Abstract
Chlamidial organisms are obligate intracellular pathogens containing highly antigenic porin-like major outer membrane proteins (MOMPs). MOMP epitopes are of substantial medical interest, and they cluster within four relatively short variable (VS) domains. If MOMPs adopt a β-barrel fold, like bacterial porins, the VS domains may form extramembranous loops and the conserved regions of the protein may correspond to predicted membrane-located β-strands. However, molecular studies on native MOMPs have been hampered by the need to culture chlamydiae in eukaryotic host cells and purification and reconstitution remain problematic. In addition, the organisms are difficult to manipulate genetically, and it has also been difficult to functionally reconstitute recombinant MOMPs. To help overcome these problems and improve our understanding of MOMP structure and function, we cloned and expressed C. trachomatis and C. psittaci MOMPs and functionally reconstituted them at the single-channel level. We measured significant functional differences between the two proteins, and by removing and exchanging VS4, we tested the hypothesis that the largest variable domain forms an extramembranous loop that contributes to these differences. Proteins in which VS4 was deleted continued to form functional ion channels, consistent with the idea that the domain forms an extramembranous protein loop and incompatible with models in which it contributes to predicted membrane-located β-strands. Additionally, the properties of the chimeric proteins strongly suggested that the VS4 domain interacts closely with other regions of the protein to form the channel entrance or vestibule. Our approach can be used to probe structure-function relationships in chlamydial MOMPs and may have implications for the generation of effective antichlamydial vaccines.
Chlamydiaceae members are obligate intracellular pathogens responsible for a broad spectrum of human and animal diseases (25, 26, 31). They include Chlamydia psittaci and Chlamydia pecorum, which mainly infect animals, and the human pathogens Chlamydia pneumoniae and Chlamydia trachomatis. The long-term complications of C. trachomatis infection, including trachoma-related blindness, ectopic pregnancy, and female infertility, have a very substantial impact on human well-being (40). The respiratory tract pathogen C. pneumoniae has also been linked to coronary thrombosis (12, 37), although a recent epidemiological study failed to confirm an association (39). Finally, C. psittaci can also infect humans, leading to respiratory disorders and abortion.
Previous studies have shown that chlamydial organisms carry epitopes recognized by both T cells (for examples, see references 1, 22, 32, and 33) and B cells (for examples, see references 2, 6, 9, 10, 29, 30, and 47), and following extensive serotyping, they have been classified according to a range of partly overlapping serogroup- and species-specific epitopes (2, 30, 45). These epitopes elicit both neutralizing (20, 21) and nonneutralizing (18) antibodies, some of which protect against infection (19, 23, 24, 46, 47). Many of the epitopes have been attributed to the major outer membrane protein (MOMP), which comprises about 60% of the total outer membrane protein of the infectious elementary body (EB) (5). As a result, efforts have been focused on delineating the role this protein has to play in the pathogenicity of Chlamydia spp. and on determining whether it might form the basis for useful antichlamydial vaccines. However, molecular studies have been severely limited by technical problems. The organism is difficult to grow in large amounts, and native MOMPs are particularly difficult to purify and reconstitute. In addition, chlamydiae cannot yet be routinely subjected to genetic manipulation, and the functional reconstitution of heterologously expressed recombinant MOMPs has proven to be problematic.
Despite these challenging technical problems, it has been discovered that MOMPs have important structural and functional roles in the complex chlamydial life cycle, in which noninfectious, metabolically active reticulate bodies (RBs) alternate with the spore-like EB. Functionally, MOMPs behave like bacterial porins (17, 21), forming a pathway for the passage of ions and small solutes through the outer membrane of the organism (4). However, unlike other porins, MOMPs contain a large number of cysteine residues (up to nine in C. trachomatis). These are relatively well conserved throughout the genus, and the ability of MOMP to form intramolecular or intermolecular disulfide bonds (or both) may contribute to the rigidity of the free-living EB, which lacks the peptidoglycan layer found in gram-negative bacteria (3).
Every known MOMP contains four domains of variable sequence (VS1 to VS4) interspersed between five highly conserved regions or sequences (CS1 to CS5) (29). As anticipated, the variable domains have been shown to underpin much of the observed serological diversity and represent major neutralization determinants that contain many antigenic epitopes (2, 27, 47) and also serovar-specific T-cell epitopes (in VS1, VS2 and VS4 [1]). Proteolytic digestion studies (2, 8, 20, 36) strongly support models in which the highly immunogenic variable domains are surface exposed and imply that the poorly immunogenic, conserved regions of MOMPs corresponding to CS1 to CS5 contain transmembrane domains. However, apart from studies such as these, we currently know very little about the structure and function of chlamydial MOMPs at the molecular level, because of the technical challenges mentioned previously.
We have speculated that chlamydial MOMPs assemble as β-barrels, like bacterial porins, and contain predicted transmembrane β-strands joined by protein turns and loops. In support of this, we purified wild-type C. psittaci MOMP and showed by circular dichroism that it had a porin-like secondary structure, with the high β-sheet and low α-helix content typical of other porins (43). We also confirmed that it formed poorly selective, high-conductance porin-like ion channels in planar lipid bilayers (43). The reconstitution of functional channels extended the liposome swelling studies of Bavoil et al. (4), who first suggested that MOMP was a porin, and opened up a new route to structure-function studies. The present study is the first investigation of structure-function relationships in chlamydial MOMPs to combine cDNA mutagenesis and functional reconstitution.
MATERIALS AND METHODS
Cloning of full-length C. trachomatis MOMP cDNA.
The MOMP gene from C. trachomatis was amplified by PCR from genomic DNA kindly provided by the Chlamydia Genome Project. The primer pairs used were the following: sense primer, 5′-GTCGACATGCTGCCTGTGGGGAATCCTGCTGAACC-3′; antisense primer, 5′-CCATGGTTTGCAAAAAAAACTGGACCCGACCG-3′. SalI and NcoI restriction sites (shown in boldface type) were engineered into the sense and antisense primers, respectively. The sense primer also contained an engineered methionine start codon (underlined). Each reaction was carried out in a 50-μl volume containing 100 ng of template, 200 μM deoxynucleoside triphosphates, a 0.5 μM concentration of each primer, 200 mM Tris-HCl (pH 8.8 at 25°C), 100 mM KCl, 100 mM (NH4)2SO4, 20 mM MgSO4, 1% (vol/vol) Triton X-100, a 1-mg/ml concentration of nuclease-free bovine serum albumin, and 2.5 U of Pfu DNA polymerase. Amplification of the target DNA was accomplished by an initial denaturation step of 5 min at 94°C, followed by a thermal cycle of 30 s at 94°C, 30 s at 65°C, and 1 min at 72°C with a final strand extension step of 72°C for 7 min. Twenty-five cycles of amplification were carried out. In order to reduce the number of mutations introduced by PCR, Pfu DNA polymerase (Promega) was used throughout. All PCR products were purified using the QIAEX II gel extraction kit (Qiagen).
