Abstract
Nematode cuticles are extracellular matrices (ECMs) that function as structural support and permeability barriers. Genetic disruption of specific cuticle collagen structures or secreted epidermal proteins in C. elegans activates stress response genes in epithelial cells suggesting the presence of an extracellular damage signaling mechanism. Cuticles are replaced during development via molting but investigations of extracellular signaling to stress responses have focused on adults. In our current study, we measured cuticle phenotypes and stress response gene expression in all post-embryonic stages of mutant strains for a collagen and two secreted epidermal proteins to gain insights into developmental patterns.
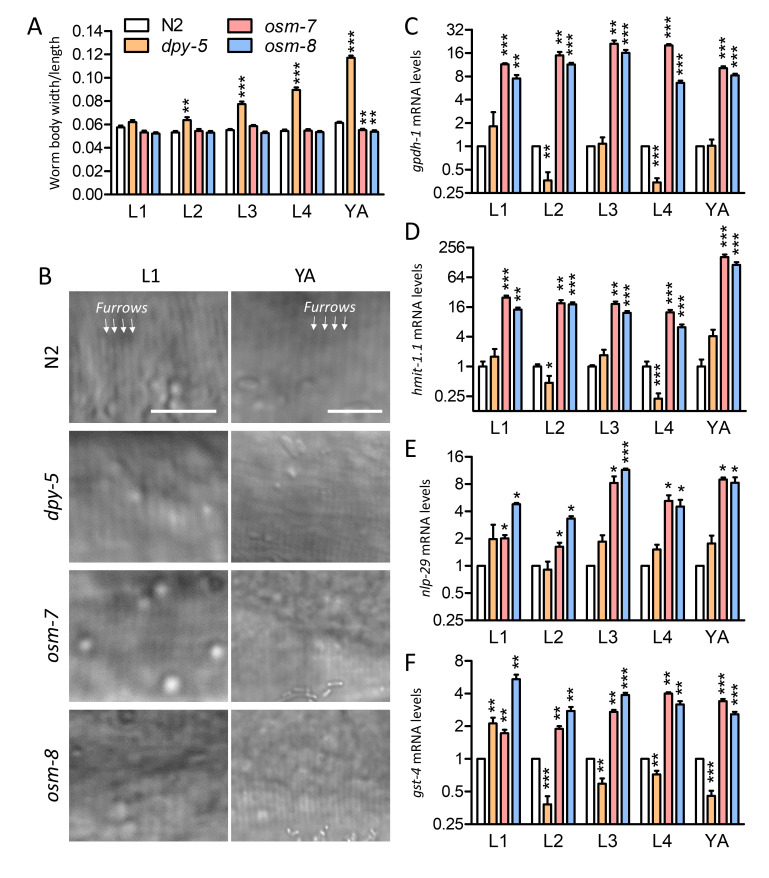
Figure 1. Cuticle phenotypes and stress response gene expression in dpy-5 , osm-7 , and osm-8 worms.
Mean plus standard error for worm body shape (A) and mRNA levels of representative stress response genes (C-F). *P<0.05, **P<0.01, and ***P<0.001; N = 10-17 individual worms in A and 4-6 replicate populations of worms in C-D. Representative high magnification DIC images of the cuticle are shown in B and were similar in all stages and individual worms. Scale bars are 5 μm in B.
Description
Extracellular matrices (ECMs) act as barriers to environmental stress on the outer surface of animals. Within cells, well-studied molecular pathways sense stress and regulate cytoprotective gene expression (Choe et al. 2009, Blackwell et al. 2015, Urso and Lamitina 2021, Pujol and Ewbank 2022). Despite being in direct contact with the environment, little is known about the role of barrier ECMs in sensing environmental conditions and regulating stress response genes in underlying cells.
