Abstract
Purpose
Convolutional neural networks (CNNs) have rapidly emerged as one of the most promising artificial intelligence methods in the field of medical and dental research. CNNs can provide an effective diagnostic methodology allowing for the detection of early-staged diseases. Therefore, this study aimed to evaluate the performance of a deep CNN algorithm for apical lesion segmentation from panoramic radiographs.
Materials and Methods
A total of 1000 panoramic images showing apical lesions were separated into training (n=800, 80%), validation (n=100, 10%), and test (n=100, 10%) datasets. The performance of identifying apical lesions was evaluated by calculating the precision, recall, and F1-score.
Results
In the test group of 180 apical lesions, 147 lesions were segmented from panoramic radiographs with an intersection over union (IoU) threshold of 0.3. The F1-score values, as a measure of performance, were 0.828, 0.815, and 0.742, respectively, with IoU thresholds of 0.3, 0.4, and 0.5.
Conclusion
This study showed the potential utility of a deep learning-guided approach for the segmentation of apical lesions. The deep CNN algorithm using U-Net demonstrated considerably high performance in detecting apical lesions.
Keywords: Artificial Intelligence; Deep Learning; Periapical Periodontitis; Radiography, Panoramic
Introduction
Apical periodontitis manifests radiographically as apical radiolucencies on various radiographic modalities such as periapical or panoramic radiography and cone-beam computed tomography (CBCT). Although CBCT has a considerably higher discriminatory ability than conventional radiography,1 its application is limited due to its cost and relatively high radiation dose. Both periapical and panoramic radiographs are common imaging modalities for the diagnosis of apical lesions. Panoramic radiography is routinely taken to screen patients for dental caries, periapical lesions, periodontal conditions, infrabony defects, and other dental diseases.2
Regardless of their ability to distinguish various entities, radiographic examinations are prone to suffering from inter- and intra-examiner reliability.3,4 The reliability of these modalities further depends on the experience of a trained examiner.5 Current empirical approaches for prediction and analysis of quantitative and qualitative dental disease, therefore, substantially rely on the combination of dental practitioners’ experience and subjective evaluations. These clinical approaches and methodologies are inefficient for achieving early detection and accurate prediction of dental diseases, as they require a large amount of time and effort in the screening process. These methods are further limited in their ability to provide highly reliable and standardized clinical evaluations. For instance, in the field of radiographic imaging, the detection of landmarks on cephalograms, detection of maxillary sinusitis, and early-stage diagnosis of dental caries and periodontitis require highly time-consuming processes, involving an interconnected set of detection tasks, which are mostly performed through manual evaluation requiring human labor.
With the aid of recently emerging artificial intelligence, recognition guided by deep learning (defined as a subset of machine learning) has successfully demonstrated its excellent ability and performance in medical and dental applications.6,7 The inherent problems of empirical approaches can be efficiently overcome, particularly in toral and maxillofacial imaging, through deep-learning approaches to classify and convert 2-dimensional panoramic radiographs, while simultaneously diagnosing diseases on the converted radiographs.8
This study performed convolutional neural network (CNN)-based apical lesion segmentation for the purpose of screening apical lesions on panoramic radiographs. Rather than relying on manually cropped image segments of each tooth as the initial training dataset,9 this study utilized whole panoramic images in the training dataset to provide an efficient method of screening and detecting early-stage apical lesions. The rapid screening and automatic prediction of dental diseases enabled by the deep-learning process facilitates more efficient diagnoses and rapid detection of apical lesions with high precision and sensitivity. The novel deep-learning approach and framework developed in this study provide fundamental insights into more efficient ways to detect and classify dental diseases using data augmentation, which can also be applied in other medical fields.
A U-Net network was adopted for apical lesion segmentation in this study, although previous studies have used YOLO-based networks for detecting dental diseases.10,11 Semantic segmentation is a form of pixel-level prediction and seemed more appropriate for identifying apical lesions with small pixels. The purpose of this study was to evaluate the performance of a pre-trained U-Net model for apical lesion segmentation on panoramic radiographs.
Materials and Methods
Data preparation
The dataset consisted of a total of 1000 panoramic radiograph samples of patients who visited Seoul National University Dental Hospital from 2018 to 2019. These radiographs included 1691 apical lesions. Panoramic radiographs were obtained from adult patients without mixed dentition, and only 1 radiograph was used from each patient. Radiographic images with severe noise and blurring were excluded. The study was approved by the Institutional Review Board (IRB) of Seoul National University Dental Hospital (ERI19010) with a waiver of informed consent. The data collection and all experiments were performed in accordance with the relevant guidelines and regulations.