Cloning of full-length C. psittaci MOMP cDNA.
A truncated MOMP gene previously cloned from C. psittaci genomic DNA, which does not encode the first 16 amino acids of the mature protein (44), was extended by PCR using a three-step heminested procedure. The sense primers were the following: T1(ps), 5′-TTAATCCATGGTACTATGTGGGAAGGAGCTTCAGGAGATCC-3′; T2(ps), 5′-CCAGCTGAACCAAGTTTATTAATCGATGGCACTATGTGGGAAG-3′; and T3(ps), 5′-CATATGTTGCCTGTGGGGAACCCAGCTGAACCAAGTTTATTAATCG-3′. We used the antisense primer T4(ps), 5′-CCATGGCAGAGATTCCTAGGTTCTGATAGCGGGACAA-3′. The T3 sense and T4 antisense primers contained the engineered restriction sites NdeI (to provide a methionine start codon) and NcoI, respectively, which are shown in boldface type. Amplification of the target DNA was accomplished using the protocol described above.
Amplification of recombinant cDNAs.
Both full-length inserts were cloned into the EcoRV site of pSTBlue-1 (Novagen), using the Perfectly Blunt cloning kit (Novagen). The nomenclature adopted here is pSTB-ps and pSTB-tra for C. psittaci and C. trachomatis constructs, respectively. These constructs were then transformed into Novablue cells (Novagen), which lack the gene for T7 RNA polymerase, to amplify and recover DNA for examination of the construct sequences. All subsequent mutations were carried out in this nonexpressing vector prior to subcloning for protein expression, and all the clones were sequenced prior to subcloning (Oswell DNA). After verification, full-length constructs were subcloned into the expression vector pET22b(+) (Novagen) using corresponding restriction sites present in the multiple cloning site of pET22b(+) (to give pET-ps and pET-tra). Ligations were carried out overnight at 16°C, and the plasmids were transformed into Novablue cells.
In vitro site-directed mutagenesis.
Site-directed mutagenesis was carried out to introduce an AatII restriction site into the 5′ and 3′ ends of the VS4-encoding domains of the C. trachomatis and C. psittaci MOMP cDNAs. The mutations were generated by the method described by Chen and Przybyla (7), using two overlapping primers containing the desired mutation. The primers used for C. psittaci MOMP cDNA were the following: 5′ sense, 5′-CGAGCAACTTTTGAC(T)GT(C)C(T)GACGCTATCCGCATC-3′; 5′antisense, 5′-GATGCGGATAGCGTCGACGTCAAAAGTTGCTCG-3′; 3′sense, 5′-AACAAATTCGCTGACG(T)TCTTGCAAATTGCTTCG-3′; and 3′ antisense, 5′-CGAAGCAATTTGCAAGACGTCAGCGAATTTGTT-3′.
For C. trachomatis MOMP cDNA, the primers were the following: 5′ sense, 5′-CGAGCAAGCTTTGAC(T)GT(C)CGATACGATTCGTATAGC-3′; 5′ antisense, 5′-GCTATACGAATCGTATCGACGTCAAAGCTTGCTCG-3′; 3′ sense, 5′-GGTCAGCTCGGAGACG(A)T(C)C(A)ATGCAAATCGTTTCC-3′; and 3′ antisense, 5′-GGAAACGATTTGCATGACGTCTCCGAGCTGACC-3′. The introduced restriction site is shown in bold, with original nucleotides in parentheses. Each PCR was carried out in a 50-μl volume containing 20 ng of plasmid, 200 μM deoxynucleoside triphosphates, 125 ng of each primer, 200 mM Tris-HCl (pH 8.8 at 25°C), 100 mM KCl, 100 mM (NH4)2SO4, 20 mM MgSO4, 1% (vol/vol) Triton X-100, a 1-mg/ml concentration of nuclease-free bovine serum albumin, and 2.5 U of Pfu DNA polymerase. Amplification was accomplished by an initial denaturation step of 1 min at 94°C followed by a thermal cycle of 94°C for 30 s, 65°C for 1 min, 72°C for 8 min (i.e., 2 min/kb), and a final strand extension step at 72°C for 7 min. Each template was subjected to 18 cycles of amplification. Mutations were first introduced into the 5′ end of the VS4-encoding region, and the DNA was then digested with DpnI (to remove the methylated, template DNA) and transformed into DH5α cells to repair the nicks in the recombinant DNA. 3′ Mutations were then introduced in the same way, using the first product as a template. Final digestion with DpnI and passage through DH5α cells produced constructs containing AatII restriction sites at each end of the VS4 region [pSTB-ps(VS4) and pSTB-tra(VS4)]. Both constructs were sequenced prior to subcloning.
VS4 domain swapping and deletion.
The mutated clones pSTB-ps(VS4) and pSTB-tra(VS4) were digested with AatII for 2 h at 37°C to yield four fragments: pSTB-ps minus VS4, ps(VS4), pSTB-tra minus VS4, and tra(VS4). The fragments were gel-purified using the QIAEX II gel extraction kit (Qiagen). ps(VS4) was ligated into pSTB-tra minus (VS4), and tra(VS4) was ligated into pSTB-ps minus (VS4). The ligation products were transformed into Novablue cells to isolate mutated constructs pSTB-tra minus VS4 plus ps(VS4) and pSTB-ps minus VS4 plus tra(VS4). Finally, these chimeric constructs and the two deletion constructs, pSTB-ps minus VS4 and pSTB-tr minus VS4, were subcloned into pET22b(+), as previously described. Domain orientation in the chimeric constructs was verified by PCR.
Protein expression and isolation of inclusion bodies (IBs).
Subcloned plasmid constructs isolated from Novablue cells were used to transform BL21 (DE3) cells to express recombinant protein. Conditions for expression were optimized in preliminary studies in each case. For example, for C. trachomatis MOMP, a single colony was inoculated into 5 ml of Luria broth containing ampicillin (100 μg/ml) and the culture was incubated at 37°C with shaking (250 rpm) overnight. This culture was diluted 1:100 into fresh Luria broth and incubated at 37°C with shaking to an optical density of 0.6 at 590 nm. Isopropyl-β-d-thiogalactopyranoside was then added to a final concentration of 1 mM to induce production of the recombinant protein, and the cells were incubated at 37°C for a further 3 h. Cells were harvested by centrifugation, and IBs were prepared as previously described (44). The channel proteins were extracted and solubilized using octylglucoside-dithiothreitol (octylglucoside-DTT), analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (44), and reconstituted into planar lipid bilayers either immediately or after storage at −70°C for up to 4 months.
