Abstract
Artificial intelligence (AI)-based approaches can now use electrocardiograms (ECGs) to provide expert-level performance in detecting heart abnormalities and diagnosing disease. Additionally, patient age predicted from ECGs by AI models has shown great potential as a biomarker for cardiovascular age, where recent work has found its deviation from chronological age (“delta age”) to be associated with mortality and co-morbidities. However, despite being crucial for understanding underlying individual risk, the genetic underpinning of delta age is unknown. In this work we performed a genome-wide association study using UK Biobank data (n=34,432) and identified eight loci associated with delta age (), including genes linked to cardiovascular disease (CVD) (e.g. SCN5A) and (heart) muscle development (e.g. TTN). Our results indicate that the genetic basis of cardiovascular ageing is predominantly determined by genes directly involved with the cardiovascular system rather than those connected to more general mechanisms of ageing. Our insights inform the epidemiology of CVD, with implications for preventative and precision medicine.
Subject terms: Genome-wide association studies, Cardiovascular genetics, Machine learning, Predictive markers
Introduction
For decades it has been known that a person’s electrocardiogram (ECG) changes with age1,2. Therefore, in light of its non-invasiveness, ease of obtainment, and consequential ubiquity, there is great potential in using the 12-lead ECG as a biomarker for physiological changes caused by ageing3. As these changes occur gradually and at a rate that is different between individuals, there is substantial variation in the risk of chronic disease and mortality in older populations. In order to understand the sources of this variation, several indicators for “biological age” have been investigated, including changes in telomere length4, the epigenome5, blood-derived biomarkers6, and the transcriptome7. Crucially, these markers have been shown to be only weakly correlated with each other8, suggesting that they do not describe the same underlying physiological processes but rather different aspects of ageing9. Since cardiovascular disease (CVD) is a major source of mortality and morbidity, with drastically increasing prevalence in older age10, the deep learning-enabled ECG-derived surrogate for cardiovascular age introduced by Attia et al.11 represents a valuable addition to other “ageing” metrics, with both preventative and personalised medicine benefits. Here we report the results of a genome-wide association study (GWAS) using the difference between a person’s actual age and this metric as phenotype.
Initial studies trying to link chronological age to the ECG signal mostly focused on human-defined ECG features, such as the QRS duration or the length of the PR interval12. However, the extraction of these features is not devoid of error13 and captures only a fraction of the available information. Recent developments in deep learning allowed researchers to address this limitation by adapting modern convolutional artificial neural network architectures to predict patients’ ages from their ECGs11,14,15. These models can be trained “end-to-end” on the raw ECG traces from which they learn to extract (and combine in a non-linear manner) the features most suitable for a prediction task. Thus, the impact of human bias is minimised and predictive power improved as all the information in the signal is taken into account. In fact, several studies have shown that deep learning models trained on ECG traces already match and in some cases even exceed the performance of medical professionals in diagnosing certain cardiac conditions16–18. Given the increasing prevalence of ECG data, machine learning models of such capabilities could transform predictive medicine and cardiovascular research.
In order to use ECGs for age prediction, the neural network needs to learn how the “average” ECG for a particular age group looks. Thus, when it predicts an age considerable larger than the corresponding person’s chronological age (a large “delta age ”), this might be indicative of accelerated ageing of the cardiovascular system – with implications for this individual’s health. Indeed, large delta age has been shown to be associated with CVD, treatment outcomes, and mortality3,11,14. This observation suggests at least two principal areas of applications for the ECG-derived age (or delta age). On one hand, it could be used in the clinic as a readily obtainable prognostic tool for screening large numbers of patients. In this capacity, delta age is conceptually similar to the “excess heart age”19, the discrepancy between a person’s chronological age and their “heart age” (the age corresponding to their risk of a CVD event), which has been devised as an easily interpretable measure for CVD risk20. However, while the excess heart age represents the increased CVD risk due to risk factors and lifestyle choices, the delta age reflects the actual functional state of the heart. Hence, in addition to clinical use cases, ECG-derived age could also complement biomarkers used in research (e.g. telomere length or the epigenetic clock, among others) for tracking ageing in general and vascular ageing in particular. One crucial advantage of the ECG-derived age over many other ageing-related biomarkers used in research is the wide-spread use of the ECG and how comparatively easy it is to obtain. This makes it especially interesting for association studies which suffer from low effective sample sizes for many disease-related phenotypes as these are usually relatively rare or can remain undiagnosed, diluting the strength of the statistical signal. Furthermore, delta age is not tied to a single type of CVD, but instead combines effects on the ECG of multiple conditions in addition to “normal” changes expected due to ageing. It might therefore lead to the discovery of genetic variants that are not associated to any individual condition.
In addition to the advances in machine learning mentioned above, the availability of genomic data (from microarrays and—more recently— whole-genome sequencing) is ever-increasing. This wealth of information has facilitated a vast number of association studies, linking biological variation in countless phenotypes to the underlying genotypes21. Some of these studies investigated the genetic basis of ECG-features (e.g. for the PR interval22 or the QRS complex23), while others sought to determine the impact of genetic variants on the shape of the ECG traces in general24 or on a more holistic representation of the cardiac state including the ECG25.
In light of these converging developments, we used a previously published convolutional neural network11 to predict the “cardiovascular age” of 36,349 participants of the UK biobank (UKB) from their 12-lead ECGs, and performed a GWAS on the difference between predicted and chronological age (i.e. delta age). We found eight loci of genome-wide significance (), many of which have been associated with cardiac or muscle development (and in extension with CVD) in the past. Functional and pathway enrichment analyses confirmed this connection to the cardiovascular system. We also explored the association of delta age with specific ECG features, risk factor-derived excess heart age, and the dynamic organism state indicator (DOSI), a complementary biomarker for ageing derived from complete blood count (CBC) data. Overall, our results elucidate the genetic underpinning of this ECG-derived biomarker for cardiovascular age and validate its utility for use in research as well as in the clinic.
Results
Predicting age from ECGs in the UK Biobank
We employed a previously described deep learning model trained on patients of the Mayo clinic11 to predict the age of 36,349 participants of the UKB from their 12-lead ECGs. On average, individuals were 64 years old, marginally more likely to be women (52%), and had high levels of education (tertiary education for more than 50%). They comprised a relatively healthy cohort (e.g. less than 6% had diagnosed cardiovascular conditions more severe than hypertension), commonly reporting lifestyle choices considered preventive of CVD (e.g. never smoked), and showing predominantly normal ranges for body mass index (BMI), lipids, and blood pressure (Table 1).
