Abstract
Photosystem II (PSII) contains many lipid molecules that are essential for the function and maintenance of PSII. Under strong light conditions, PSII complexes are dynamically modified during the repair process; however, the molecular mechanism of the dynamic changes in the PSII structure is still unclear. In the present study, we investigated the role of a lipase in the repair of PSII in Synechocystis sp. PCC 6803. We identified a protein encoded by the sll1969 gene, previously named lipase A (lipA), in the Synechocystis sp. PCC 6803 genome as a candidate for the lipase involved in PSII repair. Recombinant protein expressed in Escherichia coli cells hydrolyzed fatty acids at the sn-1 position of monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol as well as triacylglycerol esterified with stearic acids. PSII repair in a disrupted mutant of the lipA gene was suppressed by the slow degradation of damaged D1 protein under strong light. The level of the PSII dimer remained higher in lipA mutant cells than wild-type (WT) cells under strong light. LipA protein was associated with the PSII dimer in vivo, and recombinant LipA protein decomposed PSII dimers purified from WT cells to monomers by reducing MGDG content in the PSII complex. These results indicate that LipA reacts with PSII dimers, dissociates them into monomers by digesting MGDG, and enhances D1 degradation during PSII repair.
A lipase deacylates galactolipids presumably at the interface of photosystem II (PSII) dimer and induces the disassembly of the dimer that accelerates the protein degradation and repair of PSII.
Introduction
Photosynthesis uses light energy to fix carbon dioxide into organic compounds such as carbohydrates, which support the life of almost all living organisms on Earth. Understanding the mechanism of photosynthesis and taking advantage of the highly efficient energy conversion by photosynthesis are essential for our sustainable future. The energy conversion by photosynthesis occurs on the thylakoid membrane that consists of four main classes of glycerolipids: monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), sulfoquinovosyldiacylglycerol (SQDG), and phosphatidylglycerol (PG). These lipids in the thylakoid membrane perform various important functions in photosynthesis besides forming lipid bilayers of the membrane (Wada and Murata, 1998; Mizusawa and Wada, 2012).
Photosystem II (PSII) is an efficient energy converter that oxidizes water molecules by using light energy to supply electrons to the following components involved in photosynthetic-electron transport. PSII is a huge protein complex containing various cofactors such as metals, quinones, carotenoids, chlorophylls (Chls), and lipids. These cofactors are essential for the function and maintenance of PSII. Because PSII is fragile under various environmental stresses, damaged PSII is rapidly and constantly repaired in cells. During the repair, PSII complexes are dynamically changed from dimers to monomers and then CP43-less monomers followed by reconstruction of the reaction center. However, the molecular mechanism for the dynamic changes in PSII structure during the repair process is still unclear.
The crystal structure of the PSII dimer from Thermosynechococcus vulcanus has revealed several lipid molecules at the monomer/monomer interface in a PSII dimer (Umena et al., 2011). Such lipid molecules might be involved in the dimerization of PSII. In fact, applying a phospholipase A2 to PSII dimers purified from Spinacia oleracea (spinach) showed that the deacylation of PG induced monomerization of PSII dimers and the following transformation to CP43-less monomers (Kruse et al., 2000), suggesting that lipids play an important role in the dimerization of PSII. However, the results obtained from experiments with lipase treatment could not provide direct evidence for a role of lipids in PSII in vivo. Recently, Kojima et al. (2022) reported that strong-light stress activated lipid deacylation in Synechocystis sp. PCC 6803 (hereafter Synechocystis), and a mutant defective in acyl–acyl carrier protein synthetase accumulated free fatty acids because free fatty acids generated by deacylation of membrane lipids cannot be recycled in the mutant. These findings suggest that lipid molecules are remodeled by deacylation with lipases and reacylation with acyltransferases under strong light conditions, and such remodeling of lipids plays an important role in the repair process of PSII. However, no lipases involved in the repair process of PSII under strong light conditions have been identified in any photosynthetic organisms.
In the present study, we identified a lipase, previously named LipA, that hydrolyzes fatty-acyl chains at the sn-1 position of galactolipids and triacylglycerol (TAG), in Synechocystis. Biochemical analysis of recombinant LipA protein expressed in Escherichia coli cells and characterization of the lipA mutant of Synechocystis clarified the important function of LipA in the repair process of PSII. Namely, the deacylation of MGDG by LipA enhanced PSII repair under strong light by decomposing PSII dimers to monomers and enhancing degradation of damaged D1.
Results
Sll1969 cleaves C18 acyl-chains bound to galactolipids and TAG
In the genome of Synechocystis, several genes are annotated as the genes for lipases. One of the genes, sll1969, is annotated as the gene for TAG lipase in the KEGG database. A similarity search by using Gclust server 2021 (Sato, 2009) revealed that the lipase is well conserved in cyanobacterial strains and many bacterial species (Supplemental Figure S1A), however, it is not similar to a galactolipase, PGD1 found in green algae Chlamydomonas reinhardtii (Li et al., 2012). The α/β-hydrolase domain, which is usually found in the common lipases, was present in the Synechocystis Sll1969, and the domain of Sll1969 was similar to those of lipases from other bacteria and archaea; however, the amino acid sequence at the C-terminus region of Sll1969 seemed to differ from those of other lipases (Supplemental Figure S1B).
In the previous study, sll1969 was considered as a putative lipase gene and designated as lipA (Liu and Curtiss, 2012), however, the enzymatic properties of the encoded protein were not fully investigated. Moreover, Eungrasamee et al. (2019) reported sll1969 as a gene for phospholipase without any experimental evidence (Eungrasamee et al., 2019). To clarify the molecular functions of Sll1969, we expressed it as a protein with a 6×His-tag at the C-terminus in E. coli BL21 (DE3) cells harboring the pET24a (+) vector that contains the sl1969 gene (Supplemental Figure S2A) and purified the recombinant protein from the cells. About 0.5 mg of recombinant protein was obtained from 1 L cell culture, which indicates that Sll1969 protein does not show any lethality in E. coli cells, unlike another plant-type phospholipases (Wang et al., 2017).
