Abstract
Background:
Several studies have reported the impact of single nucleotide polymorphisms (SNPs) in vascular endothelial growth factor (VEGF) pathway genes on the efficacy of bevacizumab in metastatic colorectal cancer (mCRC), but results are still inconsistent. The PRODIGE 9 phase III study compared bevacizumab maintenance versus observation alone after induction chemotherapy with FOLFIRI plus bevacizumab.
Objective:
We evaluated the impact of SNPs of VEGF-A, VEGF receptors (VEGFR-1, VEGFR-2), and hypoxia inducible factor-1α (HIF-1α) on tumor control duration (TCD), overall survival (OS), progression-free survival (PFS), and duration of first chemotherapy free-intervals (CFI).
Patients and methods:
We included 314/491 patients from PRODIGE 9 with a DNA blood sample available. Nine SNPs were genotyped on germline DNA using real-time Polymerase Chain Reaction TaqMan TM (Thermo Fisher Scientific, Waltham, MA , USA 02451).
Results:
In the bevacizumab arm, patients with the VEGFR-1 rs9582036 CC genotype (n = 14) had significantly longer TCD [22.4 months (95% confidence interval (CI): 14.75-not reached)] than patients with the AA or CA genotype [14.4 months (95% CI: 11.7–17.1)] (p = 0.036), whereas there was no significant difference in the observation arm. In the bevacizumab arm, no significant difference was found between the CC, and AA or CA genotype for OS [28.2 (95% CI: 18.1–42.8) versus 22.5 (95% CI: 18.6–24.6) months, p = 0.5], PFS [9.4 (95% CI: 7.2–11.3) versus 9.2 (95% CI: 8.71–10.1)], and duration of the first CFI [4.6 (95% CI: 1.6–13.3) versus 4.14 (95% CI: 0.5–29.0) months, p = 0.3].
Conclusion:
Among mCRC patients treated with bevacizumab maintenance, those with the VEGFR-1 rs9582036 CC genotype experienced longer TCD. The presence of this genotype may thus predict a benefit of bevacizumab maintenance in mCRC.
Keywords: angiogenesis, metastatic colorectal cancer, prognostic biomarker, single nucleotide polymorphism, VEGF
Introduction
The prognosis of patients with metastatic colorectal cancer (mCRC) has been significantly improved by the combination of chemotherapy with targeted therapies, antiangiogenic agents (bevacizumab),1,2 or anti-epidermal growth factor receptor agents.3,4
Angiogenesis is a cardinal process that leads to the invasiveness and metastasis of solid tumors.5 Vascular endothelial growth factor-A (VEGF-A) is a major agent of the angiogenesis pathway. Fixation on VEGF receptor-1 and 2 (VEGFR-1 and VEGFR-2) leads to intracellular pathway activation and promotes vascular permeability, cell proliferation, survival and, migration, tubular morphogenesis and sprouting.5,6 Angiogenesis is a tumor response related to a hypoxic micro-environment. Hypoxia inducible factor-1α (HIF-1α) is a mediator of hypoxia signaling that plays a role in the VEGF-pathway.7
While the RAS mutation can predict anti-EGFR resistance,8 there are currently no predictive factors for bevacizumab’s efficacy. A pharmacogenetic study in patients with single nucleotide polymorphisms (SNPs) could be interesting to identify predictive genetic biomarkers on VEGFR genes. Germline SNPs are not dependent on tumor evolution. They are easy to identify and reliable, and can be exploited in a clinical setting with confidence.
Some studies suggest that SNPs on VEGF-A, VEGFR-1, VEGFR-2, or HIF-1α modulate protein expression, and could predict the efficacy of bevacizumab.9–15
The PRODIGE 9 study is a prospective multicenter phase III study comparing maintenance treatment with bevacizumab monotherapy to observation alone after 6 months of induction chemotherapy in mCRC. The results of the study revealed no difference between these two arms for all endpoints.16
We conducted an ancillary and exploratory study of PRODIGE 9 to evaluate the impact of SNPs on VEGF-A, VEGFR-1, VEGFR-2, and HIF-1α on tumor control duration (TCD), overall survival (OS), progression free survival (PFS), and duration of the first chemotherapy-free interval (CFI).
Patients and methods
Patients’ eligibility criteria
PRODIGE 9 was an open-label, randomized, multicenter, phase III study promoted by the Fédération Francophone de Cancérologie Digestive (FFCD) and the Partenariat de Recherche en Oncologie DIGEstive (PRODIGE) intergroup in 66 French centers. PRODIGE 9 included patients with mCRC without previous chemotherapy or antiangiogenic therapy for metastatic disease.16 PRODIGE 9 aimed to compare the TCD achieved with first-line chemotherapy followed by either bevacizumab maintenance or observation without treatment during the first CFI. Both arms received induction FOLFIRI plus bevacizumab for 12 cycles. At the end of induction, patients with disease control began a CFI. During the CFI, patients were either treated with bevacizumab monotherapy (maintenance arm) or received no antitumor treatment (observation arm). Chemotherapy was reintroduced at progression for a further eight cycles, followed by one or several new CFI. After progression, the choice of second-line and additional treatments was left to the discretion of the investigator. A blood sample was collected at enrolment of the patient in the trial. All patients from PRODIGE 9 with a DNA blood sample available were eligible for this study.
Follow-up
The tumor response was evaluated by clinicians in accordance with mRECIST 1.1 criteria every 8 weeks.
Polymorphism selection, genotyping
Genes were selected based on their involvement in VEGF-A-dependent angiogenesis. VEGF-A is directly targeted by bevacizumab. Two receptors VEGFR-1 and VEGFR-2 mediate VEGF-A signaling. HIF-1α mediates hypoxic induction of the VEGF-A gene.
All SNPs were shown in previous studies9,13–15,17–19 to play a prognostic and/or predictive role, as well as to have a functional impact and a relative frequency (minor allele frequency) over 5% (Table 1).
Table 1.
