Abstract
The rodent-borne hantavirus Puumala virus (PUUV) and related agents cause hemorrhagic fever with renal syndrome (HFRS) in humans. Other hantaviruses, including Andes virus (ANDV) and Sin Nombre virus, cause a distinct zoonotic disease, hantavirus cardiopulmonary syndrome (HCPS). Although these infections are severe and have significant case fatality rates, no FDA-approved hantavirus countermeasures are available. Recent work suggests that monoclonal antibodies may have therapeutic utility. We describe here the isolation of human neutralizing monoclonal antibodies (nAbs) against tetrameric Gn/Gc glycoprotein spikes from PUUV-experienced donors. We define a dominant class of nAbs recognizing the ‘capping loop’ of Gn that masks the hydrophobic fusion loops in Gc. A subset of nAbs in this class, including ADI-42898, bound Gn/Gc complexes but not Gn alone, strongly suggesting that they recognize a quaternary epitope encompassing both Gn and Gc. ADI-42898 blocked cell entry of seven HCPS- and HFRS-associated hantaviruses and single doses of this nAb could protect Syrian hamsters and bank voles challenged with the highly virulent HCPS-causing ANDV and HFRS-causing PUUV, respectively. ADI-42898 is a promising candidate for clinical development as a countermeasure for both HCPS and HFRS, and its mode of Gn/Gc recognition informs the development of broadly protective hantavirus vaccines.
ONE SENTENCE SUMMARY
Human neutralizing antibodies recognizing the viral glycoprotein complex are broadly protective against orthohantaviruses.
INTRODUCTION
Rodent-borne orthohantaviruses (hantaviruses) are a large, globally distributed group of enveloped viruses with segmented negative-strand RNA genomes. Hantavirus infections are associated with over 50,000 annual diagnosed cases of disease worldwide, primarily caused by rodent-to-human transmission (1) but also by direct human-to-human contact (2). The ‘New World’ hantaviruses Andes virus (ANDV) and Sin Nombre virus (SNV) are the major etiologic agents of hantavirus cardiopulmonary syndrome (HCPS) in South and North America, respectively. Case-fatality rates for both viruses are the highest among hantaviruses, reaching ~40% with cardiogenic shock and respiratory failure being the primary causes of death (3). The ‘Old World’ hantavirus Puumala virus (PUUV) and related agents, including Dobrava-Belgrade and Hantaan viruses are endemic in different regions of Europe and Asia or worldwide (Seoul virus) and cause hemorrhagic fever with renal syndrome (HFRS) with case-fatality rates of up to ~15% (4). A third of PUUV-infected individuals require hospitalization, and 5% require intensive-care stays due to shock, hemorrhage, and cardiopulmonary complications (5). Unexpectedly, large PUUV outbreaks in Sweden, Finland, and Germany between 2007–2021 have had sizeable socioeconomic impacts in endemic regions, where the incidence may exceed 50–300 cases per 100,000 residents annually (5-7). A SNV outbreak among overnight visitors in California’s Yosemite National Park in 2012 (8), and a singular ANDV super-spreader outbreak in Argentina in 2018–19 (2, 9) highlight the public health risks posed by hantaviruses. Compounding these concerns is the unknown potential for emergence of novel hantaviruses into human populations from rodent or non-rodent reservoirs with unpredictable disease and epidemic potential (10). The lack of FDA-approved or emergency use-authorized vaccines and therapeutics represents a critical gap in our preparedness for a large hantavirus outbreak and highlights the need to preposition broad anti-hantavirus countermeasures for human use.
The medium (M) segment of the hantavirus genome encodes the glycoprotein precursor, which is cotranslationally cleaved to generate N-terminal (Gn) and C-terminal (Gc) subunits that associate to form square-shaped heterotetrameric (Gn/Gc)4 spikes (11, 12). Lateral spike interactions mediated by Gc are proposed to drive (Gn/Gc)4 lattice formation and viral membrane budding. Progeny virions display a partial surface lattice of spikes (11, 13, 14). A globular membrane-distal N-terminal Gn domain is highly exposed at the viral surface and is proposed to contain the receptor-binding site (13-15). Gn also closely associates with and regulates the pre-fusion conformation of Gc, a ‘Class II’ membrane fusion protein resembling the envelope glycoproteins of other members of the order Bunyavirales (‘bunyaviruses’), flaviviruses, and alphaviruses (14, 16, 17). During cell entry, endosomal acid pH triggers Gn/Gc dissociation, allowing Gc subunits to insert into the target membrane and refold into post-fusion homotrimers, thereby driving viral membrane fusion and cytoplasmic escape (15, 18-21). Gn/Gc spikes thus provide the primary target for antibodies that mediate their neutralizing activity by blocking one or more steps in the hantavirus entry pathway.
Recent FDA approval of monoclonal antibody (mAb) countermeasures against Ebola virus and emergency use authorization (EUA) approval of mAbs for COVID-19 validate passive immunization as a strategy for viral diseases. Accumulating evidence provides rationale for the development of mAb-based hantavirus therapeutics. The kinetics and magnitude of the human neutralizing antibody (nAb) response to natural hantavirus infection can influence the severity of HCPS and HFRS (22-24), and the transfusion of convalescent hyperimmune plasma appears to be of benefit to acute HCPS patients (25). In addition, two recent studies have isolated spike-specific mAbs from ANDV- and/or SNV-experienced HCPS-convalescent donors and identified several that could protect Syrian hamsters from lethal ANDV challenge (26, 27). Most of the protective lead mAbs in these reports recognized and/or neutralized Gn/Gc in a virus-specific manner, and Engdahl and colleagues described several nAbs isolated from SNV-experienced donors with activity against members of both Old World and New World clades (hereafter, cross-clade activity; see Fig. 1A) (26). Two of these nAbs partially protected Syrian hamsters from lethal ANDV challenge, but in vivo protective efficacy against HFRS-causing hantaviruses was not evaluated. Their epitopes were not determined, and thus, the molecular bases of cross-clade Gn/Gc binding, neutralization, and in vivo protection by these rare, broadly active nAbs remain unknown.
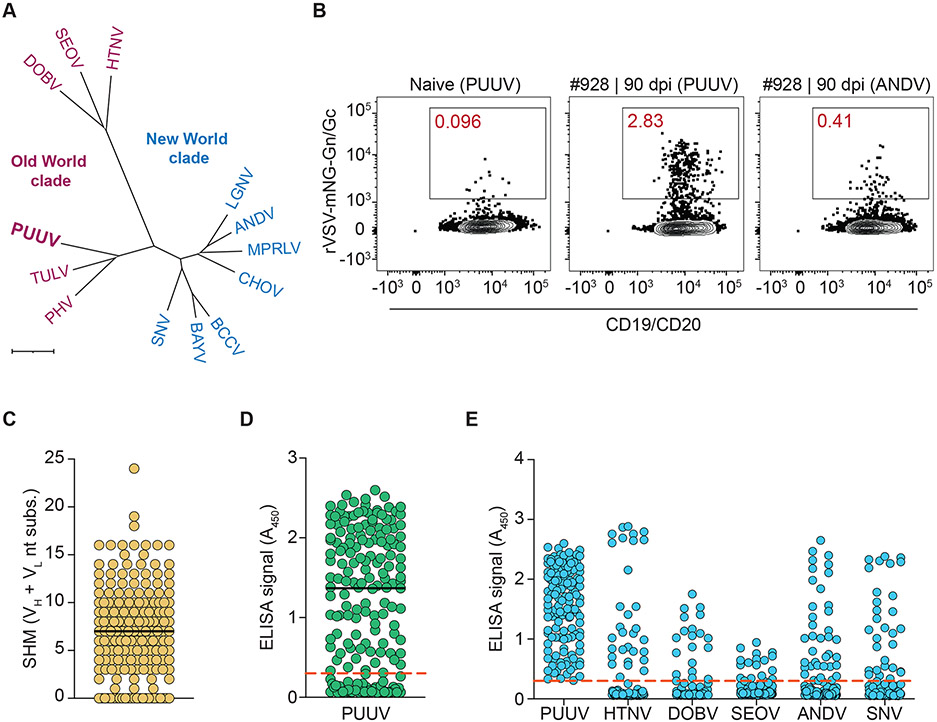
Fig. 1: Isolation and characterization of anti-Gn/Gc mAbs from PUUV-experienced donors.
(A) Phylogenetic tree of the M segment-encoded GPC sequences from Old World and New World hantaviruses (OWH/NWH) based on amino acid (aa) sequences shown in Table S1. ANDV, Andes virus; BAYV, Bayou virus; BCCV, Black Creek Canal virus; CHOV, Choclo virus; DOBV, Dobrava-Belgrade virus; HTNV, Hantaan virus; LGNV, Laguna Negra virus; MPRLV, Maporal virus; PHV, Prospect Hill virus; PUUV, Puumala virus; SEOV, Seoul virus; SNV, Sin Nombre virus; TULV, Tula virus. Scale bar represents 0.07 aa substitutions per site. (B) Frequency of hantavirus Gn/Gc-reactive B cells isolated with rVSV-mNG particles bearing PUUV or ANDV Gn/Gc from PUUV-naïve and PUUV-experienced donors. Fluorescence-activated cell sorting plots are gated on CD19+/CD20+/IgD−/IgM−/virus+ B cells. (C) Somatic hypermutation (SHM) load as determined by the number of VH and VL nucleotide substitutions away from the predicted VH and VL germline. Bar indicates median. (D) Binding reactivity of 180 isolated mAbs (25 nM) to rVSV-PUUV-Gn/Gc, as determined by ELISA. A450, absorbance at 450 nm. The red dashed line indicates the threshold for designating binders (A450=0.3). Bar indicates median. Averages for PUUV Gn/Gc binders (n=4) from two experiments. (E) Binding cross-reactivity of 135 isolated mAbs (25 nM) to rVSVs bearing Gn/Gc from the indicated viruses, as determined by ELISA. The red dashed line indicates the threshold for designating binders (A450=0.3). ANDV, SNV: averages (n=2) from one experiment. Other viruses, averages (n=4) from two experiments.
Given the cost and logistical challenges inherent in developing antiviral treatments against emerging hantaviruses, a broadly protective therapeutic is highly desirable. Accordingly, we sought to identify one or more single nAbs that could protect against divergent hantaviruses. We reasoned that the exclusive focus on New World hantavirus-experienced donors in these previous studies may have limited the discovery of nAbs that could also target Old World hantaviruses. Interestingly, a Gn/Gc sequence alignment revealed that the Old World hantavirus, PUUV, genetically bisects other Old World and New World hantaviruses (Fig. 1A and Table S1), raising the possibility that human exposure to PUUV may engender distinct, and likely cross-reactive, antibody specificities. To test this possibility, we isolated mAbs from PUUV-experienced donors from a Swedish HFRS-convalescent cohort. We discovered 135 mAbs with varying degrees of cross-clade reactivity and neutralizing activity that bound diverse epitopes in Gn/Gc. A single mAb, ADI-42898, afforded cross-clade neutralization breadth and recognized a previously unknown epitope at a Gn/Gc intersubunit interface. ADI-42898 could protect against both ANDV in Syrian hamsters and PUUV in a newly established bank vole challenge model (28) and is a promising candidate for clinical development as a broad countermeasure against HCPS and HFRS.
RESULTS
Isolation of monoclonal antibodies from PUUV-experienced donors
We screened sera from a cohort of 45 Swedish patients with clinically documented PUUV exposure to identify donors for single B cell sorting (Table S2A). Serum nAb titers were determined with recombinant vesicular stomatitis viruses (rVSV) displaying hantavirus Gn/Gc (29-31). We identified six donors with potent and broad serum nAb responses against rVSVs bearing Gn/Gc from PUUV, a divergent Old World hantavirus, Hantaan virus (HTNV), and a New World hantavirus, ANDV (Fig. S1 and Table S2B).
Recombinant hantaviral Gn/Gc proteins that recapitulate the tetrameric structure of the virion-bound glycoprotein complex are not currently available (11, 32). To sample antibody specificities recognizing conformational and quaternary epitopes, we used entry-competent rVSV-Gn/Gc particles bearing fluorescent mNeonGreen-labeled cores (rVSV-mNG-Gn/Gc) (29, 33) as baits for single-cell sorting of class-switched (swIg+), antigen-specific B cells (CD19+/IgM−/IgD−/virus+) from peripheral blood. Across donors, up to 2.8% of swIg+ B cells specifically bound rVSV-mNG-PUUV-Gn/Gc, whereas a smaller fraction (~0.4%) was reactive to rVSV-mNG-ANDV-Gn/Gc, which was concordant with our donors’ exposure history, the sequence divergence between these glycoproteins, and the lower but detectable cross-neutralizing serum activity against ANDV Gn/Gc (Figs. 1B, S1, S2 and Table S1B). Cognate antibody heavy- and light-chain pairs were recovered from 180 individual hantavirus Gn/Gc-reactive B cells and found to contain somatic hypermutations (SHMs) consistent with their memory B cell origin (Fig. 1C). Full-length immunoglobulin G subclass I (IgG1) molecules bearing these variable region sequences were expressed in an engineered strain of Saccharomyces cerevisiae (34) and screened by an enzyme-linked immunosorbent assay (ELISA) to identify 135 PUUV Gn/Gc-specific binders (Fig. 1D). The mAbs were further evaluated for their capacity to recognize rVSV particles bearing hantavirus Gn/Gc proteins from both Old World (HTNV, Seoul virus [SEOV], Dobrava-Belgrade virus [DOBV]), and New World (ANDV, SNV) hantaviruses by ELISA (29-31). Consistent with our serum activity studies (Fig. S1) and the PUUV M segment’s genetically intermediate position in Orthohantavirus, we observed a similar degree of binding cross-reactivity between PUUV and viruses belonging to either Old World or New World hantavirus clades (Fig. 1E; 14–28%).
