Abstract
The number of biomedical articles published is increasing rapidly over the years. Currently there are about 30 million articles in PubMed and over 25 million mentions in Medline. Among these fundamentals, Biomedical Named Entity Recognition (BioNER) and Biomedical Relation Extraction (BioRE) are the most essential in analysing the literature. In the biomedical domain, Knowledge Graph is used to visualize the relationships between various entities such as proteins, chemicals and diseases. Scientific publications have increased dramatically as a result of the search for treatments and potential cures for the new Coronavirus, but efficiently analysing, integrating, and utilising related sources of information remains a difficulty. In order to effectively combat the disease during pandemics like COVID-19, literature must be used quickly and effectively. In this paper, we introduced a fully automated framework consists of BERT-BiLSTM, Knowledge graph, and Representation Learning model to extract the top diseases, chemicals, and proteins related to COVID-19 from the literature. The proposed framework uses Named Entity Recognition models for disease recognition, chemical recognition, and protein recognition. Then the system uses the Chemical - Disease Relation Extraction and Chemical - Protein Relation Extraction models. And the system extracts the entities and relations from the CORD-19 dataset using the models. The system then creates a Knowledge Graph for the extracted relations and entities. The system performs Representation Learning on this KG to get the embeddings of all entities and get the top related diseases, chemicals, and proteins with respect to COVID-19.
Keywords: Biomedical Named Entity Recognition (BioNER), Relation Extraction (RE), Knowledge graph, Representation learning, BERT, BiLSTM
Graphical Abstract

1. Introduction
Biomedical Named Entity Recognition (BioNER) and Biomedical Relation Extraction (BioRE) are becoming more and more crucial for biomedical research due to the rising volume of data that has been available in the form of unstructured text articles. There are currently approximately 25 million references in Medline. Even in more specialised subjects, it is challenging to keep up with the literature at this rate (Gajendran et al., 2020). Graphs are useful tools for numerous practical applications. They have been applied to the classification of nodes and development of recommendation systems in social network mining. They have also been utilised in natural language processing to decipher basic queries and offer responses using relational data (Chen et al., 2020). Graphs have been employed in the biomedical field to undertake medication repurposing, detect drug-target interactions, and select drugs pertinent to disease (Doğan et al., 2014, Chen et al., 2020).
There are numerous approaches to build knowledge graphs using resources like text or pre-existing databases (Wang et al., 2020). Knowledge graphs are frequently built from pre-existing databases. Experts in the relevant fields create these databases using methods that range from text mining to manual curation. Manual curation requires subject-matter specialists to depth domain knowledge to claim the association between the entities. Automated methods use NLP methods to quickly identify relevant association. Rule-based systems, unsupervised, and supervised methods are the three classes into which the automated systems are divided.
The proposed system considers the Automated way of extracting knowledge graphs from text using Deep Learning and NLP techniques. The advantages of this approach include quick results and not needing ground truth information. Since its breakout, COVID-19 has been a worldwide pandemic with a high transmission rate and a substantial mortality rate that has affected millions of individuals (Gajendran et al., 2020). Scientific publications on the new Coronavirus have increased exponentially as researchers look for treatments and potential cures, but collecting, integrating, and utilising related sources of information effectively remains a challenge (Wang et al., 2020).
Scientific publications regarding COVID-19 contain various data about related diseases, proteins, chemicals and so on. The data in such publications are vastly unstructured. Most of the articles published under the title COVID-19 are gathered under the name of CORD-19. The research presents an Information Extraction (IE) method followed by Knowledge Graph building in a fully automated generic pipeline.
The proposed system creates Named Entity Recognition model using BERT-BiLSTM-CRF. Then this model is trained on multiple datasets and multiple models are generated. Then these models are used to recognize diseases, proteins, and chemicals in the prediction dataset. The Relation Extraction models are created using SciBERT with linear classifier. Then this model is trained on multiple datasets and multiple models are created. Then these models are used to extract relations such as Chemical – Protein relation and Chemical – Disease relation. Once the entities and relations are extracted from the prediction dataset, the system create the Knowledge graph with the entities as nodes and relations as edges.
2. Related work
The literature review contains all the related works relevant to the system in use such as BioNER, RE, Knowledge Graph and Representation Learning.
2.1. Named entity recognition
Identification of biomedical examples such as chemical substances, genes, proteins, viruses, diseases, DNAs, and RNAs is the goal of biomedical named entity recognition (BNER) (Gajendran and Manjula, 2020). The procedures that would be utilised to extract these things are the main issue for BNER. Dictionary-based techniques detect and extract the biomedical entities by using extensive dictionary. The interaction extraction model Polysearch (Cheng et al., 2008) provides a well-known illustration of the system. Whatizit (Rebholz-Schuhmann, 2013), an online model with distinct categories for different BioNER, is another illustration (Yoshua et al., 2003).
Machine learning techniques are now the most popular ones for named entity recognition. Support vector machines (Kazama et al., 2002), Hidden Markov models (Shen et al., 2003), Decision trees, and Naive Bayesian approaches (Nobata et al., 1999;) were the first supervised machine learning techniques to be utilised. The groundbreaking study by (Lafferty et al., 2001) on Conditional Random Fields (CRF) that considered the likelihood of word spatial interdependence redirected the researchers in a different perspective (Peters et al., 2018).
The literature has shifted toward universal deep neural network models over the past five years. For instance, different variations of convolution neural networks (CNN) and recurrent neural networks (RNN)(Kim et al., 2019) have all been applied to BioNER systems (Zhu et al., 2017). For instance, LSTM, Bi-LSTM (Yanran et al., 2015, Gajendran et al., 2020) are common RNN variations.
Bi-LSTM and CRFs models are paired with a combination of different types of embeddings in a network to produce the best results. (Gajendran et al., 2020; Giorgi et al., 2019; Luo et al., 2018; Habibi et al., 2017) Here, word embeddings are generated via a pre-trained lookup table, while character-level embeddings are generated by independent Bi-LSTMs for each word sequence (Habibi et al., 2017). These two results are then concatenated to form the word representations x1, x2,., xn. Currently, BioNER is being fine-tuned for Transformer-based models like SciBERT and BioBERT(Harnoune et al., 2021, Beltagy et al., 2019).
2.2. Relation extraction
Most of the literatures for identifying the association between the entities falls into rule based and machine learning based models. A rule-based approach heavily relies on sentence syntactic and semantic analysis for relationship extraction. In Fundel et al. (2006), for instance used the tree based structure to identify and extract the association between the entities by splitting the sentence into nouns and verbs. The nouns denotes the different entities and the verbs denotes the association between the entities.
The most popular machine learning techniques use training data from an structured dataset with labels (supervised learning). Obtaining labelled training and testing data used to be the largest barrier to implementing such machine learning algorithms for relation identification. However, this issue has been greatly mitigated by data sets produced by biomedical text mining competitions like BioCreative and BioNLP. one of the first research projects that use an SVM. The latter study, in contrast, employed a comparable SVM model to determine the polarity of food-disease relationships. According to Jensen et al. (2014), term frequency-inverse document frequency (TF-IDF) attributes were utilised to discover food-phytochemical and food-disease relationships. Whereas in (Quan et al., 2014), a CRF was employed for both entity recognition and association extraction.