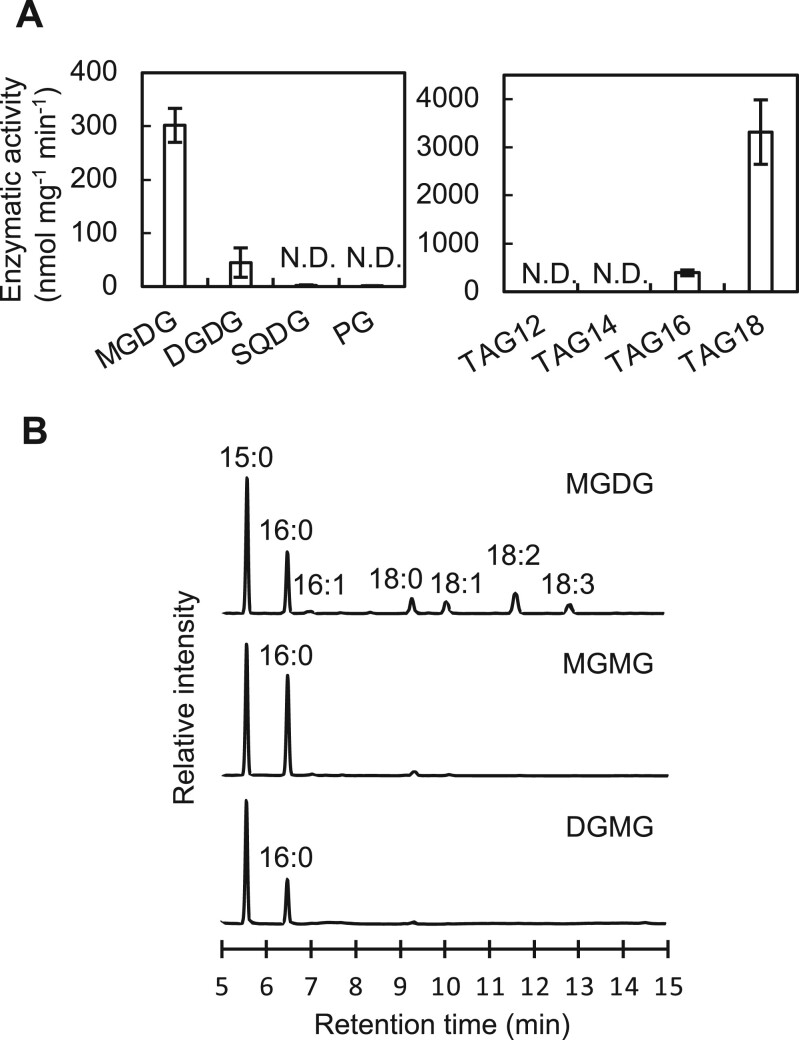
To investigate the enzymatic specificity of His-tagged Sll1969 protein, we measured its activity with p-nitrophenyl stearate as a substrate (Supplemental Figure S2). The optimal temperature and pH for the enzymic activity were determined as 35°C and pH 7.5, respectively (Supplemental Figure S2). Then we measured the activity with four glycerolipids (MGDG, DGDG, SQDG, and PG) present in Synechocystis cells. Recombinant Sll1969 protein specifically reacted with the galactolipids MGDG and DGDG but not with SQDG and PG. The mean reaction rates for MGDG and DGDG were 302 ± 32 and 45 ± 27 nmol mg−1 protein min−1, respectively (Figure 1A). The produced monogalactosylmonoacylglycerol (MGMG) and digalactosylmonoacylglycerol (DGMG) after the reaction contained mostly palmitic acid (16:0, fatty acid containing 16 carbons without double bond) (Figure 1B), which binds to the sn-2 position of glycerolipids in Synechocystis (Okazaki et al., 2006). These results suggest that Sll1969 cleaves fatty acids at the sn-1 position of galactolipids, but not SQDG and PG. The recombinant protein also reacted with TAG containing stearic (18:0) (TAG18) and palmitic acids (16:0) (TAG16). The mean reaction rates for TAG18 and TAG16 were 3,317 ± 669 and 393 ± 60 nmol mg−1 protein min−1, respectively (Figure 1A), so Sll1969 functions as a TAG lipase as well. Therefore, Sll1969 is not specific to galactolipids and cleaves fatty acids at the sn-1 position of galactolipids and TAGs. Hence, we designated it as lipA as reported previously (Liu and Curtiss, 2012). In comparison to MGDG, DGDG, and TAG, both PG and SQDG having an acidic head group were not digested by LipA, which suggests that the activity of LipA might be inhibited by the negative charge in the head groups of PG and SQDG. Since LipA cleaved C18-fatty acids at the sn-1 position of galactolipids, it seems LipA specific to fatty acids bound to the sn-1 position. However, in Synechocystis, C18-fatty acids are exclusively bound to the sn-1 position of glycerolipids, and the sn-2 position is esterified only with 16:0 (Wada and Murata, 1990), thus we cannot exclude the possibility that LipA hydrolyzes C18-fatty acids bound to uncharged lipids regardless of the sn positions.
Figure 1.
Enzymatic activity of LipA in vitro and in vivo. A, Enzymatic activity of purified Sll1969 protein with a 6× His-tag (5 μg) measured with various lipid compounds (100 nmol) in 250 μL of 50 mM Tris–HCl (pH7.5) at 35°C. Reaction times were 30 min for membrane lipids (MGDG, DGDG, SQDG, and PG) and 5 min for TAG. MGDG, DGDG, SQDG, and PG used for the assay were purified from Synechocystis cells and quantified by GC. TAG compounds were purchased from Tokyo Chemical Institute (Tokyo). Values are mean ± sd of results from three independent experiments. TAG12, tri-laureateglycerol; TAG14, tri-myristateglycerol; TAG16, tri-palmitateglycerol; TAG18, tri-stearateglycerol; and N.D., not detected. B, Gas chromatograms of fatty acid methyl-esters (MEs) from MGDG purified from Synechocystis cells, and MGMG and DGMG obtained after the reaction of LipA protein with MGDG and DGDG, respectively. 15:0, pentadecanoic acid as an internal standard; 16:0, palmitic acid; 16:1, palmitoleic acid; 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid; and 18:3, γ-linolenic acid.
To examine the roles of LipA protein in vivo, we disrupted the sll1969 gene by inserting the spectinomycin/streptomycin-resistant gene cassette (Sp/StR) into the middle of sll1969 (Supplemental Figure S3). The generated mutant of Synechocystis was named lipA. When compared with the MGDG content in wild-type (WT) cells, MGDG content in the lipA cells increased at the expense of PG content (Figure 2A). By combining thin-layer chromatography (TLC) and gas chromatography (GC), we also quantified the amount of MGMG, DGMG, and TAG, which are present in cyanobacterial cells as minor lipids. We did not find any differences in the amount of DGMG in WT and lipA cells (Figure 2B), but the amount of MGMG was lower in lipA cells than WT cells (Figure 2B), which suggests that Sll1969 deacylates MGDG and produces MGMG in vivo. The amount of TAG was eight times higher in lipA cells than in WT cells (Figure 2B). The presence of TAG in Synechocystis cells is still under discussion (Aizouq et al., 2020; Tanaka et al., 2020); however, our results support the presence of TAG and the metabolic pathway for TAG turnover in Synechocystis cells. LipA showed very high reactivity for TAG (Figure 1A), which suggests that TAG produced by TAG synthase encoded in the slr2103 gene (Aizouq et al., 2020) might be rapidly degraded in Synechocystis cells, although the physiological meaning of the rapid turnover of TAG in cyanobacteria is still unknown. In addition, because TAG is a very minor compound comparing with MGDG in Synechocystis cells, therefore LipA should react with mainly MGDG.
