Abstract
Medicago (Medicago truncatula) establishes a symbiosis with the rhizobia Sinorhizobium sp, resulting in the formation of nodules where the bacteria fix atmospheric nitrogen. The loss of immunity repression or early senescence activation compromises symbiont survival and leads to the formation of nonfunctional nodules (fix−). Despite many studies exploring an overlap between immunity and senescence responses outside the nodule context, the relationship between these processes in the nodule remains poorly understood. To investigate this phenomenon, we selected and characterized three Medicago mutants developing fix− nodules and showing senescence responses. Analysis of specific defense (PATHOGENESIS-RELATED PROTEIN) or senescence (CYSTEINE PROTEASE) marker expression demonstrated that senescence and immunity seem to be antagonistic in fix− nodules. The growth of senescence mutants on non-sterile (sand/perlite) substrate instead of sterile in vitro conditions decreased nodule senescence and enhanced defense, indicating that environment can affect the immunity/senescence balance. The application of wounding stress on wild-type (WT) fix+ nodules led to the death of intracellular rhizobia and associated with co-stimulation of defense and senescence markers, indicating that in fix+ nodules the relationship between the two processes switches from opposite to synergistic to control symbiont survival during response to the stress. Our data show that the immune response in stressed WT nodules is linked to the repression of DEFECTIVE IN NITROGEN FIXATION 2 (DNF2), Symbiotic CYSTEINE-RICH RECEPTOR-LIKE KINASE (SymCRK), and REGULATOR OF SYMBIOSOME DIFFERENTIATION (RSD), key genes involved in symbiotic immunity suppression. This study provides insight to understand the links between senescence and immunity in Medicago nodules.
Analyses of Medicago mutants with non-functional nodules highlight the relationship and mechanisms controlling the establishment of the immune and senescence programs during nodule organogenesis.
Introduction
Under nitrogen starvation, the legume plant Medicago (Medicago truncatula) is able to perform a symbiotic association with the soil nitrogen-fixing bacteria Sinorhizobium sp. During this interaction a root organ, the nodule, is formed (Oldroyd, 2013). Medicago produces indeterminate nodules characterized by the presence of a persistent meristem at the apex (zone I; ZI) responsible for nodule growth. Below ZI, in the infection zone (zone II; ZII), the rhizobia infect the plant cells. Thanks to the action of the NODULE-SPECIFIC CYSTEINE-RICH (NCR) antimicrobial peptides produced by the host plant, a differentiation process occurs in ZII leading to an increase in size and genome endoreduplication of the bacteroids (Mergaert et al., 2006; Van de Velde et al., 2010). In the fixation zone (zone III; ZIII), the differentiated bacteroids convert atmospheric nitrogen into an organic form assimilated by the plant (Paau et al., 1980).
Despite the massive invasion of the rhizobia, the symbiotic nodule cells do not produce apparent defense reactions (Gourion et al., 2015). Thanks to direct genetic studies of the Medicago–rhizobium interaction, several genes that regulate defense responses in nodules have been isolated (Kang et al., 2016; Berrabah et al., 2018b) including the DEFECTIVE IN NITROGEN FIXATION 2 (DNF2, Bourcy et al., 2013), Symbiotic CYSTEINE-RICH RECEPTOR-LIKE KINASE (SymCRK, Berrabah et al., 2014b), REGULATOR OF SYMBIOSOME DIFFERENTIATION (RSD, Sinharoy et al., 2013), and NODULES WITH ACTIVATED DEFENSE 1 (NAD1, Wang et al., 2016; Yu et al., 2018) that encode, respectively, a PHOSPHATIDYL INOSITOL SPECIFIC PHOSPHOLIPASE C-LIKE PROTEIN, a CYSTEINE-RICH RECEPTOR-LIKE KINASE, a C2H2 TRANSCRIPTION FACTOR and a protein acting positively in the maintenance of the bacteroids. Medicago mutants for these genes produce non-fixing nitrogen (fix−) nodules exhibiting necrotic tissues with typical defense features like phenolic compounds accumulation and stimulation of defense genes. The activation of these immune responses results in the death of undifferentiated bacteroids (Bourcy et al., 2013; Sinharoy et al., 2013; Berrabah et al., 2014b; Wang et al., 2016). Historically, the first up-regulated defense gene was identified in dnf2 and corresponds to the PATHOGENESIS-RELATED PROTEIN 10 (PR10, Medtr2g035150.1, Bourcy et al., 2013). This PR10 belongs to the PR gene family linked to plant–pathogen responses (Ali et al., 2018) and is stimulated in the nodules of necrotic mutants. Based on the diversity of their biochemical activities, the PR proteins can be classified into 17 groups (van Loon et al., 2006; Sels et al., 2008). Within each group, members share a specific protein domain used for the PR classification. For example, the PR1, PR5, PR8, and PR10 members include, respectively, a CYSTEINE-RICH SECRETORY PROTEIN (CAP; Schreiber et al., 1997), THAUMATIN-LIKE (Wang et al., 2010), CHITINASE TYPE III (Métraux et al., 1988), and BET V1 DOMAIN proteins (Liu and Ekramoddoullah, 2006).
During symbiosis, DNF2, SymCRK, RSD (Berrabah et al., 2015) and potentially NAD1 (Domonkos et al., 2017) act sequentially to prevent the immune response in nodules. Different factors can influence the stimulation of defenses in these mutants including the environment that can change the defense response of nodules after rhizobium internalization. For example, the Medicago dnf2 mutant grown on agarose-based medium loses the immune responses and restores nitrogen fixation (fix+). The addition of the plant defense elicitor ulvan (Jaulneau et al., 2010) to this agarose-based medium primes defense responses and dnf2 recovered the fix− phenotype (Berrabah et al., 2014a).
Nodule senescence is also an important process that controls nodule functioning and bacteroid survival. Early activation of the senescence results in bacteroid death (Berrabah et al., 2015) and nitrogen-fixing inability of the nodules (Zimmerman et al., 1983). This phenomenon can be triggered in WT nodules during developmental aging (Van de Velde et al., 2006), by the addition of a nitrogen source (e.g. nitrate) to the growth substrate (Chen and Phillips, 1977) or by the treatment of nodulated plants with the herbicide phosphinothricin that inhibits the glutamine synthase (Seabra et al., 2012). Moreover, the suppression of genes involved in essential nodule functions like iron transport (Walton et al., 2020), sulfate transport (Krusell et al., 2005), or implicated in the nitrogen fixation (Oke and Long, 1999; Maunoury et al., 2010) may also result in the formation of fix− nodules with early senescence features. During the senescence, the formation of a senescent zone (zone IV; ZIV) is observed at the base of the M. truncatula nodules in which bacteria and host cells are degraded (Van de Velde et al., 2006). In the ZIV, cellular compounds are recycled thanks to CYSTEINE PROTEASES (CPs, Wyk et al., 2014) such as CP2 to CP6 (Pérez Guerra et al., 2010) belonging to the papain cysteine protease family (Pierre et al., 2014). These CP genes are specifically expressed in senescent nodules (Fedorova et al., 2002) and the corresponding proteins are involved in proteolytic activities (Malik et al., 1981; Pladys and Vance, 1993). CPs can be inhibited by PHYTOCYSTATINS, which are proteins involved in the control of cellular proteolytic activities during various developmental processes (Martínez et al., 2012; Díaz-Mendoza et al., 2014). Members of the PHYTOCYSTATIN gene family are indeed induced during nodule senescence in soybean (Glycine max, Wyk et al., 2014) and Medicago (Lambert et al., 2020).
The interconnection between immunity and senescence in legume nodules is poorly studied, in contrast to leaves where a co-activation of these process is observed in many species (Zhang et al., 2016, 2020; Patharkar et al., 2017; Lee et al., 2018; Ma et al., 2018a; Kusch et al., 2019) including soybean in which analysis of leaf senescence revealed the expression of defense-related genes (Gupta et al., 2016).
Here, we investigated the relationship between nodule immunity and senescence using different substrates and Medicago mutants producing fix− nodules or in WT fix+ nodules exposed to wounding stress. Our results show versatile behaviors of immunity and senescence relationship between fix− and fix+ condition, opposite and co-activation of these processes are observed in, respectively, fix− nodules and during fix+ stress responses and both correspond to bacteroids suppression. Furthermore, we observed that growth substrate composition also affects defenses and senescence stimulation in fix− nodules.
Results
Medicago fix− mutants used to study the defense and senescence interaction in nodules
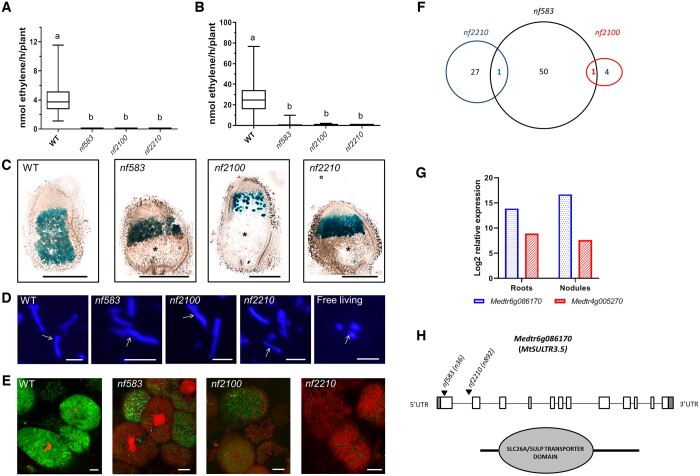
To study the relationship between defense and senescence, we use three mutants (nf583, nf2100, and nf2210) developing fix− nodules selected from a forward genetic screen of Medicago (M. truncatula) Tnt1 insertion mutant collection of the Noble Research Institute (https://medicago-mutant.dasnr.okstate.edu/mutant/index.php, Pislariu et al., 2012; Yarce et al., 2013). Nodule nitrogenase activity was measured in plants cultivated in vitro on an agar-gelified medium (Figure 1A) and in sand/perlite in a non-sterile growth chamber (Figure 1B) using the acetylene reduction assay. Nitrogenase activity was not detected in these Tnt1 mutant plants, confirming the fix− status of the nodules. To investigate the senescence feature of the symbiotic organ, 14 d post inoculation (dpi) nodule sections were prepared from plant inoculated with the S. medicae strain WSM419 constitutively expressing lacZ (Figure 1C). Bacteroids are present above a large senescence zone (ZIV) in the nodules of the mutants compared to the wild-type (WT). We will refer to these fix− mutants as senescence mutants in contrast to dnf2-4 and symCRK mutants, also used in this study and producing necrotic fix− nodules (Berrabah et al., 2015).
