Abstract
In 2022, German surveillance systems observed rapidly increasing numbers of NDM-1- and NDM-1/OXA-48-producing Klebsiella pneumoniae, which may in part reflect recurring pre-pandemic trends. Among these cases, however, a presence in Ukraine before diagnosis was frequently reported. Whole genome sequencing of 200 isolates showed a high prevalence of sequence types ST147, ST307, ST395 and ST23, including clusters corresponding to clonal dissemination and suggesting onward transmission in Germany. Screening and isolation of patients from Ukraine may help avoid onward transmission.
Keywords: Klebsiella pneumoniae, NDM-1, NDM-1/OXA-48, Ukraine, Germany, carbapenemase-producing Enterobacterales, genomic surveillance, antimicrobial resistance
Surveillance systems in Germany revealed an increase in NDM-1 and NDM-1/OXA-48-producing Klebsiella pneumoniae since March 2022. It coincided with the war in Ukraine and the arrival of refugees and evacuated patients to Germany. In a nationwide investigation, we combined epidemiological and genomic analyses to delineate transmission patterns and infer clinical care recommendations.
Epidemiology of carbapenemase-producing Enterobacterales in Germany
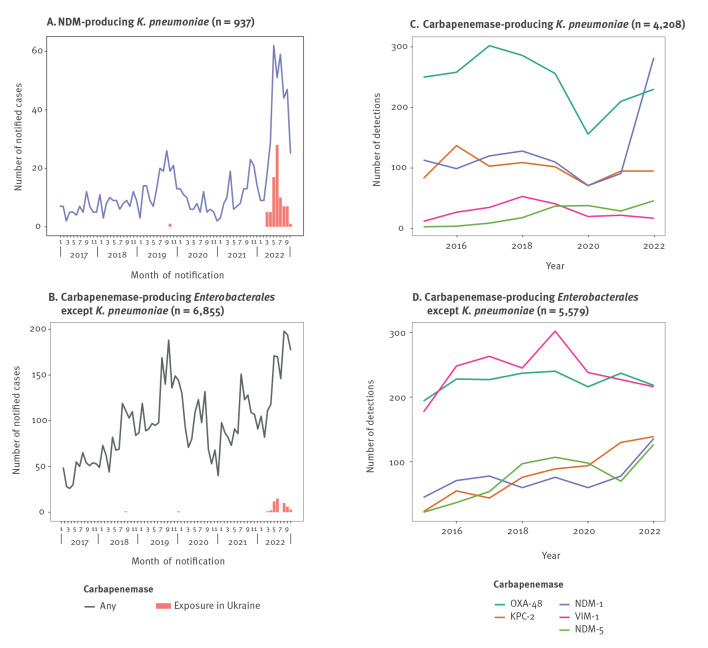
Since 2016, infections or colonisations with carbapenemase-producing Enterobacterales (CPE) have been notifiable in Germany. After a period of reduced incidence since the beginning of the coronavirus disease 2019 (COVID-19) pandemic [1,2], notifications of NDM-producing K. pneumoniae have exceeded pre-pandemic trends since March 2022, in contrast to CPE excluding K. pneumoniae (Figure 1 panels A and B). In the Supplement, part S1, we provide additional detail on the differing trends and corresponding trend analyses.
Figure 1.
Notifications of NDM-producing Klebsiella pneumoniae and other carbapenemase-producing Enterobacterales and carbapenemase detections for multidrug-resistant Gram-negative bacteria, Germany, 2022
Panels A and B: data from the national notifiable diseases surveillance system. Panels C and D: detections at the National Reference Centre.