Figure 2.
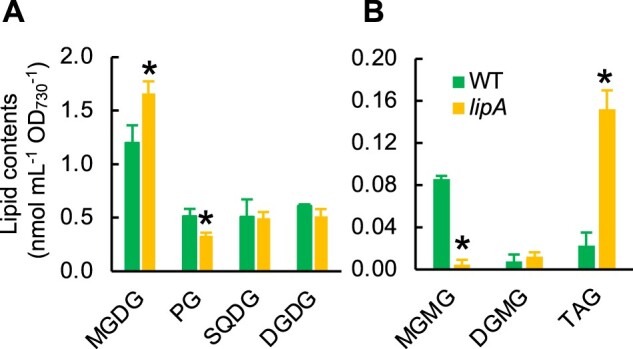
Contents of lipids in WT and lipA cells. A, Contents of major membrane lipids. Lipids are extracted from the WT and lipA cells by Bligh–Dyer method. Four major membrane lipids were separated and detected by the high-performance liquid chromatography-evaporative light scattering detector (HPLC-ELSD) system. Values are mean ± sd of results from three independent experiments. Asterisks indicate statistically significant differences (P < 0.01, Student’s t test). B, Contents of minor lipids. MGMG and DGMG were separated on the TLC plate and analyzed by GC, whereas TAG was separated and detected by HPLC-ELSD as used for analysis of major membrane lipids. Values are mean ± sd of results from three independent experiments. Asterisks indicate statistically significant differences (P < 0.01, Student’s t test).
LipA enhances the degradation of damaged D1 during PSII repair
To investigate the effect of disruption of the lipA gene on the photoinhibition of PSII, we exposed WT and lipA cells to strong light at 1,500 μmol photons m−2 s−1 at 32°C. The mean initial activities of PSII at time 0 in the WT and lipA cells were 661.4 ± 43.0 and 711.7 ± 31.5 μmol O2 mg−1 Chl h−1, respectively, so lack of LipA did not affect the activity of PSII. When the cells were exposed to strong light, the PSII activity of the WT cells decreased to 57% of the initial activity within 80 min after exposure to strong light (Figure 3A), whereas the PSII activity of lipA cells retained only 36% of the initial activity (Figure 3A). Thus, the lack of LipA extends the photoinhibition of PSII. However, the rates of photodamage to PSII in both cells, as monitored in the presence of lincomycin, did not significantly differ between the WT and lipA cells (Figure 3B). These results indicate that LipA enhances the repair of damaged PSII.
Figure 3.
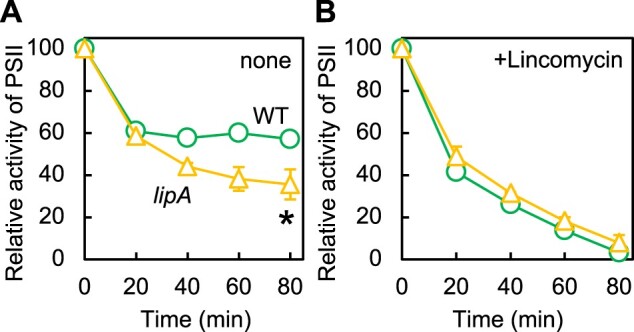
PSII photoinhibition in WT and lipA cells. WT (circles) or lipA (triangles) cells were incubated at 32°C under strong light at 1,500 μmol photons m−2 s−1 with aeration by ambient air, in the absence (A) or presence (B) of 200 μg mL−1 lincomycin. PSII activity of WT and lipA cells was measured by the evolution of oxygen in the presence of 1 mM 1,4-benzoquinone and 1 mM K3Fe(CN)6. The mean activities ± sd taken as 100% for WT and lipA cells were 661.4 ± 43.0 and 711.7 ± 31.5 μmol O2 mg−1 Chl h−1, respectively. Values are mean ± sd of results from three independent experiments. Asterisks indicate statistically significant differences (P < 0.01, Student’s t test).
Turnover of D1 has a central role in the repair of PSII (Murata and Nishiyama, 2018). To investigate the turnover rate of D1, we chased the 35S-labeled D1 under the same condition used for monitoring photoinhibition (Figure 4). In both cell types, almost the same amount of D1 protein was labeled during pulse-labeling, so the rates of de novo synthesis of D1 were similar in WT and lipA cells. By contrast, the degradation rates of labeled D1 differed between WT and lipA cells. In WT cells, only 26% of labeled D1 remained after pulse-chasing for 60 min (at 80 min in Figure 4B), whereas in lipA cells, 72% of labeled D1 remained after the pulse chase (Figure 4B). Thus, LipA enhances degradation of D1 during PSII repair. Since the compositions of membrane lipids were altered in lipA cells (Figure 2), the PSII turnover might be affected by the change in the properties of lipid phase of thylakoid membranes.
Figure 4.
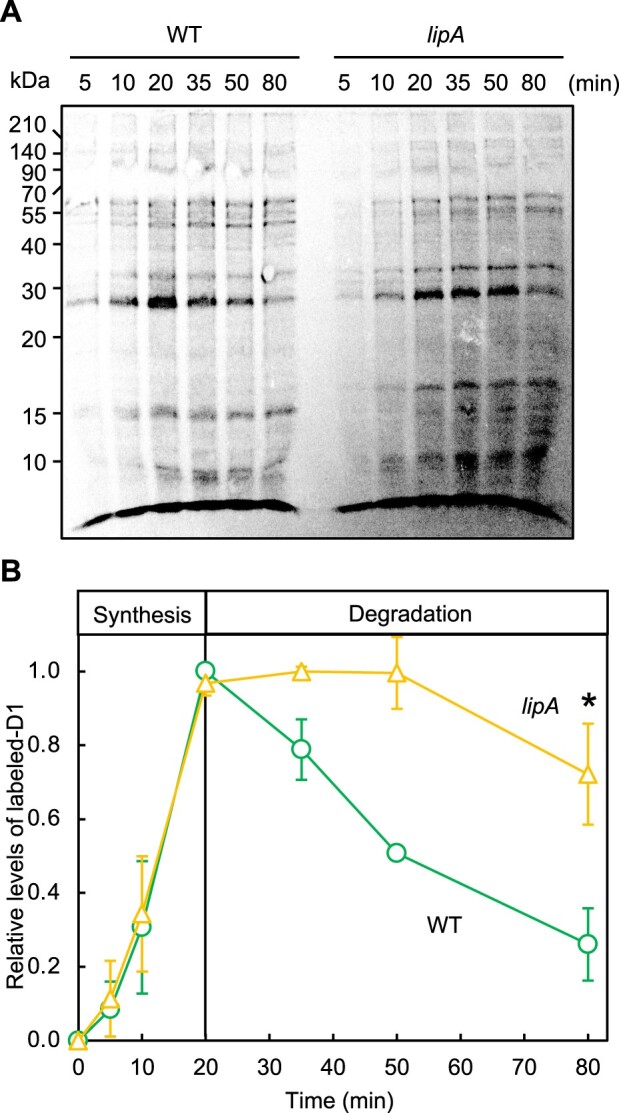
D1 degradation in WT and lipA cells. A, Proteins in WT and lipA cells were pulse-labeled by incubating cells with 35S-labeled methionine and cysteine for the indicated times under strong light at 1,500 μmol photons m−2 s−1 with aeration by ambient air. After labeling for 20 min, 1 mM each of cold methionine and cysteine was added and chased for the designated time. Thylakoid membranes were isolated, and obtained membranes corresponding to 4 μg Chl were analyzed by SDS-PAGE. A, Representative radiogram of pulse-chased proteins from thylakoid membranes. B, Quantification of the relative levels of labeled D1 protein. Values are mean ± sd of results from three independent experiments. The value for WT cells after pulse-labeling for 20 min was designated 1.0. Asterisk indicates statistically significant differences (P < 0.01, Student’s t test).
