Abstract
The KRAS gene is one of the most frequently mutated oncogenes in human cancer and gives rise to two isoforms, KRAS4A and KRAS4B. KRAS post-translational modifications (PTMs) have the potential to influence downstream signaling. However, the relationship between KRAS PTMs and oncogenic mutations remains unclear, and the extent of isoform-specific modification is unknown. Here, we present the first top–down proteomics study evaluating both KRAS4A and KRAS4B, resulting in 39 completely characterized proteoforms across colorectal cancer cell lines and primary tumor samples. We determined which KRAS PTMs are present, along with their relative abundance, and that proteoforms of KRAS4A versus KRAS4B are differentially modified. Moreover, we identified a subset of KRAS4B proteoforms lacking the C185 residue and associated C-terminal PTMs. By confocal microscopy, we confirmed that this truncated GFP-KRAS4BC185∗ proteoform is unable to associate with the plasma membrane, resulting in a decrease in mitogen-activated protein kinase signaling pathway activation. Collectively, our study provides a reference set of functionally distinct KRAS proteoforms and the colorectal cancer contexts in which they are present.
Keywords: colorectal cancer, RAS protein, proteoform, immunoprecipitation, KRAS, top–down proteomics, post-translational modification, MAPK pathway
Abbreviations: ACN, acetonitrile; AGC, automatic gain control; BU, bottom–up; CID, collision-induced dissociation; CPTAC, Clinical Proteomic Tumor Analysis Consortium; FA, formic acid; FBS, fetal bovine serum; FT–ICR, Fourier transform ion cyclotron resonance; GDC, Genomic Data Commons; ID, inner diameter; IP, immunoprecipitation; MAPK, mitogen-activated protein kinase; MEF, mouse embryonic fibroblast; MS, mass spectrometry; NCE, normalized collision energy; NCI, National Cancer Institute; 4OHT, 4-hydroxytamoxifen; PTM, post-translational modification; r.p., resolving power; RRID, Research Resource Identifier; SIM, selected ion monitoring; TDMS, top–down mass spectrometry; tMS2, targeted MS2
KRAS belongs to the RAS family of proteins, the core of which includes the genes KRAS, NRAS, and HRAS. KRAS is alternatively spliced at the fourth exon, giving rise to KRAS4A and KRAS4B isoforms. The RAS genes encode four ∼21 kDa GTPases, which play critical roles in cell signaling pathways such as those involving mitogen-activated protein kinase (MAPK) and PI3K (1). The activity of RAS isoforms is regulated by guanine nucleotide exchange factors, which promote the GTP-bound “active” state, and GTPase-activating proteins, which promote the GDP-bound “inactive” state through GTP hydrolysis (2).
KRAS is one of the most frequently mutated genes in cancer, with mutations prevalent in colorectal, pancreatic, and lung cancers. Three key sites of mutation in KRAS occur at residues G12, G13, and Q61 (3, 4, 5). Because of the critical role these residues play in coordinating the nucleotide and water molecule within the active site of KRAS, mutations at these sites “lock” KRAS into the active state, resulting in aberrant cell signaling. While there have been many attempts to develop therapeutics targeting KRAS, only a handful have been successful. The high affinity of KRAS for nucleotides and the absence of clear binding pockets for small molecules have rendered common therapeutic strategies ineffective. Attempts to block the addition of C-terminal lipid modifications required for association of KRAS with the plasma membrane have also proved unsuccessful because of compensatory modification pathways (6, 7, 8, 9). However, recent success has been achieved through covalent inhibitors like sotorasib that target the G12C mutant version of KRAS, primarily observed in cancers of the lung (10).
Previous studies have identified RAS post-translational modifications (PTMs) through methods such as immunoblotting and bottom–up (BU) proteomics. Functional assignment of these PTMs typically involves mutation of the modification site followed by experiments that examine RAS membrane association, activation of RAS-dependent signaling pathways (i.e., MAPK pathway), or protein–protein interactions (11). A critical RAS post-translational processing step occurs at the C-terminal CAAX motif, which includes C185 prenylation, subsequent proteolysis of the -AAX, and C-terminal carboxymethylation (Fig. S1). These PTMs are critical for association of RAS with the plasma membrane and thus for proper signaling through canonical pathways. In addition, there are secondary features upstream of the -CAAX motif that aid in membrane association. For KRAS4A, HRAS, and NRAS, this involves lipidation by cysteine palmitoylation, whereas KRAS4A and KRAS4B also contain a lysine-rich polybasic region, which promotes electrostatic interactions with the plasma membrane (12, 13, 14, 15, 16, 17, 18, 19, 20, 21). Other modifications that have been reported include N-terminal and lysine acetylation, cysteine nitrosylation, serine and tyrosine phosphorylation, lysine and arginine methylation, SUMOylation, and ubiquitination (22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39). The functions of many of these individual KRAS PTMs have been extensively investigated and are known to influence RAS GTPase activity, stability, subcellular localization, membrane partitioning, and protein–protein interactions (11). Despite this, the KRAS proteoform landscape, which captures the stoichiometry and dynamics of diverse and combinatorial modifications in an isoform-specific manner, remains undercharacterized.
BU relies on the tryptic digestion of proteins followed by tandem mass spectrometry (MS) sequencing of the resulting peptides, which does not allow for the complete characterization of the different protein molecular forms or proteoforms (40). For BU applications involving RAS family proteins, additional technical challenges arise because of the high sequence identity among the four RAS isoforms, a high rate of basic residues in the KRAS C-terminal domains, and separation of mutation and PTM sites within the primary sequences. Instead, we have employed top–down mass spectrometry (TDMS), which analyzes intact KRAS protein molecules, thus providing precise KRAS proteoform characterization and PTM localization (40).
Our laboratory previously reported a study employing immunoprecipitation coupled with top-down mass spectrometry (IP–TDMS) to characterize 11 KRAS4B proteoforms in isogenic colorectal cancer cell lines and 6 primary colorectal tumors (41). Here, through an improved IP–TDMS protocol with enhanced limits of detection, we characterized both KRAS4A and KRAS4B proteoforms, including those present at <5% relative abundance (41, 42). We deployed this optimized KRAS proteoform assay to a panel of 14 cell lines and 34 colorectal tumor samples. This revealed a more diverse RAS landscape with 39 completely characterized proteoforms, including a truncated form of KRAS4B lacking the C185 residue. This class of truncated proteoforms was highly abundant in the majority of primary tumors and was unable to associate with the plasma membrane or activate the RAS-dependent MAPK signaling pathway. These results offer an unprecedented level of insight into the KRAS proteoform landscape while revealing evidence for noncanonical KRAS4B-dependent signaling pathways operative in human colorectal cancers.
Results
Proteoform assay as a low bias readout of KRAS modifications
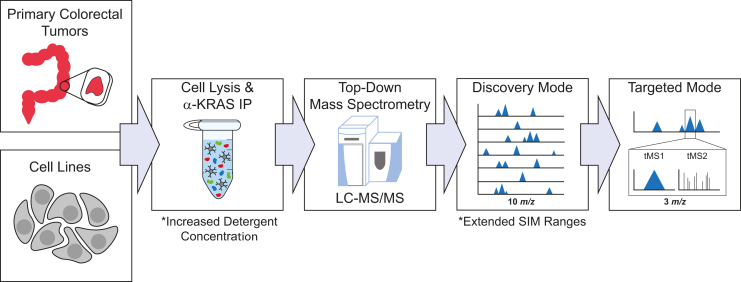
We first optimized the conditions for the IP–TDMS assay, which enabled the discovery of novel KRAS proteoforms (Fig. 1). The optimized protocol was verified to capture and enrich for all four RAS isoforms across both cell line and colorectal tumor contexts (Fig. S2). Briefly, cells or homogenized colorectal tumor tissue were lysed in a buffer containing two nonionic detergents. RAS was then immunoenriched from the lysates, with eluted proteoforms desalted by solid-phase extraction prior to LC coupled with a Q-Exactive HF mass spectrometer (42). After a survey for an initial stage of proteoform discovery for each sample type, we generated a list of proteoforms for targeted analysis and extensive characterization. A subset of samples, specified in Table S1, were analyzed in parallel on the 21 T Fourier transform ion cyclotron resonance (FT–ICR) mass spectrometer at the National High Magnetic Field Laboratory (43, 44). Use of these two platforms combined with our improved KRAS IP–TDMS assay increased the number of curated RAS proteoforms from 11 to 39, each given a proteoform record number (Figs. S3, 2, Tables S1 and S2).
Figure 1.
Diagram of the IP–TDMS workflow, which couples immunoprecipitation of KRAS with downstream top–down mass spectrometry. Proteoforms are first detected in discovery mode (intact MS1) and then further characterized in targeted mode using tandem mass spectrometry (tMS1/tMS2). Key differences in the protocol compared with those previously published (41, 42) are denoted by asterisks. IP, immunoprecipitation; TDMS, top–down mass spectrometry.
