Abstract
Mechanistically, chimeric genes result from DNA rearrangements and include parts of preexisting normal genes combined at the genomic junction site. Some rearranged genes encode pathological proteins with altered molecular functions. Those which can aberrantly promote carcinogenesis are called fusion oncogenes. Their formation is not a rare event in human cancers, and many of them were documented in numerous study reports and in specific databases. They may have various molecular peculiarities like increased stability of an oncogenic part, self-activation of tyrosine kinase receptor moiety, and altered transcriptional regulation activities. Currently, tens of low molecular mass inhibitors are approved in cancers as the drugs targeting receptor tyrosine kinase (RTK) oncogenic fusion proteins, that is, including ALK, ABL, EGFR, FGFR1-3, NTRK1-3, MET, RET, ROS1 moieties. Therein, the presence of the respective RTK fusion in the cancer genome is the diagnostic biomarker for drug prescription. However, identification of such fusion oncogenes is challenging as the breakpoint may arise in multiple sites within the gene, and the exact fusion partner is generally unknown. There is no gold standard method for RTK fusion detection, and many alternative experimental techniques are employed nowadays to solve this issue. Among them, RNA-seq-based methods offer an advantage of unbiased high-throughput analysis of only transcribed RTK fusion genes, and of simultaneous finding both fusion partners in a single RNA-seq read. Here we focus on current knowledge of biology and clinical aspects of RTK fusion genes, related databases, and laboratory detection methods.
Keywords: chimeric transcripts, fusion detection, fusion genes, genetic testing, predictive biomarker
Genomic instability and cancer evolution
Genomic instability is an exaggerated accumulation of genomic abnormalities and one of the classical cancer hallmarks.1,2 Genomic instability is associated with both the appearance of ‘driver’ and ‘passenger’ mutations and accelerated molecular evolution of the tumor. There are two main levels of genomic instability.3 At the nucleotide level, it generates mostly single nucleotide substitutions, short insertions and deletions, insertion of transposable elements,4 and variability of the microsatellite loci.5,6 In turn, chromosomal instability implies shuffling of the bigger fragments of the genome and is associated with aneuploidy,7 deletions or amplifications of genes,8 chromosomal rearrangements,7 translocations, and gene fusions.9 Chromosomal instability is currently considered one of the most common features of cancer cells. Approximately 90% of solid tumors and more than a half of hematopoietic cancers have large-scale chromosomal aberrations.10
Genomic instability generates genomic aberrations and can lead to accelerated evolution of cancers toward drug and immune resistance.11,12 The increased mutation rate can help cancer cells to control drug response by eliminating or amplifying genes related to drug efficacy mechanisms. These processes, underlaying resistance, include loss or gain of chromosomes and their fragments, as well as nucleotide-level mutations. For instance, the reversion mutations of BRCA1/2 in solid cancers make tumors more resistant to platinum or PARPi therapy.13 Oncogenic mutations can provide immune evasion, such as mutations in KRAS that promote immune suppression in colorectal cancer.14 Moreover, high level of cancer cells aneuploidy negatively correlates with the effectiveness of immunotherapy.12 On the other hand, deficient DNA repair systems can also lead to the increased mutation load in protein coding regions, thus expanding the amount of neoantigens in tumor.15 This makes tumors susceptible to recognition by immune system, thus potentially enhancing immunotherapy efficiency.16 Thus, high tumor mutation burden is one of the major biomarkers for immunotherapy prescription.17
Genomic instability is preceded by accumulation of mutations in proto-oncogenes, tumor suppressor genes, or genetic predisposition that lead to defective regulation of tightly connected mechanisms of DNA damage response and repair,18–23 cell cycle progression, senescence, and apoptosis,24 that coincide with inefficient elimination of transformed cells by the immune system.25–27 For example, negative regulators of Wnt pathway that plays a crucial role in cell proliferation, migration, polarity, and cell fate determination, are usually mutated and, consequently, downregulated in lung cancer cells.28 This results in abnormal cell proliferation and can provoke metastasis.28 Immunosuppression can be caused by various random events such as viral infection,29,30 insufficient generation and CD8 T-cell exhaustion,31 and tissue repair associated with M2 macrophage polarization.32
Chromosomal instability is associated with an ability to move and interconnect different pieces of chromosomes.33,34 During normal germ cell development, DNA is exchanged or crossed-over between chromosomes during prophase 1 of the meiosis followed by haploid cell division to generate cells with a single copy of each chromosome. These mechanisms are absent in the somatic cells and re-emerge in cancer leading to translocations and aneuploidy.35–38
One of mechanisms that promotes genomic instability is the so-called replication stress, that is, disturbances in the DNA replication process that lead to the arrest or destruction of the replication fork.39 In turn, termination of replication in the absence of timely repair can provoke DNA double-strand breaks.40
Hotspots of the DNA double-strand breaks often occur at specific ‘fragile’ sites on chromosomes.41 At these loci, there is an increased frequency of loss of homozygosity or heterozygosity in cancer cells.42 In regions of nucleotide tandem repeats, mutations also occur due to malfunctions of the replication fork.43 The system repairs DNA double-stranded breaks by the homologous recombination of adjacent genome regions. As a result, duplications of microhomologous chromosomal regions can occur, which are found in many types of cancer, such as ovarian and breast cancers.44
Mitotic disorders are another reason for the appearance of genomic instability at the chromosomal level. Errors can occur both in the early and late stages of cell division. The contribution to the development of genomic instability of sister chromatids pairing violations is well known.45 Also, mutations in the genes responsible for the passage of cell cycle checkpoints can be associated with genome instability.46 A common occurrence in the anaphase of cancer cells is chromosome lagging.47 This is usually due to abnormal attachment of chromosomes, when one kinetochore attaches to microtubules extending from different poles of the spindle. Because this phenomenon is not recognized by the spindle assembly checkpoint, this merotelic attachment and the resulting lagging of the chromosome is an important cause of genomic instability.46 Mitotic disorders can lead to aneuploidy and even genome-wide duplications.48 It is important to note that the ploidy of the genome is directly related to the ability of cells to adapt. Tetraploid cells have more mutations per genome, but not per chromosome, which increases the adaptability of the cell.49 In addition, it has been experimentally shown that genome-wide duplications increase the resistance of cells to subsequent manifestations of genomic instability.49
Telomere changes can also lead to genomic instability.50 Thus, shortening of telomeric regions is a frequent event in the early stages of tumor development.51 Normally, it occurs as a result of the accumulation of the effect of previous acts of progenitor cell replication, and when the threshold value is crossed, such cells undergo aging and/or are eliminated. However, mutations in the cancer cell, for instance, leading to loss of function of negative regulators of cell proliferation, can help to avoid apoptosis.52 Alternatively, mutations can result in function gain for the different protein families that inhibit apoptosis, for example, members of IAP and Bcl-2 protein families, or other prosurvival factors.53 This has a strong potential to evade programmed cell death.54 Telomere shortening can also be associated with various manifestations of genomic instability at the chromosomal level, from amplification or deletion of individual loci to changes in entire chromosomes.55 Late-stage tumors typically reactivate telomerase or activate an alternative telomere elongation pathway.50 The latter mechanism appears to be associated with significant genomic rearrangements and defects in DNA repair systems.56
In addition, high expression and insertions of LINE-1 and other active transposable elements within accessible chromatin regions generate multiple somatic variants, and contribute to the genomic instability in cancers.57–60 Moreover, epigenetic factors can also cause and promote genomic instability. The key epigenetic factors contributing to gene expression regulation and mutagenesis are DNA methylation, histone modification, nucleosome remodeling, and non-coding RNAs.61
Novel fusion genes in cancer
Chromosomal translocations, deletions, insertions, and inversions can lead to the formation of chimeric genes with oncogenic functions. The first chimeric gene was described within the so-called Philadelphia chromosome, which is formed as a result of the translocation of regions of chromosomes 9 and 22 [t (9;22) (q 34; q 11.2)] and occurs in approximately 90% of all cases of chronic myeloid leukemia.62,63 As a result of structural rearrangement, a chimeric gene BCR-ABL1 is formed, which encodes the corresponding chimeric protein BCR-ABL.64 It is a continuously active tyrosine kinase that permanently activates proliferation, replication, differentiation, and also renders the cell resistant to apoptosis.64 Active proliferation can lead to an increased rate of mutagenesis, which contributes to the development of drug resistance in cancer cells.65
Since the discovery of the Philadelphia chromosome, many other chimeric genes have been found that are specific to a particular type of cancer.65 For example, the chimeric EWSR1-FLI gene is characteristic for a number of sarcomas.66,67 KIAA1549-BRAF fusion oncogene is characteristic for spinal intramedullary astrocytomas,68 and fusions with JAZF1 and YWHAE genes frequently occur in leiomyosarcomas.69,70 In many cases, fusion genes are just passenger mutations accompanying carcinogenesis that do not play any significant role in cancer progression.71–73 On the other hand, some fusion genes harbor specific tumor promoting molecular activities and are, therefore, referred to as driver mutations.74 Their tumor-promoting activities may strongly differ in nature and are still most probably not completely understood.
However, even for the fusion oncogenes, only a tiny fraction of them currently serves as the targets for cancer therapeutics. All such chimeras are fusions with receptor tyrosine kinase (RTK) genes. Presence of such fusion gene in the genome can be a significant biomarker both for the diagnosis and prognosis of the disease, and for the choice of therapy. Tens of small molecular mass therapeutics that inhibit kinase activities of these gene products have been approved by the US FDA for treatment of tumors with confirmed chimeric genes (Table 1). In particular, chimeric transcripts of genes for RTKs ALK, FGFR 1-4, NTRK 1-3, RET, ROS1, and MET are used for prescription of the respective targeted therapeutics (Table 1).
Table 1.
US FDA-approved cancer drugs targeting fusion genes. Only three out of 35 drugs approved for BCR-ABL1 fusion are shown (Data collected from https://nctr-crs.fda.gov/fdalabel/ui/search).
| Cancers | Targeted kinase fusion partners | Drug | Approval, year | Reference |
|---|---|---|---|---|
| Ph+ CMLa | BCR-ABL1 | Imatinib | 2001 | Amarante-Mendes et al.75, Milojkovic and Apperle76 |
| Ph+ CMLa | BCR-ABL1 | Nilotinib | 2010 | Amarante-Mendes et al.75, Radich et al.77 |
| NSCLC ALK or ROS1 positiveb, ALCL ALK positivec | ALK, ROS1, MET | Crizotinib | 2011 | Shaw et al.,78 Solomon et al.79 |
| NSCLC ALK positived | ALK, ROS1 | Ceritinib | 2014 | Facchinetti et al.80 |
| NSCLC ALK positived | ALK, ROS1, EGFR | Brigatinib | 2017 | Descourt et al.81 |
| NSCLC ALK positive | ALK | Alectinib | 2017 | Wang et al.82 |
| Solid tumors with NTRK gene fusions | NTRK1-3 | Larotrectinib | 2018 | Rudzinski et al.83 |
| NSCLC ROS1 positivee, solid tumors with NTRK fusions | NTRK, ROS1, ALK | Entrectinib | 2019 | Dziadziuszko et al.,84 Doebele et al.85 |
| Urothelial cancer with FGFR3 fusion | FGFR3 | Erdafitinib | 2019 | Zengin et al.86 |
| Cholangiocarcinoma with FGFR2 fusion | FGFR1-3 | Pemigatinib | 2020 | Abou-Alfa et al.87 |
| RET-positive NSCLC, thyroid cancerf | RET | Pralsetinib | 2020 | Gainor et al.88 |
| RET-positive NSCLC, thyroid cancerf | RET, VEGFR1, VEGFR3 | Selpercatinib | 2020 | Subbiah et al.89 |
| NSCLC ALK positive | ALK | Lorlatinib | 2021 | Sehgal et al.,90 Descourt et al.81 |
| Cholangiocarcinoma with FGFR2 fusion | FGFR2 | Infigratinib | 2021 | Javle et al.91 |
| Ph+ CMLa | BCR-ABL1 | Asciminib | 2021 | Yeung et al.92 |
Ph+ CML – chronic myeloid leukemias with Philadelphia chromosome.
NSCLC ALK or ROS1 positive – non-small-cell lung cancers harboring translocations of ALK or ROS1 genes.
ALCL ALK positive – anaplastic large-cell lymphoma, systemic ALK-Positive.
NSCLC ALK positive – non-small-cell lung cancers with translocation of ALK gene.
NSCLC ROS1 positive – non-small-cell lung cancers with translocation of ROS1 gene.
RET-positive NSCLC – non-small-cell lung cancers with translocation of RET gene.
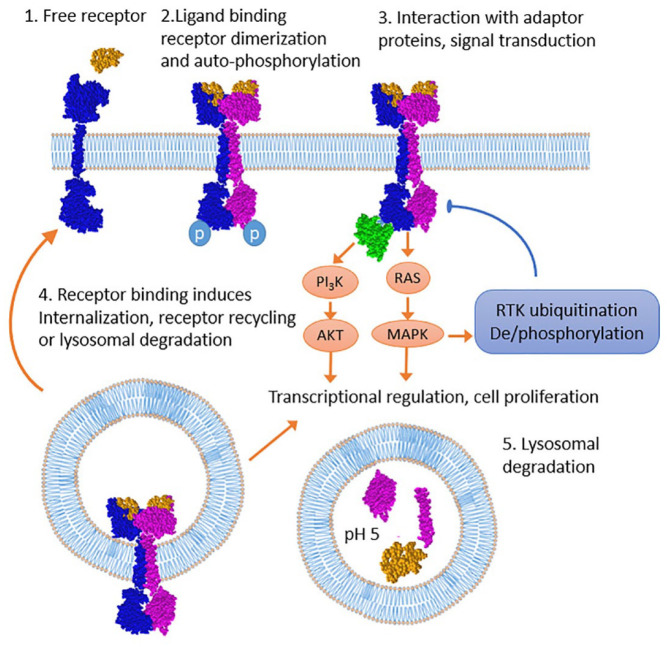
For these fusion oncogenes, all paternal tyrosine kinase receptor proteins have a set of common structural features (Figure 1). They consist of (at least) an extracellular ligand-binding domain, transmembrane domain, and cytoplasmic tyrosine kinase domains.93,94
Figure 1.
Life cycle of the tyrosine kinase receptor. Ligand binding to RTK monomer mediates receptor dimerization, autophosphorylation, and various adapter protein binding. In turn, RTK interacting proteins mediate receptor internalization and downstream RAS/MAPK cascade activation including negative and positive feedback loops regulating ubiquitination and phosphorylation. Internalized RTK continues signaling and upon ubiquitination can be recycled to the cell surface.95–98 Alternatively, RTK can be subjected to lysosomal degradation.96,98
MAPK, mitogen-activated protein kinase; RTK, receptor tyrosine kinase.
Binding of the ligand to the extracellular part of the receptor causes dimerization and transphosphorylation of tyrosine residues of the intracellular domain. This leads to activation of the kinase domain and subsequent triggering of various intracellular cascades.99 In this case, the signaling pathway of mitogen-activated protein kinase (MAPK pathway), the phosphoinositide-3-kinase/protein kinase B (PI3K/AKT/mTOR), protein kinase C (PKC), and STAT-dependent pathways are activated, which are responsible for the induction of proliferation and cell survival.100–103 Breakage and ligation during chimera formation occur in the intron region and, thus, due to splicing, the exon–intron structure of both parts of the transcript is preserved. Ligand binding leads to autophosphorylation and adaptor protein binding that further phosphorylate and ubiquitinate receptor which is subsequently internalized into lysosomes followed by either recycling or degradation,96,97 Internalized RTK continues to signal inside the cell.