SNPs studied, genetic location, ancestral allele and MAF.
| RS | SNP | Location | Ancestral allele | Variation | MAF | Ref. life technologies® |
|---|---|---|---|---|---|---|
| VEGF-A | ||||||
| rs699947 | c.-2595A > C | Promoter | A | A > C | A = 0.344/752 | C_8311602_10 |
| rs2010963 | c.-634C > G | 5′UTR | G | C > G | C = 0.343/748 | C_8311614_10 |
| rs833061 | c.-1498C > T | Promoter | C | C > T | C = 0.380/828 | C_1647381_10 |
| rs3025039 | c.*237C > T | 3′UTR | C | C > T | T = 0.149/325 | C_16198794_10 |
| VEGFR-1 | ||||||
| rs9582036 | c.3635+319G > T | Intron 3 | C | A > C | C = 0.357/778 | C_1910658_10 |
| VEGFR-2 | ||||||
| rs2071559 | c.-906T > C | Promoter | T | C > T | G = 0.459/1000 | C15869271_10 |
| rs2305948 | c.889G > A | Exon 7 | C | C > T | T = 0.131/286 | C_22271999_20 |
| rs1870377 | c.1416A > T | Exon 11 | T | A > T | A = 0.235/511 | C_11895315_20 |
| HIF-1α | ||||||
| rs11549465 | c.1744C > T | Exon 13 | C | C > T | T = 0.073/160 | C_25473074_10 |
SNPs, single nucleotide polymorphism; MAF, minor allele frequency; VEGF, vascular endothelial growth factor; VEGFR, VEGF receptor.
DNA was extracted from blood samples using an automated QIA symphony DSP DNA Mini Kit TM (Qiagen, Hilden, Germany).
DNA was quantified by fluorometry (QubitTM Thermo Fisher Scientific, Waltham, MA, USA 02451) and genotyping was performed with TaqMan® endpoint PCR in a 10 µL reaction volume (15 ng of DNA in 3 µL, 0.5 µL of probe and 5 µL of Master Mix). Negative controls were systematically added, and around 10% of samples were repeated to validate the genotyping procedure.
Outcomes
TCD was defined as the time between randomization and strategy failure, defined as tumor progression during a chemotherapy sequence.20 Patients whose tumors did not progress during a chemotherapy sequence were censored at their last follow-up within the protocol or censored at the initiation of second-line chemotherapy or another therapeutic strategy. Secondary end points were OS, defined as the time between randomization and death from any cause. Alive patients were censored at the date of the last news. PFS was defined as the time between randomization and first progression or death from any cause. Duration of the first CFI was defined as the time between the end of induction chemotherapy or the date of the last injection for patients in the maintenance arm and the first reintroduction of chemotherapy. From this cohort, all patients for whom a blood sample was available were included. The PRODIGE 9 study was approved by the Committee for the Protection of Persons Ile de France VIII on 12 July 2011 and by the French national agency (AFFSAPS) on the 25 July 2011. The trial was registered on clinicaltrials.gov with the number: NCT00952029. Written informed consent was obtained from all patients before treatment and for the recovery of tumor blocks as requested by the Helsinki declaration (1964) and its amendment (2000).
Statistical analysis
The objectives of the study were to evaluate the impact of SNPs on TCD, OS, PFS, and duration of the first CFI. All SNPs were checked for Hardy–Weinberg equilibrium (HWE) by comparing observed and expected allelic distributions using a chi-square test (χ2 < 3.84 with a significance threshold at 5% and 1 degree of freedom) or Fisher’s exact test for groups smaller than five.
Due to the exploratory design of the study, there was no pre-specified hypothesis tested regarding the effects of a specific genotype and a particular treatment outcome. Multiple statistical tests were performed and no adjustments were made.
Due to small numbers of patients (<4) in some genotype groups: HIF-1 α rs11549465 c.1744C > T and VEGF-A rs3025039 c.*237C > T, patients homozygous for the rare allele and heterozygous were combined for analyses.
Tumor response was analyzed using an univariate correlation test (Fisher’s exact or chi square with a threshold at 0.05).
The survival analysis (TCD, OS, and PFS) were performed using the Kaplan–Meier model with the log rank test (significance threshold 0.05).
We included significant genotypes (p ⩽ 0.05 from univariate analyses) in a single multivariate analysis using the Cox model taking into account independent prognostic factors recognized in the literature, that is, WHO performance status, number of metastatic sites, and age at occurrence of hypertension. As nine SNPs were tested, p values were corrected for multiple testing using the false discovery rate controlling procedure of Benjamini and Liu.21 Adjusted p values were computed using the discrete module type package for R 3.1 (R Foundation for Statistical Computing, Vienna, Austria). All other statistical tests were performed with IBM SPSS v20.0 (IBM Inc., New York, USA).
Results
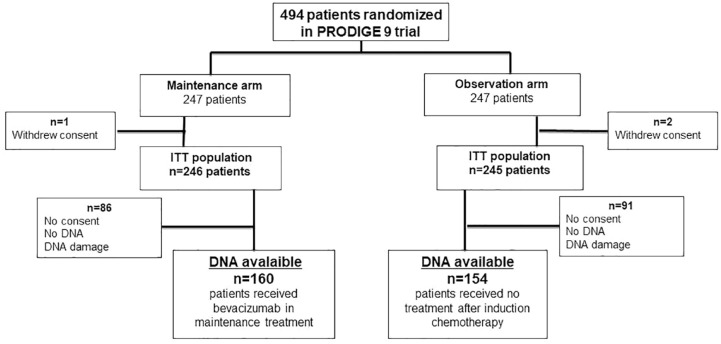
From March 2010 to July 2013, 494 patients were randomized in the PRODIGE 9 study.16 DNA was available for 314 patients: 160 in the bevacizumab maintenance arm and 154 in the observation arm during the first CFI (Figure 1). These 314 patients were comparable to the overall population of PRODIGE 9.
Figure 1.
Study flow chart.
The results are presented for the 314 patients enrolled in this study. In the overall population after the first induction chemotherapy, 156 (52.5%) patients had a complete or partial response, 133 (44.8%) patients had stable disease, and eight (2.7%) had disease progression. Patients characteristics are summarized in Table 2.
Table 2.
