Abstract
Embryonic development results in the production of distinct tissue types, and different cell types within each tissue. A major goal of developmental biology is to uncover the "parts list" of cell types that comprise each organ. Here we perform single cell RNA sequencing (scRNA-seq) of the Drosophila embryo to identify the genes that characterize different cell and tissue types during development. We assay three different timepoints, revealing a coordinated change in gene expression within each tissue. Interestingly, we find that the elav and mhc genes, whose protein products are widely used as markers for neurons and muscles, respectively, show broad pan-embryonic expression, indicating the importance of post-transcriptional regulation. We next focus on the central nervous system (CNS), where we identify genes whose expression is enriched at each stage of neuronal differentiation: from neural progenitors, called neuroblasts, to their immediate progeny ganglion mother cells (GMCs), followed by new-born neurons, young neurons, and the most mature neurons. Finally, we ask whether the clonal progeny of a single neuroblast (NB7-1) share a similar transcriptional identity. Surprisingly, we find that clonal identity does not lead to transcriptional clustering, showing that neurons within a lineage are diverse, and that neurons with a similar transcriptional profile (e.g. motor neurons, glia) are distributed among multiple neuroblast lineages. Although each lineage consists of diverse progeny, we were able to identify a previously uncharacterized gene, Fer3, as an excellent marker for the NB7-1 lineage. Within the NB7-1 lineage, neurons which share a temporal identity (e.g. Hunchback, Kruppel, Pdm, and Castor temporal transcription factors in the NB7-1 lineage) have shared transcriptional features, allowing for the identification of candidate novel temporal factors or targets of the temporal transcription factors. In conclusion, we have characterized the embryonic transcriptome for all major tissue types and for three stages of development, as well as the first transcriptomic analysis of a single, identified neuroblast lineage, finding a lineage-enriched transcription factor.
Keywords: embryonic cell type, single cell RNAseq, neuroblast, GMC, temporal transcription factor, motor neuron, temporal identity, Nkx6, HGTX, Eve
Background
Understanding how tissues such as the nervous system develop is a central goal of developmental biology. An important part of development is the generation of cell types that vary in molecular profile, cell morphology and cell function. Identifying the different cell types in a tissue is particularly important for the study of neural development, where a vast number of distinct neurons must interconnect to form a functional nervous system (Luo, 2020). Defining neural diversity at the molecular level is important for generating a "parts list" of neurons in the brain, and will ultimately generate a better understanding of how neuronal diversity is established. Understanding the mechanisms used to generate the appropriate neurons in the correct spatial location and correct time also advances the potential for neurotherapeutics to counteract injury or disease.
Single cell RNA-sequencing (scRNA-seq) is a powerful method for determining the transcriptional profile of complex tissues such as the central nervous system (CNS), including mammalian cortical excitatory and inhibitory neurons (Shen et al., 2020), hippocampal neurons (Hodge et al., 2019), zebrafish embryonic neurons (Tambalo et al., 2020), and Drosophila larval, pupal and adult neurons (Brunet Avalos and Sprecher, 2021; Davie et al., 2018; Konstantinides et al., 2018; McLaughlin et al., 2021; Nguyen et al., 2021; Vicidomini et al., 2021; Xie et al., 2021) (Velten et al., 2022). Surprisingly, there has yet to be a sc-RNAseq study of Drosophila embryonic neurogenesis; the only embryonic sc-RNAseq report was tightly focused on pre-gastrula embryos (Karaiskos et al., 2017).
Drosophila neurogenesis is ideal for the application of transcriptional analysis, as there is a wealth of cell- and tissue-specific genes that provide ground-truth information for identifying cell types through the use of sc-RNAseq. In the Drosophila embryo, neuronal diversity is generated in three steps: (1) spatial patterning cues are used to generate neural progenitor (neuroblast) identity, with each neuroblast having a unique identity based on its spatial location (Skeath and Thor, 2003); (2) temporal patterning generated by a cascade of "temporal transcription factors" (TTFs): Hunchback > Kruppel > Pdm1/2 (Flybase Nubbin/Pdm2) > Castor diversifies ganglion mother cell (GMC) identity within each neuroblast lineage (Doe, 2017); and (3) nearly all GMCs undergo a terminal asymmetric division to partition the Notch inhibitor Numb into one of the two siblings, thereby creating NotchON and NotchOFF siblings that have unique molecular and morphological identities (Mark et al., 2021; Skeath and Doe, 1998; Truman et al., 2010).
Here we present a scRNA-seq atlas of the entire embryo at three timepoints. We subsequently focus on gene expression changes within the developing nervous system: first at stage 12 when neuroblasts are maximally proliferative and only the earliest-born neurons have begun to differentiate; then at stage 14 when both neuroblasts and differentiated neurons are well represented; and finally at stage 16 where the bulk of the mature embryonic neurons are present. In addition to tracking the transcriptome of bulk neuroblasts, GMCs and neurons, we also address the question of whether sc-RNAseq can be used to identity lineage-specific gene expression. Here, we focus on the best characterized neuroblast lineage: NB7-1 which is a model for studying spatial patterning (McDonald et al., 1998), temporal patterning (Isshiki et al., 2001; Kohwi et al., 2013; Meng et al., 2019; Meng and Heckscher, 2020; Pearson and Doe, 2003; Seroka et al., 2020; Seroka and Doe, 2019), and NotchON/NotchOFF sibling specification (Mark et al., 2021; Skeath and Doe, 1998). We can identify two classes of motor neurons and interneurons known to be present in this lineage, as well as novel gene expression patterns that identify candidates for lineage-specific functions. To our knowledge, our study is the first to characterize the Drosophila post-blastoderm embryonic transcriptome and the first to transcriptional profile a single neuroblast lineage.
Results and Discussion
The transcriptome of all embryonic cell types
To create a transcriptional time course of embryonic development, we dissociated stage 12, 14, and 16 embryos using standard methods and independently performed two technical replicates for 10X Genomics scRNAseq on cells from each timepoint. This genotype contained NB7-1-Gal4 UAS-RedStinger transgenes used in the final section below to identify the NB7-1 and its progeny (Seroka and Doe, 2019). From stage 12 embryos we isolated 20,038 cells and obtained 1,234 median genes per cell; from stage 14 embryos we isolated 28,045 cells at 656 median genes per cell; and from stage 16 embryos we isolated 24,032 cells at 450 median genes per cell. The difference in median genes per cell may be due to technical differences (the way the samples were run) or biological differences (e.g. differences in viability following dissociation). We favor the latter explanation, as our technical replicates showed an absence of batch-effects between technical replicates (Supplemental Figure 1). Cells were filtered for quality in Seurat using standard methods. Following quality control, the stage 12, 14 and 16 objects contained 17,564, 24,668 and 20,328 cells respectively. We merged these datasets using Seurat to obtain a single Uniform Manifold Approximation and Projection (UMAP; https://arxiv.org/abs/1802.03426) atlas containing 62,560 cells and 96 clusters. We chose to merge the datasets as opposed to integration, as the standard Seurat integration algorithm cannot differentiate development from “batch effects” resulting from transcriptional differences between the same cell types at different ages (e.g. the algorithm might detect genes differentially expressed between stg 12 and 16 neurons as “batch effects” instead of developmental differences, and remove those genes from analysis). We used a standard cutoff of 3000 variable genes as input to PCA and utilized all computed principal components for clustering to maximize subtype discovery (Seurat FindClusters resolution 5.0, dims 1:50) (Figure 1A; Supplemental Table 1).
