Abstract
Deep learning (DL) algorithms have demonstrated a high ability to perform speedy and accurate COVID-19 diagnosis utilizing computed tomography (CT) and X-Ray scans. The spatial information in these images was used to train DL models in the majority of relevant studies. However, training these models with images generated by radiomics approaches could enhance diagnostic accuracy. Furthermore, combining information from several radiomics approaches with time-frequency representations of the COVID-19 patterns can increase performance even further. This study introduces "RADIC", an automated tool that uses three DL models that are trained using radiomics-generated images to detect COVID-19. First, four radiomics approaches are used to analyze the original CT and X-ray images. Next, each of the three DL models is trained on a different set of radiomics, X-ray, and CT images. Then, for each DL model, deep features are obtained, and their dimensions are decreased using the Fast Walsh Hadamard Transform, yielding a time-frequency representation of the COVID-19 patterns. The tool then uses the discrete cosine transform to combine these deep features. Four classification models are then used to achieve classification. In order to validate the performance of RADIC, two benchmark datasets (CT and X-Ray) for COVID-19 are employed. The final accuracy attained using RADIC is 99.4% and 99% for the first and second datasets respectively. To prove the competing ability of RADIC, its performance is compared with related studies in the literature. The results reflect that RADIC achieve superior performance compared to other studies. The results of the proposed tool prove that a DL model can be trained more effectively with images generated by radiomics techniques than the original X-Ray and CT images. Besides, the incorporation of deep features extracted from DL models trained with multiple radiomics approaches will improve diagnostic accuracy.
Keywords: Deep learning, COVID-19, Convolution neural networks (CNN), Discrete wavelet transform, Dual-tree complex wavelet transform, Texture analysis
1. Introduction
The recent coronavirus (COVID-19) has spread rapidly around the globe from its very discovery in December 2019, and its precise origin remains undetermined [1]. Generally, the COVID-19 pandemic has resulted in a string of devastating losses over the world, infecting about 287 million individuals and killing approximately 5.4 million people. COVID-19's fast proliferation continues to endanger human life and health. COVID-19 is thus not merely an epidemiological calamity, but also a psychological and emotional one. People experience tremendous anxiety, tension, and depression as a result of the pandemic's uncertainty and loss of normalcy.
Currently, COVID-19 diagnostic standard methods are real-time reverse transcription-polymerase chain reactions (RT-PCRs) tests, but they have some disadvantages. The test is vulnerable to the risks of medical professionals. In addition, it is costly, takes time, and sometimes is incorrect [2,3]. As a result, other alternatives are needed. The normal symptoms of COVID-19 are lung infections, where chest scanning techniques such as X-Ray and computed tomography (CT) may be employed to visualize the infection [4,5]. These screening methods have been proven to be more capable of diagnosing COVID-19 than RT-PCR testing [6,7]. Nevertheless, such scanning methods necessitate the involvement of a skilled specialist physician to make a diagnosis. Medical professionals analyze these images manually to determine the visual patterns of COVID-19 infections. The diagnosis of COVID-19 is challenging because of the similarity between the configurations of COVID-19 as well as other types of comparable illnesses such as different sorts of pneumonia and the various version of COVID-19 [8]. Moreover, the routine investigation requires a while, so fully automated diagnostic tools are necessary to cut inspection duration and the effort imposed by professionals to conduct the assessment and generate a precise decision. This process must therefore be fully automated to facilitate the diagnosis procedure for COVID-19 and to achieve more accurate, faster, and more effective results. Therefore, these imaging methods when utilized along with artificial intelligence (AI) capabilities, could detect COVID-19 patients more accurately, quickly, and affordably. The failure to recognize and treat COVID-19 individuals promptly raises the death rate. As a result, detecting COVID-19 instances with AI-based models using both XR and CT images has a lot of potential in healthcare [4].
In recent years, AI has been successfully used in digital health along with assistive technology to enhance the quality of life [10,11]. Furthermore, AI methods have been extensively employed along with computer-assisted diagnosis tools (CAD) to support doctors in diagnosing many diseases accurately including heart complications [12,13], cancer [[14], [15], [16], [17], [18]], stomach diseases [19], lung diseases [[20], [21], [22]], visual disorders [23], and genetic abnormalities [24]. For the purpose of detecting coronavirus disease, the AI research community has lately committed a lot of time and funds to create deep learning models (DL) based on chest radiographs. When faced with countless numbers of people arriving with disease symptoms, many medical institutions are finding themselves in challenging situations. To address this crucial demand, new diagnostic models are required. The whole area of medical image analysis has recently been impacted by DL techniques [25]. Lately, the techniques of DL, especially the convolutional neural networks (CNN) have achieved significant advancements in the quality of automated diagnostic medical imaging systems. As a result, numerous intriguing avenues are currently emerging that may eventually aid in the diagnosis of COVID-19.
Radiomics is a new field that is used to analyze medical images [26]. It is feasible to precisely identify a variety of illnesses by using radiomics and AI techniques [27]. The primary advantage of radiomics is that it can use medical scans to obtain the texture and fundamental components of illnesses and malignancies [15]. This information may help AI techniques in diagnosing diseases accurately. Numerous research papers [9,[28], [29], [30], [31], [32]] utilized CNNs to diagnose COVID-19 with CT and X-Ray scanning [[33], [34], [35], [36], [37], [38]], however, these CNNs were trained exclusively using the spatial information found in X-Ray and CT images. Nevertheless, merging spatial information with radiomics to construct these CNNs can enhance the diagnosis [39].
The goal of the current study is to ascertain whether using images created by a variety of radiomics approaches to train CNNs for the diagnosis of COVID-19 is more effective than using the original X-ray and CT images. It also investigates whether combining time-frequency and frequency data with information obtained with radiomics can improve diagnostic performance. Furthermore, it examines if combining several radiomics techniques might enhance performance even more. The novelty and contribution of this study are highlighted below.
-
•
Constructing an automated tool for COVID-19 diagnosis named “RADIC” that could be applied for both routine imaging modalities for screening COVID-19 (X-ray and CT) not a single imaging approach.
-
•
RADIC is developed via three CNNs not only one to incorporate the benefit of each architecture.
-
•
Instead of applying the raw images for learning the CNNs which is the case in existing tools, RADIC analyzes images using four radiomics approaches and employs the output of these methods as images to the DL models.
-
•
Extracting several spatial radiomics descriptors from the three CNNs after completing the training procedure using transfer learning.
-
•
Lowering the size of the extracted features by employing the Fast Walsh Hadamard Transform (FWHT) which represents the time-frequency demonstration of the features instead of depending on the textural radiomics information alone.
-
•
Incorporating the lessened spatial-time-frequency features of the three CNNs using discrete cosine transform (DCT) which attains the frequency representation of the features.
-
•
Acquiring features from several domains including spatial, time-frequency, and frequency domains is an alternative to relying on features from a single domain which correspondingly enhances performance.
The article is organized as follows; Section II presents recent works on COVID-19 diagnosis, Section III demonstrates the methodology, including the proposed tool. Section IV illustrates the results. Section V discusses the results. Finally, section V illustrates the conclusion.
2. Related works on Covid-19 diagnosis
By the beginning of 2020, a wide range of studies had been published that covered the use of AI-based models and radiological images to detect the novel coronavirus. The methods could be classified into two groups. The first group is based on singular CNNs, whereas the other group represents the ensemble-based approaches. This section will discuss related works in these two groups.
2.1. Singular cnns based approaches
Singular CNNs were employed in several articles; for example, in the study [40], the authors used DenseNet in order to identify COVID-19 via CT scans, reaching a 96.25% accuracy rate. Later, the authors in Ref. [41] used CT images to build rigorously modified CNN models based on MobileNet V2, achieving a 96.4% accuracy rate. While in the research [42], the COVID-19 diagnosis was made using the Grad-CAM and VGG-19 techniques, with an accuracy rate of 94.4%. The study [43] independently used six separate CNNs where VGG attained the greatest accuracy of 97.78%. In contrast, deep features were extracted in the study [44] using seven DL models. These deep features are fed into six classification models and their performance is tested using test photos attaining a 96.23% accuracy rate. Whereas the authors of [45] built a DL network based on AlexNet's and modified it by adding a batch normalisation to speed up training and reduce internal covariance shift. Furthermore, the authors used three classifiers to take the place of AlexNet's fully connected layer reaching 96.3% specificity. The authors of [36] proposed a new Bayesian optimization-based CNN architecture to diagnose COVID-19 from X-Ray images. The method used a Bayesian-based optimization to fine-tune the hyperparameters of the CNN relying on an objective function. The method also extracted features from this CNN and used multiple fully connected layers to perform classification attaining an accuracy of 96%. The authors of [38] employed DenseNet-169 CNN to extract deep spatial features from X-Ray scans. Next, these features were fed into the Extreme Gradient Boosting method for diagnosing COVID-19 attaining 89.7% accuracy. Conversely, the study [33] proposed an individual CNN based on the DenseNet baseline model and fine-tuned it with weights previously set from the well-known CheXNet model achieving 87.88% accuracy. While the study [46] designed a customized ResNet with transfer learning to diagnose COVID-19 using X-ray images attaining an accuracy of 95%.
