Abstract
Maize (Zea mays) originated in southern Mexico and has spread over a wide latitudinal range. Maize expansion from tropical to temperate regions has necessitated a reduction of its photoperiod sensitivity. In this study, we cloned a quantitative trait locus (QTL) regulating flowering time in maize and show that the maize ortholog of Arabidopsis thaliana EARLY FLOWERING3, ZmELF3.1, is the causal locus. We demonstrate that ZmELF3.1 and ZmELF3.2 proteins can physically interact with ZmELF4.1/4.2 and ZmLUX1/2, to form evening complex(es; ECs) in the maize circadian clock. Loss-of-function mutants for ZmELF3.1/3.2 and ZmLUX1/2 exhibited delayed flowering under long-day and short-day conditions. We show that EC directly represses the expression of several flowering suppressor genes, such as the CONSTANS, CONSTANS-LIKE, TOC1 (CCT) genes ZmCCT9 and ZmCCT10, ZmCONSTANS-LIKE 3, and the PSEUDORESPONSE REGULATOR (PRR) genes ZmPRR37a and ZmPRR73, thus alleviating their inhibition, allowing florigen gene expression and promoting flowering. Further, we identify two closely linked retrotransposons located in the ZmELF3.1 promoter that regulate the expression levels of ZmELF3.1 and may have been positively selected during postdomestication spread of maize from tropical to temperate regions during the pre-Columbian era. These findings provide insights into circadian clock-mediated regulation of photoperiodic flowering in maize and new targets of genetic improvement for breeding.
ZmELF3.1/3.2 bridge ZmELF4.1/4.2 and ZmLUX1/2 to form evening complexes and promote flowering and adaptation of maize to temperate regions.
Introduction
Maize (Zea mays) is a major staple crop worldwide, which by itself contributes ∼40% of total cereal production (FAO, http://faostat.fao.org/). Although maize originated from its wild relative (teosinte) in southern Mexico about 9,000 years ago (Matsuoka et al., 2002), it has spread to temperate regions during pre-Columbian times, and is now cultivated across a wide latitudinal range (40°S to 45°N; Buckler et al., 2009). Nowadays, ∼80% of the total worldwide maize production is produced in temperate regions (NCGA, 2021).
Flowering time is a key determinant of crop adaptation and yield productivity. As teosinte is a short-day (SD) plant, its flowering is accelerated under SD conditions, but delayed under long-day (LD) conditions. Thus, the spread of maize from tropical to temperate regions has necessitated a reduction of its photoperiod sensitivity for adaptation to LD conditions (Hung et al., 2012). A number of genes that contributed to the pre-Columbian expansion of maize to temperate regions have been cloned and functionally characterized, including a Flowering Locus T (FT) homolog, CENTRORADIALIS 8 (ZCN8) (Lazakis et al., 2011; Meng et al., 2011; Guo et al., 2018), Vgt1 (encoding an APETALA 2-like transcription factor) (Salvi et al., 2007; Ducrocq et al., 2008; Castelletti et al., 2014), ZmCCT (CONSTANS, CONSTANS-LIKE, TOC1; also named ZmCCT10) (Hung et al., 2012; Yang et al., 2013), ZmCCT9 (Huang et al., 2018), ZmMADS69 (Liang et al., 2018), Delayed flowering 1 (which encodes a basic leucine zipper transcription factor that physically interacts with ZCN8) and ZmMADS67 (Sun et al., 2020). Independently, genomic studies with nested association mapping (NAM) populations and diverse association panels have identified hundreds of candidate genes with small effects that may contribute to large-scale environmental adaptation of maize to altitude and latitude, such as DWARF8, Phytochrome C2 (ZmPHYC2), CIRCADIAN CLOCK ASSOCIATED 1 (ZmCCA1), Zea Floricaula/Leafy 1/2, Zea mays MADS-box 5 (ZMM5), Barren inflorescence 2, EARLY FLOWERING 3 (ZmELF3), PSEUDORESPONSE REGULATOR 37 (ZmPRR37), and ZmPRR73, to name a few (Thornsberry et al., 2001; Chardon et al., 2004; Salvi et al., 2007; Buckler et al., 2009; Hung et al., 2012; Bouchet et al., 2013; Li et al., 2016; Romero Navarro et al., 2017; Li et al., 2020). However, for most, functional validation on flowering regulation, evaluation of their allelic effects on regional adaptation, and identification of the causal variants remain to be carried out.
The evening complex (EC) was first identified in Arabidopsis (Arabidopsis thaliana); it is composed of three interacting proteins, ELF3, ELF4, and LUX ARRHYTHMO (LUX). ELF3 directly interacts with and bridges ELF4 and LUX to form a transcriptional repressor complex and a core component of the plant circadian clock (Nusinow et al., 2011). LUX is a single MYB domain-containing SHAQYF-type GARP transcription factor that can mediate the direct binding of the EC to the specific LUX-binding site (LBS) motif 5′-GATWCG-3′ (where W indicates A or T) in target promoters (Nusinow et al., 2011). Mutations in any of the three genes result in early flowering under SD and LD conditions and elongated hypocotyls, and thus they were believed to mediate circadian clock regulation of photoperiod-mediated flowering in Arabidopsis (Hicks et al., 1996; Zagotta et al., 1996; Doyle et al., 2002; Hazen et al., 2005). Recent studies have shown that the EC plays a conserved role in regulating photoperiod sensitivity and regional adaptation in various crops. For example, the ELF3 orthologs in rice (Oryza sativa) OsELF3.1 (also named Early heading 7 and Heading date 17; Matsubara et al., 2012; Saito et al., 2012; Zhao et al., 2012; Zhu et al., 2018), HIGH RESPONSE TO PHOTOPERIOD of pea (Pisum sativum; Weller et al., 2012), EARLY MATURITY 8 of barley (Sorghum vulgare; Faure et al., 2012; Zakhrabekova et al., 2012), and the soybean (Glycine max) J gene (Lu et al., 2017) are important targets of postdomestication breeding selection for regional adaptation of various crops. In addition, LUX orthologs in several crops (barley, einkorn wheat [Triticum monococcum], pea, and soybean) have also been shown to regulate photoperiodic flowering and regional adaptation (Mizuno et al., 2012; Campoli et al., 2013; Liew et al., 2014; Bu et al., 2021).
In this study, we report the cloning of a QTL regulating flowering time in maize and show that an ortholog of Arabidopsis ELF3, ZmELF3.1, corresponds to this QTL. We demonstrate that ZmELF3.1 and its homolog ZmELF3.2 exhibit canonical diurnal pattern of expression, and their protein products exclusively localized to the nucleus. We show that both ZmELF3.1 and ZmELF3.2 can physically interact with two maize homologs of Arabidopsis ELF4, namely ZmELF4.1 and ZmELF4.2, and two maize homologs of Arabidopsis LUX, ZmLUX1, and ZmLUX2, thus constituting the maize EC. Loss-of-function mutants of ZmELF3s and ZmLUXs exhibit a significant delay in flowering time under both LD and SD conditions, with ZmELF3.1 and ZmLUX1 playing a more predominant role. We further demonstrate that the EC can repress the expression of several known flowering suppressor genes, ZmCCT9, ZmCCT10, ZmCOL3, ZmPRR37a, and ZmPRR73, in turn, alleviating their inhibition of the expression of several maize florigen genes (ZCN8, ZCN7, and ZCN12) to promote flowering. Finally, we identified two closely linked variants (a NonLTR/L1 retrotransposon and an LTR/Gypsy retrotransposon) inserted upstream of the ZmELF3.1 coding region that regulate ZmELF3.1 expression, and that they have been positively selected during the spread of maize from tropical to temperate regions for adaptation to higher latitudes.
Results
Identification of ZmELF3.1 as a candidate QTL for flowering time and photoperiod sensitivity
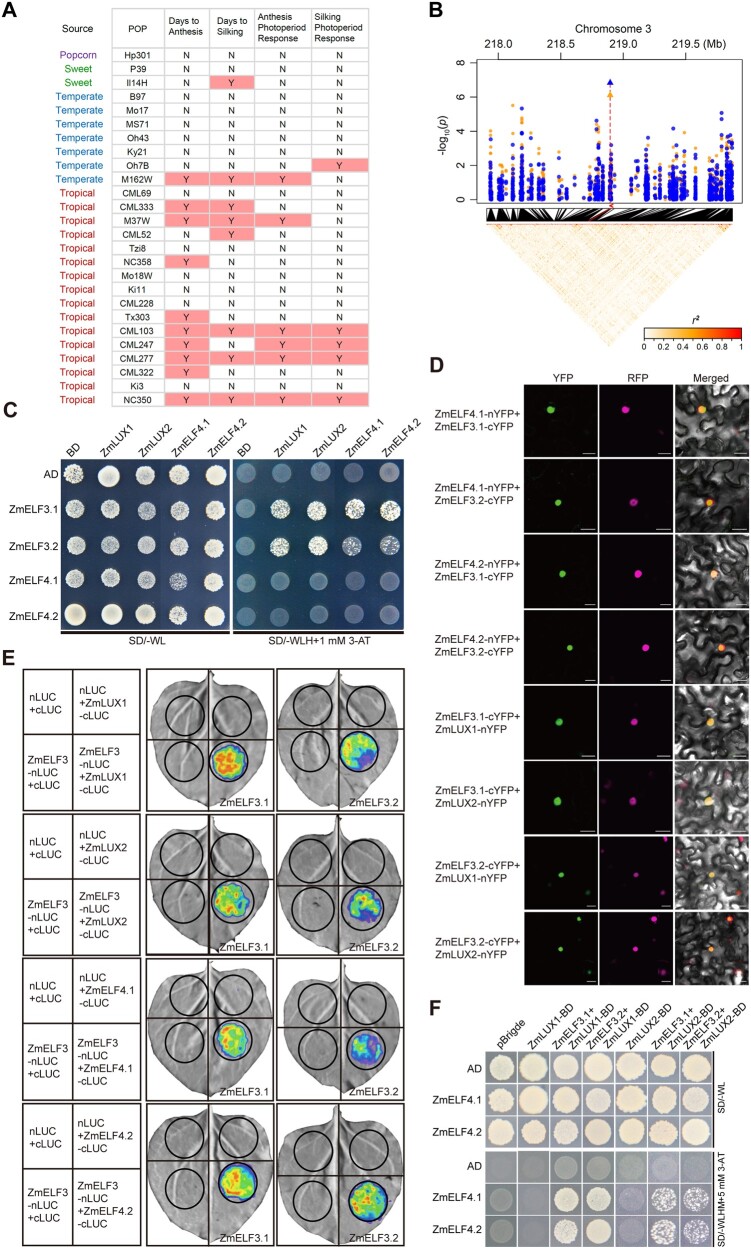
To identify new factor(s) having contributed to regional expansion of maize, we reanalyzed flowering time and photoperiod-sensitive QTLs previously identified in the maize NAM population (Buckler et al., 2009; Hung et al., 2012; Li et al., 2016). We repeatedly identified a significant QTL located at the end of chromosome 3 (216.5–221.5 Mb, centered at ∼218.4 Mb, based on the B73_v2 reference genome, thus named qFT3_218) in different NAM families (different founder lines crossed to B73), with nine families detected for days to anthesis (DTA), six families for days to silking (DTS), six families for anthesis photoperiod response, and five families for silking photoperiod response. Notably, we mostly detected this QTL in families derived from tropical founder lines crossed to B73 (a temperate inbred line) (Figure 1A). These observations indicate that qFT3_218 likely encompasses a genetic element(s) involved in the expansion of maize from tropical to temperate regions.
Figure 1.
