Abstract
Microbial targets for protective humoral immunity are typically surface-localized proteins and contain common sequence motifs related to their secretion or surface binding. Exploiting the whole genome sequence of the human bacterial pathogen Streptococcus pneumoniae, we identified 130 open reading frames encoding proteins with secretion motifs or similarity to predicted virulence factors. Mice were immunized with 108 of these proteins, and 6 conferred protection against disseminated S. pneumoniae infection. Flow cytometry confirmed the surface localization of several of these targets. Each of the six protective antigens showed broad strain distribution and immunogenicity during human infection. Our results validate the use of a genomic approach for the identification of novel microbial targets that elicit a protective immune response. These new antigens may play a role in the development of improved vaccines against S. pneumoniae.
Streptococcus pneumoniae (the pneumococcus) is the leading cause of bacterial sepsis, pneumonia, meningitis, and otitis media in young children in the United States. Annually, 7,000,000 middle-ear infections are ascribed to this organism (4). The vaccines in current use are formulations of capsular carbohydrate from the 23 serotypes responsible for 85 to 90% of infections in the United States, but these vaccines are poorly efficacious in infants and the elderly, the populations that are most at risk (1). A heptavalent-capsular-carbohydrate vaccine conjugated to the protein carrier CRM197 has been shown to be well tolerated and efficacious against invasive disease caused by the seven vaccine serotype strains (3) and has recently been approved for use in young children. However, this type of vaccine has several potential limitations, including serotype replacement by strains that are not represented (14).
The advent of whole-genome sequencing of microbes, including microbial pathogens, has revolutionized the methods by which these organisms are studied and has heightened expectations regarding the ability to predict potential targets for antimicrobial agents and vaccines (2, 12, 20). We combined sequence scanning for prediction of surface-localized proteins with an animal model which allowed us to directly screen proteins for vaccine efficacy to identify novel vaccine candidates from the genome sequence of S. pneumoniae. Here we describe the use of a clinically relevant animal model for the evaluation of the vaccine efficacy of proteins identified from the genome sequence of pneumococcus. This approach was validated by the discovery of five previously unidentified genes whose products induced immune responses that protected mice from pneumococcal infection. Similar sequence scanning methods were recently used to identify potential vaccine candidates from the genomic sequence of the gram-negative pathogen Neisseria meningitidis (21) predicted by in vitro correlates of vaccine effectiveness. Here we expand upon the use of genomics to directly demonstrate vaccine efficacy in an animal model for the important pathogen S. pneumoniae.
MATERIALS AND METHODS
Pneumococcal strains and convalescent-phase sera.
S. pneumoniae N4 was provided by Ingeborg Aaberge, National Institute of Public Health, Oslo, Norway, and is a serotype 4 strain isolated from a patient with bacteremia. Strain SJ2 (serotype 6B) was a clinical isolate from the nares of a patient at St. Jude Children's Research Hospital, Memphis, Tenn., and was the generous gift of Pat Flynn. A method involving intraperitoneal injection and isolation from the bloodstream of a mouse was used to increase the virulence of strain SJ2. Pneumococcal strains representing the following serotypes were obtained from the American Type Culture Collection, Manassas, Va.: 1, 2, 3, 4, 5, 6B, 7F, 8, 9N, 9V, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19A, 19F, 20, 22F, 23F, and 33F. Serotype 3 strains A66 and WU2, serotype 4 strain EF5668, and serotype 6A strain EF6796 were obtained courtesy of David Briles, University of Alabama, Birmingham. Pneumococci were grown in Todd-Hewitt broth (Difco, Detroit, Mich.) containing 0.5% yeast extract at 37°C in a 5% CO2 atmosphere. Convalescent-phase sera were a gift from Åke Ôrtqvist, Danderyds Sjukhus, Danderyd, Sweden. Sera were obtained from patients with culture-confirmed bacteremic pneumococcal pneumonia. The pneumococcal serogroups causing infection were determined in 14 of the cases and included types 4 (3 cases), 7 (2 cases), 8, 9, 14 (2 cases), 22 (4 cases), and 23.
Pneumococcal genome sequencing.
