Abstract
When exposed to pathogen infection or ultraviolet (UV) radiation, grapevine (Vitis vinifera) plants rapidly accumulate the stilbenoid resveratrol (Res) with concomitant increase of stilbene synthase (STS), the key enzyme in stilbene biosynthesis. Although a few transcription factors have been shown to regulate STSs, the molecular mechanism governing the regulation of STSs is not well elucidated. Our previous work showed that a VvMYB14–VvWRKY8 regulatory loop fine-tunes stilbene biosynthesis in grapevine through protein–protein interaction; overexpression of VvWRKY8 down-regulates VvMYB14 and VvSTS15/21; and application of exogenous Res up-regulates WRKY8 expression. Here, we identified an R2R3-MYB repressor, VvMYB30, which competes with the activator VvMYB14 for binding to the common binding sites in the VvSTS15/21 promoter. Similar to VvMYB14, VvMYB30 physically interacts with VvWRKY8 through their N-termini, forming a complex that does not bind DNA. Exposure to UV-B/C stress induces VvMYB14, VvWRKY8, and VvSTS15/21, but represses VvMYB30 in grapevine leaves. In addition, MYB30 expression is up-regulated by VvWRKY8-overexpression or exogenous Res. These findings suggest that the VvMYB14–VvWRKY8–VvMYB30 regulatory circuit allows grapevine to respond to UV stress by producing Res and prevents over-accumulation of Res to balance metabolic costs. Our work highlights the stress-mediated induction and feedback inhibition of stilbene biosynthesis through a complex regulatory network involving multiple positive and negative transcriptional regulators.
VvMYB30 is involved in a complex regulatory network containing multiple positive and negative transcriptional regulators that fine-tunes stilbene biosynthesis under ultraviolet stress.
Introduction
Resveratrol (Res; 3,4′,5-trihydroxystilbene) is a non-flavonoid polyphenolic compound derived from stilbene. As a phytoalexin, Res plays a crucial role in plant defense against phytopathogens and adaption to abiotic stress (Valletta et al., 2021). For example, Res inhibits the germination of conidia and sporangia of the plant pathogens Peronospora viticola and Botrytis cinerea (Adrian et al., 1997; Pezet et al., 2004). Exogenous Res can alleviate the KCl salinity stress of crabapple (Malus hupehensis) seedlings by eliminating reactive oxygen species production (Li et al., 2021). Much effort has been devoted to the study of Res for its pharmacological properties. Res reduces the risk of cancer, cardiovascular disease, and Alzheimer’s disease in humans (Diaz-Gerevini et al., 2016; Xia et al., 2017; Kiskova et al., 2020). However, Res is naturally synthesized in only a few plant species including grapevine (Vitis spp.) (Vannozzi et al., 2012). In stilbene-synthesizing plants, the stilbene biosynthetic pathway is a continuation of the shikimate pathway (Austin and Noel, 2003; Dubrovina and Kiselev, 2017; Hasan and Bae, 2017). The substrates p-coumaroyl-CoA and malony-CoA are catalyzed into Res by stilbene synthase (STS), Res is then converted into the corresponding glycoside (piceid, Pd) by the glycosyltransferases (Dubrovina and Kiselev, 2017).
Grapevine (Vitis vinifera) is currently the main source of Res worldwide because of its extensive cultivation and high production efficiency (Weiskirchen and Weiskirchen, 2016). With increasing demand for a natural source of Res, it is important to determine the gene regulatory mechanisms controlling Res production. Based on predicted amino acid sequences, 33 full-length VvSTS genes have been identified and clustered into three major groups, designated A, B, and C (Vannozzi et al., 2012). In recent years, a few transcription factors (TFs) have been demonstrated to regulate stilbene biosynthesis in grapevine. The grape MYB TFs, VvMYB14 and VvMYB15, regulate the stilbene biosynthetic pathway by activating the promoters of STS genes, resulting in increased accumulation of stilbene (Höll et al., 2013). VvMYB13 co-expresses with STSs, as well as VvMYB14 and VvMYB15, and regulates stilbene biosynthesis under biotic and abiotic stresses (Wong et al., 2016). In the Chinese wild grape Vitis davidii, VdMYB1 activates VdSTS2 and stimulates the response to pathogen infection (Yu et al., 2019). The expression of VvMYB9/14/15а/40/60/107 positively correlates with high Res content in grapevine, suggesting that these TFs are activators of stilbene biosynthesis (Kiselev et al., 2017). Recently, other TF families have been reported to participate in regulating STS. VvWRKY24 induces the expression of VvSTS29 through binding directly to its promoter (Vannozzi et al., 2018). Overexpression of the basic leucine zipper TF VqbZIP1 increases STS expression and stilbene accumulation in Vitis quinquangularis (Wang et al., 2019). Other than directly acting on the STS genes, some TFs recruit other TFs to coordinate the regulation of STS expression and stilbene accumulation. For instance, the ethylene responsive factor VqERF114 does not bind to the VqSTS promoters but interacts with VqMYB35, which binds the promoters of VqSTS15/28/44/46 to jointly promote STS transcription and stilbene accumulation (Wang and Wang, 2019). In V. quinquangularis, VqWRKY53 interacts with VqMYB14 and VqMYB15 to activate VqSTS32 and VqSTS41, and the activation of VqSTS32 and VqSTS41 by the WRKY53-MYB14/MYB15 complex is significantly higher compared with that by VqWRKY53 alone (Wang et al., 2020). Although STS genes have been found in the other stilbene-synthesizing plants, such as Japanese knotweed (Polygonum cuspidatum), Japanese red pine (Pinus densiflora), and European red pine (Pinus sylvestris), the regulatory mechanisms of non-grapevine STS genes have not been extensively investigated (Müller et al., 1999; Kodan et al., 2002; Liu et al., 2019).
The biosynthesis of stilbenes in plants can be triggered by a variety of biotic and abiotic environmental factors, such as fungal infections, ozone, wounding, and, in particular, ultraviolet (UV) radiation (Langcake and Pryce, 1976; González-Barrio et al., 2006; Cho et al., 2012; Deng et al., 2016; Yin et al., 2017; Vannozzi et al., 2018; Valletta et al., 2021). UV-B and UV-C stresses significantly increase stilbene production in different stilbene-producing plants, including grapevine, peanut, Picea jezoensis (Siebold and Zucc.) Carr., and P. cuspidatum Siebold and Zucc. (Hasan and Bae, 2017; Valletta et al., 2021). However, only a few TFs regulating STS expression have been identified, and the underlying molecular mechanisms governing UV stress-induced stilbene accumulation are not well elucidated.
Biosynthesis of specialized metabolites is often regulated by coordinated actions of transcriptional activators and repressors in response to various stimuli (Ma and Constabel, 2019). The MYB TFs are one of the largest families of transcriptional regulators in plants. Arabidopsis (Arabidopsis thaliana) R2R3 MYBs have been grouped into 25 subgroups (SGs) based on phylogeny (Dubos et al., 2010). The SG1–SG7 MYBs are known to be involved in the biosynthesis of specialized metabolites, for example, SG1 is involved in the biosynthesis of cuticular wax and stomatal control, SG2 in phytoalexins (stilbenes), SG3 in lignification, SG4 in phenylpropanoid, SG5 in proanthocyanidins, SG6 in anthocyanins, and SG7 in flavonols (Albert and Allan, 2021). Some MYBs, including SG4 of R2R3 MYBs and R3 MYBs, are transcriptional repressors of metabolic pathways (Chen et al., 2019a, 2019b; Albert and Allan, 2021). For example, AtMYB4 with a C-terminal EAR motif is a repressor of the phenylpropanoid pathway in Arabidopsis. AtMYB4 mutants show enhanced accumulation of sinapate esters functioning as UV-protecting sunscreen and are more tolerant to UV-B irradiation than the wild type (Jin et al., 2000). In Petunia, MYB27, an R2R3 MYB, represses anthocyanin biosynthetic genes (e.g. CHS, DFR, and ANS) through its C-terminal EAR motif (Albert et al., 2014). AtMYBL2, an R3-MYB protein, acts as a transcriptional repressor to modulate the expression of flavonoid pathway genes in Arabidopsis (Dubos et al., 2008; Matsui et al., 2008). CAPRICE (CPC), an R3 MYB, is a negative regulator that represses anthocyanin accumulation in Arabidopsis (Zhu et al., 2009). Besides MYBs, some WRKY TFs are also known to be negative regulators of specialized metabolism (Yokotani et al., 2013; Jiang et al., 2019). Stilbene biosynthesis may also be controlled by a complex transcriptional network that includes positive and negative regulators.
Previously, we have shown that VvWRKY8 negatively regulates VvSTS15/21 by directly interacting with the activator VvMYB14 (Jiang et al., 2019), which only partly explains why overexpression of phytoalexin genes in plants does not always result in higher phytoalexin production (Jeandet et al., 2019). As WRKY proteins are known to interact with various other TFs, we hypothesized that WRKY8 also interacts with other TFs, including additional MYB factors. We conducted a yeast two-hybrid (Y2H) screen using VvWRKY8 as bait and found the R2R3-MYB TF, VvMYB30, to be an interacting partner. In this study, we demonstrated that VvMYB30 represses VvSTS15/21 by directly binding to their promoters. Moreover, VvMYB30 does not interact with, but rather, competes with VvMYB14 for the same binding sites in the VvSTS15/21 promoter. Expression of VvMYB30 is repressed by UV-B and UV-C but induced by Res. Taken together, these results deepen our understanding of the complexity of the regulatory hub that governs balanced production of Res in grapevine in response to stresses such as UV.
Results
VvMYB30 physically interacts with VvWRKY8 through their N-termini
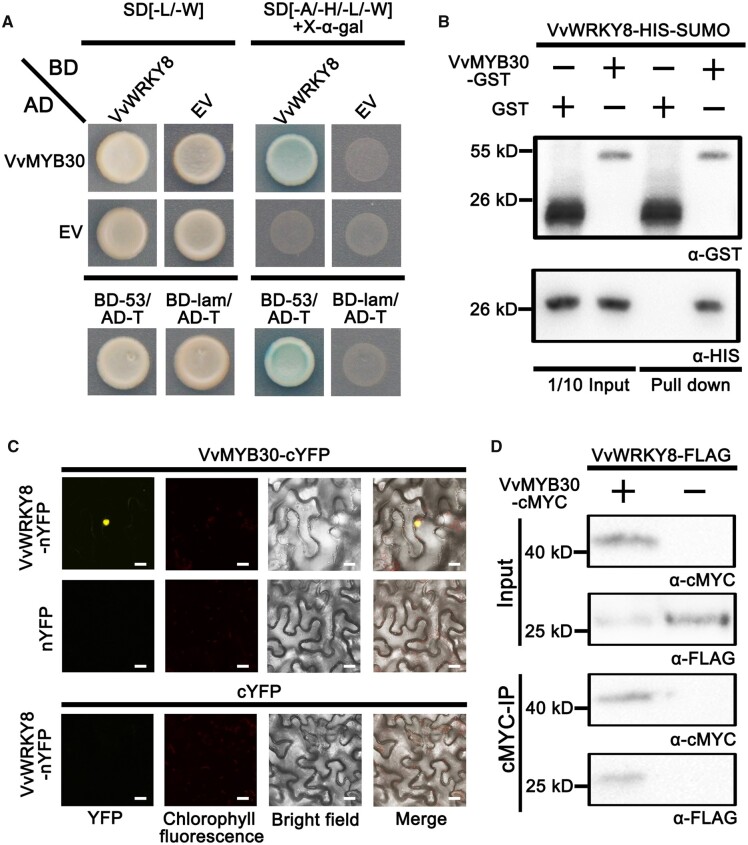
We conducted a Y2H screen of our previously described cDNA library of “Pinot Noir” grape leaf (Jiang et al., 2019), using VvWRKY8 (Gene ID: VIT_201s0010g03930) as a bait. In addition to the previously identified proteins including VvMYB14 (Gene ID: VIT_207s0005g03340), we obtained two positive clones encoding VvMYB30 (Gene ID: VIT_217s0000g06190). We carried out a follow-up Y2H assay to verify the interaction between VvWRKY8 and VvMYB30. The full-length coding sequences of VvMYB30 and VvWRKY8 were sub-cloned into the pGADT7 (AD) vector and pGBKT7 (BD) vector, respectively. After transformation, yeast cells harboring AD-VvMYB30/BD-VvWRKY8 or AD-T/BD-53 (positive control) survived on the SD/-Leu/-Trp/-His/-Ade quadruple selection medium, and the colonies turned blue when supplied with 5-bromo-4-chloro-3-indoxyl-α-d-galactopyranoside (X-α-gal), while cells transformed with AD-T/BD-Lam (negative control), AD-VvMYB30/BD-empty vector (EV), or AD-EV/BD-VvWRKY8 failed to grow on the quadruple selection medium (Figure 1A).
Figure 1.
VvMYB30 interacts physically with VvWRKY8. A, Y2H assay. Yeast cells co-transformed with VvMYB30 fused to the GAL4 activation domain and VvWRKY8 fused to the GAL4 binding domain were grown on selective media. AD-T/BD-53 was used as a positive control. VvMYB30-AD/BD, AD/VvWRKY8-BD, and AD-T/BD-lam were used as negative controls. B, In vitro pull-down assay. VvWRKY8 was fused with SUMO and HIS tags, and the molecular weights were almost 30 kD, VvMYB30 were fused with GST tag, and the molecular weight was almost 55 kD. VvMYB30 and GST were detected by GST tag monoclonal antibody, VvWRKY8 were detected by HIS tag monoclonal antibody. C, BiFC assay. VvWRKY8 was fused with the nYFP, VvMYB30 were fused with the cYFP, and the combinations of VvMYB30-cYFP/VvWRKY8-nYFP, VvMYB30-cYFP/nYFP, and cYFP/VvWRKY8-nYFP were expressed in N. benthamiana leaves, respectively. Scale bar: 20 μm. D, Co-IP assays in N. benthamiana leaves, the molecular weight of VvWRKY8 and VvMYB30 was almost 25 and 40 kD, respectively. α-cMYC, cMYC tag monoclonal antibody; α-FLAG, FLAG tag polyclonal antibody.
