 4°C, and washed twice in 1 ml of cold 1× PBS. Pellets were resuspended in 1 ml of 4% (wt/vol) paraformaldehyde (EMS; 157–4), followed by 15 min incubation at room temperature and 45 min incubation on ice in the dark. Fixed cells were centrifuged and washed three times with ice cold 1× PBS. After which cells were resuspended in 50–75 μl of Slowfade Gold antifade reagent (Invitrogen; S36936), vortexed briefly, and applied to a glass slide. A glass coverslip was gently placed onto to the slide and the samples were cured overnight at 4°C, in the dark. To visualize TADA labeling, images were taken using eFM with a Texas‐Red filter.
4.10. HaloTag labeling and epifluorescence microscopy
To determine the localization pattern of the HaloTag‐fusion proteins, cells were labeled with saturating TMR ligand and viewed by eFM as described in Perez et al. (2019); Perez, Villicana, et al. (2021). Briefly, strains with HT‐domain fusions expressed from native chromosomal loci were grown in BHI broth as described above. At 4 h of growth, 0.5 μl of working stock of TMR (500 μM TMR ligand [Promega cat #G8252] in DMSO stored at −20°C) was added to 300 μl of culture (final concentration = 0.83 μM). Tubes were inverted gently three times and then incubated for 15 min at 37°C in the dark in the absence of CO2. Cells were then collected by centrifugation at 14,000× g for 2.5 min, washed once with 500 μl of fresh BHI broth, re‐pelleted (14,000× g for 2.5 min), and resuspended in 15–20 μl of fresh BHI broth. Cell shape and fluorescence localization patterns were imaged using PCM and eFM with a Texas‐red filter.
4.11. Demograph generation
Demographs showing protein fluorescence intensity as a function of cell length were generated using Microbe J (version 5.11 s) (Ducret et al., 2016; Perez et al., 2019; Perez, Villicana, et al., 2021). Demographs were also generated from phase‐contrast images corresponding to light scattering caused by the cell body as a function of cell length. For cells displaying WT size and morphology, the following “WT” parameters were used: (area [μm2] 0.53‐max; length [μm] 0.5–3.2; width ([μm] 0.2‐max; circularity [0–1] 0‐max; curvature [0‐max] 0‐max; sinuosity [0‐max] 0‐max; angularity [rad] 0–0.38; solidity [0‐max] 0.75‐max; intensity [0‐max] 0–6200; Z‐score 2.0‐max). Within a given WT field of cells, ≈3%–5% of the population were excluded from automated selection by the program due to variables such as clustering of cells, size, shape defects, and/or out of plane of focus. These cells were not manually entered into the program. Stages of cells were classified by the degree of separation as described in Perez et al. (2019). For depletion conditions in which gross morphological changes occurred in both cell shape and size, the “WT” parameters did not fit to automated selection, leading to the exclusion of >30% of the cells from automated selection. Therefore, a separate set of “mutant” parameters were used: (area [μm2] 0.43–5; length [μm] 0.3–5; width ([μm] 0.4‐max; circularity [0–1] 0‐max; curvature [0‐max] 0–0.45; sinuosity [0‐max] 0‐max; angularity [rad] 0–0.45; solidity [0‐max] 0.75‐max; intensity [0‐max] 0–6200; Z‐score 1.0‐max). Complementation fields were analyzed using the “mutant” parameters.
4.12. Immunofluorescence microscopy of strains expressing single epitope‐tagged proteins
IFM of cells harvested after 4 h of growth was performed as described in Land et al. (2013); Tsui et al. (2014); Tsui et al. (2016). Primary antibodies were: rabbit anti‐FLAG polyclonal antibody (Sigma, F7425, 1:100 dilution) or rabbit anti‐HA polyclonal antibody (Invitrogen, 71–5500, 1:100 dilution). Secondary antibodies used were: anti‐rabbit IgG conjugated to Alexa Fluor 488 (Life Technologies; A11034, 1:100 dilution). Control experiments did not detect labeling in untagged WT IU1824 or untagged depletion IU12738 (ΔrodZ//PZn‐rodZ +) and IU12345 (ΔmreC//PZn‐mreC +) strains. Protein localization patterns were visualized and scored across multiple fields for depletion/complementation conditions and WT backgrounds. Cells were scored in accordance with the key provided in the appropriate figure legends.
4.13. Immunofluorescence microscopy of strains expressing two epitope‐tagged proteins
For co‐localization IFM studies, double epitope‐tagged strains IU7072 (rodZ‐L‐F3 ftsZ‐Myc), IU7113 (mreC‐L‐F3 rodZ‐Myc), and IU7515 (pbp1a‐L‐F3 rodZ‐Myc) were grown exponentially to OD620 ≈ 0.15–0.20 and processed as detailed in (Land et al., 2013; Tsui et al., 2014, 2016). Primary antibodies used were as follows: anti‐FLAG rabbit polyclonal antibody (dilution 1:100) and anti‐Myc mouse monoclonal antibody (dilution 1:100). Secondary antibodies used were: goat anti‐rabbit conjugated to Alexa Fluor 488 (1:100) or goat anti‐mouse‐Alexa Fluor 568 (1:100). Primary and secondary antibody incubations were for 2 h at 37°C, and 1 h at 24°C, respectively. DNA in nucleoids was stained with SlowFade Gold Antifade reagent with DAPI (Life Technologies, S36939). Image analysis was performed using a point‐and‐click IMA‐GUI organized in MATLAB (The Mathworks) as described in Land et al. (2013); Tsui et al. (2014). Pneumococcal cells were manually aligned and binned into four division stages 1 to 4: pre‐, early‐, mid‐, and late‐divisional, within the program by eye. The mean cell outline (phase‐contrast image) and fluorescence intensities of DNA or tagged‐proteins were measured and represented graphically as previously performed in Land et al. (2013), where n is the number of cells at a particular division stage averaged from two independent biological replicates. Widths of bands at midcell were measured and statistically compared as described in Tsui et al. (2014) and the legend to Figure 7.