Apical lesion labeling
Apical lesions were detected by 3 oral and maxillofacial radiologists with more than 10 years of experience in this field based on the agreement of 2 or more radiologists. Panoramic radiographs were labeled manually in red by drawing outlines of the lesions using polygon labeling tools for the training set (Fig. 1).
Fig. 1. Training software called “Deep Stack.” The lesions are labeled by outlining with polygon labeling tools.
Preprocessing and augmentation of panoramic radiographs
A total of 1000 panoramic images with a resolution of 1976×976 pixels were collected and resized to 960×480 pixels and converted into the PNG file format. The dataset was randomly divided into training (n=800 [80%]), validation (n=100, 10%), and test (n=100, 10%) datasets before data augmentation. The dataset was composed of 1511 apical lesions for training and 180 apical lesions for testing. Due to the limited number of panoramic images, data augmentation was performed to increase the amount of training data. The training dataset was randomly augmented during all phases using flip, blur, shift, scale, rotation, sharpening, emboss, contrast, brightness, grid distortion, and elastic transform through online augmentation. Online augmentation uses a specific augmentation method at each phase for optimized training, and it is transferable and more effective for models trained on limited training datasets.12 The model was optimized using PyTorch (v 1.7.1) with CUDA (v 11.0) in the Python open-source programming language (v 3.6.12; Python Software Foundation, Wilmington, DE, USA; retrieved on August 17, 2020, from https://www.python.org/). The training was processed with NVIDIA DGX Station (Nvidia Corporation, Santa Clara, CA, USA) having 4 V100 GPU cores, Intel Xeon E5-2698 v4 CPU (Intel Corp, Chandler, AZ, USA) and 256 GB of DDR4 RAM.
Architecture of the deep CNN algorithm
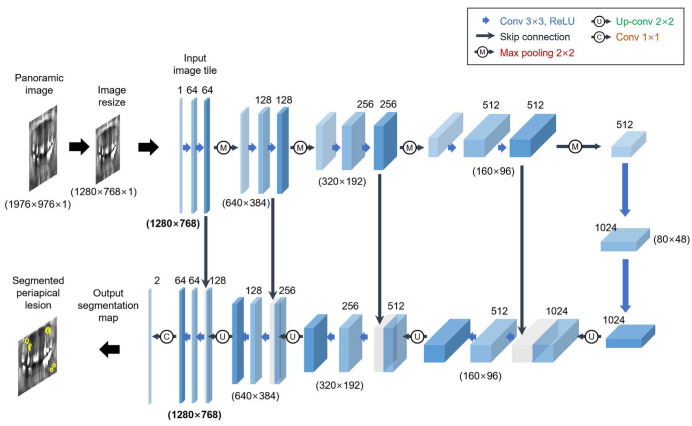
In this study, a pre-trained U-Net CNN network was used for preprocessing and transfer learning (Fig. 2). In particular, the U-Net model comprises 2 different paths: a contracting path (left) and an expansive path (right).13 In the contracting part, the original radiographic images are used as input to encode for multiple layers of convolution (3×3), rectified linear unit (ReLU) activation, max-pooling operation (2×2), and compression into a latent space. At each contracting step, the number of feature channels is doubled. The expanding part is the reverse process of the contracting part. Specifically, CNNs attempt to translate the contracted information from the latent space through an up-convolution operation (2×2) with a feature map wherein the number of feature channels is reduced to half and further to generate the segmentation mask of the image. The remaining decoding operations are the reverse of the encoding parts (cropping feature map from contracting path, 3×3 convolution, ReLU). Importantly, image cropping is required as a result of the loss of information related to border pixels during the process of every convolution. To map a 64-component feature vector to the desired number of classes, the last layer of a 1×1 convolution is finally utilized. In total, the network has 23 convolutional layers.
Fig. 2. U-Net model architecture for the detection of apical lesions (example: 32×32 pixels in the lowest resolution). Each blue box represents a multi-channel feature map. The number of channels is labeled on top of the box. The x-y-size is offered at the bottom of the box. White boxes represent duplicated feature maps. The arrows indicate the different operations.
