Abstract
Cell cycle progression and the phytohormones auxin and abscisic acid (ABA) play key roles in primary root growth, but how ABA mediates the transcription of cell cycle-related genes and the mechanism of crosstalk between ABA and auxin requires further research. Here, we report that ABA inhibits primary root growth by regulating the ABA INSENSITIVE4 (ABI4)-CYCLIN-DEPENDENT KINASE B2;2 (CDKB2;2)/CYCLIN B1;1 (CYCB1;1) module-mediated cell cycle as well as auxin biosynthesis in Arabidopsis (Arabidopsis thaliana). ABA induced ABI4 transcription in the primary root tip, and the abi4 mutant showed an ABA-insensitive phenotype in primary root growth. Compared with the wild type (WT), the meristem size and cell number of the primary root in abi4 increased in response to ABA. Further, the transcription levels of several cell-cycle positive regulator genes, including CDKB2;2 and CYCB1;1, were upregulated in abi4 primary root tips. Subsequent chromatin immunoprecipitation (ChIP)-seq, ChIP-qPCR, and biochemical analysis revealed that ABI4 repressed the expression of CDKB2;2 and CYCB1;1 by physically interacting with their promoters. Genetic analysis demonstrated that overexpression of CDKB2;2 or CYCB1;1 fully rescued the shorter primary root phenotype of ABI4-overexpression lines, and consistently, abi4/cdkb2;2-cr or abi4/cycb1;1-cr double mutations largely rescued the ABA-insensitive phenotype of abi4 with regard to primary root growth. The expression levels of DR5promoter-GFP and PIN1promoter::PIN1-GFP in abi4 primary root tips were significantly higher than those in WT after ABA treatment, with these changes being consistent with changes in auxin concentration and expression patterns of auxin biosynthesis genes. Taken together, these findings indicated that ABA inhibits primary root growth through ABI4-mediated cell cycle and auxin-related regulatory pathways.
Abscisic acid inhibits primary root elongation through ABSCISIC ACID INSENSITIVE4-mediated cell cycle progression as well as auxin biosynthesis and transport.
Introduction
The plant root system plays an indispensable role in plant development and in response to several abiotic and/or biotic stresses, providing anchorage and an interface for water and mineral nutrient absorption (Bouain et al., 2019). Root growth is precisely regulated by diverse endogenous signals, including phytohormones, and a variety of environmental cues (Pacifici et al., 2015). Among the plant hormones, the important role of auxin in the control of root development has been well documented in the past decades (Promchuea et al., 2017; Tian et al., 2017; Du and Scheres, 2018; Qin and Huang, 2018; Zhang et al., 2018; Zhao, 2018; Lv et al., 2021). Numerous elegant studies have demonstrated that auxin concentration gradient achieves crucial physiological functions in the specification and maintenance of the root stem cell niche (Dinneny and Benfey, 2008; Zhou et al., 2010; Du and Scheres, 2018; Wang et al., 2019).
Abscisic acid (ABA) is an important phytohormone which exhibits diverse roles at different plant growth stages, including seed development (Chauffour et al., 2019), seed dormancy and germination (Shu et al., 2013, 2016c; Nee et al., 2017), floral transition (Wang et al., 2013; Shu et al., 2016b), and responses to abiotic stresses such as drought and salinity (Yoshida et al., 2014; Zhu, 2016; Luo et al., 2021). For root development, low concentrations of ABA generally enhance but high concentrations inhibit primary root development (Zhang et al., 2010; Rowe et al., 2016). However, in contrast to the detailed investigations of the role of ABA in seed biology and plant–environment interactions (Shu et al., 2013, 2016b, 2016c, 2018; Wang et al., 2013; Yoshida et al., 2014; Luo et al., 2021), the relationship between ABA concentration and root system development still needs further exploration, although some important progress has been achieved in recent years.
For instance, the transcription factor OBF BINDING PROTEIN4 (OBP4)-mediated transcriptional repression of the ROOTHAIR DEFECTIVE6-LIKE2 (RSL2) gene contributes to the ABA-dependent inhibition of root hair growth in Arabidopsis (Arabidopsis thaliana) (Rymen et al., 2017). PYRABACTIN RESISTANCE1-LIKE8 (PYL8), one of the ABA receptors of the PYRABACTIN RESISTANCE 1/PYRABACTIN RESISTANCE-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR (PYR1/PYL/RCAR) protein family, is essential for the recovery of lateral root development following inhibition by exogenous ABA. The physical interaction between PYL8 and MYB DOMAIN PROTEIN 77 (MYB77) further increased the binding activity of MYB77 to the promoter fragments of its target genes, the multiple auxin-responsive genes, to augment auxin signaling (Zhao et al., 2014). Further investigations demonstrated that PYL9 is also responsible for the recovery of lateral root growth from repression by ABA through the action of MYB transcription factors (Xing et al., 2016). Another breakthrough came from analysis of the inhibitory effect of ABA on primary root development through promotion of the biosynthesis or signal transduction of the phytohormone ethylene (Beaudoin et al., 2000; Ghassemian et al., 2000; Luo et al., 2014). Our goal is to describe further regulatory cascades involved in the effect of ABA in repressing primary root development, especially with respect to the crosstalk between ABA and cell cycle progression.
The cell cycle is the ubiquitous process, leading to the production of two daughter cells (Gutierrez, 2016). Root development involves cell division in the apical meristem, with subsequent elongation of the divided cells (Zhang et al., 2010; Zhou et al., 2010). The positive regulators, cyclins, such as B-type cyclins (CYCBs), and cyclin-dependent kinases (CDKs), such as CDKBs, are central to cell cycle progression, with some negative actors, such as CDK inhibitors, like INHIBITOR/INTERACTOR WITH CKDs/KIP-RELATED PROTEINs ICKs/KRPs, also being involved in cell cycle cascades (Gutierrez, 2016). Several earlier studies had demonstrated that, during root development, ABA represses the transcription of CYCB1 at the G2/M (cell growth/mitosis) checkpoint, ultimately arresting root growth (Xu et al., 2010; Wang et al., 2011; Yang et al., 2014). In addition, ABA has also been shown to enhance the expression of ICK1/KRP1 genes encoding the CDK inhibitors, resulting in the suppression of G1/S (cell growth/DNA synthesis) transition during the cell cycle (Wang et al., 1998). Furthermore, “big data” analysis has also highlighted the observation that ABA promotes the transcription of positive regulatory genes of the cell cycle, while repressing the expression of negative regulatory genes of the cell cycle (Zhang et al., 2020). However, the details underlying the mechanisms by which ABA regulates the expression of cell cycle-related genes, such as CYCBs and CDKs, has remained elusive for a long time, in particular the identity of their direct upstream transcription factor(s).
ABA INSENSITIVE 4 (ABI4) is an APETALA 2-domain-containing transcription factor, operating as a positive regulator in the ABA signaling cascade (Soderman et al., 2000; Shu et al., 2018). Over the past decade, several groups, including our team, have revealed that ABI4 is a versatile factor which is involved in seed dormancy and germination (Shu et al., 2016a; Nee et al., 2017), flowering time control (Shu et al., 2016b), crosstalk among diverse phytohormones (Wind et al., 2013; Shu et al., 2016c, 2018; Huang et al., 2017; Chandrasekaran et al., 2020), and responses to plant abiotic stresses (Shkolnik-Inbar et al., 2013; Xie et al., 2016; Zhao et al., 2016; Luo et al., 2021). In addition, an earlier study also revealed that ABI4 negatively regulates polar auxin transport to inhibit lateral root development (Shkolnik-Inbar and Bar-Zvi, 2010). Recently, it has been shown that members of the BASIC PENTACYSTEINE (BPC) family of transcription factors can directly recruit the polycomb-repressive complex 2 (PRC2) methylase enzyme to the ABI4 locus to epigenetically repress ABI4 transcription. Ultimately, ABI4 and BPCs together coordinate the abundance of PIN1 (PIN-FORMED 1), encoding an auxin efflux carrier, and thus modulate lateral root formation (Mu et al., 2017). These investigations have identified the important regulatory roles played by ABI4 in lateral root growth, but how ABI4 performs such biological roles in ABA-mediated primary root elongation is still largely unknown.
In the current study, we show that exogenous ABA represses primary root elongation in Arabidopsis through ABI4-mediated auxin biosynthesis as well as by control of cell cycle progression. The abi4 knockout mutant exhibited an ABA-insensitive phenotype with regard to primary root growth inhibition, reflected in the upregulation of the size and cell number of the meristem zone in the abi4 primary root, compared with the wild-type (WT). Gene expression analysis revealed that the transcription level of cell cycle-related genes, including CDKB2;2 (encoding CKD2) and CYCB1;1 (encoding B2-TYPE CYCLIN), increased in the abi4 primary root tip, and the biochemical assays showed that ABI4 directly repressed the transcription of CDKB2;2 and CYCB1;1 by interacting with their promoter fragments. Subsequent genetic analysis demonstrated that overexpression of CDKB2;2 or CYCB1;1 fully rescued the shorter primary root phenotype of OE-ABI4 transgenic lines following exposure to exogenous ABA. Furthermore, we also revealed that the expression levels of the reporter genes DR5promoter-GFP and PIN1promoter::PIN1-GFP in abi4 primary root tips were significantly upregulated after ABA application, reflecting concomitant changes in auxin biosynthesis gene expression levels and the endogenous auxin concentration. Altogether, this study demonstrates that ABA inhibits primary root growth by modulating ABI4-mediated auxin biosynthesis, as well as cell cycle progression.
