Abstract
While mitochondria have long been understood to be critical to cellular function, questions remain as to how genetic variation within mitochondria may underlie variation in general metrics of organismal function. To date, studies investigating links between mitochondrial genotype and phenotype have largely focused on differences in expression of genes and physiological and life‐history traits across haplotypes. Mating display behaviours may also be sensitive to mitochondrial functionality and so may also be affected by sequence variation in mitochondrial DNA, with consequences for sexual selection and fitness. Here, we tested whether the pre‐copulatory mating success of male fruit flies (Drosophila melanogaster) varies across six different mitochondrial haplotypes expressed alongside a common nuclear genetic background. We found a significant effect of mitochondrial haplotype on our measure of competitive mating success, driven largely by the relatively poor performance of males with one particular haplotype. This haplotype, termed ‘Brownsville’, has previously been shown to have complex and sex‐specific effects, most notably including depressed fertility in males but not females. Our study extends this disproportionate effect on male reproductive success to pre‐copulatory aspects of reproduction. Our results demonstrate that mutations in mitochondrial DNA can plausibly affect pre‐copulatory mating success, with implications for future study into the subcellular underpinnings of such behaviours and the information they may communicate.
Keywords: competitive mate choice, Drosophila melanogaster, mitochondrial genetic variation, pre‐copulatory behaviour
When male fruit flies possessing one of six different focal mitochondrial haplotypes competed for copulations with females against a common strain of non‐focal males, flies with the “BRO” (or Brownsville) haplotype were significantly less successful than their counterparts.

1. INTRODUCTION
Mitochondria are central to organismal function in eukaryotes not only for harnessing energy in the form of ATP but also for signalling and serving as hubs of biosynthesis in processes as diverse as neurogenesis (Cheng et al., 2010), hormone synthesis (Bose et al., 2002) and innate immune responses (West et al., 2011). These wide‐ranging functions have sparked interest in mitochondria in the ecological, evolutionary and behavioural sciences, with increasing tests of how variation in mitochondrial genotype and phenotype may tie to variation in organismal physiological performance and life history (e.g. Dobler et al., 2014; Hill, Havird, et al., 2019).
It is exactly because mitochondria are so essential to the functionality of both cell and organism that the existence of standing variation in mitochondrial genotype and phenotype poses a puzzle. Strong purifying selection is expected to eliminate functional genetic variants affecting mitochondrial performance—particularly variation within the mitochondrial genome itself, which is streamlined in most metazoans to include a select few pivotal genes (Ballard & Kreitman, 1995; Dowling et al., 2008). Indeed, the mitochondrial genome has been expected to be under such strong selection that mitochondrial DNA sequence variation was used as a neutral mutational marker for decades (Galtier et al., 2009). However, functional variants in mitochondrial DNA have been described (e.g. Ballard & Pichaud, 2014; Dobler et al., 2014; Dowling et al., 2008; Rand, 2001), and interactions between mitochondrial genes and mitochondria‐associated nuclear genes have also been found to affect phenotypes (mito‐nuclear interactions; for example Rand et al., 2004; Wolff et al., 2014; Hill, Havird, et al., 2019). These findings have called the dogma of neutral mitochondrial genetic variation into question, altering our understanding the evolutionary forces that shape mitochondria and the populations hosting them.
Focusing on the functional effects of mitochondrial DNA variation is important to elucidating the underlying sources of variation in mitochondrial performance and the downstream processes this variation can affect—processes that are of critical importance to organismal fitness. For example, variation across mitochondrial haplotypes and mito‐nuclear combinations has been associated with expression of life‐history traits such as lifespan, juvenile development and fertility (e.g. Dobler et al., 2014; Mossman et al., 2016; Vaught & Dowling, 2018; Zhu et al., 2014). Further, the near‐exclusive maternal inheritance of mitochondria (and thereby mitochondrial DNA) in metazoans presents an unusual evolutionary context: without inheritance of male mitochondrial DNA, there is no direct means for selection to act against variants that are deleterious to males unless they are also deleterious to females (Frank & Hurst, 1996). This forms the foundation of what has become termed the ‘Mother's Curse Hypothesis’, which predicts that male‐harming mutations will accumulate in mitochondrial genomes over evolutionary time if they are benign or beneficial to females (Beekman et al., 2014; Connallon et al., 2018; Frank & Hurst, 1996; Gemmell et al., 2004). The presence and prevalence of these sex‐specific mitochondrial genetic mutations remain largely unresolved, given that predictions of the Mother's Curse Hypothesis are difficult to test, and results have been inconsistent across study designs (Dowling & Adrian, 2019).
