Abstract
The degradation products of glucosinolates (GSLs) greatly lower the nutritional value of rapeseed (Brassica napus) meal; thus, reduction of seed GSL content (SGC) has become an important objective of rapeseed breeding. In our previous study, we finely mapped a major QTL (qGSL-C2) for SGC to a 49-kb collinear region on B. rapa chromosome A2. Here, we experimentally validated that BnaC2.MYB28, encoding an R2R3-MYB transcription factor, is the causal gene of qGSL-C2. BnaC2.MYB28 is a nucleus-localized protein mainly expressed in vegetative tissues. Knockout of BnaC2.MYB28 in the high-SGC parent G120 reduced SGC to a value lower than that in the low-SGC parent ZY50, while overexpression of BnaC2.MYB28 in both parental lines (G120 and ZY50) led to extremely high SGC, indicating that BnaC2.MYB28 acts as a positive regulator of SGC in both parents. Molecular characterization revealed that BnaC2.MYB28 forms a homodimer and specifically interacts with BnaMYC3. Moreover, BnaC2.MYB28 can directly activate the expression of GSL biosynthesis genes. Differential expression abundance resulting from the polymorphic promoter sequences, in combination with the different capability in activating downstream genes involved in aliphatic GSL biosynthesis, caused the functional divergence of BnaC2.MYB28 in SGC regulation between the parents. Natural variation of BnaC2.MYB28 was highly associated with SGC in natural germplasm and has undergone artificial selection in modern low-GSL breeding. This study provides important insights into the core function of BnaC2.MYB28 in regulating SGC and a promising strategy for manipulating SGC in rapeseed.
BnaC2.MYB28, encoding an R2R3-MYB transcription factor, corresponds to a quantitative trait locus for seed glucosinolate content in rapeseed.
Introduction
Glucosinolates (GSLs), which are derived from amino acids and sugars, are known as a large group of secondary metabolites in the Brassicaceae family (Seo and Kim, 2017). In recent decades, GSLs have attracted great research attention due to the diverse biological properties of their hydrolysis products (Sonderby et al., 2010; Kittipol et al., 2019). For example, some GSLs and their hydrolysates confer Brassica vegetables with distinct taste and flavor (Halkier and Gershenzon, 2006), while others have been confirmed to be involved in plant defense reaction against pathogens and insect pests (Potter et al., 2000; Ahuja et al., 2010; Bednarek, 2012). In addition, GSL hydrolysates, particularly the well-studied sulforaphane, are considered as effective cancer-preventive agents (Keck and Finley, 2004; Li et al., 2008). Nevertheless, some GSLs such as progoitrin can be highly accumulated in seeds, and their degradation products can largely reduce the nutritional values of the cake meal and even cause diseases of the liver, thyroid, and kidney when consumed by livestock (Walker and Booth, 2001; Grubb and Abel, 2006; Tayo et al., 2012). Therefore, it is of great biological and economic importance to reveal the metabolism of GSLs in Cruciferous crops.
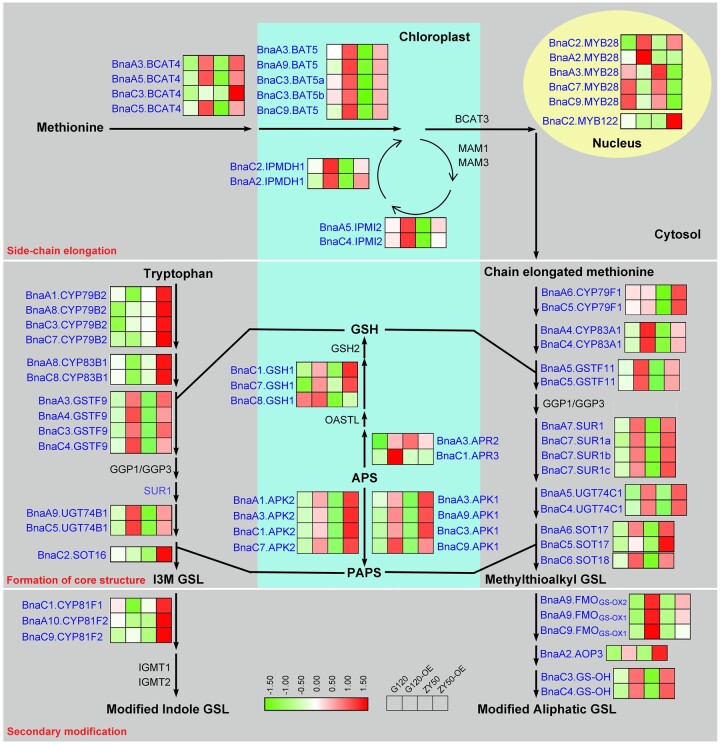
In the model plant Arabidopsis (Arabidopsis thaliana), a closely related species of Brassicaceae, the biosynthesis and metabolism pathways of GSLs have been well elucidated. Generally, the biosynthesis process of GSLs undergoes three independent stages: side chain elongation of precursor amino acids, formation of the core GSL structure, and secondary modification of the amino acid side chains (Grubb and Abel, 2006). Most genes involved in these stages have been characterized. For example, three partially redundant Methylthioalkyl Malate Synthases MAMs (MAM1, MAM2, and MAM3) were validated to be implicated in side chain elongation (Kroymann et al., 2001, 2003; Textor et al., 2007); Cytochrome P450 79B2 (CYP79B2), CYP79B3, CYP79F1, CYP79F2, CYP83A1, and CYP83B1 have been shown to participate in the formation of the core GSL structure (Mikkelsen et al., 2000; Bak and Feyereisen, 2001; Chen et al., 2003); and the Flavin-Monooxygenase GSL S-oxygenases FMOGS-OXs (FMOGS-OX1, FMOGS-OX2, FMOGS-OX3, FMOGS-OX4, and FMOGS-OX5) and Alkenyl Hydroxalkyl Producings AOPs (AOP2 and AOP3) were demonstrated to play crucial roles in the secondary modification of GSLs (Kliebenstein et al., 2001a, 2001b; Hansen et al., 2007; Li et al., 2008). Additionally, six transcription factors (TFs) belonging to the R2R3-Myeloblastosis (MYB) family, including MYB28 (HIGH ALIPHATIC GLUCOSINOLATE 1 [HAG1]), MYB29 (HAG3), MYB76 (HAG2), MYB34 (ALTERED TRYPTOPHAN REGULATION 1), MYB51 (HIGH INDOLIC GLUCOSINOLATE 1 [HIG1]) and MYB122 (HIG2), are involved in the regulation of GSL biosynthetic pathway (Hirai et al., 2007; Frerigmann and Gigolashvili, 2014). Among them, MYB28 has been recognized as the predominant regulator for the synthesis of aliphatic GSLs (Sønderby et al., 2007; Gigolashvili et al., 2008). Many critical genes involved in aliphatic GSL biosynthetic pathway have been confirmed to be positively regulated by MYB28, including side chain elongation (MAM1, MAM3, and BCAT4), core structure formation (CYP79F1, CYP79F2, CYP83A1, SOT17, and SOT18) and the secondary modification (AOP2) (Hirai et al., 2007; Gigolashvili et al., 2008).
Rapeseed (Brassica napus L.) is the second largest growing oil crop only next to soybean around the world (Carré and Pouzet, 2014). However, the high content of GSLs in the seeds not only reduces the oil quality, but also causes health problem in animals fed on the cake meal (Walker and Booth, 2001). Toward the reduction of seed GSL content (SGC) in rapeseed breeding, some quantitative trait loci (QTLs) controlling SGC have been comprehensively identified by linkage mapping (Uzunova et al., 1995; Feng et al., 2012; Wurschum et al., 2012; He et al., 2018) and genome-wide association study (GWAS) (Li et al., 2014; Lu et al., 2014; Gajardo et al., 2015; Qu et al., 2015; Wei et al., 2018; Liu et al., 2019a, 2019b). Several of these QTLs can be repeatedly detected by linkage mapping and GWAS in different environments (Liu et al., 2019a, 2019b; Zhou et al., 2021; Tan et al., 2022), and the candidate genes for them have been predicted.
Although the so-called “low-GSL breeding” has successfully lowered SGC in some elite rapeseed accessions to moderate to low levels, ranging from 30 to 50 μmol g−1 in dry seeds, the meal industry expects to further reduce SGC to a level ˂20 μmol g−1 or even lower. However, the genetic and molecular mechanisms involved in SGC regulation under low-SGC background are rarely studied. Previously, we detected a major QTL (qGSL-C2) for SGC based on a double haploid (DH) population derived from a F1 plant of the cross between G120 (SGC: 33.0 ± 2.4 μmol g−1) and ZY50 (previously named 9172; SGC:17.7 ± 1.5 μmol g−1), which showed one-fold difference in SGC but can be both classified into the low-SGC germplasm with the general standard of ˂40.0 μmol g−1 (Liu et al., 2019a, 2019b). qGSL-C2 was finally delimited to a 49-kb collinear interval on the B. rapa A2 chromosome, which contains five candidate genes (Liu et al., 2019a, 2019b). In this study, we confirmed that BnaC2.MYB28 is the underlying gene for qGSL-C2 and elucidated the molecular mechanism how the allelic variation in BnaC2.MYB28 differentially regulates SGC in rapeseed. These findings provide important clues for understanding the molecular regulation of SGC and manipulating BnaC2.MYB28 to cultivate low-GSL varieties in rapeseed.
