Abstract
Pooling structural magnetic resonance imaging (MRI) data from different imaging sites helps increase sample size to facilitate machine learning based neuroimage analysis, but usually suffers from significant cross-site and/or cross-scanner data heterogeneity. Existing studies often focus on reducing cross-site and/or cross-scanner heterogeneity at handcrafted feature level targeting specific tasks (e.g., classification or segmentation), limiting their adaptability in clinical practice. Research on image-level MRI harmonization targeting a broad range of applications is very limited. In this paper, we develop a spectrum swapping based image-level MRI harmonization (SSIMH) framework. Different from previous work, our method focuses on alleviating cross-scanner heterogeneity at raw image level. We first construct spectrum analysis to explore the influences of different frequency components on MRI harmonization. We then utilize a spectrum swapping method for the harmonization of raw MRIs acquired by different scanners. Our method does not rely on complex model training, and can be directly applied to fast real-time MRI harmonization. Experimental results on T1- and T2-weighted MRIs of phantom subjects acquired by using different scanners from the public ABCD dataset suggest the effectiveness of our method in structural MRI harmonization at the image level.
Keywords: MRI, Image-level harmonization, Spectrum analysis
1. Introduction
Structural magnetic resonance imaging (MRI) data acquired from different sites or scanners may have significant differences, which has become one of the major concerns of the medical imaging community. Those between-site or between-scanner variabilities may negatively influence the analysis results of multi-site/multi-center studies and set barrier to building large-scale publicly available medical image datasets. As for computer-aided diagnosis, since conventional machine learning theory typically assumes the training dataset and test dataset follow the same distribution, thus the distribution variability among different sites/scanners would degrade the performance of machine learning systems. A promising solution to handle the between-site/scanner variability is MR image harmonization. To this end, domain adaptation or transfer learning techniques have been extensively introduced into medical image harmonization [1–3]. However, domain adaptation-based harmonization methods generally work at the feature level which is typically associated with specific tasks, e.g., classification or segmentation [4,5]. These methods remove dataset biases in some learning modules (e.g., networks) without constructing harmonized MRIs at the whole-image level. A more general solution is image-level MRI harmonization which can be used as a universal pre-processing step to remove site or scanner bias without considering any specific downstream tasks. These methods fall into two categories: 1) non-training methods, such as intensity histogram matching (HM) [6]; and 2) deep learning methods, such as GAN [7–11]. Since only overall intensity distribution is considered without accounting for structure information, the intensity histogram matching methods may lose some detailed information within MR images. For those deep learning methods, they usually need a relatively large amount of images for complex and time-consuming model training. Also, for images from new domains/sites, they often need model re-training or fine-tuning for harmonization. These shortcomings will significantly limit their utility and flexibility in real-world applications.
In this paper, motivated by [12,13], we propose a spectrum swapping based image-level MRI harmonization (SSIMH) through spectrum analysis. Different from previous studies, our analysis and harmonization method works in the frequency domain using Discrete Cosine Transform (DCT) as the primary analysis tool. As shown in Fig. 1, an MRI is first transformed into the frequency domain by DCT. We divide the spectrum of each MRI into three frequency areas: low-frequency, middle-frequency (further divided into several sub-regions) and high-frequency. We first explore an essential problem: which is the most influential frequency region for image-level MRI harmonization. Then by swapping the corresponding frequency region of the source and target image, image-level harmonization on the raw MRIs is achieved. Our framework not only avoids complex data-driven training, but also achieves image level MRI harmonization which can offer more flexibility for clinical usage. We use T1-weighted (T1-w) and T2-weighted (T2-w) MRIs of traveling phantom MRIs acquired by using different scanners from the ABCD dataset [14] to verify the effectiveness of our methods and its ability for harmonization of multi-modality data.
Fig. 1.

Illustration of the spectrum swapping based image-level MRI harmonization (SSIMH) framework via spectrum analysis. Discrete Cosine Transform (DCT) is facilitated to source and target MRIs to project them into frequency domain. The frequency domain is divided into three spectra: low-frequency, middle-frequency, and high-frequency regions. Through swapping the key domain-variant frequency regions, harmonization can be achieved.
