Abstract
Plants acquire enhanced tolerance to intermittent abiotic stress by employing information obtained during prior exposure to an environmental disturbance, a process known as acclimation or defense priming. The capacity for stress memory is a critical feature in this process. The number of reports related to plant stress memory (PSM) has recently increased, but few studies have focused on the mechanisms that maintain PSM. Identifying the components involved in maintaining PSM is difficult due in part to the lack of clear criteria to recognize these components. In this review, based on what has been learned from genetic studies on heat acclimation memory, we propose criteria for identifying components of the regulatory networks that maintain PSM. We provide examples of the regulatory circuits formed by effectors and regulators of PSM. We also highlight strategies for assessing PSMs, update the progress in understanding the mechanisms of PSM maintenance, and provide perspectives for the further development of this exciting research field.
A review of the strategies and principles for assessing abiotic stress memory in plants, the roles of components that maintain stress memory, and the regulatory circuits they form.
Introduction
In biology, memory is defined as retained experience; when put into use, this memory changes the organism’s future behavior (Hawkins et al., 1993; Casadesús and D’Ari, 2002). In this sense, all cellular organisms including plants can “remember” experiencing various environmental stimuli, which facilitates acclimation to future changes. Multiple plant behaviors in response to environmental cues have long been associated with plant memory (Trewavas, 2003). A frequently mentioned example is the epigenetic regulation of vernalization, by which the experience of prolonged low temperatures in winter is marked or “remembered” and promotes flowering in the spring (Berry and Dean, 2015). However, such examples are scarce compared to the prevalence of human memory, so the existence and importance of plant memory are often overlooked. Despite being supported by increasing scientific evidence, plant memory remains a feature inconceivable to many scientists, including some plant biologists (Galviz et al., 2020).
An emerging area of research that illustrates the effect of memory on plant behavior is defense against biotic and abiotic stress, which is especially important, since plants are unable to move away from adverse conditions. In the past few decades, tremendous effort has focused on understanding how plants respond to and cope with various environmental stresses at the molecular level. Functional genomic studies of plant stress responses have revealed diverse mechanisms underpinning defense behaviors, including acclimation, one of the best-studied processes. Acclimation modifies plant physiology and morphology to enhance tolerance or fitness performance in response to periodic environmental changes (Taiz et al., 2015). Plants can acclimate to various abiotic stress conditions, including cold (Thomashow, 1999), heat (Hong and Vierling, 2000; Queitsch et al., 2000), drought (Harb et al., 2010), flooding (Bailey-Serres and Voesenek, 2008), salt (Xie et al., 2011), excess light (Dietz, 2015), UV (Müller-Xing et al., 2014), and mechanical loading (Martin et al., 2010). Specific and common acclimation mechanisms counteract each abiotic stress.
Different mechanisms might be invoked in response to different stress patterns, such as continuous versus intermittent stress episodes, even to the same abiotic stressor. For example, acclimation to recurring acute heat stress (HS) and prolonged moderate HS requires different genetic components (Yeh et al., 2012). Acclimation to intermittent stress involves a priming effect induced by pre-exposure to a milder stress stimulus, resulting in acquired tolerance to more severe stress challenges. This type of defense priming is typical of many plants in response to both biotic and abiotic stress (Martinez-Medina et al., 2016; Mauch-Mani et al., 2017).
According to the theory of defense priming, experiencing an external stimulus puts a plant in a primed state that persists even if the stimulus no longer exists. The primed state enables the plant to react to subsequent stress events with a faster and stronger defense response (Conrath et al., 2015). Memory capacity is defined as the ability to maintain the primed state induced by various environmental stimuli (Bruce et al., 2007; Hilker et al., 2016; Martinez-Medina et al., 2016; Oberkofler et al., 2021). The number of research articles reporting the existence of plant stress memory (PSM) is on the rise. In general, PSM, i.e. the primed state, can be divided into two parts: acquisition and maintenance.
Acquisition of PSM involves stress sensing, signal transduction, and gene regulation. These processes have been extensively studied in relation to various abiotic stresses. However, the mechanisms of PSM maintenance, as well as its physiological and ecological importance, are largely unexplored. It is crucial to understand how and to what extent plants maintain stress memories to adapt to future environmental adversities, which are likely to become more frequent and severe due to climate change (National Academies of Sciences E and Medicine, 2016).
Duration is of primary concern when it comes to the memory of a biological system. Increasing evidence shows that PSMs can have very different durations, which are classified as somatic or transgenerational based on heritability (Lämke and Bäurle, 2017). Somatic PSM endures only within the same generation of a stressed organism. By contrast, transgenerational memory is manifested when the nonstressed progeny show the stressed phenotypes of their progenitors without harboring a mutation, providing evidence of epigenetics. However, caution should be exercised in assessing PSM transmission to the first stress-free generation, as the memory effect may be induced in progeny-forming cells while they are still part of the parent plant that is exposed to stress (Pecinka and Scheid, 2012). Lämke and Bäurle (2017) proposed that memories maintained for only one stress-free generation should be referred to as intergenerational memories, and memories that are detectable after two stress-free generations should be referred to as transgenerational memories.
Transgenerational PSM is an exciting research topic that has drawn much discussion and debate, as outlined in several outstanding reviews (Chinnusamy and Zhu, 2009; Pecinka and Scheid, 2012; Weigel and Colot, 2012; Lim and Brunet, 2013). The maintenance of transgenerational PSM is primarily associated with epigenetic regulation (Chinnusamy and Zhu, 2009), whereas maintaining somatic PSM requires more diverse mechanisms. In this review, we focus on somatic abiotic stress memories, which likely contribute to within-generation plasticity to facilitate adaptation to changing environments (Auge et al., 2017). We propose guidelines for assessing PSMs based on defense priming and for recognizing components and regulatory circuits involved in PSM maintenance. We highlight the duration of PSMs during the acclimation to intermittent stress and strategies for their assessment. We also describe recent progress in elucidating the molecular mechanisms underlying the maintenance of PSMs. Finally, we provide suggestions and future perspectives on this exciting topic.
Assessment of memory during abiotic stress responses
The concept of defense priming facilitates the identification of PSMs, as illustrated by the simplified diagram in Figure 1A. In this example, plants of identical genetic makeup and growth history are divided into primed and nonprimed groups. The primed group receives an external stimulus (Stimulus I or priming), followed by the removal of the stimulus during a recovery period or memory phase (Hilker et al., 2016). A second stimulus (Stimulus II or triggering) is then applied to trigger a physiological or molecular output. The priming and triggering stimuli can be identical or different in nature and severity. The nonprimed group receives only triggering (Stimulus II) and produces output without priming. Suppose the outputs of primed and nonprimed plants are quantitatively or qualitatively different. In this case, the priming treatment is thought to modify the output of the triggering stimulus. The retainment of the primed state over time is memory, which resembles the definition of the relationship between learning and memory in animals (Hawkins et al., 1993). If the outputs of primed and nonprimed plants are not significantly different, two explanations are possible. First, perhaps the memory formed by priming was lost, leading to the loss of the primed state. Second, perhaps the priming treatment did not modify the output of the triggering stimulus, i.e. the output induced by Stimulus II is not primable by Stimulus I. Thus, the ability to be primed is a primary consideration for the assessment of PSM.
Figure 1.
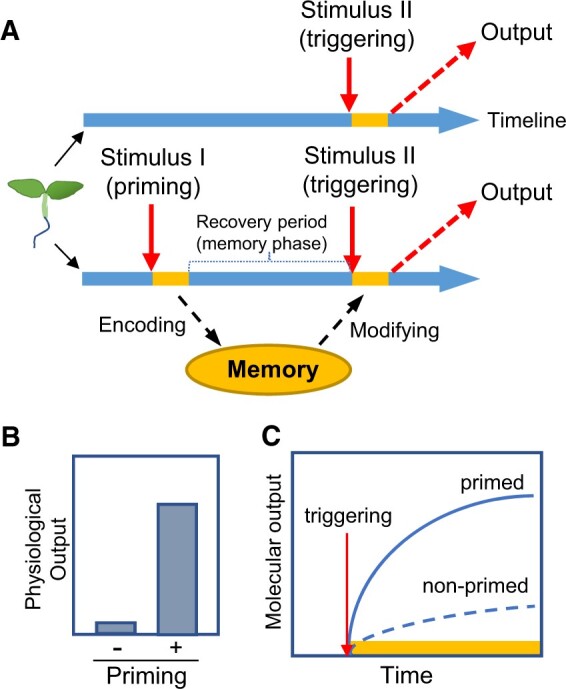
Acclimation to intermittent abiotic stress, a defense priming behavior of plants. A, A simplified diagram of the relationship between priming, triggering, memory, and output in the acclimation to recurring stress. Plants of the same genotype are subjected to two stress regimes that are run in parallel (timelines shown from left to right): nonprimed (top) and primed by Stimulus I (below). A recovery period or memory phase is introduced for the primed plants before triggering with Stimulus II. The nonprimed plants receive triggering at the same time as the primed plants. Physiological and molecular outputs are analyzed at an appropriate time point after or during triggering treatment. Memory encoded by priming is manifested by a modified output of primed versus nonprimed plants. The yellow blocks in the timelines represent the duration of stimulus application. B, Physiological outputs of primed (+) and nonprimed (−) plants subjected to the stress treatments shown in (A). The physiological outputs of primed plants are often referred to as acquired tolerance to abiotic stress. C, Molecular outputs of primed and nonprimed plants during triggering treatment (represented by the yellow bar). The primed output is stronger and occurs more rapidly than the nonprimed output.