Results
Sequence analysis of the candidate gene for qGSL-C2
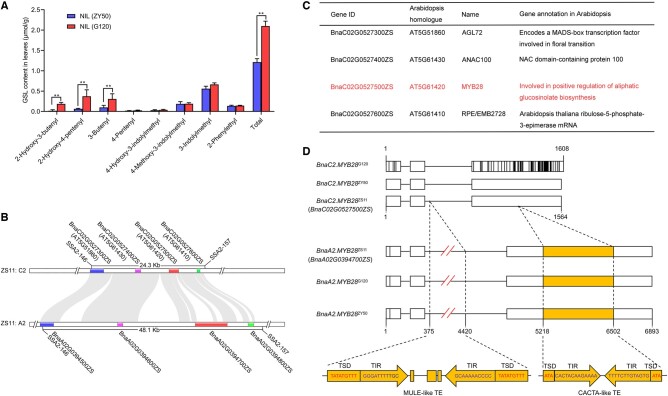
The major QTL for SGC, qGSL-C2, was delimited to a region between the markers of SSA2-146 and SSA2-157 (Liu et al., 2019a, 2019b). To clone qGSL-C2, we constructed a pair of near-isogenic lines (NILs), individually designated NIL (ZY50) and NIL (G120), from a residual heterozygous line containing a heterozygous chromosome segment encompassing the qGSL-C2 locus flanked by SSA2-146 and SSA2-157. NIL(ZY50) and NIL(G120) exhibited stable differences in SGC (Supplemental Figure S1); moreover, comparison of GSL content in leaves also revealed significant differences in three kinds of aliphatic GSLs (2-Hydroxy-3-butenyl, 2-Hydroxy-4-pentenyl, and 3-Butenyl) and total GSL content between NILs (Figure 1A). With the release of the high-quality rapeseed genome, we re-aligned the markers SSA2-146 and SSA2-157 to the ZS11 reference genome (http://cbi.hzau.edu.cn/bnapus/index.php) and obtained a target QTL region of 24.3 kb on the C2 chromosome, which showed a high collinearity with a 48.1-kb region on the A2 chromosome (Figure 1B).
Figure 1.
Sequence analysis of the candidate gene for qGSL-C2. A, GSL content of NIL(ZY50) and NIL(G120) in mature leaves. Data represent mean ± standard deviation (sd; n = 3). Asterisks indicate significant differences (t test; *P < 0.05, **P < 0.01). B, Physical region of the qGSL-C2 locus on the C2 chromosome of the ZS11 reference genome and its collinear region on the A2 chromosome. Colored bars represent gene regions. C, Genes predicted within the QTL region of qGSL-C2 and their putative orthologs in Arabidopsis. D, Genomic structure of BnaC2.MYB28 and BnaA2.MYB28 from the parents and ZS11. Horizontal lines represent introns, and boxes represent exons. The vertical lines on the boxes represent the sequence variations among genes. TSD, target site duplication; TIR, terminal inverted repeat; TE, transcription element.
Only four genes (BnaC02G0527300ZS, BnaC02G0527400ZS, BnaC02G0527500ZS, and BnaC02G0527600ZS) were identified in this target region of qGSL-C2 (Figure 1B). Putative functions of the four genes were predicted by searching the corresponding putatively orthologous genes in Arabidopsis annotation database (https://www.arabidopsis.org/) (Figure 1C). According to the re-sequencing data in the target QTL region of qGSL-C2, two (BnaC02G0527300ZS and BnaC02G0527500ZS) of the three genes with variations in the coding sequences showed differences in predicted protein sequences between parental lines (Supplemental Figure S2). BnaC02G0527500ZS encodes an R2R3-MYB TF homologous to MYB28 designated as BnaC2.MYB28. In Arabidopsis, MYB28 positively regulates the biosynthesis of aliphatic GSLs (Gigolashvili et al., 2007). The function of MYB28 may be associated with GSL content variation between the parental lines; therefore, we considered that BnaC2.MYB28 was the most promising candidate gene for qGSL-C2.
A sequence comparison of BnaC2.MYB28 from the parents and reference genome revealed that the sequences of BnaC2.MYB28ZY50 and BnaC2.MYB28ZS11 were identical, whereas the sequence of BnaC2.MYB28G120 showed great variations compared with that of BnaC2.MYB28ZS11 (Figure 1D). Sequence alignment revealed that both BnaC2.MYB28ZY50 and BnaC2.MYB28G120 contain three exons, and most of the variations, including SNPs and Indels, are scattered in the third exon (Supplemental Figure S2B). Interestingly, the sequences of BnaA2.MYB28 from G120 and ZY50 were identical to BnaA2.MYB28ZS11, and both display high similarity with BnaC2.MYB28ZS11 except for a MULE-like transposon insertion in the second intron and a CACTA-like transposon insertion in the third exon (Figure 1D). According to the RNA-seq data of ZY50, we found that the transcript of BnaA2.MYB28 has partial loss compared to that of BnaC2.MYB28 (Supplemental Figure S3). We speculated that the CACTA-like transposon inserted in the third exon may have disrupted the coding sequence of BnaC2.MYB28 and results in a dysfunctional protein.
BnaC2.MYB28 is the underlying gene of qGSL-C2 and encodes an R2R3-MYB TF
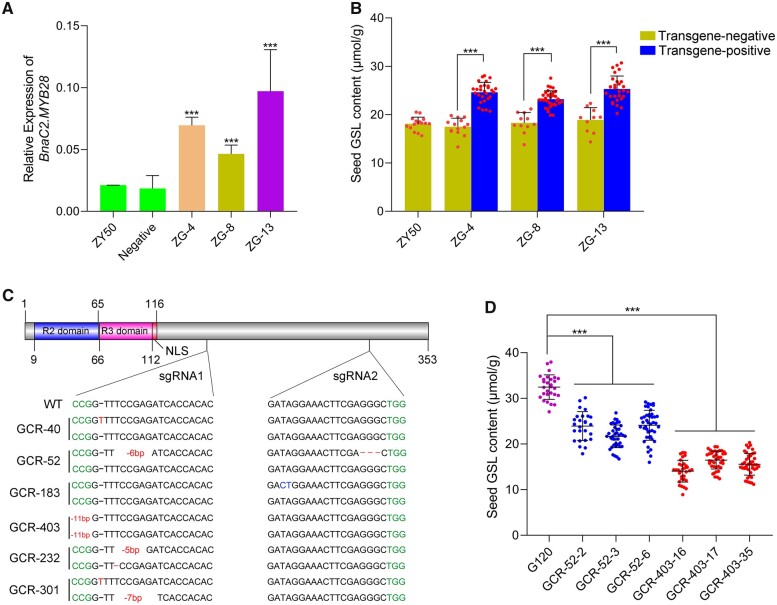
To validate that BnaC2.MYB28 is the underlying gene of the qGSL-C2 locus, a 5-kb fragment containing the 2.4-kb upstream regulatory sequence, the 1.6-kb coding sequence, and the 1-kb downstream 3′-UTR (untranslated regions) region of BnaC2.MYB28G120 was introduced into ZY50. From the 14 resultant T0 positive transformants, we randomly selected three plants (ZG−4, ZG−8, and ZG−13) and determined the expression level of BnaC2.MYB28 in their leaves of T1 generation. As shown in Figure 2A, the positive plants had significantly higher BnaC2.MYB28 expression than the negative plants and ZY50, indicating the expression of the transformed gene. A phenotypic analysis of all three T1 families demonstrated that the transgene-positive plants had significantly higher SGC than the transgene-negative plants (Figure 2B; Supplemental Table S1).
Figure 2.
Functional identification of BnaC2.MYB28. A, Relative expression of BnaC2.MYB28 in the transgenic B. napus lines. Error bars represent the sds from three independent RNA samples. BnACTIN7 was used as a control. Asterisks indicate significant differences (t test; ***P < 0.001). B, SGC in ZY50, transgene-negative and transgene-positive T1 lines (ZG−4, ZG−8, and ZG−13). Data are shown as mean ± sd (n ≥ 10). Each dot on the column represents an individual plant. Error bars represent the sds. C, Targeting of BnaC2.MYB28G120 with two (sgRNAs using the CRISPR/Cas9 system. Deletions are indicated by dashes or sequence gap lengths (bp); base substitutions are indicated by blue letters; and insertions are indicated by red letters. WT, wild-type; NLS, nuclear localization signal. D, SGC of T2 mutant lines and G120. Data are shown as mean ± sd (n ≥ 26). Each colored dot represents an individual plant. Error bars represent the sds. Two-tailed Student’s t tests were used to generate the P-values. ***P < 0.001.
To further confirm that BnaC2.MYB28 is the gene underlying the qGSL-C2 locus, the clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) system was used to generate diverse mutants of G120. Two single-guide RNAs (sgRNAs) of BnaC2.MYB28G120 were designed with the program CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). In total, six independent T0 transgenic-positive plants were confirmed to have various mutations near the target sites by sequencing, including substitution, insertion and deletion of different nucleotides (Figure 2C). To evaluate their phenotype and obtain stable mutant lines, all the T0 edited lines were progressed to T1 and T2 generations. The T1 edited lines exhibited different degrees of decrease in SGC compared with the recipient G120 plants (Supplemental Figure S4). Two T1 lines, GCR−52 and GCR−403, respectively, with amino acid deletion and frameshift mutation, were further analyzed. Their T2 lines also exhibited stable and significant decreases in SGC (Figure 2D; Supplemental Table S2). Particularly, the SGC in GCR−403 − 16 (14.1 ± 2.4 μmol g−1), GCR−403 − 17 (16.5 ± 2.0 μmol g−1), and GCR−403 − 35 (15.6 ± 2.4 μmol g−1) were even lower than that in the low-SGC parent ZY50 (17.7 ± 1.5 μmol g−1). Taken together, the genetic complementation and genome editing experiments indicated that BnaC2.MYB28 is the causal gene for qGSL-C2, and the disruption of BnaC2.MYB28 could contribute to the reduction of SGC.
