Abstract
Detection of insertions and deletions (InDels) by short-read next-generation sequencing (NGS) technology can be challenging because of frequent misaligned reads. A systematic analysis of short InDels (1 to 30 bases) and fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplications (ITDs; 6 to 183 bases) from 46 clinical cases of solid or hematologic malignancy processed with a clinical NGS assay identified misaligned reads in every case, ranging from 3% to 100% of reads with the InDel showing mismapped bases. Mismaps also increased with InDel size. As a consequence, the clinical NGS bioinformatics pipeline undercalled the variant allele frequency by 1% to 84%, incorrectly called simultaneous single-base substitutions along with InDels, or did not report an FLT3 ITD that had been detected by capillary electrophoresis. To improve the ability of the pipeline to better detect and quantify InDels, we utilized a software program called Assembly-Based ReAligner (ABRA2) to more accurately remap reads. ABRA2 was able to correct 41% to 100% of the reads with mismapped bases and led to absolute increases in the variant allele frequency from 1% to 61% along with correction of all of the single-base substitutions except for two cases. ABRA2 could also detect multiple FLT3 ITD clones except for one 183-base ITD. Our analysis has found that ABRA2 performs well on short InDels as well as FLT3 ITDs that are <100 bases.
Next-generation sequencing (NGS) has revolutionized the field of genomics for both scientific discovery and clinical care.1, 2, 3, 4, 5, 6 The affordability and efficiency of NGS has provided an opportunity for a personalized medicine approach to oncology, which has sparked widespread implementation of NGS-based approaches in clinical laboratories for cancer diagnostics.7, 8, 9 Given its applications for disease management and potential to inform patient care, the accuracy of NGS data is critical—yet, these data are highly complex and can be challenging to interpret.10 Furthermore, errors produced by sequencing and data analysis can generate erroneous artifacts, which can interfere with the detection of rare variants or even be falsely interpreted as true single-base substitutions (SBSs).11
Previous studies have revealed several sources of sequencing artifacts that can be produced using NGS approaches. Chen et al12 found that cytosine deamination occurred, as both biologic and as an artifact of thermocycling, and contributed to baseline noise in NGS data. McCall et al13 showed that off-target amplification, due to mispriming events, led to false-positive mutations during multiplex PCRs. Numerous researchers have demonstrated sequencing errors associated with GC-rich, inverted repeat, and long homopolymer regions of DNA.14, 15, 16 Many artifacts are also associated with certain sequencing platforms, largely due to unique chemistries.17 For example, the Ion Torrent platform (ThermoFisher, Waltham, MA) can produce sequencing errors within homopolymeric regions because of an inaccurate measurement of the size of the voltage pulse produced with these sequences by the semiconductor-based technology.18 Finally, in addition to errors produced from sequencing itself, artifacts can also be produced as a result of standard data analysis by bioinformatics software. For example, with short-read NGS technology, an aligner may wrongly map reads arising from a pseudogene to its paralogous functional gene or within gene families with similar sequence homology, resulting in a false-positive variant call.19,20
A critical step for NGS analysis is the process of mapping reads to the human genome, which involves converting raw sequencing data into an interpretable format for variant calling. The first step in this process is transforming raw image files into binary base call files, which represent the raw data output from a sequencing run. Base call files are then converted to FASTQ files, which is an ASCII text file format that stores both the raw sequence data and quality scores. Finally, these FASTQ files are mapped to the human reference genome, generating a SAM/BAM file, which can be visualized using software tools like Integrative Genomics Viewer (IGV)21,22 and is commonly used for variant calling. More importantly, previous studies have shown that mapping quality can be reduced because of certain features of the genome, such as repetitive and low-complexity regions.23,24
In this study, we characterize different types of mapping errors that can occur at sites of insertions and deletions (InDels). These mapping errors can result in the coexistence of InDels with artifactual single-base substitutions, a profound underestimation of InDel variant allele frequencies (VAFs), or missed detection of long InDels, such as fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplications (ITDs) by variant callers. We demonstrate that the magnitude of mapping error increases as a function of InDel size. Finally, we propose utilizing programs that can more accurately map reads to minimize these errors.
Materials and Methods
Selection of Cases
NGS reports were queried from the laboratory information system (SCC Soft Computer, Clearwater, FL) of the Molecular Diagnostics Laboratory at the Johns Hopkins Hospital (Baltimore, MD) beginning in 2017 for cases with InDels of any length. FLT3 internal tandem duplication cases were identified by searching the laboratory information system for capillary electrophoresis (CE) results of any insertion length. The total 46 cases included 26 formalin-fixed, paraffin-embedded specimens with a diagnosis of solid tumor (Supplemental Table S1) and 20 peripheral blood or bone marrow specimens with a diagnosis of acute myeloid leukemia, acute lymphoblastic leukemia, or leukopenia (Supplemental Table S2). Deletion mutations were detected in 14 formalin-fixed, paraffin-embedded specimens and 1 peripheral blood specimen, and insertion mutations were detected in 17 formalin-fixed, paraffin-embedded specimens by NGS pipeline. One or more FLT3 ITD mutations were detected by CE analysis in 19 leukemia specimens. The Johns Hopkins Medicine Institutional Review Board granted approval for this study.
Sequencing
NGS was performed using a clinically validated laboratory-developed test at the Clinical Laboratory Improvement Amendments–certified Molecular Diagnostics Laboratory at Johns Hopkins, as previously described.25,26
Sample Processing
Briefly, DNA was extracted from formalin-fixed, paraffin-embedded tissue using the Siemens Tissue Preparation System (Siemens Medical Solutions USA, Inc., Malvern, PA) and from peripheral blood or bone marrow using the Qiagen EZ1 Advanced XL (Qiagen, LLC, Germantown, MD). A total of 300 ng to 1 μg of DNA was fragmented to a size of 150 to 275 bp using a Covaris LE220-Plus sonicator (Covaris, Inc., Woburn, MA). The DNA fragments were end repaired and A tailed, then adaptors were added by ligation and the fragments were enriched by PCR. Hybrid capture DNA libraries were prepared using an Agilent SureSelect-XT (Agilent Technologies, Santa Clara, CA) target enrichment kit. Each library was hybridized to an Agilent SureSelect custom 2.8 M bait set covering mainly coding regions of 640 genes that are relevant in cancer. After stringent washing, the captured DNA was amplified with PCR, per manufacturer's protocol. The quality and quantity of the captured DNA was assessed using a Tapestation 4200 (Agilent). Final sample libraries (samples 1 to 43) were then run on an Illumina HiSeq 2500 instrument (Illumina Biotechnology, San Diego, CA) using 2 × 100 paired-end chemistry with a target read depth of 800×.
For samples 44 and 45, hybrid capture DNA libraries were prepared via a method that utilizes KAPA Hyper-prep chemistry along with custom IDT dual-indexed Illumina adapters with additional unique molecular indexes for library preparation. Each library was hybridized to the JHOPv4.2 IDT bait set, covering regions of >900 cancer-related genes, according to the manufacturer's protocol. Samples were run on an Illumina NovaSeq 6000 instrument (Illumina Biotechnology) using 2 × 100 paired-end chemistry with a target read depth of 1000×.
In terms of quality control, samples with <100 ng input DNA, specimens with <30 ng total DNA in the 100- to 700-bp range after shearing, or samples with a sequencing quality predictor ratio of ≥6.5 after shearing are reflexed to an Ion Torrent Ampliseq HotSpot panel instead. We aim to have a similar sequence depth per sample volume and do not report single-nucleotide variants or InDels with a sequencing read depth <150×.
Sequencing Pipeline
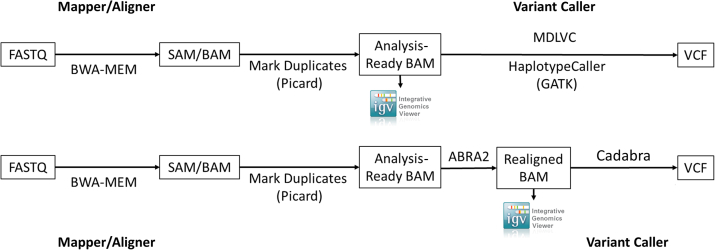
Base call files generated by the HiSeq or NovaSeq instruments were converted to FASTQ format using Illumina bcl2fastq software version 1.8.4 (Illumina Biotechnology). Sequences were aligned to the reference genome (hg19/GRCh37) using Burrows-Wheeler Aligner–maximal exact matches (BWA-MEM) version 0.7.10 (https://github.com/lh3/bwa) with the default parameters. Picard Mark Duplicates version 1.119 (Broad Institute, https://gatk.broadinstitute.org) was run on the resulting alignment files to produce analysis-ready BAM files. The BAM files were either used for variant calling by Genome Analysis Toolkit HaplotypeCaller version 3.3 (Broad Institute, https://gatk.broadinstitute.org) and an internally developed variant caller [Molecular Diagnostics Laboratory Variant Caller; short InDels: version 6, except case 45 (version 8); FLT3 ITDs: version 7, except cases 28 (version 5), 29 (version 5.6), and 46 (version 8)] (Figure 1) or processed through Assembly-Based ReAligner (ABRA2) version 2.22 followed by its variant caller Cadabra (https://github.com/mozack/abra2) (Figure 1). Molecular Diagnostics Laboratory Variant Caller will exclude variants with VAFs <5% and variants with VAFs within the mean ± 3 SDs of a reference pool of normal samples. Resulting high confidence variant calls were manually reviewed using IGV version 2.8.0 (Broad Institute, https://software.broadinstitute.org/software/igv/download).21,22
Figure 1.
Representative bioinformatics pipeline before (top row) and after (bottom row) the addition of Assembly-Based ReAligner (ABRA2). To improve the ability of the pipeline to better detect and quantify insertions and deletions, a software program called Assembly-Based ReAligner (ABRA2) was used. Cadabra is a variant caller included with ABRA2. Molecular Diagnostics Laboratory Variant Caller (MDLVC) is an in-house developed variant caller. BWA, Burrows-Wheeler Aligner; GATK, Genome Analysis Toolkit; SAM, sequence alignment/map.
Statistical Analysis
Correlation of frequencies between InDel size and percentage mismaps was examined by Spearman rank correlation coefficient (denoted as ρ) using R,27 a free software environment for statistical computing and graphics (R: The R Project for Statistical Computing, https://www.r-project.org).
