Abstract
In Nicotiana benthamiana, the expression of the Xanthomonas effector XANTHOMONAS OUTER PROTEIN Q (XopQ) triggers RECOGNITION OF XOPQ1 (ROQ1)-dependent effector-triggered immunity (ETI) responses accompanied by the accumulation of plastids around the nucleus and the formation of stromules. Both plastid clustering and stromules were proposed to contribute to ETI-related hypersensitive cell death and thereby to plant immunity. Whether these reactions are directly connected to ETI signaling events has not been tested. Here, we utilized transient expression experiments to determine whether XopQ-triggered plastid reactions are a result of XopQ perception by the immune receptor ROQ1 or a consequence of XopQ virulence activity. We found that N. benthamiana mutants lacking ROQ1, ENHANCED DISEASE SUSCEPTIBILITY 1, or the helper NUCLEOTIDE-BINDING LEUCINE-RICH REPEAT IMMUNE RECEPTORS (NLRs) N-REQUIRED GENE 1 (NRG1) and ACTIVATED DISEASE RESISTANCE GENE 1 (ADR1), fail to elicit XopQ-dependent host cell death and stromule formation. Mutants lacking only NRG1 lost XopQ-dependent cell death but retained some stromule induction that was abolished in the nrg1_adr1 double mutant. This analysis aligns XopQ-triggered stromules with the ETI signaling cascade but not to host programmed cell death. Furthermore, data reveal that XopQ-triggered plastid clustering is not strictly linked to stromule formation during ETI. Our data suggest that stromule formation, in contrast to chloroplast perinuclear dynamics, is an integral part of the N. benthamiana ETI response and that both NRG1 and ADR1 hNLRs play a role in this ETI response.
Genetic analysis aligns effector triggered immunity (ETI)-induced stromule formation with ETI signaling but not programmed cell death and questions stromule-guided perinuclear plastid clustering.
Introduction
Plastids exhibit exquisite developmental flexibility, as demonstrated by their capacity to differentiate into various plastid types with specialized functions, biochemical activities, and internal structures, depending on the plant organ, developmental stage, or environmental condition. Furthermore, plastids undergo extreme morphological changes, in some cases changing their shape within minutes or seconds (Gunning, 2005; Pyke, 2013; Delfosse et al., 2016). One highly dynamic feature of plastids is the projection of long, stroma-filled tubules formed by the two envelope membranes. These projections, also called stromules, are reliably observed when either the stroma or the envelope membranes are fluorescently labeled (reviewed in Delfosse et al., 2016). Over the last two decades, stromules have been detected by fluorescence microscopy in an increasing number of plant species throughout the Viridiplantae (“green plants”; reviewed in Gray et al., 2001), suggesting that stromule formation emerged early during plant evolution. Examination of different plant tissues revealed that while stromule frequencies may vary, stromules are a ubiquitous feature of plastids (Köhler and Hanson, 2000; Holzinger et al., 2008).
Stromules form in response to developmental cues and increase following exposure to various stresses or signaling molecules and metabolites connected to stress (Schattat and Klösgen, 2011b; Gray et al., 2012; Mathur et al., 2012; Caplan et al., 2015; Vismans et al., 2016), suggesting stromule formation is strictly controlled by the plant. These observations led to the hypothesis that stromules participate in processes that are fundamentally important for plant survival during stress, to transmit signals and/or support physiological changes.
Despite their early emergence in the evolution of Viridiplantae and their frequent observation across tissues, our knowledge of stromule function is limited. To date, mutants with defects in signaling pathways regulating stromule formation were not identified. Therefore, it remains unclear which processes or functions are carried out by stromules during stress responses and how these might be executed. As an alternative to genetic dissection of stromule formation per se, we decided instead to test the effects of mutants in defined stress responses for effects on stromule formation. Our aim is to gain insight into the role of stromules during adaptation to a specific stress and use genetic tools to decipher stromule function.
Biotic stress caused by plant interactions with recognized pathogens results in pronounced stromule formation (Krenz et al., 2012; Erickson et al., 2014; Caplan et al., 2015; Kumar et al., 2018). Many pathogenic microbes transfer virulence factors, known as effectors, into the host cell cytoplasm to promote infection, often by manipulating pattern-triggered immunity (PTI) programs (Toruño et al., 2016; Büttner, 2016). In an incompatible interaction, intracellular immune receptors recognize one or more effectors. Effector recognition triggers a robust immune response termed effector-triggered immunity (ETI), which frequently culminates in localized host-programmed cell death (a hypersensitive response = HR) at infection sites (Cui et al., 2015; Yuan et al., 2021). Most intracellular immune receptors are nucleotide binding/leucine-rich repeat (NLR) proteins. NLR proteins are represented by two major pathogen-sensing NLR receptor classes, which are defined by their N-terminal domains. The so-called TIR-NLRs (TNLs) possess a Toll/interleukin-1 domain (TIR) and CC-NLRs (CNLs) a coiled-coil (CC) domain at their N-terminus. Additionally, different families of “helper” NLRs (hNLRs) were found to function together with pathogen-detecting (sensor) NLRs (sNLRs) in Arabidopsis (Arabidopsis thaliana) and solanaceous species, thus connecting sNLRs with downstream immunity factors in ETI (Cui et al., 2015; Wu et al., 2019).
Dramatic increases in stromule frequencies were observed following the expression of effectors recognized by CC-NLRs or TIR-NLRs prior to ETI-induced cell death in Nicotiana benthamiana and Arabidopsis thaliana (Caplan et al., 2015; Erickson et al., 2018). For example, induction of ETI (resulting in HR) via the transient co-expression of the p50 helicase domain from tobacco mosaic virus and the cognate TIR-NLR immune receptor, N from tobacco (Nicotiana tabacum), in N. benthamiana results in strong stromule induction (Caplan et al., 2015). Similarly, a screen by our group revealed that the expression of XopQ from the bacterium Xanthomonas campestris pv. vesicatoria (Xcv; strain 85–10), which is recognized by the TIR-NLR immune receptor RECOGNITION OF XopQ1 (ROQ1) in N. benthamiana, also strongly enhanced stromule frequencies (Schultink et al., 2017, Erickson et al., 2018). In the case of ETI activation via N/p50, the authors reported that many stromules were in close proximity to the nucleus, and appeared to make contact. This observation suggested that plastids might directly deliver defense signals to the nucleus via stromules (Caplan et al., 2015; Kumar et al., 2018). Stromule frequency also increased during CC-NLR- and TIR-NLR-mediated ETI in A. thaliana when plants were challenged with avirulent strains of the bacterial pathogen Pseudomonas syringae (Caplan et al., 2015). Thus, it appears that stromule formation is a common response of plants during ETI.
In addition to an increase in stromule frequencies and stromule-to-nucleus contacts, the formation of plastid clusters around nuclei was observed during ETI responses in N. benthamiana (Caplan et al., 2015; Kumar et al., 2018), leading to the conclusion that plastid clusters might also support the delivery of plastid-derived defense signals to the nucleus (Ding et al., 2019; Mullineaux et al., 2020). In time-lapse experiments spanning several minutes (Kumar et al., 2018), plastid bodies moved in the direction of stromule tips/anchor points in the majority of cases, giving the impression that stromules directionally pull the plastid body with them. This observation led to the conclusion that stromules might guide plastids to the nucleus to facilitate clustering. Hence, stromules near the nucleus might have a second function in plastid positioning.
Taken together, ETI-induced stromules present a starting point for more detailed genetic analyses of stromule formation and plastid clustering following the well-defined molecular event of effector recognition. For this, we chose N. benthamiana ROQ1-mediated XopQ recognition leading to ETI as a suitable system to critically examine the functional relationship between stromule formation and immunity signaling.
The Arabidopsis immune receptor HopZ-ACTIVATED RESISTANCE 1 (ZAR1), and likely other CNLs, assembles into pentameric resistosome complexes upon activation, which may insert into membranes and function as Ca2+ influx channels (Adachi et al., 2019; Bi et al., 2021; Wang et al., 2019a). In contrast, TNLs including ROQ1 were reported to assemble into tetrameric holoenzymes with NADase activity (Ma et al., 2020; Martin et al., 2020), and cannot induce immunity directly. TNL-ETI requires at least two more components: heterodimeric complexes composed of the lipase-like protein ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and either PHYTOALEXIN DEFICIENT4 (PAD4) or SENSECENCE-ASSOCIATED GENE101 (SAG101), and hNLRs of the ACTIVATED DISEASE RESISTANCE GENE1 (ADR1) and/or N-REQUIRED GENE1 (NRG1) type. These hNLRs are characterized by an N-terminal CC domain with homology to A. thaliana RESISTANCE to POWDERY MILDEW 8 (RPW8), the so-called CCR domain, and are therefore referred to as RNLs (Collier et al., 2011; Wagner et al.; 2013; Castel et al., 2019; Jubic et al., 2019; Gantner et al., 2019; Lapin et al., 2019; Wu et al., 2019; Saile et al., 2020).
