Abstract
Powdery mildews (PM) are common and severe pathogen groups that threaten plants, and PM resistance is complex and polygenic in cucumbers. Previously mlo-based resistance was reported in various plants, including cucumber, with generated loss-of CsaMLO function mutants. However, mlo-based resistance in cucumber is also complex and involves additional mechanisms such as hypersensitive response (HR) and papillae formation. For this reason, we focused on determining the mlo-based powdery mildew resistance mechanism in cucumber. CRISPR/Cas9 was used in the present study to generate loss-of-function mutants for CsaMLO1, CsaMLO8, and CsaMLO11 of PM susceptible ADR27 cucumber inbred lines and CsaMLO mutants were obtained and validated. Trypan Blue and DAB staining were performed to detect Podosphaera xanthii germination/penetration rates and accumulation of Reactive Oxygen Species (ROS). Our results indicate that PM-susceptibility associated CsaMLOs in cucumber are negative regulators in different defense mechanisms against powdery mildew at early and late stages of infection. Further, the experiment results indicated that CsaMLO8 mutation-based resistance was associated with the pre-invasive response, while CsaMLO1 and CsaMLO11 could be negative regulators in the post-invasive defense response in cucumber against P. xanthii. Although the loss-of CsaMLO8 function confers the highest penetration resistance, CsaMLO1 and CsaMLO11 double mutations could be potential candidates for HR-based resistance against PM pathogen in cucumber. These results highlighted the crucial role of CRISPR/Cas9 to develop PM resistant cucumber cultivars, possessing strong pre-invasive defense with CsaMLO8 or post-invasive with CsaMLO1/CsaMLO11 mutations.
Keywords: cucumber, CsaMLO, defense response, plant susceptibility, powdery mildew, CRISPR/Cas9
Introduction
Podosphaera xanthii and Golovinomyces cichoracearum are the most common pathogens responsible for PM disease in cucumber (Cucumis sativus L.) Especially, P. xanthii is a serious pathogen in cucurbits, including C. sativus (Zitter et al., 1996). They are managed by fungicides in commercial agricultural production, but these chemicals affect human health and result in serious environmental problems (Xu et al., 2017). Also, PM pathogens could acquire resistance to fungicides with the excessive use of chemical fungicides which will be a more serious problem. Therefore, using PM-resistant cultivars is the most eco-friendly and durable management method for these pathogens. However, PM resistance is complex and polygenic and involves diverse plant defense mechanisms and signaling pathways (Nie et al., 2015). Generating PM-resistant cultivars with introgressed resistance (R) genes from wild form of cultivars is challenging for the breeders. Another concern is that the pathogen could overcome the resistance due to the mutations in avirulence genes even if the PM resistant cultivars are generated using long-term and tiresome backcrossing and inbreeding procedures.
Several susceptibility (S) genes were identified as an alternative for R-mediated resistance in the last decades, and it was supposed that the loss of S-gene function with controlled or naturally occurring mutations could confer durable and broad-spectrum resistance against various diseases in plants (de Almeida Engler et al., 2005). The proteins encoded by S-genes cause host recognition by the pathogen, defense suppression, and facilitate penetration (van Schie and Takken, 2014). These proteins are critical for disease caused by biotrophic pathogens such as PM and downy mildew because their vitality is directly associated with host interactions (Eckardt, 2002). MLOs are the most known S-genes responsible for PM-susceptibility in various plants. Barley mlo is one of the first identified genes associated with PM, and the loss of mlo function still confers the broad-spectrum and durable resistance against Blumeria graminis f.sp hordei (Bgh). Also, various research groups have reported mlo-mediated resistance in other plants such as Arabidopsis thaliana (Consonni et al., 2006), Triticum aestivum (Konishi et al., 2010), Solanum lycopersicum (Bai et al., 2008), Pisum sativum (Humphry et al., 2011), and Cucumis sativus (Berg et al., 2017) with phylogenetic analyses, and they revealed the fact that natural or controlled mutations on these homologs decrease PM susceptibility or confer higher resistance against PM (Jiwan et al., 2013; Zhou et al., 2013; Acevedo-Garcia et al., 2014).
mlo-based resistance contains different pathways and is associated with other defense mechanisms like R-gene-mediated PM resistance in various plants (Xu et al., 2019). For instance, papilla formation is a robust response against PM pathogens and is controlled by PENETRATION (PEN) and SNAREs VESICLE-ASSOCIATED MEM-BRANE PROTEINs (VAMPs) in A. thaliana (Assaad et al., 2004; Kwon et al., 2008; Böhlenius et al., 2010). The papilla formation is based on the accumulation of defense-related elements, such as cell-wall proteins, at the pathogen attack site (Schmelzer, 2002; Hückelhoven and Panstruga, 2011). This mechanism confers the resistance not only to PM but also to the avirulent/virulent fungi at the Microbe/Pathogen-Associated Molecular Pattern (M/PAMP) Triggered Immunity (PTI) stage, which is the host’s first layer defense response (De Wit et al., 2009). Membrane vesicles, reactive oxygen species (ROS), and phenolic compounds contribute to host cell wall reinforcement and prevent invasion (Kuhn et al., 2016). Callose is another significant compound for this pre-invasive response in plants (Ellinger and Voigt, 2014).
Hypersensitive reaction (HR) is another robust defense response against PM pathogens and occurs in Effector Triggered Immunity (ETI) as a known second layer defense response (Jones and Dangl, 2006). The intercellular branching of the PM pathogens is restricted by Programmed Cell Death (PCD) with recognition of the pathogen by host receptor proteins such as receptor-like kinase (RLK) and leucine rich repeat receptor like kinases (LRK). Phytohormones and ROS also contribute to the regulation of these defense responses to restrict pathogen growth (Micali et al., 2008; Takken and Goverse, 2012; Cao et al., 2014; Serrano et al., 2014).
R-gene-mediated PM resistance is multilayer like other pathogen resistance in the host. Different signaling pathways and proteins contribute to the regulation of defense response in plants (Kuhn et al., 2016). But how can we classify mlo-mediated resistance against PM pathogens? Recently, researchers classified the mlo-mediated resistance as a part of the non-host resistance (NHR) in A. thaliana. Because PEN proteins, the major compounds of NHR in A. thaliana against PM pathogens, are necessary for the AtMLO2-mediated resistance (Humphry et al., 2006; Appiano et al., 2015). Although mlo-mediated resistance is associated with NHR for this reason, the defense mechanism is still in ambiguity, particularly in cucumber.
C. sativus has thirteen CsaMLO genes, and three of these genes (CsaMLO1, CsaMLO8, and CsaMLO11) were classified in Clade V associated with PM-susceptibility in dicots. It has been reported that when CsaMLO1 and CsaMLO11 are minor genes for PM susceptibility, CsaMLO8 could be a major gene associated with PM susceptibility in cucumber. Also, till date there is no report on the naturally occurring mutation on CsaMLO1 or CsaMLO11 (Nie et al., 2015; Berg et al., 2015). Although several research groups have focused on understanding CsaMLO11/CsaMLO11 or CsaMLO8 mediated resistance in cucumber, the defense mechanism is still unclear. Moreover, CRISPR/Cas9 for knock-out CsaMLO1, CsaMLO8, and CsaMLO11 was suggested to determine mlo-mediated defense mechanism in cucumber (Berg et al., 2017).