Results
ABA regulates the expression pattern of cell cycle-related genes globally
From a large number of published bioinformatic databases, we found that, in response to ABA treatment, the expression of most cell cycle positive regulatory genes was downregulated (Supplemental Figure S1), while that of most negative regulatory genes was upregulated in different tissues, including leaves, roots, shoots, and entire seedlings of Arabidopsis (Supplemental Figure S2). These findings indicated that ABA globally regulates cell cycle progression by mediating the transcription of related genes, which is probably a universal mechanism in the plant kingdom. However, the detailed molecular mechanisms underlying these effects, especially the identity of the regulatory transcription factor(s) upstream of these cell cycle-related genes, need to be further investigated.
The abi4 mutant shows an ABA-insensitive phenotype with regard to inhibition of primary root growth and meristem size after ABA treatment
In our previous research, we determined that the transcription factor ABI4 mediates primary seed dormancy, seed germination, and seedling establishment as well as abiotic stress response, by modulating ABA and GA signaling pathways (Shu et al., 2013, 2016a, 2016c; Luo et al., 2021). Several other elegant studies revealed that ABI4 is also involved in the control of lateral root growth, with the root system being an excellent model for studying cell cycle progression (Shkolnik-Inbar and Bar-Zvi, 2010; Mu et al., 2017). As a result, in this study, we further explored the roles of ABI4 in cell cycle progression in response to exposure to ABA, using the primary root system.
In order to investigate the roles of both ABA and ABI4 in the regulation of primary root growth, we explored the effect of ABA on ABI4 expression in the primary root tip niche. The mRNA in the primary root tip was extracted for quantitative PCR (qPCR) assay, and the results revealed that exogenous ABA application significantly increased ABI4 transcription, with two- to three-fold increases compared with the no-ABA control (Supplemental Figure S3). The gene expression pattern suggested that ABI4 might play a key role in the control of primary root development mediated by the phytohormone ABA.
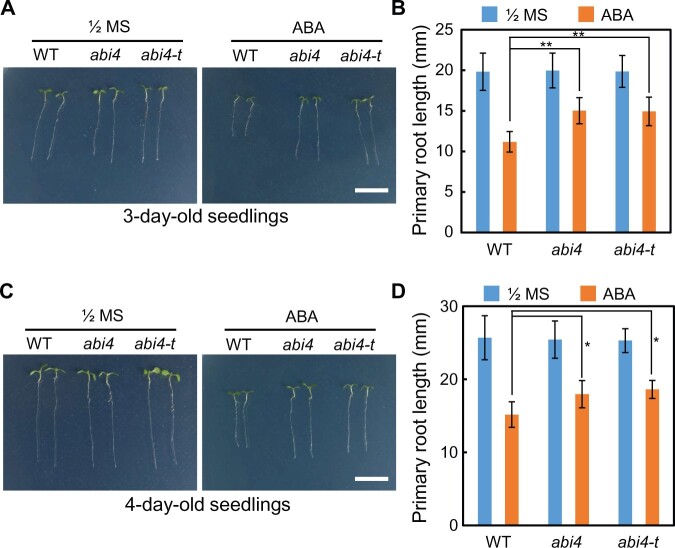
To test this hypothesis, we subsequently investigated the effect of exogenous ABA application on primary root development and the responsiveness of the abi4 knockout mutant. Two mutants carrying abi4 alleles, namely abi4 and abi4-t, which were used in our previous studies (Shu et al., 2013, 2016a, 2016b; Luo et al., 2021), were employed in this study. The 3- or 4-day-old seedlings germinated on half-strength Murashige and Skoog (1/2 MS) were transferred to 1/2 MS or ABA-containing medium, and the seedlings were harvested 2 days after transfer for analysis. The results revealed that the primary root lengths of abi4 and abi4-t were comparable with that of the WT Col-0 Arabidopsis in seedlings grown on 1/2 MS medium, whereas exogenous treatment with a high concentration of ABA (by transfer of seedlings to ABA-containing medium) markedly inhibited primary root growth in the WT (Figure 1). However, both the abi4 allelic mutants exhibited an ABA-insensitive phenotype with regard to primary root growth inhibition when grown on ABA-containing medium, compared with the WT (Figure 1).
Figure 1.
abi4 mutants show the ABA-insensitive phenotype with regard to primary root growth inhibition in response to ABA application. A, The 3-day-old seedlings germinated on 1/2 MS were transferred to 1/2 MS or ABA-containing medium, and the seedlings were harvested 2 days after transfer for photographing. B, The statistics of primary root length of the three genotypes for (A). C, The 4-day-old seedlings germinated on 1/2 MS were transferred to 1/2 MS or ABA-containing medium, and the seedlings were harvested 2 days after transfer for photographing. D, The statistics of primary root length of the three genotypes for (C). The point mutant abi4-1 (CS8104) and the T-DNA insertion mutants abi4-t (SALK_080095), previously described (Shu et al., 2013, 2016b), were employed in this assay. The values are means ± se. In each experiment at least three repeat, and in one repeat, >30 roots per genotype were used. Asterisks indicate significant difference (*P < 0.05, **P < 0.01) according to Student’s t test analysis. ABA concentration is 30 µM. Bar = 1 cm.
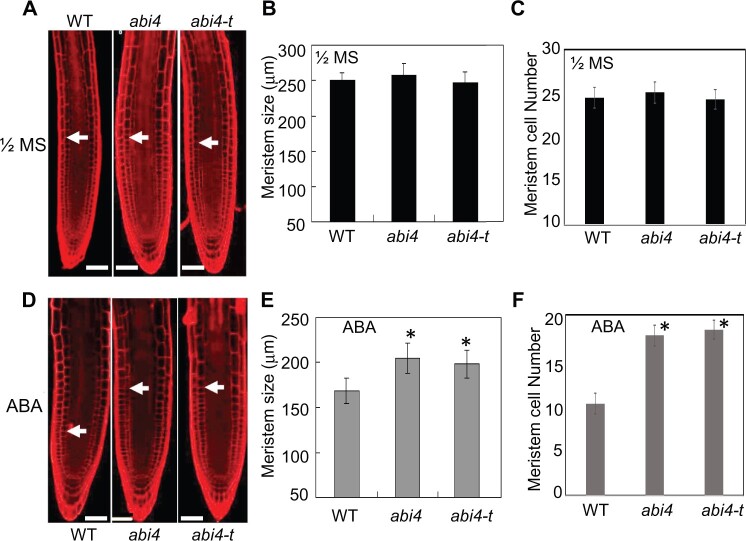
To further analyze the molecular mechanisms underlying the observed phenotypes (Figure 1), we measured root tip meristem size in the two abi4 allelic mutants and the WT, in the presence and absence of exogenous ABA exposure. On the 1/2 MS medium, the meristem traits were comparable among the three genotypes tested (Figure 2A), including meristem size (Figure 2B) and meristem zone cell number (Figure 2C). ABA treatment markedly reduced the meristem size of the WT seedlings, whereas both the abi4 allelic mutants still showed the ABA-insensitive phenotype with regard to both meristem size and meristem zone cell number (Figure 2, D–F), which is consistent with the primary root growth phenotype of abi4 and abi4-t under exogenous ABA application (Figure 1). The meristem sizes and cell numbers of both abi4 and abi4-t mutants were remarkably longer than the values for the WT (Figure 2, D–F), which might be responsible for the greater primary root length of abi4 allelic mutants, compared with the WT (Figure 1, C and D).
Figure 2.
abi4 mutant meristem size increases after ABA treatment. Phenotypic analysis of meristem cell size and cell number of abi4 and abi4-t seedlings grown on 1/2 MS or 1/2 MS+ABA media. The 4-day-old seedlings on 1/2 MS medium were harvested. Three-day-old seedlings on 1/2 MS were transferred to the ABA-containing medium with different ABA concentrations, with the seedlings being collected for analysis 2 days after transfer. A–C, Meristem cell size of seedlings on the 1/2 MS medium. D–F, Meristem cell size of seedlings on the ABA-containing medium (15 mM). Bar = 50 μm. Approximately 5–10 images were examined and at least three independent experiments were performed. The statistical significance of differences was evaluated by Student’s t test analysis. Asterisks indicate statistically significant differences from the control (CK), at the 0-h point (*P < 0.05). The values are means ± se.
It is interesting that we detected distinct responses in different ages of abi4 seedlings transferred from 1/2 MS to the ABA-containing medium. When we transferred the 3- or 4-day-old seedlings, abi4 showed the significant primary root growth arrested phenotype, compared with the WT (Figure 1). However, when the 5- or 6-day-old seedlings were employed, abi4 did not show any primary root growth inhibited phenotype, compared with the WT (Supplemental Figure S4). Indeed, we found that a previous study also documented that the older abi4 seedlings transferred from 1/2 MS to the ABA-containing medium did not have the primary root inhibition phenotype (Zhu et al., 2020), and the abi5 mutant also showed the similar trend (Ahmad et al., 2019). Thus, we suggest that ABI4 primarily functioned in the earlier developmental period with regard to ABA-mediated inhibition of primary root growth.
The transcription levels of cell cycle positive regulatory genes are increased in the abi4 primary root tip
The abi4 mutant showed a significant increase in the size of the primary root tip meristem zone (Figure 2, D–F), so we further examined the effect of ABI4 on the expression of cell cycle-related genes. The qPCR analysis revealed that the expression levels of most of the positive regulatory genes, including CYCB1;1, CYCD1;1, CYCD3;1, CYCD3;2, CYCD5;1, CDKA;1, and CDKB2;2 were markedly higher than that in the WT (Figure 3). The mRNA abundance values of CYCD2;1 and CDKB1;1 were still greater than that of the WT, although the differences from the WT were not statistically significant. The transcription levels of CYCD3;3 and CDKB2;1 were not altered or even decreased in abi4 mutant primary root tips (Figure 3) relative to WT root tips, a finding which indicated that the transcription factor ABI4 might show only a weak effect or none on the control of the expression of these genes with regard to the regulation of ABA-mediated primary root growth. However, the changing expression patterns, based on ABA response and the effects of knockout mutations on most of the cell cycle-related genes were consistent with both the primary root phenotype (Figure 1) and the meristem traits (Figure 2).