Figure 1.
New M. truncatula fix− mutants producing early senescent nodules. A, Nodules of three M. truncatula Tnt1 lines (nf583, nf2100, and nf2210) show no nitrogenase activity in vitro at 21-dpi and (B) in sand/perlite at 24-dpi growing conditions. The results are represented by boxplots showing the mean nitrogenase activity of three independent experiments with 14 plants per experiment. The central line of the box shows the median, the box limits display the upper and the lower quartiles, the whiskers show the minimum and the maximum values. The letters show statistical groups between genotypes using Student’s t test (P < 5%). C, Analysis of 17-dpi in vitro nodules infected with S. medicae WSM419 strain expressing lacZ reveals a senescent zone in the nodules of the three fix− mutants (nf583, nf2100, nf2210). The asterisk indicates the senescent zone and the scale bars represent 500 µm. D, Exploration of the bacterial differentiation with the DAPI staining of bacteroids extracted from 17-dpi in vitro nodules of WT or the isolated fix− mutants reveals an increase of the bacteroid size (corresponding to bacteroid differentiation) in nf2100, nf2210, nf583, and WT nodules compared to free living bacteria. The arrows show the bacteroids, the scale bars indicate 5 µm. E, The differentiation state is confirmed by the live and dead staining of 18-dpi in vitro nodule sections of the WT and the mutants. In this staining method, living rhizobia are stained in green with SYTO9 whereas dying cells are stained in red with PI. Death of differentiated bacteroids is observed in nf2100, nf2210, and nf583. The images were taken in the fixation zone (WT) or the putative ZIII (mutants). The scale bars indicate 10 µm. The observations in D and E are realized on plants of three independent experiments (eight plants per experiment). F, The identification of the FSTs reveals the presence of 52, 28, and 5 genes showing a Tnt1 insertion in an ORF for nf583, nf2210, and nf2100, respectively. One gene is shared between nf583 and nf2210 (Medtr6g086170) and one between nf583 and nf2100 (Medtr4g005257). G, The expression analysis of the common genes in roots and nodules of M. truncatula shows that Medtr6g086170 is induced, while Medtr4g005257 is down-regulated in the nodules compared to the roots. Expressions were determined from three independent experiments and the data are downloaded from Genevestigator. H, Medtr6g086170 encodes the sulfate transporter MtSULTR3.5 and displays Tnt1 insertions in first exon (+36) and intron (+892) in nf2210 and nf583 mutant lines, respectively.
To study the bacteroid differentiation state in the senescence mutants, we performed a DAPI staining on bacteroids extracted from WT or fix− mutant nodules (Figure 1D). Differentiated bacteroids were detected in the nodules of these senescence mutants. Furthermore, the intracellular survival of the endosymbionts was studied using the live/dead staining based on a mixture of two fluorescent probes, SYTO9 and Propidium Iodide (PI). nf583, nf2100, and nf2210 mutants displayed differentiated dead (red) bacteroids compared to the WT (Figure 1E). This staining further confirms the differentiation of the bacteroids in the senescence mutants. Altogether, our analyses indicated that nf583, nf2100, and nf2210 develop early senescent fix− nodules eliciting premature death of the differentiated bacteroids.
nf583 and nf2210 display Tnt1 insertions in the sulfate transporter MtSULTR3.5 that shows high expression in nodules
To identify the potential genes responsible for the nf583, nf2100, and nf2210 phenotypes, we searched for their Tnt1 flanking sequence tags (FSTs) in the Medicago Tnt1 mutant database (https://medicago-mutant.dasnr.okstate.edu/mutant/index.php). In order to increase the probability of selecting the genes responsible of the mutant phenotypes, we focused our analysis on the FSTs with high confidence and located in the open reading frames (ORF). Using this approach, 52, 28, and 5 tagged-genes were, respectively, identified for nf583, nf2210, and nf2100. Interestingly, the gene Medtr6g086170 (coding a SULFATE TRANSPORTER) is tagged with Tnt1 in lines nf583 and nf2210. Similarly, the gene Medtr4g005270 (coding a BETA-AMYRIN SYNTHASE) is tagged with Tnt1 in line nf2210 and nf2100 (Figure 1F; Supplemental Table S1). These two genes represent potential candidates for the symbiotic genes tagged in these mutant lines. Expression analysis using the Genevestigator database (https://genevestigator.com/; Hruz et al., 2008; Supplemental Table S1) revealed that Medtr6g086170 shows high expression in the WT nodules (Figure 1G) and especially in the zone III (https://medicago.toulouse.inrae.fr/GEA). In contrast, all the other tagged-genes with available expression data (Supplemental Table S1) including the nf2100 tagged-genes, display low variations of their expression or reduced expression in the nodule compared to the roots. Medtr6g086170 corresponds to the SULFATE TRANSPORTER MtSULTR3.5 and the sequence analysis of the associated Tnt1 insertions in the nf583 and nf2210 backgrounds reveal insertions in the first exon (+36) and first intron (+892), respectively (Figure 1H). The polymerase chain reaction genotyping confirms the Tnt1 insertion in nf583 and nf2210, moreover the mutant plants are homozygous for the mutations in MtSULTR3.5 (Supplemental Figure S1). Our data suggest that the insertions in MtSULTR3.5 are potentially responsible for the nf583 and nf2210 phenotypes.
PATHOGENESIS-RELATED (PR) genes are key markers for the assessment of nodule immunity
In order to define appropriate defense markers for the evaluation of nodule immunity, we focused our attention on the PR gene family associated with plant responses against pathogens. Genomic data mining was done using key words and blast search on two databases: phytozome and M. truncatula A17 r5.0 genome portal (see Materials and methods). This analysis revealed the presence of 106 PR genes in the Medicago genome of which PR5, PR10, and PR1 are the most represented groups with 44, 35, and 16 members, respectively (Supplemental Table S2).
To select PR candidates for nodule defense studies, the expression of the identified PR genes was analyzed using data from the M. truncatula Gene Expression Atlas (MtGEA) database (https://medicago.toulouse.inrae.fr/MtExpress) after identification of the corresponding probe sets (Supplemental Table S2). We noticed that genes showing hybridization signals (HS) values lower than 100 are usually not reproducible in the quantitative polymerase chain reaction (qPCR) analysis in our laboratory conditions. Thus, in order to select PR genes with robust expression, a filtering step was applied and the probesets displaying HS lower than 100 in both test and control conditions were excluded.
With the aim to identify PR genes potentially participating to the nodule physiology, we further selected PR genes expressed in the symbiotic organ with or without senescence stimulation. Based on the MtGEA profiles, eight different probesets (Supplemental Table S3) were selected, corresponding to PR genes induced at least two folds in the symbiotic context (WT nodules versus roots, Supplemental Figure S2A) and/or in nodules of WT plants treated with either nitrate (KNO3; Supplemental Figure S2A; Benedito et al., 2008) or phosphinothricin (Supplemental Figure S2B; Seabra et al., 2012) compared to controls. This resulted in the selection of 12 PR genes belonging to the PR5, PR8, PR10, and non-determined classes.
To check the expression of the selected PR genes in our conditions, we then examined their expression by reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis using cDNA of Medicago nodules. Expression was detected for 10 PR genes (Supplemental Table S3). The expression of these PR genes was then evaluated in nodules collected from dnf2-4 and symCRK mutants displaying exacerbated defense reactions in the symbiotic organ (Supplemental Figure S3). Five PR genes (one PR8, two PR5, and two PR10) showed a significant induction in dnf2-4 and symCRK compared to the WT and were finally selected to study defense activation in nodules. PR5.3, PR8, and PR10 are stimulated in nodules infected by the root pathogen Ralstonia solanacearum (Benezech et al., 2020), supporting the choice of these markers for defense tracking in the nodules. It is also worth noting that the sequence analysis of the identified PR10 (PR10.2 and PR10.3, Supplemental Table S3) revealed the same coding direct sequence (CDS) despite different chromosome locations (chromosomes 4 and 6, Supplemental Figure S4). As we could not discriminate PR10.2 and PR10.3 expressions by RT-qPCR, we commonly named these genes PR10 in the manuscript.
PR and CP genes show distinct expression patterns
In addition to the selected PR genes used to assess immunity activation in the nodule, the expression of four typical senescence markers (CP2, 3, 4, and 5) was monitored to follow nodule senescence stimulation. To estimate the overlap between PR and CP gene expressions in Medicago, the expression of corresponding genes was compared in different physiological contexts using the Genevestigator software. Expression analysis at different developmental stages revealed a high expression level of the PR genes in the whole plants until the beginning of the flowering stage (Supplemental Figure S5). The initiation of flowering is associated with a reduction in most of the PR gene expressions. In contrast, the CP genes show low to medium expression levels throughout the life cycle of the plants and they are less expressed than PR genes.
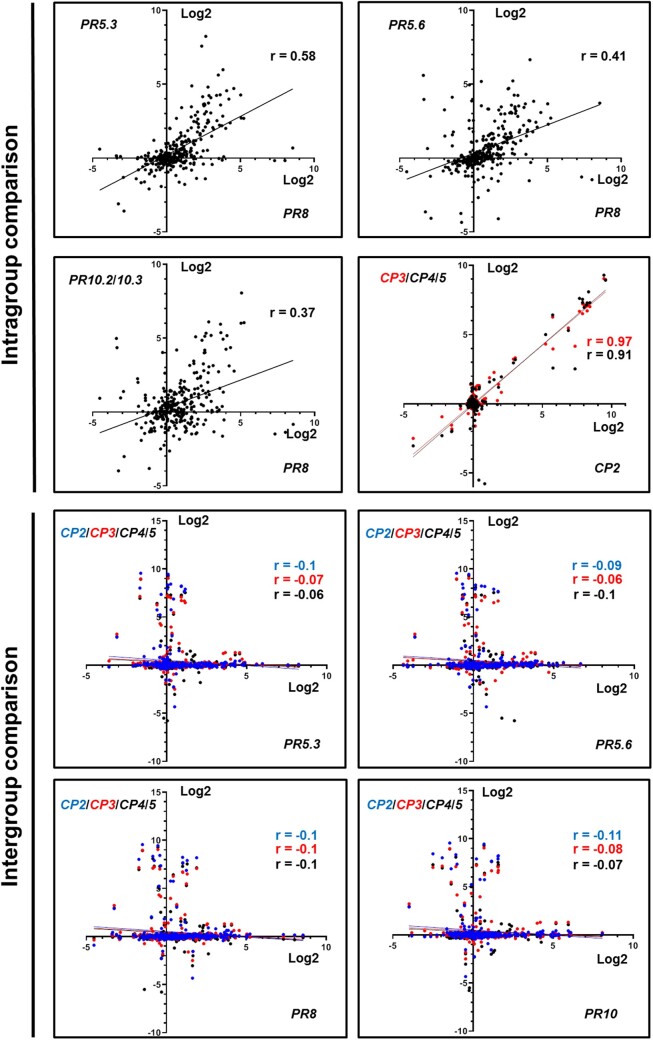
In order to compare PR and CP gene expressions in response to different biotic and abiotic elicitations (“perturbation set”, Genevestigator), we used a scatter blot analysis (Figure 2). Comparison of the PR or CP gene expression patterns revealed substantial number of conditions showing co-expression of the genes in the same group (Figure 2, intragroup comparison). In contrast, the comparison of PR to CP expressions revealed low level of expression overlap (Figure 2, intergroup comparison). Pearson analysis (Figure 2; Supplemental Table S4) showed correlation of 0.91–0.97 for CPs and 0.37–0.65 for PR genes, whereas a strong reduction of the correlations (−0.06 to −0.11) was observed when PR and CP gene expressions were compared. Together these results indicate that PR and CP genes display distinct expression patterns in Medicago.
