Abstract
Callose is a plant cell wall polysaccharide whose deposition is spatiotemporally regulated in various developmental processes and environmental stress responses. The appearance of callose in premeiotic anthers is a prominent histological hallmark for the onset of meiosis in flowering plants; however, the biological role of callose in meiosis remains unknown. Here, we show that rice (Oryza sativa) GLUCAN SYNTHASE LIKE5 (OsGSL5), a callose synthase, localizes on the plasma membrane of pollen mother cells (PMCs) and is responsible for biogenesis of callose in anther locules through premeiotic and meiotic stages. In Osgsl5 mutant anthers mostly lacking callose deposition, aberrant PMCs accompanied by aggregated, unpaired, or multivalent chromosomes were frequently observed and, furthermore, a considerable number of mutant PMCs had untimely progress into meiosis compared to that of wild-type PMCs. Immunostaining of meiosis-specific protein HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS2 in premeiotic PMCs revealed precocious meiosis entry in Osgsl5 anthers. These findings provide insights into the function of callose in controlling the timing of male meiosis initiation and progression, in addition to roles in microsporogenesis, in flowering plants.
Rice meiosis-specific callose synthase is a transmembrane protein responsible for callose accumulation in meiotic anthers and controls normal initiation and progression of male meiosis.
Introduction
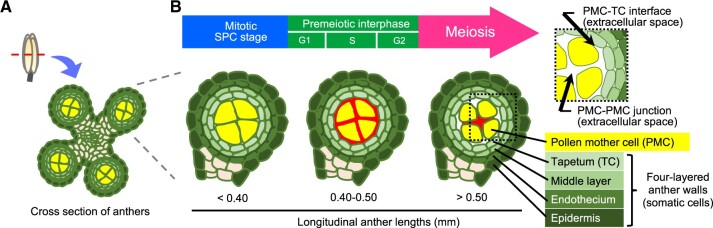
In flowering plants, successful pollen production involves a series of multiple complex steps. An important earlier step is meiosis that takes place within microsporangium or pollen sac, two pairs of which compose an anther in angiosperms (Scott et al., 2004; Zhang and Wilson, 2009). In rice (Oryza sativa) anthers, each of four microsporangia comprises central sporogenous cells (SPCs) surrounded by concentric somatic cell walls four-layered prior to male meiosis, namely tapetum, middle layer, endothecium, and epidermis from inside-out, respectively (Figure 1). After several mitotic divisions, SPCs mature into meiotically competent pollen mother cells (PMCs), which undergo meiosis following DNA replication to halve chromosome number for fertilization, while the tapetal cell (TC) layers provide metabolites and nutrients for neighboring PMCs and microspores, and eventually ends via programmed cell death (Dickinson and Bell, 1976; Steer, 1977; Lei and Liu, 2020). After anther walls are four-layered, a plant-specific carbohydrate callose fulfills extracellular spaces of a pollen sac chamber or anther locule and surrounds PMCs at premeiotic interphase, which is a prominent histological hallmark for the onset of male meiosis in flowering plants (Shivanna, 2003; Unal et al., 2013).
Figure 1.
Schematic illustration of anther development and callose accumulation during male meiosis progression. A, Illustration of a cross section of premeiotic anther with four locules. B, Enlarged view of anther locules with constituent cell types, respective anther lengths and their cell cycle status. The red regions in each locule correspond to areas for callose deposition at extracellular spaces observed in this study.
Callose is made up of linear glucose residues of β-1,3 linkages, with some having β-1,6-glucan branches, and functions in various aspects of plant growth and development spatio-temporally (Stone and Clarke, 1992; Chen and Kim, 2009; Zavaliev et al., 2011; Piršelová and Matušíková, 2013; Neduka, 2015). For instance, callose is involved in papillae cell-wall materials at bacterial and fungal contact sites (Dong et al., 2008; Voigt, 2014) and also secreted at wounded plant tissues (Jacobs et al., 2003), indicating indispensable roles of callose in defense against both biotic and abiotic stresses. In cell–cell signaling, callose regulates the conductivity of plasmodesmata (PD), forming cytoplasmic continuums in plants (Radford et al., 1998; Lucas et al., 2009; Lee and Sieburth, 2010; Zavaliev et al., 2011; Sager and Lee, 2018), and thought to permit selective diffusion of apoplastic signaling (Maltby et al., 1979; Bhalla and Slattery, 1984; Yim and Bradford, 1998). From aspects of plant development, callose is deposited transiently at dividing cell plates during cytokinesis, and aids in primary wall formation between daughter cells (Staehelin and Hepler, 1996; Hong et al., 2001; Thiele et al., 2009). It is also deposited in phloem tissue to control sieve plate development and pore size of sieve tubes (Xie et al., 2011). Toward reproduction, callose helps patterning of pollen apertures and elongation of pollen tubes (Franklin-Tong, 1999; Albert et al., 2011; Qin et al., 2012; Prieu et al. 2017).
Several reports have revealed the importance of callose accumulation during microsporogensis. In Arabidopsis (Arabidopsis thaliana), CALLOSE SYNTHASE5 (CalS5) exerts essential functions during pollen formation stages (Dong et al., 2005). Similarly, a rice gene, GLUCAN SYNTHASE LIKE5 (OsGSL5), a homolog of AtCalS5, is responsible for pollen growth and development during microsporogensis (Shi et al., 2015). Premature dissolution of callose in the rice defective callose in meiosis1 (dcm1) mutant generates abnormal pollen grains with varied size and DNA content as a result of defects in meiotic cytokinesis (Zhang et al., 2018). In rice ovary, OsGSL8 symplasmically controls carbohydrates unloading into pericarp cells of developing ovary, in addition to regulating vascular cell patterning (Song et al., 2016). In Arabidopsis, the amount of phloem-mobile green fluorescent proteins (GFPs), able to be unloaded onto all gynoecium cells before female meiosis, are extremely reduced around tetrad spores, suggesting physical isolation of meiocytes from other gynoecium cells probably by callose accumulation (Werner et al., 2011). In contrast, little is known about the roles of callose accumulation in premeiotic anther development and male meiosis, despite its noticeable amounts and cross-species conservation in land plant anthers (Musiał and Kościńska-Pająk, 2017; Sager and Lee, 2018; Seale, 2020).
Our group previously reported that MEIOSIS ARRESTED AT LEPTOTENE2 (MEL2), an RNA recognition motif protein, functions in timely transition of spore mother cells to the meiotic cycle in rice (Nonomura et al., 2011). Interestingly, one of the significantly (P < 0.05 by Student’s t test) downregulated genes in premeiotic mel2 mutant anthers was OsGSL5, detected by transcriptome (Mimura et al., 2021) and reverse transcription–quantitative polymerase chain reaction (RT–qPCR) of this study (Supplemental Figure S1), and as expected, callose accumulation was largely eliminated from mel2 anther locules during premeiosis and meiosis (Supplemental Figure S2). Among 10 rice GSL genes, OsGSL5 is the only gene expressing preferentially and abundantly in anthers during meiosis and post-meiosis (Yamaguchi et al., 2006; Shi et al., 2015; Supplemental Figure S3). These findings suggest an unknown association of callose deposition with meiotic entry control in plants.
This study demonstrates that OsGSL5 is responsible for hyper callose accumulation in extracellular spaces of anther locules at premeiosis and early meiosis, in addition to the role in late meiosis and pollen development as previously reported (Shi et al., 2015), and has an indispensable role in proper initiation of male meiosis in rice.