Planar bilayer reconstitution.
Bilayers were formed by drawing a dispersion of diphytanoyl phosphatidylcholine (Avanti) in n-decane (Sigma) across a 0.3-mm hole in a polystyrene partition separating two solution-filled chambers (35, 36, 38). The contents of both chambers were stirred (using stirbars) as required, and all the bilayers used had a final capacitance of 200 to 300 pF and a conductance of 5 to 10 pS. Spontaneous thinning of the lipid dispersion with bilayer formation was verified by monitoring the increase in membrane capacitance until it stabilized at the expected value (corresponding to a specific capacitance of 0.3 to 0.5 μF/cm2). Initially, both chambers contained 50 mM KCl, 20 mM Tris-HCl (pH 7.4), and 1 mM DTT, and the membrane was voltage-clamped at 0 mV with an Axopatch 200B amplifier (Axon Instruments). Potentials are always quoted as cis minus trans, where the cis chamber corresponds to the compartment to which protein was subsequently added. Channel incorporation was induced by adding MOMP to a final concentration of ≈1 ng/ml in the presence of 500 mM KCl in the cis chamber, with stirring. The membrane potential was repeatedly switched between +60 and –60 mV, and following the appearance of unit transmembrane currents, the cis chamber was extensively perfused with 50 mM KCl–20 mM Tris-HCl (pH 7.4) containing 1 mM DTT to limit the incorporation of additional channels. Thereafter, the salt concentration in each chamber was changed by perfusion or by direct addition of concentrated salt solutions, as required.
Single-channel recording and analysis.
Transmembrane currents were low-pass filtered (2 kHz, 8-pole Bessel response) and recorded. Voltage clamp protocols are described in detail in Results. Recordings were postfiltered and analyzed using pClamp 8 software (Axon) and PSI Plot (Polysoftware International). Positive (upgoing) currents are defined as currents in which positive ions flow cis to trans or (equivalently) negative ions flow trans to cis. The results were analyzed by nonpaired t testing, and levels of significance are given in the appropriate sections in Results.
RESULTS
Cloning and expression of full-length MOMPs.
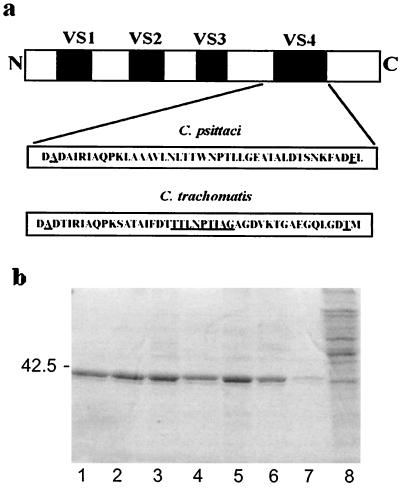
The sequence of the full-length C. trachomatis MOMP cDNA was confirmed to correspond to the published sequence of serovar Da (accession number X62918.1), and that of full-length C. psittaci MOMP corresponded to the full-length version (accession number X51859, Chlamydophila abortus) of the previously cloned ovine abortion strain S36/3 (44). Schematic representations of the full-length MOMPs, together with details of their VS4 regions, are shown in Fig. 1a. Denaturing SDS-PAGE (10% [wt/vol]) analysis of these and all the other recombinant proteins used in this study showed protein bands of the predicted sizes (≈40 kDa) (Fig. 1b).
FIG. 1.
Expression of recombinant chlamydial MOMPs. (a) Schematic representation of the approximate positions and sizes of C. psittaci and C. trachomatis MOMP VS domains. The amino acid sequences (single-letter code) include the residues (underlined) that were mutagenized to valine after AatII sites were introduced into the cDNAs to allow protein engineering. The longer underlined sequence in C. trachomatis VS4 is a species-specific conserved nonapeptide. (b) A 10% (wt/vol) Coomassie blue-stained SDS-PAGE gel of octylglucoside-dithiothreitol extracts containing the recombinant proteins used in this study. Lanes 1 to 8 contain, respectively, 10 μg of full-length C. trachomatis MOMP, C. trachomatis minus VS4, chimeric C. trachomatis MOMP containing the VS4 domain from C. psittaci MOMP, full-length C. psittaci MOMP, C. psittaci MOMP minus, chimeric C. psittaci MOMP containing the VS4 domain from C. trachomatis, uninduced transformed cells, and nontransformed cells. The position of a 42.5-kDa size marker is indicated.
Because of the high cysteine content of chlamydial MOMPs, we maintained the recombinant proteins in reducing conditions (1 mM DTT) throughout this study in order to prevent random inter- or intramolecular disulfide bond formation and protein aggregation during extraction and reconstitution.
Functional reconstitution.
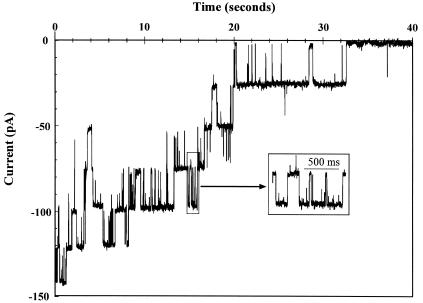
Diphytanoyl phosphatidylcholine formed exceptionally stable bilayers that routinely withstood an imposed membrane potential of ±100 mV for several minutes, and potentials of up to ±150 mV for short periods (≈1 min), without dielectric breakdown. The full-length proteins incorporated spontaneously into bilayers in the presence of high salt concentrations (500 mM) and their channel properties were consistent in over 50 independent experiments. This suggested that during membrane incorporation, the proteins folded correctly and consistently into the same structure. Full-length C. psittaci channels (Fig. 2) were similar in appearance to those previously obtained from nondenatured native MOMP (43) and from a recombinant protein containing an N-terminal 16-residue truncation (44). As detailed previously (44), we never observed channel activity attributable to contaminating Escherichia coli porins.
FIG. 2.
Functional reconstitution of C. psittaci MOMP. Typical ion channel activity obtained from recombinant full-length C. psittaci MOMP incorporated into a planar lipid bilayer. The recording was initiated by switching the membrane holding potential from 0 to −150 mV, with 50 mM KCl on both sides of the membrane. Note the successive closure of six initial unit conductances over time, with little or no activity remaining after 32 s, and the brief closures illustrated in the magnified inset (low-pass filtered at 0.2 kHz). The negative (downwards) currents are consistent with K+ flowing trans to cis and with Cl− flowing cis to trans (see Materials and Methods).
