Abstract
Delivery of messenger RNA (mRNA) using lipid nanoparticles (LNPs) is expected to be applied to various diseases following the successful clinical use of the mRNA COVID-19 vaccines. This study aimed to evaluate the effect of the cholesterol molar percentage of mRNA-LNPs on protein expression in hepatocellular carcinoma-derived cells and in the liver after intramuscular or subcutaneous administration of mRNA-LNPs in mice. For mRNA-LNPs with cholesterol molar percentages reduced to 10 mol% and 20 mol%, we formulated neutral charge particles with a diameter of approximately 100 nm and polydispersity index (PDI) <0.25. After the intramuscular or subcutaneous administration of mRNA-LNPs with different cholesterol molar percentages in mice, protein expression in the liver decreased as the cholesterol molar percentage in mRNA-LNPs decreased from 40 mol% to 20 mol% and 10 mol%, suggesting that reducing the cholesterol molar percentage in mRNA-LNPs decreases protein expression in the liver. Furthermore, in HepG2 cells, protein expression decreased as cholesterol in mRNA-LNPs was reduced by 40 mol%, 20 mol%, and 10 mol%. These results suggest that the downregulated expression of mRNA-LNPs with low cholesterol content in the liver involves degradation in systemic circulating blood and decreased protein expression after hepatocyte distribution.
Keywords: Drug delivery systems; Lipid nanoparticles (LNPs); Messenger ribonucleic acid (mRNA), Intramuscular administration
Introduction
Gene and nucleic acid therapeutics are promising drugs for treating and preventing cancer and genetic diseases. Various methods have been developed to achieve effective messenger RNA (mRNA) delivery, including mRNA conjugation, lipid nanoparticles (LNPs), and nanomicelles. Numerous designs and routes of administration of these drug delivery systems (DDSs) for mRNA delivery have been explored for their application to many targets.1 , 2 In particular, nucleic acid delivery using LNPs has attracted significant attention since the approval of Patisiran, a small interfering RNA (siRNA) medicine for hereditary variant transthyretin amyloidosis, followed by the rapid approval of COVID-19 mRNA vaccines and their mass use worldwide. Due to the ease of application of mRNA-LNPs to other mRNAs, several clinical trials are currently underway to develop mRNA vaccines against infectious diseases, cancer, and genetic diseases. In addition to intramuscular administration, various routes of mRNA-LNP administration have been studied, including intravenous, subcutaneous, intradermal, intraperitoneal, intratumoral, subretinal, intravitreal, intracerebroventricular, transpulmonary, and endonodular.3 , 4 Intramuscular and subcutaneous administrations are local administration methods that require few techniques. The intramuscular or subcutaneous administration of mRNA-LNPs is thought to occur initially at the site of administration and then through the lymphatic vessels5, 6, 7 to the vascular system.8
The liver is easily transfected by intravenous, intramuscular, and subcutaneous administration of mRNA-LNPs.3 , 9 This preference is related to the particle size8 and surface charge10 of mRNA-LNPs, the formation of protein corona11 after mRNA-LNPs enter the blood system, and uptake by apolipoprotein E (ApoE) through the low-density lipoprotein receptor (LDLR).12 When the mRNA is targeted to areas other than the liver, it should be less expressed in the liver to improve safety and reduce side effects. mRNA off-target expression may cause various side effects, including inflammation, hemolysis, and allergic reactions, as has been reported for mRNA-LNP vaccines against COVID-19.13 Cholesterol in lipids components of liposomes affects protein expression in the liver.14 Compared to cholesterol-poor or cholesterol-free liposomes, cholesterol-rich liposomes bind less protein after entering the circulating blood, resulting in delayed clearance from the circulation and greater uptake by hepatocytes than nonparenchymal cells.15 Conversely, cholesterol-poor liposomes have a higher amount of bound proteins, are taken up by the reticuloendothelial system of the liver and spleen, and show rapid clearance from the circulating blood. Furthermore, in liposome studies, cholesterol in liposome constituent lipids has also been shown to inhibit liposome aggregation and membrane stability by regulating membrane fluidity and permeability.16 Therefore, we hypothesized that decreasing the amount of cholesterol in mRNA-LNP constituent lipids may regulate its expression in the liver after intramuscular or subcutaneous administration. The relationship between the lipid composition of mRNA-LNPs and their expression distribution has been studied, including the effects of the amount17 and type18 of polyethylene glycol (PEG) lipids, type of phospholipids,19 and type of cholesterol.20 , 21 However, the effect of cholesterol content in the lipid composition of mRNA-LNPs on their expression has not yet been evaluated.
This study aimed to verify if decreasing the cholesterol content of lipid composition in mRNA-LNPs enhances the local expression of the injected site and the liver ratio in mice. In this study, we prepared mRNA-LNPs with different molar percentages of cholesterol and transfected mRNA encoding firefly luciferase into cultured cells and mice by intramuscular administration. Protein expression of the encoded luciferase was examined. We demonstrated that a lower molar percentage of cholesterol in mRNA-LNPs resulted in lower protein expression in the liver after intramuscular administration of mRNA-LNPs in mice, leading to localized protein expression in the muscle.
Material and Methods
Materials
DMG-PEG2000 (SUNBRIGHT® GM-020), DOPC (COATSOME® MC-8181), and SS-OP (COATSOME® SS-OP) were obtained from NOF CORPORATION (Tokyo, Japan). Cholesterol was purchased from Nacalai Tesque, Inc. (Kyoto, Japan); 1,2-Distearoyl-sn-glycerol-3- phosphocholine (DSPC) from Avanti Polar Lipids, Inc. (Alabaster, AL); DLin-MC3-DMA (MC3) from MedChemExpress (Monmouth Junction, NJ); DL-malic acid from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan); NucleoSpin Plasmid Transfection Grade from MACHEREY-NAGEL GmbH & Co. (KG, Germany); Sap I restriction enzyme and HiScribe T7 High Yield RNA Synthesis Kit from New England BioLabs, Inc.(Beverly, MA, USA); DNase TURBO from Life Technologies (Carlsbad, CA); CleanCap® Reagent AG from TriLink BioTechnologies (San Diego, CA); 2-(N-morpholino)ethanesulfonic acid (MES) from DOJINDO LABORATORIES (Kumamoto, Japan); and Quanti-iTTM RiboGreen RNA reagent from Molecular Probes (Eugene, OR).