Results
OsGSL5 impacts pollen viability and seed fertility
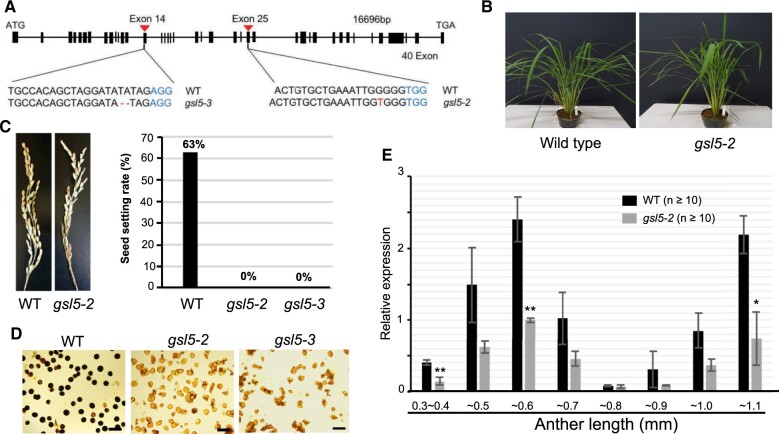
OsGSL5 is reported to be essential for pollen fertility in rice (Shi et al., 2015). Thus, to assess OsGSL5’s impact on male sporogenesis and meiosis, we exploited clustered regulatory interspaced short palindromic repeats (CRISPR)-CRISPR associated protein9 (Cas9) strategy to directly induce biallelic mutations in OsGSL5 locus. Out of the 32 CRISPR-edited plantlets screened, we obtained two independent knockout lines, Osgsl5-2 and Osgsl5-3, in which 1 bp (A) was inserted on the 25th exon and 2 bp (TA) were deleted on the 14th exon, respectively (Figure 2A). Both Osgsl5 lines were largely comparable to wild-type (WT) plants in vegetative growth and panicles and spikelets morphologies under the same condition (Figure 2, B and C), except for 2 weeks-later heading. However, both lines set no seeds, while WT plants had 63% fertility (Figure 2C;Supplemental Table S2). Pollen viability was 0.5% and 1.2% in Osgsl5-2 and Osgsl5-3, respectively, whereas it was 90.4% in the WT (Figure 2D;Supplemental Figure S4). These results were largely consistent to those previously reported (Shi et al., 2015).
Figure 2.
The Osgsl5 mutant phenotype and OsGSL5 gene expression. A, Nucleotides deletion and insertion sites in Osgsl5-2 and Osgsl5-3 knock-out mutants. B, The vegetative plant growth was comparable between WT and Osgsl5-2. C, Images of panicles at seed filling stage of WT and Osgsl5-2 (left), and the graph showing seed setting rate of WT, Osgsl5-2, and Osgsl5-3 (right). D, Pollen viability test by iodine–potassium iodide staining of WT (left), Osgsl5-2 (middle), and Osgsl5-3 (right). Darkly and faintly stained grains were categorized as viable and nonviable pollen, respectively. Bar = 40 µm. E, The relative expression levels of OsGSL5 transcripts in WT and Osgsl5-2 anthers by RT-qPCR. Errors bars each indicate the standard deviation of the mean of three biological replicates. The n value in parentheses is the number of florets used in each replicate. Asterisks indicate significant differences (*P < 0.05, **P < 0.01 by Student’s t test) between WT and Osgsl5-2.
In WT plants, the OsGSL5 transcript level was highest in anthers, while it was undetected in vegetative organs and slightly detectable in pistils (Supplemental Figure S6). It was significantly enhanced (P < 0.05 by Student’s t test) in 0.4–0.5mm anthers at premeiotic interphase, three-fold more than the level in younger 0.3–0.4mm anthers which contain SPCs proliferating mitotically (mitotic SPC stage). The expression peaked at 0.5–0.6mm anthers around leptotene to pachytene. In 0.6- to 0.7mm anthers around diplotene to telophase I, the OsGSL5 level was reduced to half or less of that at the former stage, and again elevated in 0.8–1.1mm anthers at tetrad and microspore stages (Figure 2E). Though the Osgsl5-2 mutant developed anthers with normal appearance (Supplemental Figure S7B), the OsGSL5 transcript level was lowered in all Osgsl5-2 anthers examined, and significantly reduced at premeiotic interphase, early prophase I and microspore stages (Figure 2E).
Considering the downregulation of OsGSL5 transcription and reduced fertilities of pollen and seeds similarly in two independent lines, we concluded that CRISPR–Cas9-mediated frameshift mutations within OsGSL5 coding sequence caused all above phenotypes.
The anther length is broadly utilized as a standard to assess meiotic events in many angiosperm species including rice, as it has a rough collinearity with respective meiotic stages (Itoh et al., 2005) (Figure 1). To confirm if Osgsl5 mutant anthers retain this collinearity, we examined the expression profiles of tapetum-specific genes, TDR INTERACTING PROTEIN2 (TIP2; Fu et al., 2014) and ETERNAL TAPETUM1 (EAT1, Niu et al., 2013). The expression peaks of both genes appeared at a similar level in both WT and Osgsl5-2 anthers (Supplemental Figure S7A). Noteworthily, the anther lengths where bimodal peaks of EAT1 gene (Ono et al. 2018) arose were comparable (Supplemental Figure S7A), and further, no difference in the layered structure of meiotic anthers was observed between WT and Osgsl5 plants (Supplemental Figure S7B). These results indicate that the Osgsl5 mutation unlikely affects the collinearity between anther lengths and meiotic stages, in addition to earlier anther morphogenesis. Thus, we utilized the anther length as a standard for comparison of meiotic events between WT and Osgsl5 mutants below.
OsGSL5 impacts callose accumulation in premeiotic and meiotic anther locules
We monitored the detailed pattern of callose accumulation in rice meiotic anthers by aniline blue that specifically stains β-1,3-glucan chains. The callose amount in WT anther locules, which was at an undetectable level during the mitotic SPC stage (Figure 3A), started to fulfill the extracellular spaces between cell walls and cell membranes at both PMC–PMC junctions and PMC–TC interfaces (Figure 3B). When PMCs entered into meiotic leptotene and zygotene, callose deposition was limited to PMC–PMC junctions (Figure 3, C and D). At subsequent pachytene to diakinesis stages, it can be seen enclosing PMCs with rounder shape (Figure 3, E–G). At dyad and tetrad stages, callose was detected on newly formed equatorial cell plates in addition to outer cell surfaces (Figure 3, H–I). After release of microspores to anther locules, callose can be seen fully enclosing microspores (Figure 3J). In both gsl5-2 and gsl5-3 anthers, callose signals were extremely diminished through all above stages (Figure 3, K–T).
Figure 3.
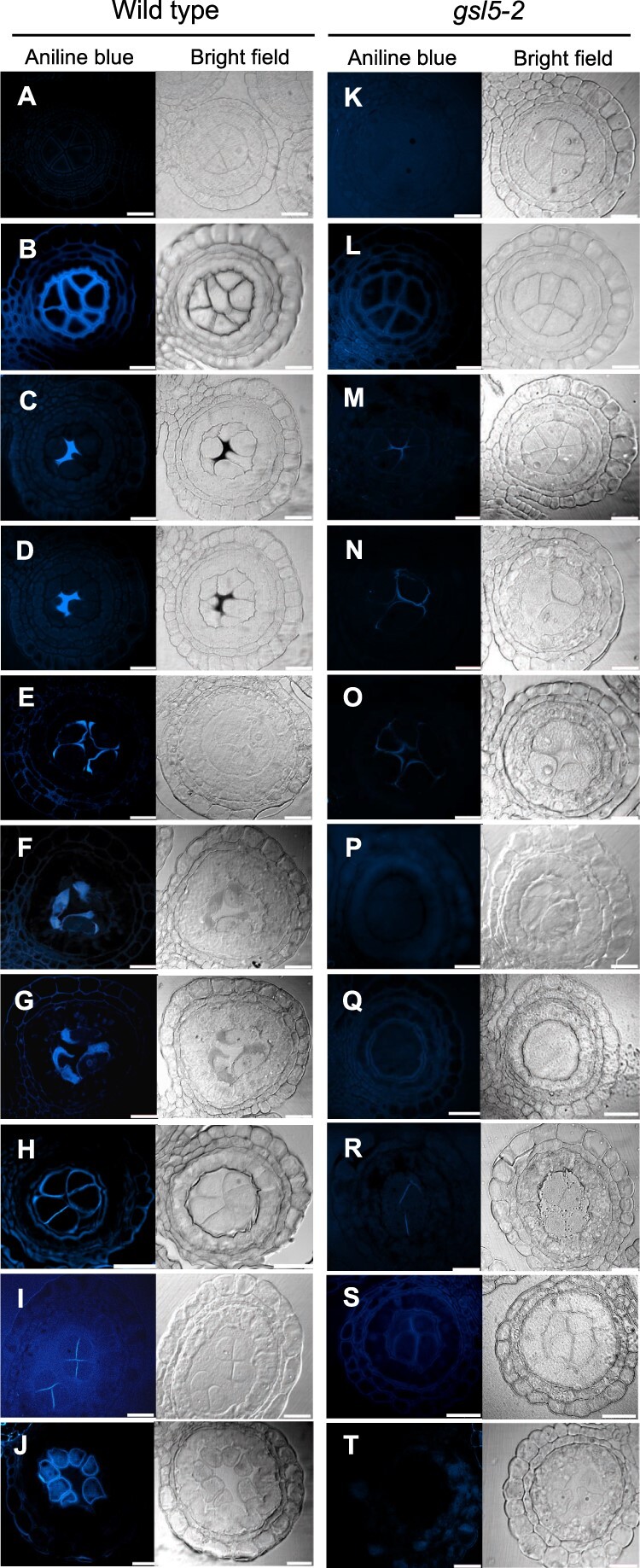
Callose accumulation during male meiosis in WT and Osgsl5-2 anthers. Callose accumulation pattern was monitored by aniline blue staining at different male meiosis substages. A and K, Mitotic SPC stage; B and L, Premeiotic interphase; C and M, Leptotene; D and N, Zygotene; E and O, Pachytene; F and P, Diplotene; G and Q, Diakinesis; H and R, Dyad; I and S, Tetrad; J and T, Microspore. An arrowhead in the inset in (B) indicates the cell wall unstained with aniline blue. Bar = 20 µm.