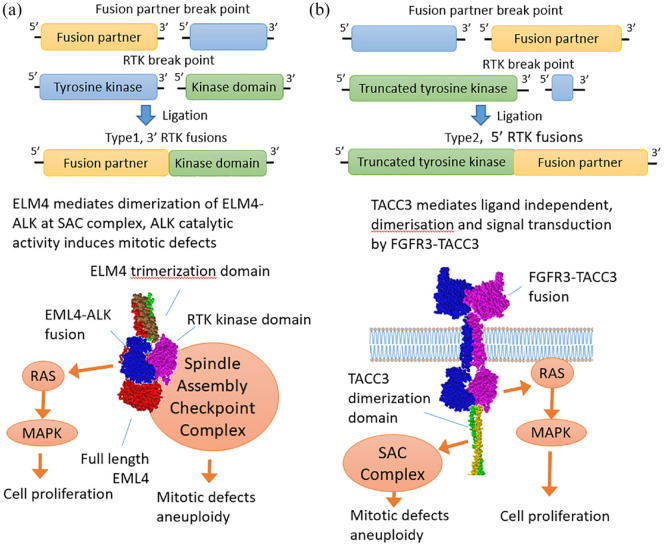
There are two main types of structure of chimeric tyrosine kinase genes (Figure 2). In the first case, the N-terminal extracellular and transmembrane domains-encoding part of a tyrosine kinase receptor gene may be replaced by a partner gene, which results in the presence of tyrosine kinase domain is the 3′-moiety of a chimeric gene. In this case, the 5′-partner domains, as a rule, contribute to the dimerization of the tyrosine kinase domain. For example, the coiled coil motif, sterile alpha domain, LIS 1-homologous, IMD domain, and caspase domain can be mentioned as the contributing structural motifs of the 5′ fusion partner.104 This can lead to ligand-independent dimerization of the tyrosine kinase domain which triggers further carcinogenic properties of such fusion oncogene.
Figure 2.
Structure and functions of the tyrosine kinase gene fusions of the first and second types.109 (a) Type 1 fusion diagram represents a fusion protein between EML4 and ALK retaining the tyrosine kinase domain, whereas the rest of the RTK including transmembrane domain is lost. The resulting chimera translocates into the cytoplasm where it signals in a RAS/MAPK-dependent manner forming lipid-independent protein granules.110,111 ELM4 is a spindle checkpoint protein112 whose trimerization domain is retained in chimeras and most likely mediates interaction with the spindle assembly checkpoint complex and mitotic defects.113,114 (b) Type 2 RTK diagram exemplifies FGFR3-TACC3 chimera in which TACC3 dimerization leucine zipper is attached to the C-terminus of FGFR3 mediating ligand-independent dimerization and signaling. In turn, TACC3 is a spindle checkpoint protein and FGFR3-TACC3 chimera causes mitotic defects.115,116
MAPK, mitogen-activated protein kinase; RTK, receptor tyrosine kinase.
In the second type, the tyrosine kinase receptor is the 5′-terminal partner, and the 3′-partner most probably can additionally stabilize the chimeric RNA or protein product.105 The breakpoint in such case is usually located after the exons encoding the tyrosine kinase domain. Thus, the partner gene is fused to the C-terminus of a nearly full-length tyrosine kinase receptor.105
Importantly, for both types, the main feature is the preservation of the active tyrosine kinase domain. This appears to be a key factor distinguishing a ‘driver’ mutation from random chimeric products arisen as a side effect of genomic instability.106 The second factor is the preservation of an open reading frame for both parts of the chimera. Both factors are important to distinguish between the ‘driver’ and ‘passenger’ gene fusion events. For example, in infant hemispheric glioma, only the patients with preserved open reading frame for a ZCCHC8-ROS1 fusion were responding to entrectinib.107 Note also the published outstanding case of heavily pretreated glioblastoma which expressed transcripts for four clinically relevant fusion transcripts: with ALK, FGFR2, NTRK2, and NTRK3 genes. Due to tumor heterogeneity, these were, however, expressed each by only a minor fraction of tumor cells, and the prescription of the corresponding targeted therapies would be most likely unsuccessful.108
Fibroblast growth factor receptor gene fusions
Fibroblast growth factor receptors 1–4 (FGFR1–4) are a highly conserved family of transmembrane tyrosine kinases that can form chimeric oncogenes in various tumors. Normally, FGFR family members play important roles in cell proliferation, embryonal development, organogenesis, maintenance of homeostasis, and tissue integrity.117 In turn, structural aberrations of FGFR family genes contribute to oncogenesis, tumor progression, and development of drug resistance. In about 8% of the cases, abnormal increase in FGFR activities in cancers is thought to be caused by their gene fusions.118
Many chimeric gene partners of the FGFR family have been described.105 The FGFR1-HOOK3 and FGFR1-TACC1 chimeras have been described for cases of the gastrointestinal stromal tumor and gliomas.119 FGFR1-ZNF703 chimeric gene is found in breast cancer.120 The products of these chimeras include the N-terminal portion of the FGFR1 protein and the coiled coil motif at the C-terminal, which induces dimerization of the tyrosine kinase triggering various signaling cascades.
Among all family members, FGFR2 gene is more likely than others to form chimeras.118 As a result of the fusion, the normally prohibited activation of FGFR2 and the launch of various signaling cascades also occur, which can stimulate oncogenesis. FGFR2 fusions are characteristic for cholangiocarcinomas, where they occur in 10–16% of the cases.121 Thus, in patients with cholangiocarcinoma, the chimeric genes FGFR2–AHCYL, FGFR2–BICC1, FGFR2–PPHLN1, and FGFR2–TACC have been identified and functionally characterized.104 The chimeric FGFR2-CCDC6 gene can initiate the proliferation of cancer cells in vivo.122 Furthermore, more than a hundred of FGFR2 chimeric partners have been identified but not studied in detail. Such partners include KIAA1217, KIAA1598, DDX21, LAMC1, NRAP, NOL4, PHC1, RABGAP1L, RASAL2, ROCK1, TFEC, AFF4, CELF2, DCTN2, DNAJC12, DZIP1, FOXP1, INA, KCTD1, LGSN, and other genes.123 In addition to cholangiocarcinoma, chimeric FGFR2 genes are also frequently found in colorectal cancer, lung cancer, and in hepatocarcinomas.124,125
In turn, FGFR3 fusions are characteristic for glioblastoma, lung, and bladder cancers.126 One of the best studied chimeras is FGFR3-TACC3.115 The product of this chimeric gene is a fusion of the N-terminal region of the FGFR3 with the coiled coil domain of TACC3.115 Coiled coil motif is located at the C-terminus of the protein and is normally involved in the formation and stabilization of the mitotic spindle.127 The FGFR3-TACC3 fusion has been found in different types of cancer: in gliomas, in cancers of the lung, bladder, head and neck, and cervix.125,128,129 The formation of this chimeric oncogene results in constitutive activation of the FGFR3 tyrosine kinase domain and, as a consequence, activation of the MEK/ERK and STAT1 signaling pathways.129 Also, the chimeric protein FGFR3-TACC3 is localized to the mitotic spindle and induces errors in the chromosome segregation process, thus resulting in the appearance of aneuploid cells in glioblastoma.115 In contrast, in bladder cancer cells, FGFR3-TACC3 chimera inhibits TACC3 localization to mitotic spindle, thereby contributing to aneuploidy.116 FGFR3 fusions with other genes have also been detected but have not been characterized in-depth. For example, the product of the FGFR3–BAIAP2L1 chimera was found in bladder and lung cancers, and the AES, ELAVL3, JAKMIP1, TNIP2, and WHSC1 genes are also known among the confirmed FGFR3 fusion partners.118,130
Fusions with FGFR genes are important prognostic molecular markers. A study was made of tumors of the bile ducts: 152 cholangiocarcinomas and 4 intraductal papillary mucinous tumors were analyzed by fluorescent in situ hybridization (FISH) for the presence of FGFR2 chimeras.131 Totally, 13 tumors carrying FGFR2 translocations were found which showed statistically significantly longer overall survival: 123 versus 37 months for the patients without translocations.131 Similar results were obtained in a study of 377 patients with biliary tract cancer, of which 63 had FGFR2 chimeras.132
Neurotrophic TRK family gene fusions
The neurotrophic TRK (NTRK) family includes TRKA, TRKB, and TRKC proteins which are encoded by the NTRK1, NTRK2, and NTRK3 genes, respectively.133 Normally, these tyrosine kinases play a pivotal role in neuronal survival and CNS plasticity.134
Various mutations of the NTRK family genes in cancer cells have been described leading to single nucleotide substitutions, amplifications, and abnormal splice isoforms.135 However, the formation of chimeric oncogenes is the most common mutation type that leads to increased kinase activity. NTRK gene sequences including a tyrosine kinase domain are usually located at the 3′ end of the chimeras, and are fused to the 5′ region of a partner gene.136 The product of the chimeric gene is an oncoprotein capable of activating the tyrosine kinase domain without the involvement of a ligand binding.137 Various NTRK chimeric 5′ partners are known, including ETV6, TPM3, and LMNA, yet their exact roles in the function of an oncoprotein have not been fully characterized.138
NTRK fusion oncogenes were found in multiple types of cancer. At the same time, their occurrence is relatively low. The analysis of 13,467 adult and pediatric tumor samples from The Cancer Genome Atlas (TCGA) and St. Jude PeCan databases showed that the frequency of chimeric NTRK genes in the most common cancer types was less than 1%.139 Another study by Rosen and colleagues showed that, when present, functional chimeric NTRK genes are expressed from the very beginning until the most advanced stages of carcinogenesis, thereby indicating a ‘driver’ rather than a ‘passenger’ nature of the mutation.140
RET gene fusions
RET mutations such as amplifications, single nucleotide substitutions, insertions and deletions, and the formation of chimeric genes have been described in cancer cells. Chimeras with the 3′ RET moiety are more frequently detected and better described. In this case, the tyrosine kinase domain part is usually placed under the control of a stronger promoter, which leads to its overexpression.106 Many 5′ fusion partners of RET have been described, of which the most common are CCDC6, NCOA4, and KIF5B.103,141,142 RET activation as a result of chimera formation can occur through different mechanisms. First, as already noted, the substitution of the 5′-part of RET for a gene fragment with a stronger promoter. Second, dimerization of RET tyrosine kinase domains due to fusion with partners providing a coiled-coil motif, as in the case of CCDC6 gene. As a result, ligand-independent kinase activation occurs.143 Also, oncogenic hyperactivation of RET in a fusion is possible due to the loss of the autoinhibitory N-terminal region.130
ROS1, ALK, and MET gene fusions
ROS1 fusions
ROS1 is a proto-oncogene encoding a tyrosine kinase with unknown physiological function. The first ROS1 chimeric gene was found in the U118MG glioblastoma cell line in 1987.144 Interestingly, the tyrosine kinase domain of ROS1 has high structural identity with another RTK, ALK.78 Impaired expression of ROS1 is known for many cancer types, and at least 55 5′-terminal partners of the ROS1 chimeras were reported.145 The frequency of occurrence of 5′-partners in the chimeras may depend on the cancer type. In particular, large heterogeneity of the 5′-partners is characteristic of glioblastoma, non-small-cell lung cancer (NSCLC), and of inflammatory myofibroblastic tumors. In NSCLC tumors, the most common 5′-partners for the ROS1 chimeras are CD74 (~44%), EZR (16%), SDC4 (14%), and SLC34A2 (10%).145
ROS1 chimeras are formed due to both intra- and interchromosomal rearrangements. For example, in glioblastoma, the chimeras are more often the result of intrachromosomal translocations, and in NSCLC, they are instead interchromosomal.146,147
The most common structure of the ROS1 chimera includes the loss of virtually the entire extracellular domain of ROS1 and the fusion with the 5′ part of the partner gene, retaining the open reading frame and a complete intracellular kinase domain.145 As before, the formation of chimeras usually results in ligand-independent, constitutive activation of the ROS1 tyrosine kinase domain. The intracellular localization of the chimeric ROS1 gene depends on the 5′-partner gene and influences the activation of specific signaling pathways. For example, the SDC4-ROS1 and SLC34A2-ROS1 chimeras are localized in endosomes and activate the MAPK pathway more efficiently than the CD74-ROS chimera localized in the endoplasmic reticulum.148 It was shown in mice models that the presence of the chimeric ROS1 gene alone may be sufficient to induce carcinogenesis. However, when combined with the loss of tumor suppressor p16Ink4a, this results in development of a more aggressive tumor.149
ALK fusions
Also, many tumors including large cell lymphoma, diffuse large B-cell lymphoma, glioma, NSCLC, colorectal cancer, breast, ovarian, and esophageal cancer have chimeras with tyrosine kinase ALK.150 More than 90 5′-partner genes of ALK have been described,150–152. ALK translocations are not uncommon and are found in approximately 8% of all NSCLCs.153 As a result of structural rearrangements, the chimeras retain a full-fledged ALK tyrosine kinase domain at the C-terminus. As in other cases, dimerization of chimeras results in aberrant persistent kinase domain activity.154 ELM4-ALK is the most frequent ALK fusion in NSCLC.155 ELM4 is a spindle checkpoint protein required for the proper chromosome alignment and attachment of microtubules to the kinetochore.112 Accordingly, ELM4-ALK fusion inhibits spindle assembly checkpoint control, that is, inhibits cell cycle arrest in response to the paclitaxel and leads to the mitotic errors (Figure 2), 113,114 This effect is partially due to the kinase activity of ALK domain and, most likely, other oncogenic ALK fusions have acquired kinase activity as well.114
MET fusions
The MET gene is another important RTK-encoding proto-oncogene. The formation of chimeras with MET is a relatively rare event, but such products are found in various cancer types. The first described chimeric gene was TPR-MET.156 The replacement of MET extracellular and juxtamembrane domains containing regulatory regions by TPR two leucine zipper domains leads to a constitutively dimerized and therefore activated Met kinase domain.156 The dimerization domains are essential for TPR-MET oncogene transforming activity, as well as the absence of MET extracellular and juxtamembrane domains.156 Similar to many other RTKs, the increased MET activity promotes activation of downstream intracellular pathways and signaling axes RAS-MAPK and PI3K-AKT.157
Structural rearrangements involving MET were detected in 0.5% of cases of NSCLC, 3% of glioblastomas, and isolated cases were described in secretory carcinoma of the salivary gland and pediatric fibrosarcoma.158–161 Several partners of MET chimeras have been described: the HLA-DRB1, KIF5B, PTPRZ1, STARD3NL, and ST7 genes.162 The stability and degradation of the MET receptor is regulated by an intracellular peri-membrane domain encoded by exon 14.163 Interestingly, the formation of TPR-MEt also resulted in the loss of exon 14 of MET without disturbing the reading frame. Mutations resulting in the loss of exon 14 in the MET mRNA occur in 3–5% of NSCLCs.164 This can be caused by both structural rearrangements and point mutations leading to splicing disorders.165 Mutations are detected both in the intron region around exon 14 and directly in the splicing sites. After pre-mRNA maturation, a mutant MET receptor with an increased lifetime is then translated.163 These mutations are characteristic of lung adenocarcinomas; however, they are also found in other types of cancer: pulmonary sarcomatoid (pleomorphic) carcinomas, squamous cell NSCLC, and less often in gliomas and other tumors.165,166
Application of protein fusion inhibitors in clinical practice
The recent findings revealed overall better response rate to the approved TKIs in patients with different target RTK fusion genes.167 Thus, development of new and broader testing of already approved drugs in more than 100 clinical trials targeting 26 gene fusions is currently underway (Table 2).
