Abstract
The Arabidopsis homeodomain protein BREVIPEDICELLUS/KNAT1 represses the expression of the gene encoding the transcription factor TCP15 to limit filament growth at late stages of stamen development.
Dear Editor,
Stamen filament elongation is a strictly controlled process, crucial for efficient self-pollination in autogamous plants. In Arabidopsis (Arabidopsis thaliana), stamen primordia emergence is followed by differentiation of filaments and anthers around Stage 7 of flower development and by fast filament elongation during Stages 10–13, when the flower opens (Smyth et al., 1990; Cardarelli and Cecchetti, 2014). The repression of genes encoding Class I KNOX homeodomain transcription factors at early stages of flower development is necessary for correct stamen development in Arabidopsis (Tabata et al., 2010; Rubio-Somoza and Weigel, 2013). A literature survey, however, indicated that the promoter of the Class I KNOX gene KNOTTED1-LIKE in ARABIDOPSIS THALIANA1 (KNAT1), also named BREVIPEDICELLUS (BP; Venglat et al., 2002), is active in elongated stamen filaments (Ori et al., 2000; Wang et al., 2006), raising the possibility of an additional role of this transcription factor during late stamen development.
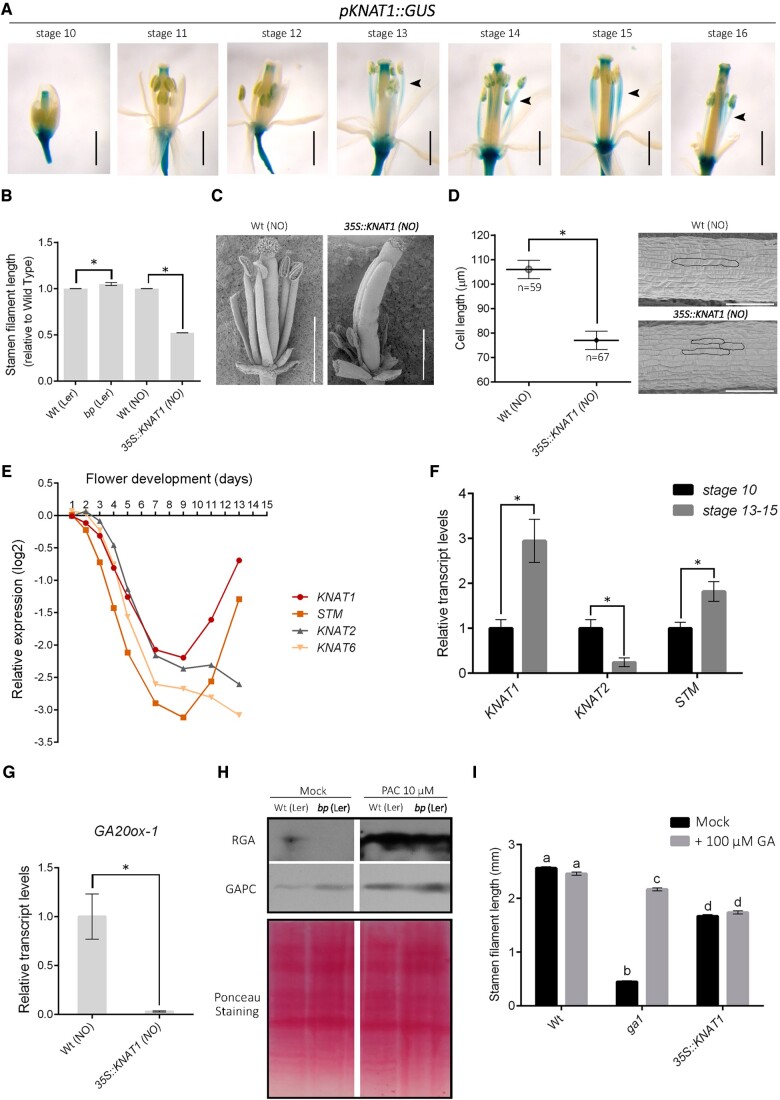
We analyzed flowers from plants that contain the pKNAT1::GUS reporter (Ori et al., 2000) and observed that KNAT1 promoter activity in elongated filaments is noticeable at flower Stage 13 or later (Figure 1A), suggesting that the repression of Class I KNOX genes at early stages of stamen development is followed by reactivation of KNAT1 around anthesis. We then analyzed stamen length in Stage 13 flowers from bp-1, a loss-of-function mutant in KNAT1, and in plants that constitutively express KNAT1 (35S::KNAT1; Ori et al., 2000). bp-1 plants showed a slight, but significant increase in stamen length, while overexpression of KNAT1 strongly affected filament elongation (Figure 1, B and C), indicating that KNAT1 is a repressor of this process. Measurement of cell length in stamens from 35S::KNAT1 plants suggested that KNAT1 affects cell elongation (Figure 1D). The mild effect of the bp-1 mutation on stamen length may be explained by functional redundancy with other Class I KNOX genes. Analysis of available global gene expression data from flowers at different developmental stages indicated that SHOOTMERISTEMLESS (STM), like KNAT1, is induced at late stages of flower development, while KNAT2 and KNAT6 remain in a repressed state (Figure 1E). Analyses of RNA levels from stamens at pre- and post-anthesis stages indicated that the induction at later stages takes place in these organs (Figure 1F). Then, KNAT1 may act redundantly with STM to limit stamen growth at post-anthesis stages. In addition, we observed an induction of STM in stamens of the bp-1 mutant relative to wild-type (Supplemental Figure S1), which may partially compensate for the absence of KNAT1 expression. Altogether, these results may explain the mild phenotype of bp-1 plants.