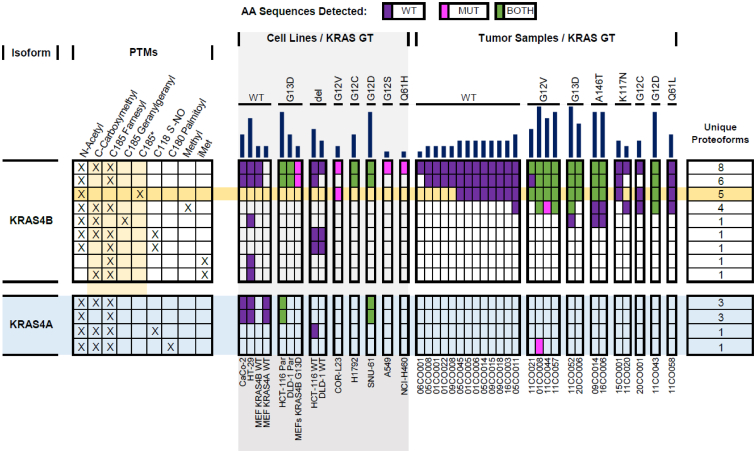
Figure 2.
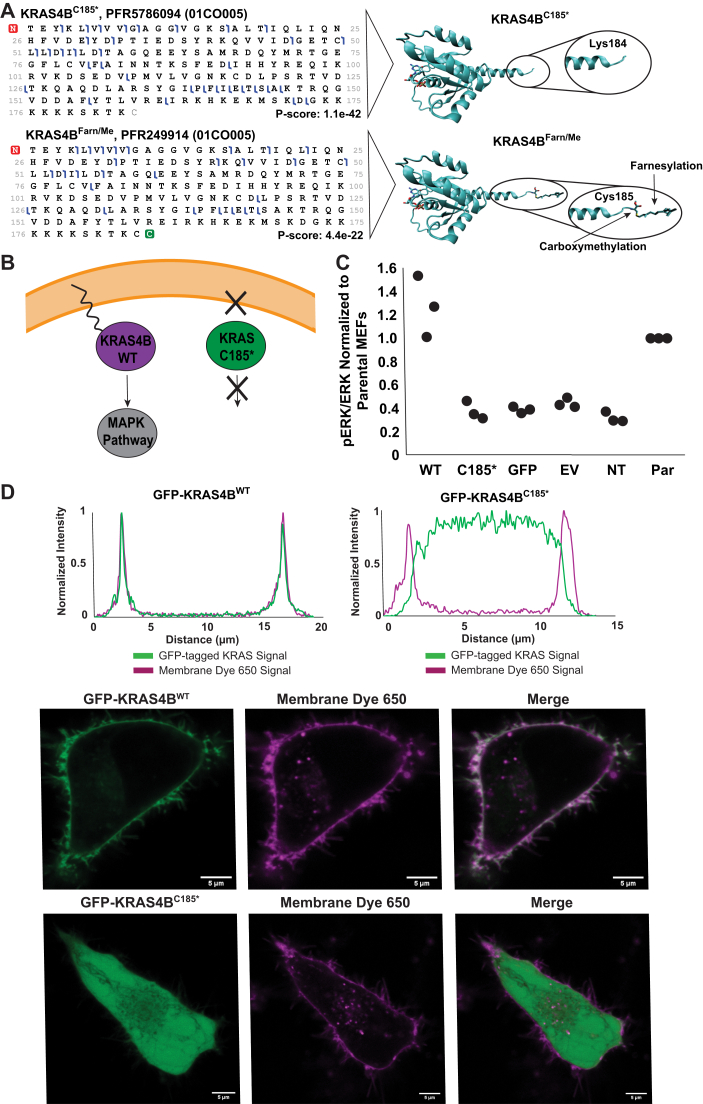
Oncoproteoform plot describing the manually validated KRAS4A (light blue) and KRAS4B proteoforms and the contexts in which they were identified. Each row represents all validated proteoforms that share the combinations of PTMs indicated with “X”s on the left. The number of unique proteoforms in each row is shown on the right. In the middle section, the presence or the absence of the proteoforms in the biological contexts (cell lines [light gray] and colorectal tumors) is displayed. The color of the box indicates the expressing allele of validated proteoforms: purple denotes WT form only, magenta is mutant form only, and green represents both WT and mutant forms. No fill indicates no proteoform was detected. The dark yellow row shows proteoforms bearing a C185∗ truncation, whereas light yellow columns in the left section highlight modifications, which involve the C-terminal residue C185. Dark blue bars on top indicate the total number of validated KRAS proteoforms in each context. PTM, post-translational modification.
KRAS4A and KRAS4B proteoform landscapes
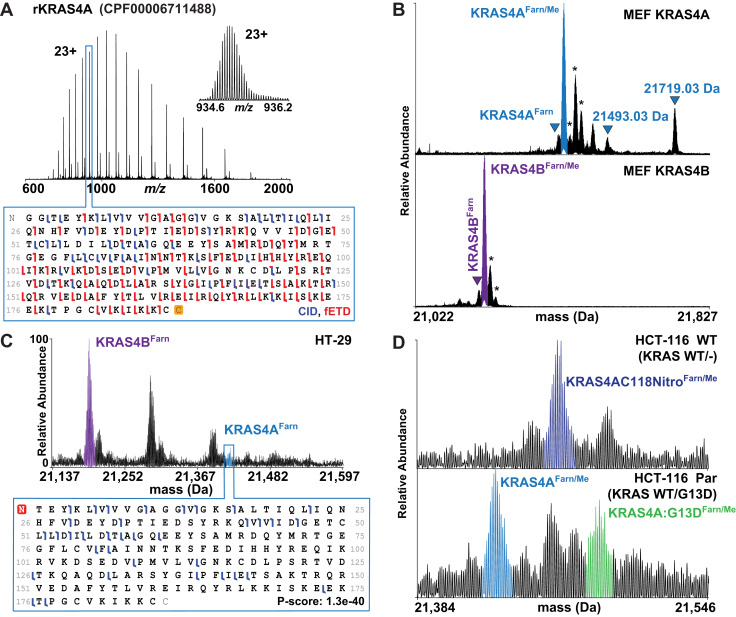
To identify KRAS4A proteoforms and compare them to those of KRAS4B, we first used recombinant protein (rKRAS4A) on both instrument platforms to optimize TDMS fragmentation parameters for optimal sequence coverage (Fig. 3A). Subsequently, we performed IP–TDMS of endogenous KRAS from two mouse embryonic fibroblast (MEF) cell lines, each expressing these specific isoforms (National Cancer Institute [NCI] RAS Initiative) (45). MEFs expressing only KRAS4A displayed proteoform distributions distinct from those in MEFs expressing only KRAS4B (Fig. 3B). The two most abundant KRAS4B proteoforms in MEFs were KRAS4BFarn/Me and KRAS4BFarn (Tables S1 and S2). While KRAS4AFarn/Me and KRAS4AFarn were present in KRAS4A MEFs, two other proteoforms were observed with intact masses consistent with KRAS4AFarn/Me plus a phosphorylation (observed mass: 21,493.03 Da) as well as a KRAS4A proteoform containing C185 geranylgeranylation in addition to Cys180 palmitoylation (observed mass: 21,719.03 Da) (Figs. 3B and 2).
Figure 3.
KRAS4AProteoforms.A, recombinant KRAS4A charge state distribution and graphical fragment map from a 21 T FT–ICR mass spectrometer. B, proteoform landscapes in MEF KRAS4A WT and MEF KRAS4B WT cell lines from a 21 T FT–ICR mass spectrometer. Peaks in the KRAS4A landscape match the masses for KRAS4A with phosphorylation (21,493.03 Da) as well as C185 geranylgeranylation with C181 palmitoylation (21,719.03 Da). C, KRAS4B versus KRAS4A proteoform relative abundances in the HT-29 cell line (top) and corresponding KRAS4A fragment ion map (bottom). D, allele-specific KRAS4A proteoforms shown in HCT-116 parental and WT isogenic cell lines. Asterisks denote oxidation. FT–ICR, Fourier transform ion cyclotron resonance; MEF, mouse embryonic fibroblast.
In all cell lines analyzed, KRAS4A proteoforms were commonly observed at <25% relative abundance of equivalent KRAS4B proteoforms (Fig. 3C and Table S1). A control experiment where IP–TDMS was performed on cell lysate with all four recombinant RAS isoforms (rRAS) spiked in at equal concentrations showed that our assay did not preferentially enrich rKRAS4B over other rRAS isoforms (Fig. S4). In addition, KRAS4B displayed a greater proteoform diversity than KRAS4A in every cancer context studied, with exception to MEFs expressing 4A (Fig. 2). While differences in modifications were observed, KRAS4A was also frequently modified like KRAS4B, including the canonical N-acetylation, C185 farnesylation, and carboxymethylation of the C terminus. Moreover, KRAS4A proteoforms are also abundantly nitrosylated for hemizygous KRAS alleles in direct alignment with prior results on KRAS4B (Fig. 3D) (41). In isogenic HCT-116 parental (KRAS WT/G13D) cells, the most abundant KRAS4A proteoforms were KRAS4AFarn/Me and KRAS4A:G13DFarn/Me, whereas in HCT-116 WT (KRAS WT/-) cells, the most abundant proteoform was KRAS4AC118NitroFarn/Me (Figs. 3D and 2).