Next, a glutathione-S-transferase (GST) pull-down assay was used to further confirm the VvWRKY8–VvMYB30 interaction using the purified recombinant GST-VvMYB30 and small ubiquitin-like modifier (SUMO)-histidine (HIS)-VvWRKY8. SUMO-HIS-VvWRKY8 was detected with a HIS-tag antibody following incubation with GST-VvMYB30 but was not detected when SUMO–HIS–VvWRKY8 was incubated with GST (Figure 1B). SUMO-HIS did not interact with GST-VvMYB30 (Supplemental Figure S1). The in vivo interaction of VvWRKY8 and VvMYB30 was also confirmed using a bimolecular fluorescence complementation (BiFC) assay in Nicotiana benthamiana leaves. VvWRKY8 and VvMYB30 were fused to the amino- and carboxy-terminus of the yellow fluorescent protein (YFP), respectively, to generate VvWRKY8-nYFP and VvMYB30-cYFP. Three days after co-infiltration of N. benthamiana leaves with VvWRKY8-nYFP and VvMYB30-cYFP, strong YFP fluorescence was observed in the nucleus. No YFP signal was detected when VvWRKY8-nYFP was co-transformed with EV-cYFP or VvMYB30-cYFP was co-transformed with EV-nYFP (Figure 1C). Finally, co-immunoprecipitation (Co-IP) was performed to verify the VvWRKY8–VvMYB30 interaction. Two tagged proteins, VvWRKY8-FLAG and VvMYB30-cMYC, were co-expressed in N. benthamiana leaves. The VvMYB30-cMYC-tagged protein was detected after precipitation using anti-cMYC antibody, whereas the VvWRKY8-FLAG-tagged protein was detected after incubation with anti-FLAG antibody (Figure 1D).
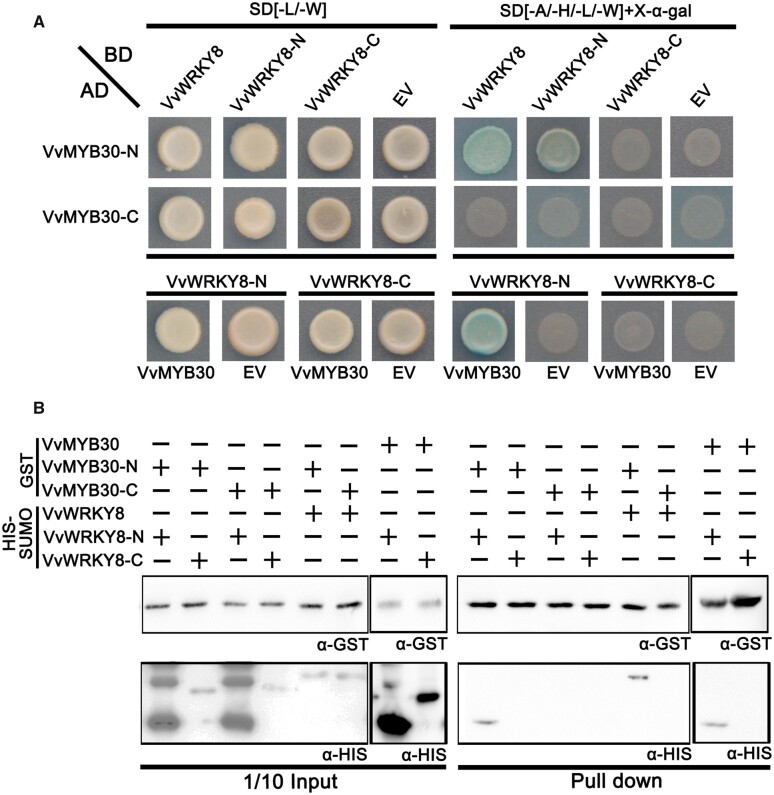
To identify the interacting domains of VvMYB30 and VvWRKY8, we divided the proteins into C-terminal (VvWRKY8-C and VvMYB30-C) and N-terminal (VvWRKY8-N and VvMYB30-N) fragments (Supplemental Figure S2). The C- and N-terminal coding sequence fragments of VvMYB30 and VvWRKY8 were cloned into the pGADT7 and pGBKT7 vectors, respectively. Y2H was carried out using the combinations of vectors harboring the full-length or fragmented coding sequences of VvMYB30 and VvWRKY8. Only the cells containing VvWRKY8/VvMYB30-N, VvWRKY8-N/VvMYB30-N, and VvWRKY8-N/VvMYB30 survived on the SD/-Leu/-Trp/-His/-Ade selection medium and appeared blue when supplied with X-α-gal (Figure 2A), indicating that VvWRKY8 and VvMYB30 interact through their N-terminal domains. To further confirm the results, we conducted a pull-down assay (Figure 2B). The purified SUMO–HIS–VvWRKY8-N and SUMO–HIS–VvWRKY8 recombinant proteins were pulled down by glutathione sepharose after incubation with purified recombinant GST-VvMYB30 or GST-VvMYB30-N (Figure 2B).
Figure 2.
Interaction between amino (N)-terminal halves of VvWRKY8 and VvMYB30. A, Y2H assay. VvMYB30-N-AD/BD, VvMYB30-C-AD/BD, AD/VvWRKY8-N-BD, and AD/VvWRKY8-C-BD were used as negative controls. B, Pull-down assay. VvWRKY8-N and VvWRKY8-C were fused with SUMO and HIS tags, and the molecular weights were almost 18 and 23 kD, respectively. VvMYB30-N and VvMYB30-C were fused with GST tag, and the molecular weight was almost 40 kD. VvMYB30 and GST were detected by GST tag monoclonal antibody, VvWRKY8 was detected by HIS tag monoclonal antibody. VvMYB30-N, N-terminal of VvMYB30; VvMYB30-C, C-terminal of VvMYB30; VvWRKY8-N, N-terminal of VvWRKY8; and VvWRKY8-C, C-terminal of VvWRKY8.
Structural and phylogenetic analysis of VvMYB30
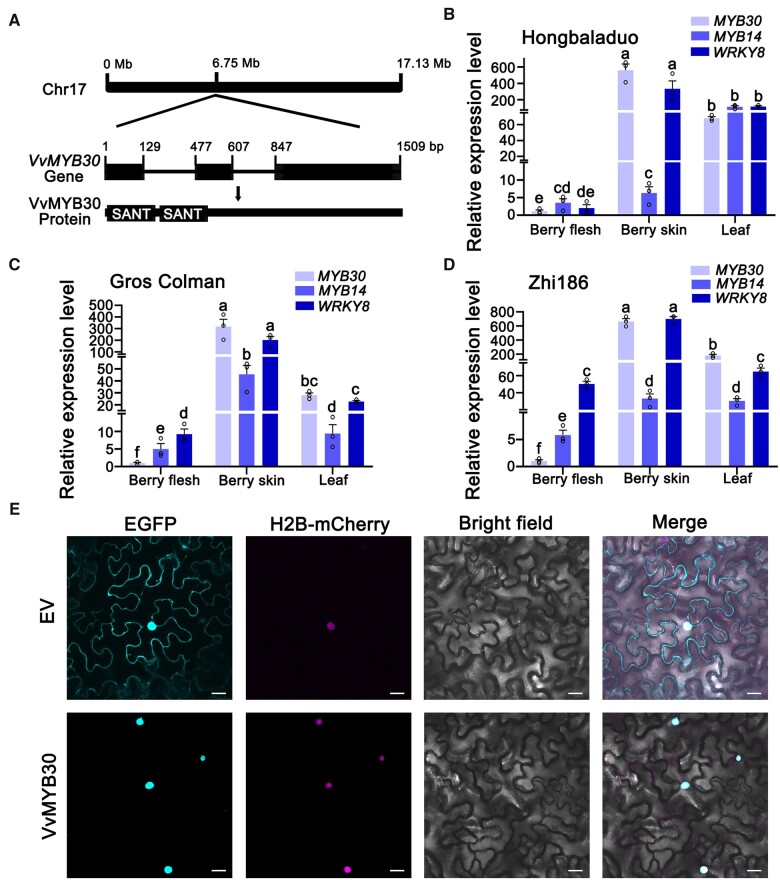
In the grapevine genome (http://www.grapegenomics.com/pages/PN40024/), the VvMYB30 gene is present in an approximately 6.75 Mb region of chromosome 17. We cloned the coding sequence and genomic DNA of VvMYB30 from “Hongbaladuo” (V. vinifera L.), and found they were identical to the corresponding sequences in the reference grapevine genome from the Pinot Noir-derived homozygous line PN40024. VvMYB30 contains three exons and two introns, encoding a 34.4-kD protein. Based on analysis using SMART (http://smart.embl-heidelberg.de/), VvMYB30 is an R2R3-MYB TF with two well-conserved N-terminal SANT domains and a variable C-terminal region (Figure 3A). The coding sequences of MYB30 were cloned from leaves of different grapevine cultivars, including “Cross Colman,” “Beihong,” and “Zhi186.” Multiple sequence alignment showed that all allelic MYB30 sequences, except for that of “Zhi186-2,” are identical (Supplemental Figure S3). To identify which SG VvMYB30 belongs to, we constructed a phylogenetic tree using Arabidopsis and grape MYB sequences. For correct classification, we selected sequences from Arabidopsis and grape genes encoding R2R3 MYBs from SG1 to SG7, which are known to be involved in specialized metabolism, and one gene encoding an R3 MYB from grape as an outlier. As expected, two previously characterized grape MYBs, VvMYB14 and VvMYB15, clustered with sequences encoding SG2 MYBs, while VvMYB30 clustered with sequences encoding Arabidopsis SG1 MYBs (Supplemental Figure S4).
Figure 3.
Genomic structure, expression profile, and subcellular localization of VvMYB30. A, Schematic diagrams of VvMYB30 gene and protein. Top panel, the solid black box (top panel) represents chromosome 17; middle panel, solid black boxes represent the exons while the lines represent the introns; bottom panel, SANT (MYB) domains at the N-terminus of VvMYB30 protein are indicated by two solid boxes. The VvMYB30 gene structure was deduced from the grape genome (http://www.grapegenomics.com/pages/PN40024/). The genomic DNA and coding sequence of VvMYB30 are from “Hongbaladuo.” B–D, Relative expression of MYB30, MYB14, and WRKY8 in berry flesh, skin, and leaf in “Hongbaladuo,” “Gros Colmam,” and “Zhi186” grapevines as measured by RT-qPCR. Data are means ± se of three biological replicates. Different letters indicate significant differences by one-way ANOVA followed by Duncan’s test (P < 0.05). E, Subcellular localization of VvMYB30. VvMYB30-EGFP and H2B-mCherry were co-transformed into N. benthamiana leaves. H2B-mCherry was used as a nuclear marker. Scale bars correspond to 20 μm.
Nucleus-localized VvMYB30 is highly expressed in berry skin and leaves
The relative expression levels of MYB30, MYB14, and WRKY8 were measured in berry flesh, berry skin, and leaves of three grapevine cultivars (Figure 3, B–D). The expression levels of MYB30 and WRKY8 were highest in berry skin, followed by leaves and flesh. The expression levels of MYB14 were higher in skin and leaves than in flesh in “Gros Colman” and “Zhi186,” while they were higher in leaves than in berry skin and flesh in “Hongbalduo.” In “Hongbalduo,” “Gros Colman,” and “Zhi186,” Res contents in the berry skin were 2.39, 43.85, and 132.57 µg g−1 FW, respectively; Res contents in the leaves were 5.17, 12.83, and 17.56 µg g−1 FW, respectively; however, Res in the berry flesh was not detectable (Supplemental Table S1). To determine the subcellular localization of VvMYB30, a VvMYB30-GFP fusion gene was transiently expressed in N. benthamiana leaves. In leaves overexpressing VvMYB30-GFP, strong fluorescence was localized in the nucleus, while in leaves expressing GFP only, fluorescence was distributed uniformly throughout the cytoplasm and nucleus (Figure 3E), suggesting that VvMYB30 is a nucleus-localized TF.
VvMYB30 negatively regulates VvSTS15/21 and stilbene biosynthesis in grapevine
The VvMYB30 interaction with VvWRKY8, a negative regulator of VvSTS15/21 (VIT_216s0100g00830/VIT_216s0100g00910) (Jiang et al., 2019), prompted us to speculate that VvMYB30 is also involved in the regulation of VvSTS genes and stilbene biosynthesis. We overexpressed and RNAi-suppressed VvMYB30 in grapevine leaves and berry skin. For overexpression, the native promoter of MYB30 was amplified from Vitis amurensis and used to express VvMYB30 (OE-VvMYB30) (Figure 4A). After agroinfiltration of the vectors into leaves from tissue culture plantlets of V. amurensis and berry skin from “Beixi” (V. vinifera “Muscat Hamburg” × V. amurensis) grapevines, expression levels of MYB30, STS15/21, and STSs were measured using RT-qPCR. Compared with the empty-vector control, the relative expression of MYB30 was significantly (P < 0.05) increased in the OE-VvMYB30-transformed leaves and skin, while that of STS15/21 and STSs was significantly (P < 0.05) down-regulated (Figure 4, B and C). Res contents were decreased in the VvMYB30-overexpressing leaves and skin (Figure 4, D and E). We also constructed an RNAi vector of VvMYB30 (Figure 4F) and transiently transformed the V. amurensis plantlets and “Kyoho” berry skin. MYB30 was down-regulated by RNAi, while the expression levels of STS15/21 and STSs (Figure 4, G and H) and the Res contents (Figure 4, I and J) were significantly (P < 0.05) increased in the leaves and skin.