Expression pattern and subcellular localization of BnaC2.MYB28
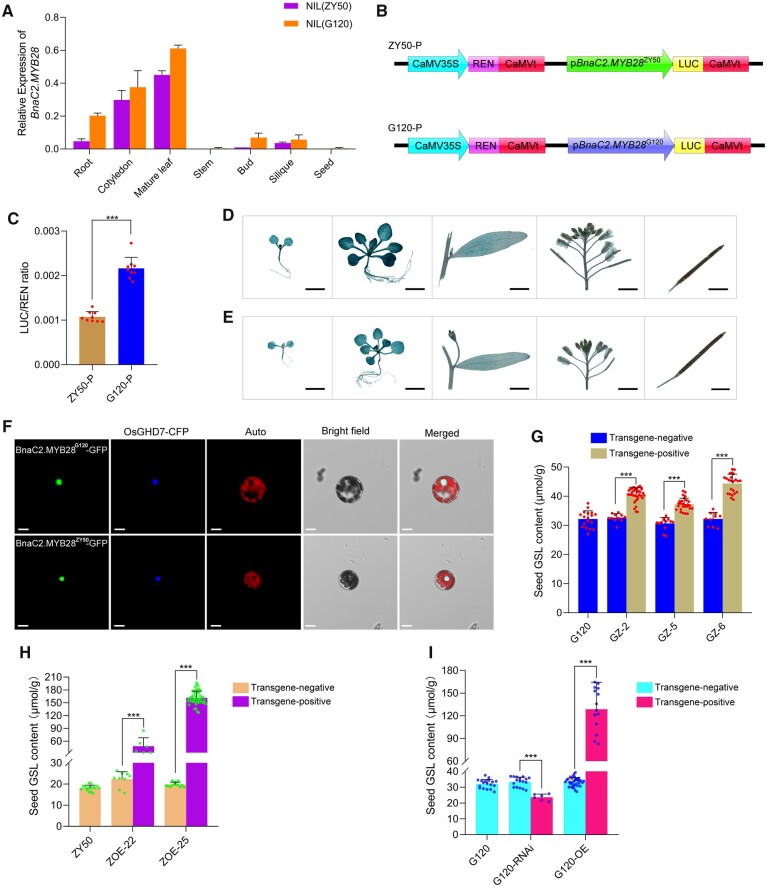
To explore the expression pattern of BnaC2.MYB28, a reverse transcription–quantitative PCR (RT–qPCR) analysis was conducted with the NILs. The results demonstrated that BnaC2.MYB28 was mainly expressed in roots, cotyledons, and mature leaves, with low transcription levels in buds and siliques and almost no transcript accumulation in stems and seeds (Figure 3A). BnaC2.MYB28G120 and BnaC2.MYB28ZY50 displayed consistent expression patterns in various tissues, with the former generally exhibiting higher abundance than the latter in the same tissue (Figure 3A). Since a great difference of promoter sequences between parental lines was observed (Supplemental Figure S5), we speculated that the promoter activity of them may be different. To quantitatively determine whether there are some differences in the promoter activity of BnaC2.MYB28G120 and BnaC2.MYB28ZY50, the pBnaC2.MYB28G120−LUC and pBnaC2.MYB28ZY50−LUC constructs (designated as G120−P and ZY50−P, respectively) with dual reporter genes were transformed into Arabidopsis protoplasts, respectively (Figure 3B). As shown in Figure 3C, the relative LUC activity of G120−P was about two-folds that of ZY50−P, further suggesting that BnaC2.MYB28G120 had a higher promoter activity than BnaC2.MYB28ZY50. To further analyze the expression pattern of BnaC2.MYB28, two constructs harboring the GUS gene, respectively, driven by BnaC2.MYB28G120 and BnaC2.MYB28ZY50 promoters were transformed into Arabidopsis wild-type (Col) plants. The results of GUS signals (Figure 3, D and E) were generally consistent with the RT–qPCR results. Collectively, BnaC2.MYB28 was mainly expressed in vegetative organs and exhibited a higher RNA accumulation in the high-SGC parent G120.
Figure 3.
Expression pattern, subcellular localization, and regulation analysis of SGC of BnaC2.MYB28. A, RT–qPCR analysis of BnaC2.MYB28 in NIL(ZY50) and NIL(G120). BnACTIN7 was used as a control. Error bars represent the sds from three independent RNA samples. B, Diagrams of the test constructs used in (C). C, Relative luciferase activity of promoters of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 in Arabidopsis protoplasts. Each dot on the column represents an independent replicate. The activities of LUC were normalized as the ratio of LUC/REN activity. Data are shown as mean ± sd (n = 9). Error bars represent the sds. GUS analysis of BnaC2.MYB28G120 (D) and BnaC2.MYB28ZY50 (E) in transgenic wild-type Arabidopsis. Bars = 2 mm. F, Subcellular localization of BnaC2.MYB28 protein in Arabidopsis leaf protoplasts. The nuclear protein OsGHD7-fused CFP was used as a nuclear marker. Bars = 10 μm. G, SGC in G120, transgene-negative and transgene-positive T1 lines (GZ−2, GZ−5, and GZ−6). Each dot on the column represents an individual plant. H, SGC in ZY50 and BnaC2.MYB28ZY50 overexpression lines (ZOE−22 and ZOE−25) in the T1 generation. Each dot on the column represents an individual plant. (I) SGC of the WT, transgene-negative and transgene-positive T0 plants of the BnaC2.MYB28G120-RNAi and BnaC2.MYB28G120-OE transformants. Each dot on the column represents an individual plant. In (G), (H), and (I), data are shown as mean ± sd (n ≥ 6); error bars represent the sds; two-tailed Student’s t tests were used to generate the P-values. ***P < 0.001.
To investigate the subcellular localization of BnaC2.MYB28, a green fluorescent protein (GFP) was fused to the N-terminus of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 driven by the CaMV35S promoter, respectively. These two constructs were then co-transformed into Arabidopsis protoplasts with a nucleus marker (OsGHD7−CFP), respectively. The fluorescence signals of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were overlapped with the signals of the nucleus marker (Figure 3F). Additionally, to further confirm the subcellular localization results, the coding sequences of BnaC2.MYB28ZY50 and BnaC2.MYB28G120 were fused with mVenus, and the fusion constructs were respectively transformed into Nicotiana benthamiana. The fluorescence signals of mVenus−BnaC2.MYB28ZY50 and mVenus−BnaC2.MYB28G120 were co-localized with the nucleus-localized OsGHD7−Turquoise2 fusion protein (Supplemental Figure S6). Taken together, both BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were normally localized to the nucleus.
BnaC2.MYB28 positively regulated GSL accumulation in seeds
Since BnaC2.MYB28ZY50 had similar expression pattern and identical protein localization to BnaC2.MYB28G120, we next attempted to explore the reasons for the functional divergence between BnaC2.MYB28ZY50 and BnaC2.MYB28G120. To this end, a construct containing the 1,525-bp upstream regulatory sequence, the 1,564-bp coding sequence, and the 767-bp downstream 3′-UTR regions of BnaC2.MYB28ZY50 was introduced into G120. As a result, eight T0 transgenic-positive plants were obtained, three of which were self-pollinated to generate T1 progeny. For these T1 families, the transgenic-positive plants exhibited significantly higher SGC than the transgenic-negative plants (Figure 3G; Supplemental Table S3), indicating that BnaC2.MYB28ZY50 is also a functional gene affecting SGC in rapeseed.
To further unravel the function of BnaC2.MYB28, two overexpression constructs of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were introduced into G120 and ZY50, respectively, and an RNA interference (RNAi) construct of BnaC2.MYB28G120 was transformed into G120. Phenotypic observation of the overexpression lines showed that compared with the transgene-negative plants, the T1 transgene-positive plants had significantly higher SGC, especially ZOE−25, which showed an extremely high value of SGC (161.3 ± 15.8 μmol g−1) (Figure 3H; Supplemental Table S4). As for BnaC2.MYB28G120, the T0 transgene-positive plants of G120−OE exhibited significantly higher SGC than the transgene-negative plants, whereas a significant decrease in SGC in the T0 transgene-positive plants of G120−RNAi was observed relative to the transgene-negative plants (Figure 3I; Supplemental Table S5). Taken together, these results suggested that both BnaC2.MYB28G120 and BnaC2.MYB28ZY50 are functional alleles with a positive effect on SGC regulation in rapeseed.
BnaC2.MYB28 formed homodimers and specifically interacted with BnaMYC3
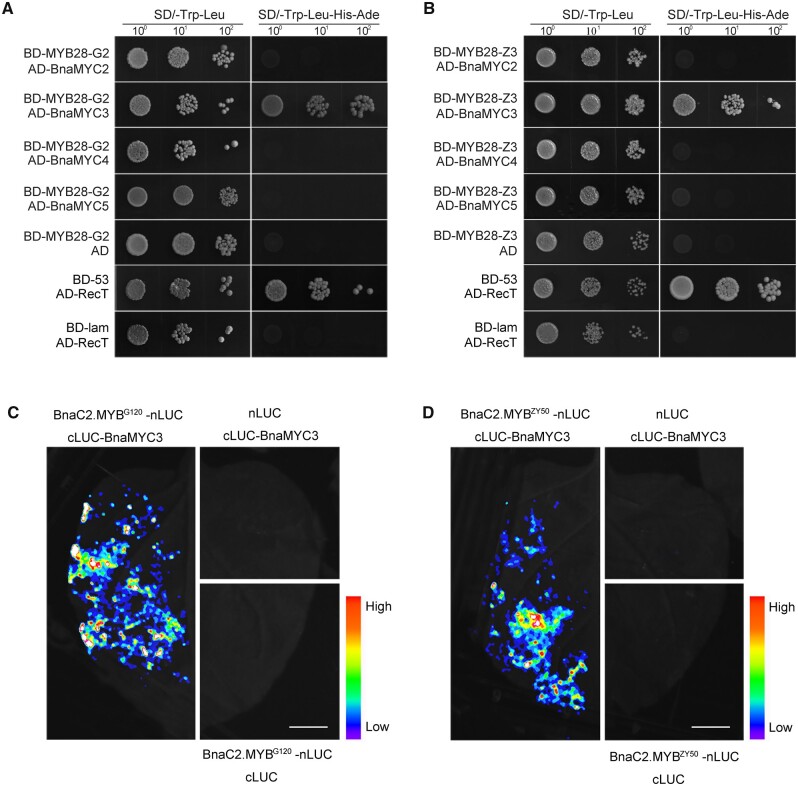
As a TF, BnaC2.MYB28 is likely to perform its function in the form of a protein complex. To explore whether there are differences in the interaction between BnaC2.MYB28 and other proteins in parent lines, we first tested the transcriptional self-activation of BnaC2.MYB28 in yeast. The results showed that both the full-length BnaC2.MYB28ZY50 and BnaC2.MYB28G120 exhibited self-activation in yeast (Supplemental Figure S7, A–C). To determine the specific transcriptional activation domain of BnaC2.MYB28, a series of fragments containing partial cDNAs of BnaC2.MYB28 were cloned into the pGBKT7 vector to construct fusion proteins for further testing the transcriptional self-activation, that is, BD-MYB28-G1/Z1, BD-MYB28-G2/Z2, BD-MYB28-G3/Z3, BD-MYB28-G4/Z4, and BD-MYB28-G5/Z5 (G and Z represent G120 and ZY50, respectively; Supplemental Figure S7A). The results showed that all the truncated proteins had transcriptional self-activation activity except for BD-MYB28-G2, BD-MYB28-G3, and BD-MYB28-Z3 (Supplemental Figure S7, B and C). According to the results of a yeast-two-hybrid assay, both BnaC2.MYB28G120 and BnaC2.MYB28ZY50 could form homodimers (Supplemental Figure S7D), which was further confirmed by the split-luciferase complementation (SLC) experiments in N. benthamiana leaves (Supplemental Figure S7E).