Table 1.
Association of anthropometric features and cardiovascular risk factors in participants of the UKB with delta age.
| Covariate | Info () |
Adjust for age, sex | Adjust for all | ||
|---|---|---|---|---|---|
| Effect size | -value | Effect size | -value | ||
| Sex (male) | 17607 (48.4%) | −0.56 (−0.71, −0.41) | 4.7e-13 | −1.15 (−1.31, −0.98) | 4.1e-42 |
| Age | 64.25 (±7.57) | −0.37 (−0.38, −0.36) | 0.0e+00 | −0.40 (−0.41, −0.39) | 0.0e+00 |
| Education | – | – | 4.6e-18 | – | 2.4e-06 |
| Secondary (ref. level) | 14437 (40.1%) | – | – | – | – |
| Tertiary | 19186 (53.3%) | −0.69 (−0.85, −0.53) | 1.7e-17 | −0.41 (−0.57, −0.25) | 9.7e-07 |
| Other | 2343 (6.5%) | 0.04 (−0.29, 0.36) | 0.83 | 0.01 (−0.33, 0.34) | 0.98 |
| History of health problems: | |||||
| Diabetes | 1979 (5.5%) | 0.81 (0.47, 1.15) | 2.2e-06 | −0.22 (−0.58, 0.14) | 0.23 |
| Hypertension | 8419 (23.2%) | 1.85 (1.67, 2.04) | 3.7e-88 | 0.77 (0.56, 0.97) | 2.1e-13 |
| Angina | 727 (2.0%) | 0.88 (0.34, 1.42) | 1.5e-03 | 0.11 (−0.48, 0.70) | 0.72 |
| Stroke | 366 (1.0%) | 1.47 (0.71, 2.23) | 1.6e-04 | 0.99 (0.20, 1.78) | 0.014 |
| Heart attack | 524 (1.4%) | 1.49 (0.85, 2.13) | 4.6e-06 | 1.43 (0.75, 2.12) | 4.1e-05 |
| Physiological measurements: | |||||
| BMI | 26.62 (±4.25) | 0.24 (0.22, 0.25) | 3.6e-149 | 0.16 (0.14, 0.18) | 3.8e-55 |
| MAP | 81.11 (±8.89) | 0.13 (0.12, 0.14) | 9.9e-173 | 0.10 (0.09, 0.11) | 3.0e-80 |
| LDL [mM] | 3.58 (±0.82) | 0.15 (0.05, 0.24) | 2.6e-03 | 0.03 (−0.07, 0.13) | 0.52 |
| Lifestyle: | |||||
| Smoking | – | – | 6.2e-11 | – | 1.6e-04 |
| Never / rarely smoked (ref. level) | 22477 (62.5%) | – | – | – | – |
| Active smoker | 1300 (3.6%) | 0.50 (0.09, 0.92) | 0.017 | 0.41 (−0.01, 0.84) | 0.056 |
| Smoked in the past | 12212 (33.9%) | 0.56 (0.40, 0.72) | 2.3e-11 | 0.34 (0.17, 0.51) | 7.9e-05 |
| Alcohol at least 3x per week | 16405 (45.2%) | 0.24 (0.09, 0.39) | 2.3e-03 | 0.33 (0.17, 0.49) | 6.3e-05 |
| Days of moderate PA per week | 3.72 (±1.87) | −0.16 (−0.20, −0.11) | 8.4e-14 | −0.020 (−0.071, 0.030) | 0.43 |
| Days of vigorous PA per week | 1.93 (±1.58) | −0.25 (−0.30, −0.20) | 1.1e-24 | −0.16 (−0.22, −0.10) | 2.0e-07 |
The “Info” column lists the number of corresponding participants for categorical features (with the percentage of the total population in parentheses) or the mean value for numerical features (with the standard deviation in parentheses). -values and effect sizes in the left double-column are adjusted for age and sex (or only sex for the age-row and vice versa). In the right double-column, the adjustment also includes all other parameters listed in the table. In the “Effect size” columns, values in parentheses denote the lower and upper bounds of the 95% confidence interval. -values smaller than the Bonferroni-corrected threshold () are highlighted in bold. BMI, body mass index; MAP, mean arterial pressure; LDL, low-density lipoprotein; PA, physical activity.
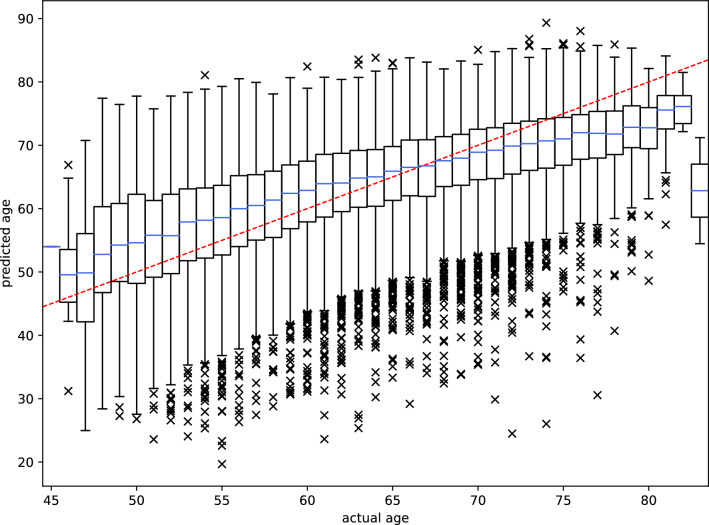
As the ECGs in the UKB were noisier than those used for training the model originally11, an initial signal filtering step was applied prior to prediction. After this pre-processing step, prediction performance on the UKB cohort was comparable to the holdout data set in the original study with a mean absolute error of 6.1 instead of 6.9 years, respectively (Fig. 1). The Pearson correlation coefficient between chronological and predicted age was =0.53.
Figure 1.
ECG-derived age vs. chronological age for 36,349 participants of the UKB. The Pearson correlation coefficient was 0.53. The red dashed line illustrates a perfect fit. Data is grouped into boxes as the chronological age at the time of recording the ECG was only available as the number of years. Note that ranges and scales are different between the x- and y-axis due to outliers in the predicted age being considerably outside the range of the chronological age in the cohort. Box plot features: blue centre lines, median; box limits, first and third quartile; whiskers, inter-quartile range; markers, outliers.