Deep learning (DL) methods have gained popularity for identifying the association between the entities in recent times because to their cutting-edge performance and reduced requirement for intricate feature processing(Percha et al., 2012). Different types of neural networks namely CNN and RNN, and combinations of CNN and RNN are the three most used DL techniques Jettakul et al. (2019). The use of different embeddings such as position, character and word level information encoded as vectors (Zeng et al., 2014) may all be feature inputs for DL models. Recently, models based on Transformers, including BioBERT and SciBERT, have been employed(Devlin et al., 2019).
2.3. Knowledge graph and representation learning
The knowledge graphs (KG) extracts information from medication, disease, and gene databases as well as their relationships to form a low dimensional structure (Zheng et al., 2020). Its advantage is that a large knowledge graph is obtained. But the data is highly generic and only takes structured data as input. The evidence text was manually encoded as a triple using the COVID-19 Knowledge Graph (source-relation-target) (Domingo-Fernandez et al., 2020). Its advantage is that extracted information is mostly correct apart from Human errors. But only a small knowledge graph is obtained and manual curation is time-consuming.
In Repke and Krestel (2021), they extracted the knowledge graphs from financial text corpus using NER and RE tasks. Its advantage is that finding financial entities is comparatively easier with rule-based approaches. In Deep Learning-based Knowledge Graph Generation for COVID-19 (Kim et al., 2021), they find entities related to COVID-19 from dictionaries and extract their relations from text corpus. Its advantage is that the Unsupervised method is devised to find entities and relations. But results vary massively in unsupervised methods.
The KG also struggles with the critical issue of information being sparse that severely impairs the ability to calculate association between items. Knowledge graph embedding, which has the ability to provide dense and low-dimensional feature space and aids in effectively calculating the semantic links between items with low computational quality, has been designed to address this issue. Translation based models is based on the idea that a triple (head, relation, tail) can be represented as a geometric principle such as h + r ≈ t (TransE) (Ji et al., 2015). Tensor Factorization-Based Models is based on the idea that all triples can be transformed to 3D binary tensor and this tensor is converted into entities and relations embeddings using Dimensionality Reduction.
Some of the problems that we found in the analysis of literature survey are Manual curation of entities and relations takes very long time and the results count will be low, Lack of ground truth information for correct and easy evaluation which results in lower quality knowledge graphs, Rule based information extraction requires manual rule setting which differs based on the information we need, usage of external knowledge base which may not be available for the concerned task and lack of structured data which will be easier to extract.
3. Proposed system
Fig. 1 displays the proposed system's overall architecture diagram. Preprocessing, Feature extraction, Named Entity Recognition, Relation Extraction, Graph construction, and Representation learning are the six modules that make up the proposed work and produces the COVID-19 knowledge graph as the outcome. In the Preprocessing module, the CORD-19 abstracts are taken and is transformed to input to the Language Model and the BERT model is finetuned to get better results for entities extraction. In the Feature extraction module, the Named Entity Recognition datasets are passed through to the BERT model to get the features from these datasets. The BERT-BiLSTM-CRF model is utilised in the Named Entity Recognition module to train on NER datasets and to predict on CORD-19. The Relation Extraction module trains the BERT model with the help of the Relations datasets, and then uses the trained models to determine whether a relation between the extracted elements from the CORD-19 exists. In the Graph Construction module, the Knowledge graph is constructed. In the Representation Learning module, the KG is used to train the TransD model to find embeddings of entities which are used to find similarity with COVID-19. Figure 3.1 shows the Complete Architecture diagram.
Fig. 1.
Overall architecture diagram.
3.1. Named entity recognition
Each sentence is tokenized using the custom tokenizer created during preprocessing. And, the sentence's special tokens [CLS] and [SEP] are added at the beginning and end, respectively. The tokenizer basically tokenizes common words as individual tokens and more rare words into meaningful sub tokens. Then the sentences are truncated or padded to accommodate the max length (256) for the BERT model. Then the mask is defined such that only words are marked as 1 and padded words are marked otherwise.
Finally, the individual sentence of size (256) is passed to the fine-tuned CORD-19 SciBERT model and the last hidden layer output is taken from the BERT model which is of the dimension 768. The output produced by the BERT model is of the shape (1,256,768). Here the 1 denotes the number of input sentences. The 256 denotes the sequence length. Each subword in the sentence is represented by a 768-dimensional vector that depends both on the subword and the words around it. Hence this type of embeddings is called Contextualized word embedding. This output is passed onto the Named Entity Recognition layers which detect the entities based on the word embedding of words in the sentence. The module takes the finetuned CORD-19 SciBERT model as the input which is used to produce the feature embeddings. The module returns the contextualized word embeddings of the input dataset regardless of whether it is a NER dataset or CORD-19 dataset.
The individual sentence features such as input_ids, attention_mask, token_type_ids are taken and are passed to the CORD-19 SciBERT. The contextualised word embedding from each token in the phrase is then extracted from the final hidden layer, which is size 768, and transferred to the bidirectional LSTM layer. The output is size 1024. (512 for forward LSTM and 512 for backward LSTM). To prevent overfitting, a different dropout values are experimented during the initial trails and finally a dropout regularisation of 30% is used since it produced better performance. The following layer is fully linked, taking an input of 1024 bytes and producing a vector with a size equal to the total number of labels in the training dataset. Then the vector is given to the Conditional Random Field (CRF) layer which learns the transitional probabilities from the input dataset and finds the best possible tag sequence for the sentence. The model uses Negative log-likelihood loss to optimise the model. It is modelled as a minimization problem.
Now there are 3 models, which are capable of finding diseases, drugs and proteins respectively. Now the Processed CORD-19 is fed into each model and the entity tags of CORD-19 dataset are found. The tags are for each word token that are encoded in BIO Scheme. The terms B-Entity, I-Entity, and O-Entity here all relate to the entity's beginning, inside, and outside, respectively. Table 1 shows sample encoding of a sentence in BIO scheme. These Tags are combined so that the final output contains CORD-19 dataset with all the diseases, chemicals and proteins mentioned.
Table 1.
Example encoding of a sentence in BIO scheme.
| Token | Encoding in BIO |
|---|---|
| Management | O |
| of | O |
| critically | O |
| ill | O |
| patients | O |
| with | O |
| Severe | B-Disease |
| Acute | I-Disease |
| Respiratory | I-Disease |
| Syndrome | I-Disease |
| . | O |
The system takes the contextualized word embeddings of the NER training datasets and CORD-19 dataset as input. Then the system returns the extracted diseases, chemicals and proteins from the CORD-19 dataset as output. The pseudocode for the BERT-BiLSTM-CRF model and extraction of biomedical entities from the CORD-19 dataset.
3.2. Relation extraction module
Fig. 2 shows the complete design diagram of the Biomedical Relation Extraction (BioRE) module. Here, BC5CDR dataset is used for extraction of chemical induced disease relations. CHEMPROT dataset is used for extraction of chemical-protein relations. The 2 datasets are preprocessed where the tokens are associated with its entities, and each sentence in the dataset is modified into the 2 sentences where the first sentence contains the two entities whose relation is in question separated by a space and the second sentence contains the actual sentence from the dataset.
Fig. 2.
Relation extraction module design diagram.
Example transformation,
Epidermal growth factor receptor gefitinib. Epidermal growth factor receptor inhibitors currently under investigation include the small molecules gefitinib (Iressa, ZD1839) and erlotinib (Tarceva, OSI-774), as well as monoclonal antibodies such as cetuximab (IMC-225, Erbitux).