Heterodimeric EDS1 complexes most likely function as receptors for TNL-derived small molecules and are essential for TNL-mediated immune responses in different dicot plants including A. thaliana and N. benthamiana (Wagner et al., 2013, Gantner et al., 2019, Lapin et al., 2020, Huang et al., 2022, Jia et al., 2022). At least in A. thaliana, small molecule binding promotes the formation of EDS1–PAD4–ADR1 and EDS1–SAG101–NRG1 complexes, which can regulate pathogen resistance and cell death, respectively, in TNL immunity (Lapin et al., 2019; Sun et al., 2021). Upon activation, ADR1 and NRG1 RNLs were reported to form a structure similar to the pentameric complex reported for the A. thaliana CNL ZAR1 (“resistosome”), which may directly integrate into membranes to function as Ca2+-permeable channels (Wang et al., 2019a, 2019b; Bi et al., 2021; Jacob et al., 2021). In N. benthamiana, immune functions are not known for EDS1–PAD4, and an EDS1–SAG101b complex appears to operate mainly through NRG1 to mediate both cell death and resistance in this species (Qi et al., 2018; Gantner et al., 2019; Lapin et al., 2019). Nicotiana benthamiana ADR1 immune functions have not been analyzed so far; however, significant XopQ-ROQ1-mediated transcriptional reprogramming in nrg1 mutant plants (which is completely abolished in eds1 mutants) in the absence of resistance and cell death suggests that ADR contributes to TNL immunity and that there is some degree of cooperativity or redundancy between the two helper NLR classes in N. benthamiana (Qi et al., 2018; Saile et al., 2020).
In this study, we capitalized on the previous characterization of XopQ–ROQ1-induced TNL immunity in N. benthamiana and positioned chloroplast stromule formation and perinuclear clustering in downstream signaling networks. Our data suggest that, although stromule formation is tightly linked to immune responses, it can be uncoupled from ETI-triggered cell death. Furthermore, our data support partially redundant functions of the NRG1 and ADR1 hNLRs in stromule formation. Intriguingly, our data indicate that plastid clustering can be largely uncoupled from ROQ1 ETI and hence is unlikely to represent an integral component of the plant’s innate immune response.
Results
Xcv-mediated stromule formation in N. benthamiana depends on XopQ
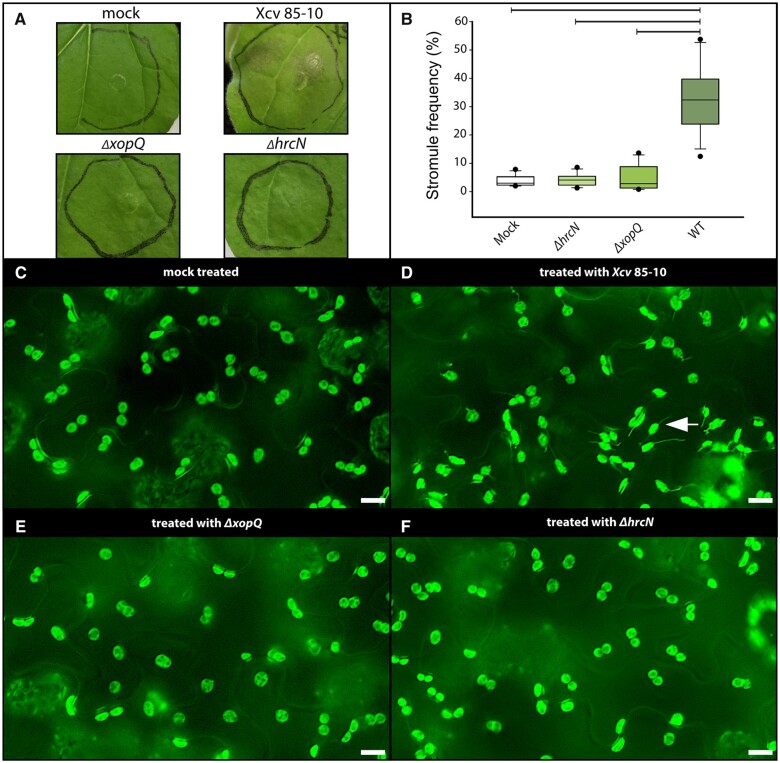
We previously reported that A. tumefaciens-mediated transient expression of XopQ from Xcv induces stromule formation in N. benthamiana (Erickson et al., 2018). During infection with Xcv strains such as Xcv 85-10, a strain that naturally delivers XopQ to host cells, XopQ is likely less abundant in infected cells than during transient expression experiments. Additionally, XopQ is translocated together with the entire type III-secreted effectome of Xcv (>30 effectors; Teper et al., 2016). In order to test the extent to which stromule frequencies measured during transient expression experiments reflect the Xcv interaction, different bacterial strains were inoculated into FNR:eGFP-expressing transgenic N. benthamiana plants. Under our greenhouse conditions, the wild-type strain Xcv 85-10 induces an ETI-associated programmed cell death response (indicating XopQ recognition), showing first signs of dead leaf tissue at 2-day post-infiltration (dpi; Adlung et al., 2016). In order to be able to observe plastids in living cells, we collected leaf samples for microscopy 43-h post-inoculation (Figure 1A). To test the role that XopQ and other effectors play in stromule response, Xcv mutant strains ΔhrcN and ΔxopQ were infiltrated on the same leaf with the wild-type strain. The ΔhrcN mutant is deficient in type III secretion and serves as a nonvirulent PTI control (Lorenz and Büttner, 2009). ΔhrcN as well as ΔxopQ mutant strains of Xcv did not induce macroscopically visual changes in infected tissues at 2 dpi and were not distinguishable from the mock infiltration (Figure 1A; Lorenz and Büttner, 2009; Adlung et al., 2016; Adlung and Bonas, 2017). When analyzing the stromule phenotype, treatments differed significantly: The Xcv 85-10 strain induced massive stromule induction in the infiltrated tissue. The ΔxopQ and ΔhrcN (no effector translocation) mutant-inoculated tissue harbored almost no stromules, with levels comparable to mock inoculations (10-mM MgCl2; see Figure 1B for stromule frequency quantification, Figure 1, C–F as well as Supplemental Figures S1 and S2 for sample images of the microscopic phenotypes and for statistical values Supplemental Table S1).
Figure 1.
Xcv inoculation experiments in wild-type FNR:eGFP transgenic plants. A, Macroscopic phenotypes of leaves infiltrated with different Xcv strains at 2 dpi. B, Stromule frequency (SF%) of Xcv-inoculated tissue at 43-h post-inoculation represented as box plots (box line = median, whiskers = 10th as well as 90th percentile, with each outlier plotted); horizontal lines above bars indicate significantly different stromule frequency values as indicated by a one way analysis of variance (ANOVA) analysis. C–F, Sample sectors of representative microscopic images used for stromule quantification. Fluorescence signals originate from the stably expressed FNR:eGFP plastid stroma marker; scale bars = 10 µm; arrow = stromule (full-frame images shown in Supplemental Figures S1 and S2).
Together with transient expression experiments using A. tumefaciens (Erickson et al., 2018), these results indicate that stromule induction at 2 dpi by the Xcv wild-type strain is strictly dependent on the presence of XopQ. No other effectors in this strain contributed measurably to stromule induction at 2 dpi when translocated at natural levels. In conclusion, inoculations with Xcv strains show that XopQ-triggered stromules appear during pathogen attack at 2 dpi, and supports the idea that stromule induction during transient assays recapitulates a physiologically relevant phenotype.
XopQ fails to induce high stromule frequency values in roq1 mutant plants
We next examined the extent to which XopQ-triggered stromule induction in the lower epidermis of N. benthamiana is a consequence of XopQ perception by the TNL receptor ROQ1, using A. tumefaciens-based transient expression. Different N. benthamiana mutant lines impaired in XopQ perception or lacking TNL downstream signaling components were co-infiltrated with Agrobacterium strains for expression of stroma-targeted eGFP (SSU:eGFP) and either xopQ:mOrange2 or mOrange2 alone, as control. Agrobacterium strains were infiltrated at a final optical density (OD)600 = 0.2, which led to only moderate Agrobacterium-dependent stromule induction (<20%, Erickson et al., 2014) well below XopQ-triggered stromule frequencies (∼60%). These experimental conditions were evaluated using N. benthamiana stably expressing stromal-targeted eGFP (FNR:eGFP; Supplemental Figure S3).