4.14. Structured‐illumination microscopy (3D‐SIM)
IFM images were taken using the Deltavision OMX Super Resolution system located in the Indiana University Bloomington Light Microscopy Imaging Center (LMIC) as detailed in (Tsui et al., 2014). Briefly, the system is equipped with four Photometrics Cascade II EMCCD cameras that allow simultaneous four‐color imaging and is controlled by DV‐OMX software, with image processing by Applied Precision softWoRx 6.0. software. For information: http://www.indiana.edu/lmic/microscopes/index.htmL#OMX.
4.15. Western blotting and immunodetection
Cell lysates were prepared by the SEDS lysis‐buffer (0.1% deoxycholate (vol/vol), 150 mM NaCl, 0.2% SDS (vol/vol), 15 mM EDTA pH 8.0) method as described in Cleverley et al. (2019). Briefly, bacteria were grown exponentially in 5 ml BHI broth to an OD620 ≈ 0.1–0.2. Aliquots of 1.0–2.0 ml were centrifuged (5 min, 16,000× g at 4°C), and cell pellets were washed once with 4°C PBS. Frozen pellets collected from 1.8 ml of cultures at OD620 = 0.16 were suspended in 80 μl of SEDS lysis buffer. Samples collected from different volumes or at different OD620 readings were resuspended in volumes of SEDS lysis buffer proportional to the culture volumes and cell densities. Cell lysis was performed by incubation at 37°C in a shaking block at 300 rpm for 15 min. Protein concentrations of lysed samples were determined with Bio‐Rad DC™ protein assay kit. Samples were denatured with 2× Laemmeli SDS loading buffer (Bio‐Rad) plus β‐mercaptoethanol (5% vol: vol) at 95°C for 10 min. 3–10 μg of total crude lysate per sample was loaded onto a 4%–15% precast gradient SDS‐PAGE gel (Bio‐Rad) and subjected to electrophoresis. Amounts of crude lysates loaded and primary antibodies are specified for individual experiments.
Sources of antibodies used for western blotting are as below. Primary antibodies used are anti‐HaloTag monoclonal antibody (Promega, G921A, 1:1000), and the following polyclonal rabbit antibodies: anti‐ FLAG (Sigma, F7425, 1:2000); anti‐HA (Invitrogen, 71–5500, 1:1000); anti‐Myc (ThermoFisher Scientific, PA1‐981, 1:1000); anti‐GFP (Invitrogen, A11122, 1:1400); anti‐StkP ([Beilharz et al., 2012], 1:10,000); anti‐PhpP ([Beilharz et al., 2012], 1:5000); anti‐MreC ((Land & Winkler, 2011), 1:5000); anti‐FtsZ ([Lara et al., 2005], 1:20,000); anti‐FtsA ([Lara et al., 2005],1:20,000); anti‐bPBP2b ((Perez, Boersma, et al., 2021), 1:10,000); anti‐bPBP2x ((Perez, Boersma, et al., 2021), 1:10,000); anti‐GpsB ((Cleverley et al., 2019), 1:2000); anti‐aPBP2a ((Cleverley et al., 2019), 1:5000); and anti‐DivIVA ([Fadda et al., 2007], 1:5000). Anti‐aPBP1a (1:5000) was generated with purified aPBP1a (aa S37 to P719) and showed no signal at 94 kDa in lysate prepared from a ∆pbp1a strain. Secondary antibodies used were anti‐mouse IgG conjugated to horseradish peroxidase (Invitrogen, SA1‐100, 1:3300), anti‐rabbit IgG conjugated to horseradish peroxidase (GE healthcare NA93AV, 1:10,000), or Licor IR Dye800 CW goat anti‐rabbit (926–32,211, 1:14,000). Chemiluminescence signals obtained with secondary HRP‐conjugated antibodies were detected using IVIS imaging system as described previously (Tsui et al., 2014). IR signals obtained with Licor IR Dye800 CW secondary antibody was detected with Azure biosystem 600.
4.16. Depletion experiments with quantitative western blotting
Depletion of proteins expressed from a Zn2+‐inducible prompter (PZn) was performed as described in Perez, Villicana, et al. (2021); Tsui et al. (2016) with the following modifications. Merodiploid strains IU12345 (ΔmreC//PZn‐mreC +) and IU10947 (ΔrodZ//PZn‐rodZ‐F) require 0.4 mM Zn + 0.04 mM Mn in BHI broth for growth and ectopic induction of MreC or RodZ‐F, respectively. To measure protein amounts during depletion of MreC, or RodZ‐F, IU1824 (WT), IU12345 (ΔmreC//PZn‐mreC +), IU14594 (rodZ‐F) and IU10947 (ΔrodZ//PZn‐rodZ‐F) were diluted from overnight cultures (IU12345 and IU10947) supplemented with Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4) and re‐suspended to an OD620 ≈ 0.003 in fresh BHI broth ± Zn inducer. Cultures were harvested at 3 or 4 h of growth, and processed for western blotting as described above. To ensure western blots were quantitative, standard curves were generated by loading a range of protein amounts on the lanes, and labeled for anti‐MreC, or anti‐FLAG for RodZ‐F (Perez, Boersma, et al., 2021). A protein amount corresponding to the mid‐range of the standard curve was loaded for each targeted protein. 3 μg of cell lysate were loaded for the detection of bPBP2x or bPBP2b, 3 or 6 μg for detection of MreC, and 10 μg for the detection of RodZ‐F. For the detection of RodZ‐F during depletion, Licor IR Dye800 CW goat anti‐rabbit secondary antibody (926–32,211)(1:14,000) was used, and IR signal was detected with Azure biosystem 600. In addition, signals obtained with anti‐F antibody were normalized with total protein stain in each lane using Totalstain Q‐NC reagent from Azure.