Particularly for case notifications of NDM-producing K. pneumoniae (Figure 1A), presence in Ukraine before diagnosis was common: 80 of 335 (24%) notifications since calendar week 10/2022, compared with one of 584 (0.2%) between 2017 and 2021. For CPE excluding K. pneumoniae (Figure 1B), respectively 68 of 2,306 (3%) and two of 12,045 (< 0.1%) cases were from Ukraine in these two time periods, (chi-squared test for exposure in Ukraine in 2022 among notifications of NDM-producing K. pneumoniae vs other CPE: p < 0.001). Notifications do not always specify a recorded presence abroad as provenance, hospitalisation or travel. However, it is likely that notified cases since March 2022 with a recorded presence in Ukraine represent predominantly refugees or evacuated patients.
At the same time, the German National Reference Centre (NRC) for multidrug-resistant Gram-negative bacteria detected NDM-1-positive K. pneumoniae isolates in unprecedented numbers compared with other carbapenemases and other years (Figure 1C) [3]. NDM-1-positive isolates of other Enterobacterales increased as well, but to a lesser extent (Figure 1D).
For 75 of 335 (22%) NDM-producing K. pneumoniae notifications since week 10/2022, the carbapenemase was specified as NDM-1. Of the remaining notifications, 250 (75%) were unspecified, eight had NDM-5, one NDM-4 and one NDM-9. Prior presence in Ukraine was reported for 24 of the 75 (32%) NDM-1 notifications (Table). Compared with other exposure locations, cases from Ukraine were younger (p < 0.01) and tended to be more often colonised, detected via screening (p = 0.24). Of all infections, wound infections predominated, in contrast to urinary tract infections among cases with exposure locations elsewhere, suggesting a considerable part of notifications related to evacuated patients [4,5]. Co-production of an OXA-48(-like) carbapenemase was found in 12 of 24 cases from Ukraine. Cases were notified from 11 of 16 German federal states and throughout the weeks 10–43 in 2022.
Table. Demographic and clinical data and sample characteristics for notified Klebsiella pneumoniae with a confirmed NDM-1 carbapenemase, Germany, weeks 10–43/2022 (n = 75).
| Patient information | Total | With reported country location of exposure | |||
|---|---|---|---|---|---|
| n | % | Germany or foreign country other than Ukraine | Ukraine | ||
| All | Co-production of NDM-1 and OXA-48(-like) carbapenemases | ||||
| Sex | 75 | 100 | 19 | 24 | 12 |
| Female | 32 | 43 | 11 | 8 | 4 |
| Male | 42 | 56 | 8 | 15 | 7 |
| Missing | 1 | 1 | 0 | 1 | 1 |
| Median age in years (IQR) | 49 (34–66) | 63 (46–72) | 41 (34–47) | 40 (29–45) | |
| Missing | 0 | 0 | 0 | 0 | |
| Sampling material a | |||||
| Blood/liquor | 7 | 9 | 2 | 1 | 1 |
| Urine | 14 | 19 | 9 | 1 | 0 |
| Wound | 17 | 23 | 1 | 9 | 4 |
| Screening/stool | 31 | 41 | 6 | 11 | 5 |
| Other or missing | 6 | 8 | 1 | 2 | 2 |
| Infections vs colonisationsb | |||||
| Infections | 39 | 52 | 13 | 11 | 5 |
| Colonisations | 34 | 45 | 6 | 13 | 7 |
| Missing | 2 | 3 | 0 | 0 | 0 |
IQR: interquartile range.
a Sampling material was documented as multiple choice but evaluated here by selecting the first material mentioned in the order: blood/liquor, urine, wound, screening/stool.
b Infection vs colonisation was inferred from the sampling material (from blood/liquor/urine/wound vs screening/stool, respectively), and from the classification as in the notification if the information on material was missing.
Whole genome sequencing and cluster detection
Between January and September 2022, the NRC received 330 non-duplicate NDM-1- and NDM-1/OXA-48-producing K. pneumoniae isolates. Of these, 200 isolates, comprising 66 NDM-1/OXA-48-producing and 134 NDM-1-producing K. pneumoniae, were subjected to Illumina (Illumina, San Diego, United States) whole genome sequencing. Sample selection prioritised NDM-1/OXA-48-producing isolates and NDM-1-producing isolates from cases from Ukraine and known hospital origin. The remaining isolates were selected randomly.