Synthesis of Messenger RNA (mRNA) by in Vitro Transcription
Plasmid DNA encoding firefly luciferase (FLuc) was constructed as previously described.22 Each plasmid DNA was amplified using E. coli DH5a and purified using NucleoSpin Plasmid Transfection-grade. Linear template DNA was produced by Sap I restriction enzyme digestion. The mRNA was synthesized using the HiScribe T7 High Yield RNA Synthesis Kit and CleanCap® Reagent AG.
Cell Culture
HepG2, a human hepatocellular carcinoma-derived cell line, was obtained from the Riken BioResource Center (Ibaraki, Japan). HepG2 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 1% penicillin-streptomycin-L-glutamine solution and 10% fetal bovine serum (FBS). The cells were maintained in a 5% CO2 atmosphere at 37°C. HepG2 cells are human hepatocellular carcinoma-derived cells commonly used for in vitro gene and nucleic acid transfection experiments,23 and we have confirmed that transfection with mRNA-LNPs is possible in HepG2 cells.24
Animals
Six-week-old male ddY mice were purchased from Japan SLC, Inc. (Shizuoka, Japan). All the animals had food and water ad libitum. All animal experiments were performed according to the regulations of the Animal Care and Use Committee of Nagasaki University.
Preparation of mRNA-Encapsulated LNPs (mRNA-LNPs)
Lipids ((SS-OP/DOPC/Cholesterol/DMG-PEG2000) or (MC3/DSPC/Cholesterol/DMG-PEG2000)) were dissolved in ethanol at a final concentration of 4.5 mM. The lipid compositions are presented in Table 1 . The mRNA was dissolved in 20 mM malic acid. The lipid-ethanol solution and mRNA solution (3:1 (v/v)) were mixed using a NanoAssemblr® Benchtop microfluidic device (Precision NanoSystems, Inc., South San Francisco, CA, USA) at a total flow rate of 4 mL/min. The SS-OP LNP solutions were dialyzed in 20 mM MES buffer (pH 6.5), and the MC3 LNP solutions were dialyzed in phosphate-buffered saline (PBS) (−) (pH 7.4) overnight at 4 °C. These LNP solutions were concentrated using a 10 kDa MWCO Amicon® Ultra15 (Merck Millipore) at 4000 × g and replaced with PBS.
Table 1.
Lipid components of LNPs. We prepared three SS-OP LNPs and three MC3 LNPs, containing 10 mol%, 20 mol% and 40 mol% of cholesterol, respectively. Here we showed the lipid components of each LNP and their mRNA encapsulation efficacy (%). Data are represented as mean ± SD (n = 3).
| SS-OP LNP# | Molar percentage (mol%) |
Encapsulation efficacy (%) | |||
|---|---|---|---|---|---|
| Cholesterol | DOPC | DMG-PEG2000 | SS-OP | ||
| 1 | 10 | 37.5 | 1.5 | 52.5 | 80.1 ± 2.12 |
| 2 | 20 | 27.5 | 1.5 | 52.5 | 87.9 ± 7.49 |
| 3 | 40 | 7.5 | 1.5 | 52.5 | 92.6 ± 9.05 |
| MC3 LNP# |
Molar percentage (mol%) |
Encapsulation efficacy (%) |
|||
| Cholesterol | DOPC | DMG-PEG2000 | MC3 | ||
| 4 | 10 | 38.5 | 1.5 | 50 | 88.6 ± 5.98 |
| 5 | 20 | 28.5 | 1.5 | 50 | 89.0 ± 8.76 |
| 6 | 40 | 8.5 | 1.5 | 50 | 97.1 ± 0.93 |
Physiological Analysis of mRNA-LNPs
The particle size (Z-average), polydispersity index (PDI), and zeta potential of the LNPs were measured using a Malvern Zetasizer Pro (Malvern Panalytical Ltd., Royston, UK). The encapsulation efficacy of mRNA was measured by the Ribogreen assay with Quanti-iT™ RiboGreen RNA Reagent, as previously described.22
In Vitro Transfection Efficiency of mRNA-LNPs
Twenty-four h before transfection, HepG2 cells were seeded in 48-well plates at 3.8×104 cells/well. Each cell line was exposed to FLuc mRNA-LNP at 0.1 mRNA/well for 24 h. Luciferase expression was analyzed using a luciferase assay, and the amount of protein was tested using a bicinchoninic acid (BCA) protein assay. For the luciferase assay, cells were washed twice with PBS, and cell membranes were disrupted in lysis buffer consisting of 0.1 M Tris-HCl (pH 7.8) containing 0.05% Triton X-100 and 2 mM EDTA. The volume of the lysis buffer was 150 μL/well. The collected cells were centrifuged at 206,000 g for 5 min. Then, 5 μL of supernatant was mixed with 50 μL of luciferase assay substrate (PicaGene®, Toyo Ink Mfg. Co., Ltd., Tokyo, Japan), and the luminescence level was measured with a luminometer (AB-2270 Luminescence Octa, ATTO Co., Ltd., Tokyo, Japan). The same supernatant was used for the BCA protein assay. The BCA protein assay was performed using a BCA protein assay kit (Thermo Fisher Scientific) and following the protocol described in the user guide.
mRNA-LNPs Administration to Mice
mRNA-LNPs at a dose of 2 µg mRNA were intramuscularly injected into mice (40 µL) with a 30G needle, or mRNA-LNPs at a dose of 2 µg mRNA were subcutaneously injected into mice (200 µL) with a 27G needle.
In Vivo Transfection Efficiency of mRNA-LNPs
For the luciferase assay, FLuc mRNA-LNPs were administered intramuscularly or subcutaneously at a dose of 2 µg mRNA per mouse. When the mRNA-LNPs were administered intramuscularly, the muscles and livers of mice were collected. When mRNA-LNPs were administered subcutaneously, the skin and livers were collected. All tissues were collected at 4.5 h after mRNA-LNP administration and homogenized with lysis buffer. The luminescence level was measured using a luminometer with a luciferase assay substrate. The amount of protein in the samples was measured using the BCA protein assay.
For in vivo imaging system (IVIS) imaging evaluation, we selected luciferase mRNA SS-OP LNPs containing 10, 20, and 40 mol% cholesterol. Naked FLuc mRNA or FLuc mRNA-LNP was administered intramuscularly at a dose of 2 µg mRNA per mouse. One hundred microliters of 30 mg/mL D-luciferin (Syd Labs, Inc., Natick, MA) solution was injected intraperitoneally at 4.5, 9, 24, 48, and 72 h after mRNA-LNP administration, and luciferase protein expression was observed by IVIS imaging (IVIS Lumina II, Caliper Life Sciences, MA) under 2% isoflurane anesthesia. The value of the total flux in the muscle to that in the liver was calculated from the imaging results. It is included as a reference value to characterize the luciferase expression among muscle and liver.
