Summary
Chromosome gains are detrimental for the development of the human embryo. As such, autosomal trisomies almost always result in spontaneous abortion, and the rare embryos surviving until live birth suffer from a plethora of pathological defects. There is no treatment currently available to ameliorate the consequences of trisomies, such as Down syndrome (trisomy of chromosome 21). Identifying the source of the phenotypes observed in cells with extra chromosomes is crucial for understanding the underlying molecular causes of trisomy syndromes. Although increased expression of the genes localized on the extra chromosome triggers several pathological phenotypes, an alternative model suggests that global, aneuploidy-associated changes in cellular physiology also contribute to the pathology. Here, we compare the molecular consequences of trisomy syndromes in vivo against engineered cell lines carrying various chromosome gains in vitro. We point out several phenotypes that are shared by variable trisomies and, therefore, might be caused by the presence of an extra chromosome per se, independent of its identity. This alternative view may provide useful insights for understanding Down syndrome pathology and open additional opportunities for diagnostics and treatments.
Keywords: aneuploidy, trisomy, Down syndrome, Autophagy, innate immune response, genome instability, interferon type I
Pathology of chromosomal trisomies
A notable category of genetic disorders is caused by the presence of an extra chromosome. The superfluous genetic material disrupts normal development, causing multiple and complex organ defects. In fact, whole organism aneuploidies of human somatic chromosomes (autosomes) cause defects in embryonic development, and only a small fraction of embryos with gains of chromosomes 13, 18, and 21 (Patau (MIM: 264480), Edwards (MIM: 601161), and Down syndrome (DS) (MIM: 190685), respectively) or even rarer mosaic trisomies of chromosomes 8, 16, 19, and 22 result in live births. A different situation is observed with gains of sex chromosomes, which generally result in less severe phenotypes characterized by infertility and reduced body growth.1,2 Of note, the loss of any single autosomal chromosome is embryonically lethal, whereas the loss of a sex chromosome is not, as in the case of monosomy for chromosome X, which is symptomatically diagnosed as Turner syndrome (MIM: 309585).3 In this review, we will focus on autosomal trisomies; readers interested in monosomies or sex chromosome aneuploidies should refer to recent papers on these topics.4,5,6
Autosomal trisomies are associated with a wide range of developmental abnormalities that often lead to early death during infancy. Recent reports determined that the 5-year survival rates were 9.7% for children with trisomy of chr. 13 (T13) and 12.3% for children with trisomy of chr. 18 (T18).7,8 This shortened life expectancy can be improved through invasive interventions; in the case of congenital heart disease, intervention can increase survival by 15 years in approximately 60% of individuals with T13 or T18.9 The trisomy that is compatible with the highest survival rates (nearly 95%) to adulthood is DS, or trisomy of chr. 21 (T21), with an average life expectancy of 60 years.10 Nevertheless, there are over 70 different pathological phenotypes associated with T21, among them most prominently intellectual disability, cardiac defects, early-onset Alzheimer disease (AD), increased autoimmune disorders and recurrent infections, as well as predisposition to acute leukemias.11 On the other hand, these individuals show a reduced incidence of solid tumors in adulthood.12 Although the etiology of these syndromes has been known for several decades, the pathogenesis remains only partially understood.13 On a cellular level, the DS-associated changes include impaired mitochondrial function, altered metabolism and epigenetic patterns, endocytic defects, altered cell fate specification during neurogenesis, increased levels of oxidative stress, and transformed proteostasis and autophagy networks.14
The prevalent hypothesis proposes that the phenotypes of trisomy syndromes arise due to the increased expression of genes located on the supernumerary chromosome through a direct effect and/or indirectly (e.g. via regulation of transcriptional factors). The phenotypes would thus be caused by a subset of genes, so-called dosage-sensitive genes (DSGs), whose abundance alters diverse cellular processes. Indeed, previous studies mapped DS phenotypes to a genomic region known as the “Down Syndrome Critical Region” (DSCR), which covers 5.4 Mb on 21q, the large arm of chr. 21 (see Box 1). The abnormalities might arise due to an indirect effect of increased abundance of chr. 21 genes, which regulate cellular processes such as splicing (U2AF1L5 [MIM: 601080], RBM1 [MIM: 400006], and U2AF1 [MIM: 191317]), DNA methylation (PRMT2 [MIM: 601961] and N6AMT1 [MIM: 614553]), and metabolism (SOD1 [MIM: 147450]). Nonetheless, linking the subtly increased expression of a given gene (on average, 50%) encoded on chromosome 21 to specific phenotypes of DS has been challenging.13,15 Additionally, mice harboring extra copies of the DSCR orthologous region do not display DS-like phenotypes.16,17,18 This evidence thus argues against the “critical region” hypothesis, suggesting rather that multiple genetic abnormalities, and not a single DS region, are responsible for the observed pathological phenotypes.
Box 1. Phenotypes associated with specific chromosome 21 genes.
Chromosome 21 carries 233 protein-coding genes, 423 non-protein-coding genes, and 188 pseudogenes.13 The leading hypothesis of DS pathology suggests that the phenotypes can be linked to increased expression of specific genes encoded on chromosome 21. For example, increased dosage of the amyloid-beta precursor protein APP promotes the susceptibility to early-onset AD in individuals with DS. Triplication of regulator of calcineurin 1 (RCAN1 [MIM: 602917]) enhances its inhibitory activity toward the phosphatase calcineurin A and thus affects endocytosis. Increased expression of the protein kinase DYRK1A causes abnormal phosphorylation status of a number of its direct targets. The increased expression of APP, RCAN1, and DYRK1A in DS contributes to AD-like neuronal malformations with abnormal synaptic plasticity and cell cycle, as well as learning and memory deficits.124,125,126 RCAN1 triplication in DS is also associated with the deregulation of NFAT signaling127,128 that underlies the increased risks of individuals with DS for inflammation and cancer.129 The elevated abundance of HMGN1 is associated with an increased incidence of B cell acute lymphoblastic leukemia.130 The MX1 gene encoding interferon-induced p78 protein (MxA) was shown to promote chronic inflammatory disease alopecia areata in individuals with DS.131 Most of these conclusions are supported, although not fully replicated, by findings in DS mouse models. For example, an overexpression of human RCAN1 in diploid control mice reproduced AD features with memory- and neuronal-specific defects associated with increased oxidative stress,132 whereas removal of one APP copy from DS mouse models partly reduced the AD-like defects.133 Overexpressed DYRK1A in the neocortex of a diploid mouse embryo compromises the mechanisms of cell proliferation and neurogenesis, contributing to cognitive impairment similar to what is observed in DS.134 Further research should focus on understanding to what degree the direct downstream effects of specific chr. 21 gene overexpression affect the phenotypes, and conversely, whether global cellular responses to chromosome gain contribute to DS pathophysiology.
Here, we present another interpretation that can complement this view, namely that cellular defects associated with trisomy syndromes are caused by the disruption of cellular homeostasis due to the presence of any extra chromosome. Indeed, analysis of model cell lines engineered to contain an additional chromosome copy revealed that low-grade, constitutive upregulation of several hundred genes triggers so-called aneuploidy-associated stresses that precipitate genome-wide gene expression changes and alter pathway networks. This, in turn, leads to phenotypes such as proliferation delay, defects in maintenance of protein homeostasis (proteostasis), metabolic changes, elevated DNA damage, and activation of innate immune response.19,20 Thus, whereas some phenotypes of trisomy syndromes may be attributed specifically to changes in DSGs on chr. 21, other pathologies may be caused by cumulative effects of chronic overexpression that lead to genome-wide deregulation of gene expression, or by some combination of both. Although this hypothesis has been proposed and discussed previously,21,22 recent progress in research using cells with engineered aneuploid karyotypes has brought additional support to these findings. It should be noted that the disrupted cellular homeostasis hypothesis is not mutually exclusive with the gene-dosage hypothesis, and we expect that it shall improve – rather than replace – the current model.
A scarcity of human primary material and strong inter-individual variations make the study of general DS phenotypes rather difficult. Therefore, several mouse models of DS, as well as model cell lines of various trisomies (including T21), were established in recent years. No mouse models for other trisomy syndromes have been established so far, and thus only data from primary material are available. Here, we compare results from various analyses of clinical samples and model systems that lend support to the idea that DS phenotypes arise mainly from chromosome gain per se and not due to the gain of chromosome 21 specifically. This perspective might improve our understanding of the pathology of T21 and allow development of novel treatments that will improve the quality of life of the affected individuals.