Table 1 lists studies that provide evidence for PSM during acclimation to various abiotic stresses. These studies used various recovery times between priming and triggering treatments and showed PSMs of different durations. Physiological outputs are mainly associated with acquired stress tolerance, in which primed plants are more tolerant to triggering stress than nonprimed plants (Figure 1B). Molecular outputs are studied in relation to transcriptional memory (TM), in which priming significantly modifies the expression of genes in response to triggering (Figure 1C). In the following sections, we highlight two primary strategies for assessing PSMs using genetic and transcriptomic approaches.
Table 1.
Studies related to PSM duration for acclimation to intermittent abiotic stress
| Stress species |
Duration of primed statea | Output | Associated molecular components | References |
|---|---|---|---|---|
| Heat | ||||
| Arabidopsis thaliana | 3 d | AT | HSA32, HSFA2, ROF1 (FKBP62), HSP101, miR156s, HSP21, FtsH6, BRU1, HLP1, JMJs | Charng et al. (2006, 2007); Meiri and Breiman (2009); Wu et al. (2013); Stief et al. (2014); Sedaghatmehr et al. (2016); Brzezinka et al. (2016); Sharma et al. (2019); Yamaguchi et al. (2021) |
| 3 d | HSA32:Hsa32-LUCIFERASE reporter activity and AT | FGT1, FGT2, and FGT3 (HSFA3) | Brzezinka et al. (2019); Urrea Castellanos et al. (2020); Friedrich et al. (2021) | |
| 6 d | TM | HSFA2 | Liu et al. (2018) | |
| 5 min | Calcium concentration | Lenzoni and Knight (2019) | ||
| Oryza sativa | 2 d | AT | HSA32 and HSP101 | Lin et al. (2014) |
| Cold/freezing | ||||
| Arabidopsis thaliana |
8–24 h 3–7 d |
TM of CBFs AT |
tAPX, AOS, and OPR3 | |
| Brachypodium distachyon | 9 d | TM | Mayer and Charron (2021) | |
| Cucumis sativus | 2 d | AT | RBOH | Di et al. (2022) |
| Dehydration/drought | ||||
| Arabidopsis thaliana | 5 d | TM | MYC2, SnRK2.2, SnRK2.3, SnRK2.6, DDE2/AOS, and COI1 | Ding et al. (2012, 2014); Liu et al. (2014); Virlouvet et al. (2014); Virlouvet and Fromm (2015); Liu et al. (2016) |
| Alopecurus pratensis | 3 weeks | AT | POX and SOD | Lukić et al. (2020) |
| Boea hygrometrica | 13 weeks | AT and TM | DNA methylation | Sun et al. (2021) |
| Salt | ||||
| Arabidopsis thaliana | 10 d | AT and TM | HKT1 | Sani et al. (2013) |
| 5 d | Proline accumulation and TM | Feng et al. (2016) | ||
| 3 d | AT and TM | bZIP17 and HRD3A | Tian et al. (2019) | |
| Lolium perenne | 46 h | AT | Hu et al. (2016) | |
| Populus alba × P. glandulosa | 3 d | AT and TM | Liu et al. (2019a) | |
| Oryza sativa | 45 d | AT and TM | do Amaral et al. (2020a, 2020b) | |
| Light/UV | ||||
| Arabidopsis thaliana | 1 d | TM | Crisp et al. (2017) | |
| 1 d | Acquired UV-C tolerance | PsbS | Gorecka et al. (2020) | |
| 3 d | Acquired UV-B tolerance | UVR8 | Xiong et al. (2021) | |
| Mechanical loading | ||||
| Mimosa pudica | 28 d | Leaf-folding habituation | Gagliano et al. (2014) | |
| Populus tremula × P. alba | 1 d | TM | Pomiès et al. (2017) |
aThe duration of the primed state was determined by measuring the length of the recovery period between priming and triggering, as indicated in Figure 1.
AT, acquired tolerance; TM, transcriptional memory.
Genetic approaches for assessing PSMs for acclimation to abiotic stress
Genetic studies have greatly improved our understanding of plant abiotic stress responses, from stress signal sensing and transduction to gene regulation at the transcriptional to post-translational levels. Recent progress highlights the power of genetic approaches for identifying components involved in maintaining PSM for heat acclimation. Here, we categorize the memory components identified using genetic approaches into effectors and their regulators. During acclimation to intermittent stress, effectors are physical substances induced by priming that directly modify the output of triggering (Figure 2A). Hence, effectors are considered to be the physical underpinnings of cellular memory, and the status of the effectors proportionally affects the output level. Thus, the maintenance of effectors equals the maintenance of memory. Depending on the primed output, effectors could be any biochemical molecules and their modified derivatives, such as methylated DNA and post-translationally modified proteins. For example, multiple effectors contribute to physiological outputs such as acquired thermotolerance (AT), most notably heat shock proteins (HSPs), which are highly induced by thermopriming (Yeh et al., 2012). We propose that these effectors persist for different amounts of time once produced, some for a short duration (usually less than a few hours) and others for a long duration (a few days). Priming–triggering tests with short and long recovery periods are instrumental in demarcating the time range covered by the effectors’ actions (Figure 2A). Mutations that fail to produce the effectors for different durations will compromise the primed output, depending on the timing of triggering. In a mutant of a short-lived effector, the primed output is only compromised if triggering is applied after a short recovery (Figure 2B).
Figure 2.
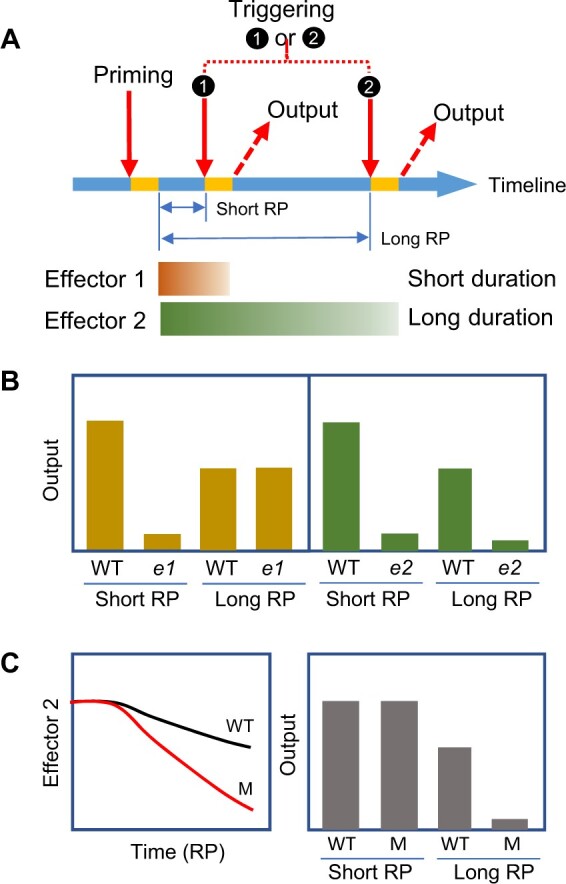
Assessment of plant abiotic stress memory using genetic approaches, and proposed criteria for short- and long-duration effectors. A, An experimental setup for priming and triggering to phenotype mutants of memory-related components. The features on the timeline are the same as in Figure 1A, except that triggering treatment is applied at either timepoint 1 or 2 by introducing a short (usually less than a few hours) or long (a few days) recovery period (RP), respectively, between priming and triggering. Priming induces the production of effectors that persist for different amounts of time (different durations). Effector 1 has a short duration, and Effector 2 has a long duration (represented by brown and green bars, respectively). B, Outputs of wild type (WT) and mutants unable to produce Effector 1 (e1) or Effector 2 (e2) after priming and triggering with a short or long RP. The output triggered after a short RP is compromised in both e1 and e2 but is compromised after a long RP only in e2. The output in the WT is lower after a long RP vs. a short one, corroborating the decay of effectors over time. C, Long-duration effectors (such as Effector 2) decay more rapidly in a maintenance mutant (M) than in WT during the RP (left panel). The mutant shows compromised output after a long RP but not a short RP (right panel), suggesting that the primed state does not persist for as long a time in the mutant as in the WT.
By contrast, a null mutation of a long-lived effector would affect the primed output triggered after both a short and long recovery (Figure 2B). Although the causal relationship between stress response genes (some encoding effectors) and acquired tolerance (primed output) has been extensively demonstrated, the duration at which effectors persist has rarely been studied. Identifying the regulators that control the durations of effectors will be critical for unraveling the mechanisms of memory maintenance during plant stress acclimation. Such regulators are not required for the induction of effectors but rather affect their duration. In the absence of a positive regulator, the decay of the effector would occur more rapidly, resulting in a compromised output triggered after a long but not short recovery (Figure 2C). Negative regulators should have the opposite effects.