ZmELF3s bridge ZmELF4s and ZmLUXs to form the EC. A, Linkage mapping using the NAM population identifies a QTL for flowering time and photoperiod response around the ZmELF3.1 region. Y or N, with or without QTL detected around the ZmELF3.1 region. B, GWAS identification of ZmELF3.1 as a candidate gene for variation in the traits DTA and DTS. The candidate genes for DTA and DTS are marked with blue points and orange points, respectively. Peak markers are shown as triangles for each trait and their positions in the linkage disequilibrium heatmap are indicated by red lines. The position of the candidate gene in the Manhattan plot is shown by the red arrow. C, Y2H assay showing that ZmELF3.1/3.2 directly interact with both ZmELF4.1/4.2 and ZmLUX1/2. Three independent experiments showed similar results. D, BiFC assay showing that ZmELF3.1/3.2 directly interact with both ZmELF4.1/4.2 and ZmLUX1/2 in N. benthamiana leaf cells. The AHL22-ERFP marker was co-infiltrated to indicate the nuclei. The interaction between nYFP and ZmELF3-cYFP or between cYFP and ZmELF4-nYFP (or ZmLUX-nYFP) is shown in Supplemental Figure S3C as the negative controls. Scale bar, 20 μm. Three independent experiments were performed, with similar results. E, LCI assay demonstrating the interaction between ZmELF3.1/3.2 with ZmELF4.1/4.2 or ZmLUX1/2 in N. benthamiana leaves. Three independent experiments were performed, with similar results. Representative images of N. benthamiana leaves 72 h after infiltration are shown. F, Yeast three-hybrid assay showing that ZmELF3.1/3.2, ZmELF4.1/4.2 and ZmLUX1/2 interact to form a tri-protein complex(es) in yeast. Two independent experiments showed similar results.
To identify the possible candidate gene(s) underlying qFT3_218, we re-performed a genome-wide association study (GWAS) for flowering time of all NAM recombinant inbred lines (RILs) using the phenotypic data across 13 environments (Buckler et al., 2009; Li et al., 2016; Figure 1B). We detected a significant signal around 218.8 Mb (B73_v2) for DTA (P-value = 1.48e–7) and DTS (P-value = 7.84e–7), with the peak single-nucleotide polymorphism (SNP; S3_218895116) being located 2.5 kb downstream of a gene (GRMZM2G045275, chromosome 3: 218,897,671–218,903,527 bp, reverse strand, B73_v2, corresponding to Zm00001d044232 in the B73_v4 genome) encoding a protein homologous to Arabidopsis ELF3 (27.9% amino acid identity). As ELF3 orthologs have been shown to regulate flowering time and photoperiod sensitivity in Arabidopsis and a number of crops, we speculated that GRMZM2G045275 (Zm00001d044232) probably represents the candidate gene for qFT3_218.
ZmELF3s form ECs with ZmELF4s and ZmLUXs
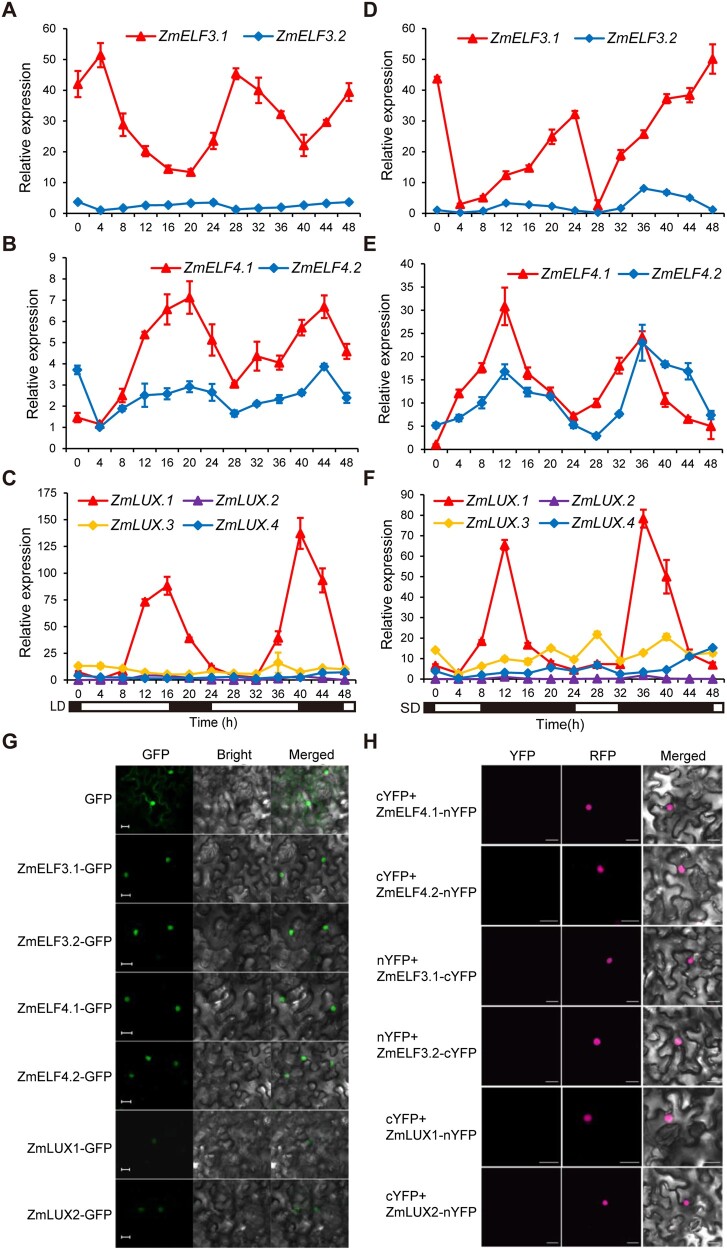
In Arabidopsis, ELF3 physically interacts with both ELF4 and LUX to form the EC, in which LUX is the transcription factor that regulates downstream gene expression (Nusinow et al., 2011; Chow et al., 2012). BLAST searches identified a gene (Zm00001d039156) highly homologous to the ELF3-like protein encoded by the qFT3_218 candidate locus GRMZM2G045275 (Zm00001d044232; 42.9% amino acid identity) in the maize B73_v4 genome. Thus, we designated these two genes ZmELF3.1 (Zm00001d044232, candidate for qFT3_218) and ZmELF3.2 (Zm00001d039156) (Supplemental Figure S1). In addition, we identified two genes homologous to Arabidopsis ELF4, designated ZmELF4.1 and ZmELF4.2, with 35.65% and 30.1% amino acid identity to Arabidopsis ELF4, respectively (Supplemental Figure S2). We also detected four homologs of Arabidopsis LUX, designated ZmLUX1, ZmLUX2, ZmLUX3, and ZmLUX4, whose encoding proteins showed 42.1%, 46.8%, 34.2%, and 32.9% amino acid identity with Arabidopsis LUX, respectively (Supplemental Figure S2). Interestingly, both ZmELF3.1 and ZmELF3.2 lack the polyglutamine (polyQ) repeats and the C-terminal prion-like domain (PrD) (Lancaster et al. 2014), which have been shown to play important roles in thermomorphogenesis for Arabidopsis ELF3 (Undurraga et al., 2012; Jung et al., 2020; Supplemental Figure S1). Yeast two-hybrid (Y2H) (Figure 1C), bimolecular fluorescence complementation (BiFC) (Figure 1D) and luciferase complementation imaging (LCI) (Figure 1E) assays all confirmed that both ZmELF3.1 and ZmELF3.2 physically interact with ZmELF4.1/4.2 and ZmLUX1/2, but we observed no direct interaction between ZmELF4.1/4.2 and ZmLUX1/2. Moreover, a yeast three-hybrid assay indicated that ZmELF3.1/3.2 bridge the interaction between ZmELF4.1/ZmELF4.2 and ZmLUX1/2 to form EC (Figure 1F), supporting the existence of conserved EC in maize. Reverse transcription-quantitative PCR (RT-qPCR) assays also showed that these genes all display distinct diurnal expression patterns under both LD and SD conditions, with ZmELF3.1, ZmELF4.1, and ZmLUX1 showing obviously higher expression levels than their respective homologs (Figure 2, A–F). Protein subcellular localization assay also showed that their green fluorescent protein (GFP) fusion proteins were all exclusively localized to the nucleus (Figure 2G).
Figure 2.
Expression pattern of ZmELF3s, ZmELF4s, and ZmLUXs and subcellular localization of their encoded proteins. A–F, ZmELF3.1/3.2, ZmELF4.1/4.2, and ZmLUX1/2/3/4 all show a rhythmic diurnal expression pattern under both LD (A–C) and SD (D–F) conditions. The sixth leaves from V6-stage plants of ZC01 (a temperate line) grown under the indicated conditions were used for RNA extraction and RT-qPCR. Values are means ± SD (n = 3 technical replicates). Two independent experiments were performed and the results were similar (leaves from six different plants were used for each biological replicate). The black bars and white bars indicate the dark period and the light period, respectively. G, ZmELF3.1/3.2, ZmELF4.1/4.2, and ZmLUX1/2 proteins localize to the nucleus. The expression constructs GFP, ZmELF3.1/3.2-GFP, ZmELF4.1/4.2-GFP, and ZmLUX1/2-GFP were individually infiltrated into N. benthamiana leaves and then incubated for 48 h in the dark prior to imaging using a confocal microscope. Scale bars, 20 µm. Three independent experiments were performed, with similar results. H, Negative controls for the BiFC assay for the interaction between ZmELF3.1/3.2, ZmELF4.1/4.2, and ZmLUX1/2 proteins. The nuclear marker AHL-RFP indicates the nuclei in N. benthamiana leaf epidermal cells. Scale bars, 20 µm. Three independent experiments were performed, with similar results.
Delayed flowering of the Zmelf3 and Zmlux mutants under both LD and SD conditions
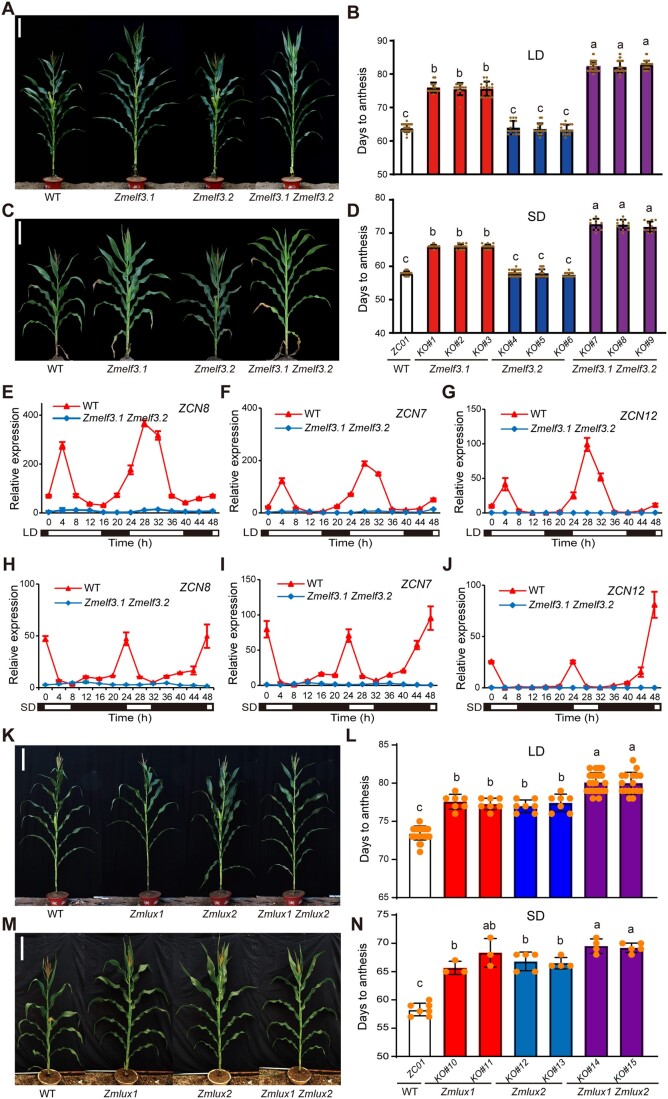
To investigate the function of maize EC components in regulating flowering time, we first overexpressed ZmELF3 genes in the Arabidopsis elf3-7 mutant. We determined that both ZmELF3.1 and ZmELF3.2 can rescue the Arabidopsis elf3-7 mutant phenotype, including long-hypocotyl and early flowering under LD conditions (Supplemental Figure S3), when overexpressed, indicating a conserved biological role of ZmELF3s in regulating flowering. We also generated both single and double knockout (ko) mutants in ZmELF3s using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated gene editing (Supplemental Figure S4). We selected three independent Zmelf3.1 single mutants (ko1#–ko#3), three independent Zmelf3.2 single mutants (ko#4–ko#6), and three independent Zmelf3.1 Zmelf3.2 double mutants (ko#7–ko#9) (Supplemental Figure S4) for analyses. We sowed all mutants together with their respective wild-type (WT) in fields at Langfang (39.53° N/116.72° E, LD) and Sanya (18.73° N/109.17° E, SD) for phenotypic examination. These field tests showed that the Zmelf3.1 single mutants, but not the Zmelf3.2 single mutants, flower significantly later than the WT plants under both LD and SD conditions (8–12 days late in anthesis and 9–15 days late in silking). Moreover, the Zmelf3.1 Zmelf3.2 double mutants flowered significantly later than the Zmelf3.1 single mutants, suggesting that ZmELF3.1 and ZmELF3.2 likely play partially redundant functions in flowering regulation, with ZmELF3.1 playing a predominant role (Figure 3, A–D). Both the Zmelf3.1 single mutants and Zmelf3.1 Zmelf3.2 double mutants displayed a more pronounced delay in flowering time under LD conditions than under SD conditions (the Zmelf3.1 single mutants delayed flowering by ∼3.8 more days under LD than under SD, and the Zmelf3.1 Zmelf3.2 double mutants by ∼4 more days under LD than under SD conditions), indicating that ZmELF3.1/ZmELF3.2 likely play a more important role in promoting flowering under LD conditions. Consistently, the Zmelf3.1 single mutants and the Zmelf3.1 Zmelf3.2 double mutants, but not the Zmelf3.2 single mutants, displayed higher plant height and ear height, and increased leaf number, under both LD and SD conditions, compared to the WT control plants (Supplemental Figure S4). Consistent with the delayed flowering phenotype of the Zmelf3.1 Zmelf3.2 double mutants, RT-qPCR analysis showed that the expression of three reported maize florigen genes (ZCN8, ZCN7, and ZCN12) (Meng et al., 2011; Castelletti et al., 2014; Mascheretti et al., 2015) is nearly not detectable in the Zmelf3.1 Zmelf3.2 double mutant under both SD and LD conditions (Figure 3, E–J).
