Abstract
Since March 2022, there has been an emergence of multidrug-resistant organisms (MDRO) in the Netherlands in patients originating from Ukraine (58 patients, 75 isolates). For about half of these patients, recent hospitalisation in Ukraine was reported. Genomic surveillance revealed that the majority of the MDRO represent globally spread epidemic lineages and that 60% contain New Delhi metallo-β-lactamase (NDM) genes. Professionals should be aware of an increase in such MDRO associated with migration and medical evacuation of people from Ukraine.
Keywords: antimicrobial resistance, patients from Ukraine, CPE, CPPA, MRSA, CRAB
The Netherlands is a country with a low prevalence of multidrug-resistant organisms (MDRO), including carbapenemase-producing Enterobacterales (CPE), carbapenemase-producing Pseudomonas aeruginosa (CPPA), carbapenem-resistant Acinetobacter baumannii complex (CRAB) and meticillin-resistant Staphylococcus aureus (MRSA) [1-4]. Active MDRO screening and infection prevention measures (e.g. isolation of suspected colonised patients) applies to all patients admitted to a Dutch hospital who have recently been admitted to a hospital abroad (< 2 months ago for > 24 h) or resided in a centre for asylum seekers. Since March 2022, there has been an emergence of MDRO in the Netherlands in persons originating from Ukraine. Persons from whom isolates were obtained were receiving hospital care in the Netherlands and are henceforth referred to as patients. Here, we describe the epidemiology and genetic analysis of MDRO from patients from Ukraine in the Netherlands.
Multidrug-resistant organisms in the Netherlands and Ukraine
In the Netherlands, the prevalence and spread of MDRO are monitored through national surveillance of CPE (since 2011), CPPA (since 2020), CRAB (since August 2022) and MRSA (since 1989), and through the mandatory notification of CPE (since July 2019). Starting from March 2022, the war in Ukraine led to migration of people from Ukraine and medical evacuation of more than 1,000 patients from Ukrainian hospitals to hospitals in other countries, including the Netherlands [5]. The European Centre for Disease Prevention and Control (ECDC) advised to pre-emptively isolate patients transferred from hospitals in Ukraine or with a history of hospital admission in Ukraine in the last 12 months and to screen for carriage of MDRO [6]. Available information from Ukrainian military and general hospitals indicates high prevalence of MDRO in Ukraine between 2014 and 2021 [3,7,8]. Percentages of resistance of 17–84% against third-generation cephalosporins and carbapenems among Enterobacterales and P. aeruginosa, and > 50% resistance to carbapenems, fluoroquinolones and aminoglycosides among Acinetobacter species have been reported [3,7,8]. European surveillance data for Ukraine indicated 18% of S. aureus to be meticillin-resistant, while 41% meticillin-resistance was reported in healthcare-associated S. aureus infections [3,8]. However, these percentages are based on isolates from hospitalised patients and may therefore not be representative for MDRO carriage in the community [3,7].
