Abstract
Retinoid X receptors (RXRs) are nuclear transcription factors that partner with other nuclear receptors to regulate numerous physiological processes. Although RXR represents a valid therapeutic target, only a few RXR-specific ligands (rexinoids) have been identified, in part due to the lack of clarity on how rexinoids selectively modulate RXR response. Previously, we showed that rexinoid UAB30 potentiates all-trans-retinoic acid (ATRA) signaling in human keratinocytes, in part by stimulating ATRA biosynthesis. Here, we examined the mechanism of action of next-generation rexinoids UAB110 and UAB111 that are more potent in vitro than UAB30 and the FDA-approved Targretin. Both UAB110 and UAB111 enhanced ATRA signaling in human organotypic epithelium at a 50-fold lower concentration than UAB30. This was consistent with the 2- to 5- fold greater increase in ATRA in organotypic epidermis treated with UAB110/111 versus UAB30. Furthermore, at 0.2 μM, UAB110/111 increased the expression of ATRA genes up to 16-fold stronger than Targretin. The less toxic and more potent UAB110 also induced more changes in differential gene expression than Targretin. Additionally, our hydrogen deuterium exchange mass spectrometry analysis showed that both ligands reduced the dynamics of the ligand-binding pocket but also induced unique dynamic responses that were indicative of higher affinity binding relative to UAB30, especially for Helix 3. UAB110 binding also showed increased dynamics towards the dimer interface through the Helix 8 and Helix 9 regions. These data suggest that UAB110 and UAB111 are potent activators of RXR–RAR signaling pathways but accomplish activation through different molecular responses to ligand binding.
Keywords: nuclear receptor, retinoic acid, retinoid X receptor, rexinoid agonists, Targretin, UAB30, UAB110, UAB111, organotypic epidermis
Abbreviations: 9cRA, 9-cis-Retinoic acid; ATRA, all-trans-retinoic acid; CPM, Counts Per Million; DMSO, dimethyl sulfoxide; DSC, differential scanning calorimetry; HDX MS, hydrogen deuterium exchange mass spectrometry; IRB, Institutional Review Board; LBD, ligand-binding domain; LBP, ligand-binding pocket; NR, nuclear receptor; RXR, retinoid X receptor
Nuclear Receptors (NR) are ligand-activated transcription factors that are key targets for drug discovery and development due to their capabilities to directly modulate gene expression (1, 2, 3, 4). There are 48 NRs and approximately half of them require heterodimerization with retinoid X receptor (RXR) to perform their function (2, 5).
The partners of RXR can be classified as permissive (PPARs, LXR, and FXR) or conditionally permissive (RAR, TR, VDR) (6, 7, 8, 9, 10, 11). RXR heterodimers with permissive partners can be activated by either RXR agonists or its partner’s specific ligands, and when both partners are activated, they act in an additive or synergistic manner. In contrast, in conditionally permissive heterodimers, the ligand-dependent transcriptional activity of RXR appears “subordinated” to the binding of the ligand to its partner (12, 13). NRs have a DNA-binding domain and a ligand-binding domain (LBD). The DNA-binding domain binds to the promoter region of DNA sequences of target genes (14). Rexinoids bind the LBD and induce a significant change in conformation of the domain, often referred to as the transconformation that involves the formation of the ligand-binding pocket (LBP) and a surface site for coactivator binding that promotes the recruitment of coactivator proteins. An example of RXR coactivators are the steroid receptor coactivator family of coactivators (15, 16). Rexinoids bind in an L-shape geometry to the LBP which consists of helices 3, 5, 7, and 11 (17, 18). The coactivator peptide has a LXXLL motif which interacts with helixes 3, 4, and 12, often referred to as the AF-2 site (19).
Considering that RXRs regulate numerous physiological processes such as differentiation and development, metabolism, apoptosis, and inflammation, RXRs represent a potential therapeutic target in many diseases (20). However, the development of RXR-selective agonists has been somewhat stifled due to the lack of understanding of the molecular mechanism of rexinoid actions in the cells and the structural basis for rexinoid-mediated modulation of RXR function.
9-cis-Retinoic acid (9cRA) was the first RXR agonist discovered. 9cRA is clinically used for Kaposi Sarcoma but treatment induces severe grade three toxicities due to its pan-agonist activity for both RAR and RXR (21, 22, 23). Targretin (Targretin) is a rexinoid agonist that targets RXR selectively and is considerably less toxic than 9cRA (24, 25, 26, 27). Even though Targretin is selective for RXR, human patients treated with this rexinoid develop hypertriglyceridemia (79%), hypercholesterolemia (48%), and hypothyroidism (40%). Our group designed the RXR-selective rexinoid, UAB30, based on the structure of 9cRA. UAB30 did not stimulate the biosynthesis of serum triglycerides in a rat model due to its low agonist effects in the liver (28, 29, 30, 31). UAB30 has been demonstrated to be effective in several model systems of cancer (28, 31).
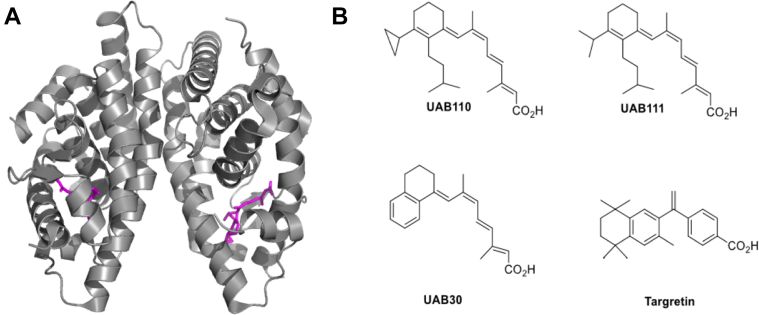
Our previous study focused on understanding the mechanism of action of UAB30 in a model of organotypic human epithelium (32). We have found that treatment of human organotypic epithelial skin raft cultures with UAB30 resulted in increased levels of all-trans-retinoic acid (ATRA) and upregulation of ATRA target genes. However, UAB30 was significantly less potent as a transcriptional regulator than Targretin, at least in this model. To further improve the potency of rexinoids, we identified two next-generation rexinoids, UAB110 and UAB111 (Fig. 1). These rexinoids were found to be effective in preventing breast cancers in an in vivo rat model alone or in combination with tamoxifen (28). Their design was facilitated by use of X-ray crystallography to define the LBP and geometry of the rexinoid bound to RXRα-LBD (33, 34, 35). The crystal structures show that UAB110 and UAB111 fill the ligand pocket well while still preserving the L-shaped geometry that renders the LBP. While crucial for the design process, crystallographic structures are static snapshots of ligand-bound proteins that exist in solution with dynamic molecular motions.
Figure 1.
RXR homodimer ligand-binding domain bound to UAB110.A, crystal structure of RXR-LBD in complex with UAB110 (magenta). B, structures of rexinoids UAB110, UAB111, UAB30, and Targretin. LBD, ligand-binding domain; RXR, retinoid X receptor.
We and others have made use of hydrogen deuterium exchange mass spectrometry (HDX MS) to examine rexinoid-induced structural dynamics in RXR (7, 18, 36, 37, 38). Through this technique, we have been able to analyze rexinoid-binding sites and binding-associated structural dynamics. We have shown that the binding of UAB30 and Targretin reduces deuterium incorporation and stabilizes peptides in the LBP of the RXRα-LBD (17, 18, 34, 35). Furthermore, our studies showed addition of coactivator LXXLL-binding motif peptide further reduces dynamics in the LBP and at the AF-2 site.
Here, to evaluate the biological efficacy of UAB110 and UAB111 relative to Targretin and UAB30, we examined their effects on gene transcription in a model of stratified human epidermis and compared their biophysical properties by HDX MS. Our results suggest that UAB110 and UAB111 are significantly more potent than either UAB30 or Targretin and reveal distinct responses of the RXRα-LBD to the four rexinoids.
Results
The effects of UAB110 and UAB111 on epithelial gene expression
To compare the effects of UAB110 and UAB111 on gene expression with those of previously characterized rexinoids, UAB30 and Targretin, we employed organotypic skin raft culture as a model. Skin raft cultures allow for the monitoring of rexinoid effects on proliferation and differentiation of the human keratinocytes through morphological changes in the three-dimensional architecture of the epidermis. Our previous results indicated that 2 μM UAB30 induced a visible reduction in the cornified and granular layers of skin rafts consistent with increased ATRA signaling (32). Therefore, we performed the initial evaluation of UAB110 and UAB111 at 2 μM. However, at this concentration of UAB110 and UAB111, the keratinocytes failed to differentiate and form stratified epithelium (data not shown), suggesting a greater potency of the new rexinoids.
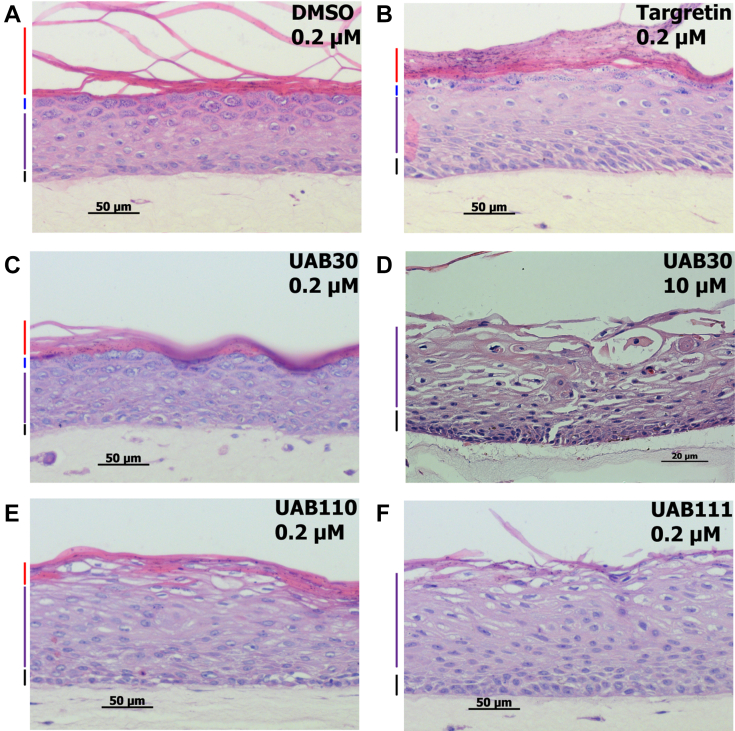
Indeed, reducing the concentration of UAB110 and UAB111 in the skin raft culture medium to 0.2 μM allowed for the growth and differentiation of keratinocytes into spinous, granular, and cornified layers of skin rafts (Fig. 2). Notably, UAB30 at 0.2 μM had little or no effect on proliferation and differentiation of keratinocytes (Fig. 2C) compared to rafts treated with dimethyl sulfoxide (DMSO) as indicated by H&E staining of sectioned rafts (Fig. 2A). In contrast, skin rafts treated with UAB110 and UAB111 displayed significantly reduced cornified and granular layers (Fig. 2, E and F), resembling the morphology of skin rafts treated with 10 μM UAB30 (Fig. 2D). Interestingly, at 0.2 μM, Targretin appeared to be less potent in triggering the changes in morphology than either UAB110 or UAB111 (Fig. 2B).