Detection of FLT3 ITD Mutations by Capillary Electrophoresis
FLT3 ITD mutations were detected as described previously.28 PCR products were analyzed by capillary electrophoresis using ABI 3130 genetic analyzer (Thermo Fisher Scientific, Waltham, MA). ITD size was calculated by subtracting the mutant peak size with the wild type peak size (329 ± 1 base). Sizing of the PCR amplicons, although precise, is not accurate compared with that determined by NGS. VAF of ITD was calculated by dividing the mutant peak height with the sum of wild-type and mutant peak heights. The amplification efficiency of the wild-type and mutant alleles may not be comparable because of their differences in length. Therefore, percentage VAF may be underestimated in larger ITDs. The limit of detection of this assay is approximately 1% VAF.
Results
Analysis of Mapping Errors Associated with InDels
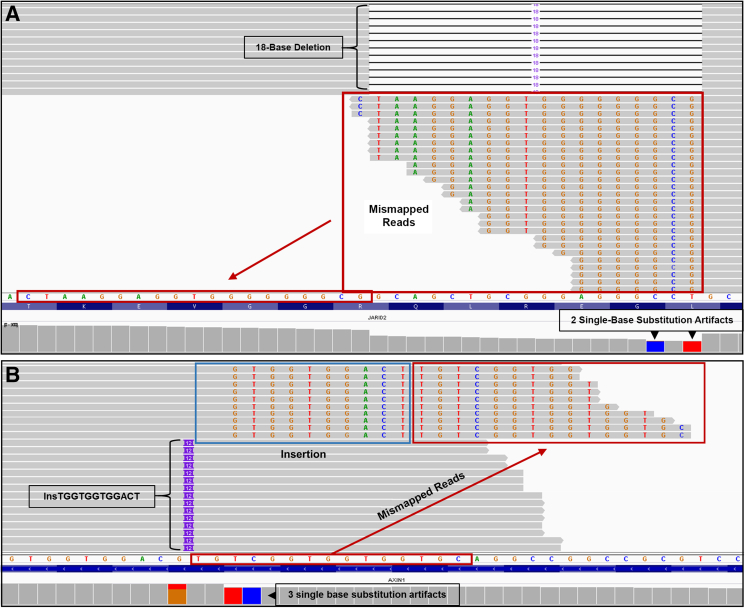
During routine clinical signout, we found a case with the co-existence of a deletion with single-base substitutions in the middle of the deleted region (Figure 2A). The question at the time was whether this was co-existence of two mutation types at a single site, or whether one was an artifact of the other. On further investigation, we recognized that the reads contributing to the single-base substitutions always had the SBSs at the ends of the reads (contrast Figure 3A vs Figure 3B; and Supplemental Figure S1) and fell within a location identified as a deletion. In addition to SBSs, it was also observed that there were multiple reads present with soft clipped bases at the ends of the reads (Figure 3B and Supplemental Figure S1), which also fell within a location of a deletion.
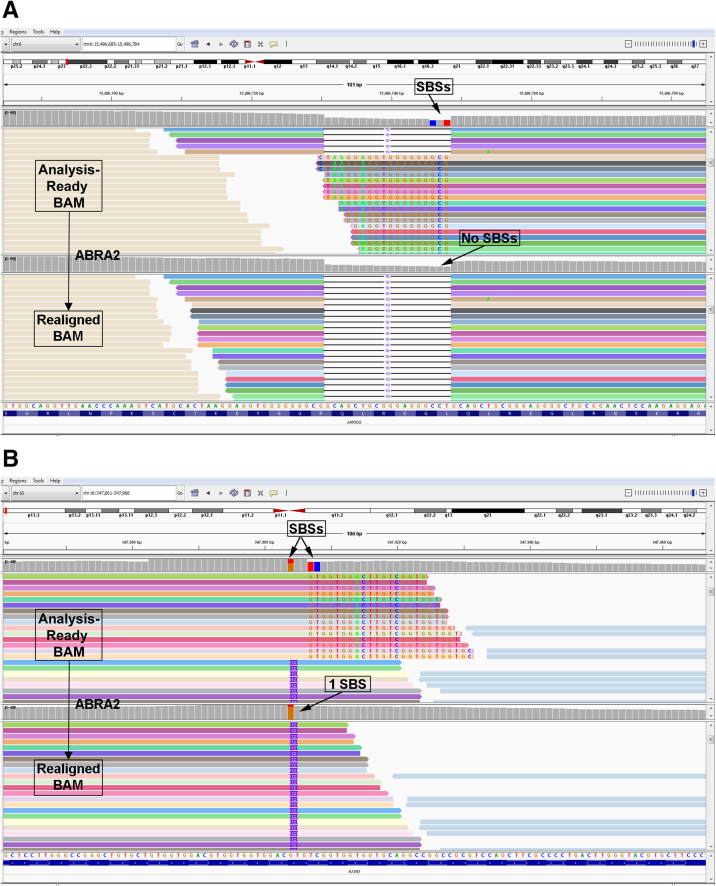
Figure 2.
Insertion and deletion mapping errors. A: Representative screenshots from Integrative Genomics Viewer (IGV) for an 18-base JARID2 deletion. Reads containing the 18-base deletion are shown at the top, because they are anchored to both the left and right of the deletion, resulting in a correctly mapped deletion. The reads at the bottom all terminate at one end within the 18-base deletion. Because these reads have the deletion, the bases to the left of the deletion (arrow) are mapped within the site of the deletion (red boxed area) and ignored (soft clipped) by the aligner. Additional reads (not shown; see Supplemental Figure S1 for a schematic) that terminate within the 18-base deletion have single-base substitutions instead of soft clips and contribute to a call of single-base substitutions (SBSs) at the first and third bases at the right edge of the deletion (blue and red bars) in IGV. B: An AXIN1 12-base insertion. The AXIN1 12-base insertion is correctly mapped for the reads at the bottom because they are anchored to both the left and right of the insertion. Because the reads at the top end within or close to the end of the insertion, they are not correctly anchored to the right. The 12-base insertion (blue rectangle) is mapped onto the reference sequence and the subsequent sequence (red rectangle) derives from the bases to the right of the insertion. Because the bases within the blue rectangle were not correctly identified as an insertion, the bases designated within the blue and red rectangles are ignored (soft clipped) by the aligner. Additional reads mapped to the reference sequence in this location (not shown; see Supplemental Figure S1 for a schematic) have single-base substitutions instead of the soft clips and result in three SBS artifacts (orange/red, red, and blue bars) in IGV.
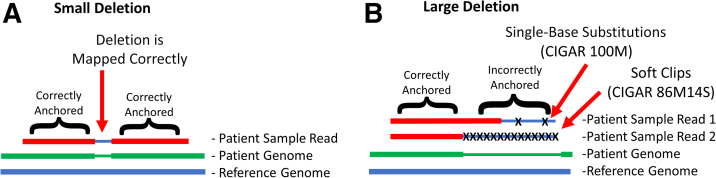
Figure 3.
Larger insertions and deletions result in incorrect anchoring of a read by the aligner. A: For a small deletion in a short read, the aligner can correctly anchor the read on both sides of the deletion. This results in a correctly mapped small deletion. B: For larger deletions, because a given read ends within the deletion, it is correctly anchored on the left side, whereas the aligner is unable to anchor the right side of the read and maps it to the reference bases within the deletion. This results in erroneous single-base substitutions (Patient Sample Read 1) or soft-clipped bases (Patient Sample Read 2). Assuming Patient Sample Reads 1 and 2 are 100 bases long each, reads with single-base substitutions are represented by the M operation in the Concise Idiosyncratic Gapped Alignment Report (CIGAR) string (which stands for match/mismatch), whereas soft clipped bases are represented by the S operation.
Soft clipped bases are bases that are unaligned (essentially skipped/hidden/ignored bases). In the sequence alignment/map format, this type of alignment is described with a Concise Idiosyncratic Gapped Alignment Report (CIGAR) string using the S operation.29 For example, an aligned 100-base read with 14 soft clipped bases at the end of the read would be described with the CIGAR string 86M14S (Figure 3B). The M operation stands for match/mismatch.29 Therefore, alternatively, an aligned 100-base read with two SBSs would appear with the CIGAR string 100M (Figure 3B). SBSs and soft clipped bases never occurred on the same read and were mutually exclusive.
We hypothesized that both these types of reads were not recognized as deletion-containing reads, and instead resulted in artifactually generated SBSs (Figure 3B and Supplemental Figure S1) or soft clipped bases (Figure 3B and Supplemental Figure S1) in the region containing the deletion. Furthermore, we realized that these mismaps (SBSs or soft clips) arose from the ends of the reads from either direction.
To determine if these mapping errors were ubiquitous at sites of InDels, 19 deletions, 21 insertions, and 24 FLT3 ITDs (two cases with multiple ITDs) were systematically analyzed (Figure 2 and Supplemental Tables S1–S6). The InDels ranged in length from 1 to 30 bases, whereas the FLT3 ITDs ranged in length from 6 to 183 bases. Examination of the sequence reads around InDel loci in IGV showed mapping errors in every case, with 3% to 100% of the reads with the InDel showing mismaps [deletions (Table 1), insertions (Table 2), FLT3 ITDs single clone (Table 3), and FLT3 ITDs multiple clones (Table 4)]. Specifically, these reads had mismapped regions of varying length occurring near the 5′ or 3′ end of the read. Careful inspection revealed that these reads actually contain the InDel and should have been mapped accordingly. Instead, they were erroneously mismapped, which led to SBS artifacts in some cases (Figure 2).
Table 1.