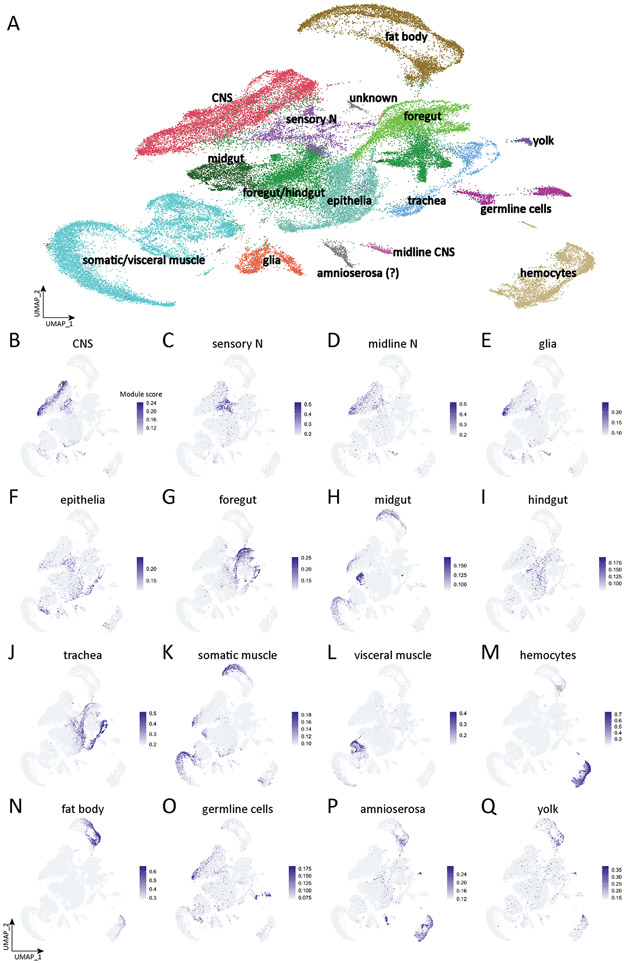
Figure 1. Tissue type atlas of whole Drosophila embryo.
(A) Integrated single-cell atlas of whole Drosophila embryos. Cluster identity is determined by the module scores based on the tissue-specific genes.
(B-Q) Plots of module scores of tissue-specific genes for each individual tissue in UMAP space. The colors encode the module scores computed by AddModuleScore function in Seurat. Tissue defining genes listed in Supplemental Table 2.
We observed clusters representing all expected embryonic cell types, identified by tissue-specific annotations in Flybase (www.flybase.org) and BDGP in situ atlas (https://insitu.fruitfly.org/cgi-bin/ex/insitu.pl) (Tomancak et al., 2007). For example, the expression of genes annotated as "glial" were used to identify cells in the UMAP object that we assign to the "glial" distribution shown in Figure 1E, and the scores were used to assign the “glial” identity in the UMAP object; a similar process was done for all tissue types. The UMAP for each annotated pool of genes is shown in Figure 1B-Q, and the list of genes in each pool is given in Supplemental Table 2.
Clusters included neural cell types in the central nervous system (Figure 1B), ciliated sensory neurons (Figure 1C), midline cells (Figure 1D), and glia (Figure 1E). These neural cell types will be further subdivided and characterized in more depth below.
We also observed clusters representing epithelia (Figure 1F); foregut (Figure 1G); midgut (Figure 1H); hindgut (Figure 1I); trachea (Figure 1J); somatic/visceral muscle (Figure 1K,1L); hemocytes (Figure 1M); fatbody (Figure 1N); germline cells (Figure 1O); amnioserosa (Figure 1P); and yolk (Figure 1Q). In addition to identifying all expected embryonic cell types, we also observe clusters that do not express tissue-specific genes (unknown, Figure 1A); these could be previously uncharacterized cell types or a mixture of cell types that were not well clustered. We conclude that we have identified transcriptional profiles for all major embryonic cell types.
Tissue-specific proteins can be widely transcribed
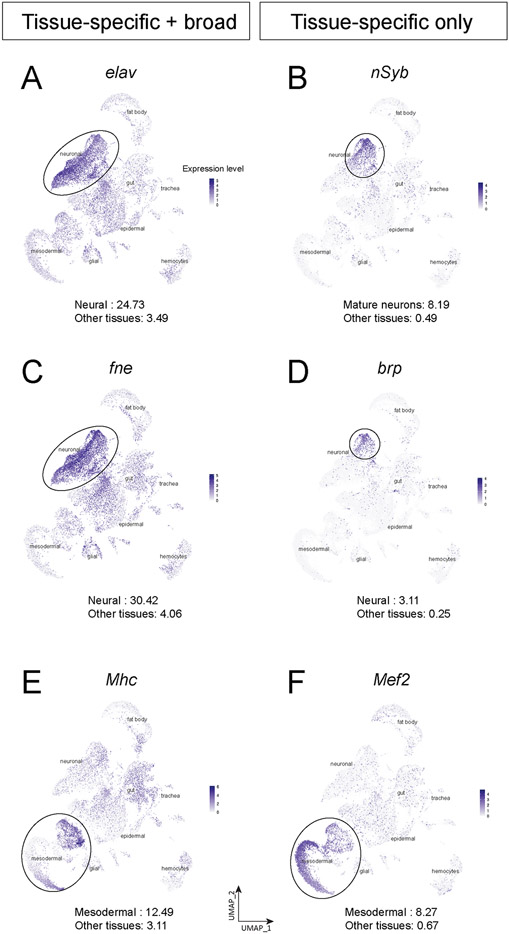
We were surprised to see widespread non-neural expression of the embryonic lethal abnormal vision (elav) gene, whose protein product Elav is widely used as a marker for post-mitotic neurons in Drosophila (Robinow and White, 1991) and vertebrates (Park et al., 2000). It has previously been noted that elav is transcribed in proliferating neuroblasts but not translated (Berger et al., 2007); we confirm here strong elav transcription in the neuroblast clusters (Figure 2A). Surprisingly, we also found elav broadly transcribed at lower levels in all tissue types of the embryo, including mesodermal derivatives, glia, trachea, gut, and fat body (Figure 2A). An elav paralog, found in neurons (fne), also shows the same pattern of high-level expression in neuronal clusters and broad, lower-level expression in all cell types (Figure 2C). This low level expression was not observed for other neuron-specific genes such as brp or nSyb (Figure 2B,D). This suggests that only post-mitotic neurons have a mechanism for translating the elav and fne transcripts. Similarly, the C. elegans single ortholog of Elav, named Exc-7, is also expressed in non-neuronal cell types including muscle and hypoderm (Pham and Hobert, 2019). In contrast, none of the zebrafish orthologs show noticeable transcription outside the CNS (Farnsworth et al., 2020).
Figure 2. Tissue-specific proteins can be widely transcribed.
(A-F) Plots of elav (A), nSyb (B), fne (C), brp (D), Mhc (E), and Mef2 (F) expression level in whole embryo single-cell atlas. The number at the bottom of each panel is the average expression level of the gene in the specific tissue (top) or other tissues (bottom) computed by the AverageExpression function in Seurat.
To see if this mechanism could be generalized to another tissue, we examined several pan-mesodermal genes, and while most showed narrow expression in some or all mesodermal derivatives (Figure 1C), the myosin heavy chain (Mhc) gene showed broad expression in all cell types (Figure 2E), even though Mhc protein is only detected in mesodermal lineages. This low level expression was not observed for other muscle-specific genes such as Mef2 (Figure 2F). We conclude that some genes are transcribed widely followed by tissue-specific translation. Candidates for positive regulators of this process would be RNA-binding proteins or long non-coding RNAs enriched specifically in mesodermal derivatives (Supplemental Table 1). It will be of interest to understand the global mechanisms regulating mRNA translation that refine these broad expression patterns to unique cell types. We conclude that some tissue-specific or cell type-specific proteins are widely transcribed, revealing a major role for post-transcriptional regulation. It will be interesting to determine the mechanism of the post-transcriptional regulation either via RNA-binding proteins or microRNAs.