Evaluation of the detection and classification performance of the deep CNN model
The performance of the developed CNN model was evaluated only using the test dataset, which was constructed independently from the training dataset. Precision, recall, and F1-score were calculated as demonstrated in the following equations (1-3). These parameters are commonly calculated to evaluate the performance of CNN models.
| (1) |
| (2) |
| (3) |
where TP represents the number of true positives, FP represents the number of false positives, and FN represents the number of false negatives.
Intensity histogram analysis
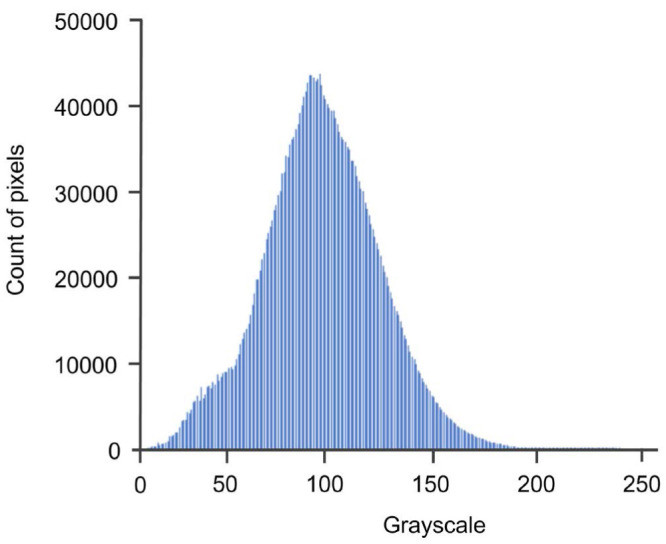
During image processing, the intensity histogram of an image is generated by converting the image to a 256-pixel scale based on the grayscale values (Fig. 3). A lower pixel number reflects darker images and a higher pixel number corresponds to lighter images. In this histogram, the pixel numbers in the range of 81-91 showed the highest probable density populations.
Fig. 3. Intensity histogram. Overall consistency was identified, without jagged edges for a specific labeled lesion.
Results
With an intersection over union (IoU) threshold of 0.3, the pre-trained CNN model segmented 147 lesions from the test group of 180 apical lesions (true positives). Thirty-one lesions were not segmented (false negatives), while 30 lesions that were not labeled manually were segmented (false positives). With an IoU threshold of 0.4, the pretrained CNN model segmented 143 lesions from the test group of 180 apical lesions (true positives). Thirty-four lesions were not segmented (false negatives), and 31 lesions that were not labeled manually were segmented (false positives). With an IoU threshold of 0.5, the pre-trained CNN model segmented 125 lesions from the test group of 180 apical lesions (true positives). Forty-four lesions were not segmented (false negatives), but 43 lesions that were not labeled manually were segmented (false positives) (Table 1).
Table 1. The number of apical lesions segmented by U-Net.
IoU: intersection over union
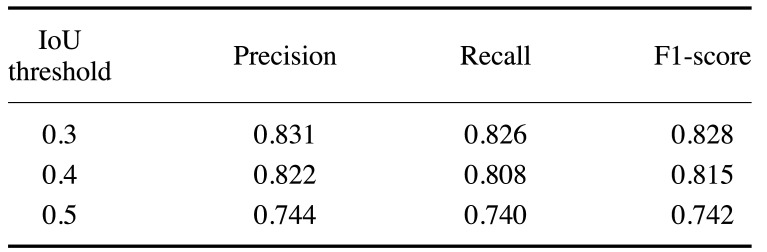
The performance of the U-Net model for apical lesion segmentation was analyzed while adjusting the IoU threshold (IoU) from 0.3 to 0.5 t. All of the detection performance metrics (precision, recall, and F1-score) generally decreased from 0.826-0.831 (at an IoU of 0.3) to 0.740-0.744 (at an IoU of 0.5) as the IoU threshold was gradually increased. The F1-score, which reflects both precision and recall, was higher with a lower IoU threshold, suggesting improved detection and classification performance (Table 2).
Table 2. Measurements of precision (positive predictive value), recall (sensitivity), and F1-score for apical lesion segmentation by U-Net.
IoU: intersection over union
The precision, recall, and F1-score were 0.831, 0.826, and 0.828 for U-Net model, respectively, at the lowest IoU threshold of 0.3. The corresponding values were 0.822, 0.808, and 0.815 with an IoU threshold of 0.4 and 0.744, 0.740, and 0.742 with an IoU threshold of 0.5, respectively (Table 2).