The participants’ chronological ages were then subtracted from the predicted ages to obtain the delta age (median 0.27; interquartile range −4.81–5.15 years). It was strongly associated with certain anthropometric features and cardiovascular conditions (Table 1), consistent with previous studies3,11,14. When adjusting for age and sex, tertiary education and physical activity were associated with a lower delta age (). BMI, mean arterial pressure (MAP), and low density lipoprotein (LDL), on the other hand, as well as classic cardiovascular risk factors and outcomes, such as frequently drinking alcohol, history of smoking, diagnosed diabetes, hypertension, angina, stroke, or heart attack were associated with higher delta age (). These findings were predominantly robust to multivariate analysis when including all mentioned variables in the model (Table 1). Interestingly, men had a lower delta age than women and the negative association with male sex increased when more covariates were taken into account.
Modern ECG machines automatically determine certain human-derived ECG features (e.g., PQ interval, QRS duration) when taking measurements. In the UKB data, many of these features were strongly associated with chronological age, predicted age, or both (Supplementary Table S1). However, only a small fraction of the variance in age could be explained by these human-derived features ( for a linear regression of age on the ECG features). The Pearson correlation coefficient between the age predicted from the ECG features and the chronological age was =0.28 (compared to =0.53 for the neural network). Interestingly, for the ages predicted by the neural network, this fraction increased almost three-fold (), indicating that the model relies on information retained in these features. This insight has also been shown in a recent study, which found that some features extracted by the convolutional layers of the neural net were strongly correlated with those defined by humans26.
GWAS on delta age
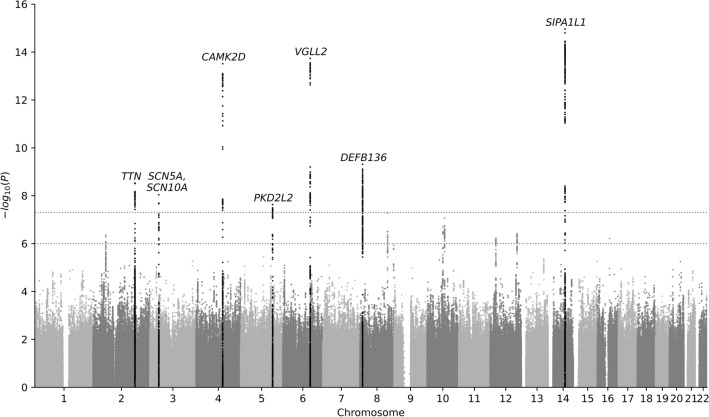
To understand the genetic underpinning of delta age, association tests were performed on 6.4 million autosomal variants in 34,432 individuals (after filtering and quality control) while adjusting for age, sex, genotyping array, and UKB assessment centre (Fig. 2). This analysis revealed eight loci of genome-wide significance () and another seven loci of suggestive significance (; Table 2).
Figure 2.
Manhattan plot. Association tests (n=34,432) were adjusted for age, sex, genotyping array, and UKB assessment centre. Horizontal lines mark the thresholds of genome-wide and suggestive significance ( and , respectively).
Table 2.
Fifteen loci were found to be associated with delta age with at least suggestive significance ().
| Chr. | Gene | rsID | Pos. | Ref. | Alt. | AF | Effect size | -value |
|---|---|---|---|---|---|---|---|---|
| 14 | SIPA1L1 | rs35866366 | 71849185 | A | G | 0.25 | 0.52 (0.39, 0.64) | 1.1e-15 |
| 6 | VGLL2 | rs6901720 | 117510203 | G | T | 0.47 | 0.43 (0.32, 0.54) | 2.8e-14 |
| 4 | CAMK2D | rs35430511 | 114387138 | T | C | 0.26 | 0.49 (0.36, 0.61) | 3.1e-14 |
| 8 | DEFB136 | rs4240678 | 11802426 | C | T | 0.40 | 0.47 (0.32, 0.62) | 4.9e-10 |
| 2 | TTN | rs11902709 | 179608207 | C | T | 0.05 | 0.78 (0.52, 1.03) | 3.0e-09 |
| 3 | SCN5A | rs6773331 | 38684397 | A | T | 0.98 | 1.24 (0.82, 1.66) | 9.1e-09 |
| 3 | SCN10A | rs6801957 | 38767315 | T | C | 0.59 | −0.32 (−0.43, −0.21) | 2.1e-08 |
| 5 | PKD2L2 | rs10076361 | 137252940 | G | A | 0.18 | 0.41 (0.27, 0.55) | 2.3e-08 |
| 8 | EXT1 | rs57237854 | 118860126 | ATCTTG | A | 0.18 | 0.40 (0.25, 0.54) | 5.3e-08 |
| 10 | AGAP5 | rs147790633 | 75447582 | T | C | 0.14 | −0.43 (−0.59, −0.27) | 8.7e-08 |
| 10 | CTNNA3 | rs72799115 | 68008504 | G | A | 0.21 | 0.35 (0.22, 0.49) | 2.0e-07 |
| 12 | TBX3 | rs1896329 | 115357432 | C | T | 0.69 | −0.31 (−0.42, −0.19) | 3.9e-07 |
| 2 | SPTBN1 | rs1802889 | 54756740 | C | T | 0.68 | −0.30 (−0.42, −0.19) | 4.4e-07 |
| 12 | SOX5 | rs12826024 | 24776799 | G | A | 0.15 | −0.39 (−0.54, −0.24) | 6.1e-07 |
| 16 | CHD9 | rs75778953 | 52906677 | C | T | 0.01 | −1.25 (−1.74, −0.76) | 6.2e-07 |
The second column lists the protein-coding gene closest to the respective lead variant. Positions correspond to the GRCh37 human genome assembly81. Values in parentheses denote the lower and upper bounds of the 95% confidence interval of the effect size estimate. -values with genome-wide significance () are highlighted in bold. Chr., Chromosome; Pos., Position; Ref., Reference allele; Alt., Alternative allele; AF, frequency of the alternative allele.