Developmental timeline of all embryonic cell types
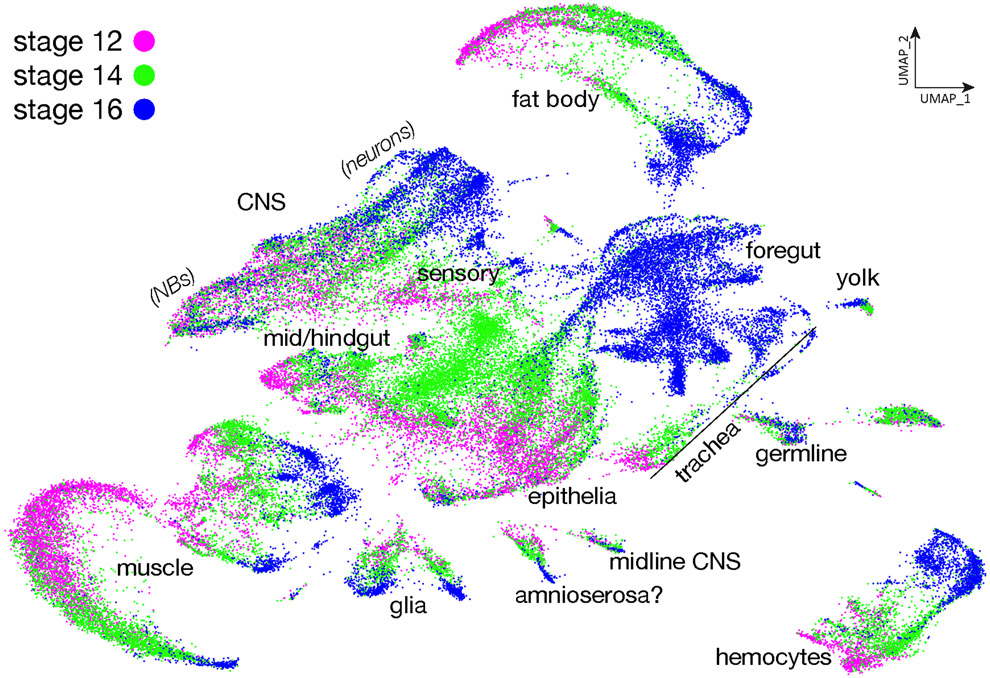
In order to visualize the developmental trajectories of each identified cell type in the atlas, we plotted the timepoint of origin of each cell (Figure 3). We observe that unbiased clustering orders most cells of each identity along a maturation axis from stage 12 to stage 16. In some cases (CNS neuroblasts, yolk) we observe less transcriptional differences over time, with cells from each timepoint clustering together instead of separating along a developmental axis; the fact that not all cell types show stage-specific clustering makes it unlikely for it to be due to batch effects. Furthermore, we see excellent correlation between the two technical replicates at stages 12, 14, or 16 (Supplemental Figure 1). We draw three conclusions from these data. Firstly, most tissue types are established early in embryogenesis and change their transcriptional programs as they mature over time. Secondly, cell types such as CNS and glial progenitors continually produce progeny and show less variability along a developmental axis, as their core transcriptional identity as progenitors is maintained from stages 12-16. Thirdly, cells in the same tissues may develop synchronously, like fat body or hemocytes, so the cells at different developmental stages cluster together to form developmental trajectories. In contrast, the cells that develop asynchronously such CNS or epithelia do not cluster together during development. Lastly, some cell types such as germline and yolk cells are established early in development and their transcriptomes stay constant across time with almost no change (Figure 3).
Figure 3. Developmental trajectory of all embryonic cells.
Plot of integrated single cell atlas based on their developmental stages in UMAP space. Stage 12, magenta; stage 14, green; stage 16, blue.
Neural cell type atlas
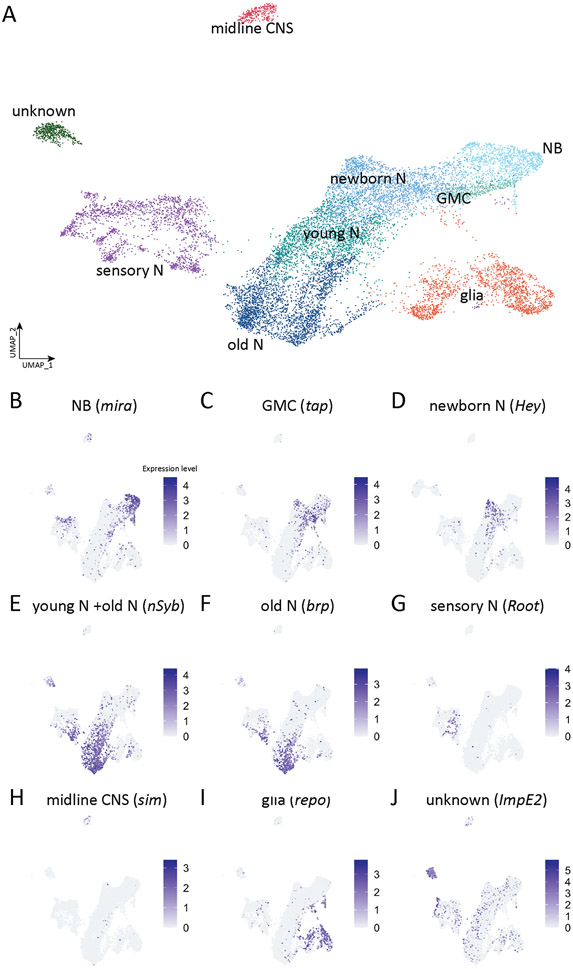
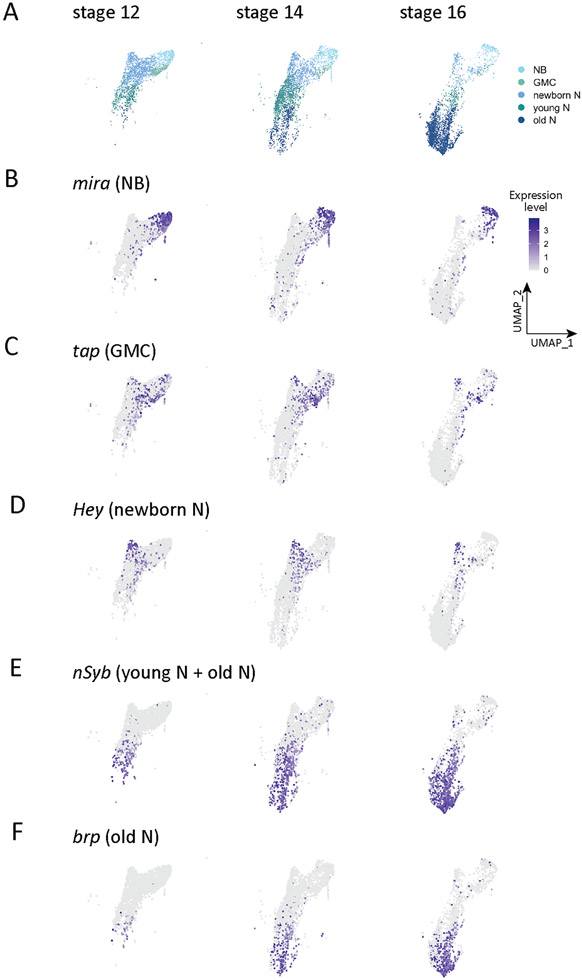
We next wanted to characterize the neural transcriptomes in more detail, so we extracted the neuronal clusters (4, 13, 14, 22, 23, 25, 27, 28, 37, 39, 45, 47, 53, 59, 61, 67, 71, 73, 75, 85) from the merged all cells data set shown in Figure 1A and generated a new "embryonic neural cell type" atlas containing 13,917 cells distributed into 32 clusters (Seurat FindClusters resolution 2.0, dims 1:50) (Supplemental Table 3). We then manually assigned different CNS cell types (Figure 4) based on the following experimentally validated marker genes (Table 1): neuroblasts, miranda (mira) (Figure 4B); GMCs, tap (Figure 4C); new-born NotchON neurons, Hey (Figure 4D); young neurons, neuronal synaptobrevin (nSyb)+ bruchpilot (brp)− (Figure 4E); mature neurons (or old neurons), brp (Figure 4F); midline cells, single-minded (sim) (Figure 4G); sensory neurons, Root (Figure 4H); glia, reverse potential (repo) (Figure 4I); and one unknown cluster (Figure 4J). We conclude that our merged stage 12, 14, and 16 atlas has representation from all major neural cell types.
Figure 4. Atlas of reclustered embryonic nervous system.
(A) Integrated single-cell atlas of Drosophila embryonic nervous system. Cluster identity is determined by the expression of ground-truth genes as indicated.
(B-I) Plots of expression level of ground-truth genes for each individual neural cell type in UMAP space. Colors encode logarithm-transformed expression levels.
Table 1.