A beginning stage of callose accumulation was further observed with the anti-β-1,3-glucan (callose)-directed antibody, to trace differences in callose amount more sensitively. In young 0.4mm or less anthers, four microsporangia of a same anther sometimes show different callose patterns with each other (Supplemental Figure S8), suggesting it being at the very beginning of premeiotic callose accumulation. In this stage, locules frequently retained callose limited to PMC–PMC junctions (Supplemental Figure S8), implicating that premeiotic callose accumulation initiates around PMC–PMC junctions, but not at PMC–TC interfaces.
Localization of callose and OsGSL5 protein in anther locules
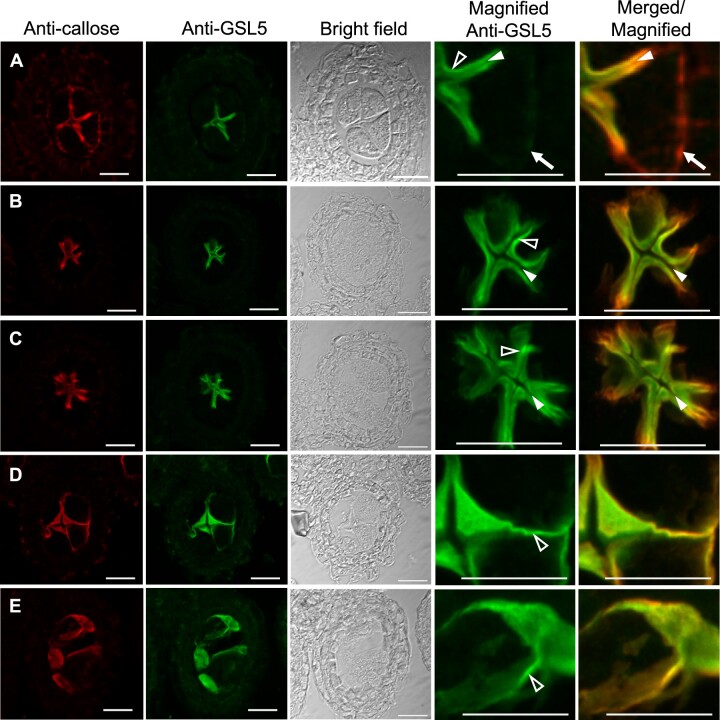
To investigate spatio-temporal localization of OsGSL5 protein, we produced anti-OsGSL5 antiserum (Supplemental Figure S5) to perform co-immunostaining with the anti-callose antibody on anther sections. After the initiation of callose accumulation at PMC–PMC junctions (Supplemental Figure S8), callose fulfilled extracellular spaces of both PMC–PMC and PMC–TC regions in anther locules (Figure 4A), as observed in aniline blue staining (Figure 3B). In contrast to callose staining, OsGSL5 localization was limited only at PMC–PMC junctions in the same anther section (Figure 4A). In subsequent leptotene, the thick callose signal declined again at PMC–TC interfaces and became almost overlapped with OsGSL5 localization at PMC–PMC junctions (Figure 4, B and C). Through all stages observed, the strongest linear OsGSL5 signals were always observed at optically sectioned edges of PMCs (open arrowheads in Figure 4), suggesting their association with the plasma membrane, well consistent to the fact that OsGSL5 and its orthologs are membrane-anchored proteins containing 15 transmembrane domains (Yamaguchi et al., 2006; Shi et al., 2015; Supplemental Figure S5A). This trend became more obvious at subsequent pachytene–diakinesis stages, where PMCs gradually took a spherical shape (Figure 4, D and E). During these stages, OsGSL5 localization (Figure 4, D and E) was gradually corresponding to callose deposition on PMC surfaces (Figures 3, E, F, 4, D and E). The OsGSL5 signal was at undetectable level in Osgsl5 mutant anthers (Supplemental Figure S9).
Figure 4.
Co-immunostaining of callose and OsGSL5 protein in WT anthers. A, Premeiotic interphase; B, Leptotene; C, Zygotene; D, Pachytene; E, Diplotene–Diakinesis. Bar = 20 µm. In (A), white arrows show the region retaining callose deposition, but lacking OsGSL5 localization, at the PMC–TC interface. The closed and open arrowheads indicate unstained cell wall regions and linearly aligned OsGSL5 signals on PMC plasma membranes, respectively.
These results strongly support that OsGSL5 takes part in callose biosynthesis during meiosis stages, and taken together with RT–qPCR results, reconfirm that the Osgsl5 mutations are null alleles.
OsGSL5 impacts male meiosis progression and chromosome behaviors
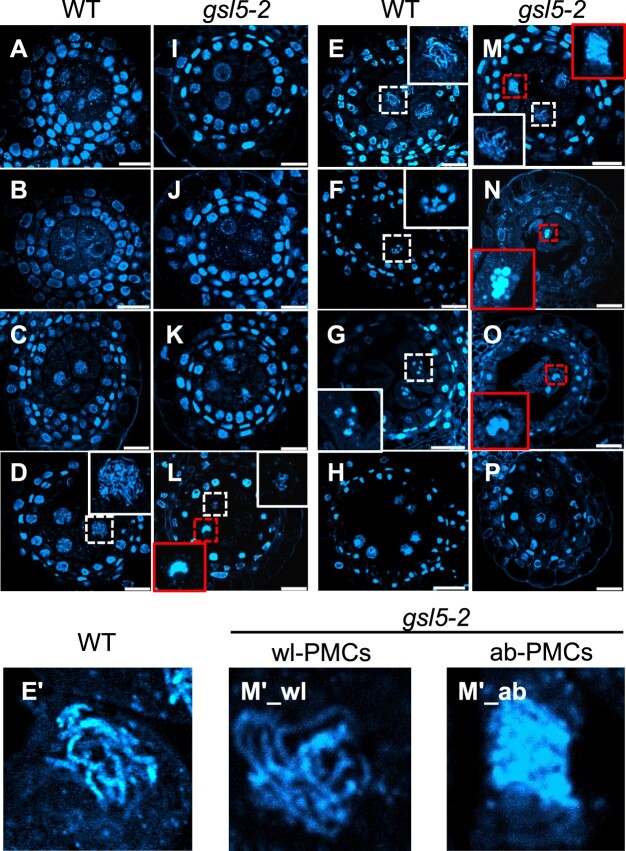
The chromosome behavior was assessed at respective meiotic stages determined by both anther lengths and chromosomal morphologies on plastic-embedded sections. In the WT, PMCs began meiotic chromosome condensation and displayed thin thread-like appearance by leptotene (Figure 5, A–C). Chromosomes were further condensed during zygotene (Figure 5D), at which homologous chromosomes begin to be synapsed. At pachytene when synapsis is completed, PMCs displayed thick-threads of homologous pairs (Figure 5, E and E′). Bivalent chromosomes were further condensed through diplotene and diakinesis (Figure 5, F and G), and either of homologous pair was delivered to opposite poles during meta/anaphase I, and eventually to either cell of the dyad (Figure 5H). In both gsl5-2 and gsl5-3 anthers, a conspicuous abnormality on male meiotic chromosomes emerged during prophase I (Figure 5, I–O), in which two different types of PMCs were contained together within a same anther—one type carrying meiotic chromosomes with seemingly normal appearance, named wildtype-like PMC (wl-PMC; Figure 5, L, M, M′_wl), and another displaying aberrant behaviors of meiotic chromosomes, such as tight aggregations and impaired homologous pairings, named aberrant PMC (ab-PMC; Figure 5, L–O, M′_ab). The chromosome spreading method further enabled to clarify the abnormality in Osgsl5 ab-PMCs (Supplemental Figure S10).
Figure 5.