LipA decomposes PSII dimers into monomers
To analyze the cellular localization, LipA protein was detected with a specific antibody against synthetic oligopeptides corresponding to the partial LipA protein. LipA protein was detected in the thylakoid membrane fraction from WT but not lipA cells (Figure 5A), although photosystem I subunit A (PsaA) protein used as a marker protein present in the thylakoid membrane was detected in thylakoid membrane fractions from both WT and lipA cells. We also analyzed the distribution of LipA protein in photosynthetic complexes separated by BN-PAGE. LipA protein was detected only in the PSII dimer separated by BN-PAGE following SDS-PAGE (Figure 5B). These results indicate that LipA protein is preferentially associated with PSII dimer. We analyzed the effect of disruption of the lipA gene on the PSII complexes in vivo and in vitro by blue native PAGE (BN-PAGE) (Figure 6A). During PSII repair, PSII dimers become monomers. In a diatom, Chaetoceros gracilis, the conversion of PSII dimers to monomers can be observed under strong light when PSII repair is inhibited by an inhibitor of protein synthesis (Nagao et al., 2016). Here, the level of PSII dimers decreased in WT cells but was unchanged in lipA cells with 30-min exposure to strong light (Figure 6B), which suggests that LipA dissociates PSII dimers into monomers. Other photosynthetic complexes, such as PSI trimers and monomers, did not differ in content between WT and lipA cells.
Figure 5.
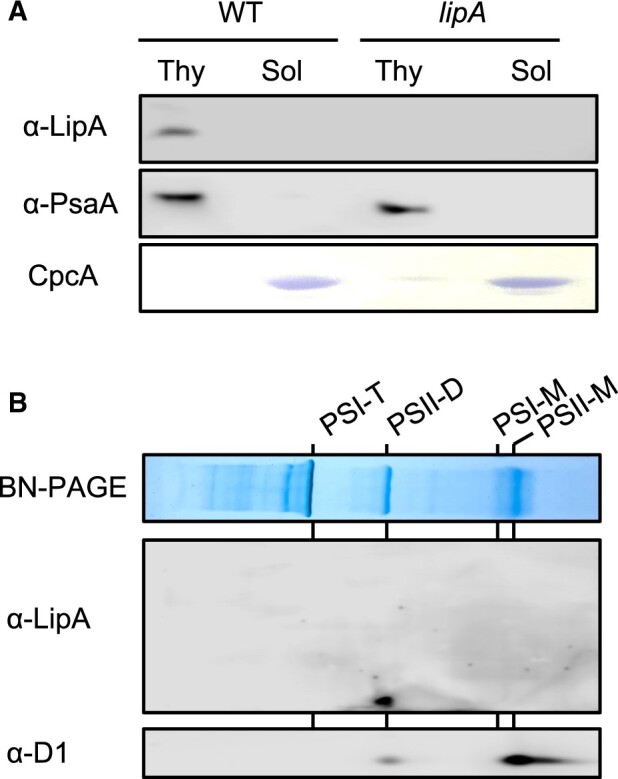
Localization of LipA protein. A, Distribution of LipA protein in fractions of thylakoid membranes or soluble proteins from WT and lipA cells analyzed with a specific antibody. PsaA and C-phycocyanin alpha chain (CpcA) proteins were used as the controls for thylakoid and soluble proteins, respectively. B, Localization of LipA protein in photosynthetic complexes in thylakoid membranes. Photosynthetic complexes in thylakoid membranes (corresponding to 8 μg chlorophyll) prepared from WT cells exposed to strong light condition for 20 min were separated on BN-PAGE, and the distribution of LipA protein in photosynthetic complexes was analyzed with specific antibodies. PSI-T, PSI trimer; PSII-D, PSII dimer; PSI-M, PSI monomer; and PSII-M, PSII monomer.
Figure 6.
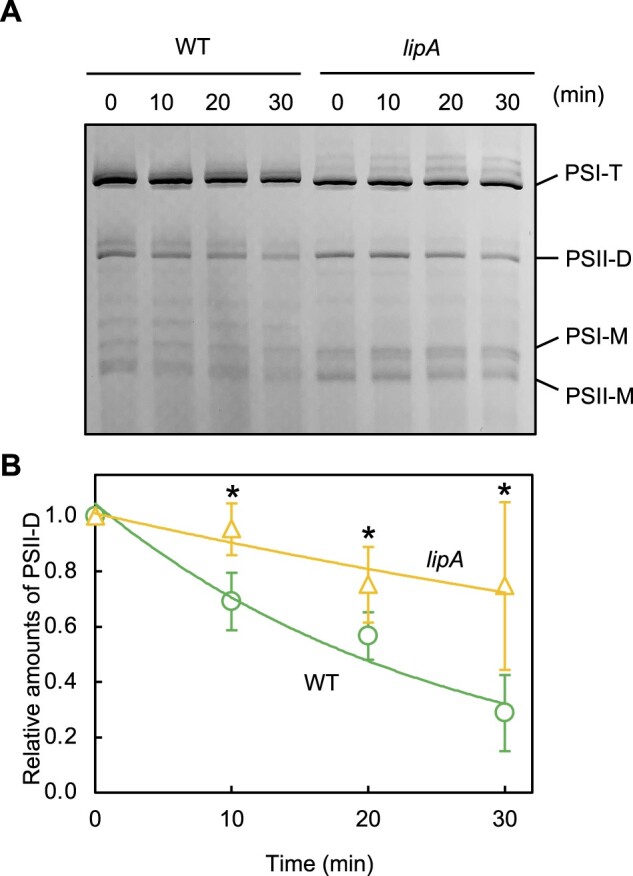
Roles of LipA in PSII complexes in vivo. A, Photosynthetic complexes in WT and lipA cells analyzed by BN-PAGE. Thylakoid membranes were prepared from WT and lipA cells that were exposed to strong light at 1,500 μmol photons m−2 s−1 (SL) at 32°C in the presence of 200 μg mL−1 lincomycin. Photosynthetic complexes were separated on BN-PAGE. PSI-T, PSI trimer; PSII-D, PSII dimer; PSI-M, PSI monomer; and PSII-M, PSII monomer. B, The relative level of PSII-D was quantified densitometrically. Asterisks indicate statistically significant differences between WT and lipA cells at the point of 30 min (P < 0.01, Student’s t test).