Figure 2.
H&E staining of skin raft cultures treated with rexinoid agonists. (A) DMSO, solvent control, human organotypic skin raft cultures were treated with rexinoid agonists: (B) 0.2 μM Targretin, (C) 0.2 μM UAB30, (D) 10 μM UAB30, (E) 0.2 μM UAB110, and (F) 0.2 μM UAB111. The rexinoids were added to the culture medium at the onset of organotypic skin raft formation, and the rafts were allowed to grow for 11 days. Colored lines at the left side of each panel demarcate the layers of the epidermis: cornified (red), granular (blue), spinous (green), and basal (black).
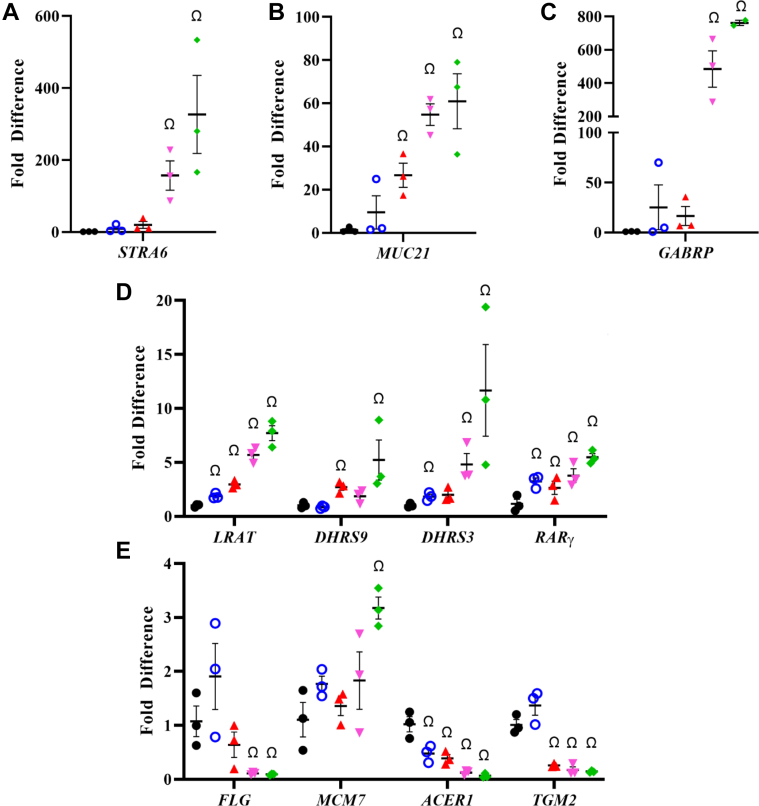
Our previous study indicated that treatment with UAB30 altered the expression of genes regulated by ATRA (32). To determine whether UAB110 and UAB111 targeted the same genes, we examined the gene expression pattern in skin rafts by quantitative PCR (qPCR) (Figs. 3 and S1). The series of targets included genes involved in ATRA biosynthesis, several known RAR- and RXR-regulated genes, as well as known markers of keratinocyte differentiation (FLG, SPINK5, ACER, TGM2). Treatment with 0.2 μM UAB110 and UAB111 resulted in strong upregulation of ATRA-sensitive genes LRAT, DHRS3, DHRS9, RARγ, STRA6, and GABRP. Transcripts that have been previously reported as downregulated by ATRA (FLG, RDHE2) were also significantly downregulated after treatment with UAB110 and UAB111. Importantly, the effects elicited by UAB110 and UAB111 at 0.2 μM were comparable to the effects produced by UAB30 at 2 μM. Thus, the next-generation rexinoids were at least ∼10-fold more potent in regulating gene expression than UAB30 and several-fold more potent than Targretin.
Figure 3.
QPCR analysis of gene expression in skin rafts treated with rexinoids.A–E, quantitative PCR analyses of gene expression in human skin raft cultures were done in triplicate. Conditions used were as follows: DMSO (black circle), UAB30 (hollow blue circle), Targretin (red triangle), UAB110 (magenta downward triangle), UAB111 (green diamond); n = 3 independent skin rafts, Ω p < 0.05 compared to DMSO control.
RXR heterodimers regulate the expression of numerous other genes in addition to those regulated by the RXR–RAR pathway. To obtain a more global picture of the gene expression changes induced by rexinoids in skin rafts, we performed RNA-seq analysis (Fig. 4). These experiments focused on comparing the effects of UAB30 and Targretin to those of UAB110. Rexinoid UAB110 was shown to act as a full and potent RXR agonist in the HEK293T cell line (33), yet the increase in serum triglycerides in rats treated by UAB110 was very similar to the low toxicity UAB30. In contrast, while UAB111 was an even more potent agonist than UAB110, its administration to rats raised triglycerides levels by 280% or more over controls. Since UAB111 did not appear as a viable drug candidate, it was not included in the RNA-seq study.
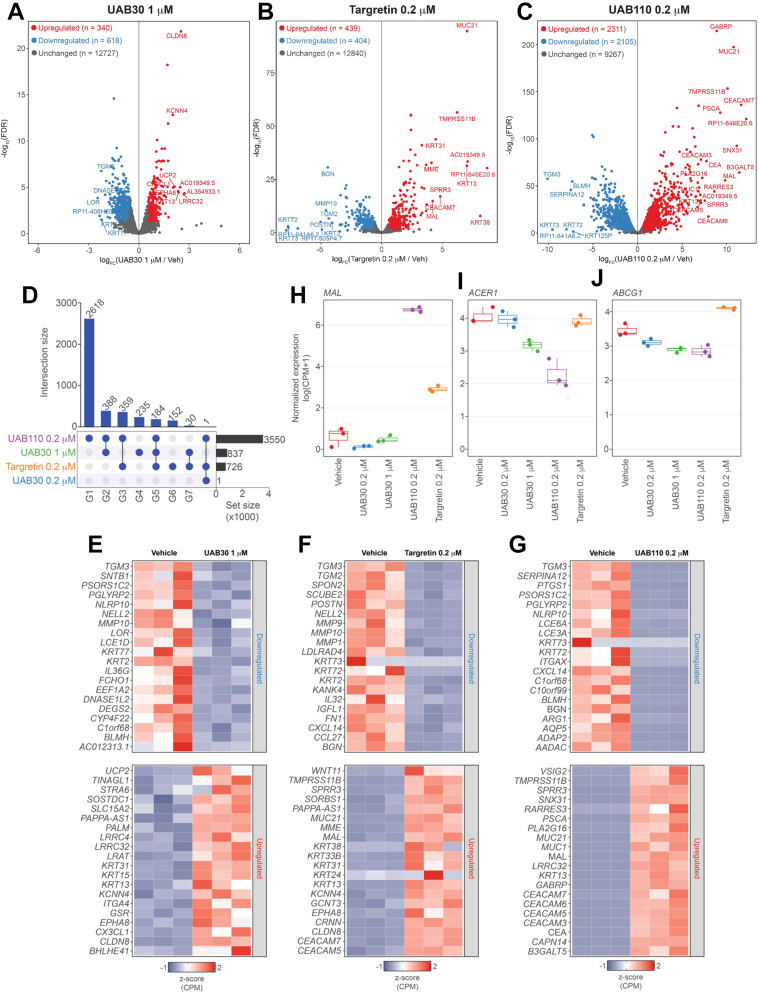
Figure 4.
RNA-seq analysis of skin rafts treated with rexinoids. RNA-seq analysis of skin raft cultures treated with rexinoids for 11 days. A–C, volcano plots showing significantly changing genes (CPM > 5, FDR < 0.05, FC ≥ 1.5). (A) 1 μM UAB30, (B) 0.2 μM Targretin, (C) 0.2 μM UAB110. D, UpSet plot showing the overlaps of significantly changing genes (CPM > 5, FDR < 0.05). Intersection size represents the number of genes unique to the overlap of the rexinoid treatments. Set size represents total size of the significantly changing genes for each rexinoid treatment. E–G, heatmaps of top 20 upregulated and downregulated genes (CPM > 5, FDR < 0.05, FC ≥ 1.5). (E) 1 μM UAB30, (F) 0.2 μM Targretin, (G) 0.2 μM UAB110. H–J, boxplots of individual gene examples of normalized logCPM expression. (H) MAL, (I) ACER1, (J) ABCG1. CPM, Counts Per Million.
Principal component analysis of the biological replicates revealed that the rexinoid treatments resulted in distinct gene expression patterns (Fig. S2). Comparing gene expression profiles between rafts treated with 0.2 μM UAB30, Targretin, and UAB110 revealed many similarities and differences between the induced gene expression changes (Fig. 4A). Due to the lower potency of UAB30, an additional dose of 1 μM UAB30 was included as a reference point. As suggested by the lack of changes in skin raft morphology (Fig. 2C), treatment with 0.2 μM UAB30 induced a significant change (FDR < 0.05) in the expression of only one gene, MAL (Fig. 4, A and B), which encodes an integral membrane protein implicated in the apical transport of proteins in polarized epithelial cells. The higher dose of 1 μM UAB30 induced much more robust changes in gene expression, upregulating 340 genes and downregulating 618 genes (Counts Per Million, CPM > 5, FDR < 0.05, FC ≥ 1.5) (Fig. 4E). ATRA-sensitive genes LRAT and STRA6 were greatly upregulated, while TGM3 was significantly downregulated (Fig. 4H). UAB30 at 1 μM uniquely altered the expression of 235 genes (Fig. 4A).
