Abstract
The EGFR/RAS/MEK/ERK signaling pathway (ERK/MAPK) is hyperactivated in most colorectal cancers. A current limitation of inhibitors of this pathway is that they primarily induce cytostatic effects in colorectal cancer cells. Nevertheless, these drugs do induce expression of proapoptotic factors, suggesting they may prime colorectal cancer cells to undergo apoptosis. As histone deacetylase inhibitors (HDACis) induce expression of multiple proapoptotic proteins, we examined whether they could synergize with ERK/MAPK inhibitors to trigger colorectal cancer cell apoptosis. Combined MEK/ERK and HDAC inhibition synergistically induced apoptosis in colorectal cancer cell lines and patient-derived tumor organoids in vitro, and attenuated Apc-initiated adenoma formation in vivo. Mechanistically, combined MAPK/HDAC inhibition enhanced expression of the BH3-only proapoptotic proteins BIM and BMF, and their knockdown significantly attenuated MAPK/HDAC inhibitor–induced apoptosis. Importantly, we demonstrate that the paradigm of combined MAPK/HDAC inhibitor treatment to induce apoptosis can be tailored to specific MAPK genotypes in colorectal cancers, by combining an HDAC inhibitor with either an EGFR, KRASG12C or BRAFV600 inhibitor in KRAS/BRAFWT; KRASG12C, BRAFV600E colorectal cancer cell lines, respectively. These findings identify a series of ERK/MAPK genotype-tailored treatment strategies that can readily undergo clinical testing for the treatment of colorectal cancer.
Introduction
The ERK/MAPK signaling pathway is constitutively activated in a high percentage of colorectal cancers. This is driven by activating mutations in KRAS, NRAS, or BRAF that collectively occur in approximately 55% of cases (1) or as a result of upregulated canonical activation of receptor tyrosine kinases such as EGFR (2).
Activation of ERK/MAPK signaling in cancer cells drives cell proliferation and is associated with loss of cell differentiation and promotion of cell survival (3). The prosurvival effects of the pathway are illustrated by the transcriptional repression of proapoptotic BH3-only proteins such as BIM (4) and PUMA (5). ERK-mediated phosphorylation of BIMEL also promotes its ubiquitination and degradation (6–8), as well as its dissociation from the prosurvival proteins BCL-XL, BCL-2, and MCL1 (8). ERK also phosphorylates BMF, which reduces BMF-dependent cell-killing activity (9) and induces RSK-mediated phosphorylation of BAD, which sequesters BAD to the cytosol (10, 11). Finally, ERK signaling promotes expression of the prosurvival protein MCL1 through ERK-mediated phosphorylation of Thr163 promoting its stabilization (12). Conversely, ERK/MAPK inhibitors (MAPKis) induce the opposite effect in cancer cell lines, increasing expression of BIM and BMF and downregulating MCL1 (13–15).
Despite these effects, preclinical studies have shown that ERK/MAPK signaling inhibitors (ERK/MAPKis) primarily induce cytostatic effects in tumor cells, including colorectal cancer cells (16, 17). This is also supported by clinical observations where agents such as cetuximab and panitumumab that inhibit ERK/MAPK signaling by inhibiting EGFR, or BRAF inhibitors, both increase progression-free survival in patients despite relatively low objective response rates (18, 19). Collectively, these findings suggest that ERK/MAPK inhibitors may be unable to alter the apoptotic “rheostat” to sufficient levels needed to trigger apoptosis but may induce a “primed” apoptotic state in tumor cells, which may be exploited through combination drug treatments.
Histone deacetylase inhibitors (HDACis) are a class of epigenetic modulators approved for treatment of the hematological malignancies cutaneous T-cell lymphoma and multiple myeloma. Several HDACis are in routine clinical use, including vorinostat (Zolinza, Merck), panobinostat (Farydak, Novartis), and romidepsin (Istodax, Celgene). Although HDACis induce multiple effects on tumor cells, preclinical studies indicate that their antitumor activity is predominantly driven through the induction of apoptosis (20). Depending on the cell type, these proapoptotic effects are triggered, at least in part, through the transcriptional activation of proapoptotic mediators, including BAD (21), BIK (21), NOXA (21), BMF (22–24), BIM (25), and BAK (26) as well as the repression of prosurvival proteins, including BCL-XL (21, 27), BFL-1 (21), BCL-w (21), and BCL-2 (28).
Although HDACis have shown limited single-agent activity in solid tumors, their unique mechanism of action makes them well suited for use in rational combination regimens and represents a potential class of drugs that may enhance the apoptotic activity of MAPK pathway inhibitors.
We therefore sought to investigate whether HDACis could enhance the apoptotic activity of MAPK pathway inhibitors in colorectal cancer cells and elucidate the underlying mechanism of action. In addition, we sought to determine whether this paradigm could be tailored to target colorectal cancers with different MAPK pathway lesions, including KRAS G12C and BRAF mutant tumors, as well as colorectal cancers in which MAPK signaling is driven by EGFR activation.
Materials and Methods
Cell culture
The following source of the cell lines were COLO 201, HT29, HCT 116, SW837, SW620, and SW1463 (ATCC); LIM2551, LIM1215, and LIM2099 (Ludwig Institute for Cancer Research, Melbourne, Australia), GEO (29), DIFI (30), and V9P (31). All cell lines were maintained in DMEM/F12 supplemented with 5% FBS (v/v; all from Thermo Fisher Scientific) at 37°C with 5% CO2. Cell lines were authenticated by short tandem repeat profiling using the GenePrint 10 system (Promega), and all were found to be exact matches to published profiles. All cell lines were frozen down as large batches of master stocks within five passages of their purchase from these commercial vendors and confirmed to be Mycoplasma negative at the point of freezing. Experiments were then performed using these master stocks for up to 20 passages. Mycoplasma testing was performed every 3 to 6 months as part of routine monitoring in our laboratory.
Generation of BIM knockout cells using CRISPR-Cas9
BIM knockout colorectal cancer cell lines were generated using a previously described doxycycline-inducible CRISPR-Cas9 lentiviral platform with the following sgRNA sequence: hBim exon 3: 5′-GCCCAAGAGTTGCGGCGTAT (32). Independent lentiviruses-expressing Cas9-mCherry and hBIM-guide RNA-GFP were generated by transfecting lentiviral components into 293T packaging cells and transduced into COLO 201 colorectal cancer cells. GPF/mCherry double-positive cells were subsequently sorted using a BD FACSAria II (BD Biosciences). Once expanded, expression of the BIM sgRNA was induced by treatment with 1-μg doxycycline-hyclate (Sigma-Aldrich), and BIM knockout was confirmed by Western blot.
Chemicals
Trametinib, panobinostat, vorinostat, mocetinostat (33), Entinostat (34), romidepsin, and vemurafenib (Selleck Chemicals), AMG 510 (Active Biochem; refs. 35, 36), and Q-VD-OPh (MP Biomedicals) were dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich). Valproic acid (Sigma-Aldrich), dissolved in water. Cetuximab (Erbitux) was obtained from the Austin Health Pharmacy.
Cell viability
Cells were seeded in 96-well plates at densities optimized to ensure control cells did not reach confluence during the experimental period. Cell viability was determined using the MTS assay (Promega), by incubating cells for 90 minutes at 37°C and measuring absorbance at 490 nm (MTS) and 630 nm (background absorbance) on a SPECTROstar Nano Spectrophotometer (BMG LabTech).