In vitro analysis of PSII monomers and dimers purified from WT cells showed that LipA protein specifically reacted with PSII dimers but not monomers and decomposed the dimers into monomers (Figure 7A). After the reaction of PSII dimers and monomers with LipA protein, the content of MGDG in PSII dimers but not monomers was reduced (Figure 7B). Therefore, LipA protein associates with PSII dimers and induces monomerization of dimers by deacylating MGDG molecules in dimers.
Figure 7.
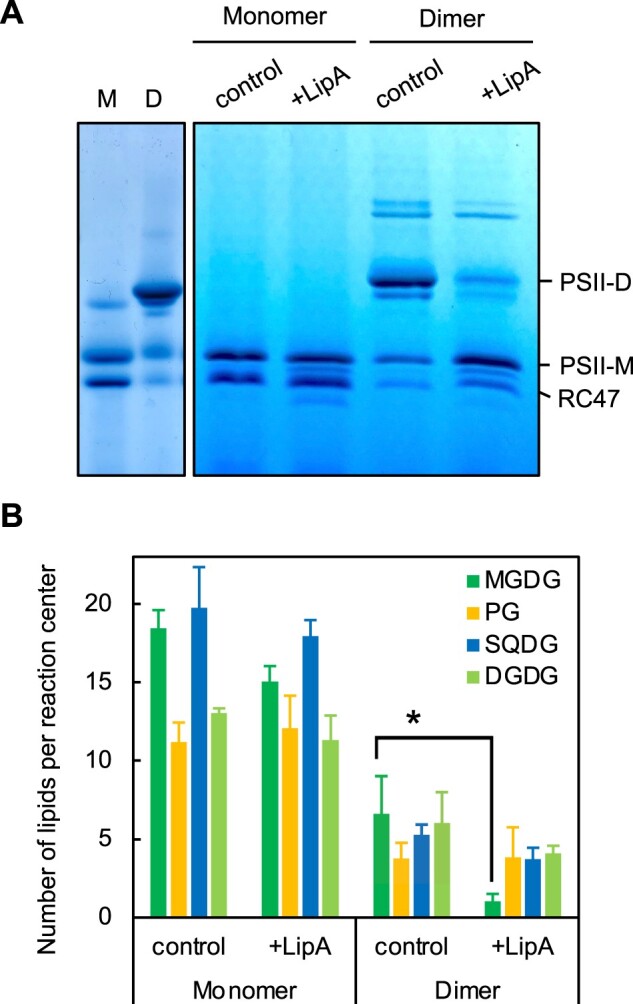
Roles of LipA in PSII complexes in vitro. Purified PSII monomers and dimers (corresponding to 10 μg chlorophyll) were incubated with recombinant LipA protein (5 μg proteins) at 32°C for 10 min, and PSII complexes (A) and lipid contents (B) were analyzed by BN-PAGE and high-performance liquid chromatography-evaporative light scattering detector system, respectively. D, dimer; M, monomer; and RC47, CP43-less monomer. Values for level of PSII-D are mean ± sd of results from three independent experiments. Asterisks indicate statistically significant differences between WT and lipA cells (P < 0.01, Student’s t test).
Discussion
Role of LipA in the repair of PSII
To reveal the role of lipids in PSII complexes, previous studies have used treatments of purified PSII complexes with commercially available lipases and cyanobacterial mutants defective in the biosynthesis of thylakoid lipids (Kruse et al., 2000; Laczko-Dobos et al., 2008; Endo et al., 2016). However, the findings obtained from the studies with lipase treatments do not provide direct evidence for a role of lipids in PSII and cannot be directly applied to the function of lipids in vivo. Also, findings obtained with mutants may include indirect effects on PSII because lack of lipids in mutant cells affects many physiological processes. In the present study, we demonstrated that a lipase, LipA, encoded in sll1969 gene in the Synechocystis genome has a role in the monomerization of PSII dimers in vivo, which is required for the efficient degradation of D1 during PSII repair under strong light conditions. In the crystal structure of the PSII dimer from T. vulcanus, two SQDG and one MGDG molecules are located at the monomer/monomer interface of the PSII dimer (Umena et al., 2011; Yoshihara and Kobayashi, 2022). Thus, the MGDG molecule at the interface might be a target of LipA to trigger the monomerization of PSII dimers. How does LipA recognize PSII dimers to be monomerized? When the purified PSII dimers from T. vulcanus were treated with a commercially available lipase, MGDG molecules in the PSII dimers were partially digested, but the dimers were not monomerized (Leng et al., 2008), which suggests that the MGDG molecules, presumably located at the monomer/monomer interface of PSII dimers and involved in the dimerization, cannot be accessed by the non-native lipase. The phylogenic analysis showed that lipA genes were well conserved in cyanobacteria, and consist of different cyanobacterial clades from other bacterial lipases (Supplemental Figure S1A), which suggests a specific function in photosynthesis. Thus, the difference of amino acids at C-terminal domain in LipA proteins may give a specificity and regulation of the lipase activity.
Land plants and cyanobacteria exhibit monomerization of PSII dimers under strong light (Nickelsen and Rengstl, 2013). Purified PSII monomers show a much lower oxygen-evolving rate than do purified PSII dimers (Sakurai et al., 2007). Thus, the monomerization of PSII can be considered as a photoprotective mechanism to reduce the electron supply for the following components in photosynthetic-electron transport. In Chlamydomonas, a galactolipase named plastid galactolipid degradation 1 (PGD1) deacylates MGDG to supply fatty acids for TAG production and decreases accepter-side limitation in photosynthetic-electron flow (Li et al., 2012; Du et al., 2018). Therefore, deacylation of MGDG by LipA and the monomerization of PSII might be important to prevent overreduction of photosynthetic-electron transport under strong light as well. In cyanobacterial cells and chloroplasts, MGDG contains a high amount of polyunsaturated fatty acids (PUFAs) such as linolenic acid (Wada and Murata, 1998). In Synechocystis, PUFAs are predominantly bound to the sn-1 position of glycerolipids (Okazaki et al., 2006). Under strong light, reactive oxygen species oxidize these PUFA molecules after the production of malondialdehydes as the end products (called lipid peroxidation) (Chan et al., 2012). The lipid peroxidation accelerates photoinhibition of PSII (Maeda et al., 2005; Pospisil and Yamamoto, 2017). Therefore, the turnover of fatty acids in MGDG might prevent the accumulation of lipid peroxidation and further photoinhibition of PSII under strong light.