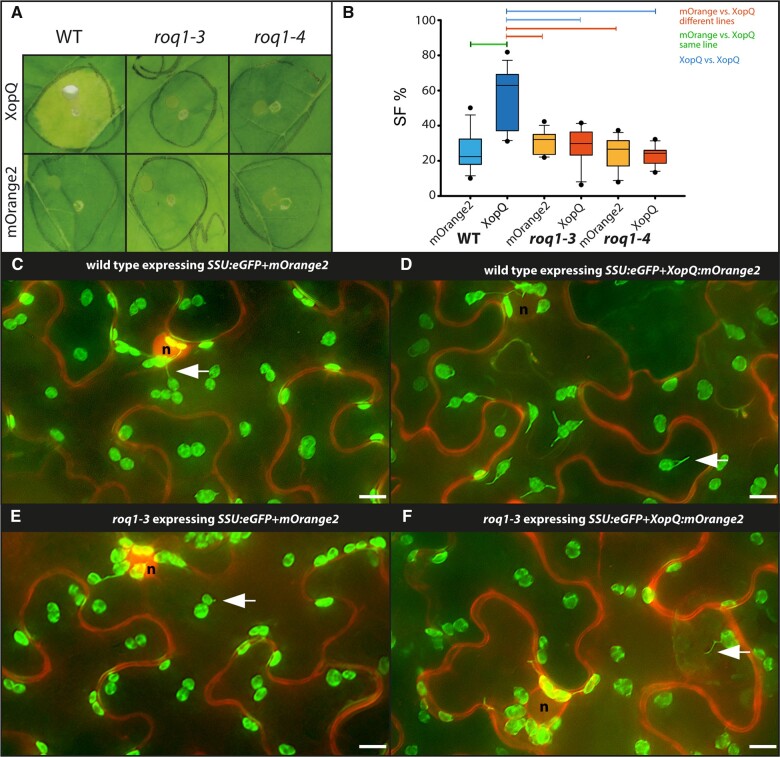
We first tested XopQ-triggered stromule frequencies in roq1 mutant plants (roq1-3 and -4). ROQ1-deficient plants fail to recognize XopQ and therefore lack the typical ETI-induced yellowing and chlorosis exhibited by wild-type plants following transient XopQ expression (Schultink et al., 2017; Gantner et al., 2019). xopQ:mOrange2 or the mOrange2 control were co-infiltrated with a stroma-targeted GFP (SSU:eGFP) into wild-type and mutant plants to allow for the visualization of plastids and stromules. As a control for XopQ recognition, macroscopic phenotypes of the co-infiltrated leaves were recorded at 10 dpi, a time point when symptoms are clear despite the low optical densities used for infiltration (Figure 2A). As expected, xopQ:mOrange2 expression in wild-type plants resulted in chlorosis of the infiltration spot, indicative of the XopQ-triggered ETI response (Adlung et al., 2016). In roq1-3 and roq1-4 plants, there was no visible chlorosis, and tissues were indistinguishable from control infiltrations. In all plant lines, mOrange2 controls showed stromule frequency values characteristic of leaves infiltrated with “empty” GV3101 (pMP90) bacteria (compare Figure 2B with Supplemental Figure S3), indicating that the roq1 mutation does not alter basal stromule frequencies. Compared to mOrange2, xopQ:mOrange2 expression resulted in significantly higher stromule induction in wild-type plants, as previously described (Erickson et al., 2018). In contrast, xopQ:mOrange2 expression failed to induce stromules beyond GV3101 (pMP90) basal levels in the roq1-3 and roq1-4 mutant plants (Figure 2B;Supplemental Table S2). Average stromule frequencies in mutants expressing xopQ:mOrange2 were equal to or less than mOrange2 controls. While mORANGE2 accumulates in the cytoplasm as well as in the nucleoplasm the fusion protein XopQ-mORANGE2 accumulates only in the cytoplasm (Figure 2, C–F; for full frame images, see Supplemental Figures S4 and S5). The full loss of XopQ-triggered stromule formation in roq1 mutant plants shows that XopQ recognition by ROQ1 is required for stromule induction and that nonrecognized XopQ activity does not generate a stromule-inducing signal.
Figure 2.
Test for XopQ-mediated stromule formation in lower leaf epidermis cells of roq1 mutants (N. benthamiana). A, Macroscopic phenotypes of Agrobacterium-mediated xopQ:mOrange2 and mOrange2-expression in wild-type, roq1-3, and roq1-4 leaves 10 dpi. B, Results of stromule quantification expressed as stromule frequency (SF%) represented as box plots (box line = median, whiskers = 10th as well as 90th percentile, with each outlier plotted); horizontal lines above bars indicate significantly different stromule frequency values as indicated by a one-way ANOVA analysis. C–F, Sample sectors of representative microscopic images used for stromule quantification. Plastid localized fluorescence originates from the SSU:eGFP plastid stroma marker; cytosolic fluorescence originates from the mORANGE2 fluorescence protein (C and E = mOrange2 controls; d and f mOrange2 fused to xopQ); nuclei = “n”; arrow = stromule; scale bars = 10 µm. (full-frame images shown in Supplemental Figures S4 and S5).
XopQ–ROQ1-dependent stromule induction requires EDS1
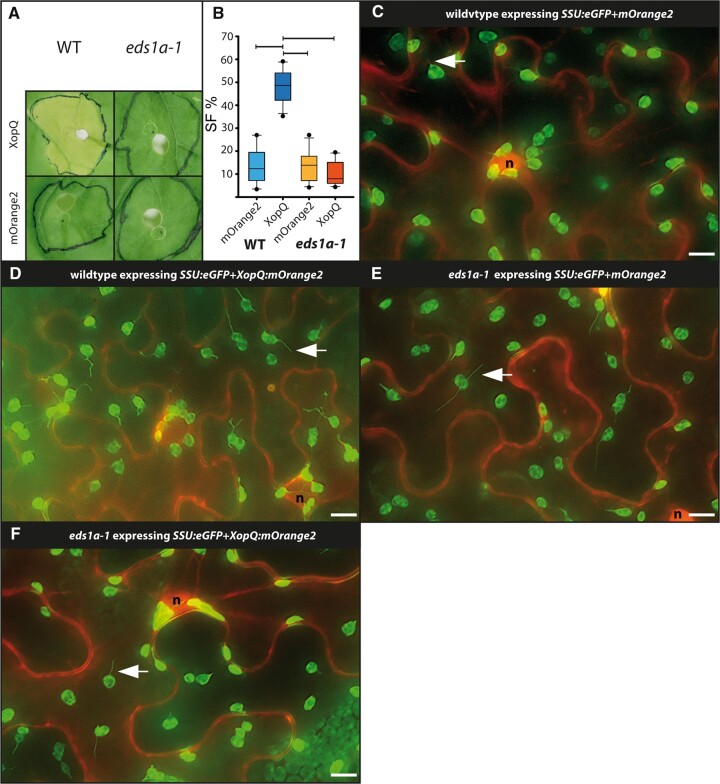
EDS1 is essential for resistance and cell death mediated by TNL-type immune receptor ROQ1 (Adlung et al., 2016; Gantner et al., 2019). In order to test if XopQ-triggered stromule formation is dependent on EDS1, co-infiltrations were repeated in eds1a-1 knockout and wild-type plants. With respect to stromule frequency and macroscopic phenotype, the wild-type plants responded as seen in previous experiments (Figure 3, A and B; Supplemental Table S2). In contrast, the wild-type eds1a-1 plants did not show signs of chlorosis at 10 dpi in response to xopQ:mOrange2 expression, which is consistent with literature reports (Figure 3A; Adlung et al., 2016; Gantner et al., 2019). As was the case for roq1 mutant plants, XopQ-triggered stromules were not observed in eds1a-1 tissues (Figure 3, C–F; for full frame images, see Supplemental Figure S6 and Supplemental Table S2). These results indicate that stromule formation in response to XopQ occurs downstream of EDS1 signaling, suggesting that XopQ–ROQ1 interaction and ROQ1 tetramerization (“resistosome” formation; Schultink et al., 2017, Martin et al., 2020) are not sufficient to induce stromules.
Figure 3.
Test for XopQ-triggered stromule formation in the lower leaf epidermis of eds1a-1 mutants (N. benthamiana). A, Macroscopic phenotypes of xopQ:mOrange2 and mOrange2 infiltrated wild-type and eds1a-1 leaves at 10 dpi. B, Results of stromule quantification expressed as stromule frequency (SF%) represented as box plots (box line = median, whiskers = 10th as well as 90th percentile, with each outlier plotted); horizontal lines above bars indicate significantly different stromule frequency values as indicated by a one-way ANOVA analysis. C–F, sample sectors of representative microscopic images from the data set used for stromule quantification. Plastid localized fluorescence originates from the SSU:eGFP plastid stroma marker; cytosolic fluorescence originates from the mORANGE2 fluorescence protein (C and E mORANGE2; D and F mORANGE2 fused to XopQ); nuclei = “n”; arrow = stromule; scale bars = 10 µm (full-frame images shown in Supplemental Figure S6.)
XopQ-triggered stromule induction depends on RNLs but is not a consequence of host cell death
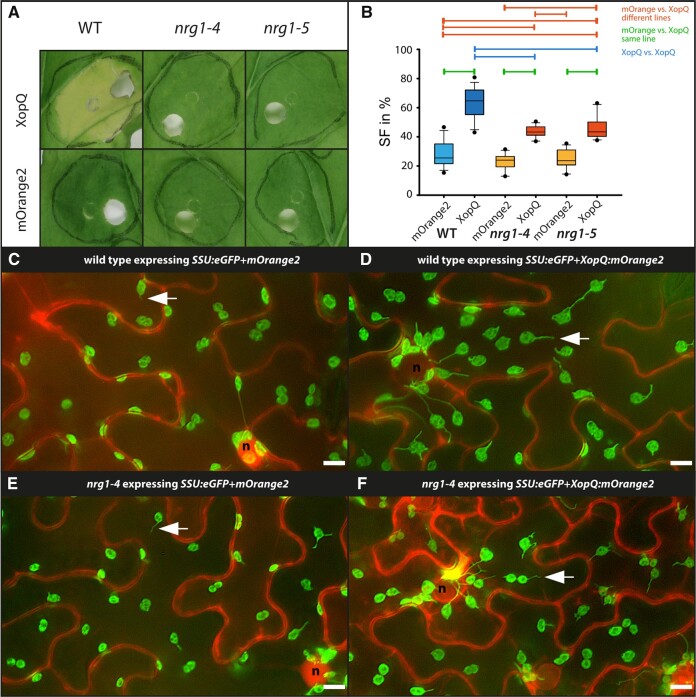
In A. thaliana, RNL-type NLRs of the ADR1 and NRG1 subfamilies contribute to TNL immunity (Castel et al., 2019; Lapin et al., 2019; Wu et al., 2019; Saile et al., 2020). In N. benthamiana nrg1 mutant plants, resistance and cell death induced by several TNLs was fully abolished, suggesting NRG1 as the major RNL in TNL immunity in this species (Qi et al., 2018). To test if XopQ-triggered stromule formation is NRG1-dependent, two mutant lines with different genomic deletions, nrg1-4 and nrg1-5 (Ordon et al., 2021), were analyzed. The nrg1 mutants did not show signs of yellowing in response to A. tumefaciens-mediated XopQ expression, and infiltration spots were macroscopically indistinguishable from control infiltrations (Figure 4A), as expected (Qi et al., 2018; Ordon et al., 2021). All three plant lines responded similarly to the mOrange2 control expression, with stromule frequencies reaching approximately 25% (Figure 4B;Supplemental Table S2). In contrast, the response to xopQ:mOrange2 was markedly different between wild-type and nrg1 mutant lines (Figure 4B). xopQ:mOrange2 expression in the nrg1 background induced stromule frequencies values which were intermediate between mOrange2 and xopQ:mOrange2 expressing wild-type plants (Figure 3, C–F; for full frame images see Supplemental Figure S7). Although roq1, eds1, and nrg1 mutants were equally deficient in XopQ-triggered cell death (necrosis), stromule formation did not strictly require NRG1 and is thus uncoupled from NRG1-mediated cell death. In support of this notion, the transient overexpression of NRG1 under the control of the native promoter (pNRG1) in FNR:eGFP plants failed to induce cell death, but significantly induced stromule formation (Supplemental Figure S8). This was in contrast to NRG1 under the control of mannopine synthase or ubiquitin promoter fragments, which resulted in strong cell-death phenotypes (Supplemental Figure S9A).