Figure 1.
KNAT1 is expressed during late stamen development and inhibits stamen filament elongation. A, GUS histochemical staining of flowers from plants carrying the pKNAT1::GUS construct at different developmental stages. Flowers were incubated in 1 mM 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid, 50 mM sodium phosphate (pH 7.0), and 0.1% (v/v) Triton X-100 at 37°C during 24 h. Representative images are shown. Scale bars: 1 mm. B, Stamen length of bp-1 and 35S::KNAT1 flowers at Stage 13, compared with their corresponding wild-type lines (ecotypes Ler and Nossen [NO], respectively). C, Scanning electron microscope (SEM) images of 35S::KNAT1 and wild-type (Nossen) flowers, obtained as described in Gastaldi et al. (2020). Sepals and petals were removed to allow visualization of the stamens. Scale bars: 1 mm. D, Cell length in stamens of 35S::KNAT1 flowers at Stage 13, compared with the corresponding wild-type line. Cell length was analyzed in SEM images of the central portion of stamens (mean±SE; n = 59–67 cells from 7 stamens from different flowers). A representative image is shown on the right. Scale bars: 100 µm. E, Relative transcript levels of Class I KNOX genes in flowers at different developmental stages. Data were obtained from Ryan et al. (2015). F, Relative KNAT1, STM, and KNAT2 transcript levels in stamens from flowers at pre- and post-anthesis stages. KNAT6 expression could not be efficiently detected. G, Relative GA20OX1 transcript levels in 35S::KNAT1 and wild-type flowers at Stage 13, determined by RT-qPCR. H, Immunoblot analysis of protein extracts from Stage 13 bp-1 and wild-type (Ler) Stage 13 flowers with anti-RGA antibodies (Agrisera, Vännäs, Sweden; dilution 1:1,000). Flowers were treated with 10 µM PAC or mock solution 4 h before collecting them from the plants. Antibodies against cytosolic glyceraldehyde-3-phosphate dehydrogenase and Ponceau S staining were used for the control of loading. I, Stamen length of wild-type, 35S::KNAT1 and ga1 (SALK_109115) flowers at Stage 13 after treatment with either 100 µM GA3 or mock solution. In B and F, flowers were photographed using a Leica MZ10F stereomicroscope equipped with a digital camera and filament length was determined using Image J software. Only the four long stamens in each flower were considered. Bars indicate the mean±SE (n = 46–123 stamens, depending on the line). In F and G, bars indicate the mean±SE of three biological replicates. In B, D, F, and G, asterisks indicate significant differences with wild type (B, D, G) or with Stage 10 flowers (F) (P < 0.05; Student’s t test). In I, different letters indicate significant differences (P < 0.05; ANOVA).