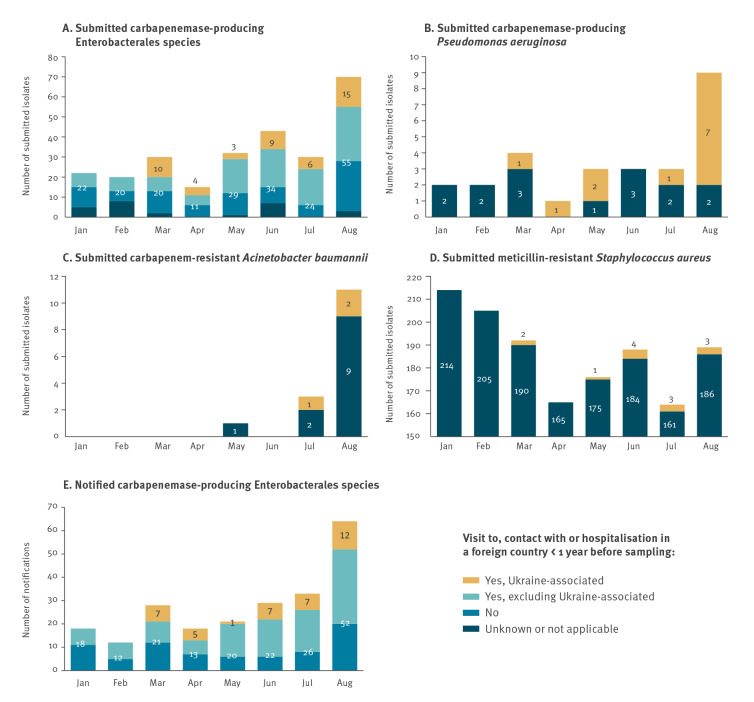
Up to March 2022, the Dutch national surveillance did not detect any MDRO in patients from Ukraine. From 1 March to 31 August 2022, 47 CPE, 12 CPPA, three CRAB and 13 MRSA isolates from 56 patients originating from Ukraine were submitted to the national surveillance (Figure, panels A-D), representing 21% (CPE), 52% (CPPA) and 1.2% (MRSA) of the total number of submitted isolates for each of these species during this period. In the mandatory CPE notification system, 39 patients from Ukraine were reported (Figure, panel E); for 37 of them, isolates had been submitted to the Dutch national surveillance for characterisation. Adding the two extra patients without submitted isolate, the total number of patients was 58. Isolates were characterised phenotypically (matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-ToF), carbapenem inactivation method and Etest for meropenem) [2,9] and genetically (whole genome (wg) sequencing, multilocus sequence typing (MLST) and wgMLST) [10], and metadata were collected during submission for CPPA, CRAB and MRSA, or through the national mandatory notification system for CPE [1,4]. Isolates and notifications were considered Ukraine-associated when the epidemiological questionnaire indicated that the patient had recently been hospitalised in Ukraine (< 2 months ago for > 24 h) or had contact with a Ukrainian hospital or visited Ukraine < 1 year before sampling.
Figure.
Number of submitted CPE (n = 262), CPPA (n = 27), CRAB (n = 15) and MRSA (n = 1,493) isolates, and mandatory CPE notifications (n = 223), the Netherlands, 1 January–31 August 2022
Numbers in the stacked bars indicate the total number of submitted isolates (panels A to D) or mandatory notifications (panel E) in a given category. For CPE submissions and notifications (panels A and E), isolates/notifications not associated with Ukraine are further stratified based on availability of data about recent travel/hospitalisation abroad in the year before sampling. The national CRAB surveillance (panel C) started on 1 Aug 2022.
Epidemiology of patients from Ukraine with multidrug-resistant organisms
In the Netherlands, a risk factor for acquisition of MDRO is recent hospitalisation abroad [1,4]. We included a figure on mandatory CPE notifications from 2019 to 2022 in Supplementary Figure S1. In general, an increase in CPE notifications is observed in the third and part of the fourth quarter of the year, both in travel and non-travel-related CPE notifications. Also in 2022, CPE notifications increased, especially in August: 58 notifications compared with an average of 23 in the previous months (Figure, panel E) and compared with 28, 12 and 28 notifications in August 2019, 2020 and 2021, respectively. The observed increase in August 2022 was in part related to a larger number of patients for whom travel and/or hospitalisation abroad was reported (n = 38), of whom nine were patients from Ukraine, but also attributable to a larger number of patients with no record of travel or hospitalisation abroad (n = 20). There were no indications of geographical clustering or a local outbreak.
Characteristics of patients from Ukraine with MDRO are shown in Table 1. The median age of patients was 33 years, and most were male. Except for CPPA and CRAB, the majority of MDRO were found in samples obtained for screening of patients considered at risk for MDRO because of recent hospitalisation or residing in a centre for asylum seekers. Twenty-one of the 39 patients with CPE had recently been hospitalised abroad. For CPPA, complete metadata were available for seven of 12 patients and six of them had been hospitalised abroad. For MRSA, complete metadata were available for 12 of 13 patients from Ukraine and for two patients, recent hospitalisation abroad was reported.
