Abstract
A recent study found that group A Streptococcus (GAS) expresses a cell surface protein with similarity to human collagen (S. Lukomski, K. Nakashima, I. Abdi, V. J. Cipriano, R. M. Ireland, S. R. Reid, G. G. Adams, and J. M. Musser, Infect. Immun. 68:6542–6553, 2000). This streptococcal collagen-like protein (Scl) contains a long region of Gly-X-X motifs and was produced by serotype M1 GAS strains. In the present study, a second member of the scl gene family was identified and designated scl2. The Scl2 protein also has a collagen-like region, which in M1 strains is composed of 38 contiguous Gly-X-X triplet motifs. The scl2 gene was present in all 50 genetically diverse GAS strains studied. The Scl2 protein is highly polymorphic, and the number of Gly-X-X motifs in the 50 strains studied ranged from 31 in one serotype M1 strain to 79 in serotype M28 and M77 isolates. The scl1 and scl2 genes were simultaneously transcribed in the exponential phase, and the Scl proteins were also produced. Scl1 and Scl2 were identified in a cell-associated form and free in culture supernatants. Production of Scl1 is regulated by Mga, a positive transcriptional regulator that controls expression of several GAS virulence factors. In contrast, production of Scl2 is controlled at the level of translation by variation in the number of short-sequence pentanucleotide repeats (CAAAA) located immediately downstream of the GTG (Val) start codon. Control of protein production by this molecular mechanism has not been identified previously in GAS. Together, the data indicate that GAS simultaneously produces two extracellular human collagen-like proteins in a regulated fashion.
Group A Streptococcus (GAS) causes human infections of the throat and soft tissue and systemic diseases (39). This broad spectrum of infected tissues indicates that GAS can adapt to changing environments during pathogen-host interactions. In addition, the wide range of infections caused by GAS suggests that virulence factor expression is a very complex regulated process. Indeed, transcriptional and posttranscriptional mechanisms that control expression of virulence genes have been described (1, 9, 17, 21, 31). For example, several streptococcal cell surface proteins expressed in exponential growth are regulated by Mga, a positive transcriptional activator protein (3, 23, 27, 32).
Microbial extracellular molecules interact with host proteins and often mediate adherence (26). Several cell surface proteins of gram-positive bacteria have structural similarities that include a variable amino terminus, a central region composed of repeating units, and a carboxy-terminal cell-associated region with an LPXTG cell wall anchor motif (8). GAS cell surface proteins have been identified as proven or potential virulence factors and include M protein (7), immunoglobulin (14) and fibronectin (13) binding proteins, serum opacity factor (4), C5a peptidase (44), and GRAB (34).
Recently, we identified a new GAS cell surface protein that contains a central region composed of variable numbers of Gly-X-X (GXX) collagen-like motifs (20). The gene (scl) encoding this streptococcal collagen-like (Scl) protein was present in all 50 GAS strains studied and was preferentially transcribed in the logarithmic phase of growth by a serotype M1 GAS strain. Although the exact role of Scl in human pathogenesis is not understood, an isogenic scl mutant had decreased adherence to human fibroblasts grown in culture and was attenuated for virulence in mice, as assessed by subcutaneous inoculation (20).
In this study, we characterized a second gene (scl2) encoding a collagen-like protein. The scl2 gene also was present in all 50 genetically diverse strains studied, together representing 21 distinct M protein serotypes. Expression of scl1 is controlled transcriptionally by Mga. In contrast, production of Scl2 is controlled at the level of translation by the number of CAAAA pentanucleotide repeats located immediately downstream from a GTG (Val) start codon. This form of regulation has not been described in GAS or other gram-positive pathogens.
MATERIALS AND METHODS
Bacterial strains and growth.
Fifty GAS strains isolated worldwide were used. The strain collection was described in a recent analysis of the molecular population genetics and virulence role of Scl1 (20). The 50 GAS strains represented 21 different M types, as verified by sequencing the emm gene fragment encoding the hypervariable amino terminus. MGAS6708 is identical to SF370, the serotype M1 strain used in a genome sequencing project (http://www.genome.ou.edu/strep.html). Isogenic M1 GAS strains JRS301 (wild type) and JRS403 (mga mutant) (28) were kindly provided by June R. Scott (Emory University).
GAS strains were grown at 37°C in 5% CO2–20% O2 in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.2% yeast extract (THY medium) or on tryptose agar with 5% sheep blood (Becton Dickinson, Cockeysville, Md.).
Cloning experiments were performed with Escherichia coli XL-1 Blue (Stratagene, La Jolla, Calif.) grown in Luria-Bertani media (Difco Laboratories). E. coli TB1 (New England Biolabs, Inc., Beverly, Mass.) was used for experiments with scl2-phoZ fusions.
Construction of scl2-phoZ fusions.
Plasmid pDC123 (obtained from C. E. Rubens, University of Washington) and the scl2 genes from MGAS5005 (serotype M1) and MGAS6274 (serotype M28) were used. Shuttle vector pDC123 (2) contains the phoZ gene (16) transcribed from constitutively expressed tetM and cat tandem promoters. The phoZ gene present in pDC123 confers a blue-colony phenotype to E. coli and GAS grown on media supplemented with 5-bromo-4-chloro-3-indolylphosphate (XP or BCIP) (2). Intracellular alkaline phosphatase (AP) activity was minimal or negligible but increased substantially when AP was secreted, indicating that abundant AP activity was export dependent.
Plasmid pDC123 was digested with restriction endonucleases Eco47III and SphI, which flank the DNA fragment containing the phoZ gene Shine-Dalgarno box, signal sequence, and multiple cloning site located at the 5′ end of phoZ. The entire promoter region and the complete signal sequence of the scl2 gene were amplified with primers scl2-SmaI and scl2-SphI from MGAS5005 (serotype M1) and MGAS6274 (serotype M28). These PCR fragments were cleaved with SmaI and SphI and directionally cloned between the Eco47III and SphI sites of digested pDC123. The new plasmids contained the phoZ structural gene fused to the 5′ region of scl2 that encodes the Scl2 signal sequence. These scl2-phoZ constructs also contain the scl2 promoter region.
DNA methods.