Table 2.
Drugs tested against neoplasms with gene fusions that are currently in clinical trials (clinicaltrials.org). Please refer to the website for complete information regarding treatment modalities and specific groups of patients in a specific clinical trial.
| Cancer type | Targeted drug(s) | Fusion gene partner(s)* | Clinical trial ID |
|---|---|---|---|
| AML | Bosutinib monohydrate, decitabine, enasidenib mesylate, gilteritinib fumarate, glasdegib maleate, ivosidenib, venetoclax | BCR-ABL1 | NCT04655391 |
| Bladder cancer | Neratinib | HER2-GRB7 | NCT01956253 |
| Bladder cancer with mutation in FGFR3 gene | Erdafitinib | FGFR3 | NCT04917809 |
| Solid cancers with fusions of NTRK, ROS1, ALK genes | Entrectinib | NTRK genes, ROS1, ALK | NCT03066661 |
| Cholangiocarcinoma | E79 | FGFR genes | NCT04238715 |
| Cholangiocarcinoma with mutations of FGF or FGFR genes | Futibatinib | FGFR genes | NCT02052778 |
| Cholangiocarcinoma with mutations in FGFR2 gene | Infigratinib (BGJ398) | FGFR2 | NCT02150967 |
| Cholangiocarcinoma with mutations in FGFR2 gene | Infigratinib (BGJ398), gemcitabine, cisplatin | FGFR2 | NCT03773302 |
| CML | Imatinib withdrawal, dasatinib, nilotinib | BCR-ABL1 | NCT04147533 |
| CML | Dasatinib, nilotinib | BCR-ABL1 | NCT03079505 |
| CML | LBH589 | BCR-ABL1 | NCT00451035 |
| CML | PF-114 | BCR-ABL1 | NCT02885766 |
| CML, ALL | Nilotinib | BCR-ABL1 | NCT01077544 |
| Clear cell sarcoma | AMG 337 | EWSR1-ATF1 | NCT03132155 |
| Epithelioid hemangioendothelioma | Trametinib | TAZ-CAMTA1 | NCT03148275 |
| Fibrosarcoma with NTRK fusion | Larotrectinib, standard of care | NTRK genes | NCT05236257 |
| Glioblastoma, IDH-wildtype | Metformin and radiation therapy, temozolomide | FGFR3-TACC3 | NCT04945148 |
| Glioma | PLB11 | PTPRZ1-MET | NCT02978261 |
| Intrahepatic cholangiocarcinoma | Derazantinib | FGFR genes | NCT03230318 |
| Malignant hepatobiliary neoplasm | Ponatinib hydrochloride | FGFR2 | NCT02265341 |
| Myeloproliferative disorders | Imatinib | PDGFR | NCT00038675 |
| Non squamous lung cancer | Crizotinib | ALK | NCT01154140 |
| Non-squamous, NSCLC | Afatinib | NRG1 | NCT04750824 |
| NRG1-rearranged malignancies | Afatinib | NRG1 | NCT04410653 |
| NSCLC | TPX-131 | ALK | NCT04849273 |
| NSCLC | Tarloxotinib bromide | ERBB | NCT03805841 |
| NSCLC | Pemigatinib | FGFR genes | NCT05210946 |
| NSCLC | Toripalimab injection combined with axitinib | negative: EGFR mutation, ALK, and ROS1 fusions | NCT04459663 |
| NSCLC | Tarloxotinib bromide | NRG1, ERBB | NCT03805841 |
| NSCLC | XZP-5955 tablets | NTRK genes | NCT04996121 |
| NSCLC | Apatinib single agent arm | RET | NCT02540824 |
| NSCLC | Vandetanib | RET | NCT01823068 |
| NSCLC | Cabozantinib | RET, ROS1, NTRK genes | NCT01639508 |
| NSCLC | AB-16 | ROS1 | NCT04395677 |
| NSCLC with ALK and ROS1 aberrations | Carboplatin + Pemetrexed + Atezolizumab + Bevacizumab, Carboplatin + Pemetrexed + Atezolizumab | ALK, ROS1 | NCT04042558 |
| NSCLC | PF-234166 | ALK | NCT00932451 |
| NSCLC | Selpercatinib, placebo | RET | NCT04819100 |
| NSCLC with ALK rearrangement | Crizotinib | ALK | NCT02201992 |
| NSCLC and thyroid neoplasms | LOXO-26 | RET | NCT05241834 |
| NSCLC and thyroid neoplasms | LOXO-26 | RET | NCT05225259 |
| Ovarian cancer and carcinosarcoma | Pamiparib | ABCB1 | NCT03933761 |
| Pancreatic cancer | Seribantumab | NRG1 | NCT04790695 |
| Prostate cancer | Cytarabine | ETS | NCT00480090 |
| Prostate cancer | ESK981 | ETS | NCT03456804 |
| Prostate cancer | Androgen deprivation therapy | TMPRSS2-ETS | NCT02303327 |
| Prostate cancer | Enzalutamide | TMPRSS2-ETS | NCT02288936 |
| Prostate cancer | Enzalutamide, abiraterone, carboplatin, cabazitaxel, docetaxel, radium chloride Ra-223, niraparib plus abiraterone acetate plus prednisone | RET | NCT03903835 |
| Recurrent IDH-wildtype glioma with FGFR(1-3)-TACC3 fusion | AZD4547 | FGFR(1–3)-TACC3 | NCT02824133 |
| Renal cell carcinoma with Xp11.2 translocation and TFE3 fusion | Cabozantinib, sunitinib | TFE3 | NCT03541902 |
| Renal cell carcinoma with Xp11.2 translocation and TFE3 fusion | Axitinib, nivolumab | TFE3 | NCT03595124 |
| Solid tumors | Debio 1347 | EGFR | NCT03834220 |
| Solid tumors | Derazantinib | EGFR | NCT01752920 |
| Solid tumors | Erdafitinib | FGFR | NCT04083976 |
| Solid tumors | HMBD-1 | NRG1 | NCT05057013 |
| Solid tumors | Seribantumab | NRG1 | NCT04383210 |
| Solid tumors | AB-16 | NTRK genes | NCT04617054 |
| Solid tumors | Entrectinib | NTRK genes | NCT02568267 |
| Solid tumors | ONO-7579 | NTRK genes | NCT03182257 |
| Solid tumors | DS-651b | NTRK genes, ROS1 | NCT02675491 |
| Solid tumors | BOS172738 | RET | NCT03780517 |
| Solid tumors | KL59586 | RET | NCT05265091 |
| Cholangiocarcinoma | Infigratinib | EGFR | NCT04233567 |
| NSCLC with RET rearrangements | Pralsetinib | RET | NCT03037385 |
| Solid tumors | Pembrolizumab | EGFR, ALK, ROS1 | NCT03049618 |
| Solid tumors | Afatinib | NRG1 | NCT05107193 |
| Solid tumors with NTRK gene fusions | Larotrectinib | NTRK genes | NCT02576431 |
| Solid tumors with NTRK, ROS1, ALK gene fusions | SIM183-1A | NTRK genes | NCT04671849 |
| NF2-deficient solid tumors with YAP1, TAZ gene fusions | IK-93 | YAP1, TAZ genes | NCT05228015 |
| CNS tumors | Entrectinib | FGFR genes | NCT02650401 |
| Solid tumors with FGFR alterations | E79 | FGFR genes | NCT04962867 |
| Solid tumors with NTRK gene fusions | Larotrectinib | NTRK genes | NCT03025360, NCT04945330, NCT02637687, NCT02122913, NCT04142437, NCT05192642 |
| Solid tumors with NTRK gene fusions | Selitrectinib | NTRK genes | NCT03206931, NCT04275960, NCT03215511 |
| Thyroid cancers with PAX8-PPARG fusion | Pioglitazone | PAX8-PPARG | NCT01655719 |
| Upper tract urothelial carcinomas, urothelial bladder cancer | Infigratinib | FGFR genes | NCT04197986 |
| Urothelial carcinoma | AZD4547 | FGFR2/3 | NCT05086666 |
Some rows of the column contain only one fusion gene in case the clinical trial was conducted regardless the second fusion partner gene.
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; FGFR, fibroblast growth factor receptor; NSCLC, non-small-cell lung cancer.
Accordingly, the presence of chimeric genes for RTKs is one of the key biomarkers facilitating the choice of therapy. For example, the drug larotrectinib is approved by the US FDA for tumors harboring chimeric NTRK genes.168 Enterectinib is another drug with a similar profile of action, with somewhat broader activity85: it can also inhibit ROS1 and ALK chimeric products.130 In addition, broad-spectrum tyrosine kinase inhibitors were the first drugs to treat tumors with FGFR chimeras: dovitinib, lenvatinib, lucitanib, nintedanib, derazantinib, and ponatinib. In addition to their activity against FGFR, they can also inhibit VEGFR, RET, KIT, and PDGFR.
However, drugs with low specificity may cause more serious side effects.169 Therefore, more precise inhibitors have been developed that act specifically against mutations of FGFR family members, including fusion formations. Two selective inhibitors for the treatment of tumors with FGFR chimeric genes have been approved by the US FDA. Erdafitinib is approved for the treatment of urothelial carcinomas with FGFR2 and FGFR3 mutations, which included the chimeric gene FGFR3-TACC3.170
Selective RET inhibitors pralsetinib (BLU-667, NCT03037385) and selpercatinib (LOXO-292, NCT03157128) in early-stage clinical trials of NSCLC with RET fusions resulted in 56% overall response rate for pralsetinib.88,89 LOXO-292 was recently approved by the US FDA for the treatment of lung and thyroid cancers with RET driver mutations, with respect to fusions.171 In addition, cases have been described of the acquisition of drug resistance to tyrosine kinase inhibitors in tumors with EGFR mutations, which occurs as a result of structural rearrangements of RET.172 This also highlights the importance of RET analysis for selecting the optimal treatment strategy.
For the treatment of NSCLC with ROS1 rearrangements, crizotinib has been approved in multiple countries, with an overall response rate of 65–80%.78 Crizotinib is also the first targeted drug for the treatment of tumors with ALK rearrangements.79
Entrectinib is an US FDA-approved drug for the treatment of NSCLC with ROS1 rearrangements.84 Entrectinib is able to penetrate the blood–brain barrier, which is especially important in the treatment of tumors that have metastasized to the brain. Currently, clinical trials investigate the activity of entrectinib against tumors with ROS1 fusions regardless of the cancer type. It was possible to observe objective responses, for example, even for the cases of melanoma and high-grade gliomas.173,174
Crizotinib, ceritinib, alectinib, brigatinib, and lorlatinib are approved by the US FDA for the treatment of ALK chimeric lung cancer. Response to ALK-inhibiting therapy has also been described in studies on renal cell and colon cancers.173 Of note, a retrospective clinical study showed that progression-free survival after treatment of ALK-positive NSCLC depends on the 5′-partner of the chimera with the 3′-part of ALK.175 However, the partner genes of the ALK chimeras are not currently routinely taken into consideration when prescribing therapy.
In addition, the drugs capmatinib and tepotinib have recently been approved for the treatment of NSCLC with MET mutations resulting in exon 14 skipping.175
Experimental methods for identification of fusion oncogenes
Identification of fusion oncogenes is a non-trivial task because there are many alternative methods available that can produce results which may contradict each other (Table 3). It is important, therefore, to carefully select method(s) of choice for the experimental testing considering availability of biomaterials, costs, equipment, high throughput, time of analysis, and even complexity of bioinformatic data interpretation.
Table 3.
Summary of advantages and disadvantages of common methods for detection of gene fusions.
| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| FISH | Diagnostic gold standard, high sensitivity and specificity | Laborious. Visual quantification, difficult to automate, requires pre-defined knowledge of fusions. Low resolution, problematic detection of fusions within the same chromosome. Cannot detect fusions with novel partners | Thompson et al.,176 Nguyen et al.,177 Ali et al.,178 Cruz-Rico et al.,180 De Luca et al.,184 |
| IHC | Common detection method | Detects protein overexpression, but not fusions | Thorne-Nuzzo et al.,185 Boyle et al.,186 Shia.256 Zhang.257 |
| RT-PCR methods | Diagnostic gold standard, excellent sensitivity and specificity, widely accepted and cheap. Easy to multiplex and automate. Straightforward interpretation, best suited for clinical laboratory analysis | Requires information about location of fusion breakpoint. Cannot detect novel fusions | Ali et al.,178 Cruz-Rico et al.,180 Lyu et al.,192 Sorber et al.,194 Abbou et al.,195 Shelton et al.196 |
| DNA sequencing of tumors | Allows unbiased detection of mutations and fusions. Covers both transcribed and non-coding regulatory elements of the genome. For example, allows detection of an active promoter – oncogenic transcript fusions | All NGS methods require bioinformatics support. Analytical methods are not standardized yet. Detects expressed and non-expressed fusions. The whole genome deep sequencing is still expensive. Lower sensitivity compared to RNA-seq-based assays | Gerstung et al.,258 ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium,72 Zhang et al.259 |
| Circulating cell-free DNA sequencing | Minimally invasive analysis of tumor DNA for monitoring of the disease. Straightforward DNA extraction from serum | Relatively high frequency of false-negative results. Accurate sample preparation is needed. Plus, the above disadvantages of DNA sequencing methods | Hofman,212 Dagogo-Jack et al.,162 Blaquier et al.,213 Leighl et al.214 |
| Targeted DNA/RNA sequencing | Relatively high-throughput with high coverage of target fusion sites | Collects only information about tested regions and/or fusion types | Heydt et al.,208 Gasc et al.,219 Zheng et al.220 |
| Bulk RNA-seq | Allows detection of known and novel fusions and the presence of an ORF for possible protein expression. Filters out passenger mutations. Permit analysis of archived FFPE samples. Relatively cheap and achieve high coverage. Allows high throughput. Simultaneously measures level of gene expression | May require further standardization. Can be technically challenging | Sorokin et al.,5 Sorokin et al.,197 Winters et al.,225 Peng et al.240 |
| Single-cell RNA-seq | Measures clonality of tumors. Independently measures gene expression and T-cell receptor heterogeneity of immune cells and quantity and gene expression of other non-cancerous cells | Requires additional equipment for single-cell library preparation. Relatively expensive and requires extensive bioinformatics support. Low coverage in terms of reads per single cell, so very low sensitivity of fusion detection. Clinical benefit is questionable | Rozenblatt-Rosen et al.,241 Zeng et al.,242 Nieto et al.,243 Amir et al.,260 Becht et al.,261 Maynard et al.244 |
FFPE, formalin-fixed paraffin-embedded; NGS, next-generation sequencing; ORF, open reading frame.