Figure 2.
Selected PR and CP display distinct pattern expressions. Scatterplot analysis of PR or CP (intragroup comparison) and PR versus CP (intergroup comparison) expression. The results display the average of the gene expressions in log2 of ratio between the test and the control obtained for 290 conditions of perturbations (response to abiotic and biotic stress, symbiosis, elicitors, defense hormones, seeds development, effect of the nutrients, and the genotypes). The data are downloaded and analyzed using the Genevestigator database. The r numbers show the Pearson correlation values between the genes. PRs are represented in black. CP2, CP3 and CP4/5 are represented in blue, red, and black.
Expression analysis of PR and CP genes reveals an opposite behavior between senescence and immunity in nodules of nf583 and nf2210 mutants
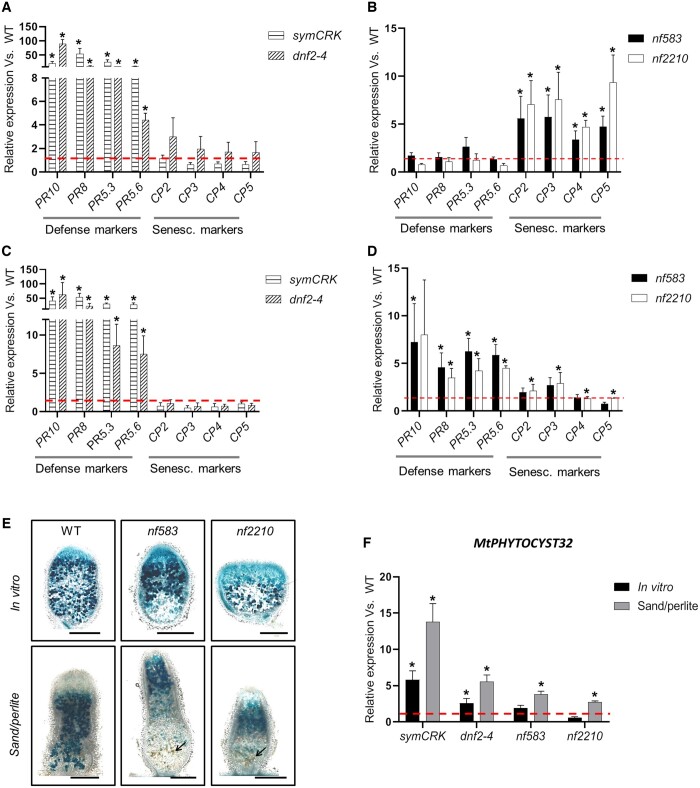
To evaluate the interconnection between immunity and senescence in nodules, the expression of the selected PRs and CP 2 to 5 genes, was evaluated in mutants cultivated in vitro and producing nodules with exacerbated defenses (dnf2-4 and symCRK; Figure 3A) or displaying early senescence (nf583 and nf2210; Figure 3B). Due to a distinct behavior, nf2100 is discussed in a dedicated section. The PR genes were highly expressed in symCRK and dnf2-4 nodules compared to the WT (Figure 3A). In the nodules of the nf583 and nf2210 early senescence mutants, the PR expressions remained low (Figure 3B). Unlike the defense markers, the CP genes were expressed at low level in nodules of the necrotic mutants compared to the WT even if a slight but not statistically significant induction of CP genes was observed in dnf2-4 (Figure 3A). In contrast, the expression of all CP genes was induced in nf583 and nf2210 compared to the control (Figure 3B).
Figure 3.
Senescence and immunity activation in fix− mutants. A, Analyses of 21-dpi nodules of plant cultivated in vitro reveal induction of all selected PRs in dnf2-4 and symCRK compared to the WT. In contrast, no significant variation of CP expressions is observed in the same condition. B, In the opposite to dnf2-4 and symCRK, PR expression is largely reduced in the nodules of the nf583 and nf2210 senescence mutants, while expression of used CPs is increased in these mutants compared to the WT. C, Analysis of 24-dpi nodules from fix− plants cultivated in sand/perlite reveals the stimulation of PRs and reduction of CPs expression in dnf2-4, symCRK. D, In the same way, nf583 shows upregulation of all PRs and nf2210 displays significant stimulation of PR8, PR5.3, PR5.6. CPs expression is reduced in the senescence mutants, nf583 and nf2210, which show stimulation of CP2/CP3 and CP4/CP5, respectively. E, Analysis of 21-dpi in vitro and sand/perlite nodules induced by S. medicae WSM419 lacZ reveals that nf583 and nf2210 produce senescent nodules without necrosis in vitro. In contrast, in sand/perlite a slight necrosis is observed and the arrows show necrotic cells. The scale bars represent 500 µm. F, Expression analysis of the PHYTOCYSTATIN32 (MtPHYTOCYST32) CP inhibitor in fix− mutant nodules compared to the WT cultivated, respectively, in vitro (21-dpi) or in sand/perlite (24-dpi) reveals induction of PHYTOCYSTATIN32 in dnf2-4 and symCRK in vitro, whereas the cultivation of plants in sand/perlite shows upregulation of PHYTOCYSTATIN32 in the necrotic mutants as well as in nf583 and nf2210. Values represent the mean of induction folds in the tested mutants compared to the WT, dashed lines represent the value of gene expression in the WT. The RT-qPCR analyses in (A)–(D) and (F) were made on three biological repetitions with two technical replicates. For each experiment, 16 and 5 plants were analyzed for in vitro and sand/perlite conditions, respectively. The actin housekeeping gene was used for expression normalization. Error bars indicate se and the asterisks represent significant variations compared to the WT using Mann–Whitney statistical test (P < 2.5%).
Together these data indicate an opposite behavior between immunity and senescence markers in nodules of in vitro cultured mutants. No overexpression of defense genes was observed during senescence while no induction of senescence markers occurred in nodules showing defense responses.
The balance between defense and senescence is influenced by the environment
To assess whether more complex conditions can have an impact on the immune and/or senescence status of nodules, expressions of PR and CP genes were analyzed in fix− mutants cultivated on sand/perlite (Figure 3, C and D). This non-sterile substrate displays more elicitors than the cleaner agar-jellified medium used for in vitro culture (Berrabah et al., 2014a). PR gene induction levels were similar in nodules of symCRK and dnf2-4 cultivated in sand/perlite compared to those observed during in vitro culture (Figure 3A versus Figure 3C). Interestingly, nf583 and nf2210 showed an increased expression of all PR genes in the sand/perlite contrary to that of agar-jellified media (Figure 3B versus Figure 3D). In contrast to PR gene induction, CP gene expression levels were reduced in the nodules of the senescence mutants grown on sand/perlite (Figure 3D) compared to in vitro conditions (Figure 3B). In addition, the analysis of nf583 and nf2210 nodules inoculated with the S. medicae lacZ strain revealed some necrotic cells at 21-dpi in the sand/perlite conditions whereas no necrosis was observed in vitro (Figure 3E).
Together these data suggest that, in contrast to in vitro conditions, when fix− mutants are cultivated on a non-sterile sand/perlite substrate, plant defense responses are activated in the nodules rather than senescence.
Expression pattern of MtPHYTOCYST32 supports the hypothesis of the opposite relationship between defense and senescence in fix− nodules
To test the hypothesis of a reduction of the senescence during defense activation in fix− nodules, we identified potential CP inhibitors acting during nodule defense response. For this purpose, co-expressed genes with PR5.3, PR5.6, PR8, and PR10 were isolated using the Phytomine tools of the Phytozome database (https://phytozome.jgi.doe.gov/phytomine/begin.do) and the genes with a Pearson correlation higher than 0.85 were selected. This analysis uncovered two PHYTOCYSTATINS encoded by the Medtr2g026040 (MtPHYTOCYST5) and Medtr5g088770 (MtPHYTOCYST32; PHYTOCYST32) genes co-expressed with PR5.3/PR5.6/PR10 and PR5.3/PR10, respectively, (Supplemental Table S5). Expression analysis of these PHYTOCYSTATINS in the nodules of fix− mutants revealed that MtPHYTOCYST5 was weakly but significantly down-regulated in symCRK and dnf2-4 in vitro (Supplemental Figure S6A) and in sand/perlite (Supplemental Figure S6B). In addition, no significant variation in the senescence mutants was observed in vitro and in sand/perlite, except for nf2210 showing a small repression of MtPHYTOCYST5 expression in vitro (Supplemental Figure S6A). In contrast, MtPHYTOCYST32 was induced in symCRK and dnf2-4 cultivated in vitro, whereas low or no induction was detected in nf583 and nf2210, respectively, compared to the reference (Figure 3F). Nodule MtPHYTOCYST32 expression increased in all fix− mutants compared to the WT (Figure 3F) when plants were grown on sand/perlite. The MtPHYTOCYST32 expression pattern agrees with the hypothesis of a reduction of senescence during the defense activation in fix− nodules. Furthermore, the behaviors of MtPHYTOCYST5 suggest that only some members of the PHYTOCYSTATINS family are stimulated during the nodule immunity.