A small fragment (1.6 to 2.0 kb) library of total genomic DNA was generated in pUC18 from strain N4. An approximately 8-fold genome coverage was achieved by generating 41,900 random sequences from this library, with an edited average length of 388 bp. A large-insert lambda library was generated to obtain a sequence scaffold. Individual sequences were assembled using the TIGR Assembler as described by Fleischmann et al. (9) to obtain 829 contiguous sequences containing a total of 2,687 predicted open reading frames (ORFs). The current version of the DNA sequence is available electronically for BLAST analysis from The Institute for Genomic Research (Rockville, Md.) at http://www.tigr.org/tdb/mdb/mdbinprogress.html.
To identify potential vaccine candidate genes we first used GeneMark software (Gene Pro, Inc., Atlanta, Ga.) to predict ORFs. ORFs encoding putative surface proteins were identified through public and custom sequence-specific search algorithms based upon their possession of sequence motifs involved in transport or binding of proteins to cell surfaces, including signal peptidase (SPase) I or II (lipoprotein) cleavage sites, type IV prepilin signal sequences, choline-binding domains, or cell wall anchor sequences characteristic of gram-positive organisms (Table 1). In addition, we searched the genome for proteins homologous to putative surface protein virulence factors previously characterized in other organisms.
TABLE 1.
Selection of ORFs from the S. pneumoniae serotype 4 predicted amino acid sequence
| Motif name (reference) | Sequence motifa | No. of ORFs selected for expression |
|---|---|---|
| SPase I signal sequenceb | 30 | |
| SPase II signal sequenceb | LXXC | 26 |
| (D,E,R,K)X6(L,G)XX(V,A)-C | ||
| Cell wall anchorc | LPXTG, LPXAG, LPXTN | 34 |
| Choline-binding domaind | 11 | |
| Homology to virulence factors | 22 | |
| Integrin-binding domain (22) | RGD | 2 |
| Type IV prepilin signal sequencee | 5 | |
| Total | 130 |
Amino acid sequences are represented by single-letter designations; “X” indicates any amino acid; any amino acid in parentheses may occupy the position indicated.
Proteins were analyzed for SPase I signal sequences using P-sort (17) or SignalP (19) algorithms and were predicted to have either cleavable or noncleavable SPase recognition sites. ORFs containing putative SPase I or II signal sequences (25) were further evaluated for the presence of a methionine start codon preceding a sequence encoding a short (<30 amino acids) hydrophobic region as predicted by Kyte-Doolittle analysis using the LaserGene DNAStar software package.
In addition to the listed sequence residues, proteins with cell wall-anchoring motifs were also examined for a characteristic N-terminal hydrophobic region followed by at least one basic residue (R or K) after the anchoring motif (8, 18).
Choline-binding proteins were predicted by comparison to the C-terminal repeat region of PspA and to the consensus domain previously described for other gram-positive bacteria (26, 28).
Type IV prepilin signal sequences were identified by comparison to the ComG locus of Bacillus subtilis (7).
Cloning and expression of vaccine candidates.
ORFs were amplified by PCR with specific oligonucleotide primers encoding restriction endonuclease recognition sites for cloning into the prokaryotic expression vector pQE10 (Qiagen, Chatsworth, Calif.). Proteins were cloned without putative signal sequences. Proteins predicted to be larger than 100 kDa were cloned in smaller subfragments to facilitate expression. Constructs were confirmed by sequencing and transformed into Escherichia coli M15 pREP4 (Qiagen) for expression of recombinant proteins. Proteins were affinity purified from guanidine-solubilized cell pellets using Ni-nitrilotriacetic acid column chromatography and dialyzed gradually against phosphate-buffered saline (PBS) to promote refolding essentially as described in The Qiaexpressionist: a Handbook for High-Level Expression and Purification of 6xHis-Tagged Proteins (Qiagen, 1999).
Mouse immunization and challenge.