KRAS proteoforms in colorectal tumors
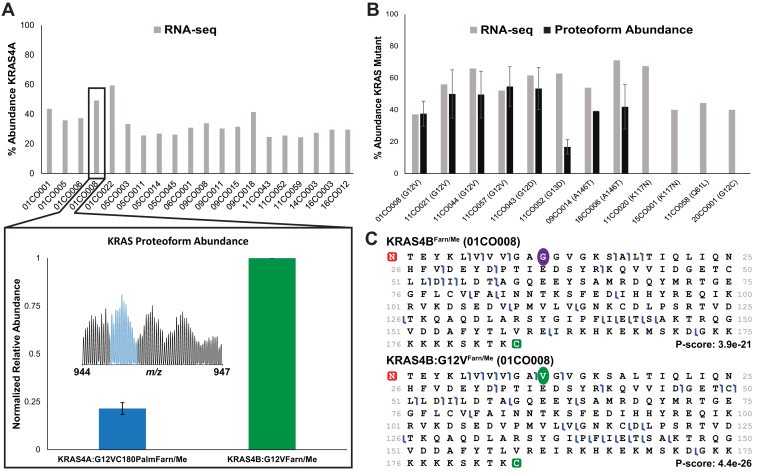
After detailed characterization of proteoforms in cell lines, next we analyzed KRAS derived from a cohort of 34 primary colorectal tumor samples procured by the Clinical Proteomic Tumor Analysis Consortium (CPTAC). Portions of these same tumors had previously undergone DNA sequencing, RNA-Seq analysis, and BU proteomics as part of a 2019 CPTAC study (46). KRAS4B proteoforms were identified in 26 of the 34 tumors. However, KRAS4A proteoforms were detectable in just a single sample (01CO008). Given that KRAS4A can be detected by the proteoform assay if present at >10% abundance relative to KRAS4B, this result stands in contrast to what one would infer from RNA-level measurements that indicated parity between KRAS4A and KRAS4B transcripts (Fig. 4A, Tables S1 and S3) (46).
Figure 4.
KRASinprimarycolorectaltumors.A, RNA-Seq data collected from tumors showing percent abundance of KRAS4A transcripts out of total KRAS (top). Only the tumor sample 01CO008 displayed a KRAS4A proteoform above the threshold for detection by TDMS. The relative abundance of this proteoform (KRAS4A:G12VC180PalmFarn/Me) was very low compared with the canonical KRAS4B:G12VFarn/Me proteoform (bottom). B, abundance of mutant KRAS4B in tumors as determined by either RNA-Seq (46) and TDMS. C, fragment ion maps showing that TDMS can distinguish mutant from WT proteoforms within the same tumor. Site of mutation is depicted by colored circles. TDMS, top–down mass spectrometry.
Thirteen of the 26 tumors positive for KRAS4B proteoforms were WT for KRAS, whereas the remaining 13 were heterozygous for KRAS mutations (Table S4) (46). Proteoforms of KRAS4B containing a driving mutation (oncoproteoforms) were detected in nine of these latter 13 tumors, many at relative abundances similar to their WT counterparts (Fig. 4B) (41). Moreover, close inspection of the RNA-Seq data from portions of these same tumors allowed calculation of the relative expression for the WT and mutant KRAS alleles (Fig. 4B and Table S4) (46) and comparison with respective proteoform relative abundances. The subset of tumors that lacked detectable KRAS oncoproteoforms (11CO020, 15CO001, 11CO058, and 20CO001) reported having mutant allele expression between 40 and 67% at the RNA level. Tumors that had detectable levels of oncoproteoforms reported having mutant allele expression between 37 and 71% (Fig. 4B and Table S4) (46). Therefore, the lack of observable proteoforms in a subset of the colorectal tumors containing the mutant allele appears not to be driven by lower expression of the mutant allele at the RNA level.
Because of the nature of TDMS, the linkage between KRAS4B mutations and PTMs could also be characterized, a feat not possible with BU once proteins are protease digested (Fig. 4C). Using these TDMS data, we were able to generate a profile of validated KRAS4B proteoforms associated with each KRAS mutation (Fig. 2, Tables S1 and S2). We also searched extensively for KRAS mutant proteoforms with unexpected modifications by manual inspection as well as using a tailored database, which returned no significant hits. In-gel trypsin digestion and subsequent BU also did not detect peptides from mutant KRAS within the flow-through, elution, and bead IP fractions, implying that the IP did not leave behind mutant KRAS proteoforms.
KRAS4B proteoforms in tumors exhibited differential abundance of PTMs critical for membrane association, including prenylation and carboxymethylation. KRAS4BFarn/Me was present in all tumor samples, irrespective of mutational status. KRAS4BFarn was present in most tumor samples, although in the majority of cases, it was at ≤50% of the relative abundance of the canonical form (KRAS4BFarn/Me). Only two tumors (05CO008 and 09CO015) showed relative abundance levels of KRAS4BFarn (the form without a C-terminal methylation) similar to KRAS4BFarn/Me. KRAS4BGeranyl/Me was only observed in three of the tumors, two of which contained A146T mutations and one which contained a G13D mutation (Fig. 2 and Table S1). Unlike KRAS4BFarn, there was no evidence to support the presence of a KRAS4BGeranyl form. Geranylgeranylation was also not observed on mutant KRAS4B proteoforms in the tumors, even within samples containing KRAS4BGeranyl/Me (Fig. 2). Finally, C185 farnesylation was observed in higher relative abundance than C185 geranylgeranylation in the three cases in which geranylgeranylation was observed (Table S1).
Finally, one of the most striking findings was the presence of novel KRAS4B proteoforms lacking the C185 residue along with the associated farnesylation and carboxymethylation, which were present at high abundance in the COR-L23 cell line and many primary colorectal tumor samples (Figs. 5, 2 and S5). These truncated proteoforms were present in the WT and the G12D, G12V, G13D, and A146T mutant variants of KRAS4B (KRAS4BC185∗, KRAS4B:G12DC185∗, KRAS4B:G12VC185∗, KRAS4B:G13DC185∗, and KRAS4B:A146TC185∗, respectively). Eighteen tumor samples contained truncated proteoforms and, in 11 cases, they were greater than twofold more abundant than canonical KRAS4BFarn/Me (Figs. 2, S5A and Table S1). After careful review of RNA-Seq data from these same tumors, we saw no evidence that the absence of KRAS4B C185 originates at the transcript level (46). We also spiked rKRAS4B into cell lysates and showed by IP–TDMS that the truncation was not because of artifactual cleavage of the Lys184–Cys185 peptide bond after cell lysis (Fig. S6).
Figure 5.
Novel KRAS4B Truncation.A, graphical fragment maps from TDMS of KRAS4BFarn/Me and KRAS4BC185∗ proteoforms and their corresponding structural models (Protein Data Bank ID: 5TAR). B, scheme depicting hypothesized differences in membrane association and cell signaling between KRAS4BFarn/Me and KRAS4BC185∗. C, scatter plot showing pERK/ERK levels in MEFs transfected with different plasmids (WT = GFP-KRAS4BWT; no Cys185 = GFP-KRAS4BC185∗; GFP = GFP-only vector; EV = empty vector; NT = no transfection control) normalized to pERK/ERK levels in parental (PAR) MEFs (no 4OHT, no transfection). Densitometry measurements were performed by Fiji ImageJ (73). All three replicates are displayed. D, intensity traces of GFP-KRAS and Membrane Dye 650 signal versus distance across a cell as determined by Fiji ImageJ (micrometer) (top) and live-cell images of HeLa cells expressing KRAS4BFarn/Me or KRAS4BC185∗ plasmids (bottom) (bar represents 5 μm). 4OHT, 4-hydroxytamoxifen; MEF, mouse embryonic fibroblast; TDMS, top–down mass spectrometry.
Functional characterization of truncated KRAS4B proteoforms
Since KRAS4BC185∗ proteoforms are neither prenylated nor carboxymethylated, we hypothesized that they would be unable to associate with the plasma membrane, thus be unable to activate the MAPK signaling pathway (Fig. 5B). To test this, HeLa and MEF cells absent of all four RAS isoforms were transfected with plasmid encoding N-terminally tagged KRAS WT bearing a premature stop codon at C185 (45). The localization and function of KRAS in these cells were then examined by phosphoimmunoblotting and live-cell imaging (Figs. 5, C and D and S7–S9).
In “Rasless” MEFs, GFP-KRAS4BC185∗ was unable to activate the MAPK pathway in contrast to GFP-KRAS4BWT (Figs. 5C and S7). There were no significant differences in p-ERK levels observed among GFP-KRAS4BC185∗, GFP only, empty vector, and nontransfected “Rasless” MEF conditions. Cells expressing GFP-KRAS4BWT exhibited p-ERK levels above all other conditions and had similar levels as those seen in the parental MEFs (no 4-hydroxytamoxifen [4OHT] treatment) (Figs. 5C and S7).
Live-cell images of HeLa cells transiently expressing the GFP-KRAS construct were taken 24 h post-transfection and in combination with CellBrite Steady 650 membrane dye. Clear differences in membrane localization were observed between GFP-KRAS4BWT and GFP-KRAS4BC185∗ (Figs. 5D and S8). Intensity profile plots for 30 cells from each condition confirmed that GFP-KRAS4BC185∗ was primarily within the cytoplasm, whereas the majority of GFP-KRAS4BWT is localized tightly to the plasma membrane (Figs. 5D and S8). Transfection of a vector expressing GFP alone showed that GFP was diffuse throughout the cell and also not localized to the plasma membrane (Fig. S9). In addition, GFP-KRAS4BC185∗ and GFP-KRAS4BWT were found to be expressed at equal abundance, suggesting that the contrast in membrane association was not driven by differences in protein expression (Fig. S10).