Treatment of skin rafts with UAB110 resulted in the largest impact on gene expression among the three rexinoids tested, upregulating 2311 and downregulating 2105 genes (CPM > 5, FDR < 0.05, FC ≥ 1.5) (Fig. 4F). Among the top altered transcripts were many ATRA-sensitive genes, such as the upregulated MUC21, GABRP, RARRES3, and MAL and downregulated TGM3 (Fig. 4F). Compared to the other rexinoid treatments, treatment with 0.2 μM UAB110 uniquely affected the expression of 2618 genes (Fig. 4A).
In comparison to UAB110, treatment of skin rafts with Targretin affected the expression of significantly fewer genes, with 152 genes uniquely regulated (FDR < 0.05) (Fig. 4A) and the overall upregulation of 439 and downregulation of 404 genes (CPM > 5, FDR < 0.05, FC ≥ 1.5) (Fig. 4G). Similar to UAB110, ATRA-sensitive genes were among the top altered transcripts, with the upregulation of MUC21 and MAL and the downregulation of TGM2 and TGM3 (Fig. 4J).
Interestingly, 184 genes were affected by all three rexinoids (Fig. 4A). Pathway analysis performed against Reactome version 78 showed that, in agreement with profound changes in the morphology of skin rafts, the common genes affected were those involved in the formation of the cornified envelope and keratinization, developmental processes, activation of matrix metalloproteases, and extracellular matrix organization.
There were also notable differences in the pathways altered by individual rexinoids. While 1 μM UAB30 primarily altered the expression of the cornified envelope genes, 0.2 μM UAB110 and Targretin also had substantial effects on endosomal/vacuolar pathway, antigen presentation, and interferon α/β signaling, with cornified envelope pathway only in the fourth position and seventh position, respectively, based on the p values.
Signaling through other RXR partners was also somewhat altered. Among the targets of the RXR–LXR pathway, the ABCG1 gene, which regulates cholesterol transportation and cellular lipid homeostasis, was down in rafts treated by UAB30 and UAB110, but it was up in rafts treated with Targretin (Fig. 4D). Similarly, the RXR-PPAR target gene ACER1, encoding a ceramidase, was downregulated after treatment with UAB30 and UAB110 but upregulated with Targretin (Fig. 4C). Thus, RNA-seq analysis provided a valuable insight into the similarities and differences in the actions of rexinoids at the level of gene transcription.
Detection of ATRA in skin rafts
Our previous study indicated that treatment with UAB30 raised the steady state levels of ATRA in organotypic skin raft cultures (32). Since the expression of ATRA-regulated genes was altered in skin rafts treated with UAB110 and UAB111, we asked whether the levels of ATRA were increased in these rafts. Skin raft cultures were treated with 0.2 μM UAB110 and UAB111 and with 2 μM UAB30, for comparison. ATRA levels were determined using multistage tandem mass spectrometry, also known as multiple reaction monitoring cubed (MRM3) which enhances selectivity in complex matrices, using an MRM3 transition of m/z 301.1 → m/z 205.1 → m/z 159. A calibration curve constructed from authentic standards was used to measure the concentration as previously described (39) (Fig. 5).
Figure 5.
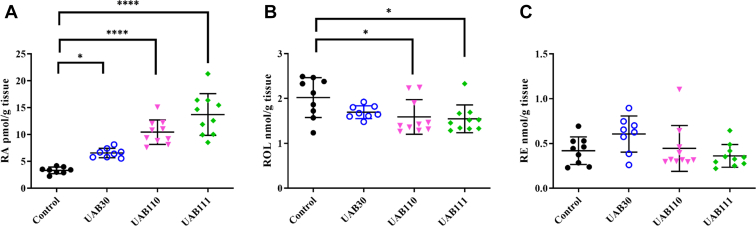
Quantification of retinoic acid, retinol, and retinyl esters. Retinoic acid (RA), retinol (ROL), and retinyl esters (RE) levels were assessed by liquid chromatography with multiple reaction monitoring mass spectrometry. Treatments of skin rafts were as follows: DMSO (black circle), UAB30 (hollow blue circle), UAB110 (magenta downward triangle), UAB111 (green diamond). RE represent the storage form of ROL. ROL is the metabolic precursor of RA. A, steady state levels of RA were significantly elevated in skin rafts treated with UAB110 or UAB111. B, skin rafts treated with UAB110 or UAB111 displayed significant reduction of retinol. C, the content of RE was similar in skin rafts treated by different rexinoids (n = 8 to 10 of skin rafts, ∗p < 0.05; ∗∗∗∗p< 0.0001 compared to DMSO control.
The basal levels of ATRA in vehicle-treated rafts were 3.33 ± 0.62 pmol/g tissue (Fig. 5A). In the 2 μM UAB30-treated rafts, the concentration of ATRA rose to 6.57 ± 0.88 pmol/g tissue. In the 0.2 μM UAB110-treated rafts, the concentration of ATRA nearly tripled to 10.45 ± 2.28 pmol/g of tissue. And the skin rafts treated with 0.2 μM UAB111 showed the highest levels of ATRA at 13.72 ± 3.88 pmol/g tissue. These results were consistent with the increasingly disordered appearance of skin rafts treated with UAB30 < UAB110 < UAB111 and with the increasing amplitude of changes in expression of ATRA-regulated genes (Fig. 3). The levels of all-trans-retinol were decreased in skin rafts treated with UAB110 and UAB111, possibly due to enhanced conversion of retinol to ATRA (Fig. 5B). There was no statistically significant difference in retinyl ester content between treated and control rafts although UAB30-treated rafts appeared to have slightly more retinyl esters (Fig. 5C). Our previous study showed a 4-fold increase in retinyl esters upon treatment with UAB30 (33), consistent with the upregulation of LRAT. The skin rafts used in the current study had 3-fold higher basal levels of retinyl esters, possibly due to individual differences among neonatal skin donors, which could mask further increase in retinyl esters somewhat. In the case of UAB110 and UAB111, the increased flux from retinol to ATRA driven by the irreversible second step of retinaldehyde oxidation to ATRA could potentially mitigate the increase in retinol esterification by LRAT. Importantly, the direct measurements of retinoids in skin rafts provided evidence that similarly to UAB30, UAB110 and UAB111 raised the cellular levels of ATRA.
HDX MS analysis of rexinoids bound to the RXRa-LBD homodimer
To examine the structural and dynamical effects of rexinoid binding to its receptor, we performed differential HDX MS where RXRα-LBD homodimer with no ligand bound (apo) was used as a reference. In-line pepsin digestions of RXRα-LBD and LC-MS analysis reliably yield 78 peptides that cover 97% of the primary sequence (Fig. S3 and Table S1). The deuterium uptake results are displayed as a difference map in Figure 6. A negative value in deuterium uptake represented a decrease in solvent accessibility and hydrogen bonding (less dynamic/more protection) in a given region of the LBD when bound to rexinoid as compared to apo. A positive value represented an increase in exchange (more dynamic/less protection). A significant change in deuterium uptake was ±0.5 deuterium (Fig. S6).
Figure 6.
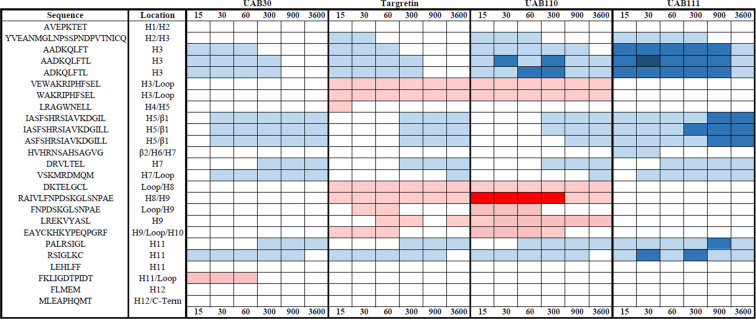
Deuterium incorporation difference maps of RXRα-LBD in complex with different rexinoids. HDX MS analysis of four rexinoids with six on-exchange time points (15, 30, 60, 300, 900, and 3600 s) organized into a difference map. Results are reported as an increase (shades of red) or decrease (shades of blue) in deuterium in different regions of the RXR–LBD–rexinoid complex relative to apo-RXR-LBD. All four complexes displayed reduced dynamics in regions of the LBP with UAB111 resulting in the greatest reduction of dynamics in H3. RXR-UAB110 and RXR-Targretin complexes revealed increased dynamics in the C-terminal end of H3, H8, and H9. Significance was established via volcano plots (Fig. S5). LBD, ligand-binding domain; LBP, ligand-binding pocket; RXR, retinoid X receptor.
The RXRα-LBD with rexinoid bound displayed overall reduced dynamics in regions that are part of the LBP, as we have previously shown (Fig. 6, shades of blue) (18). The LBP involves regions from H3, H5, β-sheet, H7, and H11. These same regions showed the greatest difference between the four rexinoids (Figs. 6, 7A, and S7). The binding of UAB111 caused the greatest reduction in dynamics for the LBP peptides in the H3, the H5/β1 region, and to a lesser extent H11 (Fig. 6 dark shades of blue). For UAB110 binding, the H3 region had reduced dynamics for all time points but to a lesser extent than UAB111. In contrast, upon UAB30 or Targretin binding, only the early time points displayed lowered dynamics. For the H5/β1 peptides, UAB111 binding to RXRα-LBD reduced dynamics in this region across all time points, whereas the other three rexinoids only displayed protection in the latter time points. The difference in the H11 region between rexinoids was more gradual moving from less protection to more protection with UAB30, Targretin, UAB110, and then UAB111.
Figure 7.