Global gene expression changes in response to trisomy
One striking consequence of chromosome gain is the global deregulation of the cellular proteome that occurs on two levels. First, the genes carried on the extra chromosome are expressed, which leads to comparatively increased mRNA and protein abundance of these specific factors. Several studies revealed a comparable upregulation of most transcripts in cells from DS-affected individuals, although the lack of proper isogenic controls and high inter-individual variability render these analyses challenging. On average, the abundance of transcripts from genes located on the trisomic chromosomes increases 1.5-fold. Recent analysis of monozygotic twins with diploid and trisomic karyotypes, respectively, confirmed differential gene expression in cells trisomic for chr. 21 compared to the diploid twin.23,24 The protein levels also increased according to gene copy number, although slightly less: 1.3 to 1.4-fold compared to the 1.5-fold observed in the transcriptome.25 Similar observations were made in several studies of gene expression changes in cells from individuals with T21 and other somatic aneuploidies.18,22,26 Importantly, comparable expression changes from the genes encoded on the extra chromosome were shown by global transcriptome and proteome profiling of human cells trisomic for other chromosomes: an increase in expression from triplicated genes, on average 1.5-fold for transcripts and 1.3-fold for proteins.25,27 The normalization of protein levels is largely due to the fact that proteins enriched for subunits of macromolecular complexes remain close to a diploid level due to post-transcriptional dosage compensation.25 Thus, the presence of a single extra chromosome leads to a modest increase in protein levels, suggesting that a potential phenotypic effect of over-expression of individual genes due to trisomy will be relatively minor. Of note, dosage compensation of chromosome gains and losses, which normalizes protein abundance to near-euploid levels, is a general phenomenon observed in aneuploid cells across various species, as well as in cancers.28
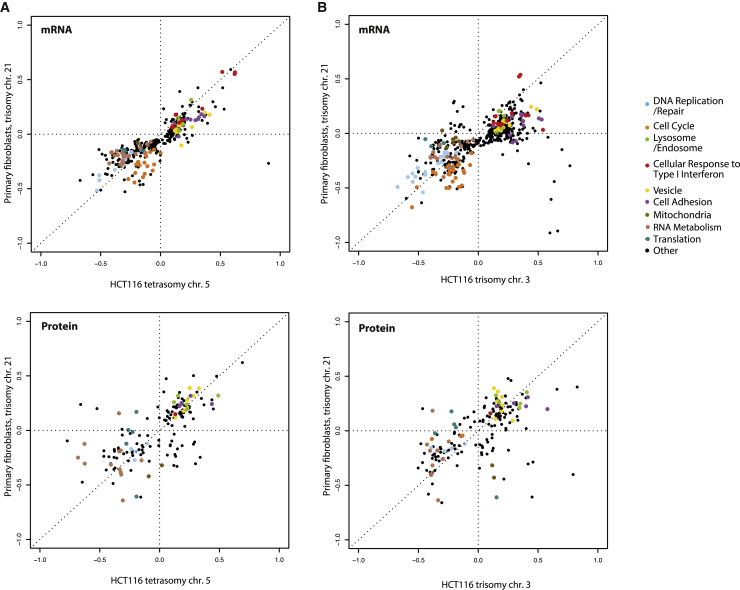
In addition to the dosage-dependent upregulation of a subset of expressed genes on chromosome 21, there is a genome-wide differential gene expression of factors encoded on other chromosomes. Some of these deregulations were proposed to occur due to an increased abundance of specific transcription factors and other regulatory proteins encoded on the trisomic chromosome, or due to chr. 21-dependent changes in global transcriptional patterns.29,30,31 However, these changes might be induced in response to chromosome gain per se. Analysis of various tissue samples from individuals with DS and mouse models revealed that T21 induces genome-wide transcriptional disruption similar to changes observed in other whole-chromosome aneuploidies.22,23,25,32,33,34,35,36,37,38,39 Indeed, human engineered trisomic cells display a differential regulation of specific pathways that are related to cell cycle regulation, lipid metabolism, mitochondrial function, autophagy and lysosomes, endosomes, and inflammatory response. This is very similar to results obtained from gene set enrichment analyses (GSEAs) of primary DS cells in which the abundance of proteins involved in cell-cycle-related functions, cell morphogenesis, lipoprotein metabolism, and cellular respiration in mitochondria are substantially altered.22,40 In addition, the expression of genes related to RNA processing, ribosome biogenesis, and translation is generally downregulated in models of chromosome gains in yeast and in human cell lines.33,35 RNA processing pathways, translation, and ribosome biogenesis are also downregulated in primary DS cells.41 These changes might be linked to altered protein homeostasis in trisomic cells and to the activation of the integrated stress response (ISR), a circuit that restores proteostasis by regulating protein synthesis rates. The ISR is activated in DS cells and various model trisomic cells.43 Direct comparison of transcriptome and proteome profiles of primary cells from individuals with DS revealed a striking similarity with model trisomic cell lines engineered to carry extra chromosome copies (Figure 1, Data S1). Thus, at least some phenotypes of DS and other trisomy syndromes might result from a general cellular response to chromosome gain.
Figure 1.
Pathway changes in human cell lines with extra chromosomes
The plots compare differentially regulated pathways based on analysis of whole-genome expression data. Two-dimensional comparative pathway analysis of transcriptomic and proteomic data from fibroblasts with trisomy of chr. 21 40,42 with data from genetically modified human colon cancer cell lines (HCT116) that were engineered to carry additional chromosomes: HCT116 with tetrasomy of chr. 5 25,41 (A) and HCT116 with trisomy of chr. 3 25(B) is shown. Respective euploid cell lines were used for the data normalization. The RNA and protein ratio calculations were previously published.25,33 Each dot represents one pathway as defined by GO category, GOCC and GOBP Slim, significantly altered with FDR 5%. Color-coding marks GO categories of frequently deregulated pathways.
Cell cycle deregulation in response to trisomy
Trisomy syndromes generally lead to reduced proliferation potential of somatic cells, which is directly reflected by changes in cell cycle. Fibroblasts from fetuses with DS show compromised proliferation rates and abnormal expression of key cell cycle regulators.43 Consequently, the growth of fetuses and children with DS is markedly slower than that of euploid counterparts. Additionally, tissue-specific proliferation and development defects, e.g. brain hypotrophy,44 are observed, supported by the observation that neurospheres of the primary DS material manifest a proliferation deficit.45,46 Histological assessment of defective prenatal lung tissues of DS-affected individuals also shows decreased proliferation in epithelial and mesenchymal cells.47
The division rates of cell lines derived from DS primary cells, e.g. lymphoblastic cell lines, are also delayed, in concordance with findings reported almost 50 years ago for trisomy 21 cells.48,49 The genome-wide expression profiles of the transcriptome and proteome of neural human induced pluripotent stem cells (iPSCs) with T21 show downregulation of the pathways responsible for cell proliferation.50 Although it is possible that the reduced proliferation in DS cells might be caused by reduced viability, current evidence instead suggests that the cells are progressing slowly through the cell cycle.51,52 Importantly, disomic murine pluripotent stem cells that were obtained by curing original trisomy of chromosome 21, showed improved proliferation and increased lifespan capacity and outgrew the trisomic cells from which they originated.53
Proliferation defects in somatic cells with trisomy are not restricted to T21. Embryos and infants with other trisomies, as well as cell lines derived from trisomic tissues, also develop significantly slower.22,54,55 Various mammalian cell lines engineered to contain extra chromosome also show delayed proliferation under optimal conditions, regardless of the identity of the extra chromosome,25,56,57,58 although interestingly, they show increased resistance to various stress conditions compared to isogenic wild-type cells.59 Whereas all cell cycle phases are affected, the proliferation defect is most apparent in the G1 phase and in the impaired transition from G1 to S phase. This fits well with transcriptome and proteome analyses, which show deregulation of cell cycle factors, thereby resembling the situation observed in DS samples.22,25,33,35,50 Additionally, proliferation of primary fibroblasts with trisomy of chr. 13, 18, or 21 display an increased dependency on serine-driven lipid synthesis, and this dependency probably reflects the need for extra membrane to accommodate the enlarged and misshapen nuclear envelope that is typical for cells with extra chromosomes.22 Thus, impaired proliferation is a general effect of chromosome gain in somatic cells and not specific to trisomy 21.
The driving factors for the observed changes in cellular proliferation remain unclear. One possible explanation – based on the observations from trisomic model cell lines –postulates that the proliferative delay arises due to increased levels of DNA damage and impaired DNA replication. Increased replication stress and subsequent genomic instability was observed in a wide range of trisomic cell lines with variable extra chromosomes.25,60 Elevated DNA damage most likely occurs due to altered expression of DNA replication factors triggered by chromosome gain. However, there is limited in vivo evidence addressing DNA integrity in trisomy syndromes. Blood cells from children with DS display increased sensitivity to DNA-damaging agents, which suggests suppressed DNA repair mechanisms.61 This correlates with the observation of increased DNA damage levels in cultured lymphocytes from DS-affected individuals.62 A recent study of mosaic DS uncovered significantly higher frequencies of micronuclei formation in trisomic versus disomic somatic cells, suggesting that trisomy drives increased chromosomal instability.63 Moreover, human lymphocytes trisomic for chr. 13, 18, or 21 show mildly elevated genomic instability and increased aneuploidy rates when forced to divide.64,65 The reduced proliferation might be due to accumulation of DNA damage and subsequent activation of the DNA damage checkpoint or the p53 pathway, as has been observed in model aneuploid cells.60,66 Decreased activity of DNA replication pathways is also observed in transcriptomic and proteomic analyses of iPSCs derived from individuals with DS.50 However, individuals with DS do not show increased frequency of chromosome gains and losses or rearranged chromosomes, suggesting that there is a strong selection against proliferation of cells with additional chromosomal changes.65,67
Other factors, such as defects in mitochondrial metabolism and changes in the nuclear envelope, might play an important role in the decreased proliferative capacity of trisomic cells.22,68 Further research will be required to determine the cause of cell cycle delays in trisomic cells, independent of the identity of the additional chromosome.