Figure 4.
VvMYB30 overexpression and knock-down in grapevine leaves and berry skin alter STS15/21 and STSs expression and Res content. The vectors were transiently transformed into leaves and berry skin to increase or reduce expression level of MYB30. For MYB30 overexpression, full-length coding sequence of VvMYB30 was inserted into pSAK277 vector with MYB30 promoter cloned from V. amurensis (A). The vectors were transiently transformed into V. amurensis leaves and “Beixi” (V. vinifera “Muscat Hamburg” × V. amurensis) berry skin. Relative expression level of MYB30 and its target gene STS15/21 in V. amurensis leaves (B) and “Beixi” berry skin (C) infiltrated with MYB30 were measured by RT-qPCR. The contents of trans/cis-Pd and trans/cis-Res in leaves (D) and berry skin (E) infiltrated with MYB30 were measured by HPLC. For knocking down the expression of MYB30, a 250-bp fragment from VvMYB30 coding sequence was inserted into pFGC5941 vector in forward and reverse (F). The vectors were transiently transformed into V. amurensis leaves and “Kyoho” (V. labrusca × V. vinifera) berry skin. Relative expression level of MYB30 and its target gene STS15/21 in leaves (G) and berry skin (H) were measured by RT-qPCR. Res contents in leaves (I) and berry skin (J) were measured by HPLC. Data are means ± se of three biological replicates. Statistical significance was calculated using Student’s t test (*P < 0.05). NS, no significant difference.
UV-B/C stress induces VvMYB14 and VvWRKY8 but represses VvMYB30
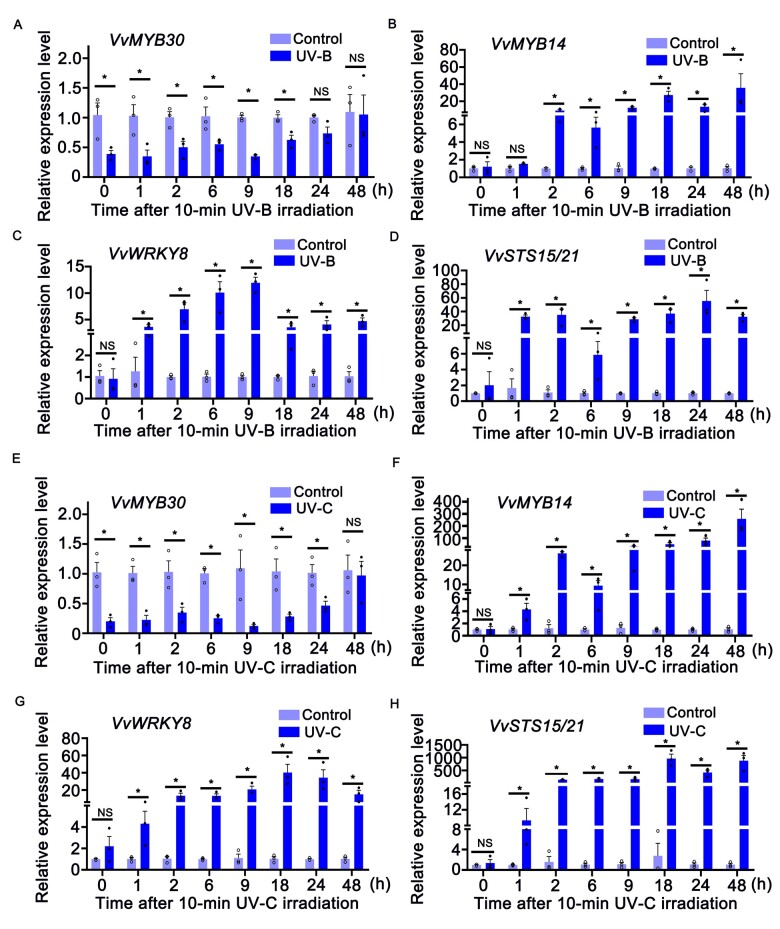
Previously, we found that UV-C treatment up-regulates VvMYB14, VvWRKY8, and VvSTS15/21 in grapevine (Jiang et al., 2019). In this study, we measured the expression of VvMYB30, VvMYB14, VvWRKY8, and VvSTS15/21 in grapevine leaves over an eight-point time course (0–48 h) after UV-B or UV-C treatment. The 10-min UV-B and UV-C treatments both caused significant repression of VvMYB30 (Supplemental Figure S5, A and B) that did not recover until 48 h later (Figure 5, A and E). In contrast, the expression levels of VvMYB14, VvWRKY8, and VvSTS15/21 were not affected immediately after UV-B/C treatment (Supplemental Figure S5, A and B) but increased 1 h after treatment despite the difference in induction profiles (Figure 5). The expression level of VvWRKY8 began to decline after 9 h of UV-B treatment and 18 h of UV-C treatment (Figure 5, C and G). For VvSTS15/21, the expression level went up after 1 h UV treatment, and quickly plateaued at 2 h (Figure 5, D and H).
Figure 5.
Relative expression of VvMYB30 (A, E), VvMYB14 (B, F), VvWRKY8 (C, G), and VvSTS15/21 (D, H) in grapevine leaves after UV-B or UV-C irradiation (6 W m−2) for 10 min as measured by RT-qPCR. Data are means ± se of three biological replicates. Statistical significance was calculated using Student’s t test (*P < 0.05). NS, no significant difference.
VvWRKY8, VvMYB30, and VvMYB14 do not mutually regulate one another
Physical interactions of VvWRKY8 with activator VvMYB14 and repressor VvMYB30 prompted us to explore the possibility of their mutual regulation. We performed a yeast one-hybrid (Y1H) assay in which the expression of the HIS3 reporter was driven by the promoter of VvMYB30 or VvWRKY8, resulting in vectors proVvMYB30-pHis2 and proVvWRKY8-pHis2, respectively. Co-transformation of proVvMYB30-pHis2 with the vector expressing VvWRKY8 (VvWRKY8-pGADT7), or proVvWRKY8-pHis2 with the vector expressing VvMYB30 (VvMYB30-pGADT7), did not produce colonies in the selection medium containing 3-amino-1,2,4-triazole (3AT) (Supplemental Figure S6), suggesting that VvMYB30 and VvWRKY8 do not mutually regulate each other. We next tested the possible mutual regulation of VvMYB14 and VvMYB30. In the Y1H assay testing the VvMYB30 promoter activation by VvMYB14 or the VvMYB14 promoter activation by VvMYB30, no yeast colonies were produced in the selection medium (Supplemental Figure S7), suggesting the absence of mutual regulation of the two MYB TFs. Together with the previous observation of the lack of mutual regulation between VvWRKY8 and VvMYB14 (Jiang et al., 2019), the results indicate that the three interacting TFs do not mutually regulate.
We also explored whether VvMYB30 binds its own promoter. Co-transformation of proVvMYB30-pHis2 and VvMYB30-pGADT7 into yeast strain Y187 did not produce colonies in the selection medium (Supplemental Figure S8), suggesting that VvMYB30 does not regulate its own expression.
VvMYB30 binds to the same cis-elements as VvMYB14 in the VvSTS15/21 promoter
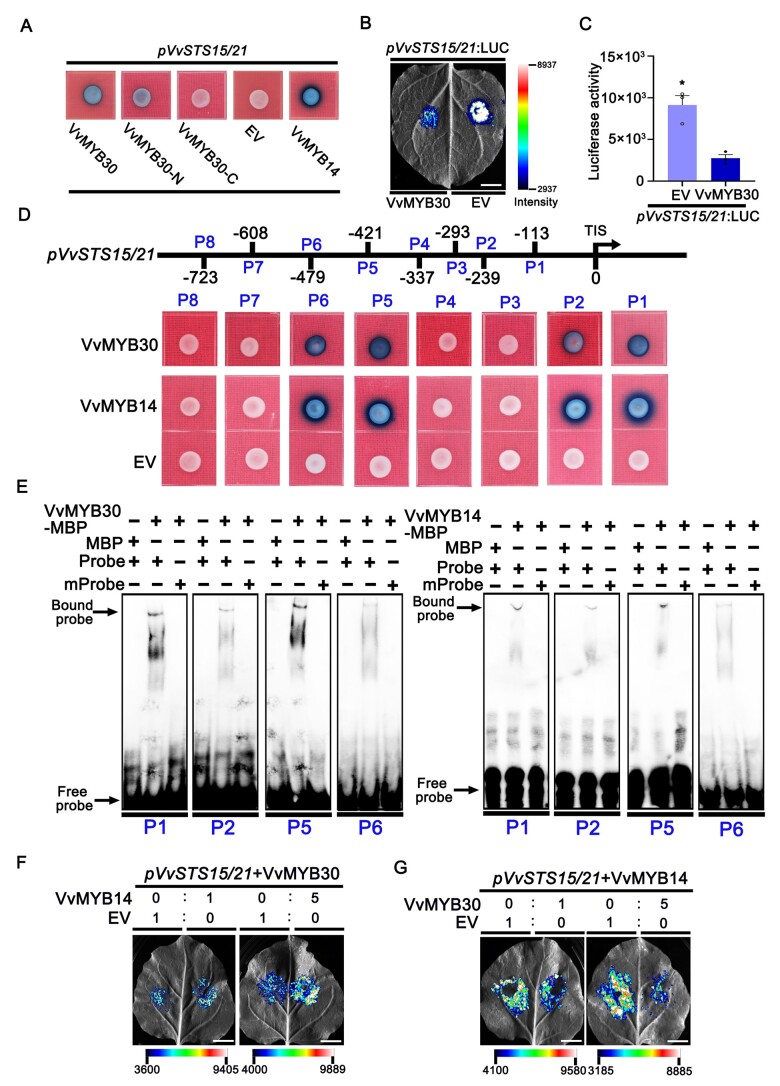
We used Y1H to demonstrate the binding of VvMYB30 to the VvSTS15/21 promoter. We generated effector vectors by cloning the full-length VvMYB30 coding sequence as well as the truncated N- and C-terminal coding sequence fragments (VvMYB30-N and VvMYB30-C) into the pGAD424 effector vector. The VvSTS15/21 promoter was cloned into the pLacZi reporter vector. The previously published VvMYB14 effector vector (Jiang et al., 2019) was used as a positive control. The Y1H results showed that the Gal4 activation-domain fusions of full-length VvMYB30, VvMYB30-N, and VvMYB14, but not VvMYB30-C, activate the VvSTS15/21 promoter (Figure 6A), suggesting that the promoter binding is mediated by the N-terminal fragment of the VvMYB30 protein.
Figure 6.
The regulation of VvSTS15/21 promoter by VvMYB30 and VvMYB14. A, Validation of the interaction of VvMYB30 with the VvSTS15/21 promoter by Y1H assay. The N-terminal, the C-terminal, and full-length coding sequences of VvMYB30 fused to the B42 activation domain (AD), respectively, and proVvSTS15/21:LacZ were co-transformed into yeast strain EGY48. Combination of pGAD424-VvMYB14 and pLacZi-proVvSTS15/21 was used as positive control. B and C, LUC activity analysis. Co-transformed the effector 35S: VvMYB30 and the reporter proVvSTS15/21: LUC into the N. benthamiana leaves. The LUC activity was measured in different combinations of effector and reporter. In (C), data are means ± se of three biological replicates. *, P < 0.05 (Student’s t test). D, Validation of the binding site on VvSTS15/21 promoter of VvMYB30. VvMYB30 fused to the B42 AD and proVvSTS15/21 (motif P1–P8): LacZ was co-transformed into yeast strain EGY48. TIS, transcription initiation site. E, Results of EMSA showing the combination of VvMYB30 or VvMYB14 to the promoter of VvSTS15/21 (motifs P1, P2, P5, and P6). VvMYB30 and VvMYB14 were all fused with MBP tag. mProbe, mutated probe. F, Transient expression assay of the proVvSTS15/21: LUC reporter in the presence of VvMYB30 or VvMYB14 effector. In the left panel, Agrobacterium containing the VvSTS15/21: LUC reporter vector (pVvSTS15/21) was infiltrated on both left and right sides of a leaf in equal amount (1:1). Agrobacterium containing VvMYB30 or VvMYB14 were co-infiltrated with pVvSTS15/21 in different ratio (1:0 and 1:1, respectively). In the right panel, the same infiltration took place except that the VvMYB30:VvMYB14 ratio is 1:5. The same experiments were performed except that the ratio of VvMYB30 and VvMYB14 is 5:1 (G). Scale bars correspond to 1 cm.
A transient promoter assay was carried out in N. benthamiana leaves. The VvSTS15/21 promoter was inserted into the pCAMBIA1302-luciferase (LUC) reporter vector to drive LUC expression. The full-length coding sequence of VvMYB30 was sub-cloned into the pSAK277 vector. The proVvSTS15/21:LUC reporter vector was infiltrated with EV pSAK277 or pSAK277-VvMYB30 into N. benthamiana leaves (Figure 6B). Firefly LUC activities were detected in the leaves infiltrated with the reporter and pSAK277 (EV), indicating that the VvSTS15/21 promoter is active in N. benthamiana leaves. In comparison, infiltration of the reporter with pSAK277-VvMYB30 resulted in lower LUC luminescence (Figure 6B). A LUC enzyme activity assay confirmed the repression of proVvSTS15/21:LUC by VvMYB30 (Figure 6C).