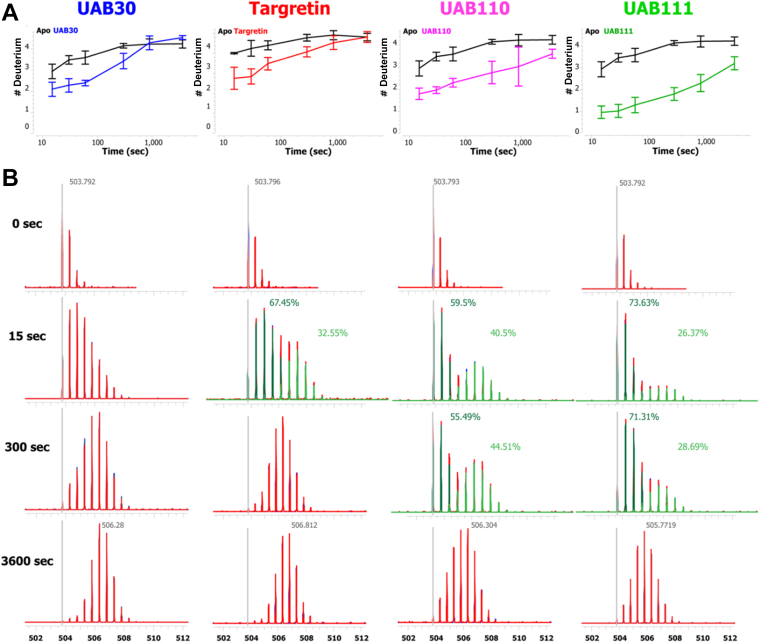
RXRα-LBD ligand-binding pocket Helix 3 demonstrates the highest extent of HDX MS variation when bound to different rexinoids. H3 is involved in both the ligand-binding pocket and the formation of the AF-2 coactivator binding site. A, Deuterium uptake versus time for the H3 peptide A271ADKQLFTL2792+ when bound to UAB30 (blue), Targretin (red), UAB110 (magenta), and UAB111 (green) relative to apo-RXRα-LBD. UAB111 binding lowers the deuterium uptake for this region the most. B, the HDX MS raw spectral data for the H3 peptide for the 0, 15, 300, and 3600 s time points. As the deuterium is incorporated over time, the isotopic envelope increases in mass. For UAB30 bound, the population of peptide ions shifts higher as a whole. When the other rexinoids are bound, a separation of the H3 peptide ion populations can be seen at the 15 s time point that indicates two populations of tightly bound versus more deuterium accessible forms of the RXR-LBD in this region. This separation in the peptide ion populations is still visible in the UAB110- and UAB111-bound raw data at the 300 s time points with both ligands still showing less overall deuterium incorporation at 3600 s (broader isotopic envelope). These HDX MS bimodal distributions for UAB110 and UAB111 demonstrate the differential interactions of the various rexinoids with H3 and H3’s “sensitivity” to differential ligand binding. Other ligand-binding pocket regions of the RXR-LBD do not show this distinction in the data when the various rexinoids are bound. A similar plot of a Helix 7 peptide D347 RVLTEL353 (1+) (also part of the ligand-binding pocket) is provided (Fig. S8). HDX MS, hydrogen deuterium exchange mass spectrometry; LBD, ligand-binding domain; RXR, retinoid X receptor.
Beyond the LPB, there were two regions of the RXRα-LBD that displayed increased dynamics for UAB110 and Targretin (Fig. 6, shades of red). These were the C-terminal portion of H3 and the H8/H9 region including the loop between the two helices. RXR in complex with UAB30 or UAB111 did not induce significant changes in deuterium incorporation for these regions (Fig. S7).
The rate of exchange was examined within the HDX MS data of the RXRα-LBD. The majority of our HDX MS data on RXRα-LBD displayed EX2 kinetics (Fig. S8). Within EX2 kinetics, the rate of exchange eventually reaches equilibrium between the open and close state and was observed through a gradual shift in the peptide isotopic distribution (40). An example of EX2 kinetics was displayed in Figure 7B in the first column and in Fig. S8. In EX1 kinetics, the rate of exchange reflects a different unfolding pattern that was identified in the H3 peptide, A271ADKQFTL279 (Fig. 7B) (41, 42). The EX1 on-exchange kinetics for H3 was manifested through two populations of isotopic distributions in several of the on-exchange time points (Fig. 7B, second, third, and fourth columns). The first population had undergone a slower opening and closing event and a second population had exposed a significant portion of the H3 region for rapid exchange. The two populations of isotopic distributions (bimodal distributions) were observed for RXRα-LBD bound to Targretin, UAB110, and UAB111. For either UAB110 or UAB111 in complex with RXRα-LBD, the bimodal distribution was seen until 3600 s. Targretin only displayed a bimodal distribution at 15 s. By 3600 s, each rexinoid bound to RXRα-LBD showed a single population of fully exchanged H3 peptides.
Based on this range in deuterium exchange for H3, we examined its correlation with the measured binding affinities for the various rexinoids. We have previously made use of differential scanning calorimetry (DSC) to determine the extent of thermostabilization caused by agonist binding at both physiological and elevated temperatures (43). UAB110 and UAB111 have been shown to stabilize the RXRα-LBD homodimer more significantly than UAB30 with differences in free energy of stabilization (ΔΔG), reaching 7 to 8 kJ/mol. In addition, the increase in thermal unfolding temperature (ΔTm) of the holo-homodimers bound with different agonists, with regard to the apo-homodimer, is directly proportional to the ΔΔG values.
Here, we measured the unfolding Tm of RXRα-LBD bound with Targretin by DSC and directly compared it with the Tm values of those bound with the UAB rexinoids Fig. S4A). Subsequently, we looked for a correlation with the H3 peptide deuterium uptake to see if the H3 region served as a leading indicator of binding affinity. When plotted (Fig. S4B), a trend between the decrease in number of deuterium incorporations by H3 and the increase in ΔTm is observed. Such correlation has been noted for other proteins (44). Overall, this analysis further confirmed the significant role of H3 in binding of rexinoids.
HDX MS analysis of rexinoids + GRIP-1 coactivator bound to the RXRa-LBD homodimer
In our previous analysis of UAB30 and Targretin, we also analyzed the RXRα-LBD by HDX MS in the presence of rexinoid and a coactivator peptide, GRIP-1. The GRIP-1–binding site is comprised of H3, H4, and H12. This work helped in the correlation between the observed dynamics and the existing X-ray crystallography structures. Thus, we performed similar HDX MS experiments with the ternary complex of RXRα-LBD-rexinoids-GRIP-1 with the two potent rexinoids, UAB110 and UAB111. The ternary complexes displayed reduced dynamics in the coactivator-binding site (Fig. 8). Compared to the HDX MS analysis of the RXRα-LBD:UAB110 or UAB111 complexes, GRIP-1 binding resulted in further suppressions in deuterium incorporation for regions involved in the LBP (Fig. 8).
Figure 8.
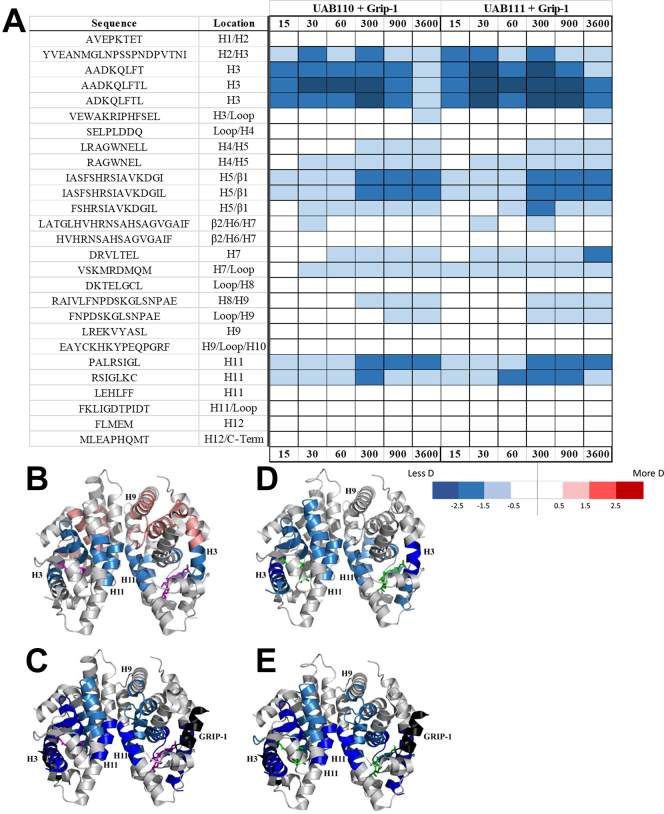
HDX MS analysis of RXR ternary complexes demonstrates potency of UAB110 and UAB111.A, the HDX MS difference map of RXRα-LBD in complex with UAB110 or UAB111 and GRIP-1 coactivator peptide. When the HDX MS results for the two potent rexinoids are painted onto X-ray crystal structures, the patterns of deuterium uptake can be visualized and shown how UAB110 binding (B, magenta) and UAB111 binding (D, green) to RXRα-LBD result in different patterns of dynamics. The regions shaded in blue show the extent of decreased dynamics for regions that are part of RXR ligand-binding pocket. There are positive dynamics observed in H3 and H8/H9 (shades of red) when UAB110 is bound. When UAB111 alone is bound (D) to the RXRα-LBD, the extent of decreased dynamics nearly matches the HDX MS results for the ternary complexes of UAB110 (C) or UAB111(E) bound plus the GRIP-1 coactivator peptide (black). Each potent rexinoid induces unique in solution dynamics for the RXRα-LBD. HDX MS, hydrogen deuterium exchange mass spectrometry; LBD, ligand-binding domain; RXR, retinoid X receptor.
These results mimicked what we saw in our previous analysis where GRIP-1 binding significantly stabilized the RXRα-LBD into the active conformation. Interestingly, the increased dynamics observed for the C-terminal end of H3 and the H8/H9 plus loop region when UAB110 alone was bound to the homodimer were not observed when GRIP-1 was added to the complex (Fig. 8, B and C). While the addition of GRIP-1 provides context for comparison of HDX MS results to existing X-ray crystal structures, the rexinoid distinguishing characteristics were best observed in complexes that did not include the coactivator peptide.