Turnover of galactolipids in Synechocystis
LipA deacylates MGDG in vitro and in vivo (Figures 1 and 2). In the transcriptomic database, the transcript level of sll1969 did not greatly change when cells were exposed to strong light (Mitschke et al., 2011), which suggests that the sll1969 transcript is stable and the enzymatic activity of LipA might be regulated at the post-transcriptional level. In lipA cells, the content of MGDG increased and that of MGMG decreased under normal conditions when compared with WT cells (Figure 2); thus, LipA is active even under normal conditions. In Arabidopsis thaliana, exposure to strong light decreased the activity of acetyl-CoA carboxylase and the biosynthesis of MGDG (Yu et al., 2021), so under normal conditions, the biosynthesis and degradation of MGDG may be balanced, and the content of MGDG becomes stable. In contrast, under strong light conditions, the biosynthesis of MGDG is suppressed and the content decreases, which might accelerate the decomposition of PSII dimers. Recent study of the large-scale CRISPERi-screening of Synechocystis revealed that knockdown of sll1969 gene increased the cellular growth rate under 1% CO2 conditions (high CO2) (Yao et al., 2020), which indicates that LipA inhibits further cellular growth under optimal conditions in Synechocystis. Synechocystis cells grown under high CO2 conditions showed increased cellular content of MGDG at the expense of PG content (Jimbo et al., 2021). Here, we found that the disruption of the lipA gene increased MGDG content (Figure 2). Therefore, under high CO2 conditions, carbon sources are utilized for cellular components other than MGDG in lipA cells, which might accelerate the cellular growth of Synechocystis.
In green alga, C. reinhardtii, MGMG seems to be a major component of membrane lipids (Iwai et al., 2021). Unlike in Chlamydomonas, in Synechocystis cells, sn-1-lyso-MGDG, a resulting product of the action of LipA, was not a major lipid (Figure 2). In Synechocystis, fatty acids from 14C-labeled lipids supplemented exogenously were transferred to MGMG (Kaczmarzyk and Fulda, 2010), which suggests that a sn-1-lysolipid acyltransferase reacylates the MGMG at the sn-1 position. Among three identified lysolipid acyltransferases, sll1848, sll1752, and slr2060, in the Synechocystis genome, only sll1752 prefers C18-fatty acids such as stearic and oleic acids as the substrates in vitro and in vivo (Okazaki et al., 2006). In Synechocystis, C18-fatty acids are predominantly bound to the sn-1 position of glycerolipids; therefore, sll1752 might be a sn-1-lysolipid acyltransferase that transfers C18-fatty acids to lyso-galactolipids. The role of reacylation of MGMG in regulating photosynthetic activity under strong light needs to be clarified in future experiments.
Materials and methods
Generation of strains and culture conditions
Glucose-tolerant Synechocystis strain (GT-I) was used as the WT cells, and all transformants were derived from the WT cells. To generate a knockout mutant of sll1969, DNA fragments containing the 5′-upstream region (0–300 nt) and 3′-downstream region (301–609 nt) of the sll1969 gene were amplified by PCR with the primers 5′-GTGGTAGCAGAATTTCCGGA-3′ (F1) and 5′-AAGGTTCTGGACCAGCGCCTAACCCGTTCCCC-3′ (R1), and 5′-GACGTCTGTATTAACGAAGCTACATCACCATTTCAGCC-3′ (F2) and 5′-TCAGGGCAACGGTTTAGCC-3′ (R2), which included 15 bp-additional sequences that are identical to Sp/StR. Another DNA fragment containing a Sp/StR gene was amplified with the primers 5′-CTGGTCCAGAACCTT-3′ (F3) and 5′-GCTTCGTTAATACAGACGTC-3′ (R3). The amplified three DNA fragments described above were combined and incubated in the NEBuilder Gibson-assembly reaction mix at 50°C for 30 min (New England Biolabs, Ipswich, Massachusetts, USA). The resulting DNA fragments were used to transform the WT cells. Segregation of the disrupted lipA gene in the genome of the transformants was checked by PCR with the same primer set F1 and R2. WT and transformant cells were grown photoautotrophically at 32°C in liquid BG-11 medium under fluorescence light at 10 μmol photons m−2 s−1 with aeration by sterile ambient air. Cultures with optical density at 730 nm (OD730) of a mean of 1.0 ± 0.1 corresponding to approximately 4.0 μg Chl mL−1 were used for assays unless indicated.
Purification of recombinant protein
The DNA fragments containing a protein coding region of sll1969 without the stop codon were amplified with a pair of primers 5′-GGACAGCAAATGGGTGTGGTAGCAGAATTTCCGGA-3′ (F4) and 5′- CTCGAGTGCGGCCGCGGGCAACGGTTTAGCC-3′ (R4), which included 15 bp-additional sequences that are identical to pET24a (+) vector. The amplified DNA fragments and XhoI-treated pET24a vector were combined and incubated in the NEBuilder Gibson-assembly reaction mix at 50°C for 30 min (New England Biolabs). The resultant DNA was used for the following purification of recombinant protein. Escherichia coli BL21 (DE3) cells harboring the pET24a (+) vector containing the sll1969 gene were grown in 100 mL LB medium at 37°C in an orbital shaker (150 rpm) overnight. The cell culture was then transferred to 1 L LB medium and incubated at 37°C until OD650 0.5. For the induction of the high expression of LipA protein, 0.1 mM isopropyl β-d-1-thiogalactopyranoside at a final concentration was added, and the cell culture was incubated at 20°C for 4 h. Cells in 10 mL breaking buffer (20 mM Tris–HCl pH 7.5, 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 0.1 mg mL−1 lysozyme) were disrupted by ultrasonic sonication (Branson). Total soluble proteins were obtained by centrifugation (22,000 g) at 4°C for 15 min. After filtration with a syringe-driven filter (Millipore, Burlington, Massachusetts, USA), recombinant protein with a 6×His-tag at the C-terminus was purified by using the AKTA start protein purification system (Cytiva, Marlborough, Massachusetts, USA) connected to the fraction collector (Frac30; Cytiva) on a system equipped with a His-trap HP column (1 mL; Cytiva). Proteins were eluted with 1 mL min−1 flow of the linear gradient of 100% buffer A (50 mM phosphate buffer pH 7.4, 500 mM NaCl, and 20 mM imidazole) to 100% buffer B (50 mM phosphate buffer pH 7.4, 500 mM NaCl, and 300 mM imidazole) within 15 min. The fractions of an identical peak monitored by absorption at 215 nm were collected into tubes and proteins were concentrated by using centrifugal filter units (Amicon Ultra-4 10K; Merck Millipore, Burlington, Massachusetts, USA). Protein concentration was determined by the Bradford method.