nf2100 displays a complex phenotype contrasting with the other senescence mutants
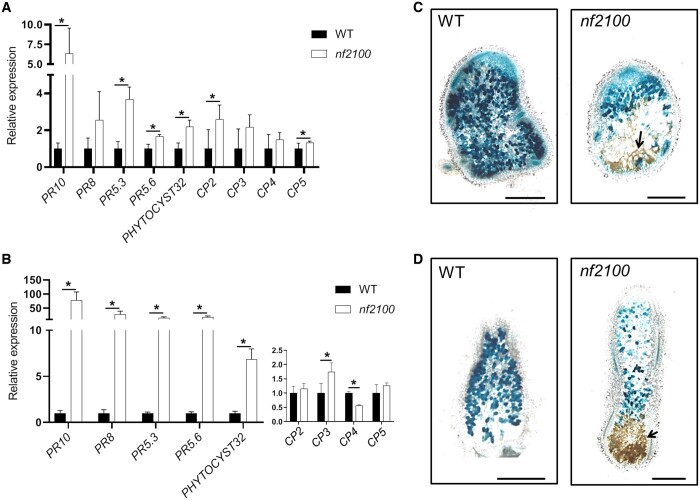
The expression of defense and senescence markers was studied on nf2100 nodules of plants cultivated in vitro (Figure 4A). Surprisingly, despite the formation of nodules with typical senescence features, nf2100 exhibits a much higher expression of defense than senescence markers. The increased expression of all PR genes together with the MtPHYTOCYSTATIN32 was observed in this mutant (Figure 4A). In contrast, CP genes showed low level of expression compared to those observed in nf583 and nf2210 (Figure 4A versus Figure 3B) with only two CP genes (CP2 and CP5) showing significant upregulation compared to the WT (Figure 4A). Cultivation of nf2100 in sand/perlite strongly increased all PRs expression in nodules (Figure 4B) with levels similar to those observed in dnf2-4 and symCRK (Figure 4B versus Figure 3C). Moreover, nf2100 displays slight induction of CP3 and repression of CP4 (Figure 4B) in sand/perlite substrate. The analysis of the necrosis in 21-dpi nodules of nf2100 grown in vitro reveals the presence of reduced necrotic zones (Figure 4C), which are greatly enlarged in sand/perlite (Figure 4D).
Figure 4.
nf2100 produces senescent nodules with higher immunity stimulation than the other senescence mutants. A, Expression analysis of PRs and CPs in 21-dpi nodules of WT or nf2100 cultivated in vitro shows the upregulation of these genes in nf2100. B, Cultivation of nf2100 or the WT on sand/perlite led to the increase of PR expression and reduction of CP expression in 24-dpi nodules of nf2100 compared to the WT. Values in (A) and (B) represent the mean of induction folds in the tested mutants compared to the WT. The RT-qPCR analyses in (A) and (B) were made on three biological repetitions with two technical replicates. For each experiment, 16 and 5 plants were analyzed for, respectively, in vitro and sand/perlite conditions. The actin housekeeping gene was used for the expression normalization. Error bars indicate se and the asterisks represent significant variations (P < 2.5%) compared to the WT using Mann–Whitney statistical test. C, Analysis of 21-dpi in vitro nodules induced by S. medicae WSM419 lacZ reveals that nf2100 produces nodules with few necrotic cells. D, In contrast, sand/perlite cultivation leads to the apparition of a large necrotic area. The arrow shows necrotic cells and the scale bars in (C) and (D) represent 500 µm.
Together these data indicate that nf2100 produces senescent nodules with more stimulation of defenses associated with reduction of the CP expression and that the growth substrate has a higher impact on nf2100 immunity than on the other senescence mutants.
Defense and senescence display simultaneous activation in nitrogen-fixing nodules under stress
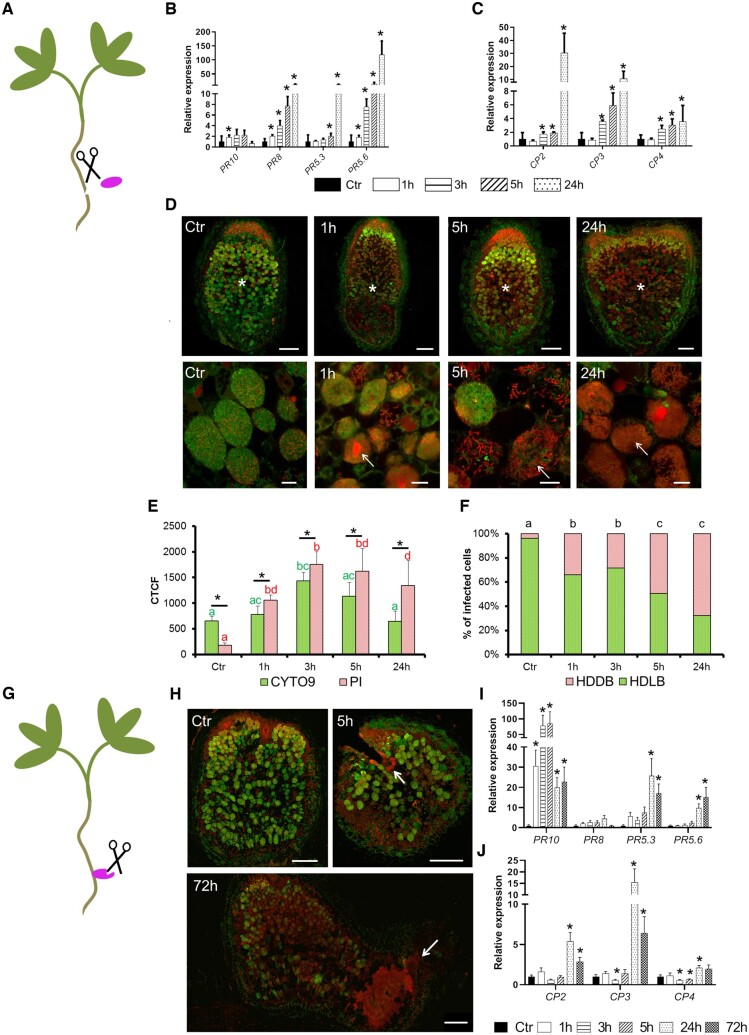
In contrast to the opposite relationship observed between immunity and senescence in the fix− mutants described above, expression data from nodules treated with nitrate (Supplemental Figure S7A) or phosphinothricin (Supplemental Figure S7B) showed concomitant induction of CP and PR genes (https://medicago.toulouse.inrae.fr/MtExpress, Benedito et al., 2008; Seabra et al., 2012) suggesting simultaneous activation of the two processes in the WT nodule upon certain circumstances. Among the main differences between the fix− mutants and the senescence induction experiments is the state of nodule development; in the former the nodules do not fix nitrogen whereas in the latter the chemical treatments were carried out on fix+ nodules. To check if co-activation of the immunity and the senescence can occur once nitrogen fixation takes place in nodule, we evaluated defense and senescence response of WT nodules during stress response. Wounding was previously shown to stimulate defense and stress responses in various plants and organs (van Loon et al., 2006; Sinha et al., 2014; Shen et al., 2018). To trigger a mechanical stress on WT nodules, we cut the nodules from the roots (Figure 5A) and we vacuum infiltrated them in liquid buffered nodulation medium (see Materials and methods).
Figure 5.
Wounding triggers defense and senescence activation in fix+ nodules associated with the death of the differentiated bacteroids. A, In the first wounding treatment the WT nodules at 21-dpi inoculated with S. medicae WSM419 were separated from the roots. Expression analysis of (B) PR and (C) CP genes after incubation of 0 (Ctr), 1, 3, 5, and 24 h revealed that PRs and CPs are, respectively, induced after 1 and 3 h. D, Observation of bacteroid survival using live (green [SYTO9]) and dead (red [PI]) staining in wounded 21-dpi nodules after 0 (Ctr), 1, 3, 5, and 24 h of incubations reveals a death of the differentiated bacteroids 1 h after incubation which increases with time. Top panel displays the nodule sections (scale bars are 200 µm) and bottom panel shows the bacteroids in the fixation zone III (scale bars are 20 µm). Asterisks indicate the nitrogen-fixation zone and the arrows show dead bacteroids. E, The CTCF of SYTO9 and PI staining calculated from nodule sections of wounded nodules reveals more PI than SYTO9 staining in 1, 3, 5, and 24 h compared to the reference (Ctr). The CTCF were calculated for each time of incubation on five to seven sections of independent nodules and error bars show the se. Asterisks show significant variation between SYTO9 and PI fluorescence and the letters show statistical groups between incubation times using Student’s t test (P < 5%). F, The percentage of nodule infected cells with HDDB or HDLB is calculated in the ZIII of sections from wounded nodules at 0 (Ctr), 1, 3, 5, and 24 h. Augmentation of HDDB cell proportion is observed as early as 1 h and increases during the time of incubation. The proportions of HDDB and HDLB were calculated on the nodule sections used in the CTCF determination. The analysis was performed on five to seven sections collected from nodules of independent plants. The letters show statistical groups between incubation times using Student’s t test (P < 5%). G, The second wounding treatment consists of cutting WT nodules attached to the roots at 21-dpi with S. medicae WSM419. H, Observation of bacteroid survival using live (green) and dead (red) staining in wounded 21-dpi nodules after 0 (Ctr), 1, 3, 5, 24, and 72 h of incubation reveals that bacteroid death starts at 5 h after incubation and is located around the wounded zones. The arrows show the wounded zones and the scale bars represent 250 µm. Expression analysis of the PRs (I) and the CPs (J) shows the upregulation of most of these genes after 24 h of incubation. The expression analysis in B, C, I, and J corresponds to the mean expression of three independent experiments (eight plants per experiment) with two to three technical replicates. The actin housekeeping gene was used for expression normalization. Error bars indicate se and asterisks represent significant variation (P < 2.5%) compared to the WT using the Mann–Whitney statistical test.
The PR and CP gene expressions were evaluated on dissected WT nodules at 0, 1, 3, 5, or 24 h of incubation (Figure 5, B and C). Induction of all PR (except PR5.3) occurred 1 h after treatment. PR8 and PR5.6 were stimulated in all the incubation times, while PR5.3 was induced at 5 and 24 h after treatment (Figure 5B). Interestingly all CPs are stimulated at 3 h and expression increased over time (Figure 5C). Altogether, these results indicate that wounding stimulates defense and senescence processes in WT mature nodules with defense stimulation taking place earlier than senescence.