Population characteristics, TCD, OS, and PFS in each arm (arm A: bevacizumab maintenance; arm B: observation arm without treatment during CFIs).
| Arm A: bevacizumab maintenance arm N = 160 | Arm B: no treatment during CFI N = 154 | Overall population N = 314 | |
|---|---|---|---|
| Age (mean, range) | 64.6 (26.9–88.8) | 64.9 (37.4–84.3) | 64.7 (26.9–88.8) |
| Sex, n.% | |||
| Males | 100 (62.5%) | 105 (68.2%) | 205 (65.3%) |
| Females | 60 (37.5%) | 49 (31.8%) | 109 (34.7%) |
| ECOG PS n.% | |||
| 0 | 77 (48.1%) | 75 (48.7%) | 152 (48.4%) |
| 1 | 75 (46.9%) | 70 (45.5%) | 145 (46.2%) |
| ⩾2 | 8 (5.0%) | 9 (5.8%) | 17 (5.4%) |
| TCD [median months (95% CI)] | 15.4 (12.5–17.42) | 17.3 (13.4–23.3) | 15.7 (13.7–18.0) |
| Overall survival [median months (95% CI)] | 22.6 (19.3–25.3) | 22.1 (230.3–25.2) | 22.6 (20.8–24.3) |
| PFS [median months (95% CI)] | 9.2 [8.9; 10.1] | 9.1 [8.0; 9.6] | 9.2 [8.9; 9.6] |
| Duration of CFI, mean. (SD) | 5.1 (4.3) | 4.7 (3.3) | 4.9 (3.9) |
CFI, chemotherapy-free interval; CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; OS, overall survival; PFS, progression-free survival; SD, standard deviation; TCD, tumor control duration.
Median TCD during CFI in the bevacizumab maintenance arm was similar to that in the observation arm. Median TCD, median OS, median PFS, and median duration of the first CFI in the 314 patients were similar to those of the main study.
The genotype distribution of the nine SNPs agreed with those predicted by HWE. Observed genotype frequencies are summarized in Table 3.
Table 3.
Frequencies of genotypes by SNPs.
| Arm A : bevacizumab maintenance arm N = 160 | Arm B: no treatment during CFI N = 154 | Overall population N = 314 | ||
|---|---|---|---|---|
| VEGF-A rs699947 | Homozygous Y | 30 (18.8%) | 31 (20.1%) | 61 (19.4%) |
| Heterozygous | 65 (40.6%) | 70 (45.5%) | 135 (43.0%) | |
| Homozygous X | 65 (40.6%) | 53 (34.4%) | 118 (37.6%) | |
| VEGF-A rs2010963 | Homozygous Y | 23 (14.4%) | 12 (7.8%) | 35 (11.1%) |
| Heterozygous | 68 (42.5%) | 71 (46.1%) | 139 (44.3%) | |
| Homozygous X | 69 (43.1%) | 71 (46.1%) | 140 (44.6%) | |
| VEGF-A rs833061 | Homozygous Y | 31 (19.4%) | 32 (20.8%) | 63 (20.1%) |
| Heterozygous | 80 (50.0%) | 82 (53.2%) | 162 (51.6%) | |
| Homozygous X | 49 (30.6%) | 40 (26.0%) | 89 (28.3%) | |
| VEGF-A rs3025039 | Homozygous Y | 121 (75.6%) | 120 (77.9%) | 241 (76.8%) |
| Heterozygous | 37 (23.1%) | 31 (20.1%) | 68 (21.7%) | |
| Homozygous X | 2 (1.3%) | 3 (1.9%) | 5 (1.6%) | |
| VEGFR-1 rs9582036 | Homozygous Y | 75 (46.9%) | 78 (50.6%) | 153 (48.7%) |
| Heterozygous | 71 (44.4%) | 60 (39.0%) | 131 (41.7%) | |
| Homozygous X | 14 (8.8%) | 16 (10.4%) | 30 (9.6%) | |
| VEGFR-2 rs2071559 | Homozygous Y | 40 (25.0%) | 45 (29.2%) | 85 (27.1%) |
| Heterozygous | 80 (50.0%) | 74 (48.1%) | 154 (49.0%) | |
| Homozygous X | 40 (25.0%) | 35 (22.7%) | 75 (23.9%) | |
| VEGFR-2 rs2305948 | Homozygous Y | 125 (78.1%) | 119 (77.3%) | 244 (77.7%) |
| Heterozygous | 32 (20.0%) | 31 (20.1%) | 63 (20.1%) | |
| Homozygous X | 2 (1.3%) | 3 (1.9%) | 5 (1.6%) | |
| D | 1 (0.6%) | 1 (0.6%) | 2 (0.6%) | |
| VEGFR-2 rs1870377 | Homozygous Y | 15 (9.4%) | 14 (9.1%) | 29 (9.2%) |
| Heterozygous | 51 (31.9%) | 57 (37.0%) | 108 (34.4%) | |
| Homozygous X | 94 (58.8%) | 81 (52.6%) | 175 (55.7%) | |
| D | . | 2 (1.3%) | 2 (0.6%) | |
| HIF-1α rs11549465 | Homozygous Y | 129 (80.6%) | 116 (75.3%) | 245 (78.0%) |
| Heterozygous | 28 (17.5%) | 35 (22.7%) | 63 (20.1%) | |
| Homozygous X | 3 (1.9%) | 3 (1.9%) | 6 (1.9%) |
CFI, chemotherapy-free interval; IQR, interquartile range; OS, overall survival; PFS, progression-free survival; TCD, tumor control duration; VEGFR, vascular endothelial growth factor receptor.
Associations between SNPs and efficacy endpoints
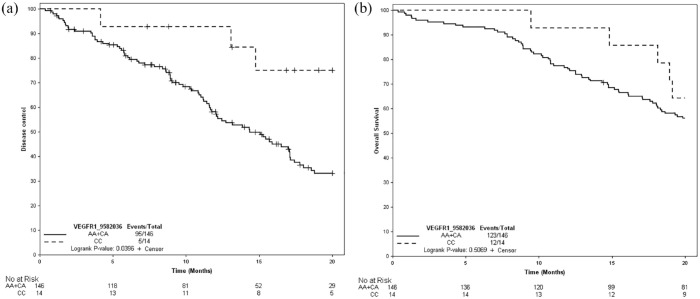
A statistically significant association was found between TCD and VEGFR-1 rs9582036. TCD was significantly longer [median 22.4 months (95% confidence interval (CI) : 14.7–NR)] in patients with the CC genotype than in patients with the AA and CA genotypes [14.4 months (95% CI : 11.7–17.1) (hazard ratio = 0.39; 95% CI : 0.16–0.97) p = 0.04] in the bevacizumab maintenance arm, as shown in Figure 2(a). In the observation arm, no significant association was found between this SNP and TCD, as shown in Table 4.