Figure 3.

Expression pattern analysis of cell cycle-related genes in abi4. qPCR analysis of expression of several cell-cycle-related genes in abi4 and WT seedling primary root tips on 1/2 MS media. The ACTIN7 mRNA was used as the reference gene. The experiments were replicated three times and the results of one typical experiment are shown. Asterisks indicate statistically significant differences from WT (**P < 0.01). The values are means ± se, n = 4. Statistical analyses were performed using Student’s t test.
Further, we also found that exogenous ABA treatment could still induce the expression of key cell cycle genes in abi4 roots with minor changes (Supplemental Figure S5), indicating that there are other factors involved in ABA-mediated cell cycle gene transcription besides ABI4. In addition, ABI3 and ABI5 also regulate the expression pattern of several key genes involved in the cell cycle cascade (Supplemental Figure S6), indicating that the regulation of cell cycle-related gene expression is under the control of several ABI transcription factors with complex mechanisms, but not restricted by only ABI4. However, it is obvious that, genes such as CDKB2;2 and CYCB1;1 showed very marked increases in expression in the abi4 primary root tip compared with the WT, with or without ABA treatment (Figure 3 and Supplemental Figure S5).
ABI4 represses CYCB1;1 and CDKB2;2 transcription by directly interacting with their promoters
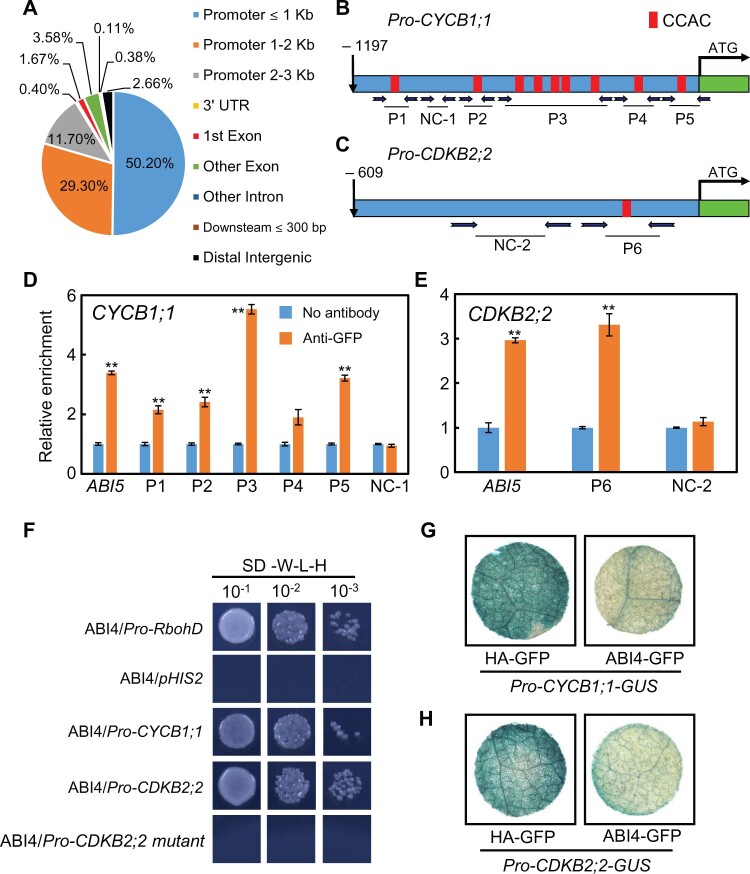
ABI4 is a versatile transcription factor, which is involved in diverse phytohormone-based signaling cascades and plant abiotic stress response networks by directly targeting the transcription of several genes (Shkolnik-Inbar et al., 2013; Shu et al., 2013, 2016a, 2018; Wind et al., 2013; Huang et al., 2017; Luo et al., 2021). Previous research had revealed that ABI4 directly binds to the CACCG or CCAC cis-element sequences in the promoters of some genes, mediating their transcription (Shu et al., 2013, 2016a, 2016b; Huang et al., 2017; Luo et al., 2021). In order to investigate whether ABI4 directly controls the transcription of the cell cycle-related genes shown in Figure 3, chromatin immunoprecipitation (ChIP)-seq assay was employed. The results showed that ABI4 could bind thousands of promoters of known genes (Supplemental Table S1), and most of the binding sites are located at 0–3,000-bp upstream of the transcription start site (Figure 4A). It is noted that dozens of potential ABI4-regulated genes are closely related to the process of cell cycle (Supplemental Table S1). Based on this, the characteristics of these gene promoters especially in Figure 3 and Supplemental Table S1 were documented. It is noted that nine CCAC motifs were detected in the CYCB1;1 promoter (Figure 4B), and one CCAC motif in the CDKB2;2 promoter (Figure 4C), indicating the potential for direct interaction between ABI4 and these promoters. The next step was to determine whether ABI4 directly binds to the respective promoters in vivo. Subsequently, a ChIP-qPCR assay was employed with the ABI4 transgenic plants which had been generated in our previous studies (Shu et al., 2013, 2016a, 2016b; Luo et al., 2021), and the ABI5 promoter element was selected as the positive control because ABI4 interacted directly with its promoter fragment (Bossi et al., 2009). Two independent OE-ABI4 transgenic lines were subjected to ChIP-qPCR analysis, and similar results were obtained for each line. As shown in Figure 4, D and E, we determined that the promoters of CYCB1;1 and CDKB2;2 were enriched with respect to the chromatin immunoprecipitated DNA, using the anti-green fluorescent protein (GFP) antibody, especially the P3 and P5 (for CYCB1;1) and P6 (for CDKB2;2) fragments (Figure 4, D and E). As expected, the subsequent yeast one-hybrid assay confirmed that ABI4 binds directly to the promoters of CYCB1;1 and CDKB2;2 genes (Figure 4F), with the Respiratory burst oxidase homolog D (RbohD) promoter fragment being used as the positive control (Luo et al., 2021). Importantly, a mutated CDKB2;2 promoter fragment, in which CCAC was changed to CCAA (Supplemental Figure S7), did not directly interact with the ABI4 transcription factor (Figure 4F). Altogether, both the ChIP-qPCR and the yeast one-hybrid assays demonstrated that ABI4 indeed physically binds directly to the promoters of CYCB1;1 and CDKB2;2.
Figure 4.
ABI4 promotes CYCB1;1 and CDKB2;2 transcription by directly binding to their promoters. A, Summary of ChIP-seq results. The pie chart shows where ABI4 binds to the promoter fragments. B and C, Promoter sequence analysis of (B) CYCB1;1 and (C) CDKB2;2. The fragment of 1,197-bp upstream of ATG of CYCB1;1 and 609-bp upstream of the start codon of CDKB2;2, respectively, was determined to be their promoters. D and E, ChIP-qPCR assay was carried out to investigate the association between the ABI4 transcription factor and the (D) CYCB1;1 and (E) CDKB2;2 promoters. Chromatin from the transgenic plant OE-ABI4 was isolated. The promoter fragment of ABI5 was chosen as a positive control, and the promoter region of ACTIN12 was employed as the internal control. Asterisks indicate statistically significant differences from no antibody (**P < 0.01). The values are means ± se, n = 4. Statistical analyses were performed using Student’s t test. F, Yeast one-hybrid assay showing the direct interaction between ABI4 and the CYCB1;1 and CDKB2;2 promoters. The full-length promoters were used in this experiment. The promoter region of RbohD was chosen as the positive control. Furthermore, the promoter of CDK2;2 was mutated (information presented in Supplemental Figure S7) and then subjected to yeast one-hybrid analysis. G and H, Nicotiana benthamiana transient expression assay revealed that ABI4 inhibited (G) CYCB1;1 and (H) CDKB2;2 expression in vivo. Representative images of the N. benthamiana leaves are shown.
Combining the gene expression analysis (Figure 3) and the biochemical evidence (Figure 4), the tobacco (Nicotiana benthamiana) transient expression system was subsequently selected to explore whether ABI4 inhibits the transcription of CYCB1;1 and CDKB2;2 in vivo. To this end, the reporter plasmids Pro-CYCB1;1-GUS and Pro-CDKB2;2-GUS, and the plasmid of pCanG-ABI4-GFP were transiently co-expressed individually in N. benthamiana leaves. As shown in Figure 4, when the Pro-CYCB1;1-GUS or Pro-CDKB2;2-GUS constructs were combined with pCanG-HA-GFP, strong β-glucuronidase (GUS) expression, visualized as staining, was detected (Figure 4, G and H); however, when the pCanG-HA-GFP control vector was replaced by an equal amount of the effector pCanG-ABI4-GFP, the GUS staining was clearly reduced (Figure 4, G and H). Consistently, the results from the quantification of GUS activity (Supplemental Figure S8) also confirmed the repressive effect of ABI4 on the expression of CYCB1;1 and CDKB2;2 in vivo. These results demonstrated that ABI4 indeed represses CYCB1;1 and CDKB2;2 transcription by directly interacting with their respective promoters.
CYCB1;1 and CDKB2;2 over-expression largely rescued the short primary root phenotype of OE-ABI4 in the presence of ABA
The results described above demonstrate that the ABI4 transcription factor directly represses transcription of the cell cycle positive regulatory genes CYCB1;1 and CDKB2;2; we hypothesized that the abi4 knockout mutant showed the ABA-insensitive phenotype because of the normal cell cycle progression in the abi4 mutant. In order to confirm this conclusion, we genetically dissected the relationship among CYCB1;1, CDKB2;2, and ABI4.