2. Materials and Experimental Settings
Materials.
We use the public ABCD dataset [14] with structural MRIs for analysis1. The ABCD dataset includes brain MRIs of nearly 12,000 youth for the research of brain development, mental health and other related topics. We use T1-w and T2-w MR images in this dataset for MRI harmonization evaluation. Five traveling phantom subjects with raw T1-weighted MRIs, and five phantom subjects with raw T2-weighted MRIs are adopted. We select three slices in the middle part of each MRI (in axial view) for harmonization. The MRIs of each traveling phantom subject were acquired by three different scanners, include GE scanner, Philips scanner and Siemens scanner. We do not perform standard brain MRI pre-processing (e.g., skull stripping or registration) and only use raw MRIs as the input of our method and each competing method.
Evaluation Metrics.
The traveling phantom data from the ABCD dataset which are acquired by using three scanners are used to assess the harmonization capability of different methods. Given a traveling phantom subject scanned at scanners A and B, we have two MR images MA and MB accordingly. Let MB be the target image (reference image). Then, MA is harmonized to MB through certain methods, and we get the harmonized MR image . For an evaluation metric ℒ(·), we calculate the value of ℒ(MA, MB) and , respectively, and compare them to check the harmonization performance. Please note that ℒ(MA, MB) is the baseline performance without any harmonization. We use the following metrics to evaluate the harmonization performance.
Image Similarity. We calculate 2D correlation coefficient (CC) between and MB. Larger CC values indicate better performance.
Peak Signal-to-Noise Ratio (PSNR). We calculate the peak signal-to-noise ratio (PSNR) between and MB. Larger PSNR values indicate better results.
Mean-Squared Error (MSE). We calculate the mean-squared error (MSE) between and MB. Smaller MSE values indicate better performance.
Visual Inspection. We also use visual inspection as a qualitative evaluation metric to evaluate the harmonization results. The original source image, harmonized source image, and the target image are visually compared in the spatial space for inspection.
Competing Methods.
We compare the proposed SSIMH method with two approaches for harmonization performance analysis in our experiment, including: (1) Baseline without harmonization. The raw phantom MRIs acquired by two different scanners are directly compared using the evaluation metrics without any harmonization. (2) Intensity histogram matching (HM) [6]. The intensity histogram of the source phantom MRI is matched with the intensity histogram of the target phantom MRI to make their intensity distribution more similar. It should be noted that we avoid using the complex model-based methods (e.g., GAN) because they need a large amount of MRI data and much training time.
3. Spectrum Analysis for Image-Level Harmonization
Discrete Cosine Transform Analysis.
The Discrete Cosine Transform (DCT) is a spectrum analysis tool which can be applied to image pixels in the spatial domain and transform them into a frequency domain. The DCT is commonly used in image compression with the advantage of information concentration and avoidance of complex number computation. Specifically, for an input image IM×N, the corresponding spectrum image via DCT is a matrix T with each entity T(i, j) calculated as:
Spectrum Analysis.
We conduct spectrum analysis for MRI harmonization. Specifically, a pair of phantom MRIs (a source MRI and a target MRI) are firstly transformed into frequency domain through DCT. Then we use five different spectrum filters to filter out different frequency components, i.e., low-frequency part, middle-frequency part (further divided into three sub-regions), and high-frequency part. As shown in Fig 2, for a M × M image (slice), we get a M × M DCT image (in frequency domain). In our experiment, M is set to 250. Then the radius of the low-frequency part is 0~5, while the radii for the three sub-regions of middle-frequency are 5~20, 20~100, 100~250, respectively. The left region is the high-frequency part. For each time, we replace one frequency part of the source image with the corresponding frequency part of the target image. Then, we use inverted DCT to transform the spectrum source image back to the spatial domain, and test the corresponding harmonization performance. The analysis results are shown in Fig. 2. From the results, we have the following findings. (1) Low-frequency part of MRI has the largest influence on the harmonization. For example, from Fig. 2, through replacing the low-frequency part of source image with the corresponding low-frequency region of target image, the harmonized source image achieves the highest similarity with the target image, and there is no structure corruption from the original source image. (2) Detailed structure information mainly lies in the middle-frequency region. As shown in the red box in Fig. 2, after replacing the second sub middle-frequency part of source image with corresponding region in target image (the 4th column), the harmonized source image has corrupted into the target domain without preserving the original anatomical structure. This indicates the anatomical structure information of MRI is mainly reflected by the middle-frequency region. (3) High-frequency region has limited contribution to harmonization. From Fig. 2, the high-frequency region has very small values which can only reflect some edges of the MRIs (see the blue box). Swapping these regions between source and target image has barely effects on the harmonization performance. It should be noted that in real-world practice, there are often noises in the MR images. Thus image quality check (QC) and image denoising need to be performed.