We used mRNA-LNPs within a day stored at 4 °C in all in vivo experiments in this study.
Statistical Analysis
Statistical analysis was performed using one-way analysis of variance (ANOVA) with Tukey's multiple comparisons test. Significant differences between groups are expressed as *P < 0.05. All statistical analyses were performed with the statistical software EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan).
Results
Physicochemical Properties of mRNA-LNPs Containing Different Amounts of Cholesterol
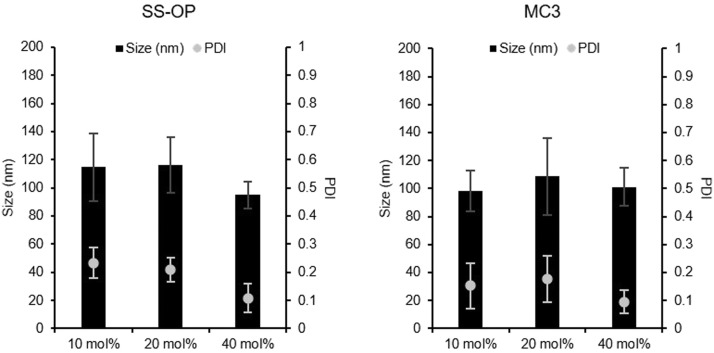
To investigate the effect of cholesterol content on the lipids constituting the mRNA-LNP formulation, 6 different mRNA-LNPs (SS-OP LNPs containing 10, 20, and 40 mol% cholesterol; MC3-LNPs containing 10, 20, and 40 mol% cholesterol) were prepared. In previous studies, mRNA-LNPs containing 40 mol% cholesterol with a standard lipid composition were used as mRNA-LNPs. Cholesterol contributes to the structural stability of mRNA-LNPs. We focused on the role of cholesterol in lipid membranes and expected that the reduction of the cholesterol molar percentage in the lipid composition of mRNA-LNPs would alter the pharmacokinetics of mRNA-LNPs. All lipids were prepared in ethanol with lipid compositions shown in Table 1, followed by microfluidic mixing with aqueous mRNA solutions at 1:3. In the mRNA encapsulation assay, all mRNA-LNPs encapsulated more than 80% of the mRNA (Fig. 1 ). We characterized the hydrodynamic size of LNPs and the zeta potential using dynamic light scattering (DLS), and all LNPs prepared with different lipid compositions showed diameters ranging from 75.5 to 140 nm and a narrow distribution (PDI < 0.25), with neutrally charged (Table 2 ). The mRNA-LNPs containing 40 mol% cholesterol showed a much narrower distribution (PDI < 0.2) than the mRNA-LNPs containing 10 mol% or 20 mol% cholesterol.
Figure 1.
Physicochemical property of mRNA-LNPs. The particle size and polydispersity index (PDI) of SS-OP LNPs containing 10 mol%, 20 mol%, and 40 mol% of cholesterol. These closed bars mean the size of mRNA-LNPs. These dots mean the PDI of mRNA-LNPs (Left). The particle size and PDI of MC3 LNPs containing 10 mol%, 20 mol%, and 40 mol% of cholesterol. These closed bars mean the size of mRNA-LNPs. These dots mean the PDI of mRNA-LNPs (Right). Data are represented as mean ± SD (n = 3–4).
Table 2.
The zeta potential of mRNA-LNPs. We prepared three SS-OP LNPs and three MC3 LNPs, containing 10 mol%, 20 mol%, and 40 mol% of cholesterol, respectively. Here we showed the zeta potential of mRNA-LNPs (mV). Data are represented as mean ± SD (n = 3).
| SS-OP LNPs | |
|---|---|
| 10 mol% Chol | −1.62 ± 1.35 mV |
| 20 mol% Chol | −2.31 ± 1.48 mV |
| 40 mol% Chol | −3.25 ± 1.76 mV |
| MC3 LNPs | |
| 10 mol% Chol | −3.68 ± 0.89 mV |
| 20 mol% Chol | −4.03 ± 0.99 mV |
| 40 mol% Chol | −3.85 ± 1.59 mV |
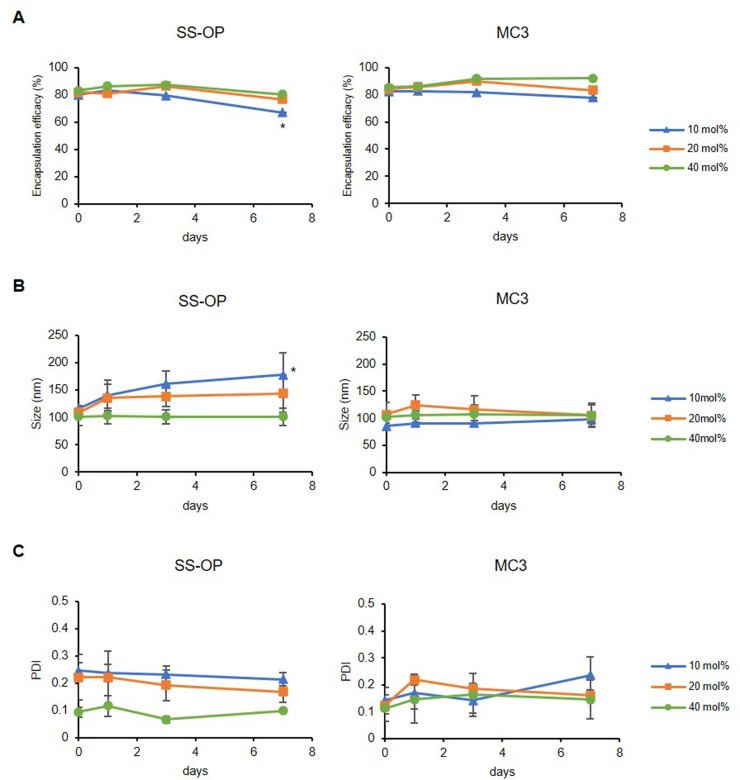
To further evaluate the differences in the structural stability of mRNA-LNPs with different cholesterol contents, physicochemical properties (particle size, PDI, and mRNA encapsulation efficiency) were analyzed by DLS and ribogreen assay at 0, 1, 3, and 7 days after mRNA-LNP preparation at 4 °C. The RNA quantification assay showed that the mRNA encapsulation efficacy in the 10 mol% cholesterol group of SS-OP LNPs decreased to 67.0% after 1 week of storage, a significant difference compared to 1 day after storage when the maximum encapsulation efficacy was reached. However, the other groups maintained an mRNA encapsulation efficacy of more than 75.2% after 1 week of storage. The particle size of the 10 mol% cholesterol group of SS-OP LNPs increased significantly after 1 week of storage compared to the day LNPs were prepared (Fig. 2 ).