The training and validation datasets were evaluated over the course of each epoch from 0-450, with each epoch representing 1 pass through the entire training dataset (Fig. 4). The loss (or accuracy in reverse) recorded for the training and validation datasets generally decreased during the course of the 450 repeated epochs. The final resulting loss value was 0.30 for the validation dataset and 0.18 for the training dataset. Both losses flattened out after 400 epochs, implying that no additional improvement was made thereafter.
Fig. 4. Training and validation curves for the pre-trained augmented model. Training and validation were performed for 450 epochs, with each epoch representing 1 pass through the entire training and validation datasets. Overfitting was minimized under 400 epochs.
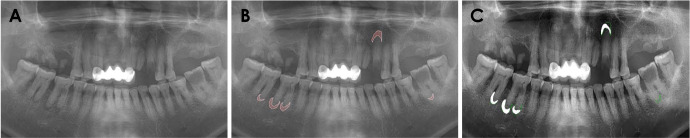
Apical lesions were segmented using the pre-trained UNet-based CNN model. Original images were prepared as the test dataset (Fig. 5A). White outlines denote the manual labeling of the ground truth to compare areas segmented by the pre-trained CNN (Fig. 5B), while white areas denote the UNet-based CNN-generated areas for apical lesions (Fig. 5C). The segmentation using the model was not significantly different from the manual labeling.
Fig. 5. An example of apical lesion segmentation from panoramic radiographs using the U-Net model. A. An original panoramic radiograph is prepared for testing. B. White outlines denote the manual labeling. C. White areas denote the CNN-generated areas by U-Net.
Discussion
To date, few studies have employed deep learning as a tool for detecting apical lesions. A deep learning-based CNN algorithm enabled the automated detection of apical lesions efficiently and effectively, minimizing the dependence on the ability of examiners. However, to the best of the authors’ knowledge, no study has yet examined the functionality of CNN for the automated diagnosis of apical lesions using entire panoramic radiographs. In this study, panoramic radiographs were used for training to detect apical lesions, and the possibility of artificial intelligence-guided diagnosis of apical lesions at early stage was confirmed.
Data augmentation is commonly used to train CNN models.11,14,15 It is an integral process of many state-of-the-art deep learning systems for image classification, object detection, and segmentation.16 Current deep neural networks have a number of parameters, tending to overfit the limited training data. Data augmentation is used to increase both the quantity and diversity of training data, thus preventing overfitting and improving generalization.17 This study adopted online data augmentation, which integrates both adversarial training and meta-learning for efficient training. This method can optimize data augmentation and target network training in an online manner. The advantages of online augmentation are the opposite features of offline methods.18 Their complementary character enables them to be applied together. The augmentation network can be applied to the target network through online training from start to finish, precluding the inconveniences of pre-training or early stopping. Offline learning methods usually rely on distributed training, as there are many parallel optimization processes, but online data augmentation is simple and easy for training.
The CNN-based model identified apical lesions in the maxilla and mandible with high performance, but it showed higher accuracy in the mandible than in the maxilla. An explanation for this may be that many anatomical structures, such as sinus floor, nasal cavity, and anterior nasal spine, were superimposed on the maxilla and interfered with the segmentation process of CNN. In contrast, there were not many overlapping anatomical structures on the mandible, facilitating better results. Therefore, further studies are needed to improve the detection performance for apical lesions in the maxilla.
All panoramic images were cropped and resized to 960×480 pixels for training and validation due to the training time, cost of computing devices, and storage space. Datasets with high-resolution images can improve the accuracy and discriminatory ability,19,20 and the lack of high-resolution images may be considered a limitation of this study. Higher-performance computing devices will enable a training process using high-resolution images. Furthermore, in future studies, panoramic images with deciduous teeth could be included, while only panoramic images with permanent teeth were included in this study.
The potential utility of deep learning for the detection and diagnosis of apical lesions was identified. The U-Net algorithm provided considerably good performance in detecting apical lesions in panoramic radiographs. CNN-based diagnosis may be a competent assistant for detecting dental and medical diseases at an early-stage.
Footnotes
This study was supported by grant (No. 04-2020-0106) from the SNUDH Research Fund and by a DDH Research Center grant funded by DDH Inc., Korea.