Figure 2.
Kaplan–Meier curves according to study treatment for VEGFR-1 rs9582036. (a) TCD. (b) OS.
OS, overall survival; TCD, tumor control duration.
Table 4.
Efficacy endpoint for the VEGFR-1 rs9582036.
| Median (95% CI) | Bevacizumab maintenance arm | Observation maintenance arm | ||||
|---|---|---|---|---|---|---|
| VEGFR-1 rs9582036 | CC N = 14 | CA + AA N = 146 | p | CC N = 16 | CA + AA N = 138 | p |
| TCD | 22.4 (14.8–NR) | 14.4 (11.7–17.1) | 0.047 | NR (9.5–NR) | 15.7 (12.7–21.9) | 0.079 |
| OS | 28.2 (18.1–42.8) | 22.5 (18.6–24.6) | 0.510 | 22.66 (17.2–NR) | 22.1 (19.5–25.2) | 0.097 |
| PFS | 9.4 (7.2–11.3) | 9.2 (8.71–10.1) | 0.59 | 9.3 (7.5–10.8) | 9.10 (7.9–9.7) | 0.785 |
| First CFI (IQR) | 4.6 (3.3–7.4) | 4.14 (2.3–6.1) | 0.303 | 3.15 (2.6–4.9) | 4 (2.7–6.0) | 0.493 |
CFI, chemotherapy-free interval; IQR, interquartile range; OS, overall survival; PFS, progression-free survival; TCD, tumor control duration; VEGFR, vascular endothelial growth factor receptor.
In the bevacizumab maintenance arm, there was a trend toward better OS in patients with the CC genotype for VEGFR-1 rs9582036 versus those with the AC + AA genotype, although the difference was not significant [28.2 (95% CI : 18.1-42.8) versus 22.5 (95% CI : 18.6–24.6) months, p = 0.5), as shown in Figure 2(b). In the observation arm, the median OS was similar in both the CC and AC + AA genotype subgroups.
Moreover, no statistically significant difference was observed for either PFS or first CFI duration for CC patients versus AC and AA patients (Table 4).
There was no significant association between any genotype and objective response after the first induction chemotherapy. However, there was a trend toward a better tumor response in patients with the VEGFR-1 rs9582036 CC genotype than that in AC + AA patients, as shown in Table 5.
Table 5.
Best response on the first induction treatment for VEGFR-1 rs9582036.
| Homozygous AA and heterozygous | Homozygous CC | Total | p | |
|---|---|---|---|---|
| Complete response or partial response | 135 (50.4%) | 21 (72.4%) | 156 (52.5%) | 0.067 |
| Stable disease | 125 (46.6%) | 8 (27.6%) | 133 (44.8%) | |
| Progression | 8 (3.0%) | 0 (0.0) | 8 (2.7%) |
VEGFR, vascular endothelial growth factor receptor.
There was no other statistically significant association between other SNPs evaluated and any endpoints, as described in Supplemental Tables 1 and 2.
Discussion
Bevacizumab is one of the main biotherapies for mCRC. In the era of ‘personalized medicine’, it has become essential to determine predictive factors of response to bevacizumab so as to identify the subgroups of patients that may benefit from this treatment without added toxicity.
In our study, we studied nine SNPs involved in the VEGF-pathway in a large series of 314 patients included in the PRODIGE 9 phase III study comparing bevacizumab maintenance versus observation alone after induction first-line chemotherapy with FOLFIRI plus bevacizumab.
Despite the randomized design of this trial set up to determine whether patients benefited or not from bevacizumab maintenance therapy, no clinical factors consistently predicted bevacizumab efficacy in this exploratory study. However, we did find that the VEGFR-1 rs9582036 genotype had an impact on survival in patients treated with maintenance bevacizumab. TCD was significantly longer in patients with the CC genotype than in those with the CA and AA genotype. We also found a trend toward better OS in patients with the CC genotype. Nonetheless, no prolongation of the CFI was observed, suggesting that the maintenance of antiangiogenic pressure could be pointless for this genotype. To date, this is the largest genotyping study of SNPs involved in the VEGF pathway in mCRC patients treated with maintenance bevacizumab as a single agent, and the first to report long-term follow-up data regarding differences in outcomes between the CC and the CA + AA genotype for VEGFR-1 rs9582036. However, the VEGFR-1 rs9582036 CC genotype was rare, found in 14 patients in the bevacizumab arm and 16 patients in the observation arm. The potential clinical value is unquestionable; but the data should still be validated in an independent series.
Hansen et al. reported a similar low frequency of the VEGFR-1 rs9582036 CC genotype, present in 22 of the 218 mCRC patients analyzed. However, they showed an inverse association, with a better response in patients with the VEGFR-1 rs9582036 AA genotype versus CC + AC.22 Chionh et al. reported no predictive association between VEGFR-1 rs9582036 and survival in patients with mCRC receiving bevacizumab plus chemotherapy. In their study, however, tumor tissue and not blood was used for the SNP analysis.23 Loupakis et al. also reported no predictive impact of the VEGFR-1 rs9582036 genotype on survival outcomes.13,24
Several studies have suggested some associations between remaining SNPs and bevacizumab efficacy in mCRC, but no correlation was strong enough to be used in clinical practice.13–15,17,22,24–27 A prognostic impact was found for VEGF-A rs1570360,15 VEGF-A rs2010963,15 VEGFR-2 rs125057758,13 and VEGFR-2 rs2305948.14 Previous data suggested the possible role of the VEGF-A rs833061 genotype in predicting the efficacy of bevacizumab,24,27 but failed to show a predictive impact.13 VEGF-A rs699947 and VEGF-A rs3025039 have been found to be associated with a lower prevalence of bevacizumab-induced hypertension.25
In other kinds of cancers, several studies have linked VEGFR-1 rs9582036 with clinical outcomes in chemotherapy plus bevacizumab-treated patients. In a retrospective study of 77 patients with pancreatic cancer, the VEGFR-1 rs9582036 CC genotype was a predictor of shorter survival in patients treated with bevacizumab, and correlated with increased VEGFR-1 expression.28 The same was shown for patients with non-small-cell lung cancer treated with bevacizumab in two small studies: shorter survival correlated with higher baseline plasma levels of VEGF-A and the presence of VEGFR-1 rs9582036 CC.29,30 Conversely, some studies showed that clinical outcomes in patients with VEGFR-1 rs9582036 AA were better than outcomes in those with CC + AC, especially in patients with breast cancer treated with bevacizumab-based therapy.31 In patients with renal cancer treated with sunitinib, a multitarget antiangiogenic agent, VEGFR-1 rs959582036 CC was associated with lower objective tumor response, PFS and OS.32
More studies are needed to understand the molecular mechanism of VEGFR-1 polymorphism in response to VEGF blockage.