4.17. Phos‐tag SDS‐PAGE and western blotting
This method is based on the mobility shift of phosphorylated proteins in SDS‐PAGE with polyacrylamide‐bound Mn2+‐Phos‐tag (Kinoshita et al., 2006). Phosphorylated proteins in gels are visualized as slower migrating bands compared with corresponding unphosphorylated proteins. Phos‐tag SDS‐PAGE and standard Western blotting were carried out as described previously with modifications (Wayne et al., 2012). Overnight BHI broth cultures were diluted and grown up to OD620 ≈ 0.2 in 30 ml of BHI. Cells were centrifuged at 14,500× g for 5 min at 4°C, and all subsequent steps were performed at 4°C. Pellets were lysed using a FastPrep homogenizer (MP biomedicals) in cold lysis buffer (20 mM Tris–HCl pH 7.0 and 1 protease inhibitor tablet [ThermoFisher Scientific] per 10 ml buffer). Cell lysates were resolved by 10% SDS‐PAGE supplemented with 75 μM Phos‐tag acrylamide (AAL‐107; Wako) and 100 μM MnCl2, and standard 10% SDS‐PAGE as control. Volumes of loaded samples were normalized to OD620 of harvested cultures (for OD620 ≈ 0.2, 30 μl of the sample was loaded). Gel electrophoresis was carried out for 3 h. RodZ‐HA3 was detected by western blotting as described above using anti‐HA as the primary antibody.
4.18. Co‐immunoprecipitation assays
Co‐immunoprecipitation (Co‐IP) assays were performed as previously described in Perez et al. (2019); Perez, Villicana, et al. (2021); Rued et al. (2017). Briefly, cultures were grown exponentially to an OD620 ≈ 0.2–0.3 in 400 ml of BHI broth, concentrated 20‐fold in 4°C PBS, and cross‐linked with 0.1% paraformaldehyde for 1 h at 37°C. After quenching with 1 M glycine and a wash with PBS, pellets were re‐suspended in 2 ml of cold lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X100 (v/v)) containing protease inhibitor, and homogenized in lysing matrix B tubes (MP Biomedicals) in a FastPrep homogenizer. One ml of lysate (input sample, normalized to ≈2–5 mg/ml) was added to 50 μl of anti‐FLAG magnetic beads (Sigma, M8823) and incubate for 2 h at 4°C with rotation. 100 μl of FLAG elution solution containing 150 ng 3× FLAG peptide/μl (Sigma, F4799) was used to elute FLAG‐tagged proteins and other associated proteins (output elution samples). Input or output samples were mixed with 2× Laemmli sample buffer (Bio‐Rad) containing 10% (vol/vol) β‐mercaptoethanol (Sigma), and were heated at 95°C for 1 h to break cross‐links, with the exception of IU8918 (ftsW‐L‐gfp) and IU16126 (ftsW‐L‐gfp rodZ‐L‐F3), when the samples were not heated. For most samples, 4–6 μl of input mixed with sample buffer was loaded onto pre‐cast SDS‐PAGE 4%–15% gels (Bio‐Rad), resulting in ≈4–9 μg of cell lysate. For the output (elutions), 15 or 25 μl of samples mixed with sample buffer were loaded. Proteins were detected using standard western blotting and immunodetection methods described above. All pair‐wise co‐IP experiments were performed independently 2–6 times.
4.19. Bacterial two‐hybrid assays
Bacterial two‐hybrid (B2H) assays were performed as described before (Cleverley et al., 2019; Perez, Villicana, et al., 2021; Rued et al., 2017) with the following modifications. The hybrid plasmids used in the B2H assays are listed in Table S3. For cloning, the target genes were amplified by PCR from S. pneumoniae D39 chromosomal DNA (or its derivatives) using the primers listed in Table S4. PCR fragments for rodZ(ΔHTH), rodZ(ΔDUF) and pbp1b were purified, digested with the appropriate restriction enzymes and cloned into the corresponding sites of the pKT25/pUT18C vectors to generate plasmids encoding the corresponding hybrid proteins fused at the C‐terminal ends of the T25/T18 fragments. E. coli DH5α or XL1‐blue transformants were selected on LB agar plates containing ampicillin (100 μg/ml) or kanamycin (50 μg/ml) and 0.4% glucose to repress leaky expression (Karimova et al., 2005). The correct sequence of each construct was verified by double‐strand sequencing, using primers listed in Table S4. B2H vectors pKT25/pUT18C containing rodZ, mreC, mpgA, pbp1a, pbp2b, rodA, pbp2a, pbp2x, ftsW, and ftsA and vectors pKNT25/pUT18 containing ftsZ, ezrA, gpsB, divIVA, stkP and mreD were previously constructed and reported (Cleverley et al., 2019; Krupka et al., 2012; Perez, Villicana, et al., 2021; Rued et al., 2017). Each pair of plasmids was co‐transformed into the E. coli cya − BTH101 strain and co‐transformation mixtures were spotted directly onto LB agar plates supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml) and X‐Gal (60 μg/ml), followed by incubation at 30°C. Plasmid pairs pKT25/pUT18C and pKT25‐zip/pUT18C‐zip were used as negative and positive controls, respectively. Plates were inspected and photographed after 24 h and 40 h. In the case of time course experiments, B2H plates were inspected for color development after 24, 30 and 36 h of incubation at 30°C and scored similarly as reported in (Bendezu et al., 2009). All the B2H experiments were performed at least twice.