Nematodes have a tough and flexible collagen-based barrier ECM called the cuticle that is re-synthesized and molted during transitions between post-embryonic developmental stages (Cox and Hirsh 1985, Johnstone 2000, Page and Johnstone 2007). We and others discovered that genetic disruption of collagens that form repeating ‘annular furrow’ structures in the superficial layer of the adult cuticle activates osmotic, detoxification, and innate immune response genes in epithelial cells (Lamitina et al. 2006, Pujol et al. 2008, Dodd et al. 2018). Manipulation of other cuticle and epidermal structures does not have the same effects on these stress responses and heat shock and organelle-specific stress responses are not activated by furrow disruption (Dodd et al. 2018). These results are consistent with a damage signaling pathway in adults that is associated with furrows. There are six collagens that are required for organization of furrows in adults and they belong to a larger class of 26 dpy genes that cause a short and wide body shape when mutated or silenced (McMahon et al. 2003).
All post-embryonic stage cuticles have furrows, but other cuticle structures, collagen gene expression, and thickness vary between stages (Cox et al. 1981, Hendriks et al. 2014). We recently demonstrated that furrow collagen gene mutations cause a progressive loss of furrow organization and body shape starting at the L3 stage (Chandler and Choe 2022); among stages, stress response gene expression was induced the strongest in adults correlated with the strongest furrow disorganization and Dpy phenotypes.
To gain further insights into extracellular stress response signaling during development, we investigated worms with mutations in a dpy collagen gene not required for furrow organization in adults (dpy-5) and non-collagen secreted protein genes osm-7 and osm-8 that are required for regulation of the same stress responses as furrow collagens without any obvious influence on body shape (Wheeler and Thomas 2006, Rohlfing et al. 2010, Wimberly and Choe 2022). Adult dpy-5 loss-of-function worms have a strong Dpy body shape phenotype but have organized furrows and do not activate stress responses (Dodd et al. 2018). Adults of osm-8 mutant worms were shown previously to have organized furrows (Rohlfing et al. 2011).
As shown in Fig. 1A, dpy-5(e61) worms developed a Dpy phenotype starting at the L2 stage that became progressively stronger with each molt. The Dpy phenotype has a similar developmental pattern in furrow collagen gene mutants albeit weaker and starting later than in dpy-5(e61) worms (Chandler and Choe 2022). These progressive body shape phenotypes are consistent with synthesis of each new cuticle being at least partially reliant on organization of the former (Chandler and Choe 2022).
Representative high magnification DIC micrographs for L1 and young adult worm cuticles are in Fig. 1B; we observed organized furrows in all individuals and stages of dpy-5 worms. We observed no Dpy phenotype for any stage of osm-7(tm2256) and osm-8(n1518) worms and slightly lower width/length ratios than wild type in adults (Fig. 1A). These worms had organized furrows at all stages (L1 and adult shown in Figs. 1B) consistent with OSM-7 and OSM-8 functioning outside or downstream of the cuticle (Wheeler and Thomas 2006).
RT-qPCR of representative canonical osmotic ( gpdh-1 and hmit-1.1 ), detoxification ( gst-4 ), and innate immune response genes ( nlp-29 ) are shown in Figures 1C-F; these genes are strongly induced in late larval and adult furrow collagen gene mutants and during exposure to hypertonicity (Dodd et al. 2018, Chandler and Choe 2022, Wimberly and Choe 2022). In dpy-5 worms, expression of these stress response genes was either not different or reduced relative to N2 at all stages except for gst-4 in L1 larvae, which was increased 2.1-fold. This lack of broad stress response gene activation indicates that a wide body shape is not sufficient to activate stress responses at any developmental stage.
In osm-7 and osm-8 worms, all four representative stress response genes were induced in all stages (Figs. 1C-F). There was no obvious developmental pattern in gpdh-1 induction in either osm-7 or osm-8 worms; hmit-1.1 was induced the greatest in adults (Figs. 1C-D). In osm-7 worms, nlp-29 and gst-4 had a trend of greater induction in late stages, but there was no obvious pattern in osm-8 worms (Figs. 1E-F). These data are consistent with the pathways responding to osm-7 and osm-8 loss functioning at all post-embryonic stages.