Table 1. Epidemiological characteristics of patients with CPE, CPPA, CRAB, MRSA and multiple MDRO originating from Ukraine, the Netherlands, sampled 1 March–31 August 2022 (n = 58).
| Characteristic | CPEa (n = 39) | CPPAb (n = 12) | CRABb (n = 3) | MRSAb (n = 13) | Multiple MDROa,b,c (n = 11) |
|---|---|---|---|---|---|
| n | n | n | n | n | |
| Median age in years (range) | 34 (0–86) | 31 (2–66) | 45 (36–66) | 27 (0–70) | 28 (16–66) |
| Male | 26 | 11 | 2 | 12 | 9 |
| Female | 13 | 1 | 1 | 1 | 2 |
| Type of material | |||||
| Swabs from nose/throat/rectum/perineum | 26 | 6 | 0 | 11 | 5 |
| Wound/pus | 10 | 6 | 3 | 2 | 5 |
| Urine | 3 | 0 | 0 | 0 | 1 |
| Additional metadata available | 39 | 7 | 0d | 12 | 11 |
| Reason for culturing | |||||
| Diagnostic/clinical indication | 5 | 4 | 0 | 3e | 2 |
| Screening for MDRO, contact tracing or belongs to risk group | 32 | 3 | 0 | 10e | 8 |
| Other/unknown | 2 | NC | NC | NC | 1 |
| Sampling location | |||||
| Outpatient clinic | 11 | 0 | 0 | 4 | 1 |
| Inpatient ward | 24 | 7 | 0 | 6 | 9 |
| Intensive care unit | 3 | 0 | 0 | 2 | 0 |
| Unknown | 1 | 0 | 0 | 0 | 1 |
| Invasive medical procedure/diagnostics | |||||
| Yes | 22 | 1 | 0 | NC | 11 |
| No | 1 | 3 | 0 | NC | 0 |
| Unknown | 16 | 3 | 0 | NC | 0 |
| Risk factor | |||||
| Hospitalisation abroad > 24 h during the previous 2 months | 21 | 6 | 0 | 2 | 10 |
CPE: carbapenemase-producing Enterobacterales; CPPA: carbapenemase-producing Pseudomonas aeruginosa; CRAB: carbapenem-resistant Acinetobacter baumannii; MDRO: multidrug-resistant organism; MRSA: meticillin-resistant Staphylococcus aureus; NC: not collected.
a Based on the notifications in the national mandatory notification system.
b Based on the metadata entered in the database of the Dutch national surveillance.
c These patients are also taken into account in columns ‘CPE’, ‘CPPA’ and ‘MRSA’, where applicable.
d For CRAB, additional epidemiological data were not available because this pilot surveillance started only in August 2022.
e One questionnaire was incomplete and only this question was available and therefore the denominator is 13 for this item.
For 11 patients (median age: 28 years; nine male), multiple MDRO isolates were submitted (Table 1). Six of the 11 patients had two different species of MDRO. Two patients carried three different MDRO: one carried one each of CPE, CPPA and CRAB and another carried two CPE and a CPPA. The remaining three patients each carried four different strains: one patient had three CPE and one CPPA, one patient had two CPE, one CPPA and one CRAB, and the final patient carried two CPE, one CPPA and one MRSA.
Genetic analysis of multidrug-resistant organisms from patients from Ukraine
All submitted CPE, CPPA and CRAB isolates (n = 62, from 44 patients from Ukraine) produced carbapenemase as assessed by the carbapenem inactivation method; the individual results are provided separately in Supplementary Table S1 [9]. The majority of isolates (39/62; 63%) were resistant to meropenem according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (minimum inhibitory concentration (MIC) > 8 mg/L [11]) as determined by Etest (bioMérieux). All submitted Escherichia coli isolates (n = 6) were susceptible with increased exposure for meropenem (MIC ≥ 2 and < 8 mg/L).
From 57 of the 62 isolates, Illumina next-generation sequencing (NGS) data were available. The majority of isolates were K. pneumoniae (37/62; 60%) and included globally spread MLST types ST307 (12/37), ST147 (10/37) and ST395 (7/37) [12,13], followed by P. aeruginosa (12/62; 19%), including ST1047 (5/12) and ST773 (4/12) [14], and E. coli (6/62; 10%) of varying STs including ST405 (2/6; Table 2) [15]. Three K. pneumoniae isolates belonged to ST23 and all CRAB isolates were from not yet assigned sequence types.