We have previously determined the PM defense mechanism in cucumber using RT-PCR and found that callose-dependent cell-wall reinforcement was a more robust response than HR in our genotypes (Tek and Calis, 2022). Therefore, we presume that the major PM-susceptibility gene CsaMLO8 could be responsible for regulating cell wall modifications negatively at pre-invasion stage. Also, we want to emphasize on the effects of the loss of function for Clade V genes in cucumber, CsaMLO1, and CsaMLO11, in defense response. Because the loss-of CsaMLO function is effect is still not clear in cucumber even if there are single CsaMLO mutant studies reported. Therefore, we used various types of loss-of CsaMLO function mutants such as triple and double mutants to determine mlo-mediated defense mechanism in cucumber. The study was carried out to reveal contribution of the CsaMLO1, CsaMLO8, and CsaMLO11 mutations for pre or post invasive responses against P. xanthii. Loss of function cucumber mutants were developed to investigate defense mechanism such as penetration resistance at early stage of the infection or hypersensitive reaction in plants by using CRISPR/Cas9. Further, ROS (Reactive Oxygen Species) accumulation such as H2O2 (Hydrogen Peroxide) associated with plant defense system against different plant pathogens was investigated to classify defense response which linked Clade V CsaMLO mutations in cucumber.
Material and methods
gRNAs selection and vector transformation
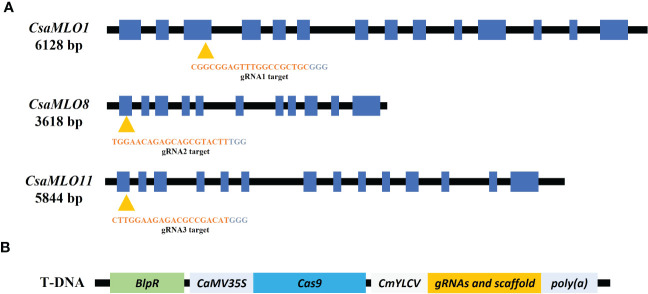
Three gRNAs have been selected for the CsaMLO1, CsaMLO8, and CsaMLO11 by using CRISPR-GE (Xie et al., 2017) for Cas9 recognition in cucumber. The gRNAs were chosen from the third exon of CsaMLO1 (Cucsa.207280) and the first exon of CsaMLO8 (Cucsa.207280) and CsaMLO11 (Cucsa.190600) sequences from Phytozome v12 ( Figure 1A ). pDIRECT_23:CmYLCV_gRNA1_gRNA2_gRNA3 plasmid ( Figure 1B ) was assembled from the described Golden Gate Assembly (Čermák et al., 2017). CmYLCV, CSY_gRNA, REP_gRNA, and csy_term oligonucleotides ( Supplementary Table 1 ) were used in PCR to amplify gRNAs and gRNA scaffolds for each gRNAs. High Fidelity Phusion Taq (Thermo) polymerase was used in reaction mixture according to the manufacturer protocol. PCR was conducted in; pre-denaturation at 98°C for 30 secs, 30 cycle; denaturation at 98°C for 10 secs, annealing at 65 °C for 30 secs, elongation at 72 °C for 15 secs, and final elongation at 72°C for 5 mins conditions to amplify gRNA:gRNA_scaffold and pDIRECT_23C was used as a template. 50 ng pDIRECT_23C ( Supplementary Figure 1 ), 0.5 µl of diluted (ten times) PCR products, 0.5 µl SapI, 0.5 µl Esp3I, 1 µl T7 DNA ligase, 10 µl 2x T7 DNA ligase buffer, and 7 µl ddH2O were used in Golden Gate reactions. The ligation was completed in 10 cycles at 37°C for 5 minutes and 25°C for 10 minutes. PmeI and PmlI restriction enzymes were used to confirm the plasmid and transferred to E. coli DH5α ( Supplementary Figure 2 ). The transformation plasmid was isolated from DH5α after validation with kanamycin (50mg/L) and ccdB selection. Transformation plasmid was transferred to electrocompetent Agrobacterium tumefaciens EHA105 by electroporation. PC primers (annealing at 57°C) were used in colony PCR to confirm transformation of EHA105.
Figure 1.
Specific gRNAs chosen from the coding site of Clade V CsaMLO (CDS) in cucumber and pDIRECT_23C plasmid. (A) gRNA1 for CsaMLO1, gRNA2 for CsaMLO8, and gRNA3 for CsaMLO11 were selected by using CRISPR-GE. (B) Selected gRNAs were amplified with PCR and cloned into pDIRECT_23C’s. Cestrum yellow leaf curling virus promoter (CmYLCV) was used for gRNAs expression and bialaphos phosphinothricin resistance (BlpR) was used to transgenic plant selection. Also, pDIRECT_23C contains Cauliflower Mosaic Virus 35S promoter (CaMV35) for Cas9.
Transformation and plant regeneration
Cotyledons of PM-susceptible cucumber line ADR27 were used as explants for A. tumefaciens EHA105-mediated transformation. The transformation and regeneration procedure were conducted as previously described (Zhang et al., 2017), with slight modifications. ADR27 inbred line’s seeds were kept in tap water for 2 hours and the seed coats were removed. Seed surfaces were sterilized using 75% ethanol for 30 seconds. Further, commercial bleach (3% sodium hypochlorite solution) was used also for sterilization for 15 minutes after the ethanol was drained out. After rinsing three times with deionized water and drying on sterile filter paper aseptically under laminar airflow cabinet, the seeds were placed in a petri dish containing plant germination medium (Murashige Skoog with 3% sucrose). Meanwhile, EHA-105 was incubated in Luria-Bertani (LB) broth containing 100 mg l-1 rifampicin and 50 mg l-1 kanamycin for 16-24 hours at 28°C and at 200 rpm. Bacterial cells were transferred to 50 ml of LB broth and incubated until the OD at 600 nm reached 0.6-0.8. After centrifugation at 4000 g for 10 minutes, the bacterial pellet was diluted in MS liquid to OD600 = 0.25. Cotyledon explants were taken from a germination medium using sterile forceps and were prepared for transformation using a sterile scalpel. Prepared explants were transferred to the EHA105-containing liquid MS, incubated for 15 minutes for agroinfiltration, and placed in a co-cultivation Medium (MS with 3% sucrose, 2 mg ml-1 BAP, 2 mg ml-1 ABA, and 200 μM acetosyringone) at 28°C for two days in the dark keeping the abaxial surface in contact with the culture medium. The explants were then transferred onto a shooting medium (MS with 3% sucrose, 2 mg ml-1 BAP, 2 mg ml-1 ABA, 150 mg l-1 Timentin) containing 1 mg ml-1 glufosinate-ammonium to select transformant plants. After 2-3 weeks of incubation at 25°C under 16-hour photoperiod, the resulting shoots were transferred to glass jars containing a rooting medium (MS with 3% sucrose, 150 mg l-1 timentin and 1 mg ml-1 glufosinate-ammonium). After 2-4 weeks, the rooted plants were transferred to pots containing wet compost, covered with a zip bag, and grown for 6-8 weeks at 25°C/18°C and 16 hours photoperiod.