Female C3H/HeJ mice, generally 6 to 8 weeks of age, were obtained from Jackson Laboratory (Bar Harbor, Maine) and were immunized subcutaneously in groups of 10 with 15 μg of protein formulated in complete Freund's adjuvant. Twenty-one days later mice were given booster immunizations in the same way with protein formulated in incomplete Freund's adjuvant. Twenty-eight days following the booster, animals were bled and immune titers were determined by an antibody capture endpoint enzyme-linked immunosorbent assay. Bacteria were diluted to approximately 35 to 100 50% lethal doses (LD50) in sterile PBS and injected intraperitoneally (i.p.) into mice in a volume of 100 μl, 35 days following the booster. Mice were monitored for 14 days for mortality. The survival rate was compared with that of a group sham immunized with PBS and adjuvant alone, and protection was evaluated using a two-sample log rank test. For passive-immunization studies, rabbit antisera were generated at Covance, Inc. (Denver, Pa.), using Freund's adjuvant and standard immunization practices. One hundred microliters of antiserum was injected i.p. into 6-week-old C3H/HeJ mice (for SJ2 challenge) or BALB/c ByJ mice (for WU2 and EF5668 challenge) 24 h prior to, and 4 h following, i.p. challenge with approximately 20 to 50 LD50 of pneumococcal strains. Animals were monitored for 14 days for mortality. All animal studies were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at MedImmune, Inc.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed using gels purchased from Novex (San Diego, Calif.). Following separation, proteins were transferred to nitrocellulose membranes and unbound membrane sites were blocked with PBS containing 0.1% Tween 20 (PBS-T) with 5% nonfat dry milk and thimerosal (0.01%). Cell lysates were prepared essentially as previously described (27). Proteins in cell lysates were detected with rabbit or mouse polyclonal antisera generated against recombinant pneumococcal proteins. Bound antibody was detected with a goat anti-rabbit or goat anti-mouse immunoglobulin G horseradish peroxidase-conjugated secondary antibody diluted 1:5,000 in PBS-T (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). Horseradish peroxidase was detected by exposure to film using ECL reagent (Amersham-Pharmacia Biotech, Inc., Piscataway, N.J.). Molecular weights of proteins were determined by comparison with a prestained molecular weight standard.
Flow cytometric analysis.
Flow cytometric analysis was performed as follows. Bacteria were grown to an A620 of between 0.4 and 0.6. The cell density was adjusted to approximately 2 × 106 CFU in 50 μl of PBS, and cells were mixed with antisera diluted 100-fold. After incubation for 1 h at 4°C, unbound antibodies were washed away by centrifugation in excess PBS. Secondary goat antibody labeled with the fluorescent dye Alexa 488 (Molecular Probes, Eugene, Oreg.) was incubated with the cells at 4°C for 1 h, the cells were washed, and bound antibody was detected using a Becton Dickinson FacStar Plus flow cytometer. Control sera included rabbit preimmune serum and rabbit polyclonal serum against E. coli GroEL (Epicentre Technologies, Madison, Wis.).
Nucleotide sequence accession numbers.
The nucleotide sequences of the genes encoding the protective antigens described here have been deposited with GenBank and are available under the accession numbers listed in Table 2.
TABLE 2.
Active immunization of C3H/HeJ mice with recombinant pneumococcal proteins in Freund's adjuvant protects mice from a lethal challenge with a serotype 6B strain of pneumococci
| Antigen | Motif | Accession no. | Subcellular locationa | Database similarityb (accession no.) | Expt | No. of survivors/total no.
|
|
|---|---|---|---|---|---|---|---|
| Active immunizationc | Sham immunization | ||||||
| Sp36 | SPase II | AF291695 | CW/M | Unknown protein from Streptococcus agalactiae (AAD13797) | 1 | 9/10 | 0/10 |
| 2 | 10/10 | 0/10 | |||||
| Sp46 | Choline-binding protein | AF291696 | M | Endo-β-N-acetylglucosaminidase (LytB) from S. pneumoniae (CAA 09078) | 1 | 6/10 | 0/10 |
| 2 | 7/10 | 0/10 | |||||
| Sp91 | Choline-binding protein | AF291697 | M | 1,4-β-N-acetylmuramidase (LytC) from S. pneumoniae (CAA08765) | 1 | 9/10 | 1/10 |
| 2 | 10/10 | 1/10 | |||||
| Sp101 | SPase II | AF291698 | CW/M | Unknown protein from Bacillus anthracis (AAF13613) | 1 | 7/10 | 0/10 |
| 2 | 9/10 | 2/10 | |||||
| Sp128 | Cell wall anchor | AF291699 | CW/M | Cell wall-associated serine proteinase from S. pneumoniae (AAD48399) | 1 | 9/10 | 0/10 |
| 2 | 6/7 | 0/10 | |||||
| Sp130 | Cell wall anchor | AF291699 | CW/M | Cell wall-associated serine proteinase from S. pneumoniae (AAD48399) | 1 | 9/10 | 0/10 |
| 2 | 10/10 | 0/10 | |||||
| PspA | Control | CW/M | 1 | 10/10 | 3/10 | ||
| 2 | 10/10 | 0/10 | |||||
CW, cell wall fraction; M, membrane fraction.
