Abstract
With the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, which are randomly mutated, the dominant strains in regions are changing globally. The development of preclinical animal models is imperative to validate vaccines and therapeutics against SARS-CoV-2 variants. The objective of this study was to develop a non-human primate (NHP) model for SARS-CoV-2 Delta variant infection. Cynomolgus macaques infected with Delta variants showed infectious viruses and viral RNA in the upper (nasal and throat) and lower respiratory (lung) tracts during the acute phase of infection. After 3 days of infection, lesions consistent with diffuse alveolar damage were observed in the lungs. For cellular immune responses, all macaques displayed transient lymphopenia and neutrophilia in the early stages of infection. SARS-CoV-2 Delta variant spike protein-specific IgM, IgG, and IgA levels were significantly increased in the plasma of these animals 14 days after infection. This new NHP Delta variant infection model can be used for comparative analysis of the difference in severity between SARS-CoV-2 variants of concern and may be useful in the efficacy evaluation of vaccines and universal therapeutic drugs for mutations.
Keywords: SARS-CoV-2, Delta variant, Interstitial pneumonia, Immunoglobulin, Primate
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), first reported in Wuhan, China in December 2019, continues to mutate and is rapidly spreading around the world (1). People with coronavirus disease 2019 (COVID-19) may develop symptoms, such as fever, runny nose, viral pneumonia, or even death. SARS-CoV-2, an RNA virus, mutates easily using a survival strategy called replication, wherein mutated viruses change in pathogenicity, infectivity, antigenicity, and transmission power (2). Thus, effective control of dominant strains is the final goal for development of vaccines and therapeutic drugs to overcome COVID-19.
Several types of preclinical animals, such as rodents, ferrets, and non-human primates (NHPs), have been used to evaluate the safety and efficacy of candidate vaccines and therapeutic drugs against COVID-19 (3,4,5). Among them, the COVID-19 pandemic has confirmed that cynomolgus and rhesus macaques, in particular, are the most suitable animal models that recapitulate human diseases because they are genetically and physiologically similar to humans. These models provide key information for the clinical application of vaccines and treatments (6,7). After the SARS-CoV-2 outbreak in Korea, we developed a preclinical model of NHPs for the S and GH clade, which were the dominant strains in April–June 2021 (8,9). These established SARS-CoV-2 infection models not only showed lymphopenia, acute interstitial pneumonia, and endotheliitis, similar to those in human COVID-19 infection, but were also consistent with the humoral immune response of convalescent patients, and were used to verify vaccine candidates and treatments made by various domestic pharmaceutical companies (10,11,12). Delta variant infection began to surge in Korea in July 2021, wherein it replaced infection by dominant strains by December 2021. Therefore, there is an urgent need to establish an animal model to verify the safety and efficacy of existing vaccines and approved therapeutics for Delta variant.
In this study, we developed a cynomolgus macaque model to evaluate the safety and efficacy of candidate vaccines and therapeutic drugs against SARS-CoV-2 Delta variant infection, and to determine potential clinical symptoms and features of immune responses through comparative analysis with previous dominant strain infection models.
MATERIALS AND METHODS
Animals and study design
A total of 6 female Cambodian-origin cynomolgus macaques (Macaca fascicularis), aged 6–7 years, were selected by institutional veterinary experts based on their general health. Animals were housed in cages in the animal biosecurity level 3 laboratory in Korea National Primate Research Center (KNPRC) at Korea Research Institute of Bioscience and Biotechnology (KRIBB). Animals were anaesthetized with a combination of ketamine hydrochloride (10 mg/kg) and tiletamine/zolazepam (5 mg/kg) for viral challenge, swabs, and blood collection. All animals were inoculated with 10.5 ml virus (2.1×106 50% tissue culture infection doses/ml [TCID50/ml]) through the mucous membranes, including the mouth, trachea, nose, and conjunctiva. Nasopharyngeal and oropharyngeal swabs were collected longitudinally at the indicated time points as shown in Fig. 1A. Swab samples in universal viral transport medium were centrifuged (1,600 g for 10 min) and filtered with 0.2-µm pore size syringe filters for virus quantification. A total of 3 animals were euthanized at days 3 and 21 after infection. Respiratory tissue samples were harvested for viral detection and microscopic analysis. Lung and bronchus tissue samples were homogenized using a Precellys homogenizer (Bertin Instruments, Montigny-le-Bretonneux, France) and centrifuged. All experimental protocols were approved by the KRIBB Institutional Animal Care and Use Committee (permit number: KRIBB-AEC-21289).