To pinpoint the VvMYB30 binding sites in the VvSTS15/21 promoter, we analyzed the VvSTS15/21 promoter using PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Eight candidate MYB-binding-site (MBS) elements were predicted: P1 (−113 to −130), P2 (−239 to −256), P3 (−293 to −310), P4 (−337 to −354), P5 (−421 to −438), P6 (−479 to −496), P7 (−608 to −625), and P8 (−723 to −740) (Figure 6D). Each potential element was individually inserted in the front of the minimum CYC1 promoter in the pLacZi reporter vector, and the resulting vectors were individually transformed into the yeast strain to generate the reporter. The reporter strains were transformed with pGAD424-VvMYB30 or pGAD424-VvMYB14 in a Y1H assay. The results showed that both VvMYB30 and VvMYB14 bind to P1, P2, P5, and P6 elements (Figure 6D).
Electrophoretic mobility shift assay (EMSA) was used to demonstrate the direct binding of the MYB proteins to the four MBS elements. Maltose binding protein (MBP)-tagged VvMYB30 and VvMYB14 recombinant proteins were produced in Escherichia coli, purified, and used for EMSA. Biotin-labeled probes (Probe), based on the sequences of the four elements, and mutant probes (mProbe), in which the binding sequences were mutated, were synthesized and incubated with VvMYB30 or VvMYB14 proteins before EMSA (Figure 6E). In all cases, up-shifted bands were observed when VvMYB30 or VvMYB14 protein was mixed with the native-sequence probes, but not with the mProbe. We also performed competition experiments for EMSA, the unlabeled sequences (cold probe) were added into the protein/probe mixture. With the increasing cold probe concentration, the up-shifted bands became weaker (Supplemental Figure S9).
We next performed a competitive promoter activation assay in N. benthamiana leaves. We first confirmed that VvMYB30 and VvMYB14 do not physically interact in a BiFC assay (Supplemental Figure S10). In the left panel of Figure 6F, Agrobacterium containing the proVvSTS15/21:LUC reporter vector (pVvSTS15/21) was infiltrated on both left and right sides of a leaf in equal amount. Agrobacterium containing the overexpression vectors, pH7WG2D-VvMYB30 and pH7WG2D-VvMYB14, was co-infiltrated with pVvSTS15/21 in different ratios (1:0 and 1:1, respectively). In the right panel, the same infiltration took place except that the VvMYB30:VvMYB14 ratio was 1:5. The results showed that, while minimal pVvSTS15/21 activation occurred when a 1:1 ratio of VvMYB30 and VvMYB14 was co-transformed, much stronger activation was observed when five times more VvMYB14 than VvMYB30 was co-transformed. The same experiments were performed except that the ratio of VvMYB30 and VvMYB14 was 5:1 (Figure 6G). When five times more VvMYB30 was present, the VvMYB14 activation of pVvSTS15/21 was significantly repressed. Together, the results suggest that VvMYB30 and VvMYB14 compete for binding to the same elements in the VvSTS15/21 promoter.
VvMYB30 and VvMYB14 compete to interact with VvWRKY8 to regulate VvSTS15/21
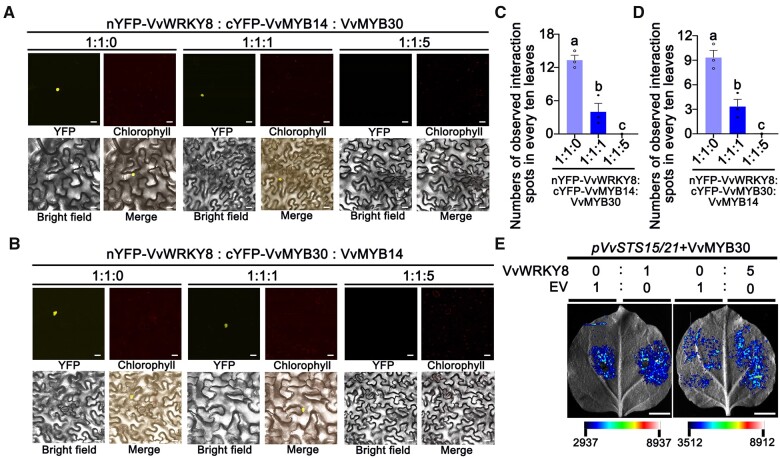
A competitive BiFC assay was used to demonstrate the competition between VvMYB30 and VvMYB14 for interaction with VvWRKY8. nYFP-VvWRKY8 and cYFP-VvMYB14 were co-transformed with or without VvMYB30 (Figure 7A). In the absence of VvMYB30 expression, nYFP-VvWRKY8 and cYFP-VvMYB14 interact (Figure 7, A and C). When VvMYB30 was co-infiltrated in a 1:1 ratio to VvMYB14, the interaction (fluorescence signal) significantly decreased. When the VvMYB30:VvMYB14 ratio was 5:1, the interaction was abolished (Figure 7, A and C). The same experiments were performed using nYFP-VvWRKY8 and cYFP-VvMYB30 with or without VvMYB14. VvMYB14 decreased the nYFP-VvWRKY8/cYFP-VvMYB30 interaction at a 1:1 ratio to VvMYB30 and demolished the interaction when the ratio was increased to 5:1 (Figure 7, B and D).
Figure 7.
VvMYB30 and VvMYB14 compete to interact with VvWRKY8 to regulate VvSTS15/21. The BiFC assay was used to demonstrate the competition between VvMYB30 and VvMYB14 for interaction with VvWRKY8. A, nYFP-VvWRKY8 and cYFP-VvMYB14 were co-expressed with or without VvMYB30. B, nYFP-VvWRKY8 and cYFP-VvMYB30 were co-expressed with or without VvMYB14. The fluorescence signals were observed by confocal microscopy. Scale bars correspond to 20 μm. Numbers of observed interaction spots were counted when nYFP-VvWRKY8 and cYFP-VvMYB14 were co-expressed with or without VvMYB30 (C) or nYFP-VvWRKY8 and cYFP-VvMYB30 were co-expressed with or without VvMYB14 (D). Data are means ± se of three biological replicates. Different letters indicate significant differences by one-way ANOVA followed by Duncan’s test (P < 0.05). E, In the left panel, Agrobacterium containing the proVvSTS15/21: LUC reporter vector (pVvSTS15/21) was infiltrated on both left and right sides of a leaf in equal amount (1:1). Agrobacterium containing VvMYB30 or VvWRKY8 was co-infiltrated with pVvSTS15/21 in different ratio (1:0 and 1:1, respectively). In the right panel, the same infiltration took place except that the VvMYB30:VvWRKY8 ratio is 1:5. Scale bars correspond to 1 cm.
In our previous work, the VvWRKY8–VvMYB14 interaction reduces the binding of VvMYB14 to the VvSTS15/21 promoter (Jiang et al., 2019). Therefore, we speculated that the VvWRKY8–VvMYB30 interaction also reduces the binding of VvMYB30 to the VvSTS15/21 promoter. In a competitive promoter activation assay, pH7WG2D-VvMYB30 or pH7WG2D-VvWRKY8 was co-transformed with pVvSTS15/21 at different concentration ratios (Figure 7E). While VvMYB30 repressed the pVvSTS15/21 activity, addition of five times more VvWRKY8 restored the pVvSTS15/21 activity. Because VvWRKY8 does not bind to pVvSTS15/21 (Jiang et al., 2019), the results suggest that the protein–protein interaction between VvWRKY8 and VvMYB30 reduces the VvMYB30 binding to the pVvSTS15/21 promoter.
Exogenous Res or VvWRKY8 overexpression induces MYB30 expression
To determine the effect of exogenous Res on MYB30 expression, RT-qPCR was conducted to measure MYB30 expression levels in grapevine suspension cells after exogenous Res treatment. The results showed that the expression of MYB30 was induced compared with that of the control (Supplemental Figure S11A). To further investigate the potential relationship between VvWRKY8 and VvMYB30, we measured the expression level of VvMYB30 in the VvWRKY8-overexpressing grapevine plants (Jiang et al., 2019) and found that VvMYB30 expression increased nearly five-folds compared with expression in the control (Supplemental Figure S11B).
Discussion
UV-B/C stress induces stilbene biosynthesis in grapevine
The solar UV radiation spectrum is subdivided into three regions, UV-A (315–400 nm), UV-B (280–315 nm), and UV-C (100–280 nm) (Björn, 2015). Naturally, plants are exposed to a little UV-B (about 5%) and little UV-C radiation, as the short wave-length radiation is effectively absorbed by stratospheric ozone (Björn, 1996; Tossi et al., 2019). However, UV-B/C has been widely used to study its effects on biosynthesis of specialized metabolites in plants (Ramani and Chelliah, 2007; Carbonell-Bejerano et al., 2014; Freitas et al., 2015; Abbasi et al., 2021; Meyer et al., 2021; Yin et al., 2022). UV-B and UV-C are efficient elicitors of stilbene biosynthesis as they intensively induce stilbene accumulation in different stilbene-producing plants including grapevine and P. cuspidatum (Adrian et al., 2000; Nishikawa et al., 2011; Suzuki et al., 2015; Guerrero et al., 2016; Sheng et al., 2018; Liu et al., 2019; Kong et al., 2020; Valletta et al., 2021). Pre-treatment of UV-B or UV-C to induce the accumulation of phytoalexin, such as stilbene, is possibly an effective strategy to increase disease resistance and immunity in grape berries and to limit the use of pesticides in vineyards (Berli et al., 2010; Gil et al., 2013; Guerrero et al., 2016). Post-harvest treatment with UV-B or UV-C has been widely used in fruit and vegetable storage, as the treatment delays fruit ripening and senescence, and activates the defenses against pathogens (Urban et al., 2016; Sheng et al., 2018). Postharvest UV treatment has been exploited in grapes to increase the content of phenolic compounds including stilbenes in berries and wines (Sheng et al., 2018; Valletta et al., 2021). The significant induction of STSs by UV-B/C as demonstrated here (Figure 5) is corroborated well with the previous reports (Versari et al., 2001; Höll et al., 2013; Xi et al., 2014; Jiang et al., 2019; Valletta et al., 2021). Additionally, we demonstrated that the TFs involved in STS regulation are differentially induced by UV treatment (Figure 5). VvMYB30 was significantly repressed, while the expression levels of VvMYB14, VvWRKY8, and VvSTS15/21 were not affected immediately after UV-B/C treatment (but increased 1 h after treatment) despite the difference in induction profiles (Figure 5 and Supplemental Figure S5, A and B), suggesting that the UV-B/C-induced Res accumulation is mainly caused by the VvMYB14–VvWRKY8–VvMYB30 regulatory module. As the UV-B or UV-C treatment used in this study is stronger than natural UV radiation, the response to the UV treatment is not likely mediated by the UV-B photoreceptor UVR8 (Brown and Jenkins, 2008; Tossi et al., 2019). Rather, UV treatment elicited a stress response that induces stilbene biosynthesis in grapevine. This approach may help us understand the changes of stilbene accumulation and gene expression under natural UV radiation.
VvMYB30 negatively regulates stilbene biosynthesis in grapevine
MYB TFs play pivotal roles in the regulation of specialized metabolism, including phenylpropanoid biosynthesis in many plant species (Dubos et al., 2010). Although many MYBs have been identified as transcriptional activators (Kobayashi et al., 2002; Walker et al., 2007; Höll et al., 2013; Wang et al., 2017; Yu et al., 2019; Sun et al., 2020), the prevalence of MYB repressors in specialized metabolism has also been recognized. The MYB repressors are mainly classified into two distinct groups, SG4 R2R3-MYB and R3 MYB repressors (Ma and Constabel, 2019). Based on the mechanism of repression, the SG4 R2R3-MYBs are again divided into two groups, one group includes MYB27 (Petunia), FaMYB1, and MtMYB2 (Aharoni et al., 2001; Albert et al., 2014; Jun et al., 2015), which do not directly bind to the promoters of target genes. Instead, they act as corepressors that bind to TF complexes (e.g. MYB-bHLH-WD40 (MBW) complex in flavonoid biosynthesis) to repress gene expression (Chen et al., 2019b). The other group includes AtMYB4 and EjMYB2 repressors (Jin et al., 2000; (Xu et al., 2014), which contain the EAR-repressor motif and bind directly to the promoters of target genes (Chen et al., 2019b). The R3 MYB repressors contain only an R3 domain (e.g. AtCPC) or an R3 domain with part of an R2 domain (e.g. AtMYBL2) (Chen et al., 2019b). AtCPC does not contain any known repression motif and competes with the R2R3 MYB for interaction with the bHLH protein, thus preventing the formation of the MBW complex (Zhu et al., 2009; Chen et al., 2019b). AtMYBL2 contains a repression motif (TLLLFR) at the C terminus and represses transcription through interacting with the MBW complex (Dubos et al., 2008; Matsui et al., 2008; Chen et al., 2019b).
In this study, we identified a previously uncharacterized R2R3-MYB, VvMYB30, as a repressor of stilbene biosynthesis (Figure 4). VvMYB30 was grouped together with SG1 Arabidopsis MYBs including AtMYB30 (Supplemental Figure S4), consistent with previous reports (Wong et al., 2016; Rodrigues et al., 2021). The Arabidopsis AtMYB30 acts as a key negative regulator of photomorphogenic development (Yan et al., 2020) but a positive regulator of the hypersensitive cell death program (Daniel et al., 1999; Vailleau et al., 2002). Our study may be the first to link the S1 SG of R2R3-MYBs to both specialized metabolism and the UV stress response. VvMYB30 does not possess any known repression motif (SID, EAR, or TLLLFR). Tamagnone et al. (1998) showed that AmMYB308 is a weak activator that competes with stronger MYB-related activators to downregulate target gene expression. This report prompted us to consider whether VvMYB30 is also a weak activator. Our GAL4-fusion experiment indicated that VvMYB30 possesses weaker transactivation activity compared with VvMYB14 (Supplemental Figure S12, A and B). Therefore, VvMYB30 is likely a weak activator that competes with VvMYB14 to control VvSTS15/21 activation. The C-terminal domains of R2R3 MYBs control transcriptional activation activity. For example, the SG6 MYBs that are involved in anthocyanin regulation include three types of activators (strong, moderate, and none-activators). These MYBs are characterized by the C-terminal motifs that are predicted to be relatively ordered compared with the flanking protein sequences (Rodrigues et al., 2021). VvMYB30 and VvMYB14 show different degrees of transcriptional activity, likely due to the large protein sequence difference in the C-terminal. However, the weaker transactivation activity of VvMYB30 requires further examination. Functionally, VvMYB30 is a negative regulator of stilbene biosynthesis.