Conflicts of Interest: The second author, Hak-Kyun Shin, invented the AI software model (called Deep Stack) and is still working for the company (DDH Inc., Seoul, South Korea) that developed it. The other authors have no conflict of interest to declare. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Leonardi Dutra K, Haas L, Porporatti AL, Flores-Mir C, Nascimento Santos J, Mezzomo LA, et al. Diagnostic accuracy of cone-beam computed tomography and conventional radiography on apical periodontitis: a systematic review and meta-analysis. J Endod. 2016;42:356–364. doi: 10.1016/j.joen.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Choi JW. Assessment of panoramic radiography as a national oral examination tool: review of the literature. Imaging Sci Dent. 2011;41:1–6. doi: 10.5624/isd.2011.41.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nardi C, Calistri L, Grazzini G, Desideri I, Lorini C, Occhipinti M, et al. Is panoramic radiography an accurate imaging technique for the detection of endodontically treated asymptomatic apical periodontitis? J Endod. 2018;44:1500–1508. doi: 10.1016/j.joen.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Nardi C, Calistri L, Pradella S, Desideri I, Lorini C, Colagrande S. Accuracy of orthopantomography for apical periodontitis without endodontic treatment. J Endod. 2017;43:1640–1646. doi: 10.1016/j.joen.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 5.Parker JM, Mol A, Rivera EM, Tawil PZ. Cone-beam computed tomography uses in clinical endodontics: observer variability in detecting periapical lesions. J Endod. 2017;43:184–187. doi: 10.1016/j.joen.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Nagi R, Aravinda K, Rakesh N, Gupta R, Pal A, Mann AK. Clinical applications and performance of intelligent systems in dental and maxillofacial radiology: a review. Imaging Sci Dent. 2020;50:81–92. doi: 10.5624/isd.2020.50.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang JJ, Jung YH, Cho BH, Heo MS. An overview of deep learning in the field of dentistry. Imaging Sci Dent. 2019;49:1–7. doi: 10.5624/isd.2019.49.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Ekert T, Krois J, Meinhold L, Elhennawy K, Emara R, Golla T, et al. Deep learning for the radiographic detection of apical lesions. J Endod. 2019;45:917–922. doi: 10.1016/j.joen.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Yüksel AE, Gültekin S, Simsar E, Özdemir ŞD, Gündoğar M, Tokgöz SB, et al. Dental enumeration and multiple treatment detection on panoramic X-rays using deep learning. Sci Rep. 2021;11:12342. doi: 10.1038/s41598-021-90386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon O, Yong TH, Kang SR, Kim JE, Huh KH, Heo MS, et al. Automatic diagnosis for cysts and tumors of both jaws on panoramic radiographs using a deep convolution neural network. Dentomaxillofac Radiol. 2020;49:20200185. doi: 10.1259/dmfr.20200185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei P, Ball JE, Anderson DT. Fusion of an ensemble of augmented image detectors for robust object detection. Sensors (Basel) 2018;18:894. doi: 10.3390/s18030894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le’Clerc Arrastia J, Heilenkötter N, Otero Baguer D, Hauberg-Lotte L, Boskamp T, Hetzer S, et al. Deeply supervised UNet for semantic segmentation to assist dermatopathological assessment of basal cell carcinoma. J Imaging. 2021;7:71. doi: 10.3390/jimaging7040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Kim DH, Jeong SN, Choi SH. Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J Dent. 2018;77:106–111. doi: 10.1016/j.jdent.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Kim DH, Jeong SN, Choi SH. Diagnosis and prediction of periodontally compromised teeth using a deep learning-based convolutional neural network algorithm. J Periodontal Implant Sci. 2018;48:114–123. doi: 10.5051/jpis.2018.48.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chlap P, Min H, Vandenberg N, Dowling J, Holloway L, Haworth A. A review of medical image data augmentation techniques for deep learning applications. J Med Imaging Radiat Oncol. 2021;65:545–563. doi: 10.1111/1754-9485.13261. [DOI] [PubMed] [Google Scholar]
- 17.Ganesan P, Rajaraman S, Long R, Ghoraani B, Antani S. Assessment of data augmentation strategies toward performance improvement of abnormality classification in chest radiographs. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:841–844. doi: 10.1109/EMBC.2019.8857516. [DOI] [PubMed] [Google Scholar]
- 18.Liu G, Zhou W, Tian L, Liu W, Liu Y, Xu H. An efficient and accurate iris recognition algorithm based on a novel condensed 2-ch deep convolutional neural network. Sensors (Basel) 2021;21:3721. doi: 10.3390/s21113721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabottke CF, Spieler BM. The effect of image resolution on deep learning in radiography. Radiol Artif Intell. 2020;2:e190015. doi: 10.1148/ryai.2019190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiol. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]