Other recent studies employed multi-agent deep learning methods such as [47]. The authors of the study [47] presented a novel multi-agent deep reinforcement learning (DRL)-based mask separation technique to reduce long-term conventional mask retrieval and improve segmentation of medical image paradigms. To allow the mask detection method to pick masks out from the image examined, this new method used a customized form of the Deep Q-Network. The authors used DRL mask retrieval methodologies on COVID-19 computed tomography (CT) images to obtain feature representations of COVID-19-affected regions and establish a reliable medical assessment.
2.2. Ensemble-based approaches
Ensemble learning was employed in other studies to carry out COVID-19 diagnosis. Some other fused multiple deep features with handcrafted attributes such as the research article [48] which combined handcrafted features along with the deep features of four CNNs trained employing CT images reaching 99% accuracy. Whereas the study [49] segmented X-Ray images using a conditional generative adversarial network. These segmented images were then used to extract significant features using two methods. The first method used the BRISK feature extraction method while the other employed deep features of several CNNs including DenseNet and VGG. These features were then fused and used as inputs to the K-NN classifier reaching 96.6% accuracy. On the other hand, others fused-only deep features like [50] where a straightforward but effective DL feature fusion model built on two customized CNNs was suggested for diagnosing COVID-19. Also, the authors of the research article [51] used 13 CNN models to diagnose COVID-19 from X-ray images. The authors combined features extracted from the 13 CNN models and used two feature selection approaches to lessen the feature space size achieving an accuracy of 99.14%. Whereas the research article [52] merged features obtained via four CNNs utilizing principal component analysis (PCA) and reached 99% accuracy. In the study [53], the authors utilized DWT to analyze images using ResNets trained with images of a private CT dataset and a public dataset. In the study [54], several fusion methods were utilized to either merge features or predictions of several CNNs including AlexNet, ResNet50, SonoNet6, Xception, Inception-Resnet-V2, and EfficientNet. These CNNs were trained with original X-ray images as well as an enhanced version of these images. The results showed that the highest accuracy of 95.57% is accomplished when fusing predictions of the ResNet-50 trained once with the original X-ray images and then with its enhanced version. The study [55] aimed to construct ensemble learning with CNN-based deep features to enhance CNN's detection performance (EL–CNN–DF). The chest X-ray pictures were first pre-processed using median filtering and image scaling. Next, the deep features were obtained from the CNN's pooling layer, and the fully connected layer is replaced with the three classifiers Support Vector Machine (SVM), Autoencoder, and Naive Bayes (NB) once the lungs have been segmented. This is known as the new EL–CNN–DF. In contrast, a completely automated approach that relies on parallel fusion and DL model optimization is proposed [56]. The presented process started with a contrast enhancement that used a top-hat and Wiener filter mixture. Next, Two DL models involving AlexNet and VGG16 were used. A parallel positive correlation ensemble approach was utilized to extract and fuse attributes. The entropy-controlled firefly optimization method was employed to choose best possible features. SVM was then used to classify images and achieved an accuracy of 98%.
While others designed end-to-end DL models such as [57] to swiftly and effectively identify COVID-19 infections in CT images. Transfer learning has been used to modify and train sixteen DL neural networks to identify COVID-19 patients [57]. The performance of COVID-19 recognition had then been enhanced utilizing the suggested DL model, which was created using three of sixteen modified pre-trained models. Alternatively, the article [58] employed an ensemble of EfficientNet CNN models based on the voting method for the diagnosis of COVID-19. Similarly, the study [32] used majority voting to merge predictions of AlexNet, GoogleNet, and ResNet CNNs achieving an accuracy of 99% accuracy using CT images. Also, majority voting is employed in Ref. [31] to merge prediction results obtained by ResNet-50 reaching 96% accuracy. The research paper [59] introduced a method based on ensemble runs of ResNet-50 reaching an accuracy that exceeded 99.63%. Alternatively, the research article [60] implemented a model called COVDC-Net, based on MobileNet and VGG-16 for COVID-19 diagnosis via X-Ray scans. The model fused the outputs of MobileNet and VGG-16 using the confidence fusion algorithm reaching 96.48% accuracy. Alternatively, other studies compared the performance of end-to-end DL models with deep feature extraction and traditional machine learning classification such as the study [61] which presented a method for detecting 15 different types of chest infections, involving COVID-19, utilizing a chest X-ray method. In the proposed framework, 2 different classification approaches were carried out. First, a CNN with a soft-max classifier was proposed. Furthermore, transfer learning is utilized to extract deep features from a presented CNN's fully-connected layer. The deep features were introduced into traditional ML classification techniques. However, the proposed framework improved COVID-19 detection accuracy and increased predictability rates for other chest diseases.
Despite the good results of earlier studies, the majority of models were constructed directly from CT or X-ray scans, relying on spatial information found in these images. Nonetheless, combining time-frequency and frequency information with radiomics information obtained from images produced by radiomics techniques may improve diagnostic performance [62]. Although earlier studies employed radiomics to detect the novel coronavirus, these methods were employed as feature extractors. Instead, various radiomics techniques are employed in this study to transform CT and X-ray images into textural images known as radiomics images. These radiomics images, together with raw CT and X-Ray scans, are fed into three CNNs. Next, deep features are obtained from the CNNs built from each radiomics image separately. Next, the fast Walsh Hadamard Transform (FWHT) is used to reduce each set of deep features, producing time-frequency representations. Discrete Cosine Transform (DCT) is then used to combine these deep features and diminish their size and represent the frequency representation of the data. Finally, these combined features are used to feed multiple machine-learning classifiers to complete the classification process.
3. Methodology
3.1. Radiomics approaches
3.1.1. Grey level run length matrix
The GLRLM method is a high-order statistical radiomics process in which the grey-level run signifies a row of pixels having similar intensities in a given orientation [63]. For an image having a dimension M x N, GLRLM computes the G which is the grey level representing a string consisting of the pairwise pixels having similar intensities of grey levels in a specific orientation and the lengthiest L run. In other words, GLRLM is a bi-dimensional matrix (G x L) of elements Q (n, m) each of which provides the number of incidences of the run having a size n of grey level m in a particular direction θ [64,65].
3.1.2. Grey level covariance matrix
GLCM is a 2nd order statistical method that depends on distributions of the grey levels between pixels. GLCM calculates the mutual frequency of a complete pairwise mixture of grey-level arrangement in each pixel of the left hemisphere (at distinct orientations) considered as reference pixels with pixels located on the right hemisphere. Consequently, several covariance matrices corresponding to each pair of pixels are generated. Subsequently, each covariance matrix is standardized to determine the relative frequency of the covariance between the grey values of mutual pixels [66].
3.1.3. Discrete wavelet transform
A potent technique for time-frequency analysis of a medical signal/image is the wavelet transform (WT) [67]. For WT, a mother wavelet is selected, and the wavelet function is scaled and translated after that. The wavelet function is multiplied by the signal and added across the time domain after scaling and translation. Throughout the signal, these processes continue. In Continuous Wavelet Transform (CWT), the wavelet function is scaled and translated over the whole time range to execute the transformation. The scaling and translation variables are typically set as discrete values (Discrete wavelet transform (DWT)) because this needs a lot of work. Operations are carried out in two dimensions when DWT is applied to the image. In the end, four matrices are created. The detail coefficient is represented by three of them (horizontal, vertical, and diagonal), whereas the approximate coefficient is represented by one of them. The low-pass filter produces the approximate coefficients, and the high-pass filter produces the detail coefficients. In the detail coefficients, the edges are hence more noticeable. Vertical, horizontal, and diagonal coefficients are represented by detail coefficients.
3.1.4. Dual-tree complex wavelet transform
DWT has shown its superior analysis performance in several domains. However, its performance is diminished in the analysis of complex and higher dimensional medical data due to the shift variance and low directional selectivity in these types of data. On the other hand, the dual-tree complex wavelet transform (DTCWT) solves these two issues as it is directionally selective and shift-invariant to high-dimensional data [68,69]. In the case of 1D input data, DTCWT applies two DWT in parallel on the same input. The filters of DTCWT should be designed in a manner that subbands of the upper DWT explain the real part of DTCWT. While the subbands of the lower DWT explain the imaginary part of DTCWT. The transformation is expanded by factor 2 and the invariant of the shift [68].
3.2. Convolution neural networks
CNNs are well-known DL techniques commonly used to analyze medical images. A CNN's main advantage over conventional artificial neural networks is that they can automatically extract images' features [70]. These networks can directly diagnose and eliminate excessive processing steps necessary by traditional machine learning techniques. The three main layers of CNNs are composed of a convolutional layer, a pooling layer, and a fully connected layer (FC). Within the convolution layer, the convolution occurs between image segments and a compact filter. Subsequently, a map called a “feature map” is created, that describes information regarding each pixel of the image segment. Because of the large dimension generated by the feature map, the pooling layer plays an important part in reducing the enormous dimension of these maps by downsampling. Lastly, FC layers collect previous layers' inputs and generate class scores [71]. The article employs three state-of-the-art CNN architectures involving MobileNet, DenseNet-201, and DarkNet-53.