Fluorescent in situ hybridization
FISH is a method based on the hybridization of labeled specific DNA probes. It allows detecting changes in the number of chromosomes, gene fusions, loss of a chromosomal locus or an entire chromosome in both fresh and fixed tumor tissue samples.176 For example, FISH is used to detect amplifications of the ERBB2 gene for Her2 protein, an important prognostic marker for breast cancer.177 FISH is also a standard method for detecting the Philadelphia chromosome in chronic myeloid leukemia.178 In particular, probes for loci that are contiguous in the absence of rearrangements are used to detect translocations. The signal from the native chromosome will be a pair of closely spaced color signals, whereas in the case of rearrangement, the two colors are separated in space.179
The chimeric EML4-ALK gene is a characteristic biomarker of lung adenocarcinoma. Routine detection of this well-known translocation can also be performed using the FISH method.180 Likewise, FISH is used to detect known rearrangements of the ETV6 gene in secretory carcinoma and pediatric fibrosarcoma specimens.181 A number of laboratories use this approach to detect NTRK gene rearrangements and other fusions such as the FGFR3-TACC3 chimeric oncogene.182,183 Nevertheless, FISH diagnostics does not provide information about the functional activity of translocations including preservation of an open reading frame and expression level and does not allow detection of the non-canonical or unknown rearrangements. In addition, this method is poorly sensitive to the detection of local intrachromosomal rearrangements that are characteristic of FGFR2 fusion genes.184
Immunohistochemistry
Immunohistochemistry ((IHC) method is simple, does not require complex equipment, is relatively affordable, and fast in execution. In addition, reliable results can be obtained using a relatively small amount of starting material. The use of IHC has been approved by the US FDA for the detection of ALK rearrangements in lung cancer samples for selection of patients for treatment with ALK-targeted therapy.185 Also, a ROS1-specific monoclonal antibody is used to detect ROS1 rearrangements by the IHC.186
Apparently, IHC method detects not the chimeric gene products themselves, but rather the expression level of the respective tyrosine kinase domain. Thus, increased IHC signal can be registered also due to stronger expression of the whole, non-fusion RTK gene which still can be targeted by the respective specific drug(s). Although this method is easy to perform and inexpensive, its accuracy may be reduced due to technical issues arising from tissue fixation, or other pre-analytical variables.187 Specifically, comparisons of IHC with FISH and next-generation sequencing (NGS) methods for screening of ALK rearrangements in lung cancer resulted in prevalence of ALK-positive samples tested by IHC.188–190 Furthermore, most of IHC-positive and FISH-negative samples were shown to be negative by NGS, thus supporting the hypothesis of frequent misleading detection of mutations other than fusions (e.g. amplifications) using IHC.189 However, some patients who were FISH negative but IHC positive for ALK fusion showed complete or partial responses to ALK-targeted therapy, thus supporting the relevance of IHC application in practice190 and clinical importance of other factors, such as the overexpression of ALK.191
PCR-based methods
PCR-based methods are widely used both for the diagnosis and for the prognosis and treatment of cancer. Also, reverse transcription PCR is a standard method for confirming the presence of chimeric transcripts resulting from structural rearrangements of chromosomes.178,180 Several kits are commercially available to detect NTRK, ALK, Ret, and ROS fusions in formalin-fixed paraffin-embedded (FFPE) RNA by the multiples real-time PCR using mutation-specific primers and/or hybridization probes with about 1% or more sensitivity and 100% specificity. Detection of 22 fusion genes in AML patients RNA using a commercial quantitative PCR kit demonstrated 99% concordance with cytogenetic analysis.192 Nonetheless, an agarose gel electrophoresis was used for measurements of multiple EWSR1-ETS and FUS-ETS fusions in Ewing sarcoma by multiplex RT-PCR (Ueno-Yokohata et al. 2021). In turn, high-resolution capillary electrophoresis was used to detect 9 fusion transcripts in the single multiplex reaction.193 Intriguingly, out of 122 patients examined, abnormal size was detected for one sample due to a rear deletion of the ETV6 exon 5 within ETV6-RUNX1 chimeric gene. Interestingly, RT-PCR was used in Idylia GeneFusion assay to detect fusion products of the NTRK gene without knowledge of the fusion partner by measuring an imbalance between 5′ and 3′ end of the gene.194 The Idylia GeneFusion assay is basically a device that automatically extracts RNA and performs expression analysis directly from the samples including fresh and FFPE samples. The data demonstrated good concordance between Idylia GeneFusion, IHC, and RNA-seq based Oncomine Focus Assay. Another method, droplet digital multiplied PCR, measures the presence of RNA in small droplets, and each of them encompasses a single RNA molecule, allowing ‘YES-NO’ quantification of PCR results. Thereby, RNA is measured by counting positive and negative droplets. This method was recently used to measure gene fusions from as little as 1 pg of RNA with great success.195,196
To conclude, traditional and novel PCR-based tools represent a gold standard for fusion detection in cancer and are widely used in clinical practice, research, and development.
NGS methods
NGS is an increasingly common method for analyzing tumors. It has a number of advantages over the approaches listed above. Thus, NGS is a more accurate and sensitive method that allows one to analyze various genetic variants in one experimental procedure, eliminating the need to conduct multiple tests in sequence to identify significant biomarkers, thereby significantly reducing the time for selecting therapy.5,197–199 It should be noted that in order to obtain informative results, NGS technologies require a minimum amount of tumor material; they are also able to capture DNA variants present in a minority of cells.200 To date, many approaches have been proposed for the detection of molecular markers of tumors based on NGS. Although whole genome studies are not widely used in clinical practice, in particular due to their high cost, whole exome analysis, gene panel analysis, and whole transcriptome technologies are increasingly used in the selection of cancer therapies. Methods based on NGS have been developed to search for single nucleotide substitutions, deletions, and duplications of genes, to assess the mutation load of tumors, structural rearrangements of chromosomes, as well as to analyze the level of expression of clinically significant genes and to detect chimeric transcripts.201,202
Several clinical trials investigating applicability of NGS for prognosis and ultimately for treatment decision can be mentioned including Strata PATH (NCT05097599), GITIC Study (NCT02013089), The MATCH Screening Trial (NCT02465060), MyTACTIC (NCT04632992), and BALLETT (NCT05058937). The methodology for detection of mutations used in these studies is based on the NGS. The aim of these studies was formulated as to determine the presence of molecular targets for the approved drugs (including gene fusions) and to verify whether patients will benefit more from the targeted treatment.
DNA sequencing
Whole genome sequencing (WGS) is the most comprehensive platform for genome profiling.203 The method has potential to identify the genomic locations of all known and currently unknown fusion events.204 However, application of WGS in tumor biology is nowadays limited to research field due to high costs, lack of standardization, and median turnaround times.
Methods based on whole exome sequencing (WES) are used to detect deletions and duplications of genes, assess the mutational load of tumors, and determine the MSI status.205 Whole exome studies mainly cover only coding regions of the genome (less than 2%), which provides a reduced cost compared to whole genome analysis with sufficient coverage. Despite technical difficulties, the method is widely used to determine various changes in the genome. For example, several approaches have been developed to detect changes in the copy numbers of specific genome regions [copy number aberration (CNA)] using sequencing data.206 Perhaps the optimal approach for the analysis of WES data for CNA is measuring of the sequencing depth of different genome regions.206 If, after normalization, an imbalance in the number of reads in certain part(s) of the genome is detected, then this can be interpreted as a marker of a copy number alteration in the corresponding chromosomal region(s).206 However, this algorithm cannot detect fusion genes. Moreover, WES as a basis for fusion detection tool has a strong technical limitation, namely its focus on gene exons, whereas most of fusion junctions are located in introns. Thus, WES capacity to detect fusion breakpoints is strongly limited. Indeed, the attempt to develop fusion detection tool based on WES data showed lower sensitivity compared to RNA-seq-based approach.207
To detect structural rearrangements of chromosomes that lead to the formation of chimeric genes, a number of targeted DNA sequencing approaches have also been developed. Thus, the panel to detect both known and not yet described chimeric genes for regulatory kinases ALK, ROS1, RET, BRAF, MET, FGFR1-3, and NTRK1-3 has been developed. The panel consists of DNA probes labeled with biotin that are complementary to the intron and exon sequences of the genome, which are known breakpoints in the formation of chimeric genes.208 Similar approaches have been used in commercial panels to search for chimeric genes from different manufacturers. Most fusions occur in intron sequences, which often contain repetitive sequences, such as insertions of transposable element, and are sequenced less efficiently than coding sequences.209 This leads to false-negative results when the presence of the chimeric gene is confirmed by another method, for example, FISH or RNA sequencing.
Davies et al. compared three approaches for detecting rearrangements of ROS1 gene in lung cancer.210 Targeted DNA sequencing did not detect 4 out of 18 chimeric genes confirmed by alternative approaches. The authors attribute false-negative results to specific structural features of specific introns.
Also, Benayed et al. analyzed the quality of chimeric genes detection using DNA sequencing panels.211 14% of tumors with confirmed chimeric genes for clinically relevant tyrosine kinases were not detected when analyzed by the US FDA approved MSK-IMPACT panel, which is based on biotinylated oligonucleotides to capture genomic sequences of interest. False-negative results are also associated with the structural features of introns.
Sequencing of circulating cell-free DNA
Sequencing of circulating cell-free DNA from plasma of cancer patients revealed the presence of mutations characteristic for tumor DNA.212 Sequencing of cell-free DNA from plasma of NSCLC patients with ALK mutation revealed acquisition of additional mutations in ALK and co-occurring amplification of MET1,162 KRAS amplification, and a PI3KCA E545K mutation.213,214 This method requires prompt isolation of plasma for DNA extraction and permits non-invasive and cheap monitoring of cancer patients.
RNA sequencing
RNA sequencing is a method that has several advantages over DNA-based approaches. This approach has been proven informative for analyzing the expression level of various genes associated with the effectiveness of the response to anticancer drugs,215 activation or inhibition of various molecular pathways.202,216,217 Approaches for the analysis of the mutational load of tumors according to full transcriptome analysis are also described.197,218 Therefore, the use of RNA sequencing makes it possible to assess the manifestation of various clinically important biomarkers in one experimental procedure.
The detection of chimeric transcripts has a number of advantages over DNA sequencing approaches. For example, when analyzing transcriptomic profile data, only transcriptionally active chimeric genes are identified, thereby filtering in only those fusions that might be the drivers of cancer progression and leaving out the passenger mutations. Second, in such a way both parts of a fusion gene can be identified at once – equally effective for both known and previously unknown fusions. Third, integrity of an open reading frame can be easily assessed, as well as the presence of a kinase domain in the chimeric gene product.
Targeted RNA sequencing
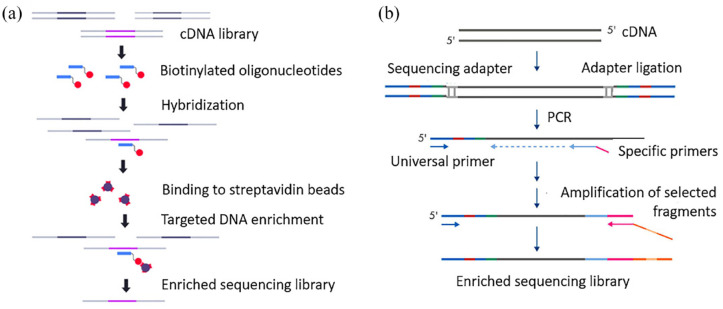
A variety of targeted panels have been proposed to search for both known and novel chimeric transcripts using RNA sequencing. Two approaches are widely used: selection of genes of interest by hybridization of cDNA with oligonucleotides labeled with biotin and complementary to exons of the target genes; or enrichment of libraries by PCR with specific primers complementary to the exon boundaries of the genes of interest and the universal adapter sequence (Figure 3).219,220
Figure 3.
Methods for enrichment of sequencing libraries with specific cDNA targets. (a) Enrichment of the library by hybridization to biotinylated oligonucleotide. (b) Enrichment of the library by PCR with a specific primer (modified, according to Kozarewa et al.221 Zheng et al.220).
In a recent study, Heydt et al. compared four RNA sequencing and one DNA-sequencing-based targeted panels for the detection of chimeric genes from cell lines and FFPE tumor samples.208 As in the studies described above, the DNA-based approach appeared to be less sensitive, producing more false negatives in the analysis of biopsies. Among the alternative RNA analytical panels, three were enriched by amplification: Archer FusionPlex Lung Panel (ArcherDX), QIAseq RNAscan Custom Panel (Qiagen), and Oncomine Focus Assay (Thermo Fisher Scientific). In turn, TruSight Tumor 170 Assay (Illumina) is based on hybridization of target sequences with biotinylated oligonucleotides. The best results were obtained for the TruSight Tumor 170 Assay (Illumina), which detected all chimeric transcripts in the samples under analysis. The single false-negative signal was observed in the results of ArcherDX and Qiagen panels; however, Qiagen panel returned more false-positive chimeras. The Thermo Fisher Scientific assay did not find 7 chimeric genes out of 18. The low sensitivity of the last panel is explained by the fact that the approach is based on classical PCR: both primers are complementary to known sequences of chimeric transcripts, which makes it impossible to detect chimeras that were never described previously and not included in the panel design. Thus, in general, RNA sequencing approaches are technically more effective than DNA analysis for the detection of chimeric oncogenes.208
Methods for generation of sequencing libraries enriched in regions close to susceptible chimerization point are highly sensitive, especially for the detection of known structural rearrangements. However, this approach is limited by the set of probes available in the targeted panels, which does not allow detecting in such a way rearrangements at previously unknown loci. Also, the use of panels limits the set of biomarkers that can be analyzed in one experimental procedure. These shortcomings are devoid of total RNA or mRNA sequencing.
Whole transcriptome RNA sequencing for detection of fusion genes
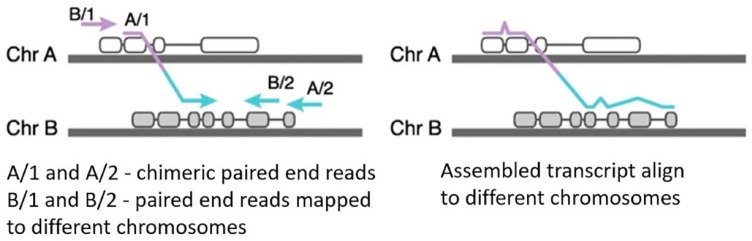
Genome-wide RNA sequencing evaluates mutations of the transcribed DNA in an unbiased manner. A number of approaches have been proposed for the detection of chimeric transcripts in the analysis of full-transcriptome data. There are two main directions to the analysis of the RNA sequencing data for the detection of chimeric transcripts: (i) alignment of reads per genome and search for those reads that map to different loci or (ii) initial assembly of reads into long transcripts followed by a search for chimeras that do not map entirely to one genome region (Figure 4).222
Figure 4.
Scheme of operation of algorithms based on alignment (left) and primary assembly (right) (according to Haas et al.,222 modified).
In search for chimeric transcripts, Haas and colleagues used both approaches to compare 23 analytic algorithms.222 The authors showed that the sensitivity of methods based on primary alignment is higher than for the approaches based on assembly of reads. The best analytic tools identified were STAR-Fusion, Arriba, and STAR-SEQR. It is important to note that algorithms for searching of the chimeric reads have been developed primarily for the analysis of cell lines and fresh frozen biopsies.223,224 Most of the work devoted to assessing their quality was carried out on artificial data, or again data obtained from cell lines and fresh frozen biopsies.225,226 However, paraffin-embedded biopsies fixed in formalin are the most common type of biomaterial in clinical practice, which is stored in collections for a long time.227,228
However, RNA in paraffin blocks is more degraded due to the fixation and storage procedures, which can lead to more false-negative results when searching for chimeric transcripts from the RNA sequencing data using existing algorithms. Validation of the work of the studied algorithms for this type of biomaterial has not yet been published.