Cell type-specific genes used for cluster annotation
| Cell type | Marker | References |
|---|---|---|
| Neuroblast | Miranda+ | (Ikeshima-Kataoka et al., 1997) |
| GMC | Target of PoxN+ | (Michki et al., 2021) |
| Neuron, new-born | Hey+ | (Monastirioti et al., 2010) |
| Neuron, immature | Brp− nSyb+ | (Deitcher et al., 1998; Wagh et al., 2006) |
| Neuron, mature | Brp+ nSyb+ | (Deitcher et al., 1998; Wagh et al., 2006) |
| Neuron, undifferentiated | Hdc+ | (Avet-Rochex et al., 2014) |
| Neuron, midline | Sim+ | (Crews et al., 1988) |
| Glial | Repo+ | (Campbell et al., 1994; Xiong et al., 1994) |
| Sensory | Root+ | (Styczynska-Soczka and Jarman, 2015) |
We next narrowed our focus to the CNS, and asked how the cell type-specific clusters changed over the three developmental stages analyzed here (stages 12, 14, 16). Stage 12 embryos are actively undergoing neuroblast divisions in this early stage of neurogenesis, while by stage 14 neurogenesis and axon outgrowth are proceeding. By stage 16 neurons are actively involved in axon guidance, dendrite outgrowth, and synaptic connectivity (Goodman and Doe, 1993). Confirming previous work, we find that there is a general shift from expression of neuroblast markers to mature neuronal markers across these timepoints (Figure 5A; Table 2). For example, the neuroblast marker miranda (mira) is expressed in many cells at stage 12, but few at stage 16 (Figure 5B; Table 2); conversely, the mature neuron marker brp is barely detected at stage 12 (these may be pioneering motor neurons (Thomas et al., 1984)), but broadly expressed at stage 16 (Figure 5F; Table 2). Genes characterizing other stages of neuronal development fall in between these extremes (Figure 5C-E; Table 2). We conclude that cell type specific clusters validated by ground truth experimental data for cluster defining genes will provide a rich resource for further identification and functional characterization that generate cell type-specific biology (e.g. neuroblast self-renewal or asymmetric division in the neuroblast cluster, or synaptogenic molecules in the mature neuron cluster). For cell types with few markers, such as GMCs or young neurons, our atlas provides the opportunity to identify additional cell type-defining genes.
Figure 5. Neural cell type atlas.
(A) Atlas of neural cells at embryonic stage 12, stage 14, and stage 16. NB, neuroblast; GMC, ganglion mother cell; newborn N, newborn neuron; young N, young neuron; old N, mature neuron.
(B-F) Distribution of ground-truth genes in the neural cell UMAP at embryonic stage 12, stage 14, and stage 16. Colors encode logarithm-transformed expression levels.
Table 2.
Cell types identified by specific gene expression across development
| Stage 12 (2589 cells) |
Stage 14 (3041 cells) |
Stage 16 (2490 cells) |
|
|---|---|---|---|
| NB | 18.7% | 12.2% | 8.5% |
| GMC | 11.6% | 3.4% | 1.2% |
| Newborn N | 49.8% | 27.3% | 9.0% |
| Young N | 18.0% | 39.6% | 10.2% |
| Old N | 2% | 17.4% | 71.1% |
Neural cell type clustering of transcription factors and cell surface molecules
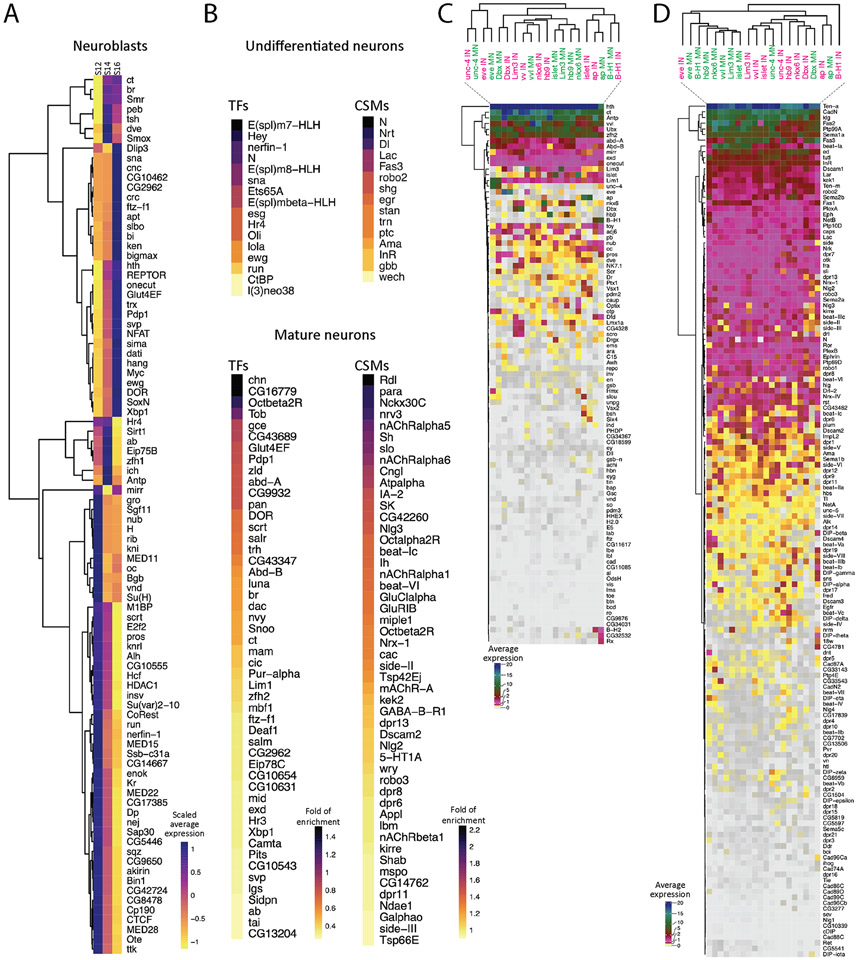
The most differentially expressed genes in the larval CNS and in other organisms are transcription factors (TFs) and cell surface molecules (CSMs) (Li et al., 2020). Here we identify all TFs differentially expressed in the neuroblasts at each developmental timepoint (Figure 6A). Although NBs express different TF subsets over time, but only Kr and pdm1 (nub) were identified as differentially expressed at stage 12 but not later stages (Figure 6A). This may be due to the TTFs Hb, Pdm2, and Cas undergoing their expression cascade at early stages (9-12) that are not fully represented in our data set. It may also be due to lineage asynchrony amongst the total NB population. Of course, it remains possible that different TFs are co-expressed with temporal TFs when examined at a single lineage level of resolution; we explore this possibility in the following section. Nevertheless, our analysis identifies TFs with enriched expression in neuroblasts at stages 12, 14, or 16 (Figure 6A); these make excellent candidates for TTFs that are expressed after the canonical embryonic neuroblast TTF cascade.
Figure 6. Gene expression profiles in neuroblasts and neurons.
(A) Heatmap of scaled average expression of pooled transcription factors statistically enriched in neuroblasts at stage 12 (S12), stage 14 (S14), and stage 16 (S16). Gene names are listed at the right side. Colors encode the levels of scaled average expression of the transcription factors at different stages. Dendrogram at the left shows the clustering of the transcription factors based on the levels of scaled average expression.
(B) Heatmap of statistically enriched transcription factors (TFs) and cell surface molecules (CSMs) in undifferentiated and mature neurons from all stages (12, 14, 16). Gene names are listed at the right side. The colors encode the logarithm-transformed folds of enrichment of average expression.
(C-D) Heatmap of homeodomain transcription factors (HDTFs) (C) and cell surface molecules (CSMs) (D) in selected motor neurons (MNs) and interneurons (INs). Gene names are listed at the right side. Colors encode the levels of average expression of HDTFs (C) and CMSs (D). Gene names are shown at the right side. Dendrogram at the top shows the clustering of different types of cells, and dendrogram at the left shows the clustering of HDTFs (C) or CSMs (D).