Characteristics of VAFs before and after ABRA2 for Deletions
| Case | Gene | Size of deletion | Estimated tumor cellularity, % | % Mismaps | % Mismaps fixed by ABRA2 | VC VAF, % | Manual calculation of |
Absolute difference between |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Before ABRA2 VAF, % |
(2) After ABRA2 VAF, % |
(3) True NGS VAF, % |
Before and after ABRA2 VAF (2) – (1), % |
After ABRA2 VAF and true NGS VAF (3) – (2), % |
Before ABRA2 VAF and true NGS VAF (3) – (1), % |
|||||||
| 1 | CDKN2A | 2 | 30 | 7.0 | 100 | 12.7 | 12.7 | 13.7 | 13.7 | 1.0 | 0 | 1.0 |
| TSC2 | 3 | 10.7 | 100 | 41.4 | 41.4 | 46.4 | 46.4 | 5.0 | 0 | 5.0 | ||
| 2 | EGFR | 18 | 20 | 38.0 | 88.9 | 12.0 | 11.9 | 18.3 | 19.1 | 6.4 | 0.8 | 7.2 |
| FOXO3 | 6 | 13.7 | 50 | 27.2 | 27.2 | 29.4 | 31.6 | 2.2 | 2.2 | 4.4 | ||
| 3 | PIK3R1 | 6 | 30 | 21.7 | 97.7 | 29.6 | 29.6 | 37.6 | 37.7 | 8.0 | 0.1 | 8.1 |
| 4 | CHD2 | 3 | 95 | 14.0 | 68.4 | 51.1 | 51.1 | 56.8 | 59.4 | 5.7 | 2.6 | 8.3 |
| 5 | NF2 | 5 | 60 | 16.4 | 66.7 | 28.6 | 28.2 | 31.9 | 33.7 | 3.7 | 1.8 | 5.5 |
| 6 | APC | 8 | 60 | 23.2 | 63.2 | 21.9 | 21.9 | 26.0 | 28.5 | 4.1 | 2.5 | 6.6 |
| ARID1A | 3 | 13.0 | 83.3 | 14.4 | 16.7 | 18.8 | 19.2 | 2.1 | 0.4 | 2.5 | ||
| 7 | APC | 1 | 60 | 5.0 | 100 | 65.5 | 65.4 | 68.9 | 68.9 | 3.5 | 0 | 3.5 |
| FANCD2 | 6 | 29.1 | 96.7 | 16.2 | 16.2 | 22.6 | 22.8 | 6.4 | 0.2 | 6.6 | ||
| PIK3R1 | 12 | 31.0 | 100 | 20.7 | 20.7 | 30.0 | 30.0 | 9.3 | 0 | 9.3 | ||
| 8 | PRKDC | 1 | 60 | 3.3 | 90.9 | 38.2 | 38.2 | 39.4 | 39.5 | 1.2 | 0.1 | 1.3 |
| 9 | HIST1H2AM | 2 | 80 | 6.6 | 96.8 | 42.5 | 42.5 | 45.4 | 45.5 | 2.9 | 0.1 | 3.0 |
| 10 | APC | 2 | 50 | 12.1 | 95 | 51.1 | 50.9 | 57.5 | 57.9 | 6.6 | 0.4 | 7.0 |
| 11 | ATRX | 1 | 90 | 5.0 | 100 | 37.7 | 37.7 | 39.7 | 39.7 | 2.0 | 0 | 2.0 |
| 12 | JARID2 | 18 | 70 | 39.8 | 100 | 27.0 | 27.0 | 44.8 | 44.8 | 17.8 | 0 | 17.8 |
| 13 | APC | 13 | 40 | 31.1 | 97.9 | 15.8 | 15.8 | 22.8 | 23.0 | 7.0 | 0.2 | 7.2 |
| 45 | KMT2D | 30 | 100 | 72.4 | 97.7 | 16.6 | 16.5 | 58.8 | 59.8 | 42.3 | 1.0 | 43.3 |
VC VAF % calculated by the VC before the use of ABRA2; (1), (2), and (3): manually calculated by visualizing reads in Integrative Genomics Viewer.
ABRA2, Assembly-Based ReAligner; NGS, next-generation sequencing; VAF, variant allele frequency; VC, variant caller.
Table 2.
Characteristics of VAFs before and after ABRA2 for Insertions
| Case | Gene | Size of insertion | Estimated tumor cellularity, % | % Mismaps | % Mismaps fixed by ABRA2 | VC VAF, % | Manual calculation of |
Absolute difference between |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Before ABRA2 VAF, % |
(2) After ABRA2 VAF, % |
(3) True NGS VAF, % |
Before and after ABRA2 VAF (2) – (1), % |
After ABRA2 VAF and true NGS VAF (3) – (2), % | Before ABRA2 VAF and true NGS VAF (3) – (1), % |
|||||||
| 6 | APC | 1 | 60 | 6.8 | 82.4 | 39.3 | 61.8 | 65.5 | 66.3 | 3.7 | 0.8 | 4.5 |
| 7 | IRS2 | 3 | 60 | 23.4 | 100 | 9.6 | 10.4 | 13.5 | 13.5 | 3.1 | 0 | 3.1 |
| RELN | 12 | 50.6 | 84.7 | 13.0 | 14.9 | 27.7 | 30.1 | 12.8 | 2.4 | 15.2 | ||
| 9 | NF1 | 1 | 80 | 10.9 | 100 | 37.9 | 58.8 | 65.9 | 65.9 | 7.1 | 0 | 7.1 |
| SMAD4 | 1 | 3.4 | 100 | 31.3 | 44.5 | 46.1 | 46.1 | 1.6 | 0 | 1.6 | ||
| 13 | ARID1A | 1 | 40 | 5.6 | 100 | 15.4 | 18.2 | 19.2 | 19.2 | 1.0 | 0 | 1.0 |
| 14 | AXIN1 | 12 | 20 | 61.2 | 73.2 | 26.6 | 26.3 | 56.7 | 67.9 | 30.4 | 11.1 | 41.5 |
| 15 | ATRX | 1 | 30 | 9.6 | 100 | 26.7 | 36.1 | 39.9 | 39.9 | 3.8 | 0 | 3.8 |
| MAPK1 | 3 | 21.2 | 100 | 25.0 | 31.7 | 40.2 | 40.2 | 8.5 | 0 | 8.5 | ||
| 16 | CIC | 1 | 90 | 8.0 | 92.3 | 39.3 | 62.5 | 67.5 | 67.9 | 5.0 | 0.4 | 5.4 |
| 17 | JAK1 | 22 | 90 | 95.7 | 69.3 | 8.7 | 1.9 | 30.5 | 43.2 | 28.6 | 12.7 | 41.3 |
| 18 | TSC1 | 1 | 30 | 6.2 | 100 | 41.6 | 69.1 | 73.7 | 73.7 | 4.6 | 0 | 4.6 |
| 19 | BRCA1 | 1 | 10 | 2.8 | 100 | 34.2 | 51.9 | 53.4 | 53.4 | 1.5 | 0 | 1.5 |
| 20 | KMT2D | 3 | 60 | 31.3 | 100 | 20.5 | 24.2 | 35.2 | 35.2 | 11.0 | 0 | 11.0 |
| 21 | TSC1 | 5 | 50 | 16.8 | 93.8 | 30.6 | 41.4 | 49.2 | 49.7 | 7.8 | 0.5 | 8.3 |
| 22 | TGFBR2 | 1 | 30 | 11.5 | 100 | 5.3 | 5.5 | 6.2 | 6.2 | 0.7 | 0 | 0.7 |
| KDM6A | 3 | 22.2 | 100 | 27.5 | 37.4 | 48.1 | 48.1 | 10.7 | 0 | 10.7 | ||
| 23 | ARID1A | 9 | 60 | 37.6 | 98.1 | 22.3 | 26.4 | 42.0 | 42.3 | 15.6 | 0.3 | 15.9 |
| 24 | NF1 | 1 | 80 | 6.6 | 94.1 | 40.5 | 67.4 | 71.9 | 72.2 | 4.5 | 0.3 | 4.8 |
| 25 | TP53 | 6 | 90 | 25.4 | 100 | 29.6 | 38.1 | 51.1 | 51.1 | 13.0 | 0 | 13.0 |
| 26 | TP53 | 4 | 30 | 21.7 | 100 | 9.6 | 10.5 | 13.4 | 13.4 | 2.9 | 0 | 2.9 |
VC VAF % calculated by the VC before the use of ABRA2; (1), (2), and (3): manually calculated from visualizing reads in Integrative Genomics Viewer.
ABRA2, Assembly-Based ReAligner; NGS, next-generation sequencing; VAF, variant allele frequency; VC, variant caller.
Table 3.
Characteristics of VAFs before and after ABRA2 for FLT3 Internal Tandem Duplications (Single Clone)
| Found by | Case | Sample type | Gene | Size of insertion (NGS) | ∼Size of insertion (CE) | % Mismaps | % Mismaps fixed by ABRA2 | VC VAF, % | Manual calculation of |
CE VAF, % | Absolute difference between |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Before ABRA2 VAF, % |
(2) After ABRA2 VAF, % |
(3) True NGS VAF, % |
Before and after ABRA2 VAF (2) – (1), % |
After ABRA2 VAF and true NGS VAF (3) – (2), % |
Before ABRA2 VAF and true NGS VAF (3) – (1), % |
||||||||||
| BWA, VC, and CE | 27 | BM | FLT3 | 6 | 6 | 28.7 | 100 | 9.0 | 10.2 | 14.4 | 14.4 | 13.6 | 4.1 | 0 | 4.1 |
| 28 | PB | FLT3 | 15 | 14 | 63.7 | 100 | 13.9 | 16.3 | 44.9 | 44.9 | 45.2 | 28.6 | 0 | 28.6 | |
| 29 | PB | FLT3 | 21 | 20 | 87.8 | 72.0 | 10.4 | 11.7 | 72.4 | 96.1 | 89.8 | 60.7 | 23.7 | 84.4 | |
| 30 | PB | FLT3 | 21 | 19 | 89.1 | 75.6 | 5.7 | 7.7 | 55.2 | 70.5 | 71.0 | 47.5 | 15.3 | 62.8 | |
| 31 | BM | FLT3 | 21 | 20 | 86.5 | 73.3 | 20.8 | 2.5 | 14.1 | 18.3 | 16.4 | 11.6 | 4.2 | 15.8 | |
| BWA and CE | 32 | PB | FLT3 | 6 | 7 | 31.0 | 100 | ND | 4.8 | 6.9 | 6.9 | 9.0 | 2.1 | 0 | 2.1 |
| VC and CE | 33 | BM | FLT3 | 27 | 25 | 100 | 98.9 | 29.7 | 0 | 28.1 | 28.5 | 28.2 | 28.1 | 0.4 | 28.5 |
| 34 | BM | FLT3 | 30 | 28 | 100 | 100 | 27.0 | 0 | 24.6 | 24.6 | 23.6 | 24.6 | 0 | 24.6 | |
| 35 | PB | FLT3 | 33 | 31 | 100 | 98.8 | 21.9 | 0 | 20.9 | 21.2 | 21.7 | 20.9 | 0.3 | 21.2 | |
| 36 | BM | FLT3 | 39 | 37 | 100 | 98.7 | 14.6 | 0 | 19.2 | 19.5 | 14.7 | 19.2 | 0.3 | 19.5 | |
| 46 | BM | FLT3 | 57 | 55 | 100 | 98.9 | 42.3 | 0 | 37.9 | 38.3 | 37.5 | 37.9 | 0.4 | 38.3 | |
| 37 | BM | FLT3 | 63 | 60 | 100 | 41.2 | 48.0 | 0 | 15.4 | 37.3 | 35.0 | 15.4 | 21.9 | 37.3 | |
| CE | 38 | BM | FLT3 | 39 | 37 | 100 | 96.6 | ND | 0 | 9.8 | 10.1 | 9.4 | 9.8 | 0.3 | 10.1 |
| 39 | BM | FLT3 | 69 | 66 | 100 | 100 | ND | 0 | 33.2 | 33.2 | 30.0 | 33.2 | 0 | 33.2 | |
| 40 | BM | FLT3 | 72 | 69 | 100 | 100 | ND | 0 | 32.0 | 32.0 | 26.5 | 32.0 | 0 | 32.0 | |
| 41 | PB | FLT3 | 84 | 82 | 100 | 99.2 | ND | 0 | 15.1 | 15.3 | 12.4 | 15.1 | 0.2 | 15.3 | |
| 42 | BM | FLT3 | 87 | 84 | 100 | 99.0 | ND | 0 | 12.1 | 12.2 | 9.3 | 12.1 | 0.1 | 12.2 | |
VC VAF % calculated by the VC before the use of ABRA2; (1), (2), and (3): manually calculated from visualizing reads in Integrative Genomics Viewer.