Identification of mAbs with potent and broad neutralizing activity
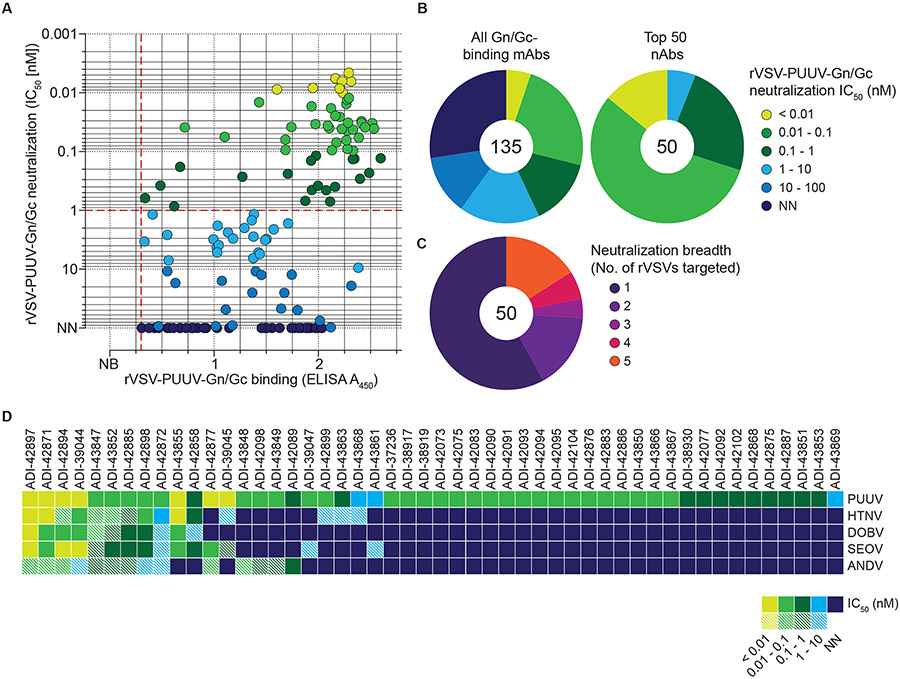
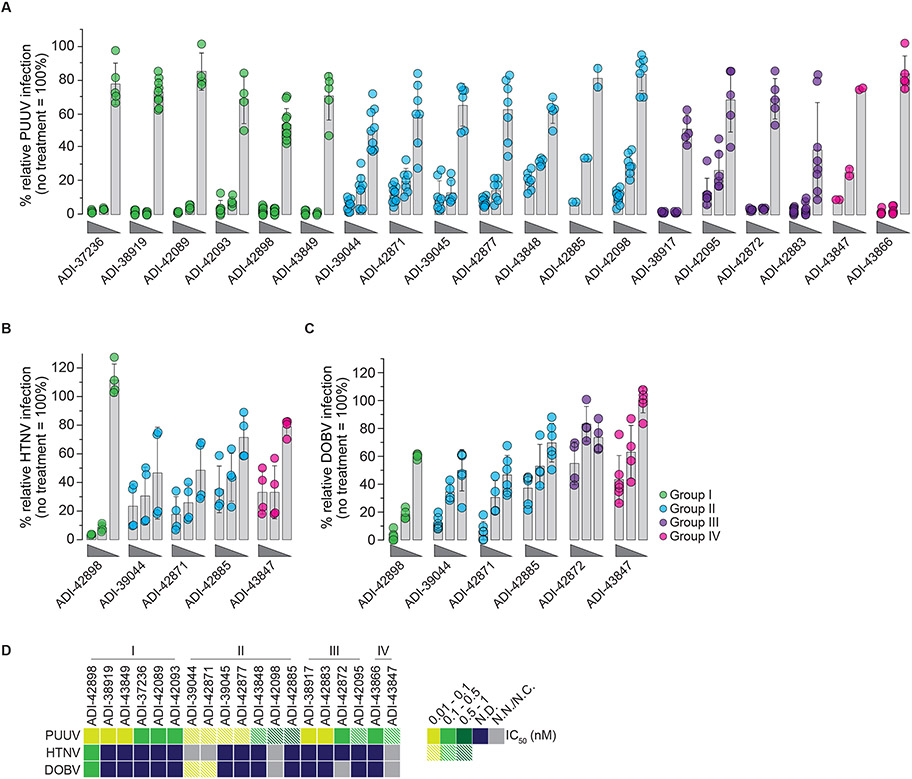
We screened the 135 PUUV Gn/Gc-reactive mAbs for neutralizing activity against rVSV-mNG-PUUV-Gn/Gc (Figs. 2A-B). 58 of these mAbs displayed half-maximal inhibitory concentration (IC50) values of <1 nM, and most bound efficiently to rVSV-PUUV-Gn/Gc particles (Fig. 2A). We evaluated the 50 most potent nAbs against rVSVs bearing divergent Gn/Gc glycoproteins from both Old World (HTNV, SEOV, DOBV) and New World hantaviruses (ANDV) (Figs. 2C-D). Although most of the nAbs (28/50) were PUUV-specific neutralizers, the remaining exhibited varying levels of neutralization breadth. 13 nAbs displayed neutralization activity against at least three hantaviruses with ten affording cross-clade neutralization, indicating that they recognize highly conserved epitopes. We noted, however, the existence of an infectious fraction of virus refractory to neutralization (‘un-neutralized fraction’ >5%, Figs. 2D and S3) that persisted even at the highest tested concentrations for some nAbs, suggesting either reduced nAb:Gn/Gc binding affinities and/or the existence of Gn/Gc subpopulations in viral particles with cryptic epitopes unavailable for antibody binding.
Fig. 2: Neutralization activity of PUUV Gn/Gc-reactive mAbs.
(A) Binding activity of mAbs (25 nM) to rVSV-PUUV-Gn/Gc, as determined by ELISA (see Fig. 1D), plotted against IC50 values calculated from rVSV-PUUV-Gn/Gc neutralization curves displayed as averages (n≥4) from at least two experiments (also see Fig. S3). Data points are colored according to neutralization activity with data points in shades of blue indicating IC50 > 1 nM and points in shade of green indicating IC50 < 1 nM. Antibodies with IC50 values >100 nM are designated as non-neutralizers (N.N.). Dashed red lines indicate the threshold for PUUV Gn/Gc binders (A450=0.3; x-axis) and potent neutralizers (IC50=1 nM; y-axis). (B) Frequency of mAbs grouped according to their neutralization activity. Number in the center of the pie chart denotes the total number of mAbs analyzed. (C) Frequency of prioritized mAbs grouped according to their neutralization activity towards rVSVs bearing divergent OWH and NWH Gn/Gc. Number in the center of the pie chart indicates the total number of mAbs analyzed. Each group is represented as a segment proportional to the group size. (D) Neutralizing activity of 50 prioritized mAbs (IC50 values) against the indicated rVSVs. mAbs with IC50>10 nM are designated non-neutralizers (N.N.), and mAbs leaving an un-neutralized virus fraction are depicted as striped squares. An un-neutralized virus fraction is defined as a residual normalized virus infection of >5% at the highest mAb concentration tested. Averages of IC50 values (n≥6) from at least two experiments.
nAbs with broad activity predominantly recognize two antigenic sites in Gn/Gc
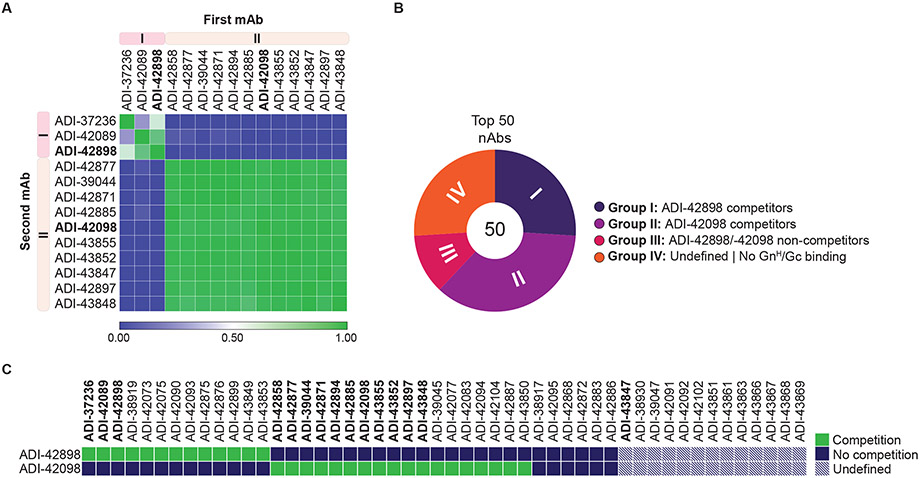
We screened a subset of nAbs with broad neutralizing activity (bnAbs) in a pairwise competition ELISA (cELISA) that employed rVSV-mNG-PUUV-Gn/Gc as antigen to begin to define the antigenic sites recognized by our nAbs (Fig. 3A). Most tested bnAbs could be assigned to one of two competition groups, which we designated Groups I and II. Group I bnAbs also competed with the PUUV-specific nAb ADI-37236, which we previously isolated from a PUUV-experienced donor by sorting with the recombinant soluble N–terminal head domain of PUUV Gn (GnH; Fig. S4), providing initial evidence that Group I nAbs recognize overlapping epitopes in the Gn subunit. We next binned top 50 nAbs against selected Group I and Group II nAbs (ADI-42898 and ADI-42098, respectively) in a high-throughput yeast display-based competition assay with a recombinant PUUV GnH/Gc heterodimer as antigen (Figs. 3B-C and S4). This second competition-binning approach largely corroborated the results from the cELISA and showed that the majority of our top 50 nAbs could be assigned to Groups I and II. Group III nAbs did not compete with ADI-42898 or ADI-42098, indicating that they bind distinct epitopes in Gn/Gc. Group IV nAbs failed to bind to soluble GnH/Gc, raising the possibility that they recognize tetrameric spike- or tetramer lattice-dependent quaternary Gn/Gc epitopes in viral particles (see Fig. 4A). Because most nAbs from Groups III–IV lacked broad neutralizing activity, we focused mostly on nAbs from Groups I–II for further characterization.
Fig. 3: Epitope binning of nAbs by competition analysis.
(A) Heatmap of competitive pairwise mAb binding studies to rVSV-PUUV-Gn/Gc. Percent mAb competition by the second mAb (columns) was normalized to the binding ELISA value for un-competed binding by the first mAb (rows). Antibodies were clustered according to their Pearson correlation-generated relatedness scores using Morpheus software (Broad Institute, Cambridge, USA). Averages (n=2-4) from one to two experiments. (B) Proportion of mAbs competing with ADI-42898 or ADI-42098 was determined in a high-throughput yeast competition assay by binding of IgG-displaying yeast cells to soluble PUUV GnH/Gc pre-complexed with F(ab)s of ADI-42898 or ADI-42098. Number in the center of the pie chart denotes the total number of mAbs analyzed. Each group (I-IV) is represented as a segment proportional to the group size. (C) Heatmap showing the capacity of selected mAbs to compete with ADI-42898 or ADI-42098 for binding to PUUV GnH/Gc from the experiments in panel B. mAbs marked in bold were tested in both competition-binding studies.
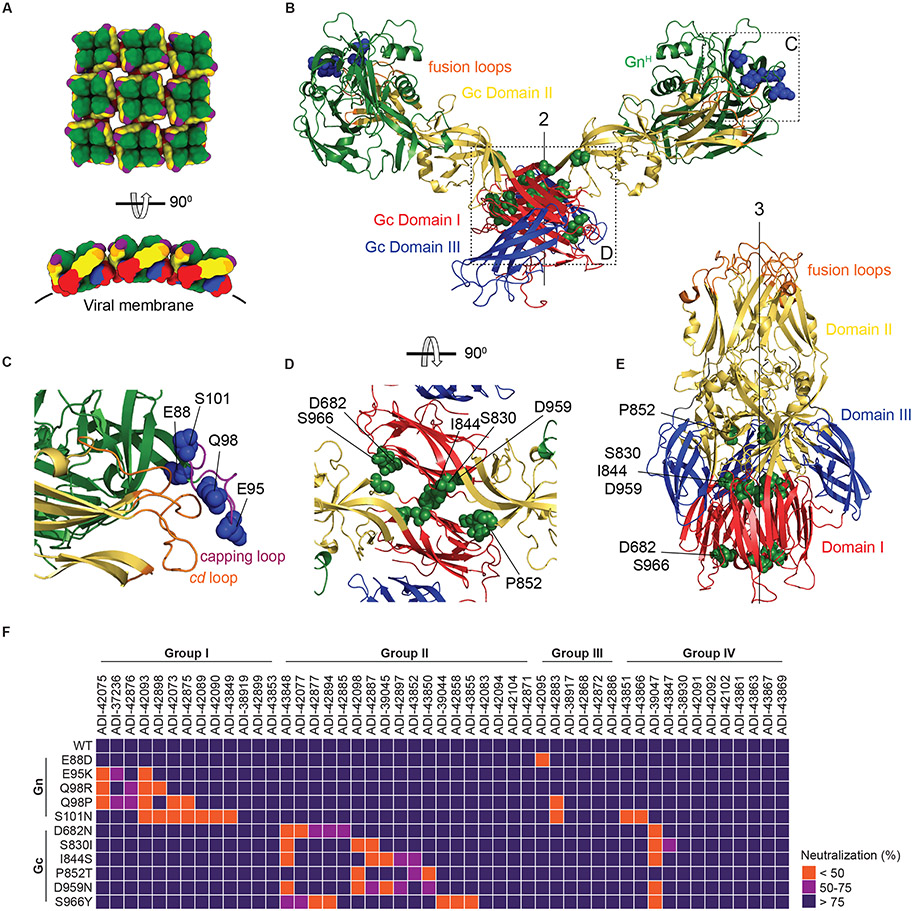
Fig. 4: Epitope mapping of nAbs by escape mutant analysis.