Activation of innate immune response as a consequence of trisomy
Innate immune response is activated in trisomic individuals and in cell culture models
A link between trisomy of chromosome 21 and increased interferon signaling was observed for the first time by Tan et al. in 1974.69 Recent analyses confirmed and elaborated this finding. Transcriptome analysis of primary material from DS-affected individuals revealed an IFN-induced response and dependence on JAK1 and TYK2 kinases.41 In fact, the entire IFN type I pathway appears to be upregulated, including the ligands (e.g. IFNA2, IFNB, and IFNG), IFN receptors (IFNAR1, IFNAR2, IFNGR2, and IL10RB), as well as the IFN-activated transcription factors (e.g. IRF3, IRF5, IRF7, and STAT1), and IFN-stimulated genes (ISG15 [MIM: 147571] and MX1 [MIM: 147150]). Fibroblasts trisomic for chr. 21 demonstrated an enhanced type I IFN response upon viral infection as compared to disomic fibroblasts.70 Proteomics data from DS blood samples further supported these observations and specifically indicated increased levels of cytokines, e.g. IL-6, IL-22, TNFα, and MCP-1, linked to IFN signaling.26 These molecular changes can be additionally linked to chronic immune system deregulation and auto-inflammation. Indeed, DS-affected individuals often present symptoms of interferonopathies.26,71,72 However, only six of the IFN-response genes upregulated in DS are located on chromosome 21; these include IFNAR1 (MIM: 107450), IFNAR2 (MIM: 602376), INGR2 (MIM: 147569), and IL10RB (MIM: 123889). Interestingly, overexpression of these factors can mimic several DS-associated phenotypes, specifically the hyperactivation of the kynurenine pathway and the associated neurological and immunological conditions in mice.73
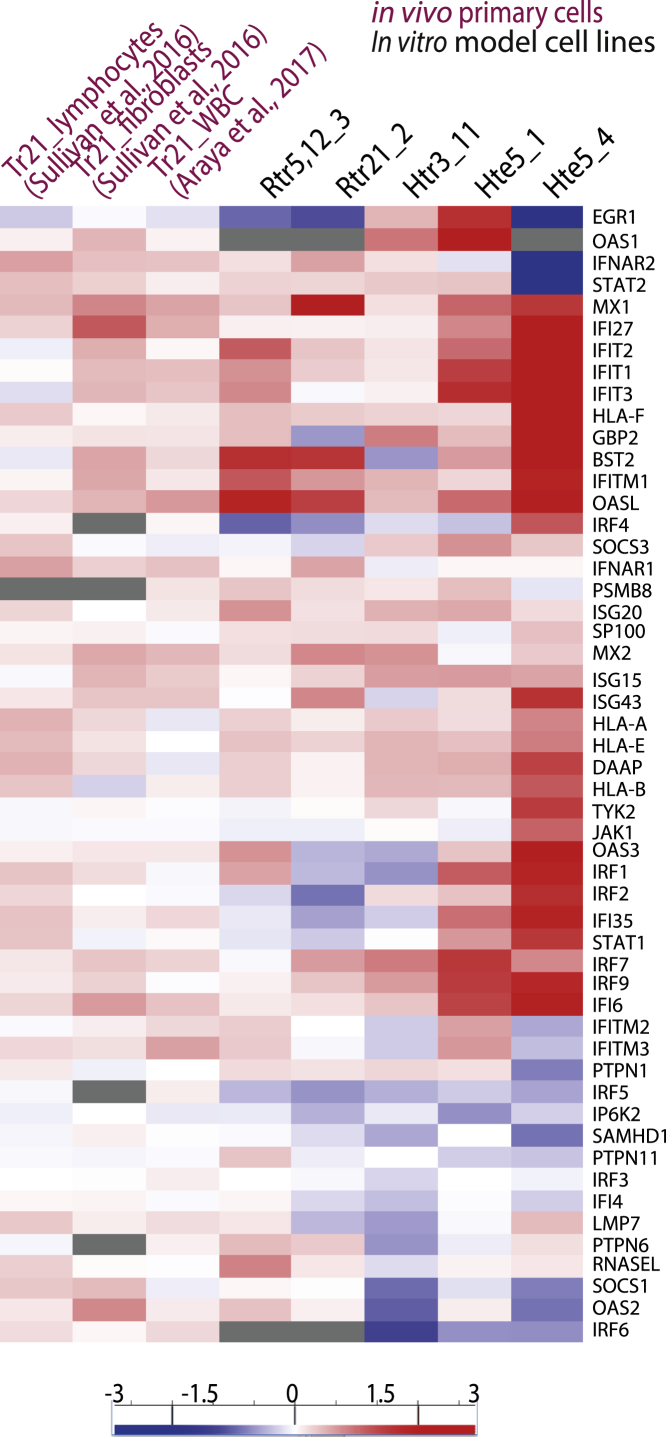
Similarly, other trisomies are also characterized by increased levels of inflammatory cytokines with damaging consequences. Early embryonic death caused by trisomy 16 has been linked to upregulation of genes (HLA-G [MIM: 142871], HLA-C [MIM: 142840], TNFα [MIM: 191160], IL18 [MIM: 600953], NCF2 [MIM: 608515], and CD16 [MIM: 146740]) required for a strong immune response, superoxide metabolism, and inflammatory reaction.24 Behçet disease, caused by trisomy of chr. 8 in bone marrow cells, is associated with myelodysplastic syndrome (MDS [MIM: 614286]) and a strong enrichment, in serum, of IFN response factors, including IL2R, IFNG, IL1b, IL6, IL8, and GMCSF,74 as well as upregulated T and B cell receptor and IL4 signaling pathways.75 Proinflammatory cytokine synthesis in T cells is typical for MDS development linked to trisomy of chr. 8.76 Trisomy 9p leads to elevated levels of IFNA, IFNB, and IFNG and to symptoms similar to the autoimmune disease systemic lupus erythematosus and interferonopathies.77 Usually, these phenotypes are explained as a consequence of an overexpression of a specific IFN type I gene cluster from an extra chromosome.78 However, upregulated IFN γ-response also is observed in trisomic human cells of variable genetic and biological backgrounds.25,33,79,80,81 Comparative transcriptome analysis of the published DS data with various model trisomic cell lines indeed shows similar activation of the type I IFN signaling (Figure 2, Data S2). Thus, the general occurrence of these changes in trisomic cells suggests that the inflammatory gene expression and IFN signaling might be a shared cellular response to chromosome gain rather than a chromosome-specific response.
Figure 2.
Transcriptome analysis reveals upregulation of type I IFN response upon chromosome gain
The GO category “Type I IFN mediated signaling pathway” was used to compare published datasets of primary trisomic chr. 21 DS lymphocytes, fibroblasts,41 and white blood cells,89 with engineered trisomic cell lines analyzed previously. Cell lines derived from RPE1 are: trisomy of chr. 5 and 12, clone 3 (Rtr5, 12_3), and trisomy chr. 21 clone 2 (Rtr21_2); cell lines derived from HCT116 are trisomy of chr. 3, clone 11 (Htr3_11) and tetrasomy chr. 5, clone 1 (Hte5_1), and clone 4 (Hte5_4) cell lines. All samples were normalized to the matching diploid controls.
What might be the reason for the general type I IFN upregulation in trisomic cells? Several factors might trigger this response, but particular attention should be given to the cGAS-STING pathway in the context of DNA damage, senescence, and cancer-associated aneuploidy.82,83 cGAS is a cytoplasmic nucleic acid receptor that stimulates an STING-dependent signaling cascade activating interferon stimulated gene expression via the IRF3 transcriptional factor.84 Strikingly, model trisomic cells, as well as cells from human embryos with trisomic phenotypes, also activated the cGAS-STING pathway, as illustrated by increased nuclear localization of IRF3 and subsequent overexpression of its targets, including IFIT1 (MIM: 147690), IFIT3 (MIM: 604650), and OAS3 (MIM: 603351).81 Immunofluorescence imaging in trisomic cells revealed increased accumulation of cytoplasmic dsDNA of nuclear origin.80,81 Although the reason for dsDNA accumulation remains unclear, it is possible that the increased DNA damage in trisomic cells contributes to this phenotype. One explanation could be the compromised integrity of the nuclear envelope, generally increased in cells with chromosomal instability, and accumulation of cGAS-positive micronuclei.55,85 Of note, increased micronuclei formation was recently reported in leukocytes from individuals with DS.63 Moreover, various DNA-damage-inducing treatments can activate the cGAS-STING pathway via increased accumulation of cytoplasmic dsDNA. This phenotype is independent of the cell type and of the identity of the extra chromosome.81 Other recent studies support the notion that IFN signaling is induced upon DNA damage in trisomic cells. For example, another cytoplasmic DNA sensor, DDX41, was connected to chr. Y and chr. 8 trisomies, which are associated with MDS and acute myeloid leukemia.86 Taken together, the available data support a model in which constitutive model trisomic human cell lines promote the expression of IRF3 targets in vitro via the cGAS-STING pathway, thereby inducing the type I IFNs. Further analysis will be required to test whether similar cGAS-STING involvement is required for the type I IFN induction in DS-affected individuals.
The deregulated immune system illustrates genome-wide changes in individuals with trisomy syndromes
Dysregulated immunity and increased autoimmunity are well-documented for individuals with trisomic genetic syndromes and have been characterized on the basis of genetic, epigenetic, and protein changes.87,88 The increased expression of various immune factors such as interleukins, cytokines, and interferons in the blood of individuals with DS has also been associated with an altered balance in post-thymic immune system. Changes in T-cell differentiation were manifested by reduced numbers of naive and increased abundance of mature – hyperactivated T-cells.89,90,91 Transcriptomic and proteomic analyses of the immune cells from blood of individuals with DS confirmed the upregulation of T-cell differentiation markers CD3, CD28, and CSF2 that correlated with increased IFN-pathway specific TBX21 (MIM: 604895), EOMES (MIM: 604615), and MKI67 (MIM: 176741) expression.89,90 Deregulation of the immune system was also observed in B cells with trisomy 21, which were underrepresented in fetal bone marrow, as well as in adults’ blood.91,92,93,94 One possible explanation for the observed phenomenon was proposed on the basis of the in vitro model of T21 iPSCs, where a reduced potential for immune cell lineage differentiation was attributed to the downregulation of endothelin receptor B.95 Alternatively, increased apoptosis rates in peripheral blood lymphocytes could cause the immune deficiency in DS.96 This hypothesis is further supported by accelerated thymic aging in children with DS.97 Similar lymphocyte imbalance was observed in individuals with chronic lymphocytic leukemia (CLL) who have mosaic trisomy for chr. 3, 8, or 12 in peripheral blood or bone marrow cells.98,99,100,101 Impaired T and NK cell development is also found in fetuses with trisomy chr. 18. Importantly, gain of chromosome 18 is associated with deregulation of lymphocyte balance, as well as with lymphomas in adults.102,103 Thus, a deregulation of the immune system is not specific to trisomy of chromosome 21, but instead might be a general cellular response triggered by chromosome gain. Together, these findings point toward elevated IFN signaling and subsequent hyperactivation of autoimmune and inflammatory responses as a general outcome of chromosome gain.
Changes in maintenance of protein homeostasis
As explained above, one of the striking cellular consequences of harboring an extra chromosome is altered protein homeostasis. The translation of the superfluous proteins from the additional chromosome leads to altered stoichiometry of multimolecular protein complexes, accumulation of protein aggregates, and increased demand for protein folding and degradation. Here we will focus on autophagy, a protein degradation process that seems particularly affected by the presence of extra chromosomes.