Stimulation of defense and senescence in fix+ nodule is associated with the death of differentiated bacteroids
To determine if the activation of defense and senescence in fix+ nodules can trigger bacteroids death, live and dead staining was performed on WT nodules at 0 (Ctr), 1, 3, 5, and 24 h after wounding (Figure 5D; Supplemental Figure S8). After 1 h, differentiated bacteroids exhibiting red staining were observed in zone III. The abundance of these dead differentiated bacteroids increased with time. At 24 h the number of dead bacteroids was increased compared to alive bacteroids. Quantification of the green/red ratio in zone III from the nodules sections was evaluated using the Corrected Total Cell Fluorescence (CTCF). It revealed a significant accumulation of red staining in wounded nodules 1, 3, 5, and 24 h after treatment compared to the control (Figure 5E). To check if infected host cells accumulated preferentially dead bacteroids during the treatment, a counting of host cells showing High Density of Live Bacteroids (HDLB) versus High Density of Dead Bacteroids (HDDB) was realized in zone III (Figure 5F). After 1 h, the proportion of the cells with HDDB raised compared to the control. The proportion of HDDB cells increased with time and reached 70% of infected cells at 24 h. Altogether these observations indicate that the stimulation of defense and senescence following wounding is associated with death of the differentiated bacteroids.
Connection of the nodules to their roots reduces wounding effects and delays defense and senescence stimulation
To test the effect of wounding on defense and senescence responses in a less destructive context, nodulated WT plants were used instead of detached nodules. To this end, the wounding treatment was applied to root-attached WT nodules (Figure 5G), which were then incubated 0 (Ctr), 1, 3, 5, 24, and 72 h. Bacteroids started to die 5 h after the incubation at the cutting site and the death increased around the treated zone upon the time of incubation (Figure 5H; Supplemental Figure S9). In this context, most PR (Figure 5I) and CP (Figure 5J) are induced after 24 h of treatment. These observations contrast with the behavior of detached nodules where a strong induction of PRs and CPs was observed already after 1 h (Figure 5, B and C). Altogether, these data confirm the observations realized on detached nodules and, reveal a delay of defense and senescence responses and moderate amplitude of bacteroid death when the wounded nodules remain attached to the plants.
Stimulation of defense in the fix+ nodules is accompanied by down-regulation of symbiotic genes repressing defense reactions
In order to investigate the mechanism controlling the activation of defense in fix+ nodules during stress response, expression of the symbiotic genes DNF2, SymCRK, and RSD was assessed in wounded detached nodules at different time points (Supplemental Figure S10). The DNF2 expression was not affected by nodule dissection (Supplemental Figure S10A), while the expressions of SymCRK and RSD were drastically reduced (Supplemental Figure S10B). Likewise, the expression of SymCRK, RSD, and DNF2 was also reduced in fix+ nodules treated with nitrate (Supplemental Figure S11A; Benedito et al., 2008) or in the nodules of plants exposed to phosphinothricin (Supplemental Figure S11B; Seabra et al., 2012), two conditions in which PR genes are upregulated (Supplemental Figure S7). These data suggest an antagonistic behavior between the genes involved in the defense repression (SymCRK, RSD, DNF2) and the PRs in fix+ nodules under stress response or senescence stimulation.
Discussion
To investigate the relationship between immunity and senescence in the nodules, we analyzed the expression of PR and CP genes in Medicago (M. truncatula) mutants forming fix− nodules with senescence or exacerbated defense. Five PR genes were identified as stimulated during nodule defense responses and were used for the tracking of defense activation. Among them, PR10 is involved in the control of the programmed cell death during plant response to pathogens (Ma et al., 2018b) and the PR5 homolog of THAUMATIN-LIKE proteins from Arabidopsis (Arabidopsis thaliana) displays an antimicrobial activity (Hu and Reddy, 1997). The soybean line displaying the loci Rj4 which carried a PR5-like gene, THAUMATIN-LIKE PROTEIN shows an arrest of the root infection with its symbiont Bradyrhizobium elkanii strain USDA61 (Hayashi et al., 2014; Tang et al., 2016; Yasuda et al., 2016). It is proposed that Rj4 restricts the nodulation of soybean through activation of defense signaling (Yasuda et al., 2016). PR8 is the last identified class and corresponds to a class III CHITINASE (Sels et al., 2008) showing homology with lysozyme, an enzyme well known for its antibacterial effect (Stintzi et al., 1993). Altogether, these observations suggest the recruitment of a wide range of defense genes by the legume probably for the control of the bacteroid persistence. Moreover, the upregulation of PRs genes (PR5.3, PR8, and PR10) in nodules infected by Ralstonia solanacearum (Benezech et al., 2020) indicates that at least a part of the described defense genes participate in nodule protection against pathogens.
Our data revealed an opposite behavior between PR and CP expressions in mutants showing nodules with early senescence or exacerbated defenses. In mutant nodules displaying typical defense responses (dnf2 or symCRK), PR genes are strongly induced in contrast to the majority of the CPs that show no stimulation. The opposite was observed in the mutants producing senescent nodules and cultivated in vitro, in which PR genes are not (nf583, nf2210) or moderately (nf2100) induced, while CP expressions are significantly increased. The defense gene induction is associated with nodule necrosis, which is absent and occasionally observed in vitro in nf583/nf2210 and nf2100, respectively. These data indicate that in the fix− mutants studied here, when the defenses are increased, generally the CPs expression is reduced. This led us to propose an opposite behavior between defense and senescence in fix− nodules and to hypothesize that these processes are preferentially stimulated in necrotic and senescent nodules, respectively.
Interestingly, the defense genes are expressed at similar levels in the nf2100, dnf2, and symCRK mutants when grown in sand/perlite and this is linked to a large necrotic zone. Likewise, the other senescence mutants also show enhanced PR expressions and reduced CP expressions with the apparition of few and disparate necrotic cells in sand/perlite compared to the in vitro growth conditions. Agar-jellified media are sufficient to induce expression of PR10 (Medtr2g035150.1) in nodules (Berrabah et al., 2014a). As agar contains agaropectin and impurities in addition to agarose, it was proposed that it displays defense elicitors that are able to prime defense reactions in the nodules. According to this hypothesis, Fukui et al. (1983) showed that agaropectin contained in the agar can stimulate defenses in Lithospermum erythrorhizon. Based on these observations, we postulate that sand/perlite contain potentially more defense elicitors than agar, enhancing the immune response and reducing senescence in nodules. In agreement with this, the behavior of the nf2100 mutant could be explained by a greater sensitivity to environmental elicitors. Its cultivation on agar-jellified media is sufficient to initiate the low level of defenses, in contrast to the behavior of the nf2210 and nf583 mutants. The CP-inhibitor MtPHYTOCYSTATIN32 shows an expression pattern similar to the PR ones in the fix− mutants, in vitro and in sand/perlite conditions, suggesting a reduction of CP activities during defense activation in fix− nodules. The contrasted results obtained in our study for fix− mutants grown in vitro or in sand/perlite suggest that in vitro studies, convenient for the control of the microbial and chemical plant environment, are clearly different from the natural situations mimicked by the sand/perlite substrate, in which plants face a more complex environment that can strongly impact their responses.
nf583 and nf2210 share FSTs in MtSULTR3.5, a sulfate transporter gene whose expression is stimulated in Medicago nodules. Both mutants display similar phenotype characteristics and defense vs. senescence responses, supporting the hypothesis of a common target gene corresponding to two mutated alleles. Interestingly, inactivation in Lotus japonicus of SYMBIOTIC SULFATE TRANSPORTER 1 (SST1), a homolog of MtSULTR3.5, also leads to the formation of fix− nodules and early senescence (Krusell et al., 2005), thereby reinforcing MtSULTR3.5 as candidate responsible for nf583 and nf2100 phenotypes. However, a potential combined effect of other mutations with MtSULTR3.5 cannot be excluded without isolation of additional alleles or complementation of the mutation. For nf2100, based on the expression pattern of the five identified tagged-genes, we failed to isolate the gene responsible for nf2100 dysfunction. Thus, the gene involved in the mutant remains to be identified.
Surprisingly, during nodule senescence induced by phosphinothricin, a co-stimulation of CP and PR genes was observed (Seabra et al., 2012). This behavior is supported by an RNA-seq analysis of Glycine max nodules which revealed the presence of PR transcripts in these organs during natural senescence (Chen et al., 2017), suggesting activation of defense in determinate as well as indeterminate nodules during induced and natural senescence. These results contrast with our observations in the fix− mutants that show an opposite pattern between expressions of defense and senescence markers in the nodules. In these previous studies, transcriptomic analyses were performed on mature fix+ nodules (Seabra et al., 2012; Chen et al., 2017), whereas in our work, the nodules of the fix− mutants are characterized by an incomplete organogenesis and early senescence. These observations prompt us to study the role of nodule development and/or the state of nitrogen fixation on the immunity and senescence relationship. To this end, we have induced a defense-like stimulation on fix+ nodules by two wounding approaches: (1) cutting of isolated nodules separated from the roots and (2) cutting the root-attached nodules. Both treatments result in bacteroid death and co-induction of PR and CP genes. Remarkably, the delay of gene iduction is accompanied by a reduction of bacteroids death when the nodules remain connected to the roots. These observations support the hypothesis that a co-occurrence between immunity and senescence activation is operating in functional nodules. In addition to the differences in nodule development between our study and previous transcriptomic analyses (Seabra et al., 2012; Chen et al., 2017), the plant genotype and the type of treatment also represent important changes. Despite these differences, the co-activation of PRs and CPs observed in all these situations indicates that the co-occurrence between immunity and senescence activations is probably an ubiquitous process rather than a specific response. Similarly, accumulation of PR transcripts takes place during leaf senescence in different plant species (Azumi and Watanabe, 1991; Hanfrey et al., 1996; Walter et al., 1996; John et al., 1997; Wingler et al., 2005). The high degree of overlap of transcriptional responses between nodule- and leaf-senescence in Medicago (Van de Velde et al., 2006) may suggest that the activation of some of the PR defense genes is a common feature between leaf and nodule senescence. We show that nodule wounding similar to nitrate or phosphinothricin treatments reduced the expression of genes that repress defense reactions in the nodule (SymCRK, RSD and DNF2) and enhanced PR expression. These data allow us to propose that defense activation in the nitrogen-fixing nodules may result from the down-regulation of SymCRK, RSD, and DNF2, that may act before and during nitrogen fixation by various ways (Sinharoy et al., 2013, Berrabah et al., 2018a).