Standard molecular-biology techniques were used (36). Plasmid DNA was purified with the UltraClean kit (Mo Bio Laboratories, Inc., Solana Beach, Calif.). GAS chromosomal DNA was isolated as described previously (25). The presence of the scl2 gene in GAS strains was assessed by PCR. The entire scl2 open reading frame (ORF) was amplified with forward primer scl2-up (5′-CTTTCAATGGATGACGATACC; nucleotides −29 to −9 upstream of the sequence shown in Fig. 1) and reverse primer scl2-rev (5′-ACTTTCCATCAGTTAGGTAGC; nucleotide positions 1160 to 1140 in Fig. 1) using Taq polymerase (Life Technologies). DNA was denatured at 94°C for 1 min. Thirty amplification cycles were performed as follows: 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min 45 s of extension at 72°C, followed by one cycle of 5 min at 72°C. The PCR products were analyzed by agarose gel electrophoresis and sequenced with internal primers and the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) with an ABI 377 instrument. The DNA sequence data were analyzed with Sequencher, version 3.1.1 (Gene Codes Corporation, Inc., Ann Arbor, Mich.) and Lasergene (DNASTAR, Inc., Madison, Wis.) software.
FIG. 1.
Nucleotide and amino acid sequences of the scl2 gene and inferred Scl2 protein in serotype M1 GAS (MGAS6708). The scl2 ORF consists of 951 bp (nucleotides 178 to 1129). The presumed scl promoter region has a predicted ribosome-binding site (RBS) and −10 and −35 regions. Dot at +1 inferred transcription start site. A potential transcription terminator (tt) consisting of two inverted repeats is located downstream of the ORF. The predicted GTG start codon (Val) and the TAA stop codon are in boldface. The inferred mature Scl2 polypeptide consists of 281 amino acids (nucleotides 284 to 1126). Four SSRs (CAAAA) located immediately after the GTG start codon would cause a frameshift in the downstream scl2 gene and result in premature termination of translation. SS, signal sequence; V, variable region; CL, collagen-like region consisting of 38 Gly-X-X triplet motifs (boxed); WM, cell wall membrane region containing the LPATG cell wall anchor motif (shaded). A T→C point mutation (dot) in the TAA stop codon of the scl2 gene was present in all serotype M3 GAS strains. This polymorphism would extend the inferred Scl2 protein by 11 amino acid residues (italicized protein sequence between stars).
RNA methods.
GAS strains were grown in THY medium and total RNA was isolated as described previously (19). Bacteria from 10-ml cultures were harvested and resuspended in Tris-EDTA buffer (10 mM Tris [pH 7.0], 1 mM EDTA). Cells were treated at 37°C for 5 min with mutanolysin (25 U) and lysozyme (1 mg/ml) in the presence of a 5 mM concentration of RNase inhibitor aurintricarboxylic acid. The cells were lysed by adding sodium dodecyl sulfate (SDS) (2% final concentration) and an equal volume of acid-phenol-chloroform at 65°C for 5 min. The samples were extracted with acid-phenol-chloroform, and RNA was precipitated with 2 volumes of ethanol in the presence of 0.2 M NaCl. DNA contamination was removed by digestion with DNase I, and the RNA was precipitated as described above.
For Northern analyses, 10 μg of total RNA was transferred onto a positively charged nylon membrane (Tropilon-Plus; Tropix, Bedford, Mass.). DNA probes were amplified by PCR using GAS genomic DNAs as templates. Since both scl1 and scl2 genes were present in GAS, the DNA probes were designed to avoid cross-hybridization in Northern blots. The DNA probes were amplified from the homologous GAS strains with the following primers: scl1 probe, 5′-GGCAAGCAGCGTTAAGGCTGA (forward) and 5′-TATGAAGACCTGCGCTTTGGTTAGCTTCTTTGTCAGCAGG (reverse); scl2 probe, 5′-TGCTGACCTTTGGAGGTGC (forward) and 5′-CGCCTGTTGCTGGCAATTGTC (reverse). The probes were biotinylated with BrightStar labeling reagents, and hybridization was performed with NorthernMax reagents (Ambion, Austin, Tex.). The hybridization signal was visualized with a chemiluminescence kit (Southern-Star; Tropix). Transcript sizes were estimated with RNA size markers (Life Technologies).
Protein methods.
The presence of the Scl1 and Scl2 proteins in culture supernatants and streptococcal cell wall fractions was studied. GAS strains were grown to exponential phase (optical density at 600 nm [OD600] of ∼0.5) in 150 ml of THY medium and pelleted by centrifugation, and total proteins in the culture supernatants were obtained by precipitation with trichloroacetic acid (TCA; 10% final concentration) on ice for 1 h. The TCA-precipitated protein samples were neutralized with saturated Tris before being loaded on an SDS–12% polyacrylamide gel electrophoresis (PAGE) gel. The cell wall-associated protein fractions were obtained from GAS cells resuspended in 2 ml of 20% sucrose with 10 mM Tris, pH 8.0, buffer containing 25 U of mutanolysin and 1 mg of lysozyme/ml. Cells were digested at 37°C for 1 h and pelleted by centrifugation, and the supernatants containing the cell wall fraction were used for subsequent analyses.
Rabbit polyclonal sera specific for Scl1 or Scl2 proteins made by several M serotype GAS strains were generated (Bethyl Laboratories, Inc., Montgomery, Tex.). The following synthetic peptides were used to raise an anti-Scl1-specific antibody: M1 GAS, TTMTSSQRESKIKEI; M28, FWGRRYFNEQEYLKS; and M52, VYQKEVEQYTKEAL. Peptides EENEKVREQEKLIQQ (serotype M1) and KLLTYLQEREQAENSW (serotype M28) were used to obtain anti-Scl2-specific sera. These peptide sequences corresponded to amino acid residues located in the amino-terminal (variable [V]) regions of mature Scl1 and Scl2 proteins. The peptides were designed to maximize antigenic and surface probability indices and minimize or avoid cross-reactivity. All immune rabbit sera had reactivity against the corresponding peptides in enzyme-linked immunosorbent assays, whereas preimmune sera from the same rabbits did not (data not shown).
Scl1 and Scl2 protein production and secretion by wild-type GAS strains were assessed by Western blot analysis. Protein samples obtained from the culture supernatants and from the cell wall fractions were separated by SDS–12% PAGE and transferred to a nitrocellulose membrane (Hybond ECL; Amersham Pharmacia Biotech, Piscataway, N.J.). Immunodetection of Scl was performed with specific rabbit antisera (1:500 dilution). Each Western blot was probed in parallel with both preimmune and immune sera to evaluate background reactivity. Horseradish peroxidase-conjugated goat anti-rabbit affinity-purified immunoglobulin G (heavy and light chains) (Bio-Rad, Hercules, Calif.) was used as the secondary antibody, and detection was done with chemiluminescence ECL reagents (Amersham Pharmacia Biotech). Prestained broad-range marker proteins (Bio-Rad) were used as molecular mass standards.