Sexually dimorphic traits are considered most likely to exhibit effects of Mother's Curse mutations because the different metabolic needs of the trait in males versus females provide the conditions in which particular mitochondrial DNA variants might confer different effects between the sexes (Dowling & Adrian, 2019). One example of a Mother's Curse mutation has been found in a naturally occurring mitochondrial haplotype of the fruit fly (Drosophila melanogaster): male flies with the ‘Brownsville’ haplotype (named after its location of origin in Texas, USA) have been found to have complete or partial sterility depending on nuclear genetic context (‘nuclear background’), while females have unchanged or even increased fertility relative to other mitochondrial haplotypes sourced from different natural populations (Camus et al., 2015; Camus & Dowling, 2018; Dowling et al., 2015; Wolff, Tompkins, et al., 2016; Yee et al., 2013). This effect has been traced to malfunctions in sperm development in the testes (Clancy et al., 2011), and adult males of this haplotype have relatively low respiratory efficiency, mitochondrial abundance and metabolic rate relative to females and to males with other haplotypes (Nagarajan‐Radha et al., 2020; Wolff, Pichaud, et al., 2016). However, males otherwise appear to have normal lifespan (Camus et al., 2012, 2015; Nagarajan‐Radha et al., 2019), and juveniles of both sexes have high viability relative to those of other haplotypes (Camus & Dowling, 2018; Wolff, Tompkins, et al., 2016). Conceivably, the highly dimorphic metabolic environments and demands of the male versus female reproductive organs and gametes are such that mitochondria with the Brownsville‐specific mutation may be specifically poorly suited for male reproductive function.
Reproductive traits are indeed expected to be especially sensitive to the accumulation of sex‐specific mitochondrial mutations—a prediction that has been supported across multiple systems with respect to male fertility (Vaught & Dowling, 2018); however, these need not be exclusively traits related to post‐copulatory aspects of reproduction (Dowling & Adrian, 2019). Traits under pre‐copulatory sexual selection are often highly sexually dimorphic as well as energetically demanding, making them potentially sensitive to Mother's Curse effects and to variation in mitochondrial performance. Recent studies have specifically predicted that expressing a pre‐copulatory display behaviour requires high‐functioning mitochondria because of links between mitochondrial processes and the physiological pathways necessary to perform complex behaviours (Koch et al., 2017; Koch & Hill, 2018). However, few studies have yet tested how mitochondrial function relates to display traits (e.g. Hill, Hood, et al., 2019), or how mitochondrial genetic variation may influence reproductive behaviour.
Investigating links between mitochondrial genetic variation and display behaviours offers an opportunity to explore the functional and potentially sex‐specific effects that are mediated by the mitochondria. The fruit fly presents a valuable system in which to explore these links because it has been well studied with respect to both mitochondrial genetic variation (including potential Mother's Curse mutations like that found in the Brownsville haplotype) and pre‐copulatory courtship behaviours (Dickson, 2008; Greenspan & Ferveur, 2000). Drosophila melanogaster pre‐copulatory behaviours involve both intra‐sexual competition (resource defence and courtship interference) and inter‐sexual courtship (Baxter et al., 2018; Greenspan & Ferveur, 2000; Kim, 2009). Male courtship behaviour, for example, features a choreographed series of acoustic and chemical displays that must be tactically adjusted in response to female receptivity, requiring finely tuned sensorimotor integration and information processing (Coen & Murthy, 2016; Dickson, 2008; Greenspan & Ferveur, 2000). These sorts of complex behaviours used in mate courtship have been hypothesized to be those that may be most sensitive to variation in mitochondrial performance (Koch & Hill, 2018), so it is possible that functional mitochondrial genetic variation may also affect pre‐copulatory mating success in this species. In this experiment, we test the relative ability of male D. melanogaster fruit flies possessing different mitochondrial haplotypes (including Brownsville) to secure copulations with females when in competition with rival males. Our simple design tests the overall outcome of intra‐sexual competition and inter‐sexual courtship, which together are behavioural determinants of pre‐copulatory mating success in this species (Baxter et al., 2018; Greenspan & Ferveur, 2000; Kim, 2009). Our study provides the first direct assessment of whether mitochondrial genetic variation influences the success of male mate acquisition.