Firstly, we employed the previously described ABI4 overexpression transgenic lines (Shu et al., 2016a, 2016b; Luo et al., 2021), and also generated the overexpression transgenic plants for CYCB1;1 and CDKB2;2. The qPCR assay revealed that CYCB1;1 and CDKB2;2 expression was higher in OE-CYCB1;1 (CYCB1;1 over-expression transgenic plants) and OE-CDKB2;2 (CDKB2;2 over-expression transgenic plants) than in the WT (Supplemental Figure S9, A and B). Phenotypic analysis demonstrated that, when grown on 1/2 MS medium, seedlings of these three genotypes showed similar primary root lengths. In the presence of exogenous ABA, however, shortened primary roots were detected in the OE-ABI4 lines, whereas the OE-CYCB1;1 and OE-CDKB2;2 transgenic plants continued to show the primary root length comparable to that of the WT, that is, the ABA-insensitive phenotype (Figure 5, A–C).
Figure 5.
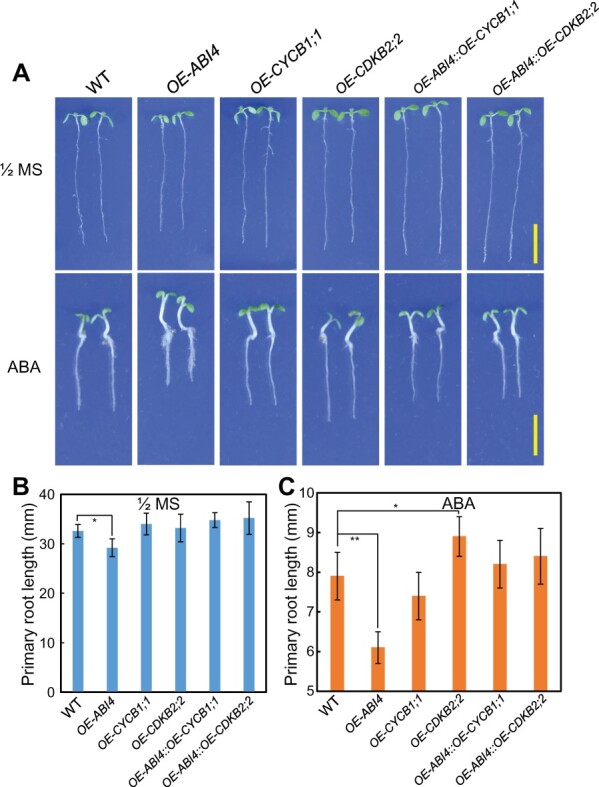
Transgenic OE-CYCB1;1 and OE-CDKB2;2 fully rescue the ABA-hypersensitive phenotype of OE-ABI4 with regard to primary root growth in response to ABA application. A, Primary root phenotype of Col-WT, OE-ABI4, OE-CYCB1;1, OE-CDKB2;2, OE-ABI4::OE-CYCB1;1, and OE-ABI4::OE-CDKB2;2 of seedlings grown on 1/2 MS medium with or without exposure to ABA (30 μM). The 3-day-old seedlings on 1/2 MS were transferred to the medium with or without ABA at different concentrations, and the seedlings were harvested 4 days after transfer. Seedlings on 1/2 MS medium, bar = 10 mm. Seedlings on ABA-containing medium, bar = 5 mm. B and C, The quantitative analysis of primary root length for (A) is shown. The values are means ± se. In one experiment at least three repeats, and in one repeat, >30 roots per genotype were used. The asterisks and indicate significant differences at the *P < 0.05 and **P < 0.01 levels, respectively, by Student’s t test analysis.
To perform the genetic analysis, we introduced the constitutively expressed 35S-ABI4-GFP construct into each of the CYCB1;1 and CDKB2;2 overexpressed transgenic line backgrounds. The OE-ABI4::OE-CYCB1;1 and OE-ABI4::OE-CDKB2;2 transgenic plants thus obtained were subjected to phenotypic analysis, in which these three genes (ABI4, CYCB1;1, and CDKB2;2) exhibited overexpression compared with the WT (Supplemental Figure S10, A and B). As shown in Figure 5, the OE-ABI4 transgenic lines showed the inhibited primary root growth phenotype following exposure to exogenous ABA (Figure 5, A–C). However, overexpression of CYCB1;1 or CDKB2;2 largely rescued the root growth inhibition phenotype of OE-ABI4 plants in the presence of exogenous ABA, as evidenced by the primary root length values (Figure 5, A–C).
Further, we also employed CRISPR–Cas9 to edit CYCB1;1 and CDKB2;2 in the background of WT and abi4. Finally, we successfully obtained the cycb1;1-cr (CRISPR/Cas9 mutant), cdkb2;2-cr (CRISPR/Cas9 mutant), abi4/cdkb2;2-cr, and abi4/cycb1;1-cr double mutants. The phenotypic analysis revealed that abi4/cdkb2;2-cr and abi4/cycb1;1-cr double mutants largely rescue the ABA-insensitive phenotype of abi4 with regard to primary root growth in response to ABA application, while under 1/2 MS media, the primary root length of those genotypes is comparable (Figure 6, A–C). Taken together, these results from genetic analysis indicated that the shorter primary root length of OE-ABI4 under ABA treatment is dependent on the ABI4-repressed expression of CYCB1;1 and CDKB2;2, and further suggested that CYCB1;1 and CDKB2;2 act genetically downstream of ABI4.
Figure 6.
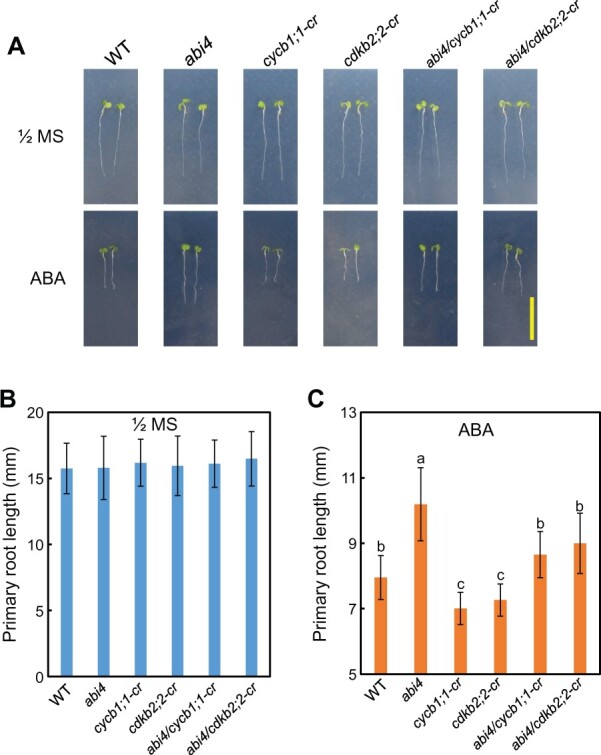
Gene editing of CYCB1;1 and CDKB2;2 (cycb1;1-cr and cdkb2;2-cr, respectively) largely rescues the ABA-insensitive phenotype of abi4 with regard to primary root growth in response to ABA application. A, Primary root phenotype of Col-WT, abi4, cycb1;1-cr, cdkb2;2-cr, abi4/cycb1;1-cr, and abi4/cdkb2;2-cr of seedlings grown on 1/2 MS medium with or without exposure to ABA (10 μM). The 3-day-old seedlings on 1/2 MS were transferred to the medium with or without ABA at different concentrations, and the seedlings were harvested 2 days after transfer. Bar = 10 mm. B and C, The quantitative analysis of primary root length for (A) is shown. The values are means ± se. In one experiment at least three repeats, and in one repeat, >30 roots per genotype were used. Duncan’s test (P < 0.05) was employed to check statistically significant means. The same letters, no significant difference; different letters, significant difference.
abi4 shows higher endogenous auxin concentrations in the primary root tip than WT after ABA application
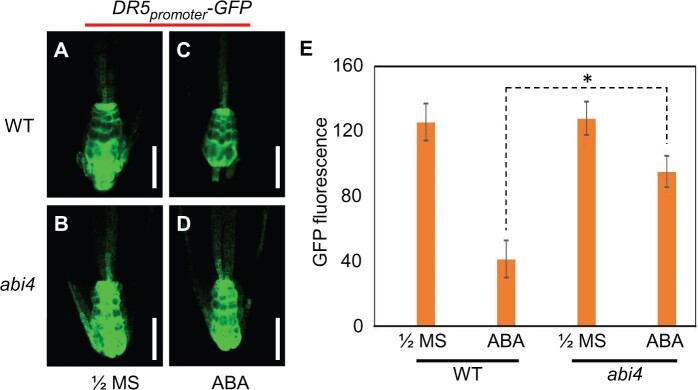
Numerous previous studies have demonstrated that an appropriate auxin concentration in the primary root tip is important for its development. We subsequently investigated any differences in auxin concentration in the primary root tip among different genotypes tested, with or without exogenous ABA application. To assess the auxin concentration, we firstly employed the DR5promoter-GFP transgenic Arabidopsis marker line, and this DR5promoter-GFP construct was crossed into the abi4 mutant background. The F2 progenies were examined by PCR, using the specified primers for abi4 point mutant genotyping. The results revealed that the fluorescence levels of DR5promoter-GFP in abi4 and WT primary root tips were comparable when the seedlings were grown on 1/2 MS medium (Figure 7, A and B), whereas, after exogenous ABA application, the fluorescence level of the GFP in the abi4 mutant was found to be significantly higher than that in WT (Figure 7, C–E).
Figure 7.
