Abstract
Leaves of shade-avoiding plants such as Arabidopsis (Arabidopsis thaliana) change their growth pattern and position in response to low red to far-red ratios (LRFRs) encountered in dense plant communities. Under LRFR, transcription factors of the phytochrome-interacting factor (PIF) family are derepressed. PIFs induce auxin production, which is required for promoting leaf hyponasty, thereby favoring access to unfiltered sunlight. Abscisic acid (ABA) has also been implicated in the control of leaf hyponasty, with gene expression patterns suggesting that LRFR regulates the ABA response. Here, we show that LRFR leads to a rapid increase in ABA levels in leaves. Changes in ABA levels depend on PIFs, which regulate the expression of genes encoding isoforms of the enzyme catalyzing a rate-limiting step in ABA biosynthesis. Interestingly, ABA biosynthesis and signaling mutants have more erect leaves than wild-type Arabidopsis under white light but respond less to LRFR. Consistent with this, ABA application decreases leaf angle under white light; however, this response is inhibited under LRFR. Tissue-specific interference with ABA signaling indicates that an ABA response is required in different cell types for LRFR-induced hyponasty. Collectively, our data indicate that LRFR triggers rapid PIF-mediated ABA production. ABA plays a different role in controlling hyponasty under white light than under LRFR. Moreover, ABA exerts its activity in multiple cell types to control leaf position.
Abscisic acid biosynthesis and signaling modulate leaf hyponasty in response to a drop in the red to far-red ratio.
Introduction
Leaf movements are controlled by the interplay of circadian and light signals (Hopkins et al., 2008; McClung, 2013; Dornbusch et al., 2014). The physiological importance of these movements is not fully understood, however, leaf position influences both photosynthesis and water loss (Hopkins et al., 2008). In open environments, light reaches intensities higher than the photosynthesis saturation point. Having more erect leaves diminishes light interception and favors cooling which is proposed to be beneficial (Hopkins et al., 2008). Consistent with this idea, Arabidopsis (Arabidopsis thaliana) accessions from lower latitudes have more erect leaf angles (Hopkins et al., 2008). Experimental evidence supports the idea that clock-controlled movements in resonance with the day/night cycle help Arabidopsis to overtop the leaves of neighbors (Woodley Of Menie et al., 2019). In addition, several environmental cues including flooding, high temperature, and shade impact on such diel leaf movements by triggering a rapid upwards leaf positioning (hyponasty; Voesenek et al., 2006; Casal and Balasubramanian, 2019). This movement is thought to help leaves acclimate to such unfavorable conditions for example by enhancing cooling capacity in warm environments (Voesenek et al., 2006; Crawford et al., 2012; Casal and Balasubramanian, 2019).
At the molecular level the mechanisms underlying hyponasty are probably best understood in response to a decrease of the red to far-red ratio (low red/far-red, abbreviated LRFR here). LRFR occurs in dense vegetational communities and is a modification of the light environment experienced by plants prior to actual shading (Casal, 2013; Ballare and Pierik, 2017). Leaf hyponasty is induced by different elements of canopy shade including both low light and LRFR (Mullen et al., 2006; Millenaar et al., 2009; Moreno et al., 2009; Casal, 2013; Ballare and Pierik, 2017). The primary photoreceptor controlling LRFR-regulated responses is phytochrome B (phyB; Casal, 2013; Ballare and Pierik, 2017). LRFR inactivates phyB leading to the derepression of several phytochrome-interacting factors (PIFs), particularly PIF4, PIF5, and PIF7 (Casal, 2013; Ballare and Pierik, 2017; Cheng et al., 2021). These PIFs promote shade-induced elongation of the hypocotyl, leaf petioles, and leaf hyponasty (Keller et al., 2011; de Wit et al., 2016; Kohnen et al., 2016; Michaud et al., 2017; Pantazopoulou et al., 2017). phyB inactivation at the leaf rim leads to PIF-mediated auxin production in the blade, followed by auxin transport and redistribution in the petiole where it promotes upwards leaf repositioning (Michaud et al., 2017; Pantazopoulou et al., 2017; Gao et al., 2020).
Beside auxin, other hormones also contribute to environmentally-controlled leaf repositioning in Arabidopsis. Ethylene is particularly important to promote upwards leaf repositioning in response to waterlogging (Millenaar et al., 2009; Rauf et al., 2013). In contrast, ethylene is not required for low light-induced hyponasty and it negatively regulates high temperature-induced hyponasty (Millenaar et al., 2009; van Zanten et al., 2009). The role of abscisic acid (ABA) in controlling leaf position is also complex and poorly understood. ABA inhibits ethylene-induced hyponasty and in white-light grown plants genetic and pharmacological experiments indicate that ABA promotes downwards leaf repositioning (Mullen et al., 2006; Benschop et al., 2007). In contrast, ABA can also positively regulate hyponasty when plants are treated with an elevated temperature (van Zanten et al., 2009). Access to water is essential for growth and reversible cellular expansion, suggesting that ABA might be an important regulator of leaf positioning (Tardieu et al., 2015; Yoshida et al., 2019). Indeed, differential petiole growth is a key contributor to upward repositioning of the leaf (Polko et al., 2012; Rauf et al., 2013). Intriguingly, shade treatments lead to higher ABA levels in tomato (Solanum lycopersicum) and based on the LRFR-induced gene expression pattern this may also be the case in Arabidopsis (Cagnola et al., 2012; Kohnen et al., 2016; Fiorucci et al., 2022). We found that LRFR leads to a rapid increase in ABA content in leaves and therefore decided to study how LRFR controls leaf ABA levels and how ABA controls hyponasty.
Results
In LRFR, PIFs enhance NCED expression and ABA production
To determine whether LRFR alters the expression of ABA biosynthesis and signaling genes in petioles, we analyzed genome-wide expression data from a recent publication (Fiorucci et al., 2022). We found that the expression of two 9-cis-epoxycarotenoid dioxygenase (NCED) genes was rapidly induced by LRFR compared with that under normal growth (high red to far-red ratio) conditions (Supplemental Figure S1). The expression of ABA signaling genes also showed an interesting pattern but changes in gene expression were delayed compared to ABA biosynthesis genes (Supplemental Figure S1). The expression of several negative regulators of the pathway from the ABI1 protein phosphatases family of ABA co-receptors was weakly induced, while we observed reduced expression of several members of the positively acting RCAR/PYR1/PYL family and SnRK2 kinases (Supplemental Figure S1). Collectively this gene expression data suggests that LRFR leads to an induction of ABA levels that is followed by reduced ABA signaling.
To test these predictions, we first determined whether LRFR rapidly changes ABA levels in Arabidopsis rosettes. 14-d-old-plants grown in long days (LDs, 16 h light, 8 h darkness), where either kept in standard conditions (white light, WL) or transferred to LRFR (WL supplemented with FR light) at ZT3. Rosettes were collected at the time of transfer to LRFR (ZT3) and 2 h later (ZT5) and ABA was quantified. This experiment showed that a 2-h LRFR treatment led to a significant increase in ABA content (Figure 1A). Since ABA levels fluctuate during the day (Lee et al., 2006; Adams et al., 2018), we also tested the effect of the LRFR treatment later in the day and found that ABA levels increase both in WL and LRFR, but at each time point there was more ABA in LRFR than in WL (Supplemental Figure S2A). LRFR led to higher ABA levels both in entire rosettes and in dissected Leaves 1 and 2, which we used to analyze the hyponastic response (Supplemental Figure S2B).
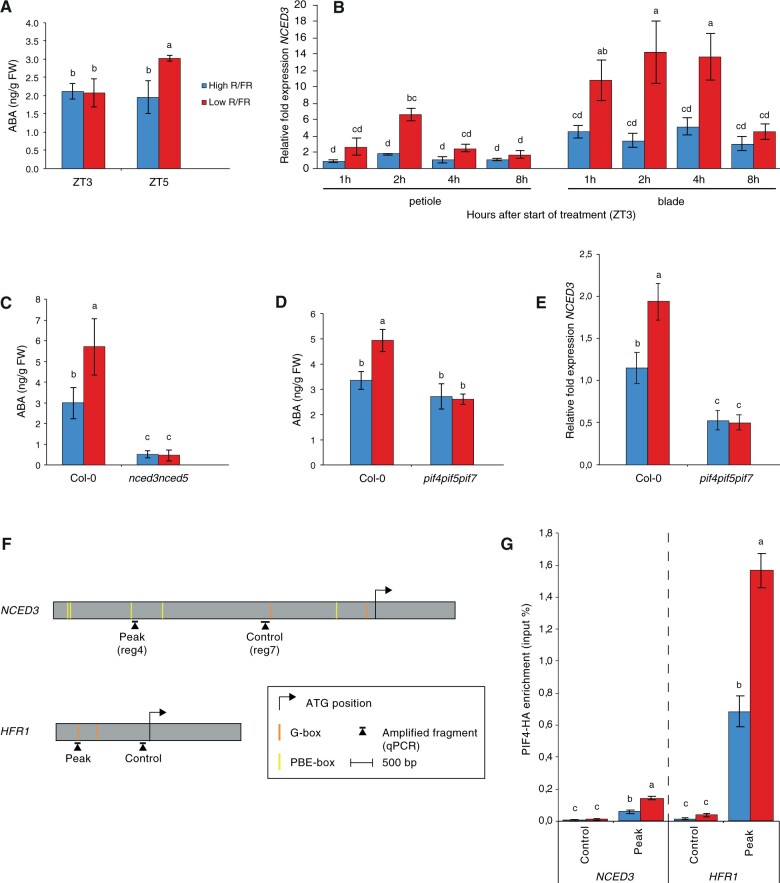
Figure 1.