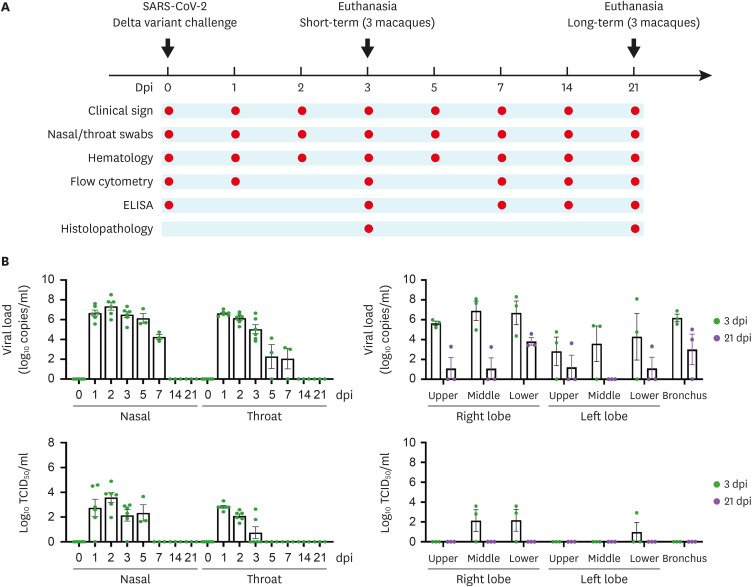
Figure 1. Scheme of experimental design and viral RNA and infectious virus in mucosal swabs and lung tissue samples. A total of 6 macaques were selected to investigate the characteristics of the SARS-CoV-2 Delta variant. (A) Baseline samples were collected on day 0 and all macaques were inoculated with SARS-CoV-2. Clinical signs, viral kinetics, and humoral and cellular immune responses to SARS-CoV-2 infection were observed at the indicated time points. (B) Viral RNA (copies/ml) and infectious virus (TCID50/ml) were determined in nasal and throat swab samples and lung tissues, and recorded at the indicated dpi. Circles indicate individual data and bar values represent the mean ± SEM.
SARS-CoV-2 virus
The SARS-CoV-2 virus (NCCP. 43390 for the GK clade [B.1.617.2 lineage]) were obtained from the National Culture Collection for Pathogens (Cheongju, Korea). The virus was propagated by inoculating VERO cells to generate viral stocks. TCID50/ml was measured in VERO cells and calculated using the Reed and Muench method.
Virus identification and quantification
VERO cells inoculated with swab samples and supernatants of tissue homogenates were incubated for 3 days at 37°C for infectious virus quantification to calculate the TCID50/ml values. To quantify viral RNA copies, RNA was extracted from the samples using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was performed with a primer/probe set to detect the ORF1b gene, as described in a previous study (9,10,13).
Histopathology and immunohistochemistry
For histological analysis, lung samples were fixed in 4% paraformaldehyde and embedded in paraffin, as described previously (8). Briefly, 4 to 5 µm sections were stained with hematoxylin and eosin and examined under a microscope. The lung sections were blindly examined and scored. The severity of lesions ranged from 0 to 4: no specific lesion, 0; alveolar wall thickening infiltrated with mononuclear cells, 1; infiltration of macrophages, mononuclear cells, and neutrophils in alveolar septa and alveolar lumina without vascular changes, 2; infiltration of inflammatory cells with vascular walls and pulmonary edema, 3; and hyaline membrane, 4 (8). For the distribution of lesions from 0 to 3: no specific lesion, 0; <30%, 1; 30%–50%, 2; and >50%, 3.
For detection of SARS-CoV-2 antigens, lung tissue sections were deparaffinized, rehydrated, and subjected to heat-induced epitope retrieval. Tissue sections were pretreated with 0.3% hydrogen peroxide, washed twice in distilled water, and blocked with 4% BSA in TBS and 1 μg dextran for 30 min at room temperature. The slides were incubated overnight at 4°C with rabbit nucleocapsid monoclonal antibodies (Sino Biological, Beijing, China) diluted 1:10,000 in TBS containing goat serum, washed twice then incubated in blocking buffer with enzyme-conjugated secondary antibody at room temperature for 1 h. Finally, the sections were developed with a chromogen and counterstained with hematoxylin.
Blood and fluorescence activated cell sorting analysis
Blood was collected from the inguinal veins at the indicated time points (Fig. 1A) and hematological examination was conducted using an autohematology analyzer (Mindray BC-5000; Mindray, Shenzhen, China). Flow cytometric analysis was performed using a BD LSR Fortessa flow cytometer (BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo v10.7.1 (BD Biosciences), as previously described (8). To exclude dead cells, blood was first stained with Fixable Viability Stain 575V (BD Biosciences) for 20 min at room temperature. For surface staining, cells were stained with the following antibodies for 30 min at 4°C: CD3 (Alexa Fluor 700; Invitrogen, Waltham, MA, USA), CD20 (APC/Cyanine7; Invitrogen), CD27 (PE/Cyanine7; Invitrogen), IgD (PE; BioLegend Inc., San Diego, CA, USA), IgM (FITC; SouthernBiotech, Birmingham, AL, USA), IgG (V450; BD Bioscience), CD4 (V500; Invitrogen), CD8 (V450; Invitrogen), CD95 (PE/Cyanine5; Invitrogen), and CD28 (ECD; Beckman Coulter, Brea, CA, USA). The cells were washed with permeabilization wash buffer and fixed with 1% paraformaldehyde. Data were acquired using an LSR Fortessa system (BD Biosciences) and analyzed using FlowJo v10.7.1 (BD Biosciences).