VEGFR-1 rs9582036 has been shown to be in high linkage disequilibrium with rs7993418. Fine-mapping experiments of the VEGFR-1 locus identified rs7993418, a synonymous SNP affecting tyrosine 1213 in the VEGFR-1 tyrosine-kinase domain, as the functional variant underlying the association. This SNP causes a shift in codon usage, leading to increased VEGFR-1 expression and downstream VEGFR-1 signaling. We could infer that CC patients in our study could be more sensitive to bevacizumab because of that variation of the VEGR-1 level. An approach with simultaneous VEGF-A dosing could be interesting to corroborate this hypothesis.
Some limitations must be considered. First, similar to other investigations in this field and due to the exploratory design, no pre-specified hypothesis was tested regarding the effects of a specific genotype and a particular treatment outcome, we conducted multiple statistical tests, and no adjustments were made.
Then, bevacizumab’s mechanisms of action are not fully known. Bevacizumab not only blocks VEGF-A, but also plays a role in the tumor microenvironment, including the inhibition of bone marrow-derived progenitors, the normalization of vessel structure, vascular ‘constriction’, disruption of the cancer stem cells niche, a direct effect on tumor cells, and interactions with the host immune system. This may explain the difficulty to identify a single ‘candidate SNP’. A broader genome-wide approach to identify more relevant SNPs could be interesting. This method was used by Kim,19 who conducted genome-wide SNP screening on high-speed DNA chips (SNP array) in blood samples from colon cancer patients. Surprisingly, genes unrelated to angiogenesis were found to be significantly linked to the bevacizumab response. However, the population studied was heterogeneous: patients were treated with bevacizumab or cetuximab, an anti-EGFR which acts via a different mechanism.
Other prognostic factors have been identified in the PRODIGE 9 study, such as the BRAF V600E mutation,33 baseline splenic volume,34 and radiomic signature,35 but these were not evaluated in correlation with SNP in this study. In recent studies, the side of the colorectal cancer, left or right, has also been reported as a prognostic factor and a predictor of response to treatment,36,37 with a worst prognosis in right-side colorectal cancer.36,38 This prognostic effect was not confirmed in other prospective trials4,39 including PRODIGE 916 and should be interpreted with caution.
It is important to note that our analyses were limited by the small number of VEGFR-1 rs9582036 CC patients, as in other studies. A larger validation cohort with more CC patients is needed to confirm the predictive value of VEGFR-1 rs9582036 for bevacizumab treatment. Moreover, the pharmacokinetics of bevacizumab was not taken into account, even though it has been suggested that clinical outcomes vary according to the concentration of bevacizumab.40 A global approach with pharmacokinetic dosing could be interesting to take into account this possible cofounding factor. In addition, a VEGFR-1 expression analysis seems necessary to understand how VEGFR-1 rs9582036 CC affects protein function.
In the PRODIGE 9 trial, bevacizumab monotherapy did not impair tumor progression during CFI. The application of this findings regarding the VEGFR-1 s9582036 CC genotype in patients treated with bevacizumab maintenance in clinical practice seems to be extremely limited. Nevertheless, a pooled analysis of several trials including PRODIGE 9. That evaluated bevacizumab monotherapy during CFI reported an improvement in PFS in patients treated with bevacizumab monotherapy.41 It would be interesting to evaluate VEGFR-1 rs9582036 in these patients to select patients that could potentially benefit from bevacizumab monotherapy during CFI.
In conclusion, our study found that VEGR-1 rs9582036 CC was associated with prolonged TCD for patients with mCRC on bevacizumab-based maintenance therapy. This result needs to be confirmed in larger studies, which should include pharmacokinetics and expression analyses to understand and strengthen the potential predictive relevance of VEGR-1 rs9582036 in patients treated with bevacizumab.
Supplemental Material
Supplemental material, sj-docx-1-tam-10.1177_17588359221141307 for Predictive value of vascular endothelial growth factor polymorphisms for maintenance bevacizumab efficacy in metastatic colorectal cancer: an ancillary study of the PRODIGE 9 phase III trial by Bernadette de Rauglaudre, Camille Sibertin-Blanc, Aurélie Fabre, Karine Le Malicot, Jaafar Bennouna, François Ghiringhelli, Julien Taïeb, Valérie Boige, Olivier Bouché, Thierry Chatellier, Roger Faroux, Eric François, Stéphane Jacquot, Dominique Genet, Claire Mulot, Sylviane Olschwang, Jean-François Seitz, Thomas Aparicio and Laetitia Dahan in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-2-tam-10.1177_17588359221141307 for Predictive value of vascular endothelial growth factor polymorphisms for maintenance bevacizumab efficacy in metastatic colorectal cancer: an ancillary study of the PRODIGE 9 phase III trial by Bernadette de Rauglaudre, Camille Sibertin-Blanc, Aurélie Fabre, Karine Le Malicot, Jaafar Bennouna, François Ghiringhelli, Julien Taïeb, Valérie Boige, Olivier Bouché, Thierry Chatellier, Roger Faroux, Eric François, Stéphane Jacquot, Dominique Genet, Claire Mulot, Sylviane Olschwang, Jean-François Seitz, Thomas Aparicio and Laetitia Dahan in Therapeutic Advances in Medical Oncology
Acknowledgments
The authors thank all participating patients and their families. We thank all the study groups, investigators from the participating countries. The authors also thank the team from the FFCD data center.