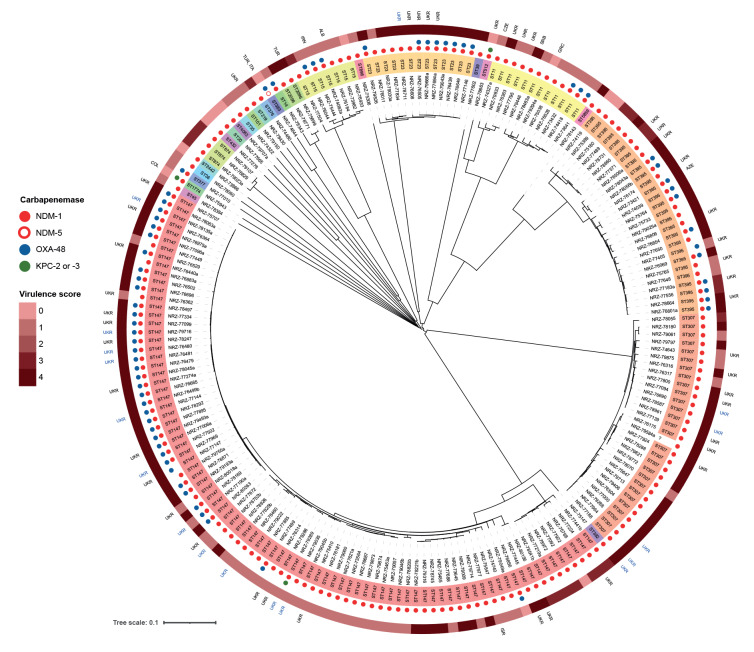
Analyses of de novo assemblies included multilocus sequence typing (MLST) and core genome (cg)MLST using the K. pneumoniae sensu lato scheme (2,358 loci) as implemented in the SeqSphere+ software version 8.4.1 (Ridom, Muenster, Germany). Among the 200 isolates, we identified 27 sequence types (ST) with a predominance of ST147 (88/200; 44%), ST307 (28/200; 14%) and ST395 (27/200; 14%) (Figure 2). Thirteen (7%) isolates belonged to ST23, often associated with hypervirulence [6,7].
Figure 2.
Phylogenetic tree of NDM-1 and NDM-1/OXA-48-producing Klebsiella pneumoniae isolates, Germany, January–September 2022 (n = 200)
We determined pairwise allelic differences between isolates and calculated a neighbour-joining tree using SeqSphere+. Metadata were annotated using Interactive Tree OF Life [20]. Sequence types (ST) are colour-coded. Coloured circles: detected carbapenemases; reddish band: Kleborate-derived virulence score [8]; 3-letter country code: presence abroad before diagnosis, as documented for isolates at the NRC (black) or matched notifications (blue).
OXA-48 was primarily identified among ST147 isolates, but also in other ST. In silico analyses revealed the presence of one or more additional extended-spectrum beta-lactamase (ESBL) genes (blaCTX-M, blaSHV, blaTEM) in almost all isolates (199/200). The detailed results of detected ESBL genes are appended in the Supplement, part S2. Kleborate [8] identified the presence of genes for the siderophores aerobactin and yersiniabactin, resulting in a virulence score of up to 4 throughout the phylogenetic tree. Colibactin-encoding genes were not found, hence, no virulence scoring of 5 was assigned.
Prior presence in Ukraine was known from case information at the NRC or from the notifications matched to isolates for 60 of 200 (30%) isolates and across all major ST: for 29 of 88 ST147, for nine of 27 ST395, for five of 13 ST23 and for 11 of 28 ST307. Overall, the diversity of ST indicates that the observed increase was not due to a single transmission cluster but rather suggests a high incidence of NDM-1 and NDM-1/OXA-48-producing K. pneumoniae in Ukraine.