The variants with the strongest association with delta age were detected on chromosome 14 in the gene SIPA1L1, which has been linked to ECG features and other cardiac traits according to the GWAS Catalog27. Recently, SIPA1L1 has also been found to be associated with heart trabeculation28 and it is involved in the regulation of water transport in the kidney29. It might thus have an impact on the cardiovascular system via kidney function or control of blood volume. However, instead of altering SIPA1L1, the causal variant in this locus could alternatively affect the expression levels of RGS6, which lies 200 kb downstream. RGS6 is listed in the GWAS Catalog as associated with systolic blood pressure, heart rate, and heart rate variability, for which there is also mechanistic evidence30.
Another strong association signal was found 30–100 kb upstream of VGLL2 on chromosome 6. VGLL2 plays a role in the development of skeletal muscle31, but, to our knowledge, has not been directly linked to CVD so far. Nonetheless, the GWAS Catalog lists associations with relevant traits like ECG morphology, blood pressure, and atrial fibrillation, but also BMI and waist circumference. Interestingly, VGLL2 has also been shown to be associated with an age-dependent response to sepsis in the hearts of mice32. However, VGLL2 is not the only protein-coding gene in the region. The next closest (100 kb) is ROS1, a variant of which has been associated with pathological vascular remodelling33.
Variants in CAMK2D also showed a strong association with delta age. CAMK2D encodes the chain of the /calmodulin-dependent protein kinase II, which phosphorylates (in addition to itself) a wide variety of targets involved in a multitude of cellular functions, including neuroplasticity and memory formation34. It also plays a role in cardiac homeostasis and constitutive activation can lead to CVD and heart failure35.
The next notable locus was found on chromosome 8 and many of the variants associated with delta age within this locus have also been associated with essential hypertension in the GWAS Catalog. It was located between a group of three genes for -defensins (DEFB136, DEFB135, DEFB134 – with DEFB136 being the closest) and CTSB. Being antimicrobial peptides, -defensins are an integral part of the innate immune system, but they also have a range of other functions36. CTSB, located 50 kb downstream of the variants associated with delta age, codes for cathepsin B, a protease relevant for proteolysis of intracellular proteins as well as constituents of the extracellular matrix37. It has been associated with a large number of diseases, including different types of cancer38, cardiac remodelling and hypertrophy39, as well as atherosclerosis40. Interestingly, cathepsin B activity has also been shown to increase with age41.
On chromosome 2, variants in TTN were associated with delta age. TTN codes for the giant protein titin, responsible for passive mechanical properties of muscle (elasticity and stiffness) and sarcomere structure42. Mutations in TTN (especially when causing truncations) have been linked to dilated cardiomyopathy (DCM)43 and the GWAS Catalog mapped a variety of cardiovascular phenotypes and ECG traits to TTN, ranging from atrial fibrillation to the PR interval and left ventricular ejection fraction.
SCN5A and the neighbouring SCN10A (both on chromosome 3) harboured two independent groups of variants at genome-wide significance. Both genes encode subunits of sodium channels (most prevalent in the myocardium44 and neurons – including intracardiac ganglia45 – respectively). Variants in SCN5A have been linked to multiple cardiac disorders and mutations in both genes can cause the arrhythmia-inducing Brugada syndrome46,47.
The last locus of genome-wide significance stretched across 400 kb and six protein-coding genes (KLHL3, HNRNPA0, MYOT, PKD2L2, FAM13B, and WNT8A) on chromosome 5. The gene product of KLHL3 causes the ubiquitination of substrate proteins and is involved in regulating kidney function48. It has been associated with a rare hereditary form of hypertension (familial hyperkalaemic hypertension)49 and other forms of congenital heart disease in the past50. FAM13B encodes a GTPase-activating protein, low expression levels of which have been linked to atrial fibrillation51. However, if we assume that there is only one causal variant at this locus, it is most likely to be found in MYOT, which codes for myotilin, a component of the Z-disc complex in skeletal and cardiac muscle52. Myotilin variants can cause myofibrillar myopathy, which sometimes also affects the heart53. We did not find any connections with cardiovascular phenotypes for the other three genes, but the GWAS Catalog lists associations with dysrhythmias and atrial fibrillation across the whole 400 kb-spanning locus and beyond.
The seven extra loci found at suggestive significance () are described in more detail in the Supplementary Results. Most of them were also in the vicinity of genes related to muscle development or the cardiovascular system, but more statistical power (e.g. through larger sample size) will be needed to confirm these associations with delta age.
To assess the robustness of our results, the GWAS was repeated with a more extensive suite of covariates (including history of CVD, exercise, and diet; for details see "Methods" section) and additionally with only those participants that reported a White British ethnic background (Supplementary Fig. S1). All three analyses showed very similar results qualitatively, with a total of 17 loci reaching at least suggestive significance in at least one analysis (Supplementary Table S2).
Heritability
The variant-based heritability () of delta age was estimated to be 12%, being robust to adjustment of cardiovascular risk factors ( for regular adjustment and for extended adjustment). This magnitude is similar to other ECG traits or cardiac phenotypes, such as PR interval (18.2%22), long QT syndrome (14.8%54), or atrial fibrillation (9.6%55). Interestingly, the 15 loci that reached at least suggestive significance only accounted for 15% of the heritability estimate ( and for regular and extended adjustment, respectively), indicating that there are likely to be many variants with lower significance that are also relevant.
Functional analysis and pathway enrichment
As described above, many loci associated with ECG-derived delta age were found in the vicinity of genes involved in cardiac development or have been linked to CVD in the past. Application of the DEPICT enrichment analysis tool56 to the 15 loci with at least suggestive significance (; see Table 2) revealed that the GO-term with the strongest signal was “intercalated discs”, which are physical connections between cardiomyocytes. The KEGG57 pathways with the strongest association were mostly linked to calcium signalling and cardiac afflictions, which was also the case with the Mammalian Phenotype Ontology58 gene sets (Supplementary Data 1). We further used DEPICT to test for tissue enrichment. All results with -values smaller than 0.05 were either connective tissues or part of the cardiovascular system (Supplementary Table S3). When including all 179 loci with , geneset and tissue enrichment were both dominated by the cardiovascular system (Supplementary Data 2, Supplementary Table S4), reinforcing the robustness of our observations. To confirm these findings with an orthologous method, we additionally employed the gProfiler functional enrichment analysis tool59, which also detected a stark overrepresentation of components of the cardiovascular system (Supplementary Table S5). Like the DEPICT analysis, the strength of the enrichment increased when more loci were included (Supplementary Table S6).