The input ids, attention mask and token types of the input sentence/text is taken and is passed into the SciBERT. The token embedding of the special token “[CLS]” is used for classification of the sentence/text to one of the relations. The 768-dimensional vector of “[CLS]” is then passed into the fully connected layer which takes this as input and produces output of size equal to the total number of types of relations in the dataset. AdamW optimizer is used to compile the model.
The BC5CDR dataset is fed into the SciBERT model which produces the Chemical Induced Disease Relation. The CHEMPROT dataset is fed into an individual SciBERT model which will be finetuned for finding relations between Chemicals and Proteins. Now the CORD-19 dataset with entity mentions is preprocessed where only the sentences with two or more entities are forwarded into the model. The cell line, cell type entities are not considered for this count. And based on the type of entities, the sentence is fed to one of two models and the model predicts whether a relation exists between two entities or not. Finally, the two models’ outputs are combined and tuples are generated of the form (Entity1, Entity2, Relation).
The system takes the relation datasets as input to train the SciBERT models. The system returns the relations between entities extracted from the CORD-19 dataset. The pseudocode for the RE training and relation extraction from CORD-19.
3.3. Graph construction
The Graph construction process takes the relation types in which each tuple is of the type (“Entity_1, Relation, Entity_2″). Then from the tuples the occurrence count of all the entities is found. Then only the tuples with either of the entities having occurrence count greater than 5 are considered for the next step. This helps in reducing the noise in the resultant Knowledge Graph.
Then a Neo4j Graph Database instance is created and all the entities from the filtered tuples are created as nodes with names and their types. Then the relations are created as directed edges from Chemical-to-Disease for Chemical Induced Disease relation and Chemical-to-Protein for Chemical Protein relation.
Mathematically, the Knowledge Graph is defined as follows,
E: a collection of nodes for diseases, proteins, and chemicals.
R: a group of labels denoting the relationships between chemicals and diseases and proteins.
G ⊆ E x R x E: a collection of connections between entity pair represented by edges.
The model takes the relation tuples extracted from the CORD-19 dataset as input. The model returns the Knowledge Graph of COVID-19 which is stored in a Neo4j Graph Database. The pseudocode for the graph construction is given in Algorithm 5.
Algorithm 5
Graph construction.
.
3.4. Representation learning module
Initially the COVID-19 Knowledge Graph is converted into triplets (head entity, relation, tail entity). Since the Knowledge Graph contains only positive relations, negative relations are generated by taking two random entities and if there is no relation between them, then these two entities are taken as negative relation.
Then these triplets are passed to train the TransD model. Each symbol (entities, relations) in the TransD model is represented by two vectors. The first one defines what an entity or relation is, while the second one is utilised to create mapping matrices. Once the TransD model is trained, the embeddings of all entities and relations are taken from the model parameters. Then only the embeddings of entities whose edges count is greater than or equal to 5 are taken are taken for comparison. Then the COVID-19 embedding is taken from the model, and is compared with all the entities from above using cosine similarity score. Then the top 25 diseases / chemicals / proteins with highest cosine similarity score is taken and is produced as output.
The model takes the COVID-19 Knowledge Graph as input in the triplet’s format. The model returns the list of top 25 diseases / chemicals / proteins related to COVID-19 along with their cosine similarity score. The pseudocode for the representation learning is given in Algorithm 6.
Algorithm 6
Representation learning.
.
4. Experimental setup
The description of each dataset utilised in the system, the hyperparameters used for training, and the evaluation metrics used to judge the suggested system are all included in this part. Our proposed method is developed in Python and makes use of the free and open-source Keras network package. With a focus on facilitating quick experimentation, Keras is capable of running on top of Theano [54], Tensorflow, and Deeplearning4j. The model is trained using the hyperparameters with the batch size of 100, entities dimension size of 400, loss function as MarginLoss, and total epochs of 700. Then the embeddings of entities are taken and only the entities with edges greater than 5 are considered and compared with coronavirus. And the diseases, chemicals and proteins with the highest cosine similarity score with coronavirus are considered and are produced as final result.
4.1. Dataset
4.1.1. NCBI-disease
To act as a research tool for the community of biomedical natural language processing, the NCBI disease corpus is completely annotated at the mention and idea levels (Doğan et al., 2014). Annotations are checked for corpus-wide consistency of annotations, and two annotators are allocated to each document (paired at random). The properties of the dataset are displayed in Table 2. The tags B-Disease, I-Disease, and O are accessible.
Table 2.
NCBI Disease dataset characteristics.
| Dataset Statistics | Dataset for Training | Dataset for Development | Dataset for Testing | Total Dataset |
|---|---|---|---|---|
| PubMed References | 593 | 100 | 100 | 793 |
| Total disease entities | 5145 | 787 | 960 | 6892 |
| Specific disease entities | 1710 | 368 | 427 | 2136 |
| Specific concept ID | 670 | 176 | 203 | 790 |
4.1.2. CHEMDNER
The CHEMDNER corpus' abstracts were chosen to be representative of all significant chemical specialties. Each mention of a chemical entity was manually categorised into one of the following classes: abbreviation, family, formula, identifier, multiple, systematic, and trivial(Krallinger et al., 2015). The properties of the dataset are displayed in Table 3. The tags B-Chemical, I-Chemical, and O are accessible.
Table 3.
CHEMDNER dataset characteristics.
| Dataset Statistics | Dataset for Training | Dataset for Development | Dataset for Testing | Total Dataset |
|---|---|---|---|---|
| Abstracts | 3500 | 3500 | 3000 | 10,000 |
| No. of Characters | 4,883,753 | 4,864,558 | 4,199,068 | 13,947,379 |
| No. of Tokens | 770,855 | 766,331 | 662,571 | 2,199,757 |
| Abstracts with classes | 2916 | 2907 | 2478 | 8301 |
| No. of Mentions | 29,478 | 29,526 | 25,351 | 84,355 |
| No. of Chemicals | 8520 | 8677 | 7563 | 19,805 |
| No. of Journals | 193 | 188 | 188 | 203 |
4.1.3. JNLPBA
The information was taken from the GENIA 3.02 corpus (Kim et al., 2003). A controlled search on MEDLINE using the MeSH terms human, blood cells, and transcription factors produced this result. The properties of the dataset are shown in Table 4. B-Protein, I-Protein, B-DNA, I-DNA, B-RNA, I-RNA, B-cell line, I-cell line, B-cell type, I-cell type, and O are the accessible tags.
Table 4.
JNLPBA dataset characteristics.
| Corpus Characteristics | Training Set | Test Set | Whole corpus |
|---|---|---|---|
| Abstracts | 2000 | 404 | 2404 |
| Sentences | 20,546 | 4260 | 24,806 |
| Words | 472,006 | 96,780 | 568,786 |
| Entities | 51,291 | 8662 | 59,953 |
4.1.4. BC5CDR
Due to their essential roles in numerous fields of biological research and healthcare, such as drug development and safety surveillance, chemicals, diseases, and their connections are among the most frequently searched subjects by PubMed users worldwide (1–3). Although creating therapeutic compounds is the ultimate goal of drug development, other objectives include identifying harmful drug interactions between substances and illnesses. The properties of the dataset are shown in Table 5. Chemical-Induced Disease is the only connection that can be made.