Enzymatic assay of the activity of purified Sll1969
In total, 10 μg purified Sll1969 protein was incubated in 240 μL reaction buffer (50 mM Tris–HCl pH 7.5 with 0.5% [w/v] Triton X-100) containing 100 nmol of various lipid compounds at 35°C. The reaction was terminated by the addition of 900 μL chloroform–methanol (1:2; v/v) followed by the lipid analysis described below.
Lipid analysis
After the enzymatic reactions, lipids were extracted by the Bligh–Dyer method (Bligh and Dyer, 1959) and applied to a TLC plate (TLC silica gel 60; Merck, Kenilworth, New Jersey, USA), and developed with chloroform:methanol:28% ammonium = 65:35:5 (v/v/v). Major membrane lipids extracted from cyanobacterial cells by the Bligh–Dyer method were analyzed by a high-performance liquid chromatography–evaporative light scattering detector (HPLC-ELSD) system as described (Jimbo et al., 2021). TAG was analyzed in the same system with the following solvent condition as described previously (Kobayashi et al., 2013). Lipids were eluted with 1 mL min−1 flow rate of the gradient of solvent A (hexane:isopropanol:acetic acid = 98.9:1:0.1 [v/v/v]) and solvent B (isopropanol:acetic acid = 99.9:0.1 [v/v]) with the following program: 0–7 min, 100% solvent A; 8 min, 5% solvent A; 9–15 min, 100% solvent B. Lyso-lipids were extracted by the Folch method (Folch et al., 1957) as briefly described below. Cyanobacterial cells in 50 mL of BG-11 culture were collected by centrifugation. After the supernatant was completely removed by wiping with paper, 1 mL chloroform–methanol (1:2; v/v) was added to the cell pellet and mixed by vortex. After incubation for 20 min at room temperature, the mixture was centrifuged at 2,800 g at room temperature, and the supernatant was transferred to a glass tube. The extraction was repeated until the color of precipitation turned bright blue. The extract was dried up by the centrifugal concentrator and dissolved in 0.02 mL chloroform. All samples were applied to TLC and developed with a solvent; chloroform:acetone:methanol:acetic acid:water = 50:20:10:15:5 (v/v/v/v/v). The spots of MGMG and DGMG identified by comparing with authentic MGMG and DGMG on the same plate were quantified by GC.
Analysis of photoinhibition of PSII and labeling of proteins in vivo
Photoinhibition of PSII and labeling of proteins in vivo were analyzed as described (Jimbo et al., 2019).
Analysis of proteins and photosynthetic complexes
Thylakoid membrane in Buffer A (KD) containing 5 mM CaCl2, 10 mM MgCl2, and 25% (v/v) glycerol buffered with 50 mM MES-NaOH pH 6.0 were isolated by breaking Synechocystis cells in a sample tube containing 0.1 μm Zirconia beads, with a beads-beater (20-s intervals on ice; Bio Medical Science, Japan), followed by centrifugations at 4°C. Thylakoid membrane (corresponding to 0.8 μg μL−1 Chl) in 10 μL of Buffer A (KD) was solubilized with 1% (w/v) n-dodecyl-β-d-maltoside on ice for 20 min, then Coomassie Brilliant Blue G-250 (CBB G-250) was added at a final concentration of 0.5% (w/v). After centrifugation (22,000 g) at 4°C for 10 min, 3.8 μL (corresponding to 3 μg Chl) of the supernatant was applied for the precast 4%–15% Native-PAGE gel (ThermoFisher, Waltham, Massachusetts, USA), and the gel was electrophoresed at 5 mA for 14 h at 4°C. Gels were visualized after excess CBB G-250 in gel was washed out by water. The level of PSII dimers was calculated from the intensity of a band representing PSII dimers by using ImageJ2. For the localization of LipA protein, proteins in the Native-PAGE gel were solubilized by SDS-PAGE buffer containing 2 M urea and separated by SDS-PAGE containing 2 M urea. Proteins in the gel were transferred to polyvinylidene difluoride membrane by electrophoresis. PsaA protein as a control protein in thylakoid membrane was detected with the PsaA-specific antibody. A specific antibody against LipA proteins was raised against an oligopeptide, CMAWQSDFLRDLNRD, which had been linked to the keyhole limpet hemocyanin as carrier protein at a cysteine residue (Eurofins Genomics, Japan), as described (Takahashi et al., 2019).
Statistical analysis
For the statistical analysis (Student’s t test), we used t test program in Excel.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries (accession no. AP012276.1) for sll1969 gene.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenic analysis of LipA proteins.
Supplemental Figure S2. Purification and characterization of recombinant LipA protein with a 6× His-tag expressed in E. coli cells.
Supplemental Figure S3. Segregation of disrupted sll1969 gene in lipA cells.
Supplementary Material
Acknowledgments
The authors thank Mr. Tsutomu Takizawa (the University of Tokyo) for the radioisotopic analysis of proteins.
Funding
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (no. 19K16161 to H.J. and no. 20K06701 to H.W.) and JST ACT-X (no. JPMJAX20B7 to H.J.).
Conflict of interest statement. None declared.
Contributor Information
Haruhiko Jimbo, Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, 3-8-1 Komaba, Meguro-ku, Tokyo 153-8902, Japan.
Hajime Wada, Department of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, 3-8-1 Komaba, Meguro-ku, Tokyo 153-8902, Japan.
H.J. designed and performed the experiments and wrote the manuscript. H.W. conceived and supervised the project, and contributed to the writing of the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Haruhiko Jimbo (h.jimbo@bio.c.u-tokyo.ac.jp).