Colony formation assays
Cells were seeded at 500 cells per well in 6-well plates and treated with drug the following day. Colonies were then allowed to form over approximately 2 weeks, at which point cells were fixed with 10% formalin for 5 minutes and stained with 0.1% crystal violet solution (Sigma-Aldrich) for 15 minutes at room temperature, washed twice using PBS and allowed to air dry.
Flow cytometry
Cells were seeded in 24-well plates and treated with drug for 24 to 72 hours. Adherent and nonadherent cells were then collected and incubated in 50 μg/mL propidium iodide (Sigma-Aldrich), 0.1% sodium citrate (w/v), and 0.1% Triton X-100 (v/v; Sigma-Aldrich) overnight at 4°C. Samples were then analyzed on an FACS Canto II flow cytometer (BD Biosciences), by analysis of 10,000 events. Apoptotic cells were defined as those with a subdiploid content and quantified using FlowJo V8.0 (FlowJo LLC).
Cytochrome c release
To assess cytochrome c release from mitochondria, cells were permeabilized in digitonin lysis buffer (20 mmol/L HEPES pH 7.2, 100 mmol/L KCl, 5 mmol/L MgCl2, 1 mmol/L EDTA, 1 mmol/L EGTA, 250 mmol/L sucrose, 0.05% (w/v) digitonin (Calbiochem), and one tablet of Roche cOmplete protease inhibitor cocktail (Roche) for 10 minutes on ice. Cell membrane permeabilization was verified by trypan blue uptake and the cells were centrifuged at 13,000 rpm for 5 minutes. The supernatant containing the soluble fraction was retained, and the pellet was lysed further using NP-40 lysis buffer (50 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mmol/L EDTA pH 8, one tablet of protease inhibitor cocktail Roche cOmplete (Roche), and one tablet of phosphatase inhibitor PhosSTOP (Roche) for 1 hour on ice. Proteins from both the soluble and pelleted fractions were resolved by Western blot analysis and probed with anti-cytochrome c (556433, BD Pharmingen).
Patient-derived tumor organoids
Patient-derived tumor organoids (PDTOs) were generated from resected tumor material from deidentified patients with colorectal cancer. All patients provided signed informed consent and the study was approved by a multi-institutional Human Research Ethics Committee (HREC 2016.249) and was conducted in accordance with the Declaration of Helsinki. Organoids were resuspended in DMEM/F-12 with Matrigel (Corning) as a single-cell suspension and seeded at 3,000 cells per well into an Optical 384-well plate (Nunc) and allowed to establish for 72 hours before the addition of drug. Drug combinations were prepared with 5 μg/mL of propidium iodide (Sigma-Aldrich) in a matrix formation using panobinostat (1, 5, 10, 25, and 50 nmol/L) and trametinib (1, 3, 5, 10, and 25 nmol/L). Organoids were imaged on a Nikon Ti2 Eclipse fluorescent microscope (Nikon, Tokyo, Japan) at ×4 magnifications at a constant temperature and CO2 levels throughout the duration of the experiment. Images were acquired at 72 hours, with a z-stack of 21 slices at 25-μm intervals. At endpoint, viability was determined using the CellTiter-Glo 3D (Promega) viability assay and luminescence read on the EnVision Plate Reader (PerkinElmer). Fluorescence activity data acquired from the Envision Plate Reader were transformed into relative activity, with DMSO as negative (vehicle) control and 1 μmol/L bortezomib as a positive (killing) control.
Apclox/lox;Cdx2-CreERT2mice
Apclox/lox;Cdx2-CreERT2mice were generated as originally described (37). Cre-recombinase–mediated Apc gene inactivation was induced in 5-week-old mice by intraperitoneal injection of 1 mg (100 μL of 10 mg/mL solution) of tamoxifen for 2 consecutive days. Mice were then left for 1 week to allow for adenoma initiation after which drug treatment was started. Mice were then randomized into four groups to receive either vehicle control, trametinib (0.3 mg/kg, oral gavage), panobinostat (5 mg/kg, intraperitoneal injection), or trametinib 0.3 mg/kg and panobinostat 5 mg/kg. Mice were treated with drug for 5 consecutive days followed by 2 days of rest and received a total of 12 treatments. The study was approved by the Austin Health Animal Ethics Committee.
Endoscopy procedure
Mice were anesthetized using (1 L O2/min and 2%–3% Isoflurane). The procedure was performed using a miniature endoscope (scope 1.9 mm outer diameter), a light source, an IMAGE 1 camera, and an air pump (all from Karl Storz, Germany), to achieve regulated inflation of the mouse colon.
Xenograft studies
For xenograft studies, 6-weeks-old female BALB/c-Foxn1nu/ARC (BALB/c nude) mice were purchased from the Animal Resources Center (Western Australia, Australia) and housed in specific pathogen free microisolators. Mice were then subcutaneously injected with 2 million LIM1215 cells into the left and right flanks in a 1:1 mixture of Matrigel matrix (75 μL; Corning): DMEM-F12 (75 μL). Once tumors became palpable, mice were randomized to receive either; vehicle control, cetuximab (40 mg/kg, intraperitoneal injection), panobinostat (5 mg/kg, intraperitoneal injection) or cetuximab 40 mg/kg and panobinostat 5 mg/kg, for five consecutive days for a total of 10 treatments. The study was approved by the Austin Health Animal Ethics Committee.
IHC
Formalin-fixed paraffin-embedded sections were de-paraffinized and rehydrated through serial washes in xylene and ethanol. Sections were rinsed in H2O and quenched in 3% H2O2 (Chem-supply) for 10 minutes. Antigen retrieval was performed by incubation in Citrate buffer (pH 6.0) in a boiling water bath for 30 minutes. Slides were probed with β-catenin (C19220, BD Transduction Laboratories) and phosho-p44/42 MAPK (ERK1/2) Thr202/Thr204 (4370, Cell Signaling Technology) at 4°C overnight, then washed and incubated with Labeled polymer HRP–anti-Rabbit and Labeled polymer HRP–anti-mouse secondary antibody (Dako) for 1 hour at room temperature. Chromagen was developed using the DAB (3, 3-diaminobenzidine) reagent (Dako). Sections were counter-stained using pre-filtered Mayer's hematoxylin (Amber Scientific) then dehydrated through serial ethanol and xylene washes before mounting using DPX mounting solution (Sigma-Aldrich).
Western blot analysis
Protein isolation was performed using NP-40 lysis buffer (50 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mmol/L EDTA pH 8 and 1 tablet of protease inhibitor cocktail Roche cOmplete (Roche) and 1 tablet of phosphatase inhibitor PhosSTOP (Roche). A total of 30 μg of protein per sample was separated on NuPAGE 4% to 12% Bis-Tris precast polyacrylamide gels (Novex, Thermo Fisher Scientific), transferred onto iBLOT2 polyvinylidene difluoride membranes (Invitrogen, Thermo Fisher Scientific) then blocked using Odyssey blocking buffer (LiCor). Primary antibodies used in Western blot analysis were cleaved caspase-3 (Asp175 No. 9661, Cell Signaling, Technology), BIMS/EL/L (ALX-804–527, Enzo Life Sciences), BMF (ALX-804–343-C100, Enzo Life Sciences), MCL-1 (5453S, Cell Signaling Technology), phosho-p44/42 MAPK (ERK1/2) Thr202/Thr204 (4370, Cell Signaling Technology), p44/42 MAPK (ERK1/2; 9107, Cell Signaling Technology), β-actin (A5316, Sigma-Aldrich), and β-tubulin (ab6046, Abcam). Secondary antibodies used include IRDye 680RD Goat anti-mouse IgG (H+L), IRDye800CW Goat anti-rabbit IgG (H+L), and IRDye680RD Goat anti-rat IgG (H+L; LiCor). Signal was detected using an Odyssey imaging system (LiCor).