(A) Schematic representation of the organization of lattices of Gn/Gc tetramers in viral particles (top, en face view; bottom, side view). Gn, green; Gn capping loop, purple; Gc domain I, red; Gc domain II, yellow; Gc domain III, blue; Gc fusion loops, orange. The approximate position of the viral membrane is indicated with a black line. (B) Ribbon representation of the ANDV GnH/Gc pre-fusion dimer (modeled from its monomer, PDB: 6Y5W; (14)) with the two-fold molecular axis of the dimeric spike indicated (“2”). Residues exchanged in the neutralization-escape mutants are shown as blue (Gn) or green (Gc) spheres. (C–D) The GnH:Gc and Gc:Gc interfaces in panel B (dotted rectangles) were enlarged for clarity and rotated as indicated to show neutralization-escape mutants in the Gn capping loop (C) and Gc (D). In panel C, the cd fusion loop in Gc associated with the capping loop in Gn is shown in orange. (E) The PUUV Gc post-fusion trimer with the three-fold molecular axis indicated (“3”) (PDB: 5J81; (20)). Labeling of Gc domains and escape mutations as in panel B. (F) Heatmap showing the capacity of mAbs to neutralize rVSV-PUUV-Gn/Gc bearing neutralization-escape Gn/Gc mutants (also see Fig. S5). mAbs were grouped according to competition Groups I-IV (see Fig 3). Group I = ADI-42898 competitors; Group II = ADI-42098 competitors; Group III = ADI-42898/ -42098 non-competitors; Group IV = undefined/ GnH/Gc non-binders. Neutralization averages (n=6) from two experiments.
Group I and II nAbs recognize epitopes in Gn and Gc, respectively
To identify Gn/Gc residues critical for viral recognition by Group I and II nAbs, we selected rVSV-mNG-PUUV-Gn/Gc mutants resistant to neutralization by ADI-42898 or ADI-42098 and sequenced the glycoprotein genes of plaque-isolated viral clones to identify escape mutations. ADI-42898 elicited an escape mutant bearing a Q98R substitution in the Gn ‘capping loop’, spanning amino acid (aa) residues 90-103 in GnH (Figs. 4B-C and S5A) (14). The capping loop associates closely with and caps the cd and bc membrane-interacting fusion loops in Gc domain II in the GnH/Gc pre-fusion heterodimer (14). Escape mutants to ADI-42098 bore two independent substitutions, S830I and P852T, in Gc’s central domain I and domain II close to the domain I-II junction, respectively, which form part of the oligomerization interfaces between adjacent pre-fusion Gn/Gc tetramers in the viral lattice and between adjacent Gc monomers in the post-fusion homotrimer (Figs. 4A, B, D, E and S5B). Concordantly, escape mutants selected with other Group I and II nAbs pinpointed additional residues in the GnH capping loop (E95K, Q98P, S101N) and Gc domain I (D682N, I844S, D959N, S966Y), respectively (Figs. 4 and S5).
Neutralization screens of our prioritized nAbs against this panel of 11 escape mutants further corroborated the competition mapping and escape mutant analyses above (Fig. 4F). Although group III–IV nAbs were largely resistant to this escape mutant panel, substitution E88D, located N-terminal to the capping loop, afforded escape by one of the Group III nAb ADI-42095 (Figs. 4C, F and S5A). Further, substitution S101N, which introduces an N-linked glycosylation (N-X-S/T sequence) at aa residue 101 in the capping loop (Fig. S6A), afforded viral escape to three of the Group III–IV nAbs (Fig. 4F), possibly through N–glycan–mediated steric blockade of nAb binding to capping loop-proximal epitopes. Taken together, these findings establish the GnH capping loop and the Gc oligomerization interface as critical sites for viral recognition and neutralization by the majority of the nAbs, including most of the bnAbs isolated herein. However, they also point to the existence of additional unmapped sites of vulnerability in hantavirus Gn/Gc, including some that only appear to be available in Gn/Gc conformers present in viral particles (Group IV).
Visualization of Gn/Gc:nAb complexes and implications for nAb breadth
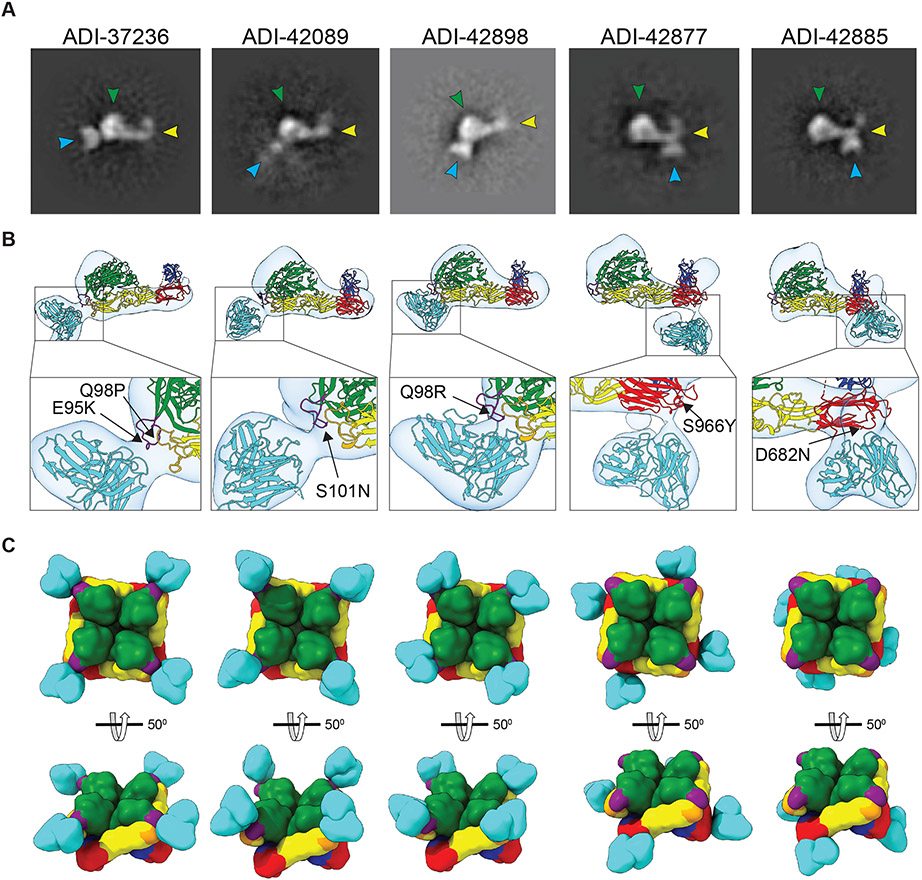
To visualize Gn/Gc recognition by Group I and II nAbs, we performed negative-stain transmission electron microscopy (nsEM) studies of the single-chain variable fragments (scFvs) of selected nAbs from Group I (ADI-37236, -42089, -42898) and Group II (ADI-42877, -42885) bound to monomeric PUUV GnH/Gc. We obtained two-dimensional classes in which the three distinctive elements of the complex, GnH, Gc, and scFvs, could be readily identified (Figs. 5A-B). We used these class averages to calculate three-dimensional maps at 20Å resolution (Fig. 5B).
Fig. 5: Negative-stain electron microscopy (nsEM) of scFv:PUUV GnH/Gc complexes.
(A) Exemplary nsEM 2D-classes of PUUV GnH/Gc bound to scFvs of the indicated mAbs. scFvs, turquoise; GnH, green with the capping loop in purple; Gc domain I, red; domain II, yellow; domain III, blue; fusion loops, orange. (B) 3D reconstructions of scFv:GnH/Gc complexes are shown in transparent surface representations (light gray) with the structure of ANDV GnH/Gc docked into the density and pseudo-colored as described in panel A. Corresponding neutralization-escape mutations are shown in the closeups. (C) Modeled interactions of the indicated scFvs with tetrameric PUUV GnH/Gc complexes are shown in surface-shaded representation and colored as in panel A. En face and side views are shown.
In agreement with our epitope assignments, all of the Group I nAbs appeared to interact with the GnH capping loop. Moreover, tetramer models of the GnH/Gc:nAb complexes suggested that these nAbs bind apically to well-exposed sequences in pre-fusion spikes at the viral surface (Fig. 5C). Although all three Group I nAbs exhibited similar binding modes at this resolution, they differ in two key functional aspects. First, ADI-37236 appears to be a ‘pure Gn binder’ that recognizes both PUUV GnH and GnH/Gc, whereas ADI-42089 and ADI-42898 only bind to the latter complex (Fig. S7). Second, ADI-37236 is PUUV-specific, whereas ADI-42089 and ADI-42898 are bnAbs (Fig. 2D). The high sequence variability of the GnH capping loop (Fig. S6A) and its proximity to the Gc fusion loops (Fig. 4B) raises the possibility that ADI-37236 contacts residues in the variable regions of GnH only, whereas ADI-42089 and ADI-42898 also make contacts with more highly conserved sequences in Gc. Reconstructions of the Group II nAbs ADI-42885 and ADI-42877 confirmed our epitope mapping to sequences in Gc domain I, near the domain I–III linker region, and in proximity to Gc:Gc homodimer interfaces between neighboring Gn/Gc tetramers (Fig. 5B) (21). The tetramer models of these GnH/Gc:nAb complexes indicate that the epitopes of Group II nAbs are more proximal to the viral membrane than those of the Group I nAbs (Fig. 5C), suggesting that the former may be less accessible in viral particles than the latter, especially in regions with a closely packed lattice of Gn/Gc tetramers (11, 19, 21). Cryo-electron tomography of PUUV virus-like particles bound to the F(ab) fragment of a bank vole-derived mAb that recognizes a similar site in Gc supports this hypothesis (32).
Group I and II nAbs specifically block viral membrane fusion
The above epitope definition and structural studies led us to postulate that nAbs from both Groups I and II target acid-triggered Gn/Gc rearrangements required for virus-cell membrane fusion, by preventing either the deployment and membrane insertion of the closely associated hydrophobic fusion loops (Group I nAbs) or the formation of a post-fusion Gc homotrimer (Group II nAbs) (19, 20). To evaluate this hypothesis, we established a ‘fusion-infection’ assay in which rVSV-PUUV-Gn/Gc particles were bound to cells at 4°C and then subjected to an extracellular acidic pulse (pH 5.5 at 37°C) to trigger virus-cell membrane fusion at the plasma membrane instead of in endosomal compartments (Fig. S8A).
All Group I and II nAbs tested in this assay mediated dose-dependent reductions in PUUV Gn/Gc-dependent fusion-infection, as also did several nAbs from Groups III and IV (Fig. S9A). Importantly, and in contrast to a Gn/Gc-specific polyclonal serum, none of these nAbs had any effect on a preceding step in viral entry, the attachment of virions to target cells (Fig. S9B). Taken together, these results indicate that many of our nAbs selected for neutralization potency and breadth directly interfere with Gn/Gc-dependent viral membrane fusion, as also observed for previously isolated nAbs (26), suggesting that they act by targeting one or more steps in the membrane fusion reaction, including Gn:Gc dissociation, and Gc target membrane insertion and homotrimerization.
Group I/II nAbs are potent and broad neutralizers of authentic Old World hantaviruses
We advanced the most potent nAbs to testing against authentic Old World hantaviruses (Fig. 6). PUUV neutralization assays largely corroborated our findings with rVSV-mNG-PUUV-Gn/Gc. However, whereas the Gn- and/or potential Gn/Gc-targeting Group I nAbs afforded complete neutralization (all nAbs in Group I and some from Groups III-IV), the Gc-targeting nAbs (all nAbs in Group II and some from Group IV) left a residual un-neutralized fraction (Figs. 6A and D). The much greater propensity of Group II nAbs to mediate incomplete neutralization in the authentic neutralization assay relative to the rVSV assay (Fig. 2D) raises the possibility that differences between authentic virions and rVSVs in the organization of the (Gn/Gc)4 spike lattice accounts for their distinct neutralization phenotypes with Gc binders.
Fig. 6: Neutralization potency of mAbs against authentic Old World hantaviruses (OWH).
Potency of mAbs to block OWH PUUV (A), HTNV (B), and DOBV (C) infection of VeroE6 cells (mAb concentrations of 100, 4, and 0.04 nM). Data points are colored according to the mAb’s competition group assignments defined in Fig. 3: Group I, green; Group II, blue; Group III, purple; Group IV, pink. Averages ± standard deviation (s.d.), n=2-10 from one to five experiments (PUUV); n=4 from two experiments (HTNV); n=4-6 from two to three experiments (DOBV). (D) Heatmap of IC50 values isolated from PUUV, HTNV, and DOBV dose-response neutralization curves (A-C) derived by non-linear regression analysis. Data points are colored according to the mAb’s neutralization potency. mAbs with IC50 values >5 nM are designated non-neutralizing mAbs (N.N.); mAbs whose neutralization curves did not adhere to a sigmoidal dose-response fit and for which an IC50 could not be computed are designated as N.C. mAbs leaving an un-neutralized virus fraction are shown as striped data points on the map. An un-neutralized virus fraction is defined as a residual normalized virus infection at the highest mAb concentration tested of >5%. N.D., not determined.