VvMYB30 and VvMYB14 compete for the same binding sites in the VvSTS15/21 promoter
It has been reported that multiple MYBs regulate the same target gene in a biological process. In loquat (Eriobotrya japonica), EjMYB1 activates the promoters of the lignin biosynthetic gene Ej4CL1, while EjMYB2, a transcriptional repressor, represses the induction of EjMYB1, resulting in the reduction of lignin production (Xu et al., 2014). Several mechanisms allow the co-regulation of a gene by both activator and repressor. They compete for the binding to the gene promoter, dimerize to form a non-DNA binding complex, or dimerize to occupy a promoter binding site (e.g. a DNA-binding activator recruits a repressor to the promoter) (Chen et al., 2019a, 2019b). In this study, we showed that VvMYB30 and VvMYB14 do not regulate each other transcriptionally (Supplemental Figure S7), nor physically interact (Supplemental Figure S10). Rather, VvMYB30 and VvMYB14 modulate stilbene biosynthesis through competing for the same MBSs in the VvSTS15/21 promoter (Figure 6, D–G and Supplemental Figure S9).
Regulation of VvSTS15/21 by VvMYB30 and VvMYB14 is modulated by VvWRKY8
The N-terminal DNA binding domains of VvMYB30 and VvMYB14 share high sequence identity (Supplemental Figure S13), and these proteins compete to interact with VvWRKY8 in a dose-dependent manner (Figure 7, A–D). Our previous study showed that the Res-induced VvWRKY8 reduces VvMYB14 activation of the VvSTS15/21 promoter by forming a complex (Jiang et al., 2019). In the current study, we showed that VvWRKY8 decreases the VvMYB30 repression of the VvSTS15/21 promoter, also in a dose-dependent manner (Figure 7E). VvWRKY8 interacts with VvMYB30 (Figures 1 and 2) and VvMYB14 (Jiang et al., 2019) through the N-termini where the MYB DNA-binding domains reside. Therefore, the VvWRKY8–VvMYB30 and VvWRKY8–VvMYB14 complexes are likely non-DNA binding because the interactions block the MYB DNA-binding domains. When VvWRKY8 expression responds to endogenous Res (Jiang et al., 2019), VvWRKY8 sequesters varied amounts of VvMYB30 and VvMYB14, thus fine-tuning VvSTS15/21 expression. As the two MYB factors do not interact (Supplemental Figure S10) and share no mutual regulation (Supplemental Figure S7), VvWRKY8 plays a central role in modulating the MYB TF-mediated VvSTS15/21 expression.
MYB and WRKY TFs often regulate the same biological process through protein interaction, transcriptional regulation, and TF recruitment. In grapevine, VvWRKY26 enhances the regulatory function of the MBW complex in flavonoids biosynthesis, vacuolar hyper-acidification, and trafficking (Amato et al., 2019). In apple, MdWRKY40 interacts with MdMYB1 to activate the anthocyanin biosynthetic genes, such as MdDFR and MdUFGT (An et al., 2019). In red-fleshed apple, MdWRKY41 inhibits the accumulation of anthocyanin and proanthocyanidin by negatively regulating MdMYB12 and interacting with MdMYB16, to down-regulate MdUFGT and MdANR (Mao et al., 2021). Our results present a prime example of a WRKY TF modulating both positive and negative MYB regulators to co-regulate the same gene in the grape stilbene biosynthetic pathway.
The VvMYB14–VvWRKY8–VvMYB30 regulatory circuit balances stilbene biosynthesis
Stilbene is quickly synthesized when grapevine is subjected to abiotic and biotic stresses; however, stilbene accumulation is dynamic and fluctuating (Versari et al., 2001; Ferri et al., 2009; Wang et al., 2016). Therefore, grapevine might have developed a dynamic regulatory mechanism that involves both positive and negative regulators to balance stilbene biosynthesis. After UV-B or UV-C treatment, the VvMYB30 expression decreased for at least 24 h before recovering by 48 h. During the 48 h, the expression levels of VvMYB14, VvWRKY8, and VvSTS15/21 were higher in treated plants than in control plants (Figure 5). The results suggest that a VvMYB14–VvWRKY8–VvMYB30 circuit dynamically regulates VvSTS15/21 expression in response to UV stress. We previously found that VvMYB14 overexpression up-regulates VvWRKY8 and increases Res accumulation in grape leaves, although VvMYB14 does not bind to the VvWRKY8 promoter (Jiang et al., 2019). In this study, overexpression and RNAi of VvMYB30 resulted in the down- and up-regulation of VvSTS15/21, respectively, and the corresponding decrease and increase of Res accumulation in grape leaves and berry skin (Figure 4). We also found that VvMYB30 does not bind to the VvWRKY8 promoter (Supplemental Figure S6). Addition of exogenous Res decreased the expression of MYB14 and STS15/21 but increased the expression of WRKY8 and MYB30 in grapevine suspension cells (Jiang et al., 2019; Supplemental Figure S11A). The specific mechanism by which exogenous Res influences VvWRKY8, VvMYB30, and VvMYB14 at the expression level remains unclear. Overexpression of VvWRKY8 resulted in the up-regulation of VvMYB30 and down-regulation of VvMYB14 (Supplemental Figure S11B; Jiang et al., 2019). However, VvWRKY8 does not directly regulate VvMYB30 and VvMYB14 expression (Supplemental Figure S6; Jiang et al., 2019). Therefore, it is likely that VvWRKY8 activates another unidentified TF in an indirect control of VvMYB14 and VvMYB30. The VvMYB14–VvWRKY8–VvMYB30 circuit allows grapevine to respond to UV by producing Res, but to prevent Res over-accumulation by feedback inhibition through formation of non-DNA binding complexes.
Such a seemingly “wasteful” complex mechanism by a regulatory circuit has previously been seen in plants. In Petunia, MYB27 represses not only anthocyanin pathway genes but also the activator AN1, the repressor MYBx, and itself (Albert et al., 2014). Similarly, VvWRKY8, although itself a repressor, transcriptionally induces the repressor VvMYB30. The mechanisms of repression are different for these factors. VvWRKY8 forms non-DNA-binding complexes with VvMYB30 and VvMYB14 while VvMYB30 directly binds to the VvSTS15/21 promoter. This regulatory mechanism enables the spatiotemporal production of Res in response to developmental and environmental cues. Concurrent expression of both activators and repressors is a hallmark of some specialized metabolic pathways. The activators (CsRuby1 and PpMYB10.1) and the repressors (CsMYB3 and PpMYB18) co-express in anthocyanin-accumulating tissues in citrus (Citrus spp.) and peach (Prunus persica) (Zhou et al., 2015, 2019; Huang et al., 2020). Although lacking mutual regulation, WRKY8, MYB30, and MYB14 co-express in grape berry skin (Figure 3, B–D) where Res is mostly accumulated (Supplemental Table S1; Jeandet et al., 1991; Ector et al., 1996; Wang et al., 2013), again implying that these three factors form a regulatory circuit.
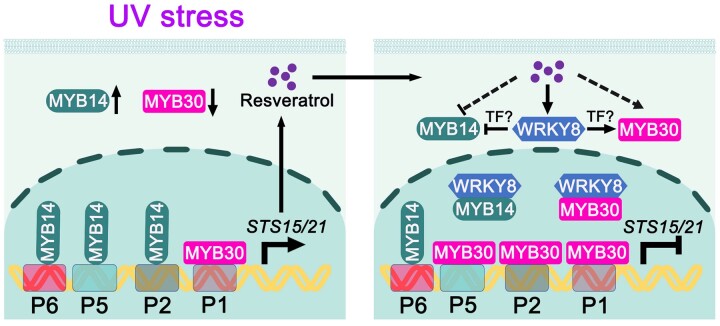
In conclusion, we identified and characterized VvMYB30, which directly binds to STS gene promoters. The repressor (VvMYB30) and activator (VvMYB14) target the same binding sites in the VvSTS15/21 promoter, and their competition to the binding sites determines the down- and up-regulation of VvSTS15/21 (Figure 8). When grapevines are exposed to UV stress, expression of VvMYB14 and VvMYB30 is activated and inhibited, respectively, resulting in a net increase of Res accumulation. After the Res content reaches a threshold level, it simultaneously activates VvWRKY8 and VvMYB30, while it represses the VvMYB14 activator. An increase in VvWRKY8 inhibits VvMYB14 expression and induces VvMYB30 expression, possibly through the activation of other TFs. VvMYB30 binds to the VvSTS15/21 promoter to reduce the activation by VvMYB14. The VvWRKY8 protein interacts with both N-termini of VvMYB30 and VvMYB14, masking the MYB DNA-binding domains, thus forming two non-DNA-binding complexes. The sequestration of the MYB factors by VvWRKY8 adds to the complexity of the VvMYB14–VvWRKY8–VvMYB30 regulatory circuit (Figure 8). Our study further paves the way for new approaches in our understanding of the feedback regulation of phytoalexin biosynthesis in plants and, through this, improved phytoalexin production in engineered stress resistance. The knowledge gained from this study will guide future research to elucidate the stilbene regulatory network that no doubt contains many more regulators.
Figure 8.
A hypothetical model of VvMYB30 regulating VvSTS15/21 through VvMYB14 and VvWRKY8 regulatory loop to control UV-induced stilbene biosynthesis in grapevine. When grapevines are exposed to UV-B/C stress, VvMYB14 and VvMYB30 expression is activated and inhibited, respectively, resulting in a net increase of Res accumulation. After the Res content reaches a threshold level, it simultaneously activates two repressors, VvWRKY8 and VvMYB30, while represses the VvMYB14 activator. Increase of VvWRKY8 reduces VvMYB14 expression and induces VvMYB30 expression, possibly through the activation of other TFs. VvMYB30 binds to VvSTS15/21 promoter to repress its expression. Simultaneously, the VvWRKY8 protein interacts with both N-termini of VvMYB30 and VvMYB14, masking the MYB DNA-binding domains, thus forms two non-DNA-binding complexes. The sequestration of the VvMYB30 by VvWRKY8 likely helps maintain the basal expression of VvSTS15/21, which adds to the complexity of the VvMYB14–VvWRKY8–VvMYB30 regulatory circuit.
Materials and methods
Plant materials and growth conditions
Experimental materials used in this study were N. benthamiana plants; plants of the grapevine cultivars “Hongbaladuo” (V. vinifera), “Cros Colman” (V. vinifera), “Beihong” (V. vinifera “Muscat Hamburg” × V. amurensis), “Beixi” (V. vinifera “Muscat Hamburg” × V. amurensis), “Zhi186” (“Zhi168” [Vitis monticola × Vitis riparia] × “Beihong”), and “Kyoho” (Vitis labrusca × V. vinifera); V. amurensis tissue culture plantlets; and “41B” (V. vinifera “Chasselas” × Vitis berlandieri) suspension cells. The grapevines were grown in the Germplasm Repository for Grapevines at the Institute of Botany, Chinese Academy of Sciences, Beijing, China. The N. benthamiana plants and V. amurensis tissue culture plantlets were grown in the greenhouse at 25°C and with 16-h light/8-h dark and a light intensity of 100 μmol m−2 s−1. The “41B” grapevine suspension cells were maintained in a tabletop shaker set at 25°C and 120 rpm.
VvMYB30 gene isolation and analysis
The nucleic acid sequence of VvMYB30 was obtained from the Grape Genome database (http://www.grapegenomics.com/pages/PN40024/) (Jaillon et al., 2007). The location of the VvMYB30 gene on the grape genome was analyzed using the On-Line BLAST tool and Genome Browser tool, which was also from the database. The primers used to amplify the full-length coding sequence and genomic DNA of VvMYB30 were designed using CE Design V1.04 software. DNA from the mature leaves of “Hongbaladuo” was extracted using a Plant Genomic DNA Extraction Kit (Aidlab Biotech, China) and the genomic DNA sequence of VvMYB30 was amplified using PCR. Total RNA extracted from the mature leaves of “Hongbaladuo,” “Gros Colman,” “Beihong,” and “Zhi186” using an RNAprep Pure Plant Kit (TIANGEN, China) was reverse transcribed using a HiScript III First Strand cDNA Synthesis Kit (Vazyme, China). The coding sequences of MYB30 were cloned from these varieties. The PCR products were cloned into the pLB-Simple vector (TIANGEN, China) and sequenced. Multiple sequence alignment of MYB30 coding sequences from the above varieties was conducted using DNAMAN. The protein domains of VvMYB30 were predicted using SMART (http://smart.embl-heidelberg.de/). Alignment of VvMYB30 and VvMYB14 amino acid sequences was also conducted using DNAMAN.
Phylogenetic analysis
Based on previous reports (Stracke et al., 2001; Wong et al., 2016; Albert and Allan, 2021; Rodrigues et al., 2021), the nucleic acid sequences of 21 and 29 SG1–SG7 R2R3 MYBs from Arabidopsis (A. thaliana) and grapevine, respectively, that are likely involved in specialized metabolism, were selected to construct a phylogenetic tree. One R3 MYB VvETC1 from grape was used as an outlier. The maximum-likelihood method was used with 1,000 bootstrap replications by MEGA11 software (Tamura et al., 2021). The substitution model is a Tamura 3-parameter model. The accession numbers of MYBs used in the tree are shown in Supplemental Table S2 and the machine-readable tree and alignment files are provided as Supplemental Files S1 and S2, respectively.