Association of variants in telomere length- and longevity-related genes
Interestingly, genes associated with other forms of biological ageing (e.g. telomere length) were mostly absent from the loci found by our analysis. In order to further investigate this surprising result, we scanned the vicinity of loci discovered by recent GWAS, which had also been performed on the UKB and used longevity60 and leukocyte telomere lengths61 as phenotypes, for variants associated with delta age. We found that none of the loci associated with longevity and only two of those associated with telomere length (rs12615793 in ACYP2 and rs12369950 close to SOX5) were within one 1 Mb of variants with at least suggestive significance according to our analysis (Supplementary Data 3). In the first case, the lead variant of the locus we discovered was located 280 kb downstream of rs12615793 and in SPTBN1, which is required for heart development62. In the second case, rs12369950 was indeed part of the same locus we found to be associated with delta age.
Further analyses
In order to further investigate the main results described above, we performed statistical tests to detect whether the effects of the genomic variants were mediated via one of the covariates most strongly associated with delta age (BMI, MAP, and diagnosed hypertension), but did not find strong evidence for mediation. Additionally, we ascertained that most of the lead variants have been shown to have a significant impact on the actual shape of the ECG in a recent study24. We also calculated the risk factor-based “heart age”20 and the whole blood counts-derived DOSI biomarker for ageing63 to contrast both with the ECG-derived cardiovascular age. We found that, while the association with delta age was substantial for the “excess” heart age (), it was weak for the “excess” DOSI (). These findings are described in greater detail in the Supplementary Results.
Discussion
We used a deep neural network to predict the age of 36,349 individuals in the UKB from their 12-lead ECGs and observed that – similar to what has been shown in other populations3,11,14 – the discrepancy to their chronological age was correlated with cardiovascular risk factors like blood pressure, BMI, and smoking status. In addition to these covariates, we also found 15 genetic loci of at least suggestive significance (), eight of which reached genome-wide significance (), in a GWAS adjusted for age, sex, genotyping array, and UKB assessment centre. We evaluated the robustness of these results by repeating the GWAS with a more extensive set of covariates including past CVD diagnoses and lifestyle variables, such as diet or the amount of physical exercise. We also carried out another round of association tests with only the subset of individuals of European ethnic origin. All three analyses yielded very similar results (Supplementary Table S7). Overall, about 12% of the variation in delta age could be explained by the genomic data, which is comparable to other cardiac phenotypes (e.g. 9.6% for atrial fibrillation55).
In order to determine whether the associations of the lead variants with the phenotype were direct and not mediated via an intermediate factor, we performed tests for mediation for the covariates most strongly associated with delta age (MAP, BMI, and diagnosed hypertension). There appeared to be weak mediating effects for some of the variants, but the signal was not strong enough to remain significant after correcting for multiple tests (). However, some metadata entries in the UKB were recorded a considerable amount of time before the imaging visit when the ECG was taken and some of the covariates might have changed in the intervening period. Because of this limitation and given the large number of (genetic and environmental) factors influencing cardiovascular health and ECG morphology, it is possible that stronger mediating effects might have been missed in the present study. More research will be required in order to disentangle the network of interactions between genetic and non-genetic variables affecting cardiovascular age and its impact on the ECG.
Most of the loci discovered in our GWAS analysis have either been associated with CVD in the past or were located in the vicinity of genes involved in cardiovascular function. Functional analyses with the DEPICT enrichment analysis tool56 found significant over-representation of gene sets related to cardiac and muscle development as well as of genes expressed in the corresponding tissues. These associations were confirmed with an alternative method (gProfiler59) and grew stronger and more robust when variants with weaker association with delta age were included in the analysis (i.e. when using -value cutoffs of or ). Similarly, only a small fraction (15%) of the heritability we found could be explained by the 15 top loci. Together, these two findings suggest that many of the variants with only moderate significance might also be potential components of the genetic basis of delta age, but larger studies will be needed to verify their signal.
In addition to their links to CVD, the lead variants in most loci of genome-wide significance have also been associated with the actual shape of the ECG in a recent study24. This is a promising sign as it might help to illuminate the “black box” character of the neural network used for age prediction. In general, the knowledge about the effects of age on the ECG and the impact of genetic variants should be combined in order to aid in the interpretation of results produced by opaque deep learning models in the medical domain.
In addition to the relatively large sample sizes possible with easily obtainable phenotypes like the ECG, another interesting aspect of using metrics like delta age (or the shape of the ECG as done in24) in association studies is that they provide a relatively “dense” signal compared to binary variables (e.g. the absence or presence of a certain type of CVD – especially when the condition is rare and / or easily misdiagnosed). Similarly, using the output of artificial intelligence (AI) models trained on diagnosing such diseases from the ECG as phenotypes might improve statistical power as their predictions need not be binary (i.e. they can – to a certain extent – quantify the severity of the condition) and they might detect diseased cases that were undiagnosed in the original data.
Several different biomarkers for ageing have been proposed in the last two decades, with telomere length and the epigenetic clock arguably receiving the most attention. Despite each being a good predictor for mortality, these metrics were shown to only correlate weakly with each other, implying that they are governed by different aspects of the mechanisms of ageing8,9. We observed something similar as we did not find a strong association of variants previously linked to ageing60 or telomere length61 with delta age. We also calculated the DOSI, a blood counts-derived marker for biological ageing and physiological resilience63, for our cohort and – as opposed to the risk factor-derived “excess” heart age – correlation of the “excess” DOSI with delta age was inconclusive. More research relating different markers of biological ageing with delta age is needed, but the available evidence suggests that genetic variants associated with more general forms of ageing (e.g. in APOE, FOXO3, TERT, LMNA) have little impact on cardiovascular age compared to genes involved in the development and function of the cardiovascular system itself.
Viewed in their entirety, our findings corroborate that the ECG-derived age reflects the physiological state of the heart and that it can be used to assess cardiovascular ageing and health. Interestingly, for two of the loci with the strongest association with delta age (SIPA1L1 and VGLL2), the connection to cardiovascular phenotypes in the literature was not as clear as for many others. They therefore represent promising targets for deeper mechanistic investigation in future work. Additionally, efforts on fine-mapping will be needed to identify individual causal variants and also to confirm relevant genes since variants in linkage disequilibrium with the lead variant spanned hundreds of kilobases for some of the loci found in this study. This raises the opportunity of narrowing down the range of potential causal variants with association studies in populations of non-European ancestry.