Footnotes
ORCID iD: Bernadette de Rauglaudre  https://orcid.org/0000-0003-2913-8129
https://orcid.org/0000-0003-2913-8129
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Bernadette de Rauglaudre, Assistance Publique Hôpitaux de Marseille – Aix-Marseille Université, CHU Timone, 264 rue Saint Pierre, Marseille Cedex 05, Marseille F-13385, France.
Camille Sibertin-Blanc, UMR S-910 INSERM, Génétique Médicale et Génomique Fonctionnelle, Aix-Marseille Université, Marseille, France; Hôpital Sainte Musse, Centre Hospitalier Intercommunal Toulon 6 La Seyne-sur-Mer, Toulon, France.
Aurélie Fabre, UMR S-910 INSERM, Génétique Médicale et Génomique Fonctionnelle, Aix-Marseille Université, Marseille, France.
Karine Le Malicot, Département de Statistique, Fédération Française de Cancérologie Digestive (FFCD), Dijon, France.
Jaafar Bennouna, Hôpital Foch, Suresne, France.
François Ghiringhelli, Centre Georges François Leclerc – Université de Dijon, Dijon, France.
Julien Taïeb, Hôpital Européen Georges Pompidou – Université Paris-Cité, SIRIC CARPEM, Paris, France.
Valérie Boige, Department of Cancer Medicine, Gustave Roussy, Villejuif, France.
Olivier Bouché, Service de Gastroentérologie et Oncologie Digestive, CHU Reims, Reims, France.
Thierry Chatellier, Centre Hospitalier de St Nazaire, St Nazaire, France.
Roger Faroux, Centre Hospitalier les Oudairies, La Roche-sur-Yon, France.
Eric François, Centre Hospitalier Antoine Lacassagne, Nice, France.
Stéphane Jacquot, Clinique Clémentville, Montpellier, France.
Dominique Genet, Clinique Privée François Chenieux, Limoges, France.
Claire Mulot, CRB EPIGENETEC, Centre de Recherche des Cordeliers, INSERM U1138 – Université de Paris, La Sorbonne, Paris, France.
Sylviane Olschwang, Hôpital Privé Clairval, Ramsay Santé, Marseille, France Medipath, Eguilles, France.
Jean-François Seitz, Hôpital la Timone, Assistance Publique Hôpitaux de Marseille – Aix-Marseille Université, Marseille, France; UMR S-910 INSERM, Génétique Médicale et Génomique Fonctionnelle, Aix-Marseille Université, Marseille, France.
Thomas Aparicio, Service de Gastroentérologie et Oncologie Digestive, Hôpital Saint Louis, Assistance Publique Hôpitaux de Paris – Université Paris Cité, Paris, France.
Laetitia Dahan, Hôpital la Timone, Assistance Publique Hôpitaux de Marseille – Aix-Marseille Université, Marseille, France; UMR S-910 INSERM, Génétique Médicale et Génomique Fonctionnelle, Aix-Marseille Université, Marseille, France.
Declarations
Ethics approval and consent to participate: The PRODIGE 9 study was approved by the Committee for the Protection of Persons Ile de France VIII on 12 July 2011 and by the French national agency (AFFSAPS) on the 25 July 2011. The trial was registered on clinicaltrials.gov with the number: NCT00952029. Written informed consent was obtained from all patients before treatment as requested by the Helsinki declaration (1964) and its amendment (2000).
Consent for publication: We confirm that informed consent for publication was provided by the participant(s) or a legally authorized representative.
Author contribution(s): Bernadette de Rauglaudre: Conceptualization; Methodology; Validation; Visualization; Writing – original draft; Writing – review & editing.
Camille Sibertin-Blanc: Conceptualization; Methodology; Resources; Validation.
Aurélie Fabre: Conceptualization; Methodology; Resources.
Karine Le Malicot: Formal analysis; Methodology; Validation; Visualization; Writing – review & editing.
Jaafar Bennouna: Investigation; Writing – review & editing.
François Ghiringhelli: Investigation; Writing – review & editing.
Julien Taïeb: Investigation; Writing – review & editing.
Valérie Boige: Investigation; Writing – review & editing.
Olivier Bouché: Investigation; Writing – review & editing.
Thierry. Chatellier: Writing – review & editing.
Roger Faroux: Investigation; Writing – review & editing.
Eric François: Investigation; Writing – review & editing.
Stéphane Jacquot: Investigation; Writing – review & editing.
Dominique Genet: Investigation; Writing – review & editing.
Claire Mulot: Conceptualization; Methodology; Resources.
Sylviane Olschwang: Conceptualization; Funding acquisition; Methodology; Project administration; Resources; Supervision.
Jean-François Seitz: Conceptualization; Funding acquisition; Investigation; Supervision; Writing – review & editing.
Thomas Aparicio: Conceptualization; Funding acquisition; Investigation; Supervision; Writing – original draft; Writing – review & editing.
Laetitia Dahan: Conceptualization; Funding acquisition; Investigation; Project administration; Supervision; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The FFCD thanks the Ligue Nationale Contre le Cancer, who provided financial support to the FFCD. Hoffmann-La Roche provided financial support for study management (Grant No. ML28232).
The authors declare the following financial interests/personal relationships that may be considered as potential competing interests: JT received honoraria as a speaker or in an advisory role from Amgen, Astellas, BMS, Merck-Serono, MSD, Novartis, Pierre Fabre, Roche, and Servier; O.B. received personal fees as a speaker and/or an advisor from Merck KGaA, Roche, Bayer, Astra-Zeneca, Grunenthal, MSD, Amgen, Sanofi, Servier, and Pierre Fabre, outside the submitted work; VB received personal fees, and non-financial support from Merck Serono, Bayer, Roche, Sanofi, Ipsen, Merck MSD, BMS, Eisai, Novartis, and Amgen outside the submitted work.