We selected a subset of these nAbs with at least some neutralization breadth against Old World hantavirus Gn/Gc in the rVSV assay (Fig. 2D) and evaluated them with authentic HTNV and DOBV (Figs. 6B-D). Concordant with our findings with authentic PUUV above, we observed a substantial narrowing in the breadth of all the Gc binders tested from Groups II and IV. Only a single GnH/Gc-specific bnAb from Group I, ADI-42898, afforded complete neutralization of HTNV and DOBV in a manner that was consistent with its exceptional breadth of activity against the corresponding rVSVs (Figs. 2D and 6B-D).
Group I/II nAbs provide pre- and post-exposure protection in a bank vole model of PUUV challenge
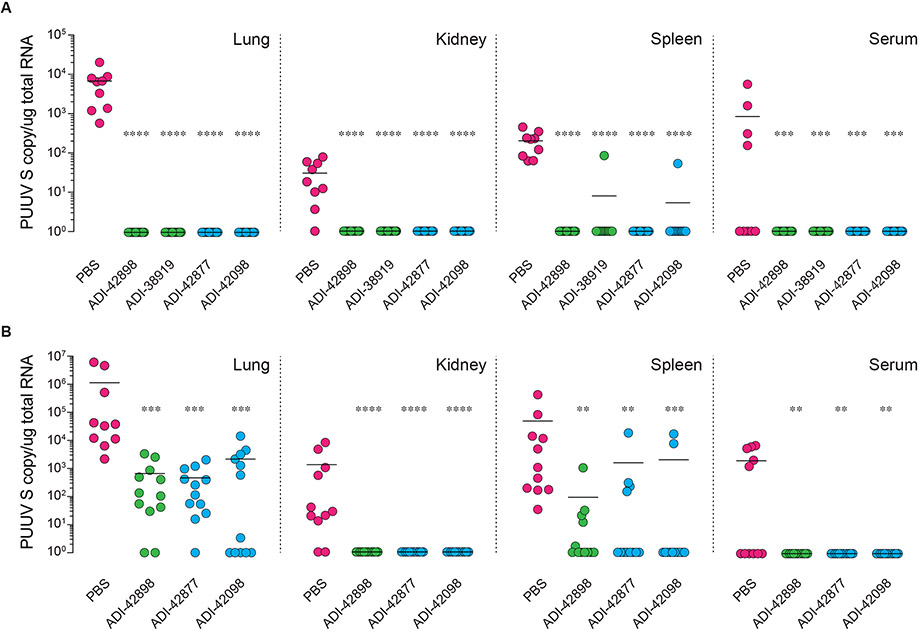
We selected two Group I (ADI-38919, -42898) and two Group II (ADI-42098, -42877) nAbs with potent activity against authentic PUUV (Fig. 6A) and assessed their protective efficacy against the PUUV strain ‘Suonenjoki’ (PUUV/Suo) in a recently described bank vole infection model (Fig. 7) (28). PUUV/Suo establishes a persistent infection in bank voles within three days after virus challenge and resembles a natural infection in these rodents in terms of viral load and tissue tropism. Virus-naïve bank voles from a laboratory-bred colony were first administered single 25-mg/kg doses of nAb intraperitoneally (i.p.) at 4 h pre-infection and then challenged with PUUV/Suo subcutaneously (s.c.). Viral loads in tissues previously associated with active PUUV replication were determined at three days post-infection. All four nAbs afforded essentially complete pre-exposure prophylactic protection against viral acquisition in the lung, kidney, spleen, and blood (Fig. 7A).
Fig. 7: In vivo protective efficacy of human nAbs in a PUUV bank vole challenge model.
(A) Bank voles were administered a single 25 mg/kg dose (i.p.) of the indicated nAbs (n=12 from two experiments) or PBS vehicle (n=9 from two experiments) followed by a challenge with PUUV/Suo (500 FFU, s.c.) at 4 h post nAb administration. Animals were sacrificed at 3 days post challenge and serum and organ viral RNA loads were determined by RT-qPCR detecting the PUUV S segment. Bars indicate median. (B) Bank voles were challenged with PUUV/Suo (500 FFU, s.c.) followed by treatment with a single 25 mg/kg dose (i.p.) of the indicated nAbs (n=12 from two experiments) or PBS vehicle (n=11 from two experiments) at 24 h post challenge. Animals were sacrificed at 3 days post challenge, and viral RNA loads were determined as above. Bars indicate median. Untreated versus nAb-treated animals, un-paired Kruskal-Wallis test: **, P=0.0021; ***, P=0.0002; ****, P<0.0001.
We next retested the nAbs as above, but in a more stringent post-exposure setting (nAb treatment 24 h post-virus challenge). Both Group I and II nAbs mediated substantial (~1,000-fold) declines in mean viral RNA levels in the lung, the organ which displayed the highest levels of viral replication, with reductions to undetectable levels in some animals. Viral RNA levels in the spleen also declined in treated animals and were uniformly reduced to undetectable levels in the kidney and in circulation (Fig. 7B).
bnAb ADI-42898 affords cross-clade neutralization and post-exposure protection
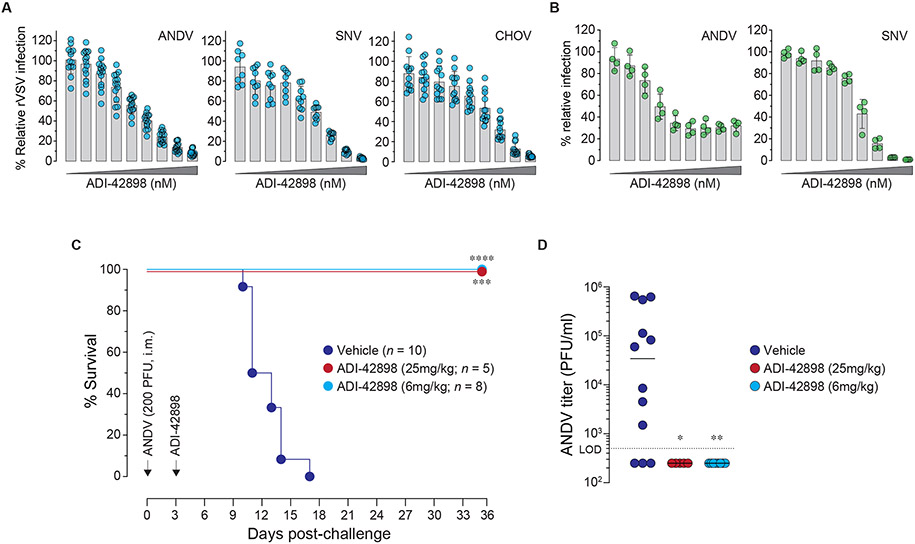
To identify bnAbs with cross-clade potential, we prioritized nAbs with activity against multiple authentic Old World hantaviruses (Fig. 6) and an rVSV bearing Gn/Gc from the New World hantavirus, ANDV (Fig. 2D). This set of nAbs, comprising a single Group I nAb, ADI-42898, and two Group II nAbs, ADI-39044 and -42871, was evaluated for neutralizing activity against rVSVs bearing New World hantavirus glycoproteins from ANDV, SNV, and Choclo virus (CHOV) as well as against authentic ANDV and SNV (Figs. 8A-B and S10). ADI-42898 completely neutralized all three rVSV-Gn/Gc and authentic SNV, and it neutralized authentic ANDV with an un-neutralized virus fraction (Figs. 8A-B). In contrast, the two Group II nAbs had little or no cross-clade activity towards authentic New World hantaviruses (Figs. S10B and D), providing another example of the discordances between the rVSV and authentic virus assays described above for Group II nAbs.
Fig. 8: Broad pan-hantavirus neutralization and protection by ADI-42898.
(A) Potency of ADI-42898 to block infection of Vero cells by rVSVs bearing Gn/Gc from the indicated NWH. Viruses were exposed to a 3-fold mAb dilution series starting at 100 nM. Averages ± s.d, n=14 from five experiments (ANDV); n=9 from three experiments (SNV); n=12 from four experiments (CHOV). (B) Potency of ADI-42898 to block ANDV (strain Chile-9717869) and SNV (strain CC107) infection of HUVECs with a 3-fold mAb dilution series starting at 300 nM. Averages ± s.d, n=4 from two experiments. (C) Syrian golden hamsters were challenged with ANDV (200 PFU, i.m.; strain Chile-9717869) followed by treatment with a single dose of ADI-42898 (~25 mg/kg or ~6 mg/kg, i.p.) at 3 days post-virus exposure. Mortality of hamsters was monitored for 35 days. Averages from two experiments, n=5 (~25 mg/kg dose); n=8 (~6 mg/kg dose), n=10 (vehicle). Untreated versus mAb-treated animals, Mantel-Cox test: ***, P=0.0003; ****, P<0.0001. (D) Serum virus titers of Syrian golden hamsters on day 8 post ANDV challenge were determined by plaque assay. Untreated versus ADI-42898-treated animals, un-paired Kruskal-Wallis test: *, P=0.03; **, P=0.002. L.O.D., limit of detection.
Finally, we evaluated ADI-42898’s protective efficacy in the gold-standard Syrian hamster model of uniformly lethal ANDV challenge (35). Accordingly, we exposed hamsters to the Chile variant of ANDV (ANDV/Chile-9717869), and then administered a single dose of ADI-42898 (4 mg; ~25-mg/kg) on day three. All nAb-treated animals survived treatment, whereas all animals in the vehicle arm succumbed to infection (Fig. 8C). Moreover, ADI-429898–mediated protection was associated with a complete reversal of viremia (Fig. 8D). Strikingly, this nAb fully retained its protective efficacy even at a fourfold lower dose (1 mg; ~6-mg/kg) (Figs. 8C-D). Thus, ADI-42898, a fully human, GnH/Gc-specific nAb with broad cross-clade activity against the major disease-associated hantaviruses displays in vivo post-exposure protective efficacy against both Old World and New World rodent-borne hantaviruses.
DISCUSSION
Previous work indicates that antibody-based therapeutics could provide urgently needed countermeasures to combat the zoonotic diseases caused by hantaviruses. Specifically, mAbs isolated from New World hantavirus ANDV- and SNV-convalescent patients or from mice vaccinated with hantavirus glycoproteins could protect against lethal ANDV challenge and development of HCPS-like disease in a Syrian hamster model (26, 27, 36). Despite this progress, however, no mAbs with demonstrated protective efficacy against less lethal but far more prevalent and globally widespread Old World hantaviruses have been described. Although previous studies suggest that vaccination can elicit cross-protective Gn/Gc-specific antibody specificities in animals, no such protective mAbs are known (37, 38). Herein, we isolated and characterized a panel of 135 human mAbs from individuals exposed to a virulent Old World hantavirus, PUUV. We identified mAbs that targeted multiple antigenic sites in membrane-bound tetramers of the Gn/Gc glycoprotein complex and displayed distinctive patterns of viral neutralization potency and breath. A single bnAb, ADI-42898, bound a quaternary epitope in the Gn/Gc heterodimer to neutralize a diverse set of virulent hantaviruses from both clades and afforded in vivo post-exposure protection against both PUUV and ANDV.
We adopted three strategies in seeking to enrich for bnAbs targeting conserved epitopes in native Gn/Gc tetramers. First, we chose PUUV as our target virus because of the genetically intermediate position of its M segment sequence (encoding Gn/Gc) relative to those of the Old World and New World hantaviruses (Fig. 1A). Second, we screened a large number of sera from PUUV-experienced donors in our Swedish cohort to identify source samples with a high degree of cross-clade serological reactivity (Fig. S1). Third, to improve our chances of retrieving nAbs targeting quaternary Gn/Gc epitopes, we panned memory B cells with fluorescently labeled rVSV particles bearing membrane-bound Gn/Gc, instead of recombinant proteins that fail to fully recapitulate the structure of the Gn/Gc tetramer (14). Of the 180 IgG VH/VL sequence pairs we captured from antigen-sorted memory B cells, 135 recognized rVSV-PUUV-Gn/Gc, attesting to the specificity of our sorting approach. Moreover, we observed some degree of cross-reactivity against Gn/Gc from five other hantaviruses and—concordant with our hypothesis—comparable cross-reactivities against both Old World and New World agents (Figs. 1E and 2D). Importantly, mAbs with broad binding activity displayed low levels of polyreactivity (Fig. S11), demonstrating that their cross-binding is not due to nonspecific cross-reactivity (39). Interestingly, mAbs from SNV-experienced donors but not ANDV-experienced donors were found to display a greater degree of cross-clade reactivity in a recent study (26), providing additional evidence that the nature of the primary antigenic exposure can influence the breadth of the antibody response against hantaviruses in humans.