Dynamics of chromosomal behavior in WT and Osgsl5-2 anthers. Plastic-embedded anthers cross-sections stained with DAPI for chromosomes observation. A and I, Mitosis; B and J, Interphase; C and K, Leptotene; D and L, Zygotene; E and M, Pachytene; F and N, Diplotene; G and O, Diakinesis; H and P, Dyad. Bar = 20 µm. Note that Osgsl5-2 mutant anthers contained two types PMCs with respect to chromosome appearance (wl-PMC and ab-PMC, see the text). The insets in each panel are magnified views of nuclei enclosed with dashed squares, in which white and red squares indicate nuclei of wl-PMCs and ab-PMCs, respectively. The insets in (E) and (M) are further magnified and shown in E′, M′1, and M′2, as examples of WT PMC, wl-PMC, and ab-PMC, respectively.
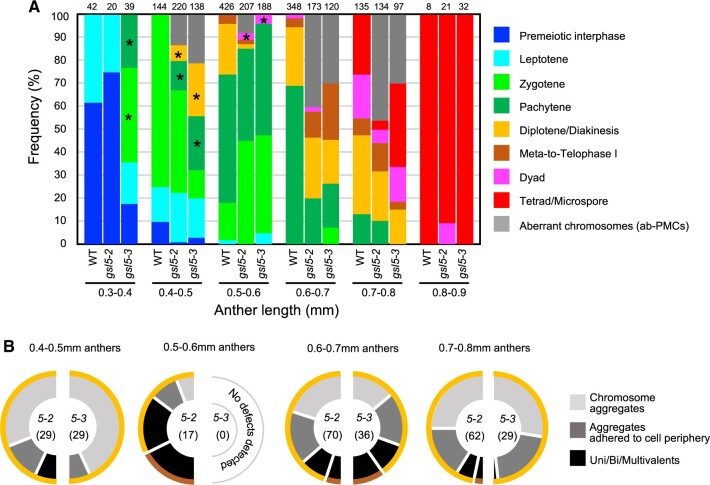
In above observations, we noticed that the Osgsl5 mutation somewhat affected the time-course progression of male meiosis, in addition to the segregation of two PMC types. Thus, to make the Osgsl5 impact on meiosis progression clearer, the frequency of each meiosis stage observed in PMCs was plotted along anther lengths. In 0.4- to 0.8mm Osgsl5 anthers, ab-PMCs appeared irregularly in the range of 8.2%–46.2% of all PMCs observed (Figure 6A;Supplemental Table S3). Another point of interest was a precocious initiation of several meiotic stages in not all, but a part of wl-PMCs, compared to WT PMCs. Interestingly, despite a subset of wl-PMCs displaying chromosomal features particularly of zygotene to dyad were observed earlier than WT PMC stages (asterisks in Figure 6A), male meiosis was completed similarly in 0.9mm anthers of both WT and Osgsl5 plants (Figure 6A;Supplemental Table S3).
Figure 6.
Male meiosis progression and ab-PMC appearance in WT and Osgsl5 anthers. A, Frequency of PMC stage observed with respect to anther lengths in WT and Osgsl5 mutants, in which the frequency of ab-PMCs in Osgsl5 anthers was shown as gray bars. The number at the top of each bar indicates the absolute number of cells counted. The wl-PMC stages marked with asterisks indicate that those stages emerged much earlier than comparable stages observed in WT anthers. B, Each half-donut graph indicates frequency of three different classes for aberrant chromosomal morphologies and behaviors in Osgsl5-2 or Osgsl5-3 ab-PMCs observed along respective anther lengths. Colored outer rims on a half donut indicate meiotic stages of wl-PMCs that concomitantly emerge with respective classes of ab-PMCs. Definition of outer rim colors is consistent to that in (A). The number in parentheses represents the absolute number of cells counted.
In 0.4 to 0.5mm Osgsl5 anthers, >85% of ab-PMCs retained chromosomal aggregates, and concomitantly appeared with diplotene/diakinesis-like wl-PMCs (Figure 6B;Supplemental Figure S10 and Supplemental Table S3). In 0.6 to 0.7mm gsl5 anthers, about >27% of ab-PMCs had more condensed univalents and/or multivalents in addition to normal bivalents, concomitant with wl-PMCs retaining diplotene/diakinesis- or meta/telophase I-like chromosomes (Figure 6B;Supplemental Figure S10 and Supplemental Table S3). In 0.7- to 0.8mm anthers, >80% ab-PMCs again displayed less-condensed aggregated chromosomes concomitantly with diplotene/diakinesis-like wl-PMCs (Figure 6B;Supplemental Figure S10 and Supplemental Table S3). The reason was ambiguous, but it may suggest that aberrant aggregations of chromosomes at early prophase I resulted in univalent/multivalent formation at later stages, and that ab-PMCs retaining univalent/multivalents were abortive and undetected during late prophase I.
Osgsl5 mutation caused precocious initiation of male meiosis
An earlier occurrence of meiotic prophase-I stages frequent in Osgsl5 anthers (Figure 6) raises a possibility that it is attributable to defects in premeiotic events. To test this hypothesis, we performed immunostaining of three meiotic initiation markers namely, HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS2 (PAIR2), PAIR3, and REC8 (Figure 7;Supplemental Figure S11). Rice PAIR2 promotes homologous chromosome synapsis, and its accumulation within the nucleus is reported to initiate during premeiotic interphase, just as following the initiation of premeiotic DNA replication (Nonomura et al., 2006). Thus, PAIR2 can be used as a maker to infer the timing of replication initiation in anthers at premeiotic interphase. PAIR3 and REC8 are a chromosome axis protein and a meiotic cohesion subunit protein, respectively, and both proteins are required for proper PAIR2 loading onto meiotic chromosome axes (Shao et al., 2011; Wang et al., 2011), thus these can serve as ideal markers for meiosis initiation time.
Figure 7.
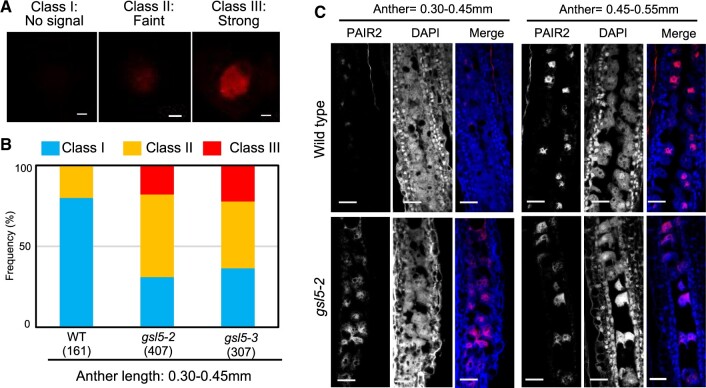
Precocious accumulation of PAIR2 in Osgsl5 PMC nuclei. A, Immunostaining of PAIR2 in isolated premeiotic PMCs. The degree of PAIR2 accumulation in the PMC nucleus was categorized into three classes based on the immunofluorescent intensity. Bar = 2.5 µm. B, The rate of three PAIR2 classes in each of WT, Osgsl5-2, and Osgsl5-3 premeiotic anthers. C, Immunostaining of PAIR2 on longitudinal premeiotic anther sections with anti-PAIR2 antibody. Bar= 50 µm.
PAIR2, PAIR3, and REC8 nucleoplasmic signals were classified into three classes based on their intensities; absent (class I), faint (class II), and strong (class III; Figure 7A;Supplemental Figure S11). The PAIR2 class II is supposed to be a stage following premeiotic replication initiation. In the 0.30- to 0.45-mm anthers observed in the WT, 80% of PMCs showed no PAIR2 signal (class I), suggesting the cells being at mitotic SPC stage or before replication, and only 20% showed class II signals. In contrast, in anthers with the same lengths, around 60%–70% of Osgsl5 PMCs showed either class II or III signals (Figure 7, B and C), suggesting the precocious initiation of meiotic S-phase in Osgsl5 PMCs. Similarly, PAIR3 and REC8 signals were quantified according to their appearance time in Osgsl5 compared to the WT (Supplemental Figure S11). In 0.40 to 0.45mm anthers observed, 65% of WT PMCs showed no PAIR3 signal (class I), and 35% showed either class II or class III signals. In contrast, in the same anther stage, 52%–100% of Osgsl5 PMCs showed either class II or III signals (Supplemental Figure S11B). In the same anther stages as above, REC8 class II or class III signals were observed in 39% WT PMCs. However, Osgsl5 PMCs displaying class II or class III signals ranged from 58% to 98% (Supplemental Figure S11B). These results clearly indicate that OsGSL5 has an impact on timely initiation of premeiotic events such as DNA replication and the formation of meiosis-specific chromosomal axes.