ELISA
The Delta variant-specific antibody in plasma was determined as previously described (8). Briefly, 96-well plates (Thermo Fisher Scientific, Waltham. MA, USA) were coated with 100 ng/well of delta variant spike (S1+S2) or receptor-binding domain (RBD) protein (all from Sino Biological) and incubated overnight at 4°C. The ELISA plates were blocked with 200 µl blocking buffer (0.05% Tween 20 and 10% FBS in PBS) for 1 h at room temperature. Diluted plasma was then added to the plate and incubated for 2 h at room temperature. After incubation, the plates were washed 6 times with 200 µl PBS-T and incubated with anti-monkey IgG and IgM-HRP conjugated antibodies (1:10,000; Rockland Immunochemicals, Gilbertsville, PA, USA) for 1 h at room temperature. After washing, the substrate tetramethylbenzidine was prepared and added to each well. After incubation, the development was stopped with 2N sulphuric acid and the absorbance at 450 nm was read using an ELISA plate reader (BioTek, Winooski, VT, USA).
Enzyme-linked immunospot (ELISPOT) assay
The membranes of high protein-binding PVDF 96-well plates (Millipore, Burlington, MA, USA) were coated with NHP-specific IFN-γ antibody (MabTech, Stockholm, Sweden) overnight at 4°C. Splenocytes (5×105/well) were stimulated for 18 h with 2 µg/ml of S peptide pool consisting of PepTivator SARS-CoV-2 Prot_S (Miltenyi Biotec, Bergisch Gladbach, Germany). After incubation, the coated plate was washed once with PBS. The spleen cells were then seeded into the coated plate with 200 μl of CTL-Test™ B Medium (Cellular Technology Ltd. [CTL], Shaker Heights, OH, USA) and incubated for 4 h in a humidified incubator providing 5% carbon dioxide at 37°C. The wells were subsequently washed twice with 200 µl PBS and twice with 200 µl PBS-T. Plates were incubated with anti-human biotin-conjugated IFN- γ antibodies (1:1,000; MabTech) in PBS-T-F (0.05% Tween 20 and 1% FBS in PBS) at room temperature for 2 h. The plates were washed twice with 200 µl PBS-T, and 80 µl of streptavidin-AP (1:1,000; CTL) in PBS-T-F was added to the wells and was incubated at room temperature for 1 h. The wells were washed twice with 200 µl PBS-T and distilled water prior to addition of developer solution (CTL), followed by incubation for 15 min at room temperature. To stop the reaction, the wells were washed with tap water and left to dry completely. Spots were scanned and counted using the immunospot CTL reader (S6 Universal Analyzer; CTL). The results were presented as spot-forming cells per 106 Spleen cells.
Statistical analysis
Statistical analyses were performed using one-way ANOVA with Tukey’s multiple comparison test using Prism version 9.3.1 (GraphPad Software, Inc., San Diego, CA, USA). All results are expressed as mean ± SEM. Statistical significance was defined as p<0.05.
RESULTS
Clinical signs and viral load kinetics of SARS-CoV-2 Delta variant infection
A total of 6 female cynomolgus macaques were challenged with SARS-CoV-2 Delta variant via multiple routes. Clinical signs of SARS-CoV-2 infection, such as changes in body temperature, body weight, and respiratory rate, were recorded. qRT-PCR was performed to assess viral kinetics in the nasal cavity, throat, and lungs throughout the experiment (Fig. 1B). None of the macaques showed notable changes in body weight or respiratory rate during infection, but the body temperature of 4 of the 6 macaques increased at 1 day post-infection (dpi) (Supplementary Fig. 1). As shown in Fig. 1B, viral loads in nasal swab samples were high at early time points (7.33±0.37 log10 copies/ml) and viral RNA was detected up to 7 dpi. Levels of viral RNA in the throat peaked on 1 dpi (6.63±0.11 log10copies/ml) and then gradually decreased over time; 7 dpi was detected in 2 subjects. Infectious virus in nasal swabs from all macaques was highest at 2 dpi (3.58±0.4 log10TCID50/ml) and sustained up to 5 dpi, whereas levels of viable virus in throat samples were peaked at 1 dpi (2.82±0.11 log10TCID50/ml) and remained up to 3 dpi. We found high viral loads in all lung lobes and bronchi of all macaques at 3 dpi, and viral particles were observed at 21 dpi as well. Infectious virus was detected in one to 2 of the 6 lobes from all subjects at 3 dpi, but could not be isolated from whole lobes and bronchi at 21 dpi. These results suggest that the SARS-CoV-2 Delta variant caused productive infection in the upper and lower respiratory tracts in the early phase of infection, and the infectious virus remained longer in the nasal cavity than in the throat.