Table 2. Carbapenemase-encoding genes and sequence types of multidrug-resistant organisms from patients from Ukraine in national surveillance, the Netherlands, sampled 1 March–31 August 2022 (n = 62).
| Species and carbapenemase allele | Total | MLST sequence type | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 23 | 39 | 46 | 147 | 231 | 307 | 395 | 405 | 654 | 773 | 1047 | 5859 | New STa | Unknownb | ||
| Acinetobacter baumannii | 3 | 3 | ||||||||||||||
| blaOXA-23, blaOXA-66 | 2 | 2 | ||||||||||||||
| bla OXA-72 | 1 | 1 | ||||||||||||||
| Enterobacter cloacae complex | 1 | 1 | ||||||||||||||
| bla NDM-1 | 1 | 1 | ||||||||||||||
| Escherichia coli | 6 | 3 | 2 | 1 | ||||||||||||
| bla NDM-5 | 4 | 3 | 1 | |||||||||||||
| bla OXA-244 | 1 | 1 | ||||||||||||||
| bla OXA-48 | 1 | 1 | ||||||||||||||
| Klebsiella pneumoniae | 37 | 1 | 3 | 1 | 10 | 12 | 7 | 2 | 1 | |||||||
| bla KPC-2 | 1 | 1 | ||||||||||||||
| bla KPC-3 | 3 | 3 | ||||||||||||||
| bla NDM-1 | 16 | 2 | 5 | 6 | 3 | |||||||||||
| blaNDM-1, blaOXA-232 | 1 | 1 | ||||||||||||||
| blaNDM-1, blaOXA-48 | 8 | 1 | 3 | 2 | 1 | 1 | ||||||||||
| bla NDM-9 | 1 | 1 | ||||||||||||||
| bla OXA-244 | 1 | 1 | ||||||||||||||
| bla OXA-48 | 3 | 1 | 2 | |||||||||||||
| bla VIM-1 | 1 | 1 | ||||||||||||||
| Unknown allele | 2 | 1 | 1 | |||||||||||||
| Proteus mirabilis | 2 | 2 | ||||||||||||||
| bla NDM-1 | 2 | 2 | ||||||||||||||
| Providencia stuartii | 1 | 1 | ||||||||||||||
| Unknown allele | 1 | 1 | ||||||||||||||
| Pseudomonas aeruginosa | 12 | 1 | 4 | 5 | 2 | |||||||||||
| blaIMP-1, blaOXA-488 | 4 | 4 | ||||||||||||||
| blaIMP-10, blaOXA-488 | 1 | 1 | ||||||||||||||
| bla NDM-1 | 4 | 4 | ||||||||||||||
| bla VIM-2 | 1 | 1 | ||||||||||||||
| Unknown allele | 2 | 2 | ||||||||||||||
| Total | 62 | 1 | 3 | 1 | 3 | 10 | 1 | 12 | 7 | 2 | 1 | 4 | 5 | 2 | 4 | 6 |
MLST: multilocus sequence typing; ST: sequence type.
a Unassigned sequence type.
b Unsequenced isolates.
The CPE, CPPA and CRAB isolates from patients from Ukraine contained blaNDM-like carbapenemase alleles (37/62; 60%), followed by blaOXA-48 (12/62; 19%), blaIMP-like (5/62; 8%) and blaKPC-like alleles (4/62; 6%; Table 2). In contrast, 31% (76/242) of the other Dutch surveillance isolates (derived from persons sampled between 1 January and 31 August 2022) carried blaNDM-like carbapenemase alleles, followed by blaOXA-48 (70/242; 29%) and blaOXA-181 (25/242; 10%) (for each isolate, the detailed results from molecular analysis are provided in Supplementary Table S1 and S2).