Osgsl5 mutation disrupts homologous synapsis
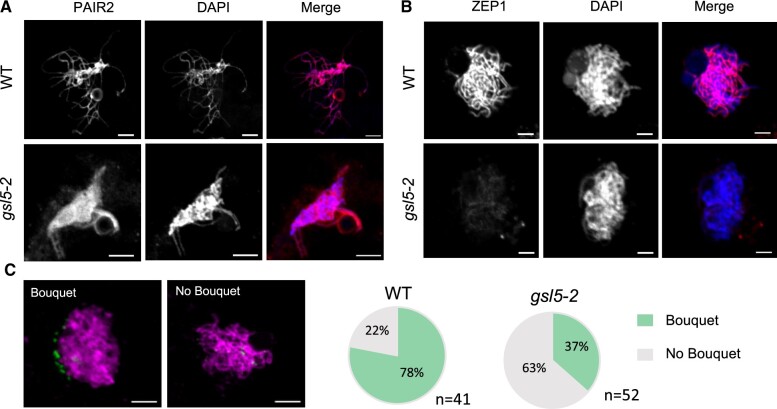
Next, we asked whether key meiotic events such as homologous chromosome synapsis was affected in Osgsl5 PMCs or not. PAIR2 was normally loaded on meiotic chromosomes in both WT PMCs (n = 168) and Osgsl5-2 PMCs observed (n = 134; Figure 8A), which further ensured that the premeiotic accumulation of PAIR2 in PMC nuclei occurs normally even in Osgsl5 mutants (Figure 7). In contrast, loading of ZEP1, which is a transverse filament component of synaptonemal complex and governs meiotic crossover numbers in rice (Wang et al., 2010), was severely diminished in all Osgsl5-2 PMCs at zygotene and pachytene (n = 85), while it was constantly observed in all WT PMCs in same stages (n = 92; Figure 8B). Though a chromosomal aggregate characteristic of ab-PMCs was hard to be distinguished at these stages, the result suggests that failed ZEP1 loading took place in both wl- and ab-PMCs in Osgsl5 anthers.
Figure 8.
ZEP1 loading to and bouquet frequency of meiotic chromosomes affected, but PAIR2 loading unaffected in Osgsl5 anthers. A, Immunostaining of PAIR2 in WT (n = 168) and Osgsl5-2 (n = 134). PAIR2 loading is unaffected in Osgsl5 mutant. B, Immunostaining of ZEP1 in WT (n = 92) and Osgsl5-2 (n = 92). ZEP1 loading onto chromosomes was inhibited in Osgsl5 mutant. C, Bouquet structures visualized by immunostaining of telomere specific OsPOT1 during leptotene–zygotene transition (0.50–0.55 mm anther) in PMCs of WT (n = 41) and Osgsl5-2 (n = 52). Bar = 5 µm.
Telomere bouquet is a chromosomal arrangement important for meiotic homologous pairing and synapsis in many eukaryotes including rice (Zhang et al., 2017). In anthers around leptotene and zygotene, only 37% PMCs displayed the bouquet in Osgsl5-2 mutant (n = 52), while 78% displayed in the WT (n = 41; Figure 8C). In 0.60 to 0.70mm anthers, no bouquet was observed both in WT and Osgsl5-2 mutant (Supplemental Figure S12), suggesting the Osgsl5 mutation restricted bouquet formation, but not dissolution.
Altered transcript levels of key meiotic and callose metabolizing genes in gsl5 anthers
The transcriptional levels of six genes having key roles in meiosis initiation and two genes involved in callose metabolism were quantified in meiotic anthers by RT–qPCR. Of the six genes, Lepoto1 and AM1 were significantly upregulated at premeiosis (0.3 to 0.4mm anthers). MEL1, SPL, and PAIR2 expression were also upregulated in young anthers compared to the WT (Supplemental Figure S13).
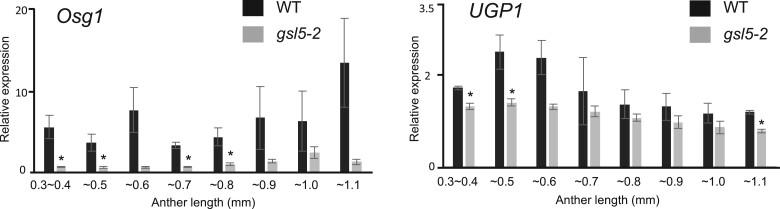
Noteworthily, two callose metabolizing genes, Osg1 gene encoding a tapetum-specific β-1,3-glucanase (Wan et al., 2011) and UGP1 gene encoding a UDP-glucose phosphorylase involved in biosynthesis of cell wall components including callose (Chen et al., 2007), were both downregulated in Osgsl5-2 anthers through all meiotic stages (Figure 9).
Figure 9.
Reduced expression of rice Osg1 and UGP1 genes in Osgsl5-2 anthers. Relative expression levels of callose metabolizing genes, Osg1 and UGP1, in WT and Osgsl5-2 anthers by RT–qPCR. Error bars indicate standard deviations of means of three biological replicates. Asterisks indicate significant differences (*P < 0.05 by Student’s t test) between WT and Osgsl5-2.
Discussion
OsGSL5-dependent callose accumulation in rice anthers during meiosis
This study gained important insights into the role of OsGSL5 callose synthase in callose deposition at extracellular spaces of anther locules during premeiotic and meiotic stages (Figure 3). Double immunostaining of OsGSL5 and callose revealed that subcellular OsGSL5 localizations are restricted on PMC plasma membranes facing PMC–PMC junctions, where multiple PMCs meet with each other along the central axis of anther locules, and served callose polysaccharides to extracellular spaces of PMC–PMC junctions through premeiotic interphase and early prophase I (Figure 4, A–D). Another important point is that during premeiosis, callose deposition was observed at PMC–TC interfaces in addition to at PMC–PMC junctions, whereas OsGSL5 localization was limited to PMC–PMC junctions (Figure 4A). Given that callose accumulation begins at PMC–PMC junctions, but not at PMC–TC interphases (Supplemental Figure S8), callose synthesized at PMC–PMC junction is likely supplied for fulfilling PMC–TC interfaces during premeiotic interphase, further affirming PMC–PMC junctions as a callose-producing center.
Fluctuation in callose levels, as above mentioned, generally involves the activities of two counteracting enzyme types: β-1,3-glucan synthases and hydrolases (Frankel et al., 1969; Stieglitz and Stern, 1973). A rice β-1,3-glucanase, Osg1, functions in timely callose degradation on pollen grains and impacts pollen fertility (Yamaguchi et al., 2006; Wan et al., 2011), and Osg1 gene expression was reduced in Osgsl5 mutant (Figure 9), likely suggesting a positive feedback regulation of Osg1 transcription by elevated callose levels to maintain callose homeostasis in anthers. Rice UGP1 is a UDP-glucose pyrophosphorylase that catalyzes the production of UDP-glucose, a substrate of glucan synthases, including GSL5. In UGP1 RNAi plants, callose accumulation in anther was substantially diminished during both meiotic and post-meiotic stages (Chen et al., 2007) which is consistent with our results (Figure 3). Thus, it is possible that UDP-glucose catalyzed by UGP1 is used for GSL5-dependent callose synthesis, leading to callose accumulation during meiosis and post-meiosis, and Osg1 probably acts in callose fluctuation antagonistically to the UGP1–GSL5 pathway.
Impact of callose accumulation on male meiosis initiation
A key insight gained from this study is the impact of OsGSL5 on male meiosis initiation and progression (Figures 6 and 7). In addition to defects in meiosis time-course, Osgsl5 mutants contained ab-PMCs that exhibited several other defects in chromosome behavior and condensation, homologous synapses, and reduced bouquet structures, along with wl-PMCs that exhibited a normal appearance of meiotic chromosomes (Figures 5 and 8;Supplemental Figure S10). Even if part of Osgsl5 PMCs passed through meiosis, callose deposition was defective in all surviving tetrad spores (Figure 3T), resulting in male sterility as previously reported (Shi et al., 2015).
Shi et al. (2015) concluded that OsGSL5 is responsible for callose accumulation at post meiosis stages, but not during early meiosis. However, this conclusion was led only by a snapshot of whole-mount staining of anthers with aniline blue, but not of sectioning, perhaps resulting in oversight of OsGSL5 impact during premeiotic interphase and prophase I stages. Similarly, a frequent appearance of survival spores, probably derived from wl-PMCs, may explain the reason why the impact of callose function during male meiosis was underestimated in previous studies using mutant plants lacking callose synthesis.