Development of T1 plants
DNA was isolated from T0 plants using the GeneJET Genomic DNA Purification Kit, and transformation was confirmed by using Cas9 primers ( Supplementary Table 1 ). After acclimatization, 0.1 M silver nitrate was sprayed on newly formed shoots to form male flowers for selfing. The transgenic T0 ADR27 male flowers, which opened on the same day, were crossed with female flowers. After the fruits ripened and matured, the T1 seeds were harvested, washed with 70% ethanol, dried, and stored at 4°C.
Analysis of mutations in T1 plants
The T1 seeds were germinated in pots containing wet compost. Genomic DNA was extracted from the T1 cotyledons using GeneJET Genomic DNA Purification Kit as per manufacturer’s instructions (Thermo Fisher, Scientific, USA). Meanwhile, WT-specific forward primers were designed from downstream and upstream of the PAM sequence in for CsaMLO1, CsaMLO8, and CsaMLO11. Reverse primers were designed from GC-rich regions of these genes with high-annealing temperatures to increase specificity. The method is previously described as an “annealing at critical temperature PCR” (ACT-PCR) by Wang and Wang, 2019. Critical temperatures were determined with the gradient-PCR (60°C to 72°C) by using DNA isolated from WT-ADR27. PCR was carried out under the following conditions: initial denaturation at 94°C for 2 minutes, denaturation at 94°C for 30 seconds, annealing at 68°C (for CsaMLO1 and CsaMLO11) and 65°C (for CsaMLO8) for 30 seconds, elongation at 72°C for 45 seconds; 30 cycle and final elongation at 72°C for 5 minutes to detect homozygous mutant. The PCR products were visualized after the agarose gel (1.5%) electrophoresis. The DNA from homozygous mutant plants that did not show amplification in the PCR using these primers could be attributed to the deletion or insertion mutation on the PAM sequence.
Determination of the CsaMLO mutation types
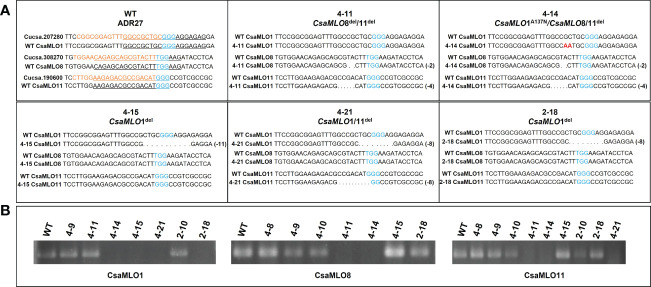
CsaMLO1, CsaMLO8, and CsaMLO11 gRNA targets of selected mutant plants accordingly ACT-PCR results were amplified with PCR by using the CsaMLO-Seq primers ( Supplementary Table 1 ). PCR were carried out; pre-denutaration at 94°C for 5 mins; denaturation at 94°C for 1 min, annealing at 58°C for 45 secs, elongation at 72°C for 45 secs and final elongation at 72°C for 5 mins. The products were then run on 1.5% agarose gel using gel electrophoresis for confirmation. Finally, the amplicon was purified and sequenced using the Sanger method by Macrogen (Madrid, Spain). The T1 plants and WT ADR27 gRNA target sequences were evaluated with alignment of WT and mutant CsaMLO genes. Mutations were detected for each sequenced plant and are presented in Figure 2 . The introns sequences were cleaned with reference CoDing Sequence (CDS) alignment. Cucsa.207280, Cucsa.308270, and Cucsa.190600 Phytozome reference sequences were used for CsaMLO1, CsaMLO8, and CsaMLO11 respectively. CDSs were converted to amino acid sequence in GeniousPrime (2021.2.2v) and protein sequences of mutant were aligned with WT-ADR27. Open Reading Frame (ORF) shifts and stop codon formations were determined according to the alignment result.
Figure 2.
Nucleotide sequence alignment of CsaMLOs. Clade V CsaMLOs of WT-ADR27 were aligned with the reference sequence to detect any genotypic differentiation, and the underlined nucleotides on the alignment shows the PCR primers for detection of homozygous mutants; (A) the sequence results indicated that 4-11 has deletion mutations on CsaMLO8 (2 bp) and CsaMLO11 (4 bp) even if there is no mutation on the CsaMLO1. Although 4-14 is triple mutant (2 bp del at CsaMLO8 and 4 bp del at CsaMLO11), there is substitution mutation on CsaMLO1 with 2 nucleotides change as “AA” rather than “GC” on the Cas9 cleavage site. 4-15 and 2-18 were determined as CsaMLO1 del with 11 bp and 8 bp deletions as a result of NHEJ. Also 8 bp deletions were detected on 4-21’s CsaMLO1 and CsaMLO11; (B) The PCR products were observed to be non-mutant genotype in terms of CsaMLO. However, there was no amplification in PCR results of mutant, because of homozygous deletion or substitution on the primer binding sites.
Inoculation and evaluation of reactions against PM
P. xanthii conidiospores were harvested from the PM infected plants with a brush and suspended in distilled water to give 2x105 spore ml-1 and sprayed three times onto T1 mutant and susceptible WT ADR27 cucumber leaves. Ten days after the inoculation, leaf samples were taken from the PM inoculated WT and mutant plants, boiled in 96% ethanol to remove the chlorophyll, and then stained with trypan blue (250 µg ml-1). The development of conidiophores and conidiospores was examined under microscope (Leica DM500). Conidiospores were counted on five leaves for each plant within 100 spores at a 0.02 mm2 area for detection of spore germination and germination/penetration rates were compared between susceptible WT-ADR27 and mutants. Determination of ROS accumulation was carried out by staining leaves with DAB combined with Trypan Blue (Thordal-Christensen et al., 1997). For this, the PM-inoculated leaves were incubated for 6 hours in DAB (1%) solution and boiled in 96% ethanol. Leaves were evaluated and scored using a modified DR (Disease Reaction) scale where 0 indicates no fungal growth and 4 indicates dense fungal growth (Adam and Somerville, 1996).
Results
Plant regeneration and development of T1 mutants
After germination of ADR27 seeds, cotyledons were removed and used as explants for transformation using A. tumefaciens EHA 105 and were then transferred to co-cultivation medium. The explants were shifted to an MS medium containing 1 mg ml-1 glufosinate-ammonium for selection of transformants. The integration of T-DNA into plant genome was confirmed by PCR using Cas9 specific primers and 46 transgenic plants were regenerated at T0. Regeneration steps and T0 plants were given in Supplementary Figure 3 . 96 T1 plants were also examined using ACT-PCR, and the result revealed that the mutation rates of the CsaMLO genes observed in the T1 generation were 25%, 2.08% and 19.79% for CsaMLO1, CsaMLO8, and CsaMLO11, respectively ( Supplementary Table 2 ).
In/dels in mutants
CsaMLO1 did not amplify in PCR that was carried out with isolated DNA samples from 4-14, 4-15, 4-21, and 2-18 lines. CsaMLO8 was amplified when the T1 plants, excluding 4-11 and 4-14 were used in PCR. Also, CsaMLO11 did not amplify in 4-11, 4-14, and 4-21. The sequences of the gRNA’s target of these lines revealed that these lines have deletion and substitution mutations on the Cas9 cleavage site. 4-14 have 2 nucleotide substitution mutations on the upstream of the PAM motif. The deletion mutations were detected in the 4-15, 4-21, and 2-18 cucumber lines. The CsaMLO1 deletion mutations in mutant lines were relatively larger than the CsaMLO8 deletions. The small deletions (2 bp) are seen upstream of the PAM in the CsaMLO8 of 4-11 and 4-14 T1 plants. Deletions were also detected in the CsaMLO11 of 4-11, 4-14, and 4-21 T1 plants. The CsaMLO11 deletions are 4 and 8 bp, and the location of the deletions differed from the PAM motif of CsaMLO1. The position of deletions ranged from downstream of the PAM to upstream in CsaMLO1, but the deletion was detected only upstream of the PAM in CsaMLO11 ( Figure 2 ).