Subcellular localization
To determine subcellular localization, the full-length VvMYB30 coding sequence was fused to the pCAMBIA2300-GFP vector (Yan et al., 2021). The nuclear localization protein H2B fused to the red fluorescent protein mCherry was used as a nuclear marker (Howe et al., 2012). The recombined vectors pCAMBIA2300-VvMYB30 and H2B-mCherry were co-transformed into the N. benthamiana leaves through Agrobacterium-mediated transformation (Sheludko et al., 2007). The combination of EV pCAMBIA2300 and H2B-mCherry was used as the control. After 72 h, localization of GFP and RFP was observed in the agro-infiltrated leaves using a Leica TCS SP5 Confocal Scanning Microscope. The excitation wavelengths of GFP and mCherry are 488 and 587 nm, respectively.
Y1H assay
Y1H assays were performed using the Matchmaker One-Hybrid System (Clontech, USA). For the Y1H assays, two different systems, pLacZi (Lin et al., 2007) and pHis2 (http://www.youbio.cn/product/vt1637), were used in this study. In the pLacZi system, the full-length coding sequence, the N-terminal/C-terminal coding sequence fragments of VvMYB30, and the full-length coding sequence of VvMYB14 were cloned into the effector vector pGAD424 (http://www.youbio.cn/product/vt1644) following the manufacturer’s instructions. The promoter of VvSTS15/21 and the predicted MYB binding motifs on VvSTS15/21 (P1: ggatgagAGTTggtgag; P2: tatgGGTAggtgaag; P3: gagaaaccGTAAgcaga; P4: atctctcTATAtaagaa; P5: cgaaGGTTggtagca; P6: attgggttGTTGagg; P7: aagtAAATacttaagtg; and P8: ttaaaaccTTTAcccga) were fused into the reporter vector pLacZi. The AD-fusion effectors were introduced into the yeast strain EGY48 with the corresponding LacZ reporters. The transformants were selected on SD/-Trp-Ura medium. The positive clones were transferred to the SC/-Trp-Ura/Gal/Raf medium supplemented with 20 mg L−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). Yeast cells were grown for 3 days at 30°C for color development.
For the pHis2 system, the full-length coding sequences of VvMYB30, VvMYB14, and VvWRKY8 were cloned into the pGADT7 effector vector. About 2,000-bp fragments upstream of the VvMYB30, VvMYB14, and VvWRKY8 start codon were cloned into the pHis2 reporter vector. Then, different combinations of effector and reporter were co-transformed into the Y187 yeast strain. The transformants were first grown on the SD/-Trp-Leu selection medium for 3 days and cells were streaked on the SD/-Trp-Leu-His selection medium supplemented with different concentrations of 3AT. Yeast cells containing the combination of pGAD53 and p53HIS were used as the positive control.
Transient LUC expression assay
For the transient LUC assay, the full-length coding sequence of VvMYB30 was ligated to the expression vector pSAK277 (Yang et al., 2022) to serve as an effector. The promoter of VvSTS15/21 was cloned into the pCAMBIA1302-LUC vector (Zhang et al., 2017). Then, the effector and the reporter plasmids were individually transformed into Agrobacterium tumefaciens EHA105 (ZOMANBIO Company, ZC142). Agrobacterium containing the effector and reporter plasmids was infiltrated into the N. benthamiana leaves with equal concentration. The combination of EV pSAK277 and proVvSTS15/21:LUC was used as the control. After 72 h, the infiltrated leaves were taken to observe the firefly luminescence using a fluorescent imager (Tanon 5200, USA). The rest of these leaves were sampled and used to measure LUC activity using the Dual-LUC Reporter Assay System (E1910, Promega, USA). According to the manufacturer’s instructions of the kit, every 0.1 g powder from six leaves (one of the three biological replicates) was added in 200 µL protein lysis buffer for total protein extraction and then the activities of firefly LUC and Renilla LUC were measured by GloMax 20/20 Luminometer (Promega). The ratio of firefly LUC/Renilla LUC was calculated to determine the effect of VvMYB30 on the transcriptional activity of VvSTS15/21. For the competitive promoter activation assay, the full-length coding sequences of VvMYB30, VvMYB14, and VvWRKY8 were ligated to the expression vector pH7WG2D (Karimi et al., 2002).
Y2H assay
Y2H assays were performed using the Matchmaker Gold Y2H System (Clontech). The two-hybrid library screening assay was conducted as described by Jiang et al. (2019). The prey cDNA library of “Pinot Noir” used was constructed by Jiang et al. (2019). The positive strains were selected on SD/–Leu/–Trp/–Ade/–His selection media supplied with 40 mg L−1 X-α-gal and 200 µg L−1 aureobasidin A (AbA).
For the Y2H assays, the full-length, the N-terminal, and the C-terminal halves of VvMYB30 coding sequences were cloned into the prey vector pGADT7 (Clontech) while the full-length, the N-terminal, and the C-terminal halves of VvWRKY8 coding sequences were fused into the bait vector pGBKT7 (Clontech). Combinations of prey and bait vectors were co-transformed into yeast strain Y2Hgold. The transformants were selected on SD/-Leu-Trp selection medium. The yeast cells were then selected on SD/-Leu/-Trp/-His/-Ade selection media with 40 mg L−1 X-α-gal to detect the interaction. Yeast strain Y2Hgold cells harboring pGADT7-T and pGBKT7-p53 were used as a positive control while cells containing pGADT7-T and pGBKT7-Lam were used as a negative control.
To determine the transcriptional activity of VvMYB14 and VvMYB30, the full-length coding sequences of VvMYB14 and VvMYB30 were fused to GAL4 BD to generate BD-VvMYB14 and BD-VvMYB30, respectively. The constructed plasmids were transferred to yeast strain Y2Hgold and selected on SD/-Trp media. Then, the yeast cells harboring BD-VvMYB14 and BD-VvMYB30 were selected on SD/-Trp media with 40 mg L−1 X-α-gal.
Yeast protein extraction and immunoblot analysis
Proteins from the yeast strain harboring BD-MYB14 or BD-MYB30 were extracted using a Yeast Total Protein Extraction Kit (Sangon Biotech Company, C500013) according to the manufacturer’s instructions. The extracted proteins were resuspended in SDS-PAGE Loading Buffer (CWBIO Company, cw0028s), boiled for 10 min, and separated on SDS–PAGE gel for immunoblot analysis. The primary antibodies used were anti-Actin (CWBIO Company, CW0096M, diluted 2,000 times) and anti-cMYC (Lablead Company, M1002, diluted 2,000 times). The gray degree of immune bands was calculated using ImageJ software.
BiFC assay
For BiFC assay, the full-length coding sequences of VvMYB30, VvMYB14, and VvWRKY8 without the stop codon were fused to the N-terminal of YFP (nYFP) and the C-terminal of YFP (cYFP) (Grefen and Blatt, 2012), respectively. The primers used to construct VvMYB30-nYFP, VvMYB30-cYFP, VvMYB14-nYFP, VvMYB14-cYFP, VvWRKY8-nYFP, and VvWRKY8-cYFP are included in Supplemental Table S3. The combination of VvMYB30-cYFP and VvMYB14-nYFP in Supplemental Figure S10 could be regarded as another negative control of VvMYB30-cYFP interaction with VvWRKY8-nYFP in Figure 1C. The nYFP and cYFP fused with corresponding proteins were co-transformed into A. tumefaciens EHA105. The Agrobacterium strains containing plasmids were infiltrated into N. benthamiana leaves. After 72 h, YFP fluorescence was observed in the infiltrated leaves using a Leica TCS SP5 Confocal Scanning Microscope.
In vitro pull-down assay and immunoprecipitation assay
For the in vitro pull-down assay, the coding sequences of VvMYB30, VvMYB30-N, and VvMYB30-C were inserted into the pGEX4T-1 (http://www.youbio.cn/product/vt1253) vector and transformed into Transetta (DE3) chemically competent cells (TransGen Biotech Company, CD801-01). The culture was induced at 16°C for 16 h and the cells were then suspended in PBS buffer (8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 in 1 L of water, pH 7.4). The E. coli suspensions were sonicated for 20 min, centrifuged (12,000 × g, 1 h, 4°C), and the supernatants were collected and incubated with glutathione sepharose 4B beads (CS20421; 25 mL) at 4°C for 1 h. After discarding the supernatants, the remaining beads were washed and eluted by PBS buffer and eluted buffer (40 mM reduced glutathione in PBS buffer). Coding sequences of VvWRKY8, VvWRKY8-N, and VvWRKY8-C were inserted into a modified pET28a vector containing a highly soluble and monomeric SUMO tag (http://www.youbio.cn/product/vt2227). The resulting vectors were transformed into BL21 (DE3) pLysS chemically competent cells (TransGen Biotech Company, CD701-01). The E. coli suspensions were sonicated for 20 min, centrifuged (12,000 × g, 1 h, 4°C), and the supernatants were incubated with Ni-NTA resin (Thermo Scientific, 88221) at 4°C for 1 h. After discarding the supernatants, the Ni-NTA resin was washed three times with 10 column volumes of wash buffer (20 mM Tris–HCl, pH 7.4, 20 mM imidazole, 500 mM NaCl), then eluted with elution buffer (20 mM Tris–HCl, pH 7.4, 500 mM imidazole, 500 mM NaCl). Purified GST-tagged VvMYB30, VvMYB30-N, and VvMYB30-C proteins were incubated with VvWRKY8, VvWRKY8-N, and VvWRKY8-C for 2 h at 4°C, respectively. The mixture was incubated with glutathione sepharose 4B beads. After discarding the supernatants, the glutathione sepharose 4B beads were rinsed three times with 500 µL of PBS buffer. The beads were then incubated with eluted buffer and the elution product was boiled with SDS-PAGE loading buffer (CWBIO Company, cw0028s) for 10 min and subjected to immunoblot analysis. The GST- and HIS-tag proteins were detected by GST (CWBIO Company, cw0084m, diluted 2,000 times) and HIS (CWBIO Company, cw0286m, diluted 2,000 times) monoclonal antibody, respectively.
For the immunoprecipitation assay, VvMYB30 with a 3× FLAG tag and VvWRKY8 fused to a 5× cMYC tag were co-transformed into N. benthamiana leaves by Agrobacterium-mediated transformation. Leaves were collected and protein was extracted using lysis buffer (50 mM PBS, pH 7.4, 1 mM DTT, 1% [v/v] NP-40, and protease inhibitor cocktail [EDTA-free, Roche, Basel, Switzerland]). Then, the proteins were incubated for 2 h at 4°C. cMYC monoclonal antibody (Sigma, M4439) (20 µL) was incubated with 20 µL (bed volume) of protein G-Sepharose beads (Roche Healthcare) for 2 h at 4°C, incubated with the protein mixture, and then centrifuged. The immunoprecipitates were washed three times with lysis buffer and the concentrates were then resuspended in SDS-PAGE loading buffer (CWBIO Company, cw0028s), boiled for 10 min, and then subjected to immunoblot analysis with FLAG antibody (Beyotime, AF0036, diluted 2,000 times). The cMYC antibody (diluted 2,000 times) was used to detect cMYC-tagged protein.
Electrophoretic mobility shift assay
The coding sequences of VvMYB14 and VvMYB30 were inserted into the pMal-c2x (http://www.addgene.org/75286/) vector and transformed into Transetta (DE3) Chemically Competent cells (TransGen Biotech Company, CD801-01), respectively. The culture was induced at 16°C for 16 h and the cells were then suspended in cell buffer (10 mM Tris–HCl pH 7.4, 30 mM NaCl). The E. coli suspensions were sonicated for 20 min and centrifuged (12,000 × g, 1 h, 4°C), and the supernatants were collected and incubated with amylose magnetic beads (NEB Corporation, E8035S) at 4°C for 1 h. After discarding the supernatants, the remaining beads were washed three times with wash buffer (20 mM Tris–HCl pH 7.4, 1 mM EDTA, 200 mM NaCl, and 1 mM DTT) and eluted with elution buffer (20 mM Tris–HCl pH 7.4, 1 mM EDTA, 200 mM NaCl, 1 mM DTT, and 10 mM maltose). The eluted protein was used to conduct EMSA. The 5′-biotin-labeled DNA probes (Supplemental Table S4) were annealed in 10× annealing buffer (100 mM Tris–HCl, pH 7.4, 10 mM EDTA, and 1 M NaCl) at 75°C for 30 min and stored at −20°C. The EMSAs were performed using a LightShift chemiluminescent EMSA kit (ThermoFisher Scientific; 20148) according to the manufacturer’s protocol.