Our work shows that genetic factors underlying cardiovascular ageing and its effect on the ECG should be incorporated into prediction models in order to improve their accuracy and interpretability. In a future of personalised medicine with readily available genomic information, the non-invasive ECG (including from wearable devices), combined with an easily obtainable measure of ECG-derived delta age, will be a valuable instrument in the clinicians’ toolkit for assessing heart health at routine examinations and monitoring treatment outcomes. Moreover, resources like the UKB, hosting an ever-increasing wealth of genomic, epigenetic, and transcriptomic data, will facilitate better comparisons as well as deeper understanding of the individual biomarkers for ageing, their underlying mechanisms, and how they complement one another. Ultimately, large-scale analysis of such data, combined with AI methodologies, will translate patient-level genomic and ECG information into preventative medicine and public health measures, leading to earlier detection of CVD and a longer healthspan.
Methods
Study population
This work has been conducted using data from the UKB, which recruited 500,000+ people aged between 40 and 69 years in 2006–2010 from across the United Kingdom64. With their informed consent, they provided detailed information about their lifestyle, had physical measures taken as well as blood, urine and saliva samples collected and stored for future analysis. We used the 10-second 12-lead ECG traces and CVD-related metadata of 37,520 participants. The ECGs were recorded during the first imaging visit (after 2014) and the metadata questionnaires were completed during the initial and first repeat assessment visits (2006–2010 and 2012–2013, respectively). All analyses were performed in accordance with relevant guidelines and regulations posed by the UKB and approved by the London School of Hygiene & Tropical Medicine ethics committee. The UKB project application reference was 54050 (www.ukbiobank.ac.uk).
Deep learning model, ECG pre-processing, and age prediction
The architecture and training procedure of the deep learning model used in this study are described in more detail in the Supplementary Methods and in the original publication11. In brief, 499,727 10-second 12-lead ECGs of patients of the Mayo clinic were used to train a convolutional neural network to predict patient age and a holdout dataset of 275,056 patients was used for testing model performance. The neural network is comprised of eight convolutional blocks in the temporal dimension, the outputs of which are combined in a single convolutional layer across the “spatial” dimension (i.e. across the 12 leads of the ECGs) with max-pooling. This is followed by two fully connected layers before being passed to the linear output layer producing the age prediction.
Due to the ECGs in the UKB being noisier than the training data, they had to undergo a filtering step prior to prediction. This was achieved using a four-pole Butterworth filter allowing frequencies from 0.5 to 100 Hz to pass. After pre-processing, ECG-derived age was predicted for 36,349 individuals in the UKB.
Metadata processing
Whenever multiple measurements of a relevant variable were available for a given sample, the mean or the value with the smallest time gap to the ECG recording was used for continuous and categorical data, respectively. MAP was calculated from systolic (SBP) and diastolic blood pressure (DBP) measurements using the equation (SBP + 2 DBP) / 3. These MAP values were then averaged with the MAP measurements derived from Pulse Wave Analysis to give the final values. The UKB contains a host of diet variables ranging from the amount of raw vegetables eaten per day to the type of fat used for cooking. We performed principal component analysis (PCA) on a selection of 24 of these variables and included the first three principal components (accounting for 25% of the total variation) as covariates in the GWAS with extended adjustment (see below).
Association testing
Pre-processing of genotype data and association testing were carried out using PLINK (v. 2.00)65. For quality control, we removed variants that either: (1) were missing in more than 1% of samples, (2) had a minor allele frequency less than 1%, (3) were not in Hardy-Weinberg Equilibrium (), or (4) had an imputation score below 0.8. Samples with more than 2% missing genotypes or that were outside of three standard deviations from the mean heterozygosity were dropped. Additionally, one sample from each closely related pair (first or second degree relations as determined by KING robust kinship inference66) was removed. The dimension of the final genotype matrix was 34,432 samples times 6,357,764 autosomal variants. PCA67 was performed on this matrix and the first 10 principal components were retained for use as covariates in the association tests.
In total, four GWAS with delta age as phenotype were carried out. The main analysis included all participants remaining after filtering and adjusted for age, sex, genotyping array, and UKB assessment centre. Additionally, in order to assess the robustness of the results, the association tests were repeated with an extended set of covariates: education (secondary, tertiary, other); smoking status (current smoker, past smoker, never / rarely smoked); alcohol consumption three or more times per week; having been diagnosed with diabetes, hypertension, angina, stroke, or heart attack in the past; BMI; MAP; LDL concentration; days of moderate exercise per week; days of vigorous exercise per week; and three principal components derived from a PCA of 24 diet variables available in the UKB. Both analyses were then repeated with the subset of participants with white British as ethnic background ().
Heritability estimation and pathway enrichment analysis
The variant-based heritability of delta age was estimated using GREML-LDMS68 implemented in GCTA (v. 1.93.2)69 while stratifying the variants based on linkage disequilibrium (four bins) and minor allele frequency (MAF) (two bins with as boundary). The analysis was carried out with both sets of covariates and later repeated with the subsets of variants found within the 15 loci of at least suggestive significance in order to also calculate the heritability of the top hits found by the GWAS. Genomic position ranges of the individual loci were calculated as part of the DEPICT workflow. DEPICT56 and gProfiler59 were used for pathway and tissue enrichment analyses. DEPICT was run on the GWAS summary statistics with and as thresholds. It uses PLINK internally to determine independent loci based on the -value threshold and a 500 kb clumping window before testing for gene set and tissue enrichment relying on data from the following databases: Gene Ontology70, KEGG57, Reactome71, InWeb72, Mouse Genome Database73, and Gene Expression Omnibus74. The coordinates of the loci found by DEPICT were additionally pasted into the gProfiler web tool, which tested for enrichment based on the Gene Ontology, KEGG, Reactome, WikiPathways75, TRANSFAC76, miRTarBase77, Human Protein Atlas78, CORUM79, and Human Phenotype Ontology80 databases.