Figure 2.
The one-week stability of mRNA-LNPs. All LNPs were tested at 0, 1, 3, and 7 days after the preparation while they were stored at 4 °C. (A) The mRNA encapsulation efficacy of SS-OP LNPs (Left) and MC3 LNPs (Right). Data are represented as mean ± SD for encapsulation efficacy (n = 3–4). (B) The particle size of SS-OP LNPs (Left) and MC3 LNPs (Right). (C) The PDI of SS-OP LNPs (Left) and MC3 LNPs (Right). Data are represented as mean ± SD for particle size and PDI (n = 3–4). Statistical analysis was performed by one-way ANOVA Tukey's multiple comparisons test. *P < 0.05, between Day 0 and Day 7 of SS-OP LNPs containing 10 mol% cholesterol groups.
Effect of Cholesterol in mRNA-LNPs on in Vitro Expression
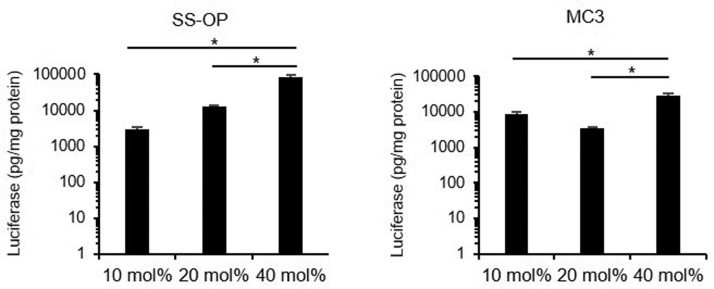
The transfection efficiency was evaluated for mRNA-LNPs with different cholesterol molar percentages. HepG2 cells were treated with LNPs encapsulating FLuc mRNA and assayed 24 h after transfection. For both SS-OP LNPs and MC3 LNPs, the cholesterol molar percentages in the 10 mol% and 20 mol% groups had significantly lower luciferase expression than the 40 mol% groups (Fig. 3 ).
Figure 3.
In vitro transfection efficiency of mRNA-LNPs. The luciferase expression levels of HepG2 cells transfected by using SS-OP LNPs containing 10 mol%, 20 mol%, and 40 mol% of cholesterol (Left) and MC3 LNPs containing 10 mol%, 20 mol%, and 40 mol% of cholesterol (Right). Data are represented as mean + SD (n = 3). Statistical analysis was performed by one-way ANOVA Tukey's multiple comparisons test (*P < 0.05).
Effect of Cholesterol in mRNA-LNPs on Luciferase Expression in Mice
Based on the in vitro results for mRNA-LNPs with varying cholesterol molar percentages, we investigated mRNA expression in vivo. SS-OP LNPs and MC3 LNPs encapsulating 2 µg of FLuc mRNA were administered intramuscularly or subcutaneously to 6-week-old male ddY mice, and luciferase expression levels and organ specificity were analyzed.
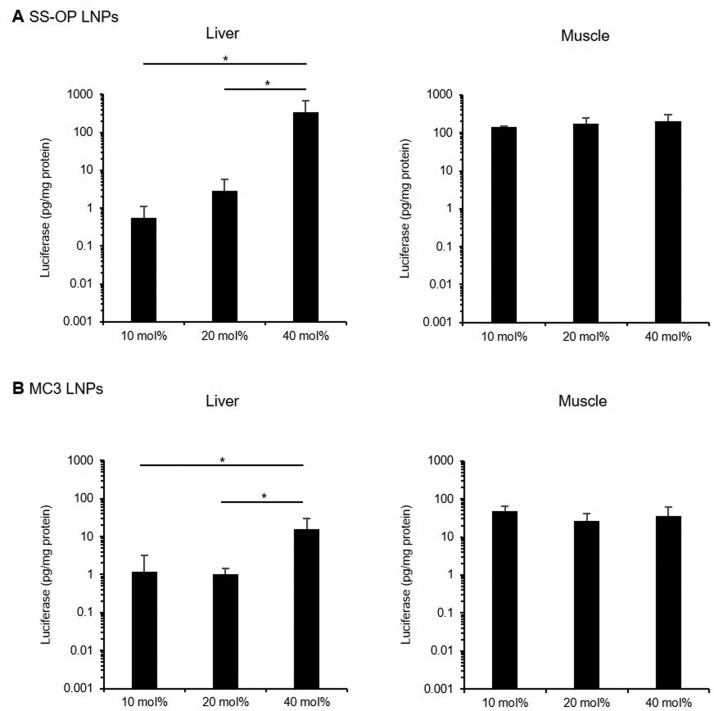
Luciferase expression levels were measured 4.5 h after administration of SS-OP LNPs containing 10 mol%, 20 mol%, and 40 mol% cholesterol and MC3 LNPs containing 10, 20, and 40 mol% cholesterol. After intramuscular administration, luciferase expression in the liver showed the same tendency as observed in vitro in the HepG2 cell line. The luciferase expression of SS-OP LNPs was significantly higher in the 40 mol% cholesterol group than in the 10 mol% (615-fold) or 20 mol% (118-fold) groups. (Fig. 4A ) For MC3 LNPs, the luciferase expression was significantly higher in the 40 mol% cholesterol group than in the 10 mol% (13.1-fold) or 20 mol% (15.3-fold) groups. (Fig. 4B) In contrast, the intramuscular luciferase expression was similar in all groups. However, intramuscular luciferase expression was significantly higher in the LNP group than in the naked mRNA intramuscular group (data not shown). The luciferase expression of muscle to that of liver in 10 mol% cholesterol of SS-OP LNPs was 434, 20 mol% was 64.7, and 40 mol% was 1.22, and the muscle-to-liver value of 10 mol% cholesterol of SS-OP LNPs was 627, 20 mol% was 34.3, and 40 mol% was 7.51.