Homology searches were performed by comparing amino acid sequences against the nonredundant protein database using the BLAST algorithm at the National Center for Biotechnology Information. The most homologous amino acid sequence and its accession number are shown here.
The number of survivors compared to that for the sham-immunized control was found to be statistically significant (P < 0.05) for all groups by using a two-sample log rank test.
RESULTS AND DISCUSSION
Identification of vaccine antigens from the pneumococcal genome.
We reasoned that successful vaccine candidates would target protein antigens accessible to antibodies at the pneumococcal surface and that such surface proteins could be identified in the genomic sequence database by one or more signature sequence motifs commonly found in known secreted proteins from other bacteria. Some of these motifs are likely to be restricted to gram-positive bacteria, like the pneumococcus. We initially scanned the genome sequence of a serotype 4 strain of pneumococci (N4) and identified 2,687 potential ORFs. These ORFs were then evaluated to determine whether the encoded gene products contained sequence motifs predictive of their localization on the surface of the bacterium (Table 1). In addition, we identified ORFs with significant homology to surface protein virulence factors of other bacteria. Altogether, this analysis identified 110 different genes contained within the pneumococcal genome from which 130 ORFs, or ORF fragments, were selected for expression (for a complete sequence listing, see the following: C. Kunsch, G. Choi, S. Johnson, and A. Hromockyj, October 1997, Patent Cooperation Treaty publication no. WO 98/18930). This analysis predicts that genes encoding surface proteins represent at least 4% of the S. pneumoniae genome, a percentage similar to that predicted for other bacteria. Proteins encoded by ORFs identified in this manner were expressed in E. coli with polyhistidine tags for ease of purification. The products of 108 of these ORFs, comprising 97 unique genes or their subfragments, were successfully expressed and purified for evaluation as vaccine candidates. Pneumococcal surface protein A (PspA) (24) from strain N4 was expressed and purified in a similar manner, as a positive control.
Evaluation of vaccine efficacy in a mouse model of lethal sepsis.
Each of the novel pneumococcal proteins was tested for the ability to induce protective antibodies against pneumococcal challenge in a mouse sepsis model wherein very low challenge doses (between 1 and 10 CFU) of virulent strains of pneumococci kill mice. Initial studies used S. pneumoniae N4 (LD50, ∼3 CFU) for the evaluation of protein vaccines in this model, but all novel antigens and the positive control (PspA) failed to protect animals from death although some antigens delayed the time to death. Earlier work by Tart et al. (24) also demonstrated that some serotype 4 strains circumvent protein-directed immunity in this model but did not identify the basis for this phenomenon. We subsequently used a different clinical isolate (strain SJ2; LD50, ∼10 CFU) with a 6B capsular serotype to evaluate the 108 vaccine candidates. Six novel antigens (Sp36, Sp46, Sp91, Sp101, Sp128, and Sp130), representing five different genes, and recombinant PspA protected against challenge with approximately 50 to 100 times the LD50 of this strain (Table 2). Of these five genes, one, which encodes a protein with a predicted mass of 220 kDa, was expressed as three fragments (Sp128, Sp129, and Sp130), but only two of these conferred protection. Four of the proteins (Sp36, Sp91, Sp128, and Sp130) also protected against challenge with a serotype 6A strain, EF6796 (data not shown). Additional experimentation revealed that antiserum raised against Sp36 protected mice against a different serotype 4 strain (Table 3). This suggests that the virulence displayed by some serotype 4 strains in this model may be due in part to the expression of additional factors beyond the capsular carbohydrate. Further optimization of expression, purification, or vaccination methods may enhance the protective efficacy of at least some of the remaining candidates. During the course of this study two of these proteins, Sp46 and Sp91, were independently identified by others as LytB (10) and LytC (11), respectively, without characterization of their vaccine efficacies. As both of these proteins contain choline-binding domains, it is possible that some of the protection conferred by these proteins may be due to cross-reactivity of antisera to these regions with other choline-binding proteins, such as CbpA or PspA. However, this possibility is unlikely, as we have found that the choline-binding domain of Sp91 is neither immunogenic nor protective in the mouse model of sepsis (unpublished data).