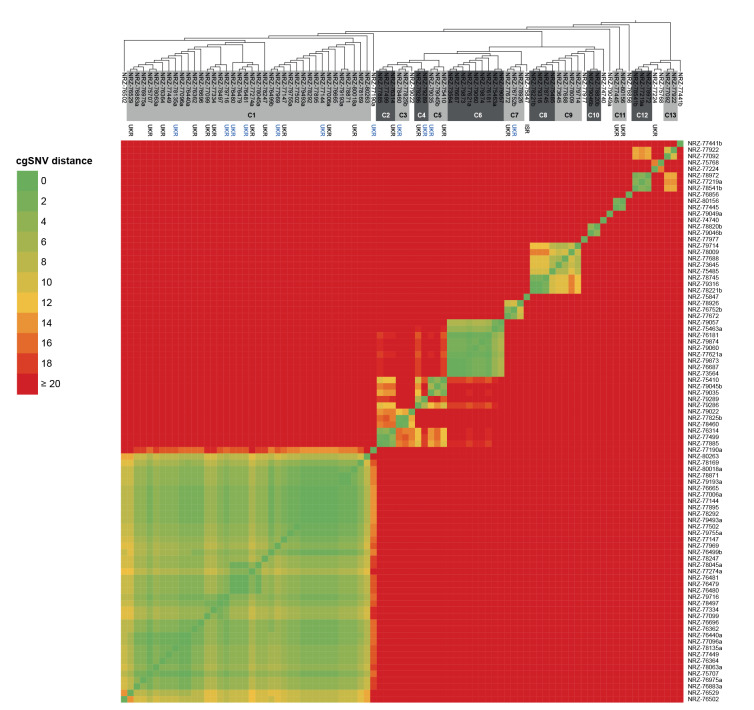
Results of cgMLST also revealed clusters of close genetic relatedness for many of the different ST, particularly among ST147. To further examine genetic relatedness, we performed single nucleotide variant (SNV)-based analyses within genes of the cgMLST scheme, as implemented in SeqSphere+. Because there is no established threshold for clonal relatedness, we performed molecular typing by XbaI-macrorestriction and pulsed-field gel electrophoresis on 14 NDM-1/OXA-48-producing K. pneumoniae ST147 isolates clustering within the phylogenetic tree of the cgMLST analysis. Following defined criteria [9], these isolates were clonally related as revealed by identical banding patterns (data not shown). In the SNV-based analysis, these 14 isolates showed a maximum of 12 cgSNV, which we therefore set as threshold to define clonality. By applying this upper limit, we identified clusters suggestive of clonal dissemination (Figure 3).
Figure 3.
Phylogenetic tree and heatmap based on pairwise single nucleotide variant (cgSNV) differences between isolates of Klebsiella pneumoniae ST147, Germany, January–September 2022 (n = 88)
Isolates of clusters differing by a maximum of 12 cgSNV in grey (see text for details). 3-letter country code: presence abroad before diagnosis, as documented for isolates at the national reference centre (black) or matched notifications (blue).
The largest cluster C1 included 38 isolates, sampled from April to September 2022. Hospital locations in Germany and from where cases were notified scattered nationwide. Prior presence in Ukraine was reported for 17 of the 38 cases, suggesting a cluster of refugees from Ukraine or evacuated patients and reflecting transmission in Ukraine. For two of the 38 cases, with sampling dates in May and September, however, reported exposure locations were exclusively in Germany, indicating potential onward transmission. In contrast, cluster C6 included cases exclusively from two neighbouring hospitals in Germany without reported links to Ukraine, potentially representing transmission within Germany. Further investigations of these clusters have begun to elucidate chains of transmission.