Table 5.
BC5CDR dataset characteristics.
| Corpus Characteristics | Training Set | Testing Set | Whole Corpus |
|---|---|---|---|
| No. of Chosen Abstracts | 1000 | 500 | 1500 |
| No. of Chemical Mentions | 10,550 | 5385 | 15,935 |
| Chemical Unique Mentions | 2973 | 1435 | 4408 |
| No. of Disease Mentions | 8426 | 4424 | 12,850 |
| Disease Unique Mentions | 3829 | 1988 | 5817 |
| Chemical Induced Disease Relations | 2050 | 1066 | 3116 |
4.1.5. CHEMPROT
The CHEMPROT corpus is a manually annotated corpus where domain experts have thoroughly categorised all references to chemicals and genes as well as any binary interactions between them that conform to a certain set of physiologically relevant relation types (CHEMPROT relation classes). The CHEMPROT track aims to encourage the creation of systems that can extract chemical-protein interactions relevant to drug discovery, precision medicine, and fundamental biomedical research. The CHEMPROT dataset's available relations are displayed in Table 6, and the dataset's corpus features are displayed in Table 7.
Table 6.
Available relations in CHEMPROT dataset.
| Group | CHEMPROT relations belonging to this group |
|---|---|
| CPR:1 | PART_OF |
| CPR:2 | REGULATOR|DIRECT_REGULATOR|INDIRECT_REGULATOR |
| CPR:3 | UPREGULATOR|ACTIVATOR|INDIRECT_UPREGULATOR |
| CPR:4 | DOWNREGULATOR|INHIBITOR|INDIRECT_DOWNREGULATOR |
| CPR:5 | AGONIST|AGONIST-ACTIVATOR|AGONIST-INHIBITOR |
| CPR:6 | ANTAGONIST |
| CPR:7 | MODULATOR|MODULATOR-REGULATOR|MODULATOR-INHIBITOR |
| CPR:8 | COFACTOR |
| CPR:9 | SUBSTRATE|PRODUCT_OF|SUBSTRATE_PRODUCT_OF |
| CPR:10 | NOT |
Table 7.
CHEMPROT dataset characteristics.
| Dataset Statistics | Dataset for Training | Dataset for Development | Dataset for Testing | Total Dataset |
|---|---|---|---|---|
| Document | 1020 | 612 | 800 | 2432 |
| Chemical | 13,017 | 8004 | 10,810 | 31,831 |
| Protein | 12,752 | 7567 | 10,019 | 30,338 |
| Positive Relation | 4157 | 2416 | 3458 | 10,031 |
| Positive relation in one sentence | 4122 | 2412 | 3444 | 9978 |
4.1.6. CORD-19
The COVID-19 Open Scientific Dataset was created in response to the COVID-19 epidemic by the White House and a consortium of top research organisations (CORD-19). The CORD-19 database contains more than 1000,000 research publications about COVID-19, SARS-CoV-2, and similar coronaviruses, including more than 350,000 full-text articles. The entire research community is given access to this freely available information so they can use cutting-edge AI approaches to produce fresh insights to aid in the ongoing battle against this contagious disease. The properties of the dataset are displayed in Table 8.
Table 8.
CORD-19 dataset characteristics.
| Subfield | Count | % Of Corpus |
|---|---|---|
| Virology | 20,116 | 42.3% |
| Immunology | 9875 | 20.7% |
| Molecular biology | 6040 | 12.7% |
| Genetics | 3783 | 8.0% |
| Intensive care medicine | 3204 | 6.7% |
| Other | 4595 | 9.6% |
4.2. Hyperparameters
For Named Entity Recognition, by training the BERT-BiLSTM-CRF model using various hyperparameters the best possible hyperparameters for the model are found which differs for the NCBI-Disease, CHEMDNER and JNLPBA dataset. Table 9 shows the Hyperparameters used for different datasets.
Table 9.
Hyperparameters for Different NER datasets.
| Dataset | No. of Labels | Epochs | Sequence Length |
|---|---|---|---|
| NCBI-Disease | 3 | 25 | 256 |
| CHEMDNER | 3 | 25 | 256 |
| JNLPBA | 11 | 32 | 256 |
For Relation Extraction, by training the SciBERT model using various hyperparameters the best possible hyperparameters for model are found which differs for the BC5CDR and CHEMPROT dataset. Table 10 shows the Hyperparameters used for different datasets.
Table 10.
Hyperparameters for Relation Extraction.
| Dataset | No. of Relations | Sequence Length | Epoch |
|---|---|---|---|
| BC5CDR | 2 | 512 | 32 |
| CHEMPROT | 9 | 512 | 39 |
4.3. Performance metrics
Here, whether or not the complete sentence is accurately detected determines the metrics of precision (P), recall (R), and F1. The following are the evaluation measures utilised in the NER and RE domains:
| (1) |
| (2) |
| (3) |
Where TP, FP and FN represents the True Positive, False Positive and False Negative respectively.
5. Results and discussion
5.1. Named entity recognition
For evaluation of the proposed systems, the NCBI-Disease dataset is considered. The proposed system is BERT-BiLSTM-CRF for Named Entity Recognition. Table 11 shows the comparison of different variants of proposed system with BiLSTM, LSTM-CRF, BiLSTM-CRF and SciBERT. As shown in the below table, the BERT-BiLSTM-CRF model has better Precision and F1 value than standard SciBERT. The BERT-BiLSTM-CRF model is used to evaluate the other datasets and the unstructured CORD-19 dataset.
Table 11.
Comparison with variants of Proposed System.
| Model | Precision | Recall | F1 |
|---|---|---|---|
| BiLSTM + Word Embedding | 84.87 | 74.11 | 79.13 |
| LSTM-CRF + Word and Char Embedding | 85.20 | 82.40 | 83.80 |
| BiLSTM-CRF + Word Embedding | 86.75 | 87.11 | 86.93 |
| SciBERT | 85.47 | 90.10 | 87.73 |
| BERT-BiLSTM-CRF (Proposed System) | 88.49 | 89.02 | 88.76 |
Initially, the CORD-19 SciBERT model is taken and the individual sentence’s features such as input ids, attention mask, token types are fed into the BERT layer and the final hidden layer of the BERT model is taken as output. The output is of size (sentence_length[256], 768). Fig. 3 shows the BERT layer output.
Fig. 3.
BERT layer output.
Then the output of the BERT layer is fed into the BiLSTM layer. It takes vectors of size 768 and output vectors of size 1024 for each word in the sentence. Since it is a bidirectional LSTM, 512 of the output is from the forward LSTM and other 512 is from the backward LSTM. (As shown in Fig. 4).
Fig. 4.
BiLSTM layer output.
The output from the LSTM layer is then given to the dropout layer where the dropout value of 0.3 is used to prevent the problem of overfitting. The output of the dropout layer is then transferred to the fully connected layer, which transforms each word's 1024 input vector into a vector with the same size as the number of label types in the NER dataset.
Then the output from the fully connected layer for each word is passed to the Conditional Random Field (CRF) layer which finds the negative log-likelihood loss and provides it as output. This loss is negated and is used as the loss function for the model and is back propagated.