The guidelines proposed above are primarily based on the findings obtained from genetic studies of PSM for heat acclimation. The simple phenotyping of Arabidopsis (Arabidopsis thaliana) seedlings grown on standard medium and the ease of HS treatment make this plant a robust system for genetic analysis of PSM. Experiments with the priming–triggering setup have long been performed in physiological studies of heat acclimation that confers AT and genetic studies on the functions of HS-induced proteins, such as HSP101, a protein disaggregase pivotal for AT (Hong and Vierling, 2000; Queitsch et al., 2000). However, these priming–triggering experiments are only associated with a short recovery period.
A critical breakthrough was made by introducing a long recovery period between priming and triggering treatments in functional studies of HSA32, a heat-stress-associated 32-kD protein (Charng et al., 2006). AT triggered after a short and long recovery are referred to as short-term and long-term AT (SAT and LAT), respectively (Yeh et al., 2012). An Arabidopsis loss-of-function mutant of HSA32 shows a defect in LAT, not SAT, suggesting that the primed state does not persist for as long a time in the mutant as the wild type. Thus, the phenotype of hsa32 represents a certain “forgetfulness” of the priming experience. Genetic screens for an hsa32-like phenotype from Arabidopsis ethane methyl sulfonate (EMS) mutant pools resulted in the isolation of defective in long-term acquired thermotolerance1-1 (dlt1-1), a missense mutant allele of HSP101 (Wu et al., 2013). The forgetful phenotype of dlt1-1, which is specifically defective in LAT, was unexpected, as the null mutation of HSP101 dramatically compromised both SAT and LAT. This finding led to the identification of a positive feedback loop between HSA32 and HSP101 (Wu et al., 2013; Lin et al., 2014). In this example, HSP101 fits the criteria of a long-lived effector, and HSA32 fits the criteria of a regulator, as illustrated in Figure 2, B and C.
Phenotyping assays of SAT and LAT were performed in reverse genetic experiments to identify other components involved in maintaining heat acclimation memory, including HSFA2 (Charng et al., 2007), ROTAMASE FKBP 1 (ROF1; Meiri and Breiman, 2009), the microRNA miR156 (Stief et al., 2014), HSP21 (also known as HSP25.3-P, Sedaghatmehr et al., 2016), BRUSHY1 (BRU1; Brzezinka et al., 2019), and HIKESHI-LIKE PROTEIN 1 (HLP1; Sharma et al., 2019). Furthermore, the forgetter mutants (fgt1, fgt2, and fgt3) were isolated by screening an EMS mutant pool generated from a transgenic HSA32:Hsa32-LUCIFERASE (LUC) line based on the phenotype of faster disappearance of LUC-derived bioluminescence after HS priming (Brzezinka et al., 2016; Urrea Castellanos et al., 2020; Friedrich et al., 2021). FGT1, FGT2, and FGT3 encode proteins with very different functions. The diverse functions of the components identified by forward and reverse genetic approaches indicate that PSM maintenance is complex. Genetic studies also identified negative regulators of PSM for heat acclimation. FtsH6, an HS-induced plastidial protease, was shown to negatively regulate HSP21, revealing a control module of PSM (Sedaghatmehr et al., 2016). We discuss these components in the context of protein and transcriptional networks below.
Thus far, components involved in maintaining PSMs during acclimation to abiotic stresses other than HS have yet to be identified or confirmed genetically. The criteria for assessing PSM for heat acclimation are likely applicable to other types of abiotic stress with rapid, irregular fluctuations and preferably a long duration of the primed state, such as freezing (Leuendorf et al., 2020) and UV-B stress (Xiong et al., 2021). The criteria may have limitations for other types of abiotic stress, which usually involve a slow process in the natural environment, such as drought or salinity stress. Intermittent salt stress may be rare, but the priming effect of mild salinity has been demonstrated in different plant species (Table 1). bZIP17, a salt-inducible membrane-bound transcription factor, was recently shown to be required for acclimation to recurring salt stress in Arabidopsis (Tian et al., 2019). The role of bZIP17 in salt acclimation memory remains to be determined. Nevertheless, studies on salt-induced memory may benefit from the criteria proposed in Figure 2.
Acclimation to intermittent water deficit has been reported in several studies. These studies employed a “training” procedure in which plants were subjected to daily dehydration and rehydration cycles (air-drying for 1–2 h followed by the replenishment of water; Table 1). In Arabidopsis, enhanced induction of the drought response genes RD29B and RAB18 was observed in plants subjected to repeated dehydration treatments (Ding et al., 2012), indicating a memory response to dehydration stress. Using the expression of RD29B and RAB18 as outputs, the primed state induced by four cycles of dehydration–rehydration was shown to be maintained for up to 5 d (Ding et al., 2012). Thus, genetic dissection of the maintenance of dehydration memory should be feasible by adopting the priming–triggering criteria proposed in Figure 2.
Identification of TM during abiotic stress responses
Analyzing the output of triggering at the molecular level is a simple way to demonstrate the priming effect and the existence of memory. This type of analysis often involves comparing the abundance of stress-induced molecules such as RNAs, proteins, and secondary metabolites in primed and nonprimed plants. For example, the enhanced nicotine accumulation in wild tobacco (Nicotiana sylvestris) roots primed by methyl jasmonate is considered to be a consequence of immunological memory (Baldwin and Schmelz, 1996). The modification of the expression of RD29B and RAB18 by the dehydration training/priming process mentioned above also serves as a good example. Another example is the desensitization of the cold-shock response of low temperature-responsive CBF genes (also known as DREB1 genes) in Arabidopsis by priming at low temperatures (Zarka et al., 2003). Since the analysis of gene transcripts is a routine molecular technique in most plant science laboratories, transcript abundance has become a prominent molecular output reported in many recent studies on PSM.
One reason for the modification of gene expression by priming is TM, a phenomenon of cellular memory first described in yeast (Saccharomyces cerevisiae) in response to priming by a specific carbon source during growth (Acar et al., 2005). Figure 1C shows a typical effect of TM on primable genes: a faster and stronger reinduction by triggering. The behaviors of RD29B and RAB18 in response to repetitive dehydration stress in Arabidopsis were shown to be regulated by TM that involves chromatin modifications and stalled RNA polymerase II at the promoters of these genes (Ding et al., 2012).
However, transcriptomic analysis of the dehydration response in Arabidopsis revealed four distinct expression patterns of primable genes (Ding et al., 2013). Priming strengthens the induction of genes that are upregulated by dehydration, such as RD29B and RAB18 (categorized as +/+), and the suppression of downregulated genes (categorized as −/−). On the other hand, priming can dampen positive and negative stress responses induced by the triggering of primable genes categorized as +/− and −/+, respectively (Ding et al., 2013). These four types of memory responses to dehydration at the gene expression level were also detected in maize (Zea mays), switchgrass (Panicum virgatum), coffee (Coffea canephora), rice (Oryza sativa), and soybean (Glycine max) by transcriptomic analyses (Ding et al., 2014; Alves de Freitas Guedes et al., 2018; Virlouvet et al., 2018; Zhang et al., 2018; Li et al., 2019; Kim et al., 2020).
Transcriptomics has also facilitated the identification of primable genes in response to other abiotic stresses, including salt (Hu et al., 2016; Liu et al., 2019a; do Amaral et al., 2020a), heat (Liu et al., 2018), cold (Zuther et al., 2019; Mayer and Charron, 2021), excess light (Crisp et al., 2017), and mechanical stress (Pomiès et al., 2017). Much information is now available for assessing PSM maintenance using the transcript abundances of primable genes as molecular outputs. The duration of PSM can be evaluated by testing the duration of TM. The durations of TM of genes induced by dehydration, salt, heat, and cold stress examined to date span several days (Hu et al., 2016; Liu et al., 2018; Mayer and Charron, 2021). For example, the TM of ASCORBATE PEROXIDASE 2 (APX2) induced by HS treatment at 37°C for 1 h could be maintained for up to 7 d (Liu et al., 2018). This information may be valuable for analyzing the maintenance of stress memory via a genetic approach using reporter genes fused to the promoters of primable genes such as RD29B and APX2 (Virlouvet et al., 2014; Liu et al., 2018).
Molecular mechanisms maintaining PSM for acclimation to abiotic stress
Plants respond to abiotic stress factors by activating molecular networks operating at different levels for processes such as signaling transduction, gene expression, protein homeostasis, and metabolic adjustment (Krasensky and Jonak, 2012). These networks are integrated to ensure the processing and retaining of environmental information perceived by plants during acclimation to intermittent stress. In Figure 2, we illustrate the temporal relationship between effectors and outputs in a priming–triggering scenario: the duration of PSM correlates with the duration of priming-induced effectors. Based on the current knowledge, we propose a schematic network that depicts the connections of the components, the effector, and their positive and negative regulators in modulating the duration of PSM (Figure 3A). Here, we focus on transcriptional and post-translational regulation, which are relatively well-understood.
Figure 3.