qRT-PCR
RNA was isolated using the ReliaPrep RNA tissue miniprep system (Promega) as per the manufacturer's protocol. cDNA was synthesized from 1 μg of RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific) as per the manufacturer's protocol. Quantitative RT-PCR was performed using SYBR green master mix (Applied Biosystems) on the Viia 7 Real-Time PCR system. A list of primers used is listed in Supplementary Table S1.
siRNA-mediated knockdown
Knockdown experiments were performed by transfecting cells when 60% confluent with a mixture of Lipofectamine RNAiMax (Invitrogen, Thermo Fisher Scientific) and 50 nmol/L Non-Targeting or BMF-targeting siRNAs (Bioneer). Cells were transfected for 24 hours before drug treatment. BMF targeting siRNAs comprised the following sequences siBMF No. 1 (5′-UAGUGAAGCUGCUAUCCU) and siBMF No. 2 (5′-UCUAAAAGUCACUUAGCU).
Statistical analysis
Statistical analysis was performed using GraphPad Prism v8.0 software (GraphPad Software), whereby experiments containing more than two groups were analyzed using one-way ANOVA, with Tukey's multiple comparison testing. Experiments containing comparison between two groups were analyzed using the Student t test with Welch's correction. In all cases, a P value of <0.05 was considered statistically significant. Synergy was analyzed using two separate models: Bliss Excess and the Highest Single Agent (HSA). All calculations and specific functions were performed in R Version 3.0.1 (R Core Team, 2012. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3–298 900051–07–0, URL http://www.R-project.org/).
Data availability statement
Raw data for this study were generated at the Olivia Newton-John Cancer Research Institute, Melbourne, Australia and Walter Eliza Hall Institute of Medical Research, Melbourne, Australia. Derived data supporting the findings of this study are available from the corresponding author upon request.
Results
Combined MEK and HDAC inhibition synergistically induces apoptosis in colorectal cancer cell lines with different MAPK pathway lesions and in PDTOs
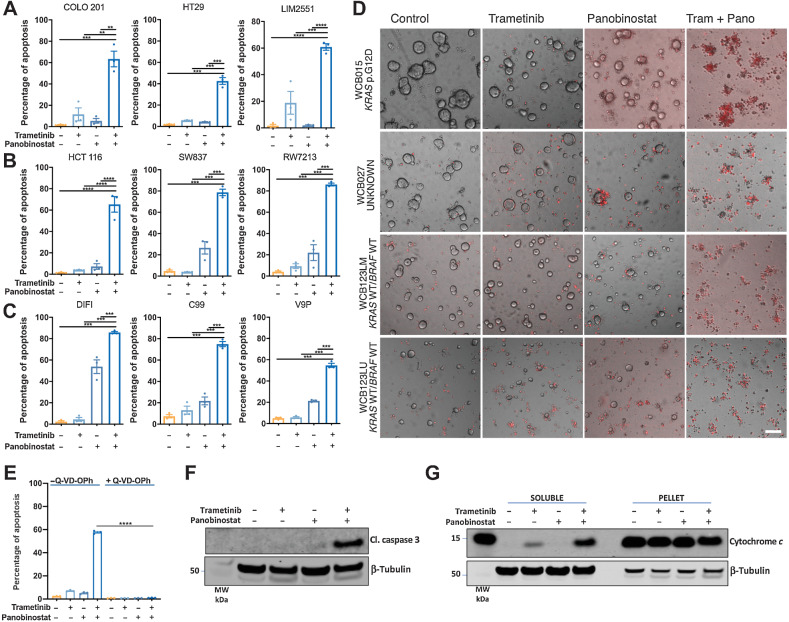
To assess the potential of combinatorial inhibition of ERK/MAPK signaling and HDAC activity as a therapeutic strategy in colorectal cancer, we treated nine colorectal cancer cell lines harboring different MAPK pathway genotypes with the MEK inhibitor (MEKi) trametinib, and the HDACi panobinostat, for 72 hours. The combination treatment induced significantly higher apoptosis compared with either agent alone in all cell lines (Fig. 1), including BRAF mutant (Fig. 1A), KRAS mutant (Fig. 1B), and KRAS/BRAF wild-type lines that also included EGFR-amplified DIFI cells (Fig. 1C). To confirm these findings in an independent model system, we tested the effects of this combination in four PDTOs of different MAPK genotypes, three of which were derived from metastatic lesions and one from a stage II tumor. As observed in colorectal cancer cell lines, the MEK/HDAC inhibitor combination induced extensive apoptosis as assessed by the uptake of propidium iodide and membrane blebbing, a key characteristic of apoptosis (Fig. 1D).
Figure 1.
Combined MEK/HDAC inhibition induces apoptosis and reduces viability of colorectal cancer cells and patient-derived colorectal tumor organoids. A,BRAFV600E mutant, (B) KRAS mutant, and (C) KRAS/BRAF wild-type colorectal cancer cell lines were treated with trametinib (10 nmol/L) and panobinostat (25 nmol/L) alone or in combination for 72 hours, and apoptosis induction was determined by propidium iodide staining and FACS analysis. The values shown are mean ± SEM from three independent experiments. Differences were compared using a one-way ANOVA, with Tukey's multiple comparison testing; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001. D, Effect of trametinib/panobinostat combination treatment on apoptosis in patient-derived colorectal tumor organoids (PDTOs). Representative images of propidium iodide–stained PDTOs treated with trametinib (10 nmol/L) and panobinostat (25 nmol/L) alone and in combination for 72 hours. Images were obtained at ×4 magnifications; scale bar, 100 μm. Cells that have incorporated propidium iodide are stained red. E, COLO 201 cells treated with trametinib (10 nmol/L) and panobinostat (25 nmol/L) alone and in combination in the presence or absence of the pan-caspase inhibitor Q-VD-OPh (10 μmol/L) for 24 hours, and apoptosis determined by PI staining and FACS analysis. The values shown are mean ± SEM from a single experiment performed in technical triplicate. Differences were compared using a one-way ANOVA, with Tukey's multiple comparison testing; ****, P ≤ 0.0001. Western blot analysis for (F) cleaved caspase 3 expression and (G) cytochrome c release in COLO 201 cells treated with trametinib (10 nmol/L) and panobinostat (25 nmol/L) for 24 hours. β-Tubulin was used as a loading control.
To determine whether the effect of the MEK/HDAC inhibitor combination was synergistic, three cell lines with different MAPK pathway genotypes (COLO 201, DIFI, and HCT116) as well the four PDTOs were treated with a 5×5 concentration matrix of trametinib and panobinostat, and synergy assessed by MTS and MTT assays, respectively, using the bliss independence and HSA excess synergy models. Synergistic activity was confirmed in all cases (Supplementary Fig. S1). In addition, combining trametinib with panobinostat also inhibited the clonogenic capacity of HCT 116 cells (Supplementary Fig. S2).