Sixteen of the 57 sequenced isolates from patients from Ukraine harboured combinations of two different carbapenemase alleles, of which blaNDM-1 with blaOXA-48 dominated (8/57). Significantly fewer (9%; 23/242; p < 0.0002) of the other Dutch surveillance isolates contained combinations of two different carbapenemase alleles, of which blaOXA-23 with blaOXA-66 (6/242; 2.5%) and blaNDM‑5 with blaOXA-48 (4/242; 1.7%) dominated. In Supplementary Figure S2, we append a resistome analysis using ResFinder v4.0 which revealed that MDRO from patients from Ukraine carried genes implicated in resistance towards more classes of antibiotics and disinfectants (p < 0.0001) than MDRO from the Netherlands.
We performed wgMLST using established in-house wgMLST schemes for K. pneumoniae, E. coli, P. aeruginosa [10,16] and A. baumannii, which indicated four potential transmission events of K. pneumoniae between patients from Ukraine and the Netherlands. Comparative wgMLST analysis of Dutch surveillance isolates with publicly available sequencing data from bacterial isolates from war-injured patients in military hospitals during the Eastern Ukraine conflict between 2014 and 2020 [7] indicates comparable sequence types (ST23, ST395, ST773), but no genetic clustering.
All MRSA isolates (n = 13, from 13 patients) carried mecA, and a small fraction harboured Panton-Valentine leukocidin (2/13). The NGS data of 12 MRSA isolates were available. Nine isolates belonged to six different MLST types and three to not yet assigned MLST types. Three isolates belonged to MLST ST5, two to ST22 and one of the isolates classified as livestock-associated MRSA (ST398). For each isolate, the detailed results from molecular analysis are provided in Supplementary Table S3. Based on wgMLST, there was no indication of MRSA transmission between patients from Ukraine and the Netherlands. No data on other MDRO are available since these are not monitored in the national surveillance.
Discussion
We observed a marked increase in Ukraine-associated CPE in August 2022, which added to the non-Ukraine-associated, travel and non-travel related increase in CPE at that time. Given the ongoing routine infection prevention measures, it is unlikely that this is a result of a change in screening practices. Potential transmission of K. pneumoniae between patients from Ukraine and the Netherlands occurred only sporadically and outbreaks were not observed, an indication that infection prevention measures performed in hospitals in the Netherlands are adequate. About half of the patients from Ukraine with MDRO and with available additional metadata had recently been hospitalised in Ukraine. Besides, in patients from Ukraine with CPE, CPPA and CRAB, combinations of two carbapenemase alleles occurred significantly more frequently than usual in the Netherlands [1,2]. This is probably attributable to the medical evacuation of patients from Ukrainian hospitals. OXA-type and NDM-1 have been reported as the most frequent alleles among carbapenemase-positive MDRO causing hospital-acquired infections in Ukraine, but carbapenemase allele combinations were not reported [8]. Limitations of our study are the lack of denominator data, including how the number of MDRO from patients from Ukraine relates to the total number of patients, the monthly number of hospitalisations in the Netherlands, and how many patients from Ukraine have been screened for MDRO or how many were not colonised with an MDRO. In addition, information on war-related injuries, other clinical data of the patients and the timing of medical evacuations is missing.
Conclusion
We observed an increase in MDRO from globally epidemic high-risk sequence types retrieved from patients from Ukraine in the Netherlands. About half of these patients had recently been hospitalised in Ukraine. Eleven patients carried multiple MDRO. The high percentage of isolates containing blaNDM-like genes in their genomes, and the observation that the isolates are potentially resistant towards multiple classes of antibiotics, imply that infections with these bacteria are difficult to treat. Additional phenotypic resistance patterns of these MDRO need to be determined, including novel combinations of antibiotics, to provide physicians with therapeutic options for treatment of infections with carbapenemase-producing Gram-negative bacteria in patients from Ukraine. Healthcare professionals should be aware of the possible presence of these microorganisms when treating hospitalised patients from Ukraine and take adequate infection prevention measures to prevent the spread of these MDRO.
Ethical statement
Ethical approval was not needed for the study since it is solely based on surveillance data. Samples from which the isolates were cultured were all collected as part of routine healthcare.
Funding statement
This study was carried out as part of the Dutch National CPE and MRSA surveillance, as part of the regular activities of the RIVM, financed by the Dutch Ministry of Health, Welfare and Sport.