Evaluating the positive RXRα-LBD dynamics and homodimer interface for the bound rexinoids
Based on the differences observed in the HDX MS profiles for each rexinoid bound to RXRα-LBD, we mapped the HDX MS deuterium incorporation results onto the X-ray crystal structures of RXRα-LBD homodimers in complex with UAB110 (PDBid: 4RMD) and UAB111 (PDBid: 4RME) (Figs. 8 and S10). By mapping the HDX MS results onto the X-ray crystal structures, we observed a connecting allosteric path of increased RXRα-LBD dynamics when UAB110 or Targretin were bound that seemed to move towards the homodimer interface based on the increased dynamics (shades of red) in the C-terminal portion of H3 and the adjacent H8-loop-H9 region. Correspondingly, the lack of these positive dynamics for UAB111 and UAB30 created a stark contrast.
Based on these observations, we then evaluated the dimer interface of all four RXRα-LBD-rexinoid-GRIP-1 X-ray structures by use of the Proteins, Interfaces, Structures, and Assemblies (PISA) server (45). Through this analysis, we identified RXRα-LBD homodimer interface contacts that are distinct when UAB111 and Targretin are bound (Fig. S9). Specifically, Lys417 in H10 forms a salt bridge with Glu390 in H9 of the opposite monomer when UAB111 is bound. There is a second salt bridge that is formed between Lys417 in H10 and Glu394 in H9 of the opposite monomer. There is a one unique Targretin-induced interaction between Lys405 in Helix 9 and Glu401 in H9 that is not seen in the other ligands. The unique and common interactions in the various homodimer interfaces identified by the PISA algorithm are listed in Fig. S9. This evaluation of the ternary complex X-ray crystal structures indicated that the rexinoids may induce unique dimer interface interactions even in the presence of a GRIP-1 coactivator.
Overall, both our biological and biophysical analyses demonstrated that UAB110 and UAB111 are more potent agonists of RXR than either UAB30 or Targretin. These studies demonstrated that the potencies of each of these rexinoids were distinct and likely followed unique RXR activation mechanisms with UAB111 being too toxic for therapeutic use. This leaves UAB110 as a uniquely potent rexinoid that does not induce toxicity and thus merits further investigations as a viable cancer chemopreventive agent.
Discussion
RXR agonists can have pleiotropic effects on gene transcription due to the role of RXR in heterodimerization with several nuclear transcription factors (46). Still, RXR remains a target for drug development based on the potential for elucidating selective RXR transcriptional response in certain tissues and different disease states such as cancer (3, 4, 20, 28). This study makes use of both an in vitro model of human epidermis and biophysical analysis to demonstrate the higher potency of two RXR-selective agonists, UAB110 and UAB111. We identify similarities and differences in (1) how these two rexinoids influence RXR ligand-mediated signaling in terms of activation of ATRA-related transcriptional profiles in the human epidermis and (2) how they influence the structural dynamics of the RXRα LBD as observed in the differential HDX MS analysis.
In our current work, at just 0.2 μM concentration, both UAB110 and UAB111 triggered changes in the epidermal stratification pattern similar to those observed in skin rafts treated with 10 μM UAB30. The differences observed in the histology of skin rafts treated with rexinoids correlated to the gene activation levels and steady state levels of ATRA. Previously, we proposed that UAB30 would raise ATRA levels in a two-step mechanism. First, UAB30 potentiates the transcriptional activity of existing cellular ATRA bound to RXR-RAR heterodimers. This upregulates the ATRA-sensitive genes which include STRA6 (47) and LRAT (48) required for the uptake and retention of retinol. Then, the increased influx of retinol promotes the biosynthesis of ATRA and increases in its cellular levels, leading to further upregulation of RXR-RAR signaling. While the increased cellular levels of ATRA provide direct support for this mechanism, it is possible that some of the stimulation of RAR-related genes could arise from low-level binding of the rexinoids to RAR itself. We and others have reported on the low-level stimulation of rexinoids in RAR reporter assays. We would presume that once stimulation of ATRA synthesis occurs, any low affinity binding of RAR by the rexinoids would be displaced by ATRA. Future studies need to include direct assessment of rexinoid binding to RAR alone or in the presence of RXR as a heterodimer as well as in the presence or absence of ATRA.
It is well known that vitamin A deficiency leads to an increased development of spontaneous and chemically induced tumors (49), whereas dietary vitamin A supplementation appears to decrease chemically induced tumor incidence. ATRA prevents tumor development by inhibiting proliferation (50, 51, 52), stimulating differentiation (53), inducing apoptosis (54, 55), or combinations of these mechanisms. We have shown that the expression of ATRA-sensitive genes is altered in UVB-irradiated mouse skin and in mouse models of UVB-induced basal cell carcinoma and squamous cell carcinoma, suggesting that exposure to UVB lowers the levels of bioactive ATRA (32). The reduced ATRA signaling can be rescued by ATRA supplementation. Additionally, organ-transplant recipients are at a higher risk of developing UVB-induced nonmelanoma skin cancers because of the drugs required to maintain their grafted tissue. Currently, ATRA is used in combination with other chemotherapeutic agents for the treatment of Acute Promyelocytic Leukemia in adults and of Neuroblastoma in children (56). Limitations in ATRAs’ therapeutic potential include rapid first-pass metabolism and poor aqueous solubility (57). One way around these limitations is to induce physiological ATRA biosynthesis through treatment with rexinoids. However, bexatorene, the only rexinoid approved by the FDA, is dose limited due to its known hyperlipidemia toxicity. A clear alternative would be UAB rexinoids. As shown in the present study, at 0.2 μM concentrations, UAB110 and UAB111 significantly upregulated the expression of STRA6 and LRAT resulting in 3- to 4-fold increase in ATRA concentrations. The high potency and low toxicity of UAB110 make it an attractive chemopreventive agent that could be used at much lower doses than UAB30 to raise ATRA levels in conditions and disease states known for reduced ATRA signaling. It should be noted that at a ten-fold lower concentration, UAB110 has a much broader impact on gene expression than UAB30. This could be due to the 1.6-fold higher concentration of ATRA in UAB110-treated skin rafts than UAB30-treated rafts, resulting in a visibly greater impact of UAB110 on epidermal stratification pattern. In addition, UAB110 could produce off-target effects, independent of RXR. This possibility will be investigated in a follow-up study.
X-ray crystal structures have provided critical details of the active conformation of the RXR-LBD in complex with rexinoids; however, these structures do not reveal dynamic responses that we have shown to be important both in ligand binding and in coactivator recruitment (17, 18, 43). In the crystal structures of RXR bound to these rexinoids, there were no significant differences in the overall structure of the LBD; however, through HDX MS, we observed how ligand binding induced rexinoid-specific dynamics in the RXRα-LBD. The HDX MS data revealed that between the four rexinoids, UAB110 and UAB111 protect the LBP structural elements from deuterium uptake to a larger extent than UAB30 and Targretin. This observed increased protection correlates well with their binding affinities and thermodynamic properties we have previously reported (33). The HDX MS data allowed for the determination of where the higher affinity binding was occurring within the RXRα-LBD. The dynamics of H3 were reduced significantly more for RXRα-LBD in complex with UAB110 or UAB111 than UAB30 or Targretin. The UAB110 in complex RXRα-LBD had a different response to binding compared to UAB111 (Fig. 6). RXR-UAB110 and RXR-Targretin complexes both displayed increased dynamics of the C-terminal end of H3 and the H8/H9 region. When mapped onto the homodimer structure, these positive dynamics appear to move in the direction of the homodimer interface. Similar dynamics profiles in the C-terminal end of H3 and the H8/H9 regions were reported for two LXR agonists–bound complexes in a HDX MS study of 17 different LXR-specific ligands (58). While this effect was not highlighted in the study, our RXRα-LBD-UAB110 biophysical data provide corroborating evidence that this can be a known response to some but not all ligands for NRs. Interestingly, H3 is the only region directly involved in both the LBP and in the formation of the coactivator-binding site. Given that RXR–UAB110 and RXR–UAB111 complexes had bimodal distributions with the largest portion displaying the slower exchange rate, it infers that UAB110 and UAB111 had higher affinity binding but also that H3 is making significant conformational changes upon ligand binding.
Upon addition of GRIP-1 coactivator peptide, the HDX MS profiles are nearly identical. Our data suggests the differences in RXR response due to UAB110 or UAB111 binding are occurring prior to coactivator binding. Based on previous reports of the toxicity of UAB111 and our current results from our in vitro organotypic skin raft cultures (morphology, qPCR, and RNA-seq), we could interpret the biophysical HDX MS results for UAB111 binding as demonstrating what a full RXRα agonist profile would look like or even some form of ligand-induced hyperactivation of RXR-mediated transcription. In fact, it is notable how similar the HDX profiles for RXRα-LBD-UAB111 and the ternary complex of RXRα-LBD-UAB110-GRIP-1 are (Figs. 6 and 8). For UAB111 bound, it is as if the coactivator is already there to lock in the active conformation of RXRα-LBD. Our HDX MS data provides a better explanation for UAB111’s toxicity than other forms of biophysical analysis can.
The two distinct RXRα-LBD biophysical responses to potent rexinoids in the context of our in vitro biological data raises questions as to how the observed LBD dynamics manifest in the molecular mechanism of RXR-mediated transcriptional signaling. Specifically, our data demonstrate these potent rexinoids are inducing RXR-RAR–mediated transcription and increased steady state levels of ATRA. In the case of UAB110 binding, do the positive RXRα-LBD dynamics in H3 and H8/H9 exert an allosteric influence on the RXR homodimer promoting dissociation for (RAR) heterodimer association? Or do they contribute an allosteric role in RAR heterodimer signaling that would not occur in other RXRα heterodimer complexes? H9 is a significant part of the RXRα-LBD homodimer interface and also a part of the RXR-RAR heterodimer interface (59). Alternatively, given H3’s dual role as part of the LBP and the AF-2 coactivator-binding site, could the increased H3 dynamics influence the extent of coactivator binding? Would increased dynamics of the AF-2 site influence full-length RXR coactivator binding? GRIP-1 (also known as TIF2) has three LXXLL motifs and has been proposed to have a cooperative binding mechanism that includes the coactivator interacting with both LBD’ AF-2 sites in an RXR-RAR heterodimer (60). In the context of a full-length GRIP-1 coactivator, the increased dynamics of UAB110 bound to RXRα could influence this dual binding. With the range of biological and biophysical responses we have demonstrated with these rexinoids in our model systems, all of these questions could be addressed going forward with focus on heterodimer and coactivator interactions. Our experiments demonstrate a unique combination of tools to understand the molecular mechanisms of differential rexinoid signaling in skin. Importantly, our work demonstrates the potential of UAB110 as a potent nontoxic anticancer agent.