Figure 3.
Maize EC regulates flowering time under both LD and SD conditions. A–D, Comparison and quantification of the flowering time of WT, Zmelf3.1 and Zmelf3.2 single mutants, and the Zmelf3.1 Zmelf3.2 double mutant under LD (A, B) or SD (C, D) conditions. Scale bars, 30 cm. Values are means ± SD (n ≥ 11 plants). Different lowercase letters indicate significant differences determined by the Duncan’s multiple-range test (P < 0.05). E–J, The relative transcript levels of three maize florigen genes (ZCN8, ZCN7, and ZCN12) are downregulated in the Zmelf3.1 Zmelf3.2 double mutant grown under LD (E–G) or SD (H–J) conditions. Leaves from V6-stage plants grown under the indicated conditions were harvested for RNA extraction. Values are means ± SD (n = 3 biological replicates). Two independent experiments were performed, with similar results. K–N, Comparison and quantification of the flowering time of WT, Zmlux1, and Zmlux2 single mutants and the Zmlux1 Zmlux2 double mutant grown under LD (K, L) or SD (M, N) conditions. Scale bars, 30 cm. Values are means ± SD (n ≥ 3 plants). Different lowercase letters indicate significant differences determined by Duncan’s multiple-range test (P < 0.05).
We also generated CRISPR/Cas9-aided ko mutants for ZmLUX1/2 (Supplemental Figure S5). We determined that the Zmlux1 and Zmlux2 single mutants and the Zmlux1 Zmlux2 double mutants all showed obviously delayed flowering under both SD and LD conditions, compared to the WT control (Figure 3, K–N). Moreover, the Zmlux1 Zmlux2 double mutants flowered significantly later than the Zmlux1 or Zmlux2 single mutants, suggesting that ZmLUX1 and ZmLUX2 also play partially redundant functions in flowering regulation (Figures 3, K–N). These results support the notion that the EC contributes to promoting flowering in maize. However, we observed no significant differences in flowering time between the ZmELF3.1 overexpressors and WT plants (Supplemental Figure S6), suggesting that the functional levels of the EC components might be optimal in WT plants. A similar observation was reported for the LUX1 and LUX2 genes in soybean (Bu et al., 2021).
ZmLUX1 directly represses the expression of a group of flowering repressor genes
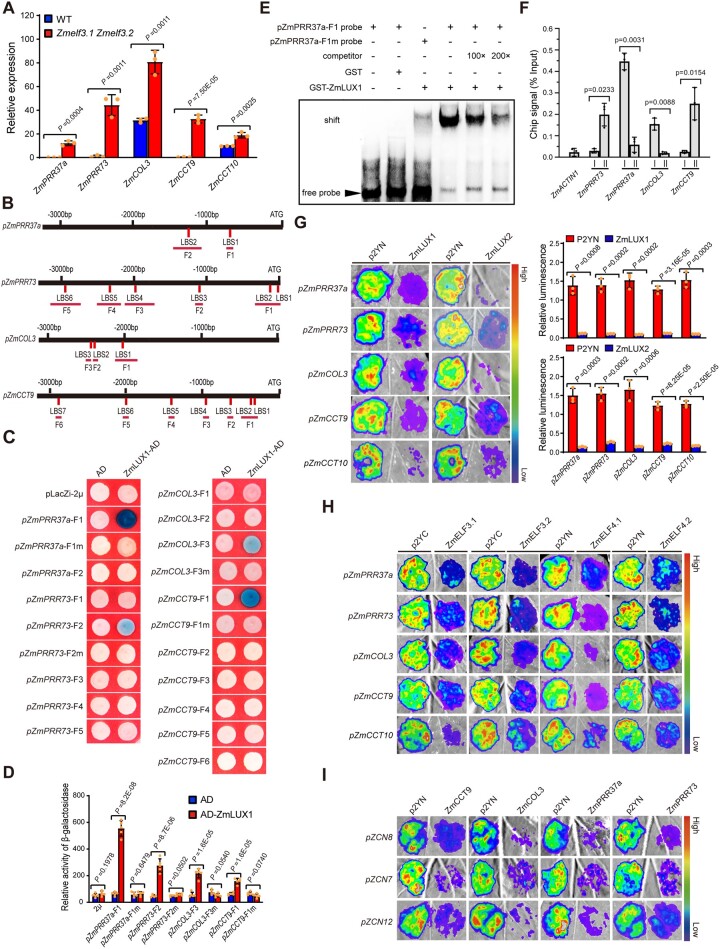
Previous studies have reported that the Arabidopsis EC directly regulates the expression of clock-regulated genes (such as PHYTOCHROME-INTERACTING FACTOR 4 [PIF4], PIF5, and PRR9) to optimize plant growth and flowering time (Nakamichi et al., 2005; Nusinow et al., 2011; Chow et al., 2012). We thus tested whether the maize EC directly regulates the expression of a set of flowering repressor genes (ZmCCT10, ZmCCT9, CONSTANS-LIKE 3 [ZmCOL3], ZmPRR37a, and ZmPRR73) (Hung et al., 2012; Li et al., 2016; Huang et al., 2018; Jin et al., 2018) by RT-qPCR. We observed that the expression of these flowering repressor genes significantly increases in the Zmelf3.1 Zmelf3.2 double mutants, compared to the WT plants (Figure 4A;Supplemental Figure S7). Sequence analysis revealed that their promoters all contain multiple copies of the LUX binding site (GATWCG; Nusinow et al., 2011; Lu et al., 2017; Figure 4B), raising the possibility that the maize EC directly regulates their expression. A yeast one-hybrid (Y1H) assay showed that indeed, ZmLUX1 binds strongly to specific promoter fragments of ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9, but only weakly to the promoter fragments of ZmCCT10 (Figures 4, C and D;Supplemental Figure S8). We verified the binding of ZmLUX1 to the ZmPRR37a promoter by electrophoretic mobility shift assay (EMSA) (Figure 4E). Further, we performed chromatin immunoprecipitation (ChIP)-qPCR assay using the leaves from V2-stage ZmELF3.1 overexpressing plants (ELF3.1 being tagged with the GFP) with an anti-GFP antibody. We again detected the binding of ZmELF3.1 to the promoter fragments of ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9 in vivo (Figure 4F). A transient expression assay in Nicotiana bentamiana leaf epidermal cells further showed that the transcription of ZmCCT9, ZmCOL3, ZmPRR37a, ZmPRR73, and ZmCCT10 (as measured by promoter constructs where each promoter individually drives firefly luciferase [LUC] expression) is significantly repressed upon co-expression of YFPn-ZmLUX1, YFPn-ZmLUX2, YFPc-ZmELF3.1, YFPc-ZmELF3.2, YFPn-ZmELF4.1, or YFPn-ZmELF4.2, compared to the control vector (p2YN [YFPn] or p2YC [YFPc]) (Figure 4, G and H), suggesting that the three components of the EC (ZmELF3s, ZmELF4s, and ZmLUXs) act together to repress the expression of downstream flowering genes.
Figure 4.
Maize EC directly represses the expression of several flowering repressor genes. A, RT-qPCR assay showing that the expression levels of several flowering repressor genes, ZmPRR37a, ZmPRR73, ZmCOL3, ZmCCT9, and ZmCCT10, are significantly upregulated in the Zmelf3.1 Zmelf3.2 double mutant compared to the WT (ZC01). Values are means ± SD (n = 3 technical replicates). Two biological replicates were performed and the results were similar (leaves from six plants were used for each biological replicate). B, The promoters of the above flowering repressor genes harbor multiple typical LUX binding sites (LBS, 5′-GATWCG-3′, 5′-GATWKG-3′, or 5′-GATWCY-3′, where W indicates A or T, K indicates C or T, and Y indicates G or T). The positions of the LBS motifs in each promoter are indicated with red vertical lines and the promoter fragments used for Y1H assay are marked with red horizontal lines. C, Y1H assay showing that ZmLUX1 directly binds to specific promoter fragments containing the LBS motif of ZmPRR37a (5′-CGTATCGTATC-3′), ZmPRR73 (5′-GATTCG-3′), ZmCOL3 (5′-GATTCG-3′), and ZmCCT9 (5′-AGAATC CATATC-3′). For mutagenesis, the LBS motif was mutated to 5′-TCCAAGTGATG-3′ in ZmPRR37apro-F1, 5′-CACACA-3′ in ZmPRR73pro-F2, 5′-CTAGGA-3′ in ZmCOL3pro-F3, and 5′-CTAGGA TCCTAG-3′ in ZmCCT9pro-F1. Three independent experiments showed similar results. D, Quantification of ZmLUX1 binding activity. Five independent yeast clones were used for activity determination. Values are means ± SD (n = 5 independent clones). Significant differences were determined by the Student’s t test. E, EMSA showing that recombinant GST-ZmLUX1 fusion protein binds to a biotin-labeled probe of the ZmPRR37a promoter. GST protein was used as a negative control. Two independent experiments were performed, with similar results. F, ChIP-qPCR assay of enrichment for ZmELF3.1 at the ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9 promoters in the leaves of V2-stage Ubipro:ZmELF3.1-EGFP transgenic seedlings grown under LD conditions. The “I” and “II” represent the individual amplified fragment of each promoter as shown in (B). Values are means ± SD (n = 3 biological replicates, leaves from six plants were used for each replicate). P-value was determined by the Student’s t test. Two independent experiments showed similar results. G, Transient expression assay showing that ZmLUX1 and ZmLUX2 repress the expression of the above flowering repressor genes. Values are means ± SD (n = 3 different infiltrated regions). At least six independent experiments were performed, with similar results. Representative images of N. benthamiana leaves 72 h after infiltration are shown. Significant differences were determined by the Student’s t test. H, Transient expression assay shows that both ZmELF3s and ZmELF4s suppress the expression of ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9. At least six independent experiments were performed, with similar results. Representative images of N. benthamiana leaves 72 h after infiltration are shown. I, Transient expression assay shows that ZmPRR37a, ZmPRR73, ZmCOL3, and ZmCCT9 suppress the expression of ZCN8, ZCN7, and ZCN12. At least six independent experiments were performed, with similar results. Representative images of N. benthamiana leaves 72 h after infiltration are shown.
ZmCOL3 was previously shown to act as a flowering repressor by directly activating the transcription of ZmCCT10, which in turn represses the expression of ZCN8 (Jin et al., 2018). Thus, we tested the effects of ZmCCT9, ZmCOL3, ZmPRR37a, and ZmPRR73 on downstream florigen gene expression. To this end, we transiently expressed ZmCCT9, ZmCOL3, ZmPRR37a, or ZmPRR73 in N. benthamiana leaf epidermal cells together with promoter:LUC reporter constructs. In all cases, the expression of ZCN8, ZCN7, or ZCN12 repressed the transcriptional output of the ZCN7, ZCN8, and ZCN12 promoters (Figure 4I). Together, these results support the notion that the maize EC promotes flowering through suppressing the expression of a set of flowering repressor genes (ZmCCT9, ZmCCT10, ZmCOL3, ZmPRR37a, and ZmPRR73).