Commonalities and differences in the KRAS proteoform landscapes between cell lines and colorectal tumors
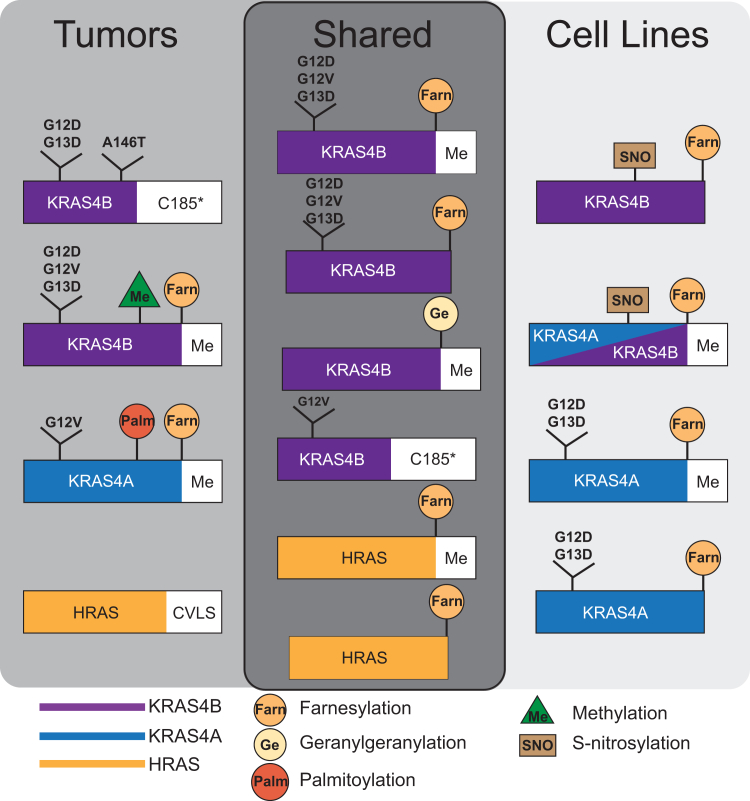
Validated proteoforms, confirmed by both intact mass measurement and targeted proteoform sequence characterization, were compared between cell lines and colorectal tumor samples to identify unique and shared proteoforms (Fig. 6). Among the proteoforms that were exclusively in cell lines were the majority of the KRAS4A proteoforms (KRAS4AFarn/Me, KRAS4AFarn, KRAS4A:G12DFarn/Me, KRAS4A:G12DFarn, KRAS4A:G13DFarn/Me, KRAS4A:G13DFarn, and KRAS4AC118NitroFarn/Me) except for KRAS4A:G12VC180PalmFarn/Me, which was found in a single tumor (01CO008) (Fig. 2). In addition, KRAS4BC118NitroFarn/Me and KRAS4AC118NitroFarn/Me were only found in isogenic cell lines that were hemizygous KRAS WT (HCT-116 WT, DLD-1 WT), as previously reported (41).
Figure 6.
Schematic diagram of RAS proteoforms either unique or shared between cell lines and colorectal tumor samples.Green triangle indicates internal methylation as distinct from carboxymethylation at the C terminus.
Proteoforms that were exclusively in the primary colorectal tumor samples included a set of KRAS4B proteoforms that lacked the C185 residue (KRAS4BC185∗, KRAS4B:G12DC185∗, KRAS4B:G13DC185∗, and KRAS4B:A146TC185∗), except for KRAS4B:G12VC185∗, which were also found in COR-L23 cells (Fig. S5B). In addition, KRAS4B proteoforms containing a single methylation within the C-terminal region of residues 147 to 184 were found only in primary tumors (KRAS4BmeFarn/Me, KRAS4B:G12DmeFarn/Me, KRAS4B:G12VmeFarn/Me, and KRAS4B:G13DmeFarn/Me) (Figs. 2, 5 and S11).
While the observed relative abundances of KRAS proteoforms were variable across samples, clear trends in the most well-validated proteoforms emerged (Table S1). The proteoforms KRAS4BFarn/Me and KRAS4BC185∗ were of consistently high abundance. Although present in the majority of contexts, KRAS4BFarn was often observed in much lower abundance than KRAS4BFarn/Me.
Notably, proteoforms containing previously reported PTMs such as S181 phosphorylation and lysine acetylation were not observed in the biological contexts examined in this study. Control experiments (Fig. S12) support that these modifications are not being lost ex vivo as a result of our IP–TDMS methodology and can be successfully characterized if in high enough relative abundance. It is possible that these previously reported modifications exist outside the contexts that have been analyzed in this study.
Discussion
With a systematic approach employing immunoenrichment and TDMS, this study provides unique data for KRAS proteoform characterization with complete molecular specificity. These analyses revealed that the RAS proteoform landscape is more diverse than previously known, with a total of 28 novel validated proteoforms harboring new types of PTMs identified (Fig. 2). This diversity of RAS modifications, as well as differential relative proteoform abundances across cell lines and tumors (Table S1), suggests that regulation of RAS activity is adaptable in multiple dimensions. Moreover, this observed KRAS proteoform complexity also reveals potential challenges for the design of targeted KRAS inhibitors, particularly those acting on a single axis.
KRAS isoforms differ in abundance and PTMs
Previous studies have found that KRAS4A acts distinctly from KRAS4B, is modified differently than KRAS4B, and plays a significant role in oncogenic signaling (17, 47, 48, 49, 50, 51, 52, 53). The new ability to detect KRAS4A proteoforms facilitated a direct examination of both KRAS4A relative abundance and PTM profile within cell lines and tumors (Fig. 3). The majority of KRAS4A proteoforms were detected only in cell lines and were observed at lower relative abundance than those of KRAS4B, including in contexts previously reported to exhibit similar KRAS4A and KRAS4B transcript levels (Fig. 6 and Table S3) (46, 52). The lack of detectable KRAS4A proteoforms in tumors was unexpected given previous reports that KRAS4A plays a prominent and specific role in cancer biology (48, 49, 52, 53). This discrepancy between proteoform and RNA-level information could be due to a sampling bias in the portion of tumor analyzed or post-transcriptional regulation events (54). It is also possible that additional KRAS4A proteoforms are present, but at an abundance so low, that it is below the levels of detection by TDMS. Furthermore, KRAS4B showed a higher diversity of PTMs than KRAS4A (Fig. 2). The notably higher relative abundance of KRAS4B, as well as the diversity in PTMs that could modulate its function, suggests the abundance-driven hypothesis that KRAS4B may be the more influential KRAS isoform within the sample types examined here. While KRAS4B was the predominant isoform in the contexts analyzed in this study, KRAS4A proteoforms may be more highly expressed in different tissues or at specific stages of cancer progression as well as participate in distinct signaling roles from KRAS4B (48, 49, 51, 52, 53).
Abundance of KRAS oncoproteoforms in tumors
The survey of colorectal tumors also revealed that KRAS4B mutant proteoforms were frequently present at higher abundance than our initial study suggested, and in some cases, were present at near equal relative abundance to their WT counterparts (Fig. 4B) (41). These results align with a study by Mageean et al. (55), which found that mutant KRAS protein represented 50% (±10%) of total KRAS expressed in SW48 isogenic colorectal cell lines. In contrast, KRAS mutant proteoforms were not detected in four of the tumors bearing KRAS mutations. While this was not because of low RNA expression levels (Fig. 4B), the lack of mutant proteoforms may have instead been because of a post-transcriptional regulatory mechanism, such as the selective regulation of KRAS mutant protein stability by SMURF2 (46, 56). In addition, there may have been sampling bias in the section of tumor that we analyzed. A specific sampling study employing techniques like MS imaging would be required to determine the protein-level dosage of mutation as a function of tumor region. With the advent of mutation-specific KRAS inhibitors, it is critical to understand the relative abundance of oncogenic KRAS4B within patients. Our findings show that oncogenic KRAS4B proteoforms represent a significant portion of the KRAS proteoform population in 9 of the 13 tumors with KRAS mutations analyzed.
KRAS PTMs in tumors
Previous studies have also found that KRAS mutant variants can lead to distinct downstream signaling targets and patient outcomes, although this has been attributed to differential nucleotide hydrolysis rates and interactions with RAS effectors (3, 5, 57). The improved KRAS IP–TDMS assay allowed us to examine whether KRAS mutant variants also exhibit differential PTMs, particularly when compared with those of WT KRAS, which could contribute to observed phenotypic differences. However, our survey of colorectal tumors did not identify any KRAS mutation- or allele-specific PTMs that could explain phenotypic differences previously seen in KRAS mutant-driven cancers. Instead, a more complex picture emerged showing that both WT and mutant KRAS proteoforms can be modified with a range of PTMs and are present at different relative abundances according to each sample (Fig. 2 and Table S4), which likely modulates their respective downstream signaling pathways. Notably, KRAS4BFarn/Me was present in every tumor sample with detectable KRAS irrespective of KRAS mutational status or percent tumor tissue (Fig. 2 and Table S4). The frequency and relatively high abundance of this proteoform underscores how critical it is to cell signaling even in the presence of oncogenic KRAS mutations.