ABRA2, Assembly-Based ReAligner; BM, bone marrow; BWA, Burrows-Wheeler Aligner; CE, capillary electrophoresis; FLT3, fms-related receptor tyrosine kinase 3; ND, not detected; NGS, next-generation sequencing; PB, peripheral blood; VAF, variant allele frequency; VC, variant caller.
Table 4.
Characteristics of VAFs before and after ABRA2 for FLT3 Internal Tandem Duplications (Multiple Clones)
| Found by | Case | Sample type | Gene | Size of insertion (NGS) | ∼Size of insertion (CE) | % Mismaps | % Mismaps fixed by ABRA2 | VC VAF, % | Manual calculation of |
CE VAF, % | Absolute difference between |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Before ABRA2 VAF, % |
(2) After ABRA2 VAF, % |
(3) True NGS VAF, % |
Before and after ABRA2 VAF (2) – (1), % |
After ABRA2 VAF and true NGS VAF (3) – (2), % |
Before ABRA2 VAF and true NGS VAF (3) – (1), % |
||||||||||
| CE | 43 | BM | FLT3 | 54 | 52 | 100 | 100 | ND | 0 | 6.1 | 6.1 | 6.0 | 6.1 | 0 | 6.1 |
| FLT3 | 30 | 29 | 100 | 100 | ND | 0 | 2.8 | 2.8 | <5 | 2.8 | 0 | 2.8 | |||
| CE and BWA | 44 | PB | FLT3 | 51 | 49 | 100 | 100 | 18.8 | 0 | 16.1 | 16.1 | 16.9 | 16.1 | 0 | 16.1 |
| FLT3 | 21 | 20 | 79.0 | 100 | ND | 0.5 | 2.2 | 2.2 | <5 | 1.7 | 0 | 1.7 | |||
| FLT3 | 24 | ND | 100 | 100 | ND | 0 | 1.0 | 1.0 | ND | 1.0 | 0 | 1.0 | |||
| FLT3 | 24 | ND | 100 | 12.0 | ND | 0 | 0.4 | 2.9 | ND | 0.4 | 2.5 | 2.9 | |||
| FLT3 | 183 | 181 | 100 | 0 | ND | 0 | 0 | 6.6 | <5 | 0 | 6.6 | 6.6 | |||
VC VAF % calculated by the VC before the use of ABRA2; (1), (2), and (3): manually calculated from visualizing reads in Integrative Genomics Viewer.
ABRA2, Assembly-Based ReAligner; BM, bone marrow; BWA, Burrows-Wheeler Aligner; CE, capillary electrophoresis; FLT3, fms-related receptor tyrosine kinase 3; ND, not detected; NGS, next-generation sequencing; PB, peripheral blood; VAF, variant allele frequency; VC, variant caller.
Using IGV to examine sequence reads near these InDel loci, we report several notable characteristics (Supplemental Figure S1 and Supplemental Tables S3–S6). First, we discovered mapping errors occurred on either side surrounding the InDel; therefore, we characterized mismapping events as occurring on the left if the sequence read had perfect homology with the reference sequence on the left side of the InDel within the IGV window, and conversely for the right side. By default, IGV will display bases in the order they appear in the FASTA file for the reference sequence, and for humans, this will be the positive strand. Although it is also possible to display the negative strand in IGV, the bases are still ordered from left to right in the window by the positive strand. Therefore, the use of left or right of the InDel in the IGV window to describe the sequencing characteristics of the mismapped reads will remain the same for all examples regardless of which strand is displayed, whether it is the sense or antisense strand for the particular gene at that location, and the direction of the reads.
On both sides of the InDel, we report the following key characteristics of the mapped reads (Supplemental Figure S1 and Supplemental Tables S3–S7): i) The minimum number of matched bases on one side of the InDel needed for a correctly mapped sequence read—we call this the minimum anchor. ii) The total number of mismapped reads due to soft-clipped bases (soft-clipped reads). A soft-clipped read is partially mapped to the reference sequence, but contains a region with unmatched bases, which are soft clipped.30 The minimum and maximum number of bases that were soft clipped among these reads was also recorded. iii) The total number of mismapped reads erroneously contributing to an SBS artifact (single-base substitutions). iv) The total number of other observed mismapped reads.
Magnitude of Mapping Errors Increase as a Function of InDel Size
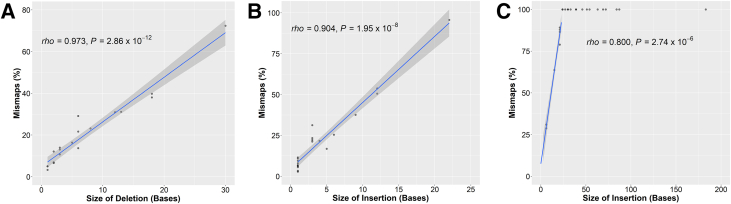
For the short deletions and insertions and the FLT3 ITDs, we determined the percentage of mismapped reads—this is calculated by summing the number of mismapped reads [soft-clipped and not soft clipped (SBSs + other)], and dividing by the sum of the number of mismapped reads and the number of correctly mapped reads that harbor the InDel. The percentage of mismapped reads and size of the InDels are positively correlated with a Spearman rank correlation coefficient of 0.973 for the deletions (P = 2.86 × 10−12) (Figure 4A), 0.904 for the insertions (P = 1.95 × 10−8) (Figure 4B), and 0.8 for the FLT3 ITDs (P = 2.74 × 10−6) (Figure 4C). All the FLT3 ITDs of ≥24 bases in length had 100% mismaps and were excluded from the correlation calculation.
Figure 4.
Mismapped reads increase with the size of the insertion and deletion. A–C: Graphed are percentage mismapped reads as a function of the size for each of the deletions (A), insertions (B), or fms-related receptor tyrosine kinase 3 internal tandem duplications (C); see Results for details. Only the data points with <100% mismaps were included for calculation of ρ (Spearman rank correlation coefficient) and P value. Above a certain size (about 25 bp), essentially 100% of the reads are mismapped. A positive correlation for each is noted. The blue lines represent linear regression lines, and the gray areas are the 95% CIs.
Mapping Errors at InDel Sites Produce SBS Artifacts
SBS artifacts were commonly discovered near sites of reported InDels; in 4 of 19 examples of deletions (21.1%) (Supplemental Table S3), in 6 of 21 examples of insertions (28.6%) (Supplemental Table S4), and in 3 of 19 cases (2 cases with multiple ITDs) of FLT3 ITDs (15.8%) (Supplemental Tables S5 and S6). These can occur within the InDel region or can be found immediately adjacent to the InDel.
For example, Figure 2A shows two SBSs associated with an 18-base deletion in JARID2. However, these SBSs were demonstrated to be artifacts due to numerous mismapped reads. Figure 2A shows mismapped reads due to soft clips (see Supplemental Figure S1 for a schematic demonstrating reads with SBSs versus soft clips), which actually have perfect homology with the reference sequence, and the correct mapping is indicated by the arrows—as such, these reads contain the deletion and should have been mapped akin to the reads at the top of the panel. In an attempt to fix the mismaps, a software program called ABRA2 was incorporated31,32 in our bioinformatics pipeline (Figure 1) to see if it could fix the mismapped reads and prevent the SBS artifacts. ABRA2 works by performing a localized de novo assembly followed by global realignment to more accurately remap reads from NGS data to the reference sequence.31,32 For JARID2, the use of ABRA2 resulted in correction of 100% of the mismapped reads (Table 1), and the two SBS artifacts were no longer present (Figure 5A). For example, one read with a single SBS had its CIGAR string updated from 101M to 1M18D100M to correctly represent the 18-base deletion in this location (Supplemental Table S7). In addition, a soft-clipped read was changed from 83M18S to 68M18D33M (Supplemental Table S7).
Figure 5.
Correction of single-base substitution (SBS) artifacts with the use of Assembly-Based ReAligner (ABRA2). A: For the JARID2 18-bp deletion (case 12; same case as shown in Figure 2A), the top panel shows the uncorrected original file, whereas the bottom panel shows the same data after ABRA2 analysis. Note that ABRA2 fixes 100% of the mismaps and corrects the two SBS artifacts. As a consequence, the variant allele frequency (VAF) is increased from 26.97% to 44.81%. Top and bottom panels: Colored reads appear in the same order. B: For the AXIN1 12-bp insertion (case 14; same case as shown in Figure 2B), ABRA2 fixes 73.2% of the mismaps and corrects two of the three SBS artifacts. The VAF is increased from 26.3% to 56.7%. Top and bottom panels: Colored reads appear in the same order.