Methods
Conceptualization, design of methodology, and preliminary assays were completed in an immersive CURE course (course-based undergraduate research experience) named “Molecular and Genetic Responses to Environmental Stress” at the University of Florida (Auchincloss et al. 2014, Wang 2017). Students from this course completed gene expression measurements and helped with body shape measurements.
Body shape was determined from images taken with a Zeiss Discovery V12 Stereo microscope and OptixCam Summit camera as a ratio of average width to total length using ImageJ version 1.53r as described previously (Schindelin et al. 2012, Chandler and Choe 2022). The superficial cuticle was imaged by DIC on an Olympus BX60 microscope with an oil 60× UPlanFL objective (numerical aperture at 1.25) and additional 2× lens in front of a Zeiss Axiocam MRm camera (Thornwood, NY) as described previously (Chandler and Choe 2022).
qRT-PCR assays were run using the delta-delta Ct method with primer efficiencies determined from standard curves as described previously (Scolaro et al. 2019, Chandler and Choe 2022) with the following modifications; after lysis, genomic DNA was removed with dsDNAse (Thermo Fisher product EN007). Numbers of worms in each replicate were adjusted to account for worm sizes as follows: 200 L1, 180 L2, 50 L3, and 15 L4 larvae for all strains; for young adults: 10 N2, osm-7 , and osm-8 and 12 dpy-5 . All RT-qPCR reactions were performed in 10 μL volumes in a Realplex2 (Eppendorf AG, Hamburg, Germany). Relative expression was normalized to a value of 1.0 for N2 within each developmental stage using the average of three reference genes ( rpl-2 , tba-1 , and cdc-42 ).
Body shapes for all mutant strains were compared to N2 with unpaired t -tests with Welch’s corrections (Ruxton 2006). All relative mRNA values were normalized within each stage to a value of 1.0 for N2 and compared to 1.0 with a one-sample t-test. P-values were false-discovery corrected with Benjamini-Hochberg adjustments. Numerical data underlying Figure 1 are in the Extended Data section.
Reagents
C. elegans strains:
N2, CB61 dpy-5(e61) , HA1857 osm-7(tm2256) , and MT3571 osm-8(n1518). Strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Primer sequences (5’ to 3’):
rpl-2 F – CTTTCCGCGACCCATACAA
rpl-2 R – CACGATGTTTCCGATTTGGAT
hmit-1.1 qF1 - TAGTGTCGGCTGCAATGCTTT
hmit-1.1 qR1 - CGCCGACCAATATAGTCGGA
gpdh-1 qPCR F - TTATGAGGCCGTGGAGCTTT
gpdh-1 qPCR R - CCAGACGGATGATAGCGGAT
nlp-29 F - CGAGGAATGTATGGAGGCTATG
nlp-29 R - TCCATGTATTTACTTTCCCCATCC
cdc-42 F3 - CGTTGACGCAGAAGGGACT
cdc-42 R3 - GAGAAGAGTGGAAGTCGGGG
tba-1 F - TCAACACTGCCATCGCCGCC
tba-1 R - TCCAAGCGAGACCAGGCTTCAG
Extended Data
Description: Extended Data - Numerical data underlying Figure 1. Resource Type: Dataset. DOI: 10.22002/d1969-eec25
Acknowledgments
Funding
This study was supported by National Science Foundation grant IOS-1452948.