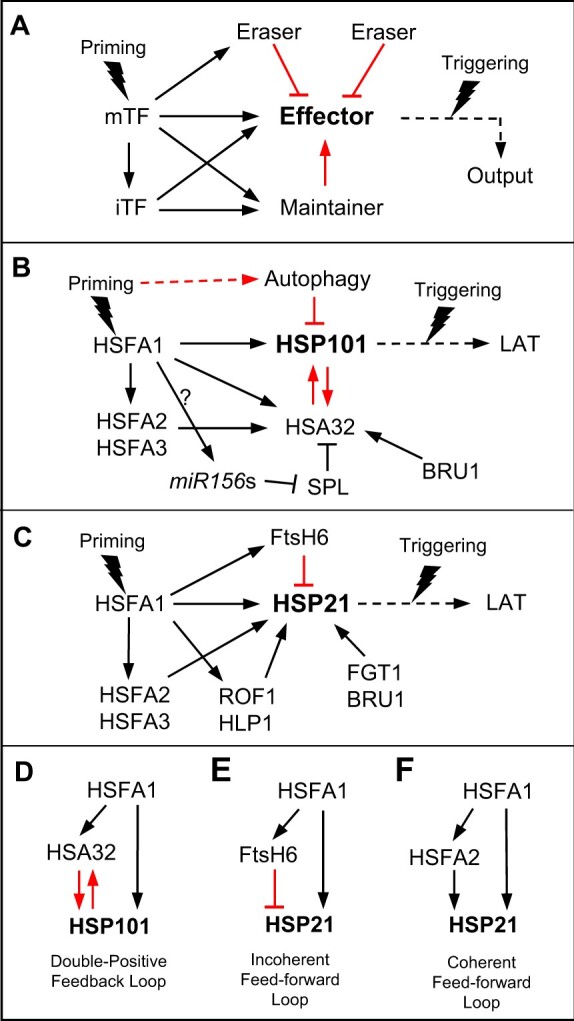
Simplified diagrams of regulatory networks and circuits for maintaining the memory of stress priming in plants. A, In response to priming, the mTFs activate downstream genes encoding priming-iTFs, effectors, and regulators (maintainers and erasers). iTF also activates the effector and regulators at the transcriptional level. At the protein level, the duration of the effectors is positively (red arrow) and negatively (red line with bar) regulated by the maintainer and erasers, respectively. The maintainer prevents the decay of the effector, while the erasers (either priming-dependent or -independent) remove the effector. B, The network that regulates the duration of HSP101, an effector that functions in heat acclimation. HSA32 acts as a maintainer by preventing the decay of HSP101, whereas autophagy functions as an eraser. HSP101 also acts as a positive regulator of HSA32 at the protein level, creating a positive feedback loop. Other factors that regulate HSA32 at the transcriptional level (indicated by black arrows) are also shown. The link between HSFA1 and miR156s has not been established (indicated by a question mark). Long-term acquired thermotolerance (LAT) is the physiological output for assessing heat acclimation memory. C, The network that regulates the duration of HSP21. The duration of HSP21 is negatively regulated at the protein level by the heat-inducible eraser FtsH6 and positively regulated by other factors at the transcriptional level. D, The double positive feedback loop that regulates HSP101 at both the transcriptional (black arrows) and protein (red arrows) levels. E, The incoherent FFL regulates HSP21 at both the transcription (black arrows) and protein (red line with bar) levels. F, The coherent FFL that regulates HSP21 at the transcriptional level (black arrows).
In response to priming, a master transcription factor (mTF) activates downstream genes encoding the effector and its regulators, including the priming-inducible transcription factor (iTF), maintainer, and erasers (Figure 3A). Like mTF, iTF also regulates the expression of the effector and the other regulators at the transcriptional level. The positive regulator that maintains the stability of the effector at the protein level is regarded as a maintainer. By contrast, the negative regulators that remove the effector protein are erasers, which can be further classified as priming-inducible or constitutive. In the following section, we use published experimental data on heat acclimation to illustrate this scheme.
Regulatory networks and circuits for maintaining heat acclimation memory
Figure 3, B and C show the networks associated with two putative effectors, HSP101 and HSP21, which are well-known molecular chaperones that confer plant thermotolerance (Härndahl et al., 1999; Hong and Vierling, 2000; Katiyar-Agarwal et al., 2003). HSP101 fulfills the criteria of a long-duration effector genetically (Figure 2). HSA32 serves as a maintainer of HSP101 by preventing its decay at the protein level during the recovery period, while HSP101 positively regulates the accumulation of HSA32 protein (Wu et al., 2013; Lin et al., 2014). In this example, HSP101 acts as a positive regulator of its maintainer, thus forming a positive feedback loop between the two players (Figure 3B). Positive feedback loops are common regulatory modules that function in cellular memory of environmental cues (Jiang and Hao, 2021). During the recovery period, HSP101 is gradually degraded along with other HSPs via an autophagy pathway activated by thermopriming (Sedaghatmehr et al., 2019). Hence, autophagy acts as an eraser of the effector HSP101. HSA32 was also shown to be positively regulated by the HS-iTFs HSFA2 and HSFA3 during the recovery period (Charng et al., 2007; Friedrich et al., 2021). On the other hand, SQUAMOSA-PROMOTER BINDING-LIKE (SPL) transcription factors, which are the targets of HS-inducible miR156s, act as negative regulators of HSA32 (Stief et al., 2014). The transcription factors responsible for the heat-induction of most microRNAs, including miR156s, have not been identified. Notably, the heat-induction of miR824 was recently shown to be HSFA1-dependent in Arabidopsis (Szaker et al., 2019).
The position of HSP21 as an effector in the network was predicted based on its chaperone function (Figure 3C). However, this is not supported by the genetic evidence provided by Sedaghatmehr et al. (2016). HSP21 reaches a high level immediately after priming and persists for a few days. However, the HSP21 knockdown mutant showed a defect in LAT but not SAT, which is not in agreement with the criteria of a long-duration effector indicated in Figure 2. Perhaps HSP21 is not immediately suppressed in the knockdown line after priming. Alternatively, perhaps HSP21 is subjected to an oxidative modification that inhibits its chaperone activity within a short recovery period window and is recovered after a long recovery period (Gustavsson et al., 2002). Nevertheless, the identification of the plastidial protease FtsH6 as a negative regulator of the duration of HSP21 provides another example of a regulatory module for PSM (Sedaghatmehr et al., 2016). FtsH6 is also an HS-inducible protease, thus functioning in negative feedback control of PSM for heat acclimation. Such feedback control found in Arabidopsis ecotype Columbia-0 is missing in ecotype N13 (Sedaghatmehr et al., 2016), suggesting that HSP21-mediated PSM is an adaptive measure. In addition, HSP21 is positively regulated by HSFA2 and HSFA3 during the recovery period (Lämke et al., 2016; Friedrich et al., 2021). ROF1 (a peptidyl-prolyl cis/trans isomerase) and HLP1, which are both responsive to HS, positively regulate the persistence of HSP21 transcript after priming (Meiri and Breiman, 2009; Sharma et al., 2019).
The two networks involving HSP101 and HSP21 operate with both unique and shared components. Both HSP101 and HSP21 and some of their regulators are induced by priming under the control of the master regulators of HS response, i.e. HSFA1a, HSFA1b, and HSFA1d, in Arabidopsis (Figure 3, B and C; Liu et al., 2011; Yoshida et al., 2011). Since many protein effectors contribute to AT (Yeh et al., 2012), the persistence/duration of other effectors downstream of HSFA1 is likely also modulated by networks similar to that for HSP101 and HSP21. Biochemical circuits formed by genes or proteins have been proposed to perform computational tasks, including the amplification, integration, and storage of information (Bray, 1995; McAdams and Shapiro, 1995). Notably, the regulatory networks involving HSP101 and HSP21 contain distinct regulatory circuits, such as the feed-forward loop (FFL), which is implicated in retaining memories of external stimuli in bacteria (Alon, 2007; Jiang and Hao, 2021). The FFL is a common computational framework in biological and technological networks for information processing for gene regulation and in neurons and electronic circuits (Milo et al., 2002). Although gene regulation circuits such as FFL function in plant growth and development (Zhong and Ye, 2012; Verweij et al., 2016; Li et al., 2022), their function in modulating PSM has not been explored. In the following section, we highlight the regulatory circuits that might be responsible for modulating the duration of heat acclimation memory.
Three regulatory circuits, each composed of three components, can be identified in the networks shown in Figure 3, B and C. These circuits include a double-positive feedback loop (DPFL, with two down-stream components under a positive regulator positively regulate each other), an incoherent FFL (in which the indirect path has the opposite effect of the direct path), and a coherent FFL (in which the indirect path has the same effect as the direct path; Figure 3, D–F) according to the definition of network motifs that initially referred to recurring patterns found in transcriptional regulatory networks (Alon, 2007). However, the regulatory circuits illustrated here are not limited to transcriptional regulation. In the case of heat acclimation, HSFA1 can form these three regulatory circuits in combination with the effectors (HSP101 and HSP21) and regulators (HSA32, HSFA2, and FtsH6).
First, HSFA1 can form a DPFL with HSP101 and HSA32 to retain the primed state or AT during the memory phase (Figure 3D). Of note, this circuit combines two regulatory layers that control the transcript and protein levels of the components (Figure 3D). A DPFL with solely transcriptional connections between its components tends to implement developmental memory and is not commonly found in networks for stress responses (Alon, 2007). Thus, the hybrid circuit depicted in Figure 3D likely represents a DPFL variant for prolonging the duration of the priming-induced effector. In this example, the reciprocally positive regulation between HSP101 and HSA32 is critical, as a missense mutation in HSP101 disrupted its action on HSA32, but not its effector function, and led to the faster decay of HSP101 during the memory phase (Wu et al., 2013). HSA32, which does not resemble any known protein chaperone and is ubiquitously present in land plants, shares a distant homology with an archaeal enzyme, phosphosulfolactate synthase (Liu et al., 2006). Its mode of action remains to be elucidated.