Our epitope-binning studies on a prioritized subset of potent nAbs uncovered at least four distinct antigenic sites in Gn/Gc that we fine-mapped through rVSV-based selections of neutralization escape mutants (Figs. 3-4). We identified a major class of nAbs (Group I) whose members engage the capping loop in the membrane-distal N–terminal head domain of Gn (GnH) (Figs. 4-5) (14). The preponderance of Group I nAbs in our panel, together with recent descriptions of nAbs elicited against HTNV Gn/Gc that recognize overlapping or adjacent sequences in GnH (40), suggest that this region of Gn, well-exposed on viral particles, is a major target of the antibody response to Gn/Gc in mammals, although it has not been described in earlier work aimed at identifying neutralizing epitopes (41). Here, we present new evidence that such mAbs exhibit at least two distinct binding modes, exemplified by ADI-37236 and previously reported mAbs (40) on the one hand and ADI-42898 on the other. Despite escaping binding and neutralization via similar mutations in the capping loop, ADI-37236 could bind both monomeric GnH and heterodimeric GnH/Gc proteins, whereas ADI-42898 could only bind GnH/Gc (Fig. S7). We propose that the former nAbs exclusively contact sequences in GnH, whereas the latter contact both GnH and residues in the closely associated Gc cd loop that forms part of Gc’s membrane-interacting fusion loops (14). This hypothesis rationalizes the observed relationship between nAb binding and breadth: ADI-37236 and other “pure” GnH binders that recognize the highly variable capping loop are virus-specific nAbs, whereas ADI-42898 and other GnH/Gc-binders whose epitopes include more conserved Gc residues are bnAbs (Figs. 2D, 4C and 4F). Our failure to obtain escape mutations in Gc with ADI-42898 may simply reflect the possibilities that such amino acid exchanges exact higher costs on viral fitness or that the energetics of mAb binding is driven by its contacts with Gn and not Gc. Finally, both Gc-dependent and Gc-independent capping-loop binders could act downstream of virus-cell attachment to block viral membrane fusion (Fig. S9), indicating that the direct engagement of Gc by Group I nAbs may not be required to prevent fusion loop deployment and/or Gc rearrangement during membrane fusion.
Members of the second major class of nAbs identified and mapped in this study (Group II) recognize central sequences of the Gc subunit, including epitopes (i) in domain I near the flexible domain I–III linker region; (ii) at or near the domain I–II junction; and (iii) the ‘inner sheets’ of domain I, which mediate Gc:Gc contacts between adjacent (Gn/Gc)4 tetramers stabilizing the spike lattice (Figs. 4-5) (19, 21). Similar nAbs, elicited through a variety of approaches for different hantaviruses, have been described previously (32, 36, 42-44), suggesting that these Gc sequences may be immunodominant despite their reduced accessibility to mAbs in intact viral particles. The behavior of Group II nAbs in neutralization assays was consistent with their mode of Gn/Gc recognition. First, we found that they tended to exhibit greater neutralization breadth than did the Group I nAbs (Fig. 2D), in accordance with the greater conservation of their target sequences (Fig. S6). Second, Group II nAbs often failed to completely neutralize authentic viral infection (Figs. 6 and S10), as also reported recently for ANDV- and SNV-targeted Gc binders (26). Cryo-EM tomographic analysis of a Gc domain I/II-targeting bank vole mAb bound to PUUV virus-like particles provides a likely structural explanation for the un-neutralized fraction: the close apposition of glycoprotein tetramers at Gc:Gc two-fold axes of symmetry permits mAb binding only at lattice-free sites in virions (11, 21, 32, 45). Together with differences in their intrinsic binding affinities for Gc from different hantaviruses, this observation may also explain the discordant behavior of Group II nAbs against rVSVs and authentic viruses (Figs. 2D, 6 and S10); lattice-free binding sites may be more available in rVSV particles due to their distinct curvature, lower levels of Gn/Gc incorporation, or both. All Gc-specific nAbs tested afforded the potent inhibition of viral membrane fusion (Fig. S9), most likely through the prevention of Gc homotrimerization and/or domain III foldback onto domain I-II trimers (19, 20, 32). The capacity of domain I inner sheet binders to block formation of the Gc:Gc interface, at least in principle, also raises the intriguing possibility that some Gc binders may act in part by interfering with glycoprotein lattice formation during Old World hantavirus assembly (21). Finally, despite their failure to fully neutralize viral infectivity, two Group II nAbs, ADI-42877 and -42098, afforded complete protection against PUUV challenge in naïve bank voles (28) (Fig. 7). Therefore, it is conceivable that Gc-targeting mAbs contribute to viral clearance through mechanisms other than entry neutralization, including Fc-mediated effector functions, as previously suggested (36). Given their non-competing epitopes and potentially distinct mechanisms of neutralization and protection, we are exploring the possibility that specific combinations of Group II and Group I nAbs could afford enhanced protection and/or robustness to variations in Gn/Gc sequence and viral neutralization escape.
We identified a single Group I nAb, ADI-42898, with remarkably broad neutralizing activity against virulent hantaviruses from both Old World and New World clades (Figs. 6 and 8) and successfully evaluated it in both pre- and post-exposure settings in a recently described bank vole model of PUUV challenge (28) (Fig. 7). ADI-42898 also afforded complete protection of Syrian hamsters from lethal ANDV challenge when administered as single 25-mg/kg or 6-mg/kg doses at three days post-virus challenge (Fig. 8), raising the possibility that it could be dosed down further (below 6-mg/kg) and still retain protective activity. By contrast, ANDV-specific nAbs previously reported to be fully protective in this model were either tested with two 5-mg/kg doses per animal (26) or with a substantially higher two-dose regimen (25–50-mg/kg) (27, 36). Moreover, recently described bnAbs only afforded partial protection against ANDV challenge, even when animals were dosed twice, and these bnAbs were not evaluated against Old World hantaviruses in vivo (26).
ADI-42898 is an antibody-based treatment that can protect against both Old World and New World hantaviruses and is a promising candidate for clinical development as a broad countermeasure against HFRS- and HCPS-causing hantaviruses. Our ongoing and future work is aimed at extending these studies of protective breadth to additional rodent and nonhuman primate models of orthohantavirus challenge (46). The studies in nonhuman primates would greatly facilitate the identification of key correlates of protection that could be used to design human clinical trials and seek drug licensure. Finally, the properties of ADI-42898 and other Group I nAbs suggest strategies for the design of broadly protective vaccines that preferentially elicit bnAbs targeting the Gn/Gc interface at the expense of virus-specific nAbs targeting only Gn.
MATERIAL AND METHODS
Study design
We initiated this study to identify, isolate and characterize human mAbs broadly reactive to hantaviral Gn/Gc, with the potential to neutralize Old World and New World hantavirus infections and prevent HCPS/HFRS. We screened sera from a cohort of 45 PUUV-experienced patients for PUUV, HTNV and ANDV Gn/Gc-specific nAb titers and identified six donors presenting potent and broad neutralization activity. PUUV and ANDV Gn/Gc-reactive memory B cells were isolated by single-cell sorting, and antibody heavy- and light-chain variable regions were cloned as full-length human IgG1 and expressed in S. cerevisiae. We characterized 135 mAbs for their cross-clade binding and neutralization activity as well as epitope specificity. We evaluated down-selected mAbs for protective efficacy in a bank vole PUUV infection model for HFRS and a Syrian hamster ANDV lethal challenge model for HCPS.
Peripheral blood mononuclear cells (PBMC) and matching serum and clinical parameters from PUUV-exposed individuals were collected at Umeå University Hospital, Sweden, 19-78 days after disease onset. Patients with laboratory-confirmed PUUV infection during acute HFRS were recruited to a prospective and longitudinal study of HFRS. The study protocol was approved by the Ethical Review Board at Umeå University, Umeå, Sweden (Dnr. 04-113M and Dnr. 07-162M). Signed informed consent was obtained from all study participants.
Bank vole (Myodes glareolus) animal protocols were approved by the Animal Experimental Board of Finland (license number ESAVI/28410/2019). Laboratory-bred 5-8 months old bank voles were housed under Biosafety level 3 conditions in an IsoCage biocontainment system (Scanbur) in groups of 3-5 animals and provided standard rodent pellets and water ad libitum. Groups of six animals (three males, three females) were used per condition to provide >80% power in a one-tailed Fisher’s exact test. This sample size would allow the experimenter to detect a minimum efficacy rate of 88% (5/6 protected) in the treated group compared to 0% (0/6 not protected) in the control group at a 95% confidence level. Randomization and blinding were not performed. All bank voles from two independent experiments were included in the data analysis; no outliers were excluded. Upon completion of the experiment 72 h post virus inoculation, bank voles were euthanized by cervical dislocation.
Syrian golden hamster (Mesocricetus auratus) animal protocols were conducted under IACUC-approved protocols in compliance with the Animal Welfare Act, Public Health Service Policy, and other applicable federal statutes and regulations relating to animals and experiments involving animals. The animal BSL-4 facility where these studies was conducted (USAMRIID) is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council. Further, all studies were performed in accordance with institutional policies for biosafety and biosecurity. Studies were powered as described above with groups of six 5-10 months old female animals per condition. Randomization and blinding were not performed. Several hamsters designated as untreated were excluded from the dataset in both independent experiments (see ‘Syrian hamster challenge studies’ section).
Single B cell sorting
B cells were purified from PBMCs using a MACS Human B Cells isolation kit (Miltenyi Biotec) and incubated for 30 min on ice with pre-titrated amounts of rVSV-mNG-P-PUUV-Gn/Gc or -ANDV-Gn/Gc to label antigen-specific B cells. The amounts of viral particles giving specific labeling of immune PBMC samples (PUUV-experienced donors) and non-specific labeling of non-immune PBMC samples (PUUV-naïve donors) were determined for each rVSV stock prior to sorting experiments. Cells were washed once with ice-cold MACS buffer, centrifuged for 10 min at 300xg at 4°C and then stained using a panel of markers to identify antigen-specific memory (class-switched) B cells for sorting: anti-human CD19 (PE-Cy7, Biolegend Cat# 302216), CD3 (PerCP-Cy5.5, Biolegend Cat# 30040), CD8 (PerCP-Cy5.5, Biolegend Cat# 344710), CD14 (PerCP-Cy5.5, Invitrogen Cat# 45-0149-42), CD16 (PerCP-Cy5.5, Biolegend Cat# 360712), propidium iodide, IgM (APC, BD Biosciences Cat#314510), IgD (BV421, Biolegend Cat# 348226) for 20 min on ice in the dark. Class-switched B cells, defined as CD19+/CD3−/CD8−/CD14−/CD16−/IgM−/IgD− cells, showing over-background reactivity to either rVSV-mNG-P-PUUV-Gn/Gc or -ANDV-Gn/Gc were single-cell sorted using BD FACS Aria II (BD Biosciences) into 96-well polystyrene microplates (Corning) containing 20 μl/well of lysis buffer (5 μl of 5x first strand Super Script IV cDNA buffer [Invitrogen], 1.25 μl dithiothreitol [Invitrogen], 0.625 μl of NP-40 [Thermo Scientific], 0.25 μl RNaseOUT [Invitrogen], and 12.8 μl dH2O). Plates were immediately stored at −80°C. Flow cytometry data was analyzed using FlowJo software. Antibody sequences were determined from sorted B cells and antibodies were produced as described in Supplementary Materials and Methods.
Cell lines
African green monkey kidney Vero cells (ATCC Cat# CCL-81) and VeroE6 cells (ATCC Cat# CRL-1586) were cultured in a humidified 37°C, 5% CO2 incubator in high-glucose Dulbecco's modified Eagle medium (DMEM, Life Technologies) supplemented with 2% fetal bovine serum (FBS, Atlanta Biologicals), 1% GlutaMAX, and 1% penicillin-streptomycin (both Life Technologies). Human hepatocarcinoma Huh.7.5.1 cells (a generous gift of Jan Carette, Stanford University, CA) were maintained in high-glucose DMEM (Life Technologies) supplemented with 10% FBS (Atlanta Biologicals), 1% GlutaMAX (Life Technologies), 1% non-essential amino acids (Life Technologies) and 1% penicillin-streptomycin (Life Technologies). Human umbilical vein endothelial cells (HUVECs, Lonza Cat# C2517A) were maintained in endothelial cell growth medium (EGM) supplemented with EGM-SingleQuots (Lonza) at 37°C with 5% CO2. Mygla.Rec.B bank vole epithelial cells (a generous gift of Isabella Eckerle, Geneva Centre for Emerging Viral Diseases, Switzerland) were cultured as described previously (28). Drosophila melanogaster S2 cells (Thermo Fisher Cat# R69007) were maintained in serum-free insect cell medium (HyClone, GE HealthCare) supplemented with 1% penicillin-streptomycin in spinner flasks in a 28°C, 150 rotation per minute (rpm) humidified shaking incubator. ExpiCHO cells (Thermo Fisher Cat# A29127) carrying a FUT8-knockout mutation (Mapp Biopharmaceuticals Inc.) were maintained in their designated growth medium (Thermo Fisher) and cultured in a 37°C, 8% CO2, 125 rpm humidified shaking incubator.
Recombinant Vesicular Stomatitis Viruses (rVSVs)
rVSVs expressing enhanced green fluorescent protein (eGFP) and bearing Gn/Gc from HTNV, SEOV, DOBV, ANDV, SNV, CHOV and VSV G were engineered, rescued and propagated following published protocols (30, 31, 47-49). rVSV carrying a phosphoprotein P fused to mNeonGreen (mNG) and bearing PUUV Gn/Gc has been described previously (29). Generation of viruses with an equivalent mNG-P backbone decorated with ANDV Gn/Gc (Chilean field strain R123, GenBank accession number NP_604472.1), SNV Gn/Gc (human field isolate, GenBank accession number NP_941974.1), and HTNV Gn/Gc (cell culture-adapted strain 76-118, GenBank accession number NP_941978.1) was performed by using standard molecular biology techniques. The identity and sequence of each virus-encoded Gn/Gc was determined by Sanger sequencing of rVSV genomic RNA-derived cDNAs. All viruses were propagated on Vero cells except rVSV-mNG-P PUUV-Gn/Gc, which was amplified on Huh.7.5.1 cells.