Since the interaction between MYB28 and MYCs plays a key role in the regulation of GSL biosynthesis in Arabidopsis, we wondered whether there is a difference in the interaction between BnaC2.MYB28 and BnaMYCs from the parental lines in rapeseed. The results of the yeast two-hybrid point-to-point assays showed that both BnaC2.MYB28G120 and BnaC2.MYB28ZY50 could interact with BnaMYC3, but not with BnaMYC2, BnaMYC4, and BnaMYC5 (Figure 4, A and B). SLC assays further confirmed the above results (Figure 4, C and D).
Figure 4.
Yeast-two-hybrid and SLC analyses of BnaC2.MYB28 interaction with BnaMYC3. Interaction of BnaC2.MYB28G120 (A) and BnaC2.MYB28ZY50 (B) with BnaMYC3 in yeast-two-hybrid assays. Interaction of BnaC2.MYB28G120 (C) and BnaC2.MYB28ZY50 (D) with BnaMYC3 in SLC assays. BD-53/AD-RecT was the positive control, and BD-Lam/AD-RecT was the negative control. AD, pGADT7/activation domain; BD, pGBKT7/binding domain; Ade, adenine; His, histidine; Lam, human lamin C; Leu, leucine; RecT, recombination of the SV40 large T antigen; SD, synthetic defined medium; Trp, tryptophan. In (C) and (D), bars = 1 cm.
BnaC2.MYB28 positively regulated GSL biosynthetic genes
To investigate the mechanism through which BnaC2.MYB28 regulates GSL accumulation, we analyzed the transcriptome profiles of mature leaves from G120, G120−OE (35S::BnaC2.MYB28G120), ZY50, and ZY50−OE (35S::BnaC2.MYB28ZY50) plants. As a result, 4,605 and 9,718 differentially expressed genes (DEGs) were identified in G120−OE and ZY50−OE relative to G120 and ZY50, respectively (Supplemental Figure S8, A and C). In the gene ontology (GO) category “biological process,” most of the DEGs were enriched in the term “glucosinolate biosynthetic process” (Supplemental Figure S8, B and D), suggesting that an increase in the expression of BnaC2.MYB28 may induce a number of GSL biosynthetic genes. Most aliphatic GSL biosynthesis genes exhibited higher expression levels in overexpression plants than in the parents (Figure 5). Four of these genes were randomly selected for RT–qPCR, and their expression levels in overexpression lines were significantly higher than those in parental lines (Supplemental Figure S9). Notably, MAM1 and MAM3, which have been confirmed to be positively regulated by MYB28 in Arabidopsis (Gigolashvili et al., 2007), showed no increase in expression in the overexpression plants (Figure 5). Interestingly, the expression of several genes involved in indole GSL biosynthesis was also substantially higher in ZY50-OE than in ZY50, suggesting that BnaC2.MYB28ZY50 may also regulate the expression of indole GSL biosynthesis genes. Taken together, BnaC2.MYB28 is a positive regulator of most GSL biosynthetic genes in rapeseed, and BnaC2.MYB28ZY50 seems to have a certain regulatory effect on indole GSL synthesis genes.
Figure 5.
Transcription levels of DEGs involved in GSL biosynthesis. The genes with black font showed no significant difference in expression level between parents and their overexpression lines (t test; significant, P < 0.05). Transcription levels are shown for two parental lines (G120 and ZY50) and corresponding overexpression lines of BnaC2.MYB28 (G120−OE and ZY50−OE). Red and green represent high and low gene expression, respectively. I3M, indolyl-3-methyl.
BnaC2.MYB28 directly activated the expression of GSL biosynthetic genes
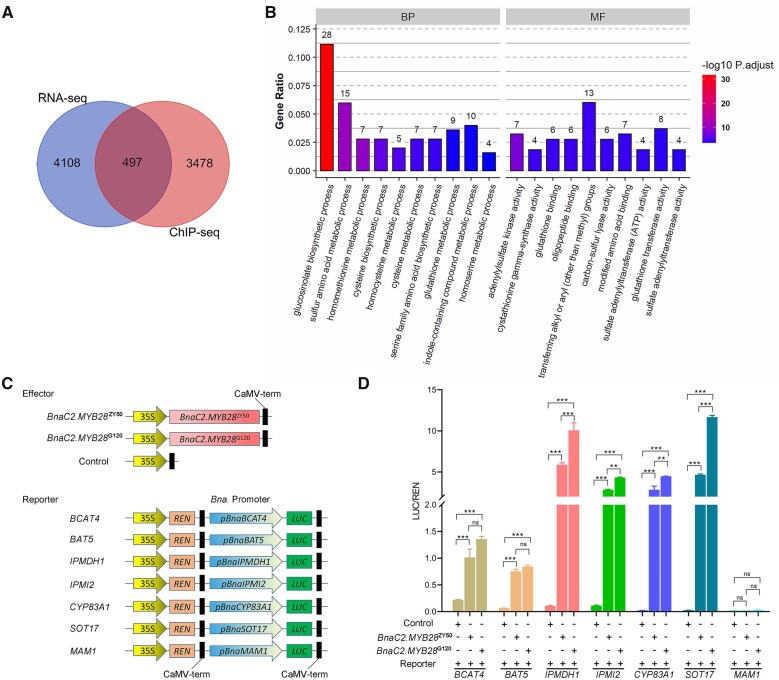
To explore the targets of BnaC2.MYB28, we performed ChIP-seq assays using BnaC2.MYB28G120-3×Flag transgenic rapeseed leaves. As a result, 3,975 genes were detected to be targeted by BnaC2.MYB28G120 (Figure 6A). A total of 4,605 DEGs were identified via RNA-seq between the overexpression lines of BnaC2.MYB28G120 and the parent G120 (Supplemental Figure S8A). We then performed an integrated analysis between the ChIP-seq and RNA-seq results and identified 497 overlapping genes (Figure 6A). GO enrichment analysis suggested that most of the overlapping genes were significantly enriched in the term of “glucosinolate biosynthetic process,” including AOPs, BAT5, BCAT4, CYP83A1, FMOGS-OXs, IPMDH1, IPMI2, SOTs, SUR1, and UTG74B1 (Figure 6B; Supplemental Table S6).
Figure 6.
Overlapping genes between ChIP-seq and RNA-seq analysis of G120 and BnaC2.MYB28G120-overexpression lines. A, Venn diagram of the DEGs. B, GO enrichment analysis of the 497 overlapping genes. The numbers above the bars indicate the number of genes for the corresponding GO term. C, Diagram of the reporter and effector constructs used in the transient dual-luciferase assays. Bna., B. napus. D, BnaC2.MYB28 activates the expression of BCAT4, BAT5, IPMDH1, IPMI2, CYP83A1, and SOT17 as assessed by the ratio of LUC/REN activity. Error bars represent the sds of three independent experiments. Two-tailed Student’s t tests were used to generate the P-values. **P < 0.01, ***P < 0.001; ns, no significance.
Considering that BnaC2.MYB28 is a transcription activator (Supplemental Figure S7, B and C), a transient dual-luciferase assay in Arabidopsis protoplasts was carried out to further determine the effect of BnaC2.MYB28 on the overlapping genes involved in the GSL biosynthetic pathway. BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were, respectively, inserted into the pGreenII 62-SK vector as effectors, and the empty pGreenII 62-SK vector was included as a control (Figure 6C). The promoters of several GSL biosynthetic genes (BCAT4, BAT5, IPMDH1, IPMI2, CYP83A1, SOT17, and MAM1) were fused with the LUC reporter, respectively. CaMV 35S-driving Renilla (REN) reporter in the same vector was used as the internal control to normalize the expression of each reporter (Figure 6C). The results showed that compared with the empty control, overexpression of BnaC2.MYB28 could significantly increase the value of the LUC/REN ratio when combined with the promoters of BCAT4, BAT5, IPMDH1, IPMI2, CYP83A1, or SOT17 (Figure 6D), indicating that the overlapping genes involved in GSL biosynthesis were activated by BnaC2.MYB28. Moreover, the LUC/REN ratios of several GSL biosynthesis genes (IPMDH1, IPMI2, CYP83A1, and SOT17) co-expressed with the effector of BnaC2.MYB28G120 were significantly higher than those of genes that co-expressed with the effector of BnaC2.MYB28ZY50 (Figure 6D). It is worth noting that overexpression of BnaC2.MYB28 had no effect on the luciferase activity driven by the promoter of MAM1 (Figure 6D). Consistently, MAM1 was not identified in the overlapping genes of ChIP-seq and RNA-seq. However, it has been confirmed that MAM1 can be activated by MYB28 in Arabidopsis (Gigolashvili et al., 2007). Collectively, these results suggested that BnaC2.MYB28 could directly activate the expression of GSL biosynthesis genes, and there is a difference in the activation ability between BnaC2.MYB28ZY50 and BnaC2.MYB28G120 on some of these genes.