Data availability
The Illumina (NGS) sequence data set generated and analysed in this study are available in the Sequence Read Archive (SRA) by the study accession number PRJNA903550 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA903550).
Acknowledgements
We thank Dr Viacheslav Kondratiuk (Department of Emergency and Military Medicine, National Pirogov Memorial Medical University, Vinnytsia, Ukraine) and Prof Dr Aidyn Salmanov (Shupyk National Healthcare University of Ukraine, Kyiv) for their support in the interpretation of the data. Dr Sebastian Haller (Department of Epidemiology, Robert Koch-Institute) exchanged data from the German observations and advised on the manuscript design. We thank the Municipal Health Services for completing the epidemiological data in the mandatory notification system Osiris, the members of the Dutch CPE and MRSA surveillance study groups and the Dutch medical microbiology laboratories for submitting isolates to the RIVM for the national surveillance program.
Members of the Dutch CPE and MRSA Surveillance Study Groups:
· A. Maijer-Reuwer, ADRZ medisch centrum, Department of Medical Microbiology, Goes
· M.A. Leversteijn-van Hall, Eurofins Clinical Diagnostics, Department of Medical Microbiology, Leiden-Leiderdorp
· W. van den Bijllaardt, Amphia Hospital, Microvida Laboratory for Microbiology, Breda
· R. van Mansfeld, Amsterdam UMC - location AMC, Department of Medical Microbiology and Infection Control, Amsterdam
· K. van Dijk, Amsterdam UMC - location Vumc, Department of Medical Microbiology and Infection Control, Amsterdam
· B. Zwart, Atalmedial, Department of Medical Microbiology, Amsterdam
· B.M.W. Diederen, Bravis Hospital/ZorgSaam Hospital Zeeuws-Vlaanderen, Department of Medical Microbiology, Roosendaal/Terneuzen
· J.W. Dorigo-Zetsma, TergooiMC, Central Bacteriology and Serology Laboratory, Hilversum
· A. Ott, Certe, Medical Microbiology Groningen, Drenthe, Groningen
· W. Ang, Comicro, Department of Medical Microbiology, Hoorn
· J. da Silva, Deventer Hospital, Department of Medical Microbiology, Deventer
· A.L.M. Vlek, Diakonessenhuis Utrecht, Department of Medical Microbiology and Immunology, Utrecht
· A.G.M. Buiting, Elisabeth-TweeSteden (ETZ) Hospital, Department of Medical Microbiology and Immunology, Tilburg
· L.G.M. Bode, Erasmus University Medical Center, Department of Medical Microbiology and Infectious Diseases, Rotterdam
· S. Paltansing, Franciscus Gasthuis & Vlietland, Department of Medical Microbiology and Infection Control, Rotterdam
· A.J. van Griethuysen, Gelderse Vallei Hospital, Department of Medical Microbiology, Ede
· M. den Reijer, Star-shl diagnostic centre, Department of Medical Microbiology, Rotterdam
· M.J.C.A. van Trijp, Groene Hart Hospital, Department of Medical Microbiology and Infection Prevention, Gouda
· M. Wong, Haga Hospital, Department of Medical Microbiology, 's-Gravenhage
· A.E. Muller, HMC Westeinde Hospital, Department of Medical Microbiology, 's-Gravenhage
· M.P.M. van der Linden, IJsselland hospital, Department of Medical Microbiology, Capelle a/d IJssel
· M. van Rijn, Ikazia Hospital, Department of Medical Microbiology, Rotterdam
· S.B. Debast, Isala Hospital, Laboratory of Medical Microbiology and Infectious Diseases, Zwolle