Fig. 2.

Influence of different spectrum/frequency of MRIs on harmonization performance.
4. Spectrum Swapping for Image-Level Harmonization
Harmonization on Phantom MRIs.
Through the spectrum analysis, we identify that the low-frequency part mainly influences harmonization. Thus we replace the low-frequency region of the source image with the corresponding low-frequency region of target image, and project the spectrum source image back into the spatial domain. We conduct harmonization on MRIs from five traveling phantom subjects in the ABCD dataset. Each phantom is acquired by two different scanners, i.e., GE scanner, Siemens scanner or Philips scanner. We use these phantoms to test the performance of different methods for cross-scanner MRI harmonization. The results in terms of three metrics are shown in Table 1. From Table 1, it can be seen that the proposed method consistently outperforms the baseline and the HM methods by significant margins. In addition, since no data-based training is required, the running speed of the proposed method is also fast, i.e., within 1 s on a Intel i-7 CPU with 16 GB memory.
Table 1.
Harmonization of T1- and T2-weighted MRIs of five traveling phantom subjects acquired by three different scanners in the ABCD dataset.
| Source→Target | Method | T1-weighted MRI | T2-weighted MRI | ||||
|---|---|---|---|---|---|---|---|
| CC | PSNR | MSE | CC | PSNR | MSE | ||
| GE→Siemens | Baseline | 0.59±0.06 | 9.61±0.29 | 0.11±0.01 | 0.73±0.06 | 11.66±0.82 | 0.07±0.01 |
| HM | 0.67±0.06 | 10.45±0.60 | 0.09±0.01 | 0.75±0.07 | 12.26±1.38 | 0.06±0.02 | |
| SSIMH | 0.84±0.02 | 13.25±0.73 | 0.05±0.01 | 0.81±0.03 | 13.37±0.75 | 0.05±0.01 | |
| Siemens→Philips | Baseline | 0.73±0.03 | 11.78±0.45 | 0.06±0.01 | 0.81±0.01 | 12.79±0.19 | 0.05±0.001 |
| HM | 0.74±0.03 | 12.00±0.47 | 0.06±0.01 | 0.82±0.002 | 12.69±0.05 | 0.05±0.002 | |
| SSIMH | 0.76±0.02 | 12.81±0.38 | 0.05±0.002 | 0.86±0.01 | 17.60±0.35 | 0.04±0.002 | |
| Philips→GE | Baseline | 0.64±0.02 | 10.70±0.17 | 0.09±0.003 | 0.75±0.02 | 11.31±0.44 | 0.07±0.01 |
| HM | 0.62±0.03 | 11.25±0.11 | 0.08±0.002 | 0.68±0.03 | 11.02±0.45 | 0.08±0.01 | |
| SSIMH | 0.70±0.03 | 12.83±0.23 | 0.05±0.002 | 0.85±0.03 | 14.43±0.82 | 0.04±0.01 | |
We also utilize visual inspection to explore the harmonization performance. A visualization result is shown in Fig. 3. The phantom is acquired by a GE scanner (source) and Siemens scanner (target), respectively. After harmonization, it can be seen that the histogram matching method loses some detailed information such as some gyrus areas in cortex (see red dotted circles), while our SSIMH method can well preserve these original anatomical structures.
Fig. 3.