Characteristically, MOMP channels activated rapidly (within 1 ms) on switching the membrane holding potential from 0 to −150 mV and gradually inactivated over the course of ≈30 to 60 s. In addition to the “staircase” effect produced by the shutting down of individual unit conductances (six in all in Fig. 2), open channels also displayed frequent short-lived closures (Fig. 2, inset). Inactivated channels could be reactivated by repolarizing or depolarizing the membrane, and similar activation was seen on switching to positive holding potentials (data not shown). The frequency of short-lived closures at negative potentials was increased compared to that at positive potentials, suggesting that the channels incorporated in a consistent orientation.
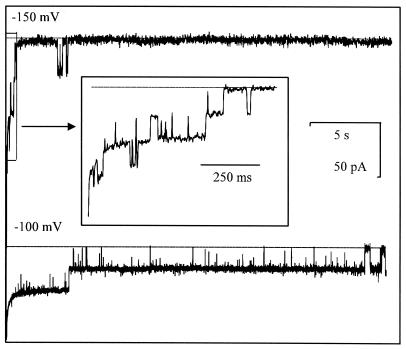
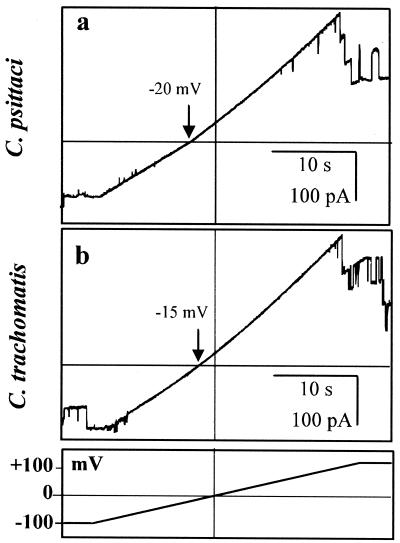
Full-length C. trachomatis MOMP channels behaved in a broadly similar way to C. psittaci MOMP (Fig. 3), except that they inactivated more rapidly at −150 mV (or at +150 mV [data not shown]). This inactivation was slow or incomplete over the course of ≈30 s at lower holding potentials (e.g., −100 mV) (Fig. 3). As previously noted for native C. psittaci MOMP (35), the number of unit conductances counted in independent experiments with both of the recombinant, full-length, wild-type proteins tended to occur in groups of three, six, or occasionally nine.
FIG. 3.
Functional reconstitution of C. trachomatis MOMP. Typical ion channel activity obtained from recombinant full-length C. trachomatis MOMP incorporated into a planar lipid bilayer. The recording was initiated by switching the membrane holding potential from 0 to −150 mV (upper trace) or to −100 mV (lower trace), with 50 mM KCl on both sides of the membrane (filtered at 0.2 kHz). The baseline bilayer currents, with no channels open, are indicated by dotted lines. The upper inset box magnifies the period of ≈1 s immediately after switching the membrane holding potential to −150 mV and illustrates three unit currents superimposed on a typical exponentially decaying bilayer membrane capacitative discharge current.
Channel conductances.
To enable a detailed analysis of the properties of reconstituted channels under reproducible experimental conditions, we used the bilayer voltage-clamp protocol summarized in Fig. 4c. Briefly, we applied a holding potential of 0 mV and then initiated repetitive sweeps consisting of 4 s at −100 mV, a voltage ramp over 32 s from −100 mV to +100 mV, 4 s at +100 mV, and finally a step return to −100 mV. This protocol was suitable for examining channel conductances over a wide range of symmetric ion concentrations, from 50 to 2,000 mM on each side of the bilayer. Repetitive sweeps (three to four) showed that the measurements were highly reproducible for each experiment.
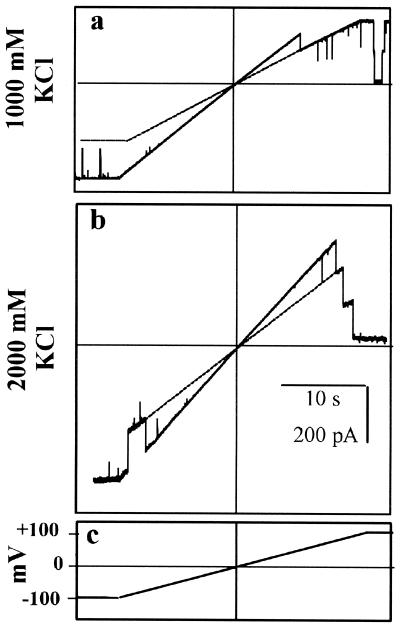
FIG. 4.
Measurement of channel conductances using voltage ramps. C. trachomatis MOMP channels (in the same experiment) were exposed to 1 M KCl (a) or 2 M KCl (b), present in each case on both sides of the incorporated proteins. The membrane potential was varied between −100 mV and +100 mV at a constant rate of 6.25 mV/s, using the voltage clamp protocol summarized in panel c. Each main panel shows three unit conductances, and the dotted lines project the “closed” levels of the top (third) conductance state. Note that all the real and projected lines are linear and intersect at the point of zero current and zero potential (the current and voltage axes are indicated by fine horizontal and vertical lines, respectively). The gradients of the channel traces (pA/V) correspond to the appropriate multiple (1, 2, or 3) of the channel slope conductance (pS).
Figure 4a and b illustrate experiments with reconstituted C. trachomatis MOMP exposed to two different KCl concentrations, 1,000 and 2,000 mM, respectively. Up to three unit conductances (single channels) are open at −100 mV. As the membrane potential is ramped towards 0 mV, the slope of the line corresponds to the total conductance of all the open channels (measured in pS, where 1 pS = 1 pA per V). The crossover point (zero current level), at 0 mV in this case, represents the point at which there is no chemical or electrical driving force for ion flux (even though all three channels are open). Finally, as the potential is ramped on towards +100 mV, the unit conductances tend to close down sequentially. It is important to note that the slopes traced by the individual unit conductances all project to 0 mV.
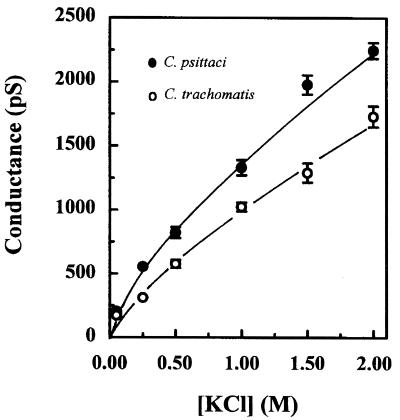
We next carried out experiments to delineate specific functional differences between C. psittaci and C. trachomatis MOMPs. Figure 5 summarizes the KCl concentration dependence of the channel conductances. Both curves are characteristic of porins, with conductances exceeding 1,000 pS at and above 1,000 mM KCl and no evidence of saturation even at 2,000 mM KCl. However, the conductances clearly differ between the two proteins, with that of C. trachomatis MOMP being consistently lower than that of C. psittaci MOMP. The value for C. trachomatis MOMP is ≈20% less than that of C. psittaci MOMP at the highest salt concentration used (1,730 ± 165 pS versus 2,250 ± 125 pS [mean ± standard deviation {SD}, respectively, n = 5). This difference is highly significant (P < 0.001).