Defects in the synthesis of gibberellins (GA) affect stamen filament elongation (Rieu et al., 2008). Class I KNOX proteins were implicated in the repression of the GA biosynthesis gene GA20OX1 in the shoot apical meristem (Hay et al., 2002). As shown in Figure 1G, KNAT1 overexpression caused a strong decrease in GA20OX1 transcript levels in flowers at Stage 13, suggesting that KNAT1 may repress stamen filament elongation by modulating GA biosynthesis. Accordingly, the amount of the DELLA protein RGA, which is negatively associated with GA levels (Silverstone et al., 2001), was lower in bp-1 than in wild-type flowers, while treatment with the GA synthesis inhibitor paclobutrazol strongly increased RGA levels in both backgrounds (Figure 1H). Notably, treatment of 35S::KNAT1 flowers with GA was ineffective in restoring stamen filament elongation, which is different from what is observed with the GA-deficient mutant ga1 (Figure 1I and Supplemental Figure S2). This implies that other factors are involved, even if a role of KNAT1 in GA metabolism during stamen development cannot be ruled out.
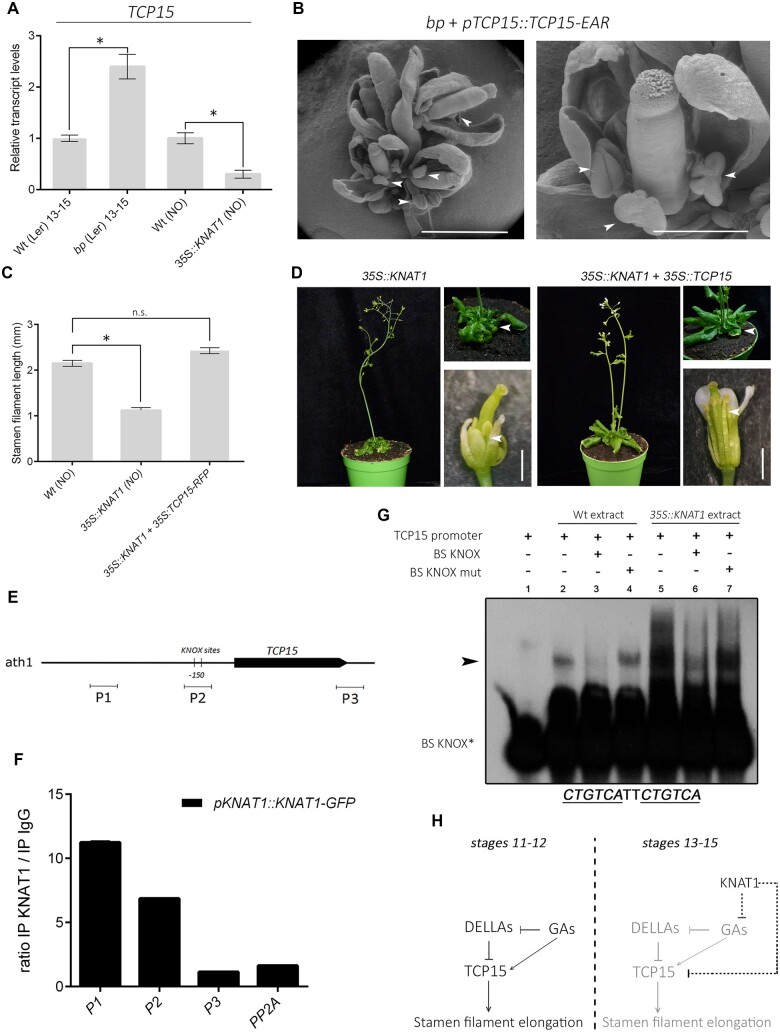
Recently, we described that members of the Class I TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR transcription factor family (TCP) stimulate late stamen filament elongation (Gastaldi et al., 2020). We then sought possible connections between KNAT1 and Class I TCPs. Transcript levels of TCP15, a Class I TCP gene, were reduced in 35S:KNAT1 plants and increased in bp-1 plants (Figure 2A). In addition, bp-1 plants that express a strong repressor form of TCP15 (TCP15-EAR; Uberti-Manassero et al., 2012) under the control of the TCP15 promoter showed short stamens (Figure 2B), while overexpression of TCP15 restored stamen filament elongation in 35S::KNAT1 plants (Figure 2, C and D). These results indicate that TCP15 most likely acts downstream of KNAT1. Interestingly, TCP15 overexpression rescued the short-stamen phenotype of 35S::KNAT1 plants but did not modify the development of crinkled leaves characteristic of these plants (Figure 2D), indicating that not all KNAT1 overexpression phenotypes depend on TCP15.
