Abstract
Motivation
Gene set enrichment analysis (GSEA) is a commonly used algorithm for characterizing gene expression changes. However, the currently available tools used to perform GSEA have a limited ability to analyze large datasets, which is particularly problematic for the analysis of single-cell data. To overcome this limitation, we developed a GSEA package in Python (GSEApy), which could efficiently analyze large single-cell datasets.
Results
We present a package (GSEApy) that performs GSEA in either the command line or Python environment. GSEApy uses a Rust implementation to enable it to calculate the same enrichment statistic as GSEA for a collection of pathways. The Rust implementation of GSEApy is 3-fold faster than the Numpy version of GSEApy (v0.10.8) and uses >4-fold less memory. GSEApy also provides an interface between Python and Enrichr web services, as well as for BioMart. The Enrichr application programming interface enables GSEApy to perform over-representation analysis for an input gene list. Furthermore, GSEApy consists of several tools, each designed to facilitate a particular type of enrichment analysis.
Availability and implementation
The new GSEApy with Rust extension is deposited in PyPI: https://pypi.org/project/gseapy/. The GSEApy source code is freely available at https://github.com/zqfang/GSEApy. Also, the documentation website is available at https://gseapy.rtfd.io/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) is a commonly used method for assessing whether a gene expression pattern measured in a test set of cells or tissues is similar to one previously found in other cell types, disease conditions or treatment responses. For example, GSEA has been used to identify genes/pathways associated with treatment response or disease prognosis (Verstockt et al., 2019; Wang et al., 2019; Labrecque et al., 2019), and to identify stem cell signatures in human cancer tissues (Merlos-Suarez et al., 2011; Corominas-Faja et al., 2013). We previously used GSEA to demonstrate that the collagen-producing myofibroblasts in mutated hepatic organoids were similar to those in fibrotic liver tissue obtained from patients with liver cancer or commonly occurring fibrotic liver diseases (Guan et al., 2021). GSEA calculates a normalized enrichment score (NES), which indicates how similar the differentially expressed gene sets are between the test and comparator datasets. However, single-cell RNA-sequencing (scRNA-Seq) is now commonly used for transcriptomic characterization of cells, organoids and tissues. When the datasets generated from scRNA-Seq are huge and provide large amounts of information, the computational challenges associated with them increase (Kiselev et al., 2019; Lakkis et al., 2019). Although there have been several different implementations can be used for GSEA analysis, such as GSEA-R (Subramanian et al., 2005), GSEA-P (Subramanian et al., 2007), fGSEA (Korotkevich et al., 2021), only GSEApy (released in early 2017) and a recently published tool, named blitzGSEA (Lachmann et al., 2022), are available for Python computing environment. Furthermore, GSEApy ships with additional features that facilitate enrichment analysis, including an application programming interface (API) to Enrichr web service (Chen et al., 2013; Kuleshov et al., 2016; Xie et al., 2021), an API to BioMart web service (Durinck et al., 2005), the single sample GSEA methodology (Barbie et al., 2009) and a utility for gene set over-representation analysis. GSEApy was implemented by Numpy with process-based parallel computing support initially. However, this implementation is not well optimized in speed and has limited utility for analyzing larger scRNA-seq datasets that require memory of more than 32 GB (Lachmann et al., 2022).
To enable enrichment analysis to be performed on large-scale data, we re-implemented GSEApy in a high-performance programming language (Rust). It provides better memory security, comparable speed to that of C/C++ and the same API as the previous version (≤ v0.10.8). Furthermore, the Rust extension of GSEApy can take full advantage of modern computers' multi-threading parallel computing capabilities because Python’s global interpreter lock was released. When small gene set libraries are analyzed (e.g. 278 pathways in this article), it runs 3-fold faster but with four times less memory usage (Supplementary Fig. S1A). For large gene set libraries (e.g. 2860 pathways in this article), it can run 80-fold faster without much more memory (Supplementary Fig. S1B).
2 Implementation
The GSEApy internal is written in Rust, and it consists of six tools:
gsea: Perform enrichment analysis using the GSEA statistical methodology.
prerank: Pre-ranked GSEA, particularly for datasets with a small number of replicates.
ssgsea: Single sample GSEA (ssGSEA) methodology.
replot: Re-generate enrichment plots from the GSEA-P output files.
enrichr: Perform an over-representation analysis on a list of genes. It supports several organisms, including human, mouse, fly, yeast, zebrafish and C.elegans.
biomart: Convert gene ids with the BioMart API.