Supplementary Information
Acknowledgements
T.G.C. was funded by the Medical Research Council UK (Grant no. MR/M01360X/1, MR/N010469/1, MR/R025576/1, MR/R020973/1, and MR/X005895/1) and a Wellcome Trust Strategic Award (Grant no. 100217/Z/12/A). D.A.L. was funded by a Wellcome Trust Strategic Award (Grant no. 100217/Z/12/A). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
D.A.L. and T.G.C. conceived the project and applied for access to the UK Biobank data. E.D.B. assisted with drafting the UK Biobank application. Z.I.A. generated the predicted age using a convolutional neural network, with the support of P.A.F. and F.L.-J. J.L.-E. and J.E.P. performed the data processing and analysis, under the supervision of T.G.C., with feedback on results from Z.I.A., S.C., F.L.-J. and D.A.L. J.L.-E. and T.G.C. wrote the first draft of the manuscript. All authors commented on versions of the manuscript and approved the final manuscript.
Data availability
All data is available from the UKB (www.ukbiobank.ac.uk).
Competing interests
P.A.F., Z.I.A., and F.L.-J. have filed intellectual property related to the AI algorithm used here to detect biological age from the ECG. The remaining authors declare no competing interests. Further information on the patent: Patent applicant: Mayo Foundation for Medical Education and Research; Names of inventors: Itzhak Zachi Attia, Paul A. Friedman, Suraj Kapa, Francisco Lopez-Jimenez; Application number: 16/960,236; Publication number: 20210361217; Status of application: pending.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-27254-z.
References
- 1.Simonson E. The effect of age on the electrocardiogram. Am. J. Cardiol. 1972;29:64–73. doi: 10.1016/0002-9149(72)90417-1. [DOI] [PubMed] [Google Scholar]
- 2.Vicent L, Martínez-Sellés M. Electrocardiogeriatrics: ECG in advanced age. J. Electrocardiol. 2017;50:698–700. doi: 10.1016/j.jelectrocard.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Ladejobi AO, et al. The 12-lead electrocardiogram as a biomarker of biological age. Eur. Heart J. Digital Health. 2021;2:379–389. doi: 10.1093/ehjdh/ztab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn EH, Epel ES, Lin J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350:1193–1198. doi: 10.1126/science.aab3389. [DOI] [PubMed] [Google Scholar]
- 5.Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: A systematic review and meta-analysis. Clin. Epigenet. 2019;11:1–17. doi: 10.1186/s13148-019-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine ME. Modeling the rate of senescence: Can estimated biological age predict mortality more accurately than chronological age? J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2013;68:667–674. doi: 10.1093/gerona/gls233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters MJ, et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 2015;6:1–14. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belsky DW, et al. Eleven telomere, epigenetic clock, and biomarker-composite quantifications of biological aging: Do they measure the same thing? Am. J. Epidemiol. 2018;187:1220–1230. doi: 10.1093/aje/kwx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yazdanyar A, Newman AB. The burden of cardiovascular disease in the elderly: Morbidity, mortality, and costs. Clin. Geriatr. Med. 2009;25:563. doi: 10.1016/j.cger.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attia ZI, et al. Age and sex estimation using artificial intelligence from standard 12-lead ECGs. Circ. Arrhythm. Electrophysiol. 2019;12:e007284. doi: 10.1161/CIRCEP.119.007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ball RL, Feiveson AH, Schlegel TT, Starc V, Dabney AR. Predicting, heart age using electrocardiography. J. Personalized Med. 2014;4:65–78. doi: 10.3390/jpm4010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah AP, Rubin SA. Errors in the computerized electrocardiogram interpretation of cardiac rhythm. J. Electrocardiol. 2007;40:385–390. doi: 10.1016/j.jelectrocard.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Lima EM, et al. Deep neural network estimated electrocardiographic-age as a mortality predictor. Nat. Commun. 2021;12:5117. doi: 10.1038/s41467-021-25351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khurshid S, et al. ECG-based deep learning and clinical risk factors to predict atrial fibrillation. Circulation. 2022;145:122–133. doi: 10.1161/CIRCULATIONAHA.121.057480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannun AY, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribeiro AH, et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-15432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon J-M, et al. Comparing the performance of artificial intelligence and conventional diagnosis criteria for detecting left ventricular hypertrophy using electrocardiography. EP Europace. 2020;22:412–419. doi: 10.1093/europace/euz324. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, et al. Vital signs: Predicted heart age and racial disparities in heart age among US adults at the state level. Morb. Mortal. Wkly Rep. 2015;64:950–958. doi: 10.15585/mmwr.mm6434a6. [DOI] [PubMed] [Google Scholar]
- 20.D’Agostino RB, Sr, et al. General cardiovascular risk profile for use in primary care: The Framingham Heart Study. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 21.Mills MC, Rahal C. A scientometric review of genome-wide association studies. Commun. Biol. 2019;2:1–11. doi: 10.1038/s42003-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ntalla I, et al. Multi-ancestry GWAS of the electrocardiographic PR interval identifies 202 loci underlying cardiac conduction. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norland K, et al. Sequence variants with large effects on cardiac electrophysiology and disease. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-12682-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verweij N, et al. The genetic makeup of the electrocardiogram. Cell Syst. 2020;11:229–238. doi: 10.1016/j.cels.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radhakrishnan, A. et al. A cross-modal autoencoder framework learns holistic representations of cardiovascular state. bioRxiv (2022). [DOI] [PMC free article] [PubMed]
- 26.Attia ZI, Lerman G, Friedman PA. Deep neural networks learn by using human-selected electrocardiogram features and novel features. Eur. Heart J. Digital Health. 2021;2:446–455. doi: 10.1093/ehjdh/ztab060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buniello A, et al. The NHGRI-EBIws GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer HV, et al. Genetic and functional insights into the fractal structure of the heart. Nature. 2020;584:589–594. doi: 10.1038/s41586-020-2635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P-J, et al. Vasopressin-induced serine 269 phosphorylation reduces Sipa1l1 (signal-induced proliferation-associated 1 like 1)-mediated aquaporin-2 endocytosis. J. Biol. Chem. 2017;292:7984–7993. doi: 10.1074/jbc.M117.779611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, et al. RGS6, a modulator of parasympathetic activation in heart. Circ. Res. 2010;107:1345–1349. doi: 10.1161/CIRCRESAHA.110.224220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda M, et al. Vestigial-like 2 contributes to normal muscle fiber type distribution in mice. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-07149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Checchia PA, et al. Myocardial transcriptional profiles in a murine model of sepsis: Evidence for the importance of age. Pediatr. Crit. Care Med. 2008;9:530–535. doi: 10.1097/PCC.0b013e3181849a2f. [DOI] [PubMed] [Google Scholar]