Insertion of two retrotransposons upstream of ZmELF3.1 confers increased expression and early flowering
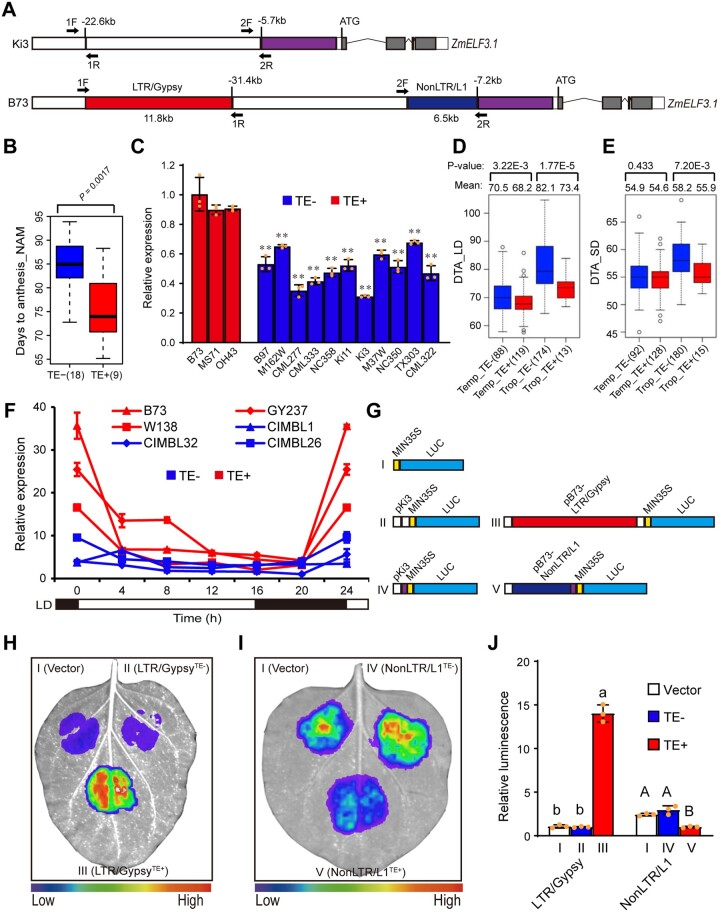
To investigate whether and how natural variation of ZmELF3.1 influences maize flowering time, we took advantage of the recently released high-quality genome assembly of 45 maize inbred lines, comprising B73 (Jiao et al., 2017), Mo17 (Sun et al., 2018), W22 (Springer et al., 2018), 4 European maize lines (Haberer et al., 2020), 26 founder lines of the NAM population (Hufford et al., 2021), and 12 founder inbred lines for modern temperate maize breeding (Wang et al., 2022; Supplemental Data Set S1). We analyzed both the ZmELF3.1 coding regions and its flanking regions up to the next neighboring gene on either side. We detected only a few nonsynonymous mutations in the coding sequences of eighty tropical inbred lines and eighty temperate inbred lines (Supplemental Data Set S2), suggesting that variation in the regulatory regions of ZmELF3.1 might play a predominant role in regulating its function. Notably, sequencing analysis identified two retrotransposon insertions: a Nonlong terminal repeat (NonLTR)/L1-type retrotransposon (6.5 kb in length) and an LTR/Gypsy type retrotransposon (11.8 kb in length), located 7 and 31 kb upstream of the ZmELF3.1 translational start site, respectively, in 22 maize inbred lines (including B73), but not another 23 inbred lines (like Ki3) (Figure 5A;Supplemental Figure S9). As structural variation in regulatory regions often influences transcription and flowering time in maize, we speculated that these retrotransposons may affect ZmELF3.1 expression, and thus flowering time. Consistent with this notion, nine NAM founder lines carrying the retrotransposon insertions flowered earlier (∼9–10 d) showed higher ZmELF3.1 expression than another 18 founder lines that lacked the retrotransposon insertions (Figure 5, B and C). Moreover, in contrast to B73 (a temperate inbred line), most families of the NAM population in which we detected qFT3_218 for DTA (all 9), DTS (5 of 6), anthesis photoperiod response (all 10) and silking photoperiod response (11 of 12) (Buckler et al., 2009; Hung et al., 2012; Li et al., 2016), their founder lines all lacked the retrotransposon insertions upstream of their ZmELF3.1 genes (except I114H for DTS, and Oh7B for silking photoperiod response) (Supplemental Data Set S3). These observations indicate that the retrotransposon insertions likely contribute to the differential expression and photoperiod flowering in the NAM founder lines.
Figure 5.
The NonLTR/L1 retrotransposon and LTR/Gypsy retrotransposon regulate ZmELF3.1 expression. A, Schematic diagram of the locations of a NonLTR/L1-type retrotransposon (6.5 kb in length) and an LTR/Gypsy type retrotransposon (11.8 kb in length) in two representative inbred lines (B73 and Ki3). 1F/1R and 2F/2R indicate the primers used to construct the vectors in Figure 5G. B, Flowering time of the NAM founder lines with or without the retrotransposons. The flowering time data were obtained from the previous NAM flowering-time mapping study (Buckler et al., 2009). C, The relative expression levels of ZmELF3.1 in the NAM founder lines with retrotransposon insertion upstream of ZmELF3.1 promoter are significantly higher than in the NAM founder lines without retrotransposon insertion. Values are means ± SD (n = 3 biological replicates, leaves from six plants were used for each replicate). D and E, Flowering time of 415 maize inbred lines (Yang et al., 2014) with or without retrotransposons in the ZmELF3.1 promoter under LD (D) or SD (E) conditions. The number of inbred lines is indicated in brackets. F, The relative expression levels of ZmELF3.1 in inbred lines with retrotransposon insertion upstream of ZmELF3.1 promoter are significantly higher than in the inbred lines without retrotransposon insertion. Three independent experiments were performed, with similar results. G, Schematic diagrams of the constructs used to test the effect of the two retrotransposon elements on gene expression in transient expression assay in N. benthamiana leaves. H and I, The LTR/Gypsy retrotransposon strongly activates ZmELF3.1 expression while the NonLTR/L1 retrotransposon mildly inhibits ZmELF3.1 expression. Representative images of N. benthamiana leaves 72 h after infiltration are shown. Three independent experiments were performed, with similar results. J, Quantification of luminescence intensity in (H) and (I). Values are means ± SD (n = 3 biological replicates). Significant differences was determined by Duncan's multiple-range test in Fig. 5J and Student's t test in the others. Three independent experiments were performed, with similar results.
To further test the effect of these two retrotransposons, we genotyped and phenotyped 415 maize inbred lines from a natural association population (Yang et al., 2014). Among them, 272 lines carried both retrotransposons while the remaining 143 lines lacked both retrotransposons (Supplemental Data Set S3), suggesting that these two retrotransposons are tightly linked. In addition, the inbred lines with the retrotransposon insertions flowered on average earlier than the inbred lines without the retrotransposon insertions under both LD (Langfang in 2015) and SD (Sanya in 2016) conditions. Notably, the differences in DTA were larger in tropical lines with and without the retrotransposon insertions than in temperate lines with and without the retrotransposon insertions, and the difference in DTA was larger under LD conditions, compared to SD conditions (Figure 5, D and E). Moreover, RT-qPCR analysis showed that three randomly selected inbred lines with the retrotransposon insertions (B73, W138, and GY237) all exhibit higher expression levels of ZmELF3.1 than three randomly selected inbred lines that do not carry the insertion (CIMBL1, CIMBL26, and CIMBL32) (Figure 5F). Consistently, the inbred lines with the retrotransposon insertions (B73, W138, and GY237) had lower expression of the EC target genes (ZmCCT9, ZmPRR37a, ZmPRR73, and ZmCOL3) than the inbred lines that do not carry the insertion (CIMBL1, CIMBL26, and CIMBL32) (Supplemental Figure S10). Collectively, these observations suggest that the two-tightly linked retrotransposons likely represent the causal variation underlying the differential expression of ZmELF3.1, and thus flowering time.
To validate this hypothesis, we cloned the B73 and Ki3 ZmELF3.1 promoter sequences that differ only at the two retrotransposons into a reporter construct upstream of a minimal promoter from the cauliflower mosaic virus (MIN35S), the LUC sequence and the CaMV poly(A) terminator (Figure 5G) for transient expression assays in N. bentamiana leaves. We established that the B73 construct containing the LTR/Gypsy element drastically enhances luciferase activity (∼14-fold), while the B73 construct containing the NonLTR/L1 element markedly represses luciferase activity (approximately 3-fold), relative to the control Ki3 construct with no insertions (Figure 5, H–J). These results suggest that in combination, the LTR/Gypsy and NonLTR1/L1 elements likely act to promote ZmELF3.1 transcription, which is consistent with the observed higher expression of ZmELF3.1 in inbred lines with the retrotransposon insertions (Figure 5, C and F).
The retrotransposon insertions were positively selected during postdomestication of maize and contributed to adaptation of maize to temperate regions
To examine the evolutionary origin of the LTR/Gypsy element, we genotyped the LTR/Gypsy and NonLTR1/L1 elements in 72 teosinte accessions (Supplemental Data Set S4). Interestingly, two accessions carried both the LTR/Gypsy and NonLTR1/L1 elements (Supplemental Figure S11), indicating that the LTR/Gypsy and NonLTR1/L1 retrotransposon insertions were already present in the maize ancestor. To determine whether selection has acted on the LTR/Gypsy and NonLTR/L1 elements during the pre-Columbian expansion of maize, we analyzed nucleotide diversity (π) and Tajima’s D across a 90-kb region centered on ZmELF3.1 in the 45 maize inbred lines (27 temperate, 18 tropical) with high-quality genome assemblies (Supplemental Data Set S1). Notably, in the region of the LTR/Gypsy and NonLTR/L1 elements, the nucleotide diversity (π) in temperate maize was much lower than that in tropical maize. Tajima’s D values of temperate maize were about –1.6 and –2.4, but Tajima’s D values of tropical maize were 1.2 and 2.0 (Supplemental Figure S11). These data indicate a strong positive selection on the LTR/Gypsy and NonLTR/L1 elements. On the contrary, we detected no significant selection signals over the ZmELF3.1 coding region.
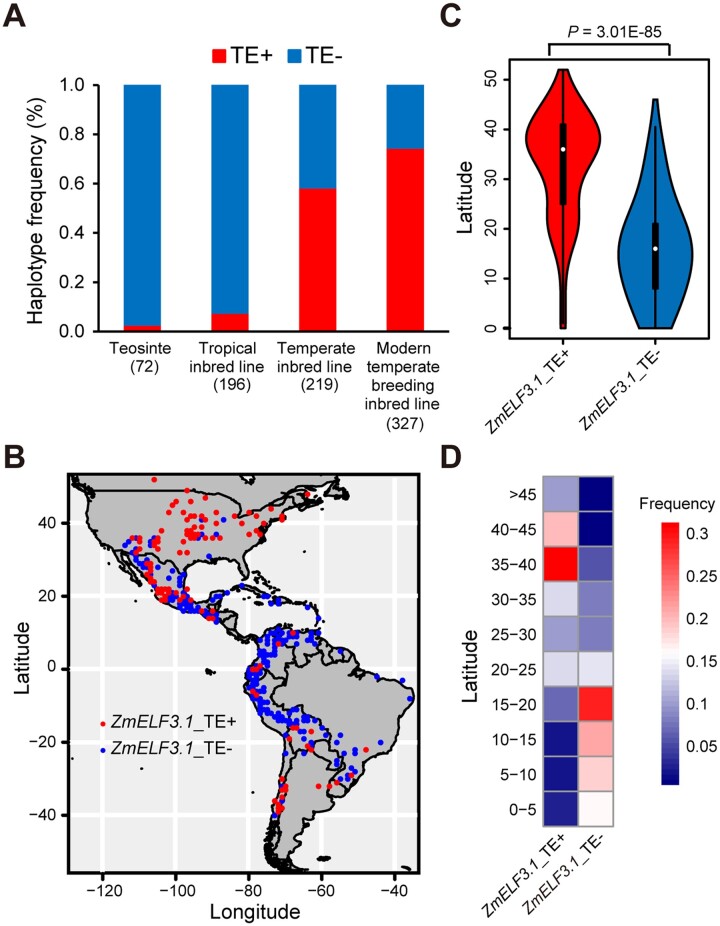
To investigate whether these two retrotransposons contributed to the expansion and adaptation of maize to temperate regions, we analyzed their distribution in 72 accessions of teosinte, 415 of a panel of 513 maize inbred lines with 196 tropical inbred lines and 219 temperate inbred lines (Yang et al., 2014; Supplemental Data Sets S3 and S4) and 327 of the 350 modern temperate maize inbred lines (Wang et al., 2020; Supplemental Data Set S5). We determined that 2.8% of teosinte, 7.7% of tropical inbred lines, 58.4% of temperate inbred lines, and 74.6% of modern maize inbred lines carry the two retrotransposons (Figure 6A), suggesting that these two retrotransposons have likely been selected during the expansion of maize from tropical to temperate regions and are still under continuous selection during modern temperate maize breeding. To substantiate this notion, we investigated the distribution of the LTR/Gypsy retrotransposon among 1,008 maize landrace accessions representing the entire pre-Columbian range of maize races native to the Americas (Huang et al., 2018; Supplemental Data Set S6). Strikingly, the LTR/Gypsy retrotransposon showed strong association with latitude, predominantly accumulating in landraces of higher latitudes. In contrast, the maize landraces without the LTR/Gypsy retrotransposon were mainly distributed at low latitudes (Figure 6, B–D).