To explore the mechanism of MEK/HDAC inhibitor-induced cell death, COLO 201 cells were treated with the drug combination in the presence of the pan-caspase inhibitor Q-VD-OPh. Drug-induced cell death was significantly attenuated by Q-VD-OPh, indicating that apoptosis induction was caspase-dependent (Fig. 1E). This was further confirmed by the robust induction of cleaved caspase 3 and cytochrome c release following MEK/HDAC inhibitor treatment (Fig. 1F and G).
As panobinostat belongs to the hydroxamic acid class of HDACis, we next assessed whether the synergistic apoptotic activity of the MEK/HDAC inhibitor combination extended to other structural classes of HDAC inhibitors. As with panobinostat, combining trametinib with another clinically used hydroxamic acid vorinostat, the cyclic tetrapeptide romidepsin, the benzamides mocetinostat or entinostat, or the short-chain fatty acid valproic acid, all significantly enhanced apoptosis compared with either agent alone (Supplementary Fig. S3).
Combined MEK/HDAC inhibition attenuates apc-initiated colon adenoma development
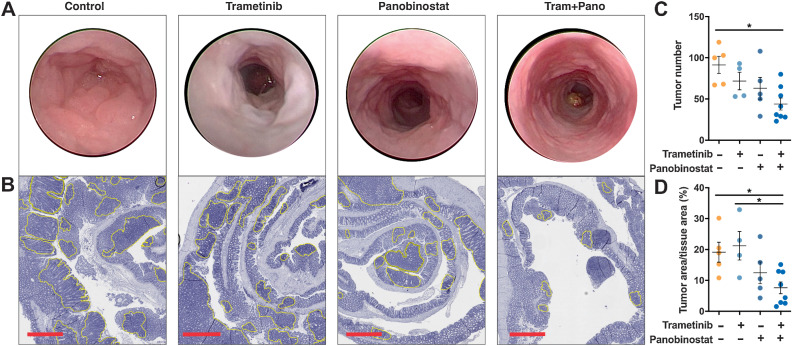
To assess the efficacy of combined MEK/HDAC inhibition in vivo as well as in earlier-stage disease, we next tested the drug combination in Apclox/lox;Cdx2-CreERT2 mice, which develop multiple colonic adenomas approximately 30 days after tamoxifen-induced inactivation of the Apc tumor-suppressor gene. Importantly, immunohistochemical staining for pERK revealed that the ERK/MAPK pathway was robustly induced in these adenomas (Supplementary Fig. S4). To test the activity of MEK/HDAC inhibition in this model, tamoxifen-induced Apclox/lox;Cdx2-CreERT2 mice were treated with vehicle, trametinib, panobinostat or the combination for 2 weeks. Endoscopic assessment of adenoma development on the day following the final drug treatment revealed fewer adenomas in mice treated with trametinib or panobinostat alone compared with control, and a further reduction in mice treated with the combination (Fig. 2A). To quantify this effect, an unbiased macroscopic count of adenomas was performed at cull that confirmed that tumor number and size was lowest in mice treated with the MEKi/HDACi combination (Fig. 2B–D). Importantly, although single-agent treatments resulted in an approximately 10% loss of overall body weight, the combination did not exacerbate these effects, indicating the tolerability of this treatment combination (Supplementary Fig. S5A).
Figure 2.
Combined MEK/HDAC inhibition attenuates colonic adenoma formation. A, Representative endoscopy images of control and drug-treated mice obtained immediately before the experimental endpoint. Mice were treated with trametinib (0.3 mg/kg), panobinostat (5 mg/kg) or the combination for five consecutive days followed by 2 days of rest and received a total of 12 treatments. B, Representative hematoxylin and eosin (H&E) stains of the entire colon from mice treated with vehicle control, trametinib, panobinostat or trametinib plus panobinostat. Yellow outlining indicates tumors; scale bar, 900 μm. C, Tumor number obtained from macroscopic counting of adenomas in resected colons. D, Tumor burden as determined by computing total tumor area relative to total tissue area from H&E-stained sections of the entire colon, from vehicle control (n = 5), trametinib (n = 4), panobinostat (n = 5), and combination (n = 8) treated mice. The values shown are mean ± SEM from the biological replicates. One-way ANOVA, with Tukey's multiple comparison testing; *, P ≤ 0.05.
Combined MEK/HDAC inhibition induces expression of the proapoptotic BH3-only proteins BMF and BIM
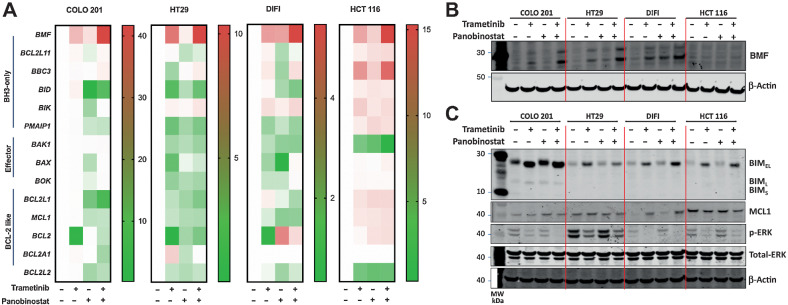
MAPKis and HDACis have been independently shown to alter expression of multiple components of the intrinsic apoptotic pathway (21, 38). We therefore sought to identify pro- and anti-apoptotic genes whose expression was altered by combined MAPK/HDAC inhibition across multiple colorectal cancer cells, with different MAPK pathway genotypes (Fig. 3A). Although expression of several apoptotic regulators was altered in individual cell lines, the only factor we found to be consistently induced across all four cell lines was the BH3-only gene BMF (Fig. 3A), which was further confirmed at the protein level in the majority of the cell lines (Fig. 3B).
Figure 3.
Induction of BMF and BIM by combined MEK/HDAC inhibition in colorectal cancer cells. Effect of trametinib (10 nmol/L) and panobinostat (25 nmol/L) treatment alone and in combination in COLO 201, HT29, DIFI, and HCT 116 cells on (A). Changes in expression of the proapoptotic genes BMF, BCL2L11, BBC3, BID, BIK, and PMAIP1; the apoptotic effectors BAK, BAX, and BOK; and the Bcl-2 only genes BCL2L1, MCL1, BCL2, BCL2A1, and BCL2L2 as determined by qRT-PCR and presented as fold change over DMSO control. B, BMF protein expression following 24 hours of treatment and (C) BIM, MCL1, pERK, and total-ERK following 6 hours of treatment as determined by Western blot. β-Actin was used as a loading control.
MAPK/ERK signaling also results in the phosphorylation and degradation of BIMEL (6–8, 39) and stabilization of MCL-1 (12). We therefore also examined expression of these proteins following combined MEK/HDAC inhibition in the same cell lines. MEKi alone increased BIM expression as well as increased the mobility of BIMEL (or decrease in the apparent molecular mass) consistent with its expected dephosphorylation following MAPK/ERK pathway inhibition, whereas HDACi treatment induced a more modest increase in BMF expression (Fig. 3C). Comparatively, expression of MCL-1 expression was not consistently changed across the four cell lines following either single-agent or combination treatment (Fig. 3C). As expected, p-ERK levels were decreased in cells treated with trametinib alone, or in combination with panobinostat, whereas panobinostat alone had no effect (Fig. 3C). Collectively, these findings show that combined MEK/HDAC inhibition consistently increased expression of BMF and BIM across multiple colorectal cancer cell lines of different MAPK pathway genotypes.