VSV infectivity measurements and neutralization assays
Confluent Vero cells were inoculated with pre-titrated amounts of pseudotyped rVSV particles bearing either PUUV, HTNV, SEOV, DOBV, ANDV, SNV, and CHOV Gn/Gc or, when indicated, rVSV-PUUV-Gn/Gc neutralization-escape mutants. Prior to infection, rVSVs were diluted in corresponding media, and infected cells were maintained at 37°C for 14 to 16 h post-infection before automated counting of eGFP+ and mNG+ cells using a Cytation 5 cell imaging multi-mode reader (BioTek Instruments) or a Cell-Insight CX5 imager (Thermo Fisher) including onboard software. For neutralization experiments, rVSV particles were incubated with increasing concentrations of mAbs or, when indicated, human serum diluted in corresponding media at room temperature for 1 h prior to addition to cell monolayers. Virus neutralization data were subjected to nonlinear regression analysis to derive IC50 values (4-parameter, variable slope sigmoidal dose-response equation constraining bottom parameters to 0-100; GraphPad Prism).
Authentic hantavirus infections and microneutralization assays
PUUV strain Suo (PUUV/Suo) was propagated in Mygla.Rec.B bank vole epithelial cells (28), whereas PUUV strain Kazan (50) and DOBV strain Ano-Poroia/AF19 (51) were grown in VeroE6 cells as described. VeroE6 cells were exposed to PUUV Kazan or DOBV at a multiplicity of infection (MOI) of 0.1-1 of fluorescent FFU following a pre-incubation with indicated mAb concentrations. Viral infectivity was calculated by immunostaining of formaldehyde-fixed cells at 20 h (PUUV) or 48 h (DOBV) post-infection using polyclonal rabbit serum targeting the PUUV nucleocapsid protein N (NR-9675, BEI Resources) or a broadly cross-reactive hantavirus N-specific antiserum (28). Plates were imaged with either the Plate RUNNER HD (Trophos) or the Opera Phenix High-Content Screening System (PerkinElmer) at the FIMM high content imaging and analysis services (University of Helsinki, Finland). Images were analyzed and individual infected cells counted using the system’s onboard software.
ANDV strain Chile-9717869, SNV strain CC107, and HTNV strain 76-118 used for all in vitro assays were propagated and viral titers determined using VeroE6 cells as previously described (30, 52-54). For neutralization assays using authentic virus, pre-titrated amounts of virus were mixed with serially diluted mAbs for 1 h before adding virus/mAb mixture to HUVECs at a MOI of 1 (ANDV and HTNV) or MOI of 2 (SNV). Viral infectivity was determined by immunostaining of formaldehyde-fixed and permeabilized cells at 48 h post-infection with polyclonal rabbit sera specific for ANDV, HTNV or SNV N (NR-9673, NR-9674, and NR-12152; BEI Resources). Images were acquired on an Operetta high-content imaging device (PerkinElmer). Images were analyzed with a customized scheme built from image-analysis functions present in the Harmonia software, and the percentage of infected cells was determined using the analysis functions. Virus neutralization data were subjected to nonlinear regression analysis to derive IC50 values (4-parameter, variable slope sigmoidal dose-response equation constraining bottom parameters to 0-100; GraphPad Prism).
Authentic hantavirus plaque assays
Serial log dilutions of serum were prepared in MEM media supplemented with 5% FBS, 2 mM L-glutamine and 1% gentamicin, and confluent monolayers of VeroE6 cells on 6-well plates were inoculated. After adsorption for 1 h at 37°C, monolayers were overlaid with Eagle basal medium (EBME) supplemented with 0.5% agarose (Seakem), 30 mM Hepes buffer, and 5% FBS. After incubation at 37°C for 8 days, a second overlay, supplemented with 5% Neutral red, was added. Plaques were counted the following day and titers were expressed as PFU/ml.
Generation of recombinant PUUV GnH and GnH/Gc antigen
To obtain the head domain of PUUV Gn (GnH, aa 20-381) and the PUUV GnH/Gc complex (both strain Mu/07/1219, GenBank code: AJC50718 (55)) we followed the approach described previously (14). In short, we generated a codon optimized sequence (GenScript) encoding GnH alone, and a single-chain construct of GnH in frame with the Gc ectodomain (aa 659-1093) in which the amphipathic C-terminal membrane proximal external region was removed. To prevent complex dissociation during purification, we joined both subunits using a flexible linker of 43-aa long (GGSGLVPRGSGGGSGGGSWSHPQFEKGGGTGGGTLVPRGSGTGG) containing two thrombin cleavage sites (underlined) at either end and a Strep-tag sequence (in bold) in the middle. Both GnH and GnH/Gc encoding sequences were cloned into a modified pMT/BiP plasmid (Invitrogen) expressing them in frame with a C-terminal double Strep-tag. This plasmid was used to generate stable Drosophila melanogaster S2 cells, which were selected and maintained in serum-free insect cell medium containing 7 μg/ml puromycin. Protein expression was induced by adding 5 μM CdCl2 and cell supernatant was harvested 5 days post induction, supplemented with 10 μg/mL avidin and 0.1 M Tris-HCl (pH 8.0). Proteins were purified via strep-tactin affinity chromatography followed by size exclusion chromatography (SEC) using a Superdex 200 10/30 column (GE Healthcare) in a buffer composed of 10 mM Tris-HCl (pH 8), 150 mM NaCl, and 1 mM EDTA.
Amplification and cloning of antibody variable genes
Human antibody variable gene transcripts (VH, Vκ, Vγ) were amplified by RT-PCR using the Super Script IV (Invitrogen) followed by nested PCRs using cocktails of IgM-, IgD-, IgA- and IgG-specific primers and the HotStarTaq Plus DNA Polymerase (Qiagen), as described previously (56). To allow cloning by homologous recombination into S. cerevisiae, the primers used in the second round of nested PCR contained 40 base pairs of 5’ and 3’ homology to the digested yeast expression vectors. Amplified variable gene transcripts were transformed into yeast using the lithium acetate method for chemical transformation (57). 200 μl of the transformed yeast cell suspension was plated on selective media and the plates were incubated at 30°C for 48 h. Following the growth of transformants on the plates, individual yeast colonies were picked for sequencing.
Expression and purification of IgGs, scFv and F(ab) fragments
IgGs were expressed in S. cerevisiae cultures grown in 24-well plates, as described previously (58). Briefly, after 6 days, the cultures were harvested by centrifugation and IgGs were purified by Protein A affinity chromatography. The bound antibodies were eluted (200 mM acetic acid/50 mM NaCl [pH 3.5]) into 1/8th volume of 2 M Hepes buffer (pH 8.0) and buffer-exchanged into phosphate buffered saline (PBS, pH 7.0).
F(ab) fragments were generated by digesting IgGs with papain for 2 h at 30°C. The digestion was terminated by the addition of iodoacetamide, and the F(ab) and Fc mixtures were passed over Protein A agarose to remove Fc fragments and undigested IgG. The flow-through of the Protein A resin was then passed over CaptureSelect™ IgG-CH1 affinity resin (Thermo Fisher) and eluted (200 mM acetic acid/50 mM NaCl [pH 3.5]) into 1/8th volume of 2 M Hepes buffer (pH 8.0). F(ab) fragments then were buffer exchanged into PBS (pH 7.0).
Single chain Fv fragments (scFv) coding sequences of ADI-37236, -42089, -42898, -42877 and -42885 were generated by joining together the sequences encoding the VH and VL chains by a flexible linker, and cloned into a modified pMT/BiP plasmid (Invitrogen). The antibody fragments were purified following the protocol described above in the ‘Generation of recombinant PUUV GnH and GnH/Gc antigen’ section.
ELISA for Gn/Gc:mAb binding
All mAbs cloned from antigen-specific B cells were tested for their reactivity to rVSV particles displaying the Gn/Gcs of PUUV, HTNV, DOBV, SEOV, ANDV and SNV in a single concentration spot ELISA. Viral particles were diluted in PBS (pH 7.4), and captured directly onto high binding half-area 96-well plates (Corning) overnight at 4°C in amounts determined using a calibration assay (designed to ensure equivalent particle loading across the antigenically distinct rVSVs). Plates were then washed with PBS and blocked with a blocking/binding buffer (PBS containing 5% non-fat dried milk) for 1 h. Blocked plates were exposed to antibodies, in duplicate, at 25 nM mAb concentration for 1 h at 37°C. Plates were washed with PBS and mAb binding was detected with anti-human IgG secondary antibody conjugated to horseradish peroxidase (HRP, Thermo Fisher) for 1 h at 37°C. Plates were washed, 1-Step™ Ultra TMB-ELISA Substrate Solution (Thermo Fisher) was added and quenched with the addition of 2 M sulfuric acid, per manufacturer recommendations. Absorbance was read at 450 nm and antibodies producing signal >0.3 were categorized as binders.
Competition ELISA
High-protein-binding 96-well ELISA plates (Corning) were coated with purified human mAbs (40 ng per well, ‘first mAb’) at 37°C and blocked with 3% (w/v) bovine serum albumin (BSA; Thermo Fisher) in PBS. The membranes of rVSV-PUUV-Gn/Gc particles were labelled with a short-chain phospholipid probe, functional-component spacer diacyl lipid conjugated to biotin (FSL-biotin, Sigma-Aldrich), as described (30). Subsequently, pre-titrated amounts of biotin-labeled rVSVs were incubated with purified human mAb (100 nM, ‘second mAb’) for 1 h at 37°C before their addition to mAb-coated plates. Bound rVSV-PUUV-Gn/Gc was detected by incubation with a streptavidin-HRP conjugate (Thermo Scientific). Competition levels were determined by reduction of rVSV binding in presence of a competitor mAb compared to rVSV binding in the absence of competition (rVSV complexed with human IgG1 [Sigma-Aldrich]). Antibodies with >75% reduction of rVSV binding were considered to be in competition with pre-complexed mAbs.
Yeast-based mAb competition assay
Antibody competition assays were performed as previously described (59). Antibody competition was measured by the ability of a competitor Group I (ADI-42898) or Group II (ADI-42098) F(ab) to inhibit binding of yeast surface-expressed anti-Gn/Gc IgGs to recombinant PUUV GnH/Gc. 50 nM biotinylated GnH/Gc was pre-incubated with 1 μM competitor F(ab) for 30 min at room temperature and then added to a suspension of yeast expressing anti-Gn/Gc IgGs in a 96-well format. Unbound antigen was removed by washing with PBS containing 0.1% BSA. After washing, bound antigen was detected using streptavidin-Alexa Fluor 633 conjugate (Life Technologies) and analyzed by flow cytometry using a FACSCanto II (BD Biosciences). The level of competition was assessed by measuring the fold reduction in PUUV GnH/Gc binding in the presence of competitor F(ab) relative to an antigen-only control. Antibodies were considered competitors when a greater than five-fold reduction was observed in the presence of control F(ab) relative to an antigen-only control.
Selection of rVSV neutralization-escape mutants
Selection of escape mutants was performed as described previously (60). In short, rVSV-PUUV-Gn/Gc was serially passaged after rVSV pre-incubation with a concentration of mAb corresponding to its IC90 value derived from neutralization assays, and then added to confluent monolayers of Huh.7.5.1 cells. Infection was allowed to proceed to completion (>90% cell death by eye), supernatants were harvested and following several passages under mAb selection, supernatants were tested for viral neutralization escape. If resistance was evident, individual viral clones were plaque-purified on Huh.7.5.1 cells, and their Gn/Gc-encoding sequences were determined.
Negative-stain electron microscopy
The PUUV GnH/Gc:scFv complexes were assembled in vitro by co-incubating the heterodimer GnH/Gc with an 1.5-fold excess of scFv for 1 h at 4°C prior to purification over a Superdex 200 gel filtration column (GE Healthcare). Complexes were diluted to a final concentration of 40 nM, deposited onto glow-discharged carbon grids (CF300, EMS), and negatively stained in 2% (w/v) uranyl acetate. The negatively stained specimens were mounted onto a transmission electron microscope holder and examined with a Tecnai F20 electron microscope operating at a voltage of 200 kV. Images were collected with a Falcon II direct electron detector (FEI) with a pixel size of 2 Å per pixel and a total accumulated dose of 20 electrons per Å2. All image processing and analyses was carried out using cryoSPARC (61). In short, the images were CTF-(contrast transfer function) corrected, the particles automatically picked, and then 2D-classified. The best 2D classes were used to generate the initial 3D model, which was further refined and filtered to 20 Å.