Nucleotide sequence accession number.
The scl2 sequence data reported here have been deposited in GenBank under accession no. AF317835.
RESULTS
Identification and analysis of the scl2 gene and inferred Scl2 protein in serotype M1 GAS.
We recently described a GAS gene encoding a presumed cell-associated protein with a long region of Gly-X-X repeats (20). The protein was named Scl for streptococcal collagen-like, and the gene was designated scl. The protein sequence corresponding to the hydrophobic cell membrane domain of cell surface protein M6 (FFTAAALTVMATAGVAAVV) (7) was used as the search query. With the exception of a small part of the scl gene sequence with similarity to the emm6 gene sequence encoding the carboxy-terminal transmembrane domain, there was no homology between scl and other GAS genes encoding cell surface proteins. This result suggested that Scl represented a new class of GAS extracellular protein. Therefore, the M1 genome database was searched again with protein sequences corresponding to the signal peptide (amino-terminal 37 amino acids) and cell wall region (carboxy-terminal 82 amino acids) of Scl (20). One highly homologous region was identified for each query. The regions of homology were located 1 kb apart on the opposite side of the GAS chromosome relative to the location of the scl gene. Analysis of this region of the GAS chromosome identified a second gene encoding a collagen-like protein with a long region of Gly-X-X repeats. To avoid confusion, the original gene was renamed scl1 and the new gene was designated scl2.
The scl2 ORF is 951 bp long (nucleotides 178 to 1129) (Fig. 1). A potential promoter located upstream of this ORF includes a −10 region (TATAAT; perfect match of the consensus sequence) and a −35 region (TTTACA; five of six bases [boldface] identical to the consensus sequence TTGACA) (35). A potential ribosome-binding site (AAAAGAGG; the consensus sequence is TAAGGAGG) is located 11 nucleotides upstream from a putative GTG (Val) start codon (11, 37). The putative scl2 gene would encode a signal sequence (ss; nucleotides 178 to 283), a variable region (nucleotides 284 to 484), a collagen-like region containing Gly-X-X motifs (CL; nucleotides 485 to 826), and a cell wall and cell membrane region containing an LPATG cell wall anchor motif (WM; nucleotides 827 to 1129). The presumed GTG (Val) start codon was out of frame with DNA located immediately downstream. Four CAAAA nucleotide sequence repeats were identified between the presumed GTG start codon and a CAT (histidine) codon adjacent to the CAAAA repeat region. The inferred amino terminus of Scl2 has structural features characteristic of signal sequences, including a short amino-terminal hydrophilic region followed by a hydrophobic transmembrane segment and a small amino acid residue at the cleavage site (29). Control of gene expression by short-sequence nucleotide repeats (SSRs) is well documented in gram-negative bacteria (40). Hence, Scl2 production could be controlled at the translation level by variation in the number of CAAAA repeats.
The predicted molecular mass of the mature Scl2 protein (residues 1 to 281) is ∼29.4 kDa, and the predicted isoelectric point is 6.22. Except for the hydrophobic transmembrane domain at the C terminus, the inferred mature Scl2 protein is hydrophilic (15). The variable region (residues 1 to 67) has a predicted α-helical structure, whereas the CL region (residues 68 to 181) has a predicted coiled structure (10).
Distribution and variation of the scl2 gene among GAS strains.
The scl2 gene was amplified from strain MGAS6708 (identical to strain SF370 used for a streptococcal genome sequencing project) and was sequenced to verify the available genome data. The scl2 gene was present in all 50 GAS strains representing the breadth of species genetic diversity as assessed by multilocus enzyme electrophoresis (24). The size of the scl2 gene varied among strains representing different M serotypes. In addition, size variation in the scl2 gene was common among GAS strains with the same M serotype. This observation suggested that variation in the scl2 gene exceeded that found in the scl1 gene. For example, no scl1 sequence variation among serotype M3 GAS strains was identified (20), whereas the scl2 gene varied in size for all five M3 strains. Hence, the scl2 gene was commonly found in GAS and was polymorphic in size.
The entire scl2 gene was sequenced in 25 GAS strains expressing 13 M types to determine the nature and extent of allelic variation (Table 1). The signal sequence region of the scl2 gene was conserved among diverse GAS strains. The 28 carboxy-terminal amino acids of the presumed Scl2 protein signal sequence (nucleotides 203 to 283) were 64% identical and 86% homologous in Scl1 and Scl2 proteins. The V regions were different in GAS strains representing different M serotypes; however, they were identical in strains of a particular M serotype. Hence, the V regions in both Scl1 and Scl2 are M type specific. The length of the V region in Scl2 varied from 61 amino acids in a serotype M9 strain to 77 residues found in M3 GAS. As identified for Scl1, the CL region of Scl2 was located C terminal to the V region. It contained a variable number of Gly-X-X motifs ranging from 33 triplet repeats in an M1 strain (MGAS252) to 116 in an M3 serotype GAS strain. The carboxy-terminal part of Scl2 (WM region) contained 100 amino acid residues that were well conserved among all 25 strains characterized. The 38 amino acid residues at the carboxy terminus of the WM region, encompassing the LPATG cell wall anchor motif and the hydrophobic transmembrane domain, were 82% identical and 92% homologous in Scl1 and Scl2. Of note, only serotype M3 GAS had a single nucleotide T→C substitution within a TAA stop codon, potentially creating an Scl2 variant extended by 11 amino acid residues (Fig. 1).
TABLE 1.