FIG. 5.
Concentration dependence of MOMP conductances. The conductances of full-length C. psittaci and C. trachomatis MOMPs were measured under symmetric ionic conditions over a range of KCl concentrations. Each point represents the average value from at least three independent experiments, and bars show ±1 SD calculated from five to eight independent experiments.
Channel selectivity.
Channel selectivities for Cl− or K+ also differed to a measurable extent. To quantify these differences, we measured reversal (equilibrium) potentials with 250 mM KCl in the cis chamber and 50 mM KCl in the trans chamber. Under these conditions, the Nernst equation predicts a reversal potential of plus or minus 38 mV for an ideally selective Cl− or K+ channel, respectively (correcting the ionic concentrations for activities [13]).
We measured reversal potentials under asymmetric ionic conditions using voltage ramps. Figure 6 shows single examples of ramps obtained from bilayers containing either C. psittaci or C. trachomatis MOMP channels. The reversal potentials are indicated. As shown here, current traces often showed a clear break at this crossover point. This represented the opening of an additional channel as the holding potential moved through the point at which the current reversed. The reversal potentials for C. psittaci and C. trachomatis MOMPs were −20.4 ± 1.5 mV (n = 8) and −14.4 ± 0.8 mV (n = 8) for n independent experiments, respectively. Thus, both channels were more selective for the cation than for the anion, but their reversal potentials (i.e., their relative anion versus cation selectivities) differed to a highly significant extent (P < 0.001).
FIG. 6.
Measurement of channel selectivities using voltage ramps. Recombinant MOMPs from C. psittaci (a) and C. trachomatis (b) were subjected to voltage ramps (lower panel) with 250 mM KCl in the cis bilayer chamber and 50 mM KCl in the trans chamber. Positive, upgoing currents at 0 mV (i.e., in the absence of an electrical driving force) indicate a net flux of K+ flowing cis to trans (i.e., channels from both species are more selective for K+ than for Cl−). The reversal (equilibrium) potentials, where negative holding potentials (applied to the cis chamber; see Materials and Methods) exactly balance the chemical driving force, are indicated by arrows.
Domain deletion.
VS4 is the most extensive variable domain in chlamydial MOMPs. It is known to be highly immunogenic and very likely to be surface exposed. Interestingly, the VS4 domain of C. trachomatis MOMP also contains a conserved nonapeptide sequence common to all serovars (see Fig. 1a). Do these VS4 domains form extramembranous protein loops that contribute to the functional differences between C. psittaci and C. trachomatis MOMPs? In order to answer this question, we first inserted unique AatII sites into regions of the MOMP cDNAs flanking the VS4 coding sequences, to facilitate manipulation of the VS4 domains. The resulting point mutations (A277V and T318V for C. trachomatis MOMP and A303V and F343V for C. psittaci MOMP) had no significant effect on channel selectivity or on channel conductances in symmetric 50 or 500 mM KCl (Table 1).
TABLE 1.
Single-channel properties of recombinant chlamydial MOMPsa
| MOMP | Reversal potential (mV) in 250 mM vs. 50 mM KCl | Conductance (pS) in symmetric KCl
|
|
|---|---|---|---|
| 50 mM | 500 mM | ||
| C. psittaci | −20.0 ± 1.5 (8) | 204 ± 19 (8) | 882 ± 88 (5) |
| C. trachomatis | −14.4 ± 0.82 (8)** | 172 ± 19 (8)NS | 575 ± 61 (6)* |
| C. psittaci (A303V/F343V) | −21.7 + 2.4 (6) | 199 ± 14 (7) | 808 ± 76 (6) |
| C. trachomatis (A277V/T318V) | −14.5 + 1.9 (5)** | 174 ± 17 (7)NS | 568 ± 80 (6)** |
| C. psittaci−VS4 | −27.7 ± 4.0 (4) | 184 ± 23 (5) | |
| C. trachomatis−VS4 | −30.0 ± 2.8 (4)NS | 193 ± 14 (4)NS | |
| C. psittaci−VS4 plus C. trachomatis VS4 | −22.1 ± 2.4 (6) | 195 ± 14 (5) | 623 ± 107 (5) |
| C. trachomatis−VS4 plus C. psittaci VS4 | −20.7 ± 2.9 (5)NS | 185 ± 27 (5)NS | 432 ± 47 (6)* |
Selectivity and conductance measurements (means ± SD for n independent experiments [n in parentheses]) for full-length wild-type MOMP channels, channels containing point mutations, deletion constructs (−VS4), and hybrid channels (VS4 domains exchanged as indicated) were carried out. The results of vertical pairwise statistical comparisons are shown (NS, not significant; *, P < 0.01; **, P < 0.001). The differences between the mean reversal potentials for C. trachomatis MOMP containing the C. psittaci VS4 domain and full-length C. trachomatis MOMP, or C. trachomatis MOMP containing point mutations, are also significant (P < 0.001 and P < 0.01, respectively). In addition, the mean reversal potentials for C. psittaci MOMP−VS4 and C. trachomatis MOMP−VS4 differ significantly from those of the corresponding full-length proteins (P < 0.01 and P < 0.001, respectively) and from the corresponding full-length proteins with point mutations (P < 0.02 and P < 0.001, respectively).
MOMPs in which the entire VS4 domain was deleted remained capable of forming functional ion channels (e.g., Fig. 7), although both deletion constructs gave rise to channels that were more K+ selective than channels formed by the full-length recombinant proteins. In each case, reversal potentials measured in 250 versus 50 mM KCl (Table 1) corresponded to a mean K+:Cl− selectivity ratio (calculated as in references 14 and 43) of ≈11:1 (correcting for ionic activities). However, the conductances of the deletion constructs measured in symmetric 50 mM KCl were not increased compared to those of full-length proteins (Table 1). This suggested that the VS4 domain does not form a partially obstructing, pore-confined loop (11, 41).
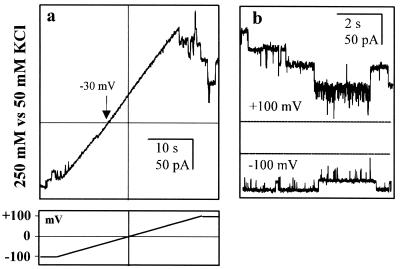
FIG. 7.