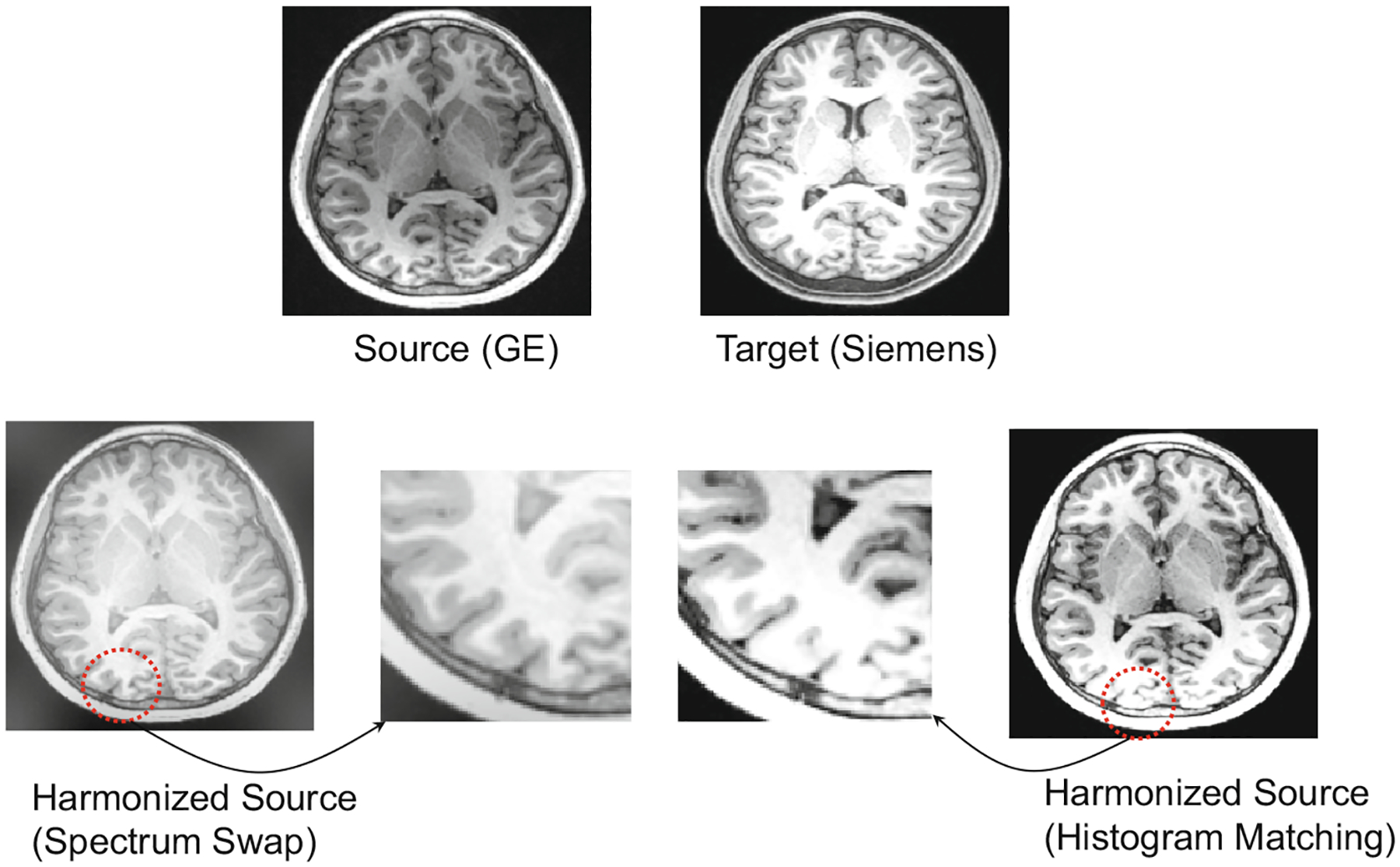
Visual inspection of MRI harmonization results achieved by two methods.
Generic Cross-Scanner Harmonization Without Phantom.