Analysis of the scl2 gene in 25 GAS strains expressing 13 M types
| M type | MGAS no. | No. of amino acids in V region | No. of GXX motifs in CL region | Scl2 protein expressiona
|
|
|---|---|---|---|---|---|
| No. of CAAAAb | GTG (Val) in-frame | ||||
| M1 | 252 | 67 | 33 | 3 | No |
| 5005 | 67 | 38 | 4 | No | |
| 5322 | 67 | 38 | 3 | No | |
| 6708 | 67 | 38 | 4 | No | |
| M2 | 3803 | 63 | 53 | 11 | Yes |
| M3 | 315 | 77 | 116 | 5 | Yes |
| M4 | 321 | 65 | 39 | 14 | Yes |
| M9 | 6159 | 64 | 58 | 17 | Yes |
| M12 | 6139 | 64 | 61 | 8 | Yes |
| 6144 | 64 | 61 | 11 | Yes | |
| 6166 | 64 | 52 | 6 | No | |
| 6198 | 64 | 61 | 6 | No | |
| 6259 | 64 | 61 | 6 | No | |
| 6276 | 64 | 72 | 5 | Yes | |
| M28 | 6141 | 73 | 73 | 16 | No |
| 6143 | 73 | 78 | 11 | Yes | |
| 6180 | 73 | 79 | 10 | No | |
| 6274 | 73 | 79 | 3c | Yes | |
| M49 | 4569 | 67 | 72 | 12 | No |
| M56 | 6146 | 72 | 37 | 17 | Yes |
| M75 | 4673 | 63 | 74 | 11 | Yes |
| M76 | 6133 | 64 | 66 | 16 | No |
| M77 | 6191 | 64 | 79 | 2 | Yes |
| 6250 | 64 | 77 | 9 | No | |
| ST2967 | 6155 | 58 | 78 | 5 | Yes |
The number of CAAAA pentanucleotide repeats downstream from the GTG start codon may cause a frameshift in the scl2 gene.
The number of CAAAA repeats also was analyzed by sequencing part of the scl2 gene in the following GAS strains (numbers of CAAAA repeats are in parentheses): M3, MGAS274 (8), MGAS335 (4), MGAS1313 (8), and AM3 (10); M18, MGAS156 (3) and MGAS300 (3); M22, MGAS6269 (5); M49, MGAS4578 (15); M52, MGAS6177 (16); M55, MGAS1863 (11); M56, MGAS4487 (11); M57, MGAS1864 (6); ST2967, MGAS1880 (17).
Three repeats in this strain do not cause a frameshift since one of the repeats is longer by one base pair, CAAAAA.
Two aspects of the scl2 gene sequence were of particular interest: (i) variation in the number of CAAAA pentanucleotide repeats located immediately downstream from the presumed GTG start codon with respect to the coding frame of the downstream sequence and (ii) the lack of a nucleotide sequence that would encode a region analogous to the linker region encoded by the scl1 gene. The number of CAAAA repeats varied greatly among the GAS strains studied, ranging from two in MGAS6191 (M77) to 17 in MGAS6159 (M9) and MGAS6146 (M56). Two CAAAA repeats is the minimal number that would permit correct translation of Scl2. Similarly, the addition of three CAAAA repeats (total of five) or multiples of three repeats (n = 8, 11, 14, or 17 repeats, etc.) should result in in-frame and full-length Scl2 protein translation. Three contiguous CAAAA nucleotide repeats would encode the pentapeptide QNKTK, whereas other numbers of CAAAA repeats should cause premature translation termination.
scl1 gene transcription and Scl production.
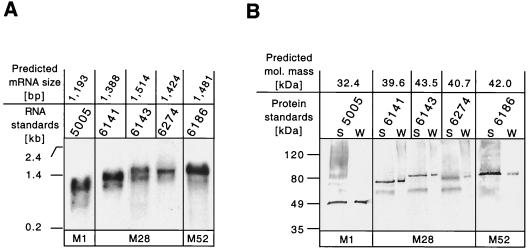
We reported recently that the scl1 gene was transcribed in two genetically distinct serotype M1 GAS strains (MGAS6708 and MGAS5005). Moreover, the Scl1 protein was present in cell wall fractions prepared from these isolates (20). To determine if the scl1 gene was transcribed by strains of GAS representing more than one M protein serotype, we studied three M28 strains (MGAS6141, MGAS6143, and MGAS6274) and an M52 strain (MGAS6186) by Northern blot analysis (Fig. 2A). The M28 strains were used because they had three distinct scl1 alleles. Total RNA was isolated from bacteria grown to logarithmic phase (OD600, ∼0.5), a time when the scl1 gene was abundantly transcribed in M1 strains (20). A single transcript was made by all strains studied, and the length of each transcript corresponded to the predicted size obtained from the gene sequence.
FIG. 2.
scl1 gene expression in GAS. (A) Northern blot analysis of total RNA isolated from cultures of GAS serotype M1, M28, and M52 strains in the logarithmic (OD600, ∼0.5) phase of growth. Total RNA (10 μg) was hybridized with biotinylated DNA probes amplified by PCR from the corresponding source strains. The blot was developed with streptavidin-AP conjugate, and the hybridizing bands were visualized by chemiluminescence. Single scl1 gene transcripts of the predicted sizes were made by all strains analyzed. RNA size markers were used to estimate the sizes of the scl1 transcripts. (B) Western blot analysis of the supernatant (S) and cell wall (W) protein samples prepared from exponential GAS cultures. Different scl1 gene alleles were translated into Scl1 protein variants. Immunoreactive bands were identified in the cell-free and cell-associated protein fractions in all strains tested. The Scl1 proteins migrated aberrantly slowly in SDS-PAGE gel. Molecular mass standards are shown.
We next determined if the Scl1 protein was produced by these M28 and M52 strains. Previously, the presence of the Scl1 protein in the cell wall-associated fraction was confirmed by analysis of two M1 GAS strains harvested in the exponential phase of growth (20). However, extracellular GAS proteins with the LPXTG cell wall anchor motif can also be present in the culture media (8). Rabbit antisera specific for the V regions of M28 and M52 strains were used. One predominant immunoreactive band was detected by Western blotting in each GAS strain (Fig. 2B). A weakly reactive band of ∼60 kDa was present in M28 GAS. This cross-reacting secreted product was most likely not related to Scl1 because it had the same size in all samples and was found in the culture supernatants only. The Scl1 protein was present in both the cell wall fraction and cell-free secreted (supernatant) form. As expected, the size of the Scl1 protein variant differed for each GAS strain. All Scl1 protein variants migrated aberrantly slowly in SDS-PAGE gels, a result confirming previous observations (20). Together, these results indicated that diverse scl1 alleles were translated into the Scl1 protein.
Transcription of the scl2 gene by GAS strains.