LRFR induces PIF-mediated ABA biosynthesis in leaves. A, ABA concentration in entire Leaves 1 and 2 of wild-type (Col-0) in high R/FR (blue) versus low R/FR (red) conditions at 0 h (ZT3) and 2 h (ZT5) after start of treatment. Plants were grown for 14 d in standard LD (16-h light, 8-h dark [16/8]) conditions. ZT0 corresponds to the beginning of the light period on Day 15. Shade treatment started on Day 15 at ZT3 by adding FR light to decrease the R/FR ratio. Each bar plot represents data from four biological replicates. Per replicate, 40 leaves (Leaves 1 and 2) from 20 plant individuals were harvested and frozen in liquid nitrogen. B, Relative fold expression of NCED3 from leaf 3 of Col-0 plants in high R/FR (blue) versus low R/FR (red) conditions over time. Gene expression values were calculated as fold induction relative to a petiole sample at time=1 h (ZT4) in high R/FR conditions. Plants were grown for 15 d in standard LD (16-h light, 8-h dark [16/8]) conditions. ZT0 corresponds to the beginning of the light period on Day 16. Shade treatment started on Day 16 at ZT3 by adding FR light to decrease the R/FR ratio. Petioles and lamina of Leaf 3 were separately pooled into three biological replicates and frozen in liquid nitrogen. C, ABA concentration in rosettes of Col-0 versus nced3nced5 double mutant plants in high R/FR (blue) versus low R/FR (red) conditions at 2 h (ZT5) after start of treatment. Plants were grown for 14 d in standard LD (16-h light, 8-h dark [16/8]) conditions. ZT0 corresponds to the beginning of the light period on Day 15. Shade treatment started on Day 15 at ZT3 by adding FR light to decrease the R/FR ratio. Each bar plot represents data from four biological replicates. Per replicate, 15 and 20 entire rosettes of Col-0 and nced3nced5 plants, respectively, were harvested and frozen in liquid nitrogen. D, ABA concentration in rosettes of Col-0 and pif4pif5pif7 triple plants in high R/FR (blue) versus low R/FR (red) conditions at 2 h (ZT5) after start of treatment. Plants were grown for 14 d in standard LD (16-h light, 8-h dark [16/8]) conditions. ZT0 corresponds to the beginning of the light period on Day 15. Shade treatment started on Day 15 at ZT3 by adding FR light to decrease the R/FR ratio. Each bar plot represents data from four biological replicates. Per replicate, 15 entire rosettes were harvested and frozen in liquid nitrogen. E, Relative fold expression of NCED3 in Col-0 and pif4pif5pif7 from Leaves 1 and 2 in high R/FR (blue) versus LRFR (red) conditions. Gene expression values were calculated as fold induction relative to a sample in high R/FR conditions. Plants were grown for 14 d in standard LD (16-h light, 8-h dark [16/8]) conditions. ZT0 corresponds to the beginning of the light period on Day 15. Shade treatment started on Day 15 at ZT3 by adding FR light to decrease the R/FR ratio. At ZT5 (±2 h shade), entire Leaves 1 and 2 were separately pooled into three biological replicates and frozen in liquid nitrogen. F, Schematic representation of the NCED3 and HFR1 genes. Regions amplified by qPCR and relative positions of G- and PBE-boxes are depicted relative to the start codon. G, PIF4-HA binding to the NCED3 and HFR1 promoter regions. Input and immunoprecipitated DNA was extracted from 10-d-old PIF4::PIF4-HA(pif4-101) seedlings exposed to ±2 h of LRFR from ZT2 and quantified by qPCR. PIF4-HA enrichment is presented as IP/input. Bars represent the mean from three technical replicates. Statistical tests were performed for each locus separately. A–E and G, Error bars represent the 2-fold SE of mean estimates. Two-way ANOVAs followed by Tukey’s Honestly Significant Difference (HSD) test were performed and different letters were assigned to significantly different groups (P-value < 0.05).
The rate-limiting step in ABA biosynthesis is catalyzed by NCED enzymes (Qin and Zeevaart, 2002). LRFR triggers a rapid increase in NCED3 and NCED5 expression in the cotyledons of young seedlings and in the petiole of rosettes (Kohnen et al., 2016; Fiorucci et al., 2022). We tested whether this is the case both in the leaf blades and the petioles of young Arabidopsis rosettes treated with LRFR. NCED3 and NCED5 were both rapidly induced in the petiole, while in the leaf blade only NCED3 expression was induced (Figure 1B, Supplemental Figure S2C). Measuring ABA levels in 2-week-old Arabidopsis rosettes showed that in nced3nced5 double mutants ABA levels were strongly reduced and LRFR did not lead to enhanced ABA accumulation (Figure 1C). Given the prevalent role of PIF transcription factors for shade-induced transcriptional reprogramming (de Wit et al., 2016), in particular PIF4, PIF5 and PIF7, we compared ABA levels in the wild type (Col-0) and the pif4pif5pif7 triple mutant. This experiment showed that ABA levels did not increase in pif4pif5pif7 in response to LRFR (Figure 1D). To determine whether this is due to PIF-mediated NCED expression, we compared the rapid LRFR-induction of NCED3 in Col-0 and pif4pif5pif7. This experiment showed that in pif4pif5pif7 NCED3 expression was reduced in WL and the rapid LRFR-induction was absent (Figure 1E).
Genome-wide analysis of PIF4 binding sites identified several PIF4-binding peaks in the NCED3 promoter of low-blue light-grown seedlings (Pedmale et al., 2016). We designed a series of amplicons along the large NCED3 promoter and found enhanced PIF4 binding in LRFR-grown plants particularly in the region corresponding to Peaks 3 and 4 identified in (Pedmale et al., 2016; Supplemental Figure S2, D and E). To determine whether the rapid LRFR-induced NCED3 expression coincides with PIF4 binding, we performed ChIP experiments in plants either maintained in WL or transferred to LRFR for 2 h and used HFR1, a well-known PIF target gene, as a control (Figure 1F). Our experiments showed enhanced PIF4-HA binding following an LRFR treatment on the promoters of HFR1 and NCED3 (Figure 1G). Collectively these results indicate that PIF transcription factors directly control NCED3 expression in response to LRFR potentially explaining the observed increase in ABA content upon transfer to LRFR.
ABA biosynthesis and signaling are required for a normal LRFR-induced hyponastic response
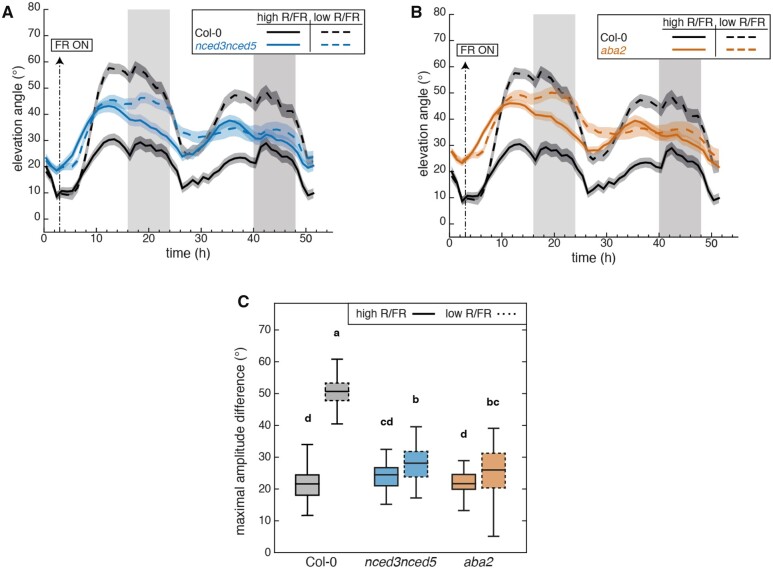
To determine the role of ABA biosynthesis during LRFR-induced hyponasty, we compared the hyponastic response of the wild type with the nced3nced5 double mutants over 2 d in plants either remaining in WL or being transferred to LRFR at ZT3 of Day 1. We focused on Leaves 1 and 2, which possess a similar hyponastic response compared to other leaves at the same developmental stage (Dornbusch et al., 2014; Michaud et al., 2017). nced3nced5 double mutants had more erect leaves than Col-0 throughout the experiment in WL (Figure 2A). However, this double mutant showed a limited response to an LRFR treatment (Figure 2A), while nced3 and nced5 single mutants had modest phenotypes (Supplemental Figure S3, A–C). We also tested the aba2 mutant which is known to have low ABA levels (Gonzalez-Guzman et al., 2002). The phenotype of aba2 was very similar to that of nced3nced5 (Figure 2, A and B), consistent with the importance of ABA for normal leaf positioning in WL and a robust hyponastic response to LRFR. We note that despite having more erect leaves, aba2 and nced3nced5 mutants displayed diel leaf movements with a normal amplitude in WL, while their ability to respond to LRFR was severely impaired (Figure 2C).
Figure 2.
Diel- and shade-induced hyponasties both require a functional ABA biosynthetic pathway. A, Leaf elevation angle of Leaves 1 and 2 in Col-0 (black) and nced3nced5 mutant (blue) plants in high R/FR (solid) versus low R/FR (dashed) conditions. Leaf elevation angles are mean values (n=44–58). B, Leaf elevation angle of Leaves 1 and 2 in Col-0 (black) and aba2 mutant (orange) plants in high R/FR (solid) versus low R/FR (dashed) conditions. Leaf elevation angles are mean values (n=44–60). A and B, Plants were grown for 14 d in standard LD (16/8) conditions. Imaging started on Day 15 at ZT0 (t=0), plants were maintained in LD. Shade treatment started at ZT3 by adding FR light to decrease the R/FR ratio. Col-0 plants analyzed in (A) and (B) are same. Opaque bands around mean lines represent the 95% confidence interval of mean estimates. Vertical gray bars represent night periods. C, Boxplots representing the amplitude of leaf movement between maximum and minimum leaf elevation angles over the time period from t=3 to t=16 and computed for each individual leaf analyzed in (A) and (B). The central mark indicates the median; the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively; and the whiskers extend to the most extreme data points. Solid and dashed plots represent data from high R/FR and low R/FR conditions, respectively. Two-way ANOVA followed by Tukey’s HSD test were performed and different letters were assigned to significantly different groups (P-value < 0.05).