Figure 2.
KNAT1 targets TCP15 to modulate stamen filament elongation. A, TCP15 transcript levels in bp-1 and 35S::KNAT1 flowers at Stages 13–15, relative to their corresponding wild-type lines, determined by RT-qPCR. The bars indicate the mean±SE of three biological replicates. Asterisks indicate significant differences with wild-type plants (P < 0.05; Student’s t test). B, SEM images of flowers from plants that express TCP15-EAR under the control of the TCP15 promoter in a bp-1 background. Arrows point to stamens. Scale bars: 2 mm (left), 0.5 mm (right). C, Stamen length of Stage 13 flowers from wild-type, 35S::KNAT1 and 35S::KNAT1 35S::TCP15 plants. Bars indicate the mean±SE (n = 10–15 stamens, depending on the line). Asterisks indicate significant differences with wild-type plants (P < 0.05; Student’s t test). KNAT1 expression levels in the different lines are shown in Supplemental Figure S5. D, Representative images of 35S::KNAT1 and 35S::KNAT1 35S::TCP15 plants. Arrows in the upper and lower panel point to stamens and leaves, respectively. Scale bars: 1 mm. E, Scheme of the TCP15 gene. The pointed rectangle represents the transcribed region. The location of the TGACAG boxes (KNOX sites) is shown. P1–P3 indicate the regions amplified by oligonucleotides used in the ChIP analysis. F, ChIP analysis of the binding of KNAT1-GFP to the TCP15 promoter region. The PP2A gene was used as a negative control. The results are expressed as the ratio of the signal obtained after immunoprecipitation with anti-GFP antibodies and anti-IgG (control) and show the mean±SE of two technical replicates. A similar result was obtained in an additional, independent experiment. G, EMSA of the binding of proteins present in extracts of wild-type and 35S::KNAT1 plants to the TCP15 promoter region that contains the TGACAG boxes. “BS KNOX*” indicates the labeled double-stranded oligonucleotide that represents the TCP15 promoter region under analysis. “BS KNOX” and “BS KNOX mut” indicate unlabeled double-stranded oligonucleotides carrying either the native or the mutated TCP15 promoter region and were added in a 100-fold molar excess where indicated to test the specificity of binding. The arrow points to a specific retarded band observed upon addition of plant extracts. H, Scheme of the role of KNAT1 during late stamen filament elongation. Before Stage 13, TCP15 induces the expression of SAUR63 and related SAUR genes involved in stamen filament elongation (Gastaldi et al., 2020; left panel). Reactivation of KNAT1 expression at later stages represses TCP15 expression, leading to filament elongation arrest (right panel). KNAT1 may also restrict stamen filament elongation acting on gibberellin (GA)-dependent processes, which would also affect TCP15 activity.