Besides using phantom data for harmonization evaluation, we also select independent subjects that acquired by using different scanners, i.e., GE, Philips, and Siemens. Each subject is harmonized to the MRI styles of the other two scanners. We conduct harmonization on two groups of MRIs. One is T1-w images, while the other group is T2-w images. The visualization of the harmonization for T1-w and T2-w MRIs are shown in Figs. 4 and 5, respectively. From the results, it can be seen that the MRIs acquired with different scanners have significantly different imaging styles, and the proposed method can harmonize MRIs to different scanner styles well. Since the proposed method does not need pre-training using numerous MRIs or phantom data, it is very flexible and convenient to use in practice.
Fig. 4.
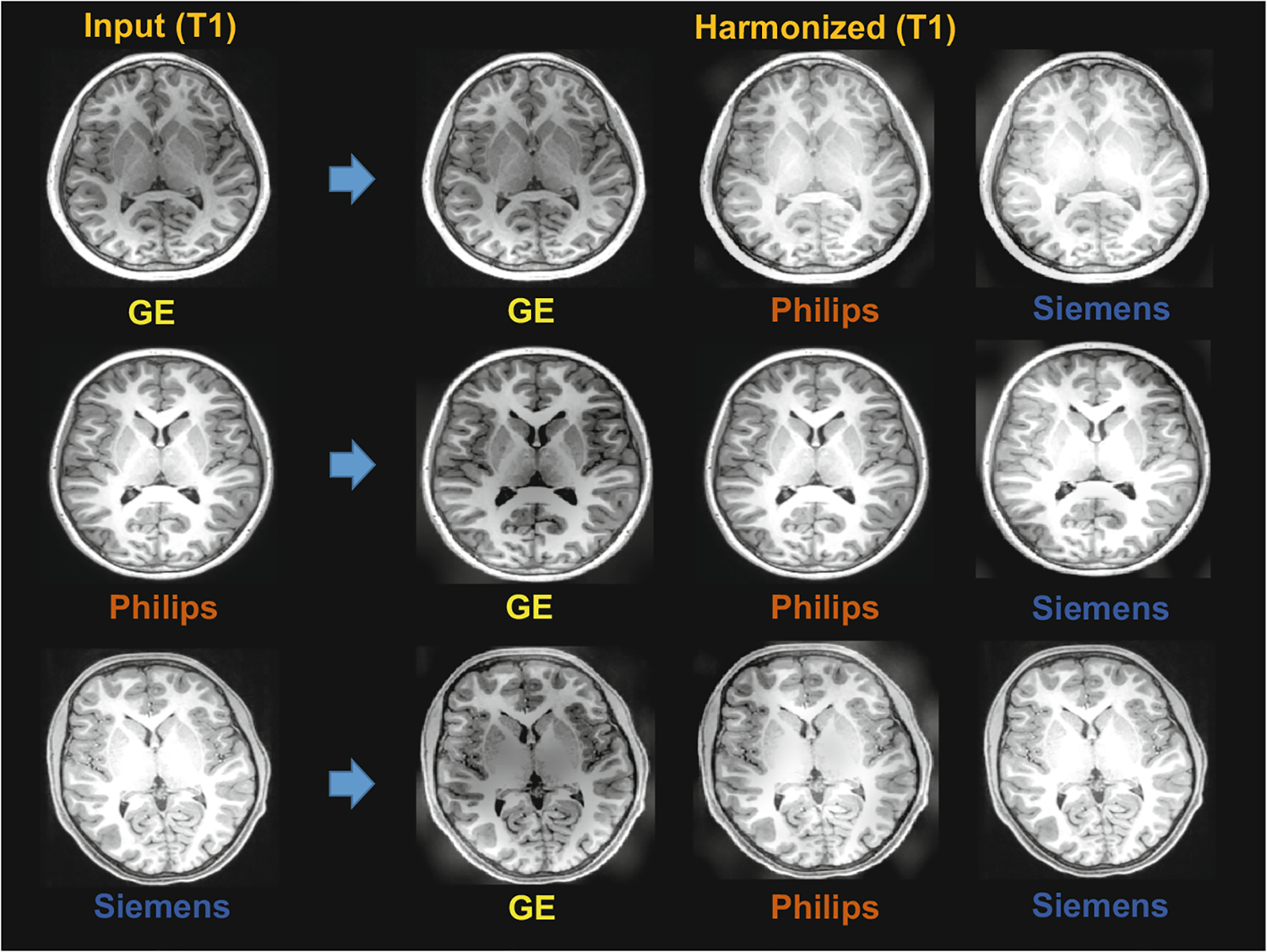
Visualization of harmonization results of our SSIMH method for T1-w MRIs acquired by different scanners (i.e., GE, Philips, Siemens).