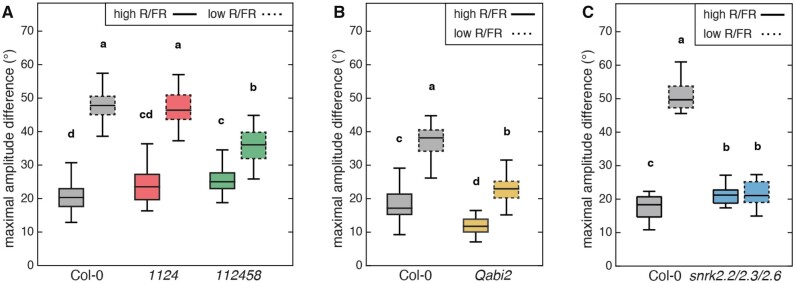
To further investigate the role of ABA in the modulation of leaf hyponasty we analyzed higher-order mutants lacking members of the major ABA receptor from the RCAR/PYR1/PYL family (Raghavendra et al., 2010; Weiner et al., 2010). While leaf movements in the quadruple mutant pyr1pyl1pyl2pyl4 were very similar to Col-0, the pyr1pyl1pyl2pyl4pyl5pyl8 septuple mutant had an interesting phenotype (Figure 3A, Supplemental Figure S4, A and B). As observed for ABA biosynthesis mutants, pyr1pyl1pyl2pyl4pyl5pyl8 had constitutively more erect leaves, a leaf movement amplitude in WL that was at least as robust as in Col-0 but a reduced response to LRFR (Figure 3A, Supplemental Figure S4, A and B). Higher-order mutants lacking protein phosphatases from the ABI1 family of ABA co-receptors (Raghavendra et al., 2010; Weiner et al., 2010) also showed altered leaf movements. This was most striking in the abi1abi2hab1pp2ca quadruple (Qabi2) mutant, which showed altered leaf movements both in WL and LRFR (Figure 3B, Supplemental Figure S4C). In contrast to the other mutants tested, Qabi2 also showed a strong increase in the leaf elevation angle at night (Supplemental Figure S4C). The hab1abi1pp2ca triple mutant had wild-type leaf movements in WL but a reduced response to LRFR (Supplemental Figure S4, D and E). We also analyzed mutants lacking the SnRK2 kinases acting directly downstream of the ABA receptor (Raghavendra et al., 2010; Weiner et al., 2010) and found that the snrk2.2snrk2.3snrk2.6 triple mutant showed a very severe phenotype (Figure 3C, Supplemental Figure S4F). This triple mutant showed constitutively erect leaves with no response to LRFR (Figure 3C, Supplemental Figure S4F). This phenotype was only observed in the triple mutant and not in the snrk2.6 or snrk2.2snrk2.3 mutants (Supplemental Figure S4G). A potential limitation of our studies with ABA biosynthesis and signaling mutants is that strong mutants have substantially smaller rosettes than the wild type (Supplemental Figure S5A). However, we found that growing such mutants in saturating humidity largely rescued this growth phenotype (Supplemental Figure S5, A and B). We then compared the leaf hyponasty phenotype in response to LRFR in saturating humidity of the WT and the snrk triple mutant previously grown in medium humidity (approx. 80%, snrk triple small) or snrk triple mutant grown in saturating humidity (snrk triple large; Supplemental Figure S5, C–E). The snrk triple mutant showed a reduced ability to increase its leaf angle in response to LRFR in both cases (Supplemental Figure S5, C–E). Collectively our data indicate that mutants affecting the positive regulators of ABA signaling (PYR1/PYL and Snrk2 kinases) have very similar phenotypes to ABA biosynthesis mutants with a reduced hyponasty amplitude in response to LRFR. In contrast, mutants affecting members of the ABI1 protein phosphatase family also alter leaf movements and the ability to respond to LRFR but the diel leaf movement patterns were different (Supplemental Figure S4).
Figure 3.
Functional ABA signaling is required for diel- and shade-induced hyponasties. A, Boxplots representing the amplitude of leaf movement between maximum and minimum leaf elevation angles over the time period from t=3 to t=16 of Col-0 (black), pyr1pyl1pyl2pyl4 quadruple mutant (1124, red) and pyr1pyl1pyl2pyl4pyl5pyl8 sextuple mutant (112458, green). Data were computed for each individual leaf analyzed in Supplemental Figure S4, A and B. Solid and dashed plots represent data from high R/FR and low R/FR conditions, respectively. B and C, Boxplots representing the amplitude of leaf movement between maximum and minimum leaf elevation angles over the time period from t=3 to t=16 (solid plots, high R/FR) or from t=27 to t=40 (dashed plots, low R/FR) for Col-0 (black), abi1abi2hab1pp2ca mutant (B, brown, Qabi2) or snrk2.2snrk2.3snrk2.6 mutant (C, blue) plants and computed for each individual leaf analyzed in Supplemental Figure S4C (B) or in Supplemental Figure S4F (C). The central mark indicates the median; the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively; and the whiskers extend to the most extreme data points. A–C, Two-way ANOVA followed by Tukey’s HSD test were performed and different letters were assigned to significantly different groups (P-value < 0.05).
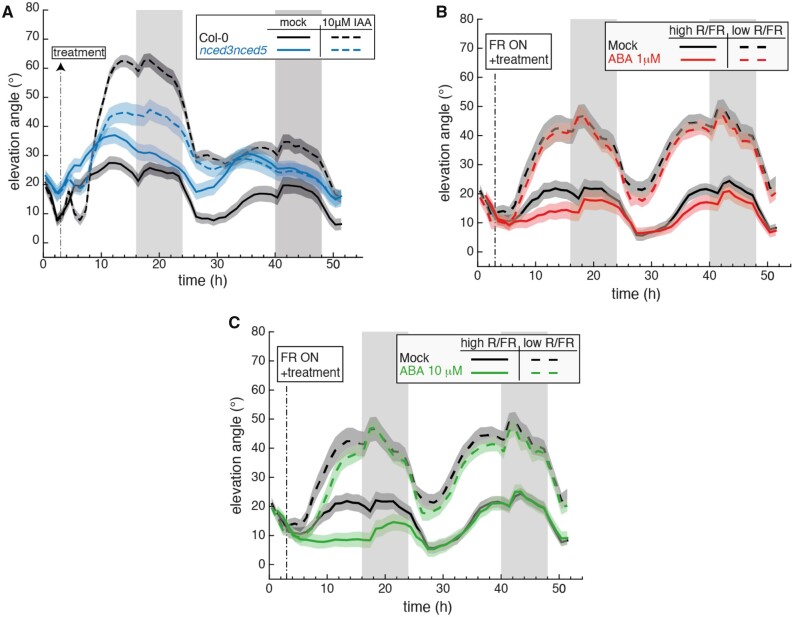
Exogenous auxin (Indole-3-acetic acid, IAA) application to the leaf tip leads to a similar leaf hyponastic response as transferring plants into LRFR (Michaud et al., 2017; Pantazopoulou et al., 2017). We therefore tested whether ABA biosynthesis mutants can respond to auxin application. These experiments showed that nced3nced5 and aba2 mutants showed modest responses to IAA application (Figure 4A, Supplemental Figure S6, A and B). The IAA and LRFR responses of these mutants were very similar (Figures 2–4, Supplemental Figure S6, A and B) suggesting that normal ABA levels are required to respond to the LRFR-induced IAA production required to induce hyponasty. Collectively our data indicate that ABA and ABA signaling modulate diel leaf hyponastic movement and are required for a robust LRFR-induced hyponastic response acting downstream of IAA production.
Figure 4.
Hormone treatments reveal dysfunctional auxin-induced hyponasty in ABA mutants and reduced sensitivity to ABA in LRFR. A, Leaf elevation angle of Leaves 1 and 2 in Col-0 (black) and nced3nced5 mutant (blue) plants treated with mock solution (solid lines) or 10 μM IAA (dashed lines). At ZT3 on Day 15 (t=3) a 1-μL drop of the corresponding solution was applied to the leaf tip (adaxial side). Leaf elevation angles are mean values (n=22–30). B and C, Leaf elevation angle of Leaves 1 and 2 in Col-0 plants treated with mock (black), 1 μM ABA (B, red), and 10 μM ABA (C, green) solutions in high R/FR (solid) versus low R/FR (dashed) conditions. Leaf elevation angles are mean values (n=26–30). At ZT3 on Day 15 (t=3), the corresponding solution was sprayed on the entire leaves (adaxial side). Shade treatment started at t=3 by adding FR light to decrease the R/FR ratio. Col-0 plants analyzed in (B) and (C) are same. A–C, Plants were grown for 14 d in standard LD (16/8) conditions. Imaging started on Day 15 at ZT0 (t=0), plants were maintained in LD. Opaque bands around mean lines represent the 95% confidence interval of mean estimates. Vertical gray bars represent night periods.
To determine whether ABA application alters leaf hyponasty we sprayed rosettes with different concentrations of ABA and followed diel leaf movements over 2 d either in WL or LRFR. Consistent with the constitutively high leaf position of WL-grown ABA biosynthesis mutants (Figure 2), ABA application reduced the leaf angle during the first day after the application in WL in a dose dependent manner (compare Figure 4, B and C). The effect of ABA application was transient as leaf movements of treated and untreated plants were very similar the second day (Figure 4, B and C). Interestingly, in LRFR the effect of exogenous ABA application was largely canceled (Figure 4, B and C). We observed no substantial effect following a treatment with 1 µM ABA and a delay in the hyponastic response when we applied 10 µM ABA (Figure 4, B and C). Our data show that ABA application inhibits leaf elevation happening during diel leaf movements in WL while in LRFR the effect of exogenous ABA was largely abolished suggesting a reduced sensitivity to ABA in shade-mimicking conditions. This change in ABA sensitivity might be due to the LRFR-induced modifications in expression of the early components of ABA sensing and response (Supplemental Figure S1).