Figure 4.
Test for XopQ-mediated stromule formation in lower leaf epidermis cells of nrg1 mutants (N. benthamiana). A, Macroscopic phenotypes of XopQ:mOrange2 and mOrange2-expressing wild-type, nrg1-4 and nrg1-5 leaves at 10 dpi. B, Results of stromule quantification expressed as stromule frequency (SF%) represented as box plots (box line = median, whiskers = 10th as well as 90th percentile, with each outlier plotted); horizontal lines above bars indicate significantly different stromule frequency values as indicated by a one-way ANOVA analysis. C–F, Sample sectors of representative microscopic images used for stromule quantification. Plastid localized fluorescence originates from the SSU:eGFP plastid stroma marker; cytosolic fluorescence originates from the mORANGE2 fluorescence protein (C and E = mOrange2 control; d and f = mOrange2 fused to XopQ); nuclei = “n”; arrow = stromule; scale bars = 10 µm. (full-frame images shown in Supplemental Figure S7).
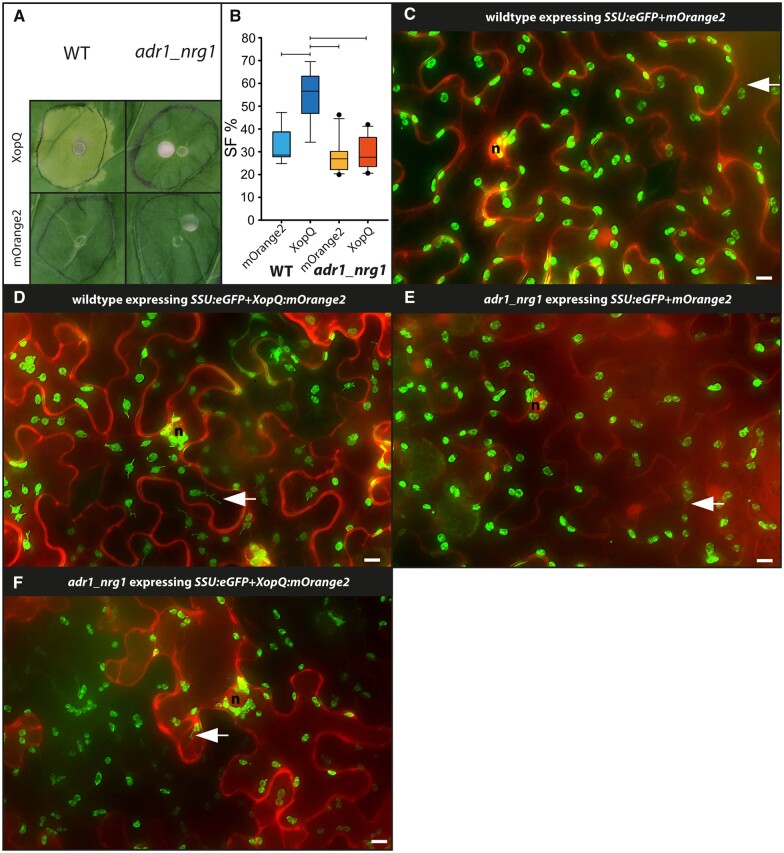
So far, a function of ADR1 was not identified in N. benthamiana but varied contributions of these RNLs to Arabidopsis TNL ETI suggested that N. benthamiana ADR1 might steer residual stromule formation in N. benthamiana nrg1 lines (Lapin et al., 2019; Saile et al., 2020; Sun et al., 2021). Therefore, we tested stromule formation in response to XopQ in a recently generated adr1_nrg1 double mutant line using co-infiltrations as before. As in the nrg1 single mutants, xopQ:mOrange2 expression did not induce yellowing or cell death in adr1_nrg1 plants (Figure 5A). In contrast to our observations in nrg1 single mutants, xopQ:mOrange2 did not induce the formation of stromules beyond the mOrange2 control in adr1_nrg1 plants (see Figure 5B for box plots and Figure 5, C–F for representative images; for full frame images, see Supplemental Figure S9; for statistical values, see Supplemental Table S2). Accordingly, the adr1_nrg1 mutant exhibited stromule and cell death phenotypes similar to the roq1 (Figure 2A) and eds1 (Figure 3A) mutant lines. Overall, these results show that not only NRG1 but also ADR1 contributes to stromule formation in N. benthamiana. Hence, these results uncover that the ADR1 family of RNLs exhibits not only ETI signaling functions in A. thaliana ETI but, at least in the absence of NRG1, also functions in the ETI response of N. benthamiana plants.
Figure 5.
Test for XopQ mediated stromule formation in lower leaf epidermis cells of adr1_nrg1 double mutants (N. benthamiana). A, Macroscopic phenotypes of xopQ:mOrange2 and mOrange2-infiltrated wild-type and adr1_nrg1-5 leaves at 10 dpi. B, Results of stromule quantification expressed as stromule frequency (SF%) represented as box plots (box line = median, whiskers = 10th as well as 90th percentile, with each outlier plotted); horizontal lines above bars indicate significantly different stromule frequency values as indicated by a one-way ANOVA analysis; wild-type three plants and adr1_nrg1 five plants for each of three repeats. C–F, Cropped images used for stromule quantification. Plastid localized fluorescence originates from the SSU:eGFP plastid stroma marker; cytosolic fluorescence originates from the mORANGE2 fluorescence protein (C and E are mOrange2 controls; d and f show mOrange2 translation fusion with XopQ); nuclei = “n”; arrow = stromule; scale bars = 10 µm (full-frame images shown in Supplemental Figure S8).
XopQ-triggered perinuclear plastid clustering does not require TNL immune signaling
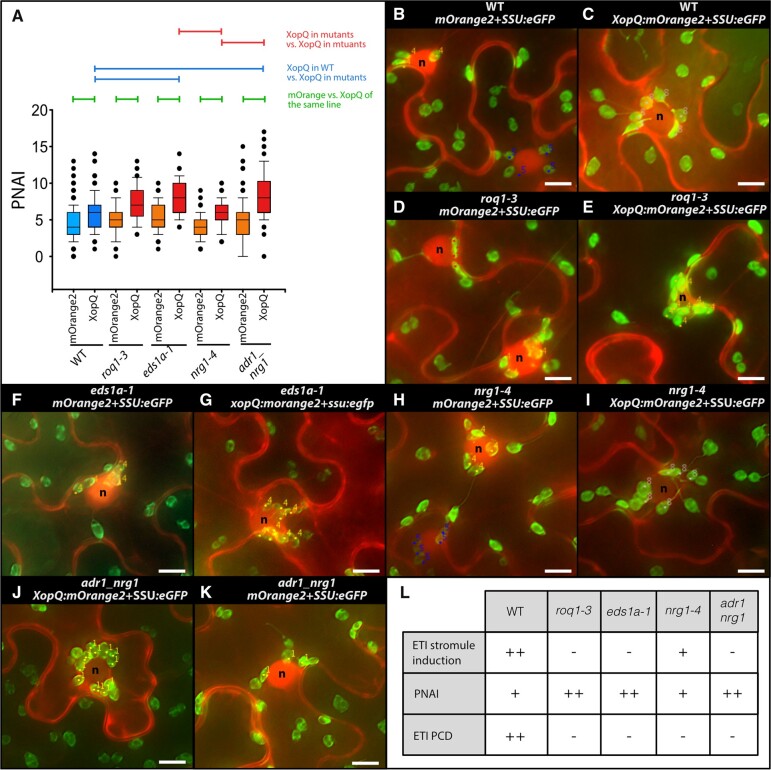
In order to test if XopQ-triggered ETI facilitates the formation of chloroplast clusters, and whether this has the same genetic dependencies as found for stromule frequencies, chloroplast clustering was quantified in wild-type, roq1-3, eds1a-1, nrg1-4, and adr1_nrg1 lines. As a measure for plastid clustering, the number of plastids in close proximity (up to one plastid in diameter) to the nucleus was counted and expressed as the plastid–nucleus association index (PNAI; see Erickson et al., 2014, 2018). In these experiments, mORANGE2 or XopQ-mORANGE2 fluorescence, respectively, served to highlight the position of nuclei (see Figure 6, B–K). In control infiltrations (mOrange2), wild-type and all four mutants produced similar numbers of plastids around the nucleus (Figure 6A;Supplemental Table S3); no significant differences in PNAI were detected. When challenged with XopQ, plastid clustering increased in wild-type plants (Figure 6A;Supplemental Table S3). Although plastid clustering has been considered an ETI response (Caplan et al., 2015; Kumar et al., 2018; Ding et al., 2019), we found that XopQ-triggered plastid clustering was not diminished in mutant lines impaired in XopQ recognition or downstream signaling (Figure 6A;Supplemental Table S3). Upon expression of xopQ:mOrange2, nrg1-4 plants showed wild-type levels of plastid clustering, while eds1 and adr1_nrg1 mutants actually had significantly higher PNAI values (Figure 6A;Supplemental Table S3). Figure 6, B–K shows representative images of the different plant lines expressing SSU:eGFP+xopQ:mOrange2 or SSU:eGFP+mOrange2. This is further supported by the NRG1 over-expression experiments (Supplemental Figure S9) where despite the induction of stromules, pNRG1::NRG1 expression does not increase PNAI values; as would be expected if NRG1-dependent signaling events contribute to perinuclear plastid clustering.