Role of BMF and BIM in MEK/HDAC inhibitor–induced apoptosis
To determine whether the induction of BMF and BIM is required for MEKi/HDACi-induced apoptosis, we first deleted BIM in COLO 201 colorectal cancer cells using CRISPR/Cas9. BMF was subsequently downregulated in BIM knockout cells by transient transfection with BMF-targeting siRNAs. Efficient inactivation of BMF and BIM under basal conditions, or following MEKi/HDACi treatment, was confirmed by qRT-PCR and Western blot, respectively (Fig. 4A and B). Although inactivation of either BMF or BIM alone resulted in partial attenuation of MEKi/HDACi-induced apoptosis, combined inactivation of both factors resulted in a significantly greater attenuation in apoptosis (Fig. 4C), demonstrating that BMF and BIM play a significant role in MEK/HDAC inhibitor–induced apoptosis.
Figure 4.
Effect of dual inactivation of BIM and BMF on trametinib and panobinostat-induced apoptosis. COLO 201 BIM deletion CRISPR cells [minus doxycycline (−DOX) and plus doxycycline (+DOX)] were transiently transfected with nontargeting or BMF-targeting siRNAs for 24 hours. Cells were then treated with trametinib (10 nmol/L) and panobinostat (25 nmol/L) alone and in combination for 24 hours. A, Knockdown efficiency of BMF as determined by qRT-PCR. B, Validation of CRISPR-Cas9–mediated BIM deletion by Western blot. β-Tubulin was used as a loading control. C, Apoptosis determined by propidium iodide staining and FACS analysis. The values shown are mean ± SEM from a single experiment performed in technical triplicate. Similar results were obtained in a second independent experiment. One-way ANOVA, with Tukey's multiple comparison testing; ****, P ≤ 0.0001.
Tailoring combined MAPK/HDAC inhibition to the MAPK pathway genotype of colorectal cancer cells
ERK/MAPK signaling is constitutively activated in colorectal cancer by multiple mechanisms, including canonical EGFR-driven signaling as well as RAS and BRAF mutations. Multiple therapies have been developed to selectively inhibit the pathway at specific nodes. These include EGFR inhibitors, BRAF inhibitors, and most recently G12C mutant KRAS inhibitors. We therefore sought to determine the efficacy of combining an HDAC inhibitor with an MAPK pathway inhibitor tailored to the “MAPK genotype” of the tumor.
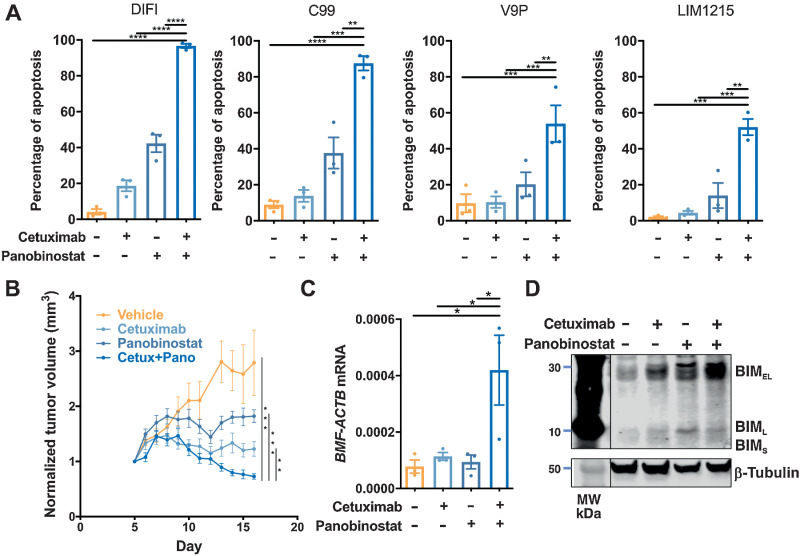
To address this, we first tested the effect of combining panobinostat with the EGFR-targeting antibody (cetuximab) in four KRAS/BRAF wild-type colorectal cancer cell lines. Combined EGFR/HDAC inhibition significantly enhanced apoptosis compared with treatment with vehicle or either agent alone, in all cell lines (Fig. 5A). To validate this finding in vivo, we tested the efficacy of combining cetuximab and panobinostat on growth of LIM1215 xenografts. Although both cetuximab and panobinostat alone significantly reduced tumor volume compared with vehicle control, a further reduction was observed in mice treated with the drug combination (Fig. 5B). Importantly, the cetuximab/panobinostat combination was well tolerated, as indicated by stable maintenance of body weight for the duration of the experiment in all treatment groups (Supplementary Fig. S5B). Similarly, to the effects induced by MEK/HDAC inhibition, EGFR/HDAC inhibition additively induced BMF mRNA (Fig. 5C), and BIM protein expression in resected tumor samples (Fig. 5D).
Figure 5.
Combined EGFR/HDAC inhibition induces apoptosis in colorectal cancer cells. A,KRAS/BRAF wild-type colorectal cancer cell lines treated with cetuximab (10 μg/mL) and panobinostat (25 nmol/L) alone or in combination for 72 hours and apoptosis induction determined by propidium iodide staining and FACS analysis. The values shown are mean ± SEM from three independent experiments. One-way ANOVA, with Tukey's multiple comparison testing; *, P ≤0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001. B, Effect of cetuximab and panobinostat on growth of LIM1215 tumor xenografts. BALB/c nude mice per group were subcutaneously injected with 2×106 LIM1215 cells into the right and left flanks (n = 8–12). Mice were then randomized to receive either vehicle control, cetuximab (n = 5, 40 mg/kg), panobinostat (n = 5, 5 mg/kg), or the combination for a total of 12 treatments. Tumor volume was normalized to day 1 of treatment. The values shown are mean ± SEM. C, Expression of BMF mRNA expression in excised tumors (n = 3, all groups). The values shown are mean ± SEM. One-way ANOVA, with Tukey's multiple comparison testing; * P ≤ 0.05. D, Expression of BIM in representative excised tumors from each treatment group as determined by Western blot analysis. β-Tubulin was used as a loading control.
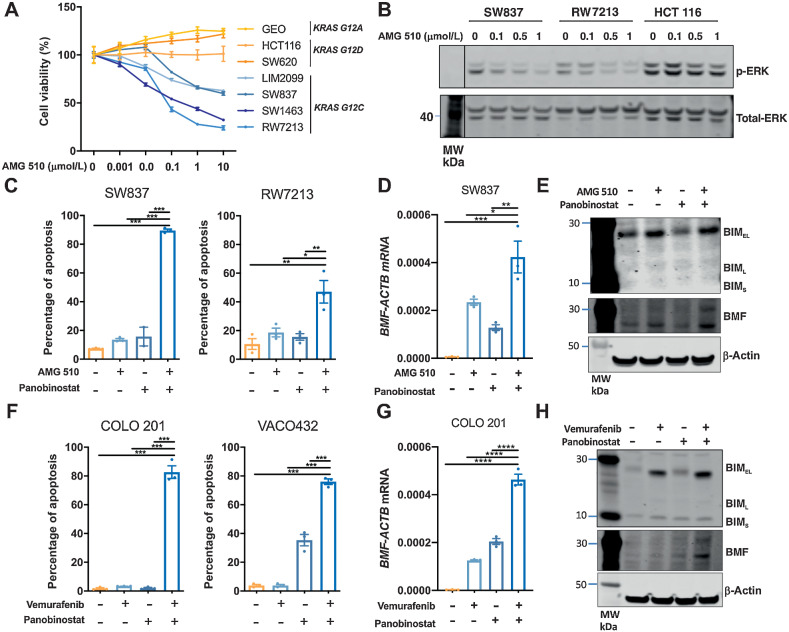
We next assessed the effect of combining panobinostat with the KRASG12C inhibitor AMG 510 in KRASG12C mutant cell lines. The selectivity of AMG510 for KRASG12C was first confirmed by demonstrating its ability to selectively inhibit cell proliferation and ERK/MAPK signaling in KRASG12C mutant colorectal cancer cells (Fig. 6A and B). As observed for the previous MAPK/HDAC inhibitor combinations, KRASG12C/HDAC inhibition significantly increased apoptosis induction compared with either agent alone in two KRASG12C cell lines (Fig. 6C). The combination also increased expression of BMF (Fig. 6D and E).