BnaC2.MYB28 differentially regulates several key genes involved in aliphatic GSL biosynthesis between NILs
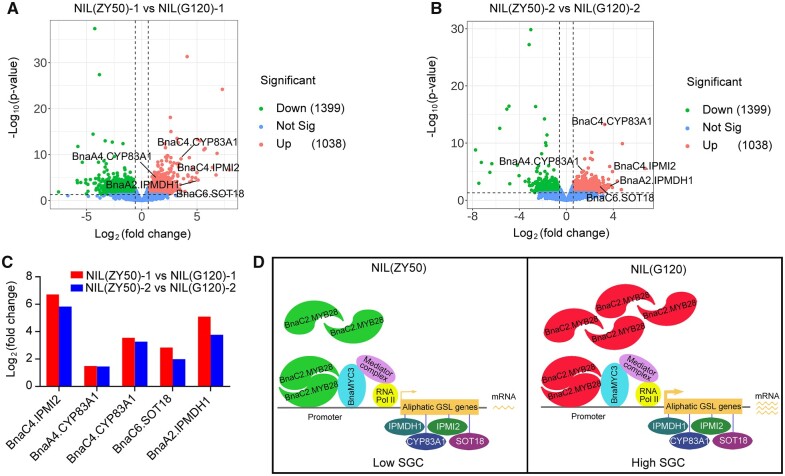
To further explore the regulation of different alleles of BnaC2.MYB28 on SGC in the same genetic background, we performed RNA-seq analysis on mature leaves of NIL(G120) and NIL(ZY50) lines. In the leaves of NIL(G120)-1 (60 days after sowing), 1,038 genes were upregulated, and 1,399 genes were downregulated (Figure 7A). Compared with the leaves of NIL(ZY50)-2 (90 days after sowing), 712 genes were upregulated, and 581 genes were downregulated in NIL(G120)-2 (Figure 7B). Notably, both of the upregulated genes in NIL(G120)-1 and NIL(G120)-2 contained five GSL biosynthesis genes, that is, BnaA4.CYP83A1, BnaC4.CYP83A1, BnaC6.SOT18, BnaA2.IPMDH1, and BnaC4.IPMI2 (Figure 7, A–C). Among these genes, BnaA2.IPMDH1 and BnaC4.IPMI2 participate in the side chain elongation of amino acids, while BnaA4.CYP83A1, BnaC4.CYP83A1, and BnaC6.SOT18 participate in the development of the core structure. All of them are involved in aliphatic GSL biosynthesis. These results are consistent with the fact that the difference in total GSL content in mature leaves between NIL(ZY50) and NIL(G120) is mainly due to the difference in aliphatic GSL content (Figure 1B). Since all of the above DEGs involved in GSL biosynthesis were regulated by BnaC2.MYB28, we considered that the difference of SGC between NIL(ZY50) and NIL(G120) was caused by the differential regulation of several aliphatic GSL biosynthesis genes by the corresponding alleles of BnaC2.MYB28.
Figure 7.
Alleles of BnaC2.MYB28 in NILs differentially regulate GSL biosynthesis genes. A, Volcano plot showing DEGs from leaves of plant 60 days after sowing between NIL(ZY50) and NIL(G120). B, Volcano plot showing DEGs from leaves of plant 90 days after sowing between NIL(ZY50) and NIL(G120). C. Log2(fold change) (n = 3) of DEGs involved in GSL biosynthesis. D, Model for the explaining of the difference of SGC between NIL(ZY50) and NIL(G120). Sig, significant.
Selection and haplotype analyses of BnaC2.MYB28 in natural population
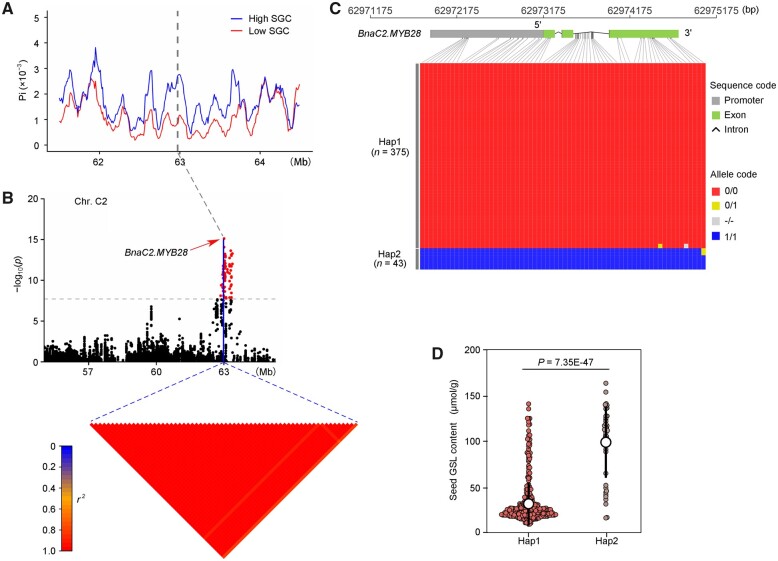
A sequence analysis of BnaC2.MYB28 was performed based on a natural population of 505 rapeseed accessions, including semi-winter (SW) and spring ecotypes. The population structure was analyzed using ADMIXTURE (Alexander et al., 2009) with ancestry kinship (K) set from 2 to 7. Based on the phenotypic distribution of accessions within subpopulation, linkage disequilibrium (LD) decay, and Fst between subpopulations, K value was set to 3 according to a previous study (Tang et al., 2021). With the K value set to 3, and the SW ecotypes could be divided into two subpopulations (SW1 and SW2). Interestingly, SW1 primarily consisted of the accessions with a low erucic acid and GSL content, while SW2 majorly included the ones with a high erucic acid and GSL content (Tang et al., 2021). Genetic diversity (π) was calculated in each subpopulation. The results showed that the genetic diversity (2.69 × 10−3) around the region of BnaC2.MYB28 was higher in the subpopulation with high SGC than that (0.87 × 10−3) with low SGC (Figure 8A), indicating the presence of an artificial selection of the region harboring BnaC2.MYB28 in low SGC accessions.
Figure 8.
Selection analysis and haplotype analysis of BnaC2.MYB28 in natural population. A, Nucleotide diversity (π) around BnaC2.MYB28. Vertical dashed line indicates the physical position of BnaC2.MYB28 on ZS11 genome. Curves in the panel are π of sliding regions (100-kb window sliding in 10 kb steps) colored by the two accession groups. B, GWAS signal of MLM around BnaC2.MYB28. The dots above horizontal dash line indicate the SNPs significantly associated with SGC (MLM, P < 7.96 × 10−7). The vertical line indicates the physical position of BnaC2.MYB28 on ZS11 genome. Heat maps spanning the SNP markers in LD with the most strongly associated SNPs in BnaC2.MYB28 gene regions. C, Two major haplotypes were identified in the BnaC2.MYB28 gene regions. D, SGC of the two haplotypes. Data are shown as mean ± sd (n ≥ 43). Error bars represent the sds. The empty circles and vertical lines indicate means and error bars, respectively. A two-tailed Student’s t test was used to generate the P-value. Chr, chromosome; bp, base pair.
Based on the natural population, we used a reference genome of “ZS11” to perform regional GWAS for SGC. Notably, numerous SNPs in the region of BnaC2.MYB28 were observed to be significantly (P < 7.96 × 10−7) associated with SGC using the mixed linear model (MLM) (Figure 8B). Since there were a number of variations in the promoter and genomic region of BnaC2.MYB28, we performed a haplotype analysis of BnaC2.MYB28 in 505 rapeseed accessions. As a result, these accessions could be mainly divided into two haplotypes designated as Hap1 and Hap2 (Figure 8C). The genotypes of BnaC2.MYB28 from the parental lines ZY50 and G120 were highly similar to Hap1 and Hap2, respectively. The average SGC of Hap1 (38.3 ± 6.8 μmol g−1) was significantly lower (P = 7.35E-47) than that of Hap2 (96.2 ± 16.2 μmol g−1) (Figure 8D). Interestingly, the number of accessions carrying Hap1 (375) was far larger than that of accessions carrying Hap2 (43), which is consistent with the history of reducing SGC in the development of modern rapeseed cultivars (Tan et al., 2022). Taken together, these results indicated that natural variations of BnaC2.MYB28 are significantly associated with SGC in natural population, and BnaC2.MYB28 has undergone artificial selection in the history of rapeseed breeding.
Surprisingly, compared with the relatively minor difference (around 12 μmol g−1) between the parental lines, the difference in average SGC (around 58 μmol g−1) between the Hap1 and Hap2 of BnaC2.MYB28 was much greater (Figure 8D). Since BnaC2.MYB28 (62.97 Mb) is physically close to BnaC02.GTR2 (63.43 Mb), the target gene underlying qGSL.C02.1 (Tan et al., 2022), we speculated that BnaC2.MYB28 may have undergone selective sweep together with BnaC02.GTR2 in the breeding process of low-SGC rapeseed. LD analysis showed that a strong LD was existed between BnaC2.MYB28 and BnaC2.GTR2 on chromosome C02 (Supplemental Figure S10A). Haplotype analysis of BnaC2.GTR2 in the 505 rapeseed accessions mentioned above showed that the accessions could be mainly divided into two haplotypes designated as Types 1 and 2 (Supplemental Figure S10B). The SGC of Type 1 (95.4 ± 21.8 μmol g−1) was significantly higher (P = 1.88E-74) than that of Type 2 (39.4 ± 11.7 μmol g−1) (Supplemental Figure S10C). Notably, most of the accessions (367/375) carrying Hap1 (Low SGC haplotype) overlapped with those carrying Type 2, and nearly all the accessions (41/43) carrying Hap2 (High SGC haplotype) included by those carrying Type 1 (Supplemental Figure S10D). These results indicated that the great phenotypic difference between Hap1 and Hap2 may be attributed to not only the genetic effect of BnaC2.MYB28, but also the contribution of BnaC2.GTR2.
Discussion
As an important trait affecting the seed quality, SGC has been widely studied in recent years. Although a number of QTLs for SGC have been reported in rapeseed (Wurschum et al., 2012; Zou et al., 2016; Liu et al., 2019a, 2019b; Zhou et al., 2021), few QTLs have been cloned and functionally verified. Here, we experimentally confirmed BnaC2.MYB28 as the target gene underlying qGSL-C2, a major QTL controlling SGC. BnaC2.MYB28 is homologous to MYB28, which encodes an R2R3-MYB TF acting as a positive regulator of aliphatic GSL biosynthesis in Arabidopsis (Gigolashvili et al., 2007). The BnaC2.MYB28 overexpression plants showed a significant increase in SGC (Figure 3, H and I), whereas an obvious decrease in SGC was observed in its null mutants (Figure 2D), suggesting that BnaC2.MYB28 can positively regulate SGC in rapeseed with a similar function to AtMYB28. In fact, MYB28 homologs in other Brassicaceae species like Brassica rapa (BrMYB28), Brassica oleracea (BoaMYB28), and Brassica juncea (BjuMYB28) have also been demonstrated to have similar functions to AtMYB28 (Augustine et al., 2013; Seo et al., 2016; Yin et al., 2017; Yang et al., 2021), indicating that MYB28 genes may play a conserved role in regulating SGC.