Ion channels formed by C. trachomatis MOMP lacking the VS4 domain. (a) With 250 mM KCl cis versus 50 mM KCl trans, the reversal potential for C. trachomatis MOMP lacking the VS4 domain is −30 mV (arrow). (b) Longer recordings at membrane potentials of +100 mV and −100 mV (low-pass filtered at 0.2 kHz, with the zero-current baseline, i.e., no open channels, indicated in each case by a dotted line). The standard voltage-clamp protocol is also shown.
Domain swapping.
Does the VS4 domain influence channel function by forming a nonessential but functionally significant part of the MOMP channel entrance or vestibule? To test this idea, we used the engineered restriction sites in our MOMP cDNAs to interchange the VS4 domain in C. psittaci MOMP (see Fig. 1a) with the VS4 domain of C. trachomatis MOMP. We then reexamined the single-channel properties previously found to be statistically different between the two full-length, recombinant proteins (including proteins containing the point mutations introduced by the restriction sites).
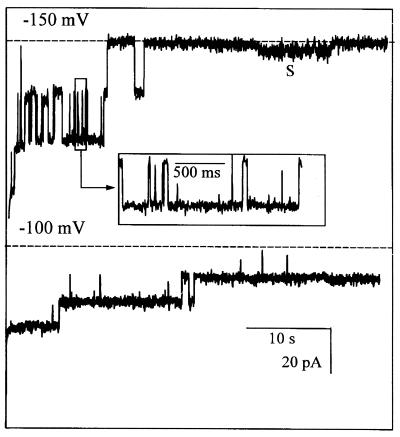
The ion channels formed from the chimeric C. trachomatis protein containing C. psittaci VS4 (Fig. 8) tended to inactivate within ≈20 s at −150 mV but remained active for much longer at −100 mV, similar to the behavior of full-length C. trachomatis MOMP. Strikingly, the mean reversal potential of the chimera in 250 versus 50 mM KCl was indistinguishable from that of full-length C. psittaci MOMP, or C. psittaci MOMP containing point mutations, but was significantly different from that of the corresponding C. trachomatis MOMPs (Table 1). Figure 8 also shows a subconductance level (S) that was not detected in other recordings.
FIG. 8.
Functional reconstitution of C. trachomatis MOMP containing C. psittaci VS4. Typical ion channel activity obtained from recombinant C. trachomatis MOMP containing the C. psittaci VS4 domain in place of C. trachomatis VS4 is shown. As in Fig. 3, the recording was initiated by switching the membrane holding potential from 0 to −150 mV (upper trace) or to −100 mV (lower trace), with 50 mM KCl on both sides of the membrane. The baseline bilayer currents, with no channels open, are indicated by the dotted lines. The upper inset box details gating behavior on an expanded time scale (filtered at 0.2 kHz), and S indicates a region of prominent substate behavior.
The chimeric C. psittaci protein containing C. trachomatis VS4 also formed fully functional ion channels. These tended to behave like full-length C. psittaci channels in their activation and conductance behavior, and their selectivity was indistinguishable from that of full-length C. psittaci MOMP, including C. psittaci MOMP containing point mutations (Table 1). Finally, the conductances of the two chimeras were not significantly different in relatively low concentrations of KCl (symmetric 50 mM KCl), but like the full-length proteins, their conductances differed significantly in symmetric 500 mM KCl (Table 1).
DISCUSSION
MOMPs as porins.
The β-sheet-rich secondary structure and ion channel functions of chlamydial MOMPs (43, 44) strongly resemble the properties of bacterial porins (11, 17, 41). Porin β-barrels have turns and loops connecting transmembrane β-strands, and our general model for MOMP encompasses the idea that some of its extramembranous loops and turns are formed by the variable domains. In this paper, we combined channel engineering and functional analysis by single-channel recording to test the idea that VS4, the largest variable domain, contributes an extramembranous loop. The VS4 domain of C. trachomatis is also of particular interest because it contains a species-specific epitope comprising nine amino acids that is conserved between all the known serovars (28, 34, 35).
When used in tandem with protein mutagenesis and engineering, single-channel recording can provide a powerful assay to test specific structural hypotheses, even in the absence of a crystal structure. A particular advantage of single-molecule studies is that they do not rely on the availability of large amounts of refolded proteins but can proceed at the rate of just one refolded molecule at a time, provided the molecules fold consistently and operate in the same way. The full-length recombinant MOMPs reconstituted in the present study displayed highly reproducible functional properties, similar to purified C. psittaci MOMP channels (43). This strongly suggests that during reconstitution, the proteins fold consistently and adopt a native-like conformation. In addition, the channels appeared to insert into the bilayer in the same orientation, because they retained the same asymmetry in voltage-dependent gating.
The VS4 domain is not essential for pore formation.
Typical of general diffusion porins, the channels formed by full-length C. psittaci and C. trachomatis MOMPs were of high conductance and showed little evidence of saturation, even at KCl concentrations of 2,000 mM. However, we were able to detect significant differences in conductance after increasing the salt concentration to 500 mM. Although both channels were poorly selective, we noted that they were both more selective for K+ than for Cl−, similar to many bacterial porins and to recombinant truncated MOMPs (44). We had previously noted that purified native C. psittaci MOMP was slightly anion selective (43), although it had a conductance similar to that of the recombinant proteins (44). The native preparation may have retained associated proteins or small molecules that bind to the channel and modify its selectivity. By varying the experimental conditions, we also found a difference in anion versus cation selectivity between C. psittaci and C. trachomatis MOMPs, and this was statistically significant in 250 mM versus 50 mM KCl.
To investigate the role of the VS4 domain, we deleted the regions encoding it from the respective genes and expressed and functionally reconstituted the corresponding channels. Both recombinant MOMPs formed functional ion channels. This provides very strong support for the idea that VS4 is not involved in the formation of predicted transmembrane β-strands, because in that case proteins lacking VS4 would be very unlikely to be able to fold into a β-barrel (i.e., a rolled-up β-sheet) (16). In addition, the conductances of the channels in which VS4 was deleted were no higher than those of either the full-length or the chimeric proteins, suggesting that VS4 does not form a pore-confined “eyelet” loop (11). In other porins, eyelet loops substantially limit the flow of ions, and simple geometric calculations suggest that porin β-barrels in which such an obstruction was removed would increase their conductance by at least an order of magnitude (15).
The VS4 domain may contribute to the channel vestibule.
If VS4 domains do not form transmembrane β-strands or pore-confined loops, an alternative role would be to contribute to the formation of the channel entrance or vestibule. In this context, the domain might exert a particularly strong influence on ion selectivity. In particular, the addition of fixed negative charges might increase the local activity of cations and promote the permeation of K+ rather than Cl−. However, despite losing a substantial net negative charge of 2 or 3 U, the MOMP channels in which the VS4 domain was removed became markedly more K+ selective, rather than more Cl− selective.