Pathological changes
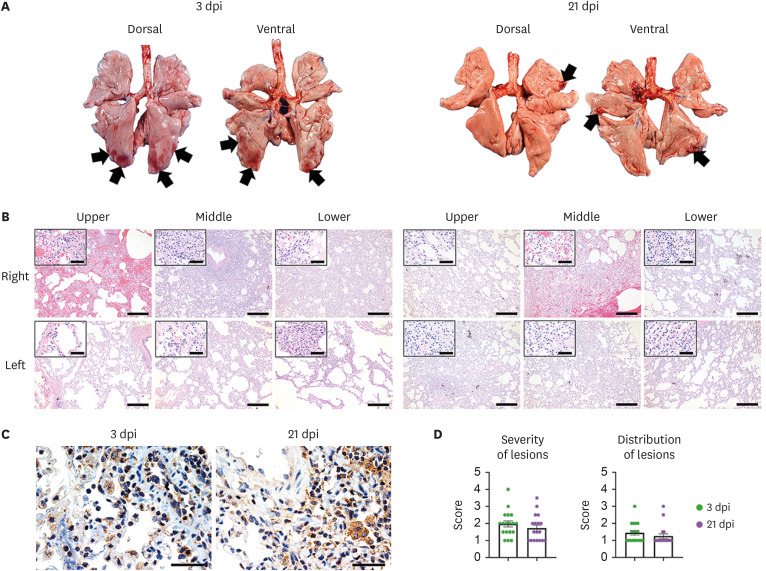
A total of 2 out of the 6 macaque groups (short- and long-term groups) were euthanized at the indicated time points, and necropsies were performed (Fig. 1A). Grossly, consolidations were observed in 2–3 lobes of the lung at 3 dpi, and the size of the lesions varied from lobe to lobe and individual to individual (Fig. 2A). At 21 dpi, consolidations were noted at the edges of the lung, and the size and distribution of lesions tended to decrease compared to those at 3 dpi. As shown in Fig. 2B, acute diffuse alveolar damage (DAD) was the major histologic lesion in the pulmonary tissue of infected monkeys. Neutrophils, alveolar macrophages, lymphocytes, fibrin, and eosinophilic edema were observed in the lumen of alveoli. The alveolar wall was dissociated due to edema and pulmonary necrosis, and monocytes and neutrophils were also observed infiltrating the wall. In one of 3 cases, a large number of neutrophils migrated around the alveolar lumen and wall. A hyaline membrane was seen in some damaged alveoli. Degeneration of vascular endothelial cells was observed, and neutrophils and monocytes aggregated around them. In the lesions except severe pulmonary edema, there was no alveolar lumen edema, but the alveolar wall was moderately thickened with infiltration of neutrophils and mononuclear leukocytes. One of 3 macaques showed pulmonary edema at 3 and 21 dpi. There was no significant difference in the severity and distribution of lesions between days 3 and 21 post-infection (Fig. 2D). The SARS-CoV-2 viral nucleocapsid protein antigen was detected in type 1 and 2 pneumocytes and alveolar macrophages in the lungs of macaques at 3 and 21 dpi (Fig. 2C). These results revealed that infection with the SARS-CoV-2 Delta variant could infect the lung, resulting in DAD, and could last for 21 dpi without alleviation of lung damage.
Figure 2. Histopathological changes in macaques during SARS-CoV-2 Delta variant infection. Macaques of the short-term (n=3) and long-term (n=3) groups were euthanized at 3 and 21 dpi, respectively. (A) Lungs from macaques presented local pulmonary consolidation (black arrow) at 3 and 21 dpi. (B) Histological changes were examined in the lung tissues at 3 and 21 dpi. The thickened alveolar walls and lumina were filled with immune cells. Scale bar, 400 µm; inlet, 50 µm. (C) Immunohistochemistry showing detection of SARS-CoV-2 antigens in the lungs. Scale bar, 40 µm. (D) Semi-quantitative analyses of the severity and distribution of lung lesions in macaques at 3 and 21 dpi. Data are presented as the scores of the 6 lung lobes of each animal per group.
Cellular immune responses to SARS-CoV-2 Delta variant
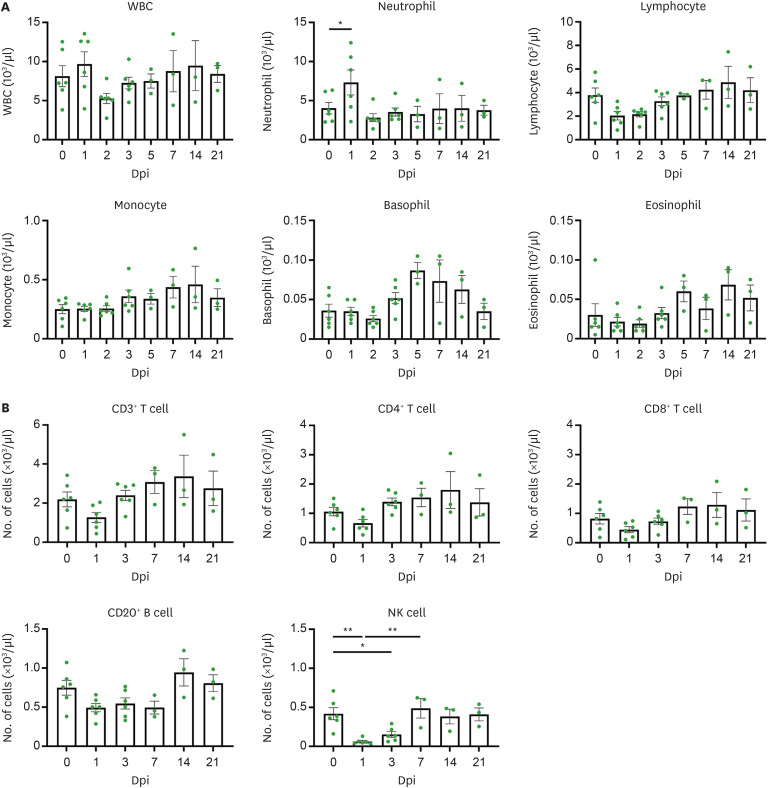
Cellular immune responses and hematological changes induced by the SARS-CoV-2 Delta variant were assessed using flow cytometry and a hematological analyzer. As shown in Fig. 3A, neutrophils, an important component of innate immunity against early viral infection, significantly increased at 1 dpi and returned to normal levels at 2 dpi. All macaques exhibited transient lymphopenia at 1 or 2 dpi. We observed a significant increase in basophils and eosinophils at 5 dpi, which gradually decreased over time. No remarkable changes in white blood cells and monocytes were observed during the infection.