ABA signaling in multiple tissues is required to control hyponasty in LRFR
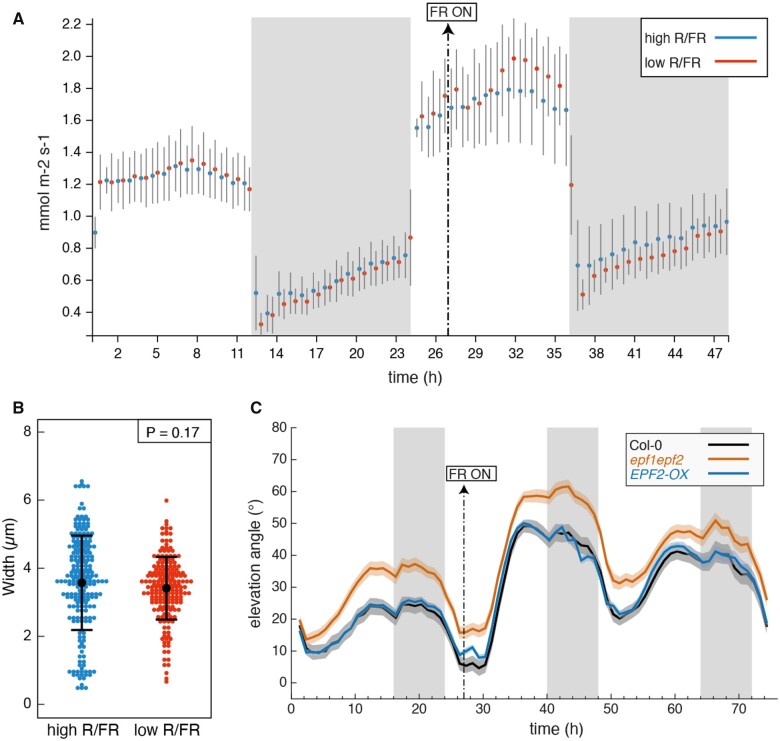
Given the established importance of ABA in controlling stomata aperture, we first tested whether neighbor-proximity induced changes in leaf transpiration. Young rosettes were transferred from WL to LRFR at ZT3 and a slight increase in transpiration was observed late in the day but the difference between WL and LRFR was not significant (t tests with P-value > 0.05; Figure 5A). We also compared stomata aperture at ZT8 from plants that either remained in WL or were transferred to LRFR at ZT3 and observed no change (Figure 5B). In contrast, treating plants with ABA or comparing plants during the day with plants that remained in darkness (night extension) validated our stomata opening measuring method as both treatments led to previously documented stomatal closure (Supplemental Figure S7, A and B). To determine the importance of stomata for diel leaf movements and the response to LRFR we used epf1epf2 double mutants and EPF2 over-expressing (EPF2-OX) plants which maintain a normal stomata morphology but have respectively higher and lower stomata density consistent with higher and lower transpiration (Hepworth et al., 2015). While the epf1epf2 double mutant had more erect leaves in WL, it showed a robust response to LRFR (Figure 5C, Supplemental Figure S7C). Leaf movements in EPF2-OX plants were similar but not identical to Col-0 (Figure 5C, Supplemental Figure S7C). Collectively, these data suggest that LRFR-regulated changes in transpiration and stomata aperture are not a major control point for LRFR-induced leaf hyponasty.
Figure 5.
Transpiration and stomata opening during LRFR-induced hyponasty. A, Transpiration of the aerial parts of 21-d-old Col-0 plants grown in hydroponics system in equinoctial (12:12) conditions, measured by infrared gas analysis (illumination at 150 μmol m−2 s−1, 20°C, 60% relative humidity, 380 ppm CO2). Mean values from three and four biological replicates (±SD) are given for high R/FR and low R/FR treatments, respectively. Vertical gray bars represent night periods. Supplementation with FR light 3 h into the second day is indicated. B, Stomatal pore width in high and low R/FR at ZT8 (low R/FR from ZT3), from dental paste imprints of 2-week-old plants grown in LD. P-value is given by a t test without assumption of equal variance. The smaller dots represent individual measurements. The big dot in the middle is the mean. Error bars represent SD. C, Leaf elevation angle of Leaves 1 and 2 in Col-0 (black), epf1epf2 double mutant (orange), EPF2-OX mutant (blue) plants in high R/FR then low R/FR conditions. Shade treatment started on Day 16 at t=27 (ZT3) by adding FR light to decrease the R/FR ratio. Leaf elevation angles are mean values (n=29–60). Plants were grown for 14 d in standard LD (16/8) conditions. Imaging started on Day 15 at ZT0 (t=0), plants were maintained in LD. Opaque bands around mean lines represent the 95% confidence interval of mean estimates. Vertical gray bars represent night periods.
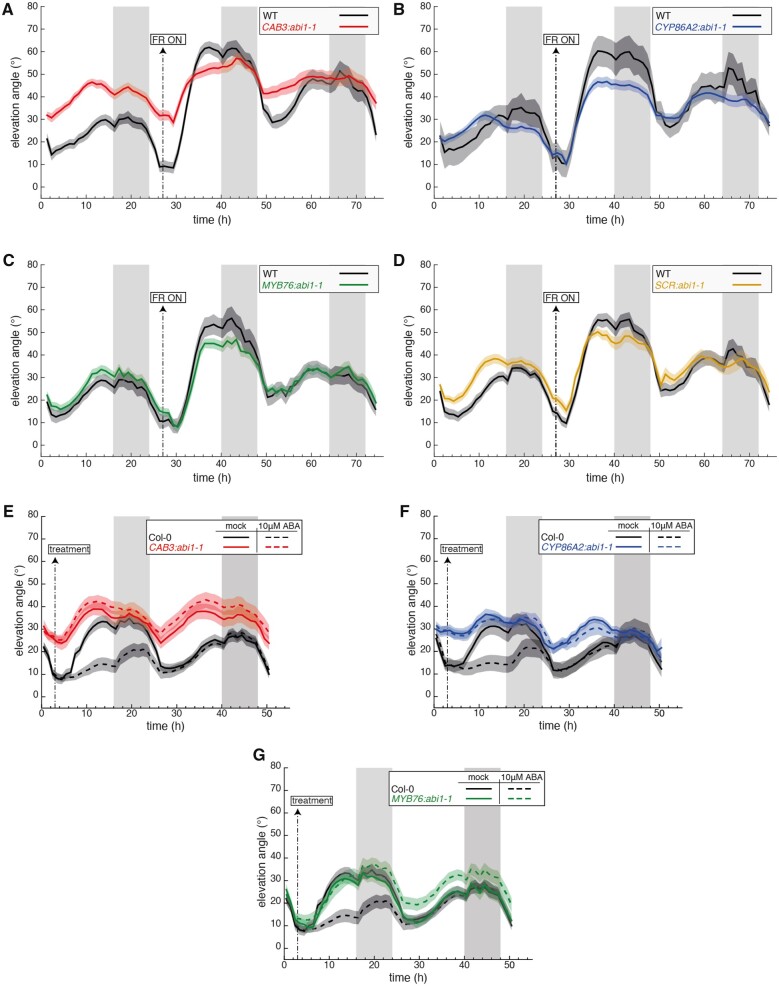
The use of auxin reporter genes was instrumental to define the role of IAA in LRFR-induced leaf hyponasty (Michaud et al., 2017; Pantazopoulou et al., 2017). We attempted an analogous approach using ABA reporter genes encouraged by the finding that RD29B and MAPKKK18 expression, two previously characterized ABA response genes (Christmann et al., 2005; Okamoto et al., 2013), was slightly induced in the petiole of LRFR-treated rosettes (Supplemental Figure S1). While RD29B and MAPKKK18 GUS reporter lines showed clear induction in response to exogenous ABA application, we observed no change in response to LRFR, thereby limiting the potential of this approach (Supplemental Figure S8). We therefore decided to disrupt ABA signaling in different cell types to better understand the role of ABA in LRFR-regulated hyponasty. We generated lines expressing the dominant negative allele abi1-1 in stomata (CYP86A2 promoter), mesophyll (CAB3 promoter) and bundle sheath cells (SCR and MYB76 promoters; Dickinson et al., 2020). We compared the leaf movements of these plants over 1 LD in WL and then transferred them into LRFR at ZT3 on Day 2 and continued imaging for 48 h. All the lines had a reduced capacity to elevate their leaves in response to LRFR (Figure 6, A–D, Supplemental Figure S9, A–D). The leaf movement pattern of the previously described pCOR:abi1-1:RFP line (Duan et al., 2013) was similar to our pCAB3:abi1-1 lines (Figure 6A, Supplemental Figure S9E), consistent with a role of ABA in mesophyll cells to control leaf movements. We also tested the ability of these lines to respond to exogenously applied ABA and found that disrupting ABA signaling in the stomata, mesophyll or bundle sheath cells largely prevented ABA-induced leaf repositioning (Figure 6, E–G, Supplemental Figure S9, F and G). Collectively, our results indicate that different cell types are required for shade-regulated leaf hyponasty and for ABA-induced downward positioning of leaves grown in WL.
Figure 6.