Multiple optimizers are tried and the optimizer that is finalised is Stochastic Gradient Descent (SGD). The system uses momentum to avoid oscillations and to converge faster than traditional gradient descent algorithms. The individual sentence features are passed to the three models and based on the predicted tags the entities are extracted by matching the tags with the words. After running the procedure for all the sentences, the extracted entities are stored in a MongoDB database for future reference. Table 12 shows the results obtained for different datasets for biomedical named entity recognition. And Table 13 shows the complete statistics of the entities found from CORD-19 based on the training.
Table 12.
Named Entity Recognition evaluation metrics.
| Dataset | Precision | Recall | F1 |
|---|---|---|---|
| NCBI-Disease | 88.49 | 89.02 | 88.76 |
| JNLPBA | 71.25 | 81.5 | 76.06 |
| CHEMDNER | 90.88 | 92.25 | 91.56 |
Table 13.
Overall entities extracted from CORD-19.
| Entity Type | Total no. of instances | Unique no. of instances |
|---|---|---|
| Disease | 1,00,441 | 17,672 |
| Chemical | 66,212 | 7841 |
| Protein | 3,66,963 | 1,29,972 |
In this section, we are going to compare the results of the proposed system for the three gold standard datasets with the other existing systems. The comparison of the results of the proposed system with the existing system is shown in Table 14. In general, the proposed framework performed poorly in JNLPBA corpus. The model can readily reach the local optimal during JNLPBA training, and investigation has not revealed a technique to enhance the effect. Except JNLPBA, the proposed framework performed marginally good when comparing with the other existing models. Hierarchical shared learning model achieved more than single task model through further improvement with multi task models and exceeded the proposed framework in CHEMDNER dataset. Hunflair model is better than the hierarchical shared learning model in NCBI disease. However, the proposed framework outperformed all the models including the multi task models and massively pretrained models in NCBI disease dataset. One may claim that prior knowledge training in biological disciplines can greatly enhance entity recognition and entity identification of genes and proteins. But our proposed framework performed marginally well in all the datasets without any massive pretraining in medical data.
Table 14.
NER model performance of F1 score with existing systems.
| NCBI | CHEMDNER | JNLPBA | |
|---|---|---|---|
| SOTA | 88.60 | 91.14 | 77.39 |
| Yoon et. al (Yoon et al., 2019) | 84.69 | 88.19 | 77.39 |
| Bert (Lee et al., 2020) | 85.63 | 90.04 | 74.94 |
| Hunflair (Weber et al., 2019) | 88.26 | - | 77.60 |
| Chai et. al (Chai et al., 2022) | 88.26 | 91.70 | - |
| Proposed System | 88.76 | 91.56 | 76.06 |
We list the scores of the state-of-the-art (SOTA) models on different datasets as follows: scores of Xu et al. (2019) on NCBI Disease, scores of Luo et al. (2018) on CHEMDNER, and scores of Yoon et al. (2019) on JNLPBA.
We define a new experiment for testing speed and accuracy metrics of our implementation with the default Keras framework while maintaining the hyper parameters constant across all datasets because the experiment in the previous section involved hyper parameter tuning. Additionally, we employ the same biomedical datasets and contextual word embeddings indicated above to ensure that the studies can be replicated. We have used learning rate as 0.01, LSTM state size as 300, batch size as 32, 10 epochs, and SGD optimizer as the hyperparameters for testing the speed.
Results shows that the CHEMDNER dataset takes more time comparing with the other biomedical datasets. The possible reason for the CHEMNDER dataset taking more time could be the size of the dataset. Another interesting thing from the results is that there is a large difference between the tuned results and this result in JNLPBA corpus. There is almost more than 5% difference in the F1 score as shown in the Table 15. This might be due to the smaller number of epochs used and because of this, there is a huge variation in terms of convergence of the neural network between the JNLPBA corpus and other two corpus. Also, the number of labels in JNLPBA corpus is higher than the other two corpus. So, there is a higher chance of identifying the labels wrong in JNLPBA corpus because of the low epoch value.
Table 15.
Performance evaluation on Biomedical datasets.
| Dataset | Time (Sec) | macro – F1 |
|---|---|---|
| NCBI-Disease | 301 | 0.865 |
| JNLPBA | 510 | 0.719 |
| CHEMDNER | 2864 | 0.884 |
5.2. Relation extraction
For evaluation of the relation extraction with the different variants of the proposed system, the BC5CDR dataset is considered. Table 16 shows the results of the comparison analysis with different variants like 1d-CNN, LSTM-SVM, LSTM-CNN, LSTM-CRF, and SciBERT. The SciBERT model obtained the better results when compared with the other variants and it is used to evaluate the CHEMPORT dataset.
Table 16.
Comparison with variants of Proposed System for RE.
| Model | Precision | Recall | F1 |
|---|---|---|---|
| 1d-CNN + Glove | 60.85 | 56.42 | 58.55 |
| LSTM-SVM | 64.90 | 49.30 | 56.00 |
| LSTM-CNN | 54.30 | 65.90 | 59.50 |
| LSTM-CRF | 60.00 | 67.50 | 63.50 |
| SciBERT (Proposed System) | 74.00 | 73.00 | 73.00 |
In the BC5CDR dataset, only positive relations are given. To create negative relations to equal the dataset for better training of the model and to prevent overfitting. Two random entities from the text that are not in relation are taken and are marked as negative relations. For example, in abstract 227,508, the found entities are shown in Fig. 5. Here only D007022 and D008750 are in positive relation. So, the negative relation is created by combining D007022 and D003000 or D006973 or D009270 and so on. In CHEMPROT dataset classes CPR:8 and CPR:0 is left out because there are not enough samples to evaluate them correctly in the development and test dataset.
Fig. 5.
Chemicals and Diseases from sample BC5CDR row.
The dataset is taken and is read into the memory. It is converted into the formation of [text, entity1, entity2, relation type]. Then the entity1 and entity2 of the relation is concatenated using space and \t is added to the end to mark a single sentence in the SciBERT input. The actual text is designed as the second sentence in the SciBERT input. Figure 4.23 shows a sample BC5CDR dataset row after transformation. Then the text is passed to the SciBERT sub word tokenizer to get input ids corresponding to the words. Then the attention mask is calculated using the input ids where there is a pad token the mask value is 0 otherwise 1. Then the token type ids are calculated using the concept of sentence 1 has a value of 0 and sentence 1 has a value of 1 and pad tokens have a value of 0. And finally, the labels are converted to tensor form as shown in Fig. 6.
Fig. 6.
Input ids of sample BC5CDR row.
The above-mentioned output is passed to the SciBERT model. The output will be of size 512 × 768 where 512 is the length of the input padded or truncated. A vector with a size equal to the number of relation types in the dataset is produced by the fully connected layer in this case using the output of the [CLS] token. The model is fine-tuned by back propagating the errors. The ultimate outcomes of fine-tuning SciBERT are displayed in Fig. 7.
Fig. 7.
Fine tuning SciBERT for BC5CDR.
Once the entities in a sentence are found in Named Entity Recognition, if there are both chemicals and diseases in a sentence then Chemical-Disease Relation extraction is carried out. If there are both chemicals and proteins in a sentence then Chemical-Protein relation extraction is carried out. Initially for each pair of entity_1, entity_2 in a sentence, they are added to the start of the text and is passed to the SciBERT tokenizer to get the input, attention mask and token type for each text. Then they are passed to the corresponding Relation Extraction model based on the type of entities to predict the relation between them. Table 17 shows the results obtained for relation extraction for different datasets. And Table 18 shows the statistics of relations extracted from CORD-19 based on the RE training.