In a simplistic model, the net loss of negative charge on removing the VS4 domains would be expected to reduce the negative surface potential at or near the pore entrance and reduce rather than enhance cation permeability. Also, transplanting the VS4 domain from C. psittaci to C. trachomatis MOMP made the latter more K+ selective, not less, despite the fact that this actually decreased the net negative charge (though only by one unit). In contrast to their selectivities, the conductances of the hybrid channels continued to show a significant difference (with slightly more variability for the C. psittaci protein containing the C. trachomatis VS4 domain). These findings suggest that the VS4 domain influences selectivity by complex interactions with other parts of the protein, including other loops and turns in the channel vestibule, and its role cannot be considered in isolation from the rest of the protein, with which it must interact. However, it is clear that the structure of this loop is not important for proper assembly of the putative β-barrel.
Conclusions.
The functional reconstitution of wild-type and mutagenized chlamydial MOMPs in planar lipid bilayers provides a new way to probe MOMP structure-function relationships. We have shown that the VS4 domain is not required for pore formation, strongly implying (for a β-barrel model) that it does not contribute to predicted transmembrane β-strands. It does not appear to be a pore-confined loop, and instead our results suggest that it may help to form the channel vestibule, where it interacts closely with other protein loops or turns. The conformations of these surface-exposed regions will be of particular interest when the crystal structures of MOMPs from different species become available.
Our findings may have implications for the design of anti-MOMP vaccines. First, it appears to be likely that the utility of subunit vaccines based on the VS4 domain and, by implication, other VS domains will be limited by the dependence of protein folding on the local protein environment. With the possible exception of further work based on the highly conserved nonapeptide in the C. trachomatis VS4 domain, this suggests that only linear epitopes (rather than conformational epitopes) may be of value. On the other hand, we have shown that MOMPs can be substantially modified yet still remain functional. Organisms in which MOMPs are attenuated could have a valuable role in future vaccination strategies.
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust.
We thank the Chlamydia Genome Project for genomic DNA.
REFERENCES
- 1.Arno J N, Xie C, Jones R B, van der Pol B. Identification of T cells that respond to serovar-specific regions of the Chlamydia trachomatis major outer membrane protein in persons with serovar E infection. J Infect Dis. 1998;178:1713–1718. doi: 10.1086/314478. [DOI] [PubMed] [Google Scholar]
- 2.Baehr W, Zhang Y-X, Joseph T, Su H, Nano F E, Everett K D E, Caldwell H D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci USA. 1988;85:4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour A G, Amano K-I, Hackstadt T, Perry L, Caldwell H D. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J Bacteriol. 1982;151:420–428. doi: 10.1128/jb.151.1.420-428.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bavoil P, Ohlin A, Schachter J. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect Immun. 1984;44:479–485. doi: 10.1128/iai.44.2.479-485.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell H D, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell H D, Schachter J. Antigenic analysis of the major outer membrane protein of Chlamydia spp. Infect Immun. 1982;35:1024–1031. doi: 10.1128/iai.35.3.1024-1031.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:567–659. [PubMed] [Google Scholar]
- 8.Collet R A, Newhall W J, Jersild R A, Jones R B. Detection of surface-exposed epitopes on Chlamydia trachomatis by immune electron microscopy. J Gen Microbiol. 1989;135:85–94. doi: 10.1099/00221287-135-1-85. [DOI] [PubMed] [Google Scholar]
- 9.Conlan J W, Clarke I N, Ward M E. Epitope mapping with solid-phase peptides: identification of type-, subspecies-, species- and genus-reactive antibody binding domains on the major outer membrane protein of Chlamydia trachomatis. Mol Microbiol. 1988;2:673–679. doi: 10.1111/j.1365-2958.1988.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 10.Conlan J W, Kajbaf M, Clarke I N, Chantler S, Ward M E. The major outer membrane protein of Chlamydia trachomatis: critical binding site and conformation determine the specificity of antibody binding to viable Chlamydiae. Mol Microbiol. 1989;3:311–318. doi: 10.1111/j.1365-2958.1989.tb00176.x. [DOI] [PubMed] [Google Scholar]
- 11.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 12.Grayston J T, Campbell L A, Kuo C-C, Mordhorst C H, Saikku P, Thom D H, Wang S-P. A new respiratory tract pathogen: Chlamydia pneumoniae strain TWAR. J Infect Dis. 1990;161:618–625. doi: 10.1093/infdis/161.4.618. [DOI] [PubMed] [Google Scholar]
- 13.Harned H S, Owen B B. The physical chemistry of electrolyte solutions. New York, N.Y: Reinhold; 1957. [Google Scholar]
- 14.Hayman K A, Ashley R H. Structural features of a multisubstate cardiac mitoplast anion channel: inferences from single channel recording. J Membr Biol. 1993;136:191–197. doi: 10.1007/BF02505763. [DOI] [PubMed] [Google Scholar]
- 15.Hille B. Ionic channels of excitable membranes. 2nd ed. Sunderland, Mass: Sinauer; 1994. pp. 294–296. [Google Scholar]
- 16.Huang H, Jeanteur D, Pattus F, Hancock R E W. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol Microbiol. 1995;16:931–941. doi: 10.1111/j.1365-2958.1995.tb02319.x. [DOI] [PubMed] [Google Scholar]
- 17.Jap B K, Walian P J. Biophysics of the stucture and function of porins. Q Rev Biophys. 1990;23:367–403. doi: 10.1017/s003358350000559x. [DOI] [PubMed] [Google Scholar]
- 18.Lampe M F, Wong K G, Kuehl L M, Stamm W E. Chlamydia trachomatis major outer membrane protein variants escape neutralization by both monoclonal antibodies and human immune sera. Infect Immun. 1997;65:317–319. doi: 10.1128/iai.65.1.317-319.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCafferty M C, Herring A J, Andersen A A, Jones G E. Electrophoretic analysis of the major outer membrane protein of Chlamydia psittaci reveals multimers which are recognized by protective monoclonal antibodies. Infect Immun. 1995;63:2387–2389. doi: 10.1128/iai.63.6.2387-2389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newhall W J. Antigenic structure of surface-exposed regions of the major outer membrane protein of Chlamydia trachomatis. Rev Infect Dis. 1988;10:S386–S390. doi: 10.1093/cid/10.supplement_2.s386. [DOI] [PubMed] [Google Scholar]