2. MATERIALS AND METHODS
2.1. Experimental subjects
The flies used in this study were sourced from a set of strains with varying mitochondrial DNA haplotypes, but a controlled nuclear background, created through selective introgression of mitochondrial haplotypes into a targeted nuclear background and maintained as near‐isogenic. Each strain possesses one of six distinct mitochondrial haplotypes (originally sourced from D. melanogaster populations around the world; Clancy, 2008; Camus et al., 2012) expressed alongside one common nuclear background (MAD; originally sourced from Madang, Papua New Guinea). All strains are maintained as near‐isogenic by crossing females from each strain with males from a MAD stock population, that is propagated via full‐sibling crosses to prevent maintenance or accumulation of genetic variation (hereafter referred to as ‘pure MAD’); as a result of more than 60 generations of such a process, we expect little to no nuclear genetic variation among strains. The six mitochondrial haplotypes represented in the strains used in this experiment are ALS (Alstonville, New South Wales, Australia), BAR (Barcelona, Spain), BRO (Brownsville, Texas, USA), HAW (Hawaii, USA), ISR (Israel) and JAP (Japan); altogether, they cover a substantial portion of the mitochondrial genetic variation that has been detected across a wider set of strains (see Wolff, Pichaud, et al., 2016). Each of these strains is maintained in duplicate, resulting in a set of 12 distinct strain replicates (i.e. ALS1, ALS2, BAR1 and BAR2) for testing. All files are maintained in 40 mL vials containing 6 ml food media (mixed from potato, dextrose, yeast and agar, sprinkled with ad lib live yeast granules and treated to prevent mould) and sealed with foam plugs.
2.2. Competitive mating success experimental protocol
The experiment took place over five non‐consecutive generations (five ‘blocks’; January and September–December 2020). To source flies to test in each block, we first collected vials of ‘parental’ flies (five pairs per vial) from each strain replicate and from the pure MAD population. The parental flies oviposited in these vials, with the eggs producing the focal flies used in the experiment. We transferred parents into fresh vials of media every 24 h 1–4 times per block to provide multiple cohorts of focal flies born to the same group of mothers over a particular 24‐h time period (termed ‘lay groups’ hereafter). Flies for testing were collected from these vials as virgins under CO2 anaesthesia and placed in same‐sex cohorts of up to 30 flies per vial. We collected one vial for each of the 12 mitochondrial strain replicates and 12–15 vials each of pure MAD males and females per lay group per block, and all flies collected from a lay group were tested on the same day (at 3–7 days of age since eclosion). In summary, all flies within any given experimental trial had matching ages and parental ages.
Each experimental trial comprised five males from a mitochondrial strain replicate (‘focal’ males) competing against five pure MAD males (‘tester’ males) for copulations with five pure MAD females. Because focal and tester males are visually indistinguishable, we dusted the males with brightly coloured green or pink powder (Barnes Products DayGlo powder), respectively, 24 h before testing. We coloured each vial of focal or tester males by transferring them into a vial containing small granules of coloured powder, shaking the vial briefly to distribute the powder, then transferring the coated flies back into the original vial. After colouring, flies were observed to groom vigorously such that most powder was removed and normal behaviours were observed by the following day, but green‐ or pink‐coloured males were still distinguishable by faint coloration on their thoraces (Figure 1). Focal males were always coloured green and tester males were always coloured pink in order to provide a consistent basis of comparison among trials, since our focus was on comparing the performance of different focal strains (i.e. comparing across different strains of green flies) rather than the performance of focal vs. tester strains (i.e. comparing green vs. pink flies). While this methodology cannot rule out that coloration and mitochondrial strain interact to affect mating success, colouring all focal strains green provided greater experimental power to compare among strains within the constraints of our sample size. We note there is no a priori basis for why we would predict an interaction between mitochondrial genotype and colour of powder shaping male reproductive outcomes.
FIGURE 1.

Green‐coloured focal male and a pink‐coloured tester male, pictured 24 h after dusting with coloured powder
For each experimental trial, we first used an aspirator to place five focal males and five tester males into each of 12 substrate‐free vials (one per mitochondrial strain replicate). We then placed five females each in 12 separate substrate‐free vials. To start the trial, we added one vial of five females into each vial of males, then allowed males and females to interact undisturbed for 15 min, at which point we assessed the coloration of copulating males (Figure 1). We chose 15 min as the duration of our trials because, during careful observation in tests of this procedure with these fly strains and personal observations from work with these and similar inbred strains in our lab, we found this time to be sufficient for copulations to be initiated, but not so long that successful copulations would be completed and pairs separated. While our 15‐min snapshot will not capture unsuccessful copulation attempts, we focus in this study specifically on successful copulations, which we believe our chosen time frame sufficiently captures for these strains; at 15 min, at least three pairs were currently copulating in 75% of our trials, and at least one pair was copulating in nearly 95% of the trials (Appendix S1).
We repeated this process (up to five times per day) with new male and female flies such that no fly was tested more than once; all trials took place over the same time of day (10 AM to 2 PM). The structure of our data therefore is as follows: five blocks (separated by one or more generations), with one to five lay groups per block (each lay group comprising flies from the same mothers of a particular age) and up to five trials performed on a given day. Not every trial featured the full group of 12 mitochondrial strain replicates due to human error and limits to the numbers of flies available for testing in any given block.