· K. Waar, Certe, Medical Microbiology Friesland, Noordoostpolder, Leeuwarden
· E. Kolwijck, Jeroen Bosch Hospital, Department of Medical Microbiology and Infection Control, 's-Hertogenbosch
· N. Al Naiemi, LabMicTA, Regional Laboratory of Microbiology Twente Achterhoek, Hengelo
· T. Schulin, Laurentius Hospital, Department of Medical Microbiology, Roermond
· S. Dinant, Maasstad Hospital, Department of Medical Microbiology, Rotterdam
· S.P. van Mens, Maastricht University Medical Centre, Department of Medical Microbiology, Infectious Diseases & Infection Prevention, Maastricht
· D.C. Melles, Meander Medical Center, Department of Medical Microbiology, Amersfoort
· M.P.A. van Meer, Rijnstate Hospital, Laboratory for Medical Microbiology and Immunology, Velp
· J.W.T. Cohen Stuart, Noordwest Ziekenhuisgroep, Department of Medical Microbiology, Alkmaar
· P. Gruteke, OLVG Lab BV, Department of Medical Microbiology, Amsterdam
· I.T.M.A. Overdevest, Eurofins PAMM, Department of Medical Microbiology, Veldhoven
· A. van Dam, Public Health Service, Public Health Laboratory, Amsterdam
· I. Maat, Radboud University Medical Center, Department of Medical Microbiology, Nijmegen
· B. Maraha, Albert Schweitzer Hospital, Department of Medical Microbiology, Dordrecht
· J.C. Sinnige, Regional Laboratory of Public Health, Department of Medical Microbiology, Haarlem
· E.E. Mattsson, Reinier de Graaf Groep, Department of Medical Microbiology, Delft
· N. van Maarseveen, Saltro Diagnostic Centre, Department of Medical Microbiology, Utrecht
· E. de Jong, Slingeland Hospital, Department of Medical Microbiology, Doetinchem
· S.J. Vainio, St Antonius Hospital, Department of Medical Microbiology and Immunology, Nieuwegein
· E. Heikens, St Jansdal Hospital, Department of Medical Microbiology, Harderwijk
· R. Steingrover, St. Maarten Laboratory Services, Department of Medical Microbiology, Cay Hill (St. Maarten)
· A. Troelstra, University Medical Center Utrecht, Department of Medical Microbiology, Utrecht
· E. Bathoorn, University of Groningen, Department of Medical Microbiology, Groningen
· J. de Vries, VieCuri Medical Center, Department of Medical Microbiology, Venlo
· D.W. van Dam, Zuyderland Medical Centre, Department of Medical Microbiology and Infection Control, Sittard-Geleen
· E.I.G.B. de Brauwer, Zuyderland Medical Centre, Department of Medical Microbiology and Infection Control, Heerlen
· NN, Analytical Diagnostic Center N.V. Curaçao, Department of Medical Microbiology, Willemstad (Curaçao)
· H. Berkhout, Canisius Wilhelmina Hospital, Department of Medical Microbiology and Infectious Diseases, Nijmegen
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: Conceptualisation, RDZ, CCHW, DWN, NJV, AFS, CS-vdL, EJK, SCG and APAH; data curation, SW, AH, JeB and JaB; processing of NGS data, SW, VAG, and AH; formal analysis, RDZ, CCHW and APAH; visualisation, RDZ, CCHW and APAH; funding, not applicable; sample collection, Dutch CPE and MRSA surveillance study Groups; laboratory experiments, JeB and AH; supervision, EJK, SCG and APAH; manuscript preparation – original draft, RDZ, CCHW and APAH; review and editing, all authors; review and approval of final manuscript, all authors.