ABA signaling in multiple tissues modulates the hyponastic response. A–D, Leaf elevation angle of Leaves 1 and 2 in Col-0 (A-D, black), CAB3::abi1-1 mutant (A, red, line #MT36-11), pCYP86A2::abi1-1 mutant (B, blue, line #MT39-03), pMYB76::abi1-1 mutant (C, green, line #MT41-13), pSCR::abi1-1 mutant (D, orange, line #MT37-24) plants in high R/FR then low R/FR conditions. Shade treatment started on Day 16 at t=27 (ZT3) by adding FR light to decrease the R/FR ratio. Leaf elevation angles are mean values (A, n=18–24; B, n=6–30; C, n=16–28; D, n=16–26). E–G, Leaf elevation angle of Leaves 1 and 2 in Col-0 (E-G, black), pCAB3::abi1-1 mutant (E, red, line #MT36-11), pCYP86A2::abi1-1 mutant (F, blue, line #MT39-03), pMYB76::abi1-1 mutant (G, green, line #MT41-13) plants sprayed with mock solution (solid lines) or 10 μM ABA (dashed lines). At ZT3 on Day 15 (t=3) mock or ABA solutions were sprayed on the entire rosette (adaxial side). Col-0 plants analyzed in (E) and (G) are same. Leaf elevation angles are mean values (E, n=26–28; F, n=25–28; G, n=21–27). A–G, Plants were grown for 14 d in standard LD (16/8) conditions. Imaging started on Day 15 at ZT0 (t=0), plants were maintained in LD. Opaque bands around mean lines represent the 95% confidence interval of mean estimates. Vertical gray bars represent night periods.
Discussion
We showed that upon transfer to LRFR ABA content rapidly increases in leaves (Figure 1, Supplemental Figure S2). Higher ABA levels were also reported in shade treated tomato and in Arabidopsis phyB mutants, which display a constitutive shade avoidance response (Cagnola et al., 2012; Gonzalez et al., 2012). Moreover, LRFR leads to increased ABA levels in Arabidopsis buds to prevent lateral shoot development (Gonzalez-Grandio et al., 2013; Holalu and Finlayson, 2017). In buds PIFs rapidly induce BRANCHED1 expression, which was proposed to enhance NCED3 expression, thereby increasing ABA production (Gonzalez-Grandio et al., 2013; Holalu et al., 2020). In contrast to most previous studies, we looked at a rapid effect of LRFR and detect an increase in NCED3 and NCED5 expression within 1–2 h and higher ABA levels within 2 h of the shade-simulating treatment (Figure 1). LRFR promotion of NCED3 expression and ABA levels depend on PIF4, PIF5, and/or PIF7 and LRFR-induced ABA levels is lost in the nced3nced5 double mutant (Figure 1). Consistent with a direct role of PIFs in controlling ABA biosynthesis, we find increased PIF4 binding on the NCED3 promoter in response to LRFR (Figure 1, Supplemental Figure S1). Collectively, our data indicate that in response to LRFR PIFs enhance ABA levels in leaves. In response to LRFR PIFs also mediate rapid IAA production, which, upon asymmetric redistribution in the petiole, controls leaf repositioning (Michaud et al., 2017; Pantazopoulou et al., 2017). Based on IAA application experiments it appears that ABA acts downstream of IAA (Figure 4, Supplemental Figure S4). ABA plays a major role in the regulation of water movements (Tardieu et al., 2015; Yoshida et al., 2019). The response to LRFR or temperature elevation triggers rapid IAA production (Quint et al., 2016; Casal and Questa, 2018), which promotes growth and repositioning of leaves (van Zanten et al., 2009; Michaud et al., 2017; Pantazopoulou et al., 2017). Whether these responses are due to reversible or irreversible cellular expansion (growth) is not fully understood but in both cases sufficient water supply is required. We propose that in this context, PIF-regulated ABA levels contributes to a normal hyponastic response.
The role of ABA in controlling plant growth and morphology is context-dependent and best understood in response to stress. Various abiotic stresses (e.g. drought, salinity) enhance ABA biosynthesis, which limits gas exchange and growth to promote tolerance to unfavorable environments (Yoshida et al., 2019; Kinoshita et al., 2021). For example, when a shade treatment occurs in the presence of high sodium chloride, hypocotyl elongation is inhibited in an ABA-dependent manner (Hayes et al., 2019). The relationship between ABA and growth in non-stressful conditions is complex and ABA acts both as an activator and a suppressor of growth (Yoshida et al., 2019). One study on the role of ABA in LRFR-induced hypocotyl elongation concluded that ABA rather prevents growth (Ortiz-Alcaide et al., 2019). Similarly, the role of ABA in leaf positioning also appears to be complex. In standard light conditions application of ABA prevents hyponasty, while decreasing ABA levels genetically of pharmacologically leads to plants with more erect leaves (Mullen et al., 2006; Benschop et al., 2007; Figures 2 and 4). In contrast, during high temperature-induced hyponasty ABA was proposed to promote leaf hyponasty (van Zanten et al., 2009). Similarly, our data indicate that in unstressed conditions (WL), ABA limits leaf hyponasty (Figures 2–4). We note that having more erect leaves in WL largely is accompanied by high transpiration. This is true for mutants affecting the PYR/PYL ABA receptors (Gonzalez-Guzman et al., 2012), SnRK2 protein kinases (Fujii and Zhu, 2009), ABA biosynthesis mutants (Merlot et al., 2002) or epf1epf2 mutants with more stomata (Hepworth et al., 2015; Figures 3 and 5). Moreover, PP2C ABA-co-receptor mutants have lower amplitude leaf oscillations during the day in WL (Figure 3B) and reduced transpiration (Rubio et al., 2009). Collectively this suggests that in WL, transpiration, which is controlled by ABA, contributes to leaf position. The role of ABA appears to change during the LRFR response, as ABA biosynthesis and the response are required for full leaf repositioning, suggesting that in LRFR ABA promotes hyponasty (Figures 2 and 3). The different involvement of ABA in controlling hyponasty in high versus low R/FR is also illustrated by the strongly reduced response to applied ABA in LRFR-grown plants (Figure 4). Similarly, when ABA was applied to heat-treated plants to trigger hyponasty, it did not inhibit upwards repositioning of leaves (van Zanten et al., 2009). Intriguingly, wounding also transiently inhibits hyponasty in WL but not in low R/FR, although this is a Jasmonic Acid (JA) response (Fiorucci et al., 2022). Collectively, these experiments show a context-dependent role of ABA (and JA) in controlling leaf hyponasty. In standard growth conditions low ABA levels or signaling leads to a higher minimal leaf angle with a normal or slightly higher amplitude. In contrast, in response to stresses (warm temperature or shade cues) ABA is required for a strong hyponastic response.
One difficulty in studying the role of ABA in controlling leaf hyponasty is the strong phenotype of ABA biosynthesis or signaling mutants. We found that growing snrk triple mutants in saturating humidity largely rescued the growth phenotype without rescuing the ability of the mutant to fully erect its leaves in response to LRFR (Supplemental Figure S5). This indicates that the growth defects of this mutant cannot account for its hyponastic defect. Moreover, the high stomata density mutant epf1epf2 which, like ABA biosynthesis or ABA response mutants, has smaller rosettes and more erect leaves in WL, was able to fully respond to LRFR (Figure 5). This suggests that the inability to respond to LRFR in ABA biosynthesis or signaling mutants is not due to their constitutively erect leaves. Moreover, it suggests that having a normal stomata response despite a high or a low transpiration baseline (epf1epf2 and EPF2-OX respectively; Hepworth et al., 2015), does not prevent LRFR-induced hyponasty (Figure 5). We conclude that despite the aforementioned limitations of our genetic experiments ABA biosynthesis and response are required for a robust LRFR-induced hyponastic response.
The mechanisms underlying the control of hyponasty by ABA remain poorly understood. In day-night conditions, stomata open rapidly at dawn leading to CO2 fixation and enhanced transpiration. However, leaves only start to move upwards several hours later when leaves grow fast (Dornbusch et al., 2014). Hence, stomata opening at dawn does not trigger a rapid leaf elevation in WL conditions. When transferred to LRFR, we did not observe stomatal opening or substantial changes in transpiration accompanied by leaf elevation (although the data indicate a tendency for slightly enhanced transpiration; Figure 5, Supplemental Figure S7). However, based on ABA application experiments (Figure 4; Mullen et al., 2006; Benschop et al., 2007) it is reasonable to conclude that regulated stomata opening is required for leaf hyponasty. This is also supported by our finding that abi1-1 expression from a stomata-specific promoter alters diel leaf movements (Figure 6, Supplemental Figure S9). Tissue-specific abi1-1 expression indicates that ABA signaling is required in multiple tissues for normal leaf positioning (Figure 6, Supplemental Figure S9). By controlling transpiration, stomata are a well-known control point of water movements. However, water fluxes are also controlled in other cells including bundle sheath and mesophyll (Negin et al., 2019; Grunwald et al., 2021). In LDs, leaf hydraulic conductivity peaks before mid-day (Prado et al., 2019). This pattern in hydraulic conductivity does not directly relate to leaf position in LDs (see e.g. Figure 2). In WL, all the abi1-1 expressing lines were largely unresponsive to ABA application indicating that ABA signaling is required in stomata, mesophyll and bundle sheath to control leaf hyponasty (Supplemental Figure S9). Expression of abi1-1 in mesophyll and stomata clearly altered leaf position in WL, while this was not obvious in the bundle sheath expressing lines (Figure 6, Supplemental Figure S9). However, the hyponastic response in plants transferred to LRFR was reduced in all abi1-1 expressing lines indicating that ABA signaling is required in stomata, mesophyll, and bundle sheath for shade-induced leaf hyponasty (Figure 6, Supplemental Figure S9). This might be due to altered regulation of the leaf water potential as ABA inhibits H+ ATPases in bundle sheath cells similar to what happens in stomata (Grunwald et al., 2021). It is noteworthy that LRFR leads to IAA production and a strong auxin response (Michaud et al., 2017; Pantazopoulou et al., 2017). While ABA inhibits H+ ATPases, auxin activates these proton pumps (Miao et al., 2022). Hence, we hypothesize that in LRFR the correct balance between IAA and ABA-regulated water movements is required to control leaf positioning. Another striking feature of the LRFR response is the reduced sensitivity to applied ABA (Figure 4). Reduced ABA sensitivity in LRFR might be due to changes in gene expression. Indeed, in petioles and cotyledons multiple members of the positively acting PYR/PYL and SNRK2 genes are downregulated, while some of the negatively acting PP2C ABA co-receptors genes are upregulated (Kohnen et al., 2016; Supplemental Figure S1). In petioles, these changes might be triggered by the rapid induction of NCED gene expression and ABA biosynthesis, which precede strong regulation of ABA sensing and response genes (Figure 1, Supplemental Figures S1 and S2). A similar gene expression signature was observed in high light which also triggers ABA production (Huang et al., 2019). That said, high light leads to 4–5 times higher ABA levels typical of a stress response (Huang et al., 2019), while LRFR only led to a 30% increase that we could not detect using typical ABA response marker lines (Figure 1, Supplemental Figure S8). We propose that the mechanisms by which ABA controls leaf position depends on the co-occurrence of an IAA response that is induced both by LRFR or following an increased temperature. However, the cellular mechanisms by which ABA controls environmentally regulated leaf positioning require future studies.