Table 17.
Relation Extraction evaluation metrics.
| Dataset | Precision | Recall | F1 |
|---|---|---|---|
| BC5CDR | 74.00 | 73.00 | 73.00 |
| CHEMPROT | 72.00 | 71.00 | 71.00 |
Table 18.
Overall relations extracted from CORD-19.
| Relation Type | Total no. of Instances | Unique no. of Instances |
|---|---|---|
| Chemical Induced Disease | 5425 | 4071 |
| Chemical Protein Relation | 87,794 | 72,891 |
5.3. Graph construction
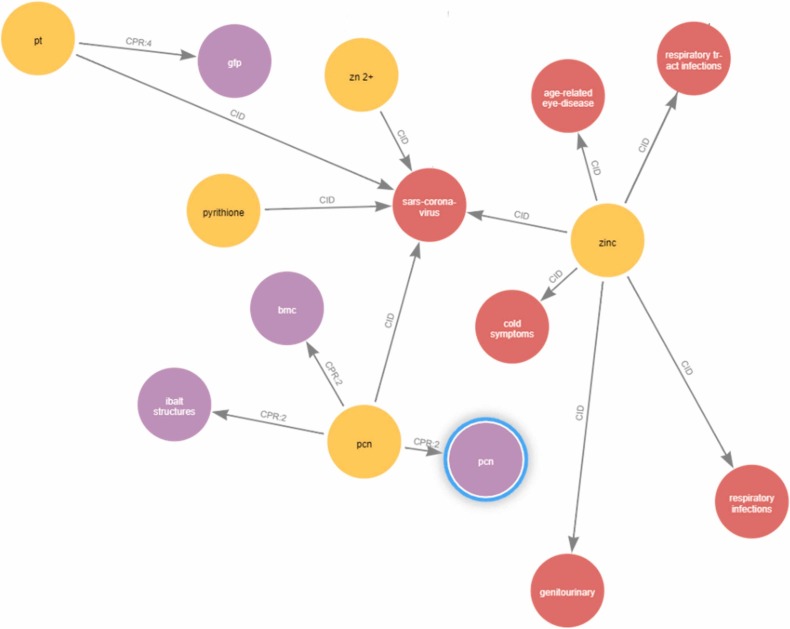
The relations extracted from the CORD-19 dataset is taken from the database and assigned with entity 1, entity 2, relation. Only the relations whose entities occurs more than 5 times are taken into consideration. The knowledge graph is built using the final list of relationships. The database has a total of 76962 relations, 5237 entries that have occurred more than five times, and 69,782 relations that contain at least one entity with five occurrences. A Neo4j graph database is initiated. The nodes are entities and the edges are relations. And all the relations are inserted into the database and the knowledge graph is created. Fig. 8 shows a portion of Knowledge Graph focused on coronavirus since there are more than 75,000 relations. Here Red colour nodes denotes Disease, Yellow for Chemical and Purple for Protein. The edges in the figures represents the diseases, chemicals, and protein closely related to COVID-19 from the extracted relations. The figure also shows the inter-relation between the entities. For example, Zinc is the chemical which is closely related to corona virus as well as age related eye diseases. Since we are interested in extracting the interaction of these entities only with the corona virus, the representation learning framework is used to extract the top chemicals, diseases, and proteins related to COVID-19.
Fig. 8.
Portion of knowledge graph focused on COVID-19.
5.4. Representation learning
Initially the relations list is taken and from them negative relations are created by taking random entities from different relations and combining them. Fig. 9, Fig. 10 and Fig. 11 shows the top diseases, chemicals and proteins related to COVID-19 respectively. Without any prior domain knowledge, our proposed methodology identified the most important drugs, diseases, and proteins associated with COVID-19. The biomedical research community could benefit from these derived entities by having a better grasp of COVID-19. In these figures thicker edges and darker colours indicates strong relationship. For example, ARDS (Acute respiratory distress syndrome), CRD (Chronic respiratory diseases), Hepatitis C, and HSV-2 (Herpes Simplex Virus) are a few diseases that patients with COVID-19 are likely to develop. Entities like Polyvinylidene fluoride, Poly adp ribose, and Succinate are likely to be considered as drugs for COVID-19. Proteins like Trypsin 1, IL-1β (Interleukin-1), Kinase, DICER, and Cyclo Oxygenase 2 have direct association with COVID-19.
Fig. 9.
Top Diseases related to COVID-19.
Fig. 10.
Top chemicals related to COVID-19.
Fig. 11.
Top proteins related to COVID-19.
Since there is no gold standard data available on the CORD-19 dataset. The system uses the Relation Extraction metrics as the output of the system. To ensure the validity of the formed Knowledge Graph by performing one of the downstream tasks of representation learning and the output of the representation learning is verified manually by checking the relation between the mentioned entity and COVID-19. The relation between entities obtained by the representation learning module needs to be verified manually. Table 19 showcases some of the evidences. The limitations of our approach is that we have evaluated only on CORD-19 dataset and no additional resources have been employed. However, due to the lack of gold standard data we have manually verified some of the randomly identified associations. Another limitation in the proposed framework is that the entities identified as closely related to COVID-19 might have a opposite association. For example, entity like PGE2 (prostaglandin E2) is a drug identified as top chemical for COIVD-19 but it increases the severity of COVID-19 disease. After the discovery of entities, we intend to develop a normalisation and abbreviation expansion mechanism as a future scope. The domain experts can better comprehend the many associations and relationships these top anticipated entities display with respect to COVID-19 by studying these entities.
Table 19.
Evidences for certain highly COVID-19 related entities.