Expression pattern of DR5promoter-GFP in the WT and abi4 mutant roots. A–D, Expression patterns of the DR5promoter-GFP reporter gene in both genotypes grown on 1/2 MS (A and B), and ABA-containing media (C and D). E, Statistical analysis of GFP fluorescence levels for (A)–(D). The 4-day-old seedlings grown on 1/2 MS medium were sampled. Three-day-old seedlings on 1/2 MS medium were transferred to the ABA-containing medium (30 μM ABA), with the seedlings being harvested 2 days after transfer, for analysis. Bar = 50 mm. Five–ten images were examined per genotype, and at least three independent experiments were performed. The statistical significance of differences was evaluated by Student’s t test analysis. The values are means ± se. Asterisks indicate statistically significant differences from the control (CK), 0-h point (*P < 0.05).
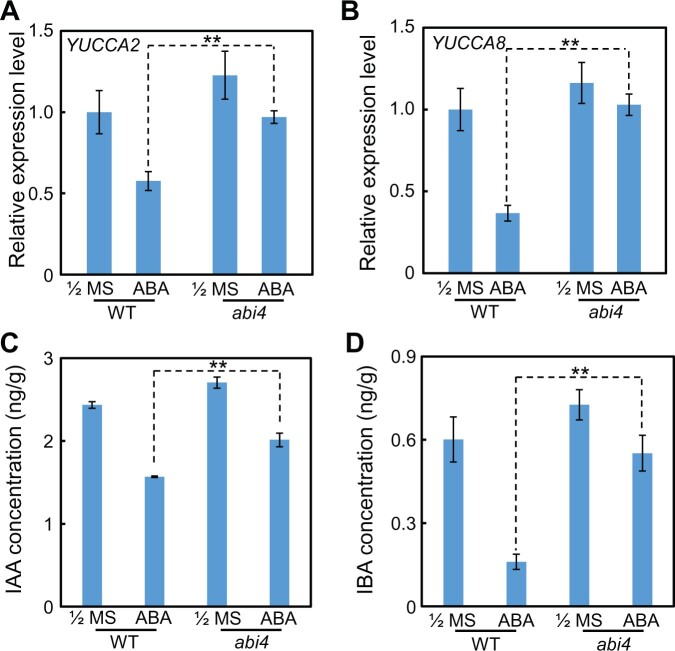
To further explore the effect of ABI4 on auxin biosynthesis in the primary root tip under ABA treatment, we subsequently examined the transcription patterns of auxin biosynthesis genes. The results revealed that, in the WT background, exogenous ABA application significantly down-regulated expression levels of YUCCA2 and YUCCA8, two key genes responsible for auxin biosynthesis (Figure 8, A and B). However, in the abi4 mutant genetic background, the transcription levels of YUCCA2 and YUCCA8 after ABA treatment were similar to those in seedlings grown on the 1/2 MS medium (Figure 8, A and B). ABI3 and ABI5 showed a weak effect on YUCCA2 and YUCCA8 expression (Supplemental Figure S11, A–D). Collectively, after ABA application, the expression levels of YUCCA2 and YUCCA8 in the abi4 primary root tip were higher than that in the WT, which further suggests that the abi4 primary root probably contains higher auxin concentrations, compared with the WT, in the presence of exogenous ABA.
Figure 8.
ABI4 decreases the auxin concentration in primary roots after exposure to exogenous ABA application. Expression of auxin biosynthesis genes (A) YUCCA2, (B) YUCCA8, and concentration of (C) IAA and (D) IBA in WT and abi4 mutant seedlings grown on the 1/2 MS or ABA-application media. The 4-day-old seedlings were treated with or without 30-μM ABA for 2 h, and then the mRNA was extracted or the IAA and IBA concentrations were determined. The asterisks indicate the significant differences at the **P < 0.01 level by Student’s t test analysis. The values are means ± se.
To test this hypothesis described above, we next examined the endogenous auxin concentrations, namely indole-3-acetic acid (IAA) and indole-3-butyric acid (IBA), in the abi4 primary root tip with or without exogenous ABA treatment. The results showed that the IAA and IBA concentrations in the abi4 primary root tip were significantly higher than those in the WT after ABA treatment, whereas on the 1/2 MS medium, the auxin concentrations were similar between the isogenic abi4 and WT lines (Figure 8, C and D). Subsequently, we further demonstrated that exogenous IAA application could largely rescue the inhibited primary root growth phenotype of OE-ABI4 plants in the presence of exogenous ABA (Figure 9, A–C), which confirmed that the shorter root phenotype of OE-ABI4 under ABA treatment is caused at least partially by the decreased auxin concentration.
Figure 9.
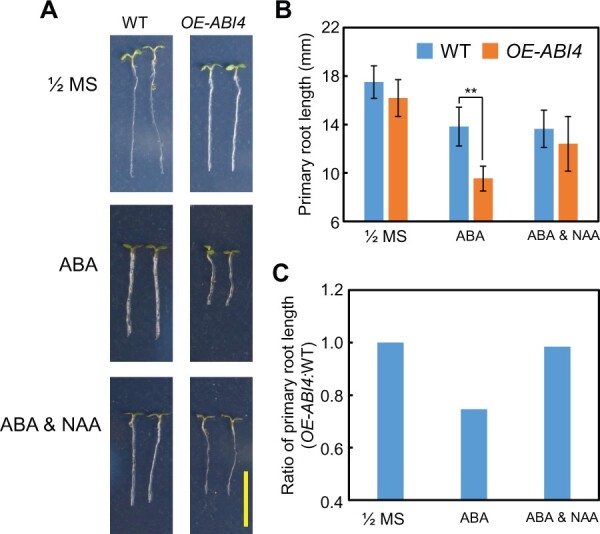
Exogenous application of auxin largely rescues the primary root growth inhibition phenotype of OE-ABI4 under ABA treatment. A, Primary root phenotype of WT and OE-ABI4 lines on 1/2 MS medium with or without ABA (30 μM) or NAA (1-Naphthylacetic acid, 1 nM) treatment. The 4-day-old seedlings on 1/2 MS medium were transferred to the medium with or without ABA or NAA, and the seedlings were harvested 1.5 days after transfer. Bar = 10 mm. B, The quantitative analysis of the primary root length for (A). The asterisks indicate the significant differences at the **P < 0.01 level by Student’s t test analysis. The values are means ± se, one experiment at least three repeats, and in one repeat, n >30 per genotype. C, The ratio of OE-ABI4:WT under each treatment in (B). Calculated as the average value in (B).
To better understand the changing pattern of auxin concentration in the abi4 primary root tip in response to exogenous ABA treatment, we explored the transcription level, under different conditions, of PIN1 (PIN-FORMED1), which is a key gene encoding an auxin efflux transporter (Zhou et al., 2010). PIN1 expression was monitored using the reporter construct PIN1promoter::PIN1-GFP, which was crossed into the abi4 mutant background. The results showed that the expression level of PIN1promoter::PIN1-GFP in the abi4 primary root tip after exogenous ABA application was significantly higher than that in the WT (Supplemental Figure S12, A and B). Altogether, the upregulation of auxin concentration in the abi4 primary root tip is responsible for its greater root length compared with the WT in response to exogenous ABA treatment.
Discussion
As a phytohormone widely known to be involved in abiotic stress signaling, ABA plays many important roles in plant–environment interaction networks, including stresses such as drought and salinity. Recently published findings have demonstrated that ABA is also involved in plant development, including seed maturation, seed dormancy, and germination processes (Chauffour et al., 2019). In support of this, the current study focused mainly on the inhibitory effect of ABA on primary root growth. An earlier study had demonstrated that a low level of ABA was essential for the maintenance of the quiescent center cells and suppression of stem cell differentiation (Zhang et al., 2010). The roles of ABA in root branching have recently been recognized and there is some evidence for the involvement of high concentrations of ABA in the repression of primary and lateral root initiation and growth (Luo et al., 2014; Rowe et al., 2016; Mu et al., 2017; Sun et al., 2018), but the detailed regulatory mechanisms of ABA on primary root growth need to be further explored, especially with respect to the mechanisms by which ABA affects cell cycle progression and the interaction between ABA and the auxin signaling cascade.
ABI4-CYCB1;1/CDKB2;2 module-mediated cell cycles play a key role in primary root elongation in response to ABA application
The root meristem is not only a major determinant of longitudinal growth but is also important for root initiation and elongation (Van Norman et al., 2013). It is noteworthy that downregulation of PLT1 and PLT2, key regulatory genes in root elongation, which encode members of the PLETHORA family of transcription factors, was responsible for meristem size reduction (Mu et al., 2017); indeed, plant growth and development is closely associated with cell cycle progression (Polyn et al., 2015). In this study, we have shown that ABI4 plays a key role with regard to the repressive effect of exogenous ABA on primary root growth. The numerous studies have demonstrated that ABA represses the expression of positive regulatory genes of the cell cycle, including CYCB1s, and induces transcription of negative regulatory genes, such as the ICK/KRP CKD inhibitor genes, of the cell cycle (Wang et al., 1998, 2011; Xu et al., 2010; Yang et al., 2014; Zhang et al., 2020). However, little is known about the direct upstream regulator(s) encoded by these cell cycle-related genes at both the transcriptional and translational levels, especially during primary root elongation under the control of ABA.
In the current study, we detected that the repressive effect of exogenous ABA on primary root development was largely dependent on the ABI4 transcription factor (Figure 1), and the evidence, including investigation of meristem size and the number of meristem zone cells, was consistent with the phenotypic description (Figure 2). Further, we showed that in the abi4 mutant primary root tip, transcription of several cell cycle-related genes was altered (Figure 3), with the subsequent biochemical and genetic analysis demonstrating that transcription factor ABI4 acted upstream of CYCB1;1 and CDKB2;2, directly repressing their expression by physical interaction with their promoters (Figures 4–6). Collectively, this body of evidence demonstrated that ABA-repressed CYCB1;1 and CDKB2;2 expression is largely dependent on the transcription factor ABI4, and that the ABI4-CYCB1;1/CDKB2;2 regulatory module precisely regulates cell cycle progression, ultimately mediating cell cycle progression and primary root elongation.