Autophagy impairment in individuals with trisomy syndromes
Most research in cells from individuals with DS reports impaired autophagy and accumulation of waste products, which might contribute to deleterious phenotypes, such as the early senescence or neurodegeneration observed in DS.104 Genome-wide defects in chromosomal segregation and the nuclear envelope are also associated with severe senescence phenotypes in iPSC-derived neural progenitor cells.105 Additionally, thymus epithelial cells, thymocytes, and T-cells with T21 show increased reactive oxygen species, DNA damage markers, cell-cycle arrest, and telomere length regulation, contributing to the aging phenotype.97 Primary fibroblasts derived from individuals with trisomies of chr. 13, 18, and 21 similarly show early signs of senescence, which can be successfully rescued by reduction of accumulated protein aggregates from the cytoplasm, a finding that connects aneuploidy and senescence with defective proteostasis.106 Protein aggregates are primarily removed by the autophagy pathway, and impaired autophagy activation is often observed in cells from individuals with DS. The subsequent accumulation of aggregates is usually associated with enhanced gene expression of proteins encoded on chr. 21. The triplication of amyloid precursor protein (APP [MIM: 104760]) from chr. 21 and a consequent increase of amyloid load were proposed to cause the autophagy impairment observed in vivo and in vitro.107,108 However, the data suggest that three copies of APP are necessary, but not sufficient, for AD in individuals with DS.18,108,109 Normalized expression of another chr. 21 gene, encoding the kinase DYRK1A (MIM: 600855), together with APP was sufficient to rescue neuronal differentiation mainly dependent on axonal transport in DS, as well as in AD.110 Similarly, balancing an aberrant O-GlcNAcylation reversed the AD-like neural phenotypes in DS mice, and this was associated with positive effects on autophagy.111
On the other hand, genome-wide expression analysis in DS also revealed downregulated autophagy. One of the general molecular mechanisms behind suppressed autophagy could be a hyperstimulation of the negative regulator of autophagy mTORC1, revealed by the transcriptome analysis in human primary fibroblasts and brain neurons of individuals with DS.112,113 Active mTORC1 inhibits TFEB, a key transcription factor promoting expression of autophagy and lysosomal-specific genes. As a consequence, DS primary fibroblasts show low autophagosome formation and reduced synthesis of ATG7, ATG3, and SQSTM1, the major factors regulating autophagosome formation.112 Additionally, the SNARE protein family members required for autophagosome-lysosome fusion were diminished.40,114 The reduced fusion prevents autophagosomes from recycling, leading to their accumulation, which is documented by an increase of autophagy markers LC3-II, SQSTM1, and NBR1 in DS cells upon autophagy activation by starvation. Finally, an excess of APP and its cleavage product directly triggered multiple lysosomal defects in DS cell lines.115,116
However, evaluation of autophagy, an extremely dynamic and multifaceted process, is difficult, particularly in primary material. In addition to mTORC1, a master regulator of autophagy, other factors play important roles, including the recently discovered function of cGAS-STING-dependent TFEB-transcriptional activation.81,117
Elevated autophagy activity in trisomic model cell lines
Model cell lines with constitutive trisomies also suffer from impaired protein homeostasis, which is manifested by accumulation of cytoplasmic protein aggregates, increased sensitivity to heat shock and to inhibitors of protein folding and translation.19,118 Additionally, the cells often show deregulated expression of proteins involved in autophagy. The dynamics of autophagy in aneuploid cells in vitro have been studied in two different settings. First, induced chromosome segregation errors allow for the assessment of the immediate response. In this model system, the lysosomal stress response was observed, manifested by upregulated activity of TFEB and reduced autophagosome-lysosome fusion.19 Although TFEB upregulates autophagy and lysosomal-specific gene expression, cargo to be degraded is stuck in the autophagosome, leading to reduced autophagy flux. These observations agree with the findings in DS cells following starvation-induced autophagy.
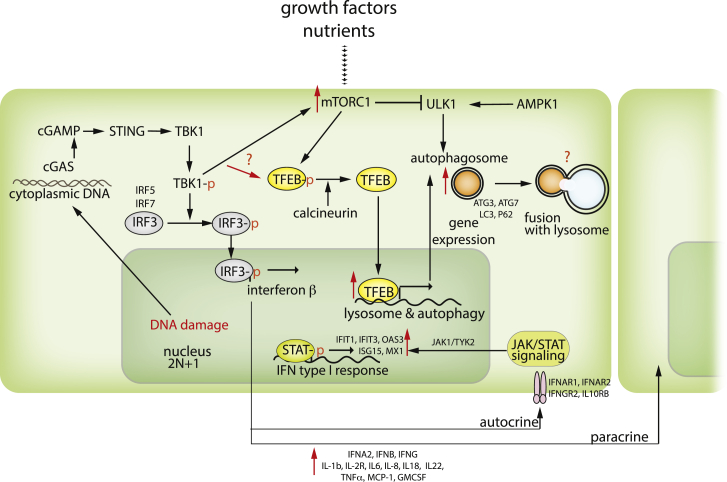
In contrast, in the second model, constitutive aneuploid cell lines with variable trisomies appear to adapt to the stress conditions triggered by chromosome gain. These cells upregulate the expression of autophagy and lysosomal-specific genes, a signature that resembles lysosomal stress.33 However, the autophagy flux is not impaired.81,118 Importantly, trisomic human mammary epithelial cell lines with extra chr. 8 or 22 activate autophagy, probably to cope with the increased oxidative stress and DNA damage induced by aneuploidy.119 The autophagy inhibitor chloroquine impairs proliferation of murine trisomic cells.120 Together, the available research suggests an alternative interpretation of the autophagy changes in trisomic cells. The basal levels of autophagy appear to be increased even under standard conditions in trisomic cells. Additional activation, e.g. via starvation or mTORC1 inhibition, leads only to a small degree of further activation, probably because the autophagy levels are near the maximal cellular capacity, achieved by mTORC1-independent pathways in trisomic cells. Of note, autophagy can be activated via the cGAS-STING pathway, linking it to the type I IFN response and DNA damage.81,121 Increased demands for the maintenance of protein homeostasis in aneuploid cells put further strain on the autophagy pathway, leading to cumulative defects in this essential recycling process (Figure 3).
Figure 3.
Schematic depiction of autophagy activation in the context of trisomy
Upon activation via the ULK1 kinase, the phagophore membrane engulfs the cytoplasmic components to be degraded and fuses with lysosomes to enable degradation. mTORC1 kinase, a key nutrient sensor, regulates autophagosome assembly via inhibitory phosphorylations. Additionally, the transcription factors of the MIT/TFEB family, which are also mTORC1 targets, regulate the expression of autophagy- and lysosomal-specific genes. Autophagy is activated upon starvation, oxidative stress, unfolded protein response, or DNA damage via the cGAS/STING pathway.
TFEB-dependent gene expression is increased in triploid model systems
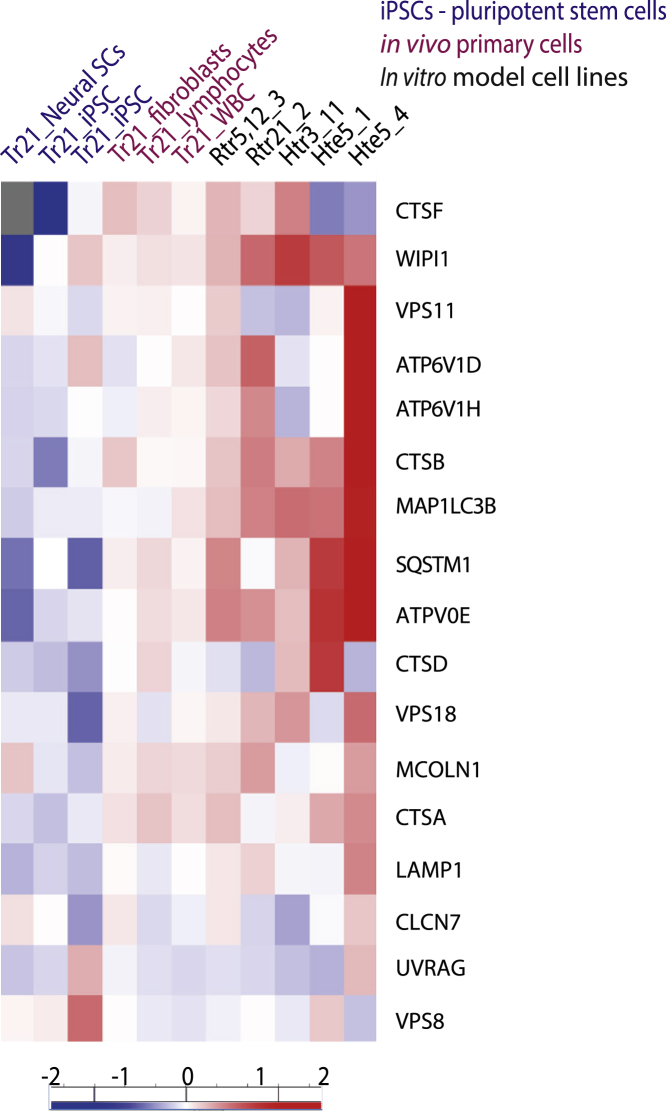
The molecular mechanisms of autophagy regulation in trisomic cells are poorly understood. An autophagy regulator, TFEB, and its behavior in the context of aneuploidy-induced proteotoxic and genotoxic stress is probably a key factor. Analysis of TFEB-target gene expression from DS samples and cell line models with constitutive trisomy shows that stimulation of TFEB activity, and subsequent upregulation of autophagy and lysosomal-specific gene expression, is the critical step in all aneuploid somatic cells (Figure 4, Data S3). It should be noted that this phenotype does not seem to hold true for stem cells. This is in line with the observation that DS iPSCs do not show reduced proliferation after treatment with autophagy inhibitor chloroquine.122 This might reflect an ability of undifferentiated stem cells to proliferate without active autophagy, which is dispensable until the onset of differentiation during embryonal development.123 Indeed, the analysis revealed downregulation of TFEB-dependent expression, suggesting that autophagy is dispensable for pluripotent stem cells even when they are trisomic. Therefore, pluripotent stem cells might not be suitable to address the role of autophagy in cellular response to aneuploidy.
Figure 4.