Microscopic observations of Osgsl5 mutant PMCs implicated that wl-PMCs with meiotic chromosomes lacked ZEP1 loading (Figure 8B), but achieved seemingly normal bivalent formation and disjunction of homologous pairs in meiosis I (Supplemental Figure S10, F and I). It is not surprising, because in rice zep1 mutants, bivalents were formed at the normal level and a considerable number of tetrads were formed, while the viability of gametes greatly reduced probably due to aberrant chromosomal condensation in microspores (Wang et al., 2010). Rather, it is a wonder why meiotic chromosomes of Osgsl5 wl-PMCs lose the capacity for ZEP1 loading. Furthermore, the frequent appearance of ab-PMCs is difficult to be explained only by the loss of ZEP1, because no chromosomal aggregate was reported during prophase I in zep1 mutants (Wang et al., 2010). A similar loss of ZEP1 loading is observed in mel2 mutant PMCs (Nonomura et al., 2011), in which OsGSL5 expression is significantly reduced (Supplemental Figure S1). Furthermore, PMCs at various cell cycle stages are segregated in the same mel2 anther due to asynchronous initiation of DNA replication (Nonomura et al., 2011). This observation may account for the appearance of two different PMC types in Osgsl5 anthers (Figures 5 and 6), occurring together with the precocious male-meiosis entry (Figure 7). Rice LEPTOTENE1 (LEPTO1) is a type-B response regulator that participates in establishing key features of meiotic leptotene chromosomes. In lepto1 mutant anthers, the expression levels of both OsGSL5 and UGP1 genes were reduced significantly and callose was depleted during meiosis (Zhao et al., 2018). Interestingly, the loading of important meiotic chromosome elements, such as OsREC8, OsAM1, and ZEP1, was also defective in lepto1 (Zhao et al., 2018). The past observations and findings of this study together strongly implicate that callose filling of the extracellular spaces of anther locules is an important step for proper PMC differentiation and/or male meiosis initiation, while the underlying mechanisms remain elusive.
Recent studies often suggest the role of callose accumulation in male meiosis via cross-talking among anther locular cells (Plackett et al., 2014; Zhai et al., 2015; Liu et al., 2017; Huang et al., 2019; Lei and Liu, 2020). PMCs are interconnected with each other and with the surrounding TCs through PD or cytomictic channels at early meiosis (Heslop-Harrison, 1964; Mamun et al., 2005; Mursalimov et al., 2010; Mursalimov et al., 2013). However, during the transition to meiosis, such intercellular connections are solved/blocked likely by hyper callose accumulation in anther locules (Sager and Lee, 2018). In addition to controlling the symplastic pathway, callose accumulation is thought to function as a molecular filter for signaling from TCs to PMCs via apoplastic pathway (Clement and Audran, 1995; Roschzttardtz et al., 2013). Biochemical evidence further suggests that callose deposition can alter the permeability and plasticity of cell walls in coexistence with cell wall components like cellulose (Abou-Saleh et al., 2018). The above findings imply that hyper callose accumulation has the potential to bring dramatic microenvironmental changes to its surrounding PMCs by controlling symplastic or apoplastic pathways or both, which entails being confirmed in future studies.
In summary, this study demonstrates that GSL5-dependent callose deposition during meiosis is crucial for proper timing of meiosis initiation and subsequent progression, and upon callose depletion at this point of time perturbs normal meiosis onset and consequently poses severe impact on several meiotic events. This study sheds light on the importance of callose in meiosis of flowering plants which is an important progress in the field of plant reproductive biology, and is a step toward understanding the mechanistic basis of GSL5 and callose function in meiosis initiation.
Materials and methods
Plant materials and growth conditions
For target mutagenesis of OsGSL5 gene (Os06g0182300), potential CRISPR guide-RNA sequences were designed using CRISPR-P v2.0 software (Liu et al., 2017). Double-stranded DNAs were produced from oligo DNA pairs of gRNAF1/gRNAR1 and gRNAF2/gRNAR2 for Osgsl5-2 and Osgsl5-3 (Supplemental Table S2), respectively, by annealing. After cloning into pU6 vector, the pU6 promoter-fused double-stranded DNA was transferred to pZD shuttle vector (Mikami et al., 2015), and introduced into seed-derived calli of rice (Oryza sativa japonica “Nipponbare”) by the method previously reported (Hiei et al., 1994). All plants were grown in growth chambers at 30°C day and 25°C night temperature and 70% relative humidity with a daylength of 12 h.
Pollen and seed fertility tests
For pollen fertility, anthers extracted from fixed panicles with Carnoy’s fluid were squashed in iodine–potassium iodide solution and viable pollen grains stained were counted under a light microscope (BX50; Olympus). For seed fertility, the ratio of fertile spikelet numbers was counted in each of the five panicles and averaged.
Cytological observations
For observations of chromosome behaviors and callose deposition on anther sections, anthers were fixed in 4% (w/v) paraformaldehyde (PFA)/1× PBS. After the removal of lemma and palea of florets, anthers were dehydrated in ethanol-graded series for 30–60 min each, followed by infiltration in embedder with hardener1 of Technovit 7100 (Kulzer Technique), overnight on rotor at 4°C. The solution was replaced with fresh Technovit with hardener1 every 6 h and incubated overnight on rotor at 4°C. Then, anthers were transferred to a cryo-dish with Technovit polymerized by the addition of harderner2 and placed at 50°C–60°C for hardening. Plastic-embedded sections with 4- to 6-µm thickness were taken using the microtome R2255 (Leica), and air dried at room temperature. The section was stained for 25–30 min with 0.01% (v/v) aniline blue (Sigma Aldrich) in 0.1 M K3PO4 (pH 12) for callose, or with a drop of 1.5 µg/mL 4',6-diamidino-2-phenylindole (DAPI)/Vector shield (Vectorlabs) for chromosome observation. The images were captured under a confocal laser scanning microscope system (FV300; Olympus), and processed with ImageJ (https://imagej.nih.gov/ij/docs/intro.html).
For chromosomal spreads, whole panicles were fixed in Carnoy’s fluid and stored at 4°C until use. Fixed anthers incubated with 0.1% (w/v) FeCl2 overnight were squashed in acetocarmine solution (1% (w/v) carmine (Merk)/45% (v/v) acetic acid) with forceps on a clean glass slide. After the quick removal of anther-wall remnants, a suspension of released PMCs was covered with a cover slip, and gently heat-treated followed by gentle thumb compression. The images of chromosomal spreads were captured under a light microscope (BX50) with a DP2-SAL CCD camera system (Olympus). The number of PMCs each classified by certain phenotypes in chromosome behaviors was counted and used for quantification together with those observed in plastic-embedded sections.
RT–qPCR
To quantify the transcript levels of rice genes, anthers and tissues were collected, immediately frozen in liquid nitrogen in a 2-mL tube with 2-mm beads, and homogenized on an automated shaker (BMS-A20TP; Bio Medical Science) at 1,100 rpm for 2 min. RNAs were extracted using a TRIZOL RNA extraction kit according to the manufacturer’s instructions (Invitrogen) and supplied to Super Script III First-Strand synthesis system (Invitrogen) for cDNA library construction. RT–qPCR was performed using a Real Time System (TP800; TAKARA biosystems) according to the manufacturer’s instructions. All primer sequences used for RT–qPCR are shown in Supplemental Table S2. The Rice Actin1 (RAc1) gene was used as an internal control to normalize the expression levels at all meiosis stages quantified.
Antibody production
To produce an antibody specific for OsGSL5 of 1910 amino acids (aa), the cDNA sequence encoding the 1,009–1,260 aa position was amplified using the above cDNA library as a template (Supplemental Figure S5 and Supplemental Table S2), and cloned into pDEST17 vectors (Invitrogen). The His-tagged protein expressed in Escherichia coli strain BL21-AI (Invitrogen) was purified using Ni-NTA agarose resin (FUJIFILM) and immunized to rabbits and guinea pigs.
To observe telomere behaviors in PMCs, the antibody was raised against rice PROTECTION OF TELOMERE1 (OsPOT1), which is encoded by a single-gene locus Os04g0467800 (LOC_Os04g39280.1), while the Arabidopsis (A. thaliana) genome has two putatively paralogous loci, POT1a and POT1b (Shakirov et al., 2005). Procedures to raise antisera are the same as above.