The second regulatory circuit is the incoherent FFL constituted by HSFA1, HSP21, and FtsH6. This circuit combines two regulatory layers at the transcriptional and post-transcriptional levels (Figure 3E). HSFA1 serves as the transcriptional activator of both HSP21 and FtsH6, and FtsH6 suppresses the accumulation of HSP21, presumably via its protease activity (Sedaghatmehr et al., 2016). This circuit shortens the duration of the effector HSP21 during the memory phase. FtsH6 transcription is induced immediately after thermopriming, but the accumulation of FtsH6 protein is substantially delayed (Sedaghatmehr et al., 2016). This phenomenon points to post-transcriptional regulation of the eraser. The delay prevents the quick decay of HSP21 from occurring within a short recovery period after priming. How FtsH6 is regulated post-transcriptionally is unclear.
The coherent FFL between HSFA1, HSFA2, and HSP21 occurs at the transcriptional level (Figure 3F). Either HSFA1 or HSFA2 can regulate HSP21 in response to thermopriming (Liu and Charng, 2013; Sedaghatmehr et al., 2016), and the expression of HSFA2 is HSFA1-dependent (Liu et al., 2011). In addition to HSP21, HSFA1 and HSFA2 also form a coherent FFL with HSA32 (Figure 3B). This type of coherent FFL has been shown to prolong expression of genes encoding components of the flagellum in Escherichia coli after signal removal (Kalir et al., 2005). Similar to the function of the FFL in E. coli flagella, the HSFA1-HSFA2-HSP21/HSA32 FFLs prolong the expression of HSP21 and HSA32 in Arabidopsis during the memory phase after priming. Deleting HSFA2 abolished the prolonged expression of HSP21 and HSA32 but not their induction by thermopriming (Charng et al., 2007; Lämke et al., 2016).
It can be seen that the maintenance of PSM involves the integration of multiple regulatory circuits. For example, HSA32 is regulated by at least two different circuits, a DPFL and a coherent FFL, and HSP21 by incoherent and coherent FFLs. Why a component of PSM maintenance is controlled by multiple regulatory circuits is unclear. One possibility is that the wiring of a memory component to different circuits provides better flexibility for fine-tuning PSM in response to different stress intensities. Additionally, a memory component may be involved in different circuits to generate different outputs. For example, in Arabidopsis, priming-induced HSFA2 was shown to regulate the expression of genes exhibiting different types of TM, as discussed below.
Maintenance of TM
Sustained transcriptional activity and modification of the expression of stress response genes induced by priming are both critical for abiotic stress acclimation. Two types of TM have been proposed to distinguish the different effects of priming on gene expression behavior (Lämke et al., 2016; Oberkofler et al., 2021). Type I TM enables the persistent expression of TM genes during the recovery period and is usually manifested by physiological outputs (Figure 4A, top panel). Type II TM modifies the expression of TM genes during the triggering session, resulting in stronger and faster expression (Figure 4A, bottom panel), which serves as a molecular output in studies of PSM duration in response to intermittent abiotic stress (Table 1). The priming and triggering treatments for Type II TM are usually identical in nature and severity to avoid the secondary effect caused by triggering under a condition much harsher than priming. As more components of PSM are identified, and their relationships become more complicated, it is becoming important to develop a systemic classification of TM.
Figure 4.
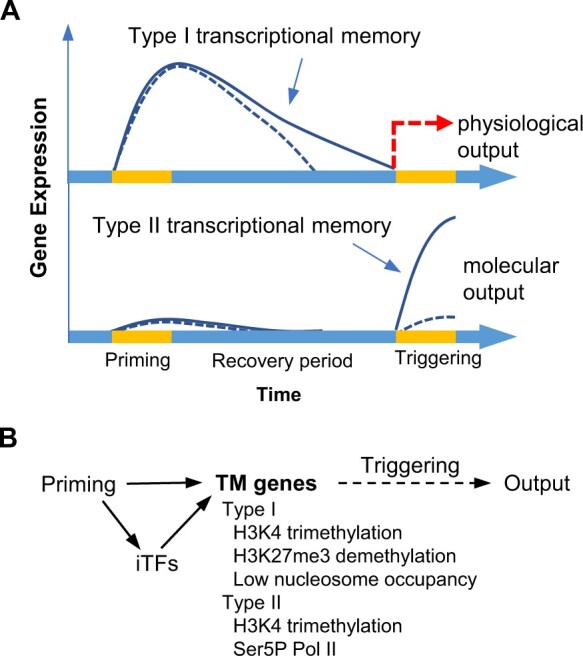
Transcriptional memories formed by abiotic stress priming and transcriptional networks. A, Two types of TM formed by priming. Type I TM enables the persistent expression of TM genes during the recovery period (top panel). Type II TM modifies the expression of TM genes during the triggering session, resulting in stronger and faster expression (bottom panel). The expression behaviors of TM genes in mutants that fail to maintain TM are represented by a dashed line, whereas that of the wild type is represented by a solid line. B, A simplified transcriptional network for the maintenance of TM. Stress-iTFs bind to the promoters of TM genes, resulting in the tri-methylation of histone H3 lysine 4 (H3K4me3) in both types of TM. The paused form of RNA polymerase II (Ser5P Pol II) accumulates at the promoter regions of Type II TM genes. Both H3K4me3 and Ser5P Pol II are associated with the primed state of TM-II genes. Demethylation of H3K27me3 and low nucleosome occupancy are associated with the sustained expression of Type I TM genes.
Type I TM was first demonstrated for LAT mediated by HSFA2. HSFA2 was shown to be required for maintaining the duration of AT and the sustained expression of a group of HS response genes, hereafter called TM-I genes (Charng et al., 2007; Lämke et al., 2016). Interestingly, HSFA2 is also required for Type II TM of a subset of HS response genes, hereafter called TM-II genes (Liu et al., 2018). Some TM-I genes, such as HSA32 and HSP21, do not show expression behavior like TM-II genes, suggesting that these types of TM regulation are specific despite involving the same iTF (HSFA2). It is unclear how HSFA2 distinguishes TM-I from TM-II genes. Since HSFA2 is under the control of HSFA1, both transcription factors play critical roles in Type I and Type II TM in response to thermopriming, but they may have different preferences for TM-I and TM-II genes. HSA32 and HSP21 (TM-I genes) could be activated by either HSFA1 or HSFA2, while the heat-induced expression of APX2 and MIPS2 (TM-II genes) is preferentially activated by HSFA2 (Liu and Charng, 2013). Thus, unlike TM-I genes, TM-II genes may not form a typical coherent FFL with HSFA1 and HSFA2 (Figure 3F). The Arabidopsis fgt3 mutant, which was isolated based on its compromised Type I TM, was recently shown to harbor a mutation in HSFA3 (Friedrich et al., 2021). Intriguingly, HSFA3 is not required for Type II TM even though HSFA3 and HSFA2 can form a heteromeric complex and redundantly maintain Type I TM (Friedrich et al., 2021). These findings reveal a complex relationship between HS-induced transcription factors and different types of TM in heat acclimation.
Epigenetic regulation has been the primary focus of studies on the TM of abiotic stress. Based on current knowledge, we propose a generalized diagram of the regulatory network to explain the mechanisms for the formation and maintenance of TM (Figure 4B). Specific transcription factors are induced by priming to mark the promoters of both TM-I and TM-II genes. The most prevalent marker associated with the primed state of TM genes identified to date is the tri-methylation of histone H3 lysine 4 (H3K4me3; reviewed by Oberkofler et al., 2021, and the references therein). Hyper-methylation of H3K4 is thought to be critical for both Type I and Type II TM. However, there is no conclusive evidence for a causal relationship. Moreover, it appears that this modification is not the only requirement for Type II TM because H3K4me3 is enriched at the promoters of TM-I genes (such as HSP21, HSP22, and HSP18.2) after thermopriming, but these TM-I genes do not show Type II TM (Lämke et al., 2016; Liu et al., 2018).
An attempt was recently made to transiently remove H3K4me3 in Arabidopsis by inducible CRISPR-based epigenomics editing using HS-inducible transgenes encoding APX2-targeting small guide RNA and the JMJ18 (H3K4 demethylase) active domain and inactive Cas9 fusion protein (dCAS9-JMJ; Oberkofler and Bäurle, 2022). Unfortunately, the results were not conclusive, as dCAS9 fused to both an active and inactivated JMJ18 domain compromised Type II TM of APX2 (a typical TM-II gene) in a similar manner. Perhaps the HS-inducible inactivated fusion protein bound to the APX2 promoter hindered the tri-methylation of H3K4 induced by priming. Alternatively, the inactive JMJ domain may recruit a complex that contains endogenous active demethylases (Oberkofler and Bäurle, 2022). A chemically inducible promoter might be favored to drive the editing process during the recovery period. Nevertheless, these findings will be valuable for other studies aimed at epigenomic editing.