Materials and methods
Plant material and plasmid construction
All Arabidopsis (A. thaliana) lines used in this study are in the Col-0 background, nced3nced5, nced3, nced5 (Frey et al., 2012), aba2 (Leon-Kloosterziel et al., 1996), pyr1pyl1pyl2pyl4 (Park et al., 2009), pyr1pyl1pyl2pyl4pyl5pyl8 (Gonzalez-Guzman et al., 2012), hab1abi1pp2ca (Rubio et al., 2009), hab1abi1abi2pp2ca (named Qabi2; Antoni et al., 2013), snrk2.2snrk2.3, snrk2.6, snrk2.2snrk2.3snrk2.6 (Fujii and Zhu, 2009), pif4pif5pif7 (de Wit et al., 2015), yuc2yuc5yuc8yuc9 (Nozue et al., 2015), pPIF4::PIF4-3HA (pif4-101; Zhang et al., 2017), epf1epf2, EPF2-OX (Hepworth et al., 2015), pCOR:abi1-1:RFP (Duan et al., 2013), pRD29B:GUS (Christmann et al., 2005), pMAPKKK18:GUS (Okamoto et al., 2013) were previously described.
All constructs were obtained with the In-Fusion HD cloning kit (Takara) and sequence verified. pCAB3:abi1-1-citrine (pMT36) was obtained by replacing the PHOT1 coding sequence from pCAB3:PHOT1-citrine (Preuten et al., 2013) with abi1-1 which was amplified with oligonucleotides MT49 and MT50 using plasmid pEN-1-abi1-1-2 as a template (Barberon et al., 2016). pSCR:abi1-1citrine (pMT37) was obtained by replacing the PHOT1 coding sequence from pSCR:PHOT1-citrine (Preuten et al., 2013) with abi1-1 which was amplified with oligonucleotides MT49 and MT51 using plasmid pEN-1-abi1-1-2 as a template (Barberon et al., 2016). pCYP86A2:abi1-1citrine (pMT39) was obtained by replacing pSCR promoter from pMT37 with the CYP86A2 promoter which was amplified with oligonucleotides MT57 and MT58 using plasmid proCYP86A2:GFP-PYR1MANDI as a template (Park et al., 2015). pMYB76:abi1-1citrine (pMT41, 2xDH-35S promoter) was obtained by replacing pSCR promoter from pMT37 with the 2xDH-35S promoter which was amplified with oligonucleotides MT61 and MT62 using plasmid pMYB76-2xDH-35Smin:GUS as a template (Dickinson et al., 2020). Transgenic plants were generated by introduction of the plant expression constructs in an Agrobacterium tumefaciens strain GV3101/2. Transformation of A. thaliana Columbia (Col-0) accession was done by floral dipping. Plasmids contained Basta resistance for plant selection. Oligonucleotides sequences are listed in Supplemental Table S1.
Growth conditions and phenotyping analyses
Seeds were stratified at 4°C for 3 d in darkness and then sown on soil saturated with deionized water in a Percival CU-36L4 incubator (Percival Scientific) at 21°C, 85% relative humidity, and PAR =175 μmol·m−2·s−1 under LD (16:8) conditions. After 13 d plants were transferred to the ScanAlyzer HTS (LemnaTec) for acclimation (with day–night cycles and light conditions as in the incubator) 24 h before scanning. Experiments were performed under LD conditions (16-h day/8-h night). To grow and image plants in saturating humidity pots were placed in clear plexiglass boxes. For shade treatments, the R/FR was decreased from 4.2 to 0.2 using FR-emitting diodes positioned on the ceiling of the ScanAlyzer HTS. Further experimental details, spectral composition of light, computation of the R/FR ratio, and technical specifications of the phenotyping device are described in detail in Dornbusch et al. (2012). For gene expression experiments in Figure 1B and Supplemental Figure S1C, Col-0 plants were grown as described in de Wit et al. (2015). In brief, seeds were directly sown on soil and stratified at 4°C for 3 d in darkness. Plants were then grown for 14 d at 20°C, 70% relative humidity, and PAR =220 μmol·m−2·s−1 under LD (16:8) conditions. Afterwards, plants were divided over two Percival I-66L incubators (Percival Scientific) at PAR =130 μmol·m−2·s−1 24 h before the start of the experiment. Experiment was performed the following day (Day 16). At ZT3 on Day 16, R/FR was decreased in one of the incubators from 1.4 to 0.2 using FR-emitting diodes positioned on the ceiling.
Pharmacological treatments
IAA (10 µM; Sigma-Aldrich) and (±)-ABA (1–10 µM; Sigma-Aldrich) solutions were freshly prepared from concentrated dimethylsulfoxide (DMSO) and ethanol (EtOH) stocks, respectively, before each application. Mock solutions were similarly prepared to contain 0.15% (v/v) Tween-20, 0.1% (v/v) DMSO and 0.1% (v/v) EtOH, respectively.
Hormone quantification
ABA measurements were performed as previously described in Glauser et al. (2014). In brief, fresh frozen samples were ground to a fine powder using mortars and pestles under liquid nitrogen and about 40 mg of powder was weighed in 2.0 mL Eppendorf tubes. To the tubes were added 5–6 glass beads (2 mm diameter), 990 μL of extraction solvent (ethyl acetate/formic acid, 99.5: 0.5, v/v), and 10 μL of internal standard solution containing D6-ABA at 100 ng/mL. The tubes were shaken for 4 min at 30 Hz in a tissue lyser, centrifuged at 14,000 g at room temperature for 3 min, the supernatant was recovered and the pellet re-extracted with 500 μL of extraction solvent. Both solutions were then combined, evaporated, and reconstituted in 100 μL of methanol 70% (v/v). The final extracts were analyzed by UHPLC-MS/MS using an Ultimate 3000 RSLC (Thermo Scientific Dionex) coupled with a 4,000 QTRAP (AB Sciex). To quantify ABA in plant samples, a calibration curve based on calibration points at 0.2, 2, 10, 50, and 200 ng/mL, all containing D6-ABA at a fixed concentration of 10 ng/mL, and weighted by 1/x was used (x refers to the concentration of the corresponding calibration point).
Stomata opening
Stomatal aperture was measured in Leaves 1 and 2 of 2-week-old rosettes at ZT8, either in high R/FR or after 5 h in LRFR, using dental paste imprints. We applied dental paste (Take 1 Advanced Light Body Wash, Kerr™, 34149) to the lower leaf epidermis. After solidification, imprints were removed from the leaf and covered with nail polish (60 Seconds Super Shine 740 Clear, Rimmel, France) which was left to dry and peeled off for visualization by light microscopy. For quantification, ellipses were fit into the opening in ImageJ and the length of the minor axis was used as a measure of stomatal aperture.
Leaf transpiration
Col-0 plants were grown for 21 d in hydroponics system in equinoctial (12:12) conditions at PAR = 150 µmol photons m−2 s−1, 20°C and 60% relative humidity. Growth medium contained 1.5 mM KNO3, 0.75 mM Ca(NO3)2, 1 mM MgSO4, 1 mM KH2PO4, 0.5 mM K2SO4, 0.35 mM CaCl2, 12.25 µM MnSO4, 61.25 µM H3BO3, 875 nM ZnSO4, 437.5 nM CuSO4 175 nM (NH4)6Mo7O24 and 9 mg/L iron-EDTA (adapted from Orsel et al., 2004). Plants were mounted into custom-built gas exchange chambers (Kolling et al., 2015). Three plants were measured in a growth cabinet compartment with a high red to far-red light ratio, i.e. normal growth conditions. Four plants were measured in a growth cabinet compartment with far-red light supplementation by FR-emitting diodes (λ = 740 nm). Far-red supplementation was started 3 h after onset of light on the second day. Gas exchange of the aerial parts of the plants was measured continuously for 48 h at 380 ppm CO2 using an infrared gas analyzer (LI-7000 CO2/H2O Gas Analyzer, LI-COR Biosciences GmbH). After measurement, leaf area size was extracted from photographs of individual plants and used to normalize the transpiration data. Data were processed, analyzed, and plotted using custom-built software (George et al., 2018).
Analysis of leaf position
For time-lapse experiments, plants were scanned at intervals of 60 min with the ScanAlyzer HTS (LemnaTec). As output, we obtained time-lapse images in which the distance of measured plant surface points from a reference plane was color-coded. These images were then transformed into 3D point clouds that yield a precise representation of plant surfaces over time as previously described in Dornbusch et al. (2012). Leaf elevation angle (tip elevation angle) was delineated by the vector taking as origin the position of the basal end of the petiole organ and as extremity the position of the tip of the blade organ. A detailed description of the geometric definitions of leaf elevation angle (ϕtip) as well as image and data processing are available in Dornbusch et al. (2012, 2014).