Antimicrobial resistance
In phenotypical tests, nine isolates representative of ST147, ST395, ST307 and ST23 were resistant to meropenem and imipenem, piperacillin/tazobactam, ceftazidime, aztreonam, and to the inhibitor combinations ceftazidime/avibactam, meropenem/vaborbactam and imipenem/relebactam (testing and interpretations was done according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) v12.0 [10]). They were also resistant to fluoroquinolones and cefiderocol (or within the area of technical uncertainty), which in case of cefiderocol has been previously reported for several NDM producers [11,12]. For many isolates, we detected low minimum inhibitory concentration (MIC) of colistin (MIC ≤ 2 mg/L) and tigecycline (MIC ≤ 0.5 mg/L); however, neither of these drugs is recommended in serious infections [13]. Checkerboard titrations with aztreonam (0.5–32 mg/L) and ceftazidime/avibactam (0.25–64 mg/L and 4 mg/L, respectively) were done and the addition of ceftazidime/avibactam to aztreonam resulted in markedly reduced MIC to aztreonam (MIC of aztreonam in combination ≤ 0.5 mg/L), suggesting synergism and a reduction into the therapeutically usable range as reported previously [14,15].
Discussion
Since March 2022, NDM-1-producing K. pneumoniae increased rapidly in Germany and in these cases, presence in Ukraine before diagnosis was frequently reported. Ukraine reported high AMR proportions in Gram-negative bacterial isolates previously (mostly bloodstream infections) to the Central Asian and European Surveillance of Antimicrobial Resistance (CAESAR) network, including K. pneumoniae with resistance to carbapenems in 64% and Acinetobacter spp. with resistance to carbapenems in 73% in 2021 [16]. As a result, the European Centre for Disease Prevention and Control (ECDC) recommended in March 2022 to screen for carriage of multidrug-resistant organisms (MDRO) in patients transferred from hospitals in Ukraine or with a history of hospital admission in Ukraine and to implement multimodal infection prevention and control (IPC) measures [17]. The IPC guidelines in Germany generally recommend screening for MDRO in individuals with reported hospitalisation in countries with high antimicrobial resistance rates, including Ukraine [18,19]. However, the increasing incidence and proportion of NDM-producing K. pneumoniae in cases from Ukraine have not occurred for other CPE, suggesting that the increase does not only stem from intensified screening of patients from Ukraine. Diagnostic laboratories in Germany should voluntarily send carbapenem-resistant isolates to the NRC for carbapenemase specification and notify all cases to the mandatory notification system. But there is a known under-ascertainment in both data sources and so far, isolates and notifications cannot be linked by a common identifier. Linking the information of the two used data sources for cluster analyses as we present here is challenged by the lack of a unique common identifier. We used several variables for matching, which is less reliable.
Whole genome sequencing revealed the predominance of K. pneumoniae sequence types ST147 and ST307, both of which are known to be successful epidemic clones. Although detailed genomic analyses indicate the clonal relatedness of NDM-1 and NDM-1/OXA-48-producing K. pneumoniae ST147 isolates, a comparison with international ST147 isolates will evaluate whether the close genetic relatedness reflects transmissions of strains or simply is a characteristic of the epidemic clone.
Conclusions
NDM-1-producing K. pneumoniae has become more frequent in Germany, particularly in 2022. Our findings suggest an elevated proportion of NDM-1 and NDM-1/OXA-48-producing K. pneumoniae among refugees or evacuated patients from Ukraine, adding to the baseline incidence in Germany. We need to further investigate if high transmission occurs in single or multiple treatment centres in Germany or Ukraine to specifically assess how best to support IPC and outbreak control. Adherence to recommended IPC measures in Germany and other European countries needs to be stressed to prevent onward transmission. The combination of aztreonam with ceftazidime-avibactam may be considered for the treatment of severe infections caused by these K. pneumoniae strains.
Ethical statement
Ethical approval was not necessary for this work as it was performed as part of routine surveillance activities in Germany, in accordance with the German Infection Protection Act.