TABLE 3.
Protection of mice from a lethal challenge with pneumococci following passive immunization with rabbit polyclonal antisera raised against recombinant pneumococcal proteins
| Antigen | Motif | No. of survivors/total no.
|
|||||
|---|---|---|---|---|---|---|---|
| SJ2 (type 6B)
|
WU2 (type
3)
|
EF5668 (type 4)
|
|||||
| Specific serum | Nonspecific serum | Specific serum | Nonspecific serum | Specific serum | Nonspecific serum | ||
| Sp36 | SPase II | 10/10a | 1/10 | 9/10a | 0/10 | 5/10a | 0/10 |
| Sp46 | Choline-binding protein | 5/10a | 1/10 | NDb | ND | ||
| Sp91 | Choline-binding protein | 2/10 | 1/10 | 7/10a | 0/10 | ND | |
| Sp130 | Cell wall anchor | 2/10 | 2/10 | 7/10a | 0/10 | ND | |
Significantly different from the value for the nonspecific serum group (P < 0.05).
ND, not determined.
To confirm that protection was antibody mediated, mice were passively immunized with rabbit hyperimmune sera raised against a subset of the recombinant proteins that had conferred protection by active immunization. Antisera raised against proteins Sp36 and Sp46 significantly protected mice from a lethal challenge with pneumococcal strain SJ2 (Table 3), although antiserum against pneumococcal proteins Sp91 and Sp130 did not passively confer protection against this challenge dose. Antisera recognizing Sp36, Sp91, and Sp130 also protected against lethal challenge with a serotype 3 strain of pneumococci (WU2). These data indicate that protection with these antigens is not capsular serotype restricted. Indeed, protection mediated by Sp36 extends to at least four different capsular types (types 3, 4, 6A, and 6B).
Conservation of vaccine antigens among diverse pneumococcal serotypes.
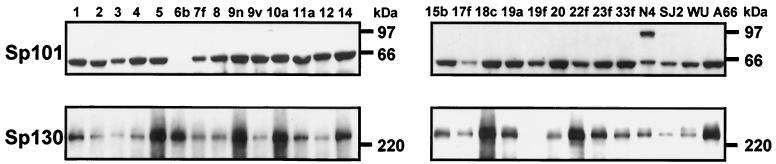
For the vaccine antigens described to be clinically useful, they must be expressed by the majority of the most prevalent pneumococcal strains. Immunoblot analysis was performed with cell lysates prepared from pneumococcal strains representative of the 23 major capsular serotypes and probed with polyclonal antisera specific for the six protective antigens. The results demonstrated a high degree of serological cross-reactivity for all the antigens among the majority of capsular serotypes (Fig. 1). In particular, proteins of the predicted molecular masses that were reactive with antisera to Sp36, Sp46, and Sp128 were expressed in 23 of 23 isolates, Sp91 was detected in all serotypes except a type 3 strain, Sp101 was detected in all serotypes except a type 6B strain, and Sp130 was detected in all except serotype 19F. These results were similar to those reported by Crain et al. (5), who demonstrated that rabbit polyclonal antiserum against PspA recognized that protein from 16 different capsular serotypes, indicating that some PspA epitopes are broadly cross-reactive.
FIG. 1.
Seroconservation of proteins Sp101 and Sp130 among pneumococcal strains. Whole-cell lysates were prepared from strains with serotypes representative of those contained in current 23-valent carbohydrate vaccines (serotypes 1, 2, 3, 4, 5, 6B, 7F, 8, 9N, 9V, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19A, 19F, 20, 22F, 23F, and 33F) or from clinical isolates N4 (serotype 4 strain; source of genomic DNA used in sequencing), SJ2 (serotype 6B), and WU2 and A66 (serotype 3). Proteins in lysates were separated by electrophoresis and immunoblots were probed with rabbit antisera raised against either recombinant Sp101 or Sp130.