Figure 3. Cellular immune response in SARS-CoV-2 Delta variant infected cynomolgus macaques. (A) Hematologic features of SARS-CoV-2 infection in whole blood of all macaques following SARS-CoV-2 infection up to 21 days (n=6 at 0, 1, 2, 3 dpi; n=3 at 5, 7, 14, 21 dpi). (B) Immune cell dynamics in whole blood of macaques after SARS-CoV-2 infection up to 21 days (n=6 at 0, 1, 3 dpi; n=3 at 7, 14, 21 dpi). Circles indicate individual data measured from macaques and data are presented as mean±SEM. One-way ANOVA with Tukey’s multiple comparison test.
WBC, white blood cell.
*p<0.05, **p<0.01.
As shown in Supplementary Fig. 2, the population of immune cells (CD3+ T cells, CD4+ T cells, CD8+ T cells, CD20+ B cells, and NK cells) decreased in the acute phase of infection (1 and 2 dpi) and then gradually recovered thereafter. Notably, NK cells were significantly reduced at 1 dpi, maintained up to 3 dpi, and then increased to normal levels (Fig. 3B). More specifically, the number of CD4+T subsets (CD4+ naïve, central memory, effective memory) rapidly returned to baseline after 1 day of SARS-CoV-2 infection, and the CD8+ T subset (CD8+ naïve, central memory, effective memory) showed the same pattern. No statistically significant changes were observed in the CD20+ B subset (naïve, double-negative, and switched memory) cells. Among monocyte subtypes, changes in classic monocytes were not observed before and after SARS-CoV-2 inoculation, but the number of intermediate monocytes (3 of 6 macaques) increased and decreased at 3 and 21 dpi, respectively. The number of non-classic monocytes significantly increased at 7 dpi and then decreased to normal levels. Natural killer T cell and plasma blast frequencies decreased at early infection time points and returned to baseline thereafter. We did not observe significant changes in hematological parameters, including red blood cell, hemoglobin, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, red cell distribution width, platelet, mean platelet volume, platelet distribution width, and plateletcrit after the SARS-CoV-2 Delta variant challenge (Supplementary Fig. 3). SARS-CoV-2-specific T cell responses increased at 21 dpi compared to those at 3 dpi (3dpi: 2±1.1, 21dpi: 80.67±36) (Supplementary Fig. 4).
Antibody responses to SARS-CoV-2 Delta variant infection
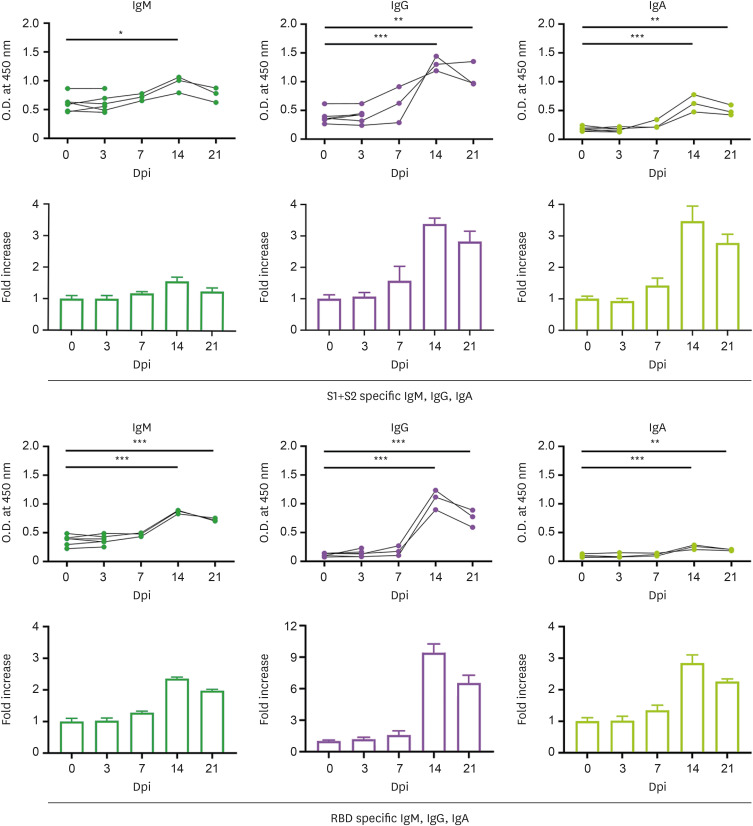
Spike protein (comprising the S1 and S2 domains) of SARS-CoV-2 plays a major role in direct viral attachment to membrane receptors and its fusion, and entry into host cells (14). RBD of the S1 domain initiates viral infection by binding to angiotensin-converting enzyme 2 in host cells (14). To evaluate humoral immune responses to SARS-CoV-2 Delta variant infection, we measured the plasma levels of SARS-CoV-2 spike antigen (S1+S2) and RBD-specific IgM, IgG, and IgA by ELISA, following infection. As shown in Fig. 4, the levels of IgM specific for the spike and RBD were significantly increased at 14 dpi (1.57-fold and 2.53-fold, respectively). The switched immunoglobulins, including IgG and IgA, against SARS-CoV-2 Delta variant were gradually elevated compared to that before the virus challenge, and peaked at 14 dpi (S1+S2 specific IgG: 3.38-fold, RBD specific IgG: 9.4-fold, S1+S2 specific IgA: 3.47-fold, RBD specific IgA: 2.84-fold). These results suggest that all macaques seroconverted to spike and RBD proteins of the SARS-CoV-2 Delta variant.