References
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ. NaN NaN NaN;13(1):29–40. doi: 10.1187/cbe.14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med. 2015 Aug 5;88(Pt B):290–301. doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler LM, Choe KP. Extracellular matrix regulation of stress response genes during larval development in Caenorhabditis elegans. G3 (Bethesda) 2022 Nov 4;12(11) doi: 10.1093/g3journal/jkac221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. 2009 Mar 9;29(10):2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GN, Hirsh D. Stage-specific patterns of collagen gene expression during development of Caenorhabditis elegans. Mol Cell Biol. 1985 Feb 1;5(2):363–372. doi: 10.1128/mcb.5.2.363-372.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GN, Staprans S, Edgar RS. The cuticle of Caenorhabditis elegans. II. Stage-specific changes in ultrastructure and protein composition during postembryonic development. Dev Biol. 1981 Sep 1;86(2):456–470. doi: 10.1016/0012-1606(81)90204-9. [DOI] [PubMed] [Google Scholar]
- Dodd W, Tang L, Lone JC, Wimberly K, Wu CW, Consalvo C, Wright JE, Pujol N, Choe KP. A Damage Sensor Associated with the Cuticle Coordinates Three Core Environmental Stress Responses in Caenorhabditis elegans . . Genetics. 2018 Feb 27;208(4):1467–1482. doi: 10.1534/genetics.118.300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks GJ, Gaidatzis D, Aeschimann F, Großhans H. Extensive oscillatory gene expression during C. elegans larval development. Mol Cell. 2014 Jan 16;53(3):380–392. doi: 10.1016/j.molcel.2013.12.013. [DOI] [PubMed] [Google Scholar]
- Johnstone IL. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 2000 Jan 1;16(1):21–27. doi: 10.1016/s0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- Lamitina T, Huang CG, Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc Natl Acad Sci U S A. 2006 Jul 31;103(32):12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon L, Muriel JM, Roberts B, Quinn M, Johnstone IL. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol Biol Cell. 2003 Apr 1;14(4):1366–1378. doi: 10.1091/mbc.e02-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A. P. and I. L. Johnstone (2007). The cuticle. WormBook. C. e. r. community, C. elegans Research Community. DOI: doi/10.1895/wormbook.1.138.1
- Pujol N, Ewbank JJ. C. elegans: out on an evolutionary limb. Immunogenetics. 2021 Nov 10;74(1):63–73. doi: 10.1007/s00251-021-01231-8. [DOI] [PubMed] [Google Scholar]
- Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, Ewbank JJ. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008 Jul 18;4(7):e1000105–e1000105. doi: 10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing AK, Miteva Y, Hannenhalli S, Lamitina T. Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS One. 2010 Feb 2;5(2):e9010–e9010. doi: 10.1371/journal.pone.0009010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing AK, Miteva Y, Moronetti L, He L, Lamitina T. The Caenorhabditis elegans mucin-like protein OSM-8 negatively regulates osmosensitive physiology via the transmembrane protein PTR-23. PLoS Genet. 2011 Jan 6;7(1):e1001267–e1001267. doi: 10.1371/journal.pgen.1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruxton, G. (2006). The unequal variance t-test is an underused alternative to Student’s t-test and the Mann–Whitney U test. Behavioral Ecology 17(4): 688-690. DOI: doi.org/10.1093/beheco/ark016
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jun 28;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolaro G, Bridges K, Curry S, Jacobson S, LoPresti M, Pappas K, Ramirez N, Savigne L, Sherman S, Upshaw K, Walsh E, Choe K. Increased expression of pgph-1, T23F2.4, and cyp-14A5 in C. elegans dpy-7 mutants and by high salt. MicroPubl Biol. 2019 Aug 29;2019 doi: 10.17912/micropub.biology.000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urso SJ, Lamitina T. The C. elegans Hypertonic Stress Response: Big Insights from Shrinking Worms. Cell Physiol Biochem. 2021 Feb 25;55(S1):89–105. doi: 10.33594/000000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JTH. Course-based undergraduate research experiences in molecular biosciences-patterns, trends, and faculty support. FEMS Microbiol Lett. 2017 Aug 15;364(15) doi: 10.1093/femsle/fnx157. [DOI] [PubMed] [Google Scholar]
- Wheeler JM, Thomas JH. Identification of a novel gene family involved in osmotic stress response in Caenorhabditis elegans. Genetics. 2006 Sep 15;174(3):1327–1336. doi: 10.1534/genetics.106.059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly K, Choe KP. An extracellular matrix damage sensor signals through membrane-associated kinase DRL-1 to mediate cytoprotective responses in Caenorhabditis elegans. Genetics. 2022 Mar 3;220(3) doi: 10.1093/genetics/iyab217. [DOI] [PMC free article] [PubMed] [Google Scholar]