Analysis of leaf length
Leaf length (tip length) was determined by the vector taking as origin the position of the basal end of the petiole organ and as extremity the position of the tip of the blade organ. A detailed description of the geometric definitions of tip length (ltip) as well as image and data processing are available in Dornbusch et al. (2014).
RT-qPCR
RNA extraction, cDNA reverse transcription and RT-qPCR were performed as previously described (de Wit et al., 2015). In brief, entire Leaves 1 and 2 (Figure 1E) or petioles/lamina of Leaves 3 (Figure 1B, Supplemental Figure S2C, samples from de Wit et al., 2015) were separately pooled into three biological replicates and frozen in liquid nitrogen. After consecutive RNA extraction and reverse transcription, RT-qPCR was performed in three technical replicates for each sample using QuantStudio 6 Flex Real-Time PCR sequence detection system (Applied Biosystems) and FastStart Universal SYBR green Master mix (Roche). Data were normalized against two reference genes (YELLOW-LEAF-SPECIFIC GENE 8 [YLS8] and UBIQUITIN-CONJUGATING ENZYME 21 [UBC21]) using the Biogazelle qbase software. Gene-specific oligonucleotides used for RT-qPCR reactions are listed in Supplemental Table S1.
ChIP-qPCR
Seedlings were harvested and cross-linked as described in (Bourbousse et al., 2012). Subsequent steps of chromatin immune-precipitation were performed as described in (Fiorucci et al., 2020) using an anti-HA antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA; sc-7392 X). For the long-term LRFR treatment (Supplementary Figure S2) 4-d-old LD grown p35S-PIF4-3XHA (Lorrain et al., 2008) seedlings were either kept in the same conditions for an additional 3 d or transferred for 3 d into LRFR before harvesting. For the short-term LRFR treatment (Figure 1), 10-d-old pPIF4p::PIF4-3XHA (in pif4-101) seedlings were grown in LDs and either kept in high R/FR or shifted at ZT2 to LRFR for 2 h before harvesting in liquid nitrogen. Oligonucleotides used for ChIP-qPCR reactions are listed in Supplemental Table S1.
GUS histochemical staining
For ABA reporter GUS staining, samples were grown in the same conditions as in the laser scanner experiments. On Day 15, plants were subjected to one of four different treatments at ZT3: mock spray (0.015% [v/v] Tween, 0.1% DMSO), ABA spray (10 µM), high R/FR, or low R/FR. At ZT8.5, plants were harvested into ice cold 90% (v/v) acetone and vacuum infiltrated. Acetone was washed off twice with 50 mM sodium phosphate buffer pH7.2 and plants were vacuum infiltrated twice with staining solution (50 mM sodium phosphate pH 7.2, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 0.1% (v/v) TritonX-100, 2 mM X-GlucA). After overnight incubation in staining solution at 37°C, samples were cleared with 70% (v/v) EtOH at 4°C three times over 24 h and imaged on a stereomicroscope with a Leica DFC7000 T camera.
Accession numbers
The Arabidopsis Genome Initiative numbers for the genes mentioned in this article are as follows: AT2G18790 (PHYB), AT2G43010 (PIF4), AT3G59060 (PIF5), AT5G61270 (PIF7), AT3G14440 (NCED3), AT1G30100 (NCED5), AT1G52340 (ABA2), AT4G17870 (PYR1), AT5G46790 (PYL1), AT2G26040 (PYL2), AT2G38310 (PYL4), AT5G05440 (PYL5), AT5G53160 (PYL8), AT4G26080 (ABI1), AT5G57050 (ABI2), AT1G72770 (HAB1), AT3G11410 (PP2CA), AT3G50500 (SNRK2.2), AT5G66880 (SNRK2.3), AT4G33950 (SNRK2.6), AT2G20875 (EPF1), AT1G34245 (EPF2), AT4G00360 (CYP86A2), AT1G29910 (CAB3), AT3G54220 (SCR), AT5G07700 (MYB76), AT5G52300 (RD29B), AT1G05100 (MAPKKK18).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcriptional regulation of ABA biosynthetic and signaling genes in leaf petioles by LRFR.
Supplemental Figure S2. Detailed analysis of LRFR-induced and PIF-mediated ABA biosynthesis in leaves.
Supplemental Figure S3. Diel- and shade-induced hyponasties in nced single mutants.
Supplemental Figure S4. Functional ABA signaling is required for diel- and shade-induced hyponasties.
Supplemental Figure S5. ABA effects on LRFR-induced leaf hyponasty are independent of leaf size.
Supplemental Figure S6. Detailed analysis of auxin-induced hyponasty in ABA biosynthetic mutants.
Supplemental Figure S7. Validation of stomatal measurement methodology.
Supplemental Figure S8. ABA response in leaves treated with ABA versus LRFR.
Supplemental Figure S9. Dysfunctional ABA signaling in internal leaf tissues affects light-modulated hyponastic responses.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Gaetan Glauser (Neuchâtel Platform of Analytical Chemistry) for determining ABA content. We thank Annie Marion-Poll (Université Paris-Saclay), Hiroaki Fujii (University of Turku), Julian Schroeder (UCSD), Julie Gray (University of Sheffield), Pedro Rodriguez (IBMCP), José Dinneny (Stanford), Sean Cutler (UC Riverside), Diana Santelia (ETH Zurich), Luis Lopez-Molina (University of Geneva), and Niko Geldner (University of Lausanne) for seeds or plasmids.
Funding
This work was supported by the University of Lausanne, ETH Zurich, the Swiss National Science Foundation (grant no. 310030B_179558 and 310030_200318 to C.F.), A Velux Foundation Grant (Project 1455 to C.F. and J.K.), ERC (RG80867 Revolution to J.M.H.), the BBSRC (BBP0031171 to J.M.H.), and a Derek Brewer Ph.D. studentship to J.K.
Conflict of interest statement. None declared.
Contributor Information
Olivier Michaud, Faculty of Biology and Medicine, Centre for Integrative Genomics, University of Lausanne, Génopode Building, Lausanne CH-1015, Switzerland.
Johanna Krahmer, Faculty of Biology and Medicine, Centre for Integrative Genomics, University of Lausanne, Génopode Building, Lausanne CH-1015, Switzerland.
Florian Galbier, Plant Biochemistry, Department of Biology, ETH Zürich, Universität-Str. 2, CH-8092 Zürich, Switzerland.
Maud Lagier, Faculty of Biology and Medicine, Centre for Integrative Genomics, University of Lausanne, Génopode Building, Lausanne CH-1015, Switzerland.
Vinicius Costa Galvão, Faculty of Biology and Medicine, Centre for Integrative Genomics, University of Lausanne, Génopode Building, Lausanne CH-1015, Switzerland.
Yetkin Çaka Ince, Faculty of Biology and Medicine, Centre for Integrative Genomics, University of Lausanne, Génopode Building, Lausanne CH-1015, Switzerland.
Martine Trevisan, Faculty of Biology and Medicine, Centre for Integrative Genomics, University of Lausanne, Génopode Building, Lausanne CH-1015, Switzerland.
Jana Knerova, Department of Plant Sciences, Downing Street, Cambridge, University of Cambridge, CB2 3EA, UK.
Patrick Dickinson, Department of Plant Sciences, Downing Street, Cambridge, University of Cambridge, CB2 3EA, UK.
Julian M Hibberd, Department of Plant Sciences, Downing Street, Cambridge, University of Cambridge, CB2 3EA, UK.
Samuel C Zeeman, Plant Biochemistry, Department of Biology, ETH Zürich, Universität-Str. 2, CH-8092 Zürich, Switzerland.
Christian Fankhauser, Faculty of Biology and Medicine, Centre for Integrative Genomics, University of Lausanne, Génopode Building, Lausanne CH-1015, Switzerland.
O.M. and C.F conceived the original research plan; O.M., Jo.K., F.G., M.L., Y.C.I., V.C.G., S.C.Z., and C.F. conceived experiments; O.M., Jo.K., F.G., M.L., M.T., Y.C.I., V.C.G., S.C.Z., and C.F. performed experiments and/or analyzed the data; Ja.K., P.D., and J.M.H. provided unpublished material; C.F. wrote the article with contributions of all the authors; C.F. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Christian Fankhauser (Christian.fankhauser@unil.ch).