Most strikingly, we observed two novel classes of KRAS proteoforms within colorectal tumors. The first class comprised WT and mutant KRAS4B proteoforms bearing a monomethylation site within the C-terminal region spanning residues 147 to 184 (Figs. 6, green triangle, S11 and 2). While the specific modified residue(s) could not be site localized using the proteoform fragmentation data collected here because of the high frequency of candidate Lys residues within this region, this PTM could be analogous to HRAS K147me1 (58). The functional relevance of this monomethylation is unknown but may present an interesting avenue for future investigations. The second class comprised WT and mutant KRAS4B proteoforms lacking the C185 residue and associated PTMs (farnesylation and carboxymethylation) (Figs. 5A and S5). These novel truncated KRAS4B proteoforms were detected at high relative abundance within the majority of the tumor samples analyzed. In addition, the detection of mutant truncated KRAS4B proteoforms indicated that these originated from within the tumor tissue and are not merely artefacts from the surrounding healthy tissue.
KRAS4B undergoes a post-translational processing at a C-terminal CAAX motif, which involves farnesylation at C185, proteolysis of the AAX, and carboxymethylation of the resultant C terminus (Fig. S1) (12, 14, 18, 21). These modifications, along with a lysine-rich region, facilitate KRAS4B association with the plasma membrane (13, 21). Previous studies investigating this process employed a C181S mutation, resulting in a C-terminal sequence of SAAX lacking farnesylation and carboxymethylation (13, 59). However, functional analyses of KRAS4B lacking C185, the AAX motif, and associated PTMs have not been reported.
Fully processed KRAS4B interacts with RAF at the plasma membrane, leading to activation of the downstream MAPK signaling pathway (20, 60, 61). Given the loss of membrane association exhibited by GFP-KRAS4BC185∗ in our live-cell imaging experiments, we hypothesized that this proteoform would be unable to activate the MAPK pathway and induce phosphorylation of ERK1/2 at Thr202/Tyr204 (Fig. 5B) (62). Indeed, GFP-KRAS4BC185∗ was unable to induce ERK1/2 phosphorylation above the basal levels seen within the controls (Fig. 5C). As activation of the MAPK pathway is associated with cell growth and proliferation, KRAS4BC185∗ may therefore be truncated as part of an antiproliferative regulatory mechanism (62). In addition, oncogenic KRAS4BC185∗ may act as a dominant negative inhibitor since malignant transformation requires RAS prenylation (63, 64). No clear trends were observed by principal component analysis between the presence of WT or mutant KRAS4BC185∗ proteoforms and the cancer stage (I–IV), subsite, or vital status (deceased/living) of the patients from which the tumors originated (data not shown). The heterogenous nature of the colorectal tumors analyzed in the current study (e.g., treatment status, subsite, percent tumor tissue, mutational status, etc.), along with sample cohort size, made it challenging to identify clear associations between a specific proteoform and patient phenotype. Our generation of a reference set of KRAS proteoforms enables for future controlled TDMS studies to test for correlations to cancer stage or other patient metrics of high clinical utility.
The mechanism by which the novel KRAS4BC185∗ proteoforms are generated is unclear. RNA-Seq data from the tumor cohort showed no evidence of genetic or transcriptional alteration that would give rise to these proteoforms (46). Instead, an enzyme such as Ste24, a metalloprotease that cleaves farnesylated proteins at both CAAX sites and N-terminal distal sites, could be responsible (65, 66). The elucidation of the responsible mechanism for the novel KRAS4BC185∗ truncation could shed insight into why this proteoform is highly abundant in some biological contexts, yet not in others. This will be critically important for understanding how the entire KRAS proteoform landscape within a given patient may play a role in both cancer severity and progression. Understanding the generative mechanism of KRAS4BC185∗ and resulting modulation of plasma membrane association could also pave the way for the development of anti-KRAS therapeutics, as prior strategies targeting KRAS membrane localization have yielded disappointing results (6, 7, 9). Finally, systematic discovery of KRAS proteoforms highlights the ability of TDMS to provide a unique perspective on RAS modifications and inspire new lines of investigation into RAS biology.
Experimental procedures
Cell culture and primary colorectal tumor tissue
HCT-116 and DLD-1 parental (KRAS WT/G13D) and WT (KRAS WT/-) cells were purchased from Horizon Discovery. Cells were maintained at 37 °C and 5% CO2 in RPMI1640 supplemented with 10% fetal bovine serum (FBS) and 5% antibiotic–antimycotic (Anti-Anti; Thermo Fisher Scientific). HT-29 (KRAS WT/WT) cells were purchased from American Type Culture Collection. HeLa cells are a kind gift from S.J. Flint and maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 5% antibiotic–antimycotic. “Rasless” MEFs expressing KRAS4A WT, KRAS4B WT, or KRAS4B G13D were generously donated by the NCI RAS Initiative. Cells were maintained at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 5% penicillin–streptomycin, and 4 μg/ml blasticidin. Confluent cells were washed twice in ice-cold PBS, harvested by scraping, resuspended in ice-cold PBS, divided into aliquots containing 1 × 108 cells, and pelleted by centrifugation. The resulting cell pellets were processed directly or snap-frozen in liquid nitrogen for storage at −80 °C. Identities of cell lines were confirmed by short tandem repeat profiling performed by the NU SeqCore. SNU-61 (KRAS WT/G12D), CaCo-2 (KRAS WT/WT), COR-L23 (KRAS G12V/G12V), and NCI-H1792 (KRAS WT/G12C) cell pellets were generously donated by the NCI RAS Initiative. Cells tested negative for mycoplasma (American Type Culture Collection, catalog no.: 30-1012K).
The 34 primary colorectal tumor samples were obtained from the US NCI’s CPTAC, National Institutes of Health. Tumors were collected, quality control approved, and processed according to CPTAC standard operating procedures, shipped on dry ice, and maintained in liquid nitrogen until the time of analysis.
IP
IP with anti-v-HRAS agarose IP beads (OP01A; MilliporeSigma, Research Resource Identifier [RRID]: AB_437743) was performed as previously reported (42), with the exception of the lysis buffer composition, which now contained 50 mM Tris (pH 7.5), 150 mM NaCl, 1% NP-40 (MilliporeSigma), 1% Triton X-100 (Thermo Fisher Scientific), and 1× final concentration of HALT Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). IP reactions were performed in triplicate for cell lines. Each replicate had four LC injections sample. A single IP reaction was performed for each tumor, followed by four LC injections per sample if proteoforms were detected.
LC and MS
QE-HF mass spectrometer (Thermo Fisher Scientific) parameters
Immunopurified RAS proteins were further resolved by reverse-phase nanocapillary LC delivered by a Dionex UltiMate 3000 system (Thermo Fisher Scientific) prior to introduction into a Q-Exactive HF BioPharma mass spectrometer (Thermo Fisher Scientific). Injections (5 μl) of sample were loaded onto a trap column (150 μm inner diameter [ID]; 3 cm length, L) and washed in HPLC solvent A (5% Optima acetonitrile [ACN], 95% Optima H2O, 0.2% MS grade formic acid (FA); all Thermo Fisher Scientific) for 10 min at a flow rate of 2.5 μl/min. Samples were then resolved on a nanocapillary analytical column (75 μm ID, 25 cm L) coupled to a vented tee setup and a nanospray emitter (New Objective FS3605015N20). Trap columns, analytical columns, and spray emitters were packed in-house with PLRP-S resin (5 μm particle size, 1000 Å pore size; Agilent Technologies) and maintained at 45 °C during LC/MS analysis. RAS proteins were eluted into the mass spectrometer at a flow rate of 0.3 μl/min by the following gradient: 5% HPLC solvent B (95% ACN with 0.2% FA) at 0 min, 30% solvent B at 5 min, 45% solvent B at 25 min, 95% solvent B from 28 to 31 min, and 5% solvent B from 34 to 50 min.
Intact mass (MS1) spectra were acquired using a full scan method covering m/z 800 to 1000 to capture any abundant untargeted RAS species or a selected ion monitoring (SIM) method covering m/z 900 to 970 or 910 to 970 in 6 or 7 × 10 m/z windows, the range within which the 23+ charge states of all RAS proteoforms of interest were expected to fall. Both MS1 scan methods were performed in “protein mode” at a resolving power (r.p.) of 120,000 (at 200 m/z), with an average of four microscans, an automatic gain control (AGC) target of 1E + 06 (full) or 3E + 06 (SIM), and a maximum ion injection time of 50 ms (full) or 600 ms (SIM). Fragment ion (MS2) spectra, which can confirm proteoform sequence and localize PTMs, were acquired in either a data-dependent method targeting the two most abundant species within each MS1 scan (top2dd) or by targeting a list of preselected values with increasingly narrow m/z windows to provide diagnostic fragment ions to be used in proteoform quantitation and comparison (targeted MS2 [tMS2]). MS2 scans were acquired at an r.p. of 60,000 (at 200 m/z), with an isolation window of 4 m/z (full) or 3 m/z (tMS2), an AGC target of 1E + 06, and a maximum ion injection time of 800 ms. Fragmentation was triggered by high-energy collisional dissociation, with a normalized collision energy (NCE) applied in 2% steps between 19 and 25%. Additional MS parameters included a heated transfer capillary temperature of 320 °C, an S-lens radiofrequency amplitude of 50%, and 15 eV in-source dissociation to facilitate protein desolvation and adduct removal.