Figure 4.
In vivo transfection efficiency of mRNA-LNPs by intramuscular administration. Transfection efficiency of SS-OP LNPs (A) and MC3 LNPs (B) by intramuscular administration. Mice were intramuscularly administrated with FLuc mRNA LNPs, containing 10 mol%, 20 mol%, 40 mol% cholesterol. Four and a half h after administration, luciferase expression levels were analyzed in the muscle and the liver (n=3–4). Statistical analysis was performed by one-way ANOVA Tukey's multiple comparisons test (*P < 0.05). Data are represented as mean + SD (n=3–4).
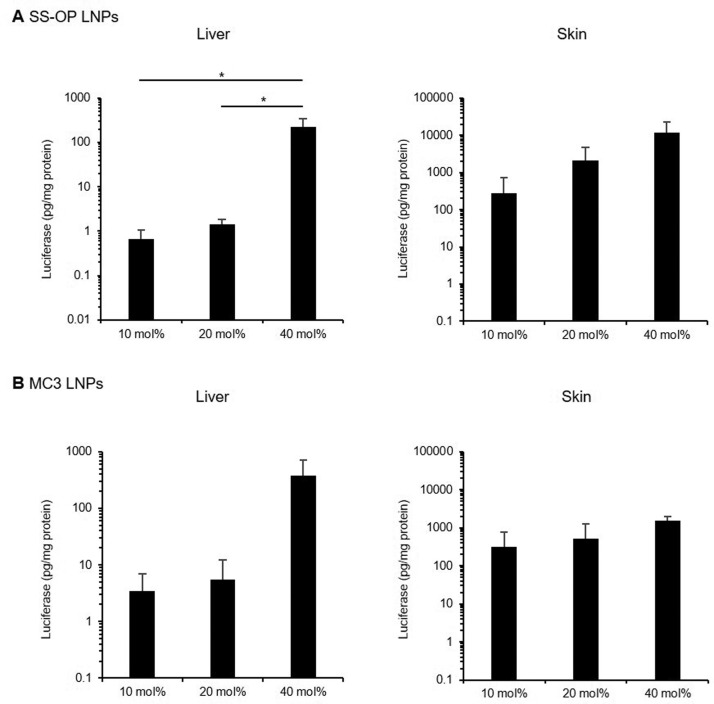
In terms of luciferase expression in the liver after subcutaneous administration, the luciferase expression of SS-OP LNPs was significantly higher in the 40 mol% cholesterol group than that in the 10 mol% (333-fold) or 20 mol% (157-fold) groups. (Fig. 5A ) For MC3 LNPs, luciferase expression was 107- and 67.9- fold higher in the 40 mol% cholesterol group than in the 10 mol% or 20 mol% groups, respectively. (Fig. 5B) There was no significant difference in luciferase expression in the skin near the administration site. However, the trend of mRNA-LNP transfection was 40 mol% > 20 mol% > 10 mol%.
Figure 5.
In vivo transfection efficiency of mRNA-LNPs by subcutaneous administration. Transfection efficiency of SS-OP LNPs (A) and MC3 LNPs (B) by subcutaneous administration. Mice were subcutaneously administrated with FLuc mRNA-LNPs, containing 10 mol%, 20 mol%, 40 mol% cholesterol. Four and a half h after administration, luciferase expression levels were analyzed in the muscle and the liver (n=3–4). Statistical analysis was performed by one-way ANOVA Tukey's multiple comparisons test (*P < 0.05). Data are represented as mean + SD (n=3–4).
The Kinetics of SS-OP LNPs
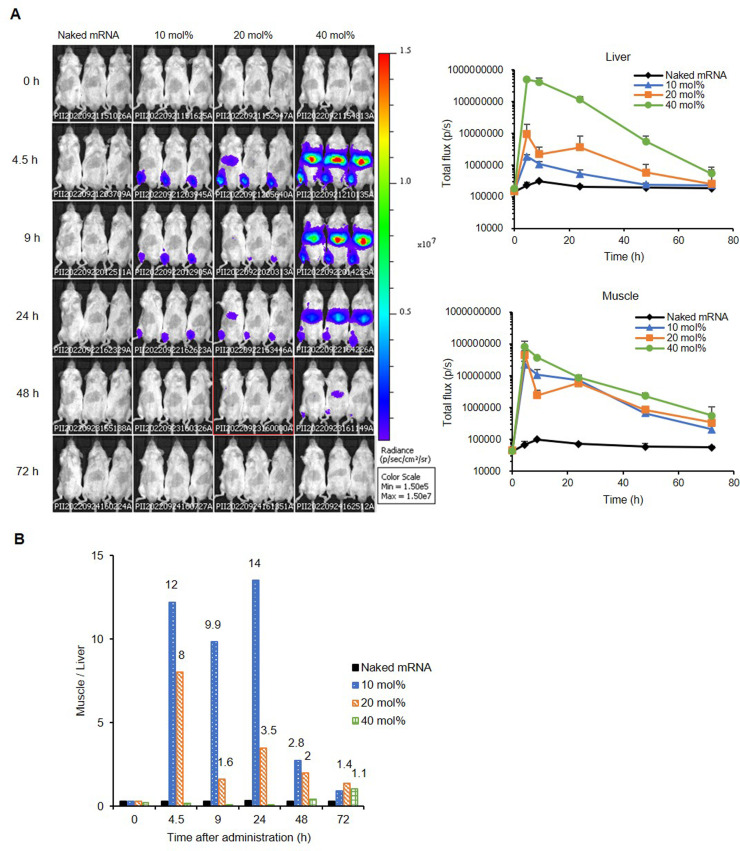
Based on the results of in vivo transfection, we selected intramuscular administration of SS-OP LNPs because they have high luciferase mRNA expression efficiency. We evaluated the effect of cholesterol in the mRNA-LNP constituent lipids on the kinetics of mRNA translation of mRNA-LNPs in mice. Mice were intramuscularly administered naked FLuc mRNA or SS-OP LNPs containing 10 mol%, 20, or 40 mol% cholesterol. Mice were intraperitoneally administered 3 mg of D-luciferin at 0, 4.5, 9, 24, 48, and 72 h after transfection. Luminescence imaging was performed using the IVIS imaging system at each time point (Fig. 6A and B). We then analyzed the luminescence levels of the muscle and liver from these images. The levels of muscle to those of the liver are shown in the graph of Fig. 6B as values to demonstrate the transfer of mRNA-LNP to the liver when administered intramuscularly under the same conditions.