Increased RNA expression of TFEB targets in cells carrying an extra chromosome
Transcriptomic data showing TFEB-response genes based on the published gene set.19 Published datasets of primary cells with trisomy chr. 21 of DS, including pluripotent stem cells,23,34 lymphocytes, fibroblasts,41 and white blood cells,89 were compared with our engineered trisomic RPE1 with trisomy of chr. 5 and 12, clone 3 (Rtr5, 12_3) and chr. 21, clone 2 (Rtr21_2), as well as HCT116 with trisomy of chr. 3. clone 11 (Htr3_11) and tetrasomy of chr. 5, clone 1 (Hte5_1), and clone 4 (Hte5_4) cell lines, normalized to diploid isogenic controls. Trisomies of chromosomes are generally lethal, and rare survivors must cope with multiple pathologies. Down syndrome (trisomy chr. 21) pathological phenotypes are connected to increased expression of Hsa 21 genes. We present an alternative model describing global, aneuploidy-associated changes, similar in trisomy syndromes and cell lines with different chromosome gains.
Taken together, autophagy and proteostasis are deregulated in trisomic model cells, as well as in cells from individuals with DS. Although the details remain unclear, this deregulation contributes to the observed phenotypes, as well as to pathophysiological changes, and is linked to TFEB-dependent transcription. Autophagy evidently plays an important role in aneuploid cell survival and adaptation to stress associated with extra chromosome presence. The discrepancy between observations of samples from individuals with DS and in vitro aneuploid cell line data might reflect the ability of cells in culture to increase autophagy to cope with the detrimental outcomes of aneuploidy. Understanding the mechanisms that trisomic cells use to upregulate autophagy in culture could provide insights into possible routes to mitigate the deleterious consequences of aneuploidy in vivo.
Conclusion
The phenotypes observed in individuals with DS and other trisomy syndromes are complex and variable. The data collected so far indicate that trisomy syndromes are not just a collection of independent single-gene phenotypes. Therefore, it has been proposed that these phenotypes might arise from a synergy of overexpression of multiple genes. Here we discuss evidence showing that a general cellular response to aneuploidy, in addition to the deregulation of specific genes on chromosome 21, contributes to the observed DS phenotypes. This notion has been supported by recently accumulated data from engineered aneuploid human cells that were created to model cancer karyotypes. Strikingly, several of the physiological changes observed in these cells overlap with the changes found in cells from individuals with trisomy syndromes. Although we focused only on some common cellular phenotypes, such as the activation in the interferon type I response, changes in autophagy, and altered proliferation, we also observed other notable cellular changes, including abnormalities of the nuclear envelope and changes in mitochondrial metabolism. Taken together, the current evidence suggests that the deregulation of cellular networks observed in trisomy syndromes is largely independent of the identity of the extra chromosome and might reflect the cellular response to aneuploidy-induced stresses. Future research should focus on addressing both of these aspects – the gene-specific response and the general response to aneuploidy – to improve the quality of life of individuals with trisomy syndromes.
Acknowledgments
This work was funded by the Rhineland-Palatine Research Initiative BioComp to Z.S. and the budget of the medical faculty of Heidelberg University Hospital to M.K.
Declaration of interests
The authors declare no conflicts of interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.10.014.
Supplemental information
References
- 1.Hong D.S., Reiss A.L. Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol. 2014;13:306–318. doi: 10.1016/S1474-4422(13)70302-8. [DOI] [PubMed] [Google Scholar]
- 2.Raznahan A., Parikshak N.N., Chandran V., Blumenthal J.D., Clasen L.S., Alexander-Bloch A.F., Zinn A.R., Wangsa D., Wise J., Murphy D.G.M., et al. Sex-chromosome dosage effects on gene expression in humans. Proc. Natl. Acad. Sci. USA. 2018;115:7398–7403. doi: 10.1073/pnas.1802889115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ford C., Jones K.W., Polani P.E., De Almeida J.C., Briggs J.H. A Sex-chromosome anomaly in a case of gonadal dysgenesis (Turner’s syndrome) Lancet. 1959;273:711–713. doi: 10.1016/S0140-6736(59)91893-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X., Hong D., Ma S., Ward T., Ho M., Pattni R., Duren Z., Stankov A., Bade Shrestha S., Hallmayer J., et al. Integrated functional genomic analyses of Klinefelter and Turner syndromes reveal global network effects of altered X chromosome dosage. Proc. Natl. Acad. Sci. USA. 2020;117:4864–4873. doi: 10.1073/PNAS.1910003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chunduri N.K., Barthel K., Storchova Z. Consequences of chromosome loss: why do cells need each chromosome twice? Cells. 2022;11:1530. doi: 10.3390/CELLS11091530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krivega M., Zimmer J., Slezko A., Frank-Herrmann P., Rehnitz J., Hohenfellner M., Betterndorf M., Luzarowski M., Strowitzki T. Genomic instability in patients with sex determination defects and germ cell cancer. Preprint at bioRxiv. 2022 doi: 10.1101/2022.06.08.495249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costello J.P., Weiderhold A., Louis C., Shaughnessy C., Peer S.M., Zurakowski D., Jonas R.A., Nath D.S. A Contemporary, single-institutional experience of surgical versus expectant management of congenital heart disease in trisomy 13 and 18 patients. Pediatr. Cardiol. 2015;36:987–992. doi: 10.1007/S00246-015-1109-5/TABLES/5. [DOI] [PubMed] [Google Scholar]
- 8.Lebedoff A.N., Carey J.C. Parent-reported histories of adults with trisomy 13 syndrome. Am. J. Med. Genet. 2021;185:1743–1756. doi: 10.1002/AJMG.A.62165. [DOI] [PubMed] [Google Scholar]
- 9.Peterson J.K., Kochilas L.K., Catton K.G., Moller J.H., Setty S.P. Long-term outcomes of children with trisomy 13 and 18 after congenital heart disease interventions. Ann. Thorac. Surg. 2017;103:1941–1949. doi: 10.1016/J.ATHORACSUR.2017.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arumugam A., Raja K., Venugopalan M., Chandrasekaran B., Kovanur Sampath K., Muthusamy H., Shanmugam N. Down syndrome-A narrative review with a focus on anatomical features. Clin. Anat. 2016;29:568–577. doi: 10.1002/CA.22672. [DOI] [PubMed] [Google Scholar]
- 11.Carfì A., Antocicco M., Brandi V., Cipriani C., Fiore F., Mascia D., Settanni S., Vetrano D.L., Bernabei R., Onder G. Characteristics of adults with down syndrome: prevalence of age-related conditions. Front. Med. 2014;1:51. doi: 10.3389/fmed.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tweats D., Eastmond D.A., Lynch A.M., Elhajouji A., Froetschl R., Kirsch-Volders M., Marchetti F., Masumura K., Pacchierotti F., Schuler M. Role of aneuploidy in the carcinogenic process: Part 3 of the report of the 2017 IWGT workgroup on assessing the risk of aneugens for carcinogenesis and hereditary diseases. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019;847:403032. doi: 10.1016/J.MRGENTOX.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Antonarakis S.E., Skotko B.G., Rafii M.S., Strydom A., Pape S.E., Bianchi D.W., Sherman S.L., Reeves R.H. Down syndrome. Nat. Rev. Dis. Prim. 2020;6:9–20. doi: 10.1038/s41572-019-0143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanzillotta C., Di Domenico F. Stress responses in down syndrome neurodegeneration: state of the art and therapeutic molecules. Biomolecules. 2021;11:266–334. doi: 10.3390/BIOM11020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonarakis S.E., Lyle R., Dermitzakis E.T., Reymond A., Deutsch S. Chromosome 21 and Down syndrome: From genomics to pathophysiology. Nat. Rev. Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 16.Olson L.E., Richtsmeier J.T., Leszl J., Reeves R.H. A chromosome 21 critical region does not cause specific down syndrome phenotypes. Science. 2004;306:687–690. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyle R., Béna F., Gagos S., Gehrig C., Lopez G., Schinzel A., Lespinasse J., Bottani A., Dahoun S., Taine L., et al. Genotype–phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet. 2008;17:454–466. doi: 10.1038/ejhg.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korbel J.O., Tirosh-Wagner T., Urban A.E., Chen X.N., Kasowski M., Dai L., Grubert F., Erdman C., Gao M.C., Lange K., et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. USA. 2009;106:12031–12036. doi: 10.1073/PNAS.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santaguida S., Vasile E., White E., Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015;29:2010–2021. doi: 10.1101/gad.269118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chunduri N.K., Storchová Z. The diverse consequences of aneuploidy. Nat. Cell Biol. 2019;21:54–62. doi: 10.1038/s41556-018-0243-8. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro B.L. Down syndrome--a disruption of homeostasis. Am. J. Med. Genet. 1983;14:241–269. doi: 10.1002/AJMG.1320140206. [DOI] [PubMed] [Google Scholar]
- 22.Hwang S., Cavaliere P., Li R., Zhu L.J., Dephoure N., Torres E.M. Consequences of aneuploidy in human fibroblasts with trisomy 21. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/PNAS.2014723118/-/DCSUPPLEMENTAL. e2014723118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letourneau A., Santoni F.A., Bonilla X., Sailani M.R., Gonzalez D., Kind J., Chevalier C., Thurman R., Sandstrom R.S., Hibaoui Y., et al. Domains of genome-wide gene expression dysregulation in Down’s syndrome. Nature. 2014;508:345–350. doi: 10.1038/nature13200. [DOI] [PubMed] [Google Scholar]