The main cause of using Darknet-53 is its effectiveness as it could ensure both accuracy and speed [72]. Given the same processing conditions, Redmon et al. [73] demonstrated that Darknet-53 is more efficient than ResNet-152, Darknet-19, ResNet-50, and state-of-the-art DL models. Darknet-53 outperforms state-of-the-art methods because of its rapid speed and low amount of floating-point processes. Furthermore, Darknet-53 includes small-scale successive convolutional filters of 3x3 and 1x1, that assist in the detection of objects or patterns of varying sizes [74].
The reason for choosing DenseNet is its unique structure which depends on shorter links among layers which makes the network more accurate and has better and more effective training capability [75]. DenseNet connects every layer in the feed-forward procedure to all other layers. Moreover, the DenseNet structure has many remarkable benefits. First, it solves the vanishing-gradient issue. Also, it empowers feature reusing and improves feature progression. Furthermore, DenseNet decreases the sum of parameters significantly [[76], [77], [78]].
Finally, MobileNet is used in this study because it is a lightweight CNN that has lesser parameters, lower deep layers, and could achieve greater accuracy even being compact. The foundation of the MobileNet design is depth-wise separable convolution [79]. A conventional convolution is factorized into a depth-wise convolution and a 1x1 point-wise convolution by the depth-wise separable convolution. The primary advantage of depth separable convolution over regular convolution is the necessity for less computational effort for large and complicated convolutional networks [80].
3.3. COVID-19 datasets
CT Dataset: The study uses the benchmark COVID-19 dataset commonly used in literature, SARS-COV-2 CT [81] referred to as Dataset I. It includes 2482 CT scans, 1230 individuals without corona virus and 1252 individuals with corona virus. The size of these CT scans differs from 119 × 104 to 416 × 512. Fig. 1 shows the sample images of this dataset.
Fig. 1.
Samples of Dataset I for COVID-19 and Non-COVID-19 cases.
X-Ray Dataset: This study used another dataset referred to as Dataset II containing 4883 X-Ray scans having 3 classes which are normal, COVID-19, and pneumonia. This dataset can be found in the Kaggle COVID-19 radiology database [82]. It contains images from different sources. The dataset consists of 1784 COVID-19 X-Ray images, 1754 healthy X-Ray scans, and 1345 X-Ray scans containing pneumonia. Samples of scans of these three classes are shown in Fig. 2 .
Fig. 2.
Samples of Dataset II for COVID-19, Normal, and Pneumonia cases.
3.4. Proposed tool
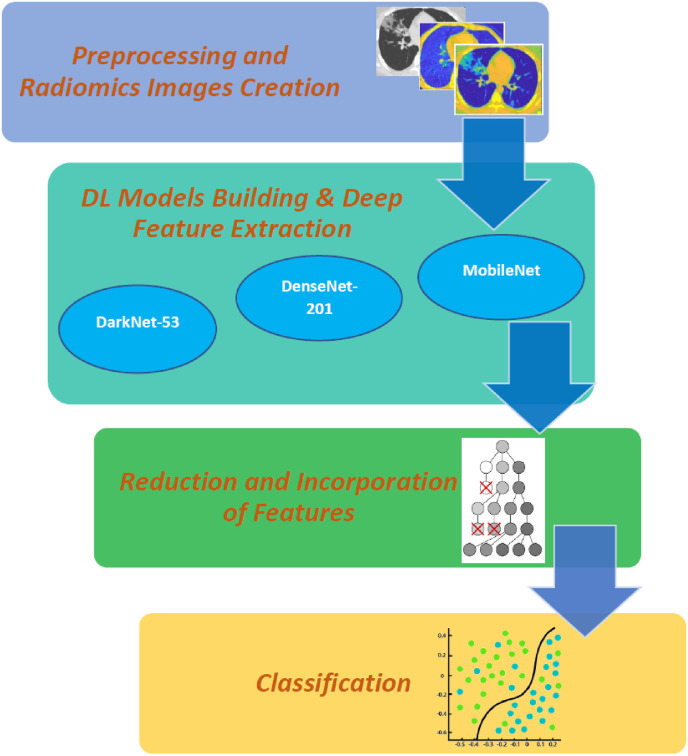
The proposed tool is called “RADIC”. It includes four steps: preprocessing and radiomics images creation, DL models building and deep feature extraction, reduction and incorporation of features, and finally classification. A flowchart of the proposed tool is presented in Fig. 3 .
Fig. 3.
A flowchart of the proposed RADIC tool.
3.4.1. Preprocessing and radiomics images creation
This step analyzes CT and X-Ray images using four radiomics methods, involving GLRLM, GLCM, DWT, and DTCWT. Afterward, the outputs of these methods are converted to heatmaps images. Then the size of these images is adjusted to 224x224x3, equivalent to MobileNet and DenseNet-201 input layer size, and 226x226x3 equal to the DarkNet-53 input layer's dimension. Subsequently, these images are divided into training and test sets with a 70%–30% ratio. The training images are augmented using several techniques to increase the number of images available during the CNNs training stage to avoid over-fitting. These techniques are scaling, rotation, flipping, and shearing. Fig. 4 and Fig. 5 show samples of the radiomics images generated using the four radiomics methods (GLRLM, GLCM, DWT, and DTCWT) for Dataset I and II respectively.
Fig. 4.
Radiomic images samples for Dataset I (a) DWT, (b) DTCWT, (c) GLCM, and (d) GLRLM.
Fig. 5.
Radiomic images samples of Dataset II (a) DWT, (b) DTCWT, (c) GLCM, and (d) GLRLM.
3.4.2. DL models building & deep feature extraction
Three pre-trained CNNs involving DenseNet-201, DarkNet-53, and MobileNet are created via transfer learning (TL). TL is a machine learning technique applied to transfer the knowledge acquired in the training procedure of CNNs in order to resolve a comparable learning problem, but with a dataset that has few training data. TL helps avoid problems of overfitting and convergence. This process generally increases diagnostic accuracy. Thus, the study used three pre-trained CNNs. Before the three pre-trained CNNs are re-trained, the size of the output layer is altered to two or three, corresponding to the number of labels of Dataset I and Dataset II respectively. Next, the retraining of the CNNs begins. Deep features are acquired via the final CNNs' batch normalisation layers upon the completion of the retraining procedure. Extracted deep features’ size is 65536 for DarkNet-53, 94080 for DenseNet-201, and 62720 for MobileNet.
3.4.3. Reduction and incorporation of features
As noticed from the previous step, the size of extracted features is huge, thus feature reduction procedure is essential to lower the complexity and training time of the classification models. Consequently, after the deep feature extraction stage, each CNN lowers these features by employing the Fast Walsh Hadamard Transform (FWHT). FWHT is an efficient technique that examines input data and transforms it into Walsh basic functions which are sets of perpendicular waveforms. It breaks down the input data of length 2n into 2n coefficients, which are equivalent to the input data's discrete WHT. The primary benefits of FWHT include its simplicity, speed, and minimal storage space requirement for decomposition coefficients [83,84]. FWHT can also be used for feature reduction. In order to obtain a time-frequency representation of the COVID-19 patterns, it is employed to reduce the dimension of the spatial features that were recovered in the previous phase. For each CNN, the size of the features after the FWHT step is 500.
Later, for each CNN, the reduced features of each radiomics are incorporated using discrete cosine transform (DCT). DCT is often used to convert the input data into the basic frequency components. Input data is represented by DCT as the sum of the oscillating cosine functions at distinct frequencies [85]. Typically, DCTs are applied to input data in order to obtain DCT coefficients. These coefficients are split into two groups, known as low frequencies namely (DC coefficients), and high frequencies namely (AC coefficients). The latter describes low noise and deviations (details). While low frequencies are associated with brightness conditions. DCT itself does not reduce data dimensions but reduces most image information by a few coefficients using another reduction phase [86]. This reduction phase usually uses zigzag scanning, where a few coefficients are selected to create feature vectors. For each CNN, the size of radiomics incorporated features is 500 (deep features extracted from the CNN trained with radiomics scans).
3.4.4. Classification
Several classification models, such as the linear support vector machine (L-SVM), quadratic-SVM, linear discriminant analysis (LDA), and ensemble subspace discriminant (ESD), are used in the classification process.In this step, images are classified either as COVID-19 or as non-COVID-19 in the case of Dataset I, and COVID-19, normal, and pneumonia for Dataset II. The proposed tool's classification process is carried out in three stages. The first stage is a complete DL classification with three CNNs to accomplish the classification process. This stage aims to investigate whether CNNs learned with radiomics images perform better than those constructed with CT and X-Ray images. In the second stage, the deep features attained from these CNNs learned with radiomics images and reduced using FWHT are fed to the classification models. In the last stage, for each CNN, features obtained using multiple radiomics after being reduced and incorporated using DCT are fed to the classifiers. These classifiers' performance is evaluated in comparison to classifiers learnt using features generated using CNNs trained on CT and X-Ray images. The target of this stage is to examine if incorporating multiple radiomics information can boost performance. To evaluate the effectiveness of the suggested tool, five-fold cross-validation techniques is employed.