Analysis of gene fusions from the FFPE material by RNA-seq
The use of DNA and RNA from archival material for genome-wide studies attract researchers’ attention for a long time and it was initially demonstrated that about 80% of genes, expressed in the fresh tissue can be detected in the FFPE samples by microarrays.229,230 Since then, we have witnessed considerable progress in the field and more recent investigations of RNA from paraffin blocks for gene expression analysis using microarrays231 and sequencing228,232–234 revealed sufficient quality of RNA obtained from the FFPE samples to generate reproducible data consistent with RNA from the unfixed material. Furthermore, specific features such as conservation of open reading frame in both fusion partners, presence of RTK domain, and finding of several non-duplicate transcript reads for a fusion were shown as the efficient criteria for discriminating true versus artifact fusion reads in FFPE-derived RNA sequencing data.235
Indeed, despite the absolute gene expression levels being not necessarily the same, very similar pathways were overrepresented within dysregulated genes obtained from the FFPE and freshly extracted RNA,232,233 thus demonstrating consistency with IHC studies.236,237
Fusion transcript detection by RNA-seq from FFPE samples was recently reviewed, focusing on the experimental variables; however, the difference in bioinformatics approaches of fusion transcript detention was not discussed.238 Gene fusions were detected by RNA sequencing in 7 out of 8 cases of DNA-fusion positive fibrous histiocytomas.239 Interestingly, comparison of ChimeraScan with TopHat software used in this study revealed better sensitivity of the former (9 versus 5 fusion transcripts detected) suggesting that detailed analysis of the software applications for the fusion transcript detection is needed.239 It was shown that RNA-seq from the FFPE clinical material detects fusions with 94% (43 out of 46 fusions) concordance with DNA fusions, and one ST7-MET fusion was detected only by RNA-seq.240
Analysis of single-cell RNA sequencing
High-throughput sequencing technology made single-cell RNA (scRNA) or DNA analysis possible at an unprecedented scale. Lately, several consortiums published aggregations and the analysis of the scRNA sequencing data.241–243
Functional enrichment analysis distinguishes different cell types as well as cancer cells, which also can be distinguished by the mutations and copy number variations typically observed in cancer,244
Importantly, single-cell sequencing characterizes not only cancer cells, but also immune cells infiltrating the tumor. And, analysis of this data might reveal information which is relevant to tumor progression and treatment strategies. For example, sequencing of the T-cell receptor repertoire from glioblastomas treated by vaccination with heat shock protein peptide complex-96 identified dominant T-cell clones that reside in glioblastomas before treatment and stratify patients that are more sensitive to therapy.245
Various methods have been developed to analyze gene expression of single cancer cells and to dissect their molecular subgroups.246 Finding of gene fusions at single cell level can potentially shed light on specific features of cells and their subtypes. Several algorithms for fusion identification in bulk RNA-seq data have been developed,222,247 but detection of fusions on single cell level is still largely unsolved task. Indeed, the ambiguous and complicated library preparation steps result in generation of artificial chimeric reads and significant increase in the number of false-positive results. Moreover, the probability to detect fusion present in many cells is higher using bulk methods. Thus, several approaches both for sample preparation and for data analyses stages have been proposed. For example, using full-length scRNA-seq method enabled to detected and experimentally verify more gene fusions in scRNA-seq data than in bulk RNA-seq data for HeLa S3 cells.248 These results were congruent with the finding published by another group of authors who could identify well-known as well as potential new fusion in colon cancer samples solely on single cell level.249 However, experimental verification could not be performed here; thus, the increased number of fusions might at least partly represent artifacts of library preparation.249 Another attempt to increase sensitivity of fusion detection at single cell level is to include specific primers targeted for the genes of interest.250 This approach increased sensitivity of BCR-ABL1 detection in chronic myeloid leukemia cells.250 However, this method is suitable only for the analysis of known fusions.
A more recent algorithm called scFusion was published for improving data analysis step.251 This method utilizes both statistics and a deep learning model to exclude false-positive results. The algorithm also relies on the hypothesis that cells collected from one sample are more likely to contain the same gene fusions.251 This limits its ability to detect rare or low expressed fusions, especially in highly heterogenous samples. Overall, scFusion requires greater sequencing depth as well as sequencing of larger amounts of cells from the same sample for obtaining reliable results. Analysis of the single-cell DNA sequencing of the NSCLC cohort enrolled in the MATCH-R (NCT0251782) trial that developed osimertinib resistance revealed heterogeneity of acquired mutations in the cells including FGFR3-TACC3, KIF5B-RE, and STRN-ALK fusions that can be treated by existing drugs, thus suggesting possible treatments to overcome osimertinib resistance.252 Similarly, RNA sequencing revealed that ALK junctional heterogeneity in NSCLC may predict resistance to crizotinib.253 Likewise scRNA-seq of chemoresistant cervical cancer revealed induction of the (PI3K)/AKT pathway.254 Thus, it is possible to infer the mechanisms of acquired resistance and to monitor clonal changes of tumors in response to therapy.
Detection of fusions at single cell level can improve distinguishing cells subpopulations, thus shedding light on drug-resistant subclones in a tumor. However, this type of analysis still has strong limitations such as low coverage per individual cells, high PCR amplification bias and lack of standardization in data analysis.
Fusion oncogene databases
The fast growth of gene fusion data necessitates major organizational effort to gather them in the databases, and there is currently nearly a dozen of published databases of cancer fusion genes. Table 4 summarizes the common databases that are specified for fusion genes. One of the first databases designed to catalog gene fusions is the Mitelman database of chromosome aberrations and gene fusions in cancer that was first published in 1994. This database is supplemented with clinical association information that relate cytogenetic and genomic abnormalities, in particular gene fusions, to tumor characteristics or patient prognosis, based either on individual cases or associations. The database is searchable by a wide variety of fields, such as patient age, publication authors, gene, tumor histology, tissue type, mutation recurrence, associated clinical features, and cancer types.255
Table 4.
Databases of gene fusions.
| Database name and URL | Description | Database size | Source of data | Reference |
|---|---|---|---|---|
| Mitelman https://mitelmandatabase.isb-cgc.org | Relates gene fusions and other chromosomal aberrations to tumor characteristics, based either on individual cases or associations | Total number of unique gene fusions 32,962, Total number of genes involved 14,016 as for April 2022 | Manual curation of literature | Denomy et al.255 |
| COSMIC: the Catalogue Of Somatic Mutations In Cancer https://cancer.sanger.ac.uk | Catalog of translocations and fusions between gene pairs supplemented with extensive clinical data | 19 368 Gene Fusions as for August 2018 | Manual curation of literature | Tate et al.,262 |
| TumorFusions: TCGA Fusion Gene Data portal https://tumorfusions.org/ | An integrative resource for cancer-associated transcript fusions giving a landscape of cancer-associated fusions using the pipeline for RNA sequencing data analysis | Total fusion called across multiple cancer types is 20731 as of July 2017 | Integrated analysis of RNA-seq and DNA-seq data from TCGA | Hu et al.,263 Yoshihara et al.264 |
| ChimerDB http://www.kobic.re.kr/chimerdb/ | Fusion transcripts across multiple species knowledgebase and information on cancer breakpoints | 67 610 fusion gene pairs, of which 1597 fusion genes with publication support as for November 2019 | Sequencing data from TCGA and PubMed publications curation | Jang et al.265 |
| ChiTaRS http://chitars.md.biu.ac.il/ | Catalogue of fusion transcripts in humans, mice, fruit flies, zebrafishes, cows, rats, pig and yeast using RNA-seq data | 111,582 fusion transcripts in humans, mice, fruit flies, rats, zebrafishes, cows, pigs, and yeast as for 2019 | EST and mRNA sequences analysis from NCBI/GenBank and review of publications by Mertens et al. and from the Mitelman collection | Balamurali et al.266 |
| FusionCancer http://donglab.ecnu.edu.cn/databases/FusionCancer/ | Fusion gene database derived exclusively from cancer RNA-seq data | 11 839 gene fusions as for 2015 | Analysis of 591 RNA-seq cancer datasets | Wang et al.267 |
| FARE-CAFÉ http://ppi.bioinfo.asia.edu.tw/FARE-CAFE | Database of functional and regulatory elements in gene fusion events related to cancer | 1587 gene fusions as for 2015 | Combination of information from different sources, including TICdb, dbCRID, NCBI, and miRTarBase | Korla et al.268 |
| HYBRIDdb http://www.primate.or.kr/hybriddb | Database of hybrid genes in the human genome | 3404 gene fusions as for 2007 | Data from INSDC about mRNA, EST, cDNA, and genomic DNA sequences | Kim et al.269 |
| FusionGDB 2.0 https://compbio.uth.edu/FusionGDB2/ | Fusion Gene annotation DataBase | 102 645 fusion genes with 16 146 in-frame as for 2021 | Combined data from ChiTaRS 5.0 and ChimerDB 4.0 and manual curation of PubMed articles for the most frequent fusions | Kim et al.270 |
| dbCRID http://dbCRID.biolead.org | Curated database of human chromosomal rearrangements, associated diseases and clinical symptoms | 2643 chromosome rearrangements as for 2010 | Manual curation of literature | Kong et al.271 |
| TICdb http://www.unav.es/genetica/TICdb/ | Finely mapped translocation breakpoints in cancer | 1374 fusion sequences from 431 different genes as for 2007 | Databases analysis and publications review | Novo et al.272 |
| KuNG FU (KiNase Gene FUsion) http://www.kungfudb.org/ | Database containing in-frame kinase gene fusions with intact kinase domain in cell lines | 108 total fusions as for January 2021 | Manual curation of literature | Kim et al.269 |
EST, expressed sequence tag; FISH, fluorescent in situ hybridization; TCGS, The Cancer Genome Atlas.
The Mitelman database was followed by COSMIC database in 2004, that started with only four genes,273 then evolved to contain more than 5 million somatic mutations and more than 19 thousand gene fusions.262
RNA-seq data is a major source of mining fusion transcripts. Several fusion databases were generated from transcript sequences available datasets, such as TCGA dataset. The Fusion Analysis Working Group identified 25 664 fusion events using RNA-seq data from tumor and normal samples from TCGA using multiple fusion calling tools.223 Similarly, TumorFusions database is a searchable portal that catalogues over two thousand gene fusions detected in cancer and normal samples from TCGA.263,264
ChimerDB is another knowledgebase database of fusion genes265,274; this database contains fusions identified using bioinformatics analysis of transcript sequences compiled from GenBank and various other well-known public fusion databases and PubMed articles reporting fusion genes. ChimerDB is composed of three modules dealing with the analysis of deep sequencing data (ChimerSeq) and text mining of publications (ChimerPub) with extensive manual annotations (ChimerKB).265
Newer gene fusion database is the ChiTaRS, generated by performing a bioinformatics analysis of transcript sequences and expressed sequence tags (ESTs) for multiple organisms in GenBank, starting from three organisms in the first version,275 then extended to include eight organisms: human, mouse, fruit fly, rats, zebrafishes, cows, pigs, and yeast in the most recent version.266 The latest version includes an extended information about fusions features, as well as 3D chromatin contact maps. In addition, FusionCancer database contains cancer fusion genes deduced from RNA-seq data.267
Other databases combine fusion genes functional, regulatory, and genomic information. Example of such is the FARE-CAFE database that collect various aspects and data concerning fusion genes and proteins as protein domains, domain–domain interactions, protein–protein interactions, transcription factors and microRNAs. FARE-CAFE database incorporates chimeric transcripts and their Genomic information from different resources including Mitelman’s, dbCRID and the Trans location in Cancer (TICdb) databases.268
dbCRID is a comprehensive database of human chromosomal rearrangements events and their associated diseases that documents the type of each event with the related disease or symptoms, the breakpoint positions and other genomic information.271 Similarly, TICdb documents the precise location of each breakpoint inside a fusion gene. TICdb gather molecular information on gene fusions resulting from reciprocal translocation events associated with tumors.272
Aside from cataloging gene fusions, numerous algorithms have been developed to predict fusion candidates from transcriptome data. Data aggregation and functional annotation with visualization support are necessary to assess the reliability, functional significance, and biological roles of predicted fusions. Programs such as GFusion, FusionScan, and STAR-Fusion are thought to be highly sensitive in detecting fusions with less false positives.265 More recently, FusionHub introduced an integrated web platform that supports both annotation and visualization for the largest collection of fusion gene datasets aggregated from 24 resources.276
HYBRIDdb is one of the earliest databases of hybrid genes to use a bioinformatics analysis for identifying gene fusions. HybridDB identified more than 3000 fusions from mRNA, EST, cDNA, and transcript sequences in the NCBI database.269
Kim and Zhou built FusionGDB (Fusion Gene annotation DataBase) that gathers more than 40,000 fusion genes from fusion gene public resources such as TumorFusions and ChiTaRS 3.1. FusionGDB provides extensive functional annotations for these collected fusion events. Most importantly, the gene assessment across pan-cancer fusion genes, open reading frame (ORF) assignment and retention search of protein features.277 FusionGDB was recently updated using deep learning techniques to provide eight categories of annotations: Fusion Gene Summary, Fusion Gene ORF analysis, Fusion Gene Genomic Features, Fusion Protein Features, Fusion Gene Sequence, Fusion Gene PPI analysis, Related Drugs, and Related Diseases.270
Since kinase gene fusions are valuable biomarkers and promising drug targets, specific databases of such fusions have been developed. KuNG FU (KiNase Gene FUsion) is one of the largest curated databases, containing precise annotations on solely in-frame kinase gene fusions with intact kinase domain, which parameters were investigated in cancer cell lines. KuNG FU database is an available and informative tool for facilitating drug development and diagnostic studying.270
Conclusions and further perspectives
Gene rearrangements stand among the major driver mutations in cancer. Recent developments unravel several therapeutically actionable fusion genes with approved targeted drugs effective against hematological and solid tumors. Many additional drugs targeting these and other fusion genes are in clinical trials and in development. Altogether, this gives physicians more options for the choice of therapy. However, cancer genomic instability leads to clonal heterogeneity and selection of cells with resistance to treatment, immune evasion, and metastatic potential, in many cases leading to failure of the therapy. Metastasis is the major problem in oncology. In particular, the picture becomes more complicated by the fact that each metastasis loci represents an individual clone with different mutational profile and drug sensitivity. To choose a proper therapy, it is critically important to provide a physician with actionable information about emerging clones. Thus, the future clinical oncology will utilize methods that will be able to measure mutations in the individual clones and assess the status of immune cells regulating tumor microenvironment. While single-cell sequencing methods that can measure clonal heterogeneity become available for academic research, targeted sequencing, FISH, and IHC remain the working horse of oncology.
To conclude, NGS-based methods provide several advantages for fusion detection both in clinical practice and in research studies. First, they combine fusion detection with finding of other clinically relevant biomarkers, thus providing more relevant information than ‘classical’ methods in just one test, which requires minute amounts of biosample. NGS approaches are suitable for the investigation of different sample types, including cell lines, fresh frozen, and FFPE biopsies. Different computational algorithms have been developed for the analyses of DNA and RNA NGS derived data and obtaining of highly reliable results. Further improvements in fusion detection approaches at both sample preparation and data analysis stages will expand the current knowledge of fusion frequencies among different cancer types, and of their particular impact on tumorigenesis, drug sensitivity, and resistance development. The latter has a strong potential of increasing efficacy of cancer treatment.
Acknowledgments
We thank Drs. Xinmin Li (UCLA), Ira Skvortsova (University of Innsbruck), and Ella Kim (University of Mainz) for fruitful discussion and insightful comments.
Footnotes
ORCID iD: Tharaa Mohammad  https://orcid.org/0000-0003-4237-0846
https://orcid.org/0000-0003-4237-0846
Contributor Information
Maksim Sorokin, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia; I.M. Sechenov First Moscow State Medical University, Trubetskaia street, 8-2, Moscow 119991, Russia; Omicsway Corp., Walnut, CA, USA.
Elizaveta Rabushko, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia; I.M. Sechenov First Moscow State Medical University, Moscow, Russia.