Funding statement
The work of the NRC was supported by the Robert Koch Institute with funds provided by the German Ministry of Health (grant no. 1369-402).
Acknowledgements
We thank Anke Albrecht, Nadine Frey, Svenja Hirle, Anja Kaminski, Kirsten Krengel, Ulrike Maduch, Marion Schmidt, Laura Suppa, and Joanna Waniczek (NRC) for excellent technical assistance. We thank Sibylle Müller-Bertling and Kirstin Ganske for excellent technical assistance. We thank Navina Sarma for feedback to the manuscript. We thank Andrea Thürmer and the sequencing core facility at the RKI for fast processing of samples and data generation. We thank Ed J. Kuijper and his colleagues for the exchange of findings and trustful communication.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: MS, FR, GW, SG, SH, NP initially conceived the work. MS, FR, AH, SH processed and analysed the notification data. JBH, MAF, MC, JE, YP performed the confirmatory diagnostics and sample preparation. JBH, MAF performed the sequence data analyses. MS, JBH, MAF, FR, MC, JE, YP, TE, GW, SG, SH, NP contributed to the interpretation of the data. MS, JBH, MAF drafted the manuscript. All authors critically revised the manuscript and approved the final version to be published.
References
- 1.Schranz M, Ullrich A, Rexroth U, Hamouda O, Schaade L, Diercke M, et al. Die Auswirkungen der COVID-19-Pandemie und assoziierter Public-Health-Maßnahmen auf andere meldepflichtige Infektionskrankheiten in Deutschland (MW 1/2016 – 32/2020). [The effects of the COVID-19 pandemic and associated public health measures on other notifiable infectious diseases in Germany]. Epid Bull. 2021;7:3-7. German. 10.25646/8011 [DOI]
- 2.Reuss A, Klingeberg A, Schmidt N, Eckmanns T, Zacher B. Einfluss der COVID-19-Pandemie auf die Anzahl der gemäß IfSG meldepflichtigen Nachweise von Erregern mit Antibiotikaresistenzen und C. difficile-Infektionen. [Impact of the COVID-19 pandemic on the number of confirmations of pathogens with antibiotic resistance and C. difficile infections notifiable under the Infection Protection Act]. Epid Bull. 2021;7:8-11. German. 10.25646/8026 [DOI]
- 3.Pfennigwerth N. Schauer J. Bericht des Nationalen Referenzzentrums für gramnegative Krankenhauserreger – Zeitraum 1. Januar 2021 bis 31. Dezember 2021. [Report of the National Reference Centre for Gram-negative hospital pathogens - period 1 January 2021 to 31 December 2021]. Epid Bull. 2022;19:3-9. German. Available from: http://edoc.rki.de/176904/9709
- 4.Kondratiuk V, Jones BT, Kovalchuk V, Kovalenko I, Ganiuk V, Kondratiuk O, et al. Phenotypic and genotypic characterization of antibiotic resistance in military hospital-associated bacteria from war injuries in the Eastern Ukraine conflict between 2014 and 2020. J Hosp Infect. 2021;112:69-76. 10.1016/j.jhin.2021.03.020 [DOI] [PubMed] [Google Scholar]
- 5.Dietze N, Trawinski H, Schönherr SG, Lippmann N, Ranft D, Fichtner F, et al. Infektionsmedizinische und chirurgische Herausforderungen durch Carbapenem-resistente bakterielle Erreger bei der Versorgung Kriegsverletzter aus der Ukraine. [Challenges in infection medicine and surgery caused by carbapenem-resistant bacterial pathogens in the care for war casualties from Ukraine]. Epid Bull. 2022;36:3-10. German. Available from: http://edoc.rki.de/176904/10214
- 6.European Centre for Disease Prevention and Control (ECDC). Risk assessment: Emergence of hypervirulent Klebsiella pneumoniae ST23 carrying carbapenemase genes in EU/EEA countries. Stockholm: ECDC; 2022. Available from: https://www.ecdc.europa.eu/en/ publications-data/risk-assessment-emergence-hypervirulent-klebsiella-pneumoniae-eu-eea