2.3. Data analysis
We used a generalized linear mixed model with a binomial error distribution to assess whether males possessing different mitochondrial haplotypes differed in competitive mating success, using the lme4 package (v. 1.1–26; Bates et al., 2015) in R (v. 3.6.1; R Core Team, 2021), with the sum of contrasts set to zero. To account for the structure inherent to our data, we first ran a full model with a vector of the numbers of green males and pink males copulating as the response variable, fixed effects of mitochondrial haplotype (six levels), block (five levels), order a trial was performed within a day (five levels), and lay group (five levels), and a random intercept effect of strain replicate (12 levels). While strain replicate represents an important level of biological replication in our data, the random intercept of strain replicate explained no variance in the model, and removing the term did not affect the results (Table 1, Appendix S1); however, we report the results of the full model here to most accurately represent the structure inherent to our dataset. We used the car package (v. 3.0–10; Fox & Weisberg, 2019) to provide global estimates of fixed effects using Type III Wald chi‐square tests, and the emmeans package (v. 1.7.0; Lenth, 2021) to calculate pairwise contrasts between strains using estimated marginal means from our full model, adjusted for multiple comparisons. We also used the tidyverse (v. 1.3.0; Wickham et al., 2019) and viridis (v. 0.5.1; Garnier, 2018) packages in data visualization.
TABLE 1.
Results of a global estimate of fixed effects from the full binomial mixed model, using Wald type III chi‐square tests
| Effect | Chi‐square | df | p |
|---|---|---|---|
| Intercept | 0.742 | 1 | 0.389 |
| Strain | 13.763 | 5 | 0.017 |
| Day order | 1.480 | 4 | 0.830 |
| Lay group | 0.990 | 4 | 0.925 |
| Block | 14.465 | 4 | 0.006 |
Note: Bold values indicate p < 0.05.
3. RESULTS AND DISCUSSION
In this study, we used a newly developed set of D. melanogaster fruit fly strains to test how mitochondrial genetic variation affects male pre‐copulatory mating success when in competition with standard males. We found significant variation among haplotypes in competitive mating success (Table 1) that derives primarily from the reduced performance of males possessing the Brownsville (BRO) mitochondrial haplotype (Table 2, Figure 2, Appendix S1).
TABLE 2.
Results of a binomial mixed model testing the effects of mitochondrial haplotype on competitive mating success (i.e. the proportion of copulating pairs featuring focal males vs. standard tester males), including a random intercept effect of strain replicate
| Effect | Estimate | SE | Z value | p |
|---|---|---|---|---|
| Intercept | −0.078 | 0.091 | −0.861 | 0.389 |
| ALS | 0.117 | 0.106 | 1.108 | 0.268 |
| BAR | 0.191 | 0.101 | 1.884 | 0.060 |
| BRO | −0.338 | 0.103 | −3.272 | 0.001 |
| HAW | 0.090 | 0.101 | 0.893 | 0.372 |
| ISR | −0.050 | 0.101 | −0.491 | 0.623 |
| JAP | −0.011 | 0.103 | −0.103 | 0.918 |
| Day order 1 | 0.025 | 0.087 | 0.288 | 0.773 |
| Day order 2 | −0.007 | 0.087 | −0.082 | 0.935 |
| Day order 3 | 0.100 | 0.090 | 1.116 | 0.264 |
| Day order 4 | −0.038 | 0.099 | −0.389 | 0.697 |
| Day order 5 | −0.080 | 0.125 | −0.635 | 0.525 |
| Block 1 | −0.408 | 0.113 | −3.607 | <0.001 |
| Block 2 | −0.006 | 0.115 | −0.055 | 0.956 |
| Block 3 | 0.101 | 0.106 | 0.949 | 0.343 |
| Block 4 | 0.207 | 0.125 | 1.664 | 0.096 |
| Block 5 | 0.106 | 0.104 | 1.023 | 0.307 |
| Lay group 1 | −0.008 | 0.110 | −0.070 | 0.944 |
| Lay group 2 | 0.044 | 0.098 | 0.454 | 0.650 |
| Lay group 3 | 0.022 | 0.159 | 0.137 | 0.891 |
| Lay group 4 | 0.046 | 0.133 | 0.345 | 0.730 |
| Lay group 5 | −0.104 | 0.131 | −0.799 | 0.424 |
Note: The results presented here are identical for both the binomial mixed model (including a random intercept of strain duplicate to account for data structure) and a binomial model without the random effect. The sum of contrasts was set to zero, so effect estimates are relative to the global mean rather than a reference group.