Fig. 5.
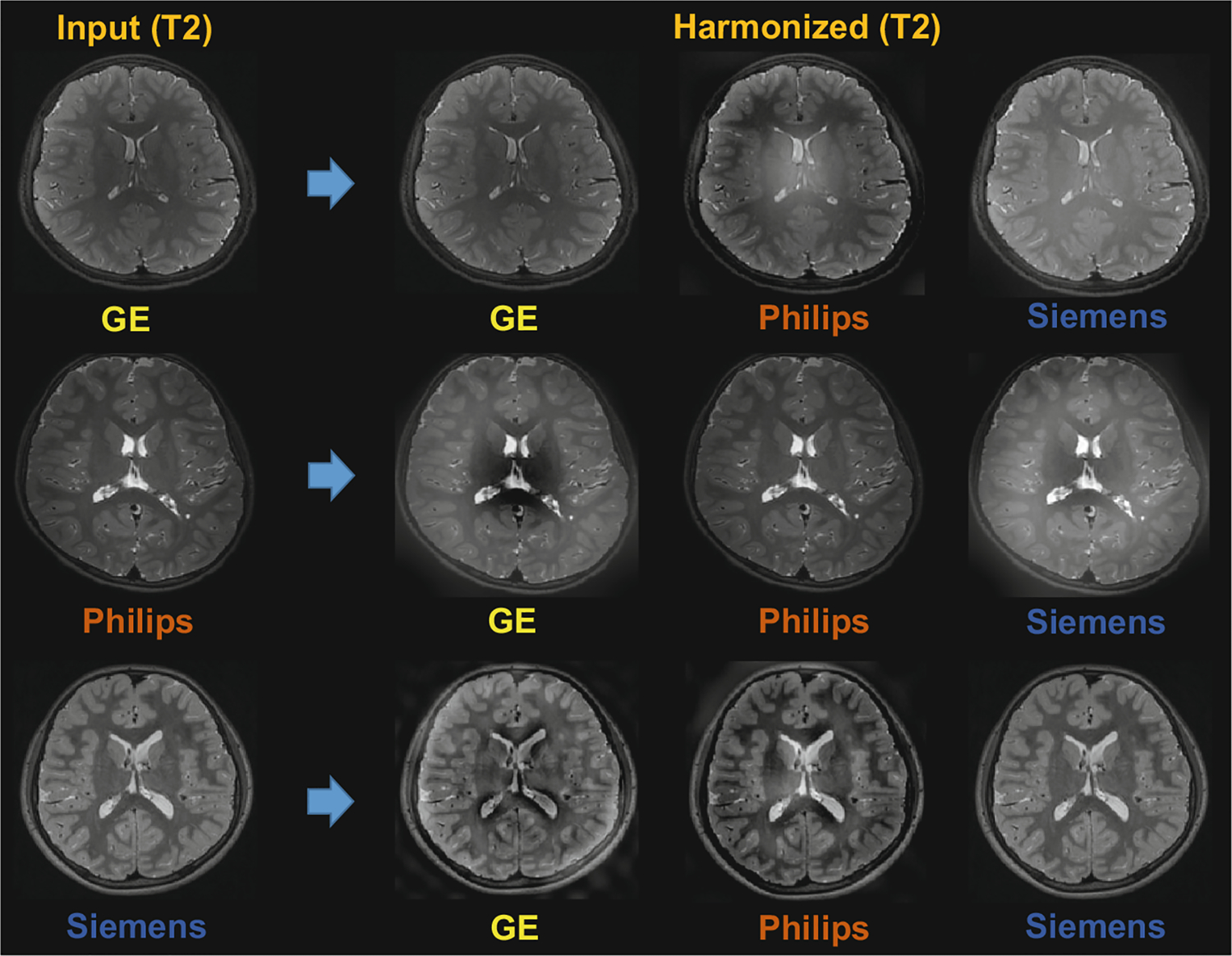
Visualization of harmonization results of our SSIMH method for T2-w MRIs acquired by different scanners (i.e., GE, Philips, Siemens).