References
- Aizouq M, Peisker H, Gutbrod K, Melzer M, Holzl G, Dormann P (2020) Triacylglycerol and phytyl ester synthesis in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 117: 6216–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Chan T, Shimizu Y, Pospisil P, Nijo N, Fujiwara A, Taninaka Y, Ishikawa T, Hori H, Nanba D, Imai A, et al. (2012) Quality control of photosystem II: lipid peroxidation accelerates photoinhibition under excessive illumination. PLoS ONE 7: e52100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZY, Lucker BF, Zienkiewicz K, Miller TE, Zienkiewicz A, Sears BB, Kramer DM, Benning C (2018) Galactoglycerolipid lipase PGD1 is involved in thylakoid membrane remodeling in response to adverse environmental conditions in Chlamydomonas. Plant Cell 30: 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Kobayashi K, Wada H (2016) Sulfoquinovosyldiacylglycerol has an essential role in Thermosynechococcus elongatus BP-1 under phosphate-deficient conditions. Plant Cell Physiol 57: 2461–2471 [DOI] [PubMed] [Google Scholar]
- Eungrasamee K, Miao R, Incharoensakdi A, Lindblad P, Jantaro S (2019) Improved lipid production via fatty acid biosynthesis and free fatty acid recycling in engineered Synechocystis sp. PCC 6803. Biotechnol Biofuels 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509 [PubMed] [Google Scholar]
- Iwai M, Yamada-Oshima Y, Asami K, Kanamori T, Yuasa H, Shimojima M, Ohta H (2021) Recycling of the major thylakoid lipid MGDG and its role in lipid homeostasis in Chlamydomonas reinhardtii. Plant Physiol 187: 1341–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimbo H, Izuhara T, Hihara Y, Hisabori T, Nishiyama Y (2019) Light-inducible expression of translation factor EF-Tu during acclimation to strong light enhances the repair of photosystem II. Proc Natl Acad Sci USA 116: 21268–21273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimbo H, Izuhara T, Hirashima T, Endo K, Nakamura Y, Wada H (2021) Membrane lipid remodeling is required for photosystem II function under low CO2. Plant J 105: 245–253 [DOI] [PubMed] [Google Scholar]
- Kaczmarzyk D, Fulda M (2010) Fatty acid activation in cyanobacteria mediated by acyl–acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol 152: 1598–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Noel EA, Barnes A, Rosenberg J, DiRusso C, Black P, Oyler GA (2013) Rapid detection and quantification of triacylglycerol by HPLC-ELSD in Chlamydomonas reinhardtii and Chlorella strains. Lipids 48: 1035–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Matsumoto U, Keta S, Nakahigashi K, Ikeda K, Takatani N, Omata T, Aichi M (2022) High-light-induced stress activates lipid deacylation at the sn-2 position in the Cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 63: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse O, Hankamer B, Konczak C, Gerle C, Morris E, Radunz A, Schmid GH, Barber J (2000) Phosphatidylglycerol is involved in the dimerization of photosystem II. J Biol Chem 275: 6509–6514 [DOI] [PubMed] [Google Scholar]
- Laczko-Dobos H, Ughy B, Toth SZ, Komenda J, Zsiros O, Domonkos I, Parducz A, Bogos B, Komura M, Itoh S, et al. (2008) Role of phosphatidylglycerol in the function and assembly of photosystem II reaction center, studied in a cdsA-inactivated PAL mutant strain of Synechocystis sp. PCC6803 that lacks phycobilisomes. Biochim Biophys Acta 1777: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Leng J, Sakurai I, Wada H, Shen JR (2008) Effects of phospholipase and lipase treatments on photosystem II core dimer from a thermophilic cyanobacterium. Photosynth Res 98: 469–478 [DOI] [PubMed] [Google Scholar]
- Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo MH, Benning C (2012) A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Curtiss R 3rd (2012) Thermorecovery of cyanobacterial fatty acids at elevated temperatures. J Biotechnol 161: 445–449 [DOI] [PubMed] [Google Scholar]
- Maeda H, Sakuragi Y, Bryant DA, Dellapenna D (2005) Tocopherols protect Synechocystis sp. strain PCC 6803 from lipid peroxidation. Plant Physiol 138: 1422–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, et al. (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 108: 2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa N, Wada H (2012) The role of lipids in photosystem II. Biochim Biophys Acta 1817: 194–208 [DOI] [PubMed] [Google Scholar]
- Murata N, Nishiyama Y (2018) ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ 41: 285–299 [DOI] [PubMed] [Google Scholar]
- Nagao R, Tomo T, Narikawa R, Enami I, Ikeuchi M (2016) Conversion of photosystem II dimer to monomers during photoinhibition is tightly coupled with decrease in oxygen-evolving activity in the diatom Chaetoceros gracilis. Photosynth Res 130: 83–91 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Okazaki K, Sato N, Tsuji N, Tsuzuki M, Nishida I (2006) The significance of C16 fatty acids in the sn-2 positions of glycerolipids in the photosynthetic growth of Synechocystis sp. PCC 6803. Plant Physiol 141: 546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisil P, Yamamoto Y (2017) Damage to photosystem II by lipid peroxidation products. Biochim Biophys Acta 1861: 457–466 [DOI] [PubMed] [Google Scholar]
- Sakurai I, Mizusawa N, Wada H, Sato N (2007) Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol 145: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N (2009) Gclust: trans-kingdom classification of proteins using automatic individual threshold setting. Bioinformatics 25: 599–605 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kusama Y, Li X, Takaichi S, Nishiyama Y (2019) Overexpression of orange carotenoid protein protects the repair of PSII under strong light in Synechocystis sp. PCC 6803. Plant Cell Physiol 60: 367–375 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Ishikawa T, Tamura S, Saito Y, Kawai-Yamada M, Hihara Y (2020) Quantitative and qualitative analyses of triacylglycerol production in the wild-type cyanobacterium Synechocystis sp. PCC 6803 and the strain expressing AtfA from Acinetobacter baylyi ADP1. Plant Cell Physiol 61: 1537–1547 [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Wada H, Murata N (1990) Temperature-induced changes in the fatty acid composition of the cyanobacterium, Synechocystis PCC 6803. Plant Physiol 92: 1062–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Murata N (1998) Membrane lipids in cyanobacteria. In PA Siegenthaler, N Murata, eds, Lipids in Photosynthesis: Structure, Function and Genetics. Kluwer Academic Publishers, Dourdrecht, Netherland, pp 65–81 [Google Scholar]
- Wang K, Froehlich JE, Zienkiewicz A, Hersh HL, Benning C (2017) A plastid phosphatidylglycerol lipase contributes to the export of acyl groups from plastids for seed oil biosynthesis. Plant Cell 29: 1678–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Shabestary K, Bjork SM, Asplund-Samuelsson J, Joensson HN, Jahn M, Hudson EP (2020) Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nat Commun 11: 1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara A, Kobayashi K (2022) Lipids in photosynthetic protein complexes in the thylakoid membrane of plants, algae, and cyanobacteria. J Exp Bot 73: 2735–2750 [DOI] [PubMed] [Google Scholar]
- Yu L, Fan J, Zhou C, Xu C (2021) Chloroplast lipid biosynthesis is fine-tuned to thylakoid membrane remodeling during light acclimation. Plant Physiol 185: 94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.