Bold values indicate p < 0.05.
FIGURE 2.
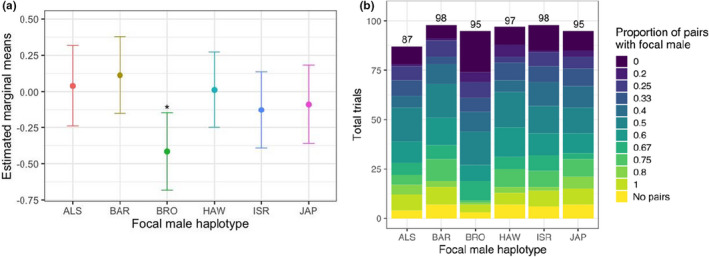
(a) Estimated marginal means (± 95% confidence limits; logit scale) for each strain based on the full binomial mixed model; star indicates the haplotype that differs significantly from the global mean (p < 0.05; see Table 2 and Appendix S1). (b) The height of each bar represents the total number of trials performed with focal males of a given mitochondrial haplotype (also numbered above each bar), and the colours within each column correspond to the proportion of mating pairs within a trial that featured a focal male. Darker blue colours indicate trials with few successful focal males, while lighter green colours indicate a higher proportion of successful focal males; yellow colours indicate trials in which no mating pairs were present. In both (a) and (b), the results for each mitochondrial haplotype include data from both duplicate populations (both strain replicates) possessing that type
It is notable that we found evidence of depressed performance specifically in male flies possessing the Brownsville haplotype, which has previously been shown to harbour a point mutation in the mitochondrial cytochrome b gene associated with reduced post‐copulatory reproductive success in males. While ours is the first study to explore flies with the Brownsville haplotype expressed alongside this particular nuclear genetic background, males with the Brownsville haplotype have previously been found to have depressed fertility or complete infertility in all other nuclear backgrounds studied (Camus & Dowling, 2018; Dowling et al., 2015; Wolff, Tompkins, et al., 2016; Yee et al., 2013). This decreased fertility found in Brownsville males has been linked to dysfunction in sperm development in the testes, at least in flies with the w 1118 nuclear background in which most research on this haplotype has been conducted (Clancy et al., 2011). The results of our study suggest that males with the Brownsville haplotype also feature depressed performance in pre‐copulatory reproductive behaviours. Whether or not this BRO‐mediated effect is general across different nuclear backgrounds will require further testing; because we tested strains of flies with varied mitochondrial haplotype expressed alongside one common nuclear background, it is not possible for us to determine whether the effect we detected is specific to the BRO mitochondrial haplotype interacting with the MAD nuclear genome or a general effect of the BRO haplotype. Despite not being able to disentangle these two possibilities, we note that our design was sufficient to have enabled us to achieve our primary aim of testing whether mtDNA variation can affect pre‐copulatory mating success. Given that male fruit flies with the BRO haplotype have previously been found to feature altered phenotype from males with other haplotypes across several other nuclear contexts; however, we expect that repeating our experiment in flies with varying nuclear backgrounds would yield similar results.
As described above, the Brownsville haplotype of D. melanogaster has been well studied because it appears to demonstrate a Mother's Curse effect: males, but not females, with the haplotype show greatly impaired fertility relative to males harbouring other naturally occurring haplotypes (Camus & Dowling, 2018). The Brownsville haplotype is distinguished from other, fully fertile mitochondrial haplotypes by a single nonsynonymous mutation leading to an amino acid change in the cytochrome B subunit of complex III of the mitochondrial electron transport chain (Clancy et al., 2011), and flies with the Brownsville haplotype have been found to also have decreased expression of the cytochrome B gene (Camus et al., 2015); however, it remains uncertain how such a change would cause noticeable effects only in males, and only in some processes. Mitochondrial function itself is difficult to quantify into a single metric that represents ‘best’ or ‘worst’ performance, as the bioenergetic processes central to mitochondrial aerobic respiration are flexibly modulated across tissues and time in response to changing environmental conditions and demands (Koch et al., 2021). Yet, a study of mitochondrial respiratory parameters in both male and female D. melanogaster of varying mitochondrial haplotypes did detect significant variation across haplotypes (as well as across age and sex groups), and results suggested that Brownsville males had lower quantities of mitochondria and performed comparatively poorly on a measure of respiratory capacity at the younger of two age groups studied (Wolff, Pichaud, et al., 2016). Similarly, in a study investigating mitochondrial variation in metabolic rate (measured through CO2 production) across these strains, Brownsville males appeared to have one of the lowest metabolic rates (Nagarajan‐Radha et al., 2020). However, males with the Brownsville haplotype have been reported to live as long or longer than males harbouring many other naturally occurring haplotypes (Camus et al., 2015; Nagarajan‐Radha et al., 2019), and both male and female juveniles exhibit high viability during development (Camus & Dowling, 2018; Wolff, Tompkins, et al., 2016), for example—so males do not exhibit poor performance across all energetically demanding traits. Further study of the density, morphology and bioenergetics of mitochondria across sexes, tissues and nuclear contexts in Brownsville flies will be necessary to further elucidate how this mitochondrial mutation affects male reproductive traits.