Immunofluorescent staining of PMCs and tissue sections
Young rice panicles were fixed with 4% (w/v) PFA/1× PMEG (25mM PIPES, 5mM EGTA, 2.5mM MgSO4, 4% (v/v) glycerol, and 0.2% (v/v) dimethyl sulfoxide (DMSO), pH 6.8), followed by washing six times with 1× PMEG, and stored at 4°C until use (Nonomura et al., 2006). For immunofluorescence of PMCs, fixed anthers were treated with the enzyme cocktail of 2% (w/v) cellulase Onozuka-RS (Yakult)/0.3% (w/v) pectolyase Y-23/0.5% (w/v) macerozyme-R10 (FUJIFILM Wako)/0.375% (w/v) Cytohelicase (Sigma-Aldrich) in 1× PME (same as 1× PMEG except without glycerol) for permeabilization on MAS-coating glass slide MAS-02 (Matsunami Glass), squashed by forceps and used for immunostaining, as described in Nonomura et al. (2006).
Immunostaining of anther sections was done as described by Tsuda and Chuck (2019) with minor modifications. Briefly, anthers were dehydrated in graded ethanol series and Histo-Clear (Cosmo bio co. Ltd), embedded into Paraplast paraffin wax (McCormick Scientific). Paraffin blocks containing anther samples were trimmed and stored at 4°C until use. The blocks were sectioned into 8- to 10-µm-thick sections by the microtome. Dewaxed and rehydrated samples were incubated with primary antibodies. The rabbit anti-OsGSL5, the rabbit anti-OsPOT1 (this study), the rabbit anti-PAIR2 (Nonomura et al., 2006), guinea pig anti-PAIR3 (Wang et al., 2011), anti-mice-REC8 (Shao et al., 2011), and rat anti-ZEP1 antibodies (Nonomura et al., 2011) were diluted to 1/100, 1/3,000, 1/3,000, 1/200, 1/200, and 1/1,000, respectively, with 3% (w/v) BSA/1× PMEG and used as primary antibodies. For callose immunostaining, monoclonal antibody specific to β-1,3-glucan (callose; Biosupplies Australia) was diluted to 1/1,000 and used as a primary antibody.
In both squash and sectioning methods, secondary antibodies Alexa fluor 488 (Abcam) and Cy3-conjugated IgG (Merck) of 1/200 dilution were used for detection. Immunofluorescent images were captured by the Fluoview FV300 CSLM system (Olympus). UV filters (380–420 nm) were used for DAPI and aniline blue stain, eGFP filters (505–525 nm) used for green dyes, BA 560–600 nm and BA 660If were used for red dyes. 1× Gain was used in all imaging conditions. Images were processed with ImageJ.
Accession numbers
Sequence data from this article can be found in the RAP-DB (Rice Annotation Project-database; https://rapdb.dna.affrc.go.jp) data libraries under accession numbers given in Supplemental Table 4.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcript levels of OsGSL5 gene quantified by RT–qPCR in WT and mel2 anthers.
Supplemental Figure S2. Callose accumulation in WT and mel2 anthers.
Supplemental Figure S3. Expression profile of OsGSL5 gene at various tissue developmental stages in WT rice plants.
Supplemental Figure S4. Pollen viability in WT and Osgsl5 mutant.
Supplemental Figure S5. OsGSL5 protein structure.
Supplemental Figure S6. The expression level of OsGSL5 transcripts in various vegetative tissues including male and female organs in WT and Osgsl5-2 plants.
Supplemental Figure S7. Anther development is unaffected in Osgsl5 mutants.
Supplemental Figure S8. Callose deposition at its beginning stage during premeiotic interphase in WT anthers.
Supplemental Figure S9. Immunostaining of OsGSL5 protein.
Supplemental Figure S10. Aberrant chromosome behaviors detected in osgsl5 mutant anthers by chromosome spreading technique.
Supplemental Figure S11. Precocious accumulation of PAIR3 and REC8 in Osgsl5 PMCs.
Supplemental Figure S12. Telomere bouquet structures visualized at late prophase stage.
Supplemental Figure S13. Transcript levels of various genes during mitosis to meiosis transition in WT and Osgsl5-2 anthers.
Supplemental Table S1. Oligo DNA sequences for guide RNA constructions and PCR primers employed in this study.
Supplemental Table S2. Seed fertility of WT, Osgsl5-2, and Osgsl5-3 plants.
Supplemental Table S3. PMC counting data on the basis of the graphs in Figure 6.
Supplemental Table S4. Gene accession numbers used in this study
Supplementary Material
Acknowledgments
We thank Dr Norio Komeda (NIG, SOKENDAI) for helping the production of anti-OsPOT1 antisera, and Dr Yoshihisa Oda (NIG) for reading manuscript and giving useful comments. The mel2 mutant used in this study was provided by NIG with support from National BioResource Project (NBRP) Rice, AMED, Japan. We also thank Drs Zhukuan Cheng and Wenqing Shi (CAS) for kindly providing us the PAIR3 and REC8 antibodies.
Contributor Information
Harsha Somashekar, Plant Cytogenetics Laboratory, Department of Gene Function and Phenomics, National Institute of Genetics, Mishima, Shizuoka 411-8540, Japan; Department of Genetics, School of Life Science, The Graduate University of Advanced Studies (SOKENDAI), Mishima, Shizuoka 411-8540, Japan.
Manaki Mimura, Plant Cytogenetics Laboratory, Department of Gene Function and Phenomics, National Institute of Genetics, Mishima, Shizuoka 411-8540, Japan.
Katsutoshi Tsuda, Plant Cytogenetics Laboratory, Department of Gene Function and Phenomics, National Institute of Genetics, Mishima, Shizuoka 411-8540, Japan; Department of Genetics, School of Life Science, The Graduate University of Advanced Studies (SOKENDAI), Mishima, Shizuoka 411-8540, Japan.
Ken-Ichi Nonomura, Plant Cytogenetics Laboratory, Department of Gene Function and Phenomics, National Institute of Genetics, Mishima, Shizuoka 411-8540, Japan; Department of Genetics, School of Life Science, The Graduate University of Advanced Studies (SOKENDAI), Mishima, Shizuoka 411-8540, Japan.
Funding
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant No. 21H04729 and 18H02181 (to K.I.N.), Bilateral Programs Grant No. JPJSBP120213510 (to K.I.N), and partly by SOKENDAI (to H.S.).
Conflict of interest statement. None declared.
H.S., M.M., and K.I.N. designed the research work. H.S. carried out most of the experiments. K.T. helped in the production of antibody against GSL5. M.M. guided in immunostaining experiments. H.S. and K.I.N. wrote the manuscript. M.M. and K.T. helped in drafting the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Ken-Ichi Nonomura (knonomur@nig.ac.jp).