Figure 6.
ZmELF3.1 was selected during the spread of maize from tropical to temperate regions. A, Frequency of the retrotransposons in the ZmELF3.1 promoter across teosinte, tropical maize, temperate inbred lines, and modern temperate breeding inbred lines. The number of various germplasms is indicated in parentheses. B, Geographic distribution of 1,008 maize landraces native to the Americas with or without the retrotransposon insertions in the ZmELF3.1 promoter (Huang et al., 2018). C, Accessions with retrotransposons insertion at ZmELF3.1 predominantly accumulate at higher latitudes, whereas accessions without the retrotransposon insertions at ZmELF3.1 mainly distribute at low latitudes. Significant differences were determined by the Student’s t test. D, Frequency of accessions with or without the retrotransposon insertions at ZmELF3.1 along a latitudinal gradient.
Discussion
The circadian clock is an internal time-keeping mechanism that enables plants to anticipate changes in external environmental conditions (such as dawn, dusk, and changes in temperature driven by daily light/dark cycles) and adjust their development and physiology accordingly (Hsu and Harmer, 2014). One major output of the circadian clock is flowering time, which is a critical determinant of regional adaptation for plants. Recent studies have demonstrated that the EC plays a conserved role in regulating photoperiod flowering in both SD and LD plants, and thus, its components have often been targets of artificial selection for crop domestication and improvement (Liu et al., 2020; McClung, 2021). For example, orthologs of ELF3 in rice, pea, barley, wheat, and soybean (Faure et al., 2012; Matsubara et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012; Lu et al., 2017; Zhu et al., 2018), orthologs of LUX in barley, einkorn wheat, pea, soybean, and rice (Mizuno et al., 2012; Campoli et al., 2013; Liew et al., 2014; Bu et al., 2021; Cai et al., 2022), orthologs of PRR7 in sugar beet (Beta vulgaris), rice, barley, wheat, sorghum, and soybean (Murakami et al., 2005; Turner et al., 2005; Beales et al., 2007; Murphy et al., 2011; Pin et al., 2012; Bu et al., 2021; Liang et al., 2021) regulate photoperiodic flowering and regional adaptation of various crops.
In this study, we showed that ZmELF3.1 corresponds to qFT3_218, a QTL regulating flowering time in maize. In addition, we established that the expression of ZmELF3.1/3.2, ZmELF4.1/4.2, and ZmLUX1/2 follows a diurnal rhythm and that ZmELF3.1/3.2 can physically interact with ZmELF4.1/4.2 and ZmLUX1/2, to form the ECs of the maize circadian clock. Similar to other SD plants with defective EC (like rice and soybean), loss-of-function mutants of ZmELF3.1/3.2 and ZmLUX1/2 exhibited delayed flowering time under both LD and SD conditions. Moreover, overexpression of either ZmELF3.1 or ZmELF3.2 largely rescued the long-hypocotyl and early flowering phenotypes of the Arabidopsis elf3-7 mutant (Supplemental Figure S3). These observations indicate that ZmELF3.1/3.2 and the EC play a conserved role in regulating flowering time in maize.
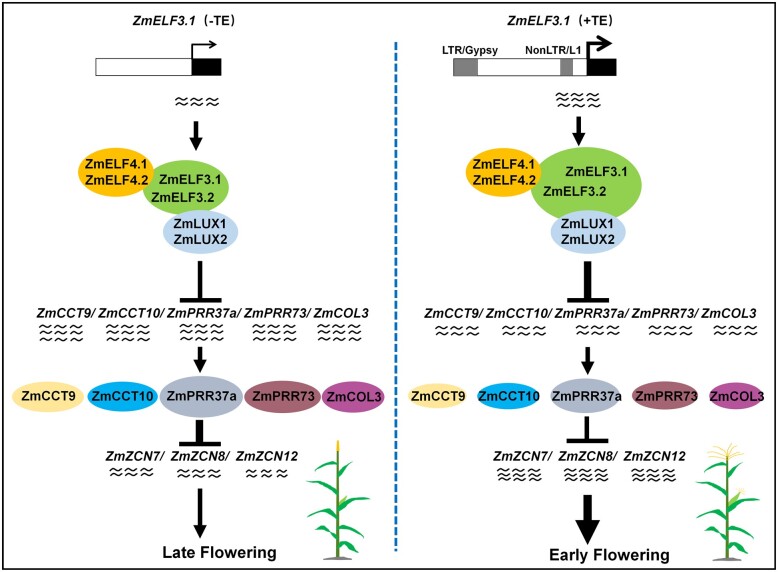
At the mechanistic level, previous studies have shown that the EC mainly acts as a transcriptional repressor to suppress the expression of downstream flowering regulatory genes. For example, in rice (an SD plant), an ortholog of ELF3 (OsELF3-1) negatively regulates the expression of the major flowering repressor Grain number, plant height, and heading date7 (an ortholog of Arabidopsis PRR7), and consequently upregulates the expression of the downstream genes Ehd1 and FT-like to promote flowering under both SD and LD conditions (Saito et al., 2012; Zhao et al., 2012). In soybean (an SD plant), the EC suppresses the transcription of a key flowering repressor, the legume-specific E1 gene (which encodes a B3 superfamily member), thereby relieving the suppression of FT2a and FT5a imposed by E1, and promoting flowering (Lu et al., 2017; Bu et al., 2021). In Arabidopsis (an LD plant), ELF3 can be recruited to the PRR9 promoter through LUX to repress PRR9 expression, thus delaying flowering (Nakamichi et al., 2005). We showed here that in maize, the EC directly represses the expression of the flowering repressor genes ZmCCT9, ZmCCT10, ZmCOL3, ZmPRR37a, and ZmPRR73, thereby alleviating the suppression of the expression of ZCN8, ZCN7, and ZCN12 mediated by their encoding proteins, to promote flowering under both SD and LD conditions (Figure 7). Thus, it appears that the switch in EC function (promoting flowering in SD plants and repressing flowering in LD plants) resides in the downstream signaling pathway in SD and LD plants.
Figure 7.
Schematic model illustrating a role of the EC in regulating flowering time in maize. ZmELF3.1/3.2 bridge ZmELF4.1/4.2 and ZmLUX1/2 to form ECs, and then the EC directly represses the expression of a set of flowering repressor genes: ZmCCT9, ZmCCT10, ZmCOL3, ZmPRR37a, and ZmPRR73, thereby relieving their suppression on expression of three maize florigen genes (ZCN8, ZCN7, and ZCN12), to promote flowering. Insertion of an LTR/Gypsy and a NonLTR/L1 retrotransposon upstream of ZmELF3.1 enhances the expression of ZmELF3.1, thus promoting flowering. Arrow: activation; Bar: repression.
The circadian clock is also implicated in temperature regulation of flowering time through temperature entrainment, which allows plants to perceive and respond to temperature cues, and adjust clock gene expression accordingly. In addition, the circadian clock can help maintain clock gene expression over a wide range of temperature through a mechanism termed temperature compensation (Salome et al., 2010). It is notable that recent studies have shown that the polyQ repeats and PrD of Arabidopsis ELF3 play a role in mediating thermal responsiveness (Undurraga et al., 2012; Jung et al., 2020). In addition, Arabidopsis ELF3 was shown to physically interact with and suppress the transcriptional activation activity of PIF4 in an EC-independent manner to regulate hypocotyl growth (Nieto et al., 2015). Moreover, ELF3 was recently shown to directly regulate the expression of PIF4 in thermoresponsive growth (Raschke et al., 2015), while ELF3 regulates circadian gating of the shade avoidance response in Arabidopsis through physically interacting with and repressing the DNA-binding activity of PIF7, thus antagonizing PIF7-induced gene expression and hypocotyl growth (Jiang et al., 2019). Intriguingly, we found that ZmELF3s in all the analyzed tropical and temperate maize lines lack the polyQ repeats and a typical PrD (Supplemental Data Set S7), hinting that ZmELF3s might not be involved temperature responsiveness. However, a recent study reported that the expression of a number of core clock genes (CCA1, GIGANTEA, PRR59, PRR73, PRR95, and LUX) in barley responds rapidly to changes in temperature and that this response is lost in elf3 mutants, indicating that the temperature response in dependent on a functional ELF3 protein (which also lacks the polyQ repeats and typical PrD domain) in barley (Ford et al., 2016). Thus, it will be an interesting research avenue to further clarify whether ZmELF3s play a role in temperature regulation of flowering time and shade avoidance response in future studies.
Previous studies have suggested that the spread of maize from tropical to temperate regions entailed stepwise regulatory changes in multiple flowering time genes. Available data suggest that SNP-1245 in the promoter of ZCN8, which is associated with differential binding of the flowering activator ZmMADS1, was first selected and nearly fixed in early domestication of maize. Then, another variant in the ZCN8 promoter, InDel-2339, which most likely originated from Z. mays ssp. Mexicana, was introgressed into maize and further selected during the spread of maize from tropical to temperate regions (Guo et al., 2018). Similarly, insertion of a MITE upstream of the vgt1 region, the CATCA-like transposable element upstream of ZmCCT10 and the Harbinger-like transposable element upstream of ZmCCT9 likely occurred after the initial domestication of maize, as they are not observed in surveyed teosinte accessions and the frequencies of their early flowering alleles rose when maize spread into the Northern USA (but not South America) (Yang et al., 2013; Huang et al., 2018). In this study, we discovered that two closely linked retrotransposons located in the ZmELF3.1 promoter upregulate ZmELF3.1 expression, and that they were already present in some accessions of teosinte, thus representing preexisting standing variations that were selected postdomestication for adaptation to temperate regions during the pre-Columbian era, both Northward and Southward (Figure 6, B–D). Analogous to our finding, a recent study reported that ancestral Puebloan people selected temperate-adapted maize for 2,000 years in situ in the southwestern USA and the early flowering alleles were from preexisting standing variation in teosinte (Swarts et al., 2017). In addition, the increased frequency of the retrotransposon-harboring haplotype in modern temperate inbred lines suggests that ZmELF3.1 is still under selection during modern temperate maize breeding (Figure 6A). Moreover, the finding that the LTR/Gypsy retrotransposon strongly activates, while the NonLTR/L1 retrotransposon mildly represses the expression of ZmELF3.1 (Figure 5, G and J) suggests a possibility of breaking their linkage or deleting the inhibitory retrotransposon using genome editing to create novel alleles of ZmELF3.1 for earlier flowering and adapting to even higher altitudes. Thus, the identification of more flowering genes and their allelic variation via tapping into the vast diversity of maize germplasm offers the promise to breed maize cultivars that adapt more efficiently to the next century of changing environments through harnessing standing natural variation or creating new variants via genome editing.