Our findings have implications for future study not only into the mechanisms linking mitochondrial genotype to sex‐specific phenotype but also to a recent hypothesis that posits that pre‐copulatory mating behaviours should reflect variation in mitochondrial genotypic quality (Hill, 2018). The results of our study suggest that links between mitochondrial haplotype and mating success are plausible. The mechanism by which variation at the level of the mitochondrial genome might affect the success of a male in acquiring a mate remains a matter of speculation, but the central role of mitochondria in a myriad of processes (including but not limited to bioenergetics) provides possible links between mitochondrial genotype and pre‐copulatory behavioural phenotype. For example, one possibility is that the low metabolic rate and decreased cytochrome B expression in male fruit flies with the Brownsville haplotype (but alongside other nuclear contexts; Camus et al., 2015; Nagarajan‐Radha et al., 2020) impairs the ability of such males to energetically support prolonged scramble competition with other males. It is also possible that slight alterations to Brownsville mitochondria could affect male performance in more subtle ways, such as by affecting biosynthesis of chemical signals or sensorimotor processes related to female courtship. While such mechanisms remain to be determined, our study suggests that functional mutations within the mitochondrial DNA sequence may affect the outcomes of pre‐copulatory sexual selection on male traits, which is intriguing given that this genetic variation is maternally transmitted and thus unable to directly respond to sexual selection in males.
AUTHOR CONTRIBUTIONS
Rebecca E. Koch: Conceptualization (equal); data curation (lead); formal analysis (lead); funding acquisition (equal); investigation (lead); methodology (lead); project administration (supporting); resources (supporting); software (lead); supervision (supporting); validation (equal); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Damian K. Dowling: Conceptualization (equal); data curation (supporting); formal analysis (supporting); funding acquisition (equal); investigation (supporting); methodology (supporting); project administration (lead); resources (lead); software (supporting); supervision (lead); validation (equal); visualization (supporting); writing – original draft (supporting); writing – review and editing (supporting).
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/jeb.14080.
Supporting information
Appendix S1
ACKNOWLEDGEMENTS
We thank James Wiles (Monash University) for contributing to the study design. This work was supported by the Australian Research Council (DE190100831 to R.E.K. and DP170100165 and DP200100892 to D.K.D.). Open access publishing facilitated by Monash University, as part of the Wiley ‐ Monash University agreement via the Council of Australian University Librarians.
Koch, R. E. , & Dowling, D. K. (2022). Effects of mitochondrial haplotype on pre‐copulatory mating success in male fruit flies (Drosophila melanogaster). Journal of Evolutionary Biology, 35, 1396–1402. 10.1111/jeb.14080
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.mgqnk9929.
REFERENCES
- Ballard, J. W. O. , & Kreitman, M. (1995). Is mitochondrial DNA a strictly neutral marker? Trends in Ecology & Evolution, 10, 485–488. [DOI] [PubMed] [Google Scholar]
- Ballard, J. W. O. , & Pichaud, N. (2014). Mitochondrial DNA: More than an evolutionary bystander. Functional Ecology, 28, 218–231. [Google Scholar]
- Bates, D. , Maechler, M. , & Walker, S. (2015). Fitting linear mixed‐effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- Baxter, C. , Mentlik, J. , Shams, I. , & Dukas, R. (2018). Mating success in fruit flies: Courtship interference versus female choice. Animal Behaviour, 138, 101–108. [Google Scholar]