Similarly, for a two-base deletion in HIST1H2AM, ABRA2 fixed 96.8% of the mismapped reads and corrected an SBS artifact (Supplemental Figure S2A). As an example, a read with an SBS showed a change in its CIGAR string from 101M to 100M2D1M (Supplemental Table S7). A single mismapped SBS read in this case went uncorrected and retained its 101M CIGAR string instead of being correctly updated to 4M2D97M (Supplemental Table S7).
In an example involving an insertion, three SBSs were associated with a 12-base insertion in AXIN1 (Figure 2B). Once again, several reads were mismapped, generating SBS artifacts. Figure 2B shows mismapped reads due to soft clips (see Supplemental Figure S1 for a schematic demonstrating reads with SBSs versus soft clips). It is apparent that these reads harbor the insertion, have perfect homology with the reference sequence, and should have been mapped as indicated by the arrow (Figure 2B). For AXIN1, ABRA2 fixed 73.2% of the mismapped reads (Table 2), resulting in a correction of two of the three SBS artifacts (Figure 5B). Supplemental Table S7 highlights the CIGAR strings of some of the uncorrected reads. Enough SBS reads remained uncorrected such that IGV still indicated that one SBS remained (Figure 5B). Similarly, for a three-base insertion in KMT2D, ABRA2 fixed 100% of the mismapped reads and corrected two SBS artifacts (Supplemental Figure S2B).
Although all the identified SBSs were highlighted by IGV when visualizing the InDel area, only two of them (case 14, AXIN1; and case 30, FLT3) (Supplemental Tables S4 and S5) were actually called by our pipeline concurrently with the InDel. This is because they were the only erroneous SBSs with a VAF of >5%, and our internally developed variant caller (Molecular Diagnostics Laboratory Variant Caller) uses a VAF cutoff of 5%, whereas IGV uses 2%. The other variant caller utilized in our pipeline, HaplotypeCaller, uses a VAF cutoff of 10%. Interestingly, ABRA2 was able to fix all the SBSs (Supplemental Tables S3–S5) except for these cases (Figure 5B and Supplemental Figure S3). In case 14, an SBS tended to occur in many of the reads rather than soft clips because the insertion sequence showed similarity to the reference sequence in 11 of the 12 bases. In case 30, ABRA2 was only able to fix 75.6% of the mismaps (Table 3), and none of those were the reads with the SBS (Supplemental Table S7 provides some examples).
Mapping Errors at InDel Sites Result in Undercalled VAFs
One consequence of mismapped reads at locations of InDels is that they do not get included in the calculation of the VAF, and thus, the VAF is erroneously low. In these cases, the true VAF is calculated by adding the correctly mapped reads with the mismapped reads and dividing by the total number of reads at that location, as indicated by the coverage. This is termed the true NGS VAF. The before and after ABRA2 VAFs were similarly calculated, using only the correctly mapped reads or the correctly mapped reads plus the ABRA2 corrected reads in the numerator, respectively. All comparisons between the VAFs were made using these calculated values for consistency, as sometimes the final VAF reported by the variant callers would be slightly different due to additional processing done by the algorithms to try to discover variants. For example, for several FLT3 ITD cases (cases 33 to 37 and 46) (Table 3), an insertion was found and reported with a VAF by the variant callers, but if one looks at the data in IGV after the alignment step, 0 insertions are identified and thus, the before ABRA2 VAF would be 0% (Table 3).
Using the aforementioned calculations, the VAF was undercalled by an absolute difference anywhere from 1.0% to 43.3% for deletions (Table 1), 1.0% to 41.5% for insertions (Table 2), and 1.0% to 84.4% for the FLT3 ITDs (Tables 3 and 4). To put this in context, for a JAK1 22-base insertion, a VAF of 1.9% was calculated using the correctly mapped reads with the insertion over the total number of reads at that location, whereas the true NGS VAF should have been 43.2% after accounting for the mismapped reads, an absolute difference of 41.3% (Table 2). Similarly, for a 21-base FLT3 ITD for case 29, the calculated VAF was 11.7%, whereas the true NGS VAF should have been 96.1%, an absolute difference of 84.4% (Table 3).
In cases where ABRA2 fixed 100% of the mismaps, the true NGS VAF would then be realized. However, ABRA2 was only able to achieve this for 6 of the 19 deletions (31.6%), 13 of the 21 insertions (61.9%), and 7 of the 19 FLT3 ITDs (36.8%). However, although some cases did not reach 100% correction, many had >90% of their mismaps corrected. In general, save for a few exceptions, the correction rate was >60% for deletions, >80% for insertions, and >70% for the single-clone FLT3 ITDs.
Therefore, when looking at the absolute amount of VAF that remained uncorrected by ABRA2, this ranged up to 2.6% for the deletions, 12.7% for the insertions, and 23.7% for the FLT3 ITDs. For example, the 22-base insertion in JAK1 saw a change in VAF from 1.9% to 30.5% after ABRA2 corrected 69.3% of the mismaps, leaving a 12.7% shortfall from the true NGS VAF of 43.2% (case 17) (Table 2). In addition, a 21-base FLT3 ITD went from a VAF of 11.7% to 72.4% after ABRA2 fixed 87.8% of the mismaps, leaving a 23.7% shortfall from the true NGS VAF of 96.1% (case 29) (Table 3).
For the FLT3 ITDs, we also have the advantage of having an additional assay, CE, which was run concurrently with the NGS and can give us a sense of the true VAF by use of an alternative method (termed CE VAF) (Tables 3 and 4). Overall, the CE VAF did not differ by >6.3% from the true NGS VAF. The approximate insertion size also did not differ by more than three bases.
Mapping Errors at InDel Sites Result in Unreported FLT3 ITDs
Although all the short InDels (1 to 30 bases) analyzed in this study were still detected by the variant callers despite the mismaps, 12 of the 24 (50.0%) FLT3 ITDs (includes 6 single-clone cases, 2 clones from case 43, and 4 clones from case 44) analyzed were not detected by our pipeline (Tables 3 and 4). However, all the single clones and several of the multiple clones were detected by capillary electrophoresis. Except for one 6-bp insertion, all of the single clone FLT3 ITDs missed by the pipeline tended to be much longer, anywhere from 39 to 87 bases. Moreover, in these cases, 100% of the reads were mismapped and >96% of the mismapped reads were corrected by ABRA2. For the rest of the single-clone cases, ABRA2 corrected >70% of the mismaps, except for one case at 41.2% (Supplemental Figure S4).
In two cases where the FLT3 ITD was missed (cases 43 and 44), multiple clones with different insertion sizes were present (Table 4). For case 43, both clones were detected by capillary electrophoresis, and they were also identifiable by NGS after manual review in IGV or with the use of ABRA2 (Figure 6). For case 44, three clones of sizes 20, 49, and 181 were identified by capillary electrophoresis, whereas manual review in IGV of the NGS data identified clones of sizes 21, 24, 24, 51, and 183. ABRA2 was able to identify all these clones except for the one with an insertion size of 183 bases (Supplemental Figure S5A). Most of these NGS clones were likely attributable to the CE peaks at 20, 49, and 181 (Supplemental Figure S5B). Supplemental Table S7 indicates how the CIGAR string for some of the mismapped reads in this case should have been corrected.
Figure 6.
Identification of fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplications (ITDs) with the use of Assembly-Based ReAligner (ABRA2; case 43). A: A FLT3 ITD with two separate clones is shown. Top panel: The original BAM file. Bottom panel: ABRA2 fixes 100% of the mismaps and identifies both clones with insertion sizes of 54 and 30 bases. The variant allele frequencies (VAFs) after ABRA2 analysis are 6.1% and 2.8%, respectively. Top and bottom panels: Colored reads appear in the same order. The red bar at the top represents the fragment amplified for capillary electrophoresis. B: Both FLT3 ITD clones are also identifiable via the use of capillary electrophoresis with similar insertion sizes of 52 and 29 bases (shorter green/blue peaks; amplicon generated from dual-labeled primers). The VAFs were reported as 6.0% and <5%, respectively. The wild-type (WT) fragment size is 329 bases (highest green/blue peak).
To determine if a variant caller could detect the missed FLT3 ITDs now that ABRA2 had been used, ABRA2's included variant caller, Cadabra, was run on the realigned BAM files. Cadabra could now identify all of the missed single-clone ITDs (cases 32 and 38 to 42), both ITD clones in case 43, and three of the five clones in case 44. In case 44, it only highlighted one of the 24-base ITD clones and did not find the 183-base ITD that was also uncorrected by ABRA2.
Mapping Errors at Sites of InDels Are Not Specific to the Platform Used
Although most of the examples used in this study came from the Illumina HiSeq platform (cases 1 to 44), the mapping errors described in this study are not specific to the platform used and continue to occur even with the use of the NovaSeq platform (cases 45 and 46). Supplemental Figure S6 shows an example from the NovaSeq platform where several reads aligned to the location of the KMT2D gene harbor multiple SBSs combined with a 3-bp insertion instead of a 30-bp deletion after the use of ABRA2. Similarly, from the NovaSeq platform, Supplemental Figure S7 shows a 57-bp FLT3 ITD identified in several reads with soft clips after the use of ABRA2.
Compute Resource
Because the clinical turnaround time for NGS assays can take time, it is important to consider the increased run time of integrating new tools into a clinical bioinformatics pipeline. Our bioinformatics pipeline is run on an approximately 500-core high-performance compute cluster consisting of Intel and AMD architecture, which is located on premise. We determined that an ABRA2-based pipeline required up to 10 GB of virtual memory and could take from 1 to 5 hours of processing time, depending on the complexity of the data.
Discussion
In the course of our clinical sign out of molecular NGS cases of solid or hematologic malignancy, the presence of misaligned reads around the location of InDels was frequently noted. Overall, it is known that InDel detection by NGS can be challenging with the use of short-read technology.33 This is because as the InDel gets larger, fewer and fewer bases from a read in that area will map to the reference sequence, and as a consequence, the aligner will often soft clip the read (hide/ignore portions of a read) instead of flagging it as a potential InDel (Figure 3).34 In this study, we further explore this phenomenon and its consequences using several examples from clinical cases. In addition, we utilize a recently developed tool, ABRA2,31,32 to improve InDel detection and avoid false SBS calls in our bioinformatics pipeline.