Early stop codon formation and amino acid substitution
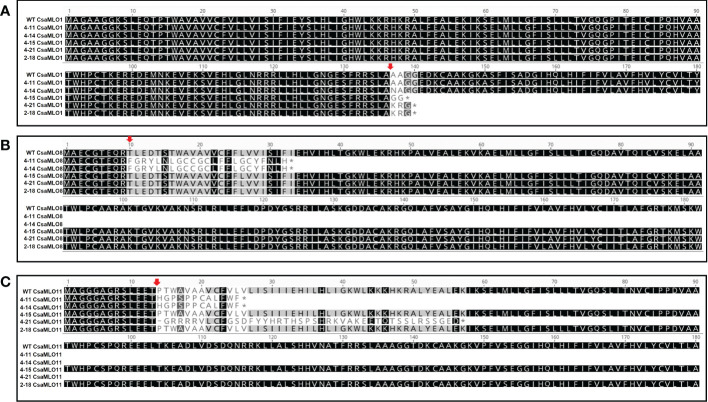
Most of the mutations occurred in different mutant plants caused early stop codon formation at the amino acid sequence except that in 4-14 mutant plant, two nucleotide mutations (GC to AA) in CsaMLO1 lead to alanine to asparagine change at the 137th amino acid as a result NHEJ (non-homologous end joining) after the Cas9 cleavage. The alanine was changed asparagine at the 137th amino acid of CsaMLO1 as a result NHEJ (non-homologous end joining) after the Cas9 cleavage. The stop codon formation was detected at the 140th amino acid of 4-21 and 2-18’s CsaMLO1 caused by 8 bp deletion, while the stop codon was also detected on 139th amino acid of 4-15’s CsaMLO1 caused by 11 bp deletion. The ORF shifts were detected on the CsaMLO8 mutants 4-11 and 4-14 at Cas9 cleavage site and stop codon formation was detected at the 33rd amino acid. ORF shift and stop codons also detected in CsaMLO11 as a result of NHEJ. The stop codons were at the 26th amino acid of 4-11 and 4-14 CsaMLO11, while stop codon formation appeared at 57th amino acid of 4-21’s CsaMLO11. 4-11 (CsaMLO8/11 del), 4-14 (CsaMLO1 A137N/CsaMLO8/11del), 4-15 (CsaMLO1 del), 4-21 (CsaMLO1/11del), and 2-18 (CsaMLO1 del) were determined as loss of function mutant ( Figure 3 ).
Figure 3.
Amino acid alignment using sequences of mutants and WT plants. (A) When the deletions on the Cas9 cleavage (red arrow) cause stop codon formation on the 4-15’s CsaMLO1 139th amino acid; the stop codon formation was detected on 2-18 and 4-21 CsaMLO1’s 140th amino acid (3rd exon). The Alanine (A) was altered with Asparagine (N) at the 137th amino acid as a result of the NHEJ caused by Cas9 cleavage at the gRNA1 target. (B) 2 bp deletions cause the unexpected stop codon formation on the 33rd amino acid of CsaMLO8 in 4-11 and 4-14. The target site of gRNA2 was on the first exon for CsaMLO8 and the changed amino acid structures are at the Cas9 cleavage (10th aa) site for this gene. The early stop codon formation caused the loss-of CsaMLO8 function in 4-11 and 4-14. (C) The mutations seem to be responsible for the stop codon formation in CsaMLO11 and stop codon’s positions of 4-11 and 4-14’s CsaMLO11 closer to that gRNA3 than 4-15 and 2-18 CsaMLO11. However, ORF shifts are initiated at gRNA3 position in whole CsaMLO11 del.
Evaluation of mutants for P. xanthii resistance
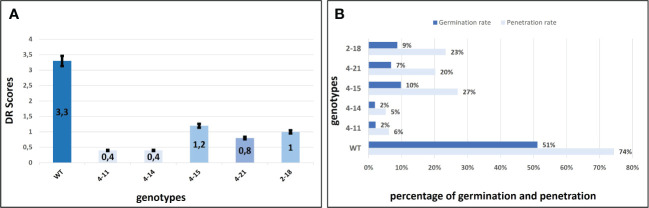
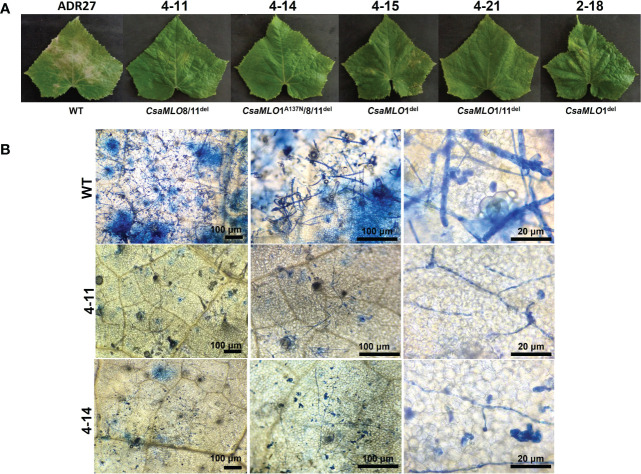
The P. xanthii was inoculated on 4-11 (CsaMLO8/11del), 4-14 (CsaMLO1 A137N/CsaMLO8/11del), 4-15 (CsaMLO1 del), 4-21 (CsaMLO1/11del), 2-18 (CsaMLO1 del), and WT cucumber plants after the determination of the mutation and mutation types. After ten days of inoculation, inoculated five leaves were examined closely for inoculated WT and mutant plants to determine DR scores ( Table 1 ). Further, leaves were also harvested from the WT and mutant plants, chlorophyll was removed, and staining was done with Trypan Blue. Typical PM symptoms were apparent on WT ADR27 leaves. 4-15 and 2-18 mutants showed fewer symptoms on the inoculated leaves. The PM symptoms on 4-21 and 4-11 plants were less severe than in CsaMLO1 del plants. Furthermore, there were no symptoms on the 4-14 plant. The microscopic observation results confirmed that the conidiospores had germinated massively and developed many new spores on the WT ADR27 plant. However, no new conidiophores or conidiospores were observed on the inoculated CsaMLO8/11del and CsaMLO1 A137N/CsamMLO8/11del ( Figure 4 ). Low spore germination and limited mycelial growth was observed on the 4-11 and 4-14 plants.
Table 1.