Production of human IgGs for animal studies
Antibody VH and VL coding sequences were ordered as gBlocks (Integrated DNA Technologies) with a 15 base pair 5’ overlap to a mouse IgKVIII secretion signal coding sequence and a 15 base pair 3’ overlap to the appropriate sequences encoding for a constant region (human kappa, human lambda or human IgG1). The variable regions coding sequences were cloned into a pcDNA3.4 (Thermo Fisher) expression vector modified to contain coding sequences of a mouse IgKVIII signal sequence and the appropriate constant regions. The gBlocks were then inserted into the vector between the secretion signal and the constant region coding sequences using In-Fusion enzyme (Takara Bio). Antibodies were transiently expressed in ExpiCHO cells (Thermo Fisher) carrying a FUT8-knockout mutation (Mapp Biopharmaceuticals Inc.) utilizing the high titer protocol for CHO Expifectamine (Thermo Fisher). Approximately 10 days post transfection, cell culture supernatants were clarified by spinning at 3,200xg for 10 min, filtered and loaded onto a HiTrap MabSelect SuRe affinity column (Cytiva) using an ÄKTA pure 25 fast protein liquid chromatography (FPLC) system. Once loaded, the column was washed with 10 column volumes of PBS (pH 7.2), followed by elution of the target antibody with Pierce IgG elution buffer (Thermo Fisher). Elution fractions containing the target antibody were pooled and neutralized to ~pH 7 with 1.0 M Tris, pH 7.8.
Bank vole challenge studies
The neutralizing potential of human mAbs against PUUV/Suo challenge in vivo was investigated in an established experimental PUUV reservoir host model system (28). The bank vole colony at the Friedrich-Loeffler-Institut, Germany was established from six breeding pairs as founder animals obtained from an existing breeding colony at the Federal Environment Agency of Germany (62). The initially introduced bank voles were tested for pathogens according to FELASA recommendation using C57/Bl6 mice as sentinels (63). Bank voles were challenged with 500 focus-forming units (FFU) of PUUV/Suo by s.c. inoculation. Group sizes for each condition were six animals with a 50/50 gender distribution. In a prophylactic treatment regimen, voles were inoculated i.p. with ~25-mg/kg of mAbs (or PBS as vehicle control) 4 h prior to challenge; in a post-exposure treatment scheme, voles were inoculated with ~25-mg/kg of mAbs 24 h post virus inoculation. The animals were euthanized 72 h post virus inoculation by cervical dislocation, after which RNA was isolated from lungs, spleen, kidneys, and serum; virus RNA loads were assessed by PUUV S-segment RT-qPCR.
Quantitative Real-Time PCR (RT-qPCR) assay
Following euthanasia and dissections, RNA was extracted from bank vole tissues using Trizol Reagent (Thermo Fisher) according to the manufacturer’s instructions. In case of serum, RNA was isolated using the QIAamp Viral RNA Mini Kit (Qiagen). For RT-qPCR, RNA was either analyzed with the qPCRBIO SyGreen 1-Step Go Lo-Rox kit (PCR Biosystems) or reverse transcribed using RevertAid reverse transcriptase (Thermo Fisher) and random hexamer primers (Thermo Fisher) followed by PCR analysis with Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher). Protocol, primers and standard for PUUV S-segment RT-qPCR analyses were carried out as previously described (64) and the analyses were performed using AriaMx instrumentation (Agilent). The results were normalized to total RNA concentration of the samples and presented as S segment copy number/μg total RNA.
Syrian hamster challenge studies
Groups (n=6) of Syrian golden hamsters (Envigo Cat# HsdHan AURA 089), 100-200 grams, were exposed i.m. to 200 plaque-forming units (PFU) of ANDV strain Chile-9717869 diluted in 0.1 ml PBS. On day 3 post-exposure, hamsters were treated i.p. with indicated amounts (~25- or ~6-mg/kg) of mAb diluted in 0.8 ml of PBS. Hamsters were monitored daily for clinical signs of disease, morbidity, and mortality for 35 days. Moribund animals, defined as those experiencing severe respiratory distress or showing unresponsiveness, were humanely euthanized according to IACUC-approved criteria. On day 8 post-exposure, blood samples were collected by venipuncture of the anterior vena cava and the serum fraction was collected following centrifugation.
Viral titers of serum collected at day 8 post-exposure were determined by plaque assay, as described in Supplementary Materials and Methods.
Human IgG concentrations in hamster serum collected at day 8 post-exposure were determined by quantitative capture ELISA, as described in Supplementary Materials and Methods, in order to exclude any hamsters that remained human IgG-negative following antibody treatment, a phenomenon that has been described previously (65). Hamsters that were human IgG negative at day 8 were deemed untreated and removed from the dataset.
Statistical analysis
Statistical details for each experiment can be found in the respective figure legends. These include the number of replicates (n), measures of precision, and the statistical test used. Dose-response neutralization curves were subjected to nonlinear regression analysis to derive IC50 values (4-parameter, variable slope sigmoidal dose-response equation constraining bottom parameters to 0-100). Viral load in animal tissue or serum (Figs. 7 and 8) was analyzed using a Kruskal-Wallis test with Dunn’s correction for multiple comparisons. Survival curves from the hamster challenge study (Fig. 8) were compared by a Mantel-Cox (log-rank) test. All statistical analyses were carried out in GraphPad Prism (alpha, 0.05).
Supplementary Material
Neutralization activity of PUUV convalescent patient sera.
Fig. S2. Flow gating strategy for isolation of Gn/Gc-reactive memory B cells.
Fig. S3. Curves for selected mAbs with different neutralization patterns.
Fig. S4. Domain organization of PUUV Gn/Gc, GnH/Gc and GnH.
Fig. S5. Characterization of rVSV-PUUV-Gn/Gc neutralization-escape mutants.
Fig. S6. Locations of viral neutralization escape mutations in PUUV Gn/Gc.
Fig. S7. Binding activity of mAbs to PUUV Gn/Gc subdomain constructs.
Fig. S8. Establishment of fusion-infection assay with rVSV-PUUV-Gn/Gc.
Fig. S9. Mechanistic basis of mAb-dependent inhibition of virus infection
Fig. S10. Activity of Group II mAbs against NWH Gn/Gc-mediated infection.
Fig. S11. Polyreactivity of mAbs isolated from PUUV-experienced donors.
Table S1. GPC amino acid sequence analyses related to Fig. 1A.
Table S2. Demographic and laboratory characteristics of PUUV cohort donors during acute phase of disease.
ACKNOWLEDGEMENTS
Research was supported by NIAID of the National Institutes of Health (NIH) under award number U19AI142777 (Centers of Excellence in Translational Research) to K.C., L.M.W., J.M.D., Z.A.B., A.S.H., S.B.B., M.N.E.F. and L.Z. M.N.E.F. was supported by a Consolidator grant from the Swedish Science Council (#2020-06235). C.A. was supported by a Cutting edge research grant from Region Västerbotten (VLL-579011). T.S. was supported by the Academy of Finland (#321809). P.G. was supported by a grant of the National French Research Agency (ANR-18-CE11-0011). A.S. was supported by a fellowship of the French Foundation pour la Recherche Médicale (FRM; fellowship FDM20170638040). F.A.R. and P.G. were supported by Labex IBEID (ANR-10-LABX-62-IBEID). R.G.U. was supported by the Bundesministerium für Bildung und Forschung (BMBF) within the Research Network Zoonotic Infectious Diseases (01KI1721A and 01KI2004A). The content is solely the responsibility of the authors and does not necessarily represent the official views of our institutions or funders. Opinions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Department of the Army and the U.S. Department of Defense.
Footnotes
Competing interests
K.C. is a scientific advisory board member of Integrum Scientific, LLC, Biovaxys Technology Corp., and the Pandemic Security Initiative of Celdara Medical, LLC. M.S., J.C.G. and L.M.W. are employees and shareholders of Adimab, LLC. L.M.W. is an employee and shareholder of Adagio Therapeutics, Inc. A.Z.W is an employee and shareholder of Dyno Therapeutics Inc., and an equity holder in Adimab, LLC. Z.A.B., D.M.A. and L.Z. are shareholders and employees of Mapp Biopharmaceutical, Inc. All other authors have no competing interests.
Data and materials availability
Data are available on request. Materials are available through material transfer agreement by contacting K.C.
REFERENCES
- 1.Kruger DH, Figueiredo LTM, Song J-W, Klempa B, Hantaviruses-globally emerging pathogens., J. Clin. Virol 64, 128–136 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Valdebenito C, Calvo M, Vial C, Mansilla R, Marco C, Palma RE, Vial PA, Valdivieso F, Mertz G, Ferrés M, Person-to-person household and nosocomial transmission of andes hantavirus, Southern Chile, 2011., Emerging Infect. Dis 20, 1629–1636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson DC, Sargianou M, Papa A, Chra P, Starakis I, Panos G, Epidemiology of Hantavirus infections in humans: a comprehensive, global overview., Crit Rev Microbiol 40, 261–272 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Jonsson CB, Figueiredo LTM, Vapalahti O, A global perspective on hantavirus ecology, epidemiology, and disease., Clin. Microbiol. Rev 23, 412–441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pettersson L, Boman J, Juto P, Evander M, Ahlm C, Outbreak of Puumala virus infection, Sweden., Emerging Infect. Dis 14, 808–810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binder F, Drewes S, Imholt C, Saathoff M, Below DA, Bendl E, Conraths FJ, Tenhaken P, Mylius M, Brockmann S, Oehme R, Freise J, Jacob J, Ulrich RG, Heterogeneous Puumala orthohantavirus situation in endemic regions in Germany in summer 2019., Transbound Emerg Dis 67, 502–509 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Makary P, Kanerva M, Ollgren J, Virtanen MJ, Vapalahti O, Lyytikäinen O, Disease burden of Puumala virus infections, 1995-2008., Epidemiol. Infect 138, 1484–1492 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Núñez JJ, Fritz CL, Knust B, Buttke D, Enge B, Novak MG, Kramer V, Osadebe L, Messenger S, Albariño CG, Ströher U, Niemela M, Amman BR, Wong D, Manning CR, Nichol ST, Rollin PE, Xia D, Watt JP, Vugia DJ, Yosemite Hantavirus Outbreak Investigation Team, Hantavirus infections among overnight visitors to Yosemite National Park, California, USA, 2012., Emerging Infect. Dis 20, 386–393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez VP, Di Paola N, Alonso DO, Pérez-Sautu U, Bellomo CM, Iglesias AA, Coelho RM, López B, Periolo N, Larson PA, Nagle ER, Chitty JA, Pratt CB, Díaz J, Cisterna D, Campos J, Sharma H, Dighero-Kemp B, Biondo E, Lewis L, Anselmo C, Olivera CP, Pontoriero F, Lavarra E, Kuhn JH, Strella T, Edelstein A, Burgos MI, Kaler M, Rubinstein A, Kugelman JR, Sanchez-Lockhart M, Perandones C, Palacios G, “Super-Spreaders” and Person-to-Person Transmission of Andes Virus in Argentina., N. Engl. J. Med 383, 2230–2241 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Klempa B, Hantaviruses and climate change., Clin. Microbiol. Infect 15, 518–523 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Huiskonen JT, Hepojoki J, Laurinmäki P, Vaheri A, Lankinen H, Butcher SJ, Grünewald K, Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses., J. Virol 84, 4889–4897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittler E, Dieterle ME, Kleinfelter LM, Slough MM, Chandran K, Jangra RK, Hantavirus entry: Perspectives and recent advances., Adv. Virus Res 104, 185–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Rissanen I, Zeltina A, Hepojoki J, Raghwani J, Harlos K, Pybus OG, Huiskonen JT, Bowden TA, A Molecular-Level Account of the Antigenic Hantaviral Surface., Cell Rep. 15, 959–967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serris A, Stass R, Bignon EA, Muena NA, Manuguerra J-C, Jangra RK, Li S, Chandran K, Tischler ND, Huiskonen JT, Rey FA, Guardado-Calvo P, The hantavirus surface glycoprotein lattice and its fusion control mechanism., Cell 183, 442–456.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rissanen I, Stass R, Zeltina A, Li S, Hepojoki J, Harlos K, Gilbert RJC, Huiskonen JT, Bowden TA, Structural transitions of the conserved and metastable hantaviral glycoprotein envelope., J. Virol 91 (2017), doi: 10.1128/JVI.00378-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guardado-Calvo P, Rey FA, The envelope proteins of the bunyavirales., Adv. Virus Res 98, 83–118 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Guardado-Calvo P, Rey FA, The surface glycoproteins of hantaviruses., Curr Opin Virol 50, 87–94 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Acuña R, Bignon EA, Mancini R, Lozach P-Y, Tischler ND, Acidification triggers Andes hantavirus membrane fusion and rearrangement of Gc into a stable post-fusion homotrimer., J. Gen. Virol 96, 3192–3197 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Guardado-Calvo P, Bignon EA, Stettner E, Jeffers SA, Préz-Vargas J, Pehau-Arnaudet G, Tortorici MA, Jestin J-L, England P, Tischler ND, Rey FA, Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc., PLoS Pathog. 12, e1005813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willensky S, Bar-Rogovsky H, Bignon EA, Tischler ND, Modis Y, Dessau M, Crystal Structure of Glycoprotein C from a Hantavirus in the Post-fusion Conformation., PLoS Pathog. 