InDels can be discovered by various algorithmic methods.35,36 In an alignment-based method, variant callers utilize aligned reads as input and are optimized for detecting small InDels.34 However, once an InDel becomes >15% of the read length,33 these callers start to degrade in performance.33,34 Therefore, tools that utilize other algorithmic methods (eg, localized assembly, split read mapping, paired end mapping, read coverage depth analysis, haplotype based, and machine learning)34,37,38 or target certain InDel lengths39,40 have been developed to address this problem. Although many comparative studies on the performance of different variant callers for the detection of InDels have been done,30,38,41, 42, 43, 44, 45, 46, 47 concordance rates among the different callers is low,30,38 and the choice of software may vary depending on the size of the InDel, sequencing parameters like coverage and read length, or tumor purity.42,46,47
We decided to use ABRA2 in this study for a few reasons. It was fairly new at the time, and other groups besides the developers had not previously published a focused evaluation tool, although some groups, such as Memorial Sloan Kettering Cancer Center, had indicated that they were utilizing a version of tool in their pipeline.48, 49, 50 Using both simulated and real tumor data, the developers demonstrated that it improves InDel detection when used with downstream variant callers.32 This was ideal for our use, because it does not call variants itself, but instead produces a realigned BAM file (Figure 1), which could be used to produce a visual result of how it changed the mismapped reads from the examples described in this article.
Our pipeline currently uses an internally developed variant caller (Molecular Diagnostics Laboratory Variant Caller) for identifying single-nucleotide variants and explicit InDels at lower allele frequencies and the Genome Analysis Toolkit HaplotypeCaller for detecting single-nucleotide variants and InDels at higher allele frequencies.51, 52, 53 HaplotypeCaller has been shown to perform well at InDel detection in several comparative studies,30,42,43,46 although most of these studies have not evaluated it in the somatic setting. HaplotypeCaller is similar to ABRA2 in that it uses a localized assembly-based method to detect InDels, although it is intended for germline variant discovery and its use is not currently recommended for somatic variants. Our use of the tool predates development of Genome Analysis Toolkit's latest somatic caller, Mutect2, which now incorporates the assembly-based machinery of HaplotypeCaller into its algorithm.
Our systematic analysis of short InDels and FLT3 ITDs showed that mapping errors by the aligner occurred in every case, with 3% to 100% of reads with the InDel showing a mismap. The mismap in most cases consisted of soft-clipped bases or an SBS (Supplemental Figure S1 and Supplemental Table S7). Although soft-clipped reads appear in the alignment file (BAM) and can be displayed in IGV, it is also possible for reads to be hard clipped, where the hard-clipped bases are trimmed from the read and do not appear in the alignment file. BWA-MEM by default will soft clip the primary alignment of a read while hard clipping any supplementary alignments. For several examples, it was observed that although ABRA2 can correct mismaps with soft-clipped bases or SBSs, hard-clipped reads are also present around the location of InDels. The developers of ABRA2 indicate that hard-clipped reads are eligible for realignment, although this was not the case in the version we used for this study. However, these reads will still be less likely to be realigned because the hard-clipped bases have been removed and do not appear in the alignment file. To rectify this issue to make the full read sequence of the supplementary alignment available for realignment would require changing an option when BWA is run to force soft clipping instead of hard clipping. This may help optimize realignment of a read by making the full alternative alignments available.
The percentage of mismapped reads also correlated positively with the size of the InDel with all of the 100% mismap cases consisting of FLT3 ITDs with insertion sizes of ≥24 bases. Despite these mismaps, our variant callers were still able to detect all of the short InDels and many of the FLT3 ITDs, although several FLT3 ITDs with large insertion sizes (≥39) or multiple clones were not detected.
For the short InDels, it was found that ABRA2 could correct >80% of the mismaps in many of the cases, but there were a few cases with correction rates only in the 50% or 60% range. On further inspection of the cases with lower correction rates, it was found that the uncorrected reads with a FOXO3 deletion (case 2) had a low-quality score, whereas there was an extraneous sequencing artifact within many of the reads in the vicinity of the deletions for CHD2 (case 4), NF2 (case 5), APC (case 6), and ARID1A (case 6). For the insertions, two of the cases with low correction rates, 82.4% for APC (case 6) and 69.3% for JAK1 (case 17), also had the exact same artifact present within many of the reads in the vicinity of the insertions, including those reads with the insertion (Supplemental Figure S8). The sequencing artifact present in all these cases (5′-TCTTTCCCTACACGACGCTCTTCCGATCT-3′ or its reverse complement) does not map to the human genome and might represent a mistake introduced at some point during the sequencing process. This 29-base sequence was soft clipped by the aligner (case 17, JAK1, CIGAR strings 10S61M29S, 46M54S, and 29S36M35S) (Supplemental Table S7). Taken as a whole, except for cases with this sequencing artifact, ABRA2 performed well at correcting short InDels.
For the FLT3 ITDs, we found that ABRA2 could correct >70% of the mismaps in the single clone case (6 to 87 bases) except for one case (Supplemental Figure S4), and only failed to correct one 183-base insertion in a multiple clone case (Supplemental Figure S5). Use of ABRA2's variant caller Cadabra resulted in identification of all of the FLT3 ITDs previously missed by the pipeline except for two clones in a multiple clone case. One comparative study done before the development of ABRA2 identified Pindel as the best program for detecting FLT3 ITDs using samples with insertion sizes up to 185 bases.38 Therefore, if laboratories plan to abandon traditional capillary electrophoresis–based methods for detecting FLT3 ITDs in favor of NGS, a multisoftware approach may be necessary utilizing tools optimized for the detection of different insertion sizes.
Although our variant callers were able to detect all the short InDels and many of the FLT3 ITDs, additional consequences of the mismaps include SBS artifacts at the location of the InDels and erroneously low VAFs. Although SBS artifacts occurred on occasion in our examples using IGV's VAF cutoff of 2%, they rarely showed up in our final lists of variants because of a more stringent VAF cutoff of 5% by our variant caller. For laboratories with a lower cutoff, the aberrant SBSs in the final lists of variants would necessitate more time during the tertiary analysis for discovery and removal from the final report. ABRA2 was able to correct all SBSs that appeared at a VAF of at least 2%, except for two cases (Figure 5B and Supplemental Figure S3).
Last, where ABRA2 made a big impact was on bringing the VAF equal to or closer to the true VAF (either by NGS or CE). Accounting for the mismaps, it was determined that the VAF was being undercalled anywhere from 1.0% to 84.4% and that the absolute amount of remaining uncorrected VAF after the use of ABRA2 ranged up to 12.7% for the short InDels and up to 23.7% for the FLT3 ITDs. Clinically, in many cases, such differences in the VAF may not make a difference in a patient's treatment course. However, in cases where targeted therapies do exist to identified variants, such as with epidermal growth factor receptor tyrosine kinase inhibitors for lung adenocarcinomas with epidermal growth factor receptor exon 19 deletions, as found in case 2,54 studies have shown an association with improved progression-free survival and overall survival in patients with higher adjusted tumoral VAFs treated with epidermal growth factor receptor tyrosine kinase inhibitors.55 The importance of defining the VAF accurately is highlighted with its use in comparing the VAFs of various mutations to determine whether a given mutation was present as an initiating or early event in contrast to those acquired late and may be subclonal and thereby poorly targetable. Moreover, for laboratories that might make use of the VAF as a means to track disease progression over time, especially in hematologic malignancies, because a laboratory's choice of software will vary, a reported VAF should not be considered comparable across institutions or different NGS assays.
In this study, we try to bring attention to a phenomenon that frequently occurs with the alignment of short reads around InDels (Table 5). Because of this, it is important for clinical molecular laboratories to utilize software specifically designed for detecting InDels, with an awareness that different algorithms may perform better within certain size ranges. We have found ABRA2 performs well on a wide variety of insertions and deletions as well as for FLT3 ITDs that are <100 bases, although sequencing artifacts may limit the percentage of reads it is able to correct. This study is meant to be a proof of concept demonstrating detailed results for many different real-world examples and not an exhaustive comparison of many different variant callers at a high level, as has already been done elsewhere.30,38,41, 42, 43, 44, 45, 46, 47 If we were to incorporate ABRA2 into our pipeline, more rigorous testing, including the use of additional software, such as Pindel,56 additional real-world data, and possibly simulated data would be utilized. Moreover, we have found that our compute resource may be a limiting factor to its use, as the increased processing time could significantly delay result generation.
Table 5.
Critical Features of InDel Artifact
|
InDel, insertion and deletion; SBS, single-base substitution; VAF, variant allele frequency.
Acknowledgments
We thank Rena R. Xian and Christopher D. Gocke for helpful discussions, constructive feedback, and input throughout the study; and Suping Chen, Lisa Haley, Emily Adams, and Katie Beierl for helpful discussions and expert technical assistance.
Footnotes
Supported in part by The Sol Goldman Pancreatic Cancer Research Center.
Disclosures: None declared.
Current address of K.E.C., Department of Pathology and Laboratory Medicine, Memorial Sloan Kettering Cancer Center, New York, NY; of C.G.F., Division of Molecular Genetics and Pathology, US Food and Drug Administration, Silver Spring, MD; of L.J., The American Genome Center, Murtha Cancer Center Research Program, Uniformed Services University of the Health Sciences, Bethesda, MD, and Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2022.08.006.
Supplemental Data
Schematic identifying key parameters for analysis. A and B: Representative reads either correctly mapped with the deletion or incorrectly mapped due to soft clips or single-base substitutions are shown. The minimum anchor is the minimum number of matched bases on one side of the insertion and deletion needed for a correctly mapped sequence read. For soft-clipped reads, the minimum and maximum number of soft-clipped bases was recorded. A: Parameters of the reads corresponding to the left side of the deletion are highlighted. B: Parameters of the reads corresponding to the right side of the deletion are highlighted.
Correction of single-base substitution (SBS) artifacts with the use of Assembly-Based ReAligner (ABRA2). A: For a HIST1H2AM 2-bp deletion (case 9; top panel), ABRA2 fixes 96.8% of the mismaps and corrects an SBS artifact (bottom panel). As a consequence, the variant allele frequency (VAF) is increased from 42.5% to 45.4%. Top and bottom panels: Colored reads appear in the same order. B: For a KMT2D 3-bp insertion (case 20; top panel), ABRA2 fixes 100% of the mismaps and corrects the two SBS artifacts (bottom panel). The VAF is increased from 24.2% to 35.2%. Top and bottom panels: Colored reads appear in the same order.