Figure 6.
Combined KRAS/HDAC and BRAF/HDAC inhibition induces apoptosis in colorectal cancer cells. A, Effect of AMG 510 on cell viability in colorectal cancer cells. KRASG12C mutant (LIM2099, RW7213, SW837, and SW1463), KRASG12D mutant (HCT116 and SW620), and KRASG12A mutant (GEO) colorectal cancer cells were treated with escalating doses of AMG 510 for 72 hours, and cell viability assessed by MTS assay. The values shown are mean ± SD from a single experiment performed in technical triplicate. Similar results were obtained in a second independent experiment. B, SW837, RW7213, and HCT 116 cells treated with AMG 510 for 1 hour and p-ERK and total ERK levels determined by Western blot analysis. C, Apoptosis analysis as determined by propidium iodide staining and FACS analysis in SW837 and RW7213 cells treated with AMG 510 (0.1 μmol/L) and panobinostat (25 nmol/L) alone and in combination for 72 hours. The values shown are mean ± SEM from a single representative experiment performed in technical triplicate. Similar results were obtained in two additional independent experiments. One-way ANOVA, with Tukey's multiple comparison testing; *, P ≤0.05; **, P ≤0.01; and ***, P ≤0.001. D,BMF mRNA expression as determined by qRT-PCR in SW837 cells treated with AMG 510 (0.1 μmol/L) and panobinostat (25 nmol/L) for 12 hours. The values shown are mean ± SEM from a single representative experiment performed in technical triplicate; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). E, Corresponding Western blot analysis for BIM and BMF expressions. β-Actin was used as a loading control. F, Apoptosis analysis as determined by propidium iodide staining and FACS analysis in COLO 201 and VACO432 cells treated with vemurafenib (2.5 μmol/L) and panobinostat (25 nmol/L) alone and in combination for 72 hours. The values shown are mean ± SEM from a single representative experiment performed in technical triplicate. Similar results were obtained in two additional independent experiments. One-way ANOVA, with Tukey's multiple comparison testing; ***, P ≤ 0.001; ****, P ≤ 0.0001. G,BMF mRNA expression as determined by qRT-PCR in COLO 201 cells treated with vemurafenib (2.5 μmol/L) and panobinostat (25 nmol/L) for 12 hours. The values shown are mean ± SEM from a single representative experiment performed in technical triplicate; ****, P ≤ 0.0001. H, Corresponding Western blot analysis for BIM and BMF expressions. β-Actin was used as a loading control.
Finally, we assessed the effect of combining panobinostat with the BRAFV600 inhibitor vemurafenib in BRAFV600E cell lines. As observed for the other MAPK genotype-tailored combinations, BRAFV600/HDAC inhibition significantly enhanced apoptosis compared with either agent alone (Fig. 6F) and significantly increased BMF mRNA and BIM protein expression (Fig. 6G and H).
Discussion
Herein, we show that combined inhibition of ERK/MAPK signaling and HDACs synergistically induces apoptosis in multiple colorectal cancer cell lines. Notably, this effect could be generically induced in colorectal cancer cell lines independent of their KRAS or BRAF mutation status by combining an MEK inhibitor with an HDAC inhibitor, or it could be selectively tailored to the ERK/MAPK genotype of colorectal cancer cells by combining EGFR, KRASG12C or BRAF inhibitors with an HDAC inhibitor, in RAS/BRAFWT, KRASG12C or BRAFV600 colorectal cancer cell lines, respectively. The latter approach may provide greater tumor-specific targeting and a better therapeutic window compared with MEK inhibition where on-target toxicities in normal cells may reduce clinical activity (40).
The robust proapoptotic activity of MEK/HDAC inhibition was also observed in PDTOs generated from four patients with colorectal cancer, three of which were derived from metastatic sites. The combination also effectively inhibited colonic adenoma growth in an Apc mutant mouse model, illustrating the potential for this regimen to be effective at multiple stages in the adenoma–carcinoma sequence.
Mechanistically, we show that the synergistic apoptotic activity of the combination is at least partially driven by the induction of the proapoptotic BH-3–only proteins, BMF and BIM. BIM has a strong binding affinity to all prosurvival BCL-2 proteins (BCL-2, BCL-XL, MCL-1, BCL-W, and BFL-1), whereas BMF primarily binds to BCL-XL, BCL-W, and BCL-2, and only weakly binds to MCL-1 and BFL-1 (41). As colorectal cancers express high levels of BCL-XL (42), we postulate that the combined induction of both BIM and BMF may be sufficient to bind and inhibit the high levels of these prosurvival factors, enabling BIM to bind and activate BAX/BAK or to inhibit other prosurvival proteins and trigger apoptosis.
Despite previous publications demonstrating BIM mRNA induction following MAPK or HDAC inhibitor treatment (25, 38), we did not observe this to be the case in all cell lines in this study. Comparatively, we observed a consistent increase in BIM protein expression, particularly when treated with the MEK inhibitor trametinib. BIM is expressed as three isoforms (BIMEL, BIML, and BIMS) of which BIMEL has been previously shown to be regulated by MAPK signaling (6–8, 39). Specifically, ERK-mediated phosphorylation of BIMEL at serine-69 promotes its ubiquitination and proteasomal degradation (6, 8), whereas BIMEL phosphorylation has also been shown to induce its rapid dissociation from the prosurvival proteins, BCL-XL and MCL-1 (39). Consistent with these reports, trametinib-mediated inhibition of MAPK/ERK signaling induced the strongest increase in protein expression of the BIMEL isoform and induced a decrease in the band size of BIMEL, consistent with a loss of phosphorylation event.
In addition, consistent induction of BMF mRNA was observed following both MAPK pathway and HDAC inhibition. Notably, the BMF promoter harbors multiple SP1/SP3-binding sites and localization of SP3 to the BMF promoter has been shown in chromatin immunoprecipitation studies (22, 24). HDACis are known to preferentially regulate SP1/SP3 target genes, either through inhibition of HDAC enzymes that are recruited by these factors, or by direct acetylation of SP1 or SP3 (43). MAPK signaling has also been shown to modulate SP1 activity through direct ERK-mediated phosphorylation of SP1 at Thr-453 and Thr-739 (44), raising the potential that MAPK pathway and HDAC inhibition converge to regulate BMF expression by modulating the activity of SP1/SP3 transcription factors.