Experimental procedures
Materials
UAB30, UAB110, and UAB111 were synthesized at the University of Alabama at Birmingham according to previous methods (33). Dr Clinton Grubbs at UAB provided Targretin. The coactivator, GRIP-1, was synthesized by Thermo Fisher Scientific (686KHKILHRLLQDSS698) with a molecular mass of 1574.86 Da. The purity and structures of the rexinoids and coactivator were confirmed by LC-MS and NMR.
Preparation of organotypic skin rafts and treatment with rexinoids
Neonatal foreskins were obtained from the Newborn Nursery of the University of Alabama at Birmingham Hospital in compliance with the University of Alabama at Birmingham Institutional Review Board (IRB) regulations. As determined by the institutional IRB, the use of discarded unidentifiable foreskin tissue met the requirements for an exemption from IRB approval. Epidermal raft cultures were prepared as described previously (61, 62). Briefly, primary human keratinocytes were isolated from freshly collected neonatal foreskins and cultured in DermaLife calcium-free medium (Lifeline Cell Technology). Primary human keratinocytes were seeded onto a dermal equivalent consisting of collagen with embedded Swiss 3T3 J2 fibroblasts. After 3 days, skin equivalents were lifted onto stainless steel grids and cultured at the medium-air interface using raft culture medium prepared from Dulbecco's modified Eagle's medium, Ham's F12 medium, and 10% fetal bovine serum (Atlanta Biologicals), which was supplemented with cholera toxin, insulin, apo-transferrin, hydrocortisone-21, and human epidermal growth factor, as described previously (61, 62). The raft culture medium was supplemented with rexinoids at specified concentrations from DMSO stocks, beginning from the day the skin equivalents were lifted onto the grids until harvest. The raft cultures were allowed to stratify and differentiate for 11 days, whereupon they were harvested for analysis.
H&E staining
The rafts were fixed in 10% buffered formalin for 45 min and embedded in paraffin. Paraffin-embedded skin rafts were cut into 5-μm sections, mounted on Superfrost/Plus slides (Thermo Fisher Scientific). Sections were deparaffinized and rehydrated by a series of washes in decreasing concentrations of ethanol (95, 85, 70, 50, and 30%), processed for Harris hematoxylin (Thermo Fisher Scientific) and eosin (Thermo Fisher Scientific), and mounted with Permount (Thermo Fisher Scientific). All sections were analyzed using an AxioImager A2 microscope equipped with an AxioCam camera and AxioVision image capture software (Carl Zeiss MicroImaging, Inc).
Quantitative analysis of gene expression
Each rexinoid treatment group included three rafts. Gene expression in skin rafts was analyzed by qPCR. RNA from mouse tissues was extracted with TRIzol (Life Technologies), treated with DNase I (Promega), and re-extracted with TRIzol according to the manufacturer's protocols. Two micrograms of RNA was reverse-transcribed in 20 μl reactions using the Superscript III kit (Life Technologies). Real-time PCR was performed in duplicates for each sample in a LightCycler 480 instrument (Roche Diagnostics) using LightCycler 480 SYBR Green I Master Mix (Roche Applied Science) with 0.5 μm primers and 2.5 μl of 5- or 15-fold dilution of reverse transcription reactions in the final volume of 10 μl. Levels of transcripts were determined using a relative quantification method (63) and normalized to the geometric mean of transcript levels of three reference genes (β-Actin, Gapdh, and Hprt and Ppia). Sequences of the primers are available by request. PCR without complementary DNA (cDNA) templates did not produce significant amplification products. The specificity of the primers was verified by amplification of a single PCR product, which was determined by observing a single dissociation curve from each tissue. The primer sequences are available upon request.
Analysis of endogenous retinoid content
Skin raft samples were frozen upon collection and kept at −80 °C until assay. Extraction of retinoids was carried out under yellow light and all retinoids were handled with glass syringes and containers during extraction and quantification procedures. For retinoid quantification, one skin raft per replicate was assayed. Skin rafts were homogenized in 0.9% normal saline as described previously prior to extraction (64, 65). A two-step liquid-liquid extraction procedure was used to extract retinoids as described previously in detail (39, 64, 65, 66, 67, 68). Briefly, the first extraction was done by mixing skin raft lysates with 1 ml 0.025 M KOH in ethanol, followed by 10 ml hexane, respectively. The mixture was vortexed and subjected to centrifugation at 1000g for 30 s. The hexane phase containing retinol and retinyl esters was transferred to a new tube and put on ice. For the second extraction, the tissue lysates were mixed with 85 μl 4 M HCl followed by 10 ml hexane, respectively. The mixture was vortexed and centrifuged at 1000g for 30 s. The solvent from the second hexane phase containing retinoic acid was transferred to a new tube and put on ice. All samples were then placed under a gentle stream of nitrogen with heating at 30 °C until dry. Samples were then resuspended in 60 μl acetonitrile, placed in HPLC vials outfitted with low-volume deactivated glass inserts, and stored at −20 °C until analyzed. As internal standards, each cell or tissue lysate sample was mixed with all-trans-4,4-dimethyl-RA (internal standard for RA) and retinyl acetate (internal standard for retinol and retinyl ester).
ATRA levels were quantified using liquid chromatography-multistage-tandem mass spectrometry using a Shimadzu Prominence UFLC XR liquid chromatography system (Shimadzu) coupled to an AB Sciex 6500 QTRAP hybrid triple quadrupole mass spectrometer (AB Sciex) using atmospheric pressure chemical ionization operated in positive ion mode as described previously in detail (39). RA quantification was performed using an injection volume of 20 μl, and the chromatographic column was maintained at 25 °C. Data were analyzed using Analyst v 1.7.2 (AB Sciex), and RA content in each sample was normalized to skin raft weight. Levels of retinol and RE were quantified by HPLC-UV using a 30 μl injection volume on a Waters H-Class UPLC system equipped with a photodiode array detector operated in single wavelength detection mode at 325 nm according to previously published methodology (32, 65, 67). HPLC-UV chromatograms were acquired and analyzed using Waters Empower Software v3. Total retinol and RE were normalized to skin raft weight.
RNA isolation, library construction and sequencing
The quality of total RNA samples was checked on Agilent BioAnalyzer using Eukaryotic Total RNA Nano Kit according to the manufacturer’s protocol. Samples with RNA integrity number value >5 were accepted for library preparation process. RNA-Seq libraries were prepared from total RNA using Ultra II RNA Sample Prep kit (New England BioLabs) according to the manufacturer’s protocol. Briefly, poly-A RNAs were captured by oligo-dT–conjugated magnetic beads, then the mRNAs were eluted and fragmented at 94-Celsius degree. First strand cDNA was generated by random priming reverse transcription and after second strand synthesis step, double stranded cDNA was generated. After repairing ends, A-tailing, adapter ligation steps, and adapter ligated fragments were amplified in enrichment PCR and sequencing libraries were generated. Sequencing runs were executed on Illumina NextSeq 500 instrument using single-end 75 cycles sequencing.
RNA-seq mapping and gene expression quantification
Sequencing quality of the single-ended mRNA reads was evaluated by FastQC software and were aligned to the human reference genome (hg19) using HISAT2 (69) with default parameters. Genes were quantified using featureCounts (70). Genes with at least 5 CPM mapped read were considered expressed. Statistically significant difference was considered with FDR < 0.05 and fold change > 1.5 using edgeR (71) package in R. Visualization of the results were performed in R.
Protein expression and purification
hRXRα-LBD (Thr223 – Thr462) was expressed and purified according to Moras and Egea (72) and Xia et al. (17). The expression of the His6-tagged hRXRα-LBD fusion protein was done in BL21 (DE3) Escherichia coli (Invitrogen). The bacteria were grown in LB medium at 20 °C. One millimolar IPTG was used to induce protein expression. The cells were first treated with 0.5 M NaCl, 20 mM Tris (pH 8.0), cOmplete ULTRA tablets, and DNAse. A French press was used to lyse the cell (1500 p.s.i.) and then centrifuged at 25,000 rpm for 30 min. The His6-tagged hRXRα-LBD was eluted from a nickel-chelating column (GE Healthcare) using a 10 mM Tris (pH 8.0) buffer containing 500 mM NaCl and 300 mM imidazole. The fractions with hRXRα-LBD were then dialyzed in 10 mM Tris buffer (pH 8) containing 2 mM DTT, 0.5 mM EDTA, and 50 mM NaCl. α-thrombin (Novagen) hydrolyzed the His6-tag at 4 °C. The hRXRα-LBD homodimers and tetramers were separated at 4 °C using a hiLoad Superdex 75 gel filtration column (SEC, GE Healthcare) with 1.0 ml/min flow rate. MALDI mass spectrometry and SDS-PAGE were used to establish a purity of >97% and mass of the monomers (m/z = 26,433.1 Da). Native PAGE and UV confirmed that the isolated fractions were hRXRα-LBD homodimers and the concentration of the fractions. Samples were then flash frozen and stored at −80 °C.
Differential scanning calorimetry
Calorimetry experiments were performed in a VP-Capillary DSC (Malvern Instruments) in 0.130 ml cells at a heating rate of 2 °C/min. An external pressure of 2.0 atm was maintained using a nitrogen tank to prevent possible degassing of the samples on heating.
For the protein, the dimeric peak freshly eluted from SEC was used, and the SEC buffer was used as the matching buffer. In the cases where a ligand was used, a 10 mM stock of the ligand in degassed DMSO was prepared and stored at −80 °C protected from light. Prior to each set of experiments, the ligand stock was diluted in DMSO to 100 times the final desired concentration and then diluted 100-fold into the protein sample or matching buffer. The samples were incubated at 22 °C for 10 to 15 min and then transferred into the DSC sample loading plate. The sample was stored at 5 °C at least 1 h prior to the DSC runs depending on their position in the queue. DSC data analysis was carried out using the built-in analysis modules in Origin 7 (OriginLab) provided by the DSC manufacturer as described (43).