| Entity | Entity Type | Evidence |
|---|---|---|
| ARDS | Disease | Aslan, Anolin et al. “Acute respiratory distress syndrome in COVID-19: possible mechanisms and therapeutic management.” Pneumonia (Nathan Qld.) vol. 13,1 14. 6 Dec. 2021, doi:10.1186/s41479–021–00092–9 |
| CRD | Disease | Beltramo, Guillaume et al. “Chronic respiratory diseases are predictors of severe outcome in COVID-19 hospitalised patients: a nationwide study.” The European respiratory journal vol. 58,6 2004474. 9 Dec. 2021, doi:10.1183/13993003.04474–2020 |
| Hepatitis C | Disease | Ronderos, Diana et al. “Chronic hepatitis-C infection in COVID-19 patients is associated with in-hospital mortality.” World journal of clinical cases vol. 9,29 (2021): 8749–8762. |
| HSV-2 | Disease | Shanshal, Mohammed, and Hayder Saad Ahmed. “COVID-19 and Herpes Simplex Virus Infection: A Cross-Sectional Study.” Cureus vol. 13,9 e18022. 16 Sep. 2021, doi:10.7759/cureus.18022 |
| PGE2 | Chemical | Ricke-Hoch, Melanie et al. “Impaired immune response mediated by prostaglandin E2 promotes severe COVID-19 disease.” PloS one vol. 16,8 e0255335. 4 Aug. 2021, doi:10.1371/journal.pone.0255335 |
| Polyvinylidene fluoride | Chemical | Nageh, Hassan et al. “Zinc Oxide Nanoparticle-Loaded Electrospun Polyvinylidene Fluoride Nanofibers as a Potential Face Protector against Respiratory Viral Infections.” ACS omega vol. 7,17 14887–14896. 22 Apr. 2022, doi:10.1021/acsomega.2c00458 |
| Poly adp ribose | Chemical | Badawy, Abdulla A-B. “Immunotherapy of COVID-19 with poly (ADP-ribose) polymerase inhibitors: starting with nicotinamide.” Bioscience reports vol. 40,10 (2020): BSR20202856. doi:10.1042/BSR20202856 |
| Succinate | Chemical | Pacl, Hayden T et al. “Water-soluble tocopherol derivatives inhibit SARS-CoV-2 RNA-dependent RNA polymerase.” bioRxiv: the preprint server for biology 2021.07.13.449251. 27 Jul. 2021, doi:10.1101/2021.07.13.449251. Preprint. |
| SER | Chemical | Rahbar Saadat, Yalda et al. “Host Serine Proteases: A Potential Targeted Therapy for COVID-19 and Influenza.” Frontiers in molecular biosciences vol. 8 725528. 30 Aug. 2021, doi:10.3389/fmolb.2021.725528 |
| Trypsin 1 | Protein | Kim, Yeeun et al. “Trypsin enhances SARS-CoV-2 infection by facilitating viral entry.” Archives of virology vol. 167,2 (2022): 441–458. doi:10.1007/s00705–021–05343–0 |
| IL-1β | Protein | Mardi, Amirhossein et al. “Interleukin-1 in COVID-19 Infection: Immunopathogenesis and Possible Therapeutic Perspective.” Viral immunology vol. 34,10 (2021): 679–688. doi:10.1089/vim.2021.0071 |
| Kinase | Protein | Pillaiyar, Thanigaimalai, and Stefan Laufer. “Kinases as Potential Therapeutic Targets for Anti-coronaviral Therapy.” Journal of medicinal chemistry vol. 65,2 (2022): 955–982. doi: 10.1021/acs.jmedchem.1c00335 |
| DICER | Protein | Mousavi, Seyyed Reza et al. “Dysregulation of RNA interference components in COVID-19 patients.” BMC research notes vol. 14,1 401. 29 Oct. 2021, doi:10.1186/s13104–021–05816–0 |
| Cyclo Oxygenase 2 | Protein | Baghaki, Semih et al. “COX2 inhibition in the treatment of COVID-19: Review of literature to propose repositioning of celecoxib for randomized controlled studies.” Int J Infect Dis. 2020;101:29–32. doi:10.1016/j.ijid.2020.09.1466 |
6. Conclusion and future work
In this study, a general pipeline for association analysis of an unstructured dataset with regard to a certain entity is proposed. Humans are kept out of the pipeline's integration of IE and KG construction. The system also suggests modifying the SciBERT on the CORD-19 abstracts to create unique SciBERT for word embeddings. For BioNER, the system also suggests the BERT-BiLSTM-CRF model, which, according to the test results, outperformed SciBERT. The SciBERT model is also used by the system to extract relationships between entities. The final collection of relations is utilised to create the Knowledge Graph after the entities are filtered to remove extraneous data. The system makes use of the TransD Knowledge Graph Embedding Method in order to learn the latent representation of the created Knowledge Graph. The system's methodology is exclusively assessed using the CORD-19 dataset; no other resources have been used. However, the system uses the metrics from Relation Extraction as the system's final metrics because there aren't any gold standard data available. Additionally, the final Knowledge graph is assessed by contrasting COVID-19 embeddings with those of all other entities to identify the top COVID-19-related illnesses, substances, and proteins.
CRediT authorship contribution statement
Sudhakaran Gajendran: Conceptualization, Methodology, Software, Writing – original draft. Manjula D: Visualization, Supervision. Vijayan Sugumaran: Writing – review & editing, Validation. R. Hema: Data curation, Investigation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.compbiolchem.2022.107808.
Appendix A. Supplementary material
Supplementary material
.
References
- Beltagy, Iz, Lo, Kyle, Cohan, Arman, 2019. Scibert: A pretrained language model for scientific text. In EMNLP/IJCNLP.
- Chai Z., Jin H., Shi S., et al. Hierarchical shared transfer learning for biomedical named entity recognition. BMC Bioinforma. 2022;23:8. doi: 10.1186/s12859-021-04551-4. https://doi.org/10.1186/s12859-021-04551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Akef Ebeid I., Bu Y., Ding Y. Coronavirus knowledge graph: a case study. arXiv e-prints. 2020 [Google Scholar]
- Cheng D., Knox C., Young N., Stothard P., Damaraju S., Wishart D.S. PolySearch: a web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn296. Jul 1;36(Web Server issue):W399-W405. doi: 10.1093/nar/gkn296. Epub 2008 May 16. PMID: 18487273; PMCID: PMC2447794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J., Chang M., Lee K., Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. ArXiv, abs/1810. 2019:04805. [Google Scholar]
- Doğan R.I., Leaman R., Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. J. BiomedInform. 2014:2014. doi: 10.1016/j.jbi.2013.12.006. Feb;47:1-10. doi: 10.1016/j.jbi.2013.12.006. Epub2014 Jan 3. PMID: 24393765; PMCID: PMC3951655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Fernandez, Daniel, Baksi, Shounak, Schultz, Bruce´, Gadiya, Yojana, Karki, Reagon, Raschka, Tamara, Ebeling, Christian, Hofmann, Martin, Alpha, Apitius, Kodamullil, Tom, 2020. Covid19 knowledge graph: a computable, multimodal, cause-and-effect knowledge model of covid-19 pathophysiology. bioRxiv. [DOI] [PMC free article] [PubMed]
- Fundel K., Küffner R., Zimmer R. RelEx—relation extraction using dependency parse trees. Bioinformatics. 2006;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- Gajendran S., Manjula D. Biomedical named entity recognition (Bner) using word representation features based on Crf. Int. J. Creat. Res. Thoughts (IJCRT) 2020:89120. [Google Scholar]
- Gajendran S., Manjula D., Sugumaran V. character level and word level embedding with bidirectional LSTM – Dynamic recurrent neural network for biomedical named entity recognition from literature. J. Biomed. Inform. 2020;Volume 112 doi: 10.1016/j.jbi.2020.103609. ISSN 1532-0464. [DOI] [PubMed] [Google Scholar]
- Giorgi J., Wang X., Sahar N., Shin W.Y., Bader G.D., Wang B. End-to-end named entity recognition and relation extraction using pre-trained language models. arXiv Prepr. arXiv. 2019;1912:13415. [Google Scholar]
- Habibi M., Weber L., Neves M., Wiegandt D.L., Leser U. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics. 2017;33(14):i37–i48. doi: 10.1093/bioinformatics/btx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnoune A., Rhanoui M., Mikram M., Yousfi S., Elkaimbillah Z., Asri B.E. BERT based clinical knowledge extraction for biomedical knowledge graph construction and analysis. Comput. Methods Prog. Biomed. Update. 2021;Volume 1(2021) ISSN2666-9900. [Google Scholar]
- Jensen K., Panagiotou G., Kouskoumvekaki I. Integrated text mining and chemoinformatics analysis associates diet to health benefit at molecular level. PLoS Comput. Biol. 2014;10(1) doi: 10.1371/journal.pcbi.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jettakul A., Wichadakul D., Vateekul P. Relation extraction between bacteria and biotopes from biomedical texts with attention mechanisms and domain-specific contextual representations. BMC Bioinforma. 2019;20(1):1–17. doi: 10.1186/s12859-019-3217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guoliang Ji, Shizhu He, Liheng Xu, Kang Liu, and Jun Zhao. (2015) Knowledge Graph Embedding via Dynamic Mapping Matrix. In Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pages 687–696, Beijing, China. Association for Computational Linguistics.