Figure 6.
PNAI—Analysis of plastid clustering in response to XopQ expression (N. benthamiana). PNAI at 3 dpi following mOrange2 and xopQ:mOrange2 expression in wild-type as well as mutant plant lines (roq1-3, eds1a, nrg1-4, and adr1_nrg1). A, PNAI values represented in box plots (box line = median, whiskers = 10th as well as 90th percentile, with each outlier plotted); horizontal lines above box plots indicate significantly different stromule frequency values as indicated by a one-way ANOVA on ranks analysis. B–K, Sample images of nuclei with associated plastids (labeled with a dot a number as they were during original plastid scoring); “n” = nuclei; scale bars = 10 µm. L, Summary of observed ETI stromule, PNAI and cell death phenotypes where “−” = no change compared to control, “+” = visible but moderate increase; “++” = strong increase.
We concluded that when the XopQ-triggered ETI signal cascade is blocked (roq1, eds1, and adr1_nrg1 mutants), the tendency of plastids to cluster around the nucleus remains, and is even enhanced compared to wild-type plants or the nrg1 line with residual ETI signaling (Figure 6H). These data suggest that one feature of ROQ1–XopQ-triggered ETI is suppression of plastid clustering.
Discussion
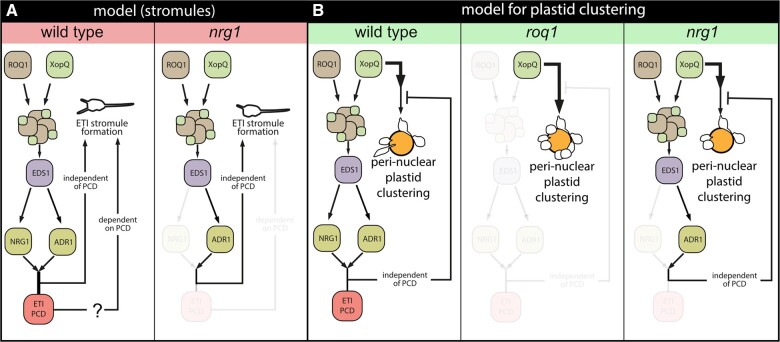
Here, we set out to understand how ETI-associated stromule formation aligns with signaling processes downstream of immune receptor activation, using recognition of the effector XopQ by the TNL ROQ1 in N. benthamiana as a case study. A quantitative analysis of stromule formation and perinuclear plastid clustering in XopQ recognition and ETI signaling mutants produced several important insights. First, complete absence of a XopQ-related stromule response in roq1 and eds1 mutants shows that XopQ-triggered stromules are not a result of its virulence/effector activity, but result from effector recognition by the ROQ1 immune receptor and a resulting EDS1-dependent ETI response. Second, residual induction of stromules in the nrg1 mutant in the absence of macroscopic cell death suggests that induced stromule formation is not a consequence of NRG1-mediated host cell death but is more closely related to ETI signaling. Third, analysis of the nrg1_adr1 double mutant line reveals that residual XopQ-triggered ETI and stromule frequency in nrg1 (but not eds1) is conferred by ADR1 in N. benthamiana. This reveals an ADR1 contribution to TNL ETI processes in a solanaceous plant in the absence of NRG1, suggesting usage of both RNLs NRG1 and ADR1 branches in ETI, as observed in A. thaliana. Finally, ROQ1-XopQ-triggered plastid clustering does not relate to ETI induction and stromule formation and is thus likely to be a direct or indirect consequence of XopQ virulence activity during infection. A model summarizing these findings is presented in Figure 7, A and B.
Figure 7.
Model of XopQ-mediated stromule induction and perinuclear plastid clustering. A, Model for ETI-induced stromule formation by XopQ in wild-type plants (left): XopQ (Xanthomonas outer protein Q) is recognized by the TIR-NB-LRR (TOLL INTERLEUKIN 1 RECEPTOR NUCLEOTIDE-BINDING LEUCINE-RICH REPEAT) resistance protein ROQ1 (RECOCNITION OF XopQ 1), which forms with XopQ a heteromeric protein complex, the so-called “resistosome”. From the “resistosome”, the information of XopQ recognition is transferred with the help of EDS1 to the RNL class helper NLRs NRG1 and ADR1, which culminates in programmed cell death (PCD) as well as stromule formation. In nrg1 mutants (right), XopQ fails to induce cell death but at the same time still shows significant ETI stromule induction mediated most likely by ADR1. This indicates the existence of a PCD-independent ETI stromule induction pathway. However, at this point, a role for PCD in stromule induction or a role for stromules in PCD is not ruled out (?). B, Model for perinuclear plastid clustering following xopQ expression: In wild-type plants (left), an intact ETI signal chain partially suppresses the strong perinuclear plastid clustering induced by XopQ presence. When ETI signal transduction is blocked (eds1, roq1, and adr_nrg1 mutants) this suppression does not take place and plastid clustering is enhanced (middle). The restricted ETI signal chain in nrg1 mutants (right) is sufficient for suppression of induced perinuclear plastid clustering to wild-type levels, thus suppressing signals likely originate from this pathway. Both models only consider genes, which were analyzed as part of this study. The role of the different proteins forming distinct complexes with EDS1 will have to be elucidated in the future.
Uncoupling immune signaling and HR-induced stromules
Stromules have been proposed to transmit retrograde signals to the nucleus, and to amplify programmed cell death responses as part of ETI (Caplan et al., 2015). More recently, it was suggested that stromules have the added function of locating and pulling plastid bodies to the nucleus (Kumar et al., 2018). So far, all reported microbial effectors that induce stromules also provoked programmed cell death (Caplan et al., 2015; Erickson et al., 2018). Therefore, it remained unclear whether stromule formation accompanies the cascade of events contributing to host cell death or is a by-product of physiological changes in cells as they die. The observed stromule induction upon XopQ expression in nrg1 mutant plants without cell death or measurable resistance (Qi et al., 2018) (Figure 5A) suggests stromules represent events upstream of ETI-related pathogen resistance and host cell death. This is supported by clear stromule induction and simultaneous lack of cell death in response to pNRG1::NRG1 expression in wild-type plants expressing FNR:eGFP (see Supplemental Figure S9, A–H). Hence, ETI-induced stromule formation is not coupled to cellular destruction but is more likely an integral part of the ETI response, as suggested previously (Caplan et al., 2015).
ADR1 contributes to TNL immunity and stromule formation in N. benthamiana
Dicot genomes encoding TNL receptors generally also encode RNL-type NLRs of the ADR1 and NRG1 classes (Collier et al., 2011; Lapin et al., 2020). In A. thaliana, both classes of RNLs contribute to immunity to different extents. Three A. thaliana ADR1 putative paralogs have functions in basal immunity to virulent pathogens related to salicylic acid, and they also contribute to PTI (Bonardi et al., 2011; Jubic et al., 2019; Pruitt et al., 2021; Tian et al., 2021). ADR1s and NRG1s function in A. thaliana as distinct modules, respectively, with EDS1–PAD4 and EDS1–SAG101 dimers regulating pathogen resistance and cell death (Lapin et al., 2019; Lapin et al., 2020; Saile et al., 2020; Sun et al., 2021). In N. benthamiana, only EDS1-SAG101 appears to execute TNL ETI, and NRG1 was identified as a major RNL required for cell death and resistance mediated by several tested TNLs (Qi et al., 2018; Gantner et al., 2019). A role for N. benthamiana ADR1 in immunity was so far not detected, although Qi et al. (2018) reported residual transcriptional reprogramming occurring upon ROQ1 activation in nrg1, but not eds1 mutant plants. Notably, ADR1 was among the upregulated genes of an NRG1-independent regulon (Qi et al, 2018). Residual stromule formation in nrg1, but not eds1 or nrg1_adr1 (compare panel “(A)” in Supplemental Figures S4, S5, and S6), supports an ADR1 contribution to TNL-ETI in N. benthamiana. Hence, stromule formation appears to be a highly sensitive read-out for ETI induction, occurring in the absence of cell death or measurable pathogen resistance which are blocked in N. benthamiana nrg1 and eds1 mutants (Qi et al., 2018). Current evidence suggests that RNLs, similar to the CNL ZAR1, can assemble into pore-forming resistosome complexes and function as Ca2+ permeable cation channels (Wang et al., 2019a; Jacob et al., 2021; Bi et al., 2021). Induced Ca2+ influx into host cells would then amplify ROS generation and salicylic acid signaling as well as transcriptional reprogramming (Lu and Tsuda, 2021; Yuan et al., 2021). In future work, it will be interesting to examine whether Ca2+ levels inside cells influence stromule formation, as stromule-to-nucleus connections were found to contribute to ROS formation (Caplan et al., 2015).