The GSEApy will automatically obtain gene set libraries from the Enrichr web service for analysis. To facilitate the interpretation of enrichment results, GSEApy provides several visualization methods (gseaplot, heatmap, dotplot, barplot and ringplot). We also provide a complete documentation website, including a user guide describing all of these tools, examples and frequently asked questions.
3 Results
3.1 Computational efficiency improvement by Rust
The most computationally expensive part of GSEA is generating the null distribution of Enrichment Scores (ES) for the P-value calculation. GSEA performs random permutations of either the samples or the gene labels to obtain the null distribution. One thousand permutations are set as the default parameter, which produces 1000 ES. An observed ES is then compared with the 1000 shuffled ES to calculate a P-value. We previously implemented a Numpy version that performed the GSEA. However, the Numpy version consumed a huge amount of memory while using multi-CPUs, an issue reported by many GSEApy users. To address this, we re-implemented the GSEA algorithm in Rust. When we compared the time and memory cost between the Numpy (v0.10.8) and Rust version with a gene permutation experiment (22 922 genes, 278 or 2860 pathways, 1000 permutations), we found that the Rust implementation was 3- to 80-fold faster in run time with one thread (Supplementary Fig. S1A). The run time with eight threads was decreased to a few minutes for both versions, but the Rust version was still 2-fold faster, and the memory cost was reduced from 50 GB to 1.4 GB (for the 2860 pathways) (Supplementary Fig. S1B). We also compared the execution speed of fGSEA, blitzGSEA, GSEA-P, GSEA-R and GSEApy on a single thread (Supplementary Fig. S1C). While it took over 1 hour for GSEA-R to calculate the statistics for 2860 pathways, GSEA-P and GSEApy took 3.7 and 5.1 minutes, respectively. fGSEA and blitzGSEA outperformed the other tools since their execution time was <1 minute. In a sample permutation experiment, the Rust binding version was almost 3-fold faster than GSEA-R (Supplementary Fig. S1D), and the Numpy and Rust versions had similar memory costs for small datasets (Supplementary Fig. S1E).
3.2 Enrichment analysis and data visualization
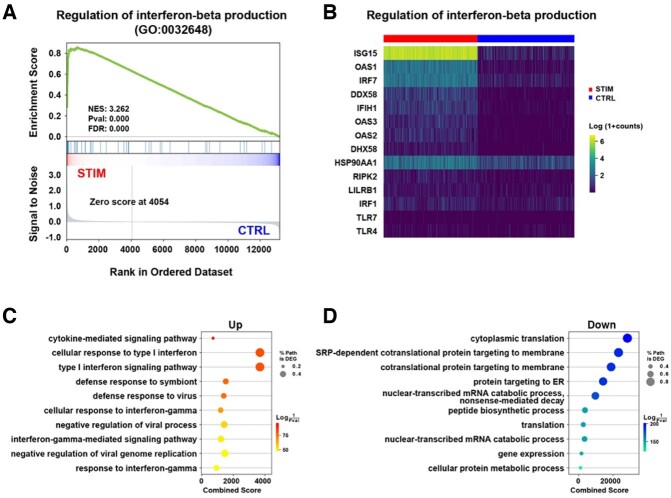
The improvement in computational efficiency enables GSEApy to perform enrichment analysis for large-scale samples. To illustrate how GSEApy could be used for biological discovery, we analyzed a publicly available scRNA-seq dataset (GSE96583) of peripheral blood mononuclear cells (PBMCs) obtained from control and interferon β-treated subjects (Kang et al., 2018). After generating the log normalized counts using Scanpy (Wolf et al., 2018), we analyzed the annotated subset of CD14+ monocytes (2215 control and 2147 stimulated cells). We performed GSEA (1000 permutations, sample permutation and 6036 pathways) on the 4362 transcriptomes of the CD14+ monocytes. The interferon β production pathway was significantly enriched (P-value < 0.01, False Discovery Rate (FDR) < 0.01, NES = 3.262) in the stimulated CD14+ monocytes (Fig. 1A and B). Next, we identified 1717 differentially expressed genes (DEGs) (Wilcoxon test with FDR < 0.05) using the rank_genes_groups function in Scanpy. An over-representation analysis (GSEApy’s Enrichr API) was performed on the 687 up-regulated and the 1030 down-regulated DEGs. The up-regulated genes indicated that the interferon signature was activated in the interferon β-stimulated cells (Fig. 1C), while the down-regulated genes were associated with general cellular functions, such as protein translation (Fig. 1D). This analysis confirms the effect of interferon treatment, which indicates that this dataset can be used for subsequent investigations.