- 21.Nikaido H. Porins and specific channels of bacterial outer membranes. Mol Microbiol. 1992;6:435–442. doi: 10.1111/j.1365-2958.1992.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz L, Angevine M, Kim S-K, Watkins D, DeMars R. T-cell epitopes in variable segments of Chlamydia trachomatis major outer membrane protein elicit serovar-specific immune responses in infected humans. Infect Immun. 2000;68:1719–1723. doi: 10.1128/iai.68.3.1719-1723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peeling R, Maclean I W, Brunham R C. In vitro neutralization of Chlamydia trachomatis with monoclonal antibody to an epitope on the major outer membrane protein. Infect Immun. 1984;46:484–488. doi: 10.1128/iai.46.2.484-488.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sa S, Souriau A, Bernard F, Salinas J, Rodolakis A. An oligomer of the major outer membrane protein of Chlamydia psittaci is recognized by monoclonal antibodies which protect mice from abortion. Infect Immun. 1995;63:4912–4916. doi: 10.1128/iai.63.12.4912-4916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schachter J. Overview of human diseases. In: Barron A L, editor. Microbiology of Chlamydia. Boca Raton, Fla: CRC Press; 1988. pp. 153–166. [Google Scholar]
- 26.Schachter J. The intracellular life of Chlamydia. Curr Top Microbiol Immunol. 1988;138:109–139. [PubMed] [Google Scholar]
- 27.Stephens S P, Wagar E A, Schoolnik G K. High-resolution mapping of serovar-specific and common antigenic determinants of the major outer membrane protein of Chlamydia trachomatis. J Exp Med. 1988;167:817–831. doi: 10.1084/jem.167.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens R S, Fawaz F S, Kennedy K A, Koshiyama K, Nichols B, Ooij C, Engel J N. Eukaryotic cell uptake of heparin-coated microspheres: a model of host cell invasion by Chlamydia trachomatis. Infect Immun. 2000;68:1080–1085. doi: 10.1128/iai.68.3.1080-1085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens R S, Sanchez-Pescador R, Wagar E A, Inouye C, Urdea M S. Diversity of Chlamydia trachomatis major outer membrane protein genes. J Bacteriol. 1987;169:3879–3885. doi: 10.1128/jb.169.9.3879-3885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens R S, Tam M R, Kuo C C, Nowinski R C. Monoclonal antibodies to Chlamydia trachomatis: antibody specificities and antigen characterization. J Immunol. 1982;128:1083–1089. [PubMed] [Google Scholar]
- 31.Storz J. Overview of chlamydial diseases. In: Barron A L, editor. Microbiology of Chlamydia. Boca Raton, Fla: CRC Press; 1988. pp. 167–192. [Google Scholar]
- 32.Su H, Caldwell H D. Immunogenicity of a chimeric peptide corresponding to T helper and B cell epitopes of the Chlamydia trachomatis major outer membrane protein. J Exp Med. 1992;175:227–235. doi: 10.1084/jem.175.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su H, Caldwell H D. Immunogenicity of a synthetic oligopeptide corresponding to antigenically common T-helper and B cell neutralizing epitopes of the major outer membrane protein of Chlamydia trachomatis. Vaccine. 1993;11:1159–1166. doi: 10.1016/0264-410x(93)90080-h. [DOI] [PubMed] [Google Scholar]
- 34.Su H, Raymond L, Rockey D D, Fischer E, Hackstadt T, Caldwell H D. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc Natl Acad Sci USA. 1996;93:11143–11148. doi: 10.1073/pnas.93.20.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su H, Watkins N G, Zhang Y-X, Caldwell H D. Chlamydia trachomatis-host cell interactions: role of the chlamydial major outer membrane protein as an adhesin. Infect Immun. 1990;58:1017–1025. doi: 10.1128/iai.58.4.1017-1025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su H, Zhang Y-X. Differential effect of trypsin on infectivity of Chlamydia trachomatis: loss of infectivity requires cleavage of major outer membrane protein variable domains II and IV. Infect Immun. 1988;56:2094–2100. doi: 10.1128/iai.56.8.2094-2100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valtonen V V. Infection as a risk factor for infarction and atherosclerosis. Ann Med. 1991;23:539–543. doi: 10.3109/07853899109150515. [DOI] [PubMed] [Google Scholar]
- 38.Villeneuve A, Brossay L, Paradis G, Hebert J. Determination of neutralizing epitopes in variable domains I and IV of the major outer membrane protein from Chlamydia trachomatis serovar K. Microbiology. 1994;140:2481–2487. doi: 10.1099/13500872-140-9-2481. [DOI] [PubMed] [Google Scholar]
- 39.Wald N J, Law M R, Morris J K, Zhou X, Wong Y, Ward M E. Chlamydia pneumoniae infection and mortality from ischaemic heart disease: large prospective study. Br Med J. 2000;321:204–207. doi: 10.1136/bmj.321.7255.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward M E. Chlamydial vaccines—future trends. J Infect. 1992;25:S11–S26. doi: 10.1016/0163-4453(92)91882-c. [DOI] [PubMed] [Google Scholar]
- 41.Weiss M S, Kreusch A, Schiltz E, Nestel V, Wette W, Weckesser J, Schultz G E. The structure of porin from Rhodobacter capsulatus at 1.8 A resolution. FEBS Lett. 1991;280:379–382. doi: 10.1016/0014-5793(91)80336-2. [DOI] [PubMed] [Google Scholar]
- 42.Williams A J. The measurement of the function of ion channels reconstituted into artificial membranes. In: Ashley R H, editor. Ion channels, a practical approach. Oxford, United Kingdom: IRL/OUP; 1995. pp. 43–67. [Google Scholar]
- 43.Wyllie S, Ashley R A, Longbottom D, Herring A J. The major outer membrane protein in Chlamydia psittaci functions as a porin-like ion channel. Infect Immun. 1998;66:5202–5207. doi: 10.1128/iai.66.11.5202-5207.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyllie S, Longbottom D, Herring A J, Ashley R H. Single channel analysis of recombinant major outer membrane protein porins from Chlamydia psittaci and Chlamydia pneumoniae. FEBS Lett. 1999;445:192–196. doi: 10.1016/s0014-5793(99)00121-0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y-U, Stewart S J, Caldwell H D. Protective monoclonal antibodies to Chlamydia trachomatis serovar- and serogroup-specific major outer membrane protein determinants. Infect Immun. 1989;57:636–638. doi: 10.1128/iai.57.2.636-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y-X, Stewart S, Joseph T, Taylor H R, Caldwell H D. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J Immunol. 1987;138:575–581. [PubMed] [Google Scholar]
- 47.Zhong G, Reid R E, Brunham R C. Mapping antigenic sites on the major outer membrane protein of Chlamydia trachomatis with synthetic peptides. Infect Immun. 1990;58:1450–1455. doi: 10.1128/iai.58.5.1450-1455.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]