In Arabidopsis, MYB28 specifically regulates the expression of aliphatic GSL biosynthetic genes by interacting with MYCs (MYC2, MYC3, and MYC4) (Gigolashvili et al., 2007; Schweizer et al., 2013). However, in this study, in addition to a number of aliphatic GSL biosynthetic genes, we also observed increases in the expression of several indole GSL biosynthetic genes in BnaC2.MYB28 overexpression plants (Figure 5), suggesting that BnaC2.MYB28 may also regulate the biosynthesis of indole GSL. Moreover, protein interaction analysis indicated that BnaC2.MYB28 specifically interacts with BnaMYC3 but not with BnaMYC2, BnaMYC4, and BnaMYC5 (Figure 4). Additionally, the transient dual-luciferase assays suggested that BnaC2.MYB28 could not activate the expression of MAM1. Taken together, it can be speculated that BnaC2.MYB28 may have some functional divergence from its putative ortholog AtMYB28 in Arabidopsis to some extent.
In the present study, sequence comparison indicated extensive variations in both promoter and CDS regions between BnaC2.MYB28G120 and BnaC2.MYB28ZY50. Expression pattern and promoter activity analysis showed that the promoter activity of BnaC2.MYB28G120 was stronger than that of BnaC2.MYB28ZY50 (Figure 3). Transcriptional activation experiments revealed that BnaC2.MYB28G120 has a stronger activation activity on GSL biosynthesis genes including CYP83A1, IPMDH1, and IPMI2 than BnaC2.MYB28ZY50 (Figure 6D). Thus, we propose a possible model for the difference in SGC between NIL(ZY50) and NIL(G120). Both BnaC2.MYB28ZY50 and BnaC2.MYB28G120 can normally form homodimers and interact with BnaMYC3 to regulate the expression of aliphatic GSL biosynthesis genes (Figure 7D). However, the expression level of BnaC2.MYB28 in NIL(G120) is higher than that in NIL(ZY50), and the transcriptional activation ability of BnaC2.MYB28 in NIL(G120) is stronger than that in NIL(ZY50), thus the expression level of aliphatic GSL biosynthesis genes (IPMDH1, IPMI2, CYP83A1, and SOT18) positively regulated by BnaC2.MYB28 in NIL(G120) is higher than those in NIL(ZY50) (Figure 7D). The higher expression of these aliphatic GSL biosynthesis genes in NIL(G120) contributed more accumulation of GSL in leaves, resulting in higher SGC in the seeds of NIL(G120). According to the model, the sequence variation in both promoter and CDS regions contributes to the functional divergence between BnaC2.MYB28G120 and BnaC2.MYB28ZY50. Since the DEGs involved in GSL biosynthesis between NIL(ZY50) and NIL(G120) were predominantly regulated by BnaC2.MYB28, we speculated that both of the differences in expression level and activation ability between the BnaC2.MYB28 alleles are responsible for the difference in SGC between NIL(ZY50) and NIL(G120). However, which is the main factor contributes to the difference in SGC between NIL(ZY50) and NIL(G120) needs to be further verified.
As SGC is an important parameter to evaluate seed quality, reduction of SGC has been an important breeding goal in rapeseed. Generally, it is much more difficult to reduce SGC from a medium level to a low level (<30 μmol g−1) than from a high level (>100 μmol g−1) to a medium level (30–100 μmol g−1). In this study, we successfully reduced the SGC of the parent G120 from a medium level (33.0 ± 2.4 μmol g−1) to a low level (14.1 ± 2.4 μmol g−1) by disrupting BnaC2.MYB28, suggesting that artificial mutation of BnaC2.MYB28 may be a promising approach for further reduction of SGC in the future. Besides, since extremely high SGC (>150 μmol g−1) can be induced by overexpression of BnaC2.MYB28, novel rapeseed accessions with low GSL in seeds but high GSL in vegetable tissues can be generated by artificial design of optimal genotype combinations of BnaC2.MYB28 and Bna.GTR2s.
Materials and methods
Plant materials, growth conditions, and phenotypic measurement
Two SW rapeseed (B. napus) inbred lines G120 and 9172 (hereafter designated as ZY50) together with their NILs were used in the present study as described previously (Liu et al., 2019a, 2019b). All rapeseed plants were grown in the experimental base of Huazhong Agricultural University, Wuhan, China in autumn. The planting density was 20 cm between plants and 25 cm between rows. The field management followed standard agricultural practice. For the measurement of SGC in each plant, open-pollinated mature seeds from the whole plant were harvested for near-infrared reflectance spectroscopy. The SGC of all the parental lines, NILs and transgenic lines was estimated as the average of three measurements. Arabidopsis (A. thaliana) plants, N. benthamiana plants, and a part of rapeseed plants were grown in a plant growth chamber at 22°C–24°C and 70% humidity under a 16-h/8-h light/dark photoperiod. The same natural population that was once used for analysis with BnaC02.GTR2 was explored for the haplotype effect of BnaC2.MYB28 in this study (Tang et al., 2021; Tan et al., 2022), with the SGC phenotype collected previously (Tan et al., 2022).
Vector construction and genetic transformation
To validate the function of BnaC2.MYB28, a 5,032-bp fragment containing the 2,406-bp sequence upstream of the translation start site, the 1,608-bp coding sequence, and the 1,018-bp downstream 3′-UTR region was amplified from G120. Then, the fragment confirmed by sequencing was cloned into the EcoRI/BamHI site of the binary vector pCAMBIA1301 to construct PC-BnaC2.MYB28G120. To knockout BnaC2.MYB28, a CRISPR/Cas9 vector was constructed. Two sgRNAs were designed using the CRISPR-P database (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). Then, the CRISPR/Cas9 binary vector pKSE401 containing the two sgRNAs was constructed according to previous descriptions (Xing et al., 2014).
To investigate the effects of different expression levels of BnaC2.MYB28G120 on SGC, an overexpression and an RNAi vector were constructed. To construct the RNAi vector, a 217-bp fragment amplified from the third exon of BnaC2.MYB28G120 was cloned into the AscI/SwaI site and BamHI/SmaI site of the pFGC5941 vector with sense and antisense orientations, respectively. To construct the overexpression vector, a double 35S promoter was cloned into the HindI/PstI site and a terminator connected with 3×Flag fragment was cloned into the EcoRI site of the binary vector pCAMBIA2300. Subsequently, a 1,605-bp full-length genomic DNA fragment without the stop codon of BnaC2.MYB28G120 was cloned into the KpnI/BamHI site of the modified pCAMBIA2300 vector to construct OE-BnaC2.MYB28G120.
To determine whether BnaC2.MYB28ZY50 is a functionally defective allele, a 3,856-bp fragment containing the 1,525-bp sequence upstream of the translation start site, the 1,564-bp coding sequence and the 767-bp downstream 3′-UTR region amplified from ZY50 was cloned into the EcoRI/BamHI site of the binary vector pCAMBIA1301. Additionally, a 1,561-bp genomic sequence without the stop codon of BnaC2.MYB28ZY50 was cloned into the KpnI/BamHI site of the modified pCAMBIA2300 vector to construct an overexpression construct OE-BnaC2.MYB28ZY50. All the recombinant plasmids used for genetic transformation were transformed into Agrobacterium tumefaciens strain GV3101. The PC-BnaC2.MYB28ZY50, CRISPR/Cas9, RNAi, and OE-BnaC2.MYB28G120 vectors were transformed into G120, whereas the PC-BnaC2.MYB28G120 and OE-BnaC2.MYB28ZY50 vectors were introduced into ZY50 via Agrobacterium-mediated genetic transformation (Cardoza and Stewart, 2003).
Expression analysis
Total RNA was extracted from various plant tissues using the Eastep Super RNA extraction kit (LS1040, Promega, Madison, WI, USA; https://www.promega.com/). First-strand cDNA was synthesized using the GoScript Reverse Transcription Mix (A2790; Promega). RT–qPCR reactions were performed on a CFX96 Touch Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA; http://www.bio-rad.com/) with the following program: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 25 s. Each sample was represented by three biological and three technical repeats. BnACTIN7 was used to normalize the expression levels of the target genes, which were calculated with the 2−ΔΔCt method. All the primer sequences for RT–qPCR are listed in Supplemental Table S7.
Histochemical GUS staining
Two BnaC2.MYB28 promoter sequences from G120 and ZY50, which respectively contained the 1,881-bp and 1,525-bp sequence upstream of the translation start site, were amplified to generate the pBnaC2.MYB28-GUS constructs. Then, the two promoter sequences of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were cloned into the PstI/BamHI sites of the binary vector pCAMBIA2300-GUS derived from a modified pCAMBIA2300 according to the method described previously (Xu et al., 2017). Subsequently, the two constructs were transformed into wild-type Arabidopsis (Col-0) by floral dipping (Clough and Bent, 1998). Fifteen and Eighteen independent T2 transgenic lines of pBnaC2.MYB28-GUS, respectively, from G120 and ZY50 were used for GUS staining according to previous descriptions (Li et al., 2015).
Subcellular localization
The full-length CDS of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 without termination codon (TGA) was amplified with the primer pair of G120-pM999F/R and ZY50-pM999F/R (Supplemental Table S7). The amplified CDS fragments were cloned into the SmaI/BamHI sites of the pM999-GFP vector. The rice (Oryza sativa) nuclear protein GHD7 fused with CFP was used as a nuclear marker (Xue et al., 2008). The BnaC2.MYB28G120/ZY50−GFP fusion construct and nuclear marker (GHD7−CFP) were co-transformed into Arabidopsis protoplasts by polyethylene glycol/calcium-mediated transformation (Yoo et al., 2007). Fluorescence signals in the transformed protoplasts were detected with a confocal laser scanning microscope and visualized using a Leica Microsystem LAS AF. Excitation wavelengths and emission filters were 405 nm/bandpass 420–485 nm for CFP, 488 nm/bandpass 495–542 nm for GFP, and 594 nm/bandpass 600–650 nm for red fluorescent protein (RFP). Additionally, the CDS fragments of BnaC2.MYB28 amplified from G120 and ZY50 were cloned into the pDOE-20 vector to be fused with the sequence encoding mVenus protein. The coding sequence of rice nuclear protein GHD7 was also cloned into the pDOE-20 vector to be fused with the sequence encoding mTurquoise2. The fusion construct with dual fluorescent reporter genes was infiltrated into N. benthamiana leaves as previously described (Yao et al., 2015). Fluorescence signals in the N. benthamiana leaves were detected under a confocal laser scanning system (SP2; Leica Microsystem, Wetzlar, Germany). Argon laser line excitation wavelength and emission bandpass filter wavelengths for mTurquoise2 and mVenus were 458 nm and 480–520 nm, and 488 nm and 500–550 nm, respectively.