21 T FT–ICR instrument parameters for top–down LC–MS/MS
Immunopurified KRAS proteoforms were further resolved by reversed-phase nano-LC delivered by an ACQUITY M-Class chromatographic system (Waters) prior to introduction into a custom 21 T FT–ICR mass spectrometer (National High Magnetic Field Laboratory) (43, 44). Injections (5 μl) of sample were loaded onto a trap column (150 μm ID; 3 cm length, L) and washed in HPLC solvent A for 10 min at a flow rate of 2.5 μl/min. Samples were then separated using a nanocapillary analytical column (75 μm ID, 15 cm L) coupled to a 15 μm nanospray emitter (New Objective). Trap columns, analytical columns, and spray emitters were packed in-house with PLRP-S resin (5 μm particle size, 1000 Å pore size; Agilent Technologies) and maintained at room tempearture during LC–MS/MS analysis. KRAS proteoforms were eluted and directly electrosprayed into the mass spectrometer at a flow rate of 0.3 μl/min by use of the following gradient: 5% HPLC solvent B (45% ACN, 45% LCMS-grade isopropanol [Honeywell], and 0.3% FA) at 0 min, 30% solvent B at 5 min, 45% solvent B at 35 min, 75% solvent B at 45 min, 75% solvent B from 45 to 48 min, and 5% solvent B from 48 to 70 min.
For all experiments, the electrospray ionization source was biased at 2.75 kV, and the inlet capillary was heated to 325 °C. MS1 spectra were recorded from m/z 300 to 2000, 700 to 1500, or 800 to 1000 at an r.p. of 150,000 or 300,000 (at 400 m/z) as the sum of four microscans, with an AGC target of 1E + 06 charges, and a maximum ion injection time of 500 ms. tMS2 spectra utilized the same r.p. settings but were recorded from m/z 300 to 2000 as the sum of two microscans. Fragmentation was performed by collision-induced dissociation (CID) or front-end electron-transfer dissociation (67) in the high-pressure cell of a modified Velos Pro linear ion trap assembly (Thermo Fisher Scientific) with precursor isolation windows ranging from 10 to 50 Th and 300 to 400 ms maximum precursor injection times. MSN AGC targets were 3E + 05 for ETD or 4E + 05 for CID, and an external multipole storage device was used to store multiple accumulations of product ions prior to high-resolution mass analysis in the ICR cell such that cumulative MS2 AGC targets were >2E + 06 charges. For fragmentation by CID, an NCE of 0% (isolation only for improved S/N of precursors) or 35% and activation q of 0.250 were employed for 10 ms. For fragmentation by front-end electron-transfer dissociation, the reagent AGC target was 6E + 05, and precursors were allowed to react for 15 ms. Spectra were stored in .raw file format in reduced profile mode (i.e., noise baseline-subtracted).
Data processing
Data were processed using Xcalibur QualBrowser (Thermo Fisher Scientific), ProSight Lite 1.4 (http://prosightlite.northwestern.edu/), ProSight PD 4.0 (Thermo Fisher Scientific), and TDValidator 1.0 (Proteinaceous) (68, 69). For ProSight PD searches, a custom RAS proteoform sequence and PTM database was created with Protein Annotator (http://proteinannotator.kelleher.northwestern.edu/) and was deposited on MassIVE. RAS isoform sequences were downloaded from UniProt, and mutations, SNPs, and PTMs were manually annotated. Raw files were run through ProSight PD with subsequent manual validation by ProSight Light or TDValidator (70). Observed masses, error, and p-scores of completely characterized RAS proteoforms are reported in Table S5. The top two most abundant RAS proteoforms as determined by protein ion relative ratios and fragment ion relative ratios for Figure 4 were quantified by the method described in the study by Pesavento et al. (71). Fragment ion relative ratios were taken from 3 m/z isolation windows for the canonical forms of KRAS WT and mutant (Table S6). Incompletely characterized RAS proteoforms are reported in Table S7. Raw files and the custom ProSight PD database are deposited on MassIVE (MSV000088748). Proteoform record numbers can be searched in a database provided by the Consortium for Top–Down Proteomics.
RNA-Seq data analysis
The mutational status and relative expression of RAS variants were determined by processing data from the CPTAC Proteogenomic Confirmatory Study of Breast, Colon, Lung, and Ovarian Tumors (dbGaP Study Accession: phs000892.v6.p1) deposited in the NCI Genomic Data Commons (GDC). GDC data were downloaded using the GDC Data Transfer Tool Client (version 1.6.1). Variants in KRAS, NRAS, and HRAS were cataloged by searching masked MAF files from whole exome sequencing. BAM files from RNA-Seq experiments were downloaded from GDC and inspected with Integrative Genomics Viewer (version 2.9.4; Broad Institute) (72). The relative expression of variants was estimated as the uncorrected ratio of read counts at each variant locus. Manifest files as well as sample sheets containing accession numbers for the BAM and MAF files can be found at https://github.com/bdrown/ras-cptac-analysis.
Plasmids and site-directed mutagenesis
Plasmids expressing GFP-tagged KRAS4B (HG12259-ANG), pCMV3-N-GFPSpark control vector (CV027), or pCMV3-untagged negative control vector (CV011) were purchased from Sino Biological. PCR site-directed mutagenesis was performed on the GFP-tagged KRAS4B plasmid to convert the Q61H mutation to Q61 WT (IDT primers: 5′-TGCACTGTACTCCTCTTGACCTGCTGTGTCG-3′ and 5′-CGACACAGCAGGTCA-AGAGGAGTACAGTGCA-3′), and a two-base mutation was introduced for a stop codon in place of cysteine codon at C185 (5′-GGCCGCTCTAGATTTACATAATTA-CTTACTTTGTCTTTGACTTCTTTTTCTT-3′ and 5′-AAGAAAAAGAAGTCAAAGACAAA-GTAAGTAATTATGTAAATCTAGAGCGGCC-3′). Mutations were confirmed by Sanger sequencing by ACGT.
Generation of Rasless MEFs
Parent MEFs (KRAS fl/fl, HRAS-null, and NRAS-null) were provided by the Ras Reagent Group from the NCI Ras Initiative at the Fredrick National Laboratory for Cancer Research and were made “Rasless” by addition of 4OHT (Sigma–Aldrich) as previously described (45). Cell lines were validated by Western blot and sulforhodamine B assay (Abcam; catalog no.: ab235935) to ensure that the MEFs were viable and “Rasless” after 4OHT treatment.
Western blots
Protein samples were run on NuPAGE 4 to 12% Bis–Tris protein gels (Invitrogen), with 1× Mes buffer, at constant 120 V or 150 V. Transfers were performed using an iBlot 2 with polyvinylidene difluoride transfer stacks (Thermo Fisher Scientific). Membranes were passivated with 5% milk/1× Tris-buffered saline with Tween-20 or bovine serum albumin. Antibodies for Western blots included anti-Pan RAS (catalog no.: ab52939; Abcam; RRID: AB_2121042), anti-Vinculin (catalog no.: 13901T; Cell Signaling Technologies), anti-P44/42 MAPK (Erk1/2) (catalog no.: 4695S; Cell Signaling Technologies; RRID: AB_390779), anti-phospho p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP (catalog no.: 4370S; Cell Signaling Technologies; RRID: AB_2315112), and anti-GFP (catalog no.: MA515256; Thermo Fisher Scientific; RRID: AB_10979281).
For MAPK Western blots, “Rasless” MEFs were transfected with 2.5 μg of plasmids using Lipofectamine 3000 (Thermo Fisher Scientific). After 48 h of incubation, cells were washed with 1× Dulbecco’s PBS and lysed with the same ice-cold 1× radioimmunoprecipitation assay buffer for 15 min. Cell lysate was scraped, transferred into a LoBind tube, and centrifuged at 16,000g, 4 °C for 15 min. Supernatants were transferred to clean LoBind tubes, and protein concentration was determined by Pierce bicinchoninic acid protein assay (Thermo Fisher Scientific). A total protein concentration of 30 μg for each sample was added to sample buffer, boiled at 95 °C for 5 min, and then loaded into a gel as described previously. Antibodies were probed in the following order starting with pErk1/2, Erk1/2, GFP, and vinculin. Between each antibody probing, the membrane was stripped with Restore stripping buffer (Thermo Fisher Scientific) following product protocols. Experiments were performed in triplicate. Densitometry measurements were done using Fiji ImageJ (73). The ratios of p-Erk/Erk were calculated for each sample and then normalized to the p-Erk/Erk ratio of the MEF parental cell line. Individual data points from all three replicates are depicted in Figures 5C and S7.
Live-cell imaging
HeLa cells on poly-l-lysine–treated coverslips in 12-well dishes were transfected with 1000 ng of plasmid DNA using Lipofectamine 3000. Cells expressed plasmids for 24 h prior to staining with CellBrite Steady 650 membrane dye for 30 min. Live cells were imaged using a Zeiss Axio Observer 7 confocal microscope with LSM800 GaAsP-PMT detectors and a Plan-Apochromat 40× objective (Zeiss, 1.3 numerical aperture, oil immersion) with a pixel size of 0.077 μm × 0.077 μm. About 488 and 650 nm lasers were used to excite GFP and membrane dye. Three technical replicates were performed, and z-stacks for n = 30 cells were analyzed per sample type. Images were analyzed by Fiji ImageJ using the Plot Profiles function (73). Intensity values were normalized to the highest signal intensity. Signal intensity plots are reported for each individual cell in Fig. S8. No averaging between samples was performed.