4. Results
4.1. Setting the experiments
The amount of epochs (20), the frequency of validation (173), the learning rate (0.001), and the mini-batch dimension (10) are among the CNN's parameters that are set. The three CNNs are trained with the technique of stochastic gradient with a moment. For the SVM classifier, linear and quadratic kernels are used with a box constraint level adjusted to 1 all other parameters are kept with their default values. In the case of the ensemble subspace discriminate classifier, the number of learners is 30, the learning rate is 0.1, the subspace dimension is 5, and the maximum number of splitting is 20. For the LDA, the full size of the covariance structure is used. Precision, sensitivity, F1-score, accuracy, Mathew correlation coefficient (MCC), area under the operating characteristic curve (AUC), and specificity are some of the assessment indicators used to evaluate the effectiveness of the presented tool. To determine these indications, the following (1), (2), (3), (5), (6) are being used.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Where TN is consistent with the total number of non-COVID-19 scans, it is incorrectly diagnosed. TP represents the total number of COVID-19 images that were perfectly identified. FN is the total number of COVID-19 scans that are incorrectly recognized as non-COVID-19 scans. FP represents the total number of non-COVID-19 images that have been incorrectly identified as COVID-19.
Certain CNNs’ parameters, such as batch normalisation, which can reduce overfitting, are chosen to avoid overfitting [87]. Additionally, the augmentation procedure is used to increase the size of the training dataset and avert overfitting [88]. Besides that, because the huge amount of features used to learn machine learning classifiers could result in overfitting, we used the fast Hadamard transform to diminish the dimension of deep features obtained from each CNN. Furthermore, discrete cosine transform was then employed to decrease the dimension of attributes after the fusion step was accomplished to merge the deep features of the multiple CNNs.Thereby, feature reduction methods are necessary to prevent overfitting.
4.2. Stage I results
The outcomes of the DL classification process (end-to-end) are presented here. The three CNNs are constructed independently with CT images, X-Ray images, and the four radiomics methods involving GLRLM, GLCM, DWT, and DTCWT. The accuracies achieved using these CNN are displayed in Fig. 6 . This figure indicates that the performance of the three CNNs learned with radiomics images (GLRLM, GLCM, DWT, and DTCWT) is greater than those constructed directly with CT or X-Ray scans. This is obvious from Fig. 6(a), as for MobileNet constructed with CT scans, the accuracies when training with GLRLM, GLCM, DWT, and DTCWT images are 81.74%,83.36%,82.82%, and 86.44% respectively which are higher than that obtained by the raw CT image. Similarly, for DarkNet-53 learned using GLRLM, GLCM, DWT, and DTCWT photos, the accuracies range from 73.02% to 80.54% which is better than the 71.01% accuracy achieved with the genuine CT scan. Likewise, for DenseNet-201, the accuracy attained with radiomics based generated images of GLRLM, GLCM, DWT, and DTCWT are 81.48%, 84.83%,79.87%, 81% which is superior to the 73.02% accomplished with the raw CT photos. Furthermore, the same scenario appears again in Fig. 6 (b) for the case of X-ray images, as for MobileNet, the accuracy varies from 95.29% to 96.31% utilizing the GLRLM, GLCM, DWT, and DTCWT photos. These accuracies are greater than the 94.81% obtained by the original X-ray scans. Also, for DarkNet-53, the radiomics generated images via GLRLM, GLCM, DWT, and DTCWT photos resulted in accuracies that alter from 91% to 93.79% which is more than that reached by the raw X-ray images. Comparably, the accuracies of DenseNet-201 realized 94.2%,95.5%,96.38%, and 95.7% employing the GLRLM, GLCM, DWT, and DTCWT pictures which exceed that attained by the authentic X-ray image (86.21%). These findings verify that making use of radiomics pictures to feed CNNs could be preferable to directly using X-Ray or CT scans.
Fig. 6.
The accuracies (%) achieved using the three CNNs trained separately using CT images and radiomics images 'of (a) Dataset I, (b) Dataset II.
4.3. Stage II results
The classification findings of the machine learning classifiers, which were trained using deep features derived from the CNNs, are described in this section. These CNNs were trained using radiomics-generated images. The classification accuracy of stage II is displayed in Table 1, Table 2 for Dataset I and II respectively. Table 1 shows that the classification accuracy of four classification models (stage II) is higher than the end-to-end DL classification (stage I) for Dataset I as shown in Fig. 6 (a). This is because the classification accuracy for DarkNet-53 features trained with radiomics-based generated images of the CT method ranges from 93.3% to 97.5% which is greater than that obtained by the end-to-end DL classification of DarkNet-53 in Fig. 6 (a), The similar case occurs for MobileNet and DenseNet-201 attributes trained with radiomics-based generated images of CT method. As MobileNet and DenseNet features reach an accuracy that fluctuates between 96.8%- 98.7% and 93.0%–98.5% which beats that achieved in Fig. 6(b). This demonstrates that TL has enhanced the proposed tool's diagnostic performance. For DarkNet-53, the highest accuracy is achieved by using the quadratic-SVM classifier that is trained with deep features obtained with DWT radiomics-generated images. Whereas for MobileNet the greatest accuracy of 98.7% is attained using the quadratic-SVM classifier fed via deep features obtained with GLRLM radiomics images. In contrast, the quadratic-SVM classifier trained with deep features collected from GLRLM radiomics-generated images for DenseNet-201 has the highest accuracy (98.5%).
Table 1.
The SVM classification accuracy (%) achieved with deep feature obtained from the three CNNs constructed with radiomics images of dataset I.
| Kernel |
LDA | Linear-SVM | Quadratic-SVM | ESD |
|---|---|---|---|---|
| Radiomics | ||||
| DarkNet-53 | ||||
| GLCM | 96.2 | 94.6 | 97.2 | 96.2 |
| GLRLM | 94.3 | 93.3 | 95.4 | 94.0 |
| DWT | 96.7 | 94.6 | 97.3 | 96.6 |
| DTCWT | 94.9 | 93.3 | 95.5 | 94.4 |
| MobileNet | ||||
| GLCM | 97.7 | 97.3 | 98.5 | 98.3 |
| GLRLM | 96.8 | 97.0 | 98.6 | 97.0 |
| DWT | 97.5 | 97.5 | 98.7 | 97.5 |
| DTCWT | 97.6 | 96.9 | 98.6 | 96.9 |
| DenseNet-201 | ||||
| GLCM | 96.7 | 96.2 | 98.1 | 96.1 |
| GLRLM | 94.2 | 93.0 | 95.1 | 94.0 |
| DWT | 96.3 | 96.0 | 96.6 | 96.0 |
| DTCWT | 98.3 | 98.0 | 98.5 | 97.8 |
Table 2.
The SVM classification accuracy (%) achieved with deep feature obtained from the three CNNs constructed with radiomics images of Dataset II.
| Kernel |
LDA | Linear-SVM | Quadratic-SVM | ESD |
|---|---|---|---|---|
| Radiomics | ||||
| DarkNet-53 | ||||
| GLCM | 96 | 96.3 | 96.5 | 96 |
| GLRLM | 94.7 | 95.2 | 94.9 | 95.2 |
| DWT | 95.3 | 95.8 | 95.6 | 95.5 |
| DTCWT | 94.1 | 94.6 | 94.6 | 94.6 |
| MobileNet | ||||
| GLCM | 98 | 98.4 | 98.4 | 98.2 |
| GLRLM | 97.7 | 97.7 | 97.9 | 97.8 |
| DWT | 97.6 | 97.7 | 97.7 | 97.8 |
| DTCWT | 97.5 | 97.7 | 98 | 97.6 |
| DenseNet-201 | ||||
| GLCM | 98.2 | 98.6 | 98.3 | 98.1 |
| GLRLM | 97.9 | 98.3 | 98.3 | 97.9 |
| DWT | 97.9 | 98.4 | 98.4 | 98.0 |
| DTCWT | 97.9 | 98 | 98.1 | 97.8 |
Similarly, Table 2 shows that the diagnostic accuracy of the four classifiers (stage II) is greater than that of the DL end-to-end classification (stage I) for Dataset II as demonstrated in Fig. 6 (b). Table 2 demonstrates that the accuracy of the classification for DarkNet-53 attributes learned with radiomics-based generated images of the X-ray method varies from 94.1% to 96.5%, which is higher than the end-to-end DL classification of DarkNet-53 displayed in Fig. 6 (b). A similar situation exists for MobileNet and DenseNet-201 features learned with radiomics-based generated images of the X-Ray technique. The accuracy of MobileNet and DenseNet features varies between 97.7%-98.4% and 97.8%–98.6%, respectively, exceeding that of Fig. 6. (b). These results also confirm that TL can improve the diagnostic accuracy of the proposed tool. The accuracy obtained for data Dataset II varies between 94.1% and 98.6%. The DarkNet-53 provides the highest accuracy (96.5%) using the quadratic-SVM classifier, which is trained with deep features taken from GLCM radiomics images. While MobileNet has reached 98.4% accuracy with the quadratic-SVM classifier trained using deep features obtained from the GLCM radiomics images. On the other hand, DenseNet-201 achieves maximum accuracy (98.6%) with a linear-SVM classifier trained in the GLCM radiomics-generated images.