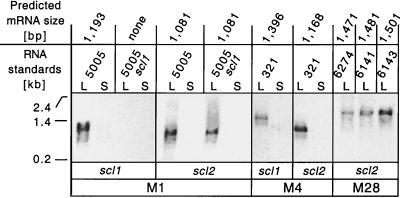
Sequence analysis (Fig. 1) identified a presumed promoter region upstream of the scl2 gene and a potential transcription terminator with two inverted repeats ∼40 bp downstream of a TAA stop codon, suggesting that the scl2 gene encoded a monocistronic mRNA. Many streptococcal genes are temporally expressed during growth, including scl1. Therefore, transcription of the scl2 gene was studied with RNA samples extracted from GAS cells harvested in exponential (OD600, ∼0.5) and stationary (OD600, ∼0.9) phases. Two controls were included to test the stringency of hybridization with the scl1- and scl2-specific probes. First, MGAS321 was included on the basis of DNA sequence data predicting that the scl2 gene transcript would be more than 200 bp shorter than the mRNA for the scl1 gene. Therefore, we assumed that hybridizing bands would be easily resolved by electrophoresis. Second, an isogenic MGAS5005 scl mutant (20) was included as a negative control for the presence of the scl1-specific transcript.
The scl1 and scl2 genes were transcribed simultaneously in the logarithmic phase by MGAS5005 (M1) (Fig. 3). The scl2 gene produced a monocistronic transcript that was ∼100 bp shorter than the scl1 transcript. The scl2 mRNA was made by MGAS5005 and its scl1 isogenic mutant (MGAS5005 scl). In contrast, the scl1 gene transcript was detected only in wild-type strain MGAS5005. MGAS321 (M4) also expressed both scl genes simultaneously and only during exponential growth. The transcript sizes corresponded to the predicted lengths based on DNA sequence analysis of the genes. The three serotype M28 strains studied also simultaneously transcribed scl1 and scl2 in the logarithmic phase. In summary, Northern blot analyses showed that both scl genes were transcribed in the exponential phase by genetically unrelated GAS strains. No evidence that gene transcription occurred in the stationary phase was obtained.
FIG. 3.
Transcription of the scl2 gene in GAS. (A) Northern blot analysis of the total RNA isolated from cultures of GAS serotype M1, M4, and M28 strains in the logarithmic (L; OD600, ∼0.5) or stationary (S; OD600, ∼0.9) phase of growth. Ten micrograms of total RNA was hybridized with biotinylated DNA probes prepared from the corresponding source strains by PCR. Hybridization was identified with streptavidin-AP conjugate with a chemiluminescence detection method. The scl2 gene was transcribed by all GAS strains in the exponential phase. RNA size markers were used to estimate the sizes of the scl2 transcripts.
Transcription of scl1 is regulated by mga.
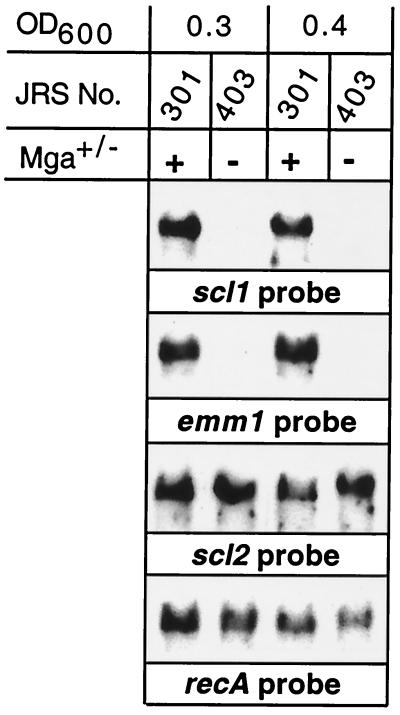
Several GAS cell surface proteins expressed in exponential growth are regulated by Mga, a positive transcriptional activator protein (1, 3, 30). The scl1 gene promoter region has a potential Mga binding site (20). To directly test the hypothesis that transcription of scl1 is regulated by Mga, we used an isogenic M1 mutant strain in which mga had been insertionally inactivated (28). Northern blot analysis showed that an scl1 transcript was made by wild-type strain JRS301 but not by the isogenic mutant strain, JRS403 (Fig. 4). No difference between early- and mid-logarithmic-phase cultures was identified. In contrast, an scl2 transcript was made by the wild-type and mutant organisms, a result indicating that this gene is not regulated by Mga.
FIG. 4.
Control of scl1 gene expression by Mga. The effect of inactivation of the mga gene on transcription of scl1 and scl2 was studied. Shown is a Northern blot analysis of the total RNA isolated during the early (OD600, ∼0.3) and middle (OD600, ∼0.4) logarithmic phases of growth from wild-type strain JRS301 and the isogenic mga mutant, JRS403. scl1 and control emm1 genes were transcribed in an mga-dependent fashion. In contrast, transcription of scl2 and the control gene recA did not require a functional mga gene.
Scl2 protein production is controlled by the number of CAAAA nucleotide repeats.
The sequence of the scl2 gene in serotype M1 GAS strains indicated that the full-length Scl2 protein would not be produced due to premature translation termination. We also observed that the number of CAAAA pentanucleotide repeats located next to the presumed GTG start codon varied among the 25 GAS strains sequenced for scl2 (Table 1). In principle, variation in the number of these repeats would either permit full-length translation of the scl2 transcript or cause premature termination. Control of gene expression at the level of translation by variation in the number of SSRs has been reported for many gram-negative bacterial species (40), but this mechanism of gene regulation has not been described for GAS or gram-positive organisms.
Western blot analysis with antibodies raised against synthetic peptides specific for the V region of Scl2 was used to test if protein production was associated with the number of CAAAA nucleotide repeats (Fig. 5). All M28 strains transcribed the scl2 gene. Supernatant and cell wall protein fractions were prepared from bacteria grown to the logarithmic phase. Scl1 was present in these protein fractions (Fig. 2B). Two M28 strains, MGAS6143 containing 11 CAAAA repeats and MGAS6274 with 3 CAAAA repeats (1 of the CAAAA repeats in this strain is longer by 1 bp, CAAAAA, and therefore 3 repeats in this strain do not cause the frameshift), were expected to produce Scl2. In contrast, MGAS6141 (16 CAAAA repeats) and MGAS6180 (10 CAAAA repeats) should not produce the Scl2 protein due to premature translation termination (Fig. 5A). Protein samples prepared from MGAS6143 and MGAS6274 had single immunoreactive bands (Fig. 5B). In contrast, only background immunoreactivity was detected in the protein samples obtained from MGAS6141 and MGAS6180. A similar background level of immunoreactivity was observed when preimmune rabbit serum was used (data not shown). Protein samples prepared from MGAS5005 contained positive immunoreactivity for the anti-Scl1 serum (Fig. 2B). As expected, the same protein samples did not contain material that reacted with the anti-Scl2 serum (data not shown). A positive control was not available for M1 strains because none was expected to produce Scl2 on the basis of DNA sequence data.