Sequence analysis of pneumococcal DNA from strains representing 4 capsular serotypes (types 1, 2, 5, and 6B) demonstrated that the amino acid sequence of Sp36 from these strains was >97% identical to the sequence of this protein from strain N4 (data not shown). However, evaluation of additional strains revealed some degree of heterogeneity among several of the other proteins. For example, Sp91 was detected in only two of three serotype 3 strains tested, while Sp101 was not detected in one serotype 6B isolate but was cross-reactive with the serotype 6B isolate used for murine challenge.
Surface accessibility of vaccine antigens and expression during infection of the human host.
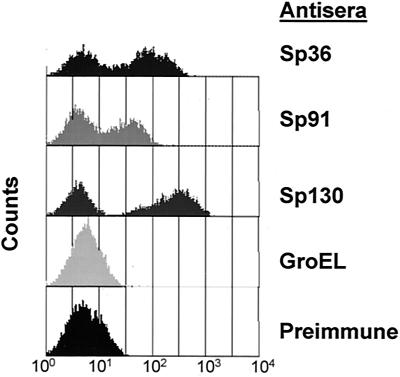
The fact that the immune response elicited by the vaccine antigens described here was protective is presumptive evidence of the antigens' surface localization in pneumococci. Fractionation of pneumococci into subcellular compartments (28), followed by Western blot analysis with antisera specific for 97 of the vaccine candidates, demonstrated that 54 of these proteins were associated with either the cell wall or membrane of the bacterium (results not shown). The products of the genes encoding the six protective antigens and of pspA also fractionated into similar compartments (Table 2). However, to demonstrate surface accessibility directly, we developed an assay based on flow cytometry for detection of antibodies bound to intact pneumococcal cells. Several of the proteins that demonstrated vaccine efficacy were stained by antibodies on intact bacteria (Fig. 2), although only a subpopulation of the bacteria were stained, as indicated by the detection of two peaks. This phenomenon may be a result of differential expression of the gene products during the growth of the bacterium or partial binding inhibition of the antibodies caused by other surface molecules, such as the capsule. Although the flow cytometry data presented here were obtained with the homologous serotype 4 strain used for sequencing, we have observed similar results with other strains which were protected by immunization with these antigens. Thus, the subpopulation of pneumococci that stains more weakly by flow cytometry is still vulnerable to protein-directed antibodies in vivo. Antiserum specific for the E. coli cytosolic chaperone GroEL reacted with a protein (∼70 kDa) in a cytosolic extract of S. pneumoniae by immunoblot analysis but not with intact pneumococci in the flow cytometry assay, providing evidence that labeling of cells was specific for surface-exposed proteins (data not shown). Two antigens (Sp46 and Sp101) that were able to confer protection on mice following active immunization were not detected by flow cytometry. This may be due to an inherent insensitivity of this assay for molecules present in limited quantities on the surface of the pneumococcus, blockage of the antibody binding sites by more abundant surface molecules during in vitro growth, or enhanced in vivo expression of these antigens. For Sp36 we extended this analysis to include the 23 serotypes contained in the currently available pneumococcal polysaccharide vaccines and determined that it could be detected on the surfaces of all serotypes tested (results not shown).
FIG. 2.
Immune detection of proteins on the surface of a serotype 4 pneumococcal strain by flow cytometry. Intact pneumococcal cells were probed with antisera generated against recombinant proteins Sp36, Sp91, and Sp130 or with preimmune serum or antiserum specific for the cytosolic chaperone GroEL as negative controls.