The exogenous application of a high concentration of ABA represses cell cycle progression (Wang et al., 1998, 2011; Xu et al., 2010; Yang et al., 2014; Zhang et al., 2020), and this is in line with the character of ABA as a stress phytohormone, which arrests plant growth, especially under one of several abiotic stress conditions. Consistently, a recent study also showed that the shoot-derived exogenous application of ABA enhances primary root cell division and elongation (Xie et al., 2020), suggesting that a very low concentration of ABA was transported from the shoot to the primary root tip. However, under stress conditions, such as drought or salinity, the increased endogenous ABA level shows the opposite effect on the cell cycle and thus arrests plant development, ultimately increasing the stress tolerance of plants. Indeed, several other phytohormones have also shown different physiological roles at high and low concentrations, such as the effect of different levels of auxin on cell cycle progression (Cao et al., 2019), although the detailed regulatory mechanism by which ABA impairs the cell cycle is still largely unknown. This study employed the primary root tip as a research target system and demonstrated that ABA represses cell cycle progression by inhibiting the transcription of the ABI4-CYCB1;1/CDKB2;2 regulatory module (Figure 10).
Figure 10.
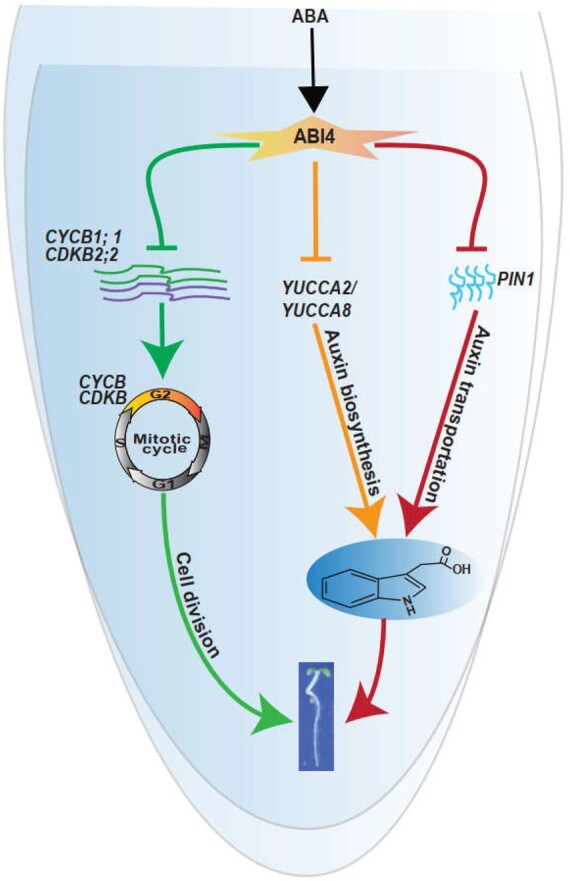
Proposed working model in which ABA represses primary root elongation by impairing the transcription of the ABI4-CYCB1;1/CDKB2;2 regulatory module and auxin biosynthesis. This simplified model showed that exogenous ABA application induces ABI4 transcription in the primary root tip, and the increased level of ABI4 further inhibits the cell cycle cascade by directly reducing CYCB1;1 and CDKB2;2 expression. Concurrently, ABI4 decreases the auxin concentration in the primary root tip by inhibiting the expression of the auxin biosynthesis genes YUCCA2 and YUCCA8, and the auxin transporter encoding gene PIN1. Altogether, the phytohormone ABA inhibits primary root growth through its effects on ABI4-mediated auxin and cell cycle pathways.
ABA inhibits primary root growth by impairing ABI4-mediated auxin biosynthesis and transport
Molecular genetic studies using Arabidopsis mutants have revealed that the auxin transport system, with a balance of influx and efflux, is important for primary root initiation and development (Du and Scheres, 2018). A previous study demonstrated that ABI4 is involved in plant lateral root development (Shkolnik-Inbar and Bar-Zvi, 2010). The number of lateral roots is higher in the abi4 mutant and lower in the ABI4-overexpressing transgenic plants, and this effect of ABI4 on lateral root development is possibly mediated through repression of PIN1, which encodes the dominant auxin-efflux carrier, thereby affecting auxin distribution (Shkolnik-Inbar and Bar-Zvi, 2010). The molecular evidence to support this hypothesis includes the observation that the transport rates of auxin were elevated in the abi4 mutant but reduced in OE-ABI4 lines (Shkolnik-Inbar and Bar-Zvi, 2010).
In addition to the ABA–auxin interaction during the primary root growth stage, it has been noted that previous studies showed that ABA also represses primary root elongation by enhancing ethylene signaling (Beaudoin et al., 2000; Ghassemian et al., 2000) and biosynthesis pathways (Luo et al., 2014). Thus, the current study opens up the possibility that ABA inhibits primary root growth by impairing ABI4-PIN1-mediated auxin biosynthesis and transport cascades. The present study revealed that ABI4 negatively controls PIN1 expression in the presence of exogenous ABA (Figure 9;Supplemental Figure S12), and that ABI4 represses the transcription of auxin biosynthesis genes (Figure 8, A and B), ultimately downregulating auxin levels (Figures 7 and 8, C and D). This decrease in the auxin level indicates the direct role of ABI4 in interfering with the threshold hormonal balance in the primary root tip.
Taken together, and combined with the previous published observations, the current study demonstrates the existence of a pivotal regulatory module, ABI4-CYCB1;1/CDKB2;2, which precisely mediates cell cycle progression, as well as auxin biosynthesis and transport, and eventually represses primary root elongation in the presence of high level of ABA (Figure 10). It is believed that these findings provide the insights into the mechanisms of ABA repression of cell cycle progression and auxin biosynthesis and transport beyond the already-known ABA–ethylene interaction cascade.
Materials and methods
Plant materials and growth conditions
The Arabidopsis ecotype Col-0 was employed as WT in this research. The point mutant abi4 (CS8104) and the T-DNA insertion mutant abi4-t (SALK_080095) were supplied by the Arabidopsis Biological Resource Center (ABRC; Ohio State University, Columbus, Ohio, USA). abi3 (abi3-8) and abi5 (abi5-7) mutant lines were provided by Dr. Diqiu Yu (State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Kunming, China). The OE-ABI4 (pCanG-ABI4-HA-GFP) transgenic line used in this study was previously generated in our laboratory (Shu et al., 2013, 2016a, 2016b; Luo et al., 2021). To produce 35S::CYCB1;1 and 35S::CDKB2;2 transgenic plants, the full-length CDS (coding sequence) fragments of both genes were amplified by PCR and then cloned into the vector pCambia-3301-HA, in which CYCB1;1 and CDKB2;2 were expressed under the control of the CaMV 35S promoter. Using the Agrobacterium tumefaciens strain GV3101 dip WT or OE-ABI4 plants separately. In order to perform genetic analysis, cycb1;1-cr, cdkb2;2-cr, abi4/cycb1;1-cr, and abi4/cdkb2;2-cr construction vector of materials was generated as previously described (Yan et al., 2015), and then using the A. tumefaciens strain GV3101 dip WT or abi4 plants separately.
Using 10% bleach (v/v), Arabidopsis seeds were surface-sterilized and then washed with sterile water three times. Seeds were suspended in 0.2% agar (w/v) and sown on 1/2 MS solid medium supplemented with sucrose (1%, w/v). Seeds were stratified under darkness by incubating the plates for 3 days at 4°C, after which they were transferred to 22°C under a 16-h light/8-h dark photoperiod. Then, the seedlings were treated by ABA (product number D8942, Sigma-Aldrich, USA) and NAA (product number N0640, Sigma-Aldrich, USA) when needed according to the requirements of the different experiments. Primers and restriction sites used in this study are shown in Supplemental Table S2.
Measurement of the primary root length
The primary root lengths of the WT, abi4, abi4-t, OE-ABI4, OE-CYCB1;1, OE-CDKB2;2, OE-ABI4:: OE-CYCB1;1, OE-ABI4::OE-CYCB1;1, cycb1;1-cr, cdkb1;1-cr, abi4/cycb1;1-cr, and abi4/cdkb2;2-cr genotypes were measured with or without ABA treatment, according to the requirements of the different experiments, using software Image J (National Institute of Health, http://imagej.nih.gov/ij/). The statistical significance of differences between samples was evaluated by Student’s t test analysis.
Mutant or transgenic line genotyping
The abi4 and abi4-t allelic mutants had been used in our previous studies (Shu et al., 2013; Luo et al., 2021). DR5promoter-GFP and PIN1promoter::PIN1-GFP marker lines, kindly provided by Dr. Chuanyou Li and Dr. Klaus Palme (Zhou et al., 2010), were crossed into the abi4 background, and plants homozygous for the abi4 mutations and transgene markers were identified from the F2 population and confirmed in the next generation. Parental transgenic marker lines were employed to compare with those in the abi4 background.
Microscopy of primary root
For confocal microscopy, seedlings on 1/2 MS medium were observed from 3 to 4 days using a Leica TCS SP5 confocal laser scanning microscope. Plants were stained with 10 mg mL−1 propidium iodide for 5 min and washed once in water. Propidium iodide was visualized using wavelengths of 600–640 nm, whereas the wavelengths to visualize GFP were 500–540 nm. GFP fluorescence was quantified with the LAS AF Lite program on confocal sections acquired with the same microscope settings (Zhou et al., 2010). Approximately 5–10 images were examined, and at least three independent experiments were performed. The statistical significance of differences was evaluated by Student’s t test analysis.