References
- Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S, Carre IA (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol 220: 893–907 [DOI] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, et al. (2013) PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol 161: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare CL, Pierik R (2017) The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ 40: 2530–2543 [DOI] [PubMed] [Google Scholar]
- Barberon M, Vermeer JE, De Bellis D, Wang P, Naseer S, Andersen TG, Humbel BM, Nawrath C, Takano J, Salt DE, et al. (2016) Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164: 447–459 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Millenaar FF, Smeets ME, van Zanten M, Voesenek LA, Peeters AJ (2007) Abscisic acid antagonizes ethylene-induced hyponastic growth in Arabidopsis. Plant Physiol 143: 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F (2012) Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet 8:e1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnola JI, Ploschuk E, Benech-Arnold T, Finlayson SA, Casal JJ (2012) Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol 160: 1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S (2019) Thermomorphogenesis. Annu Rev Plant Biol 70: 321–346 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Questa JI (2018) Light and temperature cues: multitasking receptors and transcriptional integrators. New Phytol 217: 1029–1034 [DOI] [PubMed] [Google Scholar]
- Cheng MC, Kathare PK, Paik I, Huq E (2021) Phytochrome signaling networks. Annu Rev Plant Biol 72: 217–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann A, Hoffmann T, Teplova I, Grill E, Muller A (2005) Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol 137: 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AJ, McLachlan DH, Hetherington AM, Franklin KA (2012) High temperature exposure increases plant cooling capacity. Curr Biol 22:R396–R–397. [DOI] [PubMed] [Google Scholar]
- de Wit M, Keuskamp DH, Bongers FJ, Hornitschek P, Gommers CMM, Reinen E, Martinez-Ceron C, Fankhauser C, Pierik R (2016) Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr Biol 26: 3320–3326 [DOI] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C (2015) Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol 208: 198–209 [DOI] [PubMed] [Google Scholar]
- Dickinson PJ, Knerova J, Szecowka M, Stevenson SR, Burgess SJ, Mulvey H, Bagman AM, Gaudinier A, Brady SM, Hibberd JM (2020) A bipartite transcription factor module controlling expression in the bundle sheath of Arabidopsis thaliana. Nat Plants 6: 1468–1479 [DOI] [PubMed] [Google Scholar]
- Dornbusch T, Lorrain SV, Kuznetsov D, Fortier A, Liechti R, Xenarios I, Fankhauser C (2012) Measuring the diurnal pattern of leaf hyponasty and growth in Arabidopsis – a novel phenotyping approach using laser scanning. Funct Plant Biol 39: 860–869 [DOI] [PubMed] [Google Scholar]
- Dornbusch T, Michaud O, Xenarios I, Fankhauser C (2014) Differentially phased leaf growth and movements in Arabidopsis depend on coordinated circadian and light regulation. Plant Cell 26: 3911–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PM, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci AS, Galvao VC, Ince YC, Boccaccini A, Goyal A, Allenbach Petrolati L, Trevisan M, Fankhauser C (2020) PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol 226: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci AS, Michaud O, Schmid-Siegert E, Trevisan M, Allenbach Petrolati L, Caka Ince Y, Fankhauser C (2022) Shade suppresses wound-induced leaf repositioning through a mechanism involving PHYTOCHROME KINASE SUBSTRATE (PKS) genes. PloS Genet 18:e1010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, Berger A, Sechet J, To A, North HM, Marion-Poll A (2012) Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J 70: 501–512 [DOI] [PubMed] [Google Scholar]
- Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Liu X, De Storme N, Jensen KH, Xu Q, Yang J, Liu X, Chen S, Martens HJ, Schulz A, et al. (2020) Directionality of plasmodesmata-mediated transport in Arabidopsis leaves supports auxin channeling. Curr Biol 30: 1970–1977 e1974 [DOI] [PubMed] [Google Scholar]
- George GM, Kolling K, Kuenzli R, Hirsch-Hoffmann M, Flutsch P, Zeeman SC (2018) Design and use of a digitally controlled device for accurate, multiplexed gas exchange measurements of the complete foliar parts of plants. Methods Mol Biol 1770: 45–68 [DOI] [PubMed] [Google Scholar]
- Glauser G, Vallat A, Balmer D (2014) Hormone profiling. Methods Mol Biol 1062: 597–608 [DOI] [PubMed] [Google Scholar]
- Gonzalez CV, Ibarra SE, Piccoli PN, Botto JF, Boccalandro HE (2012) Phytochrome B increases drought tolerance by enhancing ABA sensitivity in Arabidopsis thaliana. Plant Cell Environ 35: 1958–1968 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Grandio E, Poza-Carrion C, Sorzano CO, Cubas P (2013) BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, Vera-Sirera F, Merilo E, Bassel GW, Fernandez MA, Holdsworth MJ, Perez-Amador MA, Kollist H, et al. (2012) Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald Y, Wigoda N, Sade N, Yaaran A, Torne T, Gosa SC, Moran N, Moshelion M (2021) Arabidopsis leaf hydraulic conductance is regulated by xylem sap pH, controlled, in turn, by a P-type H(+) -ATPase of vascular bundle sheath cells. Plant J 106: 301–313 [DOI] [PubMed] [Google Scholar]
- Hayes S, Pantazopoulou CK, van Gelderen K, Reinen E, Tween AL, Sharma A, de Vries M, Prat S, Schuurink RC, Testerink C, et al. (2019) Soil salinity limits plant shade avoidance. Curr Biol 29: 1669–1676 e1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth C, Doheny-Adams T, Hunt L, Cameron DD, Gray JE (2015) Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol 208: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holalu SV, Finlayson SA (2017) The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J Exp Bot 68: 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holalu SV, Reddy SK, Blackman BK, Finlayson SA (2020) Phytochrome interacting factors 4 and 5 regulate axillary branching via bud abscisic acid and stem auxin signalling. Plant Cell Environ 43: 2224–2238 [DOI] [PubMed] [Google Scholar]
- Hopkins R, Schmitt J, Stinchcombe JR (2008) A latitudinal cline and response to vernalization in leaf angle and morphology in Arabidopsis thaliana (Brassicaceae). New Phytol 179: 155–164 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao X, Chory J (2019) The Arabidopsis transcriptome responds specifically and dynamically to high light stress. Cell Rep 29: 4186–4199 e4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, Ballare CL (2011) Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J 67: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Toh S, Torii KU (2021) Chemical control of stomatal function and development. Curr Opin Plant Biol 60: 102010. [DOI] [PubMed] [Google Scholar]
- Kohnen MV, Schmid-Siegert E, Trevisan M, Petrolati LA, Senechal F, Muller-Moule P, Maloof J, Xenarios I, Fankhauser C (2016) Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling K, George GM, Kunzli R, Flutsch P, Zeeman SC (2015) A whole-plant chamber system for parallel gas exchange measurements of Arabidopsis and other herbaceous species. Plant Methods 11: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- McClung CR (2013) Beyond Arabidopsis: the circadian clock in non-model plant species. Semin Cell Dev Biol 24: 430–436 [DOI] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J (2002) Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J 30: 601–609 [DOI] [PubMed] [Google Scholar]
- Miao R, Russinova E, Rodriguez PL (2022) Tripartite hormonal regulation of plasma membrane H(+)-ATPase activity. Trends Plant Sci 10.1016/j.tplants.2021.12.011 [DOI] [PubMed] [Google Scholar]
- Michaud O, Fiorucci AS, Xenarios I, Fankhauser C (2017) Local auxin production underlies a spatially restricted neighbor-detection response in Arabidopsis. Proc Natl Acad Sci USA 114: 7444–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar FF, Van Zanten M, Cox MCH, Pierik R, Voesenek L, Peeters AJM (2009) Differential petiole growth in Arabidopsis thaliana: photocontrol and hormonal regulation. New Phytol 184: 141–152 [DOI] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballare CL (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JL, Weinig C, Hangarter RP (2006) Shade avoidance and the regulation of leaf inclination in Arabidopsis. Plant Cell Environ 29: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Negin B, Yaaran A, Kelly G, Zait Y, Moshelion M (2019) Mesophyll abscisic acid restrains early growth and flowering but does not directly suppress photosynthesis. Plant Physiol 180: 910–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Tat AV, Kumar Devisetty U, Robinson M, Mumbach MR, Ichihashi Y, Lekkala S, Maloof JN (2015) Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PloS Genet 11:e1004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR (2013) Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA 110: 12132–12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F (2004) Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219: 714–721 [DOI] [PubMed] [Google Scholar]
- Ortiz-Alcaide M, Llamas E, Gomez-Cadenas A, Nagatani A, Martinez-Garcia JF, Rodriguez-Concepcion M (2019) Chloroplasts modulate elongation responses to canopy shade by retrograde pathways involving HY5 and abscisic acid. Plant Cell 31: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulou CK, Bongers FJ, Kupers JJ, Reinen E, Das D, Evers JB, Anten NPR, Pierik R (2017) Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc Natl Acad Sci USA 114: 7450–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Peterson FC, Mosquna A, Yao J, Volkman BF, Cutler SR (2015) Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 520: 545–548 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko JK, van Zanten M, van Rooij JA, Maree AF, Voesenek LA, Peeters AJ, Pierik R (2012) Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol 193: 339–348 [DOI] [PubMed] [Google Scholar]
- Prado K, Cotelle V, Li G, Bellati J, Tang N, Tournaire-Roux C, Martiniere A, Santoni V, Maurel C (2019) Oscillating aquaporin phosphorylation and 14-3-3 proteins mediate the circadian regulation of leaf hydraulics. Plant Cell 31: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuten T, Hohm T, Bergmann S, Fankhauser C (2013) Defining the site of light perception and initiation of phototropism in Arabidopsis. Curr Biol 23: 1934–1938 [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JA (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B (2013) NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 25: 4941–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Simonneau T, Parent B (2015) Modelling the coordination of the controls of stomatal aperture, transpiration, leaf growth, and abscisic acid: update and extension of the Tardieu-Davies model. J Exp Bot 66: 2227–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Voesenek LA, Peeters AJ, Millenaar FF (2009) Hormone- and light-mediated regulation of heat-induced differential petiole growth in Arabidopsis. Plant Physiol 151: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek LA, Colmer TD, Pierik R, Millenaar FF, Peeters AJ (2006) How plants cope with complete submergence. New Phytol 170: 213–226 [DOI] [PubMed] [Google Scholar]
- Weiner JJ, Peterson FC, Volkman BF, Cutler SR (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodley Of Menie MA, Pawlik P, Webb MT, Bruce KD, Devlin PF (2019) Circadian leaf movements facilitate overtopping of neighbors. Prog Biophys Mol Biol 146: 104–111 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Christmann A, Yamaguchi-Shinozaki K, Grill E, Fernie AR (2019) Revisiting the basal role of ABA - roles outside of stress. Trends Plant Sci 24: 625–635 [DOI] [PubMed] [Google Scholar]
- Zhang B, Holmlund M, Lorrain S, Norberg M, Bako L, Fankhauser C, Nilsson O (2017) BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. Elife 6: e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.