Julian Markovich Rozenberg, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia.
Tharaa Mohammad, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia.
Aleksander Seryakov, Medical Holding SM-Clinic, Moscow, Russia.
Marina Sekacheva, I.M. Sechenov First Moscow State Medical University, Moscow, Russia.
Anton Buzdin, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia; I.M. Sechenov First Moscow State Medical University, Moscow, Russia; Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia; PathoBiology Group, European Organization for Research and Treatment of Cancer (EORTC), Brussels, Belgium.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contribution(s): Maksim Sorokin: Writing – original draft.
Elizaveta Rabushko: Writing – review & editing.
Julian Markovich Rozenberg: Writing – original draft.
Tharaa Mohammad: Writing – review & editing.
Aleksander Seryakov: Writing – review & editing.
Marina Sekacheva: Project administration; Writing – review & editing.
Anton Buzdin: Conceptualization; Project administration; Writing – original draft.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Russian Science Foundation grant 22-24-00682.
Authors Maxim Sorokin and Anton Buzdin were employed by the company OmicsWay Corp. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Availability of data and material: Not applicable.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 2. Schmitt MW, Prindle MJ, Loeb LA. Implications of genetic heterogeneity in cancer. Ann NY Acad Sci 2012; 1267: 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee J-K, Choi Y-L, Kwon M, et al. Mechanisms and consequences of cancer genome instability: lessons from genome sequencing studies. Annu Rev Pathol 2016; 11: 283–312. [DOI] [PubMed] [Google Scholar]
- 4. Schneider MA, Buzdin AA, Weber A, et al. Combination of antiretroviral drugs zidovudine and efavirenz impairs tumor growths in a mouse model of cancer. Viruses 2021; 13: 2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sorokin M, Rabushko E, Efimov V, et al. Experimental and meta-analytic validation of RNA sequencing signatures for predicting status of microsatellite instability. Front Mol Biosci 2021; 8: 737821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kibriya MG, Raza M, Kamal M, et al. Relative telomere length change in colorectal carcinoma and its association with tumor characteristics, gene expression and microsatellite instability. Cancers (Basel) 2022; 14: 2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bolhaqueiro ACF, Ponsioen B, Bakker B, et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat Genet 2019; 51: 824–834. [DOI] [PubMed] [Google Scholar]
- 8. Ohshima K, Hatakeyama K, Nagashima T, et al. Integrated analysis of gene expression and copy number identified potential cancer driver genes with amplification-dependent overexpression in 1,454 solid tumors. Sci Rep 2017; 7: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng J. Oncogenic chromosomal translocations and human cancer (review). Oncol Rep 2013; 30: 2011–2019. [DOI] [PubMed] [Google Scholar]
- 10. Gagos S, Irminger-Finger I. Chromosome instability in neoplasia: chaotic roots to continuous growth. Int J Biochem Cell Biol 2005; 37(5): 1014–1033. [DOI] [PubMed] [Google Scholar]
- 11. Bakhoum SF, Landau DA. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb Perspect Med 2017; 7: a029611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017; 355: eaaf839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tobalina L, Armenia J, Irving E, et al. A meta-analysis of reversion mutations in BRCA genes identifies signatures of DNA end-joining repair mechanisms driving therapy resistance. Ann Oncol 2021; 32: 103–112. [DOI] [PubMed] [Google Scholar]
- 14. Liao W, Overman MJ, Boutin AT, et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 2019; 35: 559.e7–572.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015; 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Litchfield K, Reading JL, Puttick C, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021; 184: 596.e14–614.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klempner SJ, Fabrizio D, Bane S, et al. Tumor mutational burden as a predictive biomarker for response to immune checkpoint inhibitors: A review of current evidence. Oncologist 2020; 25: e147–e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. LaDuca H, Polley EC, Yussuf A, et al. A clinical guide to hereditary cancer panel testing: evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet Med 2020; 22: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamamoto H, Hirasawa A. Homologous recombination deficiencies and hereditary tumors. Int J Mol Sci 2021; 23: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zolotovskaia MA, Tkachev VS, Seryakov AP, et al. Mutation enrichment and transcriptomic activation signatures of 419 molecular pathways in cancer. Cancers (Basel) 2020; 12: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saadatzadeh MR, Elmi AN, Pandya PH, et al. The role of MDM2 in promoting genome stability versus instability. Int J Mol Sci 2017; 18: 2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rozenberg JM, Zvereva S, Dalina A, et al. The p53 family member p73 in the regulation of cell stress response. Biol Direct 2021; 16: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vladimirova U, Rumiantsev P, Zolotovskaia M, et al. DNA repair pathway activation features in follicular and papillary thyroid tumors, interrogated using 95 experimental RNA sequencing profiles. Heliyon 2021; 7: e06408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science 2008; 319: 1352–1355. [DOI] [PubMed] [Google Scholar]
- 25. Chen M, Linstra R, van Vugt MATM. Genomic instability, inflammatory signaling and response to cancer immunotherapy. Biochim Biophys Acta Rev Cancer 2022; 1877: 188661. [DOI] [PubMed] [Google Scholar]
- 26. Li P, Chen C, Li J, et al. Homologous recombination related signatures predict prognosis and immunotherapy response in metastatic urothelial carcinoma. Front Genet 2022; 13: 875128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rozenberg JM, Zvereva S, Dalina A, et al. Dual role of p73 in cancer microenvironment and DNA damage response. Cells 2021; 10: 3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koni M, Pinnarò V, Brizzi MF. The wnt signalling pathway: A tailored target in cancer. Int J Mol Sci 2020; 21: 7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tutaeva VV, Bobin AN, Ovsiannikova MR, et al. Disseminated form of the Kaposi sarcoma in HIV-negative patient associated with Hodgkin’s lymphoma. Oxf Med Case Reports 2020; 2020: omaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ock C-Y, Keam B, Kim S, et al. Pan-cancer immunogenomic perspective on the tumor microenvironment based on PD-L1 and CD8 T-cell infiltration. Clin Cancer Res 2016; 22: 2261–2270. [DOI] [PubMed] [Google Scholar]
- 31. Chang C-M, Wu C-L, Lu Y-T. Cancer-associated immune deficiency: a form of accelerated immunosenescence? In: Mohan R. (ed.) Topics in cancer survivorship. London: IntechOpen, 2012; pp. 95–108. [Google Scholar]
- 32. Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev 2018; 32: 1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ribeiro IP, Melo JB, Carreira IM. Cytogenetics and cytogenomics evaluation in cancer. Int J Mol Sci 2019; 20: 4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ried T, Liyanage M, du Manoir S, et al. Tumor cytogenetics revisited: comparative genomic hybridization and spectral karyotyping. J Mol Med 1997; 75(11–12): 801–814. [DOI] [PubMed] [Google Scholar]
- 35. McFarlane RJ, Wakeman JA. Meiosis-like functions in oncogenesis: a new view of cancer. Cancer Res 2017; 77: 5712–5716. [DOI] [PubMed] [Google Scholar]
- 36. Gantchev J, Ramchatesingh B, Berman-Rosa M, et al. Tools used to assay genomic instability in cancers and cancer meiomitosis. J Cell Commun Signal 2022; 16: 159–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lingg L, Rottenberg S, Francica P. Meiotic genes and DNA double strand break repair in cancer. Front Genet 2022; 13: 831620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindsey SF, Byrnes DM, Eller MS, et al. Potential role of meiosis proteins in melanoma chromosomal instability. J Skin Cancer 2013; 2013: 190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol 2015; 10: 425–448. [DOI] [PubMed] [Google Scholar]
- 40. Petermann E, Orta ML, Issaeva N, et al. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 2010; 37: 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boteva L, Nozawa R-S, Naughton C, et al. Common fragile sites are characterized by faulty condensin loading after replication stress. Cell Rep 2020; 32: 108177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sarni D, Kerem B. The complex nature of fragile site plasticity and its importance in cancer. Curr Opin Cell Biol 2016; 40: 131–136. [DOI] [PubMed] [Google Scholar]
- 43. Costantino L, Sotiriou SK, Rantala JK, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 2014; 343: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McBride DJ, Etemadmoghadam D, Cooke SL, et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J Pathol 2012; 227: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manning AL, Yazinski SA, Nicolay B, et al. Suppression of genome instability in pRB-deficient cells by enhancement of chromosome cohesion. Mol Cell 2014; 53: 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 2009; 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 2008; 180: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci 2008; 121: 3859–3866. [DOI] [PubMed] [Google Scholar]
- 49. Dewhurst SM, McGranahan N, Burrell RA, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 2014; 4: 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science 2002; 297: 565–569. [DOI] [PubMed] [Google Scholar]
- 51. Roger L, Jones RE, Heppel NH, et al. Extensive telomere erosion in the initiation of colorectal adenomas and its association with chromosomal instability. J Natl Cancer Inst 2013; 105: 1202–1211. [DOI] [PubMed] [Google Scholar]
- 52. Rivlin N, Brosh R, Oren M, et al. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2011; 2: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wong RSY. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res 2011; 30: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iyer S, Bell F, Westphal D, et al. Bak apoptotic pores involve a flexible C-terminal region and juxtaposition of the C-terminal transmembrane domains. Cell Death Differ 2015; 22: 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 2017; 18: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lovejoy CA, Li W, Reisenweber S, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 2012; 8: e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet 2020; 52: 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McKerrow W, Wang X, Mendez-Dorantes C, et al. LINE-1 expression in cancer correlates with p53 mutation, copy number alteration, and S phase checkpoint. Proc Natl Acad Sci USA 2022; 119: e2115999119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang C, Liang C. The insertion and dysregulation of transposable elements in osteosarcoma and their association with patient event-free survival. Sci Rep 2022; 12: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schumann GG, Gogvadze EV, Osanai-Futahashi M, et al. Unique functions of repetitive transcriptomes. Int Rev Cell Mol Biol 2010; 285: 115–188. [DOI] [PubMed] [Google Scholar]
- 61. Reis AHO, Vargas FR, Lemos B. Biomarkers of genome instability and cancer epigenetics. Tumour Biol 2016; 37: 13029–13038. [DOI] [PubMed] [Google Scholar]
- 62. Zhou T, Medeiros LJ, Hu S. Chronic myeloid leukemia: Beyond BCR-ABL1. Curr Hematol Malig Rep 2018; 13: 435–445. [DOI] [PubMed] [Google Scholar]
- 63. Pagani IS, Dang P, Kommers IO, et al. BCR-ABL1 genomic DNA PCR response kinetics during first-line imatinib treatment of chronic myeloid leukemia. Haematologica 2018; 103: 2026–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cilloni D, Saglio G. Molecular pathways: BCR-ABL. Clin Cancer Res 2012; 18: 930–937. [DOI] [PubMed] [Google Scholar]
- 65. Mertens F, Johansson B, Fioretos T, et al. The emerging complexity of gene fusions in cancer. Nat Rev Cancer 2015; 15: 371–381. [DOI] [PubMed] [Google Scholar]
- 66. Crompton BD, Stewart C, Taylor-Weiner A, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 2014; 4: 1326–1341. [DOI] [PubMed] [Google Scholar]
- 67. Boone MA, Taslim C, Crow JC, et al. The FLI portion of EWS/FLI contributes a transcriptional regulatory function that is distinct and separable from its DNA-binding function in Ewing sarcoma. Oncogene 2021; 40: 4759–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Konovalov N, Timonin S, Asyutin D, et al. Transcriptomic portraits and molecular pathway activation features of adult spinal intramedullary astrocytomas. Front Oncol 2022; 12: 837570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brahmi M, Franceschi T, Treilleux I, et al. Molecular classification of endometrial stromal sarcomas using RNA sequencing defines nosological and prognostic subgroups with different natural history. Cancers (Basel) 2020; 12: 2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Seryakov A, Magomedova Z, Suntsova M, et al. RNA sequencing for personalized treatment of metastatic leiomyosarcoma: case report. Front Oncol 2021; 11: 666001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020; 578: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020; 578: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kalyana-Sundaram S, Shankar S, Deroo S, et al. Gene fusions associated with recurrent amplicons represent a class of passenger aberrations in breast cancer. Neoplasia 2012; 14: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Spirin PV, Lebedev TD, Orlova NN, et al. Silencing AML1-ETO gene expression leads to simultaneous activation of both pro-apoptotic and proliferation signaling. Leukemia 2014; 28: 2222–2228. [DOI] [PubMed] [Google Scholar]
- 75. Amarante-Mendes GP, Rana A, Datoguia TS, et al. BCR-ABL1 tyrosine kinase complex signaling transduction: challenges to overcome resistance in chronic myeloid leukemia. Pharmaceutics 2022; 14: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Milojkovic D, Apperley J. Mechanisms of resistance to imatinib and second-generation tyrosine inhibitors in chronic myeloid leukemia. Clin Cancer Res 2009; 15: 7519–7527. [DOI] [PubMed] [Google Scholar]
- 77. Radich JP, Hochhaus A, Masszi T, et al. Treatment-free remission following frontline nilotinib in patients with chronic phase chronic myeloid leukemia: 5-year update of the ENESTfreedom trial. Leukemia 2021; 35: 1344–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shaw AT, Ou S-HI, Bang Y-J, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014; 371: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Solomon BJ, Mok T, Kim D-W, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014; 371: 2167–2177. [DOI] [PubMed] [Google Scholar]
- 80. Facchinetti F, Caramella C, Auger N, et al. Crizotinib primary resistance overcome by ceritinib in a patient with ALK-rearranged non-small cell lung cancer. Tumori 2016; 102: S46–S49. [DOI] [PubMed] [Google Scholar]
- 81. Descourt R, Pérol M, Rousseau-Bussac G, et al. Brigatinib for pretreated, ALK-positive, advanced non-small-cell lung cancers: long-term follow-up and focus on post-brigatinib lorlatinib efficacy in the multicenter, real-world brigALK2 study. Cancers (Basel) 2022; 14: 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang Y, Shen S, Hu P, et al. Alectinib versus crizotinib in ALK-positive advanced non-small cell lung cancer and comparison of next-generation TKIs after crizotinib failure: real-world evidence. Cancer Med 2022; 11: 4491–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rudzinski ER, Hechtman J, Roy-Chowdhuri S, et al. Diagnostic testing approaches for the identification of patients with TRK fusion cancer prior to enrollment in clinical trials investigating larotrectinib. Cancer Genet 2022; 260–261: 46–52. [DOI] [PubMed] [Google Scholar]
- 84. Dziadziuszko R, Krebs MG, De Braud F, et al. Updated integrated analysis of the efficacy and safety of entrectinib in locally advanced or metastatic ROS1 fusion-positive non-small-cell lung cancer. J Clin Oncol 2021; 39: 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Doebele RC, Drilon A, Paz-Ares L, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol 2020; 21: 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zengin ZB, Chehrazi-Raffle A, Salgia NJ, et al. Targeted therapies: expanding the role of FGFR3 inhibition in urothelial carcinoma. Urol Oncol 2022; 40: 25–36. [DOI] [PubMed] [Google Scholar]
- 87. Abou-Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020; 21: 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gainor JF, Lee DH, Curigliano G, et al. Clinical activity and tolerability of BLU-667, a highly potent and selective RET inhibitor, in patients (pts) with advanced RET-fusion+ non-small cell lung cancer (NSCLC). JCO 2019; 37: 9008–9008. [Google Scholar]