Our finding that combined MAPK/HDAC inhibition enhances cell death in colorectal cancer cells is reminiscent of studies in other tumor types. In triple-negative and inflammatory breast cancer, combined MEK/HDAC inhibition enhanced apoptosis by increasing expression of the proapoptotic factor NOXA and subsequent inhibition of MCL-1 (45). Notably, NOXA (PMAIP1) expression was not consistently altered in our analysis of the intrinsic apoptotic pathway across the four colorectal cancer cell lines, which may reflect differences in tumor type. Several studies have also demonstrated increased cell killing by combined MAPK/HDAC inhibition in melanoma cells, with multiple mechanisms implicated, including attenuation of PI3K/AKT signaling and YAP activation (46), suppression of ELK, suppression of the homologous DNA repair and nonhomologous end-joining pathways (47), and increased ROS production (48). These findings emphasize that in addition to the induction of BIM and BMF that we demonstrate in this study, several other mechanisms are also likely to be involved in driving MAPKi/HDACi-induced apoptosis.
In summary, our finding that combining MAPK pathway inhibitors with HDACis can synergistically induce apoptosis in multiple preclinical models of colorectal cancer, suggests this concept is worthy of clinical investigation. Furthermore, we identify strategies in which this can be achieved in a highly targeted manner by tailoring treatment combinations to the specific MAPK genotype of colorectal cancers.
Supplementary Material
Supplementary table 1
Supplementary Figure 1 shows the synergistic effect of trametinib plus panobinostat in CRC cell lines and PDTO's
Supplementary figure 2 shows the effect of trametinib plus panobinostat on colony formation in HCT 116 cells
Supplementary figure 3 shows combining trametinib with different classes of HDAC inhibitors in COLO 201 cells
Supplementary figure 4 shows phosphorylated ERK staining in an adenoma from a ACDX2 mouse
Supplementary figure 5 shows mouse weight during treatment in two of the mouse studies
Acknowledgments
This project was supported by the Tour de Cure Senior Research grant (17-ONJC-RS-05) and the Operational Infrastructure Support Program, Victorian Government, Australia. L.J. Jenkins was supported by La Trobe University Australian Postgraduate Awards. G. Iatropoulos, I. Ng, and Z. Alam were supported by La Trove University Postgraduate Research Scholarship. The patient-derived tumor organoid study was supported by the Stafford Fox Medical Research Foundation. J.M. Mariadason (1046092) and O.M. Sieber (1136119) were supported by National Health and Medical Research Council (NHMRC) Senior Research Fellowships.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
J. Desai reports grants, personal fees, and nonfinancial support from Beigene and Roche/Genentech, as well as grants and nonfinancial support from Bristol Myers Squibb and personal fees and nonfinancial support from Amgen, and grants, personal fees, and nonfinancial support from Pierre-Fabre, and grants and nonfinancial support from GlaxoSmithKline, as well as personal fees from Merck KGaA, and personal fees and nonfinancial support from Boehringer Ingelheim outside the submitted work. O.M. Sieber reports grants from Stafford Fox Medical Research Foundation and National Health and Medical Research Council during the conduct of the study. N.C. Tebbutt reports personal fees from Amgen, Bristol Myers Squibb, AstraZeneca, Merck & Co Inc., and Servier outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
L.J. Jenkins: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. I.Y. Luk: Data curation, validation. W.D. Fairlie: Resources, writing–review and editing. E.F. Lee: Resources, writing–review and editing. M. Palmieri: Data curation, formal analysis. K.L. Schoffer: Formal analysis, investigation, methodology. T. Tan: Data curation. I. Ng: Data curation, validation. N. Vukelic: Data curation, validation. S. Tran: Data curation, methodology. J.W.T. Tse: Data curation. R. Nightingale: Data curation. Z. Alam: Data curation. F. Chionh: Data curation. G. Iatropoulos: Data curation. M. Ernst: Resources, writing–review and editing. S. Afshar-Sterle: Data curation. J. Desai: Conceptualization. P. Gibbs: Resources. O.M. Sieber: Resources. A.S. Dhillon: Conceptualization, supervision, writing–review and editing. N.C. Tebbutt: Conceptualization, supervision. J.M. Mariadason: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McKay JA, Murray LJ, Curran S, Ross VG, Clark C, Murray GI, et al. Evaluation of the epidermal growth factor receptor (EGFR) in colorectal tumours and lymph node metastases. Eur J Cancer 2002;38:2258–64. [DOI] [PubMed] [Google Scholar]
- 3. Lavoie H, Gagnon J, Therrien M. ERK signalling: a master regulator of cell behaviour, life, and fate. Nat Rev Mol Cell Biol 2020;21:607–32. [DOI] [PubMed] [Google Scholar]
- 4. Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol 2008;10:138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin L, Ding D, Jiang Y, Li Y, Li S. MEK inhibitors induce apoptosis via FoxO3a-dependent PUMA induction in colorectal cancer cells. Oncogenesis 2018;7:67. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6. Marani M, Hancock D, Lopes R, Tenev T, Downward J, Lemoine NR. Role of Bim in the survival pathway induced by Raf in epithelial cells. Oncogene 2004;23:2431–41. [DOI] [PubMed] [Google Scholar]
- 7. Hubner A, Barrett T, Flavell RA, Davis RJ. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell 2008;30:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 2003;278:18811–6. [DOI] [PubMed] [Google Scholar]
- 9. Shao Y, Aplin AE. ERK2 phosphorylation of serine 77 regulates Bmf proapoptotic activity. Cell Death Dis 2012;3:e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14–3-3 not BCL-X(L). Cell 1996;87:619–28. [DOI] [PubMed] [Google Scholar]
- 11. Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 1999;286:1358–62. [DOI] [PubMed] [Google Scholar]
- 12. Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 2004;23:5301–15. [DOI] [PubMed] [Google Scholar]
- 13. Sale MJ, Minihane E, Monks NR, Gilley R, Richards FM, Schifferli KP, et al. Targeting melanoma's MCL1 bias unleashes the apoptotic potential of BRAF and ERK1/2 pathway inhibitors. Nat Commun 2019;10:5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nangia V, Siddiqui FM, Caenepeel S, Timonina D, Bilton SJ, Phan N, et al. Exploiting MCL1 dependency with combination MEK + MCL1 inhibitors leads to induction of apoptosis and tumor regression in KRAS-mutant non–small cell lung cancer. Cancer Discov 2018;8:1598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawakami H, Huang S, Pal K, Dutta SK, Mukhopadhyay D, Sinicrope FA. Mutant BRAF upregulates MCL-1 to confer apoptosis resistance that is reversed by MCL-1 antagonism and cobimetinib in colorectal cancer. Mol Cancer Ther 2016;15:3015–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res 2008;68:1953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau DK, Luk IY, Jenkins LJ, Martin A, Williams DS, Schoffer KL, et al. Rapid resistance of FGFR-driven gastric cancers to regorafenib and targeted FGFR inhibitors can be overcome by parallel inhibition of MEK. Mol Cancer Ther 2021;20:704–15. [DOI] [PubMed] [Google Scholar]
- 18. Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007;357:2040–8. [DOI] [PubMed] [Google Scholar]