Protoplast transformation and transient expression assays
To quantitatively detect the activity of the promoter, the promoter sequences of BnaC2.MYB28 amplified from G120 and ZY50 were cloned into the KpnI/BamHI sites of the pGreenII0800−LUC vector to create the pBnaC2.MYB28G120−LUC and pBnaC2.MYB28ZY50−LUC constructs, which were then transformed into Arabidopsis protoplasts for transient expression assays. In these vectors, an REN-LUC gene under the control of the CaMV35S promoter was used as an internal control (Hellens et al., 2005).
To detect the impact of BnaC2.MYB28 on the expression of the GSL biosynthetic genes, the promoters (−2,000 bp) of several GSL biosynthetic genes amplified from G120 were cloned into pGreenII0800-LUC vector to construct the reporters. The coding sequences of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were, respectively, linked to the pGreen II 62-SK vector to construct the effectors. Then, each of the reporters was co-transformed into Arabidopsis protoplasts along with an effector, respectively. The activities of firefly LUC and REN LUC were assayed using the dual luciferase assay reagents (Catalog No. E1910; Promega, USA). Each experiment was performed in three independent transfections and in triplicate. The activity was expressed as the ratio of LUC to REN activity.
RNA-seq analysis
Total RNA was extracted from the mature leaves of parental lines, NILs and BnaC2.MYB28 overexpression individuals for transcriptome analysis. Three biological replicates were carried out for each sample. Library construction and sequencing of the RNA-seq were performed using an Illumina Hiseq2000 (San Diego, CA, USA) by Nanjing Personal Gene Technology Co., Ltd. The low-quality reads were removed according to Shi et al. (2019), and the clean raw reads were aligned to the B. napus reference genome (http://cbi.hzau.edu.cn/bnapus/index.php) (Sun et al., 2017). The bioinformatics analysis of RNA-seq data was performed according to previous descriptions (Li et al., 2019).
Yeast-two-hybrid assays
The cDNA of G120 and ZY50 was used as the template to amplify BnaC2.MYB28, BnaMYC2, BnaMYC3, BnaMYC4, and BnaMYC5. The full-length CDS of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 was cloned into the pGBKT7 vector as bait plasmids, and the pGADT7 vector was used as prey plasmids. The CDS of BnaMYCs from G120 and ZY50 was individually fused to the pGADT7 vector. To test the self-activation of the truncated protein, a series of incomplete fragments amplified from BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were, respectively, cloned into the pGBKT7 vector. The bait plasmids and prey plasmids were co-transformed into AH109 yeast cells. In all the experiments, pGBKT7-53 and pGADT7-T served as the positive controls, while pGBKT7-lam and pGADT7-T were used as the negative controls.
SLC
The coding sequence fragments of BnaC2.MYB28G120 and BnaC2.MYB28ZY50 were amplified and cloned into the JW771-nLUC and JW772-cLUC vector. The coding regions of BnaMYC2, BnaMYC3, BnaMYC4, and BnaMYC5 derived from G120 and ZY50 were amplified and cloned into JW771-nLUC vector. All the constructs were transformed into A. tumefaciens strain GV3101. The mixed bacterial suspensions were injected into N. benthamiana leaves with a needleless syringe according to a previous study (Chen et al., 2008). The LUC signal was detected under the CCD imaging apparatus (Lumazone Pylon2048B).
ChIP-Seq library construction and data analysis
To explore the target genes regulated by BnaC2.MYB28, ChIP assays were performed as previously described (Zhao et al., 2020). Young leaves of overexpression plants were cross-linked with 1% (v/v) formaldehyde and quenched using 0.2-M glycine. For each experiment, ∼1 g of leaf sample was ground into fine power in liquid nitrogen and resuspended in 1.5× volume of Buffer S (50-mM HEPES-KOH, 150-mM NaCl, 1-mM EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) sodium deoxycholate, 1% (w/v) sodium dodecyl sulphate (SDS), and protease inhibitor cocktail). Next, 6× volume of Buffer S without SDS was added for each sample, and the chromatin was fragmented by sonication in a bioruptor (Diagenode). The chromatin sample was then centrifuged at max speed for 10 min at 4°C, and the supernatant was used for immunoprecipitation (IP). ChIP was performed using FLAG antibody (Sigma, catalog no. F3165). After washing the bead-IP complexes and purifying the ChIP DNA, the library was prepared according to the manufacturer’s guidelines. Finally, the DNA fragments were sequenced by Illumina HiSeq X Ten system (paired-end 150-bp reads, Annoroad Gene Technology).
The ChIP-Seq sequences were mapped to the rapeseed “Darmor-bzh” genome using the BWA-MEM algorithm (Li and Durbin, 2009). The ChIP-Seq data (accession number PRJNA831305) were visualized using an integrative genomics viewer (IGV). The aligned reads were used for peak identification, and the binding peaks of BnaC2.MYB28 were obtained by model-based analysis of ChIP-Seq with default parameters (Zhang et al., 2008). The peak summits in the promoter region (2-kb upstream of transcription start site) of the target genes were subjected to a cutoff of peak fold-enrichment >2 and –log10(P) ≥ 2 (Li et al., 2017). The binding sites of BnaC2.MYB28 were identified using HOMER version 4.11 (http://homer.ucsd.edu/homer/).
Analysis of GSL in leaves
The GSL content of mature leaves of NILs were analyzed; three biological replications were included. At approximately 90 days after sowing, the third leaf from the top was harvested from each plant as a biological replication. The samples were placed into a 50-mL tube that contained 20 mL 90% (v/v) methanol, then heated in a water bath and centrifuged and the supernatant was transferred to a 12-mL column containing 1-mL DEAE Sephadex A-25. After the liquid was drained, 100 μL 2 mg mL−1 sulphatase solution was added to desulphurize the GSL. The 2-propenyl GSL (Sinigrin; Sigma-Aldrich, St. Louis, MO, USA) was used as an internal standard. GSLs were analyzed via HPLC in accordance with previously described methods (Kliebenstein et al., 2001a, 2001b).
GWAS for GSL content in seeds
The genome sequences of rapeseed “ZS11” were downloaded from BnPIR (http://cbi.hzau.edu.cn/bnapus/index.php). The reads from resequencing (Tang et al., 2021) were aligned to the reference genome using the BWA software (Li, 2013) with the command “mem -M -k 32 -t 4.” According to previous descriptions (Tang et al., 2021), a series of strict filtrations were conducted, and all high-quality SNPs (MAF > 0.05) in the 505 accessions were used for GWAS with a significant threshold set to 7.96 × 10−7. The GEMMA software package (Zhou and Stephens, 2012) was used for correlation analysis of BLUP trait data of SGC using a mixed linear model. When performing haplotype analysis, we only analyzed the accessions with identical haplotypes as the parental accessions (G120 and ZY50), while other accessions with different haplotypes were not further analyzed.
Statistical analysis
All statistical analyses were performed using Student’s t test (P, 0.05) of Microsoft Office Excel for ANOVA or other analyses mentioned in the text. For the number of experimental repetitions and specific samples, please refer to figure and table captions.
Accession numbers
Sequence data from this article can be found from https://www.genoscope.cns.fr/brassicanapus/ or the GenBank/EMBL data libraries under accession numbers: BnaC2.MYB28 (BnaC02G0527500ZS), BnaA2.MYB28 (BnaA02G0394700ZS), BnaC4.IPMI2 (BnaC04G0039400ZS), BnaA4.CYP83A1 (BnaA04G0074700ZS), BnaC4.CYP83A1 (BnaC04G0359300ZS), BnaC6.SOT18 (BnaC06G0404300ZS), BnaA2.IPMDH1 (BnaA02G0052200ZS), and GHD7 (EU286800).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of seed GSL content between NIL(ZY50) and NIL(G120).
Supplemental Figure S2. Comparison of coding sequences and amino acid sequences of candidate genes between parental lines.
Supplemental Figure S3. IGV displays the transcripts of BnaC2.MYB28ZY50 and BnaA2.MYB28ZY50.
Supplemental Figure S4. SGC of BnaC2.MYB28G120-sgRNA plants in the T1 generation.
Supplemental Figure S5. Comparison of the promoter sequences of BnaC2.MYB28 between the parental lines.
Supplemental Figure S6. Subcellular localization of BnaC2.MYB28 in tobacco (N. benthamiana) leaves.
Supplemental Figure S7. Functional regions of BnaC2.MYB28.
Supplemental Figure S8. RNA-seq analysis between parental lines and BnaC2.MYB28 overexpressing lines.
Supplemental Figure S9. Quantitative RT–qPCR confirmation of the DEGs involved in GSL biosynthesis obtained by RNA-seq.
Supplemental Figure S10. Co-selection analysis of BnaC2.MYB28 and BnaC2.GTR2 in natural population.
Supplemental Table S1. SGC in BnaC2.MYB28G120-transgenic T1 lines.
Supplemental Table S2. SGC in BnaC2.MYB28-CRISPR T2 lines.
Supplemental Table S3. SGC in BnaC2.MYB28ZY50-transgenic T1 lines.
Supplemental Table S4. SGC in two p35S::BnaC2.MYB28ZY50-transgenic T1 lines.
Supplemental Table S5. SGC in RNAi and overexpressing of BnaC2.MYB28G120-transgenic T0 lines.
Supplemental Table S6. Overlapping genes involved in GSL biosynthesis of ChIP-seq and RNA-seq.
Supplemental Table S7. Summary of primers used in this study.
Supplementary Material
Acknowledgments
We thank professor Liang Guo for kindly providing genotype and phenotype data collected from 505 natural germplasms. We would like to thank Dr. Qiang Xin for providing many valuable comments for the revision of the manuscript.
Funding
This research was supported by the Program for Modern Agricultural Industrial Technology System (CARS-12), the Wuhan Applied Foundational Frontier Project (2019020701011446), and the Open Fund of the National Key Laboratory of Crop Genetic Improvement (ZK201909).
Conflict of interest statement. The authors declare no competing interests.
Contributor Information
Xianming Zhou, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China; Sanya Nanfan Research Institute of Hainan University, Hainan Yazhou Bay Seed Laboratory, Sanya 572025, China; College of Tropical Crops Hainan University, Hainan University, Haikou 570288, China.
Haiyan Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China; Sanya Nanfan Research Institute of Hainan University, Hainan Yazhou Bay Seed Laboratory, Sanya 572025, China; College of Tropical Crops Hainan University, Hainan University, Haikou 570288, China.