- 89. Subbiah V, Velcheti V, Tuch BB, et al. Selective RET kinase inhibition for patients with RET-altered cancers. Ann Oncol 2018; 29: 1869–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sehgal K, Piper-Vallillo AJ, Viray H, et al. Cases of ROS1-rearranged lung cancer: when to use crizotinib, entrectinib, lorlatinib, and beyond? Precis Cancer Med 2020; 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Javle M, Roychowdhury S, Kelley RK, et al. Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: mature results from a multicentre, open-label, single-arm, phase 2 study. Lancet Gastroenterol Hepatol 2021; 6: 803–815. [DOI] [PubMed] [Google Scholar]
- 92. Yeung DT, Shanmuganathan N, Hughes TP. Asciminib: new therapeutic option in chronic phase CML with treatment failure. Blood 2022; 139: 3474–3479. [DOI] [PubMed] [Google Scholar]
- 93. Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov 2009; 8: 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Maruyama IN. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cells 2014; 3: 304–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Weddell JC, Imoukhuede PI. Integrative meta-modeling identifies endocytic vesicles, late endosome and the nucleus as the cellular compartments primarily directing RTK signaling. Integr Biol (Camb) 2017; 9: 464–484. [DOI] [PubMed] [Google Scholar]
- 96. Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol 2013; 5: a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Miaczynska M. Effects of membrane trafficking on signaling by receptor tyrosine kinases. Cold Spring Harb Perspect Biol 2013; 5: a009035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Critchley WR, Pellet-Many C, Ringham-Terry B, et al. Receptor tyrosine kinase ubiquitination and de-ubiquitination in signal transduction and receptor trafficking. Cells 2018; 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer 2017; 17: 318–332. [DOI] [PubMed] [Google Scholar]
- 100. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005; 16: 139–149. [DOI] [PubMed] [Google Scholar]
- 101. Ibáñez CF. Structure and physiology of the RET receptor tyrosine kinase. Cold Spring Harb Perspect Biol 2013; 5: a009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol 2000; 10: 381–391. [DOI] [PubMed] [Google Scholar]
- 103. Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012; 18: 378–381. [DOI] [PubMed] [Google Scholar]
- 104. Parker BC, Engels M, Annala M, et al. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J Pathol 2014; 232: 4–15. [DOI] [PubMed] [Google Scholar]
- 105. Gallo LH, Nelson KN, Meyer AN, et al. Functions of fibroblast growth factor receptors in cancer defined by novel translocations and mutations. Cytokine Growth Factor Rev 2015; 26: 425–449. [DOI] [PubMed] [Google Scholar]
- 106. Kim P, Jia P, Zhao Z. Kinase impact assessment in the landscape of fusion genes that retain kinase domains: a pan-cancer study. Brief Bioinformatics 2018; 19: 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Papusha L, Zaytseva M, Panferova A, et al. Two clinically distinct cases of infant hemispheric glioma carrying ZCCHC8: ROS1 fusion and responding to entrectinib. Neuro Oncol 2022; 24: 1029–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Samii A, Sorokin M, Kar S, et al. Case of multifocal glioblastoma with four fusion transcripts of ALK, FGFR2, NTRK2, and NTRK3 genes stresses the need for tumor tissue multisampling for transcriptomic analysis. Cold Spring Harb Mol Case Stud 2021; 7: a006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chen L, Zhang Y, Yin L, et al. Fibroblast growth factor receptor fusions in cancer: opportunities and challenges. J Exp Clin Cancer Res 2021; 40: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hrustanovic G, Olivas V, Pazarentzos E, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med 2015; 21: 1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tulpule A, Guan J, Neel DS, et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell 2021; 184: 2649.e18–2664.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Chen D, Ito S, Yuan H, et al. EML4 promotes the loading of NUDC to the spindle for mitotic progression. Cell Cycle 2015; 14: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lucken K, O’Regan L, Choi J, et al. EML4-ALK variant 3 promotes mitotic errors and spindle assembly checkpoint deficiency leading to increased microtubule poison sensitivity. Mol Cancer Res 2022; 20: 854–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bayliss R, Choi J, Fennell DA, et al. Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell Mol Life Sci 2016; 73: 1209–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012; 337: 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sarkar S, Ryan EL, Royle SJ. FGFR3-TACC3 cancer gene fusions cause mitotic defects by removal of endogenous TACC3 from the mitotic spindle. Open Biol 2017; 7: 170080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Biol 2013; 14: 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Helsten T, Elkin S, Arthur E, et al. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res 2016; 22: 259–267. [DOI] [PubMed] [Google Scholar]
- 119. Shi E, Chmielecki J, Tang C-M, et al. FGFR1 and NTRK3 actionable alterations in “Wild-Type” gastrointestinal stromal tumors. J Transl Med 2016; 14: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Stephens PJ, McBride DJ, Lin M-L, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 2009; 462: 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Farshidfar F, Zheng S, Gingras M-C, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep 2017; 18: 2780–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wang Y, Ding X, Wang S, et al. Antitumor effect of FGFR inhibitors on a novel cholangiocarcinoma patient derived xenograft mouse model endogenously expressing an FGFR2-CCDC6 fusion protein. Cancer Lett 2016; 380: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res 2018; 24: 4154–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Arai Y, Totoki Y, Hosoda F, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2014; 59: 1427–1434. [DOI] [PubMed] [Google Scholar]
- 125. Qin A, Johnson A, Ross JS, et al. Detection of known and novel FGFR fusions in non-small cell lung cancer by comprehensive genomic profiling. J Thorac Oncol 2019; 14: 54–62. [DOI] [PubMed] [Google Scholar]
- 126. AACR Project GENIE Consortium. AACR project GENIE: Powering precision medicine through an international consortium. Cancer Discov 2017; 7: 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hood FE, Royle SJ. Pulling it together: the mitotic function of TACC3. Bioarchitecture 2011; 1: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Di Stefano AL, Fucci A, Frattini V, et al. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res 2015; 21: 3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wu Y-M, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013; 3: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Schram AM, Chang MT, Jonsson P, et al. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 2017; 14: 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Graham RP, Barr Fritcher EG, Pestova E, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol 2014; 45: 1630–1638. [DOI] [PubMed] [Google Scholar]
- 132. Jain A, Borad MJ, Kelley RK, et al. Cholangiocarcinoma with FGFR genetic aberrations: a unique clinical phenotype. JCO Precis Oncol 2018; 2: 1–12. [DOI] [PubMed] [Google Scholar]
- 133. Snider WD. Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell 1994; 77: 627–638. [DOI] [PubMed] [Google Scholar]
- 134. Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett 2001; 169: 107–114. [DOI] [PubMed] [Google Scholar]
- 135. Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018; 15: 731–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014; 5: 4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol 2017; 41: 1547–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hong DS, DuBois SG, Kummar S, et al. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol 2020; 21: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Okamura R, Boichard A, Kato S, et al. Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis Oncol 2018; 2: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Rosen EY, Goldman DA, Hechtman JF, et al. TRK fusions are enriched in cancers with uncommon histologies and the absence of canonical driver mutations. Clin Cancer Res 2020; 26: 1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012; 18: 375–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012; 18: 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Monaco C, Visconti R, Barone MV, et al. The RFG oligomerization domain mediates kinase activation and re-localization of the RET/PTC3 oncoprotein to the plasma membrane. Oncogene 2001; 20: 599–608. [DOI] [PubMed] [Google Scholar]
- 144. Sharma S, Birchmeier C, Nikawa J, et al. Characterization of the ros1-gene products expressed in human glioblastoma cell lines. Oncogene Res 1989; 5: 91–100. [PubMed] [Google Scholar]
- 145. Drilon A, Jenkins C, Iyer S, et al. ROS1-dependent cancers-biology, diagnostics and therapeutics. Nat Rev Clin Oncol 2021; 18: 35–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Davare MA, Henderson JJ, Agarwal A, et al. Rare but recurrent ROS1 fusions resulting from chromosome 6q22 microdeletions are targetable oncogenes in glioma. Clin Cancer Res 2018; 24: 6471–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Park S, Ahn B-C, Lim SW, et al. Characteristics and outcome of ROS1-positive non-small cell lung cancer patients in routine clinical practice. J Thorac Oncol 2018; 13: 1373–1382. [DOI] [PubMed] [Google Scholar]
- 148. Neel DS, Allegakoen DV, Olivas V, et al. Differential subcellular localization regulates oncogenic signaling by ROS1 kinase fusion proteins. Cancer Res 2019; 79: 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Charest A, Wilker EW, McLaughlin ME, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res 2006; 66: 7473–7481. [DOI] [PubMed] [Google Scholar]
- 150. Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013; 13: 685–700. [DOI] [PubMed] [Google Scholar]
- 151. Lovly CM, Gupta A, Lipson D, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov 2014; 4: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ignatius Ou S-H, Zhu VW, Nagasaka M. Catalog of 5’ fusion partners in ALK+ NSCLC circa 2020. JTO Clin Res Rep 2020; 1: 100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014; 311: 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Mariño-Enríquez A, Dal Cin P. ALK as a paradigm of oncogenic promiscuity: different mechanisms of activation and different fusion partners drive tumors of different lineages. Cancer Genet 2013; 206: 357–373. [DOI] [PubMed] [Google Scholar]
- 155. Tao H, Shi L, Zhou A, et al. Distribution of EML4-ALK fusion variants and clinical outcomes in patients with resected non-small cell lung cancer. Lung Cancer 2020; 149: 154–161. [DOI] [PubMed] [Google Scholar]
- 156. Vigna E, Gramaglia D, Longati P, et al. Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET. Oncogene 1999; 18: 4275–4281. [DOI] [PubMed] [Google Scholar]
- 157. Garajová I, Giovannetti E, Biasco G, et al. c-Met as a target for personalized therapy. Transl Oncogenomics 2015; 7: 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. International Cancer Genome Consortium PedBrain Tumor Project. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med 2016; 22: 1314–1320. [DOI] [PubMed] [Google Scholar]
- 159. Flucke U, van Noesel MM, Wijnen M, et al. TFG-MET fusion in an infantile spindle cell sarcoma with neural features. Genes Chromosomes Cancer 2017; 56: 663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Plenker D, Bertrand M, de Langen AJ, et al. Structural alterations of MET trigger response to MET kinase inhibition in lung adenocarcinoma patients. Clin Cancer Res 2018; 24: 1337–1343. [DOI] [PubMed] [Google Scholar]
- 161. Rooper LM, Karantanos T, Ning Y, et al. Salivary secretory carcinoma with a novel ETV6-MET fusion: expanding the molecular spectrum of a recently described entity. Am J Surg Pathol 2018; 42: 1121–1126. [DOI] [PubMed] [Google Scholar]
- 162. Dagogo-Jack I, Yoda S, Lennerz JK, et al. MEt alterations are a recurring and actionable resistance mechanism in ALK-positive lung cancer. Clin Cancer Res 2020; 26: 2535–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Abella JV, Peschard P, Naujokas MA, et al. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol Cell Biol 2005; 25: 9632–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Fujino T, Suda K, Mitsudomi T. Lung cancer with MET exon 14 skipping mutation: genetic feature, current treatments, and future challenges. Lung Cancer (Auckl). 2021; 12: 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015; 5: 850–859. [DOI] [PubMed] [Google Scholar]
- 166. Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol 2016; 11: 1493–1502. [DOI] [PubMed] [Google Scholar]
- 167. Nikanjam M, Okamura R, Barkauskas DA, et al. Targeting fusions for improved outcomes in oncology treatment. Cancer 2020; 126: 1315–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 2018; 378: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Shah RR, Morganroth J. Update on cardiovascular safety of tyrosine kinase inhibitors: with a special focus on QT interval, left ventricular dysfunction and overall risk/benefit. Drug Saf 2015; 38: 693–710. [DOI] [PubMed] [Google Scholar]
- 170. Haura EB, Hicks JK, Boyle TA. Erdafitinib overcomes FGFR3-TACC3-mediated resistance to osimertinib. J Thorac Oncol 2020; 15: e154–e156. [DOI] [PubMed] [Google Scholar]
- 171. Markham A. Correction to: selpercatinib: first approval. Drugs 2021; 81: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and ret inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov 2018; 8: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Drilon A, Siena S, Ou S-HI, et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov 2017; 7: 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Robinson GW, Gajjar AJ, Gauvain KM, et al. Phase 1/1B trial to assess the activity of entrectinib in children and adolescents with recurrent or refractory solid tumors including central nervous system (CNS) tumors. J Clin Oncol 2019; 37: 10009–10009. [Google Scholar]
- 175. Childress MA, Himmelberg SM, Chen H, et al. ALK fusion partners impact response to ALK inhibition: differential effects on sensitivity, cellular phenotypes, and biochemical properties. Mol Cancer Res 2018; 16: 1724–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Thompson LL, Jeusset LM-P, Lepage CC, et al. Evolving therapeutic strategies to exploit chromosome instability in cancer. Cancers (Basel) 2017; 9: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Nguyen HT, Dupont LN, Cuttaz EA, et al. Breast cancer HER2 analysis by extra-short incubation microfluidics-assisted fluorescence in situ hybridization (ESIMA FISH). Microelectron Eng 2018; 189: 33–38. [Google Scholar]
- 178. Ali J, Khan SA, Rauf S-E-, et al. Comparative analysis of fluorescence in situ hybridization and real time polymerase chain reaction in diagnosis of chronic myeloid leukemia. J Coll Physicians Surg Pak 2017; 27: 26–29. [PubMed] [Google Scholar]
- 179. Cheng L, Zhang S, Wang L, et al. Fluorescence in situ hybridization in surgical pathology: principles and applications. J Pathol Clin Res 2017; 3: 73–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Cruz-Rico G, Avilés-Salas A, Segura-González M, et al. Diagnosis of EML4-ALK translocation with FISH, immunohistochemistry, and real-time polymerase chain reaction in patients with non-small cell lung cancer. Am J Clin Oncol 2017; 40: 631–638. [DOI] [PubMed] [Google Scholar]
- 181. Connor A, Perez-Ordoñez B, Shago M, et al. Mammary analog secretory carcinoma of salivary gland origin with the ETV6 gene rearrangement by FISH: expanded morphologic and immunohistochemical spectrum of a recently described entity. Am J Surg Pathol 2012; 36: 27–34. [DOI] [PubMed] [Google Scholar]
- 182. Chiang S, Cotzia P, Hyman DM, et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol 2018; 42: 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Kurobe M, Kojima T, Nishimura K, et al. Development of RNA-FISH assay for detection of oncogenic FGFR3-TACC3 fusion genes in FFPE samples. PLoS One 2016; 11: e0165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. De Luca A, Esposito Abate R, Rachiglio AM, et al. FGFR fusions in cancer: from diagnostic approaches to therapeutic intervention. Int J Mol Sci 2020; 21: 6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Thorne-Nuzzo T, Williams C, Catallini A, et al. A Sensitive ALK immunohistochemistry companion diagnostic test identifies patients eligible for treatment with crizotinib. J Thorac Oncol 2017; 12: 804–813. [DOI] [PubMed] [Google Scholar]
- 186. Boyle TA, Masago K, Ellison KE, et al. ROS1 immunohistochemistry among major genotypes of non-small-cell lung cancer. Clin Lung Cancer 2015; 16: 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Engel KB, Moore HM. Effects of preanalytical variables on the detection of proteins by immunohistochemistry in formalin-fixed, paraffin-embedded tissue. Arch Pathol Lab Med 2011; 135: 537–543. [DOI] [PubMed] [Google Scholar]
- 188. Batra U, Nathany S, Sharma M, et al. IHC versus FISH versus NGS to detect ALK gene rearrangement in NSCLC: all questions answered? J Clin Pathol 2022; 75: 405–409. [DOI] [PubMed] [Google Scholar]