DR scores at 10 dpi on different PM-inoculated leaves.
| Leaf | WT | 4-11 | 4-14 | 4-15 | 4-21 | 2-18 |
|---|---|---|---|---|---|---|
| 1 | 3.50 | 0.50 | 0.50 | 1.50 | 1 | 1 |
| 2 | 3.50 | 0.50 | 0.50 | 1.00 | 0.50 | 1 |
| 3 | 3.50 | 0.50 | 0.00 | 1.50 | 0.50 | 1 |
| 4 | 30 | 0.50 | 0.50 | 1.00 | 1 | 1 |
| 5 | 30 | 0.00 | 0.50 | 1.00 | 1 | 1 |
| Average | 3.30 | 0.4 | 0.40 | 1.20 | 0.80 | 1 |
Figure 4.
Evaluation of the DR scores against PM and P. xanthi penetration/germination rates at ten-day post-inoculation. (A) DR scores of wild-type ADR27 and mutant plants were generated means of the inoculated five leaves for each genotypes at 10 dpi ( Table 1 ) and same colors indicated that there is no difference for DR scores statistically according to standard deviation bar. The lowest DR scores were observed in 4-11 (CsaMLO8/11del) and 4-14 (CsaMLO1 A137N/8/11del). The DR scores were statistically similar for 4-15 and 2-18 plants having CsaMLO1 mutation. 4-21 was determined as CsaMLO1/11 del mutant and the DR score was lower than CsaMLO1 del even if higher DR score than 4-11 and 4-14. (B) The strong penetration resistance is defined in 4-11 and 4-14 plants having loss of CsaMLO8 function. Also, P. xanthii germination rate was lowest in these mutants. 4-15 and 2-18 (CsaMLO1 del) penetration and germination rates were similar with the CsaMLO1/11del. Percentages of the germination and penetration rates were created according to average of germinated and penetrated spores’ numbers from each genotype’s five leaves.
ROS accumulation on CsaMLO8/11del and CsaMLO1 A137N/CsaMLO8/11del
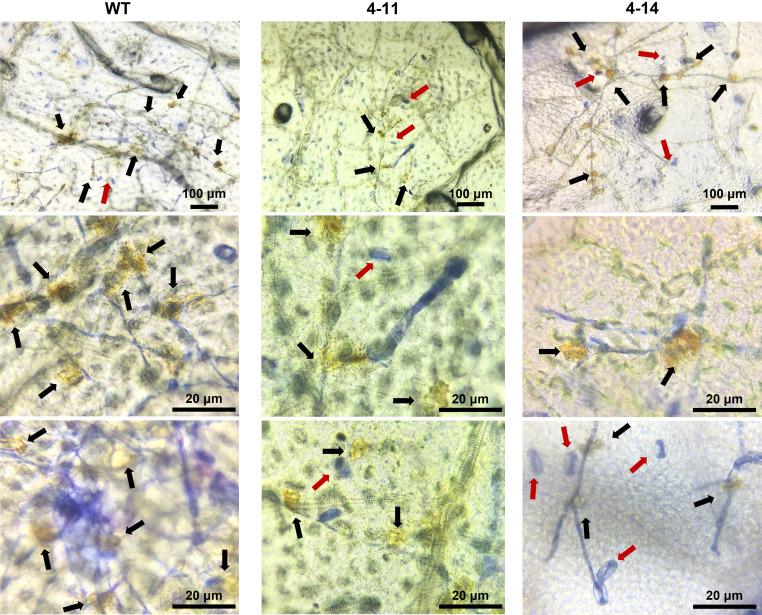
Trypan blue staining revealed massive mesophyll cell death on ADR27 associated with PM infection. However, there were restricted death cells in 4-11 and 4-14 plants ( Figure 5 ). The DAB and Trypan Blue combined staining also shows ROS accumulation on ADR27 and mutant plants. ROS accumulation was observed near the P. xanthii mycelium growth area of the leaves in both mutant and WT plants. Notably, on 4-14 mesophyll cells, the pathogen conidiospores that were unable to germinate were quite visible, even though there was no ROS accumulation and cell death. Combined staining also revealed that the mycelial growth was on wild-type ADR27 leaves, and ROS was accumulated in this growth area. The maximum ROS accumulation and P. xanthii development were on the WT ADR27 when compared with 4-11 and 4-14 ( Figure 6 ).
Figure 5.
Powdery mildew symptoms and visualization of P. xanthii spores on WT/mutant leaves using Trypan Blue. (A) Powdery mildew symptoms on leaf surfaces of WT-ADR27 and CsaMLO mutants at 10 dpi. Typical symptoms could be seen on the WT leaves as a result massive propagation caused by P. xanthii after ten-days post inoculation. Restricted symptoms were detected on P. xanthii inoculated leaves of 4-15, 4-21, and 2-18 plants. Moreover, the hypersensitive response associated lesions were apparent on the leaves of CsaMLO1 del and CsaMLO1/11del plants. However, no symptoms appeared on the leaves of 4-11 and 4-14 plants after ten days of inoculation. (B) The epidermal surface of WT-ADR27 was covered with P. xanthii conidiophore and conidiospores that was newly formed from inoculated spores. Also, massive cell deaths were observed on the leaf mesophyll as a result of pathogen activity apparent from the blue color. However only mycelial growth was detected on the leaves of 4-11 and 4-14 plants, even if there was restricted germination rate.
Figure 6.
Combined DAB and Trypan blue staining to determine ROS accumulation on leaves of inoculated WT and mutant plants at 10 dpi. Although susceptible WT-ADR27 had highest P. xanthi germination rate, ROS accumulation (black arrow) was found more frequently on the leaves of these plant as compared 4-11 and 4-14. ROS accumulation was also found in 4-11 and 4-14 which are CsaMLO8/11del and CsaMLO1 A137N/8/11del mutants. However, ROS were accumulated in the P. xanthii mycelial growth site was detected in 4-11 and 4-14. Also, non-germinated spores (red arrow) were detected on mutant leaves on the contrary to WT.
Discussion
Podosphaera xanthii is one of the important pathogens responsible for powdery mildew, which is thought to be affecting more than ten thousand plant species worldwide. The PM caused by P. xanthii is the most common and severe disease in cucurbits, including C. sativus (Zitter et al., 1996). Understanding the PM defense mechanism is the first and critical step to develop PM resistant cultivars. However, the generation of resistant cultivars is challenging because PM resistance is controlled by several genes (polygenic in nature) and involves complex defense pathways (Chen et al., 2009).
Cell wall reinforcement and HR are strong responses against PM pathogens. Significantly callose-associated cell wall reinforcement is one of the robust basal defense mechanisms found in plants against incompatible plant-microbe interaction (Ahmed et al., 2016). M/PAMPs are recognized by host receptors such as PRR and trigger the PTI, the first line of defense in plants, a defense mechanism activated to restrict the infection. However, PTI pressures the pathogen for mutation on effectors, and only successful pathogens could overcome the first obstacle (Thordal-Christensen, 2003). For this reason, we need to find new resistance sources to generate new resistant cultivars. The new sources (R-genes) could confer resistance against successful pathogens (overcame the first line) that trigger the ETI, and a hypersensitive reaction generally occurs at this stage (Jones and Dangl, 2006). However, R-gene-mediated resistance is broken, and new pathogen strains could overcome the resistance in a short time with a mutation on avirulence genes. Therefore, NHR is strongly suggested against plant pathogens to solve this infinity loop because of it is durable and broad-spectrum resistance.