A role for H3K4 tri-methylation enzymes in TM has not yet been demonstrated. The H3K4 methyltransferase ATX1 has been shown to be required for an optimal dehydration response but not for the formation of Type II TM (Ding et al., 2012). ATX1 and its homolog SDG25 were shown to be required for acclimation to continuous HS, with functional redundancy (Song et al., 2021). Disrupting both genes led to the loss of H3K4me3 at several loci (primarily transposable elements) after prolonged HS. It would be interesting to learn whether ATX1 and SDG25 are involved in forming Type I and Type II TM.
Besides H3K4, the methylation status of H3K27 has also been implicated in modulating HS memory in Arabidopsis. REF6/JMJ12, a gene from the JUMONJI (JMJ) family encoding H3K27me3 demethylase, forms a positive feedback loop with HSFA2 under prolonged moderate HS, conferring transgenerational memory to the nonstressed progeny (Liu et al., 2019b). A quadruple mutant (jmjq) of REF6/JMJ12, ELF6/JMJ11, JMJ30, and JMJ32 was recently shown to be defective in LAT, and Type I TM of HSP22 and HSP17.6C is compromised in jmjq, like it is in hsfa2 (Yamaguchi et al., 2021). Hence, JMJ proteins appear to act as positive regulators for the sustained expression of TM-I genes. However, SAT, another criterion for assessing heat acclimation memory, was not tested for jmjq. Interestingly, a high H3K27me3 level in jmjq during the recovery period is associated with low H3K4me3 levels at TM-I genes, including HSP21 (Yamaguchi et al., 2021). It remains to be clarified whether the effect on Type I TM in jmjq is indirect and is due to the reduced H3K4 trimethylation at TM-I loci. It would also be interesting to determine whether JMJ proteins are recruited to form Type II TM.
In a study of Type II TM of dehydration stress, Ding et al. (2012) showed that high levels of phosphorylated RNA polymerase II at serine 5 (Ser5P Pol II) are associated with RD29B and RAB18 (TM-II genes) during the recovery period. This phenomenon was not found in the non-TM genes RD29A and COR15A. Ser5P Pol II is associated with stalled transcriptional activity, which can be reactivated by the transition from Ser5P to Ser2P (phosphorylation at serine 2) during exposure to stress (Nechaev and Adelman, 2011). This characteristic makes Ser5P Pol II a good candidate as an effector of the output for Type II TM (Figure 4B). It remains to be seen whether Ser5P Pol II levels increase at TM-II genes in response to priming with other abiotic stress factors.
In addition to histone modifications, nucleosome occupancy is also associated with maintaining heat acclimation memory. Forward genetics facilitated the identification of FGT1 as a positive regulator of Type I TM (Brzezinka et al., 2016). FGT1 was shown to form a complex with the chromatin remodelers BRAHMA (BRM) and CHR11/17 to maintain low nucleosome occupancy at TM-I genes during the recovery period (Brzezinka et al., 2016). Thus, components that positively and negatively regulate nucleosome occupancy may form regulatory circuits that maintain PSM under various abiotic stress treatments. Comprehensive reviews on epigenetic regulation of TM include (Kinoshita and Seki, 2014; Avramova, 2015; D’Urso and Brickner, 2017; Oberkofler et al., 2021).
Perspectives and open questions
Acclimation is essential for plants to cope with recurring stress conditions in the natural environment. Memory plays a role as a defense priming behavior underpinning acclimation to intermittent abiotic stress, as evidenced by a recent flurry of reports. However, the maintenance mechanisms of PSMs are unclear due to the difficulty in identifying the underlying components and the lack of clarity surrounding the definition of memory components. This review examined the regulatory networks of heat acclimation memory components identified by phenotype-based genetic screening. We proposed criteria for identifying the effectors responsible for triggered outputs and the regulators that modulate the durations of the effectors (Figure 2), which serve as proxies of PSM. We defined the relationships between the effectors and their upstream transcription factors, maintainers, and erasers to assess and interpret the results of PSM studies (Figure 3A). We also described regulatory circuits that are known to be involved in information processing and retainment (Figure 3, D–F).
So far, relatively few distinct regulatory circuits have been found to maintain the primed state of plants. However, we believe that well-characterized regulatory circuits observed in plant abiotic stress responses are overlooked with regard to their possible roles in PSM due to an unaccounted for connection with the duration of the primed state (Vogel et al., 2005; Seo et al., 2012). The criteria regarding the duration of effectors discussed in this review may assist in uncovering new regulatory circuits for memory maintenance. For example, effectors under tight negative control for a short duration could be recognized (Figure 2). Regulatory circuits that control the stability of stress-induced molecules, such as RNAs and proteins, may provide a primed defense against a second attack within a short recovery period and allow for quick recovery from stress (Crisp et al., 2016). Identification of memory components that enforce the short persistence (short duration) of effectors might be facilitated by genetic screening for mutants that lose acquired stress tolerance within a short but not long recovery time (Figure 2B). Although this phenotype has rarely been reported, an Arabidopsis null mutant of a mitochondrial metal protease was shown to be defective in SAT but not LAT (Charng et al., 2007). The underlying mechanism, however, awaits elucidation.
Increasing studies aimed at identifying TM (mostly Type II TM) of abiotic stress have been performed using a transcriptomic approach. The maintenance of Type I TM could be achieved by a coherent FFL formed by memory components (Figure 3F) and the modulation of chromatin (Figure 4B). Efforts are needed to uncover the mechanisms that maintain Type II TM. In addition to transcriptome profiling, proteome profiling may add a new dimension through which to identify memory components in primable events at the protein level that function in abiotic stress responses (Schulze et al., 2021). Other new omics approaches, such as epitranscriptomics, are also expected to achieve the same goal. N6-methyladenosine (m6A) methylation of RNA functions in memory formation in animals (Leonetti et al., 2020). m6A methylation was recently shown to be involved in salt-stress tolerance in Arabidopsis (Hu et al., 2021). It would therefore be interesting to investigate whether m6A methylation plays a role in PSM.
Protein conformation is capable of storing biological information. For example, the conformations of self-replicating proteins such as prions are considered to be molecular memories that transmit genetic information (Shorter and Lindquist, 2005). The prion-like protein CPEB is associated with the formation of long-term memory in animals (Si et al., 2003). EARLY FLOWERING 3 (ELF3), a scaffold protein with a prion domain, functions as a thermosensor (Jung et al., 2020) that forms and stores warm temperature memory in Arabidopsis (Murcia et al., 2022). Intrinsically disordered proteins, including prion-like proteins, can adopt new conformations with significant biological functions (Wright and Dyson, 2015). Intrinsically disordered sequences might play a similar role in forming or maintaining PSM during acclimation to abiotic stress.
Multiple environmental factors likely affect the formation and duration of PSM. Indeed, PSM is unlikely to be as simple in the natural environment as in a laboratory setting. For example, light was shown to affect TM in response to salt stress (Feng et al., 2016). Sugar level is also a critical factor in heat acclimation (Sharma et al., 2019). Root endophytes can induce thermotolerance by tapping into HSFA2-mediated HS memory (Shekhawat et al., 2021). A better understanding of the fundamental mechanisms at play in PSM will be instrumental for addressing the more complex situations. Perspectives from evolution and ecology on the components for PSM maintenance are also crucial, as the cost of memory maintenance may affect fitness in organisms (Hoedjes et al., 2011; Hilker et al., 2016). It remains to be seen whether regulatory circuits are involved in ecological stress memory to help plants adapt to repeated exposure to extreme conditions (Walter et al., 2013; Ahrens et al., 2021).
Acknowledgments
We thank Chia-chen Chang for inspiration on the concept of the maintenance of plant stress memory, Tzyy-Jen Chiou for critical reading of the manuscript and giving feedback, and Miranda Loney and Nancy Eckardt for English editing. We also thank three anonymous reviewers for their constructive comments and suggestions.
Funding
This work was supported by the National Science and Technology Council, ROC (110-2311-B-001-034-MY3).
Conflict of interest statement. None declared.
Contributor Information
Yee-yung Charng, Agricultural Biotechnology Research Center, Academia Sinica, Taiwan, ROC; Molecular and Biological Agricultural Sciences Program, TIGP, Academia Sinica, Taiwan, ROC; Biotechnology Center, National Chung-Hsing University, Taichung, Taiwan, ROC; Department of Biochemical Sciences and Technology, National Taiwan University, Taipei, Taiwan, ROC.
Suma Mitra, Agricultural Biotechnology Research Center, Academia Sinica, Taiwan, ROC; Molecular and Biological Agricultural Sciences Program, TIGP, Academia Sinica, Taiwan, ROC; Graduate Institute of Biotechnology, National Chung-Hsing University, Taichung, Taiwan, ROC.
Shih-Jiun Yu, Agricultural Biotechnology Research Center, Academia Sinica, Taiwan, ROC; Department of Biochemical Sciences and Technology, National Taiwan University, Taipei, Taiwan, ROC.
Y.-y.C. wrote the article. S.M. and S.-J.Y. generated Table 1.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yee-yung Charng (yycharng@sinica.edu.tw).