Automated HDX and data analysis
Solution phase HDX experiments were performed using an automated LEAP Technologies Twin PAL RTC LEAP Technologies). Experiments were performed as previously described. Thirty picomole of hRXRα-LBD homodimer is mixed with 10 times the concentration of ligand or vehicle (DMSO) with or without coactivator peptide and incubated at room temperature for 1 hour (18). The protein mix is then dispensed into a mixing tray where it is then diluted with 54 μl of deuterated buffer (or protonated buffer for control experiments). Following on-exchange time points (20 °C), 50 μl of the sample was aspirated and dispensed into 50 μl of quench buffer (3 M Urea, 1% TFA, 50 mM TCEP, 2 °C). Samples were taken at 15 s, 30 s, 60 s, 300 s, 900 s, and 3600 s and performed in triplicate. Samples were digested for 120s on an enzymatic Waters pepsin column (2 °C). Sample loading, digestion, and desalting were driven with a Shimadzu UPLC pump at 100 μl min-1. Peptic peptides were desalted on an Acclaim PepMap C18 trap column (Thermo Fisher Scientific), followed by an Acquity BEH C18 reverse-phase column (1 mm x 50 mm; Waters). A gradient of 5 to 90% acetonitrile, 0.1% formic acid over 16 min was used to elute the peptides directly into the SYNAPT G2-Si at 100 μl min-1 with a dual electrospray ionization source. Spectra were acquired over the scan range of 300 to 1500 m/z.
PLGS program (Waters) was used for peptide selection. Seven MsE runs were collected and peptides that were replicated in five of the seven MsE runs were used for analysis. HDX MS raw files were processed and visualized by the use of HDExaminer software (Sierra Analytics). In total, 78 peptides represent 96.7% amino acid sequence coverage of the RXRα-LBD. The average length of peptides was 11.2 amino acids with an average redundancy of 3.6. Differential HDX perturbation values were calculated by taking the observed number of deuterium incorporated from ligand binding in the RXRα-LBD:rexinoid at a specific peptide and subtracting it by the number of deuterium incorporated in the apo state from that same peptide, at each of the seven time points. HDX perturbations greater than ± 0.5 deuteriums were considered significant as described previously (41). Observed RXRα-LBD HDX MS peptides were manually curated across all time points to ensure consistent T0 peptide ion identification and subsequent shifting peptide ions as deuterium is incorporated across the time points. The deuterium versus time plots for all RXRα-LBD peptides were visualized in HDExaminer to ensure reproducibility across time points. Each RXRα–LBD complex evaluated was performed at least twice. HDX MS results were then collated into the plots shown in Figures 6 and 8 based off of changes in deuterium incorporation relative to apo-RXRα-LBD. The results were also painted onto existing X-ray crystallographic structures and described in Results section.
Data availability
Sequencing data sets performed in this study are available at the NCBI GEO under accession numbers: GSE199381. GEO token ‘yxyrwwcylbmblwd’ is generated for reviewer access.
Supporting information
This article contains 10 Supplemental Figures and 1 Supplemental Table.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the UAB O’Neil Comprehensive Cancer Center and the National Cancer Institute, NIH grant CA210946.
Author contributions
N. M., O. V. B., W. K. B., L. H., J. Y., N. P., Z. Y., A. V. K., V. A., D. D. M., M. A. K., L. N., N. Y. K., and M. B. R. investigation; N. M., O. V. B., W. K. B., L. H., N. Y. K., and M. B. R. data curation; N. M., O. V. B., D. D. M., N. Y. K., and M. B. R. writing–original draft; N. M., N. Y. K., and M. B. R. visualization; W. K. B. and L. H. formal analysis; C. E., V. A., D. D. M., N. Y. K., and M. B. R. funding acquisition; C. E., D. D. M., L. N., N. Y. K., and M. B. R. conceptualization; V. A., L. N., N. Y. K., and M. B. R. resources; M. A. K. methodology; M. A. K. validation; L. N., N. Y. K., and M. B. R. supervision; N. Y. K. and M. B. R. project administration; N. M., O. V. B., W. K. B., L. H., J. Y., N. P., Z. Y., A. V. K., C. E., V. A., D. D. M., M. A. K., L. N., N. Y. K., and M. B. R. writing–review and editing.
Funding and additional information
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. N. M. was supported by an NCI diversity supplement to CA210946. L. N. was also supported by DK115924; National Institutes of Health, R01AR074846 (support to M. A. K.); University of Maryland, School of Pharmacy, P23474 (support to M. A. K.); University of Maryland, School of Pharmacy Mass Spectrometry Center, SOP1841-IQB2014 (support to M. A. K.). N. Y. K. was also supported by R01AR076924.
Edited by Donita Brady
Contributor Information
Natalia Y. Kedishvili, Email: nkedishvili@uab.edu.
Matthew B. Renfrow, Email: renfrow@uab.edu.
Supporting information
References
- 1.Bookout A.L., Jeong Y., Downes M., Ruth T.Y., Evans R.M., Mangelsdorf D.J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 3.Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ottow E., Weinmann H. Nuclear receptors as drug targets: a historical perspective of modern drug discovery. Nucl. Receptors as Drug Targets. 2008;7:679–684. [Google Scholar]
- 5.Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altucci L., Leibowitz M.D., Ogilvie K.M., de Lera A.R., Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat. Rev. Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 7.Kojetin D.J., Matta-Camacho E., Hughes T.S., Srinivasan S., Nwachukwu J.C., Cavett V., et al. Structural mechanism for signal transduction in RXR nuclear receptor heterodimers. Nat. Commun. 2015;6:1–14. doi: 10.1038/ncomms9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesario R.M., Stone J., Yen W.-C., Bissonnette R.P., Lamph W.W. Differentiation and growth inhibition mediated via the RXR: PPARγ heterodimer in colon cancer. Cancer Lett. 2006;240:225–233. doi: 10.1016/j.canlet.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Potter G.B., Beaudoin G.M., DeRenzo C.L., Zarach J.M., Chen S.H., Thompson C.C. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev. 2001;15:2687–2701. doi: 10.1101/gad.916701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturm E., Wagner M., Trauner M. Nuclear receptor ligands in therapy of cholestatic liver disease. Front. Biosci. 2009;14:4299–4325. doi: 10.2741/3529. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez-Martínez R., Castillo A.I., Steinmeyer A., Aranda A. The retinoid X receptor ligand restores defective signalling by the vitamin D receptor. EMBO Rep. 2006;7:1030–1034. doi: 10.1038/sj.embor.7400776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mark M., Ghyselinck N.B., Chambon P. Function of retinoid nuclear receptors: Lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu. Rev. Pharmacol. Toxicol. 2006;46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 13.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 14.Zhao Q., Chasse S.A., Devarakonda S., Sierk M.L., Ahvazi B., Rastinejad F. Structural basis of RXR-DNA interactions. J. Mol. Biol. 2000;296:509–520. doi: 10.1006/jmbi.1999.3457. [DOI] [PubMed] [Google Scholar]
- 15.Jeyakumar M., Tanen M.R., Bagchi M.K. Analysis of the functional role of steroid receptor coactivator-1 in ligand-induced transactivation by thyroid hormone receptor. Mol. Endocrinol. 1997;11:755–767. doi: 10.1210/mend.11.6.0003. [DOI] [PubMed] [Google Scholar]
- 16.Han J.S., Crowe D.L. Steroid receptor coactivator 1 deficiency increases MMTV-neu mediated tumor latency and differentiation specific gene expression, decreases metastasis, and inhibits response to PPAR ligands. BMC cancer. 2010;10:1–14. doi: 10.1186/1471-2407-10-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia G., Boerma L.J., Cox B.D., Qiu C., Kang S., Smith C.D., et al. Structure, energetics, and dynamics of binding coactivator peptide to the human retinoid X receptor α ligand binding domain complex with 9-cis-retinoic acid. Biochemistry. 2011;50:93–105. doi: 10.1021/bi101288y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boerma L.J., Xia G., Qui C., Cox B.D., Chalmers M.J., Smith C.D., et al. Defining the communication between agonist and coactivator binding in the retinoid X receptor α ligand binding domain. J. Biol. Chem. 2014;289:814–826. doi: 10.1074/jbc.M113.476861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heery D.M., Kalkhoven E., Hoare S., Parker M.G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 20.Overington J.P., Al-Lazikani B., Hopkins A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 21.Levin A.A., Sturzenbecker L.J., Kazmer S., Bosakowski T., Huselton C., Allenby G., et al. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXRα. Nature. 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 22.Heyman R.A., Mangelsdorf D.J., Dyck J.A., Stein R.B., Eichele G., Evans R.M., et al. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 23.Mangelsdorf D.J., Borgmeyer U., Heyman R.A., Zhou J.Y., Ong E.S., Oro A.E., et al. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 24.Dawson M. Synthetic retinoids and their nuclear receptors. Curr. Med. Chemistry-Anti-Cancer Agents. 2004;4:199–230. doi: 10.2174/1568011043352975. [DOI] [PubMed] [Google Scholar]
- 25.Gottardis M.M., Bischoff E.D., Shirley M.A., Wagoner M.A., Lamph W.W., Heyman R.A. Chemoprevention of mammary carcinoma by LGD1069 (Targretin): An RXR-selective ligand. Cancer Res. 1996;56:5566–5570. [PubMed] [Google Scholar]
- 26.Duvic M., Hymes K., Heald P., Breneman D., Martin A.G., Myskowski P., et al. Targretin is effective and safe for treatment of refractory advanced-stage cutaneous T-cell lymphoma: multinational phase II-III trial results. J. Clin. Oncol. 2001;19:2456–2471. doi: 10.1200/JCO.2001.19.9.2456. [DOI] [PubMed] [Google Scholar]
- 27.Duvic M., Martin A.G., Kim Y., Olsen E., Wood G.S., Crowley C.A., et al. Phase 2 and 3 clinical trial of oral Targretin ( capsules) for the treatment of refractory or persistent early-stage cutaneous T-cell lymphoma. Arch. Dermatol. 2001;137:581–593. [PubMed] [Google Scholar]