12, e1005948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bignon EA, Albornoz A, Guardado-Calvo P, Rey FA, Tischler ND, Molecular organization and dynamics of the fusion protein Gc at the hantavirus surface., Elife 8 (2019), doi: 10.7554/eLife.46028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bharadwaj M, Nofchissey R, Goade D, Koster F, Hjelle B, Humoral immune responses in the hantavirus cardiopulmonary syndrome., J. Infect. Dis 182, 43–48 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Pettersson L, Thunberg T, Rocklöv J, Klingström J, Evander M, Ahlm C, Viral load and humoral immune response in association with disease severity in Puumala hantavirus-infected patients--implications for treatment., Clin. Microbiol. Infect 20, 235–241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valdivieso F, Vial P, Ferres M, Ye C, Goade D, Cuiza A, Hjelle B, Neutralizing antibodies in survivors of Sin Nombre and Andes hantavirus infection., Emerging Infect. Dis 12, 166–168 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vial PA, Valdivieso F, Calvo M, Rioseco ML, Riquelme R, Araneda A, Tomicic V, Graf J, Paredes L, Florenzano M, Bidart T, Cuiza A, Marco C, Hjelle B, Ye C, Hanfelt-Goade D, Vial C, Rivera JC, Delgado I, Mertz GJ, Hantavirus Study Group in Chile, A non-randomized multicentre trial of human immune plasma for treatment of hantavirus cardiopulmonary syndrome caused by Andes virus., Antivir Ther (Lond) 20, 377–386 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Engdahl TB, Kuzmina NA, Ronk AJ, Mire CE, Hyde MA, Kose N, Josleyn MD, Sutton RE, Mehta A, Wolters RM, Lloyd NM, Valdivieso FR, Ksiazek TG, Hooper JW, Bukreyev A, Crowe JE, Broad and potently neutralizing monoclonal antibodies isolated from human survivors of New World hantavirus infection., Cell Rep. 35, 109086 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrido JL, Prescott J, Calvo M, Bravo F, Alvarez R, Salas A, Riquelme R, Rioseco ML, Williamson BN, Haddock E, Feldmann H, Barria MI, Two recombinant human monoclonal antibodies that protect against lethal Andes hantavirus infection in vivo., Sci. Transl. Med 10 (2018), doi: 10.1126/scitranslmed.aat6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strandin T, Smura T, Ahola P, Aaltonen K, Sironen T, Hepojoki J, Eckerle I, Ulrich RG, Vapalahti O, Kipar A, Forbes KM, Orthohantavirus Isolated in Reservoir Host Cells Displays Minimal Genetic Changes and Retains Wild-Type Infection Properties., Viruses 12 (2020), doi: 10.3390/v12040457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerkman PF, Dernstedt A, Tadala L, Mittler E, Dannborg M, Sundling C, Maleki KT, Tauriainen J, Tuiskunen-Bäck A, Wigren Byström J, Ocaya P, Thunberg T, Jangra RK, Román-Sosa G, Guardado-Calvo P, Rey FA, Klingström J, Chandran K, Puhar A, Ahlm C, Forsell MN, Generation of plasma cells and CD27-IgD- B cells during hantavirus infection is associated with distinct pathological findings., Clin. Transl. Immunology 10, e1313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinfelter LM, Jangra RK, Jae LT, Herbert AS, Mittler E, Stiles KM, Wirchnianski AS, Kielian M, Brummelkamp TR, Dye JM, Chandran K, Haploid genetic screen reveals a profound and direct dependence on cholesterol for hantavirus membrane fusion., MBio 6, e00801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slough MM, Chandran K, Jangra RK, Two point mutations in old world hantavirus glycoproteins afford the generation of highly infectious recombinant vesicular stomatitis virus vectors., MBio 10 (2019), doi: 10.1128/mBio.02372-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rissanen I, Stass R, Krumm SA, Seow J, Hulswit RJ, Paesen GC, Hepojoki J, Vapalahti O, Lundkvist Å, Reynard O, Volchkov V, Doores KJ, Huiskonen JT, Bowden TA, Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein., Elife 9 (2020), doi: 10.7554/eLife.58242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia M, Liberatore RA, Guo Y, Chan K-W, Pan R, Lu H, Waltari E, Mittler E, Chandran K, Finzi A, Kaufmann DE, Seaman MS, Ho DD, Shapiro L, Sheng Z, Kong X-P, Bieniasz PD, Wu X, VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA., Cell Host Microbe 27, 963–975.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu T-Y, Torrey J, Thomas J, Bobrowicz P, Vásquez M, Wittrup KD, Krauland E, Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool., Protein Eng Des Sel 26, 663–670 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Safronetz D, Zivcec M, Lacasse R, Feldmann F, Rosenke R, Long D, Haddock E, Brining D, Gardner D, Feldmann H, Ebihara H, Pathogenesis and host response in Syrian hamsters following intranasal infection with Andes virus., PLoS Pathog. 7, e1002426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duehr J, McMahon M, Williamson B, Amanat F, Durbin A, Hawman DW, Noack D, Uhl S, Tan GS, Feldmann H, Krammer F, Neutralizing Monoclonal Antibodies against the Gn and the Gc of the Andes Virus Glycoprotein Spike Complex Protect from Virus Challenge in a Preclinical Hamster Model., MBio 11 (2020), doi: 10.1128/mBio.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brocato RL, Josleyn MJ, Wahl-Jensen V, Schmaljohn CS, Hooper JW, Construction and nonclinical testing of a Puumala virus synthetic M gene-based DNA vaccine., Clin. Vaccine Immunol 20, 218–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hooper JW, Custer DM, Thompson E, Schmaljohn CS, DNA vaccination with the Hantaan virus M gene protects Hamsters against three of four HFRS hantaviruses and elicits a high-titer neutralizing antibody response in Rhesus monkeys., J. Virol 75, 8469–8477 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shehata L, Maurer DP, Wec AZ, Lilov A, Champney E, Sun T, Archambault K, Burnina I, Lynaugh H, Zhi X, Xu Y, Walker LM, Affinity maturation enhances antibody specificity but compromises conformational stability., Cell Rep. 28, 3300–3308.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Rissanen I, Krumm SA, Stass R, Whitaker A, Voss JE, Bruce EA, Rothenberger S, Kunz S, Burton DR, Huiskonen JT, Botten JW, Bowden TA, Doores KJ, Structural basis for a neutralizing antibody response elicited by a recombinant hantaan virus gn immunogen., MBio 12, e0253120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tischler ND, Galeno H, Rosemblatt M, Valenzuela PDT, Human and rodent humoral immune responses to Andes virus structural proteins., Virology 334, 319–326 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Arikawa J, Schmaljohn AL, Dalrymple JM, Schmaljohn CS, Characterization of Hantaan virus envelope glycoprotein antigenic determinants defined by monoclonal antibodies., J. Gen. Virol 70 (Pt 3), 615–624 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Lundkvist A, Hörling J, Niklasson B, The humoral response to Puumala virus infection (nephropathia epidemica) investigated by viral protein specific immunoassays., Arch. Virol 130, 121–130 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Wang M, Pennock DG, Spik KW, Schmaljohn CS, Epitope mapping studies with neutralizing and non-neutralizing monoclonal antibodies to the G1 and G2 envelope glycoproteins of Hantaan virus., Virology 197, 757–766 (1993). [DOI] [PubMed] [Google Scholar]
- 45.Parvate A, Williams EP, Taylor MK, Chu Y-K, Lanman J, Saphire EO, Jonsson CB, Diverse morphology and structural features of old and new world hantaviruses., Viruses 11 (2019), doi: 10.3390/v11090862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golden JW, Hammerbeck CD, Mucker EM, Brocato RL, Animal Models for the Study of Rodent-Borne Hemorrhagic Fever Viruses: Arenaviruses and Hantaviruses., Biomed Res. Int 2015, 793257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jangra RK, Herbert AS, Li R, Jae LT, Kleinfelter LM, Slough MM, Barker SL, Guardado-Calvo P, Román-Sosa G, Dieterle ME, Kuehne AI, Muena NA, Wirchnianski AS, Nyakatura EK, Fels JM, Ng M, Mittler E, Pan J, Bharrhan S, Wec AZ, Lai JR, Sidhu SS, Tischler ND, Rey FA, Moffat J, Brummelkamp TR, Wang Z, Dye JM, Chandran K, Protocadherin-1 is essential for cell entry by New World hantaviruses., Nature 563, 559–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K, A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry., J. Virol 84, 163–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dieterle ME, Solà-Riera C, Ye C, Goodfellow SM, Mittler E, Kasikci E, Bradfute SB, Klingström J, Jangra RK, Chandran K, Genetic depletion studies inform receptor usage by virulent hantaviruses in human endothelial cells., Elife 10 (2021), doi: 10.7554/eLife.69708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lundkvist A, Cheng Y, Sjölander KB, Niklasson B, Vaheri A, Plyusnin A, Cell culture adaptation of Puumala hantavirus changes the infectivity for its natural reservoir, Clethrionomys glareolus, and leads to accumulation of mutants with altered genomic RNA S segment., J. Virol 71, 9515–9523 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papa A, Nemirov K, Henttonen H, Niemimaa J, Antoniadis A, Vaheri A, Plyusnin A, Vapalahti O, Isolation of Dobrava virus from Apodemus flavicollis in Greece., J. Clin. Microbiol 39, 2291–2293 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper JW, Larsen T, Custer DM, Schmaljohn CS, A lethal disease model for hantavirus pulmonary syndrome., Virology 289, 6–14 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Lee HW, Lee PW, Johnson KM, Isolation of the etiologic agent of Korean Hemorrhagic fever., J. Infect. Dis 137, 298–308 (1978). [DOI] [PubMed] [Google Scholar]
- 54.Schmaljohn AL, Li D, Negley DL, Bressler DS, Turell MJ, Korch GW, Ascher MS, Schmaljohn CS, Isolation and initial characterization of a newfound hantavirus from California., Virology 206, 963–972 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Ali HS, Drewes S, Weber de Melo V, Schlegel M, Freise J, Groschup MH, Heckel G, Ulrich RG, Complete genome of a Puumala virus strain from Central Europe., Virus Genes 50, 292–298 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H, Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning., J. Immunol. Methods 329, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gietz RD, Woods RA, Yeast transformation by the LiAc/SS Carrier DNA/PEG method., Methods Mol. Biol 313, 107–120 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, Jangra RK, Dieterle ME, Lilov A, Huang D, Tse LV, Johnson NV, Hsieh C-L, Wang N, Nett JH, Champney E, Burnina I, Brown M, Lin S, Sinclair M, Johnson C, Pudi S, Bortz R, Wirchnianski AS, Laudermilch E, Florez C, Fels JM, O’Brien CM, Graham BS, Nemazee D, Burton DR, Baric RS, Voss JE, Chandran K, Dye JM, McLellan JS, Walker LM, Broad neutralization of SARS-related viruses by human monoclonal antibodies, Science (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bornholdt ZA, Turner HL, Murin CD, Li W, Sok D, Souders CA, Piper AE, Goff A, Shamblin JD, Wollen SE, Sprague TR, Fusco ML, Pommert KBJ, Cavacini LA, Smith HL, Klempner M, Reimann KA, Krauland E, Gerngross TU, Wittrup KD, Saphire EO, Burton DR, Glass PJ, Ward AB, Walker LM, Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak., Science 351, 1078–1083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wec AZ, Herbert AS, Murin CD, Nyakatura EK, Abelson DM, Fels JM, He S, James RM, de La Vega M-A, Zhu W, Bakken RR, Goodwin E, Turner HL, Jangra RK, Zeitlin L, Qiu X, Lai JR, Walker LM, Ward AB, Dye JM, Chandran K, Bornholdt ZA, Antibodies from a Human Survivor Define Sites of Vulnerability for Broad Protection against Ebolaviruses., Cell 169, 878–890.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination., Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Röhrs S, Begeman L, Straub BK, Boadella M, Hanke D, Wernike K, Drewes S, Hoffmann B, Keller M, Drexler JF, Drosten C, Höper D, Kuiken T, Ulrich RG, Beer M, The Bank Vole (Clethrionomys glareolus)—Small Animal Model for Hepacivirus Infection, Viruses 13, 2421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.FELASA working group on revision of guidelines for health monitoring of rodents and rabbits, Mähler Convenor M, Berard M, Feinstein R, Gallagher A, Illgen-Wilcke B, Pritchett-Corning K, Raspa M, FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units., Lab Anim 48, 178–192 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Wigren Byström J, Näslund J, Trulsson F, Evander M, Wesula Lwande O, Ahlm C, Bucht G, Quantification and kinetics of viral RNA transcripts produced in Orthohantavirus infected cells., Virol. J 15, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roberts A, Thomas WD, Guarner J, Lamirande EW, Babcock GJ, Greenough TC, Vogel L, Hayes N, Sullivan JL, Zaki S, Subbarao K, Ambrosino DM, Therapy with a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody reduces disease severity and viral burden in golden Syrian hamsters., J. Infect. Dis 193, 685–692 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neutralization activity of PUUV convalescent patient sera.
Fig. S2. Flow gating strategy for isolation of Gn/Gc-reactive memory B cells.
Fig. S3. Curves for selected mAbs with different neutralization patterns.
Fig. S4. Domain organization of PUUV Gn/Gc, GnH/Gc and GnH.
Fig. S5. Characterization of rVSV-PUUV-Gn/Gc neutralization-escape mutants.
Fig. S6. Locations of viral neutralization escape mutations in PUUV Gn/Gc.
Fig. S7. Binding activity of mAbs to PUUV Gn/Gc subdomain constructs.
Fig. S8. Establishment of fusion-infection assay with rVSV-PUUV-Gn/Gc.
Fig. S9. Mechanistic basis of mAb-dependent inhibition of virus infection
Fig. S10. Activity of Group II mAbs against NWH Gn/Gc-mediated infection.
Fig. S11. Polyreactivity of mAbs isolated from PUUV-experienced donors.
Table S1. GPC amino acid sequence analyses related to Fig. 1A.
Table S2. Demographic and laboratory characteristics of PUUV cohort donors during acute phase of disease.