Transient transformation of VvMYB30 in grapevine leaves and berry skin
When we attempted to overexpress MYB30 in grape leaves using the 35S promoter, VvMYB30 expression was down-regulated. We suspected this phenomenon may be co-suppression, which was firstly found in Petunia when CHS was overexpressed (Napoli et al., 1990). For transient overexpression, the CaMV35S of the vector pSAK277 was replaced by the V. amurensis MYB30 promoter (2,099 bp upstream of the MYB30 translation start codon) and the coding sequence of VvMYB30 was inserted into the pSAK277 vector containing the V. amurensis MYB30 promoter to generate pSAK277-proMYB30:MYB30. For VvMYB30 knock-down, a 250-bp fragment (301–551 bp) from the coding sequence of VvMYB30 was inserted into pFGC5941 (Kerschen et al., 2004) in sense and antisense orientation. The binary constructs were transformed into A. tumefaciens strain GV3101 (ZOMANBIO Company, ZC141). The method of transient transformation in grapevine reported by Jelly et al. (2014) was used with slight modifications. The Agrobacterium cells were grown in 10 mL YEP medium containing 10 μM acetosyringone at 28°C and 180 rpm for 12–16 h until OD600 = 1. The cells were then spun down and resuspended in liquid medium (10 mM MES-KOH, pH 5.2, 10 mM MgCl2, and 100 μM acetosyringone) and incubated for 2 h before use. For berry skin transformation, we chose three clusters of “Beixi” or “Kyoho” as three biological replicates; ten mature berries with approximate size were sampled in each biological replicate. Berries were put into Agrobacterium suspension and vacuum (−90 kPa) infiltrated for 15 min. Then, these berries were washed with ddH2O and dried with blotting paper. Berry pedicels were put into MS medium and cultured in a growth chamber at 25°C under fluorescent lights (100 μmol m−2 s−1) with a 16-h photoperiod. The constructs were infiltrated into the berry skin of “Beixi” for MYB30 overexpression or “Kyoho” for MYB30 silence. For the leaf transformation assay, we used V. amurensis tissue culture plantlets. Forty-five-day-old plantlets were used for the MYB30 overexpression and 30-day-old plantlets for MYB30 silence. Each treatment included three biological replicates, and each biological replicate had three plantlets. The plantlets were vacuum (−90 kPa) infiltrated for 20 min. Then, these plantlets were washed with ddH2O and dried with blotting paper, and the roots were inserted into MS medium. The growth conditions were the same as those mentioned above. Five days after Agrobacterium infiltration, skin and leaves were sampled.
UV-B/C treatment of grapevine leaves
In this experiment, we used UV-B and UV-C lamps to establish UV-B and UV-C treatments. The length and power of each UV lamp was 1,200 mm and 40 W, respectively. The emission peak wavelengths of UV-B and UV-C lamps were 313 and 254 nm, respectively. All lamps were bought from Beijing Zhongyi Boteng Technology Company. Before the experiment, the illumination intensity was detected using a UV radiometer (HANDY, Optical Instrument Factory of Beijing Normal University). UV-B and UV-C radiation of grapevine leaves were conducted based on Jiang et al. (2019). The UV lamps (15 tubes) were fastened tightly to the top of a big box. An adjustable platform was placed under the lamps that allows distance adjustment to the lamps. Sample flasks were secured on the platform. The grape petioles were inserted into the flasks with water. The illumination intensity was modulated by the distance between the grape leaf surfaces to the UV lamp. Healthy, 30-day-old leaves were collected from V. vinifera “Hongbaladuo” grapevines. After half an hour adaptation in the dark at 25°C, the leaves were treated with UV-B or UV-C radiation with an intensity of 6 W m−2 supplied from the UV-B/C lamp for 10 min. The petiole was kept below the water surface in the bottle until the sample was taken. Leaves were taken before treatment and at 0, 1, 2, 6, 9, 18, 24, and 48 h after 10 min treatment, and leaves without UV treatment were used as controls. Three biological replicates were set at each treatment and control point, and every similar six leaves were collected for one biological replicate. The leaves were ground into powder in liquid nitrogen and stored at −80°C for future experiments.
RNA extraction and RT-qPCR
Total RNA was isolated from grapevine leaves, berry skin, berry flesh, and suspension cells using an RNAprep Pure Plant Kit (polysaccharides and polyphenolics-rich) (TIANGEN, China) following the manufacturer’s protocol. Approximately 1 µg of total RNA was used for cDNA synthesis using the HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China). qPCR was performed in 96-well plates (Bio-Rad, USA) using AceQ qPCR SYBR Green Master (Vazyme, China). The primers used to quantify the expression level of VvMYB30, VvMYB14, VvWRKY8, VvSTS15/21, and VvSTSs were designed using NCBI Primer-Blast and are listed in Supplemental Table S3. The primers designed for VvSTSs can detect 25 VvSTSs including VvSTS15/21 (Jiang et al., 2019). All reactions were conducted in CFX96 (Bio-Rad, USA). RT-qPCR analysis was performed with three independent biological replicates, with each biological replicate containing three technical replicates. For the experiments to detect the expression level of MYB30, MYB14, and WRKY8 in “Hongbaladuo,” “Cros Colman,” and “Zhi186” grapevines, we chose three grapevines of each cultivar as three biological replicates, six 30-day-old leaves or ten mature berries with approximate size were sampled in each biological replicate.
Res extraction and content determination
Res extraction from leaves, berry skin, and berry flesh was performed following the method described by Jiang et al. (2019). Firstly, 1 g of frozen samples was ground into powder. The powder was transferred to a 50-mL centrifuge tube containing 15 mL of methanol and ethyl acetate with a 1:1 mixture (v/v). The mixture was placed in the dark at room temperature for 24 h. After extraction for 24 h, the supernatant was collected by centrifugation at 13,400 × g at 4°C for 10 min. After centrifugation, the extraction solution was added to the precipitate again for extraction, and this process was repeated two to three times to collect all the extraction solution. The collected extract was vacuum dried at 40°C in the rotary evaporator and then dissolved in 2 mL methanol. The extract was filtered through a 0.45-µm disposable filter for future analysis.
The content of Res was determined using the Waters Alliance HPLC System (Waters 2695, Waters, USA) with a photo-diode array (PDA, Waters 2998, Waters) as described by Jiang et al. (2019). The detection temperature was 30°C, the injection volume was 10 µL, and the flow rate was 1.0 mL min−1. Two detection wavelengths, 288 and 306 nm, were set to detect cis-isomers (cis-Res and cis-Pd) and trans-isomers (trans-Res and trans-Pd), respectively.
Statistical analysis
The differences between two groups were analyzed by Student’s t tests using GraphPad Prism 8 software. Asterisks (*) above columns indicate significant differences (P < 0.05) between the two groups underlined. One-way ANOVAs with Duncan’s tests were conducted to determine significant differences among more than two groups using SPSS software (IBM SPSS Statistics 21). Different letters above columns indicate significant differences (P < 0.05) among the different samples. Statistical data are shown in Supplemental Data Set S1 and complete data are provided in Supplemental Data Set S2.
Accession numbers
Sequence data from this article can be found in the V. vinifera PN40024 (12x.V2) data libraries under the following accession numbers: VIT_201s0010g03930 (VvWRKY8); VIT_207s0005g03340 (VvMYB14); VIT_217s0000g06190 (VvMYB30); VIT_216s0100g00830 (VvSTS15); and VIT_216s0100g00910 (VvSTS21).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Validation of the interaction between VvMYB30-GST and HIS-SUMO tag in vitro by pull-down assay.
Supplemental Figure S2. Schematic diagrams of VvWRKY8 and VvMYB30 showing the amino (N)- and carboxy (C)-terminal halves of VvWRKY8 (VvWRKY8-N, VvWRKY8-C) and the two halves of VvMYB30 (VvMYB30-N and VvMYB30-C).
Supplemental Figure S3. Nucleotide sequence alignment of MYB30 from the grapevine cultivars “PN40024,” “Hongbaladuo,” “Cros Colman,” “Beihong,” and “Zhi186.”
Supplemental Figure S4. Phylogenetic analysis of A. thaliana (At) and V. vinifera (Vv) R2R3 MYBs.
Supplemental Figure S5. Relative expression levels of VvMYB30, VvMYB14, VvWRKY8, and VvSTS15/21 in grapevine leaves before and immediately after UV-B and UV-C treatments.
Supplemental Figure S6. VvWRKY8 and VvMYB30 do not mutually regulate each other.
Supplemental Figure S7. VvMYB14 and VvMYB30 do not mutually regulate each other.
Supplemental Figure S8. VvMYB30 does not bind to its own promoter.
Supplemental Figure S9. EMSA showing the binding of VvMYB30 or VvMYB14 to the VvSTS15/21 promoter.
Supplemental Figure S10. VvMYB30 does not physically interact with VvMYB14.
Supplemental Figure S11. Relative expression level of MYB30 in grapevine suspension cells after exogenous supply of trans-Res and in VvWRKY8-overexpressed grapevine leaves.
Supplemental Figure S12. Transcriptional activity analysis of VvMYB30.
Supplemental Figure S13. Amino acid sequence alignment of VvMYB30 and VvMYB14.
Supplemental Table S1. Res contents in diverse tissues of different grape cultivars.
Supplemental Table S2. Gene names and the corresponding gene IDs used in this study.
Supplemental Table S3. Primers used in this study.
Supplemental Table S4. Probes used in this study.
Supplemental Data Set S1. Summary of statistical analyses.
Supplemental Data Set S2. Complete data for statistical analyses.
Supplemental File S1. Machine-readable tree files for phylogenetic analysis.
Supplemental File S2. Alignments used for phylogenetic analysis.
Funding
This work was funded by the National Key R&D Program of China (Grant No. 2019YFD1000101), Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA23080602), and the National Natural Science Foundation of China (Grant No. 31672120).
Conflict of interest statement. The authors declare that they have no competing interests.
Supplementary Material
Contributor Information
Huayuan Mu, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; Chinese National Botany Garden, Beijing 100093, China; LIA INNOGRAPE International Associated Laboratory, Beijing 100093, China.
Yang Li, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; Chinese National Botany Garden, Beijing 100093, China; LIA INNOGRAPE International Associated Laboratory, Beijing 100093, China.
Ling Yuan, Department of Plant and Soil Sciences, University of Kentucky, Lexington, Kentucky 40546, USA; Key Laboratory of South China Agricultural Plant Molecular Analysis and Genetic Improvement and Guangdong Provincial Key Laboratory of Applied Botany, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China.
Jinzhu Jiang, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Yongzan Wei, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Wei Duan, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; Chinese National Botany Garden, Beijing 100093, China; LIA INNOGRAPE International Associated Laboratory, Beijing 100093, China.
Peige Fan, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; Chinese National Botany Garden, Beijing 100093, China; LIA INNOGRAPE International Associated Laboratory, Beijing 100093, China.
Shaohua Li, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; Chinese National Botany Garden, Beijing 100093, China; LIA INNOGRAPE International Associated Laboratory, Beijing 100093, China.
Zhenchang Liang, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; Chinese National Botany Garden, Beijing 100093, China; LIA INNOGRAPE International Associated Laboratory, Beijing 100093, China.
Lijun Wang, Beijing Key Laboratory of Grape Sciences and Enology, CAS Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; Chinese National Botany Garden, Beijing 100093, China; LIA INNOGRAPE International Associated Laboratory, Beijing 100093, China.
L.W. conceived and supervised the study. H.M. and Y.L. performed the experiments and analyzed the data. J.J., Y.W., W.D., P.F., S.L., Z.L., and L.Y. helped design the experiment. H.M., Y.L., and L.W. wrote the article. L.Y. and L.W. did the final editing.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Lijun Wang (ljwang@ibcas.ac.cn).