It has been suggested that S-gene mutation mediated resistance as a novel approach for developing resistant cultivars to control plant disease. One of the most recognized and first discovered S-gene is the barley mlo. Naturally occurring mutations on this gene confer broad-spectrum and durable resistance against many Bgh isolates (Jørgensen, 1992). Unfortunately, mlo-mediated resistance is as complex as R-mediated PM resistance in various plants, till date very little is known about the mlo-mediated defense mechanism in various plants (Acevedo-Garcia et al., 2014). However, it has been shown that a calmodulin-binding domain of barley mlo increases susceptibility to PM, particularly in barley. Furthermore, the MLO have seven transmembrane domains, and one of these (the calmodulin-binding domain) is responsible for calmodulin-binding. The function of this protein is to block the Ca+2 and calmodulin interaction. The Ca+2 signal plays an essential role in recognizing the host by the PM pathogen at the early stage of the infection (Kim et al., 2002).
Phylogenetic analyses and transcriptomic studies have been focused on revealing MLO gene families in cucumber. Differentially Expressed Genes (DEGs) were determined in the loss-of-function mutants in cucumber. However, mlo-mediated resistance is still poorly understood in cucumber because of different defense responses that could occur against PM pathogens in cucumber, contrary to other plant species (Berg et al., 2015; Nie et al., 2015). Contrary to the CsaMLO1 and CsaMLO11 mutants in cucumber, mutant cucumber for CsaMLO8 was found in nature. Further, using the latest gene-editing techniques was suggested to determine mlo-mediated resistance in cucumber by obtaining different loss-of-CsaMLO function mutants such as triple or double mutants (Berg et al., 2017). Therefore, CRISPR/Cas9 was used to generate triple (CsaMLO1 A137N/CsaMLO8/11del), double (CsaMLO1/11del and CsaMLO8/11del), and single mutant (CsaMLO1 del) cucumber lines and these lines were used in this study to determine mlo-based defense mechanisms in cucumber.
The T0 lines obtained after transformation were further tested with Cas9 primers to control T-DNA integration and 46 plants determined as transgenic. We screened the mutants which are obtained from transgenic T0 with a PCR-based approach (ACT-PCR) to determine homozygous mutants in T1 generations. The mutation rates in 96 T1 plants were 25%, 2.08% and 19.79% for CsaMLO1, CsaMLO8, and CsaMLO11 respectively. The PCR-based approach could be the reason for low mutation rates in T1 reactions because the method allows the detection of only homozygous mutant plants.
The deletions mutations were detected on the CsaMLO1 target sequence of 4-15 (11 del), 4-21 (8 del), and 2-18 (8 del), and detected deletions cause loss-of CsaMLO1 function due to the formation of the unexpected stop codon formation at 139th amino acid of 4-15’s CsaMLO1, and 140th amino acid of 4-21 and 2-18’s CsaMLO1. Interestingly, two nucleotide substitution mutations at the Cas9 cleavage site of 4-14’s CsaMLO1 caused amino acid substitution. The result is similar to amino acid substitution on barley, causing the previously reported loss-of-function mutation for barley’s mlo (Reinstädler et al., 2010). However, there was no difference between DR scores of 4-11 and 4-14 plants post PM inoculation. Two deletions were detected on CsaMLO8 of 4-11 and 4-14, hence, these deletions resulted in the stop codon formation at 33rd amino acid. Also, 4 and 8 bp deletions were seen on CsaMLO11 of 4-11, 4-14, and 4-21 lines. To sum up, we report for the first-time triple mutants 4-14 (CsaMLO1 A137N/CsaMLO8/11del), double mutants 4-11 (CsaMLO8/11del), and 4-21 (CsaMLO1/11del) in cucumber lines using CRISPR/Cas9.
Although various research groups have generated CsaMLO8 mutant cucumber lines using different methods and DEGs were determined with transcriptomic approaches, but the underlying mechanism of mlo-mediated resistance in cucumber is still unclear. Therefore, we performed this study to reveal the defense mechanism involved in the mlo-mediated resistance in cucumber. In this study, we focused on the mlo-mediated resistance mechanism, and PM reactions of 4-11 (CsaMLO8/11del), 4-14 (CsaMLO1 A137N/CsaMLO8/11del), 4-15 (CsaMLO1 del), 4-21 (CsaMLO1/11del), and 2-18 (CsaMLO1 del) were evaluated post-inoculation. The results from the symptoms caused by PM on mutant plants indicated that the loss of CsaMLO8 function confers resistance against the PM. The results from the symptoms caused by PM on mutant plants indicated that the loss of CsaMLO8 function confers resistance against the PM. DR scores were lowest in 4-11 and 4-14, with 0.4 DR scores among mutant plants, there is no statistical difference in terms of DR scores between 4-15 and 2-18, which are single CsaMLO1 del, and their DR scores were higher than CsaMLO1/11 del 4-21’s DR score (0.8). The reason of the identical DR scores between 4-11 and 4-14 could associated with the early CsaMLO8 mutation-mediated response against PM or CsaMLO1 substitution mutation in 4-11 did not affect the resistance level in CsaMLO8 del plants. Although, this result is not astonishing, the evaluation of the PM reactions shed light on the mlo-mediated defense mechanism in cucumber using the DAB and Trypan Blue staining.
Hypersensitive reaction-associated lesions were seen on the leaf surface 4-15 and 2-18 plants at ten days post-inoculation, and these lines also exhibited the least deletion mutations on CsaMLO1. Deletion mutations were also observed on CsaMLO1 and CsaMLO11 in 4-21. The PM symptoms were restricted with the HR in CsaMLO1 del and CsaMLO1/11del. Although P. xanthii germination rate was higher than 4-11 and 4-14, our results have revealed that CsaMLO1 and CsaMLO11 could be negative regulators in post-invasive defense response against PM pathogen in cucumber. 4-11 and 4-14 plants recorded the lowest DR score after the PM inoculation, as expected, because of the CsaMLO8 function, a major susceptibility gene for PM. Interestingly, there was no HR observed on the adaxial leaf surface of 4-11 and 4-14 plants, and there appeared no symptoms even after ten days of inoculation. From the microscopic observation it was quite visible that attached ungerminated P. xanthii spores on epidermal cells were staining blue after Trypan blue assay. Only a few spores germinated, and only mycelial growth was detected under the cucumber leaf cells. To our surprise, accumulation of ROS was detected during DAB staining, even in the absence of HR. However, ROS such as H2O2 are not only associated with HR but have been reported to be linked with callose-dependent cell-wall reinforcement, known as papilla formation (Kuźniak and Urbaneki, 2000). The localized ROS accumulation was also detected on inoculated and non-germinated P. xanthii conidiospores. The DAB combined Trypan blue staining result indicated that accumulated ROS could be associated with PM-triggered cell-wall reinforcement in 4-11 and 4-14 plants, but the accumulated ROS did not reduce the PM infection rate in WT. The lower P. xanthii germination and restricted mycelial growth indicate that the CsaMLO8 could be a negative regulator in pre-invasive defense response. Previously, CsaMLO8 and callose interaction has been demonstrated. However, the loss-of-CsaMLO8 function provides strong penetration resistance in cucumbers at early stage of infection even if there are loss-of-function mutations on CsaMLO1 or CsaMLO11.