5. Conclusion
We present a spectrum swapping based image-level MRI harmonization (SSIMH) framework based on spectrum analysis. The Discrete Cosine Transform (DCT) is used as the tool to project the source and target images into the frequency domain. We divide the frequency domain into low-frequency, middle-frequency and high-frequency regions. We firstly analyze the influence of each region on MRI harmonization, and find that the low-frequency region plays a more important role in effective harmonization, while the middle-frequency helps keep the detailed structure information in MRIs. Through replacing the low-frequency region of source images with the corresponding low-frequency region of target image, we conduct harmonization of MRIs acquired by using different scanners. Experiments on T1- and T2-weighted MRIs from the ABCD dataset suggest that the proposed method achieves satisfactory image-level harmonization results. Another advantage of the spectrum-based method is the avoidance of complex training which relies on a large amount of training data and is time-consuming.
Acknowledgement.
This work was partly supported by NIH grants (Nos. AG073297 and AG041721).
Footnotes
References
- 1.Cheng B, Liu M, Zhang D, Shen D: Robust multi-label transfer feature learning for early diagnosis of Alzheimer’s disease. Brain Imaging Behav. 13(1), 138–153 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheplygina V, de Bruijne M, Pluim JP: Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal 54, 280–296 (2019) [DOI] [PubMed] [Google Scholar]
- 3.Guan H, Liu M: Domain adaptation for medical image analysis: a survey. IEEETrans. Biomed. Eng 69(3), 1173–1185 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinsdale NK, Jenkinson M, Namburete AI: Deep learning-based unlearningof dataset bias for MRI harmonisation and confound removal. Neuroimage 228, 117689 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan H, et al. : Multi-site MRI harmonization via attention-guided deep domainadaptation for brain disorder identification. Med. Image Anal 71, 102076 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinohara RT, et al. : Statistical normalization techniques for magnetic resonanceimaging. NeuroImage. Clin 6, 9–19 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu JY, Park T, Isola P, Efros AA: Unpaired image-to-image translationusing cycle-consistent adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2223–2232 (2017) [Google Scholar]
- 8.Zuo L, et al. : Information-based disentangled representation learning for unsupervised MR harmonization. In: Feragen A, Sommer S, Schnabel J, Nielsen M (eds.) IPMI 2021. LNCS, vol. 12729, pp. 346–359. Springer, Cham: (2021). 10.1007/978-3-030-78191-027 [DOI] [Google Scholar]
- 9.Dewey BE, et al. : Deepharmony: a deep learning approach to contrast harmonization across scanner changes. Magn. Reson. Imaging 64, 160–170 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha S, Thomopoulos SI, Lam P, Muir A, Thompson PM: Alzheimer’s disease classification accuracy is improved by MRI harmonization based on attention-guided generative adversarial networks. In: 17th International Symposium on Medical Information Processing and Analysis, vol. 12088, pp. 180–189. SPIE; (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cackowski S, Barbier EL, Dojat M, Christen T: Imunity: a generalizableVAE-GAN solution for multicenter MR image harmonization. arXiv preprint arXiv:2109.06756 (2021) [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Guan D, Xiao A, Lu S: FSDR: Frequency space domain randomization for domain generalization. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 6891–6902 (2021) [Google Scholar]
- 13.Yang Y, Soatto S: FDA: Fourier domain adaptation for semantic segmentation.In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4085–4095 (2020) [Google Scholar]
- 14.Volkow ND, et al. : The conception of the ABCD study: from substance use to abroad NIH collaboration. Dev. Cogn. Neurosci 32, 4–7 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]