References
- Abbasi BH, Khan T, Khurshid R, Nadeem M, Drouet S, Hano C (2021) UV-C mediated accumulation of pharmacologically significant phytochemicals under light regimes in in vitro culture of Fagonia indica (L.). Sci Rep 11: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian M, Jeandet P, Douillet-Breuil AC, Tesson L, Bessis R (2000) Stilbene content of mature Vitis vinifera berries in response to UV-C elicitation. J Agric Food Chem 48: 6103–6105 [DOI] [PubMed] [Google Scholar]
- Adrian M, Jeandet P, Veneau J, Weston LA, Bessis R (1997) Biological activity of resveratrol, a stilbenic compound from grapevines, against Botrytis cinerea, the causal agent for gray mold. J Chem Ecol 23: 1689–1702 [Google Scholar]
- Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O’Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 [DOI] [PubMed] [Google Scholar]
- Albert NW, Allan AC (2021) MYB genes involved in domestication and crop improvement. Ann Plant Rev Online 4: 199–242 [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Cavallini E, Walker AR, Pezzotti M, Bliek M, Quattrocchio F, Koes R, Ruperti B, Bertini E, Zenoni S, et al. (2019) The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J 99: 1220–1241 [DOI] [PubMed] [Google Scholar]
- An J, Zhang X, You C, Bi S, Wang X, Hao Y (2019) MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol 224: 380–395 [DOI] [PubMed] [Google Scholar]
- Austin MB, Noel JP (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 20: 79–110 [DOI] [PubMed] [Google Scholar]
- Berli FJ, Moreno D, Piccoli P, Hespanhol-Viana L, Silva MF, Bressan-Smith R, Cavagnaro JB, Bottini R (2010) Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet-B radiation by enhancing ultraviolet-absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ 33: 1–10 [DOI] [PubMed] [Google Scholar]
- Björn LO (1996) Effects of ozone depletion and increased UV-B on terrestrial ecosystems. Int J Environ Stud 51: 217–243 [Google Scholar]
- Björn LO (2015) Ultraviolet-A, B and C. UV4Plants Bull 1: 17–18 [Google Scholar]
- Brown BA, Jenkins GI (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146: 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Diago MP, Martínez-Abaigar J, Martínez-Zapater JM, Tardáguila J, Núñez-Olivera E (2014) Solar ultraviolet radiation is necessary to enhance grapevine fruit ripening transcriptional and phenolic responses. BMC Plant Biol 14: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang K, Khurshid M, Li J, He M, Georgiev M, Zhang X, Zhou M (2019a) MYB transcription repressors regulate plant secondary metabolism. Crit Rev Plant Sci 38: 159–170 [Google Scholar]
- Chen L, Hu B, Qin Y, Hu G, Zhao J (2019b) Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol Biochem 136: 178–187 [DOI] [PubMed] [Google Scholar]
- Cho YJ, Kim N, Kim CT, Maeng JS, Pyee J (2012) Quantitative evaluation of resveratrol enrichment induced by UV stimulus in harvested grapes. Food Sci Biotechnol 21: 597–601 [Google Scholar]
- Daniel X, Lacomme C, Morel JB, Roby D (1999) A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J 20: 57–66 [DOI] [PubMed] [Google Scholar]
- Deng N, Chang E, Li M, Ji J, Yao X, Bartish IV, Liu J, Ma J, Chen L, Jiang Z (2016) Transcriptome characterization of Gnetum parvifolium reveals candidate genes involved in important secondary metabolic pathways of flavonoids and stilbenoids. Front Plant Sci 7: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Gerevini GT, Repossi G, Dain A, Tarres MC, Das UN, Eynard AR (2016) Beneficial action of resveratrol: how and why? Nutrition 32: 174–178 [DOI] [PubMed] [Google Scholar]
- Dubos C, Gourrierec JL, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Dubrovina AS, Kiselev KV (2017) Regulation of stilbene biosynthesis in plants. Planta 246: 597–623 [DOI] [PubMed] [Google Scholar]
- Ector B, Magee JB, Hegwood CP, Coing MJ (1996) Resveratrol concentration in Muscadine berries, juice, pomace, purees, seeds, and wines. Am J Enol Vitic 47: 57–62 [Google Scholar]
- Ferri M, Tassoni A, Franceschetti M, Righetti L, Naldrett MJ, Bagni N (2009) Chitosan treatment induces changes of protein expression profile and stilbene distribution in Vitis vinifera cell suspensions. Proteomics 9: 610–624 [DOI] [PubMed] [Google Scholar]
- Freitas A, Martins MM, Costa HS, Albuquerque TG, Valentea A, Silva AS (2015) Effect of UV-C radiation on bioactive compounds of pineapple (Ananas comosus L. Merr.) by-products. J Sci Food Agric 95: 44–52 [DOI] [PubMed] [Google Scholar]
- Gil M, Bottini R, Berli F, Pontin M, Silva MF, Piccoli P (2013) Volatile organic compounds characterized from grapevine (Vitis vinifera L. cv. Malbec) berries increase at pre-harvest and in response to UV-B radiation. Phytochemistry 96: 148–157 [DOI] [PubMed] [Google Scholar]
- González-Barrio R, Beltrán D, Cantos E, Gil MI, Espín JC, Tomás-Barberán FA (2006) Comparison of ozone and UV-C treatments on the postharvest stilbenoid monomer, dimer, and trimer induction in var. ‘Superior’ white table grapes. J Agric Food Chem 54: 4222–4228 [DOI] [PubMed] [Google Scholar]
- Grefen C, Blatt MR (2012) A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 53: 311–314 [DOI] [PubMed] [Google Scholar]
- Guerrero RF, Villar EC, Puertas B, Richard T (2016) Daily preharvest UV-C light maintains the high stilbenoid concentration in grapes. J Agric Food Chem 64: 5139–5147 [DOI] [PubMed] [Google Scholar]
- Hasan MM, Bae H (2017) An overview of stress-induced resveratrol synthesis in grapes: perspectives for resveratrol-enriched grape products. Molecules 22: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J (2013) The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25: 4135–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe ES, Clemente TE, Bass HW (2012) Maize histone H2B-mCherry: a new fluorescent chromatin marker for somatic and meiotic chromosome research. DNA Cell Biol 6: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Tang Z, Fu J, Yuan Y, Deng X, Xu Q (2020) CsMYB3 and CsRuby1 form an ‘activator-and-repressor’ loop for the regulation of anthocyanin biosynthesis in Citrus. Plant Cell Physiol 61: 318–330 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jeandet P, Bessis R, Gautheron B (1991) The production of resveratrol (3,5,4′-trihydroxystilbene) by grape berries in different developmental stages. Am J Enol Vitic 42: 41–46 [Google Scholar]
- Jeandet P, Clément C, Cordelier S (2019) Regulation of resveratrol biosynthesis in grapevine: new approaches for disease resistance? J Exp Bot 70: 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelly NS, Valat L, Walter B, Maillot P (2014) Transient expression assays in grapevine: a step towards genetic improvement. Plant Biotechnol J 12: 1231–1245 [DOI] [PubMed] [Google Scholar]
- Jiang J, Xi H, Dai Z, Lecourieux F, Yuan L, Liu X, Patra B, Wei Y, Li S, Wang L (2019) VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J Exp Bot 70: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Liu C, Xiao X, Dixon RA (2015) The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell 27: 2860–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Muller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566: 223–228 [DOI] [PubMed] [Google Scholar]
- Kiselev KV, Aleynova OA, Tyunin AP (2017) Expression of the R2R3 MYB transcription factors in Vitis amurensis Rupr. plants and cell cultures with different resveratrol content. Russ J Genet 53: 465–471 [Google Scholar]
- Kiskova T, Kubatka P, Büsselberg D, Kassayova M (2020) The plant-derived compound resveratrol in brain cancer: a review. Biomolecules 10: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215: 924–933 [DOI] [PubMed] [Google Scholar]
- Kodan A, Kuroda H, Sakai F (2002) A stilbene synthase from Japanese red pine (Pinus densiflora): implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Proc Natl Acad Sci USA 99: 3335–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Deng R, Li X, Zeng Q, Zhang X, Yu X, Ren X (2020) Based on RNA-seq analysis identification and expression analysis of Trans-scripusinA synthesize-related genes of UV-treatment in postharvest grape fruit. Arch Biochem Biophys 15: 108471. [DOI] [PubMed] [Google Scholar]
- Langcake P, Pryce RJ (1976) Production of resveratrol by Vitis vinifera and other members of Vitaceae as a response to infection or injury. Physiol Plant Pathol 9: 77–86 [Google Scholar]
- Li T, Li Y, Sun Z, Xi X, Sha G, Ma C, Tian Y, Wang C, Zheng X (2021) Resveratrol alleviates the KCl salinity stress of Malus hupehensis Rhed. Front Plant Sci 12: 650485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xu J, Wu X, Wang Y, Lin Y, Wu D, Zhang H, Qin J (2019) Molecular analysis of UV-C induced resveratrol accumulation in Polygonum cuspidatum leaves. Int J Mol Sci 20: 6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Constabel CP (2019) MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci 24: 275–289 [DOI] [PubMed] [Google Scholar]
- Mao Z, Jiang H, Wang S, Wang Y, Yu L, Zou Q, Liu W, Jiang S, Wang N, Zhang Z, et al. (2021) The MdHY5–MdWRKY41–MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci 306: 110848. [DOI] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Takagi MO (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Meyer P, Van de Poel B, Coninck BD (2021) UV-B light and its application potential to reduce disease and pest incidence in crops. Hortic Res 8: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller RP, Schwekendiek A, Brehm I, Reif HJ, Kindl H (1999) Characterization of a pine multigene family containing elicitor-responsive stilbene synthase genes. Plant Mol Biol 39: 221–229 [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa Y, Tomimori S, Wada K, Kondo H (2011) Effect of cultivation practices on resveratrol content in grape berry skins. Hortic Res 10: 249–253 [Google Scholar]
- Pezet R, Gindro K, Viret O, Spring JL (2004) Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol Mol Plant Pathol 65: 297–303 [Google Scholar]
- Ramani S, Chelliah J (2007) UV-B-induced signaling events leading to enhanced-production of catharanthine in Catharanthus roseus cell suspension cultures. BMC Plant Biol 7: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JA, Espley RV, Allan AC (2021) Genomic analysis uncovers functional variation in the C-terminus of anthocyanin-activating MYB transcription factors. Hortic Res 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheludko YV, Sindarovska YR, Gerasymenko IM, Bannikova MA, Kuchuk NV (2007) Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol Bioeng 96: 608–614 [DOI] [PubMed] [Google Scholar]
- Sheng K, Zheng H, Shui S, Yan L, Liu C, Zheng L (2018) Comparison of postharvest UV-B and UV-C treatments on table grape: changes in phenolic compounds and their transcription of biosynthetic genes during storage. Postharvest Biol Technol 138: 74–81 [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Sun B, Zhou X, Chen C, Chen C, Chen K, Chen M, Liu S, Chen G, Cao B, Cao F, et al. (2020) Coexpression network analysis reveals an MYB transcriptional activator involved in capsaicinoid biosynthesis in hot peppers. Hortic Res 7: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Nakabayashi R, Ogata Y, Sakurai N, Tokimatsu T, Goto S, Suzuki M, Jasinski M, Martinoia E, Otagaki S, et al. (2015) Multiomics in grape berry skin revealed specific induction of the stilbene synthetic pathway by ultraviolet-C irradiation. Plant Physiol 168: 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38: 3022–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossi VE, Regalado JJ, Lannicelli J, Laino LE, Burrieza HP, Escandón AS, Pitta-Álvarez SL (2019) Beyond Arabidopsis: differential UV-B response mediated by UVR8 in diverse species. Front Plant Sci 10: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban L, Charles F, Miranda MRA, Aarrouf J (2016) Understanding the physiological effects of UV-C light and exploiting its agronomic potential before and after harvest. Plant Physiol Biochem 105: 1–11 [DOI] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valletta A, Iozia LM, Leonelli F (2021) Impact of environmental factors on stilbene biosynthesis. Plants 10: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M (2012) Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol 12: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannozzi A, Wong DCJ, Höll J, Hmmam I, Matus JT, Bogs J, Ziegler T, Dry I, Barcaccia G, Lucchin M (2018) Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol 59: 1043–1059 [DOI] [PubMed] [Google Scholar]
- Versari A, Parpinello GP, Tornielli GB, Ferrarini R, Giulivo C (2001) Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J Agric Food Chem 49: 5531–5536 [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Wang D, Jiang C, Li R, Wang Y (2019) VqbZIP1 isolated from Chinese wild Vitis quinquangularis is involved in the ABA signaling pathway and regulates stilbene synthesis. Plant Sci 287: 110202. [DOI] [PubMed] [Google Scholar]
- Wang D, Jiang C, Liu W, Wang Y (2020) The WRKY53 transcription factor enhances stilbene synthesis and disease resistance by interacting with MYB14 and MYB15 in Chinese wild grape. J Exp Bot 71: 3211–3226 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun Y, Wang H, Guan X, Wang L (2016) Resveratrol synthesis under natural conditions and after ultraviolet-C irradiation in grape leaves at different leaf developmental stages. Hortscience 51: 727–731 [Google Scholar]
- Wang L, Wang Y (2019) Transcription factor VqERF114 regulates stilbene synthesis in Chinese wild Vitis quinquangularis by interacting with VqMYB35. Plant Cell Rep 38: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Wang L, Xu M, Liu C, Wang J, Xi H, Wu B, Loescher W, Duan W, Fan P, Li S (2013) Resveratrols in grape berry skins and leaves in Vitis Germplasm. PLoS ONE 8: e61642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Xu H, Jiang S, Zhang Z, Lu N, Qiu H, Qu C, Wang Y, Wu S, Chen X (2017) MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J 90: 276–292 [DOI] [PubMed] [Google Scholar]
- Weiskirchen S, Weiskirchen R (2016) Resveratrol: how much wine do you have to drink to stay healthy? Adv Nutr 7: 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DCJ, Schlechter R, Vannozzi A, Höll J, Hmmam I, Bogs J, Tornielli GB, Castellarin SD, Matus JT (2016) A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res 23: 451–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia N, Daiber A, Förstermann U, Li H (2017) Antioxidant effects of resveratrol in the cardiovascular system. Br J Pharmacol 174: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Ma L, Liu G, Wang N, Wang J, Wang L, Dai Z, Li S, Wang L (2014) Transcriptomic analysis of grape (Vitis vinifera L.) leaves after exposure to ultraviolet C irradiation. PLoS ONE 9: e113772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Yin X, Zeng J, Ge H, Song M, Xu C, Li X, Ferguson IB, Chen K (2014) Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J Exp Bot 65: 4349–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Yang N, Wang X, Wang Y (2021) VqBGH40a isolated from Chinese wild Vitis quinquangularis degrades trans-piceid and enhances trans-resveratrol. Plant Sci 310: 110989. [DOI] [PubMed] [Google Scholar]
- Yang Q, Yang X, Wang L, Zheng B, Cai Y, Ogutu CO, Zhao L, Peng Q, Liao L, Zhao Y, et al. (2022) Two R2R3-MYB genes cooperatively control trichome development and cuticular wax biosynthesis in Prunus persica. New Phytol 234: 179–196 [DOI] [PubMed] [Google Scholar]
- Yan Y, Li C, Dong X, Li H, Zhang D, Zhou Y, Jiang B, Peng J, Qin X, Cheng J, et al. (2020) MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 32: 2196–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Huang L, Zhang X, Guo C, Wang H, Li Z, Wang X (2017) Expression patterns and promoter characteristics of the Vitis quinquangularis VqSTS36 gene involved in abiotic and biotic stress response. Protoplasma 254: 2247–2261 [DOI] [PubMed] [Google Scholar]
- Yin Y, Tian X, Yang J, Yang Z, Tao J, Fang W (2022) Melatonin mediates isoflavone accumulation in germinated soybeans (Glycine max L.) under ultraviolet-B stress. Plant Physiol Biochem 175: 23–32 [DOI] [PubMed] [Google Scholar]
- Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, et al. (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64: 5085–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Guo D, Li G, Yang Y, Zhang G, Li S, Liang Z (2019) The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (VdSTS2). BMC Plant Biol 19: 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huai J, Shang F, Xu G, Tang W, Jing Y, Lin R (2017) A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol 174: 2487–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin-Wang K, Wang H, Gu C, Dare AP, Espley RV, He H, Allan AC, Han Y (2015) Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J 82: 105–121 [DOI] [PubMed] [Google Scholar]
- Zhou H, Wang K, Wang F, Espley RV, Ren F, Zhao J, Ogutu C, He H, Jiang Q, Allan AC, et al. (2019) Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol 221: 1919–1934 [DOI] [PubMed] [Google Scholar]
- Zhu H, Fitzsimmons K, Khandelwal A, Kranz RG (2009) CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant 2: 790–802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.