Class I TCPs are inhibited by DELLA proteins and participate in the regulation of several processes by GA (Davière et al., 2014; Resentini et al., 2015; Lucero et al., 2017). TCP15 acts downstream of GA to stimulate stamen filament elongation, suggesting that GA may link KNAT1 to TCP15 action (Gastaldi et al., 2020). However, the fact that GA treatment did not rescue the short-stamen phenotype of 35S::KNAT1 plants while TCP15 overexpression did, led us to evaluate the existence of additional mechanisms, like the regulation of TCP15 expression at the transcriptional level. KNAT1 and TCP15 are expressed in similar stamen tissues, predominantly in the vascular system of filaments (Supplemental Figure S3). In addition, the TCP15 promoter has two adjacent putative KNOX binding sites (TGACAG) at about −500 bp from the transcription start site (Figure 2E). Similar sequences were previously shown to be essential for regulation of the maize (Zea mays L.) GA catabolism gene GA2ox1 and the Arabidopsis organ boundary gene CUC1 by Class I KNOX proteins (Bolduc and Hake, 2009; Spinelli et al., 2011), raising the possibility that TCP15 is a direct KNAT1 target. Therefore, we assessed binding of KNAT1 to the TCP15 promoter in vivo by chromatin immunoprecipitation (ChIP) using pKNAT1::KNAT1-GFP plants (Zhao et al., 2015). A TCP15 promoter fragment that includes the TGACAG sites (−537/−406) and an additional fragment located further upstream (−1806/−1685) were preferentially immunoprecipitated by anti-GFP antibodies, unlike fragments located downstream of the TCP15 coding region or within the PP2A gene, used as negative controls (Figure 2F). Further analysis of the TCP15 promoter region showed the presence of sequences with similarity to TGACAG at −1905 (TGACTG), −1801, and −1579 (both TGACAA), which may explain the observed interaction with the upstream promoter region. Nevertheless, the results show that KNAT1 binds to the TCP15 promoter in vivo. In addition, we performed an electrophoretic mobility shift assay (EMSA) with a fragment representing the region containing the TGACAG sites at −500 and extracts prepared from wild-type or 35S::KNAT1 plants. A specific retarded band (Figure 2G) was observed when using extracts from wild-type and 35S::KNAT1 plants. Moreover, the intensity of the retarded band was substantially enhanced with extracts from 35S::KNAT1 plants (Figure 2G). This implies that either KNAT1 itself or a transcription factor that is induced or activated by KNAT1 overexpression binds to the TCP15 promoter. Further analysis indicated that binding was efficiently competed by an oligonucleotide containing the KNOX binding sites but not by a similar oligonucleotide in which these sites were mutated (Figure 2G), strongly suggesting that KNAT1 is the protein that binds to this region of the TCP15 promoter. Altogether, the ChIP and EMSA results indicate that TCP15 is directly regulated by KNAT1.
In summary, we uncovered a role of KNAT1 during late stages of stamen filament elongation and identified TCP15 as a direct KNAT1 target. Reactivation of KNAT1 expression in elongated filaments probably plays a role in limiting growth during post-anthesis phases. Tashiro et al. (2009) showed that stamen growth is accelerated about 10-fold during late pre-anthesis stages and then slows down to accommodate pistil growth when the stamen reaches the top of the pistil. We postulate that KNAT1 participates in this process through direct repression of TCP15 and, possibly, through changes in GA and DELLA protein levels (Figure 2H). The relevance of limiting stamen growth after anthesis remains unclear. We analyzed the number of seeds per silique in the bp-1 mutant along two different experiments and found a slight but significant decrease in this parameter in the bp-1 mutant relative to wild type in one of the experiments (Supplemental Figure S4A). Considering that the bp-1 mutant shows slight phenotypic alterations probably due to redundancy with other Class I KNOX genes, we also analyzed two lines of 35S::TCP15 plants, which also show increased stamen growth (Gastaldi et al., 2020). Differences in the mean number of seeds per silique were more pronounced in this case (Supplemental Figure S4B), suggesting that limiting stamen growth may have an effect on seed production. Further analysis is required to support this hypothesis and to evaluate if the differences in seed production are relevant for plant fitness and reproductive efficiency.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression levels of Class I KNOX genes in stamens from bp-1 flowers.
Supplemental Figure S2. Flowers of wild-type, 35S::KNAT1 and ga1 plants after treatment with either gibberellin (GA3) or mock solution.
Supplemental Figure S3. Histochemical analysis of KNAT1 and TCP15 expression in stamen filaments.
Supplemental Figure S4. Seed production in siliques from bp-1 and 35S::TCP15 plants.