We next switched to analyzing the TF and CSM transcriptomes of undifferentiated cells (hdc+) and mature neurons (brp+) to understand the differences between immature and fully differentiated neurons (Figure 6B). We found undifferentiated neurons are enriched for Notch signaling pathway genes (Dl, Hey, E(spl)m7-HLH, E(spl)m8-HLH, E(spl)mβ-HLH, and N) (Supplemental Figure 2), or neuroblast-related genes (esg, l(3)neo38, run, and sna). This is consistent with previous findings showing that Notch signaling is important for specifying sibling neurons following GMC division (Skeath and Doe, 1998). In contrast, mature neurons are enriched for TFs promoting cell fate (ab, br, ct, dac, ftz-f1, and zfh2) and CSMs for synapse formation (beat IIc, beat-VI, dpr6, and dpr8,) and physiological functions (nAChRα1, nAChRα5, nAChRα6, Octα2R) (Figure 6B). We conclude that Notch signaling is important in early neurogenesis, whereas some TFs and CSMs play a greater role in mature neurons.
We next characterized functionally important homeodomain TFs (HDTFs) expressed in motor neurons as well as a less well characterized subset of interneurons. We chose this subset of TFs because (a) they are known to regulate aspects of motor neuron identity; (b) working with all 800+ TFs in the genome would be unwieldy; and (3) because recent work from the Hobert lab has highlighted a role for HDTFs in specifying neuronal identity (Reilly et al., 2020), and we wanted to test the generalizability of this model. We identified motor neurons as zfh1+ twit+ double positive or VGlut+ single positive; and we identified interneurons as single negative for either zfh1− twit− or VGlut− (Supplemental Figure 3). Interestingly, we found that ventral muscle motor neurons (hb9+, islet+, Lim3+, and nkx6+) express similar set of HDTFs, distinct from the HDTFs expressed by dorsal muscle motor neurons (eve+) or lateral muscle motor neurons (B-H1+) (Figure 6C). This suggests that motor neurons targeting ventral, lateral, or dorsal muscles express a pool-specific array of HDTFs. In contrast, each ventral muscle motor neuron HDTF is also expressed in a subset of interneurons; these interneurons do not cluster together like the motor neurons (Figure 6C). This suggests that interneurons are transcriptionally more distinctive than motor neurons. Interestingly, some motor neurons and interneurons cluster together, such as those having common expression of ap, B-H1, Dbx, eve, unc-4, or vvl (Figure 6C), suggesting that these motor neurons and interneurons are not very different from each other. Furthermore, all motor neurons and interneurons express more than one HDTF (Figure 6C), suggesting that each neuron may expresses a unique set of HDTFs to specify their identity, similar to neurons in C. elegans (Reilly et al., 2020).
We next wanted to identify the CSMs that may be regulated by, and thus co-clustered with, motor neuron expressed HDTFs (Figure 6D). We focused on the CSMs reported in Özkan et al. (Özkan et al., 2013) and the phosphotyrosine kinases/phosphatases. First, we found motor neurons cluster with a similar set of CSMs, regardless of their muscle targets (Figure 6D). Despite this observation, ventral muscle motor neurons (hb9+, islet+, Lim3+, and nkx6+) remain clustered independently of the dorsal muscle MNs (eve+) and lateral muscle MNs (B-H1+) (Figure 6D). In addition, interneurons express similar set of CSMs, and cluster together distinctly from motoneurons, with the exception of eve+ and B-H1+ interneurons, which cluster away from other interneurons (Figure 6D). Secondly, different motor neurons and interneurons use different combination of CSMs (Figure 6D). It remains unclear if the activation of a CSM requires only one HDTF or a unique combination of HDTFs.
NB7-1 single lineage gene expression profiles across embryonic development
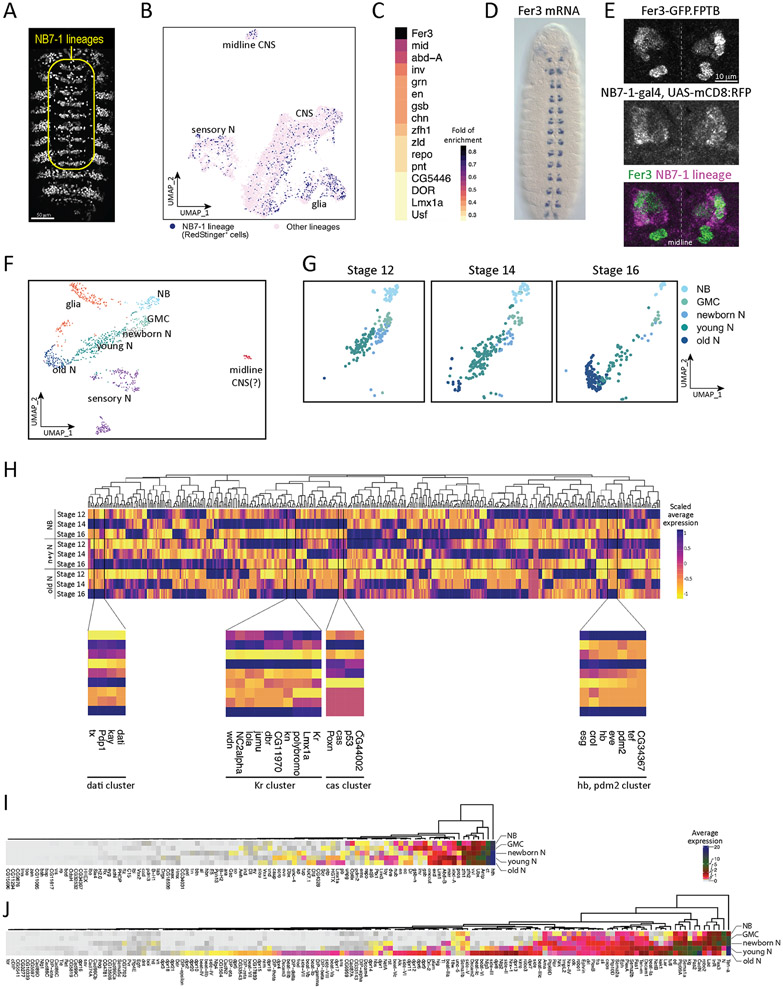
To our knowledge, transcriptional profiling of an individual neuroblast lineage has not yet been performed. Here we identify the NB7-1 lineage by using NB7-1-gal4 to drive the expression of a RedStinger reporter, which is identifiable in-silico by subsetting cells expressing the RedStinger transcript (searchable as RedstingerNLS in the atlas) (Figure 7A). We use this lineage-specific transcriptome to address these questions. (1) Do the cells of the NB7-1 lineage share a gene expression profile and thus cluster together? (2) Can we identify lineage-specific genes, that could maintain the spatial identity of the NB throughout development? (3) Does NB7-1 undergo the same pattern of embryonic stage-specific differentiation? (4) Is it possible to detect genes co-clustered with the temporal TFs within a single NB lineage?
Figure 7. Identification of NB7-1 lineage specific markers and candidate temporal transcription factors.
(A) NB7-1-gal4 drives the expression of RedStinger in the whole embryo; yellow outline shows the segmentally repeated NB7-1 lineage, whereas expression outside the outline are epidermal cells that are not part of the neural population.
(B) Distribution of NB7-1 lineage (RedStinger+) cells in the whole embryo CNS atlas.
(C) Heatmap of statistically enriched transcription factors in the NB7-1 lineage. Colors encode the logarithm-transformed folds of enrichment of average expression.
(D-E) Expression of Fer 3 mRNA (D) at stage 13 embryo (rotated from https://insitu.fruitfly.org/insitu_image_storage/img_dir_118/insitu118808.jpe ) and GFP-tagged Fer3 (E). Fer3 (E, top panel) and NB7-1-gal4 driven expression of membrane-bound RFP (E, middle panel) overlaps in NB7-1 lineage (E, bottom panel).
(F) UMAP of reclustered NB7-1 lineage cells.
(G) UMAP of NB7-1 neural cells at stage 12, stage 14, and stage 16. NB, neuroblast; GMC, ganglion mother cell; N, neuron.
(H) Scaled average expression of transcription factors in neuroblasts (NB), newborn and young neuron (newborn & young N), and mature neuron (old N) at stage 12, stage 14, and stage 16. Color encodes the levels of scaled average expression of each transcription factor at different stages. The bottom panels show the magnified branches of cluster tree clustered with known temporal identify factors (hb, Kr, pdm2, and cas), and Ptx1, and each cluster is bordered by the vertical black lines. Gene names are listed at the bottom. Note that it is not surprising that Hb and Pdm2 are in the same cluster, as they are both detected in the neuroectoderm and also in some early-born Hb+ neurons (Bhat et al., 1995).