Finally, we can develop novel resistant cultivars by using state-of-the-art gene editing techniques without understanding the underlying mechanism of this type of resistance. However, we need to figure out the defense mechanism sourced by S-gene mutations in the plant to develop broad-spectrum and durable resistance. It is not possible that the pathogen can overcome the resistance provided by S-gene mutation because this resistance is similar to NHR, especially mlo-mediated resistance. However, it is difficult to claim that S-mediated resistance will not break in the near future. Therefore, investigating the defense mechanism with novel and diverse approaches could be effective for protecting agricultural production against yield and quality losses caused by plant pathogens.
Conclusion
Three types of mutants i.e., triple mutant 4-14 (CsaMLO1 A137N/CsaMLO8/11del), double mutants 4-11 (CsaMLO8/11 del) and 4-21 (CsaMLO1/CsaMLO11 del), single mutants 4-15 and 2-18 (CsaMLO1 del) were generated in this study using the CRISPR/Cas9. Loss-of Clade V functions with diverse combinations were investigated to determine mlo-mediated resistance in cucumber. The results revealed that CsaMLO1/CsaMLO11 mutations confer resistance with HR for post-invasive defense response. Loss of CsaMLO8 function conferred higher resistance than other mutants as described previously, but we report here for the first time, that CsaMLO8 is strongly associated with pre-invasive defense response even if there are other Clade V mutants such as in triple mutant. Hence, CRISPR/Cas9 could be used for the generation of PM-resistant cucumber cultivars, which have a strong pre-invasive defense with CsaMLO8 or post-invasive with CsaMLO1/CsaMLO11 mutations.
Data availability statement
Sequence data of the cucumber CsaMLO1 (Cusca.207280), CsaMLO8 (Cusca.308270), and CsaMLO11 (Cusca.190600) from Phytozome (https://phytozome.jgi.doe.gov/) has been used for selection of the gRNAs. CsaMLO sequences of the mutant and wild type were uploaded to NCBI GenBank (ON528937-ON528951); ON528941 for WT-CsaMLO1, ON528942 for 4-14’s CsaMLO1, ON528943 for 4-15’s CsaMLO1, ON528944 for 4-21’s CsaMLO1, ON528946 for 2-12’s CsaMLO1, ON528937 for WT-CsaMLO8, ON528938 for 4-11’s CsaMLO8, ON528940 for 4-14’s CsaMLO8, ON528948 for WT-CsaMLO11, ON528949 for 4-11’s CsaMLO11, ON528950 for 4-14’s CsaMLO11, and ON528951 for 4-21’s CsaMLO11.
Author contributions
Conceptualization: OC and SW. Data curation: MT, MS, and SC. Formal analysis: MT, MS, and SC. Funding acquisition: OC. Methodology: MT. Project administration: OC and HF. Resources: SW. Supervision: OC, HF. Writing – original draft: MT. Writing – review & editing: HF, SW. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors thank to Prof. Dr. John G. Turner (Emeritus Professor, University of East Anglia, UK) for technical reading and suggestions on the manuscript. MS, and SW acknowledge the overseas fellowship grant award provided by SKAUST Kashmir, India.
Funding
This study was supported by The Scientific Research Projects Coordination Unit of Akdeniz University (Project Number: FBA-2020-5344).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1081506/full#supplementary-material
References
- Acevedo-Garcia J., Kusch S., Panstruga R. (2014). Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 204 (2), 273–281. doi: 10.1111/nph.12889 [DOI] [PubMed] [Google Scholar]
- Adam L., Somerville S. C. (1996). Genetic characterization of five powdery mildew disease resistance loci in arabidopsis thaliana. Plant journal: Cell Mol. Biol. 9 (3), 341–356. doi: 10.1046/j.1365-313x.1996.09030341.x [DOI] [PubMed] [Google Scholar]
- Ahmed A. A., McLellan H., Aguilar G. B., Hein I., Thordal-Christensen H., Birch P. R. (2016). “Engineering barriers to infection by undermining pathogen effector function or by gaining effector recognition,” in Plant pathogen resistance biotechnology Ed. Collinge D. B. Hoboken, New Jersey: John Wiley & Sons, Inc. doi: 10.1002/9781118867716.ch2 [DOI] [Google Scholar]
- Appiano M., Catalano D., Santillán Martínez M., Lotti C., Zheng Z., Visser R. G., et al. (2015). Monocot and dicot MLO powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant Biol. 15, 257. doi: 10.1186/s12870-015-0639-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad F. F., Qiu J. L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., et al. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15 (11), 5118–5129. doi: 10.1091/mbc.e04-02-0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Pavan S., Zheng Z., Zappel N. F., Reinstädler A., Lotti C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of mlo function. Mol. Plant-Microbe interactions: MPMI 21 (1), 30–39. doi: 10.1094/MPMI-21-1-0030 [DOI] [PubMed] [Google Scholar]
- Berg J. A., Appiano M., Bijsterbosch G., Visser R., Schouten H. J., Bai Y. (2017). Functional characterization of cucumber (Cucumis sativus l.) clade V MLO genes. BMC Plant Biol. 17 (1), 80. doi: 10.1186/s12870-017-1029-z. BioMed Central Ltd.: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. A., Appiano M., Santillán Martínez M., Hermans F. W., Vriezen W. H., Visser R. G., et al. (2015). A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 15, 243. doi: 10.1186/s12870-015-0635-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H., Mørch S. M., Godfrey D., Nielsen M. E., Thordal-Christensen H. (2010). The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 22 (11), 3831–3844. doi: 10.1105/tpc.110.078063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Liang Y., Tanaka K., Nguyen C. T., Jedrzejczak R. P., Joachimiak A., et al. (2014). The kinase LYK5 is a major chitin receptor in arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766. doi: 10.7554/eLife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T., Curtin S. J., Gil-Humanes J., Čegan R., Kono T. J. Y., Konečná E., et al. (2017). A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29 (6), 1196–1217. doi: 10.1105/tpc.16.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Noir S., Kwaaitaal M., Hartmann H. A., Wu M. J., Mudgil Y., et al. (2009). Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in arabidopsis root thigmomorphogenesis. Plant Cell 21 (7), 1972–1991. doi: 10.1105/tpc.108.062653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni C., Humphry M. E., Hartmann H. A., Livaja M., Durner J., Westphal L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38 (6), 716–720. doi: 10.1038/ng1806 [DOI] [PubMed] [Google Scholar]
- de Almeida Engler J., Favery B., Engler G., Abad P. (2005). Loss of susceptibility as an alternative for nematode resistance. Curr. Opin. Biotechnol. 16 (2), 112–117. doi: 10.1016/j.copbio.2005.01.009 [DOI] [PubMed] [Google Scholar]
- De Wit P. J.G.M., Joosten M. H.A.J., Thomma B. H.P.J., Ioannis S. (2009). “Gene for gene models and beyond: The cladosporium fulvum-tomato pathosystem,” in The mycota (Berlin, Heidelberg: Springer; ). doi: 10.1007/978-3-540-87407-2_7 [DOI] [Google Scholar]