Methods
Plant materials and growth conditions
All maize (Z. mays) lines used in this study were in the B73 or ZC01 inbred line backgrounds, unless other specified. For gene cloning, the seeds of B73 were directly sown on soil (Pindstrup Substrate No 2, Pindstrup Mosebrug A/S, Ryomgaard Denmark) and grown in a growth chamber under LD conditions (28°C 16-h light/22°C 8-h dark). For analysis of gene expression, maize seeds were directly sown on soil and grown in a growth chamber under either LD (16-h light/8-h dark) or SD (8-h light/16-h dark) conditions with 300 µmol m−2 s−1 light intensity provided by full-spectrum white fluorescent light tubes. The humidity was set to 60%, and the daytime and nighttime temperature was set to 28°C and 22°C, respectively. For phenotypic analysis, seeds of the mutant and WT control (ZC01) were sown in the field either under natural LD (Langfang, Hebei Province, China; 39.53°N, 116.72°E, 15-h light/9-h dark) or natural SD (Sanya, Hainan Province, China; 18.73°N, 109.17°E, 11-h light/13-h dark) conditions.
The WT and elf3-7 mutant of Arabidopsis (A. thaliana) used in this study were in the Columbia-0 accession (Reed et al., 2000). All Arabidopsis transgenic plants used in this study were generated in the elf3-7 background. Arabidopsis seeds were surface-sterilized with 5% [v/v] NaClO solution for 15 min and washed with sterile water for 3 times, then plated on half-strength Murashige and Skoog solid medium (1% [w/v] sucrose and 0.8% [w/v] agar, pH 5.8) and incubated in the dark at 4°C for 3 d for stratification. Thereafter, they were moved to the culture room under white light with 200 µmol m−2 s−1 light intensity provided by full-spectrum white fluorescent light tubes at 23°C (16-h light/8-h dark) for 10 d, then transplanted to soil.
Nicotiana benthamiana seeds were directly sown on soil and grown in the same culture room as Arabidopsis for about 1 month before use.
GWAS
A NAM population comprising 5,000 RILs was developed from crosses between B73 as a common parent and 25 other various inbred lines (Buckler et al., 2009). This NAM population was evaluated across 13 environments, consisting of eight environments in the USA (Buckler et al., 2009) and five environments in China (Li et al., 2016). The best linear unbiased predictions for flowering time for all NAM RILs, derived across these 13 environments, were calculated using a mixed model in SAS (Version 9.2, SAS Institute; Flint et al., 2011). The SNP markers around the target gene for the NAM population (http://panzea.org.com/) were selected for inclusion in the regional association mapping. The association analysis was conducted using TASSEL version 5.0 software (Bradbury et al., 2007), in which the mixed linear model with population structure (3 PCAs) and pair-kinship (K matrix) treated as covariates was applied to test for association between segregating sites and phenotypes (Yu et al., 2005). Linkage disequilibrium analysis within the target region was conducted using Haploview software (Barrett et al., 2004).
Phylogenetic and conserved motifs analysis
The homologous protein sequences for ELF3, ELF4, and LUX were retrieved from TAIR (https://www.arabidopsis.org/, for Arabidopsis), or gramene (http://ensembl.gramene.org/genome_browser/index.html, for maize and rice). The full-length amino acid sequences were aligned using the MUSCLE multiple sequence alignment program (Edgar, 2004), and the phylogenetic trees were constructed using the neighbor-joining method of MEGA version 7. The bootstrap method was used for statistical support for nodes in the phylogenetic tree. The number of bootstrap replications was 500. The Poisson model was used for substitutions model analysis to obtain tree branch lengths (Kumar et al., 2016). The alignments for the phylogenetic analysis are provided as Supplemental Files S1–S6. Conserved motifs were identified using MEME software (Bailey et al., 2006).
RNA extraction and RT-qPCR analysis
Various plant samples were collected and immediately frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted using Trizol reagent (Invitrogen, Waltham, MA, USA) from three independent pools of tissues, and first-strand cDNAs were synthesized using a reverse transcription kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. The cDNA was diluted 1:5 and RT-qPCR was performed using SuperReal PreMix Plus (TIANGEN) with a 7500 real-time PCR detection system (Applied Biosystems, Waltham, MA, USA). ZmTubulin5 and AtACT2 were used as the internal controls for maize and Arabidopsis genes, respectively. The 2−△△CT method was used to determine the relative expression levels of target genes (Schmittgen and Livak, 2008).
Protein subcellular localization assay
The coding sequences without stop codon of ZmELF3s, ZmELF4s, and ZmLUXs were obtained by PCR using maize B73 cDNA as template, and cloned into the pCAMBIA1305-EGFP binary vector digested with XbaI to generate the E35Spro:ZmELF3s-EGFP-TNos, E35Spro::ZmELF4s-EGFP-TNos, and E35Spro:ZmLUXs-EGFP-TNos constructs, respectively (primers are listed in Supplemental Data Set S8). The constructs were then introduced into Agrobacterium (Agrobacterium tumefaciens) strain EHA105 by electroporation (BIORAD, Hercules, CA, USA). To determine the subcellular localization of ZmELF3s, ZmELF4s, and ZmLUXs proteins, Agrobacterium EHA105 colonies individually carrying each of the above constructs or the nuclear marker E35Spro:AHL22-ERFP-TNos (Xiao et al., 2009) were harvested by quick centrifugation and resuspended to a cell density (OD600) of 1.0 in infiltration solution (10 mM MgCl2, 10 mM MES pH 5.7, and 150 μM acetosyringone; Wydro et al., 2006), and then co-infiltrated into N. benthamiana leaves in an equal volume. Two days after infiltration, GFP and RFP signals were observed under a confocal microscope using a 488-nm laser for excitation and an emission wavelength of 526 nm with 30% power (Zeiss LSM710).
Creation of transgenic lines in Arabidopsis and maize
PCR-amplified coding sequences without stop codon of the ZmELF3 genes were cloned into the CPB-Ubi-EGFP binary vector digested with PstI and BamHI to be placed under the control of the maize Ubiquitin promoter and the TNos terminator to obtain the Ubipro:ZmELF3s-EGFP-TNos expression cassettes (primers are listed in Supplemental Data Set S8). The constructs were then introduced into Agrobacterium strain GV3101 and separately transformed into the Arabidopsis elf3-7 mutant using the floral dip method (Clough and Bent, 1998) to obtain elf3-7 ZmELF3-OE lines. More than 10 independent transgenic lines for each transformation were obtained by selection on glufosinate ammonium and two independent lines were selected for further studies.
The above Ubipro::ZmELF3s-EGFP constructs were also introduced into Agrobacterium strain EHA105 and transformed into the maize inbred line ZC01 using immature embryos (Ishida et al., 2007). Two and three independent transgenic lines were obtained for ZmELF3.1 and ZmELF3.2, respectively.
Generation of the CRISPR/Cas9 ko lines in maize
To obtain the ko mutants of ZmELF3.1/3.2 and ZmLUX1/2, a CRISPR/Cas9 ko vector was constructed according to our previously described method with minor modifications: the E35S promoter and the AtU6-26 promoter were replaced with the maize Ubiquitin promoter and the ZmU6-6 promoter, respectively (Zhao et al., 2016). Multiple target sites were identified with SnapGene Viewer version 2.4.3 software based on the criteria of 5' G-(N)19-NGG 3'. All putative target sequences were then checked against the B73 genomic sequence at the gramene database by BLAST for specificity. Specific target sequences without PAM together with the universal scaffold sequence were used for prediction of the single-guide RNA (sgRNA) secondary structure with the program RNA Folding Form (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form2.3; Zuker, 2003) to select those target sites that did not affect the secondary structure of the universal scaffold sequence. Since the target sites were designed based on the B73 genome, and the inbred line ZC01 was used for transformation, we designed a pair of primers to amplify fragments containing the target sites and sequenced the PCR products amplified from ZC01 (primers are shown in Supplemental Data Set S8). Sequence alignment showed that target 1 of ZmELF3.1 has one base mismatch between B73 and ZC01, while the other three targets had identical sequences between B73 and ZC01 (Supplemental Data Set S1). The four target sequences of ZC01 were separately introduced into four sgRNA expression cassettes (pZmU6-6+target+universal scaffold sequence) by overlapping PCR (Urban et al., 1997). The four sgRNA expression cassettes were then sequentially cloned into the pCPB-ZmUbi::hSpCas9 vector (Zhao et al., 2016) using the HindIII restriction site. Each clone was verified by PCR and sequencing. The resulting final vector was introduced into Agrobacterium strain EHA105 and transformed into the maize inbred line ZC01 using immature embryo. Eighteen independent transgenic lines were obtained. Through sequencing analysis, we selected three independent lines for Zmelf3.1 single mutants (ko#1, ko#2, and ko#3), three independent lines of Zmelf3.2 single mutants (ko#4, ko#5, and ko#6), and three independent lines of Zmelf3.1 Zmelf3.2 double mutants (ko#7, ko#8, and ko#9) for self-pollination to obtain various homozygous mutants. For ZmLUX1/2, two independent transgenic lines each for Zmlux1, Zmlux2, and Zmlux1 Zmlux2, were selected for further study. The produced T2 generation progenies together with the WT (ZC01) were planted under natural LD and SD conditions for observation of flowering time and other phenotypes.
Y1H assay
The Y1H was performed using the pJG4-5 vector (Clontech, Mountain View, CA, USA) encoding the GAL4 activation domain and pLacZi-2µ vector harboring the bacterial lacZ reporter gene as described previously (Lin et al., 2007). The full-length coding region of ZmLUX1 was cloned into the pJG4-5 vector digested with XhoI to generate AD-ZmLUX1. To obtain the ZmCCT9pro::LacZ, ZmCOL3pro::LacZ, ZmPRR37apro::LacZ, and ZmPRR73pro::LacZ reporter constructs, the promoter fragments of ZmCCT9, ZmCOL3, ZmPRR37a, and ZmPRR73 were individually PCR amplified from B73 genomic DNA and then inserted into the pLacZi-2µ vector digested with EcoRI and XhoI (primers are listed in Supplemental Data Set S8). For mutagenesis of the LBS motif in the promoter fragments above, primer pairs were designed according to Agilent Technologies (http://www.genomics.agilent.com) and used to generate plasmids containing mutated LBS motif following the manufacturer’s instructions. The indicated plasmid pairs were then co-transformed into the yeast strain EGY48 with PEG3350-LiAc-mediated transformation. The positive transformants screened on synthetic defined medium lacking Trp and Ura (SD–Trp –Ura, Clontech) were transferred to chromogenic medium containing raffinose, galactose, and 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (Amresco) for blue color development. To quantify β-galactosidase activity, the positive clones were grown in liquid SD –Trp –Ura medium overnight, harvested by centrifugation and resuspended in Z-buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 10 mM KCl, pH 7.0, 50 mM β-mercaptoethanol) and then lysed with chloroform and 0.1% (w/v) sodium dodecyl sulfate. The substrate ONPG was added to the supernatants and incubated at 30°C for 5 min and then 1 M Na2CO3 was immediately added to terminate the reaction. After a brief centrifugation to settle cell debris, the values of OD420 and OD600 were measured from the supernatants and the original liquid cultures, respectively.
Y2H assay
The complete coding sequences of ZmELF3.1, ZmELF3.2, ZmELF4.1, ZmELF4.2, ZmLUX1, and ZmLUX2 were obtained by PCR from a cDNA library prepared from 2-week-old B73 seedlings (primers are listed in Supplemental Data Set S8). The cDNAs of ZmELF3.1, ZmELF3.2, ZmELF4.1, and ZmELF4.2 were then cloned into the pGADT7 vector (Clontech) to produce the AD-ZmELF3 and AD-ZmELF4 fusion proteins, while the cDNAs of ZmELF4.1, ZmELF4.2, ZmLUX1, and ZmLUX2 were cloned into the pGBKT7 vector (Clontech) to produce the BD-ZmELF4 and BD-ZmLUXs fusion proteins. The plasmid combinations between AD, AD-ZmELF3, AD-ZmELF4, and BD, BD-ZmLUXs, and BD-ZmELF4 were co-transformed into the yeast strain AH109 following the manufacturer’s instructions (Clontech). Positive yeast clones were screened on SD –Leu –Trp medium and then spotted onto plates containing SD –Leu –Trp and SD –Leu –Trp –His with 5 mM 3-amino-1,2,4-triazole (3-AT) for growth.