- 19. Tabernero J, Grothey A, Van Cutsem E, Yaeger R, Wasan H, Yoshino T, et al. Encorafenib Plus Cetuximab as a new standard of care for previously treated BRAF V600E-mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON study. J Clin Oncol 2021;39:273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014;124:30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bolden JE, Shi W, Jankowski K, Kan CY, Cluse L, Martin BP, et al. HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis 2013;4:e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang Y, Nian H, Rajendran P, Kim E, Dashwood WM, Pinto JT, et al. HDAC8 and STAT3 repress BMF gene activity in colon cancer cells. Cell Death Dis 2014;5:e1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiegmans AP, Alsop AE, Bots M, Cluse LA, Williams SP, Banks KM, et al. Deciphering the molecular events necessary for synergistic tumor cell apoptosis mediated by the histone deacetylase inhibitor vorinostat and the BH3 mimetic ABT-737. Cancer Res 2011;71:3603–15. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y, Adachi M, Kawamura R, Imai K. Bmf is a possible mediator in histone deacetylase inhibitors FK228 and CBHA-induced apoptosis. Cell Death Differ 2006;13:129–40. [DOI] [PubMed] [Google Scholar]
- 25. Zhao Y, Tan J, Zhuang L, Jiang X, Liu ET, Yu Q. Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc Natl Acad Sci U S A 2005;102:16090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rahmani M, Reese E, Dai Y, Bauer C, Payne SG, Dent P, et al. Coadministration of histone deacetylase inhibitors and perifosine synergistically induces apoptosis in human leukemia cells through Akt and ERK1/2 inactivation and the generation of ceramide and reactive oxygen species. Cancer Res 2005;65:2422–32. [DOI] [PubMed] [Google Scholar]
- 27. Chueh AC, Tse JWT, Dickinson M, Ioannidis P, Jenkins L, Togel L, et al. ATF3 repression of BCL-XL determines apoptotic sensitivity to HDAC inhibitors across tumor types. Clin Cancer Res 2017;23:5573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duan H, Heckman CA, Boxer LM. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol Cell Biol 2005;25:1608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ciardiello F, Bianco R, Damiano V, De Lorenzo S, Pepe S, De Placido S, et al. Antitumor activity of sequential treatment with topotecan and anti-epidermal growth factor receptor monoclonal antibody C225. Clin Cancer Res 1999;5:909–16. [PubMed] [Google Scholar]
- 30. Wu X, Fan Z, Masui H, Rosen N, Mendelsohn J. Apoptosis induced by an anti-epidermal growth factor receptor monoclonal antibody in a human colorectal carcinoma cell line and its delay by insulin. J Clin Invest 1995;95:1897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleivi K, Teixeira MR, Eknaes M, Diep CB, Jakobsen KS, Hamelin R, et al. Genome signatures of colon carcinoma cell lines. Cancer Genet Cytogenet 2004;155:119–31. [DOI] [PubMed] [Google Scholar]
- 32. Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O'Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep 2015;10:1422–32. [DOI] [PubMed] [Google Scholar]
- 33. Zhou N, Moradei O, Raeppel S, Leit S, Frechette S, Gaudette F, et al. Discovery of N-(2-aminophenyl)-4-[(4-pyridin-3-ylpyrimidin-2-ylamino)methyl]benzamide (MGCD0103), an orally active histone deacetylase inhibitor. J Med Chem 2008;51:4072–5. [DOI] [PubMed] [Google Scholar]
- 34. Suzuki T, Ando T, Tsuchiya K, Fukazawa N, Saito A, Mariko Y, et al. Synthesis and histone deacetylase inhibitory activity of new benzamide derivatives. J Med Chem 1999;42:3001–3. [DOI] [PubMed] [Google Scholar]
- 35. Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives antitumour immunity. Nature 2019;575:217–23. [DOI] [PubMed] [Google Scholar]
- 36. Lanman BA, Allen JR, Allen JG, Amegadzie AK, Ashton KS, Booker SK, et al. Discovery of a covalent inhibitor of KRAS(G12C; AMG 510) for the treatment of solid tumors. J Med Chem 2020;63:52–65. [DOI] [PubMed] [Google Scholar]
- 37. Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res 2007;67:9721–30. [DOI] [PubMed] [Google Scholar]
- 38. Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med 2007;4:1681–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ewings KE, Hadfield-Moorhouse K, Wiggins CM, Wickenden JA, Balmanno K, Gilley R, et al. ERK1/2-dependent phosphorylation of BimEL promotes its rapid dissociation from Mcl-1 and Bcl-xL. EMBO J 2007;26:2856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer 2015;15:577–92. [DOI] [PubMed] [Google Scholar]
- 41. Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005;17:393–403. [DOI] [PubMed] [Google Scholar]
- 42. Krajewska M, Moss SF, Krajewski S, Song K, Holt PR, Reed JC. Elevated expression of Bcl-X and reduced Bak in primary colorectal adenocarcinomas. Cancer Res 1996;56:2422–7. [PubMed] [Google Scholar]
- 43. Chueh AC, Tse JW, Togel L, Mariadason JM. Mechanisms of histone deacetylase inhibitor-regulated gene expression in cancer cells. Antioxid Redox Signal 2015;23:66–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Milanini-Mongiat J, Pouyssegur J, Pages G. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J Biol Chem 2002;277:20631–9. [DOI] [PubMed] [Google Scholar]
- 45. Torres-Adorno AM, Lee J, Kogawa T, Ordentlich P, Tripathy D, Lim B, et al. Histone deacetylase inhibitor enhances the efficacy of MEK inhibitor through NOXA-mediated MCL1 degradation in triple-negative and inflammatory breast cancer. Clin Cancer Res 2017;23:4780–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Faiao-Flores F, Emmons MF, Durante MA, Kinose F, Saha B, Fang B, et al. HDAC inhibition enhances the in vivo efficacy of MEK inhibitor therapy in uveal melanoma. Clin Cancer Res 2019;25:5686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maertens O, Kuzmickas R, Manchester HE, Emerson CE, Gavin AG, Guild CJ, et al. MAPK pathway suppression unmasks latent DNA repair defects and confers a chemical synthetic vulnerability in BRAF-, NRAS-, and NF1-mutant melanomas. Cancer Discov 2019;9:526–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang L, Leite de Oliveira R, Huijberts S, Bosdriesz E, Pencheva N, Brunen D, et al. An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell 2018;173:1413–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1
Supplementary Figure 1 shows the synergistic effect of trametinib plus panobinostat in CRC cell lines and PDTO's
Supplementary figure 2 shows the effect of trametinib plus panobinostat on colony formation in HCT 116 cells
Supplementary figure 3 shows combining trametinib with different classes of HDAC inhibitors in COLO 201 cells
Supplementary figure 4 shows phosphorylated ERK staining in an adenoma from a ACDX2 mouse
Supplementary figure 5 shows mouse weight during treatment in two of the mouse studies
Data Availability Statement
Raw data for this study were generated at the Olivia Newton-John Cancer Research Institute, Melbourne, Australia and Walter Eliza Hall Institute of Medical Research, Melbourne, Australia. Derived data supporting the findings of this study are available from the corresponding author upon request.



![Figure 4. Effect of dual inactivation of BIM and BMF on trametinib and panobinostat induced apoptosis. COLO 201 BIM deletion CRISPR cells [minus doxycycline (−DOX) and plus doxycycline (+DOX)] were transiently transfected with non-targeting or BMF-targeting siRNAs for 24 hours. Cells were then treated with trametinib (10 nmol/L) and panobinostat (25 nmol/L) alone and in combination for 24 hours. A, Knockdown efficiency of BMF as determined by qRT-PCR. B, Validation of CRISPR-Cas9–mediated BIM deletion by Western blot. β-Tubulin was used as a loading control. C, Apoptosis determined by propidium iodide staining and FACS analysis. The values shown are mean ± SEM from a single experiment performed in technical triplicate. Similar results were obtained in a second independent experiment. One-way ANOVA, with Tukey's multiple comparison testing; ****, P ≤ 0.0001)](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/3ad7/9808369/f7467f55271a/52fig4.jpg)