- Eckardt N. A. (2002). Plant disease susceptibility genes? Plant Cell 14 (9), 1983–1986. doi: 10.1105/tpc.140910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D., Voigt C. A. (2014). “Callose biosynthesis in arabidopsis with a focus on pathogen response: What we have learned within the last decade,” in Annals of botany, United Kingdom: Oxford University Press; vol. 114. , 1349–1358. doi: 10.1093/AOB/MCU120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R., Panstruga R. (2011). “Cell biology of the plant-powdery mildew interaction,” in Current opinion in plant biology, United Kingdom: Elsevier; 14. 738–746. doi: 10.1016/J.PBI.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Humphry M., Consonni C., Panstruga R. (2006). Mlo-based powdery mildew immunity: silver bullet or simply non-host resistance? Mol. Plant Pathol. 76, 605–610. doi: 10.1111/J.1364-3703.2006.00362.X [DOI] [PubMed] [Google Scholar]
- Humphry M., Reinstädler A., Ivanov S., Bisseling T., Panstruga R. (2011). Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant Pathol. 12 (9), 866–878. doi: 10.1111/j.1364-3703.2011.00718.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen I. H. (1992). Discovery, characterization and exploitation of mlo powdery mildew resistance in barley. Euphytica 63, 141–152. doi: 10.1007/BF00023919 [DOI] [Google Scholar]
- Jiwan D., Roalson E. H., Main D., Dhingra A. (2013). Antisense expression of peach mildew resistance locus O (PpMlo1) gene confers cross-species resistance to powdery mildew in fragaria x ananassa. Transgenic Res. 22 (6), 1119–1131. doi: 10.1007/s11248-013-9715-6 [DOI] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. doi: 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kim M. C., Panstruga R., Elliott C., Müller J., Devoto A., Yoon H. W., et al. (2002). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416 (6879), 447–451. doi: 10.1038/416447a [DOI] [PubMed] [Google Scholar]
- Konishi S., Sasakuma T., Sasanuma T. (2010). Identification of novel mlo family members in wheat and their genetic characterization. Genes Genet. Syst. 85 (3), 167–175. doi: 10.1266/ggs.85.167 [DOI] [PubMed] [Google Scholar]
- Kuhn H., Kwaaitaal M., Kusch S., Acevedo-Garcia J., Wu H., Panstruga R. (2016). Biotrophy at its best: Novel findings and unsolved mysteries of the arabidopsis-powdery mildew pathosystem. arabidopsis book 14, e0184. doi: 10.1199/tab.0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuźniak E., Urbaneki H. (2000). “The involvement of hydrogen peroxide in plant responses to stresses,” in Acta physiologiae plantarum, vol. 22. (Polish Academy of Sciences, Kraków, Poland: Springer; ), 195–203. doi: 10.1007/S11738-000-0076-4 [DOI] [Google Scholar]
- Kwon C., Neu C., Pajonk S., Yun H. S., Lipka U., Humphry M., et al. (2008). Co-Option of a default secretory pathway for plant immune responses. Nature 451 (7180), 835–840. doi: 10.1038/nature06545 [DOI] [PubMed] [Google Scholar]
- Micali C., Göllner K., Humphry M., Consonni C., Panstruga R. (2008). The powdery mildew disease of arabidopsis: A paradigm for the interaction between plants and biotrophic fungi. arabidopsis book 6, e0115. doi: 10.1199/tab.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Wang Y., He H., Guo C., Zhu W., Pan J., et al. (2015). Loss-of-Function mutations in CsMLO1 confer durable powdery mildew resistance in cucumber (Cucumis sativus l.). Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinstädler A., Müller J., Czembor J. H., Piffanelli P., Panstruga R. (2010). Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley mlo protein. BMC Plant Biol. 10, 31. doi: 10.1186/1471-2229-10-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E. (2002). Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 7 (9), 411–415. doi: 10.1016/S1360-1385(02)02307-5 [DOI] [PubMed] [Google Scholar]
- Serrano I., Gu Y., Qi D., Dubiella U., Innes R. W. (2014). The arabidopsis EDR1 protein kinase negatively regulates the ATL1 E3 ubiquitin ligase to suppress cell death. Plant Cell 26 (11), 4532–4546. doi: 10.1105/tpc.114.131540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F. L., Goverse A. (2012). How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15 (4), 375–384. doi: 10.1016/j.pbi.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Tek M. I., Calis O. (2022). “Mechanisms of resistance to powdery mildew in cucumber,” in Phytopathologia mediterranea, vol. 61. (Firenze University Press, Florence, Italy : Firenze University Press; ), 119–127. doi: 10.36253/PHYTO-13313 [DOI] [Google Scholar]
- Thordal-Christensen H. (2003). “Fresh insights into processes of nonhost resistance,” in Current opinion in plant biology, vol. 6. (United Kingdom: Elsevier Ltd; ), 351–357. doi: 10.1016/S1369-5266(03)00063-3 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/J.1365-313X.1997.11061187.X [DOI] [Google Scholar]
- van Schie C. C., Takken F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854 [DOI] [PubMed] [Google Scholar]
- Wang C., Wang K. (2019). Rapid screening of CRISPR/Cas9-induced mutants using the ACT-PCR method. Methods Mol. Biol. (Clifton N.J.) 1917, 27–32. doi: 10.1007/978-1-4939-8991-1_2 [DOI] [PubMed] [Google Scholar]
- Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y. G. (2017). CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 10 (9), 1246–1249. doi: 10.1016/j.molp.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Xu X., Liu X., Yan Y., Wang W., Gebretsadik K., Qi X., et al. (2019). “Comparative proteomic analysis of cucumber powdery mildew resistance between a single-segment substitution line and its recurrent parent,” in Horticulture research, vol. 6. (United Kingdom: Nature Publishing Group; ), 1–13. doi: 10.1038/s41438-019-0198-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Xu X., Shi Y., Qi X., Chen X. (2017). Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5.1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genomics 18 (1), 21. doi: 10.1186/s12864-016-3438-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li X., Ma S., Shan N., Zhang X., Sui X., et al. (2017). “A protocol for agrobacterium-mediated transformation of cucumber (Cucumis sativus l.) from cotyledon explants,” in Protocol exchange (United Kingdom: Springer Nature; ). doi: 10.1038/protex.2017.107 [DOI] [Google Scholar]
- Zhou S. J., Jing Z., Shi. J. L. (2013). Genome-wide identification, characterization, and expression analysis of the MLO gene family in cucumis sativus. Genet. Mol. Res. 12 (4), 6565–6578. doi: 10.4238/2013.December.11.8 [DOI] [PubMed] [Google Scholar]
- Zitter T. A., Hopkins D. L., Thomas C. E. (1996). Compendium of cucurbits diseases (Saint Paul: American Phytopathological Society; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data of the cucumber CsaMLO1 (Cusca.207280), CsaMLO8 (Cusca.308270), and CsaMLO11 (Cusca.190600) from Phytozome (https://phytozome.jgi.doe.gov/) has been used for selection of the gRNAs. CsaMLO sequences of the mutant and wild type were uploaded to NCBI GenBank (ON528937-ON528951); ON528941 for WT-CsaMLO1, ON528942 for 4-14’s CsaMLO1, ON528943 for 4-15’s CsaMLO1, ON528944 for 4-21’s CsaMLO1, ON528946 for 2-12’s CsaMLO1, ON528937 for WT-CsaMLO8, ON528938 for 4-11’s CsaMLO8, ON528940 for 4-14’s CsaMLO8, ON528948 for WT-CsaMLO11, ON528949 for 4-11’s CsaMLO11, ON528950 for 4-14’s CsaMLO11, and ON528951 for 4-21’s CsaMLO11.