BDR, CSB, AF, KLM, JB, FG, TC, RF, EF, SJ, DG, CM, SO, JFS, TA, and LD declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
JT has received honoraria as a speaker or an advisor from Amgen, Astellas, BMS, Merck-Serono, MSD, Novartis, Pierre Fabre, Roche, and Servier.
VB received personal fees and non-financial support from Merck Serono, Bayer, Roche, Ipsen, Merck MSD, Amgen, non-financial support from Sanofi, personal fees from BMS, Eisai, Novartis, outside the submitted work.
O.B. received personal fees as a speaker and/or advisor from Merck KGaA, Roche, Bayer, Astra-Zeneca, Grunenthal, MSD, Amgen, Sanofi, Servier, and Pierre Fabre, outside the submitted work.
Availability of data and materials: The PRODIGE 9 database is open to the scientific and medical community upon request to the cohort steering committee. All data will be made available (de-identified participant data, participant data with identifiers, data dictionary, or other specified data set) depending on the collaboration in place. The study protocol, statistical analysis plan, informed consent form, and consortium status are available upon request. Proposals should be addressed to Ms Cecile Girault, project manager of the study at: cecile.girault@u-bourgogne.fr. Cecile Girault will then prepare the dossiers for the steering committee. The steering committee of the FFCD will evaluate the pertinence of the request before sending the database to any academic partners. After agreement of the Steering Committee, data requestors will have to sign a data access agreement to gain access to the database. The FFCD, as the sponsor, will be vigilant regarding compliance of the requestors with the General Data Protection Regulation (GDPR).
References
- 1. Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 2004; 350: 2335–2342. [DOI] [PubMed] [Google Scholar]
- 2. Venook AP, Niedzwiecki D, Lenz H-J, et al. CALGB/SWOG 80405: phase III trial of irinotecan/5-FU/leucovorin (FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS wild-type (wt) untreated metastatic adenocarcinoma of the colon or rectum (MCRC). J Clin Oncol 2014; 32: LBA3–LBA3. [Google Scholar]
- 3. Van Cutsem E, Köhne C-H, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009; 360: 1408–1417. [DOI] [PubMed] [Google Scholar]
- 4. Heinemann V, von Weikersthal LF, Decker T, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol 2014; 15: 1065–1075. [DOI] [PubMed] [Google Scholar]
- 5. Kerbel RS. Tumor angiogenesis. N Engl J Med 2008; 358: 2039–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011; 473: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shweiki D, Itin A, Soffer D, et al. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature 1992; 359: 843–845. [DOI] [PubMed] [Google Scholar]
- 8. Lièvre A, Bachet J-B, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006; 66: 3992–3995. [DOI] [PubMed] [Google Scholar]
- 9. Jain L, Vargo CA, Danesi R, et al. The role of vascular endothelial growth factor SNPs as predictive and prognostic markers for major solid tumors. Mol Cancer Ther 2009; 8: 2496–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koukourakis MI, Papazoglou D, Giatromanolaki A, et al. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer Amst Neth 2004; 46: 293–298. [DOI] [PubMed] [Google Scholar]
- 11. Renner W, Kotschan S, Hoffmann C, et al. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res 2000; 37: 443–448. [DOI] [PubMed] [Google Scholar]
- 12. Kim HO, Jo YH, Lee J, et al. The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep 2008; 20: 1181–1187. [PubMed] [Google Scholar]
- 13. Loupakis F, Cremolini C, Yang D, et al. Prospective validation of candidate SNPs of VEGF/VEGFR pathway in metastatic colorectal cancer patients treated with first-line FOLFIRI plus bevacizumab. PloS One 2013; 8: e66774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gerger A, El-Khoueiry A, Zhang W, et al. Pharmacogenetic angiogenesis profiling for first-line Bevacizumab plus oxaliplatin-based chemotherapy in patients with metastatic colorectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res 2011; 17: 5783–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Formica V, Palmirotta R, Del Monte G, et al. Predictive value of VEGF gene polymorphisms for metastatic colorectal cancer patients receiving first-line treatment including fluorouracil, irinotecan, and bevacizumab. Int J Colorectal Dis 2011; 26: 143–151. [DOI] [PubMed] [Google Scholar]
- 16. Aparicio T, Ghiringhelli F, Boige V, et al. Bevacizumab maintenance versus no maintenance during chemotherapy-free intervals in metastatic colorectal cancer: a randomized phase III trial (PRODIGE 9). J Clin Oncol Off J Am Soc Clin Oncol 2018; 36: 674–681. [DOI] [PubMed] [Google Scholar]
- 17. Koutras A, Kotoula V, Fountzilas G. Prognostic and predictive role of vascular endothelial growth factor polymorphisms in breast cancer. Pharmacogenomics 2015; 16: 79–94. [DOI] [PubMed] [Google Scholar]
- 18. Sullivan I, Riera P, Andrés M, et al. Prognostic effect of VEGF gene variants in metastatic non-small-cell lung cancer patients. Angiogenesis 2019; 22: 433–440. [DOI] [PubMed] [Google Scholar]
- 19. Kim JG, Chae YS, Sohn SK, et al. Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with colorectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res 2008; 14: 62–66. [DOI] [PubMed] [Google Scholar]
- 20. Aparicio T, Linot B, Le Malicot K, et al. FOLFIRI+bevacizumab induction chemotherapy followed by bevacizumab or observation in metastatic colorectal cancer, a phase III trial (PRODIGE 9–FFCD 0802). Dig Liver Dis Off J Ital Soc Gastroenterol Ital Assoc Study Liver 2015; 47: 271–272. [DOI] [PubMed] [Google Scholar]
- 21. Benjamini Y, Liu W. A step-down multiple hypotheses testing procedure that controls the false discovery rate under independence. J Stat Plan Inference 1999; 82: 163–170. [Google Scholar]
- 22. Hansen TF, dePont Christensen R, Andersen RF, et al. The predictive value of single nucleotide polymorphisms in the VEGF system to the efficacy of first-line treatment with bevacizumab plus chemotherapy in patients with metastatic colorectal cancer: results from the Nordic ACT trial. Int J Colorectal Dis 2012; 27: 715–720. [DOI] [PubMed] [Google Scholar]