References
- Acar M, Becskei A, van Oudenaarden A (2005) Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232 [DOI] [PubMed] [Google Scholar]
- Ahrens CW, Challis A, Byrne M, Leigh A, Nicotra AB, Tissue D, Rymer P (2021) Repeated extreme heatwaves result in higher leaf thermal tolerances and greater safety margins. New Phytologist 232:1212–1225 [DOI] [PubMed] [Google Scholar]
- Alon U (2007) Network motifs: theory and experimental approaches. Nat Rev Genet 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Alves de Freitas Guedes F, Nobres P, Ferreira DCR, Menezes-Silva PE, Ribeiro-Alves M, Correa RL, DaMatta FM, Alves-Ferreira M (2018) Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ Exp Bot 147: 220–233 [Google Scholar]
- Auge GA, Leverett LD, Edwards BR, Donohue K (2017) Adjusting phenotypes via within- and across-generational plasticity. New Phytol 216: 343–349 [DOI] [PubMed] [Google Scholar]
- Avramova Z (2015) Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J 83: 149–159 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Schmelz EA (1996) Immunological ‘memory’ in the induced accumulation of nicotine in wild tobacco. Ecology 77: 236–246 [Google Scholar]
- Berry S, Dean C (2015) Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J 83: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner A, Hause B, Baier M (2021) Cold-priming causes dampening of oxylipin biosynthesis and signalling during the early cold- and light-triggering response of Arabidopsis thaliana. J Exp Bot 72: 7163–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D (1995) Protein molecules as computational elements in living cells. Nature 376: 307–312 [DOI] [PubMed] [Google Scholar]
- Bruce TJA, Matthes MC, Napier JAA, Pickett JA (2007) Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173: 603–608 [Google Scholar]
- Brzezinka K, Altmann S, Baeurle I (2019) BRUSHY1/TONSOKU/MGOUN3 is required for heat stress memory. Plant Cell Environ 42: 771–781 [DOI] [PubMed] [Google Scholar]
- Brzezinka K, Altmann S, Czesnick HA, Nicolas P, Gorka M, Benke E, Kabelitz T, Jaehne F, Graf A, Kappel C, Baeurle I (2016) Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLIFE 5: e17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J, D’Ari R (2002) Memory in bacteria and phage. BioEssays 24: 512–518 [DOI] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J-K (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53: 97–119 [DOI] [PubMed] [Google Scholar]
- Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ (2016) Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv 2: e1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Ganguly DR, Smith AB, Murray KD, Estavillo GM, Searle I, Ford E, Bogdanović O, Lister R, Borevitz JO, et al. (2017) Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29: 1836–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Q, Li Y, Li S, Shi A, Zhou M, Ren H, Yan Y, He C, Wang J, Sun M, et al. (2022) Photosynthesis mediated by RBOH-dependent signaling is essential for cold stress memory. Antioxidants 11: 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K-J (2015) Efficient high light acclimation involves rapid processes at multiple mechanistic levels. J Exp Bot 66: 2401–2414 [DOI] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z (2012) Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun 3: 740. [DOI] [PubMed] [Google Scholar]
- Ding Y, Liu N, Virlouvet L, Riethoven J-J, Fromm M, Avramova Z (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol 13: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Virlouvet L, Liu N, Riethoven J-J, Fromm M, Avramova Z (2014) Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol 14: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral MN, Arge LWP, Auler PA, Rossatto T, Milech C, Magalhães AMD, Braga EJB (2020a) Long-term transcriptional memory in rice plants submitted to salt shock. Planta 251: 1–16 [DOI] [PubMed] [Google Scholar]
- do Amaral MN, Auler PA, Rossatto T, Barros PM, Oliveira MM, Braga EJB (2020b) Long-term somatic memory of salinity unveiled from physiological, biochemical and epigenetic responses in two contrasting rice genotypes. Physiol Plantarum 170: 248–268 [DOI] [PubMed] [Google Scholar]
- D’Urso A, Brickner JH (2017) Epigenetic transcriptional memory. Curr Genet 63: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XJ, Li JR, Qi SL, Lin QFA, Jin JB, Hua XJ (2016) Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc Natl Acad Sci USA 113: E8335–E8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T, Oberkofler V, Trindade I, Altmann S, Brzezinka K, Lämke J, Gorka M, Kappel C, Sokolowska E, Skirycz A, et al. (2021) Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat Commun 12: 3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliano M, Renton M, Depczynski M, Mancuso S (2014) Experience teaches plants to learn faster and forget slower in environments where it matters. Oecologia 175: 63–72 [DOI] [PubMed] [Google Scholar]
- Galviz YCF, Ribeiro RV, Souza GM (2020) Yes, plants do have memory. Theor Exp Plant Physiol 32: 195–202 [Google Scholar]
- Gorecka M, Lewandowska M, Dabrowska-Bronk J, Bialasek M, Barczak-Brzyzek A, Kulasek M, Mielecki J, Kozlowska-Makulska A, Gawronski P, Karpinski S (2020) Photosystem II 22kDa protein level - a prerequisite for excess light-inducible memory, cross-tolerance to UV-C and regulation of electrical signalling. Plant Cell Environ 43: 649–661 [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Kokke BPA, Härndahl U, Silow M, Bechtold U, Poghosyan Z, Murphy D, Boelens WC, Sundby C (2002) A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. Plant J 29: 545–553 [DOI] [PubMed] [Google Scholar]
- Harb A, Krishnan A, Ambavaram MMR, Pereira A (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154: 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härndahl U, Hall RB, Osteryoung KW, Vierling E, Bornman JF, Sundby C (1999) The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperones 4: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Kandel ER, Siegelbaum SA (1993) Learning to modulate transmitter release: themes and variations in synaptic plasticity. Annu Rev Neurosci 16: 625–665 [DOI] [PubMed] [Google Scholar]
- Hilker M, Schwachtje J, Baier M, Balazadeh S, Baeurle I, Geiselhardt S, Hincha DK, Kunze R, Mueller-Roeber B, Rillig MG, et al. (2016) Priming and memory of stress responses in organisms lacking a nervous system. Biol Rev 91: 1118–1133 [DOI] [PubMed] [Google Scholar]
- Hoedjes KM, Kruidhof HM, Huigens ME, Dicke M, Vet LEM, Smid HM (2011) Natural variation in learning rate and memory dynamics in parasitoid wasps: opportunities for converging ecology and neuroscience. Proc R Soc B Biol Sci 278: 889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-W, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Cai J, Park SJ, Lee K, Li Y, Chen Y, Yun J-Y, Xu T, Kang H (2021) N6-Methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J 106: 1759–1775 [DOI] [PubMed] [Google Scholar]
- Hu T, Jin Y, Li H, Amombo E, Fu J (2016) Stress memory induced transcriptional and metabolic changes of perennial ryegrass (Lolium perenne) in response to salt stress. Physiol Plant 156: 54–69 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Hao N (2021) Memorizing environmental signals through feedback and feedforward loops. Curr Opin Cell Biol 69: 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-H, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585: 256–260 [DOI] [PubMed] [Google Scholar]
- Kalir S, Mangan S, Alon U (2005) A coherent feed-forward loop with a SUM input function prolongs flagella expression in Escherichia coli. Mol Syst Biol 1: 2005.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Grover A (2003) Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol 51: 677–686 [DOI] [PubMed] [Google Scholar]
- Kim Y-K, Chae S, Oh N-I, Nguyen NH, Cheong J-J (2020) Recurrent drought conditions enhance the induction of drought stress memory genes in Glycine max L. Front Genet 11: 576086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Seki M (2014) Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol 55: 1859–1863 [DOI] [PubMed] [Google Scholar]
- Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63: 1593–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämke J, Brzezinka K, Altmann S, Bäurle I (2016) A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J 35: 162–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämke J, Bäurle I (2017) Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol 18: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzoni G, Knight MR (2019) Increases in absolute temperature stimulate free calcium concentration elevations in the chloroplast. Plant Cell Physiol 60: 538–548 [DOI] [PubMed] [Google Scholar]
- Leonetti AM, Chu MY, Ramnaraign FO, Holm S, Walters BJ (2020) An emerging role of m6A in memory: a case for translational priming. Int J Mol Sci 21: 7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuendorf JE, Frank M, Schmülling T (2020) Acclimation, priming and memory in the response of Arabidopsis thaliana seedlings to cold stress. Sci Rep 10: 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Yang H, Wang L, Liu H, Huo HA, Zhang C, Liu A, Zhu A, Hu J, Lin Y, et al. (2019) Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front Genet 10: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wei X, Tong X, Zhao J, Liu X, Wang H, Tang L, Shu Y, Li G, Wang Y, et al. (2022) The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol Plant 15: 706–722 [DOI] [PubMed] [Google Scholar]