- 28.Muccio D.D., Atigadda V.R., Brouillette W.J., Bland K.I., Krontiras H., Grubbs C.J. Translation of a tissue-selective rexinoid, UAB30, to the clinic for breast cancer prevention. Curr. Top. Med. Chem. 2017;17:676–695. doi: 10.2174/1568026616666160617093604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolesar J.M., Hoel R., Pomplun M., Havighurst T., Stublaski J., Wollmer B., et al. A pilot, first-in-human, pharmacokinetic study of 9cUAB30 in healthy volunteers. Cancer Prev. Res. 2010;3:1565–1570. doi: 10.1158/1940-6207.CAPR-10-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vedell P.T., Lu Y., Grubbs C.J., Yin Y., Jiang H., Bland K.I., et al. Effects on gene expression in rat liver after administration of RXR agonists: UAB30, 4-methyl-UAB30, and Targretin. Mol. Pharmacol. 2013;83:698–708. doi: 10.1124/mol.112.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams A.P., Garner E.F., Stafman L.L., Aye J.M., Quinn C.H., Marayati R., et al. UAB30, A novel rexinoid agonist, decreases stemness in group 3 medulloblastoma human cell line xenografts. Transl. Oncol. 2019;12:1364–1374. doi: 10.1016/j.tranon.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L., Chaudhary S.C., Atigadda V.R., Belyaeva O.V., Harville S.R., Elmets C.A., et al. Retinoid X receptor agonists upregulate genes responsible for the biosynthesis of all-trans-retinoic acid in human epidermis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atigadda V.R., Xia G., Deshpande A., Wu L., Kedishvili N., Smith C.D., et al. Conformationally defined rexinoids and their efficacy in the prevention of mammary cancers. J. Med. Chem. 2015;58:7763–7774. doi: 10.1021/acs.jmedchem.5b00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atigadda V.R., Xia G., Desphande A., Boerma L.J., Lobo-Ruppert S., Grubbs C.J., et al. Methyl substitution of a rexinoid agonist improves potency and reveals site of lipid toxicity. J. Med. Chem. 2014;57:5370–5380. doi: 10.1021/jm5004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desphande A., Xia G., Boerma L.J., Vines K.K., Atigadda V.R., Lobo-Ruppert S., et al. Methyl-substituted conformationally constrained rexinoid agonists for the retinoid X receptors demonstrate improved efficacy for cancer therapy and prevention. Bioorg. Med. Chem. 2014;22:178–185. doi: 10.1016/j.bmc.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandra V., Wu D., Li S., Potluri N., Kim Y., Rastinejad F. The quaternary architecture of RARβ–RXRα heterodimer facilitates domain–domain signal transmission. Nat. Commun. 2017;8:1–9. doi: 10.1038/s41467-017-00981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayne L. Methods in enzymology. 566. Academic Press; Cambridge, MA: 2016. Hydrogen exchange mass spectrometry; pp. 335–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcsisin S.R., Engen J.R. Hydrogen exchange mass spectrometry: What is it and what can it tell us? Anal. Bioanal. Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones J.W., Pierzchalski K., Yu J., Kane M.A. Use of fast HPLC multiple reaction monitoring cubed for endogenous retinoic acid quantification in complex matrices. Anal. Chem. 2015;87:3222–3230. doi: 10.1021/ac504597q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodge E.A., Benhaim M.A., Lee K.K. Bridging protein structure, dynamics, and function using hydrogen/deuterium-exchange mass spectrometry. Protein Sci. 2020;29:843–855. doi: 10.1002/pro.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engen J.R. ACS Publications; Washington D.C.: 2009. Analysis of Protein Conformation and Dynamics by Hydrogen/deuterium Exchange MS. [Google Scholar]
- 42.Morgan C.R., Engen J.R. Investigating solution-phase protein structure and dynamics by hydrogen exchange mass spectrometry. Curr. Protoc. Protein Sci. 2009;58 doi: 10.1002/0471140864.ps1706s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z., Muccio D.D., Melo N., Atigadda V.R., Renfrow M.B. Stability of the retinoid X receptor-α homodimer in the presence and absence of rexinoid and coactivator peptide. Biochemistry. 2021;60:1165–1177. doi: 10.1021/acs.biochem.0c00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strutzenberg T.S., Garcia-Ordonez R.D., Novick S.J., Park H., Chang M.R., Doebellin C., et al. HDX-MS reveals structural determinants for RORγ hyperactivation by synthetic agonists. Elife. 2019;8 doi: 10.7554/eLife.47172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Evans R.M., Mangelsdorf D.J. Nuclear receptors, RXR, and the big bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawaguchi R., Zhong M., Kassai M., Ter-Stepanian M., Sun H. Vitamin A transport mechanism of the multitransmembrane cell-surface receptor STRA6. Membranes. 2015;5:425–453. doi: 10.3390/membranes5030425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Byrne S.M., Blaner W.S. Retinol and retinyl esters: biochemistry and physiology: thematic review series: fat-soluble vitamins: vitamin A. J. lipid Res. 2013;54:1731–1743. doi: 10.1194/jlr.R037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niles R.M. Signaling pathways in retinoid chemoprevention and treatment of cancer. Mutat. Research/Fundamental Mol. Mech. Mutagenesis. 2004;555:97–105. doi: 10.1016/j.mrfmmm.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 50.Langenfeld J., Kiyokawa H., Sekula D., Boyle J., Dmitrovsky E. Posttranslational regulation of cyclin D1 by retinoic acid: a chemoprevention mechanism. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12070–12074. doi: 10.1073/pnas.94.22.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuo T., Thiele C.J. p27 Kip1: a key mediator of retinoic acid induced growth arrest in the SMS–KCNR human neuroblastoma cell line. Oncogene. 1998;16:3337–3343. doi: 10.1038/sj.onc.1201830. [DOI] [PubMed] [Google Scholar]
- 52.Teixeira C., Pratt M.C. CDK2 is a target for retinoic acid-mediated growth inhibition in MCF-7 human breast cancer cells. Mol. Endocrinol. 1997;11:1191–1202. doi: 10.1210/mend.11.9.9977. [DOI] [PubMed] [Google Scholar]
- 53.Chen L.-C., Sly L., De Luca L.M. High dietary retinoic acid prevents malignant conversion of skin papillomas induced by a two-stage carcinogenesis protocol in female SENCAR mice. Carcinogenesis. 1994;15:2383–2386. doi: 10.1093/carcin/15.10.2383. [DOI] [PubMed] [Google Scholar]
- 54.Fujimura S., Suzumiya J., Anzai K., Ohkubo K., Hata T., Yamada Y., et al. Retinoic acids induce growth inhibition and apoptosis in adult T-cell leukemia (ATL) cell lines. Leuk. Res. 1998;22:611–618. doi: 10.1016/s0145-2126(98)00049-6. [DOI] [PubMed] [Google Scholar]
- 55.Hsu S.-L., Yin S.-C., Liu M.-C., Reichert U., Ho W.L. Involvement of cyclin-dependent kinase activities in CD437-induced apoptosis. Exp. Cell Res. 1999;252:332–341. doi: 10.1006/excr.1999.4625. [DOI] [PubMed] [Google Scholar]
- 56.Reynolds C.P., Matthay K.K., Villablanca J.G., Maurer B.J. Retinoid therapy of high-risk neuroblastoma. Cancer Lett. 2003;197:185–192. doi: 10.1016/s0304-3835(03)00108-3. [DOI] [PubMed] [Google Scholar]
- 57.Giuli M.V., Hanieh P.N., Giuliani E., Rinaldi F., Marianecci C., Screpanti I., et al. Current trends in ATRA delivery for cancer therapy. Pharmaceutics. 2020;12:707. doi: 10.3390/pharmaceutics12080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belorusova A.Y., Evertsson E., Hovdal D., Sandmark J., Bratt E., Maxvall I., et al. Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands. Commun. Biol. 2019;2:1–13. doi: 10.1038/s42003-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bourguet W., Ruff M., Chambon P., Gronemeyer H., Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 60.Senicourt L., Le Maire A., Allemand F., Carvalho J.E., Guee L., Germain P., et al. Structural insights into the interaction of the intrinsically disordered co-activator TIF2 with retinoic acid receptor heterodimer (RXR/RAR) J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.166899. [DOI] [PubMed] [Google Scholar]
- 61.Lee S.-A., Belyaeva O.V., Wu L., Kedishvili N.Y. Retinol dehydrogenase 10 but not retinol/sterol dehydrogenase (s) regulates the expression of retinoic acid-responsive genes in human transgenic skin raft culture. J. Biol. Chem. 2011;286:13550–13560. doi: 10.1074/jbc.M110.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H.-J., Chen T., Cheng L., Cheng T., Tung Y. Human keratinocyte culture using porcine pituitary extract in serum-free medium. Burns. 1995;21:503–506. doi: 10.1016/0305-4179(95)00030-f. [DOI] [PubMed] [Google Scholar]
- 63.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucl. Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Napoli J.L., Horst R.L. Quantitative analyses of naturally occurring retinoids. Met. Mol. Biol. 1998;89:29–40. doi: 10.1385/0-89603-438-0:29. [DOI] [PubMed] [Google Scholar]
- 65.Kane M.A., Napoli J.L. Retinoids. Springer; New York City, NY: 2010. Quantification of endogenous retinoids; pp. 1–54. [Google Scholar]
- 66.Kane M.A., Chen N., Sparks S., Napoli J.L. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem. J. 2005;388:363–369. doi: 10.1042/BJ20041867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kane M.A., Folias A.E., Napoli J.L. HPLC/UV quantitation of retinal, retinol, and retinyl esters in serum and tissues. Anal. Biochem. 2008;378:71–79. doi: 10.1016/j.ab.2008.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kane M.A., Folias A.E., Wang C., Napoli J.L. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal. Chem. 2008;80:1702–1708. doi: 10.1021/ac702030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mihaela P., Daehwan K., Geo M., Jeffrey T., Steven L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bates D., Mächler M., Bolker B., Walker S. Fitting linear mixed-effects models using lme4. arXiv. 2014 doi: 10.48550/arXiv.1406.5823. [preprint] [DOI] [Google Scholar]
- 71.Yu X., Liu H., Hamel K.A., Morvan M.G., Yu S., Leff J., et al. Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-019-13839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Egea P.F., Moras D. Purification and crystallization of the human RXRα ligand-binding domain–9-cisRA complex. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2001;57:434–437. doi: 10.1107/s0907444901000385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data sets performed in this study are available at the NCBI GEO under accession numbers: GSE199381. GEO token ‘yxyrwwcylbmblwd’ is generated for reviewer access.