- 23. Chionh F, Gebski V, Al-Obaidi SJ, et al. VEGF-A, VEGFR1 and VEGFR2 single nucleotide polymorphisms and outcomes from the AGITG MAX trial of capecitabine, bevacizumab and mitomycin C in metastatic colorectal cancer. Sci Rep 2022; 12: 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loupakis F, Ruzzo A, Salvatore L, et al. Retrospective exploratory analysis of VEGF polymorphisms in the prediction of benefit from first-line FOLFIRI plus bevacizumab in metastatic colorectal cancer. BMC Cancer 2011; 11: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morita S, Uehara K, Nakayama G, et al. Association between bevacizumab-related hypertension and vascular endothelial growth factor (VEGF) gene polymorphisms in Japanese patients with metastatic colorectal cancer. Cancer Chemother Pharmacol 2013; 71: 405–411. [DOI] [PubMed] [Google Scholar]
- 26. Abajo A, Rodriguez J, Bitarte N, et al. Dose-finding study and pharmacogenomic analysis of fixed-rate infusion of gemcitabine, irinotecan and bevacizumab in pretreated metastatic colorectal cancer patients. Br J Cancer 2010; 103: 1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cui W, Li F, Yuan Q, et al. Role of VEGFA gene polymorphisms in colorectal cancer patients who treated with bevacizumab. Oncotarget 2017; 8: 105472–105478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambrechts D, Claes B, Delmar P, et al. VEGF pathway genetic variants as biomarkers of treatment outcome with bevacizumab: an analysis of data from the AViTA and AVOREN randomised trials. Lancet Oncol 2012; 13: 724–733. [DOI] [PubMed] [Google Scholar]
- 29. Jantus-Lewintre E, Sureda BM, González Larriba JL, et al. Prospective exploratory analysis of angiogenic biomarkers in peripheral blood in advanced NSCLC patients treated with bevacizumab plus chemotherapy: the ANGIOMET study. Front Oncol 2021; 11: 695038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Glubb DM, Paré-Brunet L, Jantus-Lewintre E, et al. Functional FLT1 genetic variation is a prognostic factor for recurrence in stage I-III non-small-cell lung cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer 2015; 10: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gal J, Milano G, Brest P, et al. VEGF-related germinal polymorphisms may identify a subgroup of breast cancer patients with favorable outcome under bevacizumab-based therapy-a message from COMET, a French Unicancer Multicentric Study. Pharm Basel Switz 2020; 13: E414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beuselinck B, Jean-Baptiste J, Schöffski P, et al. Validation of VEGFR1 rs9582036 as predictive biomarker in metastatic clear-cell renal cell carcinoma patients treated with sunitinib. BJU Int 2016; 118: 890–901. [DOI] [PubMed] [Google Scholar]
- 33. Aparicio T, Bennouna J, Le Malicot K, et al. Predictive factors for early progression during induction chemotherapy and chemotherapy-free interval: analysis from PRODIGE 9 trial. Br J Cancer 2020; 122: 957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niogret J, Limagne E, Thibaudin M, et al. Baseline splenic volume as a prognostic biomarker of FOLFIRI efficacy and a surrogate marker of MDSC accumulation in metastatic colorectal carcinoma. Cancers 2020; 12: E1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dohan A, Gallix B, Guiu B, et al. Early evaluation using a radiomic signature of unresectable hepatic metastases to predict outcome in patients with colorectal cancer treated with FOLFIRI and bevacizumab. Gut 2020; 69: 531–539. [DOI] [PubMed] [Google Scholar]
- 36. Arnold D, Lueza B, Douillard J-Y, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol Off J Eur Soc Med Oncol 2017; 28: 1713–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He W-Z, Liao F-X, Jiang C, et al. Primary tumor location as a predictive factor for first-line bevacizumab effectiveness in metastatic colorectal cancer patients. J Cancer 2017; 8: 388–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petrelli F, Tomasello G, Borgonovo K, et al. Prognostic survival associated with left-sided vs right-sided colon cancer: a systematic review and meta-analysis. JAMA Oncol 2017; 3: 211–219. [DOI] [PubMed] [Google Scholar]
- 39. Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol 2017; 3: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caulet M, Lecomte T, Bouché O, et al. Bevacizumab pharmacokinetics influence overall and progression-free survival in metastatic colorectal cancer patients. Clin Pharmacokinet 2016; 55: 1381–1394. [DOI] [PubMed] [Google Scholar]
- 41. Salvatore L, Bria E, Sperduti I, et al. Bevacizumab as maintenance therapy in patients with metastatic colorectal cancer: a meta-analysis of individual patients’ data from 3 phase III studies. Cancer Treat Rev 2021; 97: 102202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tam-10.1177_17588359221141307 for Predictive value of vascular endothelial growth factor polymorphisms for maintenance bevacizumab efficacy in metastatic colorectal cancer: an ancillary study of the PRODIGE 9 phase III trial by Bernadette de Rauglaudre, Camille Sibertin-Blanc, Aurélie Fabre, Karine Le Malicot, Jaafar Bennouna, François Ghiringhelli, Julien Taïeb, Valérie Boige, Olivier Bouché, Thierry Chatellier, Roger Faroux, Eric François, Stéphane Jacquot, Dominique Genet, Claire Mulot, Sylviane Olschwang, Jean-François Seitz, Thomas Aparicio and Laetitia Dahan in Therapeutic Advances in Medical Oncology
Supplemental material, sj-docx-2-tam-10.1177_17588359221141307 for Predictive value of vascular endothelial growth factor polymorphisms for maintenance bevacizumab efficacy in metastatic colorectal cancer: an ancillary study of the PRODIGE 9 phase III trial by Bernadette de Rauglaudre, Camille Sibertin-Blanc, Aurélie Fabre, Karine Le Malicot, Jaafar Bennouna, François Ghiringhelli, Julien Taïeb, Valérie Boige, Olivier Bouché, Thierry Chatellier, Roger Faroux, Eric François, Stéphane Jacquot, Dominique Genet, Claire Mulot, Sylviane Olschwang, Jean-François Seitz, Thomas Aparicio and Laetitia Dahan in Therapeutic Advances in Medical Oncology