4.4. Stage III results
The following section describes the results obtained after incorporating deep features of multiple radiomics methods obtained from each CNN. As displayed in Table 3 , the results are compared to those classification models trained using features obtained with the three CNNs constructed via raw CT and X-Ray scans. Table 3 shows that the feature incorporation and reduction procedures have increased the classification accuracies for the three CNNs. This is due to the fact that for Dataset I, the DarkNet-53 achieves 97.4%,95.2%, 97.8 and 91.1% accuracies after the feature incorporation and reduction using DCT for the LDA, linear-SVM, quadratic-SVM, and ESD classifiers. Those results are greater than the 96.5%, 94.8%, 96.9, and 95.7% accuracies achieved using the exact classifiers but with features obtained using the raw CT scans. Likewise, the classification accuracies obtained with deep features of radiomics-generated images extracted from MobileNet and DenseNet-201 are improved after the feature incorporation and reduction steps compared to the classification models constructed with deep features of CT images. This is obvious as the accuracies accomplished from the classifiers using the fused and reduced deep features obtained via the quad-radiomics-generated images for DenseNet-201 and MobileNet are (98.5%, 98.5%), (98.3%,98.6%), (99.4%,99.4%), and (97.9% and 98.2%) for the LDA, linear-SVM, quadratic-SVM, and ESD classifier respectively.
Table 3.
SVM Classifiers Classification Accuracies achieved with incorporated and Reduced features of three CNNs learned with Radiomics images against CT and X-ray images.
| Model | LDA |
Linear-SVM |
Quadratic-SVM |
ESD |
||||
|---|---|---|---|---|---|---|---|---|
| Dataset I | ||||||||
| CT | Fused | CT | Fused | CT | Fused | CT | Fused | |
| Dark-53 | 96.5 | 97.4 | 94.8 | 95.3 | 96.9 | 97.8 | 95.7 | 97.1 |
| MobileNet | 96.9 | 98.5 | 96.8 | 98.3 | 98.1 | 99.4 | 96.6 | 97.9 |
|
DenseNet-201 |
96.9 |
98.5 |
96.1 |
98.6 |
97.9 |
99.4 |
95.8 |
98.2 |
|
Model |
Dataset II | |||||||
|
X-Ray |
Fused |
X-Ray |
Fused |
X-Ray |
Fused |
X-Ray |
Fused |
|
| Dark-53 | 96.7 | 97.3 | 96.4 | 96.7 | 96.6 | 97.2 | 96.6 | 97.5 |
| MobileNet | 96.8 | 97.5 | 96.5 | 97.7 | 96.8 | 98.0 | 96.6 | 97.6 |
| DenseNet-201 | 98.2 | 98.7 | 98.1 | 99.0 | 98.3 | 99.0 | 98.2 | 98.9 |
The feature fusion and reduction using DCT have also improved the classification accuracy for the three deep learning models for Dataset II, according to Table 3. The results after these procedures are compared to that achieved by the original X-Ray images and are shown in Table 3. In the case of MobileNet, the accuracies achieved after the fusion and reduction process is 97.5%, 97.7%, 98.0%, and 97.6%. These accuracies are greater than the 96.8%m 96.5%, 96.8%, and 96.6% accuracies attained using the same classifier trained with features of the original X-Ray images. Likewise, for DarkNet-53 and DenseNet-201, the classification accuracies have increased after the feature incorporation and reduction procedures compared to those obtained by the features of the original X-Ray images.
It can be noted from Table 3 that the quadratic-SVM classifier achieved the highest accuracies for the three CNNs. In Table 4 , additional evaluation indicators for the quadratic-SVM classifier are computed and shown. The table indicates that for Dataset I, the fused radiomics deep features of DenseNet-201 and MobileNet have comparable performance. The precision, specificity, F1-score, MCC, and sensitivity achieved using the quadratic SVM trained with radiomics deep features of DenseNet-201 are 99.44%, 99.43%, 99.40%, 97.79%, and 99.36% respectively. Similarly, for Dataset II, radiomics features of both MobileNet and DenseNet-101 have equivalent performance. The radiomics deep features of DenseNet-201 attained 99.01% precision, 99.49% specificity, 99.01 F1-score, 98.5% MCC, and 98.94% sensitivity. The confusion matrices for the LDA and linear-SVM classifiers trained with radiomics deep features of DenseNet-201 are displayed in Fig. 7 and Fig. 8 for Dataset I and Dataset II respectively. These Figures demonstrate that RADIC can accurately classify each class of Dataset I with a sensitivity of 97.7% and 98.2% for COVID-19 class and 99.3% and 98.9% for the Non-Covid-19 class using LDA and linear SVM respectively (Fig. 7). Similarly, for Dataset II, RADIC is able to accurately classify 98.5% and 99.0% of the COVID-19 cases, 98.5% and 98.6% of the normal cases n and finally 99.2% and 99.3% of the pneumonia cases utilizing the LDA and linear SVM classifiers. Additionally, Fig. 9, Fig. 10 for Dataset I and Dataset II, respectively, demonstrate the receiving operating characteristic curve (ROC) and the area under the ROC (AUC) for the LDA and linear-SVM classifiers trained with radiomics deep features of DenseNet-201. Fig. 9, Fig. 10 show that the LDA, and linear SVM classifiers all achieved an AUC of 1.
Table 4.
Performance metrics (%) for the quadratic SVM trained with the fused radiomics features.
| Model | Precision | Specificity | F1-score | MCC | Sensitivity |
|---|---|---|---|---|---|
| Dataset I | |||||
| Dark-53 | 97.36 | 97.34 | 97.79 | 95.57 | 98.23 |
| MobileNet | 99.60 | 99.59 | 99.40 | 98.79 | 99.20 |
| DenseNet-201 | 99.44 | 99.43 | 99.40 | 98.79 | 99.36 |
| Dataset II | |||||
| Dark-53 | 97.21 | 97.55 | 97.24 | 95.78 | 97.27 |
| MobileNet | 98.94 | 99.45 | 98.94 | 98.38 | 98.94 |
| DenseNet-201 | 99.01 | 99.49 | 99.01 | 98.5 | 99.01 |
Fig. 7.
The confusion matrices for Dataset I achieved with incorporated radiomics deep features of DenseNet-201 (a) LDA classifier, and (b) Linear-SVM classifier.
Fig. 8.
The confusion matrices for Dataset II achieved with incorporated radiomics deep features of DenseNet-201 (a) LDA classifier, and (b) Linear-SVM classifier.
Fig. 9.
The ROC curves for Dataset I obtained employing DenseNet-201's integrated radiomics deep features (a) LDA classifier and (b) Linear-SVM classifier.
Fig. 10.
The ROC curves for Dataset II obtained employing DenseNet-201's integrated radiomics deep features (a) LDA classifier and (b) Linear-SVM classifier.
5. Discussion
This article proposed a DL tool called “RADIC” for COVID-19. Rather than directly using CT or X-Ray scans to build diagnostic models, RADIC utilized multiple radiomics images generated through quad-radiomics methods involving DWT, DTCWT, GLRLM, and GLCM. RADIC operated the classification process through three stages. In stage I, an end-to-end DL classification procedure is performed, where three CNNs (DarkNet-53, MobileNet, and DenseNet-201) were trained with each type of radiomics individually. The performance of these CNNs was compared with the same CNNs trained with the raw CT and X-Ray images. The results of this comparison were displayed in Fig. 6. The results that were shown in Fig. 6 confirmed that employing radiomic-generated images as an alternative to the original X-Ray and CT-scans is capable of boosting the performance of RADIC. Next, in stage II, deep features were extracted from each CNN trained with images of each radiomics approach and reduced using FWHT, then used to train four classifiers. The results of this stage were displayed in Table 1, Table 2 for Dataset I and II respectively. These results proved that TL has increased the classification performance compared to end-to-end DL classification. The results also verified that using the time-frequency representation of COVID-19 patterns obtained with FWHT is useful in enhancing diagnostic performance. For Dataset I, deep features of both GLCM and DWT radiomics-generated images had the highest performance for the three CNNs. Whereas, for Dataset II, GLCM, GLRLM, DTCWT, and DWT radiomics-generated images had comparable performance, whereas radiomics deep features of GLCM had slightly higher performance than other radiomics.
Finally, in the last classification stage of RADIC, the four types of radiomics deep features extracted from each CNN were incorporated and reduced using DCT. The results of this stage were shown in Table 3. The results proved that the third stage of RADIC had a further improvement in diagnostic performance. These outcomes were contrasted with the effectiveness of classifiers built using deep features from the original X-Ray and CT images. This comparison indicated that using radiomics information is better than utilizing the spatial information existing in the X-Ray and CT scans.