BU analysis of select colorectal tumor samples
IP flow-through fractions, elution fractions preserved in acetone, and IP beads in sample buffer were prepared using trichloroacetic acid/acetone precipitation followed by in-gel digestion with trypsin (Promega). A single set of IP fractions was used for each individual tumor. The obtained peptides were analyzed by LC–MS/MS using a Dionex UltiMate 3000 Rapid Separation nanoLC and a Q Exactive HF Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). Samples were loaded onto a house-packed C18 column and separated with a 5 to 40% of solvent (0.1% FA in ACN) for 120 min by an analytical column (PicoChip, New Objective, Inc). Full MS scans were acquired from 300 to 2000 m/z at 60,000 r.p. using an isolation width of 2.0 m/z. The top 20 most abundant precursor ions in each full MS scan were selected for MS/MS fragmentation by higher-energy collisional dissociation at 30% NCE. MS/MS spectra were searched against a custom database, SwissProt Homo sapiens database plus the mutant sequence of a KRAS protein, using the Mascot search engine (Matrix Science; version 2.8.0). All searches included carbamidomethyl Cys as a fixed modification and oxidized Met; deamidated Asn and Gln; and acetylated N-term as variable modifications. The search result was visualized by Scaffold, version 5.0.1 (Proteome Software, Inc). Proteins were identified with a 1% false discovery rate and a minimum of two unique peptides.
Data availability
Mass spectra raw files, custom databases, and analysis result files are available on MassIVE (MSV000088748). RNA-Seq data were acquired from dbGaP Study Accession phs000892.v6.p1. Confocal microscopy images are available upon request from the corresponding author.
Supporting information
This article contains supporting information (24, 42, 73).
Conflict of interest
N. L. K. is involved in commercialization of proteomics software. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by federal funds from the NCI (Office of Cancer Clinical Proteomics Research), National Institutes of Health (NIH), and Leidos Biomedical Research under contract HHSN261200800001E and was carried out in collaboration with the National Resource for Translational and Developmental Proteomics under the NIH grant P41 GM108569. A portion of this work was performed at the National High Magnetic Field Laboratory, which is supported by the National Science Foundation Cooperative Agreement No. DMR-1644779 and the State of Florida. We thank Byoung-Kyu and Young ah Goo for their work on BU MS in this study. BU analysis was performed by the Northwestern Proteomics Core Facility, supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center, instrumentation award (S10OD025194) from the NIH Office of the Director, and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. We also thank Paul M. Thomas for helpful discussions throughout this work and Karla Satchell for valuable feedback.
Author contributions
L. M. A., C. J. D., and N. L. K. conceptualization; L. M. A., C. J. D., L. C. A., and R. V. methodology; L. M. A., C. J. D., and L. C. A. validation; L. M. A., C. J. D., and B. S. D. formal analysis; L. M. A., C. J. D., L. C. A., and R. V. investigation; E. S. B., T. M. H., C. L. H., H. R., R. V., and N. L. K. resources; L. M. A., C. J. D., and L. C. A. data curation; L. M. A. writing–original draft; L. M. A., C. J. D., B. S. D., L. C. A., and N. L. K. writing–review & editing; L. M. A., C. J. D., and M. C. visualization; W. B., C. L. H., and N. L. K. supervision; W. B., E. S. B., T. M. H., C. L. H., H. R., and N. L. K. project administration; W. B., C. L. H., and N. L. K. funding acquisition.
Funding and additional information
L. M. A. was supported by T32GM008382. B. S. D. was supported by the NCI of the NIH under award number F32CA246894. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Edited by Donita Brady
Footnotes
Emily S. Boja is now affiliated with the Office of Data Sharing at the National Cancer Institute.
Supporting information
References
- 1.Barbacid M. Ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Vigil D., Cherfils J., Rossman K.L., Der C.J. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer. 2011;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prior I.A., Hood F.E., Hartley J.L. The frequency of ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haigis K.M. KRAS alleles: the devil is in the detail. Trends Cancer. 2017;3:686–697. doi: 10.1016/j.trecan.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porru M., Pompili L., Caruso C., Biroccio A., Leonetti C. Targeting KRAS in metastatic colorectal cancer: current strategies and emerging opportunities. J. Exp. Clin. Cancer Res. 2018;37:57. doi: 10.1186/s13046-018-0719-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baranyi M., Buday L., Hegedus B. K-Ras prenylation as a potential anticancer target. Cancer Metastasis Rev. 2020;39:1127–1141. doi: 10.1007/s10555-020-09902-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toby T.K., Fornelli L., Srzentic K., DeHart C.J., Levitsky J., Friedewald J., et al. A comprehensive pipeline for translational top-down proteomics from a single blood draw. Nat. Protoc. 2019;14:119–152. doi: 10.1038/s41596-018-0085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox A.D., Der C.J., Philips M.R. Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin. Cancer Res. 2015;21:1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahearn I., Zhou M., Phillips M. Post-translational modifications of RAS proteins. Cold Spring Harb. Perspect. Med. 2018;8 doi: 10.1101/cshperspect.a031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hancock J.F., Magee A.I., Childs J.E., Marshall C.J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 13.Hancock J.F., Paterson H., Marshall C.J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localise p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 14.Hancock J.F., Cadwallader K., Paterson H., Marshall C.J. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock J.F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 16.Plowman S.J., Ariotti N., Goodall A., Parton R.G., Hancock J.F. Electrostatic interactions positively regulate K-ras nanocluster formation and function. Mol. Cell Biol. 2008;28:4377–4385. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busquets-Hernandez C., Triola G. Palmitoylation as a key regulator of ras localization and function. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.659861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang H., Abraham S.J., Chavan T.S., Hitchinson B., Khavrutskii L., Tarasova N.I., et al. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J. Biol. Chem. 2015;290:9465–9477. doi: 10.1074/jbc.M114.620724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welman A., Burger M.M., Hagmann J. Structure and function of the C-terminal hypervariable region of K-Ras4B in plasma membrane targetting and transformation. Oncogene. 2000;19:4582–4591. doi: 10.1038/sj.onc.1203818. [DOI] [PubMed] [Google Scholar]
- 20.Willumsen B.M., Christensen A., Hubbert N.L., Papageorge A.G., Lowy D.R. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984;310:583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- 21.Wright L.P., Philips M.R. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of ras. J. Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Yang M.H., Nickerson S., Kim E.T., Liot C., Laurent G., Spang R., et al. Regulation of RAS oncogenicity by acetylation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10843–10848. doi: 10.1073/pnas.1201487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M.H., Laurent G., Bause A.S., Spang R., German N., Haigis M.C., et al. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol. Cancer Res. 2013;11:1072–1077. doi: 10.1158/1541-7786.MCR-13-0040-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knyphausen P., Lang F., Baldus L., Extra A., Lammers M. Insights into K-Ras 4B regulation by post-translational lysine acetylation. Biol. Chem. 2016;397:1071–1085. doi: 10.1515/hsz-2016-0118. [DOI] [PubMed] [Google Scholar]
- 25.Lander H.M., Hajjar D.P., Hempsteadi B.L., Mirza U.A., Chait B.T., Campbell S., et al. A molecular redox switch on p21ras. J. Biol. Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- 26.Lander H.M., Ogiste J.S., Teng K.K., Novogrodsky A. p21ras as a common signaling target of reactive free radicals and cellular redox stress. J. Biol. Chem. 1995;270:21195–21198. doi: 10.1074/jbc.270.36.21195. [DOI] [PubMed] [Google Scholar]
- 27.Williams J.G., Pappu K., Campbell S.L. Structural and biochemical studies of p21Ras S-nitrosylation and nitric oxide-mediated guanine nucleotide exchange. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6376–6381. doi: 10.1073/pnas.1037299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heo J., Prutzman K.C., Mocanu V., Campbell S.L. Mechanism of free Radical nitric oxide-mediated ras guanine nucleotide dissociation. J. Mol. Biol. 2005;346:1423–1440. doi: 10.1016/j.jmb.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 29.Huang L., Carney J., Cardona D.M., Counter C.M. Decreased tumorigenesis in mice with a Kras point mutation at C118. Nat. Commun. 2014;5:5410. doi: 10.1038/ncomms6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballester R., Furth M.E., Rosen O.M. Phorbol ester- andP rotein Kinase C-mediated phosphorylation of the cellular kirsten ras gene product. J. Biol. Chem. 1987;262:2688–2695. [PubMed] [Google Scholar]
- 31.Barcelo C., Paco N., Beckett A.J., Alvarez-Moya B., Garrido E., Gelabert M., et al. Oncogenic K-ras segregates at spatially distinct plasma membrane signaling platforms according to its phosphorylation status. J. Cell Sci. 2013;126:4553–4559. doi: 10.1242/jcs.123737. [DOI] [PubMed] [Google Scholar]
- 32.Barcelo C., Paco N., Morell M., Alvarez-Moya B., Bota-Rabassedas N., Jaumot M., et al. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Res. 2013;74:1190–1199. doi: 10.1158/0008-5472.CAN-13-1750. [DOI] [PubMed] [Google Scholar]