- Lim JP, Brunet A (2013) Bridging the transgenerational gap with epigenetic memory. Trends Genet 29: 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MY, Chai KH, Ko SS, Kuang LY, Lur HS, Charng YY (2014) A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol 164: 2045–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N,, Ding Y,, Fromm M,, Avramova Z (2014) Different gene-specific mechanisms determine the ‘revised-response' memory transcription patterns of a subset of A. thaliana dehydration stress responding genes. Nucleic Acids Res 42: 5556–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Lämke J, Lin SY, Hung MJ, Liu KM, Charng YY, Bäurle I (2018) Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J 95: 401–413 [DOI] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Liu HC, Charng YY (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163: 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-G, Han X, Yang T, Cui W-H, Wu A-M, Fu C-X, Wang B-C, Liu L-J (2019a) Genome-wide transcriptional adaptation to salt stress in Populus. BMC Plant Biol 19: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Feng L, Gu X, Deng X, Qiu Q, Li Q, Zhang Y, Wang M, Deng Y, Wang E, et al. (2019b) An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res 29: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Staswick PE, Avramova Z (2016) Memory responses of jasmonic acid‐associated Arabidopsis genes to a repeated dehydration stress. Plant Cell Environ 39: 2515–2529 [DOI] [PubMed] [Google Scholar]
- Liu NY, Ko SS, Yeh KC, Charng YY (2006) Isolation and characterization of tomato Hsa32 encoding a novel heat-shock protein. Plant Sci 170: 976–985 [Google Scholar]
- Lukić N, Kukavica B, Davidović-Plavšić B, Hasanagić D, Walter J (2020) Plant stress memory is linked to high levels of anti-oxidative enzymes over several weeks. Environ Exp Bot 178: 104166 [Google Scholar]
- Martin L, Leblanc-Fournier N, Julien J-L, Moulia B, Coutand C (2010) Acclimation kinetics of physiological and molecular responses of plants to multiple mechanical loadings. J Exp Bot 61: 2403–2412 [DOI] [PubMed] [Google Scholar]
- Martinez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CMJ, Pozo MJ, Ton J, van Dam NM, Conrath U (2016) Recognizing plant defense priming. Trends Plant Sci 21: 818–822 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Baccelli I, Luna E, Flors V (2017) Defense priming: an adaptive part of induced resistance. Annu Rev Plant Biol 68: 485–512 [DOI] [PubMed] [Google Scholar]
- Mayer BF, Charron J-B (2021) Transcriptional memories mediate the plasticity of cold stress responses to enable morphological acclimation in Brachypodium distachyon. New Phytol 229: 1615–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Shapiro L (1995) Circuit simulation of genetic networks. Science 269: 650–656 [DOI] [PubMed] [Google Scholar]
- Meiri D, Breiman A (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59: 387–399 [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298: 824–827 [DOI] [PubMed] [Google Scholar]
- Müller-Xing R, Xing Q, Goodrich J (2014) Footprints of the sun: memory of UV and light stress in plants. Front Plant Sci 5: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia G, Nieto C, Sellaro R, Prat S, Casal JJ (2022) Hysteresis in PHYTOCHROME INTERACTING FACTOR 4 and EARLY-FLOWERING 3 dynamics dominates warm daytime memory in Arabidopsis. Plant Cell 34: 2188–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences E, and Medicine (2016) Attribution of extreme weather events in the context of climate change. National Academies Press, Washington [Google Scholar]
- Nechaev S, Adelman K (2011) Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberkofler V, Bäurle I (2022) Inducible epigenome editing probes for the role of histone H3K4 methylation in Arabidopsis heat stress memory. Plant Physiol 189: 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberkofler V, Pratx L, Bäurle I (2021) Epigenetic regulation of abiotic stress memory: maintaining the good things while they last. Curr Opin Plant Biol Epigenet 61: 102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A, Scheid OM (2012) Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol 53: 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomiès L, Decourteix M, Franchel J, Moulia B, Leblanc-Fournier N (2017) Poplar stem transcriptome is massively remodelled in response to single or repeated mechanical stimuli. BMC Genom 18: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong S-W, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani E, Herzyk P, Perrella G, Colot V, Amtmann A (2013) Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genom Biol 14: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze WX, Altenbuchinger M, He M, Kränzlein M, Zörb C (2021) Proteome profiling of repeated drought stress reveals genotype-specific responses and memory effects in maize. Plant Physiol Biochem 159: 67–79 [DOI] [PubMed] [Google Scholar]
- Sedaghatmehr M, Mueller-Roeber B, Balazadeh S (2016) The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat Commun 7: 12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghatmehr M, Thirumalaikumar VP, Kamranfar I, Marmagne A, Masclaux-Daubresse C, Balazadeh S (2019) A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ 42: 1054–1064 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park M-J, Lim M-H, Kim S-G, Lee M, Baldwin IT, Park C-M (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Banday ZZ, Shukla BN, Laxmi A (2019) Glucose-regulated HLP1 acts as a key molecule in governing thermomemory. Plant Physiol 180: 1081–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhawat K, Saad MM, Sheikh A, Mariappan K, Al-Mahmoudi H, Abdulhakim F, Eida AA, Jalal R, Masmoudi K, Hirt H (2021) Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep 22: e51049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2005) Prions as adaptive conduits of memory and inheritance. Nat Rev Genet 6: 435–450 [DOI] [PubMed] [Google Scholar]
- Si K, Lindquist S, Kandel ER (2003) A neuronal isoform of the Aplysia CPEB Has prion-like properties. Cell 115: 879–891 [DOI] [PubMed] [Google Scholar]
- Song Z-T, Zhang L-L, Han J-J, Zhou M, Liu J-X (2021) Histone H3K4 methyltransferases SDG25 and ATX1 maintain heat stress gene expression during recovery in Arabidopsis. Plant J 105: 1326–1338 [DOI] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant B, Datt Scheible W-R, Bäurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R-Z, Liu J, Wang Y-Y, Deng X (2021) DNA methylation-mediated modulation of rapid desiccation tolerance acquisition and dehydration stress memory in the resurrection plant Boea hygrometrica. PLOS Genet 17: e1009549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaker HM, Darkó É, Medzihradszky A, Janda T, Liu H-c, Charng Y-y, Csorba T (2019) miR824/AGAMOUS-LIKE16 module integrates recurring environmental heat stress changes to fine-tune poststress development. Front Plant Sci 10: 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Zeiger E, Møller IM, Murphy A (2015) Plant physiology and development. 6th edition. Sinauer Associates Incorporated, Sunderland, USA
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Tian L, Zhang Y, Kang E, Ma H, Zhao H, Yuan M, Zhu L, Fu Y (2019) Basic-leucine zipper 17 and Hmg-CoA reductase degradation 3A are involved in salt acclimation memory in Arabidopsis. J Integr Plant Biol 61: 1062–1084 [DOI] [PubMed] [Google Scholar]
- Trewavas A (2003) Aspects of plant intelligence. Ann Bot 92: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrea Castellanos R, Friedrich T, Petrovic N, Altmann S, Brzezinka K, Gorka M, Graf A, Bäurle I (2020) FORGETTER2 protein phosphatase and phospholipase D modulate heat stress memory in Arabidopsis. Plant J 104: 7–17 [DOI] [PubMed] [Google Scholar]
- Verweij W, Spelt CE, Bliek M, de Vries M, Wit N, Faraco M, Koes R, Quattrocchio FM (2016) Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in Arabidopsis. Plant Cell 28: 786–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlouvet L, Avenson TJ, Du Q, Zhang C, Liu N, Fromm M, Avramova Z, Russo SE (2018) Dehydration stress memory: gene networks linked to physiological responses during repeated stresses of Zea mays. Front Plant Sci 9: 1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlouvet L, Ding Y, Fujii H, Avramova Z, Fromm M (2014) ABA signaling is necessary but not sufficient for RD29B transcriptional memory during successive dehydration stresses in Arabidopsis thaliana. Plant J 79: 150–161 [DOI] [PubMed] [Google Scholar]
- Virlouvet L, Fromm M (2015) Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol 205: 596–607 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Walter J, Jentsch A, Beierkuhnlein C, Kreyling J (2013) Ecological stress memory and cross stress tolerance in plants in the face of climate extremes. Environ Exp Bot 94: 3–8 [Google Scholar]
- Weigel D, Colot V (2012) Epialleles in plant evolution. Genome Biol 13: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Juan YT, Hsu YH, Wu SH, Liao HT, Fung RW, Charng YY (2013) Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol 161: 2075–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y-J, Xu S, Han B, Wu M-Z, Yuan X-X, Han Y, Gu Q, Xu D-K, Yang Q, Shen W-B (2011) Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J 66: 280–292 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Xing Q, Müller-Xing R (2021) A novel UV-B priming system reveals an UVR8-depedent memory, which provides resistance against UV-B stress in Arabidopsis leaves. Plant Signal Behav 16: 1879533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Matsubara S, Yoshimizu K, Seki M, Hamada K, Kamitani M, Kurita Y, Nomura Y, Nagashima K, Inagaki S, et al. (2021) H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat Commun 12: 3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C-H, Kaplinsky NJ, Hu C, Charng Y-y (2012) Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci 195: 10–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim J-M, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genom 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Peng X, Guo X, Tang G, Sun F, Liu S, Xi Y (2018) Transcriptional and physiological data reveal the dehydration memory behavior in switchgrass (Panicum virgatum L.). Biotechnol Biofuels 11: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H (2012) MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53: 368–380 [DOI] [PubMed] [Google Scholar]
- Zuther E, Schaarschmidt S, Fischer A, Erban A, Pagter M, Mubeen U, Giavalisco P, Kopka J, Sprenger H, Hincha DK (2019) Molecular signatures associated with increased freezing tolerance due to low temperature memory in Arabidopsis. Plant Cell Environ 42: 854–873 [DOI] [PubMed] [Google Scholar]