In order to visualize and discuss the explainability of the results, a gradient-weighted class activation mapping (Grad-CAM) analysis is conducted. In the X-Ray or CT images, the Grad-CAM generates a heap map of the affected region of the lungs. A few examples of the results are presented in Fig. 11 and Fig. 12 for Dataset I and Dataset II respectively. This research utilizes the jet colour theme. Throughout this scheme, blue shades resemble smaller values, indicating that no attributes have been obtained for a particular category, whilst green and yellow shades demonstrate intermediate values, demonstrating significantly fewer retrieved features, and red and darker red shades comprise higher values, revealing that the attributes in the area characterize the particular class [89].
Fig. 11.
Sample images of Grad-CAM analysis on the COVID-19 CT radiomics based generated images for MobileNet, DarkNet-53, and DenseNet-201.
Fig. 12.
Sample images of Grad-CAM analysis on the COVID-19 X-Ray radiomics based generated images for MobileNet, DarkNet-53, and DenseNet-201.
5.1. Comparisons with deep learning models
In order to prove the outstanding performance of the RADIC tool, its performance is compared against baseline DL classification models. The results of this comparison are demonstrated in Fig. 13 . This figure confirms the beating performance of RADIC compared with other deep learning models. This is because RADIC has achieved an accuracy of 99.4% and 99.0% for Dataset I and Dataset II respectively which are greater than that attained by other deep learning classification models that were trained with either CT or X-Ray original images.
Fig. 13.
Comparison between the performance of RADIC and baseline deep learning classification models.
5.2. Comparisons with related studies
The proposed tool's performance is compared against related research articles that employed Dataset I. Table 5 demonstrates this comparison. Table 5 validates the competitiveness of the proposed tool. The table also demonstrates the superiority of using radiomics pictures while creating CNNs over using actual CT images. This is clear because the proposed tool results are higher than other studies employing CT scans for diagnostic procedures. Consequently, the RADIC tool can aid radiologists in making a more timely and precise diagnosis.
Table 5.
Performance metrics (%) Comparisons with other related studies based on Dataset I.
| Article | Accuracy | Precision | Sensitivity | Specificity | F1-score | MCC | AUC |
|---|---|---|---|---|---|---|---|
| [90] | 97.8 | 97.8 | 97.8 | – | 97.7 | – | 97.81 |
| [58] | 87.6 | – | – | – | 86.2 | – | 90.5 |
| [91] | 97.0 | – | – | – | 97.0 | – | 99.38 |
| [42] | 94.0 | 95.00 | 94.00 | – | 94.5 | – | – |
| [40] | 96.3 | 96.3 | 96.3 | – | 96.3 | – | – |
| [41] | 96.4 | 94.6 | 98.4 | 94.4 | 96.5 | 92.9 | – |
| Proposed RADIC | 99.4 | 99.44 | 99.36 | 99.43 | 99.40 | 98.79 | 100 |
RADIC tool performance is also compared to research studies based on the same X-ray dataset and other comparable datasets containing the same three classes. The results of this comparison are shown in Table 6 . This comparison shows that the proposed tool has a strong competing ability. This table also validates that using images created by radiomics to build a CNN is better than using an original X-ray image. This is clear because RADIC's performance is outperforming other studies that used X-Ray scans for diagnosing COVID-19. Consequently, RADIC tools can help radiologists diagnose more accurately and faster.
Table 6.
Performance metrics (%) Comparisons with other related studies based on Dataset II and other X-Ray 3- classes datasets.
5.3. Complexity analysis of RADIC tool
As stated before, the classification procedure of the proposed approach is done in three stages. In the first stage, end-to-end DL classification is performed. In this stage, the classification is achieved by three CNNs trained with radiomics-based generated images. Next, in the second stage, deep features were extracted from each CNN and reduced using FHWT which then feed machine learning classifiers. Finally, these reduced deep features are combined using DCT to further diminish the dimension of features.
According to the training complexity of the first and second stages, it is clear from Table 7 that only 500 features are required to execute classification with the LDA and SVM classifiers used in the RADIC tool, which isn't the situation for CNNs, which require the entire images to accomplish classification and go through a complex process to achieve a decision (great amount of deep layers and vast quantity of parameters). Furthermore, in the final stage of the RADIC tool, only a few training parameters are required to complete the classification procedure, as opposed to millions of parameters in each CNN classification model. Lastly, the layer structure of every CNN model (number of layers and complexity per layer) is much more complex compared to the RADIC tool's final stages, resulting in a greater computational load to complete the identification process. Consequently, when compared to end-to-end DL models, the RADIC tool was verified to be more accurate, efficient, and had the lowest computational burden (few amount of input data dimensions, training hyper-parameters, as well as classification layers).
Table 7.
The complexity analysis of the three classification stages of RADIC.
| Model | Input Data Dimesnion to the Model | Number of the model hyperparameters parameters (N) | Number of Layers | Training Complexity per Layer (O) |
|---|---|---|---|---|
| Stage I of Classification | ||||
| DenseNet-201 | Images of Size 224x224x3 | 16.5 M | 201 |
[96] k: kernel size n: sequence length (number of input data) d: representation dimension |
| DarkNet-53 | Images of Size 256x256x3 | 41.0 M | 53 | |
| MobileNet | Images of Size 224x224x3 | 3.5 M | 28 | |
| Stage II of Classification | ||||
| Deep Features Extraction from Radiomics-based Generated Images and Reduction with FWHT and SVM classifiers | 500 Features |
SVM C: Regularization parameter Gamma: width of the Kernel C: number of class labels. p: number of features |
– |
SVM ) [97] p: number of features m: number of input samples LDA ) [98] p: number of features m: number of input samples s: the average number of non-zero features of one sample t: min (m,p) |
| Stage III of Classification | ||||
| Incorporated Deep Features of the three CNNs using DCT and SVM classifiers | 500 Features |
SVM C: Regularization parameter Gamma:width of the Kernel C: number of class labels. p: number of features |
– |
SVM ) p: number of features m: number of input samples LDA ) p: number of features m: number of input samples s: the average number of non-zero features of one sample t: min (m,p) |
6. Conclusion
The study proposed a DL-based diagnostic tool called “RADIC” for COVID-19 diagnosis. The proposed tool did not use directly the actual X-Ray and CT images but utilized radiomics images generated by four radiomics approaches including GLCM, GLRLM, DWT, and DTCWT methods. The proposed tool accomplished classification in three stages. Firstly, the four radiomics methods are applied to the original images, then radiomics images are created by generating heatmaps of the outputs of the GLCM, GLRLM, DWT, and DTCWT methods. Afterward, those pictures are employed as inputs to the three CNNs including DarkNet-53, MobileNet, and DenseNet-201. This is the first classification stage of the proposed tool. Subsequently, features were attained through every CNN constructed with radiomics-generated images. These features are of high dimension. Thus they are reduced using FWHT which demonstrated the time-frequency representation of COVID-19 patterns. These reduced features are used to train four machine learning models independently. In the suggested tool, this stage serves as the second classification stage. Next, in stage III for each CNN, the deep features obtained from DL models trained using the radiomics methods are incorporated and reduced using DCT. Stage I results confirmed that radiomics images are more useful in training CNNs than X-Ray and CT images. Furthermore, stage II results have shown that TL for feature extraction of deep features can increase diagnostic performance. Additionally, it demonstrated how time-frequency representations can improve diagnostic efficiency. Finally, stage III showed that merging multiple radiomics information could further improve performance to an accuracy of 99.4% and 99.0% for the CT and X-Ray datasets respectively. As a result, the proposed tool can help medical experts to accurately and quickly diagnose COVID-19. It can be employed as a clinical diagnostic tool for numerous disorders thanks to the proposed tool's promising performance. This study does not consider methods of optimization for the selection of deep learning hyperparameters. Furthermore, the study did not take into account the uncertainties of the dataset entered into deep learning models. The noise present in real CT and X-ray scanning may lower diagnostic accuracy, but this problem has not been studied. Furthermore, the study did not address the issue of distinguishing COVID-19 from other types of pneumonia and chest diseases, which should be tackled in future research. Also, it was not evaluated for other diseases or medical abnormalities. Also, it does not provide a correlation analysis to measure the degree of association between deep features and medical interpretations. Future work will address the aforementioned limitations. Besides, upcoming work will consider the use of more feature reduction techniques and deep learning methods.
Availability of supporting data
Dataset I is available at: https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset.
Dataset II is available at: https://github.com/sharma-anubhav/COVDC-Net.
Funding
The study did not receive any sort of external funding.