Perinuclear plastid clustering is independent of ETI stromule induction
In an emerging concept, plastids are the source of important immune response signaling and defense metabolites, including precursors of salicylic acid and jasmonic acid. Many plastid-derived signals must reach the nucleus to fulfill their proposed functions (reviewed in Kretschmer et al., 2020). Thus, re-localization of plastids toward the nucleus might promote more efficient signal transmission. Indeed, when challenged with different pathogens and H2O2, plastids relocated toward the nucleus in N. benthamiana epidermis leaf cells, forming perinuclear clusters (Erickson et al., 2014; Caplan et al., 2015; Ding et al., 2019a). How plant cells regulate the re-localization of plastids to the nucleus upon different stimuli remains unknown. Based on the observations of stromule orientation often coinciding with plastid directional movement, it was proposed that stromules are initiated during ETI and extend along the microtubules network, finding anchor points on actin filaments close to the nucleus which guide plastid body movement toward nuclei (Kumar et al., 2018). Perinuclear plastid clustering, as a consequence, might enhance plastid-to-nucleus signal transfer underpinning immune responses. When we challenged wild-type plants with XopQ-mORANGE2, stromule formation as well as perinuclear plastid clustering were consistently induced in lower epidermis cells (Figure 6, A–K). Additionally, stromules facing the nucleus and seemingly anchored in the nuclear periphery were observed (see Figures 2, B, 3, B, 4, B, and 5, B for SF%; e.g. 2D, 3D, and 4D for nucleus-associated stromules). Both observations support stromules guiding plastid body movement (Kumar et al., 2018). However, based on this model we expected impaired or abolished perinuclear clustering in plant lines unable to recognize XopQ (roq1) or to initiate TNL downstream signaling (eds1, nrg1, nrg1 adr1). Despite having reduced numbers of ETI-associated stromules, the plastid clustering still occurred when xopQ:mOrange2 was expressed in respective mutant backgrounds. Notably, plastid clustering was more pronounced in the mutants compared to the wild-type. In contrast, perinuclear clustering in response to xopQ:mOrange2 expression was not reduced in nrg1 plants (Figure 6, A–K) or upon expression of pNRG1::NRG1 in wild-type plants (Supplemental Figure S9C). In summary, we observe a negative association between stromule frequency and perinuclear plastid clustering. Accordingly, in our assays, ETI induction of stromules was associated with lower plastid clustering compared to when ETI was disabled (Figure 6H). These data suggest that perinuclear plastid clustering is not facilitated by ETI-induced stromules, but instead might be due to other mechanisms, which are enhanced by XopQ virulence activity.
Is induction of perinuclear plastid clustering a part of XopQ’s function?
Stronger plastid clustering observed in the absence of ROQ1, EDS1, and ADR1 together with NRG1 RNLs suggests it may represent a consequence of undisturbed XopQ activity (Figures 6, A and 7, B). This observation partially contradicts the suggestion that perinuclear plastid clustering supports ETI responses by facilitating more efficient transfer of pro-defense signals from plastids to the nucleus (discussed in Mullineaux et al., 2020). If the sole function of perinuclear plastids is to enhance ETI, why should the bacteria facilitate perinuclear plastid clustering via XopQ in the absence of effector recognition? Conversely, why suppress clustering when ETI is induced by XopQ? Our results suggest that clustering may serve multiple functions or is the consequence of several stimuli in plant–pathogen interactions. In support of this hypothesis, while plastid clustering in N. benthamiana occurs in response to ETI-triggering stimuli (e.g. TMV-p50 and AvrRpt2 recognition Kumar et al., 2018; Ding et al., 2019), it also occurs in response to PTI stimuli (Pst DC3000 ΔhopQ1-1, flg22 and H2O2, Ding et al., 2019), which demonstrates that plastid clustering is not ETI specific. Additionally, plastid clustering is not restricted to plant–microbe interactions and has been found to be important for plastid inheritance during cell division (Sheahan et al., 2004, 2020) and has been observed following the exposure of N. benthamiana epidermis leaf cells to cytokinin (Erickson et al., 2014). In summary, although plastid accumulation at the nucleus is linked to plant–microbe interactions, it is not exclusively so and may reflect one output resulting from changes to different cell physiological parameters (i.e. altered hormone or H2O2 levels). Currently, although we see that XopQ activity induces clustering, the trigger for this phenotype remains enigmatic and it remains to be seen whether it is of any benefit to Xcv during an infection.
Does PTI play a role in the phenotypes observed in response to XopQ expression?
Hormone-triggered expression of effectors from transgenes, as recently developed by Ngou et al. (2020), allows for the induction of ETI responses without simultaneously triggering PTI via PAMPs derived from bacterial infiltration. This approach revealed that despite separate early signaling components, PTI and ETI signaling partially converge further downstream, resulting in similar outputs, suggesting PTI and ETI signaling crosstalk (reviewed in Yuan et al., 2021). Since the system chosen for this study utilizes A. tumefaciens to mediate protein expression, the ETI-induced phenotypes observed are always occurring in the presence of a basal immune response to the bacteria, as would be the case during an encounter with a pathogen. However, in the future using the stable inducible transgenic system described by Ngou et al. (2020) in EDS1-dependent signaling mutants will allow for the evaluation of the importance of PTI to XopQ-triggered responses described here.
Conclusion
The goal of this study was to test if stromule formation in response to XopQ-triggered ETI is merely the consequence of cell death and if XopQ-triggered stromules support perinuclear plastid clustering. Here we provide experimental evidence for a direct link between ETI-signal induction and stromule formation, supporting the hypothesis of Caplan et al. (2015), which suggests that stromules play a specific role during ETI. Our findings therefore encourage the enquiry of the nature of this specific role in the future. In contrast to this, our results do not support the second hypothesis, which suggested that stromules might be needed to guide plastid movement toward the nucleus (Kumar et al., 2018), highlighting the fact that there is currently no mechanistic explanation for perinuclear plastid accumulation and that an explanation for this phenomenon will require further investigation. Additionally, residual stromule formation in nrg1, but not adr1_nrg1 double mutant plants, suggests that stromules are a highly sensitive read-out for low-level ETI responses in the absence of resistance and cell death.
Materials and methods
Plant material
Nicotiana benthamiana plant lines used in this study were: wild-type, roq1-3, and roq1-4 (Gantner et al., 2019), eds1-1 (also referred to as eds1a-1; Ordon et al., 2017), nrg1-4 and nrg1-5 (Ordon et al., 2021). An adr1_nrg1 double mutant was created by genome editing using a derivative of pDGE311, a plant transformation vector containing additional counter-selection markers and an intron-optimized zCas9i gene, as recently described (Grützner et al., 2021; Stuttmann et al., 2021) (Supplemental Materials and Methods S1). For Xanthomonas inoculations transgenic N. benthamiana plants of the plant line FNR:eGFP#7-25 expressing the plastid marker FNR:eGFP (Schattat et al., 2011a) were used. Plants were grown in long-day conditions (16-h day and 8-h night) in greenhouse chambers with controlled temperature and humidity. The temperature was approximately 23°C during the day and 19°C at night. Relative humidity was kept around 55%.
Bacterial strains and cultivation
Escherichia coli Top10 cells were used for cloning and DNA propagation. Cells were cultivated at 37°C in LB with the appropriate antibiotic selection. Agrobacterium strain GV3101 (pMP90; Koncz and Schell, 1986) was grown in liquid or on solid yeast extract beef (YEB) media containing rifampicin, gentamycin, and either spectinomycin or carbenicillin, while Xcv strains were grown in NYG medium supplemented with rifampicin (30°C for both). Xanthomonas strains utilized were: Xcv 85-10 (wild-type; Thieme et al., 2005), Xcv ΔhrcN (strain deficient in an ATPase required for type III secretion of effectors; Lorenz and Büttner, 2009 and Xcv ΔxopQ; Adlung et al., 2016).
Plasmids
For visualization of plastids and stromules, a plastid organelle marker construct was created using the Modular Cloning Toolbox (Weber et al., 2011; Engler et al., 2014). The final construct consisted of the 35S promotor (pICH51277), the chloroplast transit peptide of RUBISCO (SSU in the backbone pICH41258), eGFP (in the backbone pICH41264), and the nopalin synthase (NOS) terminator (pICH41421), assembled in a Level 1 acceptor plasmid. The xopQ:mOrange2 expression construct was described previously (Erickson et al., 2018; for more details see Supplemental Materials and Methods S2).
Agrobacterium tumefaciens-mediated transient expression
Plasmids were transformed into A. tumefaciens strain GV3101 (pMP90). For transient expression experiments, strains harboring the binary vectors were grown overnight in 5-mL YEB liquid cultures (with appropriate antibiotics), harvested by centrifugation, and resuspended in agrobacterium infiltration medium (10-mM MgCl2; 5-mM MES, pH 5.6; 0.15-µM Acetosyringone) with a final optical density (OD600nm) of 0.2. Bacteria harboring the plastid marker and the effector or the mORANGE2 control were mixed in a 1:1 ratio. Using a needless syringe, bacterial suspensions were inoculated into intercostal areas of the youngest fully expanded leaves of 5- to 6-week-old N. benthamiana plants (see Supplemental Materials and Methods S3).
Xcv inoculations
Xcv NYG liquid cultures were centrifuged to harvest cells, bacteria were resuspended in 10-mM MgCl2, and suspensions were adjusted to an OD600nm of 0.1. All three strains, as well as a buffer control, were then inoculated as described for A. tumefaciens. Plastids/stromules were observed at 2 dpi using an epi-fluorescence microscope.
Imaging hardware
For image acquisition, an epi-fluorescence microscope (AxioObserver Z1) setup from Zeiss (Jena, Germany) equipped with an X-Cite fluorescence light source and an MRm monochrome camera (Zeiss, Jena, Germany) was used. GFP fluorescence was recorded using a 38 HE filter cube (Carl Zeiss AG, Jena, Germany). mORANGE2 fluorescence was recorded utilizing the 43 HE filter cube (Carl Zeiss AG. Jena, Germany). The microscope manufacturer’s software (ZenBlue, Zeiss, Germany) controlled image acquisition. All images were captured using a 40x/0.75 NA EC PLAN NEOFLUAR lens.