- Kazama Jun’ichi, Makino Takaki, Ohta Yoshihiro, Tsujii Jun’ichi. Proceedings of the ACL-02 Workshop on Natural Language Processing in the Biomedical Domain, pages. Association for Computational Linguistics,; Phildadelphia, Pennsylvania, USA: 2002. Tuning support vector machines for biomedical named entity recognition; pp. 1–8. [Google Scholar]
- Kim D., Lee J., So C.H., Jeon H., Jeong M., Choi Y., Kang J. A neural named entity recognition and multi-type normalization tool for biomedical text mining. IEEE Access. 2019;7:73729–73740. [Google Scholar]
- Kim T., Yun Y., Kim N. Deep learning-based knowledge graph generation for COVID-19. Sustainability. 2021;2021(13):2276. [Google Scholar]
- Krallinger M., Rabal O., Leitner F., Vazquez M., Salgado D., LuZ, Leaman R., Lu Y., Ji D., Lowe D.M., Sayle R.A., Batista-Navarro R.T., Rak R., Huber T., Rocktäschel T., Matos S., Campos D., TangB, XuH, Munkhdalai T., Ryu K.H., Ramanan S.V., Nathan S., Žitnik S., BajecM, Weber L., Irmer M., Akhondi S.A., Kors J.A., Xu S., An X., Sikdar U.K., Ekbal A., Yoshioka M., Dieb T.M., Choi M., Verspoor K., KhabsaM, Giles C.L., Liu H., Ravikumar K.E., Lamurias A., Couto F.M., Dai H.J., Tsai R.T., Ata C., Can T., Usié A., Alves R., Segura-Bedmar I., MartínezP, Oyarzabal J., Valencia A. The CHEMDNER corpus of chemicals and drugs and its annotation principles. J. Chemin. 2015 doi: 10.1186/1758-2946-7-S1-S2. 2015Jan19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty, J., McCallum, A., Pereira, F.C.N., 2001. Conditional random fields: Probabilistic models for segmenting and labeling sequence data.
- Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., Kang J. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4) doi: 10.1093/bioinformatics/btz682. 1234– 0. https://doi.org/10.1093/bioinformatics/btz682. arXiv:1901.08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Yang Z., Yang P., Zhang Y., Wang L., Lin H., Wang J. An attention-based BiLSTM-CRF approach to document-level chemical named entity recognition. Bioinformatics. 2018;34(8):1381–1388. doi: 10.1093/bioinformatics/btx761. [DOI] [PubMed] [Google Scholar]
- Nobata C., Collier N., Tsujii J.-I. Automatic term identification and classification in biology texts. Proc. 5th Nlprs. Citeseer. 1999:369–374. [Google Scholar]
- Percha B., Garten Y., Altman R.B. Discovery and explanation of drug-drug interactions via text mining. Biocomputing. 2012;2012:410–421. [PMC free article] [PubMed] [Google Scholar]
- Peters, Matthew E., Neumann, Mark, Iyyer, Mohit, Gardner, Matt, Clark, Christopher, Lee, Kenton, Zettlemoyer, Luke, 2018 Deep contextualized word representations. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), pages 2227–2237, New Orleans, Louisiana. Association for Computational Linguistics.
- Quan C., Wang M., Ren F. An unsupervised text mining method for relation extraction from biomedical literature. PloS one. 2014;9(7) doi: 10.1371/journal.pone.0102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebholz-Schuhmann D. In: Encyclopedia of Systems Biology. Dubitzky W., Wolkenhauer O., Cho K.H., Yokota H., editors. Springer; New York, NY: 2013. Biomedical named entity recognition, Whatizit. [DOI] [Google Scholar]
- Repke T., Krestel R. In: Data Science for Economics and Finance. Consoli S., Reforgiato Recupero D., Saisana M., editors. Springer; Cham: 2021. Extraction and representation of financial entities from text. [Google Scholar]
- Shen, Dan, Zhang, Jie, Zhou, Guodong, Su, Jian, Tan, Chew-Lim, 2003. Effective adaptation of hidden markov model-based named entity recognizer for biomedical domain. In: Proceedings of the ACL 2003 Workshop on Natural Language Processing in Biomedicine, pages 49–56, Sapporo, Japan. Association for Computational Linguistics.
- Wang Q., Li M., Wang X., Parulian N., Han G., Ma J., Tu J., Lin Y., Zhang H., Liu W., Chauhan A., Guan Y., Li B., Li R., Song X., Ji H., Han J., Chang S., Pustejovsky J., Liem D., El-Sayed A., Palmer M., Rah J., Schneider C., Onyshkevych B. Covid-19 literature knowledge graph construction and drug repurposing report generation. ArXiv, abs/2007. 2020:00576. [Google Scholar]
- Weber L., Sänger M., Münchmeyer J., Habibi M., Leser U., Akbik A. HunFlair: an easy-to-use tool for state-of-the-art biomedical named entity recognition. Bioinformatics. 2019;37(17):2792–2794. doi: 10.1093/bioinformatics/btab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Yang Z., Kang P., Wang Q., Liu W. Document-level attention-based BiLSTM-CRF incorporating disease dictionary for disease named entity recognition. Comput. Biol. Med. 2019;108(May):122–132. doi: 10.1016/j.compbiomed.2019.04.002. Epub 2019 Apr 7. PMID: 31003175. [DOI] [PubMed] [Google Scholar]
- Yanran Li, Wenjie Li, Fei Sun, Sujian Li. Component-enhanced chinese character embeddings. arXiv Prepr. arXiv. 2015;1508:06669. [Google Scholar]
- Yoon W., So C.H., Lee J., Kang J. CollaboNet: collaboration of deep neural networks for biomedical named entity recognition. BMC Bioinform. 2019 doi: 10.1186/s12859-019-2813-6. https://doi.org/10.1186/s12859-019-2813-6. arXiv:1809.07950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshua Bengio, Ducharme R., Vincent Pascal, Janvin Christian. A neural probabilistic language model. J. Mach. Learn. Res. 2003;3:1137–1155. [Google Scholar]
- Zeng, D., Liu, K., Lai, S., Zhou, G., & Zhao, J. (2014, August). Relation classification via convolutional deep neural network. In: Proceedings of COLING 2014, the 25th international conference on computational linguistics: technical papers (pp. 2335–2344).
- Zheng S., Rao J., Song Y., Zhang J., Xiao X., Fang E., Yang Y., Niu Z. PharmKG: a dedicated knowledge graph benchmark for biomedical data mining. Brief. Bioinforma. 2020 doi: 10.1093/bib/bbaa344. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Li X., Conesa A., Pereira C. GRAM-CNN: a deep learning approach with local context for named entity recognition in biomedical text. Bioinformatics. 2017;34(9):1547–1554. doi: 10.1093/bioinformatics/btx815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material