AUTHOR CONTRIBUTIONS
Melissa M. Lamanna, Ho‐Ching T. Tsui, and Malcolm E. Winkler contributed to the conception or design of this study. Melissa M. Lamanna, Irfan Manzoor, Merrin Joseph, Ziyun A. Ye, Mattia Benedet, Alessia Zanardi, Zhongqing Ren, Xindan Wang. Orietta Massidda, Ho‐Ching T. Tsui, and Malcolm E. Winkler contributed to the acquisition, analysis, and interpretation of the data. Melissa M. Lamanna, Orietta Massidda, Ho‐Ching T. Tsui, and Malcolm E. Winkler contributed to the writing of the manuscript with input from the other authors.
ETHICS STATEMENT
This work did not include animal or human experimental subjects requiring formal approval or consent. Antibodies used in this study are available commercially, were published previously, or were prepared by companies approved by the Indiana University Bloomington Institutional Animal Care andUse Committee.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interests.
Supporting information
supinfo
Appendix S1
ACKNOWLEDGMENTS
We thank Kevin Bruce, Jiaqi Zheng, and other members of the Winkler lab for discussions about this work; Jim Powers and Sidney Shaw (Indiana University Bloomington) for advice about light microscopy; Jason Rosch (St. Jude's) and Tim van Opijnen (Boston College) for Tn‐seq protocols; Mike VanNieuwenhze (Indiana University Bloomington) for TADA FDAA reagent; and Suzanne Walker and David Rudner (Harvard Medical School) and Linda Doubravova and Pavel Branny (Czech Academy of Sciences) for antibodies against pneumococcal PG synthesis and division proteins. This work was supported by NIH Grants R35GM131767 (to MEW), R01GM141242 (to XW), Predoctoral Grant F31AI138430 (to MML), and NIH Equipment Grant S10OD024988 (to the Indiana University Bloomington [IUB] Light Microscopy Imaging Center); and by institutional research funds from the CIBIO Department of the University of Trento (to OM).
Lamanna, M. M. , Manzoor, I. , Joseph, M. , Ye, Z. A. , Benedet, M. , Zanardi, A. , Ren, Z. , Wang, X. , Massidda, O. , Tsui, H‐C. & Winkler, M. E. (2022). Roles of RodZ and class A PBP1b in the assembly and regulation of the peripheral peptidoglycan elongasome in ovoid‐shaped cells of Streptococcus pneumoniae D39 . Molecular Microbiology, 118, 336–368. 10.1111/mmi.14969
Contributor Information
Ho‐Ching T. Tsui, Email: tttsui@indiana.edu.
Malcolm E. Winkler, Email: winklerm@indiana.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in Appendix S1 and from the corresponding authors upon reasonable request.
REFERENCES
- Aaron, M. , Charbon, G. , Lam, H. , Schwarz, H. , Vollmer, W. & Jacobs‐Wagner, C. (2007) The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus . Molecular Microbiology, 64, 938–952. [DOI] [PubMed] [Google Scholar]
- Ago, R. & Shiomi, D. (2019) RodZ: a key‐player in cell elongation and cell division in Escherichia coli . AIMS Microbiology, 5, 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyahya, S.A. , Alexander, R. , Costa, T. , Henriques, A.O. , Emonet, T. & Jacobs‐Wagner, C. (2009) RodZ, a component of the bacterial core morphogenic apparatus. Proceedings of the National Academy of Sciences of the United States of America, 106, 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L. , Anantharaman, V. , Balaji, S. , Babu, M.M. & Iyer, L.M. (2005) The many faces of the helix‐turn‐helix domain: transcription regulation and beyond. FEMS Microbiology Reviews, 29, 231–262. [DOI] [PubMed] [Google Scholar]
- Barendt, S.M. , Land, A.D. , Sham, L.T. , Ng, W.L. , Tsui, H.C. , Arnold, R.J. et al. (2009) Influences of capsule on cell shape and chain formation of wild‐type and pcsB mutants of serotype 2 Streptococcus pneumoniae . Journal of Bacteriology, 191, 3024–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartual, S.G. , Straume, D. , Stamsås, G.A. , Munoz, I.G. , Alfonso, C. , Martinez‐Ripoll, M. et al. (2014) Structural basis of PcsB‐mediated cell separation in Streptococcus pneumoniae . Nature Communications, 5, 3842. 10.1038/ncomms4842 [DOI] [PubMed] [Google Scholar]
- van Beilen, J. , Blohmke, C.J. , Folkerts, H. , de Boer, R. , Zakrzewska, A. , Kulik, W. et al. (2016) RodZ and PgsA play intertwined roles in membrane homeostasis of Bacillus subtilis and resistance to weak organic acid stress. Frontiers in Microbiology, 7, 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz, K. , Novakova, L. , Fadda, D. , Branny, P. , Massidda, O. & Veening, J.W. (2012) Control of cell division in Streptococcus pneumoniae by the conserved ser/Thr protein kinase StkP. Proceedings of the National Academy of Sciences, 109, E905–E913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu, F.O. & de Boer, P.A. (2008) Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli . Journal of Bacteriology, 190, 1792–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu, F.O. , Hale, C.A. , Bernhardt, T.G. & de Boer, P.A. (2009) Rodz (YfgA) is required for proper assembly of the MreB Actin cytoskeleton and cell shape in E. coli . The EMBO Journal, 28, 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, K.H. , Stamsas, G.A. , Straume, D. & Havarstein, L.S. (2013) Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. Journal of Bacteriology, 195, 4342–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen, T. , Andreu, J.M. & Monasterio, O. (2014) Bacterial cell division proteins as antibiotic targets. Bioorganic Chemistry, 55, 27–38. [DOI] [PubMed] [Google Scholar]