Assembly-Based ReAligner ABRA2) is unable to correct an aberrant single-base substitution (SBS) next to an fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) (case 30). Reads in which the 21-base FLT3 ITD was found by ABRA2 are represented by multiple colors in the middle and bottom of the image for each panel, whereas a purple vertical line represents the location of the insertion. Reads in which ABRA2 failed to find the 21-base insertion are colored in salmon. Purple reads are background reads without the insertion. Overall, ABRA2 corrected 75.6% of the mismaps. The red bar at the top represents the fragment amplified for capillary electrophoresis.
Assembly-Based ReAligner (ABRA2) is only able to correct 41.2% of the mismapped reads with a 63-base fms-related receptor tyrosine kinase 3 internal tandem duplication (case 37). The panels show reads before (top panel) and after (bottom panel) running ABRA2. Uncorrected reads are represented by a salmon color, whereas corrected reads are represented by multiple colors. Purple reads are background reads without the insertion. The red bar at the top represents the fragment amplified for capillary electrophoresis.
Failure to detect a 183-base fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) spanning more than one read length with the use of Assembly-Based ReAligner (ABRA2; case 44). A: For an FLT3 ITD clone with an insertion size of 183 bases, ABRA2 is unable to identify the clone. The soft-clipped area of the reads on the left should line up with the normal reference sequence with the reads on the right, and the remainder of the sequence constitutes the beginning of the insertion. The variant allele frequency (VAF) is 6.6%. The red bar at the top represents the fragment amplified for capillary electrophoresis. B: All clonal FLT3 ITDs from case 44 with different insertion sizes were identified after manual review of the next-generation sequencing data (21, 24, 24, 51, and 183 bases) or with ABRA2 (21, 24, 24, and 51 bases) and correspond with peaks identified by capillary electrophoresis (20, 49, and 181 bases; shorter green/blue peaks). The VAFs from the capillary electrophoresis were reported as <5%, 6.0%, and <5%, respectively. The wild-type (WT) fragment size is 329 bases (highest green/blue peak).
Correction of single-base substitution (SBS) artifacts and improved detection of a KMT2D 30-bp deletion (Novaseq platform; case 45). Top panel: The read mappings from the original BAM file. Bottom panel: The read mappings after running Assembly-Based ReAligner (ABRA2). Top and bottom panels: The multicolored reads appear in the same order. After running ABRA2, it is clear that a 30-bp deletion is present in this region of KMT2D (black horizontal lines; bottom panel) instead of multiple SBSs (blue and green vertical lines; top panel) and a small 3-bp insertion (purple boxed areas; top panel). Salmon colored reads are background reads without the deletion.
Improved detection of a 57-bp fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) with Assembly-Based ReAligner (ABRA2; Novaseq platform; case 46). Top panel: The read mappings from the original BAM file. Bottom panel: The read mappings after running ABRA2. Top and bottom panels: The multicolored reads appear in the same order. Top panel: The vertical colored lines represent soft-clipped bases at the right end of the reads. Bottom panel: A 57-base FLT3 ITD in these reads is only evident after running ABRA2 (purple boxed areas). Salmon colored reads are background reads without the insertion.
Assembly-Based ReAligner (ABRA2) is unable to completely correct reads with a JAK1 22-base insertion due to a sequencing artifact. The panels show reads before (top panel) and after (bottom panel) running ABRA2. Only 69.3% of the mismapped reads with a JAK1 22-base insertion are corrected by ABRA2. Several of the reads that remain uncorrected (salmon color) contain a sequencing artifact within the read that is soft clipped. Top and bottom panels: Reads that were corrected by ABRA2 are ordered the same and appear in the same color. Purple reads are background reads without the insertion.
References
- 1.Harismendy O., Schwab R.B., Bao L., Olson J., Rozenzhak S., Kotsopoulos S.K., Pond S., Crain B., Chee M.S., Messer K., Link D.R., Frazer K.A. Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol. 2011;12:R124. doi: 10.1186/gb-2011-12-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagle N., Berger M.F., Davis M.J., Blumenstiel B., Defelice M., Pochanard P., Ducar M., Van Hummelen P., Macconaill L.E., Hahn W.C., Meyerson M., Gabriel S.B., Garraway L.A. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singhi A.D., McGrath K., Brand R.E., Khalid A., Zeh H.J., Chennat J.S., Fasanella K.E., Papachristou G.I., Slivka A., Bartlett D.L., Dasyam A.K., Hogg M., Lee K.K., Marsh J.W., Monaco S.E., Ohori N.P., Pingpank J.F., Tsung A., Zureikat A.H., Wald A.I., Nikiforova M.N. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut. 2018;67:2131–2141. doi: 10.1136/gutjnl-2016-313586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Springer S., Wang Y., Dal Molin M., Masica D.L., Jiao Y., Kinde I., et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology. 2015;149:1501–1510. doi: 10.1053/j.gastro.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J., Jiao Y., Dal Molin M., Maitra A., de Wilde R.F., Wood L.D., Eshleman J.R., Goggins M.G., Wolfgang C.L., Canto M.I., Schulick R.D., Edil B.H., Choti M.A., Adsay V., Klimstra D.S., Offerhaus G.J., Klein A.P., Kopelovich L., Carter H., Karchin R., Allen P.J., Schmidt C.M., Naito Y., Diaz L.A., Jr., Kinzler K.W., Papadopoulos N., Hruban R.H., Vogelstein B. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci U S A. 2011;108:21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J., Matthaei H., Maitra A., Dal Molin M., Wood L.D., Eshleman J.R., Goggins M., Canto M.I., Schulick R.D., Edil B.H., Wolfgang C.L., Klein A.P., Diaz L.A., Jr., Allen P.J., Schmidt C.M., Kinzler K.W., Papadopoulos N., Hruban R.H., Vogelstein B. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garraway L.A. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 8.Lin M.T., Mosier S.L., Thiess M., Beierl K.F., Debeljak M., Tseng L.H., Chen G., Yegnasubramanian S., Ho H., Cope L., Wheelan S.J., Gocke C.D., Eshleman J.R. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. 2014;141:856–866. doi: 10.1309/AJCPMWGWGO34EGOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes D.N., Kim W.Y. The next steps in next-gen sequencing of cancer genomes. J Clin Invest. 2015;125:462–468. doi: 10.1172/JCI68339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy S., LaFramboise W.A., Nikiforov Y.E., Nikiforova M.N., Routbort M.J., Pfeifer J., Nagarajan R., Carter A.B., Pantanowitz L. Next-generation sequencing informatics: challenges and strategies for implementation in a clinical environment. Arch Pathol Lab Med. 2016;140:958–975. doi: 10.5858/arpa.2015-0507-RA. [DOI] [PubMed] [Google Scholar]
- 11.Do H., Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61:64–71. doi: 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- 12.Chen G., Mosier S., Gocke C.D., Lin M.T., Eshleman J.R. Cytosine deamination is a major cause of baseline noise in next-generation sequencing. Mol Diagn Ther. 2014;18:587–593. doi: 10.1007/s40291-014-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCall C.M., Mosier S., Thiess M., Debeljak M., Pallavajjala A., Beierl K., Deak K.L., Datto M.B., Gocke C.D., Lin M.T., Eshleman J.R. False positives in multiplex PCR-based next-generation sequencing have unique signatures. J Mol Diagn. 2014;16:541–549. doi: 10.1016/j.jmoldx.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura K., Oshima T., Morimoto T., Ikeda S., Yoshikawa H., Shiwa Y., Ishikawa S., Linak M.C., Hirai A., Takahashi H., Altaf-Ul-Amin M., Ogasawara N., Kanaya S. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross M.G., Russ C., Costello M., Hollinger A., Lennon N.J., Hegarty R., Nusbaum C., Jaffe D.B. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R., Bertoni A., Swerdlow H.P., Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laehnemann D., Borkhardt A., McHardy A.C. Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief Bioinform. 2016;17:154–179. doi: 10.1093/bib/bbv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng W., Zhao S., Xue D., Song F., Li Z., Chen D., He B., Hao Y., Wang Y., Liu Y. Improving alignment accuracy on homopolymer regions for semiconductor-based sequencing technologies. BMC Genomics. 2016;17(Suppl 7):521. doi: 10.1186/s12864-016-2894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandelker D., Schmidt R.J., Ankala A., McDonald Gibson K., Bowser M., Sharma H., Duffy E., Hegde M., Santani A., Lebo M., Funke B. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet Med. 2016;18:1282–1289. doi: 10.1038/gim.2016.58. [DOI] [PubMed] [Google Scholar]
- 20.Claes K.B.M., Rosseel T., De Leeneer K. In: Dealing with pseudogenes in molecular diagnostics in the next generation sequencing era. Pseudogenes: Functions and Protocols. Poliseno L., editor. Springer US; New York, NY: 2021. pp. 363–381. [DOI] [PubMed] [Google Scholar]
- 21.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson J.T., Thorvaldsdottir H., Wenger A.M., Zehir A., Mesirov J.P. Variant review with the integrative genomics viewer. Cancer Res. 2017;77:e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treangen T.J., Salzberg S.L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xian R.R., Xie Y., Haley L.M., Yonescu R., Pallavajjala A., Pittaluga S., Jaffe E.S., Duffield A.S., McCall C.M., Gheith S.M.F., Gocke C.D. CREBBP and STAT6 co-mutation and 16p13 and 1p36 loss define the t(14;18)-negative diffuse variant of follicular lymphoma. Blood Cancer J. 2020;10:69. doi: 10.1038/s41408-020-0335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng G., Chen P., Pallavajjalla A., Haley L., Gondek L., Dezern A., Ling H., De Marchi F., Lin M.T., Gocke C. The diagnostic utility of targeted gene panel sequencing in discriminating etiologies of cytopenia. Am J Hematol. 2019;94:1141–1148. doi: 10.1002/ajh.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 28.Murphy K.M., Levis M., Hafez M.J., Geiger T., Cooper L.C., Smith B.D., Small D., Berg K.D. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003;5:96–102. doi: 10.1016/S1525-1578(10)60458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome project data processing S: the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li D., Kim W., Wang L., Yoon K.A., Park B., Park C., Kong S.Y., Hwang Y., Baek D., Lee E.S., Won S. Comparison of INDEL calling tools with simulation data and real short-read data. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:1635–1644. doi: 10.1109/TCBB.2018.2854793. [DOI] [PubMed] [Google Scholar]