Figure 6.
Luciferase expression kinetics of SS-OP LNPs by intramuscular administration. Naked FLuc mRNA, SS-OP LNPs containing 10 mol%, 20 mol%, or 40 mol% cholesterol were administered intramuscularly to ddY male mice as a dose of 2 µg of FLuc mRNA. Then, 0, 4.5, 9, 24, 48, and 72 h after the administration, 3 mg of D-luciferin was intraperitoneally injected, and IVIS imaging was performed at each time point (A). The value of the total flux of muscle to the liver was calculated (B). More than 1 is shown in the graph as numbers.
Discussion
This study showed that mRNA-LNPs could be successfully formulated even when the cholesterol molar percentage was reduced to 10 or 20 mol% (Table 1, Figs. 1 and 2). The protein expression levels of mRNA-LNP transfection in HepG2 cells and the liver of mice after intramuscular administration were in the order of cholesterol molar percentage of 40 mol% > 20 mol% > 10 mol% (Figs. 3 and 4). To evaluate whether these results show a similar trend when administered subcutaneously, experiments were conducted with subcutaneous administration, which is used to administer various vaccines by local administration similar to intramuscular administration. The value of the expression in the injected site-to-liver in subcutaneous administration was higher with lower cholesterol levels of mRNA-LNPs, similar to what was observed in intramuscular administration (Fig. 5). These results support our hypothesis that reducing the cholesterol molar percentage of mRNA-LNPs decreases protein expression in the liver during intramuscular or subcutaneous administration of mRNA-LNPs.
Intramuscular administration of mRNA-LNPs in mice decreased its expression in the liver as the molar percentage of cholesterol in mRNA-LNPs decreased from 40 mol% to 20 mol% and 10 mol% (Figs. 4 and 6). These results are consistent with previous studies that showed that cholesterol-rich liposomes were distributed in hepatocytes compared to cholesterol-poor liposomes and cholesterol-free liposomes during intravenous administration in mice,15 and as the percentage of cholesterol in liposomes increased, blood protein binding decreased and plasma clearance was delayed.14 , 25 Considering that intramuscularly administered mRNA-LNPs finally enter the systemic circulation like liposomes having neutral charge and about 100 nm mean particle size.8 Intramuscular administration of mRNA-LNP at a lower cholesterol molar percentage of mRNA-LNP can be degraded in the systemic circulation and decrease its protein expression in the liver.
Protein expression in muscle was more sustained than the protein expression in the liver. In Fig. 6A, we evaluated the elimination of the luciferase activity in muscle and liver. Not only cellular uptake as mRNA-LNPs, but also mRNA expression leaked from unstable mRNA-LNPs may contribute to the protein expression at the injected site for intramuscular administration, as Wolff et al. reported that protein is expressed even after intramuscular administration of naked mRNA.26 For protein expression in the liver, mRNA-LNP uptake via LDLR in hepatocytes has been reported, and the contribution of LNP particle stability is assumed to be relatively large. It has also been reported that protein expression efficiency may vary from tissue to tissue because mRNA release efficiency in tissues may be different.8 The tendency is consistent with previous reports, suggesting differences in expression profiles by tissue.
To date, there have been few reports on the effect of the cholesterol content of mRNA-LNPs on protein expression in cultured hepatocytes. To estimate the effect of the cholesterol content of the lipid composition of mRNA-LNPs on protein expression after reaching the liver, we evaluated the efficiency of protein expression in HepG2 cells. Protein expression also decreased in HepG2 cells as the cholesterol molar percentage of mRNA-LNPs decreased from 40 mol% to 20 mol% and 10 mol%. (Fig. 3) It has been reported that the bending stiffness of nanoparticles varies with the cholesterol content.27 The mechanical stiffness of liposomes is often defined by the bending stiffness of the membrane, an important parameter of particle stability.28 Furthermore, The bending stiffness of liposome membranes has been reported to play a key role in cellular uptake by endocytosis.29 Even if blood protein binding is not involved, the membrane stability of the mRNA-LNPs might be involved in protein expression in HepG2 cells. We revealed the possibility that the downregulated expression of mRNA-LNPs with low cholesterol content in the liver involves not only degradation in systemic circulating blood, but also decreased protein expression after hepatocyte distribution.
In this study, we considered MC3 as the gold standard ionizable lipid for LNP because it was also used in Patisiran and was the ionizable lipid on the market at the beginning of the study. On the other hand, we chose SS-OP30 which has been reported to have superior endosomal escape ability because of the aromatic ring in its structure and show higher protein expression efficiency compared to MC3, and because it forms LNPs with a different phospholipid, DOPC. We prepared formulations of various ionizable lipids (SS-OP or MC3), phospholipids (DOPC or DSPC), DMG-PEG lipids, and cholesterol. We arranged the cholesterol molar percentages of mRNA-LNPs as we arranged the phospholipids molar percentages to assess the effect of cholesterol content on the expression of mRNA-LNPs. The cholesterol molar percentage of SS-OP LNPs was arranged from 10 mol% to 20 mol% or 40 mol%, while the DOPC molar percentage was changed from 27.5 mol% to 17.5 mol%, 7.5 mol%. The cholesterol molar percentage of MC3 LNPs was arranged from 28.5 mol% to 18.5 mol%, 8.5 mol% because ionizable lipids are responsible for the transfection efficiency of mRNA-LNPs, and PEG lipids are related to size limitations. The particle sizes of all mRNA-LNPs ranged from 75.5 to 140 nm, and the PDI of the mRNA-LNPs was <0.25. The standard cholesterol levels in mRNA-LNP are 30–50 mol% because cholesterol contributes to structural stability.31 On the other hand, cholesterol-free liposomes were formulated in some previous studies about liposomes. However, their expression characteristics differ from liposomes containing cholesterol.14 , 15 , 25 Thus, we hypothesized that it would also be possible to formulate particles in mRNA-LNPs. This study found that mRNA-LNPs can be formed even when the cholesterol molar percentage is reduced. However, we also found that the stability of the one-week formulation was inferior in mRNA-LNPs with a 10 mol% cholesterol molar percentage compared to the 20 or 40 mol% groups (Fig. 2), suggesting that the cholesterol molar percentage contributes to the structural stability of mRNA-LNPs. This information can be valuable for the rational use of mRNA-LNPs with a lower cholesterol content.