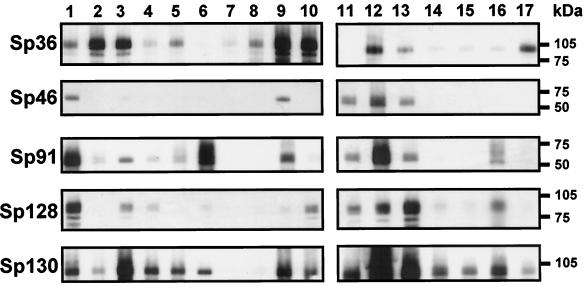
To evaluate expression of these novel pneumococcal antigens during infection in the human host, convalescent-phase sera from 17 patients recovering from bacteremic pneumococcal pneumonia were tested for reactivity with five of the recombinant pneumococcal antigens in an immunoblot assay (Fig. 3). The results demonstrated that immune sera generated during pneumococcal infection in 15 of 17 patients recognized Sp36. In addition, proteins Sp91, Sp128, and Sp130 were recognized by the majority of patient sera (76, 82, and 94%, respectively). Convalescent-phase sera from 71% of patients did not recognize protein Sp46, possibly indicating that this protein is expressed at low levels during this clinical disorder or is poorly immunogenic in its natural context. Due to limitations in quantities of convalescent-phase antisera, Sp101 was not assayed in this manner. While the presence of an immune response to these antigens is not in itself predictive of their ability to protect, the data do indicate that they are expressed in vivo, are immunogenic during infection of the host, and are serologically cross-reactive among capsular serotypes. Serial analyses of the magnitude of the immune responses to these proteins during natural infection and correlation to subsequent pneumococcal colonization and disease may disclose levels of specific antibody required for prophylactic protein vaccines.
FIG. 3.
Immune recognition of pneumococcal vaccine proteins by convalescent-phase sera from patients diagnosed with pneumococcal infections. Recombinant pneumococcal proteins were resolved by polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were probed with convalescent-phase sera obtained from patients recovering from bacteremic pneumococcal pneumonia. Bound antibody was detected with an enzyme-conjugated secondary antibody specific for human immunoglobulin.
Implications for development of bacterial vaccines.
As expected from the clinical experience with Haemophilus influenzae type b capsular conjugate vaccine (16, 23), pneumococcal capsular conjugate vaccines have been efficacious in clinical trials against invasive disease caused by vaccine serotypes. However, most morbidity from S. pneumoniae is due to disease at mucosal sites, chiefly acute otitis media. Clinical trials with heptavalent or nonavalent pneumococcal capsular conjugate vaccines in children have demonstrated a significant reduction in carriage of vaccine serotypes in the nasopharynx in vaccinees (6, 15). This benefit, however, was accompanied by an eventual increase in carriage of nonvaccine serotypes. In the mouse invasive-disease model, immunity to PspA confers protection across several pneumococcal capsular serotypes (13). The same is true of the antigens identified in the present study (Tables 2 and 3 and data not shown). Thus, protein antigens, such as those identified here, may overcome the problem of capsular serotype replacement. They also might act synergistically with capsular conjugate vaccines providing protection against both mucosal and invasive disease, particularly if they are expressed during nasopharyngeal colonization.
The results of this study demonstrate the power of a microbial genomic approach in identifying novel vaccine antigens. This approach is applicable for any microbial pathogen for which genomic sequence data are available, a suitable in vivo model exists, and humoral immunity is important in conferring protection. We are currently evaluating the utility of these proteins, either singly or in combination with each other or with capsular-carbohydrate vaccines, for prevention of mucosal infection and invasive disease caused by pneumococci.
ACKNOWLEDGMENTS
We thank Melissa Dormitzer and the vivarium staff at MedImmune for expert technical assistance, David Carlin and Harry Yang for help with statistical analysis, and Donni Leach for reviewing the manuscript. The contributions of members of the DNA sequencing facility and the Microbial Genomics Group at Human Genome Sciences are also recognized.