Yeast three-hybrid assay
For yeast three-hybrid assay, full-length coding sequences (cDNAs) of ZmLUX1 and ZmLUX2 were cloned into the pBridge vector (Clontech) at the EcoRI site to generate BD-ZmLUX1 and BD-ZmLUX2, respectively. The cDNAs of ZmELF3.1 and ZmELF3.2 were then further ligated into the generated constructs above at the BglII site to produce the BD-ZmLUX-ZmELF3 constructs (primers are listed in Supplemental Data Set S8). The combinations between AD- and BD-fused plasmids were co-transformed into yeast strain AH109 and the positive yeast colonies were screened on selective medium (SD –Leu/–Trp). The positive clones were spotted onto plates of SD –Leu –Trp or SD –Leu –Trp–His-Met with 1 mM 3-AT for growth.
Recombinant protein production
For protein production and purification, the coding sequence of ZmLUX1 was amplified from the above BD-ZmLUX1 plasmid and cloned into the pGEX-4T-1 vector at the EcoRI restriction site to generate the GST-ZmLUX1 construct, which was transformed into Escherichia coli strain Transette (TransGen Biotech, China). The production of GST-ZmLUX1 recombinant protein was induced by the addition of 0.4 mM isopropyl β-D-thiogalactopyranoside and growth at 16°C overnight before purification with glutathione sepharose resin (GE Healthcare, Chicago, IL, USA) according to the manufacturer’s protocol.
Gel mobility shift assay
The direct binding of ZmLUX1 to the ZmPRR37a promoter was assessed using an EMSA kit (Beyotime, Jiangsu, China) following the manufacturer’s protocol with probes listed in Supplemental Data Set S8. The GST (glutathione S-transferase) proteins were used as the controls.
ChIP-qPCR assay
The leaves from V2-stage UBIpro::ZmELF3.1-EGFP transgenic seedlings grown under LD conditions were crosslinked with 1% (w/v) formaldehyde and ground in liquid nitrogen. The chromatin complex was prepared following the method of Xie et al. (2020). In brief, the supernatant was precleared with 40 µL Protein-A-Agarose (16-157, EMD Millipore Corp., Burlington, MA, USA) and incubated at 4°C for 1 h. The supernatant was then incubated with 50 µL anti-GFP Magarose beads (SM038001, SMART Lifesciences) at 4°C overnight. After washing, the immune complex was eluted from the beads with elution buffer (1% SDS and 0.1 M NaHCO3) and harvested the supernatant (eluate) by centrifugation. The precipitated DNA was then recovered and quantified using quantitative PCR with the primer pairs listed in Supplemental Data Set S8. The values were normalized to input DNA to obtain the fold-enrichment. ACTIN1 was used as the internal control.
BiFC assay
The cDNAs of ZmELF3.1 and ZmELF3.2 were individually cloned into the p2YC vector (digested with PacI and SpeI) encoding C-terminal YFP fragments to generate the ZmELF3.1-cYFP and ZmELF3.2-cYFP constructs. The cDNAs of ZmELF4.1, ZmELF4.2, ZmLUX1, and ZmLUX2 were separately cloned into the similarly PacI-SpeI digested p2YN vector (Walter et al., 2004; Yang et al., 2007) encoding N-terminal YFP fragments to obtain ZmELF4.1-nYFP, ZmELF4.2-nYFP, ZmLUX1-nYFP, and ZmLUX2-nYFP, respectively (primers are listed in Supplemental Data Set S8). Various combinations of these constructs were introduced into Agrobacterium strain EHA105 and co-infiltrated into N. benthamiana leaves with the viral silencing suppressor P19 and the nuclear protein marker AHL22-ERFP. The infiltrated N. benthamiana plants were grown in the dark for 48 h, then grown in the light for 3–6 h before observation with a confocal microscope (Zeiss LSM710).
LCI assay
The vectors pCAMBIA1300-nLUC and pCAMBIA1300-cLUC were used for LCI assay according to previously described procedures (Chen et al., 2008). The coding sequences of ZmELF3.1 and ZmELF3.2 were ligated into the pCAMBIA1300-nLUC vector at the KpnI-SalI sites to generate the ZmELF3.1-nLUC and ZmELF3.2-nLUC vectors, respectively. The coding sequences of ZmELF4.1, ZmELF4.2, ZmLUX1, and ZmLUX2, were ligated into the pCAMBIA1300-cLUC vector at the KpnI-SalI sites to produce the ZmELF4.1-cLUC, ZmELF4.2-cLUC, ZmLUX1-cLUC, and ZmLUX2-cLUC vectors, respectively (primers are listed in Supplemental Data Set S8). The constructs were individually transformed into Agrobacterium strain EHA105 and co-infiltrated into N. benthamiana leaves using the indicated plasmid combinations and Agrobacterium strain EHA105 carrying the P19 vector was simultaneously infiltrated with each combination. The infiltrated plants were first grown under dark for 24 h and then in the light (16-h light/8-h dark) for 24–48 h. Luciferase activity was acquired with the Plant Imaging System of NightShade LB985 (Berthold Technologies, Bad Wildbad, Germany) after applying 20 mg/mL D-luciferin potassium salt (Gold Biotech, St. Louis, MO, USA).
Transient expression assay
Transient expression assays were carried out with the vector pGreenII 0800-LUC (Dual-LUC) as described previously (Hellens et al., 2005). To obtain the ZmCCT9pro::LUC, ZmCCT10pro::LUC, ZmCOL3pro::LUC, ZmPRR37apro::LUC, ZmPRR73pro::LUC, ZCN8pro::LUC, ZCN7pro::LUC, and ZCN12pro::LUC reporter constructs, the promoters (∼3 kb) of ZmCCT9, ZmCCT10, ZmCOL3, ZmPRR37a, ZmPRR73, ZCN8, ZCN7, and ZCN12 were amplified from B73 genomic DNA and ligated into the vector pGreenII 0800-LUC at the HindIII site (primers are listed in Supplemental Data Set S8). Agrobacterium colonies (strain EHA105) carrying the indicated plasmid combinations were co-infiltrated into N. benthamiana leaves together with Agrobacterium strain EHA105 harboring P19. Subsequent processing was the same as the LCI assay.
Statistical analysis
To assess statistical significance of the experimental data, Student’s t tests were employed to determine significant differences between two groups. If more than two groups of data are analyzed, one-way analysis of variance was adopted using the commercially available package SAS (version 9.2, SAS Institute, Cary, NC, USA) and Duncan’s multiple-range test was used for all pairwise comparisons with P-values corrected for multiple comparisons to control against type I errors (Brady et al., 2015).
Accession numbers
The genes studies in this article are under the following accession numbers: AtELF3 (At2g25930), AtACT2 (At3g18780), ZmELF3.1 (Zm00001d044232), ZmELF3.2 (Zm00001d039156), ZmTubulin5 (Zm00001d006651), ZmELF4.1 (Zm00001d047269), ZmELF4.2 (Zm00001d020364), ZmLUX1 (Zm00001d011785), ZmLUX2 (Zm00001d041960), ZmLUX3 (Zm00001d010280), ZmLUX4 (Zm00001d038191), ZmCCT9 (Zm00001d000176), ZmCCT10 (Zm00001d024909), ZmCOL3 (Zm00001d017176), ZmPRR37a (Zm00001d022590), ZmPRR73 (Zm00001d047761), ZCN7 (Zm00001d038725), ZCN8 (Zm00001d010752), ZCN12 (Zm00001d043461), OsELF3.1 (Os06g0142600), OsELF3.2 (Os01g0566050), OsELF4.1 (Os06g0142600), OsELF4.2 (Os01g0566050), OsELF4.3 (Os06g0142600), OsLUX1 (Os01g0971800), OsLUX2 (Os05g0412000) and OsLUX3 (Os01g0844900).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence comparison and phylogeny of ZmELF3.1/3.2 with AtELF3 and OsELF3.1/3.2.
Supplemental Figure S2. Amino acid sequence comparison and phylogeny of ELF4 and LUX orthologs from Arabidopsis, maize, and rice.
Supplemental Figure S3. Both ZmELF3.1 and ZmELF3.2 rescue the Arabidopsis elf3-7 mutant phenotypes when overexpressed.
Supplemental Figure S4. Loss-of-function of ZmELF3.1 results in greater plant height and ear height.
Supplemental Figure S5. Generation of Zmlux ko mutants using CRISPR/Cas9.
Supplemental Figure S6. WT plants and ZmELF3.1 overexpressors show no significant differences in flowering time.
Supplemental Figure S7. Lower expression levels of several flowering repressor genes in Zmelf3.1 Zmelf3.2 double mutants.
Supplemental Figure S8. ZmLUX1 and ZmLUX2 weakly bind to the ZmCCT10 promoter.
Supplemental Figure S9. Schematic diagram of retrotransposon insertions in 45 maize inbred lines with high-quality genome assembly.
Supplemental Figure S10. Comparison of the expression of EC target genes in the inbred lines with or without the retrotransposon insertion in ZmELF3.1.
Supplemental Figure S11. Positive selection of the NonLTR/L1 and LTR/Gypsy retrotransposons in temperate inbred lines.
Supplemental Table S1. Target genes and their two sgRNA target sequences with PAM of B73 and ZC01.
Supplemental Table S2. Detailed information for the constructs in Figure 5, A and G.
Supplemental Table S3. Mutant or transgenic lines used in this study.
Supplemental Data Set S1. Distribution of LTR/Gypsy and NonLTR/L1 retrotransposons in the 45 inbred lines with high-quality genome assembly.
Supplemental Data Set S2. Genetic variation in the cDNA sequence of ZmELF3.1 in 160 inbred lines (80 tropical and 80 temperate).
Supplemental Data Set S3. Distribution of LTR/Gypsy and NonLTR/L1 retrotransposons in 415 inbred lines (196 tropical lines and 219 temperate lines).
Supplemental Data Set S4. Distribution of LTR/Gypsy and NonLTR/L1 retrotransposons in 72 teosinte accessions.
Supplemental Data Set S5. Distribution of LTR/Gypsy and NonLTR/L1 retrotransposons in 327 modern temperate maize breeding inbred lines.
Supplemental Data Set S6. Distribution of LTR/Gypsy and NonLTR/L1 retrotransposons in 1,008 landrace accessions native to the Americas.
Supplemental Data Set S7. Amino acid sequences of ZmELF3.1 in 160 inbred lines (80 tropical and 80 temperate).
Supplemental Data Set S8. Primers used in this study.
Supplemental File S1. Alignment of ELF3 proteins in fas format.
Supplemental File S2. Phylogenetic tree of ELF3 proteins in Newick format.
Supplemental File S3. Alignment of ELF4 proteins in fas format.
Supplemental File S4. Phylogenetic tree of ELF4 proteins in Newick format.
Supplemental File S5. Alignment of LUX proteins in fas format.
Supplemental File S6. Phylogenetic tree of LUX proteins in Newick format.
Funding
This work was supported by the National Key R&D program of China (grant no. 2021YFF1000301), the National Natural Science Foundation of China (32022065, 31921004, and 31871639), the Major Program of Guangdong Basic and Applied Research (2019B030302006), the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, and a project from Hainan Yazhou Bay Seed Lab (B21HJ8101).
Conflict of interest statement. The authors declare no competing interests.
Supplementary Material
Contributor Information
Yongping Zhao, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Binbin Zhao, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Yurong Xie, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China; HainanYazhou Bay Seed Lab, Sanya, 572025, China.
Hong Jia, Department of Plant Genetics and Breeding, State Key Laboratory of Agrobiotechnology and National Maize Improvement Center, China Agricultural University, Beijing, 100193, China.
Yongxiang Li, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, 10008, China.
Miaoyun Xu, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China; HainanYazhou Bay Seed Lab, Sanya, 572025, China.
Guangxia Wu, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Xiaojing Ma, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Quanquan Li, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Mei Hou, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Changyu Li, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Zhanchao Xia, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Gang He, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Hua Xu, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Zhijing Bai, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China.
Dexin Kong, School of Life Sciences, and State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, 510642, China.
Zhigang Zheng, School of Life Sciences, and State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, 510642, China.
Qing Liu, School of Life Sciences, and State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, 510642, China.
Yuting Liu, School of Life Sciences, and State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, 510642, China.
Jinshun Zhong, School of Life Sciences, and State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, 510642, China.
Feng Tian, Department of Plant Genetics and Breeding, State Key Laboratory of Agrobiotechnology and National Maize Improvement Center, China Agricultural University, Beijing, 100193, China.
Baobao Wang, Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, 100081, China; HainanYazhou Bay Seed Lab, Sanya, 572025, China.
Haiyang Wang, School of Life Sciences, and State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, 510642, China.