Author statement
The current study was conducted by Omneya Attallah who is the single author of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Dataset I is available at: https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset Dataset II is available at: https://github.com/sharma-anubhav/COVDC-Net
References
- 1.Domingo J.L. What we know and what we need to know about the origin of SARS-CoV-2. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., et al. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int. J. Infect. Dis. 2020;94:107–109. doi: 10.1016/j.ijid.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attallah O. ECG-BiCoNet: an ECG-based pipeline for COVID-19 diagnosis using Bi-Layers of deep features integration. Comput. Biol. Med. 2022 doi: 10.1016/j.compbiomed.2022.105210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung M., et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cozzi A., et al. Chest x-ray in the COVID-19 pandemic: radiologists' real-world reader performance. Eur. J. Radiol. 2020;132 doi: 10.1016/j.ejrad.2020.109272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rousan L.A., Elobeid E., Karrar M., Khader Y. Chest x-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020;20(1):1–9. doi: 10.1186/s12890-020-01286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serrano C.O., et al. Pediatric chest x-ray in covid-19 infection. Eur. J. Radiol. 2020;131 doi: 10.1016/j.ejrad.2020.109236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alyasseri Z.A.A., et al. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expet Syst. 2021 doi: 10.1111/exsy.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Attallah O., Abougharbia J., Tamazin M., Nasser A.A. A BCI system based on motor imagery for assisting people with motor deficiencies in the limbs. Brain Sci. 2020;10(11):864–888. doi: 10.3390/brainsci10110864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohr A., Memarzadeh K. Artificial Intelligence in Healthcare. Elsevier; 2020. The rise of artificial intelligence in healthcare applications; pp. 25–60. [Google Scholar]
- 12.Karthikesalingam A., et al. An artificial neural network stratifies the risks of Reintervention and mortality after endovascular aneurysm repair; a retrospective observational study. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0129024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attallah O., Ragab D.A. Auto-MyIn: automatic diagnosis of myocardial infarction via multiple GLCMs, CNNs, and SVMs. Biomed. Signal Process Control. 2023;80 [Google Scholar]
- 14.Attallah O., Ai-His M.B.- Histopathological diagnosis of pediatric medulloblastoma and its subtypes via AI. Diagnostics. 2021;11(2):359–384. doi: 10.3390/diagnostics11020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attallah O., Sharkas M. vol. 2021. Contrast Media & Molecular Imaging; 2021. (Intelligent Dermatologist Tool for Classifying Multiple Skin Cancer Subtypes by Incorporating Manifold Radiomics Features Categories). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attallah O. CoMB-deep: composite deep learning-based pipeline for classifying childhood medulloblastoma and its classes. Front. Neuroinf. 2021;15 doi: 10.3389/fninf.2021.663592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Attallah O., Anwar F., Ghanem N.M., Ismail M.A. Histo-CADx: duo cascaded fusion stages for breast cancer diagnosis from histopathological images. PeerJ Computer Science. 2021;7:e493. doi: 10.7717/peerj-cs.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attallah O., Aslan M.F., Sabanci K. A framework for lung and colon cancer diagnosis via lightweight deep learning models and transformation methods. Diagnostics. 2022;12(12):2926. doi: 10.3390/diagnostics12122926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Attallah O., Sharkas M. GASTRO-CADx: a three stages framework for diagnosing gastrointestinal diseases. PeerJ Computer Science. 2021;7:e423. doi: 10.7717/peerj-cs.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arumugam M., Sangaiah A.K. Arrhythmia identification and classification using wavelet centered methodology in ECG signals. Concurrency Comput. Pract. Ex. 2020;32(17):e5553. [Google Scholar]
- 21.Attallah O. An intelligent ECG-based tool for diagnosing COVID-19 via ensemble deep learning techniques. Biosensors. 2022;12(5):299. doi: 10.3390/bios12050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attallah O. 2022 the 12th International Conference on Information Communication and Management. 2022. Deep learning-based CAD system for COVID-19 diagnosis via spectral-temporal images; pp. 25–33. [Google Scholar]
- 23.Attallah O. DIAROP: automated deep learning-based diagnostic tool for retinopathy of prematurity. Diagnostics. 2021;11(11):2034. doi: 10.3390/diagnostics11112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attallah O. A deep learning-based diagnostic tool for identifying various diseases via facial images. Digital Health. 2022;8 doi: 10.1177/20552076221124432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hertel R., Benlamri R. A deep learning segmentation-classification pipeline for x-ray-based covid-19 diagnosis. Biomedical Engineering Advances. 2022 doi: 10.1016/j.bea.2022.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Timmeren J.E., Cester D., Tanadini-Lang S., Alkadhi H., Baessler B. Radiomics in medical imaging—‘how-to’ guide and critical reflection. Insights into imaging. 2020;11(1):1–16. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le N.Q.K., Hung T.N.K., Do D.T., Lam L.H.T., Dang L.H., Huynh T.-T. Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104320. [DOI] [PubMed] [Google Scholar]
- 28.Alafif T., Tehame A.M., Bajaba S., Barnawi A., Zia S. Machine and deep learning towards COVID-19 diagnosis and treatment: survey, challenges, and future directions. Int. J. Environ. Res. Publ. Health. 2021;18(3):1117. doi: 10.3390/ijerph18031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., et al. Diagnosis of COVID-19 pneumonia via a novel deep learning architecture. J. Comput. Sci. Technol. 2022;37(2):330–343. doi: 10.1007/s11390-020-0679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian N., Elharrouss O., Al-Maadeed S., Chowdhury M. A review of deep learning-based detection methods for COVID-19. Comput. Biol. Med. 2022 doi: 10.1016/j.compbiomed.2022.105233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serte S., Demirel H. Deep learning for diagnosis of COVID-19 using 3D CT scans. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haghanifar A., Majdabadi M.M., Choi Y., Deivalakshmi S., Ko S. Covid-cxnet: detecting covid-19 in frontal chest x-ray images using deep learning. Multimed. Tool. Appl. 2022:1–31. doi: 10.1007/s11042-022-12156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharifrazi D., et al. Fusion of convolution neural network, support vector machine and Sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomed. Signal Process Control. 2021;68 doi: 10.1016/j.bspc.2021.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R.K., Pandey R., Babu R.N. COVIDScreen: explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Comput. Appl. 2021;33(14):8871–8892. doi: 10.1007/s00521-020-05636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loey M., El-Sappagh S., Mirjalili S. Bayesian-based optimized deep learning model to detect COVID-19 patients using chest X-ray image data. Comput. Biol. Med. 2022 doi: 10.1016/j.compbiomed.2022.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jalali S.M.J., Ahmadian M., Ahmadian S., Hedjam R., Khosravi A., Nahavandi S. X-ray image based COVID-19 detection using evolutionary deep learning approach. Expert Syst. Appl. 2022;201 doi: 10.1016/j.eswa.2022.116942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nasiri H., Hasani S. Automated detection of COVID-19 cases from chest X-ray images using deep neural network and XGBoost. Radiography. 2022;28(3):732–738. doi: 10.1016/j.radi.2022.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Attallah O. A computer-aided diagnostic framework for coronavirus diagnosis using texture-based radiomics images. Digital Health. 2022;8 doi: 10.1177/20552076221092543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2021;39(15):5682–5689. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 41.Singh V.K., Kolekar M.H. Deep learning empowered COVID-19 diagnosis using chest CT scan images for collaborative edge-cloud computing platform. Multimed. Tool. Appl. 2022;81(1):3–30. doi: 10.1007/s11042-021-11158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-scan images. Chaos, Solit. Fractals. 2020 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kogilavani S.V., et al. COVID-19 detection based on lung CT scan using deep learning techniques. Comput. Math. Methods Med. 2022;2022 doi: 10.1155/2022/7672196. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Oğuz Ç., Yağanoğlu M. Detection of COVID-19 using deep learning techniques and classification methods. Inf. Process. Manag. 2022;59(5) doi: 10.1016/j.ipm.2022.103025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., et al. Diagnosis of COVID-19 pneumonia via a novel deep learning architecture. J. Comput. Sci. Technol. 2022;37(2):330–343. doi: 10.1007/s11390-020-0679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Showkat S., Qureshi S. Efficacy of transfer learning-based ResNet models in chest X-ray image classification for detecting COVID-19 pneumonia. Chemometr. Intell. Lab. Syst. 2022;224 doi: 10.1016/j.chemolab.2022.104534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allioui H., et al. A multi-agent deep reinforcement learning approach for enhancement of COVID-19 CT image segmentation. J. Personalized Med. 2022;12(2):309. doi: 10.3390/jpm12020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ragab D.A., Attallah O. FUSI-CAD: coronavirus (COVID-19) diagnosis based on the fusion of CNNs and handcrafted features. PeerJ Computer Science. 2020;6:e306. doi: 10.7717/peerj-cs.