- 24.Yao T., Hou H., Liu G., Wu J., Qin Z., Sun Y., Jin X., Chen J., Chen Y., Xu Z. Quantitative proteomics suggest a potential link between early embryonic death and trisomy 16. Reprod. Fertil. Dev. 2019;31:1116–1126. doi: 10.1071/RD17319. [DOI] [PubMed] [Google Scholar]
- 25.Stingele S., Stoehr G., Peplowska K., Cox J., Mann M., Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan K.D., Evans D., Pandey A., Hraha T.H., Smith K.P., Markham N., Rachubinski A.L., Wolter-Warmerdam K., Hickey F., Espinosa J.M., Blumenthal T. Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation. Sci. Rep. 2017;7:14818. doi: 10.1038/s41598-017-13858-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dephoure N., Hwang S., O’Sullivan C., Dodgson S.E., Gygi S.P., Amon A., Torres E.M. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023–e03027. doi: 10.7554/ELIFE.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schukken K.M., Sheltzer J.M. Extensive protein dosage compensation in aneuploid human cancers. Genome Res. 2022;32:1254–1270. doi: 10.1101/GR.276378.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim J.H., Han Y.J., Kim H.J., Kim M.Y., Park S.Y., Cho Y.H., Ryu H.M. Integrative analyses of genes and microRNA expressions in human trisomy 21 placentas. BMC Med. Genom. 2018;11:46–47. doi: 10.1186/S12920-018-0361-Y/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muskens I.S., Li S., Jackson T., Elliot N., Hansen H.M., Myint S.S., Pandey P., Schraw J.M., Roy R., Anguiano J., et al. The genome-wide impact of trisomy 21 on DNA methylation and its implications for hematopoiesis. Nat. Commun. 2021;12:821. doi: 10.1038/S41467-021-21064-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C., Jin L., Bai Y., Chen Q., Fu L., Yang M., Xiao H., Zhao G., Wang S. Genome-wide expression analysis in down syndrome: insight into immunodeficiency. pLoS One. 2012;7:e49130. doi: 10.1371/JOURNAL.PONE.0049130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianco K., Gormley M., Farrell J., Zhou Y., Oliverio O., Tilden H., McMaster M., Fisher S.J. Placental transcriptomes in the common aneuploidies reveal critical regions on the trisomic chromosomes and genome-wide effects. Prenat. Diagn. 2016;36:812–822. doi: 10.1002/PD.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dürrbaum M., Kuznetsova A.Y., Passerini V., Stingele S., Stoehr G., Storchová Z. Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genom. 2014;15:139. doi: 10.1186/1471-2164-15-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzales P.K., Roberts C.M., Fonte V., Jacobsen C., Stein G.H., Link C.D. Transcriptome analysis of genetically matched human induced pluripotent stem cells disomic or trisomic for chromosome 21. PLoS One. 2018;13:e0194581. doi: 10.1371/journal.pone.0194581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheltzer J.M., Torres E.M., Dunham M.J., Amon A. Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. USA. 2012;109:12644–12649. doi: 10.1073/PNAS.1209227109/SUPPL_FILE/SD01.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olmos-Serrano J.L., Kang H.J., Tyler W.A., Silbereis J.C., Cheng F., Zhu Y., Pletikos M., Jankovic-Rapan L., Cramer N.P., Galdzicki Z., et al. Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron. 2016;89:1208–1222. doi: 10.1016/J.NEURON.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyler W.A., Haydar T.F. Multiplex genetic fate mapping reveals a novel route of neocortical neurogenesis, which is altered in the Ts65Dn mouse model of Down syndrome. J. Neurosci. 2013;33:5106–5119. doi: 10.1523/JNEUROSCI.5380-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walus M., Kida E., Rabe A., Albertini G., Golabek A.A. Widespread cerebellar transcriptome changes in Ts65Dn Down syndrome mouse model after lifelong running. Behav. Brain Res. 2016;296:35–46. doi: 10.1016/J.BBR.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 39.FitzPatrick D.R., Ramsay J., McGill N.I., Shade M., Carothers A.D., Hastie N.D. Transcriptome analysis of human autosomal trisomy. Hum. Mol. Genet. 2002;11:3249–3256. doi: 10.1093/HMG/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y., Borel C., Li L., Müller T., Williams E.G., Germain P.L., Buljan M., Sajic T., Boersema P.J., Shao W., et al. Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nat. Commun. 2017;8:1212. doi: 10.1038/s41467-017-01422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan K.D., Lewis H.C., Hill A.A., Pandey A., Jackson L.P., Cabral J.M., Smith K.P., Liggett L.A., Gomez E.B., Galbraith M.D., et al. Trisomy 21 consistently activates the interferon response. 2016;29:e16220. doi: 10.7554/eLife.16220.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu P.J., Khatiwada S., Cui Y., Reineke L.C., Dooling S.W., Kim J.J., Li W., Walter P., Costa-Mattioli M. Activation of the ISR mediates the behavioral and neurophysiological abnormalities in down syndrome. Science. 2019;366:843–849. doi: 10.1126/SCIENCE.AAW5185/SUPPL_FILE/AAW5185_ZHU_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gimeno A., García-Giménez J.L., Audí L., Toran N., Andaluz P., Dasí F., Viña J., Pallardó F.V. Decreased cell proliferation and higher oxidative stress in fibroblasts from down syndrome fetuses. Preliminary study. Biochim. Biophys. Acta. 2014;1842:116–125. doi: 10.1016/j.bbadis.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 44.Larsen K.B., Laursen H., Græm N., Samuelsen G.B., Bogdanovic N., Pakkenberg B. Reduced cell number in the neocortical part of the human fetal brain in Down syndrome. Ann. Anat. 2008;190:421–427. doi: 10.1016/j.aanat.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Nakano-Kobayashi A., Awaya T., Kii I., Sumida Y., Okuno Y., Yoshida S., Sumida T., Inoue H., Hosoya T., Hagiwara M. Prenatal neurogenesis induction therapy normalizes brain structure and function in Down syndrome mice. Proc. Natl. Acad. Sci. USA. 2017;114:10268–10273. doi: 10.1073/pnas.1704143114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esposito G., Imitola J., Lu J., De Filippis D., Scuderi C., Ganesh V.S., Folkerth R., Hecht J., Shin S., Iuvone T., et al. Genomic and functional profiling of human Down syndrome neural progenitors implicates S100B and aquaporin 4 in cell injury. Hum. Mol. Genet. 2008;17:440–457. doi: 10.1093/hmg/ddm322. [DOI] [PubMed] [Google Scholar]
- 47.Danopoulos S., Bhattacharya S., Deutsch G., Nih L.R., Slaunwhite C., Mariani T.J., Al Alam D. Prenatal histological, cellular, and molecular anomalies in trisomy 21 lung. J. Pathol. 2021;255:41–51. doi: 10.1002/PATH.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segal D.J., McCoy E.E. Studies on Down’s syndrome in tissue culture. I. Growth rates protein contents of fibroblast cultures. J. Cell. Physiol. 1974;83:85–90. doi: 10.1002/JCP.1040830112. [DOI] [PubMed] [Google Scholar]
- 49.Coskun P., Helguera P., Nemati Z., Bohannan R.C., Thomas J., Samuel S.E., Argueta J., Doran E., Wallace D.C., Lott I.T., Busciglio J. Metabolic and growth rate alterations in lymphoblastic cell lines discriminate between down syndrome and alzheimer’s disease. J. Alzheimers Dis. 2017;55:737–748. doi: 10.3233/JAD-160278. [DOI] [PubMed] [Google Scholar]
- 50.Sobol M., Klar J., Laan L., Shahsavani M., Schuster J., Annerén G., Konzer A., Mi J., Bergquist J., Nordlund J., et al. Transcriptome and proteome profiling of neural induced pluripotent stem cells from individuals with down syndrome disclose dynamic dysregulations of key pathways and cellular functions. Mol. Neurobiol. 2019;56:7113–7127. doi: 10.1007/s12035-019-1585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roper R.J., Reeves R.H. Understanding the basis for down syndrome phenotypes. PLoS Genet. 2006;2:e50. doi: 10.1371/JOURNAL.PGEN.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guidi S., Ciani E., Bonasoni P., Santini D., Bartesaghi R. Widespread proliferation impairment and hypocellularity in the cerebellum of fetuses with down syndrome. Brain Pathol. 2011;21:361–373. doi: 10.1111/J.1750-3639.2010.00459.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanuma-Takahashi A., Inoue M., Kajiwara K., Takagi R., Yamaguchi A., Samura O., Akutsu H., Sago H., Kiyono T., Okamoto A., Umezawa A. Restoration of keratinocytic phenotypes in autonomous trisomy-rescued cells. Stem Cell Res. Ther. 2021;12:476–511. doi: 10.1186/S13287-021-02448-W/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brewer C.M., Holloway S.H., Stone D.H., Carothers A.D., FitzPatrick D.R. Survival in trisomy 13 and trisomy 18 cases ascertained from population based registers. J. Med. Genet. 2002;39:e54. doi: 10.1136/jmg.39.9.e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang S., Williams J.F., Kneissig M., Lioudyno M., Rivera I., Helguera P., Busciglio J., Storchova Z., King M.C., Torres E.M. Suppressing aneuploidy-associated phenotypes improves the fitness of trisomy 21 cells. Cell Rep. 2019;29:2473–2488.e5. doi: 10.1016/J.CELREP.2019.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kneissig M., Keuper K., De Pagter M.S., Van Roosmalen M.J., Martin J., Otto H., Passerini V., Campos Sparr A., Renkens I., Kropveld F., et al. Micronuclei-based model system reveals functional consequences of chromothripsis in human cells. Elife. 2019;8:e50292. doi: 10.7554/eLife.50292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torres E.M., Williams B.R., Amon A. Aneuploidy: Cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams B.R., Prabhu V.R., Hunter K.E., Glazier C.M., Whittaker C.A., Housman D.E., Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/SCIENCE.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rutledge S.D., Douglas T.A., Nicholson J.M., Vila-Casadesús M., Kantzler C.L., Wangsa D., Barroso-Vilares M., Kale S.D., Logarinho E., Cimini D. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci. Rep. 2016;6:22828. doi: 10.1038/srep22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Passerini V., Ozeri-Galai E., De Pagter M.S., Donnelly N., Schmalbrock S., Kloosterman W.P., Kerem B., Storchová Z. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 2016;7:10754. doi: 10.1038/ncomms10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morawiec Z., Janik K., Kowalski M., Stetkiewicz T., Szaflik J., Morawiec-Bajda A., Sobczuk A., Blasiak J. DNA damage and repair in children with Down’s syndrome. Mutat. Res. 2008;637:118–123. doi: 10.1016/j.mrfmmm.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 62.George A., Venkatesan S., Ashok N., Saraswathy R., Hande M.P. Assessment of genomic instability and proliferation index in cultured lymphocytes of patients with Down syndrome, congenital anomalies and aplastic anaemia. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018;836:98–103. doi: 10.1016/j.mrgentox.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 63.Rafferty K., Archer K.J., Turner K., Brown R., Jackson-Cook C. Trisomy 21-associated increases in chromosomal instability are unmasked by comparing isogenic trisomic/disomic leukocytes from people with mosaic Down syndrome. PLoS One. 2021;16:e0254806. doi: 10.1371/JOURNAL.PONE.0254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reish O., Regev M., Kanesky A., Girafi S., Mashevich M. Sporadic aneuploidy in PHA-stimulated lymphocytes of trisomies 21, 18, and 13. Cytogenet. Genome Res. 2011;133:184–189. doi: 10.1159/000323504. [DOI] [PubMed] [Google Scholar]