FIG. 5.
Scl2 protein production depends on the number of CAAAA pentanucleotide repeats. (A) Schematic of the scl2 gene GTG start codon and CAAAA repeat region in four serotype M28 strains. The scl2 genes in MGAS6141 and MGAS6180 have 16 and 10 CAAAA nucleotide repeats, respectively, resulting in early translation termination. In contrast, scl2 genes in MGAS6143 and MGAS6274 have 11 and 3 CAAAA repeats, respectively, and should produce the extracellular Scl2 protein. Star, stop codon. (B) Immunoblot analysis showing the presence of Scl2 in the supernatant (S) and cell wall (W) protein samples obtained from the logarithmic GAS cultures. As anticipated, the Scl2 proteins were identified in cell-free and cell-associated fractions prepared from MGAS6143 and MGAS6274. Molecular mass standards are shown.
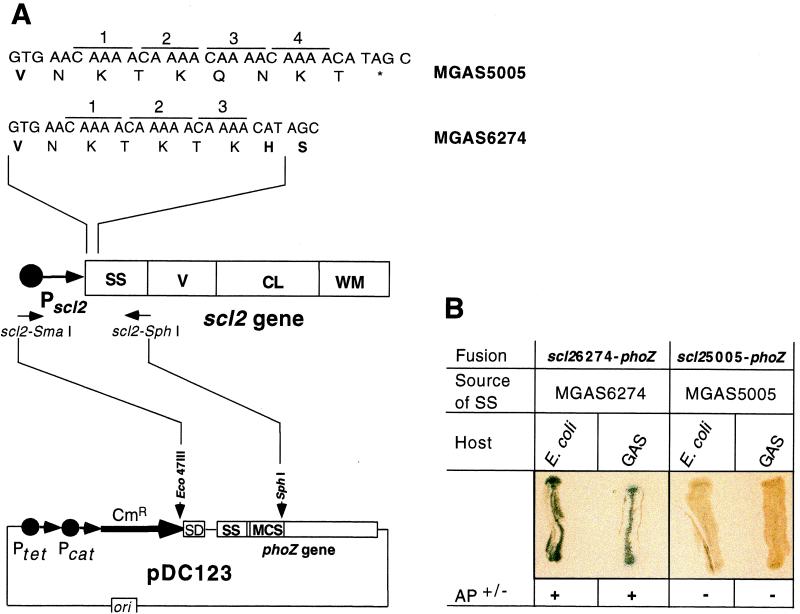
To further investigate the involvement of the CAAAA nucleotide repeats in translational control of Scl2 production, a reporter system employing the Enterococcus faecalis phoZ gene was used. Secreted, but not intracellular, PhoZ protein has AP activity and produces a blue-colony phenotype on media containing XP (see Material and Methods). This reporter system (2, 16) was used because (i) it was present in pDC123, an E. coli-GAS shuttle vector, (ii) it conferred a blue-white-colony phenotype in both bacterial species, and (iii) the PhoZ signal sequence could be replaced with the signal sequence from C5a peptidase (an extracellular GAS protein) without loss of the AP activity, indicating that the reporter system functions in GAS.
The phoZ signal sequence was replaced with part of the scl2 gene encoding the promoter region and signal sequence (Fig. 6A). Constructs were made with scl2 fragments obtained from either MGAS5005 (scl25005-phoZ) or MGAS6274 (scl26274-phoZ). The scl2 gene in the former strain had four CAAAA pentanucleotide repeats, presumably responsible for early translation termination of the Scl2 protein (Table 1). In contrast, the scl2 gene present in the latter strain had three CAAAA repeats and the full-length Scl2 protein was made (Fig. 5). As predicted only E. coli and GAS containing pDC123::scl26274-phoZ had a blue-colony phenotype, whereas colonies with pDC123::scl25005-phoZ were white on medium with XP (Fig. 6B). This important observation indicated that lack of Scl2 protein production by MGAS5005 was caused by the scl2 gene sequence, not by the genetic background of the host strain. Hence, export-dependent AP activity occurred only when the correct (in-frame) number of CAAAA nucleotide repeats was located 5′ to the phoZ gene. Taken together, the comparative sequence data, immunoblot analyses, and scl2-phoZ reporter studies strongly suggest that Scl2 protein production is regulated at the level of translation by variation in the number of CAAAA pentanucleotide repeats located immediately downstream of the GTG (Val) start codon, in the region of the scl2 gene that encodes the Scl2 signal sequence.
FIG. 6.
Analysis of translation control of Scl2 production by the number of CAAAA pentanucleotide repeats with PhoZ reporter constructs. (A) Schematic representation of the fusions between the region of the scl2 gene encoding the Scl2 signal sequence and phoZ (drawing not to scale). (Bottom) Plasmid pDC123 contains the phoZ gene under the control of the tetM and cat tandem promoters. The signal sequence (SS) and multiple cloning site (MCS) are located at the amino terminus of the functional PhoZ protein. pDC123 was digested with Eco47III and SphI restriction enzymes. (Middle) Schematic of the GAS scl2 gene, including the promoter (Pscl2) and regions encoding the signal sequence (SS), variable region (V), collagen-like region (CL), and cell wall membrane region (WM). A DNA fragment of scl2 containing the promoter region and encoding the signal sequence of Scl2 was amplified by PCR, digested with SmaI and SphI, and cloned into pDC123. In the resulting construct, the secretion signal sequence of PhoZ is replaced by the secretion signal sequence of Scl2; hence, production of the full-length Scl2-PhoZ chimeric protein is dependent on the presence of an in-frame number of CAAAA nucleotide repeats located immediately downstream of the GTG start codon. (Top) Amino acid sequences at the amino termini of the Scl2 signal sequences in two GAS isolates and associated nucleotide sequences. Different numbers of CAAAA nucleotide repeats located downstream of the GTG start codon are shown. Four CAAAA repeats cause a frameshift in the downstream DNA resulting in premature termination of translation in MGAS5005. In contrast, three CAAAA repeats present in the scl2 gene from MGAS6274 encode a functional signal sequence, resulting in Scl2 production. (B) Blue-white-colony phenotype depends on the number of CAAAA pentanucleotide repeats. AP activity was detected in both E. coli and GAS only when the phoZ reporter was fused to the scl2 signal sequence with the correct (in-frame) number of CAAAA repeats, scl26274-phoZ.