- Boersma, M.J. , Kuru, E. , Rittichier, J.T. , VanNieuwenhze, M.S. , Brun, Y.V. & Winkler, M.E. (2015) Minimal peptidoglycan (PG) turnover in wild‐type and PG hydrolase and cell division mutants of Streptococcus pneumoniae D39 growing planktonically and in host‐relevant biofilms. Journal of Bacteriology, 197, 3472–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton, B.P. , Shaevitz, J.W. , Gitai, Z. & Morgenstein, R.M. (2018) MreB polymers and curvature localization are enhanced by RodZ and predict E. coli's cylindrical uniformity. Nature Communications, 9, 2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, N.S. , Bruce, K.E. , Naskar, S. , Winkler, M.E. & Roper, D.I. (2021) The pneumococcal divisome: dynamic control of Streptococcus pneumoniae cell division. Frontiers in Microbiology, 12, 737396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, K. & Bradford, P.A. (2016) Beta‐lactams and beta‐lactamase inhibitors: an overview. Cold Spring Harbor Perspectives in Medicine, 6, a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver, T. , Harris, S.R. , Berriman, M. , Parkhill, J. & McQuillan, J.A. (2012) Artemis: an integrated platform for visualization and analysis of high‐throughput sequence‐based experimental data. Bioinformatics, 28, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . (2019) Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC. Available from: http://www.cdc.gov/drugresistance/Biggest‐Threats.html [Accessed 1st June 2022]. [Google Scholar]
- Cho, H. , Wivagg, C.N. , Kapoor, M. , Barry, Z. , Rohs, P.D.A. , Suh, H. et al. (2016) Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi‐autonomously. Nature Microbiology, 1, 16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleverley, R.M. , Rutter, Z.J. , Rismondo, J. , Corona, F. , Tsui, H.T. , Alatawi, F.A. et al. (2019) The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes. Nature Communications, 10, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavin, A. , Shi, H. & Huang, K.C. (2018) Rodz modulates geometric localization of the bacterial Actin MreB to regulate cell shape. Nature Communications, 9, 1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitch, A.K. & Goley, E.D. (2020) Uncovering unappreciated activities and niche functions of bacterial cell wall enzymes. Current Biology, 30, R1170–R1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion, M.F. , Kapoor, M. , Sun, Y. , Wilson, S. , Ryan, J. , Vigouroux, A. et al. (2019) Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nature Microbiology, 4, 1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret, A. & Grangeasse, C. (2017) Bacterial physiology: wrapping the cell in a CozE shell. Nature Microbiology, 2, 16262. [DOI] [PubMed] [Google Scholar]
- Ducret, A. , Quardokus, E.M. & Brun, Y.V. (2016) MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature Microbiology, 1, 16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, A.J.F. , Errington, J. & Vollmer, W. (2020) Regulation of peptidoglycan synthesis and remodelling. Nature Reviews. Microbiology, 18, 446–460. [DOI] [PubMed] [Google Scholar]
- van den Ent, F. , Johnson, C.M. , Persons, L. , de Boer, P. & Lowe, J. (2010) Bacterial Actin MreB assembles in complex with cell shape protein RodZ. The EMBO Journal, 29, 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington, J. & Wu, L.J. (2017) Cell cycle machinery in Bacillus subtilis . Sub‐Cellular Biochemistry, 84, 67–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, R. , O'Neill, M. , Pritzel, A. , Antropova, N. , Senior, A. , Green, T. , Žídek, A. , Bates, R. , Blackwell, S. , Yim, J. , Ronneberger, O. , Bodenstein, S. , Zielinski, M. , Bridgland, A. , Potapenko, A. , Cowie, A. , Tunyasuvunakool, K. , Jain, R. , Clancy, E. , Kohli, P. , Jumper, J. , and Hassabis, D. (2021) Protein complex prediction with AlphaFold‐Multimer. bioRxiv. 10.1101/2021.10.04.463034 [DOI] [Google Scholar]
- Fadda, D. , Santona, A. , D'Ulisse, V. , Ghelardini, P. , Ennas, M.G. , Whalen, M.B. et al. (2007) Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD‐free context. Journal of Bacteriology, 189, 1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton, A.K. , El Mortaji, L. , Lau, D.T. , Rudner, D.Z. & Bernhardt, T.G. (2016) CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae . Nature Microbiology, 2, 16237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton, A.K. , Manuse, S. , Flores‐Kim, J. , Garcia, P.S. , Mercy, C. , Grangeasse, C. et al. (2018) Phosphorylation‐dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proceedings of the National Academy of Sciences of the United States of America, 115, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie, A. , Lesterlin, C. , Manuse, S. , Zhao, C. , Cluzel, C. , Lavergne, J.P. et al. (2014) MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae . Nature, 516, 259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie, A. , Manuse, S. , Zhao, C. , Campo, N. , Cluzel, C. , Lavergne, J.‐P. et al. (2014) Interplay of the serine/threonine‐kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genetics, 10, e1004275. 10.1371/journal.pgen.1004275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutu, A.D. , Wayne, K.J. , Sham, L.T. & Winkler, M.E. (2010) Kinetic characterization of the WalRKSpn (VicRK) two‐component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. Journal of Bacteriology, 192, 2346–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenbeck, R. (2014) Discovery of beta‐lactam‐resistant variants in diverse pneumococcal populations. Genome Medicine, 6, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenbeck, R. , Bruckner, R. , Denapaite, D. & Maurer, P. (2012) Molecular mechanisms of beta‐lactam resistance in Streptococcus pneumoniae . Future Microbiology, 7, 395–410. [DOI] [PubMed] [Google Scholar]