Conclusion
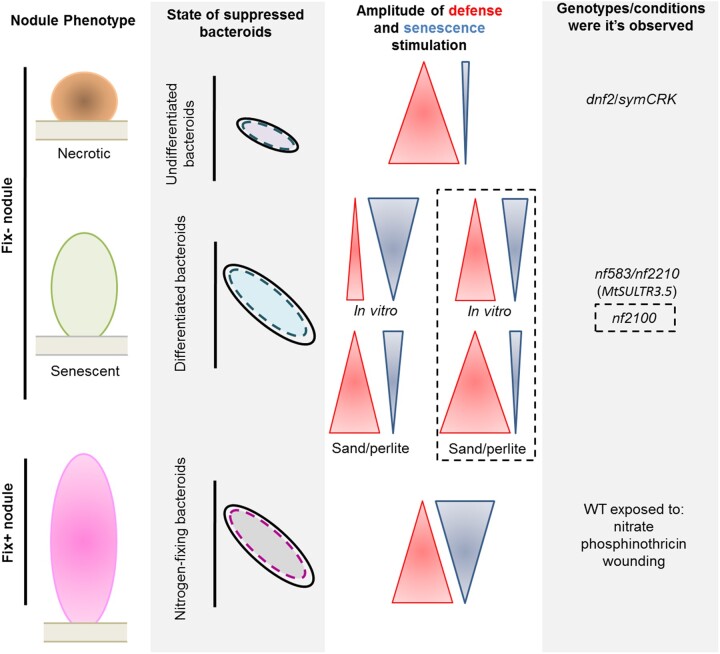
This work deciphers the relationship between immunity and senescence. The use of mutants producing non-fixing nitrogen (fix−) nodules uncovers the mechanisms controlling the dynamic of the establishment of the immune and the senescence programs during nodule organogenesis. In symCRK and dnf2 nodules, which display symbiotic arrest prior to bacteroid differentiation, defense is stimulated more than the senescence (Figure 6). In contrast, under in vitro growth conditions, senescence is more promoted than defense in the senescence mutants nf583 and nf2210 containing differentiated fix− bacteroids. The senescence mutant nf2100 in the same growth substrates shows a greater defense than senescence response. The growth of the fix− mutants in sand/perlite enhances greatly defenses and reduces senescence, pointing out the influence of the environment in the defense/senescence balance. Finally, when the nodule becomes functional (fix+), a co-activation of defense and senescence in response to stresses (wounding, phosphinothricin) or induced senescence (nitrate) is observed and is associated with suppression of the nitrogen-fixing bacteroids at least during nodule responses to wounding.
Figure 6.
Defense and senescence activation in Medicago nodules. After rhizobia internalization, failure in defense repression can lead to death of undifferentiated bacteroids in dnf2 and symCRK producing non-fixing nitrogen (fix−) nodules showing necrosis and low stimulation of senescence. In contrast, the senescence mutants show degradation of differentiated bacteroids in a fix− senescent nodule associated with senescence marker expression and low defense responses in nf583 and nf2210, two potential mutated alleles of MtSULTR3.5. Interestingly, nf2100 displays more defense stimulation than the other senescence mutants. The environment (as sand/perlite substrate) can stimulate immunity and reduce senescence stimulation in senescence mutants. In the sand/perlite conditions, nf2100 shows PR stimulation similar to that observed in dnf2 and symCRK with the accumulation of necrotic tissues. Finally, in nitrogen-fixing (fix+) nodules, stress conditions (wounding, phosphinothricin) or induction of senescence with the addition of nitrate to the growth medium, lead to a co-activation of defense and senescence and suppression of nitrogen-fixing bacteroids.
Materials and methods
Bacterial material and growth conditions
Sinorhizobium medicae strains WSM419 (Ma and Ewing, 1986) and WSM419 expressing lacZ provided by G. Endre (Horváth et al., 2015) were used. The bacteria were cultivated in yeast extract broth medium (Krall et al., 2002) supplemented with the appropriate antibiotics for 24–48 h at 30°C. The following antibiotics were added to the media: chloramphenicol at 12.5 µg·mL−1 for WSM419, chloramphenicol at 12.5 µg·mL−1, and tetracycline at 5 µg·mL−1 for WSM419 expressing lacZ.
Plant material
Medicago (M. truncatula) ecotype R108 (Hoffmann et al., 1997) and the derived Tnt1 transposon tagged-lines (provided by the Noble Research Institute) nf583, nf2100, and nf2210 isolated in a community screen (Tadege et al., 2008; Pislariu et al., 2012; Cheng et al., 2014) as well as nf737 (symCRK, Berrabah et al., 2014b) were used in this study. In addition, the MERE1 insertion mutant line ms240 (dnf2-4) corresponding to a somaclonal variant obtained by regeneration of a T-DNA-tagged Medicago line was used (Bourcy et al., 2013).
Growth conditions and plant inoculation
Medicago seeds were surface sterilized as previously described by Berrabah et al. (2015) and vernalized for at least 48 h at 4°C in the dark on solid medium (Bacto-agar 1% w/v). Seeds were then germinated for 24 h in the dark at 24°C before transfer to BNM (Ehrhardt et al., 1992) solidified with 1.5% (w/v) bacto-agar for plants cultivated in vitro or in a mixture of sand/perlite (2/1, v/v). The plants are cultivated into a growth chamber conditions at 24°C and 60% humidity under a photoperiod of 16-h light/8-h dark (150 μE intensity).
Overnight cultures of the bacterial strains were pelleted and washed twice in sterile water. OD600 nm was adjusted to 0.1 in water. Roots of eight seedlings per plate (in vitro culture) or five seedlings per pot (growth in sand/perlite) were inoculated with 1 mL or 10 mL of bacterial cell suspension, respectively.
Plant treatments
Wounding treatment
Twenty-one dpi nodules from WT plant inoculated with S. medicae WSM419 were collected using forceps and scalpel or wounded with one wound on the nodule attached to the plant and incubated in 5 mL of liquid BNM. Immediately after harvesting, to enhance the mechanical stress with nodule integrity preservation, the collected nodules or nodulated plants were vacuum-infiltrated for 15 min and collected (control) or incubated for 1, 3, 5, 24, and 72 h under checking in multiwell plates filled with BNM.
Nitrogenase activity
Acetylene reduction assays (ARA) were conducted on individual plants with a modified protocol from Koch and Evans (1966). Plants were harvested after in vitro growth at 21 dpi or in a growth chamber at 24 dpi. Individual whole plants (in vitro) or nodulated roots (growth chamber) were incubated with 500 µL of acetylene for 2 h at room temperature in 21 mL air-tight glass vials sealed with rubber septa. After incubation, 200 µL of gas samples was removed from the vial and was injected into a gas chromatography system (7820A; Agilent Technology) to determine the ethylene production. For each test 14 plants were used for ARA analysis (see “Replicates and statistical tests” part).
Histological analysis
LacZ staining
Nodules were embedded into agarose 6% (w/v, Bourcy et al., 2013) and 60 µm sections were prepared using the vibratome VT1200S (Leica Biosystems GmbH, Germany). For LacZ activity detection, the slices were incubated for 15 min in Z’ buffer (phosphate buffer pH 7 [100 mM], MgCl2 [1 mM], and KCl [10 mM]). The slices were then incubated for 2 h under darkness, at 28°C in reaction buffer (Z’ buffer, potassium ferricyanide [5 mM], potassium ferrocyanide [5 mM], 45 µm filtrated 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside [X-gal, 2.4 mM]). The samples were observed using AZ10 macroscope (Nikon).
DAPI staining
The bacteroids were purified from 17-dpi aged nodules cultivated in vitro and stained with DAPI as described in Mergaert et al. (2006). Free-living bacteria or bacteroids were incubated for 10 min in 50 μg·mL−1 of DAPI at 60°C and then observed using an epifluorescent microscope (AxioImager Z2, Zeiss) with the following setup: 365 nm and 420–470 nm, respectively, for filter excitation and emission wavelengths, 47.89% for light source intensity. The contrast and the brightness are equally adjusted between the test and the control in each experiment.
Live and dead staining
The nodules were embedded in 6% (w/v) agarose and sliced into 70 µm sections using the vibratome VT1200S. Live and dead stainings were carried out as previously described by Haag et al. (2011). Nodule sections were incubated in a 50 mM Tris-HCl buffer (pH 7.2) containing 30 µM PI and 5 µM SYTO9 (Life Technology) for 20 min. Stained sections were then mounted between slide and slip cover with a few Tris-HCl buffer drops and observed using the confocal microscope LSM880 (Zeiss) with the following setup: 561 nm and 488 nm for laser wavelengths, 594–687 nm and 508–553 nm for detection wavelengths, 550–600 V for detector gains. The images were not subjected to erasure; the contrast and the brightness are equally adjusted between the test and the control in each experiment.
Quantification of PI and SYTO9 fluorescence in nodule sections was carried out using the CTCF as described by Jakic et al. (2017) in the ImageJ software (https://imagej.net/Bio7). The following equation was used for CTCF calculation:
RNA extraction, cDNA synthesis, and expression analysis
RNA extraction, cDNA synthesis and RT-qPCR were performed as previously described (Berrabah et al., 2018a). After freezing in liquid nitrogen, the nodules collected from 16 plants (in vitro growth) or 5 plants (sand/perlite growth) per experiment were ground in a 2 mL tube with beads and the total RNA was extracted using a TRI Reagent procedure recommended by the manufacturer (Molecular Research Center). DNA was removed from the samples using the DNAse I kit (Invitrogen) as recommended by the manufacturer. The concentration and the RNA quality were checked using the NanoDrop ND-1000 (Thermo Scientific).
Reverse transcription was performed on 0.5 or 1 µg of the total RNA (DNA free) using oligo dT and SuperScript II (Life Technology) according to the supplier in a final volume of 20 µL.
For each tested gene, the primers amplified 200–300 nucleotides of the cDNA sequence and the quantification was made using qPCR on a LightCycler 480 (Roche Life Science) with the LightCycler FastStart DNA Master SYBR green I kit according to manufacturer’s instructions (Roche). The temperatures of 94°C, 58°C–62°C and 72°C were used, respectively, for denaturation, annealing, and extension steps. In all analyzed samples, expression levels were normalized using the housekeeping gene MtACT (Actin 11; Supplemental Table S6; Plet et al., 2011).
Identification of PR genes and sequence analyses
Identification of PR genes was realized using two genome databases: phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and Medicago A17 r5.0 genome portal (https://medicago.toulouse.inra.fr/MtrunA17r5.0-ANR/). Key word search was done on the used databases with the term “Pathogenesis”. The genes corresponding to PRs were then isolated. A complementary approach of identification was realized by blasting the coding DNA sequence (CDS) of identified PR. To confirm the classification of the PRs, functional domains were detected on full-length protein sequences using the NCBI prediction domain tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The identified PRs, their corresponding groups and their domains used for the classification are reported in Supplemental Table S1. Sequence comparison between PR10.2 and PR10.3 was performed on the CDS using ClustalW method in the BioEdit software (https://bioedit.software.informer.com).