(I-J) Heatmap of average expression of homeodomain transcription factors (HDTFs) and cell surface molecules (CSMs) in NB7-1 neuroblast (NB), GMC (ganglion mother cells), newborn neurons (newborn N), young neurons (young N) and mature neurons (old N). Gene names are shown at the bottom. Dendrogram at the top shows the clustering of HDTFs (I) and CSMs (J). Colors encode the levels of average expression.
(1) Surprisingly, when unsupervised clustering was performed on all CNS cells, the NB7-1 lineage did not cluster distinctly away from other lineages in UMAP space (Figure 7B). This indicates significant overlap in gene expression patterns between individual NBs.
(2) We were able to identify several genes enriched in NB7-1 (Figure 7C), including some known to be expressed in NB7-1, such as the tandem engrailed (en)/invected (inv) genes (Broadus et al., 1995), gooseberry (gsb) (Broadus et al., 1995) and neuromancer 2 (Flybase: mid) (Leal et al., 2009). In addition, the top enriched gene was Fer3, a transcription factor that has not been previously characterized in the CNS. The Fer3 RNA expression pattern shows a segmentally repeated cluster of cells adjacent to the midline consistent with expression in NB7-1 (https://insitu.fruitfly.org/cgi-bin/ex/report.pl?ftype=3&ftext=LD04689-a); Figure 7D). We use an endogenous Fer3:GFP transgene and found it overlaps with NB7-1-gal4 UAS-RFP expression, thereby validating its expression in the NB7-1 lineage (Figure 7E). Note that there is also a cluster of Fer3+ cells that is RFP-negative, which is likely to be expression in an adjacent lineage.
(3) We reclustered the 655 NB7-1 lineage cells (RedStinger+) from the whole CNS (8595 cells) (Seurat FindClusters resolution 2.0, dims 1:50), and found that the NB7-1 lineage generates multiple cell types (Figure 7F). We also found that the NB7-1 lineage shows the same pattern of differentiation seen in the general CNS population (Figure 7G; Table 3).
Table 3.
Cell types in NB7-1 lineage identified by specific gene expression across development
| Stage 12 (187 cells) |
Stage 14 (229 cells) |
Stage 16 (205 cells) |
|
|---|---|---|---|
| NB | 25.1% | 13.1% | 7.3% |
| GMC | 15.0% | 9.7% | 5.4% |
| Newborn N | 21.4% | 10.0% | 2.9% |
| Young N | 38.0% | 60.3% | 30.7% |
| Old N | 0.5% | 7.0% | 53.7% |
(4) We found genes that cluster with the temporal TFs Hb, Kr, Pdm, or Cas. The genes (crol, eve, esg, and tef) are expressed in a similar spatiotemporal pattern to hb (Figure 7H); dbr, kn, Lmx1A, and wdn, are expressed similarly to Kr (Figure 7H), and CG44002 and Poxn are similar to cas (Figure 7H). We also found a gene module (dati cluster, Figure 7H) which shows expression at later timepoints than hb, Kr, and cas; this module may include novel temporal TFs which function late in the lineage. Some of these genes are targets of temporal TFs in the NB7-1 lineage (e.g. eve) (Isshiki et al., 2001), whereas others may be acting in parallel or after the known neuroblast temporal TFs. In any case, these co-clustered genes are excellent candidates for regulating neuronal identity in the NB7-1 lineage.
Homeodomain transcription factors and cell surface molecules are upregulated from progenitors to neurons in the NB7-1 lineage
We have shown earlier that HDTFs and CSMs are differentially expressed in post-mitotic motor neurons and interneurons. It remains unclear if these molecules, essential for neuron fate specification and morphogenesis, are inherited from progenitor cells or synthesized de-novo in post-mitotic neurons. Here we use the NB7-1 lineage to address this concept. We computed the average expression of all HDTFs and selected CSMs (see Figure 6) in the NB7-1 lineage. We found that some HDTFs may initially be expressed in NBs; these include spatial factors (gsb) or neuroblast self-renewal factors (pros). Some HDTFs are modestly expressed in neuroblasts, but upregulated in newborn neurons (ems, pdm2, and repo). Some are highly expressed in young neurons (acj6, caup, Dbx, eve, and scro), while some HDTFs only expressed at significant levels in mature neurons (CG4328, Dfd, and Nk7.1) (Figure 7I, Supplemental Figure 4). Most of these CSMs have expression restricted to mature neurons consistent with a role in neuron pathfinding and synapse formation. Interestingly, some CSMs, especially that play a role in axon guidance, show early enrichment in neuroblasts (Fas3, Sema1a, Ten-a, and robo2) (Figure 7J, Supplemental Figure 4); their function in progenitors remains unknown. In conclusion, we found several HDTFs and CSMs with known roles in neuronal fate determination and morphogenesis to be enriched in the NB7-1 lineage as early as in neuroblasts.
Methods
Fly Stocks
Male and female Drosophila melanogaster were used. The chromosomes and insertion sites of transgenes (if known) are shown next to genotypes. Previously published gal4 lines, mutants and reporters used were: NB7-1-gal4KZ (II) (Seroka and Doe, 2019), called NB7-1-gal4 here; UAS-RedStinger [RRID:BDSC_8547]; UAS-mCD8:RFP[RRID:BDSC_32218]; and Fer3-GFP.FPTB[RRID:BDSC_66447].
Immunostaining and imaging
DyLight 488-conjugated goat anti-GFP antibody was used (Novus Biologicals, Centennial, CO). Embryos were fixed and stained as previously described (Seroka and Doe, 2019). The samples were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Images were captured with a Zeiss LSM 900 confocal microscope with a z-resolution of 0.5 μm. Images were processed using the open-source software FIJI. Figures were assembled in Adobe Illustrator (Adobe, San Jose, CA).
Embryo dissociation
Cell dissociates were prepared from 8-9 hr (stage 12), 10-11 hr (stage 14) and 15-16 hr (stage 16) embryos respectively. Embryos were washed in DI water, before surface sterilization in 100% bleach for 5 minutes. Embryos were homogenized in Chan-Gehring (C+G) media by six to eight strokes of a loose-fitting dounce. The cell suspension was filtered through a 40 μm Nitex mesh, and cells were pelleted in a clinical centrifuge at 4°C (setting 5, IEC). The cell pellet was washed twice by pouring off the supernatant and gently triturating the pellet in fresh C+G. Percent cell-survival was determined for each dissociate by BioRad TC-20 trypan-blue assay.
Single cell RNA-seq
Sample preparation was performed by the University of Oregon Genomics and Cell Characterization core facility (https://gc3f.uoregon.edu/) Dissociated cells were run on a 10X Chromium platform using 10X V2 chemistry targeting 10,000 cells per sample. Following cDNA library preparation, the library for each timepoint was amplified with 15 cycles of PCR before sequencing on two separate Illumina Hi-seq lanes, providing two technical replicates for each timepoint (stages 12, 14, 16). Following examination for batch effects between technical replicates (Supplemental Figure 1) the datasets were merged using the CellRanger Aggregate function prior to quality control and downstream analysis. Reads were aligned to the Drosophila genome (BDGP6.22) and protein coding reads were counted. The resulting sequencing data were analyzed using the 10X CellRanger pipeline, version 3.1.0 (Zheng et al., 2017) and the Seurat software package for R, v3.1.2 using standard quality control, normalization, and analysis steps. Cells were filtered by % expression of mitochondrial genes, indicating high stress state. Only cells expressing <20% mitochondrial reads were retained for analysis. Additionally, cells containing reads for <50 and >3000 unique genes were filtered out of downstream analysis. For each gene, expression levels were normalized by total expression, multiplied by a scale factor (10,000) and log-transformed, and the top 3000 variable genes were identified for downstream PCA. Clustering was performed using all computed principal components (common max of 50 PCs) to maximize detection of heterogeneity. Differential expression analysis was performed with the FindAllMarkers function in Seurat using Wilcoxon rank sum test. Tissue identity was determined by the expression score of tissue-specific genes (Supplemental Table 2) with the AddModuleScore function in Seurat. Cells were subsetted for further analysis based on the clustering and expression of ground-truth genes (see Results sections). Raw sequence files have been deposited at NCBI GEO (GSE202987) and R scripts used for analysis have been deposited at Github (https://github.com/AustinSeroka/2022-Doe-Drosophila-Embryo-Atlas).