- Holeckova, N. , Doubravova, L. , Massidda, O. , Molle, V. , Buriankova, K. , Benada, O. et al. (2014) LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae . mBio, 6, e01700‐01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor, J. , Garriss, G. , Di Giorgio, S. , Hack, L.M. , Vanselow, J.T. , Forstner, K.U. et al. (2020) Grad‐seq in a gram‐positive bacterium reveals exonucleolytic sRNA activation in competence control. The EMBO Journal, 39, e103852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, S. , Wivagg, C.N. , Szwedziak, P. , Wong, F. , Schaefer, K. , Izore, T. et al. (2018) MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife, 7, e32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, F.E. , Kazmierczak, K.M. , Lisher, J.P. , Winkler, M.E. & Giedroc, D.P. (2011) Interplay between manganese and zinc homeostasis in the human pathogen Streptococcus pneumoniae . Metallomics, 3, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper, J. , Evans, R. , Pritzel, A. , Green, T. , Figurnov, M. , Ronneberger, O. et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature, 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova, G. , Dautin, N. & Ladant, D. (2005) Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two‐hybrid analysis. Journal of Bacteriology, 187, 2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemege, K.E. , Hickey, J.M. , Barta, M.L. , Wickstrum, J. , Balwalli, N. , Lovell, S. et al. (2015) Chlamydia trachomatis protein CT009 is a structural and functional homolog to the key morphogenesis component RodZ and interacts with division septal plane localized MreB. Molecular Microbiology, 95, 365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, E. , Kinoshita‐Kikuta, E. , Takiyama, K. & Koike, T. (2006) Phosphate‐binding tag, a new tool to visualize phosphorylated proteins. Molecular & Cellular Proteomics, 5, 749–757. [DOI] [PubMed] [Google Scholar]
- Krupka, M. , Rivas, G. , Rico, A.I. & Vicente, M. (2012) Key role of two terminal domains in the bidirectional polymerization of FtsA protein. The Journal of Biological Chemistry, 287, 7756–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Mollo, A. , Kahne, D. & Ruiz, N. (2022) The bacterial cell wall: from lipid II flipping to polymerization. Chemical Reviews, 122, 8884–8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru, E. , Hughes, H.V. , Brown, P.J. , Hall, E. , Tekkam, S. , Cava, F. et al. (2012) In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D‐amino acids. Angewandte Chemie (International Ed. in English), 51, 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna, M.M. & Maurelli, A.T. (2022) What is motion? Recent advances in the study of molecular movement patterns of the peptidoglycan synthesis machines. Journal of Bacteriology, 204, e0059821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land, A.D. , Tsui, H.C. , Kocaoglu, O. , Vella, S.A. , Shaw, S.L. , Keen, S.K. et al. (2013) Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Molecular Microbiology, 90, 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land, A.D. & Winkler, M.E. (2011) The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. Journal of Bacteriology, 193, 4166–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanie, J.A. , Ng, W.L. , Kazmierczak, K.M. , Andrzejewski, T.M. , Davidsen, T.M. , Wayne, K.J. et al. (2007) Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. Journal of Bacteriology, 189, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara, B. , Rico, A.I. , Petruzzelli, S. , Santona, A. , Dumas, J. , Biton, J. et al. (2005) Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Molecular Microbiology, 55, 699–711. [DOI] [PubMed] [Google Scholar]
- Lewis, K. (2020) The science of antibiotic discovery. Cell, 181, 29–45. [DOI] [PubMed] [Google Scholar]
- Liechti, G. , Kuru, E. , Packiam, M. , Hsu, Y.P. , Tekkam, S. , Hall, E. et al. (2016) Pathogenic chlamydia lack a classical sacculus but synthesize a narrow, mid‐cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathogens, 12, e1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, L.L. , Schneider, T. , Peoples, A.J. , Spoering, A.L. , Engels, I. , Conlon, B.P. et al. (2015) A new antibiotic kills pathogens without detectable resistance. Nature, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, V.A. , Wacnik, K. , Turner, R.D. , Cotterell, B.E. , Walther, C.G. , Fenn, S.J. et al. (2018) Molecular coordination of Staphylococcus aureus cell division. eLife, 7, e32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Galiano, A.J. , Yuste, J. , Cercenado, M.I. & de la Campa, A.G. (2014) Inspecting the potential physiological and biomedical value of 44 conserved uncharacterised proteins of Streptococcus pneumoniae . BMC Genomics, 15, 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda, O. , Novakova, L. & Vollmer, W. (2013) From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environmental Microbiology, 15, 3133–3157. [DOI] [PubMed] [Google Scholar]
- Meeske, A.J. , Riley, E.P. , Robins, W.P. , Uehara, T. , Mekalanos, J.J. , Kahne, D. et al. (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature, 537, 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehla, J. , Liechti, G. , Morgenstein, R.M. , Caufield, J.H. , Hosseinnia, A. , Gagarinova, A. et al. (2021) ZapG (YhcB/DUF1043), a novel cell division protein in gamma‐proteobacteria linking the Z‐ring to septal peptidoglycan synthesis. The Journal of Biological Chemistry, 296, 100700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstein, R.M. , Bratton, B.P. , Nguyen, J.P. , Ouzounov, N. , Shaevitz, J.W. & Gitai, Z. (2015) RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proceedings of the National Academy of Sciences of the United States of America, 112, 12510–12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchova, K. , Chromikova, Z. & Barak, I. (2013) Control of Bacillus subtilis cell shape by RodZ. Environmental Microbiology, 15, 3259–3271. [DOI] [PubMed] [Google Scholar]