- Beekman, M. , Dowling, D. K. , & Aanen, D. K. (2014). The costs of being male: Are there sex‐specific effects of uniparental mitochondrial inheritance? Philosophical Transactions of the Royal Society B: Biological Sciences, 369, 20130440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, H. S. , Lingappa, V. R. , & Miller, W. L. (2002). Rapid regulation of steroidogenesis by mitochondrial protein import. Nature, 417, 87–91. [DOI] [PubMed] [Google Scholar]
- Camus, M. F. , Clancy, D. J. , & Dowling, D. K. (2012). Mitochondria, maternal inheritance, and male aging. Current Biology, 22, 1717–1721. [DOI] [PubMed] [Google Scholar]
- Camus, M. F. , & Dowling, D. K. (2018). Mitochondrial genetic effects on reproductive success: Signatures of positive intrasexual, but negative intersexual pleiotropy. Proceedings of the Royal Society B: Biological Sciences, 285, 20180187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus, M. F. , Wolf, J. B. W. , Morrow, E. H. , & Dowling, D. K. (2015). Single nucleotides in the mtDNA sequence modify mitochondrial molecular function and are associated with sex‐specific effects on fertility and aging. Current Biology, 25, 2717–2722. [DOI] [PubMed] [Google Scholar]
- Cheng, A. , Hou, Y. , & Mattson, M. P. (2010). Mitochondria and neuroplasticity. ASN Neuro, 2, AN20100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, D. J. (2008). Variation in mitochondrial genotype has substantial lifespan effects which may be modulated by nuclear background. Aging Cell, 7, 795–804. [DOI] [PubMed] [Google Scholar]
- Clancy, D. J. , Hime, G. R. , & Shirras, A. D. (2011). Cytoplasmic male sterility in Drosophila melanogaster associated with a mitochondrial CYTB variant. Heredity, 107, 374–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen, P. , & Murthy, M. (2016). Singing on the fly: Sensorimotor integration and acoustic communication in Drosophila . Current Opinion in Neurobiology, 38, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon, T. , Camus, M. F. , Morrow, E. H. , & Dowling, D. K. (2018). Coadaptation of mitochondrial and nuclear genes, and the cost of mother's curse. Proceedings of the Royal Society B: Biological Sciences, 285, 20172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, B. J. (2008). Wired for sex: The neurobiology of Drosophila mating decisions. Science, 322, 904–909. [DOI] [PubMed] [Google Scholar]
- Dobler, R. , Rogell, B. , Budar, F. , & Dowling, D. K. (2014). A meta‐analysis of the strength and nature of cytoplasmic genetic effects. Journal of Evolutionary Biology, 27, 2021–2034. [DOI] [PubMed] [Google Scholar]
- Dowling, D. K. , & Adrian, R. E. (2019). Challenges and prospects for testing the Mother's curse hypothesis. Integrative and Comparative Biology, 59, 875–889. [DOI] [PubMed] [Google Scholar]
- Dowling, D. K. , Friberg, U. , & Lindell, J. (2008). Evolutionary implications of non‐neutral mitochondrial genetic variation. Trends in Ecology & Evolution, 23, 546–554. [DOI] [PubMed] [Google Scholar]
- Dowling, D. K. , Tompkins, D. M. , & Gemmell, N. J. (2015). The trojan female technique for pest control: A candidate mitochondrial mutation confers low male fertility across diverse nuclear backgrounds in Drosophila melanogaster . Evolutionary Applications, 8, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, J. , & Weisberg, S. (2019). An R companion to applied regression (3rd ed.). SAGE. [Google Scholar]
- Frank, S. A. , & Hurst, L. D. (1996). Mitochondria and male disease. Nature, 383, 224. [DOI] [PubMed] [Google Scholar]
- Galtier, N. , Nabholz, B. , Glémin, S. , & Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Molecular Ecology, 18, 4541–4550. [DOI] [PubMed] [Google Scholar]
- Garnier, S. (2018). viridis: Default Color Maps from “matplotlib” .