Replicates, statistical tests, and data representation
ARA tests were analyzed using three independent experiments with 14 plants per experiment. For all microscopic analyses, at least two independent replicates with at least 10 samples were observed. Expression analyses were carried out on two to three independent experiments with two technical replicates. For each experiment 16 and 5 plants were analyzed for, respectively, in vitro and sand/perlite condition. In all RT-qPCR data, mean expression is represented with standard error (SE). For RT-qPCR data a Mann–Whitney statistical test was performed and only experiments with a P < 2.5% were considered as statistically significant. Student’s t tests were realized for ARA experiments, SYTO9 and PI fluorescence quantification and the evaluation of bacteroid death, and only variations with P < 5% were considered as significant. All graphics were generated using the Prism8 software (https://www.graphpad.com/scientific-software/prism/), with the exception of graphics in Figure 5, E and F for which an Excel software was used (https://www.microsoft.com/fr-fr/microsoft-365/excel).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: Medtr4g107930: CP3; Medtr4g079770: CP4; Medtr5g022560: CP2; Medtr4g079470: CP5; TC106667: Actine; Medtr1g099310.1: PR8; Medtr4g120970.1/Medtr6g033450.1: PR10; Medtr5g010640.1: PR5.3; Medtr8g096910.1: PR5.6; Medtr5g088770.1: PHYTOCYSTATIN32; Medtr2g026040.1: PHYTOCYSTATIN5; Medtr4g085800: DNF2; Medtr3g079850: SymCRK; MtrunA17_Chr7g0239441: RSD.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. PCR genotyping of the Tnt1 insertion in nf583 and nf2210.
Supplemental Figure S2. Expression patterns of PR candidate genes in WT nodules in response to nitrate and phosphinothricin.
Supplemental Figure S3. Validation by RT-qPCR analysis of 10 PR genes selected for defense monitoring in the Medicago nodules.
Supplemental Figure S4. Comparison of CDS sequences between PR10.2 (Medtr4g120970.1) and PR10.3 (Medtr6g033450.1).
Supplemental Figure S5. Expression analyses of PRs and CPs during the development of Medicago.
Supplemental Figure S6. Expression pattern of PHYCYST5 in Medicago fix− nodules of mutants in response to different environmental conditions.
Supplemental Figure S7. Expression pattern of senescence and defense markers in Medicago WT nodules in response to nitrate and phosphinothricin treatments.
Supplemental Figure S8. Live and dead staining of WT inoculated nodules separated from the roots.
Supplemental Figure S9. Live and dead staining of Medicago WT inoculated nodules attached to the roots.
Supplemental Figure S10. Expression patterns of DNF2, SymCRK, and RSD in Medicago WT nodules in response to wounding.
Supplemental Figure S11. Expression pattern of DNF2, SymCRK, and RSD in Medicago WT nodules in response to nitrate and phosphinothricin treatments.
Supplemental Table S1. List of nf583, nf2210, and nf2100 genes with FSTs.
Supplemental Table S2. List of the identified PR genes in the M. truncatula genome.
Supplemental Table S3. List of PR genes validated by RT-qPCR for the study.
Supplemental Table S4. Pearson correlation analysis of the PRs, CPs, and PRs versus CPs expression.
Supplemental Table S5. Co-expressed PHYTOCYSTATIN genes with the studied PRs.
Supplemental Table S6. List of primers used in this study.
Supplementary Material
Acknowledgments
The authors thank Hossein Khademian and Peter Mergaert for preliminary characterization of line NF583 and to Messaoudi Hala Selma and Souaiaia Nour El Houda for their help during PR identification.
Funding
This work was supported by the grant ANR-15-CE20 0005 (STAYPINK) and funding obtained from the invitation programs for foreign researchers from Paris Cité University and Paris Saclay University. This study contributes to the IdEX Université de Paris ANR-18-IDEX-0001. Medicago truncatula Tnt1 mutants were created through research funded, in part, by grants from the National Science Foundation, USA (DBI 0703285 and IOS-1127155).
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Fathi Berrabah, Faculty of Sciences, Department of Biology, Amar Telidji University, 03000 Laghouat, Algeria; Research Unit of Medicinal Plants (RUMP), National Center of Biotechnology Research, CRBt, 25000 Constantine, Algeria.
Gautier Bernal, Université Paris-Saclay, CNRS, INRAE, Université d’Évry, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France; Université Paris Cité, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France.
Ait-Salem Elhosseyn, Université Paris-Saclay, CNRS, INRAE, Université d’Évry, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France; Université Paris Cité, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France.
Cyrille El Kassis, Université Paris-Saclay, CNRS, INRAE, Université d’Évry, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France; Université Paris Cité, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France.
Roxane L’Horset, Pôle de Protection des Plantes, UMR PVBMT, 97410 Saint-Pierre, Réunion, France.
Farouk Benaceur, Faculty of Sciences, Department of Biology, Amar Telidji University, 03000 Laghouat, Algeria; Research Unit of Medicinal Plants (RUMP), National Center of Biotechnology Research, CRBt, 25000 Constantine, Algeria.
Jiangqi Wen, The Institute of Agricultural Biosciences, Oklahoma State University, Ardmore, Oklahoma 73401, USA.
Kirankumar S Mysore, The Institute of Agricultural Biosciences, Oklahoma State University, Ardmore, Oklahoma 73401, USA.
Marie Garmier, Université Paris-Saclay, CNRS, INRAE, Université d’Évry, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France; Université Paris Cité, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France.
Benjamin Gourion, LIPME, Université de Toulouse, INRAE, CNRS, 31320 Castanet-Tolosan, France.
Pascal Ratet, Université Paris-Saclay, CNRS, INRAE, Université d’Évry, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France; Université Paris Cité, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France.
Véronique Gruber, Université Paris-Saclay, CNRS, INRAE, Université d’Évry, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France; Université Paris Cité, Institute of Plant Sciences Paris-Saclay (IPS2), 91190 Gif-sur-Yvette, France.
Fat.B. and V.G. designed the experiments, supervised the experiments, analyzed the data, and wrote the paper. Fat.B., G.B., A-S.E., C.E.K., and R.L.H. conducted the experiments. Fat.B., Far.B., M.G., B.G., P.R., and V.G analyzed the data and discussed the paper. K.S.M. and J.W. contributed reagents and edited the manuscript. Fat.B. and V.G. agree to serve as the authors responsible for contact and ensure communication.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Véronique Gruber (veronique.gruber@u-paris.fr) and Fathi Berrabah (fa.berrabah@lagh-univ.dz).
References
- Ali S, Ahmad B, Kamili AN, Ali A, Ahmad Z, Akhter J, Tyagi A, Tajamul S, Mushtaq M, Yadav P, et al (2018) Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol Res 212–213: 29–37 [DOI] [PubMed] [Google Scholar]
- Azumi Y, Watanabe A (1991) Evidence for a senescence-associated gene induced by darkness. Plant Physiol 95: 577–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Ott T, Moreau S, Niebel A, Frickey T (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Benezech C, Berrabah F, Jardinaud MF, Le Scornet A, Milhes M, Jiang G, George J, Ratet P, Vailleau F, Gourion B (2020) Medicago-Sinorhizobium-Ralstonia co-infection reveals legume nodules as pathogen confined infection sites developing weak defenses. Curr Biol 30: 351–358 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Balliau T, A EH, George J, Zivy M, Ratet P, Gourion B (2018a) Control of the ethylene signaling pathway prevents plant defenses during intracellular accommodation of the rhizobia. New Phytol 219: 310–323 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Cayrel A, Eschstruth A, Mondy S, Ratet P, Gourion B (2014a) Growth conditions determine the DNF2 requirement for symbiosis. PLoS One 9: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, et al (2014b) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Hosseyn E, Salem A, Garmier M, Ratet P (2018b) The multiple faces of the Medicago-Sinorhizobium Symbiosis. Methods Mol Biol 1822: 241–260 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Ratet P, Gourion B (2015) Multiple steps control immunity during the intracellular accommodation of rhizobia. J Exp Bot 66: 1977–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Mickael K U, Benjamin G, Ratet P (2013) Medicago truncatula DNF2 is a PI-PLC-XD-containing protein requierd for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol 197: 1250–1261 [DOI] [PubMed] [Google Scholar]
- Chen P-C, Phillips DA (1977) Induction of root nodule senescence by combined nitrogen in Pisum sativum L. Plant Physiol 59: 440–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Shan ZH, Yang ZL, Zhang XJ, Qiu DZ, Zhou XA (2017) RNA-Seq analysis of nodule development at five different developmental stages of soybean (Glycine max) inoculated with Bradyrhizobium japonicum strain 113-2. Scient Rep 7: 42248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee HK, Tadege M, Ratet P, Udvardi M, Mysore KS, Wen J (2014) An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol 201: 1065–1076 [DOI] [PubMed] [Google Scholar]
- Díaz-Mendoza M, Velasco-Arroyo B, González-Melendi P, Martínez M, Díaz I (2014) C1A cysteine protease-cystatin interactions in leaf senescence. J Exp Bot 65: 3825–3833 [DOI] [PubMed] [Google Scholar]
- Domonkos A, Szilard K, Aniko G, Erno K, Horváth B, Gyongyi ZK, Attila F, Monika TT, Ferhan A, Karoly B, et al (2017) NAD1 Controls defense-like responses in Medicago truncatula symbiotic nitrogen fixing nodules following rhizobial colonization in a BacA-independent manner. Genes (Basel) 8: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Morrey Atkinson E, Long SR (1992) Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti nod factors. Science 256: 998–1000 [DOI] [PubMed] [Google Scholar]
- Fedorova M, Van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Yoshikawa N, Tabata M (1983) Induction of shikonin formation by agar in Lithospermum erythrorhizon cell suspension cultures. Phytochemistry 22: 2451–2453 [Google Scholar]
- Gourion B, Berrabah F, Ratet P, Stacey G (2015) Rhizobium–legume symbioses : the crucial role of plant immunity. Trends Plant Sci 20: 186–194 [DOI] [PubMed] [Google Scholar]
- Gupta R, Lee SJ, Min CW, Kim SW, Park KH, Bae DW, Lee BW, Agrawal GK, Rakwal R, Kim ST (2016) Coupling of gel-based 2-DE and 1-DE shotgun proteomics approaches to dig deep into the leaf senescence proteome of Glycine max. J Proteomics 148: 65–74 [DOI] [PubMed] [Google Scholar]
- Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Angelo SD, Kondorosi E, Longhi R, Boncompagni E, He D, et al (2011) Protection of sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol 9: e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfrey C, Fife M, Buchanan-Wollaston V (1996) Leaf senescence in Brassica napus: expression of genes encoding pathogenesis-related proteins. Plant Mol Biol 30: 597–609 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Shiro S, Kanamori H, Mori-Hosokawa S, Sasaki-Yamagata H, Sayama T, Nishioka M, Takahashi M, Ishimoto M, Katayose Y, et al (2014) A thaumatin-like protein, Rj4, controls nodule symbiotic specificity in soybean. Plant Cell Physiol 55: 1679–1689 [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Trinh TH, Leung J, Kondorosi A, Kondorosi E (1997) A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol Plant-Microbe Interact 10: 307–315 [Google Scholar]
- Horváth B, Domonkos Á, Kereszt A, Sz}ucsb A, Ábrahám E, Ayaydin F, Bóka K, Chen Y, Chen R, et al (2015) Loss of the nodule-specific cysteine riche peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci 112: 15232–15237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Reddy ASN (1997) Cloning and expression of a PR5-like protein from Arabidopsis: inhibition of fungal growth by bacterially expressed protein. Plant Mol Biol 34: 949–959 [DOI] [PubMed] [Google Scholar]
- Jakic B, Buszko M, Cappellano G, Wick G (2017) Elevated sodium leads to the increased expression of HSP60 and induces apoptosis in HUVECs. PLoS One 12: e0179383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulneau V, Lafitte C, Jacquet C, Fournier S, Salamagne S, Briand X, Esquerré-Tugayé M-T, Dumas B (2010) Ulvan, a sulfated polysaccharide from green algae, activates plant immunity through the jasmonic acid signaling pathway. J Biomed Biotechnol 2010: 525291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John I, Hackett R, Cooper W, Drake R, Farrell A, Grierson D (1997) Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol Biol 33: 641–651 [DOI] [PubMed] [Google Scholar]
- Kang Y, Li M, Sinharoy S, Verdier J (2016) A snapshot of functional genetic studies in Medicago truncatula. Front Plant Sci 7: 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B, Evans HJ (1966) Reduction of acetylene to ethylene by soybean root nodules. Plant Physiol 41: 1748–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C (2002) Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc Natl Acad Sci USA 99: 11405–11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch S, Thiery S, Reinstädler A, Gruner K, Zienkiewicz K, Feussner I, Panstruga R (2019) Arabidopsis mlo3 mutant plants exhibit spontaneous callose deposition and signs of early leaf senescence. Plant Mol Biol 101: 21–40 [DOI] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert I, Pervent M, Le Queré A, Clément G, Tauzin M, Severac D, Benezech C, Tillard P, Martin-Magniette M-L, Colella S, et al (2020) Responses of mature symbiotic nodules to the whole-plant systemic nitrogen signaling. J Exp Bot 71: 5039–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Lee G, Kim B, Jang S, Lee Y, Yu Y, Seo J, Kim S, Lee YH, Lee J, et al (2018) Identification of a spotted leaf sheath gene involved in early senescence and defense response in rice. Front Plant Sci 9: 1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Ekramoddoullah AKM (2006) The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol 68: 3–13 [Google Scholar]
- Ma H, Ewing J (1986) Acid tolerance in the Rhizobium meliloti – Medicago symbiosis. Aust J Agric Res 37: 55–64 [Google Scholar]
- Ma X, Keller B, McDonald BA, Palma-Guerrero J, Wicker T (2018a) Comparative transcriptomics reveals how wheat responds to infection by zymoseptoria tritici. MPMI 31: 420–431 [DOI] [PubMed] [Google Scholar]
- Ma H, Xiang G, Li Z, Wang Y, Dou M, Su L, Yin X, Liu R, Wang Y, Xu Y (2018b) Grapevine VpPR10.1 functions in resistance to Plasmopara viticola through triggering a cell death-like defence response by interacting with VpVDAC3. Plant Biotechnol J 16: 1488–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik NSA, Pfeiffer NE, Williams DR, Wagner FW (1981) Peptidohydrolases of soybean root nodules. Plant Physiol 68: 386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez M, Cambra I, González-Melendi P, Santamaría ME, Díaz I (2012) C1A cysteine-proteases and their inhibitors in plants. Physiol Plant 145: 85–94 [DOI] [PubMed] [Google Scholar]
- Maunoury N, Redondo-Nieto M, Bourcy M, Van De Velde W, Alunni B, Ratet P, Aggerbeck L, Kondorosi E, Mergaert P (2010) Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One 5: e9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset A-E, Barloy-Hubler F, Galibert F, Kondorosi A, et al (2006) Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc Natl Acad Sci USA 103: 5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métraux JP, Streit L, Staub T (1988) A pathogenesis-related protein in cucumber is a chitinase. Physiol Mol Plant Pathol 33: 1–9 [Google Scholar]
- Oke V, Long SR (1999) Bacteroid formation in the Rhizobium-legume symbiosis. Curr Opin Microbiol 2: 641–646 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Paau AS, Bloch CB, Brill WJ (1980) Developmental fate of Rhizobium meliloti bacteroids in alfalfa nodules. J Bacteriol 143: 1480–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar OR, Gassmann W, Walker JC (2017) Leaf shedding as an anti-bacterial defense in Arabidopsis cauline leaves. PLoS Genet 13: 849–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Guerra JC, Coussens G, De Keyser A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M (2010) Comparison of developmental and stress-induced nodule senescence in Medicago truncatula. Plant Physiol 152: 1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre O, Hopkins J, Combier M, Baldacci F, Engler G, Brouquisse R, Hérouart D, Boncompagni E (2014) Involvement of papain and legumain proteinase in the senescence process of Medicago truncatula nodules. New Phytol 202: 849–863 [DOI] [PubMed] [Google Scholar]
- Pislariu CI, Murray JD, Wen J, Cosson V, Muni RRD, Wang M, Benedito VA, Andriankaja A, Cheng X, Jerez IT, et al (2012) A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol 159: 1686–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pladys D, Vance CP (1993) Proteolysis during development and senescence of effective and plant gene-controlled ineffective alfalfa nodules. Plant Physiol 103: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F (2011) MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Schreiber MC, Karlo JC, Kovalick GE (1997) A novel cDNA from Drosophila encoding a protein with similarity to mammalian cysteine-rich secretory proteins, wasp venom antigen 5, and plant group 1 pathogenesis-related proteins. Gene 191: 135–141 [DOI] [PubMed] [Google Scholar]
- Seabra AR, Pereira PA, Becker JD, Carvalho HG (2012) Inhibition of glutamine synthetase by phosphinothricin leads to transcriptome reprograming in root nodules of Medicago truncatula. Mol Plant Microbe Interact 25: 976–992 [DOI] [PubMed] [Google Scholar]
- Sels J, Mathys J, De Coninck BMA a, Cammue BPA a, De Bolle MFCC (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46: 941–50 [DOI] [PubMed] [Google Scholar]
- Shen Q, Liu L, Wang L, Wang Q (2018) Indole primes plant defense against necrotrophic fungal pathogen infection. PLoS One 13: e0207607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Singh RP, Kushwaha GS, Iqbal N, Singh A, Kaushik S, Kaur P, Sharma S, Singh TP (2014) Current overview of allergens of plant pathogenesis related protein families. Sci World J 2014: 543195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinharoy S, Torres-Jerez I, Bandyopadhyay K, Kereszt A, Pislariu CI, Nakashima J, Benedito VA, Kondorosi E, Udvardi MK (2013) The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 25: 3584–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B (1993) Plant “pathogenesis-related” proteins and their role in defense against pathogens. Biochimie 75: 687–706 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tang F, Yang S, Liu J, Zhu H (2016) Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol 170: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JCP, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H, et al (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Van Wyk SG, Du Plessis M, Cullis CA, Kunert KJ, Vorster BJ (2014) Cysteine protease and cystatin expression and activity during soybean nodule development and senescence. BMC Plant Biol 14: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MH, Liu JW, Wünn J, Hess D (1996) Bean ribonuclease-like pathogenesis-related protein genes (Ypr10) display complex patterns of developmental, dark-induced and exogenous-stimulus-dependent expression. Eur J Biochem 239: 281–293 [DOI] [PubMed] [Google Scholar]
- Walton JH, Kontra-Kováts G, Green RT, Domonkos Á, Horváth B, Brear EM, Franceschetti M, Kaló P, Balk J (2020) The Medicago truncatula vacuolar iron transporter-like proteins VTL4 and VTL8 deliver iron to symbiotic bacteria at different stages of the infection process. New Phytol 228: 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yu H, Luo L, Duan L, Cai L, He X, Wen J, Mysore KS, Li G, Xiao A, et al (2016) NODULES WITH ACTIVATED DEFENSE 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol 212: 176–191 [DOI] [PubMed] [Google Scholar]
- Wang X, Tang C, Deng L, Cai G, Liu X, Liu B, Han Q (2010) Characterization of a pathogenesis-related thaumatin-like protein gene TaPR5 from wheat induced by stripe rust. Physiol Plant 139: 27–38 [DOI] [PubMed] [Google Scholar]
- Wingler A, Brownhill E, Pourtau N (2005) Mechanisms of the light-dependent induction of cell death in tobacco plants with delayed senescence. J Exp Bot 56: 2897–2905 [DOI] [PubMed] [Google Scholar]
- Yarce JCS, Lee HK, Tadege M, Ratet P, Mysore KS (2013) Forward genetics screening of Medicago truncatula Tnt1 insertion lines. Methods Mol Biol 1069: 93–100 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Miwa H, Masuda S, Takebayashi Y, Sakakibara H, Okazaki S (2016) Effector-triggered immunity determines host genotype-specific incompatibility in legume-rhizobium symbiosis. Plant Cell Physiol 57: 1791–1800 [DOI] [PubMed] [Google Scholar]
- Yu H, Xiao A, Dong R, Fan Y, Zhang X, Liu C, Wang C, Zhu H, Duanmu D, Cao Y, et al (2018) Suppression of innate immunity mediated by the CDPK-Rboh complex is required for rhizobial colonization in Medicago truncatula nodules. New Phytol 220: 425–434 [DOI] [PubMed] [Google Scholar]
- Zhang H, Dugé de Bernonville T, Body M, Glevarec G, Reichelt M, Unsicker S, Bruneau M, Renou JP, Huguet E, Dubreuil G, et al (2016) Leaf-mining by Phyllonorycter blancardella reprograms the host-leaf transcriptome to modulate phytohormones associated with nutrient mobilization and plant defense. J Insect Physiol 84: 114–127 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang H-L, Li Z, Guo H (2020) Genetic Network between leaf senescence and plant immunity : crucial regulatory nodes and new insights. Plants(Basel) 9: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JL, Szeto WW, Ausubel FM (1983) Molecular characterization of Tn5-induced symbiotic (Fix-) mutants of Rhizobium meliloti. J Bacteriol 156: 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.