Gene expression analyses
Total RNA was extracted from the primary root tips of 1-week-old seedlings, and first-strand cDNA synthesis, and qPCR were performed as previously described (Zhou et al., 2010). DNase I-treated total RNA (2 μg) was denatured and subjected to reverse transcription using Moloney murine leukemia virus reverse transcriptase (200 units per reaction; Promega Corporation, Madison, Wisconsin, USA). qPCR was performed using the SsoFast EvaGreen Supermix (Bio-Rad) and CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Gene expression was quantified at the logarithmic phase using the expression of the housekeeping gene ACTIN2 mRNA as an internal control. Primers used in qPCR are shown in Supplemental Table S2.
ChIP-seq and data analysis
Samples for ChIP-seq were constructed with NovoNGS CUT&Tag 3.0 High-Sensitivity Kit for Illumina (Novoprotein, product number N259-YH01) (Kaya-Okur et al., 2019; Huang and Sun, 2022). One-week-old seedlings of OE-ABI4 were selected, and then about 3 g of seedling was ground in liquid nitrogen following operation according to the manufacturer’s instructions. Anti-GFP (Proteintech, product number 66002) was used as primary antibody, goat anti-mouse IgG (H + L) (BOSTER, product number BA1038) was used as secondary antibody. The DNA fragments were extracted by phenol chloroform and used for sequencing. Paired-end sequencing by Hangzhou Lianchuan Biological Information Co., Ltd. Software FastQC for quality control of raw data, clean data were obtained by detecting and removing adapters and low-quality bases with TRIM GALORE software. Subsequently, BOWTIE2 was used to compare the sequences with the reference genome (A. thaliana reference genome TAIR10), and SAMTOOLS and BEDTOOLS were employed to complete the filtering and file format conversion. Next, MACS2 was used to conduct peak calling on the compared data, and finally CHIPSEEKER was employed to annotate peaks. The raw data have been submitted to National Center for Biotechnology Information database that can be accessed with the number of PRJNA859368.
ChIP-qPCR analysis
To explore the binding of ABI4 to the promoter of CYCB1;1 or CDKB2;2, a ChIP-qPCR procedure was performed. One-week-old seedlings of OE-ABI4 were selected; the whole seedlings were crosslinked with 1% formaldehyde (w/v) for 10 min under vacuum and the reaction stopped by the addition of 0.125 M glycine. The samples were snap-frozen in liquid nitrogen, ground, and the nuclei extracted. Using a COVARIS M220 focused-ultrasonicator (Gene Company Limited, Hong Kong, China), the chromatin was disrupted. ChIP was performed with the anti-GFP antibody and protein G beads, with samples without anti-GFP being used as the empty control (CK). Finally, the nuclear DNA was washed with 75% ethanol (v/v) and dissolved in 50 µL water. The qPCR analysis was performed using specific primers corresponding to different promoter fragments of CYCB1;1 and CDKB2;2. The ABI5 promoter was used as the positive control and TUB4 was used as the internal control in this experiment. Primers used in the ChIP-qPCR are shown in Supplemental Table S2.
Yeast one-hybrid analysis
Yeast one-hybrid analysis was performed as previously described (Fu et al., 2018). pGADT7-ABI4 vector had previously been constructed in our laboratory (Luo et al., 2021). Full-length CYCB1;1 and CDKB2;2 promoter fragments were amplified by PCR and then cloned into the vector pHIS2. The positive control pHIS2-ProRbohD vector, generated in our previous study (Luo et al., 2021), was employed. Primers and restriction sites used in the yeast one-hybrid analysis are shown in Supplemental Table S2.
Transient expression analysis of CYCB1;1 and CDKB2;2 promoter activity by ABI4 in vivo
The transient expression assay was performed in N. benthamiana leaves according to our previous methodology (Shu et al., 2016a, 2016b; Luo et al., 2021). The full-length promoter sequences of CYCB1;1 and CDKB2;2 were amplified by PCR and then cloned into the vector pCambia1300-221-GUS, by replacing the original CaMV 35S promoter. pCanG-HA-GFP is the effector construct used in this experiment. Agrobacterium cells (strain GV3101) were cultured at 30°C overnight and the pellet obtained following centrifugation was resuspended in the buffer used to infiltrate the N. benthamiana leaves. GUS activity was measured in leaf disks 2 days after infiltration into N. benthamiana leaves. The GUS staining procedure was followed using a GUS staining kit (G3061; Solarbio Corp., Beijing, China), and the GUS activity was quantified using a protocol that had been described previously (Luo et al., 2021). A digital camera (D3500; Nikon, Tokyo, Japan) was used for photography of the images. Primers used in this experiment are shown in Supplemental Table S2.
Quantification of endogenous auxin
Quantification of endogenous auxin concentrations was performed according to previously described methods (Pan and Wang, 2009; Pan et al., 2010). The primary root tips from 4-day-old seedlings were selected and snap-frozen in liquid nitrogen, ground to a fine powder, and extracted with 80% (v/v) methanol. Purification was performed using a Poly-Sery MAX SPE Cartridge (CNW, Germany) and eluted with 0.5% formic acid (v/v) in methanol. The eluate was then dried under nitrogen, reconstituted in methanol, and injected into a high-performance liquid chromatography (HPLC)-MS/MS system consisting of an Agilent 1290 HPLC system (Agilent Company, USA) and a SCIEX-6500 Qtrap MS/MS tandem mass spectrometer (AB SCIEX Company, USA). These auxin quantification experiments were performed by Wuhan ProNets Biotechnology Co., Ltd, Wuhan, China. Three biological replications were conducted.
Global gene expression analysis
Heat maps of the transcriptome data were generated from Arabidopsis RNA-Seq data resources (http://ipf.sustech.edu.cn/pub/athrna/) (Zhang et al., 2020), the project numbers including PRJNA272719, PRJNA277840, PRJNA371677, PRJNA389285, PRJNA471243, PRJNA472051, PRJNA478998, and PRJNA494179. Global gene expression profiles were visualized by heat maps via TBtools software as described previously (Chen et al., 2020).
Accession numbers
Arabidopsis Genome Initiative locus identifiers for the major genes mentioned in this article are as follows: ABI3 (AT3G24650), ABI4 (AT2G40220), ABI5 (AT2G36270), PIN1 (At1G73590), RbohD (AT5G47910), YUCCA2 (AT4G13260), YUCCA8 (AT4G28720), CDKB1;1 (AT3G54180), CDKB2;1 (AT1G76540), CDKB2;2 (AT1G20930), CDKA;1 (AT3G48750), CYCB1;1 (AT1G70210), CYCD1;1 (AT4G37490), CYCD2;1 (AT2G22490), CYCD3;3 (AT3G50070), CYCD3;1 (AT4G34160), CYCD3;2 (AT5G67260), CYCD5;1 (AT4G37630), KRP1 (AT2G23430), KRP2 (AT3G50630), KRP3 (AT5G48820), KRP4 (AT2G32710), KRP5 (AT3G24810), KRP6 (AT3G19150), and KRP7 (AT1G49620). The stock numbers of the mutants used in this study are as follows: abi4 (CS8104) and abi4-t (SALK_080095).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Published database bioinformatic analysis showed that ABA promotes the transcription of cell cycle-related positive regulatory genes.
Supplemental Figure S2. Published database bioinformatic analysis showed that ABA promotes the transcription of cell cycle-related negative regulatory genes.
Supplemental Figure S3. ABA induces ABI4 transcription in the primary root tip.
Supplemental Figure S4. Distinct response of different age of abi4 seedlings with regard to primary root growth after ABA treatment.
Supplemental Figure S5. ABA-induced cell cycle-related gene expression in abi4.
Supplemental Figure S6. Expression pattern analysis of cell cycle-related genes in abi3 and abi5.
Supplemental Figure S7. Mutation description of CDKB2;2 promoter for yeast one-hybrid assay.
Supplemental Figure S8. Quantitative analysis of GUS activity.
Supplemental Figure S9. Gene expression levels in OE-CYCB1;1 and OE-CDKB2;2 transgenic lines.
Supplemental Figure S10. Gene expression analysis in OE-ABI4::OE-CYCB1;1 and OE-ABI4::OE-CDKB2;2 transgenic lines.
Supplemental Figure S11. The effect of ABI3 and ABI5 on the auxin biosynthesis genes in primary roots after exposure to exogenous ABA application.
Supplemental Figure S12. Transcription pattern of the PIN1promoter::PIN1-GFP reporter construct in the WT and abi4 mutant roots.
Supplemental Table S1. ChIP-seq analysis database.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Klaus Palme (the University of Freiburg) and Dr. Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for kindly providing the DR5promoter-GFP and PIN1promoter::PIN1-GFP marker lines. abi3-8 and abi5-7 mutant lines were provided by Dr. Diqiu Yu (State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan University, Kunming, China). The point and T-DNA insertion abi4 mutants were provided by the ABRC.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 31872804 and 31701064) and the Innovation Foundation for Doctoral Dissertations of Northwestern Polytechnical University (CX2021040).
Conflict of interest statement. The authors declare no interests.
Contributor Information
Xiaofeng Luo, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Jiahui Xu, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China; College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Chuan Zheng, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China; College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China; Institute of Ecological Agriculture, Sichuan Agricultural University, Chengdu 611130, China.
Yingzeng Yang, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China; Institute of Ecological Agriculture, Sichuan Agricultural University, Chengdu 611130, China.
Lei Wang, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Ranran Zhang, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Xiaotong Ren, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Shaowei Wei, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Usman Aziz, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Junbo Du, Institute of Ecological Agriculture, Sichuan Agricultural University, Chengdu 611130, China.