Figure 4. Seroconversion in cynomolgus macaques infected with SARS-CoV-2 Delta variant. (A) SARS-CoV-2 spike (S1+S2) protein specific IgM, IgG, IgA antibodies were measured by ELISA at 0, 3, 7, 14, 21 dpi and fold change in IgM, IgG and IgA antibodies was determined by dividing with baseline (0 dpi). (B) SARS-CoV-2 RBD specific IgM, IgG, IgA antibodies were measured by ELISA at same time points and fold change in IgM, IgG, and IgA antibody was determined by dividing with baseline (0 dpi). n=6 at 0, 3 dpi; n=3 at 7, 14, 21 dpi. Circles indicate individual data measured from macaques and bars show the group mean±SEM. One-way ANOVA with Tukey’s multiple comparison test.
*p<0.05, **p<0.001, ***p<0.0001.
DISCUSSION
The mission of KNPRC is to establish NHP infection models for validation of vaccines and therapeutics against SARS-CoV-2 variants of concern (VOCs). In April and December 2020, the Korea Disease Control Prevention Agency declared that the S and GH clade became dominant strains in South Korea; hence we developed an NHP model of SARS-CoV-2 infection (8,9). This model was used to evaluate the efficacy of vaccine and drug candidates against COVID-19 and to support the entry of these candidates into clinical trials (10,12,15). The dominant strain changed to the SARS-CoV-2 Delta variant, which spread rapidly throughout South Korea in July 2021. Here, our goal was to develop NHP infection models that could be utilized in the evaluation of viral replication inhibition and immunological and protective efficacy of promising vaccines and treatment drugs against the SARS-CoV-2 Delta variant.
Our NHP models of SARS-CoV-2 (S and GH clade) had a high viral load in the nasal cavity and throat at early stage of infection, but showed faster removal of live virus from the throat than from the nasal cavity (8,9). Viral RNA remained in the whole lung lobes and bronchi taken from all macaques, and infectious viruses were also detected at the acute stage of infection (8,9). Cynomolgus macaques infected with the SARS-CoV-2 Delta variant had a similar pattern of high viral RNA and viable virus in the nasal cavity and throat, but displayed a tendency of removal at the late stage of infection. These findings suggest that there is no difference between the NHP infection models of the SARS-CoV-2 Delta variant and S/GH clades in terms of virus shedding and replication patterns.
Regarding the severity of lung lesions after SARS-CoV-2 infection, 75% (18 of 24 lung lobes of 4 macaques) in the SARS-CoV-2 S clade (8) and 17% (3 of 18 lung lobes of 3 macaques) in the SARS-CoV-2 Delta variant were evaluated as having a severity score of 3 or higher. As Shuai et al. (16) reported, our results were in line with histological scores that were significantly decreased in mice infected with the Delta variant than in the wild-type strain at an early time point. Additionally, computed tomography (CT) scores from pediatric patients infected with the Delta variant of SARS-CoV-2 were lower than those of children infected with the original SARS-CoV-2 (17). The CT imaging-based severity of Delta-variant pediatric cases was significantly milder than the lung damage in patients in 2020 (17). Therefore, these comparative results suggest that the severity of pulmonary lesions in the Delta variant is milder than that in the original strain in the acute phase of infection. However, further studies should be performed to investigate the immunological mechanism by which Delta variant causes less damage to the lungs compared to previous strains, as well as the virological characteristics of SARS-CoV-2 variants that have higher transmissibility and decreased pathogenicity through mutation.