Imaging procedures and image processing
For the quantification of stromule frequencies, a single leaf disc of each infiltration spot was harvested using a cork borer. Leaf discs were vacuum-infiltrated and mounted on glass slides, and three independent z-stacks of the lower epidermis were collected in transmitted light, eGFP, and mORANGE2 channels. In order to obtain 2D extended depth of field images for quantification, single images of the z-series of each channel were first exported into separate file folders and subsequently combined into single images using software and procedures described in Schattat and Klösgen (2009) (total of three images per disc).
For the quantification of stromule frequencies (SF%), we measured the proportion of plastids with at least one stromule (Erickson et al., 2014). To facilitate the faster quantification of stromule and plastid counts in N. benthamiana tissues, we expanded on the previously published MTBCellCounter (Franke et al., 2015) via a ridge detection-based stromule detection algorithm (Möller and Schattat, 2019). The extended MTBCellCounter allows for the detection of plastid bodies as described in Franke et al. (2015) and identifies subsequently plastids with stromules.
The PNAI was described previously (Erickson et al., 2014) and represents the absolute number of plastids in close association with a given nucleus. PNAI was evaluated in the 2D projected images (see image processing). Nuclei were counted as nucleus associated when either the plastid body touched, overlapped with, or was within a distance of 4 µm from the nucleus. Four micrometer corresponds to the average epidermis plastid diameter.
For sample sizes and details on statistical analysis of SF% and PNAI, see Supplemental Materials and Methods S4, Supplemental Tables S1–S5, and Supplemental Statistics S1–S8.
Naming conventions
For conventions used to name mutants, genes, proteins, and artificial DNA constructs, see Supplemental Materials and Methods S5.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AAV74206 (xopQ), AAY5460 (NRG1), AAL85347 (EDS1), Gene ID Niben101Scf02118g00018 (ADR1), ATD14363 (ROG1), and ABC66096 (mOrange2).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Full frames of stacked fluorescence images of a mock and Xcv ΔhrcN infiltrated FNR:eGFP-7-25 N. benthamiana plant.
Supplemental Figure S2. Full frames of stacked fluorescence images of a Xcv wild-type and Xanthomonas campestris pv. vesicatoria ΔxopQ infiltrated FNReGFP-7-25 N. benthamiana plant.
Supplemental Figure S3. Moderate optical densities of GV3101 (pMP90) induce moderate stromule frequencies at 3 dpi.
Supplemental Figure S4. Full-frame stacked fluorescence images of an mOrange2+SSU:eGFP and an xopQ:mOrange2+SSU:eGFP inoculated N. benthamiana wild-type plant.
Supplemental Figure S5. Full-frame stacked fluorescence images of an mOrange2+SSU:eGFP and an xopQ:mOrange2+SSU:eGFP inoculated N. benthamiana roq1 plant.
Supplemental Figure S6. Full-frame stacked fluorescence images of an mOrange2+SSU:eGFP and an xopQ:mOrange2+SSU:eGFP inoculated N. benthamiana eds1 plant.
Supplemental Figure S7. Full-frame stacked fluorescence images of an mOrange2+SSU:eGFP and an xopQ:mOrange2+SSU:eGFP inoculated N. benthamiana nrg1 plant.
Supplemental Figure S8. Macroscopic phenotype, SF%, and PNAI in response to NRG1 over-expression in FNR:eGFP-7-25 transgenic WT plants.
Supplemental Figure S9. Full-frame stacked fluorescence images of an inoculated N. benthamiana adr1_nrg1 plant.
Supplemental Table S1. Summary of SF% values used for stromule frequency bar blots in the main manuscript.
Supplemental Table S2. Summary of PNAI values used for box blots in the main manuscript.
Supplemental Table S3. Values used for SF% bar plots in Supplemental Figure S3A.
Supplemental Table S4. Values used for SF% box plots in Supplemental Figure S8B.
Supplemental Table S5. Values used for PNAI box plots in Supplemental Figure S8D.
Supplemental Statistics S1. For Figure 1 SF.
Supplemental Statistics S2. For Figure 2 SF.
Supplemental Statistics S3. For Figure 3 SF.
Supplemental Statistics S4. For Figure 4 SF.
Supplemental Statistics S5. For Figure 5 SF.
Supplemental Statistics S6. For Figure 6 PNAI.
Supplemental Statistics S7. For Supplemental Figure S3 SF.
Supplemental Statistics S8. For Supplemental Figure S8 SF.
Supplemental Statistics S9. For Supplemental Figure S8 PNAI.
Supplemental Materials and Methods S1. Generation of Nb nrg1 adr1 double mutant line.
Supplemental Materials and Methods S2. Cloning of plasmids.
Supplemental Materials and Methods S3. Experimental procedure utilized for A. tumefaciens infiltration experiments.
Supplemental Materials and Methods S4. Information on sample sizes and data analysis for stromule frequencies and PNAI values.
Supplemental Materials and Methods S5. Naming conventions.
Supplementary Material
Acknowledgments
All Xcv strains were kindly provided by Prof. Ulla Bonas, Martin-Luther-Universität Halle-Wittenberg, Germany. Dr. Sebastian Schornack, TSL, Cambridge, UK for advice on the manuscript. We thank Jaqueline Bautor, Junli Wang and Dmitry Lapin (MPIPZ) for help in developing the Cas9-free adr1_nrg1 double mutant.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–400681449/GRK2498, STU642/1-1, and SFB-1403–414786233 as well as Martin-Luther-University core funding.
Conflict of interest statement. There is no conflict of interest.
Contributor Information
Jennifer Prautsch, Biology, Plant Physiology, Martin-Luther-University Halle-Wittenberg, Halle, Germany.
Jessica Lee Erickson, Biology, Plant Genetics, Martin-Luther-University Halle-Wittenberg, Halle, Germany; Leibniz-Institut for Plant Biochemistry, Halle, Germany.
Sedef Özyürek, Biology, Plant Physiology, Martin-Luther-University Halle-Wittenberg, Halle, Germany.
Rahel Gormanns, Biology, Plant Physiology, Martin-Luther-University Halle-Wittenberg, Halle, Germany.
Lars Franke, Biochemistry and Biotechnology, Martin-Luther-University Halle-Wittenberg, Halle, Germany.
Yang Lu, Biology, Plant Physiology, Martin-Luther-University Halle-Wittenberg, Halle, Germany.
Jolina Marx, Leibniz-Institut for Plant Biochemistry, Halle, Germany.
Frederik Niemeyer, Biology, Plant Physiology, Martin-Luther-University Halle-Wittenberg, Halle, Germany.
Jane E Parker, Max Planck Institute for Plant Breeding Research, Cologne, Germany.
Johannes Stuttmann, Biology, Plant Genetics, Martin-Luther-University Halle-Wittenberg, Halle, Germany; Institute for Biosafety in Plant Biotechnology, Federal Research Centre for Cultivated Plants, Julius Kühn-Institute (JKI), Quedlinburg, Germany.
Martin Hartmut Schattat, Biology, Plant Physiology, Martin-Luther-University Halle-Wittenberg, Halle, Germany.
J.L.E., J.S., J.E.P., and M.H.S. contributed to experimental design and the written text; J.P., S.Ö., R.G., Y.L., F.N., J.M., and L.F. conducted the A. tumefaciens inoculation experiments in N. benthamiana and respective image analysis; J.L.E. conducted Xcv inoculation experiments and respective image analysis.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Martin Hartmut Schattat (martin.schattat@pflanzenphys.uni-halle.de), Jane Parker (parker@mpipz.mpg.de), and Johannes Stuttmann (jstuttmann@gmail.com).