References
- 1.de Greeff SC, Kolwijck E, Schoffelen AF, Verduin CM. NethMap 2022. Consumption of antimicrobial agents and antimicrobial resistance among medically important bacteria in the Netherlands in 2021 / MARAN 2022. Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2021. Bilthoven: Rijksinstituut voor Volksgezondheid en Milieu; 2022. Available from: https://www.rivm.nl/publicaties/nethmap-2022-consumption-of-antimicrobial-agents [Google Scholar]
- 2.van der Zwaluw K, Witteveen S, Wielders L, van Santen M, Landman F, de Haan A, et al. Molecular characteristics of carbapenemase-producing Enterobacterales in the Netherlands; results of the 2014-2018 national laboratory surveillance. Clin Microbiol Infect. 2020;26(10):1412.e7-12. 10.1016/j.cmi.2020.01.027 [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Regional Office for Europe (WHO/Europe)/European Centre for Disease Prevention and Control (ECDC). Antimicrobial resistance surveillance in Europe 2022 – 2020 data. Copenhagen: WHO/Europe; 2022. Available from: https://apps.who.int/iris/handle/10665/351141. License: CC BY-NC-SA 3.0 IGO
- 4.Wielders CCH, Schouls LM, Woudt SHS, Notermans DW, Hendrickx APA, Bakker J, et al. Epidemiology of carbapenem-resistant and carbapenemase-producing Enterobacterales in the Netherlands 2017-2019. Antimicrob Resist Infect Control. 2022;11(1):57. 10.1186/s13756-022-01097-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Commission (EC). Ukraine: 1,000 Ukrainian patients transferred to European hospitals. Press release. Brussels: EC; 2022. Available from: https://ec.europa.eu/commission/presscorner/detail/en/IP_22_4882
- 6.European Centre for Disease Prevention and Control (ECDC). Operational public health considerations for the prevention and control of infectious diseases in the context of Russia’s aggression towards Ukraine. Stockholm: ECDC; 2022. Available from: https://www.ecdc.europa.eu/en/publications-data/operational-public-health-considerations-prevention-and-control-infectious
- 7.Kondratiuk V, Jones BT, Kovalchuk V, Kovalenko I, Ganiuk V, Kondratiuk O, et al. Phenotypic and genotypic characterization of antibiotic resistance in military hospital-associated bacteria from war injuries in the Eastern Ukraine conflict between 2014 and 2020. J Hosp Infect. 2021;112:69-76. 10.1016/j.jhin.2021.03.020 [DOI] [PubMed] [Google Scholar]
- 8.Salmanov A, Shchehlov D, Svyrydiuk O, Bortnik I, Mamonova M, Korniyenko S, et al. Epidemiology of healthcare-associated infections and mechanisms of antimicrobial resistance of responsible pathogens in Ukraine: a multicentre study. J Hosp Infect. 2022;131:129-38. [DOI] [PubMed] [Google Scholar]
- 9.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One. 2015;10(3):e0123690. 10.1371/journal.pone.0123690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrickx APA, Landman F, de Haan A, Witteveen S, van Santen-Verheuvel MG, Schouls LM, et al. blaOXA-48-like genome architecture among carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the Netherlands. Microb Genom. 2021;7(5):000512. 10.1099/mgen.0.000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clinical breakpoints - breakpoints and guidance. Växjö: EUCAST; 2022 Available from: https://www.eucast.org/clinical_breakpoints
- 12.Argimón S, David S, Underwood A, Abrudan M, Wheeler NE, Kekre M, et al. Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin Infect Dis. 2021;73(Suppl_4):S325-35. 10.1093/cid/ciab784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castanheira M, Doyle TB, Collingsworth TD, Sader HS, Mendes RE. Increasing frequency of OXA-48-producing Enterobacterales worldwide and activity of ceftazidime/avibactam, meropenem/vaborbactam and comparators against these isolates. J Antimicrob Chemother. 2021;76(12):3125-34. 10.1093/jac/dkab306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor E, Jauneikaite E, Sriskandan S, Woodford N, Hopkins KL. Detection and characterisation of 16S rRNA methyltransferase-producing Pseudomonas aeruginosa from the UK and Republic of Ireland from 2003-2015. Int J Antimicrob Agents. 2022;59(3):106550. 10.1016/j.ijantimicag.2022.106550 [DOI] [PubMed] [Google Scholar]
- 15.Dadashi M, Yaslianifard S, Hajikhani B, Kabir K, Owlia P, Goudarzi M, et al. Frequency distribution, genotypes and prevalent sequence types of New Delhi metallo-β-lactamase-producing Escherichia coli among clinical isolates around the world: A review. J Glob Antimicrob Resist. 2019;19:284-93. 10.1016/j.jgar.2019.06.008 [DOI] [PubMed] [Google Scholar]
- 16.Pirzadian J, Persoon MC, Severin JA, Klaassen CHW, de Greeff SC, Mennen MG, et al. National surveillance pilot study unveils a multicenter, clonal outbreak of VIM-2-producing Pseudomonas aeruginosa ST111 in the Netherlands between 2015 and 2017. Sci Rep. 2021;11(1):21015. 10.1038/s41598-021-00205-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.