- 31.Mose L.E., Wilkerson M.D., Hayes D.N., Perou C.M., Parker J.S. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics. 2014;30:2813–2815. doi: 10.1093/bioinformatics/btu376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mose L.E., Perou C.M., Parker J.S. Improved indel detection in DNA and RNA via realignment with ABRA2. Bioinformatics. 2019;35:2966–2973. doi: 10.1093/bioinformatics/btz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abel H.J., Duncavage E.J. Detection of structural DNA variation from next generation sequencing data: a review of informatic approaches. Cancer Genet. 2013;206:432–440. doi: 10.1016/j.cancergen.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narzisi G., Schatz M.C. The challenge of small-scale repeats for indel discovery. Front Bioeng Biotechnol. 2015;3:8. doi: 10.3389/fbioe.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medvedev P., Stanciu M., Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z.D., Du J., Lam H., Abyzov A., Urban A.E., Snyder M., Gerstein M. Identification of genomic indels and structural variations using split reads. BMC Genomics. 2011;12:375. doi: 10.1186/1471-2164-12-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm D., Hagmann J., Koenig D., Weigel D., Borgwardt K. Accurate indel prediction using paired-end short reads. BMC Genomics. 2013;14:132. doi: 10.1186/1471-2164-14-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J., Guo J.T. Comparative assessments of indel annotations in healthy and cancer genomes with next-generation sequencing data. BMC Med Genomics. 2020;13:170. doi: 10.1186/s12920-020-00818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer D.H., Abel H.J., Lockwood C.M., Payton J.E., Szankasi P., Kelley T.W., Kulkarni S., Pfeifer J.D., Duncavage E.J. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn. 2013;15:81–93. doi: 10.1016/j.jmoldx.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Shigemizu D., Miya F., Akiyama S., Okuda S., Boroevich K.A., Fujimoto A., Nakagawa H., Ozaki K., Niida S., Kanemura Y., Okamoto N., Saitoh S., Kato M., Yamasaki M., Matsunaga T., Mutai H., Kosaki K., Tsunoda T. IMSindel: an accurate intermediate-size indel detection tool incorporating de novo assembly and gapped global-local alignment with split read analysis. Sci Rep. 2018;8:5608. doi: 10.1038/s41598-018-23978-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X., Han S., Wang Z., Gelernter J., Yang B.Z. Variant callers for next-generation sequencing data: a comparison study. PLoS One. 2013;8:e75619. doi: 10.1371/journal.pone.0075619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghoneim D.H., Myers J.R., Tuttle E., Paciorkowski A.R. Comparison of insertion/deletion calling algorithms on human next-generation sequencing data. BMC Res Notes. 2014;7:864. doi: 10.1186/1756-0500-7-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasan M.S., Wu X., Zhang L. Performance evaluation of indel calling tools using real short-read data. Hum Genomics. 2015;9:20. doi: 10.1186/s40246-015-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu C. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput Struct Biotechnol J. 2018;16:15–24. doi: 10.1016/j.csbj.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bian X., Zhu B., Wang M., Hu Y., Chen Q., Nguyen C., Hicks B., Meerzaman D. Comparing the performance of selected variant callers using synthetic data and genome segmentation. BMC Bioinformatics. 2018;19:429. doi: 10.1186/s12859-018-2440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang N., Lysenkov V., Orte K., Kairisto V., Aakko J., Khan S., Elo L.L. Variant calling tool evaluation for variable size indel calling from next generation whole genome and targeted sequencing data. bioRxiv. 2021 doi: 10.1101/2021.07.15.452444. [Preprint] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pei S., Liu T., Ren X., Li W., Chen C., Xie Z. Benchmarking variant callers in next-generation and third-generation sequencing analysis. Brief Bioinform. 2021;22:bbaa148. doi: 10.1093/bib/bbaa148. [DOI] [PubMed] [Google Scholar]
- 48.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., Brannon A.R., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rose Brannon A., Jayakumaran G., Diosdado M., Patel J., Razumova A., Hu Y., et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat Commun. 2021;12:3770. doi: 10.1038/s41467-021-24109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poplin R., Ruano-Rubio V., DePristo M., Fennell T., Carneiro M., Van der Auwera G., Kling DE G L.D., Levy-Moonshine A., Roazen D., Shakir K., Thibault J., Chandran S., Whelan C., Lek M., Gabriel S., Daly M., Neale B., MacArthur D., Banks E. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017 doi: 10.1101/201178. [Preprint] doi: [DOI] [Google Scholar]
- 53.Van der Auwera G., O'Connor B. ed 1. Sebastopol, CA: O'Reilly Media; 2020. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. [Google Scholar]
- 54.Thai A.A., Solomon B.J., Sequist L.V., Gainor J.F., Heist R.S. Lung cancer. Lancet. 2021;398:535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 55.Gieszer B., Megyesfalvi Z., Dulai V., Papay J., Kovalszky I., Timar J., Fillinger J., Harko T., Pipek O., Teglasi V., Regos E., Papp G., Szallasi Z., Laszlo V., Renyi-Vamos F., Galffy G., Bodor C., Dome B., Moldvay J. EGFR variant allele frequency predicts EGFR-TKI efficacy in lung adenocarcinoma: a multicenter study. Transl Lung Cancer Res. 2021;10:662–674. doi: 10.21037/tlcr-20-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic identifying key parameters for analysis. A and B: Representative reads either correctly mapped with the deletion or incorrectly mapped due to soft clips or single-base substitutions are shown. The minimum anchor is the minimum number of matched bases on one side of the insertion and deletion needed for a correctly mapped sequence read. For soft-clipped reads, the minimum and maximum number of soft-clipped bases was recorded. A: Parameters of the reads corresponding to the left side of the deletion are highlighted. B: Parameters of the reads corresponding to the right side of the deletion are highlighted.
Correction of single-base substitution (SBS) artifacts with the use of Assembly-Based ReAligner (ABRA2). A: For a HIST1H2AM 2-bp deletion (case 9; top panel), ABRA2 fixes 96.8% of the mismaps and corrects an SBS artifact (bottom panel). As a consequence, the variant allele frequency (VAF) is increased from 42.5% to 45.4%. Top and bottom panels: Colored reads appear in the same order. B: For a KMT2D 3-bp insertion (case 20; top panel), ABRA2 fixes 100% of the mismaps and corrects the two SBS artifacts (bottom panel). The VAF is increased from 24.2% to 35.2%. Top and bottom panels: Colored reads appear in the same order.
Assembly-Based ReAligner ABRA2) is unable to correct an aberrant single-base substitution (SBS) next to an fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) (case 30). Reads in which the 21-base FLT3 ITD was found by ABRA2 are represented by multiple colors in the middle and bottom of the image for each panel, whereas a purple vertical line represents the location of the insertion. Reads in which ABRA2 failed to find the 21-base insertion are colored in salmon. Purple reads are background reads without the insertion. Overall, ABRA2 corrected 75.6% of the mismaps. The red bar at the top represents the fragment amplified for capillary electrophoresis.
Assembly-Based ReAligner (ABRA2) is only able to correct 41.2% of the mismapped reads with a 63-base fms-related receptor tyrosine kinase 3 internal tandem duplication (case 37). The panels show reads before (top panel) and after (bottom panel) running ABRA2. Uncorrected reads are represented by a salmon color, whereas corrected reads are represented by multiple colors. Purple reads are background reads without the insertion. The red bar at the top represents the fragment amplified for capillary electrophoresis.
Failure to detect a 183-base fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) spanning more than one read length with the use of Assembly-Based ReAligner (ABRA2; case 44). A: For an FLT3 ITD clone with an insertion size of 183 bases, ABRA2 is unable to identify the clone. The soft-clipped area of the reads on the left should line up with the normal reference sequence with the reads on the right, and the remainder of the sequence constitutes the beginning of the insertion. The variant allele frequency (VAF) is 6.6%. The red bar at the top represents the fragment amplified for capillary electrophoresis. B: All clonal FLT3 ITDs from case 44 with different insertion sizes were identified after manual review of the next-generation sequencing data (21, 24, 24, 51, and 183 bases) or with ABRA2 (21, 24, 24, and 51 bases) and correspond with peaks identified by capillary electrophoresis (20, 49, and 181 bases; shorter green/blue peaks). The VAFs from the capillary electrophoresis were reported as <5%, 6.0%, and <5%, respectively. The wild-type (WT) fragment size is 329 bases (highest green/blue peak).
Correction of single-base substitution (SBS) artifacts and improved detection of a KMT2D 30-bp deletion (Novaseq platform; case 45). Top panel: The read mappings from the original BAM file. Bottom panel: The read mappings after running Assembly-Based ReAligner (ABRA2). Top and bottom panels: The multicolored reads appear in the same order. After running ABRA2, it is clear that a 30-bp deletion is present in this region of KMT2D (black horizontal lines; bottom panel) instead of multiple SBSs (blue and green vertical lines; top panel) and a small 3-bp insertion (purple boxed areas; top panel). Salmon colored reads are background reads without the deletion.
Improved detection of a 57-bp fms-related receptor tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) with Assembly-Based ReAligner (ABRA2; Novaseq platform; case 46). Top panel: The read mappings from the original BAM file. Bottom panel: The read mappings after running ABRA2. Top and bottom panels: The multicolored reads appear in the same order. Top panel: The vertical colored lines represent soft-clipped bases at the right end of the reads. Bottom panel: A 57-base FLT3 ITD in these reads is only evident after running ABRA2 (purple boxed areas). Salmon colored reads are background reads without the insertion.
Assembly-Based ReAligner (ABRA2) is unable to completely correct reads with a JAK1 22-base insertion due to a sequencing artifact. The panels show reads before (top panel) and after (bottom panel) running ABRA2. Only 69.3% of the mismapped reads with a JAK1 22-base insertion are corrected by ABRA2. Several of the reads that remain uncorrected (salmon color) contain a sequencing artifact within the read that is soft clipped. Top and bottom panels: Reads that were corrected by ABRA2 are ordered the same and appear in the same color. Purple reads are background reads without the insertion.