DISCUSSION
The data presented in this paper and another very recent contribution (20) indicate that GAS strains have two genes that encode collagen-like proteins (Table 2). The Scl1 and Scl2 proteins have several features in common with other GAS cell surface proteins (8), including a secretion signal sequence, variable domain at the amino terminus of the mature protein, repetitive central part, and conserved cell wall membrane domain with an LPXTG cell wall anchor motif. All 50 GAS strains tested, which together represent the breadth of species diversity in GAS, have both the scl1 and scl2 genes. This finding differs from what was found for emm and emm-like genes encoding streptococcal cell surface proteins M and M-like, respectively. All GAS strains have the emm gene encoding type-specific M protein, but other emm family members (enn, fcrA, and sph) are present only in some strains (5). The emm and emm family genes are located contiguously and their transcription is coordinately controlled by Mga. The two scl genes are expressed in the exponential phase of growth; however, the scl1 gene is regulated by Mga, whereas scl2 is not. The scl1 and mga genes are located ∼30 kb apart in the chromosome of an available serotype M1 GAS strain. At least one other gene controlled by Mga (sof, encoding serum opacity factor) is located outside of the region of the chromosome that contains mga and genes immediately downstream of mga controlled by it (23). Our study provides no insight into the molecular mechanism controlling temporal regulation of scl2 gene expression. However, Mga-independent but growth phase-dependent expression has been reported for the slo (streptolysin O) and plr genes (22).
TABLE 2.
Characteristics of two scl genes and Scl proteins in GASa
| Gene/ protein |
scl gene
|
No. of amino acids in Scl protein:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Distributionb | Locus (frame)c | Expression | Signal sequence | V region | CL region | L regiond | WM region | |
| scl1/Scl1 | All GAS | 1651591–1652637 (−3) | Mga-regulated transcription | 37 | 66–85 | 42–186 | 30–135 | 63 |
| scl2/Scl2 | All GAS | 863909–864860 (+3) | CAAAA-regulated translation | 32–57 | 61–77 | 99–348 | Not present | 100 |
scl1 scl1 genes from 50 GAS strains and scl2 genes from 25 GAS strains were sequenced.
50 GAS strains were screened by PCR for the presence of scl1 and scl2 genes.
Designated on the basis of an available serotype M1 genome sequence.
L, linker.
After this paper was submitted, Rasmussen et al. reported that the scl1 gene was under Mga control in GAS strain AP1 (serotype M1) by using a transposon-inactivated mga mutant (33).
Although we found that the scl2 gene was transcribed by all six GAS strains tested in this study, not all of these organisms produced full-length Scl2 protein. Our data indicate that failure to produce Scl2 by some or all strains was due to premature translation termination caused by variable numbers of CAAAA pentanucleotide repeats located immediately downstream from the GTG (Val) start codon. Analysis of the scl2 genes in four M28 serotype GAS strains with 3 to 16 CAAAA repeats predicted that only two of these four strains should produce the full-length Scl2 protein. Immunoblot and phoZ reporter fusion analyses fully supported this prediction. Only the correct number of CAAAA repeats in a signal sequence resulted in Scl2 protein production by GAS or in a blue-colony phenotype by phoZ fusion. In-frame expansion of the number of CAAAA repeats would result in elongation of the signal sequence, a process that could detrimentally affect Scl2 secretion. However, structural predictions made for the longest variant of the Scl2 signal sequence (made by the allele with 17 CAAAA repeats) indicated that this is not expected to be the case. Expansion of the CAAAA repeats would result in production of additional QNKTK pentapeptides and extend the charged domain of the secretion signal sequence. Of note, an scl2 gene with 11 CAAAA repeats is present in MGAS6143 and this strain secreted the Scl2 protein, a result indicating that elongation of the charged domains of the secretion signal sequence is not an impediment to extracellular production of Scl2.
Control of protein expression by variation in the number of SSRs such as CAAAA has not been reported for GAS or other gram-positive bacteria. There are several examples of genes whose expression is controlled by SSRs in gram-negative bacteria. For example, variation in the number of CAAT tetranucleotide repeats in lic1, lic2, and lc3 regulates lipopolysaccharide production by Haemophilus influenzae (42, 43). Cell surface variation in Neisseria gonorrhoeae is caused by early translation termination of the lsi-2 (lipopolysaccharide synthesis) and opa (opacity protein) genes and is due to variation in the length of a polyguanidine tract and the number of CTCTT repeats, respectively (6, 38). Phase and antigenic variation in these microorganisms affects important biological traits such as the ability of bacteria to colonize the host mucosal surface, to evade the host immune response, and to cause disease.
The frequency with which the number of CAAAA repeats varies in clonal descendants of a GAS strain is unknown. Consequently, it is not known if extracellular production of Scl2 undergoes classical, high-frequency phase variation. However, the gene sequence data provide strong indirect evidence that phase variation occurs (Table 1). Four of the six serotype M12 strains (MGAS6139, MGAS6144, MGAS6198, and MGAS6259) have scl2 genes that differ only by the number of CAAAA repeats. As a result, strains MGAS6139 (8 CAAAA repeats) and MGAS6144 (11 CAAAA repeats) are expected to express the same mature Scl2 protein variant whereas translation of Scl2 by strains MGAS6198 and MGAS6259 is expected to terminate prematurely (both strains have 6 CAAAA repeats). These four strains have the same multilocus enzyme electrophoretic type and multilocus gene sequence type (unpublished data) and hence have a recent common ancestor. Interestingly, these strains were isolated from patients in Texas during an outbreak of invasive disease that occurred in the winter of 1997 to 1998. Similarly, the scl2 genes in serotype M28 strains MGAS6274 and MGAS6180 also differed only by the number of CAAAA pentanucleotide repeats. As expected, MGAS6274 produces extracellular Scl whereas MGAS6180 does not because of premature translation termination (Fig. 5). These two strains also have a recent common ancestor, as assessed by multilocus enzyme electrophoresis and comparative sequencing of several other genes. Although these data are limited, taken together they suggest that Scl2 extracellular production can be modulated by cells that have recently descended from a common ancestor. We speculate that phase variation of Scl2 extracellular production in the course of pathogen-host interaction provides a survival advantage or enhances durability. In this regard, we note that variation in capsule production by Neisseria meningitidis is due to 1-bp deletions and insertions occurring in contiguous cytosine residues present in siaD (polysialyltransferase gene). Insertion or deletion of one cytosine residue results in a frameshift that produces premature translation termination or full-length translation of this enzyme, which participates in capsule synthesis (12). Spontaneous reversion of the capsule-deficient variant occurred in vitro at the high frequency of 10−3. Similarly, size variation in several variable-number-of-tandem-repeat loci has been reported among strains of H. influenzae isolated from patients in an outbreak of lung infections (41). Variation in the number of SSRs is presumed to occur by slipped-strand mispairing during replication (18), and there is evidence that amplification of the number of SSRs by slipped-strand mispairing increases the likelihood of subsequent slippage events. Hence, infrequent expansion of the CAAAA present in scl2 might accelerate the frequency of insertions and deletions. Therefore, it is possible that an scl2 allele with 2 to 4 CAAAA repeats would be more stable than an scl2 allele with 8 to 10 CAAAA repeats. Additional experimental and epidemiological studies are needed to fully understand molecular events controlling production of Scl2 by GAS.