- 65.Valind A., Jin Y., Baldetorp B., Gisselsson D. Whole chromosome gain does not in itself confer cancer-like chromosomal instability. Proc. Natl. Acad. Sci. USA. 2013;110:21119–21123. doi: 10.1073/PNAS.1311163110/SUPPL_FILE/PNAS.201311163SI.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson S.L., Compton D.A. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/J.AJHG.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yim A., Koti P., Bonnard A., Marchiano F., Dürrbaum M., Garcia-Perez C., Villaveces J., Gamal S., Cardone G., Perocchi F., et al. mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dynamics and mutations. Nucleic Acids Res. 2020;48:605–632. doi: 10.1093/nar/gkz1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan Y.H., Schneider E.L., Tischfield J., Epstein C.J., Ruddle F.H. Human chromosome 21 dosage: effect on the expression of the interferon induced antiviral state. Science. 1974;186:61–63. doi: 10.1126/SCIENCE.186.4158.61. [DOI] [PubMed] [Google Scholar]
- 70.Royzman K., Drews C., Maxwell T., Singh G., Keefe C., James S. Fibroblasts from individuals with Trisomy 21 have increased Type-I Interferon responses to HSV-1 compared to typical adults. J. Immunol. 2019;202 [Google Scholar]
- 71.Crow Y.J., Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 72.Malle L., Bogunovic D. Down syndrome and type I interferon: not so simple. Curr. Opin. Immunol. 2021;72:196–205. doi: 10.1016/J.COI.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Powers R.K., Culp-Hill R., Ludwig M.P., Smith K.P., Waugh K.A., Minter R., Tuttle K.D., Lewis H.C., Rachubinski A.L., Granrath R.E., et al. Trisomy 21 activates the kynurenine pathway via increased dosage of interferon receptors. Nat. Commun. 2019;10:4766. doi: 10.1038/s41467-019-12739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hasegawa H., Iwamasa K., Hatta N., Fujita S. Behçet’s disease associated with myelodysplastic syndrome with elevated levels of inflammatory cytokines. Mod. Rheumatol. 2003;13:350–355. doi: 10.1007/s10165-003-0245-6. [DOI] [PubMed] [Google Scholar]
- 75.Pellagatti A., Cazzola M., Giagounidis A., Perry J., Malcovati L., Della Porta M.G., Jädersten M., Killick S., Verma A., Norbury C.J., et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24:756–764. doi: 10.1038/leu.2010.31. [DOI] [PubMed] [Google Scholar]
- 76.Oka S., Ono K., Nohgawa M. The acquisition of trisomy 8 associated with Behçet’s-like disease in myelodysplastic syndrome. Leuk. Res. Rep. 2020;13:100196. doi: 10.1016/J.LRR.2020.100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inoue Y., Yamamoto T., Honda Y., Izawa K., Yasumi T. Partial Trisomy 9p with clinical symptoms resembling interferonopathies. J. Clin. Immunol. 2021;42:203–205. doi: 10.1007/S10875-021-01153-W. [DOI] [PubMed] [Google Scholar]
- 78.Zhuang H., Kosboth M., Lee P., Rice A., Driscoll D.J., Zori R., Narain S., Lyons R., Satoh M., Sobel E., Reeves W.H. Lupus-like disease and high interferon levels corresponding to trisomy of the type I interferon cluster on chromosome 9p. Arthritis Rheum. 2006;54:1573–1579. doi: 10.1002/art.21800. [DOI] [PubMed] [Google Scholar]
- 79.Santaguida S., Richardson A., Iyer D.R., M’Saad O., Zasadil L., Knouse K.A., Wong Y.L., Rhind N., Desai A., Amon A. Chromosome Mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell. 2017;41:638–651.e5. doi: 10.1016/j.devcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Viganó C., von Schubert C., Ahrné E., Schmidt A., Lorber T., Bubendorf L., De Vetter J.R.F., Zaman G.J.R., Storchova Z., Nigg E.A. Quantitative proteomic and phosphoproteomic comparison of human colon cancer DLD-1 cells differing in ploidy and chromosome stability. Mol. Biol. Cell. 2018;29:1031–1047. doi: 10.1091/mbc.E17-10-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krivega M., Stiefel C.M., Karbassi S., Andersen L.L., Chunduri N.K., Donnelly N., Pichlmair A., Storchová Z. Genotoxic stress in constitutive trisomies induces autophagy and the innate immune response via the cGAS-STING pathway. Commun. Biol. 2021;4:831. doi: 10.1038/s42003-021-02278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tijhuis A.E., Johnson S.C., McClelland S.E. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol. Cytogenet. 2019;12:17. doi: 10.1186/s13039-019-0429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barroso-Vilares M., Logarinho E. Chromosomal instability and pro-inflammatory response in aging. Mech. Ageing Dev. 2019;182:111118. doi: 10.1016/j.mad.2019.111118. [DOI] [PubMed] [Google Scholar]
- 84.Hopfner K.P., Hornung V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 85.MacKenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., et al. CGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wan Z., Han B. Clinical features of DDX41 mutation-related diseases: a systematic review with individual patient data. 2021. [DOI] [PMC free article] [PubMed]
- 87.Ferrari M., Stagi S. Autoimmunity and genetic syndromes: a focus on down syndrome. Genes. 2021;12:268. doi: 10.3390/GENES12020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huggard D., Doherty D.G., Molloy E.J. Immune dysregulation in children with down syndrome. Front. Pediatr. 2020;8:73. doi: 10.3389/FPED.2020.00073/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Araya P., Waugh K.A., Sullivan K.D., Núñez N.G., Roselli E., Smith K.P., Granrath R.E., Rachubinski A.L., Enriquez Estrada B., Butcher E.T., et al. Trisomy 21 dysregulates T cell lineages toward an autoimmunity-prone state associated with interferon hyperactivity. Proc. Natl. Acad. Sci. USA. 2019;116:24231–24241. doi: 10.1073/pnas.1908129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waugh K.A., Araya P., Pandey A., Jordan K.R., Smith K.P., Granrath R.E., Khanal S., Butcher E.T., Estrada B.E., Rachubinski A.L., et al. Mass cytometry reveals global immune remodeling with multi-lineage hypersensitivity to type I interferon in down syndrome. Cell Rep. 2019;29:1893–1908.e4. doi: 10.1016/j.celrep.2019.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dieudonné Y., Uring-Lambert B., Jeljeli M.M., Gies V., Alembik Y., Korganow A.S., Guffroy A. Immune defect in adults with down syndrome: insights into a complex issue. Front. Immunol. 2020;11:840. doi: 10.3389/FIMMU.2020.00840/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roy A., Bohn G., Goudevenou K., Cowan G., Bhatnagar N., O’Connor D., Tunstall O., Chan J., Bennett P., Kumar S., et al. Developmental stage specific B-progenitor expansion in normal fetal bone marrow is absent in down syndrome: implications for infant ALL. Blood. 2014;124:4331. doi: 10.1182/BLOOD.V124.21.4331.4331. [DOI] [Google Scholar]
- 93.O’Byrne S.I., Elliott N., Buck G., Rice S., O’Connor D., Oswald J., Fuchs H., Labbett E.-M., Fordham N.J., Roy A., Roberts I. Trisomy 21 driven pro-inflammatory signalling in fetal bone marrow may play a role in perturbed B-lymphopoiesis and acute lymphoblastic leukemia of down syndrome. Blood. 2019;134:1206. doi: 10.1182/BLOOD-2019-130180. [DOI] [Google Scholar]
- 94.Verstegen R.H.J., Kusters M.A.A., Gemen E.F.A., De Vries E. Down Syndrome B-Lymphocyte subpopulations, intrinsic defect or decreased T-lymphocyte help. Pediatr. Res. 2010;67:563–569. doi: 10.1203/pdr.0b013e3181d4ecc1. [DOI] [PubMed] [Google Scholar]
- 95.MacLean G.A., McEldoon J., Huang J., Allred J., Canver M.C., Orkin S.H. Downregulation of endothelin receptor B contributes to defective B cell lymphopoiesis in trisomy 21 pluripotent stem cells. Sci. Rep. 2018;8:8001. doi: 10.1038/s41598-018-26123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Elsayed S.M., Elsayed G.M. Phenotype of apoptotic lymphocytes in children with Down syndrome. Immun. Ageing. 2009;6:2–5. doi: 10.1186/1742-4933-6-2/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marcovecchio G.E., Ferrua F., Fontana E., Beretta S., Genua M., Bortolomai I., Conti A., Montin D., Cascarano M.T., Bergante S., et al. Premature senescence and increased oxidative stress in the thymus of down syndrome patients. Front. Immunol. 2021;12:1830. doi: 10.3389/FIMMU.2021.669893/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hjalmar V., Hast R., Kimby E. Cell surface expression of CD25, CD54, and CD95 on B- and T-cells in chronic lymphocytic leukaemia in relation to trisomy 12, atypical morphology and clinical course. Eur. J. Haematol. 2002;68:127–134. doi: 10.1034/j.1600-0609.2002.01515.x. [DOI] [PubMed] [Google Scholar]