Although we evaluated the effect of cholesterol content in mRNA-LNPs on protein expression in vitro and in vivo, we acknowledge that the limitations of this study should be described here. Based on the process of protein expression in tissues after local administration, various factors should contribute to the protein expression in the liver after intramuscular or subcutaneous injection of LNP, including the adsorption into the blood (via lymph), the clearance from blood circulation, the interaction with ApoE and the cholesterol of mRNA-LNP surface, uptake rate by hepatocytes including interaction with LDLR and the complex of ApoE and mRNA-LNPs, the release rate of mRNA into the cytoplasm, and transcription of mRNA in the hepatocyte. First, it is reported the particle size and surface charge of mRNA-LNPs can affect their transfer into the lymphatic system after local administration.32 When administered subcutaneously, it has been reported that LNPs smaller than 50 nm33 and negatively charged34 are more likely to be transferred to the lymphatic system. At this point, all of the mRNA-LNPs administrated to the mice in this article were neutrally charged with their particle size of approximately 100 nm, and possible to remain their distribution properties. Therefore, the surface charge of these mRNA-LNPs would not be a factor affecting their kinetics in this study. Secondly, it has been reported that an increased cholesterol molar percentage decreases protein binding in blood.15 However, there are several reports that ApoE, a blood protein, is contributed to the hepatic uptake of mRNA-LNPs via LDLR on hepatocytes.12 , 35 ApoE usually circulates in the blood as a part of chylomicron remnant, very low density lipoprotein (VLDL), intermediate-density lipoprotein (IDL), and some high density lipoprotein (HDL) and interacts with the VLDL receptor and catabolizes lipoproteins. In peripheral tissues, ApoE is responsible for cholesterol metabolism. It is unclear which factors contribute significantly to this process. It is presumed that such multiple factors influence the cholesterol content and the expression, and it would be complicated to design an experiment to identify the most prominent factor. More detailed evaluation is needed to clarify this mechanism in the future. In this study, the toxicity was not evaluated. However, the purpose of this study was to characterize the gene expression profiles. We used mRNA-LNPs with two types of ionizable lipids that are commonly used in mRNA-LNP studies. In the case of mRNA-LNPs with MC3, it has been reported that subcutaneous administration of 0.3 mg mRNA/kg in mice did not cause hepatotoxicity.36 In this experiment, we chose a sufficiently low dose, about one-fourth of the previous report, because 2 µg mRNA/mouse (about 0.08 mg mRNA/kg) was sufficient to express luciferase and to analyze the effect of cholesterol content on the protein expression in mice. However, systemic toxicity is needed to be evaluated in the future for therapeutic applications.
In this section, we discuss some future applications of our findings. We found that by reducing the cholesterol content of LNP, the expression in the liver could be reduced, and localized expression in the muscle could be achieved. However, its expression in the muscle was not increased. In Duchenne muscular dystrophy (DMD), a genetic disease of muscle, permanent protein expression can be achieved even at low expression levels at the time of transfection by combining genome editing. In addition, it is important to enabling local expression for genome editing because if expression outside the target tissue is permanent, side effects are unavoidable.
Conclusion
In this study, we verified that decreasing the cholesterol content in mRNA-LNPs suppressed protein expression in the liver during intramuscular administration of mRNA-LNPs. This study showed for the first time that mRNA-LNPs with a particle size of approximately 100 nm, PDI <0.25, and mRNA encapsulation efficacy of more than 80% can be stably generated even when the cholesterol molar percentage in LNPs is reduced. More importantly, the study demonstrated that decreasing the cholesterol molar percentage can reduce the expression level in the liver while maintaining the expression level in the muscle. We expect that the relative increase in muscle expression with cholesterol-reduced LNPs can be an effective method for the safe treatment of DMD by local in vivo genome editing using these mRNA-LNPs in the future.
Source of Funding
This work was partly supported by JSPS KAKENHI Grant Number 21H03818 (S.K.), AMED Grant Number JP22ak0101178 (H.M.).
Disclosure
Conflicts of Interest The author Shusaku Mizukami has received a research budget from 428 SHIONOGI & CO., LTD. It has been used for the employment of Shusaku Mizukami, but not for 429 other uses in this study
CRediT authorship contribution statement
Maho Kawaguchi: Conceptualization, Investigation, Methodology, Writing – original draft. Marin Noda: Investigation, Writing – review & editing. Akari Ono: Investigation, Writing – review & editing. Mariko Kamiya: Investigation, Writing – review & editing. Makoto Matsumoto: Investigation, Writing – review & editing. Masako Tsurumaru: Writing – review & editing. Shusaku Mizukami: Resources, Writing – review & editing. Hidefumi Mukai: Funding acquisition, Supervision, Writing – review & editing. Shigeru Kawakami: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was partly supported by JSPS KAKENHI Grant Number 21H03818 (S.K.), AMED Grant Number JP22ak0101178 (H.M.), and the Nagai Memorial Research Scholarship from the Pharmaceutical Society of Japan (M.K.).