- Mura, A. , Fadda, D. , Perez, A.J. , Danforth, M.L. , Musu, D. , Rico, A.I. et al. (2017) Roles of the essential protein FtsA in cell growth and division in Streptococcus pneumoniae . Journal of Bacteriology, 199, e00608–e00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.L. , Tsui, H.C. & Winkler, M.E. (2005) Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae . Journal of Bacteriology, 187, 7444–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak, M. , Jiang, X. , Basczok, M.M. & Storz, G. (2022) KH domain proteins: another family of bacterial RNA matchmakers? Molecular Microbiology, 117, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen, T. & Camilli, A. (2012) A fine scale phenotype‐genotype virulence map of a bacterial pathogen. Genome Research, 22, 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen, T. , Lazinski, D.W. & Camilli, A. (2015) Genome‐wide fitness and genetic interactions determined by Tn‐seq, a high‐throughput massively parallel sequencing method for microorganisms. Current Protocols in Microbiology, 36, 1E 3 1–1E 3 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette, S.P. , Lee, J. & Cox, J.V. (2020) Division without binary fission: cell division in the FtsZ‐less chlamydia . Journal of Bacteriology, 202, e00252–e00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbaykal, G. , Wollrab, E. , Simon, F. , Vigouroux, A. , Cordier, B. , Aristov, A. et al. (2020) The transpeptidase PBP2 governs initial localization and activity of the major cell‐wall synthesis machinery in E. coli . eLife, 9, e50629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, J. , Kern, I. , Lurz, R. & Hakenbeck, R. (1999) Mutational analysis of the Streptococcus pneumoniae bimodular class a penicillin‐binding proteins. Journal of Bacteriology, 181, 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos, M. , Peters, K. , Casanova, M. , Palacios, P. , VanNieuwenhze, M. , Breukink, E. et al. (2018) Z‐ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nature Communications, 9, 5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos, M. & Vollmer, W. (2021) Regulation and function of class a penicillin‐binding proteins. Current Opinion in Microbiology, 60, 80–87. [DOI] [PubMed] [Google Scholar]
- Perez, A.J. , Boersma, M.J. , Bruce, K.E. , Lamanna, M.M. , Shaw, S.L. , Tsui, H.T. et al. (2021) Organization of peptidoglycan synthesis in nodes and separate rings at different stages of cell division of Streptococcus pneumoniae . Molecular Microbiology, 115, 1152–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, A.J. , Cesbron, Y. , Shaw, S.L. , Bazan Villicana, J. , Tsui, H.T. , Boersma, M.J. et al. (2019) Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae . Proceedings of the National Academy of Sciences of the United States of America, 116, 3211–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, A.J. , Villicana, J.B. , Tsui, H.T. , Danforth, M.L. , Benedet, M. , Massidda, O. et al. (2021) FtsZ‐ring regulation and cell division are mediated by essential EzrA and accessory proteins ZapA and ZapJ in Streptococcus pneumoniae . Frontiers in Microbiology, 12, 780864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe, J. , Vernet, T. & Zapun, A. (2014) The elongation of ovococci. Microbial Drug Resistance, 20, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff, S. , Du, S. & Lutkenhaus, J. (2019) Roles of FtsEX in cell division. Research in Microbiology, 170, 374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal, M. & Walker, S. (2017) Envelope structures of gram‐positive bacteria. Current Topics in Microbiology and Immunology, 404, 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos‐Montanez, S. , Tsui, H.C. , Wayne, K.J. , Morris, J.L. , Peters, L.E. , Zhang, F. et al. (2008) Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS‐HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae . Molecular Microbiology, 67, 729–746. [DOI] [PubMed] [Google Scholar]
- Ranjit, D.K. , Liechti, G.W. & Maurelli, A.T. (2020) Chlamydial MreB directs cell division and peptidoglycan synthesis in Escherichia coli in the absence of FtsZ activity. mBio, 11, e03222–e03219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs, P.D.A. & Bernhardt, T.G. (2021) Growth and division of the peptidoglycan matrix. Annual Review of Microbiology, 75, 315–336. [DOI] [PubMed] [Google Scholar]
- Rohs, P.D.A. , Buss, J. , Sim, S.I. , Squyres, G.R. , Srisuknimit, V. , Smith, M. et al. (2018) A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genetics, 14, e1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs, P.D.A. , Qiu, J.M. , Torres, G. , Smith, M.D. , Fivenson, E.M. & Bernhardt, T.G. (2021) Identification of potential regulatory domains within the MreC and MreD components of the cell elongation machinery. Journal of Bacteriology, 203, e00493‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rued, B.E. , Alcorlo, M. , Edmonds, K.A. , Martínez‐Caballero, S. , Straume, D. , Fu, Y. et al. (2019) Structure of the large extracellular loop of FtsX and its interaction with the essential peptidoglycan hydrolase PcsB in Streptococcus pneumoniae . mBio, 10, e02622‐18. 10.1128/mBio.02622-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rued, B.E. , Zheng, J.J. , Mura, A. , Tsui, H.T. , Boersma, M.J. , Mazny, J.L. et al. (2017) Suppression and synthetic‐lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin‐binding protein interactions in Streptococcus pneumoniae D39. Molecular Microbiology, 103, 931–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva, B.M. , Sorg, M. , Pereira, A.R. , Ferreira, M.J. , Caulat, L.C. , Reichmann, N.T. et al. (2020) Reassessment of the distinctive geometry of Staphylococcus aureus cell division. Nature Communications, 11, 4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham, L.T. , Barendt, S.M. , Kopecky, K.E. & Winkler, M.E. (2011) Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proceedings of the National Academy of Sciences of the United States of America, 108, E1061–E1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham, L.T. , Jensen, K.R. , Bruce, K.E. & Winkler, M.E. (2013) Involvement of FtsE ATPase and FtsX extracellular loops 1 and 2 in FtsEX‐PcsB complex function in cell division of Streptococcus pneumoniae D39. mBio, 4, e00431‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham, L.T. , Tsui, H.C. , Land, A.D. , Barendt, S.M. & Winkler, M.E. (2012) Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Current Opinion in Microbiology, 15, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi, D. , Sakai, M. & Niki, H. (2008) Determination of bacterial rod shape by a novel cytoskeletal membrane protein. The EMBO Journal, 27, 3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi, D. , Toyoda, A. , Aizu, T. , Ejima, F. , Fujiyama, A. , Shini, T. et al. (2013) Mutations in cell elongation genes mreB, mrdA, and mrdB suppress the shape defect of