Weiguo Liu, Institute of Ecological Agriculture, Sichuan Agricultural University, Chengdu 611130, China.
Weiming Tan, College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
Kai Shu, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
K.S. conceived this study. K.S. and W.T. supervised the research. X.L., J.X., C.Z., Y.Y., L.W., S.W., U.A., J.D., and W.L. performed the experiments. K.S., X.L., R.Z., X.R., and W.T. analyzed the data. K.S. and X.L. wrote the manuscript. All authors approved the submission of the final draft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Kai Shu (kshu@nwpu.edu.cn).
References
- Ahmad R, Liu Y, Wang TJ, Meng Q, Yin H, Wang X, Wu Y, Nan N, Liu B, Xu ZY (2019) GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiol 179: 1844–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupre P, Mendoza MS, Roman CS, Leon P (2009) The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J 59: 359–374 [DOI] [PubMed] [Google Scholar]
- Bouain N, Krouk G, Lacombe B, Rouached H (2019) Getting to the root of plant mineral nutrition: combinatorial nutrient stresses reveal emergent properties. Trends Plant Sci 24: 542–552 [DOI] [PubMed] [Google Scholar]
- Cao M, Chen R, Li P, Yu Y, Zheng R, Ge D, Zheng W, Wang X, Gu Y, Gelova Z, et al. (2019) TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568: 240–243 [DOI] [PubMed] [Google Scholar]
- Chandrasekaran U, Luo X, Zhou W, Shu K (2020) Multifaceted signaling networks mediated by abscisic acid insensitive 4. Plant Commun 1: 100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauffour F, Bailly M, Perreau F, Cueff G, Suzuki H, Collet B, Frey A, Clement G, Soubigou-Taconnat L, Balliau T, et al. (2019) Multi-omics analysis reveals sequential roles for ABA during seed maturation. Plant Physiol 180: 1198–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13: 1194–1202 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Benfey PN (2008) Plant stem cell niches: standing the test of time. Cell 132: 553–557 [DOI] [PubMed] [Google Scholar]
- Du Y, Scheres B (2018) Lateral root formation and the multiple roles of auxin. J Exp Bot 69: 155–167 [DOI] [PubMed] [Google Scholar]
- Fu JY, Liu Q, Wang C, Liang J, Liu LJ, Wang Q (2018) ZmWRKY79 positively regulates maize phytoalexin biosynthetic gene expression and is involved in stress response. J Exp Bot 69: 497–510 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C (2016) 25 years of cell cycle research: what’s ahead? Trends Plant Sci 21: 823–833 [DOI] [PubMed] [Google Scholar]
- Huang X, Sun MX (2022) H3K27 methylation regulates the fate of two cell lineages in male gametophytes. Plant Cell 34: 2989–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XZ, Zhang XY, Gong ZZ, Yang SH, Shi YT (2017) ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J 89: 354–365 [DOI] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S (2019) CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10: 1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XF, Dai YJ, Zheng C, Yang YZ, Chen W, Wang QC, Chandrasekaran U, Du JB, Liu WG, Shu K (2021) The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol 229: 950–962 [DOI] [PubMed] [Google Scholar]
- Luo XJ, Chen ZZ, Gao JP, Gong ZZ (2014) Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J 79: 44–55 [DOI] [PubMed] [Google Scholar]
- Lv B, Wei K, Hu K, Tian T, Zhang F, Yu Z, Zhang D, Su Y, Sang Y, Zhang X, et al. (2021) MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol Plant 14: 285–297 [DOI] [PubMed] [Google Scholar]
- Mu Y, Zou MJ, Sun XW, He BY, Xu XM, Liu YN, Zhang LX, Chi W (2017) BASIC PENTACYSTEINE proteins repress ABSCISIC ACID INSENSITIVE4 expression via direct recruitment of the polycomb-repressive complex 2 in Arabidopsis root development. Plant Cell Physiol 58: 607–621 [DOI] [PubMed] [Google Scholar]
- Nee G, Kramer K, Nakabayashi K, Yuan B, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ (2017) DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat Commun 8: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifici E, Polverari L, Sabatini S (2015) Plant hormone cross-talk: the pivot of root growth. J Exp Bot 66: 1113–1121 [DOI] [PubMed] [Google Scholar]
- Pan XQ, Wang XM (2009) Profiling of plant hormones by mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 877: 2806–2813 [DOI] [PubMed] [Google Scholar]
- Pan XQ, Welti R, Wang XM (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Polyn S, Willems A, De Veylder L (2015) Cell cycle entry, maintenance, and exit during plant development. Curr Opin Plant Biol 23: 1–7 [DOI] [PubMed] [Google Scholar]
- Promchuea S, Zhu Y, Chen Z, Zhang J, Gong Z (2017) ARF2 coordinates with PLETHORAs and PINs to orchestrate ABA-mediated root meristem activity in Arabidopsis. J Integr Plant Biol 59: 30–43 [DOI] [PubMed] [Google Scholar]
- Qin H, Huang R (2018) Auxin controlled by ethylene steers root development. Int J Mol Sci 19: 3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JH, Topping JF, Liu J, Lindsey K (2016) Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol 211: 225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Schafer S, Breuer C, Iwase A, Shibata M, Ikeda M, Mitsuda N, Koncz C, Ohme-Takagi M, et al. (2017) ABA suppresses root hair growth via the OBP4 transcriptional regulator. Plant Physiol 173: 1750–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Adler G, Bar-Zvi D (2013) ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J 73: 993–1005 [DOI] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu Y, Liu R, Zhang H, Wang P, Li Y, Wang S, Tang S, Liu C, et al. (2016a) ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J 85: 348–361 [DOI] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu YR, Liu RJ, Zhang HW, Wang SF, Tang SY, Yang WY, Xie Q (2016b) ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J Exp Bot 67: 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH (2016c) Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant 9: 34–45 [DOI] [PubMed] [Google Scholar]
- Shu K, Zhang HW, Wang SF, Chen ML, Wu YR, Tang SY, Liu CY, Feng YQ, Cao XF, Xie Q (2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLos Genet 9: e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhou WG, Yang WY (2018) APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol 217: 977–983 [DOI] [PubMed] [Google Scholar]
- Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LR, Wang YB, He SB, Hao FS (2018) Mechanisms for abscisic acid inhibition of primary root growth. Plant Signal Behav 13: e1500069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Lv B, Ding T, Bai M, Ding Z (2017) Auxin-BR interaction regulates plant growth and development. Front Plant Sci 8: 2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN (2013) To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140: 4301–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qi QG, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Wang L, Hua D, He J, Duan Y, Chen Z, Hong X, Gong Z (2011) Auxin response factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet 7: e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang W, Zuo Y, Zhu L, Hastwell AH, Chen L, Tian Y, Su C, Ferguson BJ, Li X (2019) GmYUC2a is important for mediating auxin biosynthesis during root development and nodulation in soybean. J Exp Bot 70: 3165–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Li L, Ye TT, Lu YM, Chen X, Wu Y (2013) The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J Exp Bot 64: 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC (2013) ABI4: versatile activator and repressor. Trends Plant Sci 18: 125–132 [DOI] [PubMed] [Google Scholar]
- Xie Q, Essemine J, Pang X, Chen H, Cai W (2020) Exogenous application of abscisic acid to shoots promotes primary root cell division and elongation. Plant Sci 292: 110385. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Mao Y, Duan XL, Zhou H, Lai DW, Zhang YH, Shen WB (2016) Arabidopsis HY1-modulated stomatal movement: an integrative hub is functionally associated with ABI4 in dehydration-induced ABA responsiveness. Plant Physiol 170: 1699–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Zhao Y, Gao JH, Xiang CB, Zhu JK (2016) The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci Rep 6: 27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Gao GL, Du JJ, Guo Y, Yang CW (2010) Cell cycle modulation in response of the primary root of Arabidopsis to ABA. Pak J Bot 42: 2703–2710 [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q (2015) High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant 8: 1820–1823 [DOI] [PubMed] [Google Scholar]
- Yang L, Zhang J, He J, Qin Y, Hua D, Duan Y, Chen Z, Gong Z (2014) ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet 10: e1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21: 133–139 [DOI] [PubMed] [Google Scholar]
- Zhang H, Han W, De Smet I, Talboys P, Loya R, Hassan A, Rong H, Jurgens G, Paul Knox J, Wang MH (2010) ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J 64: 764–774 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang F, Yu Y, Feng L, Jia J, Liu B, Li B, Guo H, Zhai J (2020) A comprehensive online database for exploring approximately 20,000 public Arabidopsis RNA-seq libraries. Mol Plant 13: 1231–1233 [DOI] [PubMed] [Google Scholar]
- Zhang M, Lu X, Li C, Zhang B, Zhang C, Zhang XS, Ding Z (2018) Auxin efflux carrier ZmPGP1 mediates root growth inhibition under aluminum stress. Plant Physiol 177: 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y (2018) Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu Rev Plant Biol 69: 417–435 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ai XH, Wang MC, Xiao LT, Xia GM (2016) A putative pyruvate transporter TaBASS2 positively regulates salinity tolerance in wheat via modulation of ABI4 expression. BMC Plant Biol 16: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang XG, Hou YJ, Gao JH, Wang PC, Duan CG, Zhu XH, Zhu JK (2014) The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal 7: ra53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wei L, Xu J, Zhai Q, Jiang H, Chen R, Chen Q, Sun J, Chu J, Zhu L, et al. (2010) Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Hu X, Duan Y, Li S, Wang Y, Rehman AU, He J, Zhang J, Hua D, Yang L, et al. (2020) The Arabidopsis nodulin homeobox factor AtNDX interacts with AtRING1A/B and negatively regulates abscisic acid signaling. Plant Cell 32: 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.