In our previous studies, NHP infected with SARS-CoV-2 showed a significant loss of lymphocytes and an increase in neutrophils at the early stages of infection, but gradually recovered over time (8,9,18). Likewise, this Delta variant macaque model did not display notable cellular immune responses, except transient lymphopenia and neutrophilia during the acute phase of infection, suggesting common immune cell dynamics in SARS-CoV-2 wild-type and Delta variant infection. These results are consistent with a recent report that showed decreased peripheral blood lymphocytes and increased neutrophils in COVID-19 Delta variant patients compared with the original clade, supporting that NHPs infected with Delta variants could recapitulate the cellular responses in patients with COVID-19 (15).
In terms of humoral immune responses to SARS-CoV-2, NHP models of the S clade present a significant increase in switched immunoglobulin IgG and IgA following an increase in plasma IgM (9). Similarly, NHPs infected with the Delta variant had a similar seroconversion, suggesting that SARS-CoV-2 Delta variant infection forms germinal centers (microstructures in which B cells differentiate into antibody-secreting cells and memory B cells) in the lymph nodes and spleen. Therefore, further studies using isolated blood and lymphoid tissue samples would help us understand how or whether memory cells control SARS-CoV-2 variants following re-infection in future.
Whenever SARS-CoV-2 VOCs emerge, the development of NHP infection models is required to study virus replication kinetics, shedding, pathogenesis, and potential therapeutic strategies. This newly established model is expected to enable a comparative analysis of the differences in the severity of SARS-CoV-2 VOCs, including Delta mutations, and is considered to be useful in verifying the efficacy of universal treatments and vaccine candidates for SARS-CoV-2 variants. Currently, Omicron variant has become the dominant strain around the world. Therefore, NHP modeling is needed to better understand characteristics for Omicron variant.
ACKNOWLEDGEMENTS
This work was supported by the Ministry of Science and ICT of Korea (Korea Research Institute of Bioscience and Biotechnology Research Initiative Programs [grant number KGM 4572222]) and the Korea Centers for Disease Control Prevention (grant number 2021-ER1608-00).
Abbreviations
- COVID-19
coronavirus disease 2019
- CT
computed tomography
- CTL
Cellular Technology Ltd.
- DAD
diffuse alveolar damage
- dpi
day post-infection
- ELISA
enzyme-linked immunosorbent assay
- ELISPOT
enzyme-linked immunospot
- KNPRC
Korea National Primate Research Center
- KRIBB
Korea Research Institute of Bioscience and Biotechnology
- NHP
non-human primate
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RBD
receptor-binding domain
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- TCID50/ml
50% tissue culture infection doses/ml
- VOC
variant of concern
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Park JH, Hong JJ.
- Data curation: Baek SH, Oh H, Koo BS, Kim G, Hwang EH, Jung H, An YJ.
- Investigation: Oh H, Koo BS, Kim G, Hwang EH, Jung H, An YJ.
- Supervision: Park JH, Hong JJ.
- Writing - original draft: Baek SH.
- Writing - review & editing: Baek SH, Oh H, Hong JJ.
SUPPLEMENTARY MATERIALS
Clinical signs in cynomolgus macaques infected with SARS-CoV-2 Delta variant. Macaques were monitored for changes in body weight, temperature, and respiratory rate during infection after the virus challenge. Changes in body temperature were determined by subtracting the baseline temperature from each time point (n=6 at 0, 1, 2, 3 dpi; n=3 at 5, 7, 14, 21 dpi).
Cellular immune response to macaques infected with SARS-CoV-2 Delta variant. (A) Representative gating strategy for immune cell sub-population. (B) Immune cell dynamics of T cell sub-populations (CD4+ naïve, CD4+ TCM, CD4+ TEM and CD8+ naïve, CD8+ TCM, CD8+ TEM), B cell sub-populations (naïve, DN B cell, Sm B cell), PB cell, monocytes subtypes (classic/intermediate/non-classic), NKT cells in macaques infected with SARS-CoV-2 Delta variant were analyzed up to 21 days (n=6 at 0, 1, 3 dpi; n=3 at 7, 14, 21 dpi).
Changes of hematologic parameters in macaques infected with SARS-CoV-2 Delta variant. Hematological parameters were observed up to 21 days after SARS-CoV-2 Delta variant infection (n=6 at 0, 1, 2, 3 dpi; n=3 at 5, 7, 14, 21 dpi).