REFERENCES
- 1.Austrian R. The pneumococcus at the millennium: not down, not out. J Infect Dis. 1999;179(Suppl. 2):S338–S441. doi: 10.1086/513841. [DOI] [PubMed] [Google Scholar]
- 2.Baltz R H, Norris F H, Matsushima P, DeHoff B S, Rockey P, Porter G, Burgett S, Peery R, Hoskins J, Braverman L, Jenkins I, Solenberg P, Young M, McHenney M A, Skatrud P L, Rosteck P R., Jr DNA sequence sampling of the Streptococcus pneumoniaegenome to identify novel targets for antibiotic development. Microb Drug Resist. 1998;4:1–9. doi: 10.1089/mdr.1998.4.1. [DOI] [PubMed] [Google Scholar]
- 3.Black S, et al. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Pediatr Infect Dis J. 2000;19:187–195. doi: 10.1097/00006454-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP) Morb Mortal Wkly Rep. 1997;46(RR-08):1–24. [PubMed] [Google Scholar]
- 5.Crain M J, Waltman II W D, Turner J S, Yother J, Talkington D F, McDaniel L S, Gray B M, Briles D E. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect Immun. 1990;58:3293–3299. doi: 10.1128/iai.58.10.3293-3299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dagan R, Melamed R, Muallem M, Piglansky L, Greenberg D, Abramson O, Mendelman P M, Bohidar N, Yagupsky P. Reduction of nasopharyngeal carriage of pneumococci during the second year of life by a heptavalent conjugate pneumococcal vaccine. J Infect Dis. 1996;174:1271–1278. doi: 10.1093/infdis/174.6.1271. [DOI] [PubMed] [Google Scholar]
- 7.Dubnau D. Genetic exchange and homologous recombination. In: Sonenshein A L, Hoch J, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 555–584. [Google Scholar]
- 8.Fischetti V A, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from Gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 9.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzaeRD. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.García P, González M P, García E, López R, Garciá J L. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol Microbiol. 1999;31:1275–1277. doi: 10.1046/j.1365-2958.1999.01238.x. [DOI] [PubMed] [Google Scholar]
- 11.García P, González M P, García E, Garciá J L, López R. The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniaereveals evolutionary mobile domains. Mol Microbiol. 1999;33:128–138. doi: 10.1046/j.1365-2958.1999.01455.x. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman S L, Rogers W O, Carucci D J, Venter J C. From genomics to vaccines: malaria as a model system. Nat Med. 1998;4:1351–1353. doi: 10.1038/3934. [DOI] [PubMed] [Google Scholar]
- 13.Langermann S, Palaszynski S R, Burlein J E, Koenig S, Hanson M S, Briles D E, Stover C K. Protective humoral response against pneumococcal infection in mice elicited by recombinant bacille Calmette-Guerin vaccines expressing pneumococcal surface protein A. J Exp Med. 1994;180:2277–2286. doi: 10.1084/jem.180.6.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipsitch M. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg Infect Dis. 1999;5:336–345. doi: 10.3201/eid0503.990304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mbelle N, Huebner R E, Wasas A D, Kimura A, Chang I, Klugman K P. Immunogenicity and impact on nasopharyngeal carriage of a nonavalent pneumococcal conjugate vaccine. J Infect Dis. 1999;180:1171–1176. doi: 10.1086/315009. [DOI] [PubMed] [Google Scholar]
- 16.Murphy T V, Pastor P, Medley F, Osterholm M T, Granoff D M. Decreased Haemophilus colonization in children vaccinated with Haemophilus influenzae type b conjugate vaccine. J Pediatr. 1993;122:517–523. doi: 10.1016/s0022-3476(05)83529-2. [DOI] [PubMed] [Google Scholar]
- 17.Nakai K, Horton P. PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 18.Navarre W W, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Nowak R. Bacterial genome sequence bagged. Science. 1995;269:468–470. doi: 10.1126/science.7624767. [DOI] [PubMed] [Google Scholar]
- 21.Pizza M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 22.Ruoslahti E, Pierschbacher M D. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 23.Takala A K, et al. Reduction of oropharyngeal carriage of Haemophilus influenzaetype b (Hib) in children immunized with an Hib conjugate vaccine. J Infect Dis. 1991;164:982–986. doi: 10.1093/infdis/164.5.982. [DOI] [PubMed] [Google Scholar]
- 24.Tart R C, McDaniel L S, Ralph B A, Briles D E. Truncated Streptococcus pneumoniaePspA molecules elicit cross-protective immunity against pneumococcal challenge in mice. J Infect Dis. 1996;173:380–386. doi: 10.1093/infdis/173.2.380. [DOI] [PubMed] [Google Scholar]
- 25.von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 26.Wren B W. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol. 1991;5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 27.Yother J, Handsome G L, Briles D E. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspAgene. J Bacteriol. 1992;174:610–618. doi: 10.1128/jb.174.2.610-618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yother J, White J M. Novel surface attachment mechanism of the Streptococcus pneumoniaeprotein PspA. J Bacteriol. 1994;176:2976–2985. doi: 10.1128/jb.176.10.2976-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]