ACKNOWLEDGMENTS
We thank J. R. Scott and C. E. Rubens for providing bacterial strains and plasmids.
This research was supported by Public Health Service grant AI-33119 to J.M.M. and by funds from the Moran Foundation to S.L. We acknowledge a streptococcal genome sequencing project funded by USPHS/NIH grant AI38406.
REFERENCES
- 1.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaffin D O, Rubens C E. Blue/white screening of recombinant plasmids in gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Bormann N, Cleary P P. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet. 1993;241:685–693. doi: 10.1007/BF00279912. [DOI] [PubMed] [Google Scholar]
- 4.Courtney H S, Chiang H C, Thacker J L, Dale J B. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol Microbiol. 1999;32:89–98. doi: 10.1046/j.1365-2958.1999.01328.x. [DOI] [PubMed] [Google Scholar]
- 5.Courtney H S, Dale J B, Hasty D L. Strategies for preventing group A streptococcal adhesion and infection. In: An Y H, Friedman R J, editors. Handbook of bacterial adhesion: principles, methods, and applications. Totowa, N.J: Humana Press, Inc.; 2000. pp. 553–579. [Google Scholar]
- 6.Danaher R J, Levin J C, Arking D, Burch C L, Sandlin R, Stein D C. Genetic basis of Neisseria gonorrhoeae lipooligosaccharide antigenic variation. J Bacteriol. 1995;177:7275–7279. doi: 10.1128/jb.177.24.7275-7279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischetti V A. Surface proteins on gram-positive bacteria. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 11–24. [Google Scholar]
- 9.Fogg G C, Gibson C M, Caparon M G. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an mγδ-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol. 1994;11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 10.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 11.Gold L, Pribnow D, Schneider T, Shinedling S, Singer B W, Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- 12.Hammerschmidt S, Muller A, Sillmann H, Muhlenhoff M, Borrow R, Fox A, van Puten J, Zollinger W D, Gerardy-Schahn R, Frosch M. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol. 1996;20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 13.Hanski E, Caparon M. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc Natl Acad Sci USA. 1992;89:6172–6176. doi: 10.1073/pnas.89.13.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath D G, Cleary P P. Fc-receptor and M-protein genes of group A streptococci are products of gene duplication. Proc Natl Acad Sci USA. 1989;86:4741–4745. doi: 10.1073/pnas.86.12.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 16.Lee M H, Nittayajarn A, Ross R P, Rothschild C B, Parsonage D, Claiborne A, Rubens C E. Characterization of Enterococcus faecalis alkaline phosphatase and use in identifying Streptococcus agalactiae secreted proteins. J Bacteriol. 1999;181:5790–5799. doi: 10.1128/jb.181.18.5790-5799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin J C, Wessels M R. Identification of csrRlcsrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 18.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 19.Lukomski S, Hoe N P, Abdi I, Rurangirwa J, Kordari P, Liu M, Shu-Jun D, Adams G G, Musser J M. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukomski S, Nakashima K, Abdi I, Cipriano V J, Ireland R M, Reid S R, Adams G G, Musser J M. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect Immun. 2000;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyon W R, Gibson C M, Caparon M G. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIver K S, Scott J R. Role of mga in growth phase regulation of virulence genes of the group A Streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLandsborough L A, Cleary P P. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of ScpA, FcRA, OF, and M protein. FEMS Microbiol Lett. 1995;128:45–51. doi: 10.1111/j.1574-6968.1995.tb07498.x. [DOI] [PubMed] [Google Scholar]
- 24.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patti J M, Allen B L, McGavin M J, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Casal J F, Dillon H F, Husmann L K, Graham B, Scott J R. Virulence of two Streptococcus pyogenes strains (types M1 and M3) associated with toxic-shock-like syndrome depends on an intact mry-like gene. Infect Immun. 1993;61:5426–5430. doi: 10.1128/iai.61.12.5426-5430.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perlman D, Halvorson H O. A putative signal recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983;167:391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- 30.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podbielski A, Woischnik M, Leonard B A B, Schmidt K-H. Characterization of nra, a global negative regulator gene in group A streptococci. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 32.Podbielski A, Woischnik M, Pohl B, Schmidt K H. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface factors. Med Microbiol Immunol. 1996;185:171–181. doi: 10.1007/s004300050028. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen M, Eden A, Bjorck L. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect Immun. 2000;68:6370–6377. doi: 10.1128/iai.68.11.6370-6377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen M, Muller H-P, Bjorck L. Protein GRAB of Streptococcus pyogenes regulates proteolysis at bacterial surface by binding α2-macroglobulin. J Biol Chem. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg D E, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stern A, Brown M, Nickel P, Meyer T F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 39.Stevens D L. Invasive group A Streptococcus infections. Clin Infect Dis. 1992;14:2–13. doi: 10.1093/clinids/14.1.2. [DOI] [PubMed] [Google Scholar]
- 40.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Belkum A, Scherer S, van Leeuwen W, Willemse D, van Alphen L, Verbrugh H. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect Immun. 1997;65:5017–5027. doi: 10.1128/iai.65.12.5017-5027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiser J N, Love J M, Moxon E R. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 43.Weiser J N, Maskell D J, Butler P D, Lindberg A A, Moxon E R. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzae lipopolysaccharide. J Bacteriol. 1990;172:3304–3309. doi: 10.1128/jb.172.6.3304-3309.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wexler D E, Chenoweth D E, Cleary P P. Mechanism of action of the group A streptococcal C5a inactivator. Proc Natl Acad Sci USA. 1985;82:8144–8148. doi: 10.1073/pnas.82.23.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]