Zhaoqi Xie, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Ying Liu, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Pengfei Wang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Lihong Dai, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Xiaohui Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Zhaoyang Wang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Zhuanrong Wang, Institute of Crops, Wuhan Academy of Agricultural Sciences, Wuhan 430065, China.
Lili Wan, Institute of Crops, Wuhan Academy of Agricultural Sciences, Wuhan 430065, China.
Guangsheng Yang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China.
Dengfeng Hong, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
D.H. and G.Y. conceived and designed the experiments. X.Zhou and H.Z. performed most of the experiments. P.W. performed the bioinformatics analysis. H.Z., Y.L., and Z.X. participated in genetic transformation. Zhao.W., X.Zhang, and L.D. participated in phenotypic and genotypic analyses of transgene plants. L.W. and Zhuan.W. provided experimental ideas and technical guidance. X.Zhou wrote the manuscript. D.H. revised the manuscript. All the authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Dengfeng Hong (dfhong@mail.hzau.edu.cn).
References
- Ahuja I, Rohloff J, Bones AM (2010) Defence mechanisms of Brassicaceae: implications for plant-insect interactions and potential for integrated pest management: A review. Agron Sustain Dev 30: 311–348 [Google Scholar]
- Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R, Majee M, Gershenzon J, Bisht NC (2013) Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J Exp Bot 64: 4907–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Feyereisen R (2001) The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P (2012) Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr Opin Plant Biol 15: 407–414 [DOI] [PubMed] [Google Scholar]
- Cardoza V, Stewart CN (2003) Increased Agrobacterium-mediated transformation and rooting efficiencies in canola (Brassica napus L.) from hypocotyl segment explants. Plant Cell Rep 21: 599–604 [DOI] [PubMed] [Google Scholar]
- Carré P, Pouzet A (2014) Rapeseed market, worldwide and in Europe. OCL 21: D102 [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Glawischnig E, Jørgensen K, Naur PBJ, Olsen C, Hansen CH, Rasmussen H, Pickett JA, Halkier BA (2003) CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J 33: 923–937 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Feng J, Long Y, Shi L, Shi J, Barker G, Meng J (2012) Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol 193: 96–108 [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T (2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7: 814–828 [DOI] [PubMed] [Google Scholar]
- Gajardo HA, Wittkop B, Soto-Cerda B, Higgins EE, Parkin IAP, Snowdon RJ, Federico ML, Iniguez-Luy FL (2015) Association mapping of seed quality traits in Brassica napus L. using GWAS and candidate QTL approaches. Mol Breed 35: 143 [Google Scholar]
- Gigolashvili T, Berger B, Flügge UI (2008) Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem Rev 8: 3–13 [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Muller C, Flugge UI (2007) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51: 247–261 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11: 89–100 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kliebenstein DJ, Halkier BA (2007) Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J 50: 902–910 [DOI] [PubMed] [Google Scholar]
- He Y, Fu Y, Hu D, Wei D, Qian W (2018) QTL mapping of seed glucosinolate content responsible for environment in Brassica napus. Front Plant Sci 9: 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck AS, Finley JW (2004) Cruciferous vegetables: cancer protective mechanisms of glucosinolate hydrolysis products and selenium. Integr Cancer Ther 3: 5–12 [DOI] [PubMed] [Google Scholar]
- Kittipol V, He Z, Wang L, Doheny-Adams T, Langer S, Bancroft I (2019) Genetic architecture of glucosinolate variation in Brassica napus. J Plant Physiol 240: 152988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001a) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001b) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate–dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA 100: 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127: 1077–1088 [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen B, Xu K, Wu J, Song W, Bancroft I, Harper AL, Trick M, Liu S, Gao G, et al. (2014) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res 21: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. figshare. Poster. 10.6084/m9.figshare.963153.v1 [DOI]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li J, Song J, Zhao B, Guo C, Wang B, Zhang Q, Wang J, King GJ, Liu K (2019) An auxin signaling gene BnaA3.IAA7 contributes to improved plant architecture and yield heterosis in rapeseed. New Phytol 222: 837–851 [DOI] [PubMed] [Google Scholar]
- Li J, Hansen BG, Ober JA, Kliebenstein DJ, Halkier BA (2008) Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol 148: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen L, Zhang L, Li X, Liu Y, Wu Z, Dong F, Wan L, Liu K, Hong D, et al. (2015) BnaC9.SMG7b functions as a positive regulator of the number of seeds per silique in Brassica napus by regulating the formation of functional female gametophytes. Plant Physiol 169: 2744–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, et al. (2017) A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170: 114–126 e115 [DOI] [PubMed] [Google Scholar]
- Liu S, Huang H, Yi X, Zhang Y, Yang Q, Zhang C, Fan C, Zhou Y (2019a) Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome‐wide association study. Plant Biotechnol J 18: 1472–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou X, Yan M, Wang P, Wang H, Xin Q, Yang L, Hong D, Yang G (2019b) Fine mapping and candidate gene analysis of a seed glucosinolate content QTL, qGSL-C2, in rapeseed (Brassica napus L.). Theor Appl Genet 133: 479–490 [DOI] [PubMed] [Google Scholar]
- Lu G, Harper AL, Trick M, Morgan C, Fraser F, O’Neill C, Bancroft I (2014) Associative transcriptomics study dissects the genetic architecture of seed glucosinolate content in Brassica napus. DNA Res 21: 613–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem 275: 33712–33717 [DOI] [PubMed] [Google Scholar]
- Potter MJ, Vanstone VA, Davies KA, Rathjen AJ (2000) Breeding to increase the concentration of 2-phenylethyl glucosinolate in the roots of Brassica napus. J Chem Ecol 26: 1811–1820 [Google Scholar]
- Qu CM, Li SM, Duan XJ, Fan JH, Jia LD, Zhao HY, Lu K, Li JN, Xu XF, Wang R (2015) Identification of candidate genes for seed glucosinolate content using association mapping in Brassica napus L. Genes (Basel) 6: 1215–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P (2013) Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo MS, Jin M, Chun JH, Kim SJ, Park BS, Shon SH, Kim JS (2016) Functional analysis of three BrMYB28 transcription factors controlling the biosynthesis of glucosinolates in Brassica rapa. Plant Mol Biol 90: 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo MS, Kim JS (2017) Understanding of MYB transcription factors involved in glucosinolate biosynthesis in brassicaceae. Molecules 22: 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderby IE, Geu-Flores F, Halkier BA (2010) Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci 15: 283–290 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ (2007) A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS One 2: e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Fan G, Hu Q, Zhou Y, Guan M, Tong C, Li J, Du D, Qi C, Jiang L, et al. (2017) The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J 92: 452–468 [DOI] [PubMed] [Google Scholar]
- Tan Z, Xie Z, Dai L, Zhang Y, Zhao H, Tang S, Wan L, Yao X, Guo L, Hong D (2022) Genome- and transcriptome-wide association studies reveal the genetic basis and the breeding history of seed glucosinolate content in Brassica napus. Plant Biotechnol J 20: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Zhao H, Lu S, Yu L, Zhang G, Zhang Y, Yang QY, Zhou Y, Wang X, Ma W, et al. (2021) Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol Plant 14: 470–487 [DOI] [PubMed] [Google Scholar]
- Tayo T, Dutta N, Sharma K (2012) Effect of feeding canola quality rapeseed mustard meal on animal production - a review. Agric Rev 33: 114–121 [Google Scholar]
- Textor S, de Kraker JW, Hause B, Gershenzon J, Tokuhisa JG (2007) MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol 144: 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova M, Ecke W, Weissleder K, R6bbelen G (1995) Mapping the genome of rapeseed (Brassica napus L.). I. Construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor Appl Genet 90: 194–204 [DOI] [PubMed] [Google Scholar]
- Walker KC, Booth EJ (2001) Agricultural aspects of rape and other Brassica products. Eur J Lipid Sci Technol 103: 441–446 [Google Scholar]
- Wei D, Cui Y, Mei J, Qian L, Lu K, Wang ZM, Li J, Tang Q, Qian W (2018) Genome-wide identification of loci affecting seed glucosinolate contents in Brassica napus L. J Integr Plant Biol 61: 611–623 [DOI] [PubMed] [Google Scholar]
- Wurschum T, Liu W, Maurer HP, Abel S, Reif JC (2012) Dissecting the genetic architecture of agronomic traits in multiple segregating populations in rapeseed (Brassica napus L.). Theor Appl Genet 124: 153–161 [DOI] [PubMed] [Google Scholar]
- Xing H, Dong L, Wang Z, Zhang H, Han C, Liu B, Wang X, Chen Q (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Cao S, Hu K, Wang X, Huang W, Wang G, Lv Z, Liu Z, Wen J, Yi B, et al. (2017) Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci Rep 7: 3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Yang J, Wang J, Li Z, Li X, He Z, Zhang L, Sha T, Lyu X, Chen S, Gu Y, et al. (2021) Genomic signatures of vegetable and oilseed allopolyploid Brassica juncea and genetic loci controlling the accumulation of glucosinolates. Plant Biotechnol J 19: 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Li J, Liu J, Liu K (2015) An Arabidopsis mitochondria-localized RRL protein mediates abscisic acid signal transduction through mitochondrial retrograde regulation involving ABI4. J Exp Bot 66: 6431–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Chen H, Cao B, Lei J, Chen G (2017) Molecular characterization of MYB28 involved in aliphatic glucosinolate biosynthesis in Chinese Kale (Brassica oleracea var. alboglabra Bailey). Front Plant Sci 8: 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Xie L, Zhang Q, Ouyang W, Deng L, Guan P, Ma M, Li Y, Zhang Y, Xiao Q, et al. (2020) Integrative analysis of reference epigenomes in 20 rice varieties. Nat Commun 11: 2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Dai L, Wang P, Liu Y, Xie Z, Zhang H, Xin Q, Wan L, Yang L, Yang G, et al. (2021) Mining favorable alleles for five agronomic traits from the elite rapeseed cultivar zhongshuang 11 by QTL mapping and integration. Crop J 9: 1449–1459 [Google Scholar]
- Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44: 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhao Y, Liu P, Shi L, Wang X, Wang M, Meng J, Reif JC (2016) Seed quality traits can be predicted with high accuracy in Brassica napus using genomic data. PLoS One 11: e0166624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.