- 7.Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan YH, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):2703. 10.1038/s41467-018-05114-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(1):4188. 10.1038/s41467-021-24448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233-9. 10.1128/jcm.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters, Version 12.0. Växjö: EUCAST; 2022. Available from: https://www. eucast.org/clinical_breakpoints
- 11.Oueslati S, Bogaerts P, Dortet L, Bernabeu S, Ben Lakhal H, Longshaw C, et al. In vitro activity of cefiderocol and comparators against carbapenem-resistant Gram-negative pathogens from France and Belgium. Antibiotics (Basel). 2022;11(10):1352. 10.3390/antibiotics11101352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isler B, Aslan AT, Akova M, Harris P, Paterson DL. Treatment strategies for OXA-48-like and NDM producing Klebsiella pneumoniae infections. Expert Rev Anti Infect Ther. 2022;20(11):1389-400. 10.1080/14787210.2022.2128764 [DOI] [PubMed] [Google Scholar]
- 13.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST guidance documents. Växjö: EUCAST; 2022. Available from: https://www.eucast.org/eucastguidancedocuments
- 14.Falcone M, Daikos GL, Tiseo G, Bassoulis D, Giordano C, Galfo V, et al. Efficacy of ceftazidime-avibactam plus aztreonam in patients with bloodstream infections caused by metallo-β-lactamase-producing Enterobacterales. Clin Infect Dis. 2021;72(11):1871-8. 10.1093/cid/ciaa586 [DOI] [PubMed] [Google Scholar]
- 15.Shaw E, Rombauts A, Tubau F, Padullés A, Càmara J, Lozano T, et al. Clinical outcomes after combination treatment with ceftazidime/avibactam and aztreonam for NDM-1/OXA-48/CTX-M-15-producing Klebsiella pneumoniae infection. J Antimicrob Chemother. 2018;73(4):1104-6. 10.1093/jac/dkx496 [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization Regional Office for Europe (WHO/Europe). Antimicrobial Resistance Map. Acinetobacter spp. Aminoglycosides. Copenhagen: WHO/Europe. [Accessed:13 Dec 2022]. Available from: https://worldhealthorg.shinyapps.io/WHO-AMR-Dashboard/?_ga=2.172166563.1827800992.1668654440-1324205868.1668654440
- 17.European Centre for Disease Prevention and Control (ECDC). Operational public health considerations for the prevention and control of infectious diseases in the context of Russia’s aggression towards Ukraine. Stockholm: ECDC; 2022. Available from: https://www.ecdc.europa.eu/en/publications-data/operational-public-health-considerations-prevention-and-control-infectious
- 18.Kommission für Krankenhaushygiene und Infektionsprävention (KRINKO) . (Hygiene measures for infection or colonization with multidrug-resistant gram-negative bacilli. Commission recommendation for hospital hygiene and infection prevention (KRINKO) at the Robert Koch Institute (RKI)). Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012;55(10):1311-54. [DOI] [PubMed] [Google Scholar]
- 19.Kommission für Krankenhaushygiene und Infektionsprävention (KRINKO). Ergänzung zu den “Hygienemaßnahmen bei Infektionen oder Besiedlung mit multiresistenten gramnegativen Stäbchen” (2012) im Rahmen der Anpassung an die epidemiologische Situation. [Supplement to the " Hygiene measures for infection or colonization with multidrug-resistant gram-negative bacilli" (2012) as part of the adaptation to the epidemiological situation]. Epid Bull. 2014;21:183-4. German. Available from: https://www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2014/Ausgaben/21_14.pdf?__blob=publicationFile
- 20.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293-6. 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.