References
- Abou-Saleh RH, Hernandez-Gomez MC, Amsbury S, Paniagua C, Bourdon M, Miyashima S, Helariutta Y, Fuller M, Budtova T, Connell SD, et al. (2018) Interactions between callose and cellulose revealed through the analysis of biopolymer mixtures. Nat Commun 9: 4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B, Ressayre A, Nadot S (2011) Correlation between pollen aperture pattern and callose deposition in late tetrad stage in three species producing atypical pollen grains. Am J Bot 98: 189–196 [DOI] [PubMed] [Google Scholar]
- Bhalla PL, Slattery HD (1984) Callose deposits make clover seeds impermeable to water. Ann Bot 53: 125–128 [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Kim JY (2009) Callose synthesis in higher plants. Plant Signal Behav 4: 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement C, Audran JC (1995) Anther wall layers control pollen sugar nutrition in Lilium. Protoplasma 187: 172–181 [Google Scholar]
- Dickinson HG, Bell PR (1976) The changes in the tapetum of pinus banksiana accompanying formation and maturation of the pollen. Ann Bot 40: 1101–1109 10.1093/oxfordjournals.aob.a085219 [DOI] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DP (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Dong X, Hong Z, Chatterjee J, Kim S, Verma DPS (2008) Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 229: 87–98 [DOI] [PubMed] [Google Scholar]
- Frankel R, Izhar S, Nitsan J (1969) Timing of callase activity and cytoplasmic male sterility in Petunia. Biochem Genet 3: 451–455 [DOI] [PubMed] [Google Scholar]
- Franklin-Tong VE (1999) Signaling and the modulation of pollen tube growth. Plant Cell 11: 727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J (1964) Cell walls, cell membranes and protoplasmic connections during meiosis and pollen development. InLinskens HF, ed, Pollen Physiology and Fertilization. North Holland Publishers, Amsterdam, pp. 39–47 [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hong Z, Delauney AJ, Verma DP (2001) A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang C, Wang H, Lu P, Zheng B, Ma H, Copenhaver GP, Wang Y (2019) Meiocyte-specific and AtSPO11-1-dependent small RNAs and their association with meiotic gene expression and recombination. Plant Cell 31: 444–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura KI, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005) Rice plant development: from zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB (2003) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15: 2503–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Sieburth LE (2010) Plasmodesmata formation: poking holes in walls with ISE. Curr Biol 20: R488–R490 [DOI] [PubMed] [Google Scholar]
- Lei X, Liu B (2020) Tapetum-dependent male meiosis progression in plants: increasing evidence emerges. Front Plant Sci 10: 1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, De Storme N, Geelen D (2017) Gibberellin induces diploid pollen formation by interfering with meiotic cytokinesis. Plant Physiol 173: 338–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL (2017) CRISPR-P 2.0: an improved CRISPR/Cas9 tool for genome editing in plants. Mo Plant 10: 530–532 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Ham BK, Kim JY (2009) Plasmodesmata - bridging the gap between neighboring plant cells. Trends Cell Biol 19: 495–503 [DOI] [PubMed] [Google Scholar]
- Maltby D, Carpita NC, Montezinos D, Kulow C, Delmer DP (1979) Β-1,3-glucan in developing cotton fibers: structure, localization, and relationship of synthesis to that of secondary wall cellulose. Plant Physiol 63: 1158–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamun EA, Cantrill LC, Overall RL, Sutton BG (2005) Cellular organisation in meiotic and early post-meiotic rice anthers. Cell Biol Int 29: 903–913 [DOI] [PubMed] [Google Scholar]
- Mikami M, Toki S, Endo M (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88: 561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura M, Ono S, Nonomura KI (2021) Rice MEL2 regulates the timing of meiotic transition as a component of cytoplasmic RNA granules. bioRxiv. doi: 10.1101/2021.03.24.433842, Publication date: March 25, 2021 [DOI] [Google Scholar]
- Mursalimov SR, Baiborodin SI, Sidorchuk YV, Shumny VK, Deineko EV (2010) Characteristics of the cytomictic channel formation in Nicotiana tabacum L. pollen mother cells. Cytol Genet 44: 14–18 [PubMed] [Google Scholar]
- Mursalimov SR, Sidorchuk YV, Deineko EV (2013) New insights into cytomixis: specific cellular features and prevalence in higher plants. Planta 238: 415–423 [DOI] [PubMed] [Google Scholar]
- Musiał K, Kościńska-Pająk M (2017) Pattern of callose deposition during the course of meiotic diplospory in Chondrilla juncea (Asteraceae, Cichorioideae). Protoplasma 254: 1499–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedukha OM (2015) Callose: localization, functions, and synthesis in plant cells. Cytol Genet 49: 49–57 [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Nonomura KI, Nakano M, Eiguchi M, Suzuki T, Kurata N (2006) PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119 (Pt 2): 217–225 [DOI] [PubMed] [Google Scholar]
- Nonomura KI, Eiguchi M, Nakano M, Takashima K, Komeda N, Fukuchi S, Miyazaki S, Miyao A, Hirochika H, Kurata N (2011) A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet 7: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Liu H, Tsuda K, Fukai E, Tanaka K, Sasaki T, Nonomura KI (2018) EAT1 transcription factor, a non-cell-autonomous regulator of pollen production, activates meiotic small RNA biogenesis in rice anther tapetum. PLoS Genet 14: e1007238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piršelová B, Matušíková I (2013) Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol Planta 35: 635–644 [Google Scholar]
- Plackett A, Ferguson AC, Powers SJ, Wanchoo-Kohli A, Phillips AL, Wilson ZA, Hedden P, Thomas SG (2014) DELLA activity is required for successful pollen development in the Columbia ecotype of Arabidopsis. New Phytol 201: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieu C, Sauquet H, Gouyon PH, Albert B (2017) More than sixty origins of pantoporate pollen in angiosperms. Am J Bot 104: 1837–1845 [DOI] [PubMed] [Google Scholar]
- Qin P, Ting D, Shieh A, McCormick S (2012) Callose plug deposition patterns vary in pollen tubes of Arabidopsis thaliana ecotypes and tomato species. BMC Plant Biology 12: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford JE, Vesk M, Overall RL (1998) Callose deposition at plasmodesmata. Protoplasma 201: 30–37 [Google Scholar]
- Roschzttardtz H, Conéjéro G, Divol F, Alcon C, Verdeil JL, Curie C, Mari S (2013) New insights into Fe localization in plant tissues. Front Plant Sci 4: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager RE, Lee JY (2018) Plasmodesmata at a glance. J Cell Sci 131: jcs209346. [DOI] [PubMed] [Google Scholar]
- Seale M (2020) Callose deposition during pollen development. Plant Physiol 184: 564–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Surovtseva YV, Osbun N, Shippen DE (2005) The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol 25: 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T, Tang D, Wang K, Wang M, Che L, Qin B, Yu H, Li M, Gu M, Cheng Z (2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Sun X, Zhang Z, Feng D, Zhang Q, Han L, Wu J, Lu T (2015) GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol 56: 497–509 [DOI] [PubMed] [Google Scholar]
- Shivanna KR (2003) Pollen Biology and Biotechnology. Science Publishers, Plymouth [Google Scholar]
- Song L, Wang R, Zhang L, Wang Y, Yao S (2016) CRR1 encoding callose synthase functions in ovary expansion by affecting vascular cell patterning in rice. Plant J 88: 620–632 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Hepler PK (1996) Cytokinesis in higher plants. Cell 84: 821–824 [DOI] [PubMed] [Google Scholar]
- Steer MW (1977) Differentiation of the tapetum in Avena. I. The cell surface. J Cell Sci 25: 125–138 [DOI] [PubMed] [Google Scholar]
- Stone BA, Clarke AE (1992) Chemistry and Biology of (1-3)-β-D-Glucans. La Trobe University Press, Melbourne [Google Scholar]
- Stieglitz H, Stern H (1973) Regulation of β-1,3-glucanase activity in developing anthers of Lilium. Dev Biol 34: 169–173 [DOI] [PubMed] [Google Scholar]
- Thiele K, Wanner G, Kindzierski V, Jürgens G, Mayer U, Pachl F, Assaad FF (2009) The timely deposition of callose is essential for cytokinesis in Arabidopsis. Plant J 58: 13–26 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Chuck G (2019) Heat induced epitope retrieval (HIER) assisted protein immunostaining in maize. Bio-101: e3260. [Google Scholar]
- Unal M, Vardar F, Ayturk Ö (2013) Callose in plant sexual reproduction. InSilva-Opps M, ed, Current Progress in Biological Research. InTechOpen, ; 10.5772/53001 [DOI] [Google Scholar]
- Voigt CA (2014) Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front Plant Sci 5: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Zha W, Cheng X, Liu C, Lv L, Liu C, Wang Z, Du B, Chen R, Zhu L, et al. (2011) A rice β-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233: 309–323 [DOI] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Qin B, Li M, Cheng Z (2011) PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol Biol Cell 22: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z (2010) The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner D, Gerlitz N, Stadler R (2011) A dual switch in phloem unloading during ovule development in Arabidopsis. Protoplasma 248: 225–235 [DOI] [PubMed] [Google Scholar]
- Xie B, Wang X, Zhu M, Zhang Z, Hong Z (2011) CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J 65: 1–14 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Hayashi T, Nakayama K, Koike S (2006) Expression analysis of genes for callose synthases and Rho-type small GTP-binding proteins that are related to callose synthesis in rice anther. Biosci Biotechnol Biochem 70: 639–645 [DOI] [PubMed] [Google Scholar]
- Yim KO, Bradford KJ (1998) Callose deposition is responsible for apoplastic semipermeability of the endosperm envelope of muskmelon seeds1. Plant Physiol 118: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev R, Ueki S, Epel BL, Citovsky V (2011) Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan GL, Walbot V, Meyers BC (2015) Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA 112: 3146–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wilson ZA (2009) Stamen specification and anther development in rice. Chinese Sci Bull 54: 2342–2353 [Google Scholar]
- Zhang F, Tang D, Shen Y, Xue Z, Shi W, Ren L, Du G, Li Y, Cheng Z (2017) The F-box protein ZYGO1 mediates bouquet formation to promote homologous pairing, synapsis, and recombination in rice meiosis. Plant Cell 29: 2597–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Shen Y, Tang D, Shi W, Zhang D, Du G, Zhou Y, Liang G, Li Y, Cheng Z (2018) The zinc finger protein DCM1 is required for male meiotic cytokinesis by preserving callose in rice. PLoS Genet 14: e1007769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Ren L, Chen X, Yu H, Liu C, Shen Y, Shi W, Tang D, Du G, Li Y, et al. (2018) The OsRR24/LEPTO1 type-B response regulator is essential for the organization of leptotene chromosomes in rice meiosis. Plant Cell 30: 3024–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.