- 33.Ali ZA, et al. Oxido-reductive regulation of vascular remodeling by receptor tyrosine kinase ROS1. J. Clin. Investig. 2014;124:5159–5174. doi: 10.1172/JCI77484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayer KU, Schulman H. CaM kinase: Still inspiring at 40. Neuron. 2019;103:380–394. doi: 10.1016/j.neuron.2019.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattiazzi A, et al. Chasing cardiac physiology and pathology down the CaMKII cascade. Am. J. Physiol. Heart Circ. Physiol. 2015;308:H1177–H1191. doi: 10.1152/ajpheart.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shelley JR, Davidson DJ, Dorin JR. The dichotomous responses driven by -Defensins. Front. Immunol. 2020;11:1176. doi: 10.3389/fimmu.2020.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadati T, Houben T, Bitorina A, Shiri-Sverdlov R. The ins and outs of cathepsins: Physiological function and role in disease management. Cells. 2020;9:1679. doi: 10.3390/cells9071679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aggarwal N, Sloane BF. Cathepsin B: Multiple roles in cancer. PROTEOMICS Clin. Appl. 2014;8:427–437. doi: 10.1002/prca.201300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blondelle J, Lange S, Greenberg BH, Cowling RT. Cathepsins in heart disease-chewing on the heartache? Am. J. Physiol. Heart Circ. Physiol. 2015;308:H974–H976. doi: 10.1152/ajpheart.00125.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mareti A, et al. Cathepsin B expression is associated with arterial stiffening and atherosclerotic vascular disease. Eur. J. Prev. Cardiol. 2020;27:2288–2291. doi: 10.1177/2047487319893042. [DOI] [PubMed] [Google Scholar]
- 41.Wyczalkowska-Tomasik A, Paczek L. Cathepsin B and L activity in the serum during the human aging process: Cathepsin B and L in aging. Arch. Gerontol. Geriatr. 2012;55:735–738. doi: 10.1016/j.archger.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 42.LeWinter MM, Granzier H. Cardiac titin: A multifunctional giant. Circulation. 2010;121:2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tharp CA, Haywood ME, Sbaizero O, Taylor MR, Mestroni L. The giant protein titin’s role in cardiomyopathy: Genetic, transcriptional, and post-translational modifications of TTN and their contribution to cardiac disease. Front. Physiol. 2019;10:1436. doi: 10.3389/fphys.2019.01436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remme C, et al. The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res. Cardiol. 2009;104:511–522. doi: 10.1007/s00395-009-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verkerk AO, et al. Functional Nav1.8 channels in intracardiac neurons: The link between SCN10A and cardiac electrophysiology. Circ. Res. 2012;111:333–343. doi: 10.1161/CIRCRESAHA.112.274035. [DOI] [PubMed] [Google Scholar]
- 46.Li W, et al. SCN5A variants: Association with cardiac disorders. Front. Physiol. 2018;9:1372. doi: 10.3389/fphys.2018.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu D, et al. Mutations in SCN10A are responsible for a large fraction of cases of Brugada syndrome. J. Am. Coll. Cardiol. 2014;64:66–79. doi: 10.1016/j.jacc.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong Y, et al. KLHL3 regulates paracellular chloride transport in the kidney by ubiquitination of claudin-8. Proc. Natl. Acad. Sci. 2015;112:4340–4345. doi: 10.1073/pnas.1421441112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glover M, et al. Detection of mutations in KLHL3 and CUL3 in families with FHHt (familial hyperkalaemic hypertension or Gordon’s syndrome) Clin. Sci. 2014;126:721–726. doi: 10.1042/CS20130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Lai G, Chu G, Liang X, Zhao Y. cMyBP-C was decreased via KLHL3-mediated proteasomal degradation in congenital heart diseases. Exp. Cell Res. 2017;355:18–25. doi: 10.1016/j.yexcr.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 51.Hsu J, et al. Genetic control of left atrial gene expression yields insights into the genetic susceptibility for atrial fibrillation. Circ. Genomic Precis. Med. 2018;11:e002107. doi: 10.1161/CIRCGEN.118.002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Dube DK, Mittal B, Sanger JM, Sanger JW. Myotilin dynamics in cardiac and skeletal muscle cells. Cytoskeleton. 2011;68:661–670. doi: 10.1002/cm.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olivé M, Kley RA, Goldfarb LG. Myofibrillar myopathies: New developments. Curr. Opin. Neurol. 2013;26:527. doi: 10.1097/WCO.0b013e328364d6b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lahrouchi N, et al. Transethnic genome-wide association study provides insights in the genetic architecture and heritability of long QT syndrome. Circulation. 2020;142:324–338. doi: 10.1161/CIRCULATIONAHA.120.045956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen JB, et al. Genome-wide study of atrial fibrillation identifies seven risk loci and highlights biological pathways and regulatory elements involved in cardiac development. Am. J. Hum. Genet. 2018;102:103–115. doi: 10.1016/j.ajhg.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:1–9. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith CL, Eppig JT. The mammalian phenotype ontology: Enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raudvere U, et al. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pilling LC, et al. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging (Albany NY) 2017;9:2504. doi: 10.18632/aging.101334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Codd V, et al. Polygenic basis and biomedical consequences of telomere length variation. Nat. Genet. 2021;53:1425–1433. doi: 10.1038/s41588-021-00944-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang P, et al. II spectrin (SPTBN1): Biological function and clinical potential in cancer and other diseases. Int. J. Biol. Sci. 2021;17:32. doi: 10.7150/ijbs.52375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pyrkov TV, et al. Longitudinal analysis of blood markers reveals progressive loss of resilience and predicts human lifespan limit. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-021-23014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sudlow C, et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:015–s13742. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galinsky KJ, et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016;98:456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Croft D, et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2010;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 73.Blake JA, et al. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrett T, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slenter DN, et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46:D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matys V, et al. TRANSFAC®and its module TRANSCompel®: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chou C-H, et al. miRTarBase update 2018: A resource for experimentally validated microRNA- target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 79.Giurgiu M, et al. CORUM: The comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47:D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Köhler S, et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019;47:D1018–D1027. doi: 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Church DM, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available from the UKB (www.ukbiobank.ac.uk).