- Gemmell, N. J. , Metcalf, V. J. , & Allendorf, F. W. (2004). Mother's curse: The effect of mtDNA on individual fitness and population viability. Trends in Ecology & Evolution, 19, 238–244. [DOI] [PubMed] [Google Scholar]
- Greenspan, R. J. , & Ferveur, J.‐F. (2000). Courtship in Drosophila . Annual Review of Genetics, 34, 205–232. [DOI] [PubMed] [Google Scholar]
- Hill, G. E. (2018). Mitonuclear mate choice: A missing component of sexual selection theory? BioEssays, 40, 1700191. [DOI] [PubMed] [Google Scholar]
- Hill, G. E. , Havird, J. C. , Sloan, D. B. , Burton, R. S. , Greening, C. , & Dowling, D. K. (2019). Assessing the fitness consequences of mitonuclear interactions in natural populations. Biological Reviews, 94, 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, G. E. , Hood, W. R. , Ge, Z. , Grinter, R. , Greening, C. , Johnson, J. D. , Park, N. R. , Taylor, H. A. , Andreasen, V. A. , Powers, M. J. , Justyn, N. M. , Parry, H. A. , Kavazis, A. N. , & Zhang, Y. (2019). Plumage redness signals mitochondrial function in the house finch. Proceedings of the Royal Society B, 286, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.‐K. (2009). Sexual selection and aggressive behavior in Drosophila . In Kim Y.‐K. (Ed.), Handbook of behavior genetics (pp. 317–330). Springer. [Google Scholar]
- Koch, R. E. , Buchanan, K. L. , Casagrande, S. , Crino, O. , Dowling, D. K. , Hill, G. E. , Hood, W. R. , McKenzie, M. , Mariette, M. M. , Noble, D. W. A. , Pavlova, A. , Seebacher, F. , Sunnucks, P. , Udino, E. , White, C. R. , Salin, K. , & Stier, A. (2021). Integrating mitochondrial aerobic metabolism into ecology and evolution. Trends in Ecology & Evolution, 36, 321–332. [DOI] [PubMed] [Google Scholar]
- Koch, R. E. , & Hill, G. E. (2018). Behavioural mating displays depend on mitochondrial function: A potential mechanism for linking behaviour to individual condition. Biological Reviews, 93, 1387–1398. [DOI] [PubMed] [Google Scholar]
- Koch, R. E. , Josefson, C. C. , & Hill, G. E. (2017). Mitochondrial function, ornamentation, and immunocompetence. Biological Reviews, 92, 1459–1474. [DOI] [PubMed] [Google Scholar]
- Lenth, R. V. (2021). emmeans: Estimated marginal means, aka least‐squares means . https://github.com/rvlenth/emmeans
- Mossman, J. A. , Biancani, L. M. , Zhu, C.‐T. , & Rand, D. M. (2016). Mitonuclear epistasis for development time and its modification by diet in Drosophila . Genetics, 203, 463–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan‐Radha, V. , Aitkenhead, I. , Clancy, D. J. , Chown, S. L. , & Dowling, D. K. (2020). Sex‐specific effects of mitochondrial haplotype on metabolic rate in Drosophila melanogaster support predictions of the Mother's curse hypothesis. Philosophical Transactions of the Royal Society B: Biological Sciences, 375, 20190178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan‐Radha, V. , Rapkin, J. , Hunt, J. , & Dowling, D. K. (2019). Interactions between mitochondrial haplotype and dietary macronutrient ratios confer sex‐specific effects on longevity in Drosophila melanogaster . The Journals of Gerontology: Series A, 74, 1573–1581. [DOI] [PubMed] [Google Scholar]
- R Core Team . (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Rand, D. M. (2001). The units of selection on mitochondrial DNA. Annual Review of Ecology and Systematics, 32, 415–448. [Google Scholar]
- Rand, D. M. , Haney, R. A. , & Fry, A. J. (2004). Cytonuclear coevolution: The genomics of cooperation. Trends in Ecology & Evolution, 19, 645–653. [DOI] [PubMed] [Google Scholar]
- Vaught, R. C. , & Dowling, D. K. (2018). Maternal inheritance of mitochondria: Implications for male fertility? Reproduction, 155, R159–R168. [DOI] [PubMed] [Google Scholar]
- West, A. P. , Shadel, G. S. , & Ghosh, S. (2011). Mitochondria in innate immune responses. Nature Reviews Immunology, 11, 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. , Averick, M. , Bryan, J. , Chang, W. , McGowan, L. D. , François, R. , et al. (2019). Welcome to the Tidyverse. Journal of Open Source Software, 4, 1686. [Google Scholar]
- Wolff, J. N. , Ladoukakis, E. D. , Enríquez, J. A. , & Dowling, D. K. (2014). Mitonuclear interactions: Evolutionary consequences over multiple biological scales. Philosophical Transactions of the Royal Society B: Biological Sciences, 369, 20130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, J. N. , Pichaud, N. , Camus, M. F. , Côté, G. , Blier, P. U. , & Dowling, D. K. (2016). Evolutionary implications of mitochondrial genetic variation: Mitochondrial genetic effects on OXPHOS respiration and mitochondrial quantity change with age and sex in fruit flies. Journal of Evolutionary Biology, 29, 736–747. [DOI] [PubMed] [Google Scholar]
- Wolff, J. N. , Tompkins, D. M. , Gemmell, N. J. , & Dowling, D. K. (2016). Mitonuclear interactions, mtDNA‐mediated thermal plasticity and implications for the trojan female technique for pest control. Scientific Reports, 6, 30016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee, W. K. W. , Sutton, K. L. , & Dowling, D. K. (2013). In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Current Biology, 23, R55–R56. [DOI] [PubMed] [Google Scholar]
- Zhu, C.‐T. , Ingelmo, P. , & Rand, D. M. (2014). G×G×E for lifespan in Drosophila: Mitochondrial, nuclear, and dietary interactions that modify longevity. PLoS Genetics, 10, e1004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Data Availability Statement
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.mgqnk9929.