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhattacharyya A., Bhaik D., Kumar S., Thakur P., Sharma R., Pachori R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed. Signal Process Control. 2022;71 doi: 10.1016/j.bspc.2021.103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdar M., et al. UncertaintyFuseNet: robust uncertainty-aware hierarchical feature fusion model with ensemble Monte Carlo dropout for COVID-19 detection. Inf. Fusion. 2022;90:364–381. doi: 10.1016/j.inffus.2022.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aslan N., Koca G.O., Kobat M.A., Dogan S. Multi-classification deep CNN model for diagnosing COVID-19 using iterative neighborhood component analysis and iterative ReliefF feature selection techniques with X-ray images. Chemometr. Intell. Lab. Syst. 2022;224 doi: 10.1016/j.chemolab.2022.104539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Attallah O., Ragab D.A., Sharkas M. MULTI-DEEP: a novel CAD system for coronavirus (COVID-19) diagnosis from CT images using multiple convolution neural networks. PeerJ. 2020;8 doi: 10.7717/peerj.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Attallah O., Samir A. A wavelet-based deep learning pipeline for efficient COVID-19 diagnosis via CT slices. Appl. Soft Comput. 2022 doi: 10.1016/j.asoc.2022.109401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi X., Brown L.G., Foran D.J., Nosher J., Hacihaliloglu I. Chest X-ray image phase features for improved diagnosis of COVID-19 using convolutional neural network. Int. J. Comput. Assist. Radiol. Surg. 2021;16(2):197–206. doi: 10.1007/s11548-020-02305-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das A. Adaptive UNet-based lung segmentation and ensemble learning with CNN-based deep features for automated COVID-19 diagnosis. Multimed. Tool. Appl. 2022;81(4):5407–5441. doi: 10.1007/s11042-021-11787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khan M.A., et al. COVID-19 case recognition from chest CT images by deep learning, entropy-controlled firefly optimization, and parallel feature fusion. Sensors. 2021;21(21):7286. doi: 10.3390/s21217286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang L., Wang S.-H., Zhang Y.-D. Ednc: ensemble deep neural network for covid-19 recognition. Tomography. 2022;8(2):869–890. doi: 10.3390/tomography8020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva P., et al. COVID-19 detection in CT images with deep learning: a voting-based scheme and cross-datasets analysis. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gouda W., Almurafeh M., Humayun M., Jhanjhi N.Z. Detection of COVID-19 based on chest X-rays using deep learning. Healthcare. 2022;10(2):343. doi: 10.3390/healthcare10020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma A., Singh K., Koundal D. A novel fusion based convolutional neural network approach for classification of COVID-19 from chest X-ray images. Biomed. Signal Process Control. 2022 doi: 10.1016/j.bspc.2022.103778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rehman N., et al. A self-activated cnn approach for multi-class chest-related COVID-19 detection. Appl. Sci. 2021;11(19):9023. [Google Scholar]
- 62.Attallah O., Zaghlool S. AI-based pipeline for classifying pediatric medulloblastoma using histopathological and textural images. Life. 2022;12(2):232. doi: 10.3390/life12020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humeau-Heurtier A. Texture feature extraction methods: a survey. IEEE Access. 2019;7:8975–9000. [Google Scholar]
- 64.Mohanty A.K., Beberta S., Lenka S.K. Classifying benign and malignant mass using GLCM and GLRLM based texture features from mammogram. Int. J. Eng. Res. Afr. 2011;1(3):687–693. [Google Scholar]
- 65.Mishra S., Majhi B., Sa P.K. Recent Findings in Intelligent Computing Techniques. Springer; 2018. Glrlm-based feature extraction for acute lymphoblastic leukemia (all) detection; pp. 399–407. [Google Scholar]
- 66.Ming J.T.C., Noor N.M., Rijal O.M., Kassim R.M., Yunus A. Lung disease classification using GLCM and deep features from different deep learning architectures with principal component analysis. International Journal of Integrated Engineering. 2018;10(7) [Google Scholar]
- 67.Sundararajan D. John Wiley & Sons; 2016. Discrete Wavelet Transform: a Signal Processing Approach. [Google Scholar]
- 68.Raj V.N.P., Venkateswarlu T. Denoising of medical images using dual tree complex wavelet transform. Procedia Technology. 2012;4:238–244. [Google Scholar]
- 69.Vermaak H., Nsengiyumva P., Luwes N. Using the dual-tree complex wavelet transform for improved fabric defect detection. J. Sens. 2016;2016 [Google Scholar]
- 70.Anwar S.M., Majid M., Qayyum A., Awais M., Alnowami M., Khan M.K. Medical image analysis using convolutional neural networks: a review. J. Med. Syst. 2018;42(11):1–13. doi: 10.1007/s10916-018-1088-1. [DOI] [PubMed] [Google Scholar]
- 71.Liu W., Wang Z., Liu X., Zeng N., Liu Y., Alsaadi F.E. A survey of deep neural network architectures and their applications. Neurocomputing. 2017;234:11–26. [Google Scholar]
- 72.Xing J., Jia M. A convolutional neural network-based method for workpiece surface defect detection. Measurement. 2021;176 [Google Scholar]
- 73.Redmon J., Farhadi A. 2018. Yolov3: an Incremental Improvement. arXiv preprint arXiv:1804.02767. [Google Scholar]
- 74.Ullah A., Muhammad K., Ding W., Palade V., Haq I.U., Baik S.W. Efficient activity recognition using lightweight CNN and DS-GRU network for surveillance applications. Appl. Soft Comput. 2021;103 [Google Scholar]
- 75.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 76.Islam M.M., Shuvo M.M.H. 2019 5th International Conference on Advances in Electrical Engineering. ICAEE); 2019. DenseNet based speech imagery EEG signal classification using gramian angular field; pp. 149–154. [Google Scholar]
- 77.Shan L., Wang W. DenseNet-based land cover classification network with deep fusion. Geosci. Rem. Sens. Lett. IEEE. 2021;19:1–5. [Google Scholar]
- 78.Wang S.-H., Zhang Y.-D. DenseNet-201-based deep neural network with composite learning factor and precomputation for multiple sclerosis classification. ACM Trans. Multimed Comput. Commun. Appl. 2020;16(2s):1–19. [Google Scholar]
- 79.Howard A.G., et al. 2017. Mobilenets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv preprint arXiv:1704.04861. [Google Scholar]
- 80.Ahmed S., Bons M. Proceedings of the 5th International Workshop on Non-intrusive Load Monitoring. 2020. Edge computed NILM: a phone-based implementation using MobileNet compressed by tensorflow lite; pp. 44–48. [Google Scholar]
- 81.Soares E., Angelov P., Biaso S., Froes M.H., Abe D.K. SARS-CoV-2 CT-scan dataset: a large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv. 2020 [Google Scholar]
- 82.A. Sharma, K. Singh, and K. Koundal, “Dataset for COVDC-net.” Accessed: January. 4, 2022. [Online]. Available: https://github.com/sharma-anubhav/COVDC-.
- 83.Ahmed N., Rao K.R. Springer Science & Business Media; 2012. Orthogonal Transforms for Digital Signal Processing. [Google Scholar]
- 84.Subathra M.S.P., Mohammed M.A., Maashi M.S., Garcia-Zapirain B., Sairamya N.J., George S.T. Detection of focal and non-focal electroencephalogram signals using fast walsh-hadamard transform and artificial neural network. Sensors. 2020;20(17):4952. doi: 10.3390/s20174952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aydoğdu Ö., Ekinci M. An approach for streaming data feature extraction based on discrete cosine transform and particle swarm optimization. Symmetry. 2020;12(2):299. [Google Scholar]
- 86.Dabbaghchian S., Ghaemmaghami M.P., Aghagolzadeh A. Feature extraction using discrete cosine transform and discrimination power analysis with a face recognition technology. Pattern Recogn. 2010;43(4):1431–1440. [Google Scholar]
- 87.Garbin C., Zhu X., Marques O. Dropout vs. batch normalization: an empirical study of their impact to deep learning. Multimed. Tool. Appl. 2020;79(19):12777–12815. [Google Scholar]
- 88.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. Journal of Big Data. 2019;6(1):1–48. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Umair M., et al. Detection of COVID-19 using transfer learning and grad-CAM visualization on indigenously collected X-ray dataset. Sensors. 2021;21(17):5813. doi: 10.3390/s21175813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kundu R., Singh P.K., Ferrara M., Ahmadian A., Sarkar R. ET-NET: an ensemble of transfer learning models for prediction of COVID-19 infection through chest CT-scan images. Multimed. Tool. Appl. 2022;81(1):31–50. doi: 10.1007/s11042-021-11319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng B., Zhu Y., Shi Q., Yang D., Shao Y., Xu T. MA-Net: mutex attention network for COVID-19 diagnosis on CT images. Appl. Intell. 2022:1–16. doi: 10.1007/s10489-022-03431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and engineering sciences in medicine. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cengil E., Çınar A. The effect of deep feature concatenation in the classification problem: an approach on COVID-19 disease detection. Int. J. Imag. Syst. Technol. 2022;32(1):26–40. doi: 10.1002/ima.22659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khan A.I., Shah J.L., Bhat M.M. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Progr. Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaswani A., et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017;30 [Google Scholar]
- 97.Younes H., Alameh M., Ibrahim A., Rizk M., Valle M. Electronic Skin. River Publishers; 2022. Efficient algorithms for embedded tactile data processing; pp. 113–138. [Google Scholar]
- 98.Cai D., He X., Han J. 2008 IEEE 24th International Conference on Data Engineering. 2008. Training linear discriminant analysis in linear time; pp. 209–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Dataset I is available at: https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset.
Dataset II is available at: https://github.com/sharma-anubhav/COVDC-Net.
Dataset I is available at: https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset Dataset II is available at: https://github.com/sharma-anubhav/COVDC-Net