References
- Adachi H, Contreras M, Harant A, Wu C, Derevnina L, Sakai T, Duggan C, Moratto E, Bozkurt T, Maqbool A, et al. (2019) An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife 8: e49956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlung N, Bonas U (2017) Dissecting virulence function from recognition: cell death suppression in Nicotiana benthamiana by XopQ/HopQ1-family effectors relies on EDS1-dependent immunity. Plant J 91: 430–442 [DOI] [PubMed] [Google Scholar]
- Adlung N, Prochaska H, Thieme S, Banik A, Blüher D, John P, Nagel O, Schulze S, Gantner J, Delker C, et al. (2016) Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front Plant Sci 7: 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Su M, Li N, Liang Y, Dang S, Xu J, Hu M, Wang J, Zou M, Deng Y, et al. (2021) The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184: 3528–3541.e12 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108: 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D (2016) Behind the lines–actions of bacterial type III effector proteins in plant cells. FEMS Microbiol Rev 40: 894–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Kumar AS, Park E, Padmanabhan MS, Hoban K, Modla S, Czymmek K, Dinesh-Kumar SP (2015) Chloroplast stromules function during innate immunity. Dev Cell 34: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel B, Ngou P-M, Cevik V, Redkar A, Kim D-S, Yang Y, Ding P, Jones JDG (2019) Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol 222: 966–980 [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel L-P, Moffett P (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. MPMI 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Delfosse K, Wozny MR, Jaipargas E-A, Barton KA, Anderson C, Mathur J (2016) Fluorescent protein aided insights on plastids and their extensions: a critical appraisal. Front Plant Sci 6: 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Jimenez-Gongora T, Krenz B, Lozano-Duran R (2019) Chloroplast clustering around the nucleus is a general response to pathogen perception in Nicotiana benthamiana. Mol Plant Pathol 8: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843 [DOI] [PubMed] [Google Scholar]
- Erickson JL, Adlung N, Lampe C, Bonas U, Schattat MH (2018) The Xanthomonas effector XopL uncovers the role of microtubules in stromule extension and dynamics in Nicotiana benthamiana. Plant J 93: 856–870 [DOI] [PubMed] [Google Scholar]
- Erickson JL, Ziegler J, Guevara D, Abel S, Klösgen RB, Mathur J, Rothstein SJ, Schattat MH (2014) Agrobacterium-derived cytokinin influences plastid morphology and starch accumulation in Nicotiana benthamiana during transient assays. BMC Plant Biol 14: 127–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke L, Erickson JL, Rödel D, Schröter D, Storbeck B, Möller B, Schattat MH (2015) The “MTB Cell Counter” a versatile tool for the semi-automated quantification of sub-cellular phenotypes in fluorescence microscopy images. A case study on plastids, nuclei and peroxisomes. Endocytobiosis Cell Res 26: 31–42 [Google Scholar]
- Gantner J, Ordon J, Kretschmer C, Guerois R, Stuttmann J (2019) An EDS1-SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana. Plant Cell 31: 2456–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JC, Hansen MR, Shaw DJ, Graham K, Dale R, Smallman P, Natesan SKA, Newell CA (2012) Plastid stromules are induced by stress treatments acting through abscisic acid. Plant J 69: 387–398 [DOI] [PubMed] [Google Scholar]
- Gray JC, Sullivan JA, Hibberd JM, Hansen MR (2001) Stromules: mobile protrusions and interconnections between plastids. Plant Biol 3: 223–233 [Google Scholar]
- Grützner R, Martin P, Horn C, Mortensen S, Cram EJ, Lee-Parsons CWT, Stuttmann J, Marillonnet S (2021) High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun 2: 100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning BES (2005) Plastid stromules: video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging, and tip-shedding. Protoplasma 225: 33–42 [DOI] [PubMed] [Google Scholar]
- Holzinger A, Kwok EY, Hanson MR (2008) Effects of arc3, arc5 and arc6 mutations on plastid morphology and stromule formation in green and nongreen tissues of Arabidopsis thaliana. Photochem Photobiol 84: 1324–1335 [DOI] [PubMed] [Google Scholar]
- Huang S, Jia A, Song W, Hessler G, Meng Y, Sun Y, Xu L, Laessle H, Jirschitzka J, Ma S, et al. (2022) Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity. Science (New York, NY) 377: abq3297. [DOI] [PubMed] [Google Scholar]
- Jacob P, Kim NH, Wu F, Kasmi El F, Chi Y, Walton WG, Furzer OJ, Lietzan AD, Sunil S, Kempthorn K, et al. (2021) Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science (New York, NY) 373: 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia A, Huang S, Song W, Wang J, Meng Y, Sun Y, Xu L, Laessle H, Jirschitzka J, Hou J, et al. (2022) TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity. Science (New York, N.Y) 377: eabq8180. [DOI] [PubMed] [Google Scholar]
- Jubic LM, Saile S, Furzer OJ, Kasmi El F, Dangl JL (2019) Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol 50: 82–94 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of T L-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. 204: 383–396 [Google Scholar]
- Köhler RH, Hanson MR (2000) Plastid tubules of higher plants are tissue-specific and developmentally regulated. J Cell Sci 113 (Pt 1): 81–89 [DOI] [PubMed] [Google Scholar]
- Krenz B, Jeske H, Kleinow T (2012) The induction of stromule formation by a plant DNA-virus in epidermal leaf tissues suggests a novel intra- and intercellular macromolecular trafficking route. Front Plant Sci 3: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer M, Damoo D, Djamei A, Kronstad J (2020) Chloroplasts and Plant Immunity: Where Are the Fungal Effectors? Pathogens 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AS, Park E, Nedo A, Alqarni A, Ren L, Hoban K, Modla S, McDonald JH, Kambhamettu C, Dinesh-Kumar SP, et al. (2018) Stromule extension along microtubules coordinated with actin-mediated anchoring guides perinuclear chloroplast movement during innate immunity. eLife 7: e23625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D, Bhandari DD, Parker JE (2020) Origins and immunity networking functions of EDS1 family proteins. Annu Rev Phytopathol 58: 253–276 [DOI] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, Born von P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. (2019) A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31: 2430–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Büttner D (2009) Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J Bacteriol 191: 1414–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Tsuda K (2021) Intimate association of PRR- and NLR-mediated signaling in plant immunity. MPMI 34: 3–14 [DOI] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, et al. (2020) Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science (New York, NY) 370: eabe3069 [DOI] [PubMed] [Google Scholar]
- Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, Staskawicz BJ (2020) Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science (New York, NY) 370: eabd9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mammone A, Barton KA (2012) Organelle extensions in plant cells. J Integr Plant Biol 54: 851–867 [DOI] [PubMed] [Google Scholar]
- Möller B, Schattat MH (2019) Quantification of stromule frequencies in microscope images of plastids combining ridge detection and geometric criteria. In Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2019), Vol. 2: BIOIMAGING. INSTICC, SciTePress, Prague, Czech Republic, pp. 38–48
- Mullineaux PM, Selga T, Selga M, Exposito-Rodriguez M, Laissue PP, Gobiņš V, Smirnoff N, Laser AO, Park E (2020) Spatial chloroplast-to-nucleus signalling involving plastid-nuclear complexes and stromules. Philos Trans R Soc Lond B Biol Sci 375: 20190405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou BPM, Ahn H-K, Ding P, Redkar A, Brown H, Ma Y, Youles M, Tomlinson L, Jones JDG (2020) Estradiol-inducible AvrRps4 expression reveals distinct properties of TIR-NLR-mediated effector-triggered immunity. J Exp Bot 71: 2186–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordon J, Gantner J, Kemna J, Schwalgun L, Reschke M, Streubel J, Boch J, Stuttmann J (2017) Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J 89: 155–168 [DOI] [PubMed] [Google Scholar]
- Ordon J, Martin P, Erickson JL, Ferik F, Balcke G, Bonas U, Stuttmann J (2021) Disentangling cause and consequence: genetic dissection of the DANGEROUS MIX2 risk locus, and activation of the DM2h NLR in autoimmunity. Plant J 106: 1008–1023 [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, Karelina D, Hua C, Fröhlich K, Wan W-L, et al. (2021) The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598: 495–499 [DOI] [PubMed] [Google Scholar]
- Pyke KA (2013) Divide and shape: an endosymbiont in action. Planta 237: 381–387 [DOI] [PubMed] [Google Scholar]
- Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho M-J, Schultink A, Staskawicz BJ (2018) NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci USA 115: E10979–E10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile SC, Jacob P, Castel B, Jubic LM, Salas-Gonzáles I, Bäcker M, Jones JDG, Dangl JL, El Kasmi F (2020) Two unequally redundant "helper" immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol 18: e3000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattat M, Barton K, Baudisch B, Klösgen RB, Mathur J (2011a) Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol 155: 1667–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattat MH, Klösgen RB (2009) Improvement of plant cell microscope images by use of “depth of field” extending software. Endocytobiosis Cell Res 19: 11–19 [Google Scholar]
- Schattat MH, Klösgen RB (2011b) Induction of stromule formation by extracellular sucrose and glucose in epidermal leaf tissue of Arabidopsis thaliana. BMC Plant Biol 11: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz B (2017) Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J 92: 787–795 [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Collings DA, Rose RJ, McCurdy DW (2020) ACTIN7 is required for perinuclear clustering of chloroplasts during Arabidopsis protoplast culture. Plants 9: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, Rose RJ, McCurdy DW (2004) Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J 37: 379–390 [DOI] [PubMed] [Google Scholar]
- Stuttmann J, Barthel K, Martin P, Ordon J, Erickson JL, Herr R, Ferik F, Kretschmer C, Berner T, Keilwagen J, et al. (2021) Highly efficient multiplex editing: one-shot generation of 8× Nicotiana benthamiana and 12× Arabidopsis mutants. Plant J 106: 8–22 [DOI] [PubMed] [Google Scholar]
- Sun X, Lapin D, Feehan JM, Stolze SC, Kramer K, Dongus JA, Rzemieniewski J, Blanvillain-Baufumé S, Harzen A, Bautor J, et al. (2021) Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat Commun 12: 3335–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper D, Burstein D, Salomon D, Gershovitz M, Pupko T, Sessa G (2016) Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine-learning approach. Mol Plant Pathol 17: 398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme F, Koebnik R, Bekel T, Berger C, Boch J, Büttner D, Caldana C, Gaigalat L, Goesmann A, Kay S, et al. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol 187: 7254–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Wu Z, Chen S, Ao K, Huang W, Yaghmaiean H, Sun T, Xu F, Zhang Y, Wang S, et al. (2021) Activation of TIR signalling boosts pattern-triggered immunity. Nature 598: 500. [DOI] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G (2016) Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54: 419–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vismans G, van der Meer T, Langevoort O, Schreuder M, Bouwmeester H, Peisker H, Dörman P, Ketelaar T, van der Krol A (2016) Low-phosphate induction of plastidal stromules is dependent on strigolactones but not on the canonical strigolactone signaling component MAX2. Plant Physiol 172: 2235–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE (2013) Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14: 619–630 [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang H-W, Zhou J-M, Chai J (2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science (New York, NY). doi: 10.1126/science.aav5870 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang H-W, et al. (2019b) Ligand-triggered allosteric ADP release primes a plant NLR complex. Science (New York, NY). doi: 10.1126/science.aav5868 [DOI] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X (2019) Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol 222: 938–953 [DOI] [PubMed] [Google Scholar]
- Yuan M, Ngou BPM, Ding P, Xin X-F (2021) PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62: 102030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.