References
- 1.Mukai H., Ogawa K., Kato N., Kawakami S. Recent advances in lipid nanoparticles for delivery of nucleic acid, mRNA, and gene editing-based therapeutics. Drug Metab Pharmacokinet. 2022;44 doi: 10.1016/j.dmpk.2022.100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oyama N., Kawaguchi M., Itaka K., Kawakami S. Efficient messenger RNA delivery to the kidney using renal pelvis injection in mice. Pharmaceutics. 2021;13(11):1810. doi: 10.3390/pharmaceutics13111810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pardi N., Tuyishime S., Muramatsu H., et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Cont Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loughrey D., Dahlman J.E. Non-liver mRNA Delivery. Acc Chem Res. 2022;55(1):13–23. doi: 10.1021/acs.accounts.1c00601. [DOI] [PubMed] [Google Scholar]
- 5.Chen S., Tam Y.Y.C., Lin P.J.C., Leung A.K.K., Tam Y.K., Cullis P.R. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J Cont Release. 2014;196(28):106–112. doi: 10.1016/j.jconrel.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Oussoren C., Zuidema J., Crommelin D., Storm G. Lymphatic uptake and biodistribution of liposomes after subcutaneous injection.: II. Influence of liposomal size, lipid composition and lipid dose. BBA. 1997;1328(2):261–272. doi: 10.1016/S0005-2736(97)00122-3. [DOI] [PubMed] [Google Scholar]
- 7.Trevaskis N.L., Kaminskas L.M., Porter C.J.H. From sewer to saviour — targeting the lymphatic system to promote drug exposure and activity. Nature Rev Drug Discov. 2015;14:781–803. doi: 10.1038/nrd4608. [DOI] [PubMed] [Google Scholar]
- 8.Di J., Du Z., Wu K., Jin S., Wang X., Li T., Xu Y. Biodistribution and non-linear gene expression of mRNA LNPs affected by delivery route and particle size. Pharm Res. 2022;39:105–114. doi: 10.1007/s11095-022-03166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang N., Li X., Deng Y., et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5):1271–1283. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrasco M.J., Alishetty S., Alameh M., et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Nature Comm Biol. 2021;4:956. doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francia V., Schiffelers R.M., Cullis P.R., Witzigmann D. The biomolecular corona of lipid nanoparticles for gene therapy. Bioconjugate Chem. 2020;31(9):2046–2059. doi: 10.1021/acs.bioconjchem.0c00366. [DOI] [PubMed] [Google Scholar]
- 12.Akinc A., Querbes W., De S., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18(7):1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson E.J., Rouphael N.G., Widge A.T., et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semple S.C., Chonn A., Cullis P.R. Influence of cholesterol on the association of plasma proteins with liposomes. Biochemistry. 1996;35(8):2521–2525. doi: 10.1021/bi950414i. [DOI] [PubMed] [Google Scholar]
- 15.Murao A., Nishikawa M., Managit C., et al. Targeting efficiency of galactosylated liposomes to hepatocytes in vivo: effect of lipid composition. Pharm Res. 2002;19(12):1808–1814. doi: 10.1023/A:1021433206081. [DOI] [PubMed] [Google Scholar]
- 16.Nakhaei P., Margiana R., Bokov D.O., et al. Liposomes: structure, biomedical applications, and stability parameters with emphasis on cholesterol. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.705886. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Ryals R.C., Patel S., Acosta C., McKinney M., Pennesi M.E., Sahay G. The effects of PEGylation on LNP based mRNA delivery to the eye. PLoS ONE. 2020;15(10) doi: 10.1371/journal.pone.0241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J Cont Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 19.Zhang R., El-Mayta R., Murdoch T.J., et al. Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater Sci. 2021;9:1449–1463. doi: 10.1039/D0BM01609H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paunovska K., Sanchez A., Sago C.D., et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv Mater. 2019;31(14) doi: 10.1002/adma.201807748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel S., Ashwanikumar N., Robinson E., et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nature Comm. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa K., Kato N., Yoshida M., et al. Focused ultrasound/microbubbles-assisted BBB opening enhances LNP-mediated mRNA delivery to brain. J Cont Release. 2022;348:34–41. doi: 10.1016/j.jconrel.2022.05.042. [DOI] [PubMed] [Google Scholar]
- 23.Niemietz C., Nadzemova O., Zibert A., Schmidt H.H.J. APOE polymorphism in ATTR amyloidosis patients treated with lipid nanoparticle siRNA. Amyloid. 2020;27(1):45–51. doi: 10.1080/13506129.2019.1681392. [DOI] [PubMed] [Google Scholar]
- 24.Kamiya M., Matsumoto M., Yamashita K., et al. Stability study of mRNA-Lipid nanoparticles exposed to various conditions based on the evaluation between physicochemical properties and their relation with protein expression ability. pharmaceutics. 2022;14(11):2357. doi: 10.3390/pharmaceutics14112357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel H.M., Tuz̈el N.S., Ryman B.E. Inhibitory effect of cholesterol on the uptake of liposomes by liver and spleen. BBA. 1983;761(2):142–151. doi: 10.1016/0304-4165(83)90223-4. [DOI] [PubMed] [Google Scholar]
- 26.Wolff J.A., Malone R.W., Williams P., et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi-Haraya Y., Sakai-Kato K., Abe Y., Kawanishi T., Okuda H., Goda Y. Observation of liposomes of differing lipid composition in aqueous medium by means of atomic force microscopy. Microscopy. 2016;65(4):383–389. doi: 10.1093/jmicro/dfw011. [DOI] [PubMed] [Google Scholar]
- 28.Liang X., Mao G., Ng K.Y.S. Mechanical properties and stability measurement of cholesterol-containing liposome on mica by atomic force microscopy. J Colloid Interface Sci. 2004;278(1):53–62. doi: 10.1016/j.jcis.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S., Gao H., Bao G. Physical principles of nanoparticle cellular endocytosis. ACS Nano. 2015;9:8655–8671. doi: 10.1021/acsnano.5b03184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka H., Takahashi T., Konishi M., et al. Self-degradable lipid-like materials based on “hydrolysis accelerated by the intra-particle enrichment of reactant (HyPER)” for messenger RNA delivery. Adv Funct Mater. 2020 doi: 10.1002/adfm.201910575. [DOI] [Google Scholar]
- 31.Schoenmaker L., Witzigmann D., Kulkarni J.A., et al. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura T., Kawai M., Sato Y., Maeki M., Tokeshi M., Harashima H. The effect of size and charge of lipid nanoparticles prepared by microfluidic mixing on their lymph node transitivity and distribution. Mol Pharmaceutics. 2020;17(3):944–953. doi: 10.1021/acs.molpharmaceut.9b01182. [DOI] [PubMed] [Google Scholar]
- 33.Reddy S.T., Rehor A., Schmoekel H.G., Hubbell J.A., Swartz M.A. In vivo targeting of dendritic cells in lymph nodes with poly(propylene sulfide) nanoparticles. J Controlled Release. 2006;112(1):26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Rao D.A., Forrest M.L., Alani A.W.G., Kwon G.S., Robinson J.R. Biodegradable PLGA based nanoparticles for sustained regional lymphatic drug delivery. J Pharm Sci. 2010;99(4):2018–2031. doi: 10.1002/jps.21970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sebastiani F., Arteta Y.M., Lerche M., et al. Apolipoprotein E binding drives structural and compositional rearrangement of mRNA-containing lipid nanoparticles. ACS Nano. 2021;15(4):6709–6722. doi: 10.1021/acsnano.0c10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies N., Hovdal D., Edmunds N., et al. Functionalized lipid nanoparticles for subcutaneous administration of mRNA to achieve systemic exposures of a therapeutic protein. Mol Ther Nucleic Acids. 2021;24:369–384. doi: 10.1016/j.omtn.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]