- 99.Hui H.Y.L., Clarke K.M., Fuller K.A., Stanley J., Chuah H.H., Ng T.F., Cheah C., McQuillan A., Erber W.N. “Immuno-flowFISH” for the assessment of cytogenetic abnormalities in chronic lymphocytic leukemia. Cytometry A. 2019;95:521–533. doi: 10.1002/cyto.a.23769. [DOI] [PubMed] [Google Scholar]
- 100.Sloand E.M., Melenhorst J.J., Tucker Z.C.G., Pfannes L., Brenchley J.M., Yong A., Visconte V., Wu C., Gostick E., Scheinberg P., et al. T-cell immune responses to Wilms tumor 1 protein in myelodysplasia responsive to immunosuppressive therapy. Blood. 2011;117:2691–2699. doi: 10.1182/blood-2010-04-277921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schlegelberger B., Nölle I., Feller A.C., Bauer E., Grote W. Angioimmunoblastic lymphadenopathy with trisomy 3: The cells of the malignant clone are T cells. Hematol. Pathol. 1990;4:179–183. [PubMed] [Google Scholar]
- 102.Makrydimas G., Plachouras N., Nicolaides K.H. Abnormal immunological development in fetuses with trisomy 18. Prenat. Diagn. 1994;14:239–241. doi: 10.1002/pd.1970140403. [DOI] [PubMed] [Google Scholar]
- 103.Stevens W.B.C., Mendeville M., Redd R., Clear A.J., Bladergroen R., Calaminici M., Rosenwald A., Hoster E., Hiddemann W., Gaulard P., et al. Prognostic relevance of CD163 and CD8 combined with EZH2 and gain of chromosome 18 in follicular lymphoma: A study by the Lunenburg Lymphoma Biomarker Consortium. Haematologica. 2017;102:1413–1423. doi: 10.3324/haematol.2017.165415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Colacurcio D.J., Pensalfini A., Jiang Y., Nixon R.A. Dysfunction of autophagy and endosomal-lysosomal pathways: roles in pathogenesis of Down syndrome and Alzheimer’s Disease. Free Radic. Biol. Med. 2018;114:40–51. doi: 10.1016/j.freeradbiomed.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meharena H.S., Marco A., Dileep V., Lockshin E.R., Akatsu G.Y., Mullahoo J., Watson L.A., Ko T., Guerin L.N., Abdurrob F., et al. Down-syndrome-induced senescence disrupts the nuclear architecture of neural progenitors. Cell Stem Cell. 2022;29:116–130.e7. doi: 10.1016/J.STEM.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nawa N., Hirata K., Kawatani K., Nambara T., Omori S., Banno K., Kokubu C., Takeda J., Nishimura K., Ohtaka M., et al. Elimination of protein aggregates prevents premature senescence in human trisomy 21 fibroblasts. PLoS One. 2019;14:e0219592. doi: 10.1371/journal.pone.0219592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.González A.E., Muñoz V.C., Cavieres V.A., Bustamante H.A., Cornejo V.H., Januário Y.C., González I., Hetz C., Dasilva L.L., Rojas-Fernández A., et al. Autophagosomes cooperate in the degradation of intracellular C-terminal fragments of the amyloid precursor protein via the MVB/lysosomal pathway. FASEB. J. 2017;31:2446–2459. doi: 10.1096/fj.201600713R. [DOI] [PubMed] [Google Scholar]
- 108.Doran E., Keator D., Head E., Phelan M.J., Kim R., Totoiu M., Barrio J.R., Small G.W., Potkin S.G., Lott I.T. Down Syndrome, partial trisomy 21, and absence of Alzheimer’s Disease: the role of APP. J. Alzheimers Dis. 2017;56:459–470. doi: 10.3233/JAD-160836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prasher V.P., Farrer M.J., Kessling A.M., Fisher E.M., West R.J., Barber P.C., Butler A.C. Molecular mapping of alzheimer-type dementia in Down’s syndrome. Ann. Neurol. 1998;43:380–383. doi: 10.1002/ANA.410430316. [DOI] [PubMed] [Google Scholar]
- 110.Wu C.I., Vinton E.A., Pearse R.V., Heo K., Aylward A.J., Hsieh Y.C., Bi Y., Adeleye S., Fancher S., Duong D.M., et al. APP and DYRK1A regulate axonal and synaptic vesicle protein networks and mediate Alzheimer’s pathology in trisomy 21 neurons. Mol. Psychiatr. 2022;27:1970–1989. doi: 10.1038/s41380-022-01454-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zuliani I., Lanzillotta C., Tramutola A., Francioso A., Pagnotta S., Barone E., Perluigi M., Di Domenico F. The Dysregulation of OGT/OGA cycle mediates Tau and APP neuropathology in down syndrome. Neurotherapeutics. 2021;18:340–363. doi: 10.1007/S13311-020-00978-4/FIGURES/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bordi M., Darji S., Sato Y., Mellén M., Berg M.J., Kumar A., Jiang Y., Nixon R.A. mTOR hyperactivation in Down Syndrome underlies deficits in autophagy induction, autophagosome formation, and mitophagy. Cell Death Dis. 2019;10:563. doi: 10.1038/s41419-019-1752-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perluigi M., Pupo G., Tramutola A., Cini C., Coccia R., Barone E., Head E., Butterfield D.A., Di Domenico F. Neuropathological role of PI3K/Akt/mTOR axis in Down syndrome brain. Biochim. Biophys. Acta. 2014;1842:1144–1153. doi: 10.1016/j.bbadis.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aivazidis S., Jain A., Rauniyar A.K., Anderson C.C., Marentette J.O., Orlicky D.J., Fritz K.S., Harris P.S., Siegel D., Maclean K.N., Roede J.R. SNARE proteins rescue impaired autophagic flux in down syndrome. PLoS One. 2019;14:e0223254. doi: 10.1371/journal.pone.0223254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lauritzen I., Pardossi-Piquard R., Bourgeois A., Pagnotta S., Biferi M.G., Barkats M., Lacor P., Klein W., Bauer C., Checler F. Intraneuronal aggregation of the β-CTF fragment of APP (C99) induces Aβ-independent lysosomal-autophagic pathology. Acta Neuropathol. 2016;132:257–276. doi: 10.1007/s00401-016-1577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang Y., Mullaney K.A., Peterhoff C.M., Che S., Schmidt S.D., Boyer-Boiteau A., Ginsberg S.D., Cataldo A.M., Mathews P.M., Nixon R.A. Alzheimer’s-related endosome dysfunction in Down syndrome is Aβ-independent but requires APP and is reversed by BACE-1 inhibition. Proc. Natl. Acad. Sci. USA. 2010;107:1630–1635. doi: 10.1073/pnas.0908953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gui X., Yang H., Li T., Tan X., Shi P., Li M., Du F., Chen Z.J. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stingele S., Stoehr G., Storchova Z. Activation of autophagy in cells with abnormal karyotype. Autophagy. 2013;9:246–248. doi: 10.4161/auto.22558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ariyoshi K., Miura T., Kasai K., Fujishima Y., Oshimura M., Yoshida M.A. Induction of genomic instability and activation of autophagy in artificial human aneuploid cells. Mutat. Res. 2016;790:19–30. doi: 10.1016/j.mrfmmm.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 120.Tang Y.C., Williams B.R., Siegel J.J., Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nassour J., Radford R., Correia A., Fusté J.M., Schoell B., Jauch A., Shaw R.J., Karlseder J. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–663. doi: 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li L.B., Chang K.-H., Wang P.-R., Hirata R.K., Papayannopoulou T., Russell D.W. Trisomy Correction in Down Syndrome Induced Pluripotent Stem Cells. Cell Stem Cell. 2012;11:615–619. doi: 10.1016/j.stem.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 124.Martin K.R., Corlett A., Dubach D., Mustafa T., Coleman H.A., Parkington H.C., Merson T.D., Bourne J.A., Porta S., Arbonés M.L., et al. Over-expression of RCAN1 causes down syndrome-like hippocampal deficits that alter learning and memory. Hum. Mol. Genet. 2012;21:3025–3041. doi: 10.1093/hmg/dds134. [DOI] [PubMed] [Google Scholar]
- 125.Soppa U., Schumacher J., Florencio Ortiz V., Pasqualon T., Tejedor F.J., Becker W. The down syndrome-related protein kinase DYRK1A phosphorylates p27 Kip1and cyclin D1 and induces cell cycle exit and neuronal differentiation. Cell Cycle. 2014;13:2084–2100. doi: 10.4161/cc.29104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park J., Oh Y., Chung K.C. Two key genes closely implicated with the neuropathological characteristics in Down syndrome: DYRK1A and RCAN1. BMB Rep. 2009;42:6–15. doi: 10.5483/BMBRep.2009.42.1.006. [DOI] [PubMed] [Google Scholar]
- 127.Arron J.R., Winslow M.M., Polleri A., Chang C.P., Wu H., Gao X., Neilson J.R., Chen L., Heit J.J., Kim S.K., et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 128.Shin S.Y., Yang H.W., Kim J.R., Heo W.D., Cho K.H. A hidden incoherent switch regulates RCAN1 in the calcineurin-NFAT signaling network. J. Cell Sci. 2011;124:82–90. doi: 10.1242/jcs.076034. [DOI] [PubMed] [Google Scholar]
- 129.Pan M.-G., Xiong Y., Chen F. NFAT Gene Family in Inflammation and Cancer. Curr. Mol. Med. 2013;13:543–554. doi: 10.2174/1566524011313040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lane A.A., Chapuy B., Lin C.Y., Tivey T., Li H., Townsend E.C., Van Bodegom D., Day T.A., Wu S.C., Liu H., et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat. Genet. 2014;46:618–623. doi: 10.1038/ng.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tazi-Ahnini R., di Giovine F.S., McDonagh A.J., Messenger A.G., Amadou C., Cox A., Duff G.W., Cork M.J. Structure and polymorphism of the human gene for the interferon-induced p78 protein (MX1): evidence of association with alopecia areata in the Down syndrome region. Hum. Genet. 2000;106:639–645. doi: 10.1007/s004390000318. [DOI] [PubMed] [Google Scholar]
- 132.Wong H., Levenga J., Cain P., Rothermel B., Klann E., Hoeffer C. RCAN1 overexpression promotes age-dependent mitochondrial dysregulation related to neurodegeneration in Alzheimer’s disease. Acta Neuropathol. 2015;130:829–843. doi: 10.1007/s00401-015-1499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Salehi A., Delcroix J.D., Belichenko P.V., Zhan K., Wu C., Valletta J.S., Takimoto-Kimura R., Kleschevnikov A.M., Sambamurti K., Chung P.P., et al. Increased app expression in a mouse model of down’s syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 2006;51:29–42. doi: 10.1016/J.NEURON.2006.05.022/ATTACHMENT/23C4EAE1-4B9E-4912-9DB6-4941C2366E46/MMC1.PDF. [DOI] [PubMed] [Google Scholar]
- 134.Yabut O., Domogauer J., D’Arcangelo G. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J. Neurosci. 2010;30:4004–4014. doi: 10.1523/JNEUROSCI.4711-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.