H.W. and B.W. conceived and designed the project. Y.Z., B.Z., and Y.X. performed most of the experiments. M.X., H.X., Z.B., Q.Liu, Y.Liu, and J.Z. helped with some molecular experiments, G.W., Q.Li, Z.X., G.H., D.K., and Z.Z. conducted some field experiments and agronomic traits measurement. X.M. conducted Arabidopsis transformation and phenotypic analysis. M.H. and C.L. helped with bioinformatics analyses, Y.Li performed GWAS analysis of flowering time, and H.J. and F.T. analyzed the distribution of ZmELF3.1 genotypes in 1,008 accessions native to the Americas. H.W., Y.Z., B.W., and Y.X. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Haiyang Wang (whyang@scau.edu.cn).
References
- Bailey TL, Williams N, Misleh C, Li W (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2004) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265 [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115: 721–733 [DOI] [PubMed] [Google Scholar]
- Bouchet S, Servin B, Bertin P, Madur D, Combes V, Dumas F, Brunel D, Laborde J, Charcosset A, Nicolas S (2013) Adaptation of maize to temperate climates: mid-density genome-wide association genetics and diversity patterns reveal key genomic regions, with a major contribution of the Vgt2 (ZCN8) locus. PLoS One 8: e71377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Brady SM, Burow M, Busch W, Carlborg Ö, Denby KJ, Glazebrook J, Hamilton ES, Harmer SL, Haswell ES, Maloof JN, et al. (2015) Reassess the t test: interact with all your data via ANOVA. Plant Cell 27: 2088–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu T, Lu S, Wang K, Dong L, Li S, Xie Q, Xu X, Cheng Q, Chen L, Fang C, et al. (2021) A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc Natl Acad Sci USA 118: e2010241118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Cai Z, Zhang Y, Tang W, Chen X, Lin C, Liu Y, Ye Y, Wu W, Duan Y (2022) LUX ARRHYTHMO interacts with ELF3a and ELF4a to coordinate vegetative growth and photoperiodic flowering in irce. Front Plant Sci 13: 853042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M (2013) HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199: 1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelletti S, Tuberosa R, Pindo M, Salvi S (2014) A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL Vgt1. G3 (Bethesda, MD) 4: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A (2004) Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou J (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Helfer A, Nusinow DA, Kay SA (2012) ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signal Behav 7: 170–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Ducrocq S, Madur D, Veyrieras JB, Camus-Kulandaivelu L, Kloiber-Maitz M, Presterl T, Ouzunova M, Manicacci D, Charcosset A (2008) Key impact of Vgt1 on flowering time adaptation in maize: evidence from association mapping and ecogeographical information. Genetics 178: 2433–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequnce alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2021) Food and Agriculture Organization of the United Nations Agriculture Databases (FAO). http://faostat.fao.org/
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA (2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109: 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, Holland JB, Flint-Garcia S, McMullen MD, et al. (2011) Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet 7: e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford B, Deng W, Clausen J, Oliver S, Boden S, Hemming M, Trevaskis B (2016) Barley (Hordeum vulgare) circadian clock genes can respond rapidly to temperature in an EARLY FLOWERING 3-dependent manner. J Exp Bot 67: 5517–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Wang X, Zhao M, Huang C, Li C, Li D, Yang CJ, York AM, Xue W, Xu G, et al. (2018) Stepwise cis-regulatory changes in ZCN8 contribute to maize flowering-time adaptation. Curr Biol 28: 3005–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Kamal N, Bauer E, Gundlach H, Fischer I, Seidel MA, Spannagl M, Marcon C, Ruban A, Urbany C, et al. (2020) European maize genomes highlight intraspecies variation in repeat and gene content. Nat Genet 52: 950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Sun H, Xu D, Chen Q, Liang Y, Wang X, Xu G, Tian J, Wang C, Li D, et al. (2018) ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci USA 115: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, Liu J, Ricci WA, Guo T, Olson A, Qiu Y, et al. (2021) De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 373: 655–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HY, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, McMullen MD, Ware D, Buckler ES, Doebley JF, et al. (2012) ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA 109: E1913–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Hiei Y, Komari T (2007) Agrobacterium-mediated transformation of maize. Nat Protoc 2: 1614–1621 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang C, Huang S, Xie F, Xu Y, Liu C, Li L (2019) The ELF3-PIF7 interaction mediates the circadian gating of the shade response in Arabidopsis. iScience 22: 288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Liu X, Jia W, Liu H, Li W, Peng Y, Du Y, Wang Y, Yin Y, Zhang X, et al. (2018) ZmCOL3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J Integr Plant Biol 60: 465–480 [DOI] [PubMed] [Google Scholar]
- Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F, et al. (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585: 256–260 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, King OD (2014) PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30: 2501–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazakis CM, Coneva V, Colasanti J (2011) ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J Exp Bot 62: 4833–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wu G, Zhao Y, Wang B, Zhao B, Kong D, Wei H, Chen C, Wang H (2020) CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height. Plant Biotechnol J 18: 2520–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YX, Li C, Bradbury PJ, Liu X, Lu F, Romay CM, Glaubitz JC, Wu X, Peng B, Shi Y, et al. (2016) Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. Plant J 86: 391–402 [DOI] [PubMed] [Google Scholar]
- Liang Y, Liu HJ, Yan J, Tian F (2021) Natural variation in crops: realized understanding, continuing promise. Annu Rev Plant Biol 72: 357–385 [DOI] [PubMed] [Google Scholar]
- Liang Y, Liu Q, Wang X, Huang C, Xu G, Hey S, Lin HY, Li C, Xu D, Wu L, et al. (2018) ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN 8 regulatory module and contributes to maize flowering time adaptation. New Phytol 221: 2335–2347 [DOI] [PubMed] [Google Scholar]
- Liew LC, Hecht V, Sussmilch FC, Weller JL (2014) The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol 165: 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhou X, Li Q, Wang L, Xing Y (2020) CCT domain-containing genes in cereal crops: flowering time and beyond. Teor Appl Genet 133: 1385–1396 [DOI] [PubMed] [Google Scholar]
- Lu S, Zhao X, Hu Y, Liu S, Nan H, Li X, Fang C, Cao D, Shi X, Kong L, et al. (2017) Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet 49: 773–779 [DOI] [PubMed] [Google Scholar]
- Mascheretti I, Turner K, Brivio RS, Hand A, Colasanti J, Rossi V (2015) Florigen-encoding genes of day-neutral and photoperiod-densitive maize are regulated by different chromatin modifications at the floral transition. Plant Physiol 168: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M (2012) Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol 53: 709–716 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J (2002) A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99: 6080–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR (2021) Circadian clock components offer targets for crop domestication and improvement. Genes 12: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON (2011) The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23: 942–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Nitta M, Sato K, Nasuda S (2012) A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet Syst 87: 357–367 [DOI] [PubMed] [Google Scholar]
- Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T (2005) Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci Biotechnol Biochem 69: 410–414 [DOI] [PubMed] [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE (2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA 108: 16469–16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) Pseudo-response regulators, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46: 686–698 [DOI] [PubMed] [Google Scholar]
- NCGA (2021) National corn growers association, world of corn (NCGA, WOC, 2021). http://www.worldofcorn.com/#us-select-crop-value
- Nieto C, López-Salmerón V, Davière JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin PA, Zhang W, Vogt Sebastian H, Dally N, Büttner B, Schulze-Buxloh G, Jelly Noémie S, Chia Tansy YP, Mutasa-Göttgens ES, Dohm JC, et al. (2012) The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr Biol 22: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Raschke A, Ibañez C, Ullrich KK, Anwer MU, Becker S, Glöckner A, Trenner J, Denk K, Saal B, Sun X, et al. (2015) Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol 15: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Bastow RM, Solomon KS, Dowson-Day MJ, Elumalai RP, Millar AJ (2000) Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol 122: 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Navarro JA, Willcox M, Burgueño J, Romay C, Swarts K, Trachsel S, Preciado E, Terron A, Delgado HV, et al. (2017) A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nat Genet 49: 476–480 [DOI] [PubMed] [Google Scholar]
- Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, Yokoo T, Matsubara K, Hori K, Yano M, Inoue H, et al. (2012) Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol 53: 717–728 [DOI] [PubMed] [Google Scholar]
- Salome PA, Weigel D, McClung CR (2010) The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22: 3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S, Sponza G, Morgante M, Tomes D, Niu X, Fengler KA, Meeley R, Ananiev EV, Svitashev S, Bruggemann E, et al. (2007) Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci USA 104: 11376–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Springer NM, Anderson SN, Andorf CM, Ahern KR, Bai F, Barad O, Barbazuk WB, Bass HW, Baruch K, Ben-Zvi G, et al. (2018) The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat Genet 50: 1282–1288 [DOI] [PubMed] [Google Scholar]
- Sun H, Wang C, Chen X, Liu H, Huang Y, Li S, Dong Z, Zhao X, Tian F, Jin W (2020) dlf1 promotes floral transition by directly activating ZmMADS4 and ZmMADS67 in the maize shoot apex. New Phytol 228: 1386–1400 [DOI] [PubMed] [Google Scholar]
- Sun S, Zhou Y, Chen J, Shi J, Zhao H, Zhao H, Song W, Zhang M, Cui Y, Dong X, et al. (2018) Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat Genet 50: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Swarts K, Gutaker RM, Benz B, Blake M, Bukowski R, Holland J, Kruse-Peeples M, Lepak N, Prim L, Romay MC, et al. (2017) Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America. Science 357: 512–515 [DOI] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler EST (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28: 286–289 [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulatorPpd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Undurraga SF, Press MO, Legendre M, Bujdoso N, Bale J, Wang H, Davis SJ, Verstrepen KJ, Queitsch C (2012) Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc Natl Acad Sci USA 109: 19363–19367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban A, Neukirchen S, Jaeger KE (1997) A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res 25: 2227–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang B, Lin Z, Li X, Zhao Y, Zhao B, Wu G, Ma X, Wang H, Xie Y, Li Q, Song G, et al. (2020) Genome-wide selection and genetic improvement during modern maize breeding. Nat Genet 52: 565–571 [DOI] [PubMed] [Google Scholar]
- Wang B, Hou M, Shi J, Ku L, Song W, Li C, Ning Q, Li X, Li C, Zhao B, et al. (2022) De novo assembly and analyses of twelve founder inbred lines provide genomic insights into heterotic groups and heterosis of maize. Nat Genet Under review [Google Scholar]
- Weller JL, Liew LC, Hecht VF, Rajandran V, Laurie RE, Ridge S, Wenden B, Vander Schoor JK, Jaminon O, Blassiau C, et al. (2012) A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Natl Acad Sci USA 109: 21158–21163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydro M, Kozubek E, Lehmann P (2006) Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim Pol 53: 289–298 [PubMed] [Google Scholar]
- Xiao C, Chen F, Yu X, Lin C, Fu YF (2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71: 39–50 [DOI] [PubMed] [Google Scholar]
- Xie Y,, Liu Y,, Ma M,, Zhou Q,, Zhao Y,, Zhao B,, Wang B,, Wei H,, Wang H (2020) Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat Commun 11: 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Lu Y, Yang X, Huang J, Zhou Y, Ali F, Wen W, Liu J, Li J, Yan J (2014) Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 gronomic traits in an enlarged maize association panel. PLoS Genet 10: e1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Li Z, Li W, Ku L, Wang C, Ye J, Li K, Yang N, Li Y, Zhong T, et al. (2013) CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci USA 110: 16969–16974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Baliji S, Buchmann RC, Wang H, Lindbo JA, Sunter G, Bisaro DM (2007) Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J Virol 81: 11972–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. (2005) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR (1996) The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702 [DOI] [PubMed] [Google Scholar]
- Zakhrabekova S, Gough SP, Braumann I, Müller AH, Lundqvist J, Ahmann K, Dockter C, Matyszczak I, Kurowska M, Druka A, et al. (2012) Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci USA 109: 4326–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huang X, Ouyang X, Chen W, Du A, Zhu L, Wang S, Deng X, Li S (2012) OsELF3-1, an ortholog of Arabidopsisearly flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One 7: e43705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li W, Mao L, Chen B, Xu Y, et al. (2016) An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep 6: 23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Peng Q, Fu D, Zhuang D, Yu Y, Duan M, Xie W, Cai Y, Ouyang Y, Lian X, et al. (2018) The E3 ubiquitin ligase HAF1 modulates circadian accumulation of EARLY FLOWERING3 to control heading date in rice under long-day conditions. Plant Cell 30: 2352–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.