- 189. Mok T, Peters S, Camidge DR, et al. Outcomes according to ALK status determined by central immunohistochemistry or fluorescence in situ hybridization in patients with ALK-positive NSCLC enrolled in the phase 3 ALEX study. J Thorac Oncol 2021; 16: 259–268. [DOI] [PubMed] [Google Scholar]
- 190. Cabillic F, Hofman P, Ilie M, et al. ALK IHC and FISH discordant results in patients with NSCLC and treatment response: for discussion of the question-to treat or not to treat? ESMO Open 2018; 3: e000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Poddubskaya E, Bondarenko A, Boroda A, et al. Transcriptomics-guided personalized prescription of targeted therapeutics for metastatic ALK-positive lung cancer case following recurrence on ALK inhibitors. Front Oncol 2019; 9: 1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lyu X, Wang X, Zhang L, et al. Detection of 22 common leukemic fusion genes using a single-step multiplex qRT-PCR-based assay. Diagn Pathol 2017; 12: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Chen Q, Hu Z, Fang Y, et al. Development and application of a rapid molecular method for detection of fusion genes in pediatric leukemia. Int J Clin Exp Pathol 2018; 11: 1074–1087. [PMC free article] [PubMed] [Google Scholar]
- 194. Sorber L, Van Dorst B, Bellon E, et al. NTRK gene fusion detection in a pan-cancer setting using the Idylla genefusion assay. J Mol Diagn 2022; 24: 750–759. [DOI] [PubMed] [Google Scholar]
- 195. Abbou S, Finstuen-Magro S, McDannell B, et al. Rapid and highly sensitive approach for multiplexed somatic fusion detection. Mod Pathol 2022; 35: 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Shelton DN, Bhagavatula P, Sepulveda N, et al. Performance characteristics of the first food and drug administration (FDA)-cleared digital droplet PCR (ddPCR) assay for BCR::ABL1 monitoring in chronic myelogenous leukemia. PLoS One 2022; 17: e0265278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Sorokin M, Gorelyshev A, Efimov V, et al. RNA sequencing data for FFPE tumor blocks can be used for robust estimation of tumor mutation burden in individual biosamples. Front Oncol 2021; 11: 732644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Shao D, Lin Y, Liu J, et al. A targeted next-generation sequencing method for identifying clinically relevant mutation profiles in lung adenocarcinoma. Sci Rep 2016; 6: 22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Gudkov A, Shirokorad V, Kashintsev K, et al. Gene expression-based signature can predict sorafenib response in kidney cancer. Front Mol Biosci 2022; 9: 753318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Moskalev EA, Stöhr R, Rieker R, et al. Increased detection rates of EGFR and KRAS mutations in NSCLC specimens with low tumour cell content by 454 deep sequencing. Virchows Arch 2013; 462: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Borisov N, Sergeeva A, Suntsova M, et al. Machine learning applicability for classification of PAD/VCD chemotherapy response using 53 multiple myeloma RNA sequencing profiles. Front Oncol 2021; 11: 652063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Sorokin M, Poddubskaya E, Baranova M, et al. RNA sequencing profiles and diagnostic signatures linked with response to ramucirumab in gastric cancer. Cold Spring Harb Mol Case Stud 2020; 6: a004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Zhao EY, Jones M, Jones SJM. Whole-genome sequencing in cancer. Cold Spring Harb Perspect Med 2019; 9: a034579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Nakagawa H, Fujita M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci 2018; 109: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Wang Y, Tong Z, Zhang W, et al. FDA-approved and emerging next generation predictive biomarkers for immune checkpoint inhibitors in cancer patients. Front Oncol 2021; 11: 683419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Zhao M, Wang Q, Wang Q, et al. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics 2013; 14: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Deng W, Murugan S, Lindberg J, et al. Fusion gene detection using whole-exome sequencing data in cancer patients. Front Genet 2022; 13: 820493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Heydt C, Wölwer CB, Velazquez Camacho O, et al. Detection of gene fusions using targeted next-generation sequencing: a comparative evaluation. BMC Med Genomics 2021; 14: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 2011; 13: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Davies KD, Le AT, Sheren J, et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol 2018; 13: 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Benayed R, Offin M, Mullaney K, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res 2019; 25: 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Hofman P. Detecting resistance to therapeutic ALK inhibitors in tumor tissue and liquid biopsy markers: an update to a clinical routine practice. Cells 2021; 10: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Blaquier JB, Cardona AF, Russo A, et al. Next-generation sequencing using liquid biopsy in the care of patients with ALK-rearranged non-small cell lung cancer: a focus on lorlatinib. Precis Cancer Med 2021; 4: 28. [Google Scholar]
- 214. Leighl NB, Page RD, Raymond VM, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res 2019; 25: 4691–4700. [DOI] [PubMed] [Google Scholar]
- 215. Buzdin A, Tkachev V, Zolotovskaia M, et al. Using proteomic and transcriptomic data to assess activation of intracellular molecular pathways. Adv Protein Chem Struct Biol 2021; 127: 1–53. [DOI] [PubMed] [Google Scholar]
- 216. Buzdin A, Sorokin M, Garazha A, et al. Molecular pathway activation–New type of biomarkers for tumor morphology and personalized selection of target drugs. Semin Cancer Biol 2018; 53: 110–124. [DOI] [PubMed] [Google Scholar]
- 217. Sorokin M, Kholodenko I, Kalinovsky D, et al. RNA sequencing-based identification of ganglioside GD2-positive cancer phenotype. Biomedicines 2020; 8: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. DiGuardo MA, Davila JI, Jackson RA, et al. RNA-seq reveals differences in expressed tumor mutation burden in colorectal and endometrial cancers with and without defective DNA-mismatch repair. J Mol Diagn 2021; 23: 555–564. [DOI] [PubMed] [Google Scholar]
- 219. Gasc C, Peyretaillade E, Peyret P. Sequence capture by hybridization to explore modern and ancient genomic diversity in model and nonmodel organisms. Nucleic Acids Res 2016; 44: 4504–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014; 20: 1479–1484. [DOI] [PubMed] [Google Scholar]
- 221. Kozarewa I, Armisen J, Gardner AF, et al. Overview of target enrichment strategies. Curr Protoc Mol Biol 2015; 112: 7.21.1–7.21.23. [DOI] [PubMed] [Google Scholar]
- 222. Haas BJ, Dobin A, Li B, et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol 2019; 20: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Gao Q, Liang W-W, Foltz SM, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep 2018; 23: 227.e3–238.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Ghandi M, Huang FW, Jané-Valbuena J, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019; 569: 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Winters JL, Davila JI, McDonald AM, et al. Development and verification of an RNA sequencing (RNA-Seq) assay for the detection of gene fusions in tumors. J Mol Diagn 2018; 20: 495–511. [DOI] [PubMed] [Google Scholar]
- 226. López-Nieva P, Fernández-Navarro P, Graña-Castro O, et al. Detection of novel fusion-transcripts by RNA-seq in T-cell lymphoblastic lymphoma. Sci Rep 2019; 9: 5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Arreaza G, Qiu P, Pang L, et al. Pre-analytical considerations for successful next-generation sequencing (NGS): challenges and opportunities for formalin-fixed and paraffin-embedded tumor tissue (FFPE) samples. Int J Mol Sci 2016; 17: 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Suntsova M, Gaifullin N, Allina D, et al. Atlas of RNA sequencing profiles for normal human tissues. Sci Data 2019; 6: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Lewis F, Maughan NJ, Smith V, et al. Unlocking the archive–gene expression in paraffin-embedded tissue. J Pathol 2001; 195: 66–71. [DOI] [PubMed] [Google Scholar]
- 230. Zhang P, Lehmann BD, Shyr Y, et al. The utilization of formalin fixed-paraffin-embedded specimens in high throughput genomic studies. Int J Genomics 2017; 2017: 1926304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Wimmer I, Tröscher AR, Brunner F, et al. Systematic evaluation of RNA quality, microarray data reliability and pathway analysis in fresh, fresh frozen and formalin-fixed paraffin-embedded tissue samples. Sci Rep 2018; 8: 6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Marczyk M, Fu C, Lau R, et al. The impact of RNA extraction method on accurate RNA sequencing from formalin-fixed paraffin-embedded tissues. BMC Cancer 2019; 19: 1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Pennock ND, Jindal S, Horton W, et al. RNA-seq from archival FFPE breast cancer samples: molecular pathway fidelity and novel discovery. BMC Med Genomics 2019; 12: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Borisov N, Sorokin M, Zolotovskaya M, et al. Shambhala-2: a protocol for uniformly shaped harmonization of gene expression profiles of various formats. Curr Protoc 2022; 2: e444. [DOI] [PubMed] [Google Scholar]
- 235. Rabushko E, Sorokin M, Suntsova M, et al. Experimentally deduced criteria for detection of clinically relevant fusion 3’ oncogenes from FFPE bulk RNA sequencing data. Biomedicines 2022; 10: 1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Sorokin M, Ignatev K, Poddubskaya E, et al. RNA sequencing in comparison to immunohistochemistry for measuring cancer biomarkers in breast cancer and lung cancer specimens. Biomedicines 2020; 8: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Buzdin A, Sorokin M, Garazha A, et al. RNA sequencing for research and diagnostics in clinical oncology. Semin Cancer Biol 2020; 60: 311–323. [DOI] [PubMed] [Google Scholar]
- 238. Talebi A, Thiery JP, Kerachian MA. Fusion transcript discovery using RNA sequencing in formalin-fixed paraffin-embedded specimen. Crit Rev Oncol Hematol 2021; 160: 103303. [DOI] [PubMed] [Google Scholar]
- 239. Walther C, Hofvander J, Nilsson J, et al. Gene fusion detection in formalin-fixed paraffin-embedded benign fibrous histiocytomas using fluorescence in situ hybridization and RNA sequencing. Lab Invest 2015; 95: 1071–1076. [DOI] [PubMed] [Google Scholar]
- 240. Peng H, Huang R, Wang K, et al. Development and validation of an RNA sequencing assay for gene fusion detection in formalin-fixed, paraffin-embedded tumors. J Mol Diagn 2021; 23: 223–233. [DOI] [PubMed] [Google Scholar]
- 241. Rozenblatt-Rosen O, Regev A, Oberdoerffer P, et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell 2020; 181: 236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Zeng J, Zhang Y, Shang Y, et al. CancerSCEM: a database of single-cell expression map across various human cancers. Nucleic Acids Res 2022; 50: D1147–D1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Nieto P, Elosua-Bayes M, Trincado JL, et al. A single-cell tumor immune atlas for precision oncology. Genome Res 2021; 31: 1913–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Maynard A, McCoach CE, Rotow JK, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell 2020; 182: 1232.e22–1251.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Zhang Y, Mudgal P, Wang L, et al. T cell receptor repertoire as a prognosis marker for heat shock protein peptide complex-96 vaccine trial against newly diagnosed glioblastoma. Oncoimmunology 2020; 9: 1749476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Kharchenko PV. The triumphs and limitations of computational methods for scRNA-seq. Nat Methods 2021; 18: 723–732. [DOI] [PubMed] [Google Scholar]
- 247. McPherson A, Hormozdiari F, Zayed A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 2011; 7: e1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Wu L, Zhang X, Zhao Z, et al. Full-length single-cell RNA-seq applied to a viral human cancer: applications to HPV expression and splicing analysis in HeLa S3 cells. Gigascience 2015; 4: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Chen J, Zhou Q, Wang Y, et al. Single-cell SNP analyses and interpretations based on RNA-seq data for colon cancer research. Sci Rep 2016; 6: 34420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Giustacchini A, Thongjuea S, Barkas N, et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med 2017; 23: 692–702. [DOI] [PubMed] [Google Scholar]
- 251. Jin Z, Huang W, Shen N, et al. Single-cell gene fusion detection by scFusion. Nat Commun 2022; 13: 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Chen J, Facchinetti F, Braye F, et al. Single-cell DNA-seq depicts clonal evolution of multiple driver alterations in osimertinib-resistant patients. Ann Oncol 2022; 33: 434–444. [DOI] [PubMed] [Google Scholar]
- 253. Song Z, Lian S, Mak S, et al. Deep RNA sequencing revealed fusion junctional heterogeneity may predict crizotinib treatment efficacy in ALK-rearranged NSCLC. J Thorac Oncol 2022; 17: 264–276. [DOI] [PubMed] [Google Scholar]
- 254. Gu M, He T, Yuan Y, et al. Single-Cell RNA sequencing reveals multiple pathways and the tumor microenvironment could lead to chemotherapy resistance in cervical cancer. Front Oncol 2021; 11: 753386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Denomy C, Germain S, Haave B, et al. Banding together: a systematic comparison of the cancer genome atlas and the mitelman databases. Cancer Res 2019; 79: 5181–5190. [DOI] [PubMed] [Google Scholar]
- 256. Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn 2008; 10: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Zhang L. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part II. The utility of microsatellite instability testing. J Mol Diagn 2008; 10: 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Gerstung M, Jolly C, Leshchiner I, et al. The evolutionary history of 2,658 cancers. Nature 2020; 578: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Zhang Y, Chen F, Fonseca NA, et al. High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. Nat Commun 2020; 11: 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Amir ED, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013; 31: 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018; 37: 38–44. [DOI] [PubMed] [Google Scholar]
- 262. Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019; 47: D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Hu X, Wang Q, Tang M, et al. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 2018; 46: D1144–D1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Yoshihara K, Wang Q, Torres-Garcia W, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 2015; 34: 4845–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Jang YE, Jang I, Kim S, et al. ChimerDB 4.0: an updated and expanded database of fusion genes. Nucleic Acids Res 2020; 48: D817–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Balamurali D, Gorohovski A, Detroja R, et al. ChiTaRS 5.0: the comprehensive database of chimeric transcripts matched with druggable fusions and 3D chromatin maps. Nucleic Acids Res 2020; 48: D825–D834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Wang Y, Wu N, Liu J, et al. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn Pathol 2015; 10: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Korla PK, Cheng J, Huang C-H, et al. FARE-CAFE: a database of functional and regulatory elements of cancer-associated fusion events. Database (Oxford) 2015; 2015: bav086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Kim D-S, Huh J-W, Kim H-S. HYBRIDdb: a database of hybrid genes in the human genome. BMC Genomics 2007; 8: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Kim P, Tan H, Liu J, et al. FusionGDB 2.0: fusion gene annotation updates aided by deep learning. Nucleic Acids Res 2022; 50: D1221–D1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Kong F, Zhu J, Wu J, et al. dbCRID: a database of chromosomal rearrangements in human diseases. Nucleic Acids Res 2011; 39: D895–D900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Novo FJ, de Mendíbil IO, Vizmanos JL. TICdb: a collection of gene-mapped translocation breakpoints in cancer. BMC Genomics 2007; 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Bamford S, Dawson E, Forbes S, et al. The COSMIC (catalogue of somatic mutations in cancer) database and website. Br J Cancer 2004; 91: 355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Lee M, Lee K, Yu N, et al. ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res 2017; 45: D784–D789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Frenkel-Morgenstern M, Gorohovski A, Lacroix V, et al. ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res 2013; 41: D142–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Panigrahi P, Jere A, Anamika K. FusionHub: a unified web platform for annotation and visualization of gene fusion events in human cancer. PLoS One 2018; 13: e0196588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Kim P, Zhou X. FusionGDB: fusion gene annotation DataBase. Nucleic Acids Res 2019; 47: D994–D1004. [DOI] [PMC free article] [PubMed] [Google Scholar]