Fig. 1.
An example of enrichment analysis performed using GSEApy. (A) GSEA enrichment plot of the regulation of interferon-beta production pathway. (B) A heatmap shows the leading-edge genes that appear in the ranked list at or before the point at which the running ES reaches its maximum deviation from zero identified in (A). (C, D) over-representation analysis shows the top 10 significantly enriched pathways of up-regulated genes (C) and down-regulated genes (D), respectively. % Path is DEGs, the percentage of DEGs that overlapped with the pathway of interest. Combined score is defined by the Enrichr (Xie et al., 2021)
4 Conclusion
GSEApy provides a fast and straightforward way to perform enrichment analysis. It requires minimal arguments, provides clear output and operates from both the command line and Python environment, which maximizes the ease of use, accommodates novice programmers and supports large-scale dataset analysis. It also can quickly generate high-quality, publication-ready plots. GSEApy will be one of the fundamental packages for enrichment analysis in Python.
Supplementary Material
Contributor Information
Zhuoqing Fang, Department of Anesthesia, Pain and Perioperative Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Xinyuan Liu, Department of Otolaryngology-Head and Neck Surgery, Stanford University School of Medicine, Stanford, CA 94305, USA.
Gary Peltz, Department of Anesthesia, Pain and Perioperative Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Funding
This work was supported by a National Institute of Health (National Institute for Drug Addiction) award [5U01DA04439902 to G.P.]
Conflict of Interest: The authors declare that they have no competing interests.
Data availability
The GSEApy source code is freely available at https://github.com/zqfang/GSEApy.
References
- Barbie D.A. et al. (2009) Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature, 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y. et al. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas-Faja B. et al. (2013) Stem cell-like ALDH(bright) cellular states in EGFR-mutant non-small cell lung cancer: a novel mechanism of acquired resistance to erlotinib targetable with the natural polyphenol silibinin. Cell Cycle, 12, 3390–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S. et al. (2005) BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics, 21, 3439–3440. [DOI] [PubMed] [Google Scholar]
- Guan Y. et al. (2021) A human multi-lineage hepatic organoid model for liver fibrosis. Nat. Commun., 12, 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M. et al. (2018) Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol., 36, 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev V.Y. et al. (2019) Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet., 20, 273–282. [DOI] [PubMed] [Google Scholar]
- Korotkevich G. et al. (2021) Fast gene set enrichment analysis. bioRxiv. 10.1101/060012. [DOI]
- Kuleshov M.V. et al. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res., 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrecque M.P. et al. (2019) Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Invest., 129, 4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A. et al. (2022) blitzGSEA: efficient computation of gene set enrichment analysis through gamma distribution approximation. Bioinformatics, 38, 2356–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkis J. et al. (2021) A joint deep learning model enables simultaneous batch effect correction, denoising and clustering in single-cell transcriptomics. Genome Res., 31, 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suarez A. et al. (2011) The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell, 8, 511–524. [DOI] [PubMed] [Google Scholar]
- Subramanian A. et al. (2007) GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics, 23, 3251–3253. [DOI] [PubMed] [Google Scholar]
- Subramanian A. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstockt B. et al. (2019) Expression levels of 4 genes in Colon tissue might be used to predict which patients will enter endoscopic remission after vedolizumab therapy for inflammatory bowel diseases. Clin. Gastroenterol. Hepatol., 18, 1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. (2019) Identification of seven-gene signature for prediction of lung squamous cell carcinoma. Onco. Targets Ther., 12, 5979–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A. et al. (2018) SCANPY: large-scale single-cell gene expression data analysis. Genome Biol., 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. et al. (2021) Gene set knowledge discovery with Enrichr. Curr. Protoc., 1, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GSEApy source code is freely available at https://github.com/zqfang/GSEApy.