- 33.Alvarez-Moya B., Lopez-Alcala C., Drosten M., Bachs O., Agell N. K-Ras4B phosphorylation at Ser181 is inhibited by calmodulin and modulates K-Ras activity and function. Oncogene. 2010;29:5911–5922. doi: 10.1038/onc.2010.298. [DOI] [PubMed] [Google Scholar]
- 34.Kano Y., Gebregiworgis T., Marshall C.B., Radulovich N., Poon B.P.K., St-Germain J., et al. Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation. Nat. Commun. 2019;10:224. doi: 10.1038/s41467-018-08115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jura N., Scotto-Lavino E., Sobczyk A., Bar-Sagi D. Differential modification of ras proteins by ubiquitination. Mol. Cell. 2006;21:679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Baker R., Lewis S.M., Sasaki A.T., Wilkerson E.M., Locasale J.W., Cantley L.C., et al. Site–specific monoubiquitination activates ras by impeding GTPase activating protein function. Nat. Struct. Mol. Biol. 2013;20:46–52. doi: 10.1038/nsmb.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker R., Wilkerson E.M., Sumita K., Isom D.G., Sasaki A.T., Dohlman H.G., et al. Differences in the regulation of K-ras and H-ras isoforms by monoubiquitination. J. Biol. Chem. 2013;288:36856–36862. doi: 10.1074/jbc.C113.525691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki A.T., Carracedo A., Locasale J.W., Anastasiou D., Takeuchi K., Kahoud E.R., et al. Ubiquitination of K-ras enhances activation and facilitates binding to Select downstream effectors. Cell Biol. 2011;4 doi: 10.1126/scisignal.2001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell S.L., Philips M.R. Post-translational modification of RAS proteins. Curr. Opin. Struct. Biol. 2021;71:180–192. doi: 10.1016/j.sbi.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toby T.K., Fornelli L., Kelleher N.L. Progress in top-down proteomics and the analysis of proteoforms. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2016;9:499–519. doi: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ntai I., Fornelli L., DeHart C.J., Hutton J.E., Doubleday P.F., LeDuc R.D., et al. Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk. Proc. Natl. Acad. Sci. U. S. A. 2018;115:4140–4145. doi: 10.1073/pnas.1716122115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams L.M., DeHart C.J., Kelleher N.L. In: Ras Activity and Signaling: Methods and Protocols. Rubio I., Prior I., editors. Springer US; New York, NY: 2021. Precise characterization of KRAS4B proteoforms by combining immunoprecipitation with top-down mass spectrometry; pp. 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson L.C., DeHart C.J., Kaiser N.K., Fellers R.T., Smith D.F., Greer J.B., et al. Identification and characterization of human proteoforms by top-down LC-21 tesla FT-ICR mass spectrometry. J. Proteome Res. 2017;16:1087–1096. doi: 10.1021/acs.jproteome.6b00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendrickson C.L., Quinn J.P., Kaiser N.K., Smith D.F., Blakney G.T., Chen T., et al. 21 tesla fourier transform ion cyclotron resonance mass spectrometer: a national resource for ultrahigh resolution mass analysis. J. Am. Soc. Mass Spectrom. 2015;26:1626–1632. doi: 10.1007/s13361-015-1182-2. [DOI] [PubMed] [Google Scholar]
- 45.Drosten M., Dhawahir A., Sum E.Y., Urosevic J., Lechuga C.G., Esteban L.M., et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 2010;29:1091–1104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasaikar S., Huang C., Wang X., Petyuk V.A., Savage S.R., Wen B., et al. Proteogenomic analysis of human Colon cancer reveals new therapeutic opportunities. Cell. 2019;177:1035–1049.e1019. doi: 10.1016/j.cell.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nussinov R., Tsai C.J., Chakrabarti M., Jang H. A new view of ras isoforms in cancers. Cancer Res. 2016;76:18–23. doi: 10.1158/0008-5472.CAN-15-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amendola C.R., Mahaffey J.P., Parker S.J., Ahearn I.M., Chen W., Zhou M., et al. KRAS4A directly regulates hexokinase 1. Nature. 2019;576:482–486. doi: 10.1038/s41586-019-1832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W.C., To M.D., Westcott P.M.K., Delrosario R., Kim I.J., Philips M., et al. Targeting KRAS4A splicing through the RBM39/DCAF15 pathway inhibits cancer stem cells. Nat. Commun. 2021;12:4288. doi: 10.1038/s41467-021-24498-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung S.I., Moon H., Ju H., Kim D.Y., Cho K.J., Ribback S., et al. Comparison of liver oncogenic potential among human RAS isoforms. Oncotarget. 2016;7:7354–7366. doi: 10.18632/oncotarget.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jing H., Zhang X., Wisner S.A., Chen X., Spiegelman N.A., Linder M.E., et al. SIRT2 and lysine fatty acylation regulate the transforming activity of K-Ras4a. Elife. 2017;6 doi: 10.7554/eLife.32436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai F.D., Lopes M.S., Zhou M., Court H., Ponce O., Fiordalisi J.J., et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl. Acad. Sci. U. S. A. 2015;112:779–784. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X., Cao J., Miller S.P., Jing H., Lin H. Comparative nucleotide-dependent interactome analysis reveals shared and differential properties of KRas4a and KRas4b. ACS Cent. Sci. 2018;4:71–80. doi: 10.1021/acscentsci.7b00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 55.Mageean C.J., Griffiths J.R., Smith D.L., Clague M.J., Prior I.A. Absolute quantification of endogenous ras isoform abundance. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shukla S., Allam U.S., Ahsan A., Chen G., Krishnamurthy P.M., Marsh K., et al. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated beta-TrCP1 degradation. Neoplasia. 2014;16:115–128. doi: 10.1593/neo.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammond D.E., Mageean C.J., Rusilowicz E.V., Wickenden J.A., Clague M.J., Prior I.A. Differential reprogramming of isogenic colorectal cancer cells by distinct activating KRAS mutations. J. Proteome Res. 2015;14:1535–1546. doi: 10.1021/pr501191a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshino H., Yin G., Kawaguchi R., Popov K.I., Temple B., Sasaki M., et al. Identification of lysine methylation in the core GTPase domain by GoMADScan. PLoS One. 2019;14 doi: 10.1371/journal.pone.0219436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmick M., Vartak N., Papke B., Kovacevic M., Truxius D.C., Rossmannek L., et al. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 2014;157:459–471. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 60.Tran T.H., Chan A.H., Young L.C., Bindu L., Neale C., Messing S., et al. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun. 2021;12:1176. doi: 10.1038/s41467-021-21422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marshall M. Interactions between ras and Raf: key regulatory proteins in cellular transformation. Mol. Reprod. Dev. 1995;42:493–499. doi: 10.1002/mrd.1080420418. [DOI] [PubMed] [Google Scholar]
- 62.Guo Y.J., Pan W.W., Liu S.B., Shen Z.F., Xu Y., Hu L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020;19:1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiordalisi J.J., Holly S.P., Johnson R.L., 2nd, Parise L.V., Cox A.D. A distinct class of dominant negative ras mutants: cytosolic GTP-bound ras effector domain mutants that inhibit ras signaling and transformation and enhance cell adhesion. J. Biol. Chem. 2002;277:10813–10823. doi: 10.1074/jbc.M107684200. [DOI] [PubMed] [Google Scholar]
- 64.Lerner E.C., Qian Y., Blaskovich M.A., Fossum R.D., Vogt A., Sun J., et al. Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic Ras signaling by inducing cytoplasmic accumulation of inactive Ras-Raf complexes. J. Biol. Chem. 1995;270:26802–26806. doi: 10.1074/jbc.270.45.26802. [DOI] [PubMed] [Google Scholar]
- 65.Goblirsch B.R., Wiener M.C. Ste24: an integral membrane protein zinc metalloprotease with provocative structure and emergent biology. J. Mol. Biol. 2020;432:5079–5090. doi: 10.1016/j.jmb.2020.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mehmood S., Marcoux J., Gault J., Quigley A., Michaelis S., Young S.G., et al. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nat. Chem. 2016;8:1152–1158. doi: 10.1038/nchem.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weisbrod C.R., Kaiser N.K., Syka J.E.P., Early L., Mullen C., Dunyach J.J., et al. Front-end electron transfer dissociation coupled to a 21 tesla FT-ICR mass spectrometer for intact protein sequence analysis. J. Am. Soc. Mass Spectrom. 2017;28:1787–1795. doi: 10.1007/s13361-017-1702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fellers R.T., Greer J.B., Early B.P., Yu X., LeDuc R.D., Kelleher N.L., et al. ProSight lite: graphical software to analyze top-down mass spectrometry data. Proteomics. 2015;15:1235–1238. doi: 10.1002/pmic.201570050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeHart C.J., Fellers R.T., Fornelli L., Kelleher N.L., Thomas P.M. Bioinformatics analysis of top-down mass spectrometry data with ProSight lite. Met. Mol. Biol. 2017;1558:381–394. doi: 10.1007/978-1-4939-6783-4_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fornelli L., Srzentić K., Huguet R., Mullen C., Sharma S., Zabrouskov V., et al. Accurate sequence analysis of a monoclonal antibody by TopDown and middle-down orbitrap mass spectrometry applying multiple ion activation techniques. Anal. Chem. 2018;90:8421–8429. doi: 10.1021/acs.analchem.8b00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pesavento J.J., Mizzen C.A., Kelleher N.L. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human Histone H4. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 72.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. Fiji: an open-source platform for biological-image analysis. Nat. Met. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectra raw files, custom databases, and analysis result files are available on MassIVE (MSV000088748). RNA-Seq data were acquired from dbGaP Study Accession phs000892.v6.p1. Confocal microscopy images are available upon request from the corresponding author.