T cell responses specific for SARS-CoV-2 Delta variant. IFN-γ secreted by T cells were quantified in the splenocytes isolated from cynomologus macaques at 3- and 21-dpi using an ELISPOT assay. (A) Representative IFN-γ ELISPOT raw data are depicted by images of ELISPOT wells in duplicate. (B) Circles represent individual data obtained from each macaque. The bar values represent mean±SEM.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radvak P, Kwon HJ, Kosikova M, Ortega-Rodriguez U, Xiang R, Phue JN, Shen RF, Rozzelle J, Kapoor N, Rabara T, et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat Commun. 2021;12:6559. doi: 10.1038/s41467-021-26803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiuppesi F, Salazar MD, Contreras H, Nguyen VH, Martinez J, Park Y, Nguyen J, Kha M, Iniguez A, Zhou Q, et al. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat Commun. 2020;11:6121. doi: 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan KA, Bewley KR, Fotheringham SA, Slack GS, Brown P, Hall Y, Wand NI, Marriott AC, Cavell BE, Tree JA, et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat Commun. 2021;12:81. doi: 10.1038/s41467-020-20439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, Kranz LM, Walzer KC, Hein S, Güler A, et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 6.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, et al. Evaluation of the mrna-1273 vaccine against sars-cov-2 in nonhuman primates. N Engl J Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim G, Kim DH, Oh H, Bae S, Kwon J, Kim MJ, Lee E, Hwang EH, Jung H, Koo BS, et al. Germinal center-induced immunity is correlated with protection against SARS-CoV-2 reinfection but not lung damage. J Infect Dis. 2021;224:1861–1872. doi: 10.1093/infdis/jiab535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koo BS, Oh H, Kim G, Hwang EH, Jung H, Lee Y, Kang P, Park JH, Ryu CM, Hong JJ. Transient lymphopenia and interstitial pneumonia with endotheliitis in SARS-CoV-2-infected macaques. J Infect Dis. 2020;222:1596–1600. doi: 10.1093/infdis/jiaa486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong SH, Oh H, Park YW, Kwak HW, Oh EY, Park HJ, Kang KW, Kim G, Koo BS, Hwang EH, et al. Immunization with RBD-P2 and N protects against SARS-CoV-2 in nonhuman primates. Sci Adv. 2021;7:eabg7156. doi: 10.1126/sciadv.abg7156. [DOI] [PubMed] [Google Scholar]
- 11.Kim C, Ryu DK, Lee J, Kim YI, Seo JM, Kim YG, Jeong JH, Kim M, Kim JI, Kim P, et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo YB, Suh YS, Ryu JI, Jang H, Oh H, Koo BS, Seo SH, Hong JJ, Song M, Kim SJ, et al. Soluble spike DNA vaccine provides long-term protective immunity against SARS-CoV-2 in mice and nonhuman primates. Vaccines (Basel) 2021;9:307. doi: 10.3390/vaccines9040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, Liu Y, Ng DY, Wan CK, Yang P, Wang Q, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Chen R, Hu F, Lan Y, Yang Z, Zhan C, Shi J, Deng X, Jiang M, Zhong S, et al. Transmission, viral kinetics and clinical characteristics of the emergent SARS-CoV-2 Delta VOC in Guangzhou, China. EClinicalMedicine. 2021;40:101129. doi: 10.1016/j.eclinm.2021.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shuai H, Chan JF, Hu B, Chai Y, Yuen TT, Yin F, Huang X, Yoon C, Hu JC, Liu H, et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 17.Cheng QR, Fan MX, Hao J, Hu XC, Ge XH, Hu ZL, Li Z. Chest CT features of children infected by B.1.617.2 (Delta) variant of COVID-19. World J Pediatr. 2022;18:37–42. doi: 10.1007/s12519-021-00484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng H, Li H, Guo L, Liang Y, Li J, Wang X, Hu Y, Wang L, Liao Y, Yang F, et al. Virulence and pathogenesis of SARS-CoV-2 infection in rhesus macaques: a nonhuman primate model of COVID-19 progression. PLoS Pathog. 2020;16:e1008949. doi: 10.1371/journal.ppat.1008949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical signs in cynomolgus macaques infected with SARS-CoV-2 Delta variant. Macaques were monitored for changes in body weight, temperature, and respiratory rate during infection after the virus challenge. Changes in body temperature were determined by subtracting the baseline temperature from each time point (n=6 at 0, 1, 2, 3 dpi; n=3 at 5, 7, 14, 21 dpi).
Cellular immune response to macaques infected with SARS-CoV-2 Delta variant. (A) Representative gating strategy for immune cell sub-population. (B) Immune cell dynamics of T cell sub-populations (CD4+ naïve, CD4+ TCM, CD4+ TEM and CD8+ naïve, CD8+ TCM, CD8+ TEM), B cell sub-populations (naïve, DN B cell, Sm B cell), PB cell, monocytes subtypes (classic/intermediate/non-classic), NKT cells in macaques infected with SARS-CoV-2 Delta variant were analyzed up to 21 days (n=6 at 0, 1, 3 dpi; n=3 at 7, 14, 21 dpi).
Changes of hematologic parameters in macaques infected with SARS-CoV-2 Delta variant. Hematological parameters were observed up to 21 days after SARS-CoV-2 Delta variant infection (n=6 at 0, 1, 2, 3 dpi; n=3 at 5, 7, 14, 21 dpi).
T cell responses specific for SARS-CoV-2 Delta variant. IFN-γ secreted by T cells were quantified in the splenocytes isolated from cynomologus macaques at 3- and 21-dpi using an ELISPOT assay. (A) Representative IFN-γ ELISPOT raw data are depicted by images of ELISPOT wells in duplicate. (B) Circles represent individual data obtained from each macaque. The bar values represent mean±SEM.