Supplementary Material
Supplemental Table 1. Genes enriched in each embryonic tissue cluster in Figure 1.
A spreadsheet of markers of each individual cluster determined by FindAllMarkers function in Seurat. Identity of each cluster is determined by tissue-specific genes (Supplemental Table 2).
Supplemental Table 2. Genes used as tissue-specific "ground truth" in Figure 1.
Sixteen separate sheets are included. Each sheet contains a list of genes that are annotated to be expressed in the tissue based on in situ hybridization database (https://insitu.fruitfly.org/cgi-bin/ex/insitu.pl), and used to identify tissue-specific clusters in Figure 1.
Supplemental Table 3. Genes enriched in each CNS tissue cluster.
A spreadsheet of markers of each individual cluster determined by FindAllMarkers function in Seurat. Identity of each cluster is determined by ground-truth genes; see Figure 4.
Supplemental Figure 2. Atlas of undifferentiated neurons and mature neurons.
Atlas of undifferentiated neurons and mature neurons in CNS (top-left panel) based on headcase (hdc) expression (top left). The remaining genes are known Notch pathway components. Gray/navy color gradient encodes the expression level.
Supplemental Figure 3. Atlas of different types of motor neuron and interneuron in CNS.
Distribution of different types of motor neurons and interneurons in a UMAP CNS atlas.
Supplemental Figure 4. Atlas of genes expressed in NB7-1 lineage.
Atlas of NB7-1 lineage in UMAP and feature plots of genes expressed in NB7-1 lineage. Colors encode the expression level.
Supplemental Figure 1. Pairs of technical replicates show correlated expression.
UMAP plots show a high correlation between pairs of technical replicates at each stage.
Acknowledgements
We thank Noah Dillon and Dylan Farnsworth for comments on the manuscript. We thank Maggie Weitzman in the Genomics Core for library preparation and sequencing.
Funding
This work was funded by HHMI (S-LL,CQD), NIH R01 HD27056 (CQD), and T32 HD007348 (AQS).
Abbreviations
- sc-RNAseq
Single cell RNA sequencing
- Eve
Even-skipped
- TTF
Temporal transcription factor
- Hb
Hunchback
- Kr
Kruppel
- Pdm
Nubbin/Pou domain 2
- Cas
Castor
- NB7-1-gal4
NB7-1 split gal4
- CNS
central nervous system
Footnotes
Consent for publication
All authors have approved this manuscript.
Availability of data and materials
All code used for analysis will be uploaded to GitHub upon acceptance. No new fly stocks were generated, and all fly stocks are available from public stock centers or by request.
Competing interests
The authors declare no competing interests.
References
- Avet-Rochex A, Carvajal N, Christoforou CP, Yeung K, Maierbrugger KT, Hobbs C, Lalli G, Cagin U, Plachot C, McNeill H, Bateman JM, 2014. Unkempt is negatively regulated by mTOR and uncouples neuronal differentiation from growth control. PLoS Genet. 10, e1004624. 10.1371/journal.pgen.1004624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Renner S, Lüer K, Technau GM, 2007. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev. Dyn. Off. Publ. Am. Assoc. Anat 236, 3562–3568. 10.1002/dvdy.21372 [DOI] [PubMed] [Google Scholar]
- Bhat KM, Poole SJ, Schedl P, 1995. The miti-mere and pdm1 genes collaborate during specification of the RP2/sib lineage in Drosophila neurogenesis. Mol. Cell. Biol 15, 4052–4063. 10.1128/MCB.15.8.4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ, 1995. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Dev 53, 393–402. https://doi.org/0925477395004548 [pii] [DOI] [PubMed] [Google Scholar]
- Brunet Avalos C, Sprecher SG, 2021. Single-Cell Transcriptomic Reveals Dual and Multi-Transmitter Use in Neurons Across Metazoans. Front. Mol. Neurosci 14, 623148. 10.3389/fnmol.2021.623148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A, 1994. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development 120, 2957–66. [DOI] [PubMed] [Google Scholar]
- Crews ST, Thomas JB, Goodman CS, 1988. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell 52, 143–151. 10.1016/0092-8674(88)90538-7 [DOI] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, Bravo González-Blas C, Poovathingal S, Hulselmans G, Spanier KI, Moerman T, Vanspauwen B, Geurs S, Voet T, Lammertyn J, Thienpont B, Liu S, Konstantinides N, Fiers M, Verstreken P, Aerts S, 2018. A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 174, 982–998.e20. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitcher DL, Ueda A, Stewart BA, Burgess RW, Kidokoro Y, Schwarz TL, 1998. Distinct requirements for evoked and spontaneous release of neurotransmitter are revealed by mutations in the Drosophila gene neuronal-synaptobrevin. J. Neurosci. Off. J. Soc. Neurosci 18, 2028–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ, 2017. Temporal Patterning in the Drosophila CNS. Annu Rev Cell Dev Biol 33, in press. [DOI] [PubMed] [Google Scholar]
- Farnsworth DR, Saunders LM, Miller AC, 2020. A single-cell transcriptome atlas for zebrafish development. Dev. Biol 459, 100–108. 10.1016/j.ydbio.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CS, Doe CQ, 1993. Embryonic development of the Drosophila central nervous system, in: Bate M, Martinez Arias A (Eds.), The Development of Drosophila Melanogaster. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp. 1131–1207. [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, Yao Z, Eggermont J, Höllt T, Levi BP, Shehata SI, Aevermann B, Beller A, Bertagnolli D, Brouner K, Casper T, Cobbs C, Dalley R, Dee N, Ding S-L, Ellenbogen RG, Fong O, Garren E, Goldy J, Gwinn RP, Hirschstein D, Keene CD, Keshk M, Ko AL, Lathia K, Mahfouz A, Maltzer Z, McGraw M, Nguyen TN, Nyhus J, Ojemann JG, Oldre A, Parry S, Reynolds S, Rimorin C, Shapovalova NV, Somasundaram S, Szafer A, Thomsen ER, Tieu M, Quon G, Scheuermann RH, Yuste R, Sunkin SM, Lelieveldt B, Feng D, Ng L, Bernard A, Hawrylycz M, Phillips JW, Tasic B, Zeng H, Jones AR, Koch C, Lein ES, 2019. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F, 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, 625–9. 10.1038/37641 [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ, 2001. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–21. [DOI] [PubMed] [Google Scholar]
- Karaiskos N, Wahle P, Alles J, Boltengagen A, Ayoub S, Kipar C, Kocks C, Rajewsky N, Zinzen RP, 2017. The Drosophila embryo at single-cell transcriptome resolution. Science 358, 194–199. 10.1126/science.aan3235 [DOI] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ, 2013. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152, 97–108. 10.1016/j.cell.2012.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N, Kapuralin K, Fadil C, Barboza L, Satija R, Desplan C, 2018. Phenotypic Convergence: Distinct Transcription Factors Regulate Common Terminal Features. Cell 174, 622–635.e13. 10.1016/j.cell.2018.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal SM, Qian L, Lacin H, Bodmer R, Skeath JB, 2009. Neuromancer1 and Neuromancer2 regulate cell fate specification in the developing embryonic CNS of Drosophila melanogaster. Dev. Biol 325, 138–150. 10.1016/j.ydbio.2008.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li T, Horns F, Li J, Xie Q, Xu C, Wu B, Kebschull JM, McLaughlin CN, Kolluru SS, Jones RC, Vacek D, Xie A, Luginbuhl DJ, Quake SR, Luo L, 2020. Single-Cell Transcriptomes Reveal Diverse Regulatory Strategies for Olfactory Receptor Expression and Axon Targeting. Curr. Biol. CB 30, 1189–1198.e5. 10.1016/j.cub.2020.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, 2020. Principles of Neurobiology, 2nd edition. ed. Garland Science, Boca Raton, FL. [Google Scholar]