RodZ‐deficient cells. Molecular Microbiology, 87, 1029–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodt, M. , Brock, K. , Dobihal, G. , Rohs, P.D.A. , Green, A.G. , Hopf, T.A. et al. (2018) Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature, 556, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodt, M. , Rohs, P.D.A. , Gilman, M.S.A. , Erlandson, S.C. , Zheng, S. , Green, A.G. et al. (2020) Structural coordination of polymerization and crosslinking by a SEDS‐bPBP peptidoglycan synthase complex. Nature Microbiology, 5, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager, J. , Aprianto, R. & Veening, J.W. (2018) Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Research, 46, 9971–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamsas, G.A. , Straume, D. , Ruud Winther, A. , Kjos, M. , Frantzen, C.A. & Havarstein, L.S. (2017) Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae . Molecular Microbiology, 105, 954–967. [DOI] [PubMed] [Google Scholar]
- Straume, D. , Piechowiak, K.W. , Kjos, M. & Havarstein, L.S. (2021) Class A PBPs: it is time to rethink traditional paradigms. Molecular Microbiology, 116, 41–52. [DOI] [PubMed] [Google Scholar]
- Straume, D. , Stamsas, G.A. , Berg, K.H. , Salehian, Z. & Havarstein, L.S. (2017) Identification of pneumococcal proteins that are functionally linked to penicillin‐binding protein 2b (PBP2b). Molecular Microbiology, 103, 99–116. [DOI] [PubMed] [Google Scholar]
- Sun, Y. & Garner, E. (2020) PrkC modulates MreB filament density and cellular growth rate by monitoring cell wall precursors. bioRxiv. 10.1101/2020.08.28.272336 [DOI] [Google Scholar]
- Sung, C.K. , Li, H. , Claverys, J.P. & Morrison, D.A. (2001) An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae . Applied and Environmental Microbiology, 67, 5190–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, A. , Page, J.E. , Tsui, H.T. , Winkler, M.E. & Walker, S. (2021) Biochemical reconstitution defines new functions for membrane‐bound glycosidases in assembly of the bacterial cell wall. Proceedings of the National Academy of Sciences, 118, e2103740118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling, M.C.F. (2021) Elongation at midcell in preparation of cell division requires FtsZ, but not MreB nor PBP2 in Caulobacter crescentus . Frontiers in Microbiology, 12, 732031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouve, J. , Zapun, A. , Arthaud, C. , Durmort, C. , Di Guilmi, A.M. , Soderstrom, B. et al. (2021) Nanoscale dynamics of peptidoglycan assembly during the cell cycle of Streptococcus pneumoniae . Current Biology, 31, 2844–2856.e6. [DOI] [PubMed] [Google Scholar]
- Tsui, H.C. , Mukherjee, D. , Ray, V.A. , Sham, L.T. , Feig, A.L. & Winkler, M.E. (2010) Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. Journal of Bacteriology, 192, 264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, H.C. , Zheng, J.J. , Magallon, A.N. , Ryan, J.D. , Yunck, R. , Rued, B.E. et al. (2016) Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Molecular Microbiology, 100, 1039–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, H.T. , Boersma, M.J. , Vella, S.A. , Kocaoglu, O. , Kuru, E. , Peceny, J.K. et al. (2014) Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Molecular Microbiology, 94, 21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrych, A. , Fabrik, I. , Kupcik, R. , Vajrychova, M. , Doubravova, L. & Branny, P. (2021) Cell wall stress stimulates the activity of the protein kinase StkP of Streptococcus pneumoniae, leading to multiple phosphorylation. Journal of Molecular Biology, 433, 167319. [DOI] [PubMed] [Google Scholar]
- Vigouroux, A. , Cordier, B. , Aristov, A. , Alvarez, L. , Ozbaykal, G. , Chaze, T. et al. (2020) Class‐a penicillin binding proteins do not contribute to cell shape but repair cell‐wall defects. eLife, 9, e51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, W. , Massidda, O. & Tomasz, A. (2019) The cell wall of Streptococcus pneumoniae . Microbiology Spectrum, 7(3). 10.1128/microbiolspec.GPP3-0018-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, K.J. , Li, S. , Kazmierczak, K.M. , Tsui, H.C. & Winkler, M.E. (2012) Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross‐talk in Streptococcus pneumoniae D39 cells. Molecular Microbiology, 86, 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser, J.N. , Ferreira, D.M. & Paton, J.C. (2018) Streptococcus pneumoniae: transmission, colonization and invasion. Nature Reviews. Microbiology, 16, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, R. , Mesnage, S. , Boneca, I.G. , Hobbs, J.K. & Foster, S.J. (2011) Super‐resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Molecular Microbiology, 82, 1096–1109. [DOI] [PubMed] [Google Scholar]
- Winther, A.R. , Kjos, M. , Herigstad, M.L. , Havarstein, L.S. & Straume, D. (2021) EloR interacts with the lytic transglycosylase MltG at midcell in Streptococcus pneumoniae R6. Journal of Bacteriology, 203, e00691–e00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii, Y. , Niki, H. & Shiomi, D. (2019) Division‐site localization of RodZ is required for efficient Z ring formation in Escherichia coli . Molecular Microbiology, 111, 1229–1244. [DOI] [PubMed] [Google Scholar]
- Young, K.D. (2006) The selective value of bacterial shape. Microbiology and Molecular Biology Reviews, 70, 660–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun, A. , Vernet, T. & Pinho, M.G. (2008) The different shapes of cocci. FEMS Microbiology Reviews, 32, 345–360. [DOI] [PubMed] [Google Scholar]
- Zheng, J.J. , Perez, A.J. , Tsui, H.T. , Massidda, O. & Winkler, M.E. (2017) Absence of the KhpA and KhpB (JAG/EloR) RNA‐binding proteins suppresses the requirement for PBP2b by overproduction of FtsA in Streptococcus pneumoniae D39. Molecular Microbiology, 106, 793–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supinfo
Appendix S1
Data Availability Statement
The data that support the findings of this study are available in Appendix S1 and from the corresponding authors upon reasonable request.