Supplemental Figure S5. KNAT1 expression levels in wild-type, 35S::KNAT1 and 35S::KNAT1 35S::TCP15 plants.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Arabidopsis Biological Resource Center, Xuncheng Liu, Keqiang Wu, and Stephen Thomas for sending seeds of lines used in this study.
Funding
This work was supported by a grant from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, Argentina; PICT-2019-01204). F.D.A., I.L.V., L.E.L., and D.H.G. are members of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina). V.G. and A.L.A. are CONICET fellows. N.M. is an ANPCyT fellow.
Conflict of interest statement. None declared.
Contributor Information
Victoria Gastaldi, Instituto de Agrobiotecnología del Litoral (CONICET-UNL), Cátedra de Biología Celular y Molecular, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina.
Antonela L Alem, Instituto de Agrobiotecnología del Litoral (CONICET-UNL), Cátedra de Biología Celular y Molecular, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina.
Natanael Mansilla, Instituto de Agrobiotecnología del Litoral (CONICET-UNL), Cátedra de Biología Celular y Molecular, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina.
Federico D Ariel, Instituto de Agrobiotecnología del Litoral (CONICET-UNL), Cátedra de Biología Celular y Molecular, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina.
Ivana L Viola, Instituto de Agrobiotecnología del Litoral (CONICET-UNL), Cátedra de Biología Celular y Molecular, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina.
Leandro E Lucero, Instituto de Agrobiotecnología del Litoral (CONICET-UNL), Cátedra de Biología Celular y Molecular, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina.
Daniel H Gonzalez, Instituto de Agrobiotecnología del Litoral (CONICET-UNL), Cátedra de Biología Celular y Molecular, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina.
L.E.L. and D.H.G. designed experiments. V.G. and A.L.A. performed the experiments. N.M. participated in Western blot and reporter expression analysis. F.D.A. designed and supervised the ChIP experiment. I.L.V. designed and supervised the EMSA experiment. All authors analyzed data. V.G. made the figures. D.H.G. wrote the manuscript with contribution from all authors. All authors accepted the final version of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Daniel H. Gonzalez (dhgonza@fbcb.unl.edu.ar).
References
- Bolduc N, Hake S (2009) The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli M, Cecchetti V (2014) Auxin polar transport in stamen formation and development: how many actors? Front Plant Sci 5: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928 [DOI] [PubMed] [Google Scholar]
- Gastaldi V, Lucero LE, Ferrero LV, Ariel FD, Gonzalez DH (2020) Class-I TCP transcription factors activate the SAUR63 gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol 182: 2096–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Lucero LE, Manavella PA, Gras DE, Ariel FD, Gonzalez DH (2017) Class I and class II TCP transcription factors modulate SOC1-dependent flowering at multiple levels. Mol Plant 10: 1571–1574 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Resentini F, Felipo-Benavent A, Colombo L, Blázquez MA, Alabadí D, Masiero S (2015) TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol Plant 8: 482–485 [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D (2013) Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet 9: e1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PT, Ó'Maoiléidigh DS, Drost HG, Kwaśniewska K, Gabel A, Grosse I, Graciet E, Quint M, Wellmer F (2015) Patterns of gene expression during Arabidopsis flower development from the time of initiation to maturation. BMC Genomics 16: 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SV, Martin AP, Viola IL, Gonzalez DH, Palatnik JF (2011) A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol 156: 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S (2010) Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Tashiro S, Tian CE, Watahiki MK, Yamamoto KT (2009) Changes in growth kinetics of stamen filaments cause inefficient pollination in massugu2, an auxin insensitive, dominant mutant of Arabidopsis thaliana. Physiol Plant 137: 175–187 [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH (2012) The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63: 809–823 [DOI] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA 99: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Xu WH, Ma LG, Fu ZM, Deng XW, Li JY, Wang YH (2006) Requirement of KNAT1/BP for the development of abscission zones in Arabidopsis thaliana. J Integr Plant Biol 48: 15–26 [Google Scholar]
- Zhao M, Yang S, Chen CY, Li C, Shan W, Lu W, Cui Y, Liu X, Wu K (2015) Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet 11: e1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.