- Mark B, Lai S-L, Zarin AA, Manning L, Pollington HQ, Litwin-Kumar A, Cardona A, Truman JW, Doe CQ, 2021. A developmental framework linking neurogenesis and circuit formation in the Drosophila CNS. eLife 10. 10.7554/eLife.67510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JA, Holbrook S, Isshiki T, Weiss J, Doe CQ, Mellerick DM, 1998. Dorsoventral patterning in the Drosophila central nervous system: the vnd homeobox gene specifies ventral column identity. Genes Dev 12, 3603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin CN, Brbić M, Xie Q, Li T, Horns F, Kolluru SS, Kebschull JM, Vacek D, Xie A, Li J, Jones RC, Leskovec J, Quake SR, Luo L, Li H, 2021. Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. eLife 10, e63856. 10.7554/eLife.63856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng JL, Heckscher ES, 2020. Development of motor circuits: From neuronal stem cells and neuronal diversity to motor circuit assembly. Curr. Top. Dev. Biol in press. [DOI] [PubMed] [Google Scholar]
- Meng JL, Marshall ZD, Lobb-Rabe M, Heckscher ES, 2019. How prolonged expression of Hunchback, a temporal transcription factor, re-wires locomotor circuits. Elife 8. 10.7554/eLife.46089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michki NS, Li Y, Sanjasaz K, Zhao Y, Shen FY, Walker LA, Cao W, Lee C-Y, Cai D, 2021. The molecular landscape of neural differentiation in the developing Drosophila brain revealed by targeted scRNA-seq and multi-informatic analysis. Cell Rep. 35, 109039. 10.1016/j.celrep.2021.109039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastirioti M, Giagtzoglou N, Koumbanakis KA, Zacharioudaki E, Deligiannaki M, Wech I, Almeida M, Preiss A, Bray S, Delidakis C, 2010. Drosophila Hey is a target of Notch in asymmetric divisions during embryonic and larval neurogenesis. Development 137, 191–201. 10.1242/dev.043604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Vicidomini R, Choudhury SD, Coon SL, Iben J, Brody T, Serpe M, 2021. Single-Cell RNA Sequencing Analysis of the Drosophila Larval Ventral Cord. Curr. Protoc 1, e38. 10.1002/cpz1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, Johnson KG, Zinn K, Celniker SE, Garcia KC, 2013. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell 154, 228–239. 10.1016/j.cell.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Hong SK, Kim HS, Kim SH, Yoon EJ, Kim CH, Miki N, Huh TL, 2000. Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neurosci. Lett 279, 81–84. 10.1016/s0304-3940(99)00940-4 [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ, 2003. Regulation of neuroblast competence in Drosophila. Nature 425, 624–8. 10.1038/nature01910 nature01910 [pii] [DOI] [PubMed] [Google Scholar]
- Pham K, Hobert O, 2019. Unlike Drosophila elav, the C. elegans elav orthologue exc-7 is not panneuronally expressed. MicroPublication Biol. 2019, 10.17912/micropub.biology.000189. 10.17912/micropub.biology.000189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MB, Cros C, Varol E, Yemini E, Hobert O, 2020. Unique homeobox codes delineate all the neuron classes of C. elegans. Nature 584, 595–601. 10.1038/s41586-020-2618-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, White K, 1991. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol 22, 443–61. 10.1002/neu.480220503 [DOI] [PubMed] [Google Scholar]
- Seroka A, Yazejian RM, Lai S-L, Doe CQ, 2020. A novel temporal identity window generates alternating Eve+/Nkx6+ motor neuron subtypes in a single progenitor lineage. Neural Develop. 15, 9. 10.1186/s13064-020-00146-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroka AQ, Doe CQ, 2019. The Hunchback temporal transcription factor determines motor neuron axon and dendrite targeting in Drosophila. Development 137. 10.1242/dev.175570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Zeppillo T, Limon A, 2020. Regional transcriptome analysis of AMPA and GABAA receptor subunit expression generates E/I signatures of the human brain. Sci. Rep 10, 11352. 10.1038/s41598-020-68165-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath JB, Doe CQ, 1998. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125, 1857–65. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S, 2003. Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol 13, 8–15. https://doi.org/S0959438803000072 [pii] [DOI] [PubMed] [Google Scholar]
- Styczynska-Soczka K, Jarman AP, 2015. The Drosophila homologue of Rootletin is required for mechanosensory function and ciliary rootlet formation in chordotonal sensory neurons. Cilia 4, 9. 10.1186/s13630-015-0018-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambalo M, Mitter R, Wilkinson DG, 2020. A single cell transcriptome atlas of the developing zebrafish hindbrain. Dev. Camb. Engl 147, dev184143. 10.1242/dev.184143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JB, Bastiani MJ, Bate M, Goodman CS, 1984. From grasshopper to Drosophila: a common plan for neuronal development. Nature 310, 203–7. 10.1038/310203a0 [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM, 2007. Global analysis of patterns of gene expression during Drosophilaembryogenesis. Genome Biol. 8, R145. 10.1186/gb-2007-8-7-r145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW, 2010. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development 137, 53–61. 10.1242/dev.041749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten J, Gao X, Van Nierop Y Sanchez P, Domsch K, Agarwal R, Bognar L, Paulsen M, Velten L, Lohmann I, 2022. Single-cell RNA sequencing of motoneurons identifies regulators of synaptic wiring in Drosophila embryos. Mol. Syst. Biol 18, e10255. 10.15252/msb.202110255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicidomini R, Nguyen TH, Choudhury SD, Brody T, Serpe M, 2021. Assembly and Exploration of a Single Cell Atlas of the Drosophila Larval Ventral Cord. Identification of Rare Cell Types. Curr. Protoc 1, e37. 10.1002/cpz1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle M-C, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E, 2006. Bruchpilot, a Protein with Homology to ELKS/CAST, Is Required for Structural Integrity and Function of Synaptic Active Zones in Drosophila. Neuron 49, 833–844. 10.1016/j.neuron.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Xie Q, Brbic M, Horns F, Kolluru SS, Jones RC, Li J, Reddy AR, Xie A, Kohani S, Li Z, McLaughlin CN, Li T, Xu C, Vacek D, Luginbuhl DJ, Leskovec J, Quake SR, Luo L, Li H, 2021. Temporal evolution of single-cell transcriptomes of Drosophila olfactory projection neurons. eLife 10, e63450. 10.7554/eLife.63450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C, 1994. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 8, 981–94. [DOI] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Genes enriched in each embryonic tissue cluster in Figure 1.
A spreadsheet of markers of each individual cluster determined by FindAllMarkers function in Seurat. Identity of each cluster is determined by tissue-specific genes (Supplemental Table 2).
Supplemental Table 2. Genes used as tissue-specific "ground truth" in Figure 1.
Sixteen separate sheets are included. Each sheet contains a list of genes that are annotated to be expressed in the tissue based on in situ hybridization database (https://insitu.fruitfly.org/cgi-bin/ex/insitu.pl), and used to identify tissue-specific clusters in Figure 1.
Supplemental Table 3. Genes enriched in each CNS tissue cluster.
A spreadsheet of markers of each individual cluster determined by FindAllMarkers function in Seurat. Identity of each cluster is determined by ground-truth genes; see Figure 4.
Supplemental Figure 2. Atlas of undifferentiated neurons and mature neurons.
Atlas of undifferentiated neurons and mature neurons in CNS (top-left panel) based on headcase (hdc) expression (top left). The remaining genes are known Notch pathway components. Gray/navy color gradient encodes the expression level.
Supplemental Figure 3. Atlas of different types of motor neuron and interneuron in CNS.
Distribution of different types of motor neurons and interneurons in a UMAP CNS atlas.
Supplemental Figure 4. Atlas of genes expressed in NB7-1 lineage.
Atlas of NB7-1 lineage in UMAP and feature plots of genes expressed in NB7-1 lineage. Colors encode the expression level.
Supplemental Figure 1. Pairs of technical replicates show correlated expression.
UMAP plots show a high correlation between pairs of technical replicates at each stage.